Intense induction of PPsPase1 and PECP1 phosphatases impacts phosphocholine and phosphoethanolamine content in Arabidopsis.
Abstract
Phosphate starvation-mediated induction of the HAD-type phosphatases PPsPase1 (AT1G73010) and PECP1 (AT1G17710) has been reported in Arabidopsis (Arabidopsis thaliana). However, little is known about their in vivo function or impact on plant responses to nutrient deficiency. The preferences of PPsPase1 and PECP1 for different substrates have been studied in vitro but require confirmation in planta. Here, we examined the in vivo function of both enzymes using a reverse genetics approach. We demonstrated that PPsPase1 and PECP1 affect plant phosphocholine and phosphoethanolamine content, but not the pyrophosphate-related phenotypes. These observations suggest that the enzymes play a similar role in planta related to the recycling of polar heads from membrane lipids that is triggered during phosphate starvation. Altering the expression of the genes encoding these enzymes had no effect on lipid composition, possibly due to compensation by other lipid recycling pathways triggered during phosphate starvation. Furthermore, our results indicated that PPsPase1 and PECP1 do not influence phosphate homeostasis, since the inactivation of these genes had no effect on phosphate content or on the induction of molecular markers related to phosphate starvation. A combination of transcriptomics and imaging analyses revealed that PPsPase1 and PECP1 display a highly dynamic expression pattern that closely mirrors the phosphate status. This temporal dynamism, combined with the wide range of induction levels, broad expression, and lack of a direct effect on Pi content and regulation, makes PPsPase1 and PECP1 useful molecular markers of the phosphate starvation response.
Plant growth is highly sensitive to a lack of phosphate (Pi); hence, the application of Pi fertilizers has become standard practice for high-throughput crop production (Cordell et al., 2009). Most phosphorus in the soil is not available to plants, as it is combined with other minerals or parts of organic compounds (Bieleski, 1973; Raghothama and Karthikeyan, 2005). Only a small fraction of soluble Pi (usually present at a concentration of <10 µm in the soil) can be taken up by plants.
Although growth is not optimal under limiting conditions, plants can withstand changing Pi concentrations within heterogeneous soils or in the external nutrient supply that can lead to reprogramming of their metabolism and architecture (Hammond et al., 2003; Péret et al., 2011; Plaxton and Tran, 2011). Root architecture can be modified to facilitate Pi uptake by favoring the development of lateral roots (at the expense of primary root elongation in many plants including Arabidopsis), increasing the density and length of root hairs, and limiting the development of aerial parts (López-Bucio et al., 2002; Svistoonoff et al., 2007; Gruber et al., 2013). Pi uptake mechanisms are enhanced at the root/soil interface, particularly through the stimulation of Pi transport activity (Mudge et al., 2002; Shin et al., 2004; Nussaume et al., 2011; Ayadi et al., 2015). In parallel, mobilization of Pi sources from the soil is facilitated by the secretion of acid phosphatases and acidification of the soil surrounding roots (Raghothama, 1999; Plaxton and Tran, 2011; Wang et al., 2011; Zhang et al., 2014; Balzergue et al., 2017).
Pi stored within cells also represents an important nutrient source, and plants demonstrate an extraordinary capacity to mobilize these stocks during starvation (Ticconi and Abel, 2004). Vacuoles are the major storage compartment for Pi in the cell (storing about 80% of the total cellular Pi) in nonlimiting conditions (Bieleski, 1983; Poirier and Bucher, 2002). Pi incorporated in lipid membranes also represents an abundant and readily available source. Shortly after the onset of Pi deficiency, nutrient starvation leads to the replacement of phosphoglycerolipids, such as phosphatidyl-choline (PtdCho) and phosphatidyl-ethanolamine (PtdEth), by sulfolipids (sulfoquinovosyldiacylglycerol) and galactolipids (digalactosyldiacylglycerol) within the membranes (Misson et al., 2005; Nakamura, 2013; Siebers et al., 2015). The two latter lipid forms are devoid of Pi, which allows the cell to recycle this element into other pathways that require Pi. Many enzymes involved in this process are well known, and independent pathways are concomitantly triggered to ensure a fast and sizable lipid remobilization (Nakamura, 2013; Pant et al., 2015). For instance, SQD1 and SQD2 are involved in sulfolipid biosynthesis, whereas MGD2 and MGD3 are strongly induced to increase galactolipid content (Yu et al., 2002; Jouhet et al., 2004; Nakamura et al., 2005; Kobayashi et al., 2009). To achieve these substitutions, phospholipids are degraded by different types of lipases, resulting in the release of lipid subcomponents that can reenter the metabolism or be converted into other lipid forms (Nakamura, 2013).
The Pi starvation-induced reprogramming of architecture and metabolism is supported by vast changes in the plant transcriptome and has been demonstrated in several plant models (Wasaki et al., 2003; Chiou and Lin, 2011; Hu and Chu, 2011; Huang et al., 2011). In Arabidopsis, more than 600 genes are induced in response to Pi starvation (Misson et al., 2005; Thibaud et al., 2010). This includes genes that respond either locally to Pi fluctuations or systemically throughout the plant, in a manner reflecting the plant’s Pi status. Most of the systemically induced genes are under the control of PHR1 (a MYB family transcription factor) and its homolog PHL1 (Bustos et al., 2010). Furthermore, their corresponding mutants exhibit altered gene induction and classical Pi starvation responses, including diminished effects on Pi content and lipid remobilization (Pant et al., 2015). Candidates for the systemic control of Pi starvation responses have been proposed, including substantial transport of mRNAs throughout the plant (as reviewed in Puga et al., 2017), and the direct sensing of Pi concentration (or Pi-containing metabolites) by SPX-type proteins (Wang et al., 2009; Puga et al., 2014; Wild et al., 2016).
Two phosphatases belonging to the haloacid dehalogenase (HAD) superfamily were identified as some of the most strongly induced transcripts following Pi starvation (Bari et al., 2006; Müller et al., 2007; Thibaud et al., 2010). These enzymes were named PPsPase1 (for pyrophosphatase [PPi]-specific phosphatase1) and PECP1 (for phosphoethanolamine (PEth)/phosphocholine [PCho] phosphatase1) on the basis of their in vitro activity, following their heterologous expression in bacteria and subsequent purification (May et al., 2011, 2012). However, no formal demonstration of their physiological role has yet been achieved in planta. It has also been suggested that the HAD protein family plays a role in the general regulation of plant responses to Pi starvation. This is due to their extremely high levels of induction, their ability to dephosphorylate, and the observation that tomato (Solanum lycopersicum) plants overexpressing LePS2;1 (a homolog of these proteins) display enhanced Pi starvation symptoms (Baldwin et al., 2008). Chandrika et al. (2013) demonstrated the inability of a PPsPase1 KO (knockout) mutant to induce root hair elongation in response to Pi deficiency, suggesting a link between these enzymes and the classic architectural response to nutrient limitation (i.e. root hair elongation).
The third member of this family (AT4G29530) is missing in published transcriptomics studies related to Pi starvation. Nevertheless, this protein was recently shown to act as a thiamin monophosphate phosphatase when expressed in bacteria (Hasnain et al., 2016). Since this is the first plant protein to exhibit this type of enzymatic activity, we will name this enzyme ThMPase1. However, knockouts of ThMPase1 in Arabidopsis have not revealed any visible phenotype or modification of thiamin concentration, even when combined with other mutant lines affected in thiamin monophosphate content (Hasnain et al., 2016; Mimura et al., 2016).
In this report, we show that PPsPase1 and PECP1 (but not ThMPase1) constitute robust and sensitive reporters of Pi status, indicating a very dynamic expression response to Pi concentration. Using a reverse genetics approach, we altered the expression of these proteins and revealed that PPsPase1 and PECP1 are likely involved in the recycling of polar heads from membrane lipids, but do not play a role in the overall regulation of membrane lipid composition or plant responses to Pi starvation. Furthermore, both enzymes appear to target PCho and PEth in vivo with little or no effect on PPi levels, contrary to what was suggested by their in vitro substrate preferences.
RESULTS
PPsPase1 and PECP1 Are Dynamic Markers of Pi Starvation
PPsPase1 and PECP1 are listed as strongly induced in several transcriptomics studies related to phosphate starvation (Misson et al., 2005; Bari et al., 2006; Müller et al., 2007; Thibaud et al., 2010; Hirsch et al., 2011). However, very little is known about their precise expression patterns. To address this question, we developed the PromPPsPase1:GUS and PromPECP1:GUS reporter lines. We observed a strong and proportional response to Pi content in the medium of the GUS signal (reporting PPsPase1 and PECP1 expression; Figure 1; Supplemental Figs. 1 and 2). Comparison of internal Pi content and expression of PPsPase1 and PECP1 transcripts by real-time PCR showed that these genes responded to a wide range of Pi concentrations, with a maximum inhibition observed near 500 µm in the external medium (Supplemental Fig. 2). Inhibition of the genes was maintained at a very high external Pi concentration (1250 µm), while internal Pi concentration was decreased suggesting the occurrence of additional regulatory mechanisms in Pi-saturated conditions. This pattern of regulation was also observed with MGD3, a well-known marker of Pi starvation involved in lipid remodeling (Supplemental Fig. 2; Kobayashi et al., 2009).
