Summary
Approximately 15 genes have been directly associated with skin pigmentation variation in humans, leading to its characterization as a relatively simple trait. However, by assembling a global survey of quantitative skin pigmentation phenotypes, we demonstrate that pigmentation is more complex than previously assumed with genetic architecture varying by latitude. We investigate polygenicity in the KhoeSan, populations indigenous to southern Africa, who have considerably lighter skin than equatorial Africans. We demonstrate that skin pigmentation is highly heritable, but known pigmentation loci explain only a small fraction of the variance. Rather, baseline skin pigmentation is a complex, polygenic trait in the KhoeSan. Despite this, we identify canonical and non-canonical skin pigmentation loci, including near SLC24A5, TYRP1, SMARCA2/VLDLR, and SNX13 using a genome-wide association approach complemented by targeted resequencing. By considering diverse, under-studied African populations, we show how the architecture of skin pigmentation can vary across humans subject to different local evolutionary pressures.
Keywords: pigmentation, human evolution, Africa, heritability, population genetics
eTOC
The genetic architecture of skin pigmentation is highly complex and varies across human populations subjected to distinct geographical evolutionary pressures
Introduction
Skin pigmentation is one of the most strikingly variable and strongly selected phenotypes among human populations, with darker skin observed closer to the equator and lighter pigmentation observed at high latitudes (Sturm and Duffy, 2012). Researchers have hypothesized that variable exposure to ultra violet radiation (UVR) creates opposing selective forces for vitamin D production and folate protection, resulting in variable melanin production and global pigmentation differentiation (Chaplin and Jablonski, 2009; Jablonski and Chaplin, 2010). Skin pigmentation differences at similar latitudes and UV exposures indicate that additional evolutionary forces, such as assortative mating, drift, and epistasis, are also likely to have affected global skin pigmentation (Pospiech et al., 2014; Wilde et al., 2014). While ~171 genes have been implicated in variability across model organisms (e.g. the Color Genes database: http://www.espcr.org/micemut/), only ~15 genes have been associated with skin color differences in humans (Table 2). The relative paucity of loci identified from GWAS efforts has led to the characterization of pigmentation variation as relatively simple, with only a handful of SNPs being highly predictive of skin, eye, and hair color across populations (Hart et al., 2013a; Spichenok et al., 2011; Walsh et al., 2013).
Table 2. Replication of previously associated skin pigmentation variants in the joint ‡Khomani and Nama populations.
P-value indicates the joint association across all KhoeSan individuals using a linear mixed model accounting for European and Bantu admixture as well as kinship. Beta values reflect the effect size of adding one derived allele, assuming an additive model, to the distribution of M index (see Figure 1). See also Figure S5 and Table S6A.
| Gene | rsID | P-value | Beta | Derived frequency | Allele numbera | San-specific frequency | San 95% CIb | W. AFRc | N. EURd |
|---|---|---|---|---|---|---|---|---|---|
| UGT1A | rs6742078 | 0.58 | −0.44 | 0.54 | 460 | 0.60 | [0.54,0.69] | 0.47 | 0.29 |
| SLC45A2 | rs35395 | 0.98 | −0.02 | 0.32 | 882 | 0.21 | [0.18,0.25] | 0.20 | 0.99 |
| SLC45A2 | rs16891982 | 1.2E-03 | −2.84 | 0.14 | 882 | 0.00 | [0.00,0.02] | 0.00 | 0.98 |
| IRF4 | rs12203592 | 0.83 | −0.54 | 0.01 | 882 | 0.00 | [0.00,0.00] | 0.00 | 0.17 |
| IRF4 | rs12202284 | 0.51 | 0.99 | 0.04 | 824 | 0.00 | [0.00,0.01] | 0.15 | 0.21 |
| OPRM1 | rs6917661 | 0.29 | −0.71 | 0.66 | 882 | 0.71 | [0.67,0.79] | 0.61 | 0.76 |
| EGFR | rs12668421 | 0.65 | −0.49 | 0.08 | 882 | 0.02 | [0.01,0.08] | 0.06 | 0.27 |
| TYRP1 | rs13289810 | 0.61 | 0.53 | 0.19 | 882 | 0.18 | [0.11,0.25] | 0.24 | 0.34 |
| BNC2 | rs10756819 | 0.51 | 0.91 | 0.08 | 466 | 0.02 | [0.00,0.05] | 0.07 | 0.65 |
| GATA3 | rs376397 | 0.91 | 0.07 | 0.65 | 872 | 0.79 | [0.75,0.82] | 0.31 | 0.32 |
| GRM5, TYR | rs10831496 | 0.28 | −0.90 | 0.52 | 460 | 0.63 | [0.57,0.70] | 0.12 | 0.69 |
| TYR | rs1042602 | 0.74 | 0.58 | 0.06 | 466 | 0.00 | [0.00,0.02] | 0.00 | 0.38 |
| KITLG | rs12821256 | 0.02 | −5.28 | 0.02 | 882 | 0.00 | [0.00,0.01] | 0.00 | 0.17 |
| OCA2 | rs1800404 | 0.53 | −0.40 | 0.55 | 854 | 0.65 | [0.56,0.74] | 0.11 | 0.81 |
| OCA2 | rs7495174 | 0.92 | −0.07 | 0.71 | 716 | 0.61 | [0.55,0.69] | 0.26 | 0.90 |
| HERC2 | rs12913832 | 0.09 | −1.70 | 0.10 | 882 | 0.00 | [0.00,0.02] | 0.01 | 0.79 |
| APBA2 | rs4424881 | 0.25 | −1.24 | 0.18 | 440 | 0.02 | [0.00,0.06] | 0.07 | 0.86 |
| SLC24A5 | rs1426654 | 9.8E-09 | −3.58 | 0.40 | 882 | 0.24 | [0.17,0.32] | 0.05 | 1.00 |
| MC1R | rs1805007 | 0.80 | −0.64 | 0.01 | 630 | 0.00 | [0.00,0.03] | 0.00 | 0.11 |
Allele number indicates the total number of alleles genotyped or sequenced across all KhoeSan samples.
Confidence interval for the San-specific frequencies indicates the allele frequencies specifically on ‡Khomani haplotypes, assessed with local ancestry tracts.
W. AFR (western African) allele frequencies were estimated from 405 ESN, GWD, YRI, and MSL populations in the phase 3 1000 Genomes project
N. EUR (northern Europeans) allele frequencies were estimated from 190 GBR and CEU populations in the phase 3 1000 Genomes project
Despite being home to the greatest range of pigmentation globally, remarkably few genetic studies of pigmentation have been published to date in continental Africans (Jablonski and Chaplin, 2014; Relethford, 2000). Instead, the genetic basis of skin color has primarily been studied in Eurasians and admixed African Americans (Beleza et al., 2013; 2012; Candille et al., 2012; Sturm and Duffy, 2012; Sulem et al., 2008; 2007); selective sweeps in high latitude populations have been interpreted as resulting from strong environmental selection pressure. For example, the derived Ala111Thr allele (rs1426654) of SLC24A5 that swept to near fixation in western Eurasian populations confers the largest known effect on skin color variability (Beleza et al., 2013; Lamason, 2005). Loci in/near SLC45A2, GRM5/TYR, and APBA2/OCA2 also have divergent allele frequencies between Europeans and Africans, with large derived lightening effects in Europeans (Beleza et al., 2013; Norton et al., 2006). Smaller effects contribute to the relatively narrow variation within Europeans, including associations in/near MC1R, TYR, IRF4, and ASIP (Sulem et al., 2007; 2008). Several shared lightening effects derived in Eurasians arose through convergent evolution (e.g. rs1800414 in OCA2 in East Asians), whereas other shared effects resulted from similar selective sweeps (e.g. KITLG) (Miller et al., 2007; Yang et al., 2016). Because African populations have been understudied, the genetic architecture and higher variability of skin pigmentation is poorly understood.
Strong positive selection acting on skin pigmentation has resulted in large effects that explain a large fraction of heritable variation. For example, a previous study in recently admixed Cape Verdeans with European and West African ancestors showed that only 4 loci explain 35% of the variation in skin pigmentation (Beleza et al., 2013). In contrast, complex traits such as height and schizophrenia require ~10,000 independent SNPs derived from GWAS of >100,000 individuals to build predictors that explain ~29% and ~20% of the variance in independent cohorts, respectively (Ripke et al., 2014; Wood et al., 2014). Previous studies of positively selected traits such as pigmentation, high altitude adaptation, and response to pathogens have repeatedly produced larger effect sizes than complex common disease; these large effect loci have typically been discovered with relatively small sample sizes (i.e. ~100s of individuals) compared to common diseases (Genovese et al., 2010; Kenny et al., 2012a; Yi et al., 2010). While pigmentation loci are among the most diverged in the genome, it is noteworthy that effect size estimates of significant polymorphic GWAS loci tend to be directionally consistent across populations (Carlson et al., 2013), but that aggregate prediction accuracy varies across populations (Martin et al., 2017).
Striking skin pigmentation variability among African populations has been underappreciated in genetic studies (Jablonski and Chaplin, 2014; Relethford, 2000). Light skin pigmentation is observed in the far southern latitudes of Africa among KhoeSan§ hunter-gatherers and pastoralists in and near the Kalahari Desert. The KhoeSan are unique in their early divergence from other populations, likely dating back at least ~100,000 years ago (Schlebusch et al., 2012; Veeramah et al., 2011); they exhibit extraordinary levels of genetic diversity and low levels of linkage disequilibrium (LD) (Henn et al., 2011). Previous work points to southern Africa as the point of origin for modern humans (Henn et al., 2011; Tishkoff et al., 2009), but it is unknown whether moderate to light skin pigmentation in the different KhoeSan groups is an example of convergent evolution with northern Europeans and Asians, or reflects the ancestral human phenotype. Previous studies have noted different pigmentation allele frequencies between the Ju|’hoansi San and other Africans, but these have been based on n<7 individuals from the former population without measured phenotypes (Berg and Coop, 2014; Norton et al., 2006).
Here we report an evolutionary and genetic study of skin pigmentation with a total of 465 genotyped KhoeSan individuals (278 ‡Khomani San and 187 Nama), with targeted resequencing at associated pigmentation loci and matched quantitative spectrophotometric phenotype data (Table S4). The ‡Khomani San are traditionally a N|u-speaking hunter-gatherer population living in the southern Kalahari Desert, while the Nama are traditionally a Khoekhoe-speaking semi-nomadic pastoralist group of KhoeSan ancestry. We investigate: i) the degree of polygenicity and heritability of skin pigmentation, ii) the extent of pigmentation variation explained by previously associated or canonical pigmentation genes, and iii) novel pigmentation alleles contributing to variation in the ‡Khomani San and Nama populations.
Results
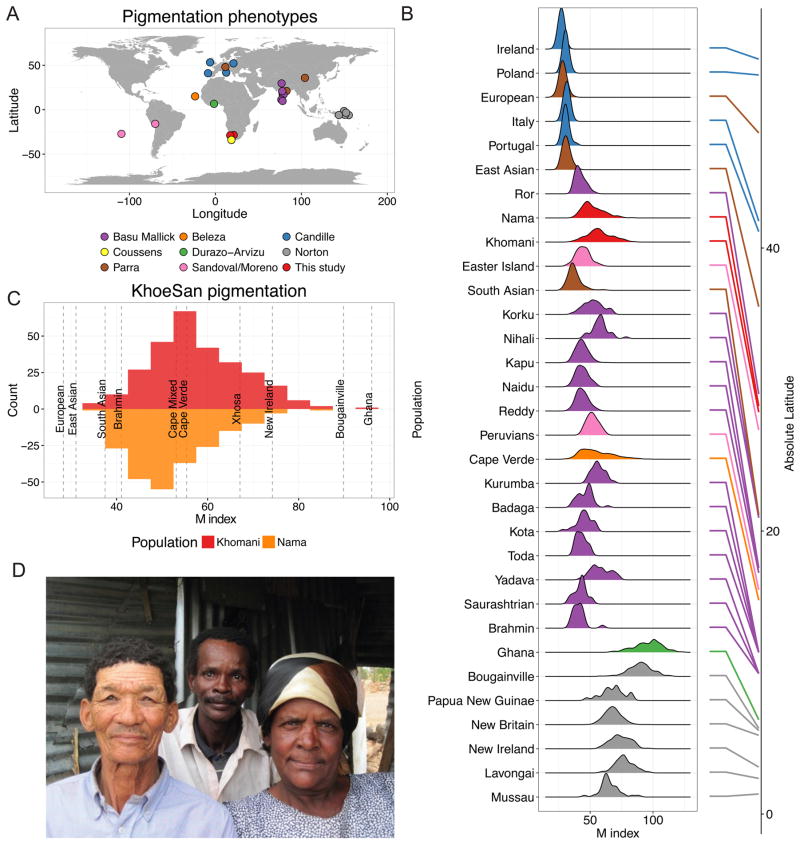
We quantitatively phenotyped baseline skin color in 479 individuals (277 ‡Khomani, 202 Nama, Table S4) via narrow-band reflectometry to measure hemoglobin and melanin of both the left and right upper inner arms (STAR Methods), with M index = log10(1 / % red reflectance). We sequenced and/or genotyped a subset of phenotyped samples (Table S4, Methods). Skin pigmentation is lighter in the KhoeSan than the majority of other African populations, with baseline upper arm M index = 57.57 ± 10.12 (mean ± sd) in the ‡Khomani San. Baseline upper arm pigmentation in the Nama is slightly lower, with M index = 52.12 ± 8.93. The ‡Khomani are on average significantly darker than the Nama (p=3.6e-10, Figure 1C), but the variance is not significantly different (p>0.05). For comparison, we aggregated quantitative skin pigmentation across 32 globally diverse populations (4,712 individuals) assayed with a Derma Spectrometer (DSMI or DSMII) (Basu Mallick et al., 2013; Beleza et al., 2013; Candille et al., 2012; Coussens et al., 2015; Durazo-Arvizu et al., 2014; Edwards et al., 2010; Norton et al., 2006) (Figure 1A–B, Table S1). Only four African populations are available for comparison; among these only the Ghanians represent an equatorial African population with minimal recent admixture. Skin color is substantially darker in equatorial Ghanaians, where M index reaches a mean of 96.04 ± 10.94; M index for Cape Verdeans, who have ~40% European admixture on average, have slightly lighter (55.39 ± 13.00, p=5.6e-3) and considerably more variable pigmentation (p=1.9e-6) than the KhoeSan. Two other populations living in South Africa, the Xhosa and admixed Coloured populations, have respectively darker (M index=67.1±7.5) and similar (M index=53.1±8.5) pigmentation compared to the KhoeSan populations (Coussens et al., 2015).
