Abstract
Amyotrophic lateral sclerosis-frontotemporal dementia (ALS-FTD) constitutes a devastating disease spectrum characterised by TDP-43 pathology. Understanding how TDP-43 contributes to neurodegeneration will help direct therapeutic efforts. Here, we have created a novel TDP-43 knock-in mouse with a human-equivalent mutation in the endogenous mouse Tardbp gene. TDP-43Q331K mice demonstrate cognitive dysfunction and a paucity of parvalbumin interneurons. Critically, TDP-43 autoregulation is perturbed leading to a gain of TDP-43 function, and altered splicing of Mapt, another pivotal dementia gene. Furthermore, a novel approach to stratify transcriptomic data by phenotype in differentially affected mutant mice reveals 471 changes linked with improved behaviour. These changes include downregulation of two known modifiers of neurodegeneration, Atxn2 and Arid4a, and upregulation of myelination and translation genes. With one base change in murine Tardbp, this study identifies TDP-43 misregulation as a pathogenic mechanism that may underpin ALS-FTD, and exploits phenotypic heterogeneity to yield candidate suppressors of neurodegenerative disease.
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are destructive neurodegenerative diseases that exist on a clinicopathological spectrum (ALS-FTD)1. ALS is characterised by motor impairment and FTD by executive dysfunction, language impairment and behavioural changes. Nearly all cases of ALS, half of FTD cases, and most hereditary forms of ALS and FTD are characterised by cytoplasmic mislocalisation and aggregation of the 43kDa TAR DNA-binding protein (TDP-43)2,3. Significantly, the identification of mutations in the gene encoding TDP-43 (TARDBP) as a cause of ALS and FTD confirmed that TDP-43 plays a mechanistic role in neurodegeneration4,5. This role remains undefined.
TDP-43 is a conserved RNA-binding protein with critical roles in splicing in the nervous system6. TDP-43 also demonstrates exquisite autoregulation by binding to its transcript, triggering alternative splicing of intron 7 within the TARDBP 3’-untranslated region (UTR) and destruction of its mRNA7. Experimentally increasing or decreasing TDP-43 levels both cause neuronal loss, but whether human neurodegeneration is caused by a gain or loss of TDP-43 function remains unclear. Modelling of mutant TDP-43 in vivo has relied on variable degrees of transgenic overexpression of TDP-43 to replicate pathological changes seen in post-mortem human tissues8. However, TDP-43 transgenic mouse models have demonstrated that TDP-43 aggregation is not necessary to cause neurodegeneration9, and whether TDP-43 aggregation is causally linked to disease onset is unclear.
A caveat of transgenic TDP-43 mouse models is that phenotypes may partly be artefacts of overexpression. Furthermore, the cell-type specific expression of single TDP-43 splice forms in transgenic models using neuronal promoters, and temporally-triggered expression of transgenes in adulthood do not reflect ubiquitous expression and alternative splicing of Tardbp, including during embryonic development10. To unravel the role of mutant TDP-43 in the disease pathogenesis we created a knock-in mouse harbouring only a human-equivalent point mutation in the endogenous mouse Tardbp gene. This model replicates the human mutant state as closely as possible, retaining the endogenous gene structure including promoters and autoregulatory 3’UTR, and maintaining the ubiquitous expression of TDP-43 during embryonic development and in adulthood. By avoiding deliberate manipulation of TDP-43 expression, this model helps elucidate both mediators and modifiers of cognitive dysfunction in ALS-FTD.
Results
TDP-43Q331K causes behavioural phenotypes and disproportionately affects male mice
Over 50 TARDBP mutations at conserved sites have been identified in ALS-FTD11. We chose to introduce the n.991C>A (p.Q331K) mutation into murine Tardbp because TDP-43Q331K is a particularly toxic species in vitro and in vivo4,9,12,13. Mutagenesis was performed using CRISPR/CAS9 methodology yielding four founders with the Q331K mutation (Fig. 1a). Mutagenesis events at predicted off-target regions and in the remainder of Tardbp were excluded by Sanger sequencing. Founder #52 was outcrossed to F4 to remove other potential off-target mutagenesis events. Heterozygous (TDP-43Q331K/+) F4 animals were intercrossed to generate mutant and wild-type mice. Homozygotes (TDP-43Q331K/Q331K) were viable (Fig. 1b, Supplementary Fig. 1a) and appeared superficially normal as juveniles. Since TDP-43 transgenic mice have not been shown to rescue TDP-43 knockout mice, TDP-43Q331K/Q331K knock-in mice represent a unique opportunity to study mutant TDP-43 in vivo in the absence of wild-type TDP-43.
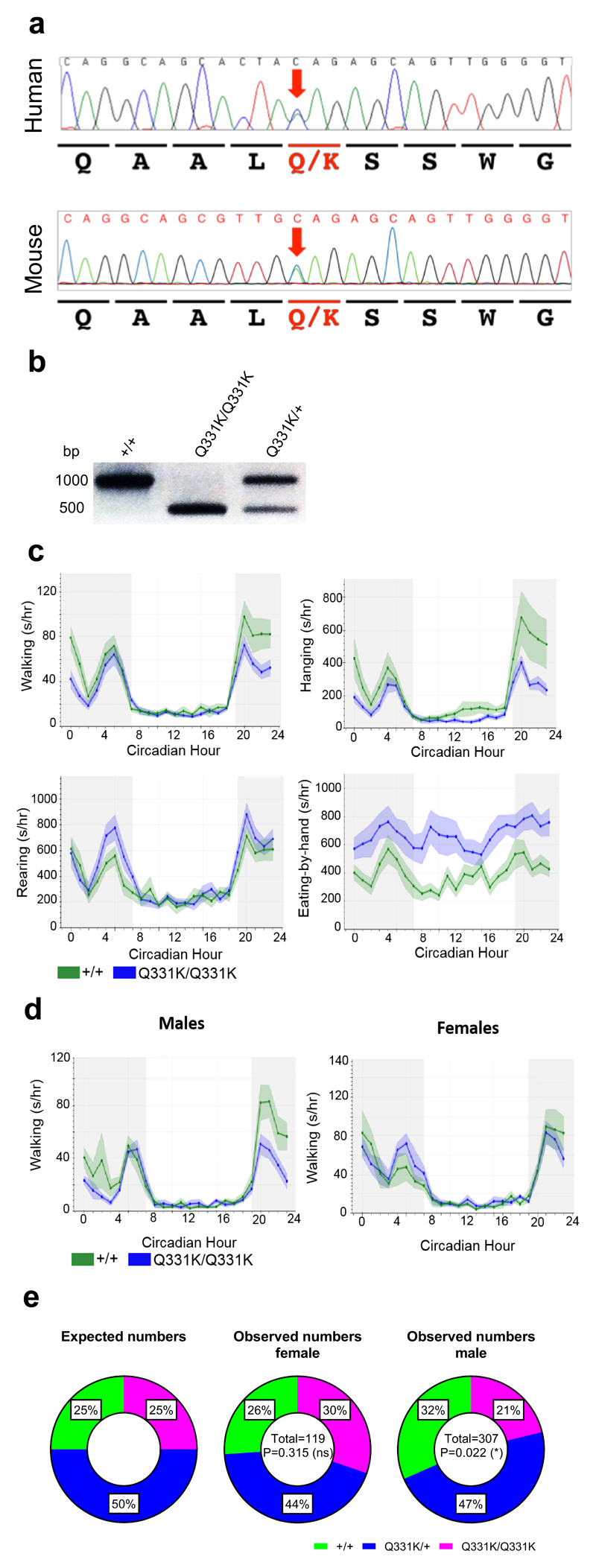
Figure 1. CRISPR mutagenesis, ACBM characterisation and breeding ratios of TDP-43Q331K mice.
(a) Chromatograms from the patient originally identified with the Q331K mutation and CRISPR/CAS9 knock-in founder mouse #52. Bases are given above the chromatograms and amino acids coded are given below. The mutation is highlighted with the red arrow.
(b) SapI restriction enzyme digestion of 1000 bp PCR products across the mutation site from representative genotyping of wild-type, TDP-43Q331K/Q331K, and TDP-43Q331K/+ mice.
(c) Automated continuous behavioural monitoring (ACBM) of 4-month-old mice (n = 10 mice per genotype; 5 males and 5 females). Significantly altered behaviours are displayed: walking: interaction P<0.0001; hanging: interaction P=0.002; rearing: interaction P=0.038; eating-by-hand: genotype P=0.008; repeated measures two-way ANOVA.
(d) Walking behaviour as assessed by ACBM in 7.5-month-old male and female mice (n = 5 mice per genotype). Walking male: interaction P<0.0001; walking female: interaction P=0.334; repeated measures two-way ANOVA.
(e) Ratios of mice genotyped at 10 days (all of which were successfully weaned) broken down by gender. Female (χ2=2.311, d.f.=2, P=0.315), Male (χ2=7.612, d.f.=2, P=0.022); Chi square test.
Error bars represent mean ± s.e.m.
We initially screened for phenotypes in a small group of wild-type and TDP-43Q331K/Q331K mice using automated continuous behavioural monitoring (ACBM)14. At ~4 months of age TDP-43Q331K/Q331K male and female mice demonstrated reduced walking and hanging, and increased rearing and eating-by-hand, but no alterations in circadian rhythmicity (Fig. 1c). The most consistent phenotype was reduced walking in males (Fig. 1d and Supplementary Fig. 1b). Further breeding revealed an under representation of male mutants, yet females were present at Mendelian ratios, further suggesting that males are more susceptible to deleterious effects of TDP-43Q331K (Fig. 1e). This is notable as sporadic ALS is more common in men, and TDP-43 mutations demonstrate greater penetrance in men than women15. We therefore focussed on males in subsequent studies, breeding two cohorts of mice: Cohort 1 for motor, pathological and transcriptomic studies; Cohort 2 for cognitive studies.
TDP-43Q331K mice display no significant motor impairment, but demonstrate weight gain due to hyperphagia and transcriptomic changes in spinal motor neurons
To identify ALS-like motor deficits we measured Rotarod performance in Cohort 1 mice. From ~6 months of age TDP-43Q331K/+ and TDP-43Q331K/Q331K mice demonstrated reduced Rotarod latencies (Fig. 2a). Interestingly, mutants demonstrated hyperphagia, a feature of FTD16, and gained more weight than wild-types (Fig. 2b,c). Increased weight could contribute to impaired Rotarod performance, so we tested Cohort 2 mice, which were weight-matched due to dietary control (Supplementary Fig. 2a). Weight-matched mutants performed similarly to wild-types up to 16 months of age (Fig. 2d), suggesting that mutant mice do not have significant impairment of motor coordination.
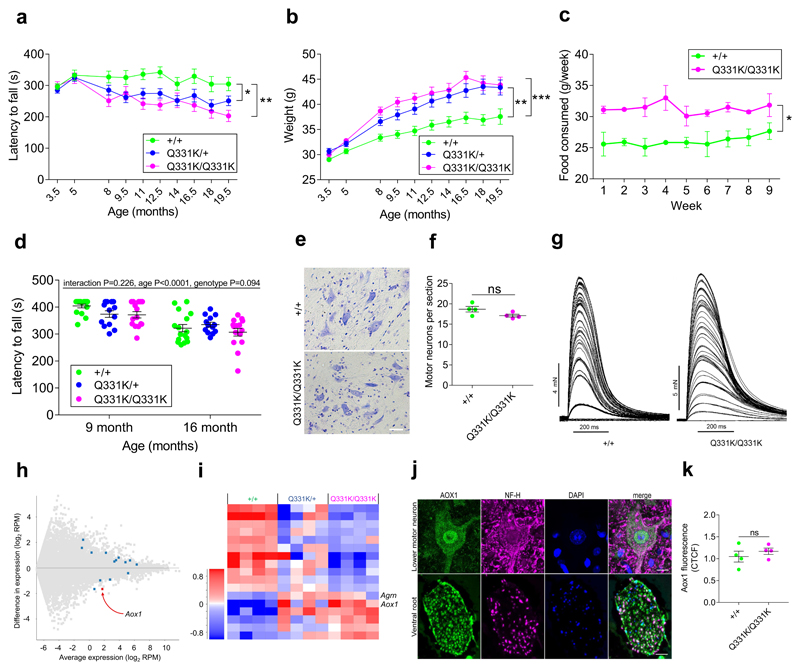
Figure 2. Motor impairment, hyperphagia and spinal motor neuronal transcriptomic changes in mutant mice.
(a) Rotarod and (b) weights of Cohort 1 mice (n = 14 wild-type, 13 TDP-43Q331K/+ and 13 TDP-43Q331K/Q331K mice). (a) Pairwise comparisons: wild-type vs. TDP-43Q331K/+: P=0.014 (*); wild-type vs. TDP-43Q331K/Q331K: P=0.0024 (**). (b) Pairwise comparisons: wild-type vs. TDP-43Q331K/+: P=0.002 (**); wild-type vs. TDP-43Q331K/Q331K: P=0.0002 (***).
(c) Weekly food consumption over 9 weeks (n = 2 cages per genotype). Comparison: Genotype: P=0.047(*).