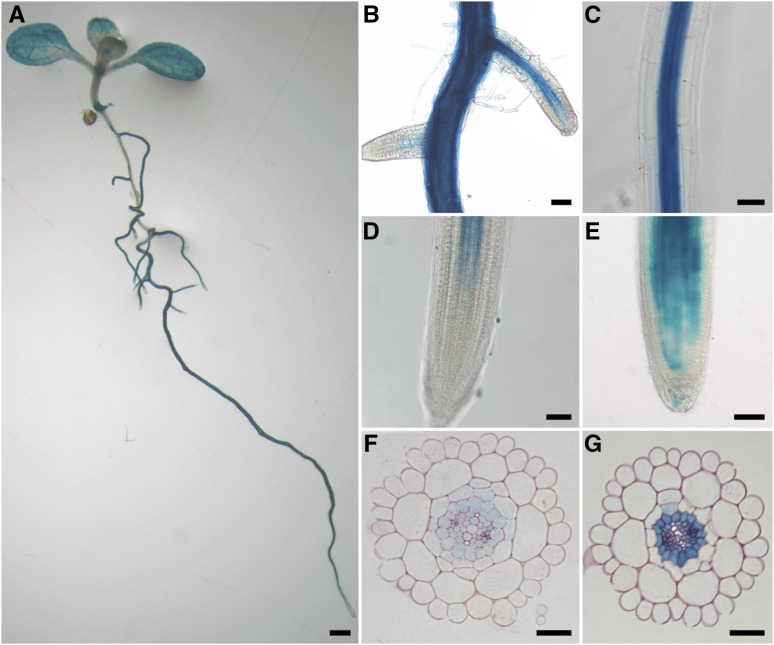
Figure 1.
PromPPsPase1:GUS expression pattern in Pi-starved plants. A, GUS expression in a whole seedling, showing heterogeneous staining along the root axis, along with vascular tissue and mesophyll staining. B, Lateral roots emerging from the primary root. C, Differentiated part of the primary root tip with strong central cylinder staining. D, Primary root tip. E, Lateral root tip. F, Cross section through a root tip. G, Cross section through a mature part of the primary root. A to E, 7-d-old seedlings (1.5 h GUS staining); F and G, 12-d-old seedlings (1 h GUS staining). Plants were grown on a medium containing 10 µm Pi. Bars = 1.5 mm in A; 80 µm in B, C, and E, 50 µm in D, and 30 µm in F and G.
Comparing low Pi and high Pi plants revealed that the overall pattern of GUS localization was maintained, with a stronger intensity in the root central cylinder, particularly in the pericycle (Fig. 1; Supplemental Fig. 1), as well as in leaf veins. Nonetheless, overall induction levels were much higher in low Pi conditions. More frequently, in low Pi conditions, expression tended to extend to other root tissues in several long patches along the root axis, up to the epidermal layer and root hairs. In low Pi, the expression also extended more broadly to the mesophyll in leaves. Expression in the root cap, hydathodes, and stomata was also generally observed in low and high Pi conditions (Fig. 1; Supplemental Fig. 1). Expression was also detected in the late stages of developing anthers (stages 13 to 14, according to Sanders et al., 1999, 2000; Supplemental Fig. 3).
To study the kinetics of transcript regulation by Pi status in PPsPase1 and PECP1, we generated reporter lines by placing the coding sequence of the firefly luciferase gene (LUC+) downstream of their promoters. We quantified the overall kinetics of gene induction during the onset of Pi starvation and then gene repression after the return to a Pi-rich solution (Fig. 2). These results were compared to a control PromMGD3:LUC+ line.
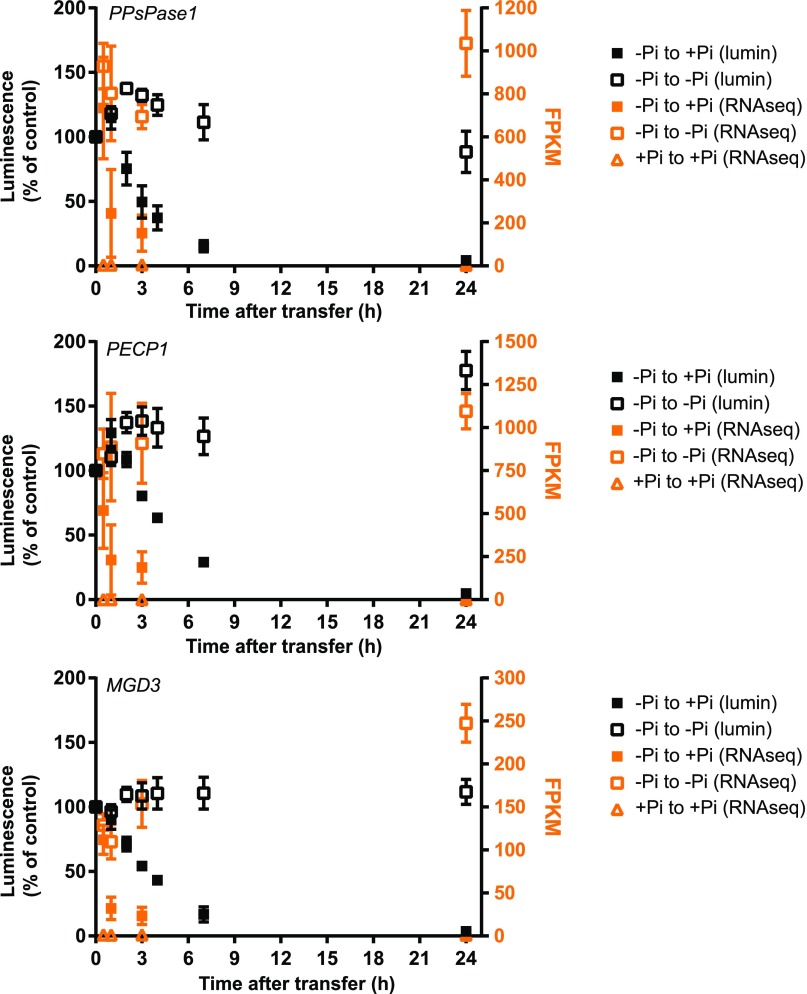
Figure 2.
Kinetics of transcript regulation following changes in Pi status for PPsPase1 (top panel) and PECP1 (middle panel). Results for MGD3 (bottom panel) were included as an example of strong regulation by Pi status. Data were acquired for the following time points: 30 min, 1 h, 3 h, and 24 h after transfer. The luminescence signal of whole seedlings (left axis, black data points) reflects the activity of corresponding Prom:LUC+ fusion and is expressed as a percentage of the luminescence detected in low Pi. Transcripts from root samples were directly quantified by RNA-seq (right axis, orange data points), with results expressed in FPKM. Samples transferred from low Pi to high Pi are represented by solid squares. Reference data (low Pi to low Pi and high Pi to high Pi transfers) are indicated by empty squares and triangles, respectively. Results shown are the mean ± sd of three biological replicates.
Luminescence quantification confirmed that PPsPase1 and PECP1 expression levels were strongly dependent on Pi content and revealed the fast decline of their expression in response to Pi replenishment (Fig. 2). After addition of Pi, a transitory peak was observed, showing a slight increase in luminescence during a 1 to 2 h period. This was followed by a rapid decrease within hours, reaching low basal values after 6 to 7 h. Similar kinetics were observed with the control PromMGD3:LUC+ line (Fig. 2).
Since this transitory initial peak was also observed after transfer to a Pi-rich solution (Fig. 2), but was not followed by the rapid decline observed in low Pi conditions, we suspected that it could be due to sample manipulation. To directly quantify the transcript levels, we performed RNA-seq analysis on roots of plants transferred from low Pi conditions to low or high Pi conditions on agar media.
Direct transcript measurements confirmed the intense transcript induction observed for PPsPase1 and PECP1, with expression values ranging from ∼ 900 FPKM (fragments per kilobase of transcript per million) in low Pi to <10 FPKM in high Pi (Fig. 2). As a comparison, MGD3 transcripts only reached ∼100 FPKM in low Pi and declined to around 2 FPKM in high Pi. The kinetics of the three genes revealed an extremely fast response to changes in Pi, with a detectable decrease of both transcript levels as early as 30 min after Pi addition to Pi-starved plants. This FPKM decrease was noticeably faster than the drop in luminescence, possibly due to the presence of regulatory elements in the transcript sequence downstream of the promoter for the three genes or, more likely, due to the fact that the luciferase transcripts were more stable than those encoded by our genes of interest.
To obtain a more precise view of gene induction dynamics throughout the plant, we built a luminescence imaging growth chamber. This chamber allowed us to grow plants in nearly constant illumination conditions, while detecting the luminescence signal in the plant during very short dark periods. Using this setup, we observed upon transfer of PromPECP1:LUC+ plants to a low Pi medium that the luminescence signal increased throughout the root system within 36 h, with an uneven induction pattern all along the root system (Fig. 3A; Supplemental Movie 1).
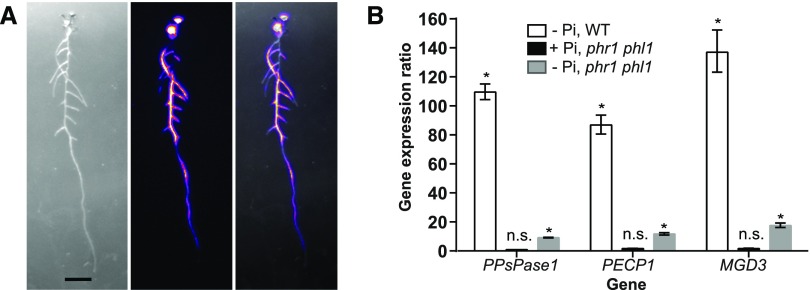
Figure 3.
PPsPase1 and PECP1 are regulated via PHR/PHL. A, The luminescence signal is rapidly induced throughout the whole plant, in 4-d-old PromPECP1:LUC+ seedlings (as shown here) and PromPPsPase1:LUC lines (Hirsch et al., 2011). Left, transmitted light image; center, luminescence signal (color-coded intensity); right, overlay. Bar = 1 cm. An animation illustrating the onset of induction is presented in Supplemental Movie 1. B, PPsPase1 and PECP1 gene induction in the wild type and the phr1 phl1 mutant in low or high Pi, compared to high Pi wild-type plants. Results for the reference gene MGD3 are also included. The expression level of genes was arbitrarily set to 1 for high Pi wild-type plants. For each sample, three reference genes (ROC3, GAPC1, and SCAMP) and three technical replicates were taken into account. Results are presented as the median ± 95% confidence interval. Asterisks indicate a statistically significant difference between the sample and the high Pi wild-type control (REST randomization test, P < 0.05). n.s., Nonsignificant.