Figure 1. Distributions of baseline pigmentation in globally diverse populations.
A) Sample locations of skin pigmentation datasets where phenotypes were measured with a DSM I or DSM II. B) Violin plots of pigmentation distributions for 32 populations from 8 studies ordered by latitude; absolute latitudes provided on the right. Corresponding datasets are colored as in A). Table S1 provides summary statistics for each population. M indices are reflectance measures that approximate melanin content. C) A comparison of skin pigmentation distributions in ‡Khomani (top) and Nama populations (bottom). Dashed grey lines and labels indicate mean M index for the indicated other global populations. D) South African individuals in a household that exemplify the substantial skin pigmentation variability in the ‡Khomani and Nama populations. Picture taken with consent for publication. See also Table S1.
Evidence of Increased Polygenicity in Skin Pigmentation Among Equatorial Populations
We tested whether the correlation between absolute latitude and pigmentation was significant with our large, quantitatively phenotyped sample of global populations. As previously observed (Byard, 1981; Jablonski and Chaplin, 2010; Zaidi et al., 2017), we find that skin pigmentation is strongly associated with absolute latitude (R2=0.53, β=−1.18 on M index scale, p<2e-16); populations further from the equator have lighter skin pigmentation. We next tested whether variance in melanin within populations also varies across populations. Skin pigmentation has primarily been studied in lightly pigmented European and East Asian populations, where skin color varies minimally among individuals (Figure 1A–B). Less-studied equatorial and admixed populations, including Melanesians, Ghanaians, Cape Verdeans, South African admixed Coloured, and South Asians vary considerably more in skin pigmentation (Figure 1B). We find that absolute latitude is also significantly negatively associated with the standard deviation in melanin (R2=0.41, p=5.0e-5). Further, melanin distributions are heteroskedastic (i.e. the variance is not constant—rather, it changes over the range of observed M index), with the coefficient of variation, a standardized metric of phenotypic dispersion, decreasing with increasing distance from the equator (cv=σ/μ, R2=0.14, p=0.03, Table S1).
A sign test comparing variances in lighter versus darker population pairs within the same study indicates that populations with lighter skin have significantly reduced phenotypic variance than expected by chance (p=2.01e-8). These results suggest that there is reduced genetic heterogeneity and/or reduced variance in the population distribution of causal effect sizes contributing to lighter versus darker pigmentation. There is more than an order of magnitude difference in variance between the lightest and darkest populations (i.e. Irish vs Ghanaian F=0.03, p=6.7e-23). Europeans and East Asians have significantly less variation than South Asians (F=0.25, p=1.06e-14 and F=0.30, p=1.27e-10, respectively, Figure 1B). Cape Verdeans with the highest quartile of European admixture have lighter, less variable skin color than individuals with the lowest quartile of European ancestry (p=4.28e-9, although notably ancestry proportions are bimodal across individuals). Among Melanesians, islands at similar latitudes with more lightly pigmented individuals on average show less variance than those with more darkly pigmented individuals (e.g. one-sided F test comparing variance among more lightly pigmented New Britain individuals versus individuals from Bougainville, p=2.89e-9, Figure 1B). Among the ‡Khomani and Nama, comparing individuals with primarily European admixture (>20%, N=124) to individuals with primarily Bantu admixture (>20%, N=91), we find significantly greater melanin variation among KhoeSan individuals with more Bantu admixture (p=1.33e-4).
Ancestry and Skin Pigmentation Variation in the KhoeSan
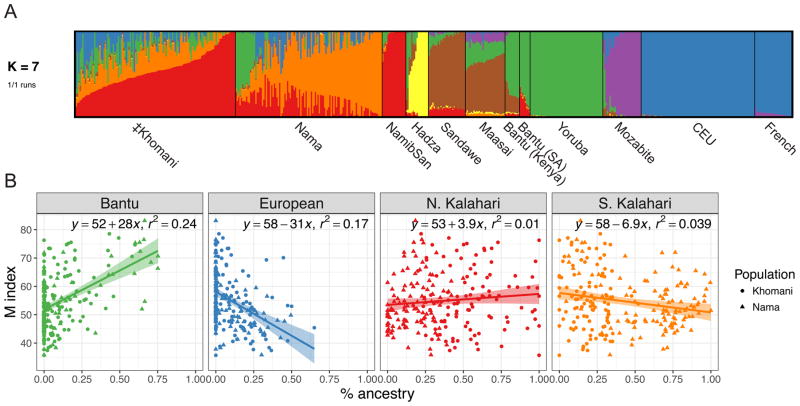
The ‡Khomani San and the Nama have both experienced admixture with neighboring darker-skinned Bantu-speaking groups beginning ~450 years ago, as well as with lighter-skinned European settlers who first arrived in the Northern Cape during the late 18th century (Uren et al., 2016a). We assessed these ancestry proportions using unsupervised allele frequency clustering with ADMIXTURE as well as principal components analysis (PCA, STAR Methods). At k=3, we observe distinct clustering between Europeans, Bantu-speaking and West African populations, and KhoeSan populations; both the Nama and the ‡Khomani have ~75–80% KhoeSan-specific ancestry. For k=7, which gives most stable ancestry estimates, we observe a partitioning of the KhoeSan ancestry into ‘northern Kalahari’ ancestry shared with Ju|’hoansi and a distinct southern or circum-Kalahari ancestry present in the Nama and the ‡Khomani. On average, in the ‡Khomani San we find 55% northern Kalahari KhoeSan ancestry, 21% southern Kalahari KhoeSan ancestry, 11% European ancestry (common in CEU and French individuals), 12% western African ancestry (common in Yoruba and Bantu-speaking populations), and 2% attributable to other African populations (Tanzanian hunter-gatherers, East African, and North African populations, Table S2, Figure 2A and Figure S2A). The Nama differ from the ‡Khomani in their proportion of northern versus southern Kalahari ancestry; they have 17% northern Kalahari ancestry, 62% southern Kalahari ancestry, 9% European ancestry, 10% western African ancestry, and 1% attributable to other African populations on average. The western African fraction in the Nama is significantly more variable among individuals (p=1.08e-5), resulting from recent Damara gene flow (Uren et al., 2016a). The partition of ancestry components occurs in the same order and is correlated between ADMIXTURE and PCA (Figure 2, Figure S2A,D–F).
Figure 2. Ancestry components in the KhoeSan and association with pigmentation.
A) ADMIXTURE proportions at k=7 for the ‡Khomani and Nama populations, using Namibian San, Hadza, Sandawe, Maasai, Kenyan Bantu, South African (SA) Bantu, Yoruba, Mozabite, Central Europeans (CEU), and French populations as a reference panel B) Associations between substantial k ancestry clusters and average melanin (M index) baseline pigmentation value in the combined ‡Khomani and Nama populations. The Bantu and European components each constitute ≥ 5% of the total KhoeSan ancestry on average and have significant associations in the best multivariate model (p <0.05). See also Figure S2 and Table S2.
In a multivariate mixed model with the significant European and Bantu admixture components, European and Bantu ancestry are strongly correlated with light (β = −18.09, p=2.9e-03) and dark skin (β = 25.60, p=1.8e-09), respectively. Together, we estimate that fixed admixture effects explained 34% of the variation in skin color (adjusted R2); by comparison, 44% of pigmentation variation in Cape Verdeans is explained by admixture effects (Beleza et al., 2013). Marginal associations are shown in Figure 2B, with pairwise ancestry correlations shown in Figure S2B. Southern Kalahari ancestry frequent in the Nama is significantly anti-correlated with Bantu ancestry and is marginally predicted to lighten skin, but not when modeled jointly with Bantu ancestry in a multivariate model. Interestingly, the mean pigmentation of Nama and ‡Khomani individuals with <90% KhoeSan ancestry is not significantly different from individuals with >90% KhoeSan ancestry (p=0.94), although the variance is significantly greater in more admixed individuals (admixture from either/both European or Bantu ancestries, p=2.2e-3). These results suggest that while admixture increases phenotypic variance, pigmentation alleles on KhoeSan haplotypes contribute more to the overall heterogeneity than those on European or Bantu haplotypes. Consistent with this result, we observe substantial skin pigmentation variation among related individuals, which, coupled with high heritability (see below) suggests a role for large effect sizes of alleles contributing to pigmentation.
Skin Pigmentation is Highly Heritable
We inferred narrow sense heritability for baseline skin pigmentation and tanning status in the KhoeSan with four methods: family pedigrees ( ), SNP array similarity matrices ( ), identity-by-descent (IBD) sharing matrices ( ), and exome sequence variation ( , Table 1). While pedigree-based heritability estimates are not based on genetic data and therefore not strongly affected by admixture, we carefully considered it for SNP-based estimates, as described previously (Beleza et al., 2013; Thornton et al., 2012a; Zaitlen et al., 2013; 2014). In each of the heritability estimates of baseline skin color and tanning status, we accounted for admixture proportions with European and Bantu ancestry as covariates, as well as familial relatedness via a kinship covariance matrix. Similarly for tanning status, we accounted for age, sex, and kinship. Previous family-based estimates for skin color heritability in other populations are high, ranging between 55–90% (Byard, 1981; Clark et al., 1981; Frisancho et al., 1981; Harrison and Owen, 1964). Interestingly, published genetic estimates of skin pigmentation heritability in Europe are low and insignificant, potentially because of reduced genetic diversity at skin pigmentation loci due to positive selection (Zaidi et al., 2017). Our heritability estimates in the KhoeSan are analogous to family-based estimates because of the elevated relatedness in our samples.
Table 1. Heritability estimates contrasting baseline skin pigmentation with tanning status.
SNP-based heritability estimates were computed with GCTA using genetic relationship matrices (GRMs) calculated from SNP gentoypes, an admixture-corrected GRM computed with REAP, and IBD segments. All models were unconstrained. See also Figure S1 and Table S3.
| Method | Dataset | SNPs | N | h2 (SE) baseline pigmentationa | h2 (SE) tanning statusb |
|---|---|---|---|---|---|
| GCTA GRM | genotype array | 286,026 | 216 | 0.90 (0.15) | 0.31 (0.19) |
| REAP GRM | genotype array | 286,026 | 216 | 0.97 (0.15) | 0.41 (0.21) |
| KIBD | genotype array | NA | 216 | 0.97 (0.16) | 0.45 (0.22) |
| GCTA GRM | exome | 117,132 | 82 | 0.95 (0.26) | 0.37 (0.37) |
| SOLAR | pedigrees | NA | 477 | 0.96 (0.12) | 0.19 (0.11) |
Bantu and European admixture proportions were included as covariates.
Age and sex were included as significant covariates for tanning status (wrist minus baseline underarm pigmentation).
We first constructed pedigrees from ethnographic interviews for individuals within the ‡Khomani and Nama populations and verified relationships where possible with genetic data. 533 individuals (including parental individuals not sampled) could be assigned to a pedigree, resulting in 354 extended pedigrees and 470 nuclear families. Via traditional pedigree-based estimation, we estimate an of 0.96 ± 0.12 for baseline skin color (STAR Methods). We then asked whether variation present on the ascertained SNP arrays or from exome sequencing could explain a similar fraction of the pigmentation variation. Genetic heritability estimates inferred from recently admixed populations have two potential problems: 1) inferred familial relationships between individuals are less accurate (Thornton et al., 2012a), and 2) environmental confounders (e.g. socioeconomic status) could be associated with the variance component attributed to additive genetic effects. In order to address the first issue, we use the proportion of KhoeSan, European and Bantu ancestry per individual to correct the SNP array genetic relatedness matrix (GRM) as described by the REAP approach (Thornton et al., 2012a). The REAP matrix is also compared to the IBS matrix inferred using default GCTA parameters that do not account for stratification (Methods). We include European and Bantu ancestry as global covariates in the heritability estimation. All further estimation of was made using the unconstrained model in GCTA. Furthermore, we contrast baseline pigmentation with tanning status (i.e. sun exposed wrist – underarm melanin pigmentation); if our estimates were inflated by environmental confounders, we would also expect inflated heritability of tanning status.
The array-based heritability point estimates are consistently but not significantly higher when using a kinship matrix from Relatedness Estimation in Admixed Populations (REAP) than GCTA’s identity-by-state (IBS) GRM, both for the joint dataset and each population separately (Table 1 and Table S3). We estimate (standard error) in an unconstrained model across both populations using the REAP GRM. We find consistent results from exome sequence data, where we estimate that in the ‡Khomani. We then used the familial relationships (Figure S1) and population-level endogamy to estimate heritability from IBD sharing among all individuals in the ‡Khomani and Nama; we obtain a similar estimate of (Methods, see also (Zaitlen et al., 2013)).
We contrast the high heritability estimates for baseline pigmentation with estimates for tanning status. Tanning status is significantly associated with both sex (male β=6.2 increase in M index, p = 4.2e-4) and age (β=0.18 increase in M index per year, p=1.8e-4), but not with admixture proportions. None of the tanning status h2 estimates, including pedigree-, IBD-, exome-, and SNP array-based estimates, are significantly greater than 0 (Table 1), consistent with previous observations that tanning status is largely environmentally determined by UV exposure (Clark et al., 1981; Nan et al., 2009). The stark contrast of the baseline pigmentation and tanning status heritability estimates, and the consistency of h2 across methods indicates that our high baseline pigmentation heritability estimates do not simply arise from pedigree and population structure and that socioeconomic factors are unlikely to have significant effect on our heritability estimates.