(d) Rotarod of weight-matched Cohort 2 mice (n = 16 wild-type, 13 TDP-43Q331K/+ and 15 TDP-43Q331K/Q331K mice).
For (a-d) repeated measures two-way ANOVA followed by Holm-Sidak post-hoc test for pairwise comparisons.
(e) Nissl-stained lumbar motor neurons of 5-month-old mice. Representative images shown. Scale bar, 40μm.
(f) Quantification of lumbar motor neurons (n = 4 mice per genotype). Comparison: P=0.089 (ns); unpaired t test.
(g) Examples of isometric twitch force recordings during graded nerve stimulation of FDB muscles from representative wild-type and TDP-43Q331K/Q331K mice. Each increment corresponds to recruitment of motor units of successively higher electrical threshold (n = 5 mice per genotype).
(h) MA plot and (i) hierarchical clustering of significantly differentially expressed genes (DEGs) in laser-captured motor neurons. In (h) blue dots indicate significant changes, red dots indicate intensity hits. In (i) Genes Aox1 and Agrin are labelled. Comparison: DESeq2 wild-type v TDP-43Q331K/Q331K
(j) Immunohistochemistry for AOX1. Representative images from a 5-month-old wild-type mouse shown. Scale bars, 10μm motor neuron, 100μm ventral root.
(k) AOX1 immunofluorescence in lumbar motor neurons. Comparison: P=0.433 (ns); unpaired t test.
For (h-k) n = 4 mice per genotype.
All error bars denote mean ± s.e.m.
To determine if mutant mice demonstrated lower motor neuron degeneration we examined spinal cords from 5-month-old mice to identify early pathological changes. Motor neurons demonstrated normal morphology and numbers with no TDP-43 aggregation or mislocalisation in TDP-43Q331K/Q331K mice (Fig. 2e,f Supplementary Fig. 2b). Quantification of neuromuscular junctions (NMJs) and succinate dehydrogenase staining in gastrocnemius muscles were normal in TDP-43Q331K/Q331K mice, suggesting no significant denervation (Supplementary Fig. 2c,d,f). Examination of 18 to 23-month-old mice similarly found no evidence of denervation (Supplementary Fig. 2e) and no electrophysiological evidence of motor unit loss (Fig. 2g, Supplementary Fig. 2g-o).
Collectively, these data indicated a remarkable resilience of neuromuscular units to TDP-43Q331K. We hypothesised that gene expression changes occurring in motor neurons of mutant mice could elucidate how these cells respond to cellular stress caused by TDP-43Q331K. We isolated RNA from laser-captured lumbar motor neurons from 5-month-old mice and performed RNASeq (Supplementary Fig. 3a,b). This yielded 31 significant expression and splicing differences between wild-type and TDP-43Q331K/Q331K mice (Fig. 2h,i Supplementary Fig. 3c-e, Supplementary Table. 1). A notable change was upregulation of Agrin. Agrin is secreted by neurons and functions through muscle specific kinase to cluster acetylcholine receptors at NMJs17. Agrin upregulation may therefore promote NMJ function in TDP-43Q331K/Q331K mice. Interestingly, the largest gene expression change was a three-fold increase in expression of aldehyde oxidase 1 (Aox1). Little is known about the neurobiological functions of AOX1 although its transcript has been observed in the anterior horn of the spinal cord18. AOX1 catalyses the conversion of retinaldehyde to retinoic acid (RA)19, which functions in neuronal maintenance in the adult nervous system and following axon injury. Thus, Aox1 upregulation may benefit motor neurons in TDP-43Q331K/Q331K mice. Immunostaining revealed expression of AOX1 in spinal motor neurons (Fig. 2j), but no difference in expression between TDP-43Q331K/Q331K and wild-type mice (Fig. 2k, Supplementary Fig. 3f). This could be because upregulated AOX1 is transported into peripheral motor axons, as we found abundant expression of AOX1 in motor axons (Fig. 2j).
TDP-43Q331K mice display executive dysfunction, memory impairment and phenotypic heterogeneity
In parallel with motor studies, to determine if TDP-43Q331K causes FTD-like cognitive dysfunction we performed neuropsychological assessments on Cohort 2 mice using touchscreen operant technology. To test if mice exhibited FTD-related deficits we conducted the 5-choice serial reaction time task (5-CSRTT; Fig. 3a), which measures frontal/executive function including attention, perseveration, impulsivity, and psychomotor speed22. At 4 months of age the number of training sessions required to reach performance criteria for probe testing was higher in TDP-43Q331K/Q331K mice than wild-types (Fig. 3b), indicating learning deficits in mutants. Following training, animals underwent probe testing at 6 and 12 months of age. Accuracy (Fig. 3c,d insets) and omission percentage were comparable between genotypes at 6 months of age (Fig. 3c). However, at 12 months of age, while accuracy remained normal, omission percentage was greater in TDP-43Q331K/+ and TDP-43Q331K/Q331K mice (Fig. 3d), suggesting attentional deficits and cognitive decline in mutants. Reward collection and response latencies, and premature and perseverative response rates were similar between genotypes (Supplementary Fig. 4a-h), arguing against visual, motivational, or significant motor deficits as causes for increased omissions. We also measured motivation using fixed (FR) and progressive-ratio (PR) schedules. No significant differences were found between genotypes, further suggesting that increased omissions in mutants were not due to motivational deficits (Fig. 3e,f). Collectively, these data indicate an inattention phenotype in TDP-43Q331K/+ and TDP-43Q331K/Q331K mice, which is consistent with frontal/executive dysfunction.
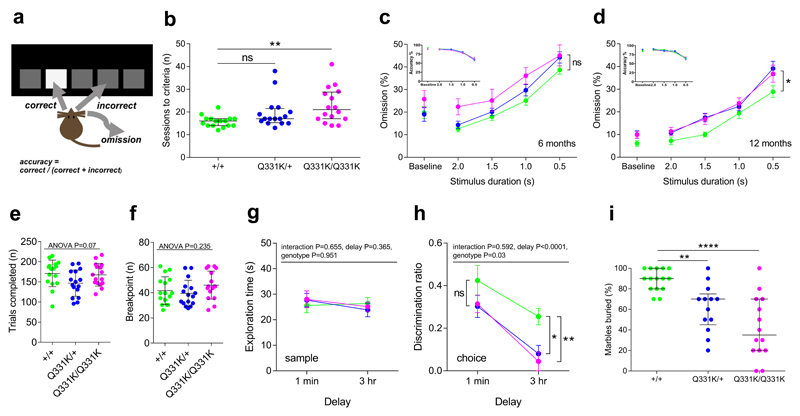
Figure 3. Cognitive testing indicates executive dysfunction, memory impairment and phenotypic heterogeneity in mutant mice.
(a) Schematic for the 5-choice serial reaction time task (5-CSRTT).
(b) Sessions required to reach performance criteria for 5-CSRTT (n = 16 per genotype). Pairwise comparisons: wild-type vs. TDP-43Q331K/+: P=0.083 (ns); wild-type vs. TDP-43Q331K/Q331K: P=0.004 (**).
(c) 5-CSRTT at 6 months of age (n = 15 wild-type, 16 TDP-43Q331K/+, 15 TDP-43Q331K/Q331K mice). Baseline session genotype effects: accuracy: P=0.109; omission: P=0.283). Stimulus duration (SD) probe test genotype effects: accuracy: P=0.833; omission: P=0.077 (ns); SD effect: accuracy and omission: P<0.001; Mixed-effects model.
(d) 5-CSRTT at 12 months of age (n = 15 wild-type, 16 TDP-43Q331K/+, 16 TDP-43Q331K/Q331K mice). Baseline session genotype effects: accuracy: P=0.487; omission: P=0.120. SD probe test genotype effects: accuracy: P=0.880; omission: P=0.044 (*); SD effect: accuracy: P<0.0001; omission: P<0.0001; genotype by SD interaction: accuracy: P=0.081; omission: P=0.271; Mixed-effects model.
(e) Mean trials completed on an unrestricted fixed-ratio schedule (n = 16 per genotype).
(f) Mean breakpoint on a progressive-ratio schedule (response increment per trial = 4; n = 16 per genotype).
(g) Novel object recognition sample and (h) choice phases (n = 8 wild-type, 9 TDP-43Q331K/+, 8 TDP-43Q331K/Q331K mice). For (h) 1 min delay pairwise comparisons: wild-type vs. TDP-43Q331K/+: P=0.158 (ns); wild-type vs. TDP-43Q331K/Q331K: P=0.158 (ns); 3 hour delay pairwise comparisons: wild-type vs. TDP-43Q331K/+: P=0.014 (*); wild-type vs. TDP-43Q331K/Q331K: P=0.009 (**).
For (b,e,f) one-way ANOVA and (g,h) two-way ANOVA, all followed by Holm-Sidak post-hoc tests for pairwise comparisons.
(i) Marbles buried in Cohort 1 at 18 months of age (n = 15 wild-type, 13 TDP-43Q331K/+, 14 TDP-43Q331K/Q331K mice). Pairwise comparisons: wild-type vs. TDP-43Q331K/+: P=0.009 (**); wild-type vs. TDP-43Q331K/Q331K: P<0.0001 (****); Kruskal-Wallis followed by Dunn’s test for pairwise comparisons.
Error bars denote s.e.m. for (c) to (h) and median and interquartile range for (b) and (i).
Next, to explore temporal lobe-dependent function, we conducted the spontaneous object recognition task, a test of declarative memory. Initial exploratory times did not differ between genotypes (Fig. 3g), but in the choice phase a deficit emerged in TDP-43Q331K/+ and TDP-43Q331K/Q331K mice (Fig. 3h), indicating memory impairment. The combination of executive dysfunction and memory impairment, together with hyperphagia in free-fed Cohort 1 mice led us to conclude that TDP-43Q331K/+ and TDP-43Q331K/Q331K mice recapitulate FTD at the behavioural level.
During touchscreen analyses we noted that some Cohort 2 mutant mice demonstrated consistently worse performance than other mutants (Fig. 3b, Supplementary Fig. 4i). This phenotypic heterogeneity was intriguing given that the mutant mice were genetically homogeneous. Furthermore, ALS-FTD is a remarkably heterogeneous disease in which patients display varying phenotypic severity and different rates of disease progression. Indeed, TARDBP mutation carriers demonstrate variable penetrance even with homozygous mutations15. We therefore looked for further evidence of phenotypic heterogeneity by examining Cohort 1 mice using the marble-burying assay, a measure of innate digging behaviour23. From 5 to 18 months of age, wild-type mice buried ~80% of marbles. Mutants demonstrated a range of digging behaviours, with some animals behaving similarly to wild-types, but others demonstrating attenuated digging behaviour (Fig. 3i, Supplementary Fig. 4j). These observations confirm the presence of phenotypic heterogeneity in genetically homogeneous groups of mutant mice, and suggest that some mutants were relatively resistant to behavioural deficits caused by TDP-43Q331K.
TDP-43Q331K mice demonstrate perturbed TDP-43 autoregulation and reduced parvalbumin-positive neurons
To obtain mechanistic insight into the cognitive dysfunction caused by TDP-43Q331K we sacrificed 5-month-old mice for pathological and transcriptomic studies. Prior to sacrifice we performed the marble-burying assay to identify animals with a range of different behaviours (Fig. 4a). Analysis of frontal cortices from wild-type and TDP-43Q331K/Q331K mice demonstrated no significant reduction in cortical thickness or cellular density in mutants (Fig. 4b, Supplementary Fig. 5a-c), and no nuclear clearing or cytoplasmic aggregation of TDP-43 (Fig. 4c). However, subcellular fractionation and immunoblotting demonstrated a ~45% increase in nuclear TDP-43 in TDP-43Q331K/Q331K compared to wild-type mice (Fig. 4d,e, Supplementary Fig. 5d).
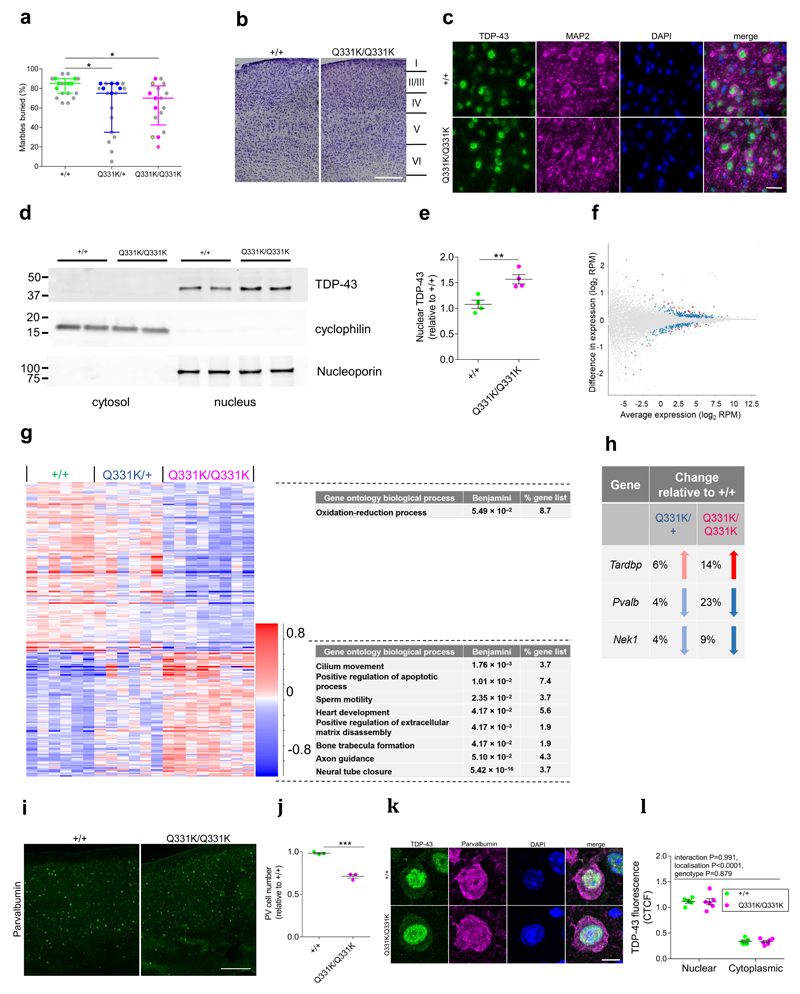
Figure 4. Perturbed TDP-43 autoregulation and loss of parvalbumin interneurons in mutant mice.