Since most regulated genes that respond to Pi starvation are under the control of the Myb transcription factor PHR1 and its close homolog PHL1 (Nilsson et al., 2007; Bustos et al., 2010; Thibaud et al., 2010; Pant et al., 2015; Sun et al., 2016), we examined the role of these factors in PPsPase1 and PECP1 induction, including MGD3 as a control. By comparing gene induction levels using RT-qPCR in the wild type and phr1 phl1 double mutants (Bustos et al., 2010), we observed that only a residual induction remained for these three genes in phr1 phl1 (Fig. 3B). Hence, these transcription factors control most of the observed transcript induction of the two phosphatases and MGD3 in response to Pi starvation.
Null Mutants of PPsPase1, PECP1, and ThMPase1 Do Not Display Growth Phenotypes
PPsPase1 KO mutants have previously been shown to display impaired root hair growth (Chandrika et al., 2013). To compare this phenotype to the PECP1 and ThMPase1 null mutants, we identified T-DNA or transposon insertional mutants for each gene (Fig. 4A), obtained homozygous mutants, and confirmed the precise T-DNA or transposon insertion site by sequencing.
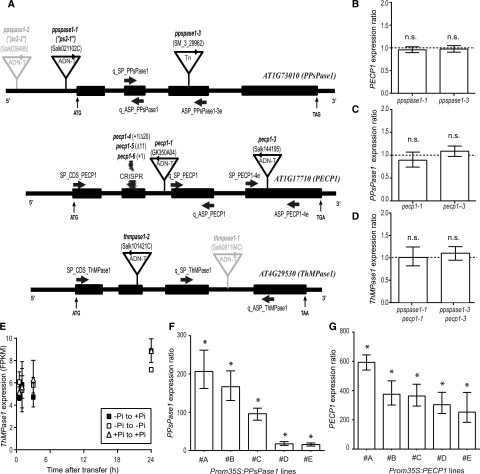
Figure 4.
Characterization of the knockout and overexpressing lines affecting PPsPase1, PECP1, and/or ThMPase1. A, T-DNA, transposon (Tn) and CRISPR-Cas9 (CRISPR) mutation localization within the targeted genes. Sequenced T-DNA and transposon borders are indicated by arrowheads. T-DNA insertions indicated in gray (ppspase1-2 and thmpase1-1) were published elsewhere and are not characterized here. Thick arrows indicate primer annealing sites. Start (ATG) and stop codons are indicated. Introns and exons are represented by thin and thick lines, respectively. The site of CRISPR mutations is also indicated. B to D, Levels of PECP1, PPsPase1, and ThMPase1 are similar in wild-type and knockout plants for PPsPase1 (B), PECP1 (C), or both (D), indicating a lack of transcriptional compensation. Results are expressed as gene induction level in mutants in comparison to wild-type plants grown under the same conditions. Wild-type control values were arbitrarily set to one (dotted line). E, ThMPase1 is expressed at low levels and does not respond to changes in Pi status for transcripts. Transcripts from root samples were quantified by RNA-seq, for the following time points: 30 min, 1 h, 3 h, and 24 h after transfer. Results are expressed in FPKM. Samples transferred from low Pi to high Pi are represented by solid squares. Reference data (low Pi to low Pi and high Pi to high Pi transfers) are indicated by empty squares and triangles, respectively. Results shown are the mean ± sd of three biological replicates. F and G, Lines that overexpress PPsPase1 (F) and PECP1 (G) show very high transcript levels for these phosphatases, even in the high Pi condition. Results are expressed as gene induction level in overexpressing lines (#A to #E) in comparison to wild-type plants grown under the same conditions. Wild-type control values were arbitrarily set to one. Results are presented as the median ± 95% confidence interval. Asterisks indicate a statistically significant difference (REST randomization test, P < 0.05). n.s., Nonsignificant. The reference genes used for the individual panels were ROC3, SCAMP, and GF14phi (B and C), or ROC3 only (D, F, and G).
During the review process of this article, Tannert et al. (2018) reported the phenotypes of three pecp1 mutant alleles (pecp1-1 to pecp1-3). To avoid confusion, pecp1 insertion lines used in this work were renamed before publication and follow the nomenclature of Tannert et al. (2018).
Two mutant alleles that knockdown gene expression were obtained for PPsPase1 and PECP1 (Supplemental Fig. 4, A–C). ppspase1-1 (identical to the ps2-1 allele from Chandrika et al. [2013]) has two face-to-face T-DNA insertions in its promoter (146 bp upstream of the ATG), which also causes a 65-bp deletion of the promoter sequence upstream of the insertion site. ppspase1-3 has a transposon insertion just upstream of the bases that encode amino acid Pro-131. pecp1-1 and pecp1-3 both have a T-DNA insertion just downstream of the bases that encode amino acid Lys-89 and Pro-201, respectively. We only identified one effective insertional mutant for ThMPase1 (Fig. 4A; Supplemental Fig. 4D) that contains a T-DNA insertion downstream of the bases that encode amino acid His-75. We named this mutant thmpase1-2, since the first ThMPase1 mutant allele was recently described (Hasnain et al., 2016).
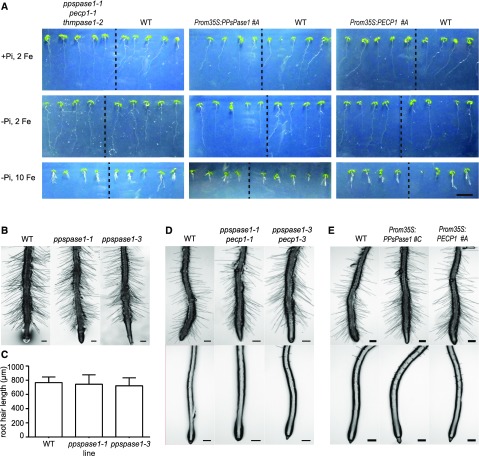
No obvious growth phenotype was observed for any of these mutants when they were grown in vitro (Fig. 5, A and B). In particular, no impairment in root hair growth was observed in any KO line, including ppspase1-1, although we used our standard low Pi/high Pi MS/10 medium as well as growth conditions described by Chandrika et al. (2013) (Fig. 5, B–E). Our inability to reproduce this phenotype may be due to a different light quality or another unidentified factor and could suggest that the phenotype is not strictly linked to Pi starvation.
Figure 5.
Growth characterization of knockout and overexpressing lines affecting PPsPase1 and/or PECP1 expression. A, Growth in mutant and overexpressing lines was similar to the control. The different conditions are: high Pi + 2 µm FeCl2 (top panel), low Pi + 2 µm FeCl2 (middle panel), and low Pi + 10 µm FeCl2 (lower panel). Plants were grown in vitro for 7 d. Each panel shows the comparison between mutant and wild-type plants, separated by a dotted line (left, triple KO; middle, PPsPase1 overexpressor; right, PECP1 overexpressor). Bar = 1 cm. B to E, Root hair growth was not affected by the expression level of PPsPase1 or PECP1, as observed in the single KO mutant (B), double KO mutants (D), and overexpressor lines (E). In low Pi, no significant differences were observed in root hair (C) length measurements (one-way ANOVA, P > 0.05; n = 32–54). Results are presented as the mean ± sd. Plants were grown in vitro in low Pi medium mimicking the conditions of Chandrika et al. (2013) for 12 d (B and C) or on our standard MS/10 low Pi (D and E, upper panel) or high Pi (D and E, lower panel) medium for 10 d. Bars = 200 µm.
When grown in soil, stems of the pecp1-3 T-DNA insertional line revealed a “glossy” phenotype that cosegregated with the pecp1 mutation, reminiscent of wax-deficient mutants (Rowland et al., 2007). The second allele (pecp1-1) did not display any such phenotype, suggesting that this phenotype was unrelated to PECP1. To confirm this hypothesis in the absence of any other satisfactory T-DNA lines, we generated new mutant alleles using CRISPR-Cas9 (CRISPR) mutagenesis (Fauser et al., 2014). Three new alleles (pecp1-4 to pecp1-6) were obtained, containing frame shifts due to single base pair insertions or deletions (up to 20 bp; Fig. 4A; Supplemental Fig. 4F). None of these three new pecp1 mutants displayed a glossy phenotype, confirming that pecp1-3 has an additional mutation genetically linked to the PECP1 gene, leading to the wax defect.
The absence of phenotype in single ppspase1 and pecp1 mutants could be caused by genetic redundancy. Hence, we produced double ppspase1 and pecp1 KO mutants and one triple KO mutant containing an additional thmpase1 mutant allele (Supplemental Fig. 4E). Our examination of the single and double KO mutants revealed that the absence of any of these three phosphatase genes did not lead to any compensatory mechanism at the transcriptional level in other members of this gene family (Fig. 4, B–D) or to any growth phenotype (Fig. 5D). RNA-seq analysis showed that the ThMPase1 transcript level was insensitive to Pi concentration and displayed an extremely low expression level (below 10 FPKM for all time points and conditions; Fig. 4E). This expression level and the lack of response to Pi changes indicate that ThMPase1 is not specifically involved in the response of plants to Pi starvation.
To complete our set of mutant lines, we generated overexpressing lines for the two genes of interest (PPsPase1 and PECP1) under the control of a 35S promoter. A preliminary screen of transformed plants allowed us to select five distinct lines (#A to #E) containing a single T-DNA insertional event (based on herbicide resistance segregation properties). The RT-qPCR analysis of transcript level in homozygous T3 lines confirmed strong overexpression in the two genes, with levels up to 200-fold for PPsPase1 and 600-fold for PECP1 (Fig. 4, F and G). For subsequent studies, we selected the two strongest overexpressing lines (#A and #B). No obvious growth phenotype was observed with any lines (Fig. 5, A and E).