A Complex Genetic Architecture in the KhoeSan
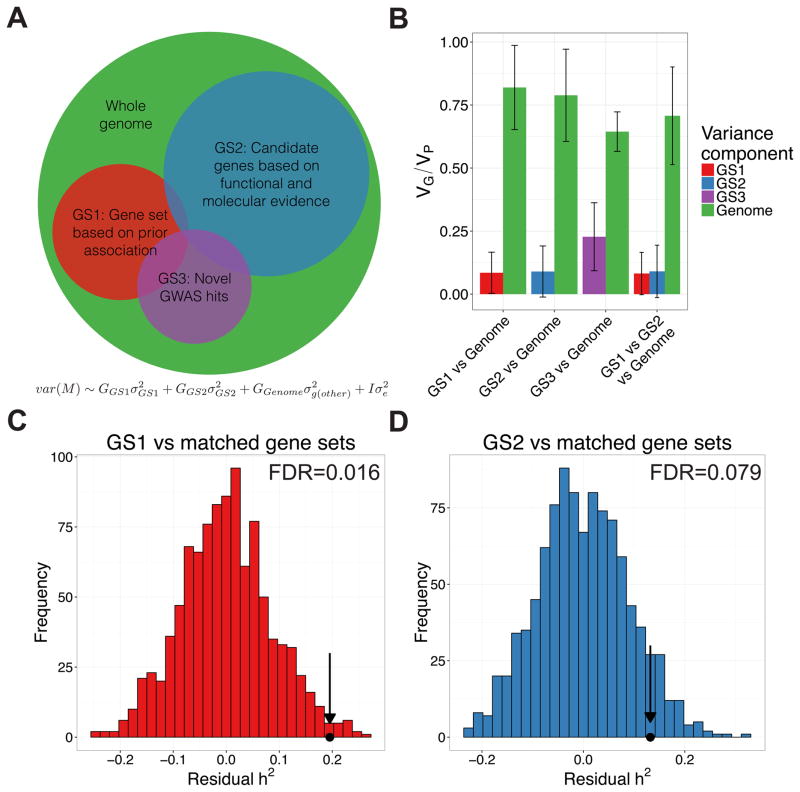
The genetic architecture of skin pigmentation has been described as simpler than many other phenotypes, for which only a few genes explain ~35% of the total variation in a given population, and average genomic ancestry explains an additional ~44% of the variation, indicating a long tail of smaller effects (Beleza et al., 2013; Candille et al., 2012). We investigated how much of the heritable variation in KhoeSan populations can be ascribed to previously annotated pigmentation gene sets (Figure 3A). The first gene set (GS1) consists of 14 genes containing or near previously discovered skin pigmentation genetic associations in Europeans, East Asians, Cape Verdeans, and Native Americans (Table 2 and Table S6A). The larger, second gene set (GS2) contains 50 genes compiled previously (Beleza et al., 2013) from human pigmentation associations, positive selection scans, and model organism pigmentation loci. The third gene set (GS3) contained 50 loci most significantly associated with pigmentation in the KhoeSan (phase 1, see novel variants section below). We partitioned the genome into GS1, GS2, GS3, and the rest of the genome and performed four comparisons, computing the variance explained by: GS1 versus the rest of the genome, GS2 versus the rest of the genome, GS3 versus the rest of the genome, and GS1 versus GS2 versus the rest of the genome. For each comparison, we performed a restricted likelihood ratio test. The GS1 and GS2 gene sets do not explain a significant fraction of the heritability; that is, the heritability estimates overlap with zero. Rather, the remainder of the genome explains the overwhelming majority of the heritability (Figure 3B, σ2GS1=0.08 vs σ2Genome=0.82, pGenome=2.7e-5; σ2GS2=0.09 vs σ2Genome=0.79, pGenome=3.3e-4; and σ2GS1=0.08 vs σ2GS2=0.09 vs σ2Genome=0.71, pGenome=2.5e-3, respectively). This result contrasts with conclusions from previous studies and indicates that the vast majority of variation in KhoeSan skin pigmentation arises from pigmentation genes yet to be discovered, providing strong evidence for a complex, polygenic architecture. GS3 explains a small but significant fraction of the heritability, as discussed below.
Figure 3. Partitioned heritability across known and novel gene sets.
Heritable variation in KhoeSan pigmentation is partially explained by previously associated loci, newly associated loci, and candidate genes discovered in divergence studies of other populations, and in animal models. A) Schema illustrating how heritability analyses were used to partition the phenotypic variance explained by candidate gene sets (GS1, GS2) and novel associations (GS3) compared to the rest of the genome. B) Variance components analysis in GCTA comparing pigmentation variability explained by GS1, GS2, and the rest of the genome. Error bars span ± 1 standard error. C) Heritability explained by estimated value observed in our data (dot and arrow) versus matched null distribution in the ‡Khomani and Nama after accounting for number of SNPs in GS1 gene sets containing 14 genes previously associated with skin pigmentation in other populations. D) As in C), where GS2 = gene set from Table S4 of (Beleza et al., 2013) compiled based on pigmentation function. See also Figure S3.
We further assessed whether GS1 and GS2 explain more of the heritable variation than a random sample of coding regions; genes tend to explain more phenotypic variation than noncoding regions (Gusev et al., 2014). After regressing out the effect of variable numbers of SNPs per gene set (STAR Methods), we find that both GS1 and GS2 explain more than random genes with a 10% false discovery rate (FDR=0.016 and FDR = 0.079, Figure 3C–D, respectively) across both KhoeSan populations. This is not significant in the Nama alone (Figure S3), likely because of ancestry heterogeneity between the two populations.
Replication of Known Pigmentation Associations in the KhoeSan
Even though previously identified pigmentation loci explain little of the phenotypic variance in our samples, it is possible that these loci simply have small effect sizes in the KhoeSan. We used SNP array and/or resequencing data in a linear mixed model with ancestry covariates (see Methods and “Novel Variants Influence Skin Pigmentation in KhoeSan Populations”) to assess both the frequencies and effect sizes of 42 previously identified eye, skin, and hair pigmentation variants, some of which have been experimentally shown to be causal (Table 2 and Table S6A). To this end, we also deconvolved recent admixture into local ancestry tracts across the genome and estimated the allele frequencies specifically on KhoeSan haplotypes via expectation-maximization. Known pigmentation allele frequencies vary considerably between the ‡Khomani San, Europeans, and West Africans (Table 2). However, most previously identified pigmentation associations do not replicate with genome-wide significance or nominally in the ‡Khomani and Nama, with a few exceptions (STAR Methods).
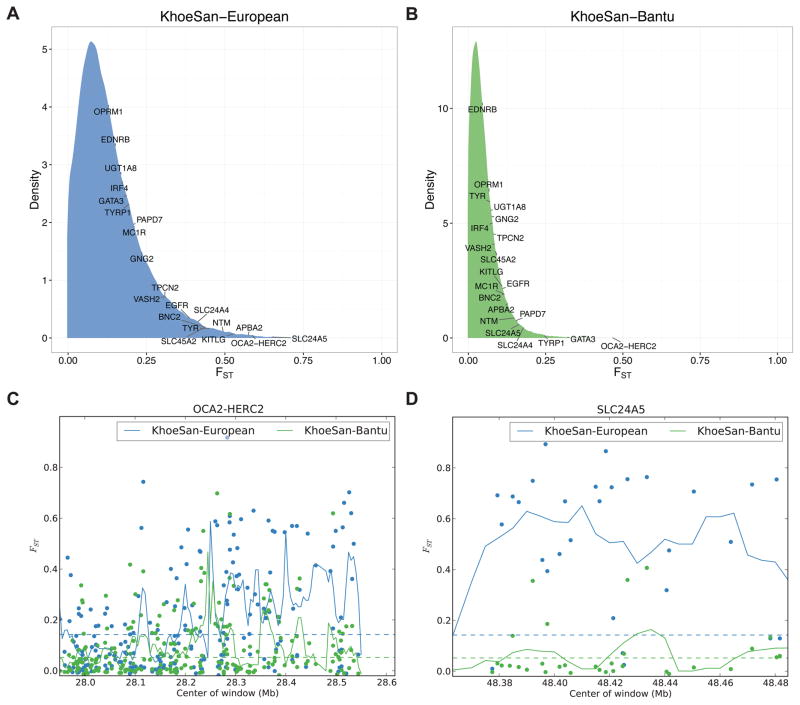
Because high divergence in a segment of the genome can be a signature of selection (e.g. XP-EHH scans), we assessed genetic divergence between KhoeSan, West African, and European populations at SNPs and in sliding windows across the genome. We find considerable divergence in many canonical pigmentation genes when comparing regions of the genome across populations (Figure 4A–B). We followed up our divergence scan by focusing on two outlier genes that were highly diverged among all three populations: SLC24A5 and OCA2 (Figure 4). The divergence in SLC24A5 is among the highest in the genome, especially between the KhoeSan and European populations (Figure 4D). Interestingly, different regions of OCA2 exhibit elevated divergence between the KhoeSan and European comparison versus the KhoeSan and West African comparison (Figure 4C). A previous study suggested that the derived, synonymous T allele of rs1800404 in OCA2 has been positively selected and is a candidate skin pigmentation variant conferring light skin in Europeans and KhoeSan populations based on its global allele frequency distribution (Norton et al., 2006). We confirm its elevated allele frequency on KhoeSan haplotypes (65%), but do not find an association with skin pigmentation (p=0.53). Variants in OCA2 explain most of the variation in human eye color (Duffy et al., 2007), and rs1800404 was later significantly associated with this phenotype (Eriksson et al., 2010); ‡Khomani and Nama individuals notably have heterogeneous eye color, with a range of brown, hazel, and green eyes. We identified a missense mutation in OCA2 (rs1800417, ns with skin pigmentation: p=0.87) with a derived allele (G) frequency of 0.32 in the KhoeSan (Table S6A) that is at low frequency in all other populations surveyed (global allele frequency = 0.016 in Phase 3 1000 Genomes and 0.0058 in the Exome Aggregation Consortium, ExAC).
Figure 4. Genetic divergence in genes previously associated with pigmentation.
A–B) Distribution of weighted FST in 20 kb moving windows of SNPs across the genome with a step size of 5 kb. Labels indicate where the maximal FST window from each canonical pigmentation gene lies in the distribution. Divergence depicted is between A) the KhoeSan and Europeans, and B) the KhoeSan and West African populations. C–D) FST in canonical pigmentation genes. Dots indicate SNPs, lines indicate moving averages over 20 kb windows with a step size of 5 kb. Canonical pigmentation loci/genes are shown as: C) the OCA2-HERC2 locus, and D) the SLC24A5 gene.
Novel Variants Influence Skin Pigmentation in KhoeSan Populations
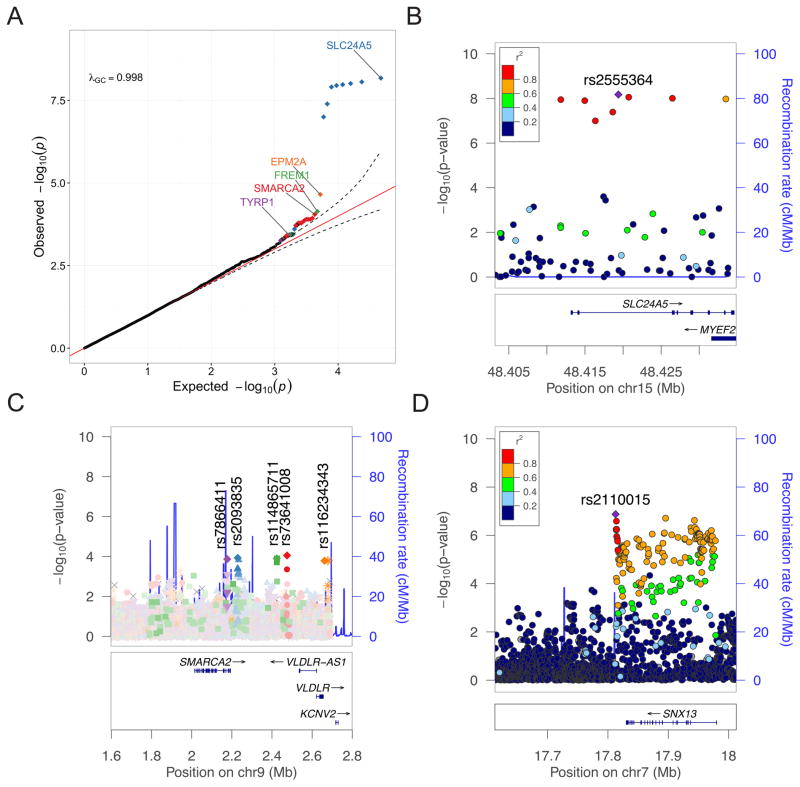
To identify novel variants associated with skin pigmentation in the ‡Khomani and Nama, we performed a 2-stage study (Figure S6A), employing a linear mixed model approach including recent admixture covariates as fixed effects and covariance matrices adjusted for admixture (akin to a GRM in GCTA) as random effects to identify associations between pigmentation and high quality imputed variants. We assessed the quality of the imputation via homozygous reference, heterozygous, and homozygous non-reference concordance with high coverage exome sequencing data (Figure S4A). We ran the initial GWAS (i.e. phase 1) with imputed variants from 107 ‡Khomani and 109 Nama individuals (Table S4, Table S5, Figure S6A–C), and the genes closest to the strongest associations (Table S5) showed a significant enrichment in multiple mammalian phenotypes related to skin pigmentation (abnormal extracutaneous pigmentation p=2.3e-3, abnormal melanocyte morphology p=5.8e-3, abnormal skin morphology p=3.5e-2). Further, the strongest signals across the genotyped ‡Khomani and Nama cohorts were near canonical pigmentation genes (e.g. TYRP1 and SLC24A5), genes associated with pigmentation-related disorders (e.g. TYRP1), or genes implicated in pigmentation in model organisms and in vitro studies (e.g. VLDLR, SMARCA2, and others) (Sturm, 2009) (Keenen et al., 2009a; Xia et al., 2013a). To assess the variation explained by the most significantly associated loci, we generated an additional gene set, referred to as “GS3”, using the 50 most significantly associated loci ± 10 kb. We find that the GS3 loci explain significantly more of the heritable variation in skin pigmentation than previously identified pigmentation candidate genes in the KhoeSan, but that the majority of heritable variation remains to be explained (Figure 3B, σ2GS3=0.23±0.13, pGS3=0.027 vs σ2Genome=0.64±0.08, pGenome<1e-5).