(a) Marbles buried by 5-month-old mice. Coloured dots indicate animals used for RNASeq analysis. Yellow dots indicate TDP-43Q331K/Q331K littermates (n = 19 wild-type, 19 TDP-43Q331K/+, 17 TDP-43Q331K/Q331K mice). Pairwise comparisons: wild-type vs. TDP-43Q331K/+: P=0.028 (*); wild-type vs. TDP-43Q331K/Q331K: P=0.013 (*); Kruskal-Wallis followed by Dunn’s test for pairwise comparisons. Error bars represent median and interquartile range.
(b) Representative Nissl staining of frontal cortex (layers indicated) (n = 5 wild-type, 6 TDP-43Q331K/Q331K mice). Scale bar, 500μm.
(c) Immunohistochemistry for TDP-43 in pyramidal neurons of motor cortex layer V. Representative images shown (n = 4 mice per genotype). Scale bar, 20μm.
(d) Immunoblot of fractionated frontal cortical tissue from 5-month-old mice (two biological replicates shown, uncropped in Supplementary Fig. 5).
(e) Immunoblot band intensity quantification (n = 4 mice per genotype). Comparison: P=0.007 (**); unpaired t test. Error bars denote s.e.m.
(f) MA plot and (g) hierarchical clustering of DEGs (n = 6 wild-type, 6 TDP-43Q331K/+, 8 TDP-43Q331K/Q331K mice) in frontal cortex. For (f) blue dots indicate significant changes, red dots indicate intensity hits. Comparison: DESeq2 wild-type v TDP-43Q331K/Q331K. For (g) gene ontology (GO) biological process and KEGG pathway enriched terms are displayed.
(h) Expression changes for parvalbumin and ALS-FTD linked genes identified by RNASeq.
(i) Immunohistochemistry for parvalbumin in cortices of 5-month-old mice. Representative images shown. Scale bar, 250μm.
(j) Quantification of parvalbumin-positive neurons (n = 3 mice per genotype). Comparison: P=0.0003 (***); unpaired t test. Error bars denote s.e.m.
(k) Immunohistochemistry for TDP-43 in parvalbumin-positive cells. Representative images shown. Scale bar, 5μm.
(l) TDP-43 expression in parvalbumin-positive cells (n=5 mice per genotype). Comparison by two-way ANOVA. Error bars denote s.e.m.
TDP-43 has critical roles in RNA processing, which may be disturbed in disease. We therefore performed transcriptomic analyses using RNASeq of frontal cortices from six wild-type, six TDP-43Q331K/+, and eight TDP-43Q331K/Q331K mice (Supplementary Fig 6a). We identified 171 genes that were upregulated and 233 that were downregulated in TDP-43Q331K/Q331K mice relative to wild-type (Fig. 4f,g). TDP-43Q331K/+ mice demonstrated changes that trended in the same direction as TDP-43Q331K/Q331K mice, suggesting a dose-dependent effect of the mutation. In particular, we noted a 14% increase in expression of Tardbp in TDP-43Q331K/Q331K mice (Fig. 4h). As nuclear TDP-43 protein expression was also raised in mutants, we conclude that the Q331K mutation disturbs TDP-43 autoregulation.
One notable gene that was downregulated in mutant mice was Nek1. This change is consistent with human data indicating that loss-of-function mutations in NEK1 cause ALS24,25. Another downregulated gene was Pvalb, which encodes the calcium buffering protein parvalbumin. Reduced parvalbumin immunopositivity is observed in patients with ALS and is linked with selective cellular vulnerability in ALS26. We therefore immunostained for parvalbumin and found a ~25% reduction in parvalbumin-positive cells in the frontal cortex of TDP-43Q331K/Q331K mice (Fig. 4i,j). Co-staining for TDP-43 in this affected subset of cortical neurons did not demonstrate TDP-43 mislocalisation (Fig. 4k,l). Notably, fast-spiking parvalbumin interneurons are GABAergic inhibitory cells that play a direct role in the control of attention27. We therefore conclude that a paucity of parvalbumin interneurons may be responsible for the attentional impairment of TDP-43Q331K mice.
Splicing analysis indicates a gain-of-function of TDP-43Q331K and links aberrant TDP-43 homeostasis with altered splicing of Mapt
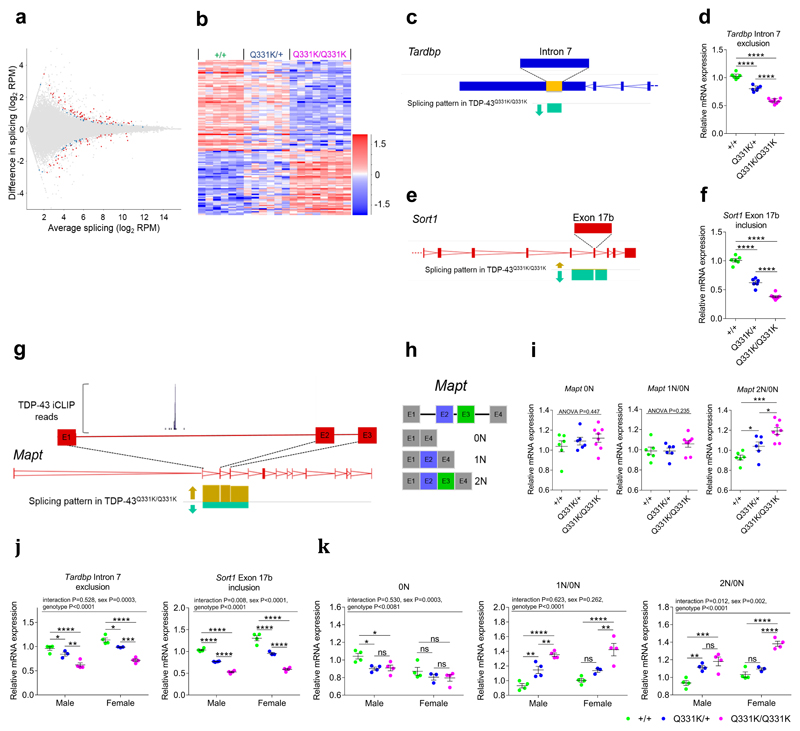
TDP-43 plays key roles in alternative splicing. We therefore interrogated the cortical transcriptomic dataset further for splicing differences between mutant and wild-type mice and identified 138 splicing changes in 106 genes (Fig. 5a,b, Supplementary Fig. 6b). This included an ~80% increase in retention of Tardbp intron 7 in TDP-43Q331K/Q331K mice (Fig. 5c,d), which will promote the production of stable mRNA species7. This confirms that TDP-43 autoregulation is perturbed in mutant mice. Another prominent change was a 2.4-fold increase in exclusion of Sort1 exon 17b, a known splicing target of TDP-43 (Fig. 5e,f). This change is consistent with a gain of function of TDP-4328.
Figure 5. Splicing analysis indicates TDP-43 misregulation, a gain of TDP-43 function and altered Mapt exon 2/3 splicing.
(a) MA plot and (b) hierarchical clustering of frontal cortical alternative splice events (n = 6 wild-type, 6 TDP-43Q331K/+, 8 TDP-43Q331K/Q331K mice). Comparison: DESeq2 wild-type v TDP-43Q331K/Q331K.
(c) Schematic of altered splicing in the 3’UTR of Tardbp. Arrow indicates reduced exclusion of intron 7 of the Tardbp transcript in TDP-43Q331K/Q331K relative to wild-type mice.
(d) Quantitative PCR (qPCR) of splicing changes in Tardbp intron 7 (n = 6 wild-type, 6 TDP-43Q331K/+, 8 TDP-43Q331K/Q331K mice).
(e) Schematic of exon 17b inclusion/exclusion in Sort1. Arrows indicate reduced inclusion of exon 17b in TDP-43Q331K/Q331K relative to wild-type mice.
(f) qPCR of splicing changes in Sort1 exon 17b (n = 6 wild-type, 6 TDP-43Q331K/+, 8 TDP-43Q331K/Q331K mice).
(g) Schematic of altered splicing of exons 2 and 3 of Mapt. Arrows indicate increased inclusion of exons 2 and 3 in the Mapt transcripts of TDP-43Q331K/Q331K relative to wild-type mice. The expanded view of exon 1 to exon 2 includes a site of TDP-43 binding as detected by iCLIP (iCount pipeline; TDP-43_CLIP_E18-brain).
(h) Schematic of N-terminal Mapt splice variants (0N, 1N and 2N).
(i) qPCR of splicing changes in Mapt exons 2 and 3 (n = 6 wild-type, 6 TDP-43Q331K/+, 8 TDP-43Q331K/Q331K mice). 2N/0N pairwise comparisons: wild-type vs. TDP-43Q331K/+: P=0.047 (*); wild-type vs. TDP-43Q331K/Q331K: P=0.0001 (***); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.013 (*).
(j-k) qPCR of hippocampal splicing changes (n = 4 wild-type, 3 TDP-43Q331K/+, 4 TDP-43Q331K/Q331K mice per gender). Pairwise comparisons: Tardbp intron 7 exclusion, male: wild-type vs. TDP-43Q331K/+: P=0.043 (*); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.002 (**); female: wild-type vs. TDP-43Q331K/+: P=0.013 (*); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.0002 (***); Mapt: 0N, male: wild-type vs. TDP-43Q331K/+: P=0.023 (*); wild-type vs. TDP-43Q331K/Q331K: P=0.023 (*); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.877 (ns); female: wild-type vs. TDP-43Q331K/+: P=0.365 (ns); wild-type vs. TDP-43Q331K/Q331K: P=0.324 (ns); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.858 (ns); 1N/0N, male: wild-type vs. TDP-43Q331K/+: P=0.008 (**); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.008 (**); female: wild-type vs. TDP-43Q331K/+: P=0.077 (ns); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.002 (**); 2N/0N, male: wild-type vs. TDP-43Q331K/+: P=0.002 (**); wild-type vs. TDP-43Q331K/Q331K: P=0.0001 (***); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.151 (ns); female: wild-type vs. TDP-43Q331K/+: P=0.202 (ns).
For (d,f,i-k) P<0.0001 (****). For (d,f,i) one-way and (j,k) two-way ANOVA, all followed by Holm-Sidak post-hoc tests for pairwise comparisons.
Error bars denote s.e.m.
We also noted altered splicing of exons 2 and 3 of Mapt, which encodes the microtubule associated protein tau and is mutated in FTD with Parkinsonism29. We detected increased inclusion of Mapt exons 2 and 3 in TDP-43Q31K/Q331K mice (Fig. 5g-i). This is notable as inclusion of exons 2 and 3 of Mapt is associated with increased somatodendritic localization and aggregation of tau30. We immunostained wild-type and mutant frontal cortices for total tau but found no difference in the localization or aggregation of tau (Supplementary Fig. 6c). Analysis of iCLIP databases (http://icount.biolab.si/groups.html) revealed that TDP-43 binds to an intronic sequence upstream of Mapt exon 2 (Fig. 5g). This confirmed that Mapt exons 2 and 3 are likely splicing targets of TDP-43. The identification of this novel splicing effect of TDP-43 on Mapt mechanistically links these two major dementia genes.
Next, to determine if TDP-43 misregulation could be responsible for temporal lobe-dependent functions we analysed hippocampal RNA extracts from male mice. We also examined hippocampi from female mice to determine if TDP-43 misregulation was restricted to male mice. Splicing analyses for Tardbp, Sort1 and Mapt were consistent with a gain of function of TDP-43 in mutant mice of both genders (Fig. 5j,k). This indicates that TDP-43 misregulation occurs beyond the frontal cortex, and in both male and female mice.