Altered Expression of PPsPase1, PECP1, or ThMPase1 Does Not Affect Phosphate Content or Molecular Markers of Phosphate Starvation
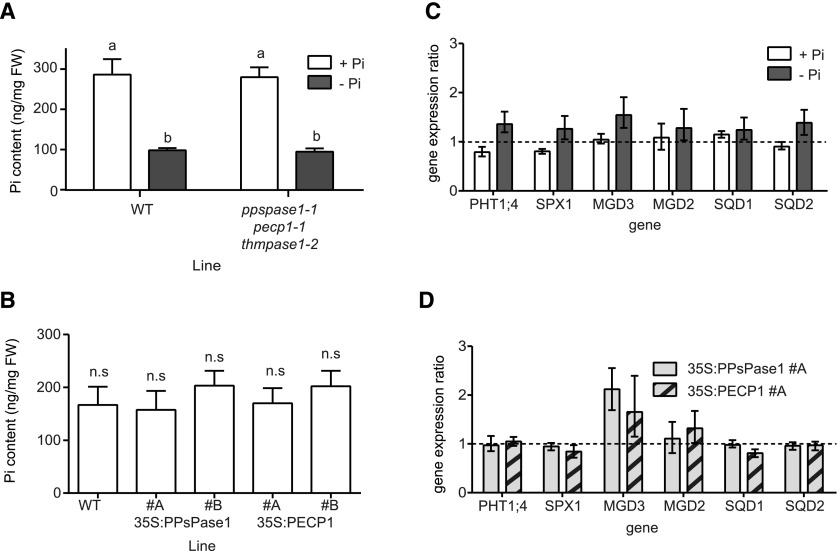
The two Pi starvation-induced HAD-type phosphatases were previously predicted to be involved in the recycling of Pi from several putative substrates including lipid polar heads, PPi, and other Pi-containing small molecules (May et al., 2011, 2012). Therefore, knocking down or increasing the expression of these genes could potentially affect the Pi content of plants. We measured Pi content in our triple KO mutant (Fig. 6A) as well as in the overexpressing lines (Fig. 6B) grown in vitro in high or low Pi conditions, but no significant difference was observed.
Figure 6.
Characterization of Pi status markers in knockout and overexpressing lines affecting PPsPase1, PECP1, and ThMPase1 expression. A, Pi quantification in triple KO mutants. Measurements were performed in triplicate on pools of 50 whole plants grown for 6 d on MS/10 high or low Pi (2 µM FeCl2). The results were confirmed in three independent experiments and are presented as the mean ± SD (n = 3). Different letters indicate significantly different Pi content (two-way ANOVA followed by Sidak’s pairwise comparison, P < 0.01). B, Quantification of Pi in overexpressing lines. Measurements were performed on the soluble fraction extracted from the leaves of individual plants grown in hydroponics on MS/10 high Pi (10 µm FeCl2). Results are presented as the mean ± SD (n = 10). Lines do not differ significantly from the wild-type control (ANOVA followed by Dunnett’s pairwise comparison, P < 0.01). C and D, Knockout or overexpressing lines have no impact on the expression of classical molecular markers of Pi starvation. C, Triple knockout and corresponding wild-type controls were grown in low Pi (white bars) or high Pi (gray bars). Wild-type control values were arbitrarily set to one for each growth condition (here represented as a dotted line). D, Overexpressing lines for PPsPase1 (light gray bars) and PPsPase1 (striped bars) were grown in high Pi along with wild-type plants. Wild-type control values were arbitrarily set to one (dotted line). Results are presented as the median ± 95% confidence interval (REST randomization test, n = 3). Reference genes used for the RT-qPCR were SCAMP and GAPC1 (C and D), and ROC3 (D).
Genes that are homologous to PPsPase1, PECP1, and ThMPase1 have also been suggested to play a regulatory role related to plant Pi starvation responses. Specifically, tomato plants overexpressing LePS2;1 (homologous to PPsPase1 and PECP1) were reported to display altered Pi content, increased anthocyanin production, and enhanced acid phosphatase activity under Pi-rich conditions (Baldwin et al., 2008). To verify this hypothesis, we quantified the induction of several robust markers of Pi starvation responses (Duan et al., 2008; Rouached et al., 2010; Thibaud et al., 2010), including known players involved in Pi uptake and lipid remobilization (PHT1;4, SPX1, MGD2, MGD3, SQD1, and SQD2), and compared them to the expression level in their respective controls (wild type, low or high Pi). The triple KO mutants (Fig. 6C) and overexpressing lines (Fig. 6D) did not show any significant modification of induction levels of these markers when compared to the wild-type plants, suggesting that these HAD-type enzymes are not essential for Pi homeostasis control in Arabidopsis.
Triple KO Mutant Does Not Mimic or Amplify Phenotypes Caused by Impaired Cytosolic PPi Regulation
Previous reports have shown that the preferred in vitro substrates of purified PPsPase1 and PECP1 proteins are PPi and PCho/PEth, respectively (May et al., 2011, 2012). We therefore performed several assays to verify these activities in planta. As PPi quantification is notoriously challenging (Gdula et al., 1998; Heinonen, 2001), we opted for a more physiological approach to compare our mutants to previously described mutant lines impaired in their cytosolic PPi concentration (Ferjani et al., 2011).
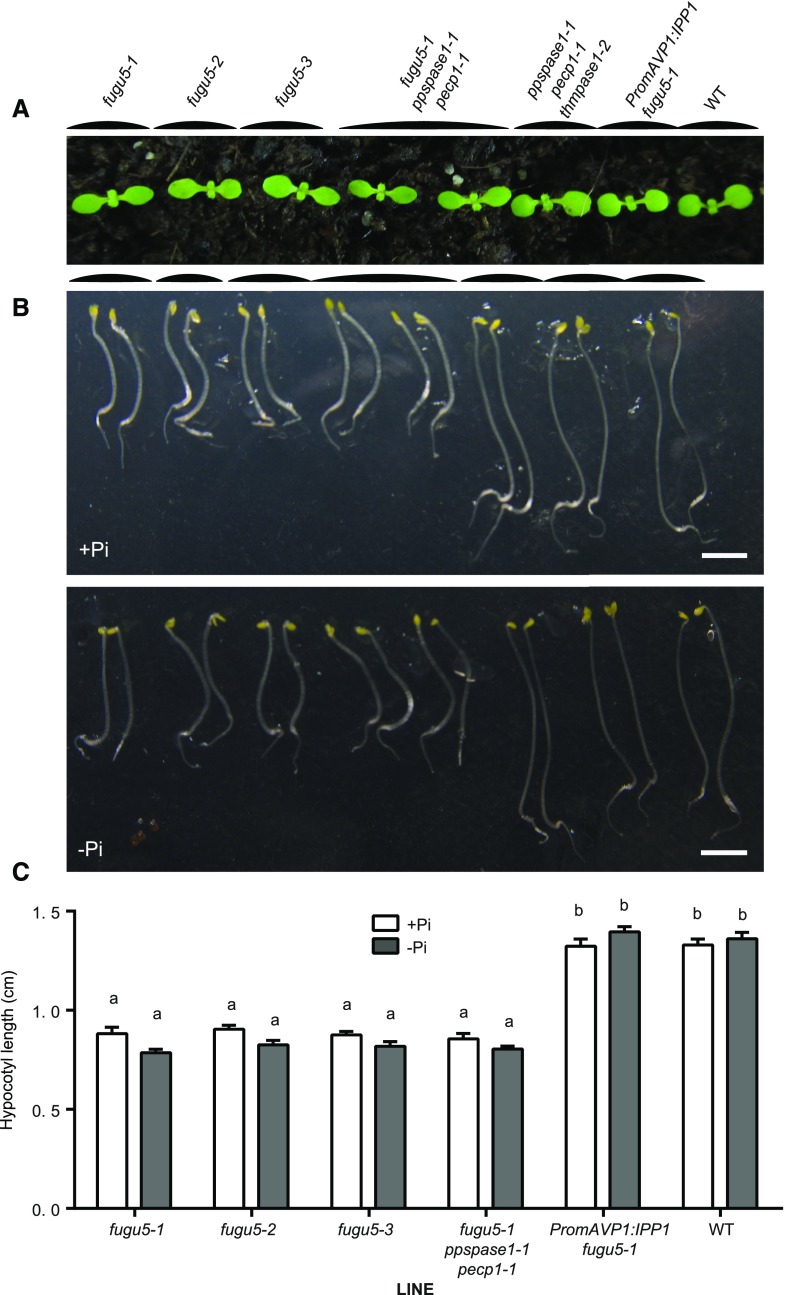
The fugu5 mutant lines are deficient in AVP1, a PPi-dependent proton pump (Ferjani et al., 2011; Pizzio et al., 2015). Previously, it was reported that the null mutants fugu5-1, fugu5-2, and fugu5-3 have an increased PPi concentration in the cytosol. This increased concentration has a visibly negative impact on the development of cotyledons and hypocotyl elongation in etiolated seedlings, due to metabolic inhibition of key enzymes by PPi overload (Ferjani et al., 2011). We compared the cotyledon growth of the triple KO mutant to the three fugu5 alleles but did not observe the typical elongated shape of cotyledons in AVP1 mutants (Fig. 7A). As previously described, the fugu5-1 line complemented with the yeast IPP1 soluble pyrophosphatase developed normal cotyledons (Ferjani et al., 2011). In addition, we combined the ppspase1-1 and pecp1-1 mutations with the fugu5-1 allele to determine whether combining the three mutations could enhance the elongated cotyledon phenotype or lead to a new phenotype (Fig. 7A). No additional effect was observed when the three mutations were combined, and the plants displayed a phenotype similar to the single fugu5-1 mutant.
Figure 7.
Knocking out PPsPase1, PECP1, or ThMPase1 does not mimic or accentuate the phenotype of fugu5 mutants. A, The classical elongated cotyledon phenotype of fugu5 mutants in comparison to other lines. Seedlings were grown for 13 d in soil. B, Hypocotyl elongation in etiolated seedlings is impaired by the fugu5 mutations, but not by the ppspase1, pecp1 or thmpase1 mutations. Seedlings were grown mostly in the dark for 3 d on high Pi (+Pi) or low Pi (−Pi) medium. Bar = 300 µm. C, Quantification of hypocotyl length in elongated seedlings. Data shown are the mean with error bars corresponding to se (n = 22 for each genotype). Different letters indicate statistically different means (two-way ANOVA followed by Tukey’s multiple comparisons, P < 0.01).