Based on initial evidence from the imputed ‡Khomani pigmentation GWAS, we designed a targeted NGS capture and successfully resequenced 36 candidate pigmentation regions (Table S7, Figure S6) across a larger set of 451 KhoeSan samples in order to improve power to detect associated loci (Table S8, STAR Methods), including 269 ‡Khomani, and 182 Nama individuals. In this larger sample, we observe more variants significantly associated with pigmentation than expected by chance in the resequencing regions (Figure 5A). The strongest signal comes from SNPs in SLC24A5, 8 of which are all in high pairwise LD (R2 > 0.6) on a high frequency haplotype (Figure 5B). We identify significant associations between lighter skin and derived SLC24A5 SNPs, including the putatively causal p.Thr111Ala rs1426654 allele (β=−3.58 on M index scale, p=9.8e-9), which has previously been associated with skin pigmentation in Eurasians. The most strongly associated SNP (rs2555364, β=−3.58 on M index scale, p=6.7e-9) is tightly linked with rs1426654 (LD R2=0.81). These variants are strongly differentiated between Europeans and Africans, with rs1426654 having derived allele frequencies of 99.7% vs 5.5% in 1000 Genomes (excluding ASW and ACB populations with recent European admixture), respectively. The derived allele of rs1426654 has previously been observed in HGDP Ju|’hoansi San samples at 7% frequency, which have no detectable recent European admixture (Norton et al., 2007). The frequency of the derived rs1426654 allele is 40% in the combined Nama + ‡Khomani dataset, which is significantly greater than expected from ~11% European admixture alone (binomial test p=7.8e-52, Table S6A, STAR Methods).
Figure 5. Associations between genetic data and baseline pigmentation.
A) Targeted resequencing QQ plot. 95% confidence interval on the QQ plot is drawn assuming the jth order statistic from a uniform sample follows a Beta(j, n − j + 1) distribution. Colors differentiate loci containing more than one variant associated more significantly than the 95% confidence interval in a region. B–C) LocusZoom plots of targeted resequencing genetic associations incorporating KhoeSan-specific LD. Recombination rates are from HapMap b37. Regions include: B) SLC24A5, and C) 5 independent signals associated with p < 1e-3 in/near SMARCA2 and VLDLR. D) LocusZoom plot of suggestive association in/near SNX13 from meta-analysis of phase 1 and phase2 imputed associations with KhoeSan-specific LD. See also Figure S6, Table S5, Table S7, and Table S6B.
Multiple low frequency (<5%) SNPs near several additional genes, including EPM2A, FREM1, SMARCA2/VLDLR, and TYRP1, are above the 95% confidence interval of expected versus observed significance (Figure 5). Two of these loci are near EPM2A and FREM1, neither of which have any known role in skin pigmentation in humans or model organisms. In contrast, there are >5 independent low frequency signals with p<1e-3 within/near SMARCA2 and VLDLR, with rs7866411 (p=8.91e-5) and rs2093835 (p=1.17e-4) being most significantly associated with skin pigmentation. We used HaploReg to infer regulatory activity in/near these peaks and identify multiple enhancer and DNAse peaks identified in skin, including melanocytes and/or keratinocytes, overlapping top tag and/or perfectly linked SNPs (Table S6B). We also identify a low frequency association (rs34803545, p=3.7e-4) ~600 kb upstream of TYRP1. This variant is perfectly linked with multiple conserved variants, one of which exhibits enhancer activity and DNAse hypersensitivity specifically in skin (Table S6B).
We performed a 2nd phase of GWAS, in which an additional 240 individuals were genotyped (Table S4, Figure S6A) and meta-analyzed with phase 1 summary statistics. While two tanning status associations met genome-wide significance, none of the loci contained linkage peaks, suggesting that they are most likely spurious as expected from a phenotype with low heritability. As expected from the resequencing study, we identified a genome-wide significant association in SLC24A5 (rs2470102 derived allele β = −3.4, p=3.6e-12) and a suggestive association upstream of TYRP1 (chr9:12088112, frequency=0.014, β = −13.6, p=1.1e-07 Figure S6B–C, Figure S6F–G). We identified an additional suggestive novel association in and near SNX13, with common derived T alleles of rs2110015 associated with light skin (β = −3.1, p=1.3e-07, Figure S6H); SNX13 regulates lysosomal degradation and G-protein signaling, but has not previously been associated with skin pigmentation.
Discussion
Pigmentation has been described previously as a relatively simple trait with few loci of large effect contributing to the phenotype (Sulem et al., 2007; Walsh et al., 2013). However, populations living in continental Africa, where humans have the greatest genetic diversity and pigmentation variability, have been largely ignored in genetic studies with quantitative phenotypes. We investigated the genetic architecture of pigmentation in two KhoeSan populations: the ‡Khomani San and Nama, where baseline melanin variation is substantial. Southern African KhoeSan populations are the most polymorphic modern human populations yet studied (Henn et al., 2011), and provide a unique glimpse into the evolution of pigmentation.
Novel Genetic Associations with Pigmentation
We have performed the first genetic discovery effort for pigmentation loci in the Nama and ‡Khomani San populations. The strongest allelic associations include previously associated variants, noncoding regions near canonical pigmentation genes, and novel genes shown in model organisms to have a role in pigmentation. The strongest association is in SLC24A5, which is a well-known pigmentation gene (Lamason, 2005) and is among the most differentiated regions of the genome between European and African populations–indicative of strong positive selection in northern Europeans (Sturm and Duffy, 2012). We find that derived variants in SLC24A5 are at high frequency in the KhoeSan, including missense mutations that influence skin and eye pigmentation (Table 2). Notably, these variants are segregating at higher frequency than expected by recent European admixture alone. Three possible evolutionary scenarios that may explain these elevated frequencies are: 1) these variants arose in southern Africa more than 100,000 years ago and were later selected for in Europeans after the out-of-Africa migration in response to northern UVR environments. Alternatively, 2) these variants arose in Europe/Near East, were introduced into KhoeSan populations via “back to Africa” migration into southern Africa predating 17th century European colonialism (Pickrell et al., 2014; Uren et al., 2016a), and have since been positively selected in the KhoeSan. Lastly, 3) a recurrent mutation (G to A transition at the CpG ancestral dinucleotide, a class of mutations shown to have elevated mutation rates) occurred. Considerable future work is needed to definitively disentangle these scenarios.
The Polygenic Architecture of Pigmentation in Africa
We assessed the heritability of baseline skin pigmentation, and find that it is virtually completely heritable in our KhoeSan sample. In contrast, tanning status is primarily environmental, with heritability estimates which are not significantly different from zero. In European populations, predictive models based on only 9 SNPs capture up to 16% of the variance in skin pigmentation (Liu et al., 2015), highlighting its relative simplicity. We applied a predictive model based on these SNPs to the Nama and ‡Khomani San populations, and find no significant association between predicted skin color and spectrophotometrically measured skin M index, showing that this estimation fails to capture the genetic variation driving the phenotype in the KhoeSan. Given the large effect sizes and high fraction of variation explained in Eurasian populations, we asked whether and how much of the phenotypic variation can be explained by previously identified genes. All gene sets, including previously associated loci, canonical pigmentation genes, and the most significantly associated variants in this study, explained a small fraction of the phenotypic variance (σ2GS1=0.08, σ2GS2=0.09, σ2GS3=0.23, respectively). As expected from previous work (Martin et al., 2017), our results indicate that genetic risk prediction is strongly affected by population structure. Most of the pigmentation variability in KhoeSan populations is not explained by previously identified loci, suggesting that more than 50 loci (and indeed, likely far more, given our genomic heritability estimates) with a distribution of mostly small effects contribute to variation in pigmentation in the KhoeSan. This suggests that skin pigmentation is a far more complex trait than previously discussed, analogous to numerous other complex traits discussed in biomedical literature.
The Evolution of Skin Pigmentation: Selection and Constraint
By aggregating a large set of quantitative skin pigmentation phenotypes (N=4,712) from globally diverse populations, we have demonstrated heteroskedasticity as a function of latitude. As observed previously, we find a strong correlation between absolute latitude and average skin pigmentation reflectance caused by melanin content. We also observe that populations with lighter skin have reduced variation within any given study: populations furthest from the equator have narrower distributions, while populations closest to the equator have wider distributions. These patterns suggest that selection is acting differently at different latitudes. In equatorial regions, strong directional selection for darker pigmentation has shifted the distribution means in some populations to M indices greater than 90, but with wide variances. This is consistent with a ‘threshold’ model (Chaplin, 2004) in which the protective benefit of melanin needs to meet some minimum threshold but with no penalty to darker pigmentation; alternatively, diversifying selection could maintain the wide variance.
In stark contrast, pigmentation in far northern European and Asian populations has been under directional selection for decreased melanin production, reflected by very narrow distributions. There may be biological constraints on the lower boundary of skin pigmentation, and/or due to the strong positive selection acting on a few large-effect alleles, there is little genetic variability left at these pigmentation loci. This would simplify the genomic architecture, with relatively few alleles of large effect driving the phenotype, particularly alleles that lighten skin at extreme northern latitudes, and could explain why prior investigations observed an almost Mendelian inheritance of large effect light pigmentation alleles.
Finally, populations at intermediate latitudes have increased variance and higher means than populations in northern Eurasia, but less than equatorial populations. The most parsimonious explanation for this pattern is that stabilizing selection affects the light and dark tails of the pigmentation distribution (Barton, 1999). The Nama and ‡Khomani San appear to be two such instances of this intermediate variation within Africa, likely attributable to their geographic distance from the equator in far southern Africa (~24–29 degrees South). The observed mean and variance differences across the full spectrum of skin pigmentation by latitude may be driven by imbalanced opposing adaptive pressures, where selective forces to produce vitamin D and protect folate from photolysis are unequal and change in response to UV radiation exposure. Given our heritability results and the observed variability in baseline pigmentation; light skin pigmentation in the KhoeSan appears to be due to a combination of many small-effect mutations as well as some large-effect variants. The evolution of the pigmentation phenotype in these populations cannot be explained in terms of only a few variants segregating in Eurasians. A fuller characterization of the genes underlying the architecture in Africans is needed before we can distinguish between the hypothesis of directional versus stabilizing selection across different latitudes (Berg and Coop, 2014).
Conclusion
Because African populations often carry the ancestral (i.e. dark) allele for skin pigmentation genes identified in Eurasians, allusions to African skin pigmentation have ignored the great variability in this phenotype across Africa. Here, we reiterate that skin pigmentation varies more in Africa than any other continent, and we show that pigmentation in African populations cannot simply be explained by the small number of large effect alleles discovered in Eurasians. Even in light to moderately pigmented KhoeSan populations, the polygenicity of skin pigmentation is much greater than Eurasians, encompassing both known pigmentation genes as well as novel loci. We argue that the distributions of skin pigmentation globally suggest different forces of selection operating at various latitudes. To better understand baseline pigmentation, one of the most rapidly-evolving traits and strongest cases for positive selection in humans, it is essential to quantitatively measure and study pigmentation in a large set of genetically diverged populations that have historically been exposed to different levels of UV radiation. As human genetics moves to ever larger studies of complex traits, the full picture of genetic architecture will remain incomplete without representation from diverse worldwide populations.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Brenna Henn (brenna.henn@stonybrook.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Sample collection and ethics approval
As described previously (Henn et al., 2011; Uren et al., 2016b), sampling of the ‡Khomani San took place in the Northern Cape of South Africa in the southern Kalahari Desert region (near Upington and neighboring villages) in 2006, 2010, 2011, 2013, and 2015. Sampling of the Nama took place in the Richtersveld in 2014 and 2015. Institutional review board (IRB) approval was obtained from Stanford University, Stony Brook University, and the University of Stellenbosch, South Africa. ‡Khomani N|u-speaking individuals, Nama individuals, local community leaders, traditional leaders, nonprofit organizations, and a legal counselor were all consulted regarding the aims of the research before collection of DNA (Henn et al., 2011). Research was conducted with the permission of the Working Group of Indigenous Minorities in Southern Africa (WIMSA) and, subsequently, the South African San Council. All individuals gave signed written and verbal consent with a witness present before participating. Individuals collected in 2006 were re-consented under an updated protocol. Ethnographic interviews of all individuals were conducted, including questions about age, language, place of birth, and ethnic group of the individual and of his/her mother, maternal grandparents, father, and paternal grandparents. All individuals included in the study were adults (age range of 18 to 94, mean=53, sd=18). Both men and women were included in the study (311 females, 190 males). We recorded the relationships between any sampled individuals if revealed during the interview. Ages of older individuals were verified with separate interviews regarding reproductive history. DNA was obtained via saliva, collected using Oragene saliva collection kits (DNAGenotek, Ontario, Canada).
METHOD DETAILS
Skin reflectance measurements
A portable reflectance spectrophotometer (DermaSpectrometer DSMII ColorMeter, Cortex Technology, Hadsund, Denmark) was used to measure skin pigmentation. Similar devices have previously been shown to measure melanin and hemoglobin (Diffey et al., 1984). The melanin content, M index is quantified as
The device was calibrated to 0 as suggested by the manufacturer twice a day while sampling. Five measurements of M index were taken on each of the left and right upper inner arms to approximate baseline constitutive skin pigmentation. We also measured the dorsal side of the left or right wrist (i.e. an area exposed to sunlight) and subtracted the baseline pigmentation for a measure of tanning status (Shriver and Parra, 2000). For the remainder of the analyses, we used the trimmed phenotype means (highest and lowest values removed); we also averaged the inner arm skin pigmentation measurements over the two arms.