Finally, to confirm that our behavioural and transcriptomic observations were caused by mutant TDP-43 and not off-target CRISPR mutagenesis effects we performed the marble-burying assay in a second line of Tardbp Q331K knock-in mice, line #3, and found a similar impairment of digging behavior to line #52 mice (Supplementary Fig. 6d). We also analysed RNA from line #3 mice and observed an increase in Tardbp expression and altered splicing of Tardbp and Sort1, which is consistent with perturbed autoregulation and a gain of function of TDP-43 (Supplementary Fig. 6e). Furthermore, line #3 TDP-43Q331K/Q331K mice also demonstrated increased inclusion of exons 2 and 3 of Mapt, and a paucity of parvalbumin-positive neurons relative to wild-type mice, replicating key splicing and pathological observations made in line #52 mice (Supplementary Fig. 6e,f),
TDP-43 misregulation in lumbar spinal cords of mutant mice further implicates interneurons in ALS-FTD pathogenesis
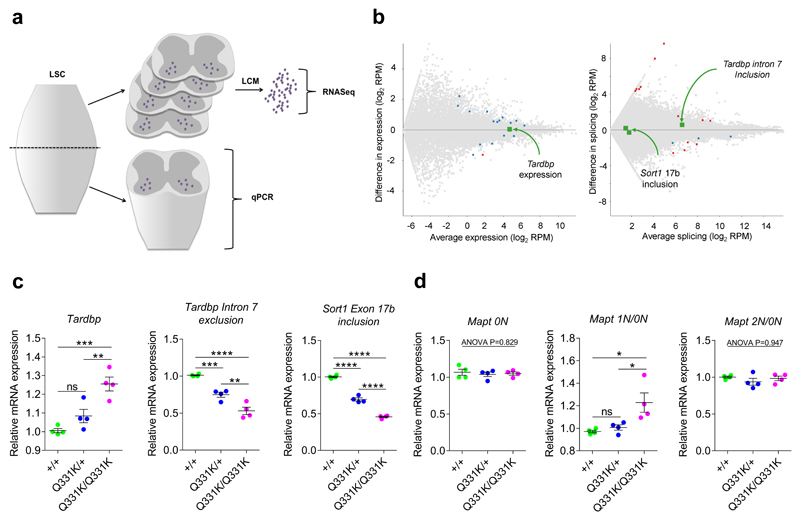
Our transcriptomic profiling of frontal cortices and hippocampi elucidated a gain of function of TDP-43 in the brains of mutant mice. By contrast, spinal motor neurons from mutants did not demonstrate TDP-43 misregulation as Tardbp, Sort1 and Mapt were not differentially expressed or spliced in these cells (Fig. 6b). However, TDP-43 misregulation could occur in other cells of the spinal cord, namely glia or interneurons. We therefore analysed RNA from homogenates of lumbar spinal cord from the mice from which we had laser captured spinal motor neurons (Fig. 6a). Interestingly, spinal cord homogenates demonstrated increased expression of Tardbp, and altered splicing of Tardbp and Sort1 consistent with a gain of function of TDP-43 in mutant mice (Fig. 6c). Furthermore, spinal cords from mutant mice also demonstrated increased inclusion of Mapt exon 2 (Fig. 6d). Given that Mapt expression is predominantly neuronal rather than glial this suggests that a gain of TDP-43 function occurs in interneurons of the spinal cord.
Figure 6. TDP-43 misregulation occurs in spinal cords of mutant mice, but not in motor neurons.
(a) Schematic detailing lumbar spinal cord (LSC) processing for transcriptomic analysis (LCM, laser capture microdissection).
(b) MA plots of lumbar motor neuronal differentially expressed and spliced genes (n = 4 mice per genotype). Comparison: DESeq2 wild-type v TDP-43Q331K/Q331K. Blue and red dots indicate significant changes. Green dots highlight Tardbp expression, Tardbp intron 7 exclusion and Sort1 exon 17b inclusion, which are not significant changes.
(c-d) Quantitative PCR of homogenised lumbar spinal cord (n = 4 wild-type, 4 TDP-43Q331K/+, 4 TDP-43Q331K/Q331K mice). Comparisons as follows:
(c) Tardbp expression: wild-type vs. TDP-43Q331K/+: P=0.103 (ns); wild-type vs. TDP-43Q331K/Q331K: P=0.0008 (***); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.007 (**). Tardbp intron 7 exclusion: wild-type vs. TDP-43Q331K/+: P=0.001 (***); wild-type vs. TDP-43Q331K/Q331K: P>0.0001 (****); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.002 (**). Sort1 exon 17b inclusion: P<0.0001 (****).
(d) 0N Mapt. 1N Mapt: wild-type vs. TDP-43Q331K/+: P=0.640 (ns); wild-type vs. TDP-43Q331K/Q331K: P=0.02 (*); TDP-43Q331K/+ vs. TDP-43Q331K/Q331K: P=0.03 (*). 2N Mapt.
(c-d) Comparisons by one-way ANOVA followed by Holm-Sidak post-hoc tests.
Error bars denote s.e.m.
Stratification of transcriptomic data from TDP-43Q331K/Q331K mice by phenotype identifies novel expression and splicing changes
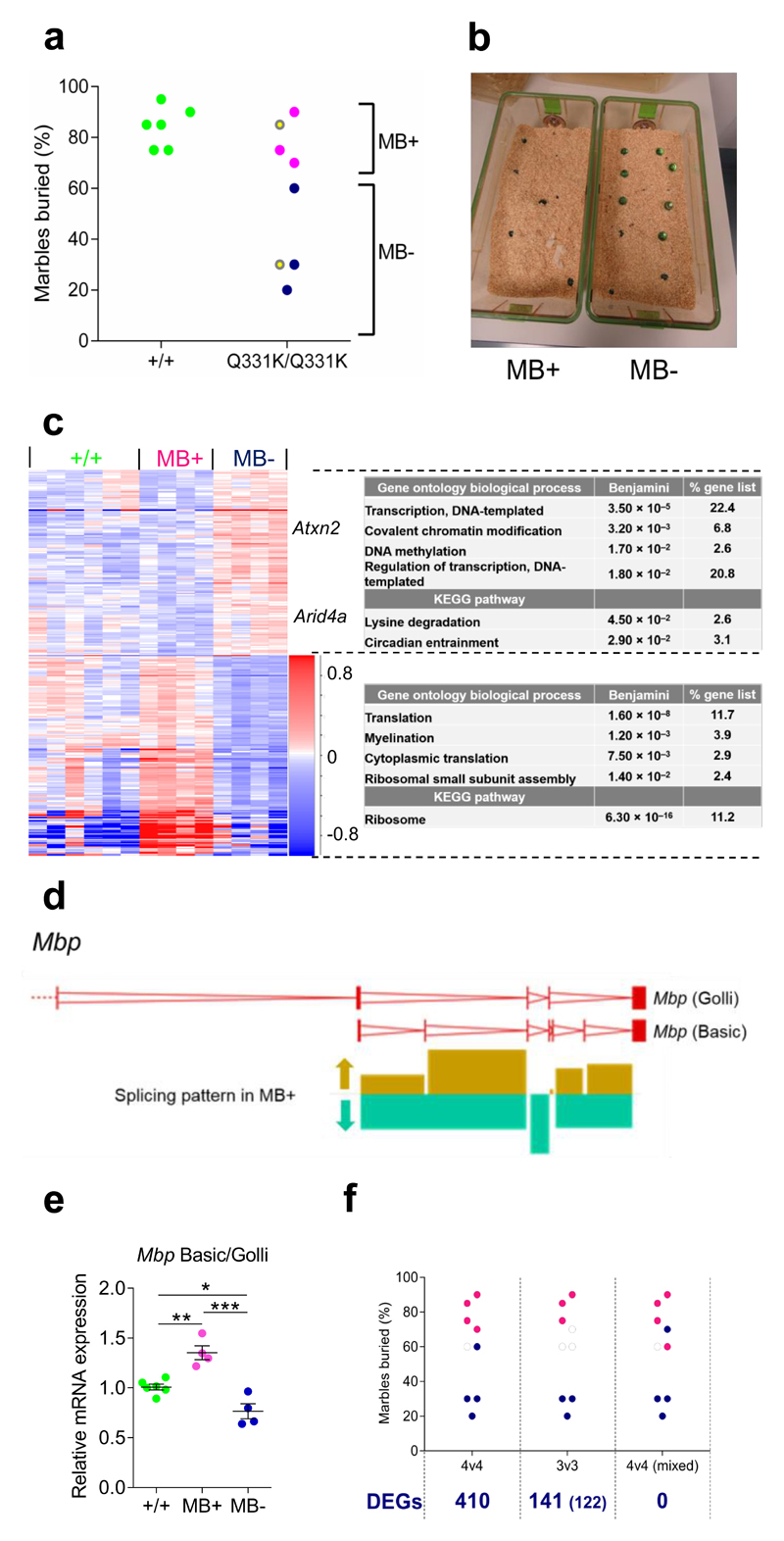
As stated earlier, some mutant mice appeared relatively resistant to the cognitive effects of the Q331K mutation. We wished to exploit this phenotypic heterogeneity in TDP-43Q331K/Q331K mice to identify potential modifiers of cognitive dysfunction. For this purpose we divided the frontal cortical transcriptomic data from the eight TDP-43Q331K/Q331K mice into two subsets according to their antemortem marble-burying behaviour. We designated this the ‘MB+/-’ comparison. TDP-43Q331K/Q331K mice that dug consistently well were designated MB+, and those that dug consistently poorly were designated MB- (Fig. 7a,b). We hypothesised that transcriptomic differences between these two genotypically homogeneous groups would indicate molecular pathways that influenced the risk of developing cognitive impairment. Using this strategy we found 410 gene-expression and 61 splicing differences between MB+ and MB- groups (Fig. 7c, Supplementary Fig. 6g,h), which were entirely different to those seen in the earlier comparison with wild-type mice when all eight TDP-43Q331K/Q331K mice were considered as one group (Fig. 4g, 5b). Interestingly, for 78% of these genes MB+ and MB- mice demonstrated opposing expression changes relative to wild-type (Fig. 7c, Supplementary Table 2 and MB+/- sections in Supplementary Table. 1). Effectively, for these genes an expression change in one direction is associated with a poor behavioural phenotype, yet an expression change in the opposite direction is associated with improved behavior. Furthermore, there was no difference in TDP-43 expression or the degree of TDP-43 gain of function as evidenced by Sort1 splicing between MB+ and MB- groups. Taken together, these data indicate that the MB+/- comparison genes could be metastable modulators of TDP-43-mediated cognitive dysfunction.
Figure 7. Phenotypic stratification of transcriptomic data from mutant mice allows the identification of putative disease modifiers.
(a) Marble-burying in 5-month-old mice prior to sacrifice. MB+ mice bury at or above the median number of marbles for the group, and MB- mice bury fewer. Yellow dots indicate TDP-43Q331K/Q331K littermates.
(b) Marble burying activity of TDP-43Q331K/Q331K littermates as described in (a).
(c) Hierarchical clustering of DEGs in frontal cortices comparing MB+ and MB- TDP-43Q331K/Q331K mice. Genes Atxn2 and Arid4a are highlighted (n = 6 wild-type, 4 MB+ TDP-43Q331K/Q331K and 4 MB- TDP-43Q331K/Q331K mice). Comparison: DESeq2 MB+ v MB-. Gene ontology (GO) biological processes and KEGG pathway enriched terms are displayed.
(d) Graphical representation of altered splicing of Mbp. Arrows indicate the altered pattern of splicing in MB+ relative to MB- TDP-43Q331K/Q331K mice.
(e) qPCR of the ratio of Mbp Basic to Mbp Golli (n = 6 wild-type, 4 TDP-43Q331K/+, 4 TDP-43Q331K/Q331K mice). Pairwise comparisons: wild-type vs. MB+: P=0.005 (**); wild-type vs. MB-: P=0.024 (*); MB+ vs. MB-: P=0.0003 (***); one-way ANOVA followed by Holm-Sidak post-hoc tests. Error bars denote s.e.m.
(f) Representative marble burying analyses: 4:4, original analysis; 3:3, comparing the three best MB+ and three worst MB- mice; 4v4 mixed, one MB- mouse swapped with one MB+ mouse. Number of DEGs identified by DESeq2 comparison of MB+ v MB- mice for each comparison is given below. For 3:3, hits common to the 4:4 stratification are shown in brackets.
Significantly, two of the genes from the MB+/- comparison have previously been linked with suppression of neurodegeneration: Atxn2 and Arid4a. Compared to wild-type mice, MB+ mice demonstrated reduced Atxn2 expression, while MB- mice demonstrated increased Atxn2 expression. This is in keeping with previous observations that Atxn2 knockdown suppresses TDP-43 toxicity in yeast, Drosophila and mouse31,32. Furthermore, intermediate expansions of Atxn2 CAG repeat length is associated with ALS disease risk in humans31. Similarly, reduced expression of the chromatin-modeling gene Arid4a in MB+ mice is notable, as we previously found that loss of function mutations in hat-trick, the Drosophila orthologue of Arid4a, suppress TDP-43-mediated neurodegeneration in flies12. It is therefore likely that reduced levels of Atxn2 and Arid4a are similarly neuroprotective in TDP-43Q331K/Q331K MB+ mice.