Similar conclusions were made when we observed and quantified hypocotyl elongation in etiolated seedlings (Fig. 7, B and C). The medium Pi content had no impact on the phenotype of the respective plant lines. Altogether, these results suggest that PPsPase1, PECP1, and ThMPase1 have a negligible, if any, impact on cytosolic PPi concentrations in planta.
Expression Levels of PPsPase1 and PECP1 Affect Phosphocholine and Phosphoethanolamine Concentration
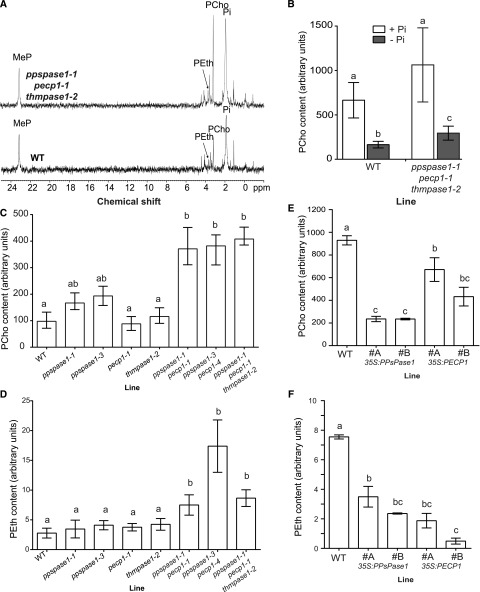
PECP1 was shown to preferentially remove Pi from PCho and PEth when the enzyme was purified and tested in vitro (May et al., 2012). The triple KO mutant and wild-type plants were analyzed by the 31P NMR technique, which can detect several phosphorus-containing compounds either in living plants or cell extracts (Bligny et al., 1990; Pratt et al., 2009). The drawback of this technique is that it requires large quantities of biological material. The triple KO mutant and wild-type plants were grown in hydroponics for several weeks and then placed in low Pi media for 12 d to enhance the Pi starvation status. In these conditions, PEth and PCho contents were found to be similarly increased in the triple mutant (Fig. 8A).
Figure 8.
PPsPase1 and PECP1 affect PCho and PEth content in plants as measured by NMR (A) and UPLC-MS/MS (B–F). A, 31P-NMR spectra of shoot extracts from the wild-type and triple KO mutant plants grown in low Pi. Peaks corresponding to the molecules of interest are labeled. MeP, Methylphosphonate (internal standard). B, PCho quantification by UPLC-MS/MS reveals that triple KO mutants contain more PCho than control plants when grown in vitro in low Pi. Contents are higher in high Pi, but similar between plant lines. Seedlings were grown for 7 d in high or low Pi. Results are presented as the mean ± sd of seven biological replicates. Different letters indicate statistically different PCho content (T-test P < 0.01). C and D, Quantification in single, double, and triple mutants reveals the importance of PPsPase1 and PECP1 expression on PCho (C) and PEth (D) content. Seedlings were grown for 8 d in low Pi. Results are presented as the mean ± sd of 4 to 11 biological replicates. Different letters indicate statistically different PCho or PEth contents (one-way ANOVA followed by Kruskal-Wallis test, P < 0.01). E and F, UPLC-MS/MS quantification shows a strong PCho (E) and PEth (F) content decrease in shoots of overexpressing lines compared to the wild-type control. Plants were grown in hydroponics in high Pi conditions. Results are presented as the mean ± se of 10 biological replicates (E) or as the range of data for two pools of five biological replicates (F). Different letters indicate statistically different PCho content (one-way ANOVA followed by Tukey’s test, P < 0.01).
To confirm this result and quantify the difference, PCho was analyzed by ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) on seedlings grown in vitro in low or high Pi in long days. Our method requires much smaller sample quantities (only 50 mg). Analysis revealed a 70 to 80% increase of PCho in the triple KO mutant compared to wild-type plants (Fig. 8B). As expected, no clear difference was noted in the high Pi condition, since these phosphatases are strongly induced in low Pi conditions. In addition, when PCho measurements were performed on mature plants grown in short days in hydroponics and transferred to short periods of Pi starvation (up to 5–6 d in low Pi conditions), no consistent difference was detected between control and KO plants.
To identify which gene could be responsible for the observed phenotype, the PCho content of wild-type seedlings grown in low Pi conditions was compared to the content of single, double, and triple KO mutants for PPsPase1, PECP1, and ThMPase1 (Fig. 8C). The PCho content in single mutants pecp1-1 and thmpase1-2 was comparable to wild-type seedlings, whereas a moderate but significant increase was detected in extracts from both ppspase1-1 and ppspase1-3 seedlings. The two ppspase1 pecp1 double mutants showed a much stronger increase (>300%) in PCho than the single mutants, confirming that these two genes function redundantly in planta (Fig. 8C). In addition, these ppspase1 pecp1 double mutants behaved similarly to the triple mutant, confirming here that ThMPase1 does not play any significant role in PCho regulation.
The quantification of PEth by UPLC-MS/MS in the same extracts did not reveal any significant difference in single mutants and yet both double and triple mutants displayed a more than 2-fold increase in PEth content (Fig. 8D).
To determine whether the overexpression of PPsPase1 or PECP1 could reciprocally diminish PCho and PEth content in plants, we quantified PCho and PEth in Prom35S:PPsPase1 and Prom35:PECP1 lines grown in hydroponics in short days and maintained in Pi-rich conditions. In these conditions, shoots had between 25 and 72% of the PCho concentration observed in the wild-type control, with the lowest amounts observed in Prom35S:PPsPase1 lines (Fig. 8E). In extracts from overexpressing lines, PEth was below the detection threshold, contrary to the wild-type extracts. However, it was possible to estimate the range of diminution in these lines by pooling and concentrating several independent biological replicates. Accordingly, PEth levels had diminished to about 50% in Prom35S:PPsPase1, and to less than 25% in Prom35:PECP1 (Fig. 8F). This result shows that both enzymes can affect PCho as well as PEth content in vivo.
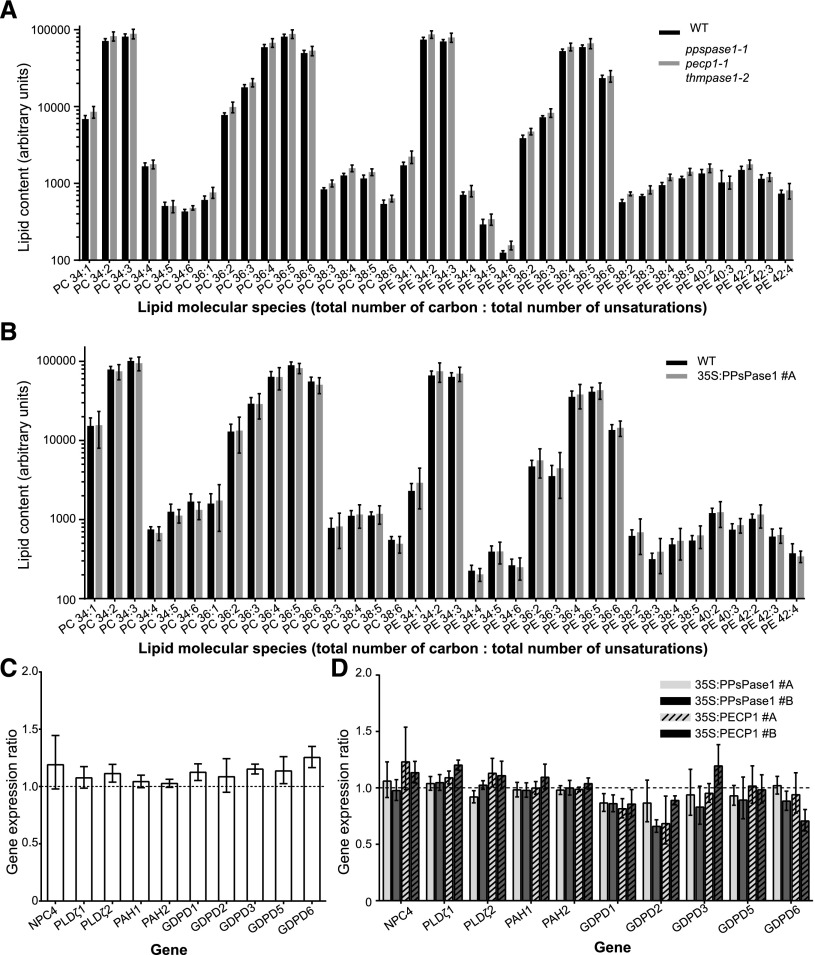
One contentious issue in the literature is whether the lipid composition of membranes, particularly lipids containing PCho and PEth polar heads (PtdCho and PtdEth), is influenced by the PCho and PEth levels in the cell (Alatorre-Cobos et al., 2012; Meï et al., 2017). To examine this issue, we evaluated if the modification of PCho and PEth observed in our mutant and overexpressing lines led to an alteration of lipid composition. For this, lipids were extracted from triple KO seedlings grown in a low-Pi condition (Fig. 9A; Supplemental Table 1), as well as overexpressing lines grown in a high-Pi condition (Fig. 9B; Supplemental Table 2). The lipid composition in both cases appeared very similar to their respective control plants, including PtdCho and PtdEth (Fig. 9A and B). Altogether, our results suggest that plants can control lipid composition independently of the cytosolic concentration of PCho and PEth, as observed with metabolic drugs that affect PCho synthesis (Meï et al., 2017).
Figure 9.