Genotyping platforms
A total of 471 KhoeSan samples were genotyped across all arrays, including the Illumina 550k array, Illumina OmniExpress and OmniExpressPlus arrays, Illumina Omni2.5 array, and the Illumina MEGA array, some of which has been described individually previously. 35 ‡Khomani, 21 Hadza, and 35 Sandawe individuals were previously genotyped on the Illumina Beadchip 550K custom v2 chip (Henn et al., 2011). 86 ‡Khomani and 13 Nama were genotyped on the Illumina OmniExpress and OmniExpressPlus arrays (same base content, additional exome content in the Plus version of the array) (Uren et al., 2016b). 105 Nama individuals were genotyped on the Illumina Omni2.5 array as part of the African Genome Diversity Project. 185 ‡Khomani and 84 Nama individuals were genotyped on the Illumina MEGA array. Table S4 indicates the number of samples genotyped on each array platform, as well as the overlapping phenotypes, exome sequencing, and targeted resequencing. A small number of individuals overlapped between multiple arrays to perform QC.
Global ancestry estimation
We derived two sets of genome-wide ancestry estimates: one from a smaller set of genotyping array data and one from a larger set of samples with targeted resequencing data. For the former set of estimates, we included genotype data from the Human Genome Diversity Project (CEPH-HGDP, sample sizes in parentheses) as reference samples, including the South African Bantu (8), Kenyan Bantu (11), Namibian San (6), Mozabites (29), and French (28). We also included genotype data from 12 Namibian San individuals from Schuster et al (2010), as well as individuals from the Hadza (17) and Sandawe (28) of Tanzania, described by Henn et al (2011). We also included individuals genotyped in the HapMap Project, including Yoruba trio parents from Ibadan, Nigeria (YRI, 55), Centre d’Etude du Polymorphisme Humain (CEPH) Utah residents with ancestry from northern and western Europe (CEU) trio parents (86), and Maasai trio parents from Kinyawa, Kenya (MKK, 30) individuals (Altshuler et al., 2010). Because of the high degree of relatedness in our dataset, we then split the merged ‡Khomani and Nama data into 11 groups of maximally unrelated KhoeSan individuals from this study based on ethnographic information, then merged in these samples as well, holding the reference panels constant. After merging the SNPs genotyped in the HapMap, CEPH-HGDP, and South African samples, and removing SNPs with any genotype missingness using PLINK 2, SNPs with minor allele frequency <1%, and SNPs in high LD (r2 > 0.9) a total of 215,607 SNPs remained. All datasets were merged to Human Genome Build hg19 as above and dbSNP v138. We ran ADMIXTURE for k = 3–7 in unsupervised mode for each of the 11 groups, then matched clusters across runs. In runs where we identified multimodality, we further split running groups of KhoeSan samples resulting in a minor mode into two sets, which resulted in unimodality across all runs. After matching clusters, we merged ancestry estimates across all 11 running groups, averaging individuals that appeared in multiple running groups. We chose k=7 as the most stable and best representation of ancestry.
For the larger resequencing data, we extracted sequence data in the targeted resequencing intervals (Table S7) for 99 CEU and 99 LWK samples from the 1000 Genomes Project bam files and generated gVCF files, then called variants jointly with HaplotypeCaller. We included unrelated KhoeSan samples estimated to have >90% KhoeSan ancestry from the genotype-based ancestry estimates as reference samples, then ran ADMIXTURE in supervised mode, projecting related and more admixed samples.
Using the same data, we also estimated ancestry estimates using PCA. Because of the elevated relatedness and admixture in our data, we applied the PC-AiR and PC-Relate approaches (Conomos et al., 2016).
Covariates
We performed forward stepwise regression using custom scripts in R to select the best multivariate mixed model of ancestry, age, and sex for pigmentation and tanning with a random effect accounting for the genetic relationships among individuals. Sex and age do not significantly correlate with baseline skin pigmentation, suggesting that our quantitative measure of underarm reflectance is not significantly affected by UV exposure. The best model fit, measured via AIC, included Bantu, European, East African, and Hadza ancestries, although the latter two components comprise ≤ 1% of individuals’ total ancestry on average and are likely imprecise.
Identity-by-descent (IBD) haplotype sharing
To estimate IBD, we phased intersected genotypes for the ‡Khomani and Nama populations both separately (number of SNPs = 300,370 in ‡Khomani, 525,934 in Nama) and jointly (number of SNPs = 241,929) using Beagle (v4.1) (Browning and Browning, 2007). Adjusting for differences in SNP density, we used a sliding window size of 600 markers with 55 overlapping SNPs between each window for the ‡Khomani, a window size of 1000 markers with 90 overlapping SNPs between each window for the Nama, and a window size of 400 markers with 39 overlapping SNPs between each window for the joint ‡Khomani and Nama intersection, with 10 iterations per run. The phased data was then used to infer haplotypes shared via IBD with length ≥ 5 cM using Germline (v.1.5.1) with the following flags for the joint haplotype calls: “-w_extend -min_m 5 -err_hom 2 -err_het 5 -bits 60” (Gusev et al., 2009). This allowed a mismatch of 2 homozygous and 5 heterozygous markers. We verified the total genomic length of the inferred cumulative IBD between pairs of individuals by comparing to pedigree relationships identified from ethnographic interviews and verified with IBD inferred here from the genotyping arrays.
Covariance matrices
To account for the considerable relatedness in our samples, which have variable degrees of admixture, we evaluated multiple covariance matrices: a Balding-Nichols matrix computed via EMMAX (Kang et al., 2010), a genetic relationship matrix (GRM) computed via GCTA (Yang et al., 2010), and a kinship matrix computed via REAP (Thornton et al., 2012b). To generate the REAP matrix, we intersected genotype data for all individuals, then included P and Q matrices obtained from an ADMIXTURE run with k=3 (described in Ancestry estimation) to construct the ancestry-corrected kinship matrix. Briefly, this approach uses individual-specific allele frequencies at SNPs that are calculated on the basis of genome-wide ancestry. We compared inferred pairwise kinship values in all covariance matrices to ethnographically and genetically validated pedigree information. We used the REAP matrix to correct for kinship in all regression models unless otherwise noted because it correlated best with true relationships (e.g. heritability analyses with different kinship matrices).
We also constructed a kinship matrix using pairwise IBD estimates (KIBD), as previously (Zaitlen et al., 2013), with haplotypes sharing calls computed as described above. We constructed a kinship covariance matrix based on IBD (KIBD), where the entry for individuals j and k are defined as follows:
where Li is the genetic length in centimorgans of ith IBD segment between individual j and k, and Lparent–offspring is the total length of IBD in centimorgans shared between a parent and an offspring (i.e. the callable length of the haploid genome).
Heritability
Heritability estimates were calculated across the full KhoeSan sample (Nama and ‡Khomani) in addition to within each population separately. We used GCTA restricted maximum likelihood (REML) analysis to compute SNP-based heritability ( ) in multiple ways with differing covariance matrices. For all heritability analyses of baseline pigmentation, we included European and Bantu ancestry proportion estimates at k=7 from ADMIXTURE as quantitative covariates. For tanning status, we included age as a quantitative covariate and sex as a binary covariate. We assessed by fitting an unconstrained linear mixed model (--reml-no-constrain) in GCTA (Yang et al., 2010), once using a covariance matrix constructed with REAP, and once with a genetic relationship matrix (GRM) generated in GCTA. We estimated heritability from the exome data ( ) similarly using a GRM generated in GCTA with exome sequencing data from 82 ‡Khomani individuals. We also estimated heritability ( ) in our study using a kinship matrix constructed from pairwise IBD estimates (KIBD), as previously (Zaitlen et al., 2013). Lastly, we estimated narrow sense heritability using pedigree relationships ( ) that were constructed from ethnographic interviews and subsequently genetically confirmed using the Sequential Oligogenic Linkage Analysis Routines (SOLAR) software (Almasy and Blangero, 1998). SOLAR employs maximum likelihood variance decomposition to determine narrow-sense h2 assuming a normal distribution. SOLAR employs maximum likelihood variance decomposition to determine narrow-sense h2 assuming a normal distribution. It calculates heritability utilizing pairwise coefficients of genetic relatedness in the full pedigree, including dummy link individuals. The “polygenic” command was used to calculate trait polygenic heritability, significance of , and the proportion of variance contributed by covariates, with the “screen” flag to assess the significance level of each covariate.
Variance partitioning
We partitioned heritability in two ways: 1) by comparing the heritability explained by candidate gene sets versus the rest of the genome, and 2) by comparing the heritability explained by candidate gene sets to randomly sampled genes. For the first type of analysis, we generated GRMs based on SNPs that fall within pigmentation candidate gene sets, including GS1 (genes in Table 2), GS2 (Table S4 from (Beleza et al., 2013)), GS3, and the rest of the genome. We performed a restricted likelihood ratio tests comparing the heritability explained by each gene set to the rest of the genome. We estimated partitioned heritability explained by different gene sets using joint linear mixed models, by including multiple genetic variance components as random effects.
For the second type of analysis, we sought to determine how likely we are to find a candidate pigmentation gene set explaining more of the heritable variation than a random gene set. To do this, we matched both candidate gene sets by number of genes, length, and number of exons and permuted these matched samples 1000 times. Specifically, we generated a GRM in GCTA based on SNPs in these candidate genes, then calculated the heritability based on this GRM. Then, we binned all genes in the genome by the natural log of their lengths (absolute value of transcription end - transcription start) and number of exons. For each gene in the gene set, we sampled with replacement from its matched length and exon bin, and constructed 1000 matched gene sets. To create an empirical null distribution, for each of the 1000 matched gene sets, we constructed a GRM, and computed heritability. We then regressed out the effect of number of SNPs on heritability explained, then generated empirical false discovery rates by comparing the residual heritability of the true candidate gene set to the residual heritability of the matched empirical null distribution.
Categorical pigmentation prediction
As described previously (Hart et al., 2013b; Spichenok et al., 2011), published categorical skin color prediction models utilize 7 SNPs (rs12913832, rs1545397, rs16891982, rs1426654, rs885479, rs6119471, rs12203592) in or nearby pigmentation genes. The model follows a bifurcating decision tree, dependent on homozygous state at each locus. At any two loci except rs6119471, if both are homozygous derived, then the phenotype is predicted as “non-dark”, i.e. medium or light. Further “light” pigmentation is confirmed if all three loci: rs12913832, rs16891982, and rs1426654 are homozygous derived. A “non-light”, i.e. medium or dark, is predicted if rs6119471 is homozygous ancestral (Figure S6).
Replication of known pigmentation loci
Few known loci replicate with genome-wide significance or even marginally in the KhoeSan populations studied here. Four SNPs in the genes SLC45A2 (rs16891982, p=1.2e-3), KITLG (rs12821256, p=0.02), and SLC24A5 (rs1426654, p=9.8e-9 and rs2470102, p=1.1e-8) marginally replicate in the ‡Khomani + Nama under an additive model. The derived allele frequencies of the associated SNPs in SLC45A2 and KITLG are low in the KhoeSan, consistent with ~10% admixture from recent European gene flow. Interestingly, however, SNPs in OCA2, SLC24A5 and GRM5/TYR are at much higher frequencies in both the ‡Khomani and Nama than expected from European admixture alone, as estimated from global ancestry (see “Global ancestry estimation”). We do not replicate the vast majority of previously observed skin pigmentation associations in our dataset, potentially due to low frequencies in the KhoeSan, power limitations, differentiated LD structure in which the tag SNPs are non-causal pigmentation alleles, or epistatic effects. It is therefore unsurprising that when we applied forensic models based on only seven SNPs that claim very high prediction accuracy of skin color across populations (>99%) (Hart et al., 2013b; Spichenok et al., 2011), we did not find a significant association with quantitatively measured M index (p=0.31, Figure S5B).
Phasing and imputation
We first ensured uniform SNP IDs by orienting all variants to dbSNP 138, then merged genotype data for KhoeSan individuals across all genotyping platforms (Illumina 550k, OmniExpress, OmniExpressPlus, and Omni2.5). We then phased all ‡Khomani (N=121) and Nama (N=112) individuals together with Shapeit2 (v2.r778) using all available genetic data. We used the full Phase 3 1000 Genomes reference panel, consisting of haplotypes from 2,535 individuals to aid phasing accuracy. Shapeit imputes missing genotypes, which can result in array-specific technical artifacts. We mitigated technical artifacts from individuals genotyped on different arrays by subsetting haplotypes to variants genotyped only the array, resulting in four sets of haplotypes. We then imputed variants in 5 Mb windows for all 4 sets using the full 1000 Genomes phase 3 reference panel as well as 53 HGDP medium coverage genomes (Henn et al., 2016) with Impute2 (v2.2.2) for all runs. After imputing each array separately, we aggregated the data across windows and runs, including only sites that were imputed with an Impute2 info metric ≥ 0.8 across all sets and subset to sites with MAF ≥ 0.01. We assessed the accuracy of the imputation in three ways. First, we assessed the homozygous reference, heterozygous, and homozygous non-reference concordance between the imputed output and two low-pass genome sequences from individuals SA1000 and SA1025 at sites that passed variant call filters and with > 5 reads. Next, we assessed concordance similarly across all 79 individuals for whom we have both genotype and high coverage exome sequencing data (see Figure S4). Finally, we ran PCA for 100,000 randomly selected imputed sites across all individuals as well as for the maximum number of unrelated individuals to test whether the primary source of aggregated imputed variation arose from technical artifacts or population/familial structure. By investigating the top PCs, we concluded the latter.
Local ancestry inference
To disentangle haplotypes specific to a given ancestry and estimate ancestry-specific allele frequencies, we inferred local ancestry along chromosomes for all the genotyped ‡Khomani (N=121) and Nama (N=112) individuals, as described in (Uren et al., 2016b). We phased haplotypes as described above. As reference panels, we defined separate classes for European, Bantu, and KhoeSan ancestries respectively using CEU, LWK, and ‡Khomani individuals from this study as well as KhoeSan individuals from a previous study (Schuster et al., 2010) with >90% KhoeSan ancestry as inferred via ADMIXTURE (see “Ancestry estimation”). We used RFMix (v1.5.4) to assess local ancestry at sites that intersected between the reference panels using an iterative expectation maximization (EM) approach with 0.2 cM windows, incorporating the reference panel throughout EM iterations and correcting potential phase errors. We used a node size of 5 to deal with class imbalances in our reference panels. For all individuals, we used calls from RFMix at the 1st iteration.