To identify the most significant pathways linked with phenotypic heterogeneity in the MB+/- comparison we cross-referenced the differential gene expression list with the Gene Ontology database for biological processes (Fig. 7c). Genes downregulated in MB+ mice were enriched for biological processes involving transcription, DNA methylation and chromatin modification. Genes upregulated in MB+ mice were enriched for processes involving protein translation and myelination, including the myelin repair gene Olig1, and Mbp, which encodes myelin basic protein (Supplementary Table 2). Furthermore, examination of the splicing gene list also identified Mbp as a candidate (Fig. 7d,e). Specifically, MB- mice demonstrated a significantly increased expression of a specific splice form, which is predicted to encode Golli-Mbp, in which three additional exons upstream of classical Mbp are normally expressed in non-myelinating cells including neurons, and in immature oligodendrocytes33. Collectively, this Gene Ontology analysis identifies an association between the upregulation of protein translation and oligodendrocyte genes and improved behaviour in TDP-43Q331K/Q331K mice, and suggests that the promotion of myelin repair pathways by oligodendrocytes in a mature state contributes to improved cognition.
To confirm the validity of MB+/- hits we deliberately swapped data from the worst performing MB+ mouse with that of the best performing MB- mouse. This resulted in all transcriptomic hits disappearing from the analysis (Fig. 7f). We also compared only the three best performing MB+ mice with the three worst performing MB- mice and found a diminished hit list, but which largely overlapped with the genes from the complete MB+/- comparison. Furthermore, we found two TDP-43Q331K/Q331K mice that were littermates yet demonstrated contrasting digging behaviour on repeated assessment (Fig. 7a,b). This indicated that transcriptomic differences between MB+ and MB- groups were not due to a genetic founder effect within our breeding program. Collectively, these data indicate that the MB+/- transcriptomic differences were genuinely reflective of two phenotypic subsets of young TDP-43Q331K/Q331K mice.
TDP-43Q331K mice demonstrate age-related deterioration of cortical transcriptomes with altered expression and splicing of other ALS-linked genes
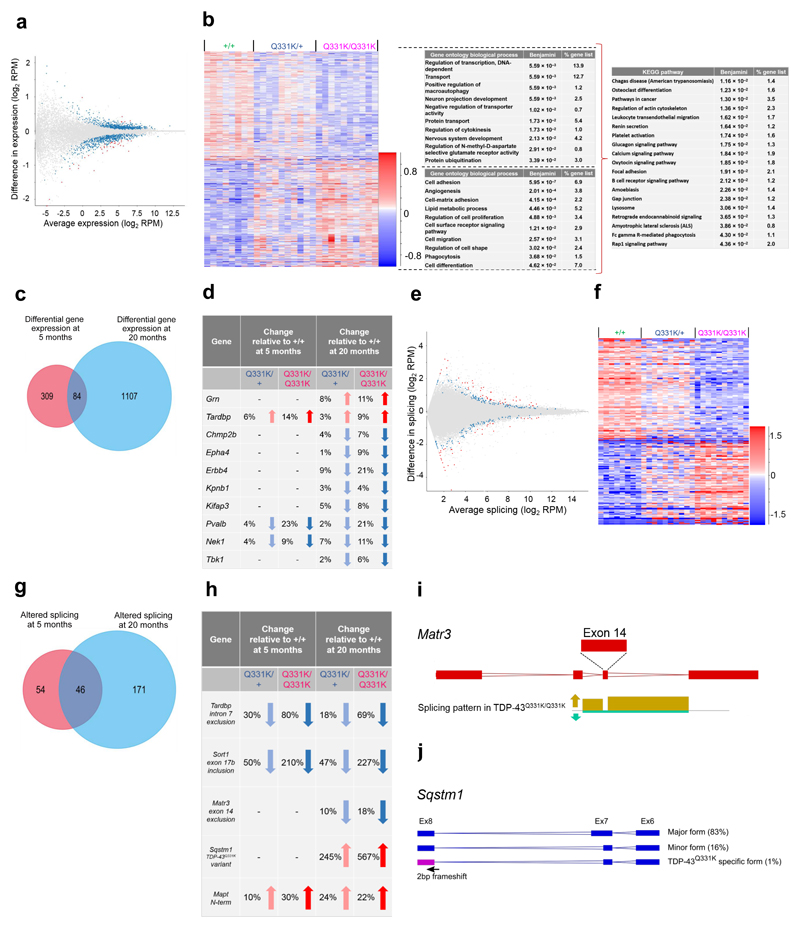
Ageing is the greatest known risk factor for sporadic ALS-FTD. To determine the effects of ageing on TDP-43Q331K mice we performed a frontal cortical RNASeq study in 20-month-old mice (Fig. 8a,b,e,f, Supplementary Fig. 7a,b). Comparison of wild-type and mutant mice revealed transcriptomic differences that partly overlapped with the 5-month-old dataset (Fig. 8c,d,g,h). Significantly, aged mutant mice still demonstrated a gain of function of TDP-43, increased retention of Mapt exons 2 and 3, and reduced Nek1 and Pvalb expression. However, a broader range of transcriptomic changes was seen, further implicating inhibitory interneuronal disturbances, including downregulation of Sirt1 and Ppargc1a, which encode proteins involved in Pvalb transcription, and downregulation of GAD1/GAD67, which encodes the GABA synthetic protein glutamate decarboxylase (Supplementary Table 1). Aged mice also demonstrated downregulation of Tbk1 (encoding Tank binding protein kinase 1) (Fig. 8d), loss of function mutations of which cause ALS and FTD34,35. Several other ALS-FTD-linked genes also demonstrated significant downregulation, including Chmp2b, mutations of which cause FTD36, Erbb4, mutations of which cause ALS37, the ALS risk-linked gene Epha438, and the TDP-43 nuclear import factor Kpnb140. We also observed altered splicing of ALS-linked genes Matr341 (decreased exclusion of exon 14, which encodes a zinc finger domain), and Sqstm142 (Fig. 8h-j, Supplementary Fig. 7f,g). For Sqstm1 two splice variants (major and minor) were detected in wild-type and mutant mice, but a third variant was present only in mutants. This TDP-43Q331K-specific variant comprises a truncated 7th exon and a 2bp frameshift in exon 8 of Sqstm1, which is predicted to introduce a premature stop codon with loss of the C-terminal ubiquitin-associated domain of sequestosome 1 (Fig. 8j). Furthermore, Gene Ontology and pathway analysis of the RNASeq dataset in 20-month-old mice revealed many more significant networks than had been identified in 5-month-old TDP-43Q331K mice. Aged mutants demonstrated changes in processes classically linked to neurodegeneration, including protein ubiquitination, autophagy, and glutamate receptor activity, while KEGG pathway analysis highlighted ‘ALS’ and immune pathways (Fig. 8b). These pathways were not invoked in young mice (Fig. 4g). Collectively, these observations in aged mutant mice validate key transcriptomic findings in young mutants, link aberrant TDP-43 homeostasis with other key ALS-FTD-linked genes, and indicate age-related progressive changes in the cortical transcriptomes of TDP-43Q331K mice.
Figure 8. TDP-43Q331K mice demonstrate age-related deterioration in cortical transcriptomes with altered expression of multiple ALS-linked genes.
(a) MA plot and (b) hierarchical clustering of DEGs in frontal cortices at 20 months of age (n = 8 wild-type, 10 TDP-43Q331K/+, 10 TDP-43Q331K/Q331K mice). For (a) blue dots indicate significant changes, red dots indicate intensity hits. Comparison: DESeq2 wild-type v TDP-43Q331K/Q331K. For (b) gene ontology (GO) biological processes and KEGG pathway enriched terms are displayed.
(c) Venn diagram highlighting DEGs between wild-type v TDP-43Q331K/Q331K mice that were common to analyses in 5 and 20-month-old mice. Known ALS-FTD linked genes within this common subset are highlighted in (d).
(e) MA plot and (f) hierarchical clustering of frontal cortical alternative splice events at 20 months of age (n = 8 wild-type, 10 TDP-43Q331K/+, 10 TDP-43Q331K/Q331K mice). Blue dots indicate significant changes, red dots indicate intensity hits. Comparison: DESeq2 wild-type v TDP-43Q331K/Q331K.
For (a,b,e,f) n = 8 wild-type, 10 TDP-43Q331K/+, 10 TDP-43Q331K/Q331K mice.
(g) Venn diagram highlighting alternative splice events between wild-type v TDP-43Q331K/Q331K mice that are common to analyses in 5 and 20-month-old mice. Known ALS-FTD linked genes within this common subset are highlighted in (h).
(i) Schematic of Matr3 exon 14 inclusion/exclusion. Arrows indicate increased inclusion of exon 14 in TDP-43Q331K/Q331K relative to wild type mice.
(j) Schematic of Sqstm1 transcript splice variants. Percentages given indicate the relative amount of each variant in TDP-43Q331K/Q331K mice. The TDP-43Q331K-specific variant is undetectable in wild-type mice.
Finally, to identify transcriptomic differences associated with long-term resistance to cognitive impairment we performed an MB+/- comparison in aged mice. As most aged TDP-43Q331K/Q331K mice had progressed to an MB- state by 20 months, we compared TDP-43Q331K/+ mice, which we were able to stratify into MB+ and MB- groups. This comparison yielded only 21 differentially expressed genes, and 45 splicing differences between TDP-43Q331K/+MB+ and MB- mice, which did not overlap with those genes identified in the MB+/- comparison of 5-month-old TDP-43Q331K/Q331K mice (Supplementary Fig. 7c-e). This suggests that aged TDP-43Q331K/+ mice are not amenable to stratification in the same way as young TDP-43Q331K/Q331K mice, and further suggests that modulation of MB+/- genes early in life has the potential to influence longer-term susceptibility to cognitive impairment secondary to aberrant TDP-43 homeostasis.
Discussion
Here, we show that with a single human disease-linked base change in murine Tardbp it is possible to replicate behavioural, pathological and transcriptomic features of the ALS-FTD spectrum. Significantly, by creating a model that mimics the human mutant state as closely as possible and in the absence of exogenous expression we elucidated that the Q331K mutation perturbs TDP-43 autoregulation. This leads to an increase in TDP-43 expression (effectively a gain of function defect). Interestingly, spinal cords from sporadic ALS patients and from TARDBP mutation carriers demonstrate increased TDP-43 mRNA expression, as do human stem cell-derived motor neurons with TARDBP mutations43,44. This indicates that TDP-43 misregulation could underpin the human disease state.
Interestingly, lumbar motor neurons of TDP-43Q331K/Q331K mice demonstrated upregulation of genes that may confer neuroprotection and did not demonstrate TDP-43 misregulation, both of which might explain why mutant mice did not demonstrate significant neuromuscular phenotypes. By contrast, the FTD-like phenotypes in mutant mice were more significant. The identification of reduced parvalbumin expression as a possible cause for cognitive impairment in ALS-FTD is intriguing as parvalbumin interneuron loss has been observed in sporadic ALS and FTD26. As parvalbumin interneurons are GABAergic a reduction in their number could increase activity of cortical projection neurons with excitotoxic consequences. Early interneuronal dysfunction may have analogous consequences in the spinal cord and is suggested by our observation that TDP-43 autoregulation is perturbed in the spinal cord, but not in motor neurons.
That TDP-43Q331K mice demonstrate a specific increase in inclusion of Mapt exons 2 and 3 is of great interest as 2N tau oligomers appear to have a greater ability to provoke tau aggregation than 0N and 1N isoforms30, and inclusion of exon 2 and 3 influence subcellular localisation and protein-protein interactions of tau45. Furthermore, in humans the H2 Mapt haplotype is associated with a greater inclusion of Mapt exon 3 and is associated with an earlier age of onset in FTD46,47. Although we did not observe clear disturbances of total tau localisation in TDP-43Q331K mice, more detailed analyses to identify specific tau isoforms are warranted. Our identification of a mechanistic link between TDP-43 and Mapt adds to growing evidence that ALS-FTD is characterised by both TDP-43 and tau pathology48. Furthermore, transcriptomic analysis of aged TDP-43Q331K mice elucidated changes in other ALS-FTD linked genes. Collectively, these findings emphasise a central role for TDP-43 in neurodegeneration.
Finally, we observed phenotypic heterogeneity among mutant mice with the same genotype and identified distinct transcriptomic profiles corresponding to differing phenotypes. This transcriptomic dataset contains genes already implicated in neurodegeneration, including Arid4a12, and Atxn231. The unbiased discovery of Atxn2 downregulation as a hit in our model is consistent with observations validating Atxn2 knockdown as a therapeutic approach for ALS-FTD32. Our data suggest a delicate balance in the transcriptome of the brain, which is metastable and can influence disease onset or progression. Identifying the environmental factors that influence this balance is a priority in future studies. Indeed, the strong representation of DNA methylation and chromatin modelling genes in the MB+/- comparison suggests a critical role for epigenetic influences in determining disease susceptibility. Genes with roles in protein translation and oligodendrocyte biology including myelination also feature in our list of putative disease modifiers, and it is encouraging that both these pathways have roles in neurodegenerative disease49,50. Our wider list of potential modifiers of disease is composed of over 450 gene-expression and splicing changes that are associated with improved behaviour in TDP-43Q331K/Q331K mice. We conclude that this list contains additional novel suppressors of neurodegeneration that will help direct efforts towards developing treatments for ALS-FTD.