Membrane lipid composition and expression of genes involved in lipid recycling do not change in response to altered PCho or PEth content. A and B, Lipid molecular species from the triple KO mutant (A) or 35S:PPsPase1 #A overexpressing line (B) do not differ from their respective controls. KO and wild-type seedlings were grown in vitro for 8 d in low Pi. Extracts from wild-type and 35S:PPsPase1 #A shoots were obtained from plants grown hydroponically in high Pi conditions. Results are presented as the mean ± sd of five biological replicates. C and D, Knockout or overexpressing lines do not show altered regulation of the genes involved in lipid recycling during Pi starvation. C, Triple knockout and corresponding wild-type controls were grown in vitro for 8 d in low Pi. Wild-type control values were arbitrarily set to one and are represented here as a dotted line. D, Overexpressing lines for PPsPase1 (#A, solid light gray bar; #B, solid dark gray bar) and PECP1 (#A, striped light gray bar; #B, striped dark gray bar) were grown in vitro for 8 d in high Pi along with wild-type plants. Wild-type control values were arbitrarily set to one (dotted line). Results are presented as the median ± 95% confidence interval of independent biological replicates (REST randomization test, n = 2 or 3). SCAMP, ROC3, and GAPC1 were used as reference genes for the RT-qPCR. No significant differences were observed.
Achieving the correct lipid composition in the triple mutant line or the overexpressing lines could involve transcriptomic and/or posttranscriptomic regulations. To verify if altered levels of PCho and PEth could lead to the transcriptomic activation or inhibition of specific lipid metabolism pathways, we selected several genes representative of membrane lipid recycling pathways triggered during Pi starvation (Nakamura, 2013). These genes included NPC4 (representative of the nonspecific PLC pathway; Nakamura et al., 2005), PLDζ1 and 2, PAH1 and 2 (for the PLD/PAP pathway; Qin and Wang, 2002; Li et al., 2006; Nakamura et al., 2009; Eastmond et al., 2010), and GDPD1, 2, 3, 5, and 6 (for the LAH/GDPD pathway; Cheng et al., 2011). In response to a lack of Pi, we observed that NPC4, PLDζ2, and GDPD1, 2, 3, 5, and 6 were induced in wild-type plants as previously reported (Supplemental Fig. 5; Nakamura et al., 2005; Li et al., 2006; Cheng et al., 2011), even if GDPD2 and 5 were only moderately induced. PLDζ1 and PAH2 did not respond to Pi deficiency, and PAH1 was moderately (yet consistently) induced in the absence of Pi. Comparing the wild type to the triple KO line grown in low Pi did not reveal any changes in transcript levels (Fig. 9C). Similarly, no statistically significant transcript level changes were detected when comparing the wild type to the overexpressing lines 35S:PPsPase1 (#A and #B) and 35S:PECP1 (#A and #B) grown in high Pi (Fig. 9D). Altogether, our results show that maintaining a correct control over the lipid composition in the presence of altered PCho or PEth content does not appear to be attributed to compensatory mechanisms that involve the transcriptional activation or repression of genes from specific lipid recycling pathways.
DISCUSSION
When plants are confronted with Pi starvation, a rapid remobilization of Pi from internal sources occurs. One of the major Pi suppliers is the membrane phospholipids, which are converted into galactolipids and sulfolipids when this nutrient is limited (Nakamura, 2013; Pant et al., 2015). Although the enzymes involved in the initial lipid degradation and the different pathways are well described, the processes involved in the recycling of subcomponents, and particularly the recycling of phospholipid polar heads, remains unknown.
The strong induction of the PPsPase1 and PECP1 HAD-type phosphatases in response to Pi starvation led us to investigate the precise role of these enzymes in the Pi recycling process triggered by phosphorus starvation. Based on their described in vitro substrate preferences (May et al., 2011, 2012), these proteins were predicted to cleave Pi from PPi sources (PPsPase1) and PCho/PEth soluble polar heads (PECP1).
Our in vivo results reveal that both enzymes, not just PECP1, can strongly affect the concentration of PCho and PEth. Surprisingly, while PPsPase1 only marginally used PCho as a substrate in vitro (contrary to PECP1), its overexpression in plants had the strongest influence on PCho content. In addition, a single mutation affecting PPsPase1 was sufficient to significantly alter PCho content, whereas inactivation of PECP1 alone had no detectable effect on this parameter. This demonstrates that although the enzymes have distinct in vitro properties, they can both efficiently use PCho as a substrate in vivo. PCho is one of the major forms of Pi transported in xylem, independent of Pi status (Martin and Tolbert, 1983). Considering that PPsPase1 and PECP1 are strongly expressed in the vascular tissues, future research should examine how these mutations affect the content of PCho in the xylem sap. Analysis of KO mutants and overexpressing lines also revealed that these enzymes strongly affect PEth content in plants, demonstrating that both PCho and PEth can be used as substrates by PPsPase1 and PECP1.
As for PPi degradation, there was no indication of PPi imbalance when our mutants were compared to or combined with known PPi-degradation defective mutants (fugu5; Ferjani et al., 2011). While it has been suggested that alternative pathways using PPi as an energy source could be an essential strategy for plants to withstand Pi starvation (Plaxton and Tran, 2011), PPsPase1 and PECP1 do not appear to significantly affect this process. Therefore, it could be appropriate to rename the enzymes PECP1 and PECP2 at this stage. However, proper quantification of PPi is necessary before concluding that PPi might not be a significant in vivo substrate for these enzymes. Furthermore, the exact importance of PPi in plant responses to Pi limitation must still be clarified, as it was recently shown that the growth of fugu5 mutants or AVP1 overexpressors was not specifically more affected when placed in low Pi conditions (Asaoka et al., 2016).
Although the expression of PPsPase1 and PECP1 is highly dynamic and can reach very high levels, impairing their normal expression levels did not have any general impact on plant responses to Pi starvation. Altogether, our overexpressors did not show any enhanced Pi starvation symptoms, in contrast to LePS2;1 overexpressing tomato plants (Baldwin et al., 2001). Indeed, in Arabidopsis, the in vivo role of these overexpressors seems more directly linked to the degradation of PCho and PEth polar heads, the by-products of phospholipid degradation triggered in response to Pi starvation. We have demonstrated that their induction is largely under the control of PHR1/PHL1, adding them to the list of proteins involved in lipid metabolism under the control of these transcription factors (Rouached et al., 2010; Pant et al., 2015).
The superfamily of HAD phosphatases is a very large and divergent group of enzymes, with very little overall homology but great diversity in substrates (Burroughs et al., 2006). These phosphatases are subdivided into different classes based on the overall structural arrangement of their active core (Burroughs et al., 2006). Interestingly, the InterPro predictive modeling software recently assigned PPsPase1, PECP1, and ThMPase1 to the PHOSPHO1/2 subgroup within the HAD-like superfamily (Stewart et al., 2003; Hunter et al., 2009; Seifried et al., 2013). This subgroup was named based on its structural homology to the human PHOSPHO1/2 proteins. PHOSPHO1, which uses PEth and PCho as substrates, is involved in releasing Pi for bone mineralization, whereas PHOSPHO2 uses pyridoxal-5-phosphate as a substrate (Roberts et al., 2005). It has also been shown that subtle differences between the PHOSPHO1 and PHOSPHO2 amino acid sequences are responsible for these substrate preferences (Roberts et al., 2005). A deeper comparative study between the Pi starvation-induced protein and the PHOSPHO1/2 proteins should provide more insight into the structural determinant of the in vitro and in vivo substrate preferences.
This project was designed to understand the link between the role of these enzymes in Pi recycling and the general plant responses to Pi starvation. We were intrigued by the previously described phenotype of several ppspase1 mutants that were unable to develop normally elongated root hairs in Pi starvation (Chandrika et al., 2013). Although we tried to reproduce the medium composition and growth conditions of this report, root hair development was normal in our mutant lines (including one line described in Chandrika et al., 2013). This impeded our efforts to associate the described root hair phenotype with a biochemical function for these enzymes. An additional factor controlling the occurrence of the previously described root hair phenotype remains to be identified and characterized.
In the past, enzymes controlling PtdCho, PCho, PEth, and choline balance were linked to altered root architecture, including root hair alteration. Results from our work and others indicate that altered PtdCho and choline contents are responsible for these phenotypes, rather than PCho or PEth, since specifically affecting their concentrations did not influence root architecture. For instance, XIPOTL1 converts PEth to PCho through a triple methylation and is strongly involved in the PtdCho balance in membranes (McNeil et al., 2001; Cruz-Ramírez et al., 2004). Furthermore, PCho has been shown to affect the expression of XIPOTL1 indirectly, through the action of an upstream regulatory µORF (Alatorre-Cobos et al., 2012). Previous studies have reported altered root architecture and epidermal cell death in a XIPOTL1/PEAMT1 KO mutant (xpl1; Cruz-Ramírez et al., 2004). However, even though PCho content was effectively diminished in this mutant, the described phenotype was more likely caused by an impaired balance of choline, PtdCho, and phosphatidic acid (Cruz-Ramírez et al., 2004; Alatorre-Cobos et al., 2012).
A connection between choline content and root hair elongation was also suggested following the mutation of PLDζ1, an enzyme responsible for the degradation of PtdCho into PA and choline (Qin and Wang, 2002; Ohashi et al., 2003). However, the double KO pldζ1 pldζ2 contradicted these results since it did not exhibit any root hair alteration (Li et al., 2006). These conflicting results resemble our attempts to reproduce the root hair phenotype described previously for ppspase1 mutants and suggest either that these phenotypes were fortuitous or that there could be another unidentified factor controlling the link between root hair formation and choline, PCho, or PtdCho contents.