Allele frequency approximation
Because the ‡Khomani San are a recently admixed population, we estimated allele frequencies with consideration to local ancestry calls. We specifically estimated allele frequencies on KhoeSan haplotypes using an expectation maximization approach (Gravel et al., 2013). Briefly, we used Bayes’ Rule to calculate the expected frequency given the observed genotype and diploid local ancestry calls. Ancestral/derived state were determined from great ape genome sequencing, where possible (Prado-Martinez et al., 2013).
Mixed-model association approach
To identify loci significantly associated with baseline skin pigmentation and tanning status, we associated high quality imputed and resequenced SNPs and indels with these pigmentation phenotypes using a linear mixed model, with a covariance matrix of relatedness as a random effect. As with the heritability analyses, we used a covariance matrix constructed using REAP to account for admixture in the construction of the covariance matrix. We included the proportion of European and Bantu ancestry estimated via ADMIXTURE as fixed effect covariates for baseline pigmentation and tanning status, as chosen in forward stepwise regression. We also included age and sex covariates for tanning, which were significantly associated with the phenotype. We performed the association analysis using EMMAX (Kang et al., 2010) for the imputed data and GCTA (Yang et al., 2010) for the resequenced data, as both employ mixed model approaches and readily support different data formats.
Meta-analysis
We performed inverse variance weighted meta-analysis of summary statistics of the phase 1 and phase 2 summary statistics from imputed data using METAL (Willer et al., 2010).
Association enrichment
Using the 50 most significant associations in the imputed dataset, we identified the closest genes using bedtools with gencode v19 gene annotations. We assessed enrichments using enrichR (Chen et al., 2013), which computes enrichment in three ways: 1) the Fisher’s exact test, the standard method implemented in most enrichment analysis tools, assuming a binomial distribution (i.e. presence/absence of a gene in a gene set) and independence of a gene belonging to any set; 2) the deviation from the expected rank by the Fisher’s exact test given many random input gene lists is computed as a z-score, providing a correction to the Fisher’s exact test; and 3) multiplying the log of the p-value from the Fisher’s exact test by the z-score computed in the second test to generate a combined score. We investigated enrichment using the Mouse Genome Informatics (MGI) Mammalian Phenotype ontology rather than the Human Phenotype ontology because pigmentation is highly diverged across populations and has not been studied thoroughly across all populations.
Across all GWAS efforts, we find a significant enrichment of genes related to melanogenesis. Specifically, we find several independent associations near SMARCA2 and VLDLR. SMARCA2 has a known role in folate biosynthesis, in vitamin D-coupled transcription regulation, and is differentially expressed across CEU and YRI populations in lymphoblastoid cell lines (Duan et al., 2009). Additionally, previous functional studies have shown that MITF, the transcription factor known as the “master regulator of melanogenesis” due to its ability to activate many melanocyte-specific genes (Praetorius et al., 2013), recruits critical components of the SWI/SNF chromatin remodeling complex (including SMARCA2), to the promoter region of its targets (Vachtenheim et al., 2010). This recruitment is required for normal expression of many MITF target genes, including TYR, TYRP1, DCT, RAB27A, BCL2, among others (Keenen et al., 2009b). Additionally, VLDLR knockout mice exhibit hypopigmented retinas (Xia et al., 2013b). We also find a suggestive association upstream of TYRP1 (Figure 5A, Figure S6G). TYRP1 mutations in humans have been associated with oculocutaneous albinism and shown to cause nearly Mendelian inheritance of blond hair in Solomon Islanders (Kenny et al., 2012b; Sarangarajan and Boissy, 2001). Thus, we observe enrichments of molecular pathways involved in pigmentation beyond those previously identified as associated with the phenotype in non-African populations.
Exome variant calling and annotation
Illumina sequencing reads from 91 KhoeSan DNA samples (of which 82 had pigmentation phenotypes) were captured with: 74 samples on an Agilent SureSelect Human All Exon V2 44Mb array (2 × 101 bp reads, sequenced at BGI on a HiSeq 2000), 8 samples on an Agilent SureSelect Human All Exon 50Mb array (2 × 101 bp reads, sequenced at BGI on a HiSeq 2000), and 8 samples on an Agilent SureSelect Human All Exon V4+UTRs 71Mb array (2 × 126 bp reads, sequenced at the New York Genome Center on a HiSeq 2500). Sequencing data was processed according to a standard pipeline informed by the 1000 Genomes Project. Briefly, we aligned reads to the hg19 reference genome using bwa-mem 0.7.10. We then sorted bam files and marked duplicate reads with Picard v1.92. We next ran RealignerTargetCreator, IndelRealigner, BaseRecalibrator, PrintReads, HaplotypeCaller, GenotypeGVCFs, and VariantRecalibrator, and ApplyRecalibration with GATK (v3.2.2). During the HaplotypeCaller step, we filtered reads down to the capture regions ± 100 bp of padding. We annotated exomic variants using the Variant Effect Prediction tool (VEP) using Ensembl version 75 annotations, which annotates variants using Gencode v19 gene set annotations. We also annotated loss-of-function variants using LOFTEE (https://github.com/konradjk/loftee). We calculated genotype concordance comparing passing variant calls to the corresponding Illumina 550k, Illumina OmniExpress, and Illumina OmniExpressPlus arrays. On target coverage was calculated using GATK’s DepthofCoverage tool.
Targeted resequencing
For 441 KhoeSan samples (N=269 ‡Khomani and N=172 Nama), we performed targeted resequencing. Older samples had a smaller quantity of DNA, so we first performed whole genome amplification (WGA) for a subset of samples. We chose resequencing targets based on the output of the ‡Khomani GWAS, enriching for strong associations, associations near genes with prior evidence for a role in pigmentation, and regions containing SNPs previously implicated in pigmentation in other populations. We chose 35 regions totaling 7.1 Mb and used the NimbleGen SeqCap EZ Choice Enrichment Kit to enrich for these loci. We barcoded then pooled 96 samples per sequencing run with the Illumina NextSeq.
Resequencing variant calling
Resequencing data was processed in the same way as exome variant calls up until the HaplotypeCaller step. Because there were not enough variants in the 7 Mb capture region to run VQSR, we applied hard filters to as quality control. We removed samples with <10× mean coverage, samples with a ≥8% contamination rate measured by verify BamID, and highly discordant samples (concordance with genotyping array <95%). We also removed variants with <1% allele frequency, sites with >50% missingness, and spanning (<*:DEL>) variants.
Resequencing QC
The resequencing design was successful for all targets, although the regions targeting SNPs previously associated with pigmentation in MC1R and TPCN2 had significantly lower coverage than the other regions. The resequencing efforts yielded large amounts of high-quality data for each pool with on average 84% of reads in target regions, achieving a median depth of coverage of 29× per sample. We compared variant calls and genotypes for samples that were genotyped on an array and achieved an average of > 99% concordance (Figure S7). We discovered a total of 46,429 SNPs and indels with a MAF > 1% that passed our quality control filters, resulting in a Bonferoni threshold of 1.08e-6.
QUANTIFICATION AND STATISTICAL ANALYSIS
All heritability analyses except for the pedigree-based analysis were performed with GCTA. Pedigree-based heritability analysis was performed with SOLAR. All mixed model association analyses with the imputed GWAS data were performed with EMMAX. All mixed model association analyses with resequenced data were performed with GCTA. All other statistical analyses, unless otherwise noted, were performed using R.
DATA AND SOFTWARE AVAILABILITY
Processed data are available here: https://data.mendeley.com/datasets/98mh8z78m3/draft?a=4a3dc606-f854-4190-9f26-e5e07110349e.
According to the newly issued San Code of Research Ethics, as published by the South African San Council, parties should first contact the South African San Council to request data access and submit a project proposal. Following local approval, Dr. Henn will release the appropriate SNP array, exome and/or phenotype data.
Contact Details: admin@sasi.org.za, South African San Institute, 4 Sanda Park, Platfontein Farm, Barkly-West / Kimberley Road, Kimberley, Northern Cape, South Africa
KEY RESOURCES TABLE
Supplementary Material
Figure S1 – Pedigrees inferred from ethnographic information. (Related to Table 1 and Methods) Ethnographically inferred pedigrees for KhoeSan individuals are shown in: A) the ‡Khomani, and B) the Nama. Different shades represent whether the samples have been genotyped. Non-phenotyped individuals are crossed in pedigrees.
Figure S2 – Ancestry estimates in ‡Khomani and Nama samples. (Related to Figure 2) A) Admixture runs across K=3–7 for the ‡Khomani and Nama populations, using Namibian San, Hadza, Sandawe, Maasai, Kenyan Bantu, South African (SA) Bantu, Yoruba, Mozabite, Central Europeans (CEU), and French populations as a reference panel, as in Figure 2A. B) Minor ancestry component associations with M index, displayed with a square root x-axis to elongate the minor contributions. C) Pairwise ancestry component correlations at k=7 from A). Upper triangular matrix shows Pearson’s correlation coefficient between pairwise ancestry estimates. Lower triangular matrix shows scatter plots with ‡Khomani shown in blue and Nama shown in red. D-F) Principal Components Analysis (PCA) biplots for the same SNPs and individuals used in the ADMIXTURE analysis adjusted for relatedness using King and the PC-AiR approach. D) PC1 vs PC2, E) PC3 vs PC4, and F) PC5 vs PC6.
Figure S3 – Partitioned heritability by population. (Related to Figure 3) Proportion of heritable variation in baseline pigmentation explained by true pigmentation gene sets (dots) versus matched null distribution after accounting for number of SNPs in gene sets in two different candidate gene sets. Results are stratified by the combined (comb) data, ‡Khomani (khm), and Nama populations. * indicates FDR < 0.1 (indicated by the top decile of the null distributions). Gene set labels are the same as in Figure 3.
Figure S4 – Imputation and targeted resequencing quality control. (Related to STAR methods) A) Imputation quality was assessed via homozygous reference (HR), heterozygous (Het), and homozygous non-reference (HN) concordance for imputed dosages with high coverage exome sequencing data. Each concordance metric was computed as the ratio of dosages for a particular genotyping class (e.g. HR) in both genotype and exome datasets to the dosages of that total class in the genotype dataset with any call in the exomes. The colors indicate the different genotyping arrays, with the 550k array in red, OmniExpress array in blue, and OmniExpressPlus array in green. B–E) Targeted resequencing quality control. B) Depth of coverage versus array concordance. Dashed line at 10× indicates minimum depth accepted for a sample’s inclusion. C) Depth of coverage by targeted resequencing region. D) Resequencing coverage by sequencing library. Solid line indicates median value, and dashed lines indicate 25% and 75% quartiles. E) Fraction of reads in targeted resequencing regions by sequencing library. Sequencing libraries (seq1, seq2, seq3, seq4, seq5, new_seq1, new_seq2, new_seq3, new_seq4, and new_seq5) consisted of barcoded and pooled samples, with most samples sequenced in multiple libraries. Libraries with “new” in the name were sequenced after the last sample collection.
Figure S5 – Pigmentation prediction from other models are inaccurate in the KhoeSan. (Related to Table 2) A) Prediction model of skin pigmentation based on seven SNPs. The model (Hart et al., 2013b; Spichenok et al., 2011) utilizes a bifurcating decision tree based on individuals’ genotypes at seven predictive loci. The condition of each step-wise decision is described, with arrows indicating the decision possibilities. Categorical predictions are shown at the bottom of the chart. B) Pigmentation prediction result is shown as individuals’ actual spectrometer measured of M index against predicted categorical pigmentation. Number of individuals assigned to each category is shown in the x axis labels. The prediction is based on a previously developed 7-SNP model (Hart et al., 2013b; Spichenok et al., 2011), with predicted output categorized as “Light”, “Medium”, “Dark”, or “No Call” if the model fails to unambiguously assign a prediction based on genotypes.
Figure S6 – Initial GWAS. (Related to Figure 5) Imputed variants were association with pigmentation and tanning status for 217 KhoeSan individuals (107 ‡Khomani and 110 Nama individuals). A) Study design overview. B) Manhattan plot for pigmentation, with LD assessed and clumped using best guess genotypes from MEGA data (i.e. largest sample size). LD clumps are shown in blue for top 25 loci and labeled with closest gene for top 5 loci. C) QQplot for pigmentation (λGC=1.018), D) LocusZoom plot for SLC24A5, among top 5 independent loci with linkage signals, E) Manhattan plot for tanning status, F) QQplot for tanning status (λGC=1.052), G) LocusZoom plot near TYRP1, among top 5 independent loci with linkage signals.
Table S1 – Summary statistics of pigmentation phenotypes for global population. (Related to Figure 1) All statistics relate to M index for baseline pigmentation. Rows with no dataset listed refer to subsets of the dataset in prior rows, e.g. >90% KhoeSan Nama refers to the Nama samples from this study. Cape Verdean samples from (Beleza et al., 2013) were broken into quantiles, labeled Q1 to Q4. μ = mean, σ = standard deviation, and cV = coefficient of variation (σ/μ), a standardized metric of dispersion.
Table S2 – Reference and KhoeSan samples included in the ADMIXTURE analysis. (Related to Figure 2)
Table S3 – Heritability estimates by population using multiple approaches. (Related to Table 1) Heritability was assessed separately and jointly in the ‡Khomani and Nama populations. Because of population structure, haplotypes were called separately, (i.e. even when populations were analyzed jointly). N = number of individuals. SNPs = number of SNPs included.
Table S4 – Number of individuals from each population that were phenotyped, genotyped, exome sequenced, or with targeted resequencing data (Related to STAR Methods). Pheno = phenotyped, Geno=genotyped, Exome = exome sequenced, Reseq = targeted resequencing. We also indicate the overlap among different genotyping arrays, which are as follows: 550k = Illumina 550k array, OmniEx = Illumina OmniExpress array, OmniExPlus = Illumina OmniExpressPlus array, Omni2.5 = Illumina Omni2.5 array, Mega = Illumina Multiethnic genotyping array. Phase 2 of the GWAS analysis consisted of samples genotyped on the MEGA array.