Online methods
CRISPR/CAS9 mutagenesis to introduce Q331K mutation
Nucleases were designed to be close to/overlap the desired point mutation. Three CRISPR-Cas9 nucleases were tested for activity using a GFP reporter plasmid. A 121 bp single-stranded DNA (ssDNA) oligonucleotide with the point mutation at the mid-point was used as a repair template. Guide RNA (gRNA) and a capped Cas9 mRNA were synthesised and injected with the donor oligonucleotide into 270 single-cell C57Bl/6J embryos. For sequences see Supplementary Table 3.
Off-targets were predicted using CRISPRseek51.
Mouse breeding and maintenance
Mouse founder #52 was outcrossed with wild-type C57Bl/6J mice through to the F3 generation. Three F3 male siblings were bred to wild-type C57Bl/6J mice to generate F4 TDP-43Q331K/+ mutants, which were intercrossed to generate animals for study.
Power calculations were based on historical rotarod and touchscreen data of wild-type mice. This indicated required group sizes of 15 animals per genotype to identify a ~20% difference in performance between genotypes. Animals were only excluded from analyses if specified in the following methods.
Mouse breeding was carried out in the UK and USA. ACBM was carried out at the Brown University Rodent Neurodevelopment Behaviour Testing Facility. All procedures were approved by the Brown University Animal Care and Use Committee. Touchscreen analysis; marble burying; object recognition; motor behaviour; food intake and weight measurement; pathology; electrophysiology and RNA sequencing all took place in the UK. All experiments were conducted in accordance with the United Kingdom Animals (Scientific Procedures) Act (1986) and the United Kingdom Animals (Scientific Procedures) Act (1986) Amendment Regulations 2012. Animals were housed in cages of up to five animals under a 12 hr light/dark cycle.
Genotyping
The Q331K mutation coincidentally introduces a SapI/EarI restriction site, which facilitates genotyping (see Supplementary Table 4).
Automated continuous behavioural monitoring
Ten TDP43Q331K/Q331K and 10 wild-type animals (5 female, 5 male of each genotype) from the same breeding campaign were obtained from the animal care facility at the University of Massachusetts Medical School. Animals were group housed between sessions, but housed individually during the 5-day ACBM recording sessions. Cages were monitored with a Firefly MV 0.3 MP Mono FireWire 1394a (Micron MT9V022) at 30 frames/s. Cameras were connected to a workstation with Ubuntu 14.04 with a firewire card to connect to all cameras. For processing by the computer vision system, all videos were down-sampled to 320×240 pixels.
The system used for ACBM was modified from that previously described and was re-implemented in Python and NVIDIA’s CUDNN to speed video analysis subroutines. All video analyses were conducted using the Brown University high-performance computer cluster. The system was retrained using data collected at the Brown Rodent Neuro-Developmental Behaviour Testing facility (~20 h of video and 40 animals total). Data were annotated by hand for 8 behaviours as previously described (drink, eat, groom, hang, rear, rest, sniff, walk). Accuracy was evaluated using by cross-validation. The average agreement with human annotations was 78% for individual behaviour and 83% overall for individual frames. Evaluation of the system was also run on a subset of the data collected for the present study, which found an overall mean agreement of 71% for individual behaviours and 82% over all video frames.
Rotarod
Motor testing was performed using Rotarod (Ugo Basile, Model 7650, Varese, Italy). At least 24 h prior to testing mice were first trained for 5 min at the slowest speed and then 7 min with acceleration. During testing mice were subjected to 7 min trials with acceleration from 3.5 to 35 rpm. In each session mice were tested 3 times with a trial separation of 30 min. The latency to fall (maximum 420 s) for each mouse was recorded and mean values for each mouse calculated. An individual mouse recording was excluded if it fell off the rod while moving backwards, accidentally slipped or jumped off at slow speed. Two consecutive passive rotations were counted as a fall and the time recorded as the end point for that mouse. Mouse weights were recorded immediately after completion of rotarod testing. All testing was conducted by operators who were blind to genotype and in a randomised order.
Feeding
Cages containing either two or three mice of the same genotype were topped up with 400g of food on Monday mornings. The following Monday the surplus food in the hopper together with any obvious lumps of food in the cage was removed and weighed. The difference from 400g was calculated and recorded as the total food consumed in seven days. This was normalised to the number of mice in a given cage. Weekly consumption was calculated for 9 consecutive weeks. Mice were 12 months of age when recording commenced. All testing was conducted while blind to genotype and in a randomised order.
Touchscreen studies
48 male mice (n = 16 per genotype) were housed in groups of 2-5 per cage under a 12 hr light/dark cycle (lights on at 7:00pm). Testing was conducted during the dark phase. To ensure sufficient levels of motivation, animals were food-restricted to ~85-90% of free-fed weights by daily provision of standard laboratory chow pellets (RM 3; Special Diet Services, Essex, UK). Drinking water was available ad libitum.
Experiments were performed in standard mouse Bussey-Saksida touchscreen chambers (Campden Instruments Ltd, Loughborough, UK). The reward for each correct trial was delivery of 20 μL of milkshake (Yazoo Strawberry milkshake®; FrieslandCampina UK, Horsham, UK). The chambers are equipped with infrared activity beams (rear beam = 3 cm from magazine port and front beam = 6 cm from screen) to monitor locomotor activity.
Following two days of habituation to touchscreen chambers, mice underwent pretraining and training. Briefly, mice were first trained to touch the correctly lit stimulus in return for a food reward, and to initiate a trial by poking and removing their nose from the magazine. Finally, mice were discouraged from making responses at non-illuminated apertures by a 5 s time-out period during which the chamber was illuminated. Investigators were blind to genotype.
5-choice serial reaction time task (5-CSRTT)
Upon completion of training at 2 s stimulus duration (baseline), mice were tested on 4 sessions of decreasing stimulus durations (2.0 s, 1.5 s, 1.0 s, 0.5 s) pseudo randomly within a session. Animals that had not reached the criterion (> 80% accuracy, < 20% omissions in two consecutive sessions in baseline training before entering the probe test, N = 1 in the first probe test) or whose body weights were below 80% of free-feeding weight (N = 1 in the first, and N = 1 in the second probe test) were excluded.
Fixed-ratio (FR) and progressive-ratio (PR) schedule
FR and PR were conducted as described elsewhere52. When performance stabilised on FR5 (completion of 30 trials within 20 min), all mice were tested on two sessions of an unrestricted FR5, which allowed an unlimited number of trials in 60 min. Next, animals underwent 3 sessions of PR4, in which animals should emit a progressively increasing number of responses (i.e. 1, 5, 9, 13, …) in each subsequent trial to obtain a single reward. PR session terminated following either 60 min or 5 min of inactivity. Breakpoint, the number of responses made to obtain the reward in the last completed trial, was recorded as an index of motivation.
Object recognition
The novel object recognition task was conducted as described elsewhere53 in a randomised order with the operator blind to genotype and under dimmed white light. Six-month-old male mice (n = 8-9 per genotype) were randomly chosen from Cohort 2. Mice were habituated to a Y-maze for 5 min. One day later mice were reintroduced to the Y-maze, which now contained two identical objects in each arm. Exploration time for each object over a 5 min period was recorded (sample phase). Mice were then removed from the maze and one of the objects replaced with a novel object. After a delay of 1 min or 3 h mice were reintroduced to the maze (choice phase) and the time spent exploring each object over a 5 min period was recorded. The memory for the familiar object was expressed as a discrimination ratio (difference in exploration of the novel and familiar objects divided by the total object exploration time).
Marble burying
All testing was conducted in the morning and blind to genotype. Cages of size 39.1cm x 19.9cm x 16.0cm height (Tecniplast) were used. Fresh bedding material (Datesand, grade 6) was placed into each cage to a height of ~6cm. Ten glass marbles (1cm) were placed evenly across the bedding. Ten cages were prepared in a single round. One mouse was placed in each of the cages and the lids replaced. Mice were left undisturbed for 30 min under white light. Mice were then removed and the number of marbles buried by at least two thirds was scored. Cages were reset using the same bedding material to test another 10 mice. In stratifying mice prior to frontal cortical RNAseq, animals were tested twice, three days apart to identify those that consistently buried high or low numbers of marbles.
Repeat behavioural studies
Cohort 1 mice underwent rotarod, weight, feeding and marble testing all under a standard light/dark cycle (lights on at 7:00am for 12h). Cohort 2 mice underwent all touchscreen, object recognition and rotarod studies under a reverse light/dark cycle.
Pathological studies
Mice were culled by cervical dislocation, decapitated and tissues processed as follows.
Brains
Right hemispheres were processed for RNA and/or protein extraction (see below). Left hemispheres were immersion fixed in 4% paraformaldehyde (PFA) at 4°C for 24 h, washed in PBS, cryoprotected in 30% sucrose in PBS at 4°C, embedded and frozen in M1 matrix (Thermo Fisher Scientific) on dry ice and sectioned coronally at 16 μm thickness on a cryostat (Leica Biosystems). Sections were mounted on Superfrost Plus charged slides (Thermo Fisher Scientific), allowed to dry overnight and stored at -80°C.
Spinal cords
Vertebral columns were dissected from culled mice, immersion fixed in 4% PFA at 4°C for 48 h, washed in PBS and dissected to extract spinal cords and nerve roots. The lumbar enlargement was sub dissected, cryoprotected in 30% sucrose at 4°C, embedded in M1 matrix in a silicon mould, frozen on dry ice and sectioned at 16 μm thickness onto charged slides, briefly air dried and stored at -80°C.
Antigen retrieval and immunostaining
Sections were thawed at R/T and briefly rinsed in distilled water. Antigen retrieval was performed by heating slides for 20 min at 95°C in antigen unmasking solution, Tris-based (Vector laboratories). Sections were cooled to R/T, washed in distilled water, and blocked and permeabilised in a solution containing 5% bovine serum albumin (BSA), 0.1% Triton X-100 and 5% serum (specific to secondary antibody species used) for 1 h at R/T. Slides were incubated with primary antibody for 2 h at R/T or 4°C overnight in 5-fold diluted blocking buffer. Secondary antibodies were applied for 1 h at R/T (Alexa Fluor conjugated, Thermo Fisher Scientific; 1:500 in diluted block). Sections were counterstained and mounted with VECTASHIELD with DAPI (Vector labs) hard-set. Alexa Fluor 568 conjugated secondary antibodies were false coloured magenta (ImageJ 1.15j).
To quantify parvalbumin-positive neurons, parvalbumin stained sections were imaged on a Nikon Ti-E live cell imager. Images were acquired using a Plan Apo lambda 10x objective with a final image dimension of 4608 x 4608 with 2x2 binning, stitched (NIS-Elements) and analysed (ImageJ 1.15j) blind to genotype. For each mouse, matching sections through the frontal cortex from Bregma 2.8 mm to 0.74 mm were analysed with a total of 10 sections quantified for 3 wild-type and TDP43Q331K/Q331K mice. Images were converted to greyscale and thresholded to produce a binary image. Consistent regions of interest were drawn around the cortex using the polygon selection tool and the ‘analyse particle’ function used to count cells.
To investigate TDP-43 in parvalbumin-positive neurons, sections were costained with antibodies against TDP-43 and parvalbumin and imaged using a Zeiss LSM 780, AxioObserver with a Plan-Apochromat 63x/1.40 Oil DIC M27 objective running Zen system software. Data analysis (ImageJ 1.15j) and imaging was carried out blind to genotype. For each cell, a maximum intensity projection of Z stacks was created and regions of interest were drawn around the nucleus and the cytoplasm using the polygon selection tool. Area, integrated density and mean grey value measurements were taken for the cytoplasm and nucleus, together with a background reading. Corrected total fluorescence for a region of interest was calculated as:
CTF = Integrated Density - (Area region of interest x background fluorescence)
Corrected fluorescence was recorded for at least 10 cells per mouse in matched sections corresponding to Bregma 1.18 mm (The Mouse Brain, compact third edition, Franklin and Paxinos).
To quantify AOX1 fluorescence in lumbar motor neurons, sections were costained with antibodies against AOX1 and neurofilament heavy and imaged on a Nikon Ti-E live cell imager with a Plan Apo VC 20x DIC N2 objective with a final image dimension of 1024 x 1022 pixels and 2x2 binning. Data analysis (ImageJ 1.15j) and imaging were carried out blind to genotype. Corrected fluorescence was recorded for at least 29 cells per mouse.
TDP-43 immunostaining in spinal cord and brain were imaged using a Nikon Ti-E live cell imager and a Plan Apo VC 100x Oil objective with a final image dimension of 1024 x 1024 pixels with 2x2 binning. Images are a maximum intensity z-stack created using ImageJ 1.15j with a z-step of 0.2µm.
Tau immunostaining in cortex was imaged using a Zeiss LSM 780, AxioObserver with a Plan-Apochromat 63x/1.40 Oil DIC M27 objective running Zen system software. Images are a maximum intensity z-stack created using ImageJ 1.15j.