The lack of architectural modification could also be explained by the fact that several independent pathways can lead to lipid degradation during Pi starvation, through phospholipase D (PLD), phosphatidate phosphatase (PAP), phospholipase C (PLC), or other types of transferases (Nakamura, 2013; Meï et al., 2017). In addition, de novo glycerolipid biosynthesis is induced to sustain root growth (Angkawijaya et al., 2017). This could explain why we did not observe any changes in membrane lipid composition despite the dramatic changes in polar head levels. This suggests that PtdCho homeostasis is not strictly dependent on the level of PCho, as reported recently by Meï et al. (2017) but in contrast to Alatorre-Cobos et al. (2012). In the ppspase1 and pecp1 mutant backgrounds, the putative compensation of the impaired degradation of PCho/PEth by other lipid recycling pathways does not seem to involve any specific transcriptional regulation. This was shown by the lack of any altered regulation of genes representative of the PLC, PLD, and GDPD pathways. Compensatory mechanisms could involve enzymes that play a dual role in metabolism and pathway regulation, such as PAH 1 and 2 (Eastmond et al., 2000; Craddock et al., 2015).
Since the PLC pathway leads directly to the generation of PCho and PEth polar heads, this pathway could be affected in our mutant lines, contrary to other pathways. To verify this hypothesis, our overexpressors and mutants will need to be combined with other lines containing mutations in different lipid degradation routes (such as the PAP and PLD pathways).
Furthermore, recent work has demonstrated that impairing choline transport, by CHER1 choline transporter inactivation, has an impact on the development of the root vasculature and plasmodesmata in particular (Dettmer et al., 2014; Kraner et al., 2017). In light of the absence of strong architectural alterations and lipid modifications caused by the greatly diminished PCho concentration in our plant lines, the CHER1 mutant phenotype is more likely linked to choline or PtdCho imbalance rather than to lower PCho content (Kraner et al., 2017).
Altogether, our results provide valuable information on the effect of PCho and PEth homeostasis on choline content and lipid composition. Future research will focus on verifying how these alterations can preserve the lipid composition in membranes while ensuring their effective recycling in Pi-starved conditions. Ultimately, this will help to identify key elements of membrane lipid control, an important topic considering the emerging regulatory role of PtdCho in the control of plant responses to the environment (Nakamura et al., 2014).
MATERIALS AND METHODS
Plant Material and Transformation
Arabidopsis (Arabidopsis thaliana) Col-0 ecotype plant lines were used for all experiments. Plant transformation was performed using Agrobacterium tumefaciens C58C1 with a simplified version of the floral dip method (Logemann et al., 2006). T-DNA or transposon insertion lines for PPsPase1 (Salk021102C and SM_3_29962), PECP1 (GK350A04 and Salk144195), and ThMPase1 (Salk101421C) were obtained from the NASC seed collection (Scholl et al., 2000). The fugu5-1 through -3 mutants as well as fugu5-1 PromAVP1:IPP1 #8-3 were previously described (Ferjani et al., 2011). The phr1 phl1 double mutant was also previously described (González et al., 2005; Bustos et al., 2010)
CRISPR Mutagenesis
To generate new pecp1 mutant alleles using CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) mutagenesis (Barrangou, 2014), the part of the second exon that is common to the two splice forms (AT1G17710.1 and AT1G17710.2) was targeted. A similar candidate target was suggested by three CRISPR designing tools (CRISPOR, CRISPR-P, and CRISPR-PLANT; Lei et al., 2014; Xie et al., 2014; Haeussler et al., 2016) and was selected for mutagenesis. A protospacer was designed using the ATTG_protospacer and AAAC_protospacer primers (Supplemental Table 3), and the annealed primers were cloned into pEn-Chimera according to Fauser et al. (2014). After sequencing, the construct was transferred to pDe-CAS9 gentaR. This final construct was used to transform Arabidopsis. Transformants were selected on Hoagland’s solution (half-strength and without Suc) with agar (0.8%) and 80 µm gentamycin and were then transferred to soil for seed collection. T2 plants carrying a CRISPR mutation were screened by high-resolution melting (HRM) using the Light Cycler 480 apparatus (Roche) and two primer pairs (HRM_PECP1_F and HRM_PECP1_R1 or R2; Supplemental Table 3; 4 µm final concentration). MgCl2 concentration was changed in the HRM reaction mix, depending on the use of the primer R1 or R2 (requiring 3.5 or 3 mm MgCl2, respectively). The targeted genomic area was sequenced to confirm the effective mutation of pecp1 and to select homozygous plants for T3 amplification. Homozygous pecp1 T3 seed batches that segregated gentamycin resistance were selected for T4 amplification, and seed batches without gentamycin resistance (i.e. batches that lost the CRISPR machinery T-DNA insertion) were selected in the following generation.
Generation of Multiple Knockout Mutants
Double and triple homozygous KO mutants in PPsPase1, PECP1, and ThMPase1 were selected by PCR screening of the T-DNA or transposon insertion (based on the presence of the mutant allele and absence of the wild-type allele) in the F2 and F3 generations. In addition, several mutant alleles were selected based on the resistance provided by the T-DNA or transposon (PPT/Basta for ppspase1-3 and sulfadiazine for pecp1-1). Plus, pecp1 CRISPR mutations were generated in a ppspase1-3 background to immediately obtain double mutants, in accordance with the procedure described in the preceding CRISPR section.
The ppspase1-1 pecp1-1 fugu5-1 triple homozygous mutant was selected in the F2 and F3 generations by combining the selection of sulfadiazine-resistant plants (provided by the T-DNA insert of the pecp1-1 allele) with PCR screening of the two T-DNA insertions (based on the presence of the mutant allele and absence of the wild-type allele). The fugu5-1 mutation (G-to-A substitution in the coding sequence leading to an A709T amino acid change) was screened by HRM using the HRM_fugu5.1_R and F primers (Supplemental Table 3). The presence of the fugu5-1 mutation and its homozygous state were confirmed by sequencing.
Generation of Reporter Lines for Promoter Activity
A site-directed mutagenesis was performed to replace LUC with LUC+ (Promega) in pBGWL7 (Karimi et al., 2002), using the QuikChange Lightning site-directed mutagenesis kit (Stratagene) combined with protocol modifications according to Liu and Naismith (2008). Specifically, the KpnI and PshAI restriction sites were introduced surrounding the LUC gene of pBGWL7 using the SDM_Kpn1_F and R and SDM_PshA1_F and R primer pairs (Supplemental Table 3). The LUC+ gene (Promega) was amplified by PCR with addition of the KpnI and PshAI restriction sites (in 5′ and 3′) using the SDM_Kpn1_F and ASP_LUC+_PshA1 primers (Supplemental Table 3), and then subcloned into pGEMT-easy (Promega) following the manufacturer’s instructions. After KpnI and PshAI digestion, LUC+ replaced the LUC gene, resulting in the new pBGWL7+ vector.
The PPsPase1 (2001 bp upstream of ATG) and PECP1 promoters (1973 bp upstream of ATG) were PCR amplified using the SP_PromPPsPase1/ASP_PromPPsPase1 and SP_PromPECP1/ASP_PromPECP1 primers, respectively (Supplemental Table 3). After amplification, the promoters were subcloned into pENTR/D-TOPO (Thermo Fisher Scientific), sequenced, and cloned via a gateway LR reaction using Gateway LR clonase II mix (Thermo Fisher Scientific) into pBGWL7+. The final constructs were sequenced and named PromPPsPase1:LUC+ and PromPECP1:LUC+. The same PPsPase1 promoter sequence was transferred into the pBGWFS7 vector (Karimi et al., 2002) using the LR Gateway reaction to create the PromPPsPase1:GFP-GUS construct.
For all the above reporter lines, T1 transformants were selected in soil using Basta, and the T2 generation was screened on plates for PPT resistance. Only those lines showing a 3:1 segregation of PPT resistance in T2 (suggesting a single T-DNA insertion event) were amplified to obtain homozygous plants in T3. The expression pattern was studied with at least four independent lines before selection of the representative line. Displayed results are from the T3 and T4 generations.
For MGD3 reporting, a 1284-bp fragment of the MGD3 (AT2g11810) 5′ sequence was amplified by PCR using the SP_PromMGD3/ASP_PromMGD3 primers (Supplemental Table 3) that added a NheI and a SalI site to the 5′ end, and a NcoI site to the 3′ end. The PCR fragment was digested with NheI and NcoI, and ligated with an equimolar amount of pSP-luc+ (Promega) that had been digested with the same enzymes. The promoter sequence was validated by Sanger sequencing on the resulting pSP-PromMGD3:LUC+. The PromMGD3:LUC+ cassette was excised with SalI and SacI, and ligated into pPTV30bar (a derivative of pGPTV-BAR) in which the nos terminator was replaced with the ocs terminator. This was introduced into A. tumefaciens and transferred into Arabidopsis by floral dip. Seven lines with single functional inserts were characterized for the absence of basal luciferase activity and the magnitude of luciferase activity increase following 6 d of growth on –Pi media. The best line was used in the experiments reported here.
Gene Overexpressing Lines
To design the overexpressing constructs under 35S promoter control, the PPsPase1 and PECP1 coding sequences (including their stop codons) were amplified from genomic DNA using the SP_CDS_PPsPase1/ASP_CDS_PPsPase1_stop and SP_CDS_PECP1/ASP_CDS_PECP1_stop primer combinations, respectively (Supplemental Table 3). PCR products were purified and cloned into pENTR/D-TOPO (Thermo Fisher Scientific), sequenced, and then recombined with pEN-L4-2-R1 and pB7m24GW (Karimi et al., 2007) via a multiple gateway LR reaction (as described above). The final constructs were sequenced and named Prom35S:PPsPase1 and Prom35S:PECP1.
After transformation, T1 transformed plants were selected in soil using Basta, and a preliminary selection of overexpressing lines was performed by RT-qPCR. Only overexpressing lines showing a 3:1 segregation of PPT resistance in T2 (suggesting a single T-DNA insertion event) were reamplified to obtain homozygous plants in T3. Quantification of the overexpression level was performed by RT-qPCR in the T3 generation.
Plant Growth Conditions
To screen the cotyledon fugu5 phenotype, seeds were vernalized for 2 d at 4°C before sowing them in soil in a short-day growth chamber (8-h-day/16-h-night photoperiod, 120–160 µmol photons m−2 s−1; 20°C day/18°C night).