Table S5 – Top 50 associations from initial GWAS. (Related to Figure 5) All associations shown are independent (R2 ≤ 0.5, window size = 250 kb), and indicate the association between genotype dosages and skin pigmentation M index across the Nama and ‡Khomani datasets. The # tagging column indicates the number of SNPs with an rsID that are tagged by the independent signal.
Table S6 – Summary statistics and regulator information for known and novel pigmentation SNPs. (Related to Table 2 and Figure 5) A) P-value, effect size and standard error, and frequency are shown for known pigmentation loci (including skin, eye, and hair color phenotypes) in the ‡Khomani, Nama, and jointly analyzed samples. B) Regulatory information including conservation, functional class, motif changes, etc, indentified via HaploReg are shown for resequencing variants in 6 of the targeted regions.
*Provided as Excel spreadsheet due to size constraints
Table S7 – Regions selected for targeted resequencing. (Related to Figure 5) Resequencing was successfully performed for a total of 441 KhoeSan individuals.
Table S8 – Power to detect loci with suggestive and genome-wide significance for effect sizes previously associated with skin pigmentation. (Related to STAR Methods) A previous study by Beleza et al, 2013 showed that 4 loci explained 35% of the variation in skin color. We show power for the approximate sample sizes in phase 1, phase 2, and the resequencing analysis.
*Provided as Excel file due to size constraints
Highlights.
Skin pigmentation in Africans is far more polygenic than light skin in Eurasians
Early diverged KhoeSan populations have light skin compared to equatorial Africans
Highly heritable KhoeSan skin color variation is poorly explained by known genes
The study of African skin color identifies novel and canonical pigmentation genes
Acknowledgments
We thank the Beijing Genome Institute and Agilent Technologies for sequencing support. We thank Sandra Beleza, Heather Norton, Esteban Parra, Chandana Basu Mallick, Andres Moreno-Estrada and Karla Sandoval, Richard Cooper, Sophie Candille, George Chaplin, and Nina Jablonski and their colleagues for providing quantitative phenotypes or summaries from their pigmentation work. We thank Catherine Guenther for discussing molecular strategies for functional followup. We also thank the Working Group of Indigenous Minorities in Southern Africa (WIMSA), community leaders, and the South African San Institute (SASI) for their advice and oversight. Finally, we thank Richard Jacobs, Wilhelmina Mondzinger, Hans Padmaker, Willem de Klerk, Hendrik Kaiman, and the communities in which we have sampled; without their support, this study would not have been possible. Funding for this project was provided by a Stanford University CDEHA seed grant (NIH, NIA P30 AG017253-12) to BMH and by a trainee research grant to ARM by the Stanford Center for Computational, Evolutionary, and Human Genomics. Funding was also provided by the Morrison Institute for Population Studies, Stanford University. Funding for genotyping additional KhoeSan individuals was provided by the Stanford Center for Computational, Evolutionary, and Human Genomics. ARM was funded by NIH Genetics and Developmental Biology Training Program T32 GM07790. EA was supported by an NIH IRACDA fellowship to Stony Brook University. CRG was supported by the Stanford Genome Training Program 5T32HG000044. CJW, MM and EGH are partly supported by the South African Medical Research Council. The content is solely the responsibility of the authors and does not necessarily represent the official views of the South African Medical Research Council.
Footnotes
Author Contributions
BMH, CRG, MWF, and CDB conceived of and designed the study. ARM and AS designed experiments. ARM, MS and AS performed experiments. ARM, JMG, ML, EGA and XL analyzed data. BMH, CRG, CJW, JWM, ARM, JMG, and MM collected samples and measured skin photometrics. EGH, CRG, DMK, MWF, CDB, and BMH supervised the study. ARM and BMH wrote the manuscript with input from ML, JMG, MM, DMK, EGH, MWF, and CRG. All authors read and approved of the final version of the manuscript.
We use the term “KhoeSan” to refer to a diverse array of indigenous populations in southern Africa that carry KhoeSan ancestry and speak Khoe, !Ui-Tuu, or Kx’a languages. “KhoeSan” is not accepted by all such communities; where possible we refer to populations by their specific ethnic name. This grouping lumps together populations of different languages, cultures, and variable genetic diversity.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almasy L, Blangero J. Multipoint Quantitative-Trait Linkage Analysis in General Pedigrees. The American Journal of Human Genetics. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Dermitzakis E, Peltonen L, Dermitzakis E, Bonnen PE, de Bakker PIW, Deloukas P, Gabriel SB, Gwilliam R, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH. Clines in polygenic traits. Genet Res. 1999;74:223–236. doi: 10.1017/s001667239900422x. [DOI] [PubMed] [Google Scholar]
- Basu Mallick C, Iliescu FM, Möls M, Hill S, Tamang R, Chaubey G, Goto R, Ho SYW, Gallego Romero I, Crivellaro F, et al. The Light Skin Allele of SLC24A5 in South Asians and Europeans Shares Identity by Descent. PLoS Genet. 2013;9:e1003912. doi: 10.1371/journal.pgen.1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleza S, Johnson NA, Candille SI, Absher DM, Coram MA, Lopes J, Campos J, Araújo II, Anderson TM, Vilhjálmsson BJ, et al. Genetic Architecture of Skin and Eye Color in an African-European Admixed Population. PLoS Genet. 2013;9:e1003372–15. doi: 10.1371/journal.pgen.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleza S, Santos AM, McEvoy B, Alves I, Martinho C, Cameron E, Shriver MD, Parra EJ, Rocha J. The Timing of Pigmentation Lightening in Europeans. Mol Biol Evol. 2012;30:24–35. doi: 10.1093/molbev/mss207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JJ, Coop G. A Population Genetic Signal of Polygenic Adaptation. PLoS Genet. 2014;10:e1004412–e1004425. doi: 10.1371/journal.pgen.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and Accurate Haplotype Phasing and Missing-Data Inference for Whole-Genome Association Studies By Use of Localized Haplotype Clustering. The American Journal of Human Genetics. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byard PJ. Quantitative genetics of human skin color. Am J Phys Anthropol. 1981;24:123–137. [Google Scholar]
- Candille SI, Absher DM, Beleza S, Bauchet M, McEvoy B, Garrison NA, Li JZ, Myers RM, Barsh GS, Tang H, et al. Genome-Wide Association Studies of Quantitatively Measured Skin, Hair, and Eye Pigmentation in Four European Populations. PLoS ONE. 2012;7:e48294–12. doi: 10.1371/journal.pone.0048294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S, Schumacher FR, Peters U, Franceschini N, Ritchie MD, et al. Generalization and Dilution of Association Results from European GWAS in Populations of Non-European Ancestry: The PAGE Study. PLoS Biol. 2013;11:e1001661–11. doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Giga Sci. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin G. Geographic distribution of environmental factors influencing human skin coloration. Am J Phys Anthropol. 2004;125:292–302. doi: 10.1002/ajpa.10263. [DOI] [PubMed] [Google Scholar]
- Chaplin G, Jablonski NG. Vitamin D and the evolution of human depigmentation. Am J Phys Anthropol. 2009;139:451–461. doi: 10.1002/ajpa.21079. [DOI] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles G, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark P, Stark AE, Walsh RJ, Jardine R, Martin NG. A twin study of skin reflectance. Annals of Human Biology. 1981;8:529–541. doi: 10.1080/03014468100005371. [DOI] [PubMed] [Google Scholar]
- Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free Estimation of Recent Genetic Relatedness. The American Journal of Human Genetics. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens AK, Naude CE, Goliath R, Chaplin G, Wilkinson RJ, Jablonski NG. High-dose vitamin D 3reduces deficiency caused by low UVB exposure and limits HIV-1 replication in urban Southern Africans. Proc Natl Acad Sci USA. 2015;112:8052–8057. doi: 10.1073/pnas.1500909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffey BL, OLIVER RJ, Farr PM. A portable instrument for quantifying erythema induced by ultraviolet radiation. British Journal of Dermatology. 1984;111:663–672. doi: 10.1111/j.1365-2133.1984.tb14149.x. [DOI] [PubMed] [Google Scholar]
- Duan S, Huang RS, Zhang W, Mi S, Bleibel WK, Kistner EO, Cox NJ, Dolan ME. Expression and alternative splicing of folate pathway genes in HapMap lymphoblastoid cell lines. Pharmacogenomics. 2009;10:549–563. doi: 10.2217/PGS.09.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DL, Montgomery GW, Chen W, Zhao ZZ, Le L, James MR, Hayward NK, Martin NG, Sturm RA. A three-single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. The American Journal of Human Genetics. 2007;80:241–252. doi: 10.1086/510885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazo-Arvizu RA, Camacho P, Bovet P, Forrester T, Lambert EV, Plange-Rhule J, Hoofnagle AN, Aloia J, Tayo B, Dugas LR, et al. 25-Hydroxyvitamin D in African-origin populations at varying latitudes challenges the construct of a physiologic norm. American Journal of Clinical Nutrition. 2014;100:908–914. doi: 10.3945/ajcn.113.066605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, Bigham A, Tan J, Li S, Gozdzik A, Ross K, Jin L, Parra EJ. Association of the OCA2 Polymorphism His615Arg with Melanin Content in East Asian Populations: Further Evidence of Convergent Evolution of Skin Pigmentation. PLoS Genet. 2010;6:e1000867. doi: 10.1371/journal.pgen.1000867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisancho AR, Wainwright R, Way A. Heritability and components of phenotypic expression in skin reflectance of Mestizos from the Peruvian Lowlands. Am J Phys Anthropol. 1981;55:203–208. doi: 10.1002/ajpa.1330550207. [DOI] [PubMed] [Google Scholar]
- Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Zakharia F, Moreno-Estrada A, Byrnes JK, Muzzio M, Rodriguez-Flores JL, Kenny EE, Gignoux CR, Maples BK, Guiblet W, et al. Reconstructing Native American Migrations from Whole-Genome and Whole-Exome Data. PLoS Genet. 2013;9:e1004023–14. doi: 10.1371/journal.pgen.1004023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Lowe JK, Stoffel M, Daly MJ, Altshuler D, Breslow JL, Friedman JM, Pe’er I. Whole population, genome-wide mapping of hidden relatedness. Genome Research. 2009;19:318–326. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Trynka G, Finucane H, Vilhjálmsson BJ, Xu H, Zang C, Stahl E, Hultman CM, Purcell SM, Daly M, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison GA, Owen JJT. Studies on the inheritance of human skin colour. Annals of Human Genetics. 1964;28:27–37. doi: 10.1111/j.1469-1809.1964.tb00457.x. [DOI] [PubMed] [Google Scholar]
- Hart KL, Kimura SL, Mushailov V, Budimlija ZM, Prinz M, Wurmbach E. Improved eye- and skin-color prediction based on 8 SNPs. Croat Med J. 2013a;54:248–256. doi: 10.3325/cmj.2013.54.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart KL, Kimura SL, Mushailov V, Budimlija ZM, Prinz M, Wurmbach E. Improved eye- and skin-color prediction based on 8 SNPs. Croat Med J. 2013b;54:248–256. doi: 10.3325/cmj.2013.54.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, Martin AR, Musharoff S, Cann H, Snyder MP, et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci USa. 2016;113:E440–E449. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Gignoux CR, Jobin M, Granka JM, Macpherson JM, Kidd JM, Rodríguez-Botigué L, Ramachandran S, Hon L, Brisbin A, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci USa. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski NG, Chaplin G. Colloquium paper: human skin pigmentation as an adaptation to UV radiation. Proc Natl Acad Sci USa. 2010;107(Suppl 2):8962–8968. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski NG, Chaplin G. The Evolution of Skin Pigmentation and Hair Texture in People of African Ancestry. Dermatologic Clinics. 2014;32:113–121. doi: 10.1016/j.det.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nature Publishing Group. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenen B, Qi H, Saladi SV, Yeung M, de la Serna IL. Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene. 2009a;29:81–92. doi: 10.1038/onc.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenen B, Qi H, Saladi SV, Yeung M, de la Serna IL. Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene. 2009b;29:81–92. doi: 10.1038/onc.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EE, Timpson NJ, Sikora M, Yee MC, Moreno-Estrada A, Eng C, Huntsman S, Burchard EG, Stoneking M, Bustamante CD, et al. Melanesian Blond Hair Is Caused by an Amino Acid Change in TYRP1. Science. 2012a;336:554–554. doi: 10.1126/science.1217849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EE, Timpson NJ, Sikora M, Yee MC, Moreno-Estrada A, Eng C, Huntsman S, Burchard EG, Stoneking M, Bustamante CD, et al. Melanesian Blond Hair Is Caused by an Amino Acid Change in TYRP1. Science. 2012b;336:554–554. doi: 10.1126/science.1217849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL. SLC24A5, a Putative Cation Exchanger, Affects Pigmentation in Zebrafish and Humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Visser M, Duffy DL, Hysi PG, Jacobs LC, Lao O, Zhong K, Walsh S, Chaitanya L, Wollstein A, et al. Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Human Genetics. 2015;134:823–835. doi: 10.1007/s00439-015-1559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: A Discriminative Modeling Approach for Rapid and Robust Local-Ancestry Inference. The American Journal of Human Genetics. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, Kenny EE. AR TICLE Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. The American Journal of Human Genetics. 2017:1–36. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016:1–14. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. cis-Regulatory Changes in Kit Ligand Expression and Parallel Evolution of Pigmentation in Sticklebacks and Humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H, Kraft P, Qureshi AA, Guo Q, Chen C, Hankinson SE, Hu FB, Thomas G, Hoover RN, Chanock S, et al. Genome-Wide Association Study of Tanning Phenotype in a Population of European Ancestry. 2009;129:2250–2257. doi: 10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton HL, Kittles RA, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD. Genetic Evidence for the Convergent Evolution of Light Skin in Europeans and East Asians. Mol Biol Evol. 2006;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- O’Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, Traglia M, Huang J, Huffman JE, Rudan I, et al. A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness. PLoS Genet. 2014;10:e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Patterson N, Loh PR, Lipson M, Berger B, Stoneking M, Pakendorf B, Reich D. Ancient west Eurasian ancestry in southern and eastern Africa. Proc Natl Acad Sci USA. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospiech E, Wojas-Pelc A, Walsh S, Liu F, Maeda H, Ishikawa T, Skowron M, Kayser M, Branicki W. The common occurrence of epistasis in the determination of human pigmentation and its impact on DNA-based pigmentation phenotype prediction. Forensic Science International: Genetics. 2014;11:64–72. doi: 10.1016/j.fsigen.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, Robinson KC, Van Otterloo E, Kim RSQ, Bergsteinsdottir K, Ogmundsdottir MH, et al. A Polymorphism in IRF4 Affects Human Pigmentation through a Tyrosinase-Dependent MITF/TFAP2A Pathway. Cell. 2013;155:1022–1033. doi: 10.1016/j.cell.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relethford JH. Human skin color diversity is highest in sub-Saharan African populations. Human Biology. 2000;72:773–780. [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh KH, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangarajan R, Boissy RE. Tyrp1 and Oculocutaneous Albinism Type 3. Pigment Cell Res. 2001;14:437–444. doi: 10.1034/j.1600-0749.2001.140603.x. [DOI] [PubMed] [Google Scholar]