For list of primary antibodies see Supplementary Table 5.
Nissl staining of spinal cord and brain
Sections were thawed at R/T, washed in distilled water then stained with cresyl etch violet (Abcam) for 5 min, briefly washed in distilled water, dehydrated in 100% ethanol, cleared in xylene, mounted (Permount, Fisher) and dried overnight at R/T. Images were taken on a Zeiss Axio Observer.Z1 running Axiovision SE64 release 4.8.3 software. Cortical images were taken with an EC Plan-Neofluar 5x/0.16 M27 objective with a total area of 4020 x 2277 pixels auto stitched within the software. Spinal cord images were acquired with an LD Plan-Neofluar 20x/0.4 korr M27 objective with an image size of 1388 x 1040 pixels.
Lumbar spinal motor neuron quantification
Motor neurons were quantified as described elsewhere54. Briefly, large motor neurons (diameter >20 μm) in the ventral horn were counted blind to genotype in 18 sections from the lumbar L3-5 levels of each animal.
Cellular quantification in brain
Data analysis using ImageJ 1.15j and imaging was carried out blind to genotype. For total frontal cortical area, matching sections through the frontal cortex from Bregma 2.8 mm to 0.74 mm were selected with a total of 10 sections quantified for six wild-type and six TDP43Q331K/Q331K mice. Matching regions of interest were drawn around the cortex and the area quantified using the measure function. To count cells within cortical sub regions, matching sections based on Bregma references were identified. Images were converted to greyscale and thresholded to produce a binary image. Consistent regions of interest were drawn around the cortex and the ‘analyse particle’ function used to count cells. A minimum size of 10 pixel units ensured that intact cells were counted and results were displayed with the overlay option selected.
Western blotting
Brain tissues were weighed to ensure equal amounts of starting material between samples, thawed on ice and processed using a modified fractional protocol55. Briefly, tissue was sequentially homogenised and centrifuged using buffers A [NaCL 150 mM, HEPES (pH 7.4) 50mM, digitonin (Sigma, D141) 25 μg/mL, Hexylene glycol (Sigma, 112100) 1 M, protease inhibitor cocktail (Sigma, P8340), 1% v:v] and B [same as buffer A except Igepal (Sigma, I7771) 1% v:v is used in place of digitonin] to extract cytoplasmic and membrane fractions respectively. The subsequent pellet was sonicated in 1% sarkosyl buffer containing 10μM Tris-Cl (pH 7.5), 10μM EDTA, 1M NaCl and centrifuged (14,000g for 30min at 4°C). The supernatant was taken as the nuclear fraction. Protein lysates were quantified (bicinchoninic acid protein assay, Pierce), electrophoresed in 4-12% or 12% SDS polyacrylamide gels, wet transferred to PVDF membranes, blocked with a 50:50 mixture of Odyssey PBS blocking buffer and PBS with 0.1% Tween20 for 1 h at R/T and then probed with primary antibodies at 4°C overnight. Secondary antibodies were either fluorescently tagged for Odyssey imaging, or HRP tagged for ECL visualisation. Western blot band intensities were quantified using Fiji (ImageJ; Version 2.0.0-rc-54/1.51h; Build: 26f53fffab) using the programs gel analysis menu option in 8-bit greyscale. Quantification was carried out by an independent user blind to genotype.
For list of primary antibodies see Supplementary Table 5.
Muscle histology
The right gastrocnemius was dissected, fixed in 4% PFA at R/T, washed in PBS for 10 min (x2) and cryoprotected and stored in 30% sucrose with 0.1% azide. Tissues were placed in a silicone mould with M1 matrix,and frozen on dry ice. Longitudinal cryosections (50 μm) were mounted onto slides (Superfrost Plus), air dried at R/T for 5 min and stored at -80°C.
To stain neuromuscular junctions (NMJs), slides were brought up to R/T and incubated in blocking solution (2% BSA, 0.2% Triton X-100, 0.1% sodium azide) for 1 h. Primary antibodies against βIII-tubulin (rabbit polyclonal, Sigma T2200) and synaptophysin (mouse monoclonal, Abcam ab8049) were applied at 1:200 dilution in blocking solution. Sections were incubated at R/T overnight. Sections were washed in PBS (x3) and incubated for 90 min with mouse and rabbit Alexa488-conjugated secondary antibodies (Thermo Fisher Scientific) diluted 1:500 in blocking solution together with TRITC-conjugated alpha bungarotoxin (Sigma, T0195) 10 μg/ml. Sections were washed in PBS and coverslipped (VECTASHIELD hardset). Confocal Z-stacks were obtained using a Zeiss LSM 780, AxioObserver with a Plan-Apochromat 20x/0.8 M27 objective running Zen system software blind to genotype.
For succinate dehydrogenase (SDH) staining, the left gastrocnemius was dissected, flash frozen in isopentane in liquid nitrogen and stored at -80°C until use. Frozen sections of 12 μm were prepared and stained using a modified version of a previously described method56. Briefly, sections were stained with freshly prepared SDH staining solution at 37°C for 3 min, washed through saline, acetone and ethanol solutions, cleared in xylene and mounted (Permount). Images were taken using an Olympus BX41 light microscope (10x objective) with Q Capture Pro 6.0.
Quantification of NMJ Innervation
NMJs from flattened z-stacks of muscle were analysed (ImageJ; Version 2.0.0-rc-54/1.51h; Build: 26f53fffab) blind to genotype. Brightness and contrast thresholds were set to optimise the signal-to-noise ratio of the presynaptic staining (anti-tubulin and anti-synaptophysin). Innervated NMJs were defined as having observed overlap of staining for pre- and post-synaptic elements. Denervated NMJs were defined as alpha-bungarotoxin signal in the absence of pre-synaptic staining. A small percentage (~5% in each genotype) of NMJs could not be scored and were excluded from this analysis.
Neuromuscular electrophysiology
Isolated FDB-tibial nerve preparations were mounted in an organ bath in HEPES-buffered MPS of the following composition (mM): Na+ (158); K+ (5); Ca2+ (2); Mg2+ (1); Cl- (169); glucose (5); HEPES (5); pH 7.2-7.4, and bubbled with air or 100% O2 for at least 20 min. The distal tendons were pinned to the base of a Sylgard-lined recording chamber and the proximal tendon connected by 6/0 silk suture to an MLT0202 force transducer (AD Instruments, Oxford, UK). The tibial nerve was aspirated into a glass suction electrode and stimuli (0.1-0.2 ms duration, nominally up to 10V) were delivered via a DS2 stimulator (Digitimer, Welwyn Garden City, UK) triggered and gated by an AD Instruments Powerlab 26T interface. Force recordings were captured and digitised at 1 kHz using the Powerlab interface and measured using Scope 4 and Labchart 7 software (AD Instruments) running on PC or Macintosh computers. For motor unit recordings, the stimulating voltage was carefully graded from threshold to saturation, to evoke the maximum number of steps in the twitch tension record. Motor unit number estimation (MUNE) was performed by inspection, counting the number of reproducible tensions steps, and by extrapolation between the average twitch tension of the four lowest threshold motor units and the maximum twitch tension. For tetanic stimulation, trains of stimuli, 1-5 s in duration were delivered at frequencies of 2-50 Hz. To measure muscle fatigue, 50 Hz stimulus trains, 1 s in duration were delivered every five seconds for about a minute. A fatigue index was calculated as the time constant of the best fitting single exponential to the decline of the maxmimum tetanic force.
Brain RNA isolation
Frontal cortices and hippocampi were subdissected in RNase free conditions (RNaseZap, Sigma Aldrich) from right hemispheres of freshly culled mice and flash frozen until further use. For RNA extraction tissue was thawed directly in TRIsure reagent (Bioline) and RNA isolated following manufacturer’s instructions. RNA was purified (RNeasy kit, Qiagen) with on-column DNase treatment and analysed on an Agilent 2100 Bioanalyzer.
Spinal motor neuron laser capture microdissection
Mice were culled by cervical dislocation and decapitation. Lumber spinal cord was rapidly dissected taking care to avoid RNase-exposure, embedded in pre-cooled M1 embedding matrix (Thermo) in a silicone mould and flash frozen in isopentane on dry ice. Samples were stored at -80°C until use. Transverse cryosections (14 μm) were taken through the lumbar enlargement and placed onto PEN membrane glass slides (Zeiss) that were kept at -20°C during sectioning. One spinal cord was processed at a time. ~50 sections were taken per mouse and placed onto two PEN slides. Slides were immediately stained in the following RNase-free, ice-cold solutions (each for 1 min): 70% ethanol, water (with gentle agitation), 1% cresyl violet in 50% ethanol, 70% ethanol, 100% ethanol (with gentle agitation), 100% ethanol (with gentle agitation). Slides were dabbed onto tissue paper to remove excess ethanol, air-dried for 1 min and taken for immediate microdissection (Zeiss PALM Microbeam). Cells were cut at x40 magnification, keeping laser power to a minimum. Motor neurons were identified by location and diameter >30 μm. ~120 cells were captured per mouse into Adhesive Cap 500 tubes (Zeiss). RNA was extracted using the Arcturus PicoPure kit (Thermofisher). 1 ul of RNA was run on an RNA 6000 Pico chip on an Agilent 2100 Bioanalyzer to evaluate RNA quality. 1ng of RNA was used as input for cDNA library preparation.
Spinal motor neuron cDNA and library preparation
Library preparation for sequencing on an Illumina HiSeq2500 sequencer was carried out using the SMART-seq v4 Ultra low Input RNA kit (Clontech) following the manufacturer’s instructions. All steps were carried out on ice unless otherwise specified. Reverse transcription, PCR cycles and incubation steps utilised a BioRad T100 Thermal Cycler. Amplification of cDNA by LC PCR used a 10-cycle protocol. After bead purification, cDNA library concentration was measured (High Sensitivity DNA kit, Agilent Technologies).
Sequencing libraries were generated using the Nextera XT DNA Library Prep Kit (Illumina) using 150 pg cDNA as input following the manufacturer’s instructions with the following modification. Following library amplification and bead purification the final fragment size was analysed and libraries quantified using the Universal KAPA Library Quantification kit (Kapa Biosystems) and a Bio-Rad C100 thermal cycler. An equal amount of cDNA was used to pool up to four samples, which were sequenced in one lane. Sequencing was carried out to a depth of 50 million 100 bp paired-end reads per library.
Frontal cortex RNAseq library preparation
Only RNA samples with RIN >8 were used for sequencing. Libraries were prepared using the TruSeq Stranded mRNA kit (Illumina) following the manufacturer’s low sample protocol with the following modification. RNA fragmentation time was reduced to 3 min at 94°C to increase median insert length. Final libraries were analysed, quantified and sequenced as above.
Bioinformatics pipeline and statistics
FastQ files were trimmed with trim galore v0.4.3 using default settings then aligned against the mouse GRCm38 genome assembly using hisat2 v2.0.5 using options --no-mixed and --no-discordant. Mapped positions with MAPQ values of <20 were discarded.
Gene expression was quantitated using the RNA-Seq quantitation pipeline in SeqMonk v1.37.0 in opposing strand specific (frontal cortex) or unstranded (motor neuron) library mode using gene models from Ensembl v67. For count based statistics, raw read counts over exons in each gene were used. For visualisation and other statistics log2 RPM (reads per million reads of library) expression values were used.
Differentially expressed genes were selected using pairwise comparisons with DESeq2 with a cut-off of P<0.05 following multiple testing correction.
Differential splice junction usage was detected by quantitating the raw observation counts for each unique splice donor/acceptor combination in all samples. Initial candidates were selected using DESeq2 with a cut-off of P<0.05 following multiple testing correction. To focus on splicing specific events hits were filtered to retain junctions whose expression change was >1.5 fold different to the overall expression change for the gene from which they derived, or which showed a significant (logistic regression P<0.05 after multiple testing correction) change in observation to another junction with the same start or end position.
A secondary intensity filter was applied to DESeq2 hits akin to a dynamic fold-change filter. DESeq2 comparisons were between wild-type and TDP43Q331K/Q331K mice or between MB+ and MB- mice. Significant expression and splicing changes between wild-type and TDP43Q331K/Q331K were used to generate hierarchical cluster plots including TDP43Q331K/+ mice to identify patterns of changes across replicates. Significant expression and splicing changes between MB+ and MB- mice were used to generate hierarchical cluster plots including wild-type mice.
GO, KEGG enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 was used for functional annotation of gene expression data in addition to the Functional Enrichment Analysis tool (FunRich v3.0) (available at:http://funrich.org). Gene ontology (GO) biological process (BP) and KEGG pathway enrichment analysis was conducted using DAVID and FunRich with a threshold Benjamini-corrected p-value=0.05.
Spinal cord RNA extraction for qPCR
Tissues were briefly washed in ice cold PBS to remove mounting media, homogenised and RNA was extracted as described above for frontal cortices and hippocampi.
Quantatitive PCR
500 ng of RNA was reverse transcribed (QuantiTect Reverse transcription kit, Qiagen) and the output volume of 20 μL diluted 10-fold in nuclease free water (Promega). Real-time PCR was performed using Brilliant-III Ultra-Fast SYBR (Agilent Technologies) on a Bio-Rad CFX96 instrument with cycle conditions based on Agilent’s quick reference guide (publication number 5990-3057, Agilent Technologies). Reaction specificity was confirmed by melt curve analysis and normalised expression (ΔΔCq) calculated using CFX Manager software 3.1 with at least four reference genes.
For qPCR primer sequence see Supplementary Table 6.
Reference genes used were: Ywhaz, Pgk1, Gapdh and Hprt1. KiCqStart SYBR Green primers for these reference genes were purchased from Sigma-Aldrich in addition to Tardbp.
Statistical analyses
Statistical analyses were conducted using Prism 6.05 (GraphPad). Graphs were plotted using Graphpad or Python. Use of parametric tests required data to be sampled from a Gaussian distribution. Homogeneity of variance between experimental groups was confirmed by the Browne-Forsythe test for ANOVA and F test for unpaired t-tests. For comparisons between genotypes or experimental groups two-tailed, unpaired t-tests or one-way ANOVA were used when comparing two or three groups respectively. Multiple comparisons by ANOVA were corrected using the Holm-Sidak test. Where the assumptions of one-way ANOVA were violated the non-parametric Kruskal-Wallis test was performed followed by Dunn’s multiple comparison test. All statistical comparisons are based on biological replicates unless stated otherwise. Where technical replication of experiments occurs, this is highlighted in the respective method.
Analyses of Rotarod performance, weights and food intake utilised repeated measures two-way ANOVA. Mice lacking measurements at any timepoint were excluded from analyses. Multiple comparisons by two-way ANOVA were corrected using the Holm-Sidak test.
TDP-43 fluorescence in the nuclear and cytoplasmic compartments of parvalbumin positive cells and cell counts in multiple regions of the cortex were compared using multiple t-tests. Multiple comparisons were corrected using the Holm-Sidak test (alpha = 5%) without assuming consistent standard deviation.
Statistical Analysis: ACBM
The ACBM system characterized each behaviour for every frame of recording and quantified the amount of time the mouse was performing a given behaviour for each hour (0-23). These data were averaged across five days of recording within each animal and then subject to statistical comparison for within-day and between-group analyses.
Statistical analysis to compare the average time spent performing a given behaviour between TDP43Q331K/Q331K and wild-type mice was conducted using repeated measures two-way ANOVA, in which the between-subjects variable was genotype and the within-subjects variable was circadian hour (0-23). We report main effects of genotype and genotype x circadian hour interactions. All statistics were calculated using IBM SPSS Statistics 24, alpha = 0.05.
Statistical analyses: Touchscreens
Data analyses for touchscreen and object recognition tasks were conducted using R version 3.3.1. Mixed-effects models were used to identify the main effects of genotype or task conditions (i.e., stimulus duration in 5-CSRTT or delay in object recognition task) and interactions between these factors. Between-genotype differences in sessions to criteria, FR, and PR outcomes were analysed by one-way ANOVA with Holm-Sidak post hoc test.
Additional statistical information
Randomisation
The order and genotype of animals and samples tested was randomized by one operator before subsequent experimental studies were conducted by a second investigator.
Reproducibility
Life Science Reporting Summary is available online.
Data availability
The authors will make all data available to readers upon request. RNAseq data have been deposited are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?accGSE99354.
Supplementary Material
Acknowledgements
We thank Babraham Institute Experimental Unit staff for technical assistance, Alexandra Weiss for technical assistance at UMMS, Michael Brodsky for assistance with CRISPR mutagenesis, the DERC morphology core at UMMS for assistance with histological preparations, and Sam Hilton for assistance with OR testing. We thank MPC lab members and J. Gallo for helpful discussions. EK is supported by a grant from the Korean Health Technology R&D Project, Korea-UK AD Collaborative Project (HI14C2173), Ministry of Health and Welfare, Republic of Korea. SY is supported by an ARUK grant (RF-2016A-1). RHB gratefully acknowledges support from the ALS Association, Project ALS, Target ALS, ALS-One, ALS Finding A Cure, and NIH grants RO1NS088689, RO1FD004127, RO1NS065847, and RO1 NS073873. JS is funded by the Motor Neuron Disease Association, the Medical Research Council UK, the Lady Edith Wolfson Fellowship Fund, and the van Geest Foundation.
Footnotes
Accession code
RNASeq data were deposited in the NCBI GEO database, number GSE99354
Author contributions
JS, MAW, MPC, RHB, TB, JF, RM, and LS designed experiments. MAW and JS performed studies on Cohort 1 mice including behavioural assessments, histology and transcriptomics, EK performed touchscreen studies on Cohort 2 mice with assistance from BUP, AD collated ACBM data and quantified NMJ innervation, RA performed spinal cord dissections for laser capture and histology, OMP and JM conducted histological studies and image analysis, JoS performed motor behavioural studies, SY and EK performed the OR assay, FM quantified motor neurons and western blots, ZL performed sequencing to exclude off-target mutagenesis events, SA and ASP assisted with analysis of RNASeq data and statistical analyses respectively, RRR performed neuromuscular electrophysiological studies, YB and TS developed ACBM software and analysed ACBM data, JS wrote the manuscript with contributions from all authors.
Competing Financial Interests
The authors declare no competing financial interests
References
- 1.Burrell JR, et al. The frontotemporal dementia-motor neuron disease continuum. Lancet. 2016;388:919–931. doi: 10.1016/S0140-6736(16)00737-6. [DOI] [PubMed] [Google Scholar]
- 2.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 3.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in fronto temporal lobar degeneration and amyotrophic lateral sclerosis. Biochemical and biophysical research communications. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 4.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benajiba L, et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–473. doi: 10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- 6.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayala YM, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philips T, Rothstein JD. Rodent Models of Amyotrophic Lateral Sclerosis. Current protocols in pharmacology. 2015;69:5 67 61–21. doi: 10.1002/0471141755.ph0567s69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold ES, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci U S A. 2013;110:E736–745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu LS, et al. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 11.Buratti E. Functional Significance of TDP-43 Mutations in Disease. Advances in genetics. 2015;91:1–53. doi: 10.1016/bs.adgen.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Sreedharan J, Neukomm LJ, Brown RH, Jr, Freeman MR. Age-Dependent TDP-43-Mediated Motor Neuron Degeneration Requires GSK3, hat-trick, and xmas-2. Current biology: CB. 2015 doi: 10.1016/j.cub.2015.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jhuang H, et al. Automated home-cage behavioural phenotyping of mice. Nature communications. 2010;1:68. doi: 10.1038/ncomms1064. [DOI] [PubMed] [Google Scholar]
- 15.Borghero G, et al. Genetic architecture of ALS in Sardinia. Neurobiol Aging. 2014;35:2882 e2887–2882 e2812. doi: 10.1016/j.neurobiolaging.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed RM, et al. Assessment of Eating Behavior Disturbance and Associated Neural Networks in Frontotemporal Dementia. JAMA Neurol. 2016;73:282–290. doi: 10.1001/jamaneurol.2015.4478. [DOI] [PubMed] [Google Scholar]
- 17.Burden SJ, Yumoto N, Zhang W. The role of MuSK in synapse formation and neuromuscular disease. Cold Spring Harb Perspect Biol. 2013;5:a009167. doi: 10.1101/cshperspect.a009167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger R, et al. Analysis of aldehyde oxidase and xanthine dehydrogenase as possible candidate genes for autosomal recessive familial amyotrophic lateral sclerosis. Human Molec Genetics. 1995;21:121–131. doi: 10.1007/BF02255787. [DOI] [PubMed] [Google Scholar]
- 19.Garattini E, Fratelli M, Terao M. The mammalian aldehyde oxidase gene family. Human genomics. 2009;4:119–130. doi: 10.1186/1479-7364-4-2-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang YM, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 21.Kolarcik CL, Bowser R. Retinoid signaling alterations in amyotrophic lateral sclerosis. American journal of neurodegenerative disease. 2012;1:130–145. [PMC free article] [PubMed] [Google Scholar]
- 22.Mar AC, et al. The touchscreen operant platform for assessing executive function in rats and mice. Nature protocols. 2013;8:1985–2005. doi: 10.1038/nprot.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas A, et al. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology. 2009;204:361–373. doi: 10.1007/s00213-009-1466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenna KP, et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2016;48:1037–1042. doi: 10.1038/ng.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenner D, et al. NEK1 mutations in familial amyotrophic lateral sclerosis. Brain. 2016;139:e28. doi: 10.1093/brain/aww033. [DOI] [PubMed] [Google Scholar]
- 26.Nihei K, McKee AC, Kowall NW. Patterns of neuronal degeneration in the motor cortex of amyotrophic lateral sclerosis patients. Acta Neuropathologica. 1993;86:55–61. doi: 10.1007/BF00454899. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, Ahrlund-Richter S, Wang X, Deisseroth K, Carlen M. Prefrontal Parvalbumin Neurons in Control of Attention. Cell. 2016;164:208–218. doi: 10.1016/j.cell.2015.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nature neuroscience. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutton M, et al. Association of missense and 5'-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 30.Swanson E, et al. Extracellular Tau Oligomers Induce Invasion of Endogenous Tau into the Somatodendritic Compartment and Axonal Transport Dysfunction. J Alzheimers Dis. 2017;58:803–820. doi: 10.3233/JAD-170168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker LA, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harauz G, Boggs JM. Myelin management by the 18.5-kDa and 21.5-kDa classic myelin basic protein isoforms. J Neurochem. 2013;125:334–361. doi: 10.1111/jnc.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freischmidt A, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 35.Cirulli ET, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skibinski G, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi Y, et al. ERBB4 mutations that disrupt the neuregulin-ErbB4 pathway cause amyotrophic lateral sclerosis type 19. Am J Hum Genet. 2013;93:900–905. doi: 10.1016/j.ajhg.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Hoecke A, et al. EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nat Med. 2012;18:1418–1422. doi: 10.1038/nm.2901. doi:nm.2901 [pii] [DOI] [PubMed] [Google Scholar]
- 39.Landers JE, et al. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0812937106. doi:0812937106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimura AL, et al. Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain. 2010;133:1763–1771. doi: 10.1093/brain/awq111. [DOI] [PubMed] [Google Scholar]
- 41.Johnson JO, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nature Neuroscience. 2014;17:664. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fecto F, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Archives of neurology. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 43.Koyama A, et al. Increased cytoplasmic TARDBP mRNA in affected spinal motor neurons in ALS caused by abnormal autoregulation of TDP-43. Nucleic acids research. 2016;44:5820–5836. doi: 10.1093/nar/gkw499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egawa N, et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Science translational medicine. 2012;4:145ra104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 45.Liu C, Song X, Nisbet R, Gotz J. Co-immunoprecipitation with Tau Isoform-specific Antibodies Reveals Distinct Protein Interactions and Highlights a Putative Role for 2N Tau in Disease. J Biol Chem. 2016;291:8173–8188. doi: 10.1074/jbc.M115.641902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trabzuni D, et al. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borroni B, et al. Association between tau H2 haplotype and age at onset in frontotemporal dementia. Archives of neurology. 2005;62:1419–1422. doi: 10.1001/archneur.62.9.1419. [DOI] [PubMed] [Google Scholar]
- 48.Behrouzi R, et al. Pathological tau deposition in Motor Neurone Disease and frontotemporal lobar degeneration associated with TDP-43 proteinopathy. Acta neuropathologica communications. 2016;4:33. doi: 10.1186/s40478-016-0301-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreno JA, et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Science translational medicine. 2013;5:206ra138. doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- 50.Kang SH, et al. Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat Neurosci. 2013;16:571–579. doi: 10.1038/nn.3357. doi:nn.3357 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu LJ, Holmes BR, Aronin N, Brodsky MH. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS One. 2014;9:e108424. doi: 10.1371/journal.pone.0108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heath CJ, Bussey TJ, Saksida LM. Motivational assessment of mice using the touchscreen operant testing system: effects of dopaminergic drugs. Psychopharmacology. 2015;232:4043–4057. doi: 10.1007/s00213-015-4009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romberg C, et al. Depletion of perineuronal nets enhances recognition memory and long-term depression in the perirhinal cortex. J Neurosci. 2013;33:7057–7065. doi: 10.1523/JNEUROSCI.6267-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu LS, Cheng WC, Shen CK. Targeted depletion of TDP-43 expression in the spinal cord motor neurons leads to the development of amyotrophic lateral sclerosis-like phenotypes in mice. J Biol Chem. 2012;287:27335–27344. doi: 10.1074/jbc.M112.359000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baghirova S, Hughes BG, Hendzel MJ, Schulz R. Sequential fractionation and isolation of subcellular proteins from tissue or cultured cells. MethodsX. 2015;2:440–445. doi: 10.1016/j.mex.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalmar B, Blanco G, Greensmith L. Determination of Muscle Fiber Type in Rodents. Current protocols in mouse biology. 2012;2:231–243. doi: 10.1002/9780470942390.mo110229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors will make all data available to readers upon request. RNAseq data have been deposited are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?accGSE99354.