- Schlebusch CM, Skoglund P, Sjodin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MGB, et al. Genomic Variation in Seven Khoe-San Groups Reveals Adaptation and Complex African History. Science. 2012;338:374–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shringarpure SS, Bustamante CD, Lange K, Alexander DH. Efficient analysis of large datasets and sex bias with ADMIXTURE. BMC Bioinformatics. 2016;17:1655. doi: 10.1186/s12859-016-1082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Parra EJ. Comparison of narrow-band reflectance spectroscopy and tristimulus colorimetry for measurements of skin and hair color in persons of different biological ancestry. Am J Phys Anthropol. 2000;112:17–27. doi: 10.1002/(SICI)1096-8644(200005)112:1<17::AID-AJPA3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Spichenok O, Budimlija ZM, Mitchell AA, Jenny A, Kovacevic L, Marjanović D, Caragine T, Prinz M, Wurmbach E. Prediction of eye and skin color in diverse populations using seven SNPs. Forensic Science International: Genetics. 2011;5:472–478. doi: 10.1016/j.fsigen.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Sturm RA. Molecular genetics of human pigmentation diversity. Human Molecular Genetics. 2009;18:R9–R17. doi: 10.1093/hmg/ddp003. [DOI] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL. Human pigmentation genes under environmental selection. Genome Biol. 2012;13:248–15. doi: 10.1186/gb-2012-13-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, Steinberg S, Gudjonsson SA, Palsson A, Thorleifsson G, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- Thornton T, Tang H, Hoffmann TJ, Ochs-Balcom HM, Caan BJ, Risch N. Estimating Kinship in Admixed Populations. The American Journal of Human Genetics. 2012a;91:122–138. doi: 10.1016/j.ajhg.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T, Tang H, Hoffmann TJ, Ochs-Balcom HM, Caan BJ, Risch N. Estimating Kinship in Admixed Populations. The American Journal of Human Genetics. 2012b;91:122–138. doi: 10.1016/j.ajhg.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, et al. The Genetic Structure and History of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren C, Kim M, Martin AR, Bobo D, Gignoux CR, van Helden PD, Möller M, Hoal EG, Henn BM. Fine-Scale Human Population Structure in Southern Africa Reflects Ecogeographic Boundaries. Genetics. 2016a;204:303–314. doi: 10.1534/genetics.116.187369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren C, Kim M, Martin AR, Bobo D, Gignoux CR, van Helden PD, Möller M, Hoal EG, Henn BM. Fine-Scale Human Population Structure in Southern Africa Reflects Ecogeographic Boundaries. Genetics. 2016b;204:303–314. doi: 10.1534/genetics.116.187369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachtenheim J, Ondrušová L, Borovanský J. SWI/SNF chromatin remodeling complex is critical for the expression of microphthalmia-associated transcription factor in melanoma cells. Biochemical and Biophysical Research Communications. 2010;392:454–459. doi: 10.1016/j.bbrc.2010.01.048. [DOI] [PubMed] [Google Scholar]
- Veeramah KR, Wegmann D, Woerner A, Mendez FL, Watkins JC, Destro-Bisol G, Soodyall H, Louie L, Hammer MF. An Early Divergence of KhoeSan Ancestors from Those of Other Modern Humans Is Supported by an ABC-Based Analysis of Autosomal Resequencing Data. Mol Biol Evol. 2011;29:617–630. doi: 10.1093/molbev/msr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh S, Liu F, Wollstein A, Kovatsi L, Ralf A, Kosiniak-Kamysz A, Branicki W, Kayser M. The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. Forensic Science International: Genetics. 2013;7:98–115. doi: 10.1016/j.fsigen.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Research. 2011;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde S, Timpson A, Kirsanow K, Kaiser E, Kayser M, Unterländer M, Hollfelder N, Potekhina ID, Schier W, Thomas MG, et al. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc Natl Acad Sci USA. 2014;111:4832–4837. doi: 10.1073/pnas.1316513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Publishing Group. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia CH, Lu E, Zeng J, Gong X. Deletion of LRP5 in VLDLR Knockout Mice Inhibits Retinal Neovascularization. PLoS ONE. 2013a;8:e75186–11. doi: 10.1371/journal.pone.0075186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia CH, Lu E, Zeng J, Gong X. Deletion of LRP5 in VLDLR Knockout Mice Inhibits Retinal Neovascularization. PLoS ONE. 2013b;8:e75186. doi: 10.1371/journal.pone.0075186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhong H, Chen J, Zhang X, Zhang H, Luo X, Xu S, Chen H, Lu D, Han Y, et al. A Genetic Mechanism for Convergent Skin Lightening during Recent Human Evolution. Mol Biol Evol. 2016;33:1177–1187. doi: 10.1093/molbev/msw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi AA, Mattern BC, Claes P, McEcoy B, Hughes C, Shriver MD. Investigating the case of human nose shape and climate adaptation. PLoS Genet. 2017;13:e1006616–e1006631. doi: 10.1371/journal.pgen.1006616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N, Kraft P, Patterson N, Pasaniuc B, Bhatia G, Pollack S, Price AL. Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits. PLoS Genet. 2013;9:e1003520–11. doi: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N, Pasaniuc B, Sankararaman S, Bhatia G, Zhang J, Gusev A, Young T, Tandon A, Pollack S, Vilhjálmsson BJ, et al. Leveraging population admixture to characterize the heritability of complex traits. Nature Publishing Group. 2014;46:1356–1362. doi: 10.1038/ng.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 – Pedigrees inferred from ethnographic information. (Related to Table 1 and Methods) Ethnographically inferred pedigrees for KhoeSan individuals are shown in: A) the ‡Khomani, and B) the Nama. Different shades represent whether the samples have been genotyped. Non-phenotyped individuals are crossed in pedigrees.
Figure S2 – Ancestry estimates in ‡Khomani and Nama samples. (Related to Figure 2) A) Admixture runs across K=3–7 for the ‡Khomani and Nama populations, using Namibian San, Hadza, Sandawe, Maasai, Kenyan Bantu, South African (SA) Bantu, Yoruba, Mozabite, Central Europeans (CEU), and French populations as a reference panel, as in Figure 2A. B) Minor ancestry component associations with M index, displayed with a square root x-axis to elongate the minor contributions. C) Pairwise ancestry component correlations at k=7 from A). Upper triangular matrix shows Pearson’s correlation coefficient between pairwise ancestry estimates. Lower triangular matrix shows scatter plots with ‡Khomani shown in blue and Nama shown in red. D-F) Principal Components Analysis (PCA) biplots for the same SNPs and individuals used in the ADMIXTURE analysis adjusted for relatedness using King and the PC-AiR approach. D) PC1 vs PC2, E) PC3 vs PC4, and F) PC5 vs PC6.
Figure S3 – Partitioned heritability by population. (Related to Figure 3) Proportion of heritable variation in baseline pigmentation explained by true pigmentation gene sets (dots) versus matched null distribution after accounting for number of SNPs in gene sets in two different candidate gene sets. Results are stratified by the combined (comb) data, ‡Khomani (khm), and Nama populations. * indicates FDR < 0.1 (indicated by the top decile of the null distributions). Gene set labels are the same as in Figure 3.
Figure S4 – Imputation and targeted resequencing quality control. (Related to STAR methods) A) Imputation quality was assessed via homozygous reference (HR), heterozygous (Het), and homozygous non-reference (HN) concordance for imputed dosages with high coverage exome sequencing data. Each concordance metric was computed as the ratio of dosages for a particular genotyping class (e.g. HR) in both genotype and exome datasets to the dosages of that total class in the genotype dataset with any call in the exomes. The colors indicate the different genotyping arrays, with the 550k array in red, OmniExpress array in blue, and OmniExpressPlus array in green. B–E) Targeted resequencing quality control. B) Depth of coverage versus array concordance. Dashed line at 10× indicates minimum depth accepted for a sample’s inclusion. C) Depth of coverage by targeted resequencing region. D) Resequencing coverage by sequencing library. Solid line indicates median value, and dashed lines indicate 25% and 75% quartiles. E) Fraction of reads in targeted resequencing regions by sequencing library. Sequencing libraries (seq1, seq2, seq3, seq4, seq5, new_seq1, new_seq2, new_seq3, new_seq4, and new_seq5) consisted of barcoded and pooled samples, with most samples sequenced in multiple libraries. Libraries with “new” in the name were sequenced after the last sample collection.
Figure S5 – Pigmentation prediction from other models are inaccurate in the KhoeSan. (Related to Table 2) A) Prediction model of skin pigmentation based on seven SNPs. The model (Hart et al., 2013b; Spichenok et al., 2011) utilizes a bifurcating decision tree based on individuals’ genotypes at seven predictive loci. The condition of each step-wise decision is described, with arrows indicating the decision possibilities. Categorical predictions are shown at the bottom of the chart. B) Pigmentation prediction result is shown as individuals’ actual spectrometer measured of M index against predicted categorical pigmentation. Number of individuals assigned to each category is shown in the x axis labels. The prediction is based on a previously developed 7-SNP model (Hart et al., 2013b; Spichenok et al., 2011), with predicted output categorized as “Light”, “Medium”, “Dark”, or “No Call” if the model fails to unambiguously assign a prediction based on genotypes.
Figure S6 – Initial GWAS. (Related to Figure 5) Imputed variants were association with pigmentation and tanning status for 217 KhoeSan individuals (107 ‡Khomani and 110 Nama individuals). A) Study design overview. B) Manhattan plot for pigmentation, with LD assessed and clumped using best guess genotypes from MEGA data (i.e. largest sample size). LD clumps are shown in blue for top 25 loci and labeled with closest gene for top 5 loci. C) QQplot for pigmentation (λGC=1.018), D) LocusZoom plot for SLC24A5, among top 5 independent loci with linkage signals, E) Manhattan plot for tanning status, F) QQplot for tanning status (λGC=1.052), G) LocusZoom plot near TYRP1, among top 5 independent loci with linkage signals.
Table S1 – Summary statistics of pigmentation phenotypes for global population. (Related to Figure 1) All statistics relate to M index for baseline pigmentation. Rows with no dataset listed refer to subsets of the dataset in prior rows, e.g. >90% KhoeSan Nama refers to the Nama samples from this study. Cape Verdean samples from (Beleza et al., 2013) were broken into quantiles, labeled Q1 to Q4. μ = mean, σ = standard deviation, and cV = coefficient of variation (σ/μ), a standardized metric of dispersion.
Table S2 – Reference and KhoeSan samples included in the ADMIXTURE analysis. (Related to Figure 2)
Table S3 – Heritability estimates by population using multiple approaches. (Related to Table 1) Heritability was assessed separately and jointly in the ‡Khomani and Nama populations. Because of population structure, haplotypes were called separately, (i.e. even when populations were analyzed jointly). N = number of individuals. SNPs = number of SNPs included.
Table S4 – Number of individuals from each population that were phenotyped, genotyped, exome sequenced, or with targeted resequencing data (Related to STAR Methods). Pheno = phenotyped, Geno=genotyped, Exome = exome sequenced, Reseq = targeted resequencing. We also indicate the overlap among different genotyping arrays, which are as follows: 550k = Illumina 550k array, OmniEx = Illumina OmniExpress array, OmniExPlus = Illumina OmniExpressPlus array, Omni2.5 = Illumina Omni2.5 array, Mega = Illumina Multiethnic genotyping array. Phase 2 of the GWAS analysis consisted of samples genotyped on the MEGA array.
Table S5 – Top 50 associations from initial GWAS. (Related to Figure 5) All associations shown are independent (R2 ≤ 0.5, window size = 250 kb), and indicate the association between genotype dosages and skin pigmentation M index across the Nama and ‡Khomani datasets. The # tagging column indicates the number of SNPs with an rsID that are tagged by the independent signal.
Table S6 – Summary statistics and regulator information for known and novel pigmentation SNPs. (Related to Table 2 and Figure 5) A) P-value, effect size and standard error, and frequency are shown for known pigmentation loci (including skin, eye, and hair color phenotypes) in the ‡Khomani, Nama, and jointly analyzed samples. B) Regulatory information including conservation, functional class, motif changes, etc, indentified via HaploReg are shown for resequencing variants in 6 of the targeted regions.
*Provided as Excel spreadsheet due to size constraints
Table S7 – Regions selected for targeted resequencing. (Related to Figure 5) Resequencing was successfully performed for a total of 441 KhoeSan individuals.
Table S8 – Power to detect loci with suggestive and genome-wide significance for effect sizes previously associated with skin pigmentation. (Related to STAR Methods) A previous study by Beleza et al, 2013 showed that 4 loci explained 35% of the variation in skin color. We show power for the approximate sample sizes in phase 1, phase 2, and the resequencing analysis.
*Provided as Excel file due to size constraints