For in vitro assays, seeds were surface sterilized for 5 min in a sterilization solution (0.3% active chlorine in 86% ethanol), followed by two rinses in 70% ethanol. Seeds were then sown on modified MS/10 agar medium (0.15 mm MgSO4, 2.07 mm NH4NO3, 1.88 mm KNO3, 0.34 mm CaCl2, 0.5 µm KI, 10 µm H3BO3, 10 µm MnSO4, 3 µm ZnSO4, 0.1 µm Na2MoO4, 0.1 µm CuSO4, 0.1 µm CoCl2, either 0.5 mm KH2PO4 or 0.5mm KCl for the low Pi version, either 2 or 10 µm FeCl2, and 3.4 mm MES, pH 5.7) supplemented with agar (0.8%) from Sigma-Aldrich (A1296 #BCBL6182V), and with or without Suc (0.5%). The low Pi version of the medium, with added KCl instead of KH2PO4, only contained Pi from the agar (∼13 µm). After ∼2 d at 4°C, plates were placed vertically in a long-day growth chamber (16-h-day/8-h-night photoperiod, 70 µmol photons m−2 s−1; 24.5°C day/21°C night). In addition, for root hair screening, plants were grown on the same medium used by Chandrika et al. (2013).
To observe etiolated seedlings, seeds were sown on either high or low Pi agar medium (with 2 µm FeCl2) without Suc. After 3 d at 4°C, the plates were placed horizontally in a 23°C growth chamber and were grown for 6 h in the light (∼ 70 µmol m−2 s−1) to induce germination, followed by 66 h in the dark for hypocotyl elongation.
For hydroponic cultures, plants were initially grown on plates for 12 to 16 d on high Pi medium in short days (8-h-day/16-h-night photoperiod, 120 µmol photons m−2 s−1; 24°C day/21°C night) before transfer to the hydroponic system. Plants were then placed on a polystyrene plate floating on 4 liters of MS/10 liquid medium supplemented with vitamins (M5519 Sigma-Aldrich) and grown in the same conditions for about 4 weeks, before being transferred to high Pi modified MS/10 liquid medium (0.15 mm MgSO4, 2.07 mm NH4NO3, 1.88 mm KNO3, 0.34 mm CaCl2, 0.5 µm KI, 10 µm H3BO3, 10 µm MnSO4, 3 µm ZnSO4, 0.1 µm Na2MoO4, 0.1 µm CuSO4, 0.1 µm CoCl2, 0.5 mm KH2PO4, and 10 µM FeCl2). Plants were maintained in this solution for at least 1 week before analysis or transfer to a low Pi condition (containing only 10 µm KH2PO4 and 0.49 mm KCl instead of 0.5 mm KH2PO4). Hydroponic solutions were replaced twice per week and were aerated constantly.
Reporter Line Signal Quantification and Imaging
To measure gene induction kinetics using the Prom:LUC+ lines, seeds were vernalized for 2 d and then germinated for 4 d on high Pi modified MS/10 medium (containing Suc and 2 µm FeCl2) in constant light (150 µmol photons m−2 s−1; 22°C). Seedlings were then individually transferred to wells in white plastic 96-well plates with 200 µL liquid/well of MS/10 medium with Suc, and supplemented with 50 µm luciferin (d-luciferin firefly potassium salt; Biosynth) under two different conditions: 500 µm KH2PO4 + 2 µm FeCl2 or 10 µm KH2PO4 + 2 µm FeCl2. Additional luciferin (50 µm) was added after 24 h. Altogether, seedlings were grown for 2 d in liquid under gentle agitation (160 rpm), and then luminescence was measured using a plate reader (Tecan Infinite M200; 3 cycles of 200 ms acquisition/well). After the measurement, 4 µL of 25 mm KH2PO4 (final concentration = 500 µm) or an equivalent volume of water was added to the wells, and further luminescence measurements were performed during ∼ 24 h.
To image luminescence, we designed a dedicated plant growth chamber (Lumalum) allowing the automated control of light phases for growth and dark phases with luminescence imaging. To achieve this, we mounted a customized light unit (RX30 Heliospectra) on top of a dark ventilated chamber. This unit was modified by the manufacturer to deactivate the internal LED controls and remove phosphor-coated LEDS to eliminate background noise. Petri plates were placed vertically inside the chamber on a carrousel, with one position of the carrousel facing the objective connected to a cooled back-illuminated CCD camera (IkonM Andor). A dedicated script was created to control the imaging setup and its carrousel through µManager (Edelstein et al., 2014), with an additional Arduino (http://www.arduino.cc) component and script for interfacing purposes. The chamber was placed in a temperature-controlled room (around 22°C) and the temperature inside the chamber was stabilized at around +24°C. The LED unit was constantly “on” (at ∼150 µmol photons m−2 s−1), except during luminescence imaging phases (2 min dark period for chlorophyll deexcitation, followed by 1 min of luminescence signal acquisition in the dark). Gene induction kinetics were imaged every hour, over 3.75 d. Plants were grown for 4 to 5 d in constant light on high Pi modified MS/10 agar medium supplemented with 50 µm luciferin and were then transferred onto low Pi medium with 50 µm luciferin immediately before the first acquisition.
GUS staining was performed as previously described (Misson et al., 2004) on plants grown in low or high Pi medium. For visualization, seedlings were either placed in water and observed under a stereomicroscope (MZ16; Leica Microsystems) or between cover slips and observed using an upright microscope (LMD6000; Leica Microsystems).
Transcript Analysis
For RNA-seq studies, sample preparation and transcript analysis were performed as previously described (Secco et al., 2015), with several exceptions. First, samples were harvested 30 min, 1 h, 3 h, or 24 h after transfer to fresh medium (from low Pi to low Pi, high Pi to high Pi, or low Pi to high Pi). Second, RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). Roots from three independent biological replicates were analyzed for each condition.
For RT-qPCR analysis, presented in Figures 3, 4, and 6, total RNA was extracted from 50 to 100 mg of frozen plant tissues using the RNeasy extraction kit (Qiagen), combined with an RNase-free DNase Set (Qiagen). After DNA quantification and quality control on agarose gel, reverse transcription of poly(dT) cDNA was performed on an initial 400 ng of total RNA, using the SuperScript VILO cDNA Synthesis kit (Invitrogen) and following the manufacturer’s instructions. For RT-qPCR analysis in Figure 9 and Supplemental Figures 2 and 5, RNA was extracted from two or three independent biological replicates (16–50 mg of frozen plant tissue per sample) using the Direct-zol RNA MiniPrep kit (Zymo Research). DNase treatment is included in the kit. Reverse transcription was performed starting with 400 ng of RNA, using the qScript XLT cDNA SuperMix (Quantabio) and following the manufacturer’s instructions. Real-time PCR was performed in 384-well plates (final volume = 5 µL/reaction) using a Lightcycler 480 (Roche) and the SYBR Green I Master 2X mix (Roche), following the manufacturer’s recommendations. Primer combinations and amplification efficiency for each gene are provided in Supplemental Table 3. Reactions were performed in triplicate.
Metabolite Quantification
Quantification of the free cellular Pi content was performed as previously described (Misson et al., 2004). Analysis of PCho and PEth content by 31P NMR was performed on shoots and roots from plants grown in hydroponics, following the procedure from Mongélard et al. (2011).
For the quantification of PCho by UPLC-MS/MS, wild-type and ppspase1-1/pecp1-1/thmpase1-2 plants were grown in vitro for 1 week on low or high Pi medium (2 µm FeCl2). For the parallel study of single, double and triple mutants, plants were grown in vitro for 8 d on low Pi medium (2 µm FeCl2). Wild-type and overexpressing lines were grown in hydroponics for 4 weeks in commercial MS/10 solution and then for 4 d in high Pi-modified MS/10 solution (10 µm FeCl2).
For each sample, 20 to 50 mg fresh weight was harvested, using whole seedlings for in vitro samples or rosettes for hydroponic plants. A direct extraction of the metabolites into hot isopropanol (without prior freezing or grinding) was favored to avoid unwanted degradation by endogenous phosphatases or lipases during the extraction procedure. Briefly, plant samples were placed in 1 or 2 mL hot isopropanol + 0.01% BHT (for in vitro and hydroponics plants, respectively), and maintained at 85°C for 30 min. After cooling to room temperature, standards were added [1 µm phosphocholine-(trimethyl-d9), 0.7 mm PE (17:0/17:0); Sigma-Aldrich] and samples were ground for 1 min using an Ultra-turrax T25 apparatus (IKA Labortechnik). A second grinding (30 s) was performed after adding 1 or 2 mL of water (for in vitro and hydroponics plants, respectively). The slurry was centrifuged for 5 min at 4,000g at 4°C to eliminate most debris. The clear supernatant was transferred to a tube and vortexed after addition of 3 or 6 mL tert-butyl methyl ether with 0.01% formic acid (for in vitro and hydroponic samples, respectively). The aqueous and lipid phases were separated by a short centrifugation (3,000g for 3 min at 4°C). The lower (aqueous) phase was placed under a gentle stream of nitrogen gas to evaporate the remaining traces of organic phase, and then stored at −20°C until analysis. The upper (organic) phase was maintained under nitrogen gas until complete evaporation and was subsequently resuspended in 200 µL acetonitrile/isopropanol/ammonium formate (65:30:5, v/v/v). The final concentration of ammonium formate was 10 mm.
PCho content (from the aqueous phase) was analyzed using an UPLC-MS/MS system (UPLC ultimate RS 3000 Dionex QTOF 5600; AB Sciex) connected to a Kinetex C18 2.1 × 150-mm column (Phenomenex). The ESI source was operated in positive mode. For in vitro samples, a binary gradient of solution A (95v:5v water/methanol with 0.01% v/v formic acid) and solution B (95v:5v methanol/isopropanol with 0.01% v/v formic acid) was applied. Enrichment of solution A was from 0 to 100% within 20 min at a speed of 0.3 mL min−1 and then at 100% for 5 min. Solution A was then decreased to a 95% enrichment during 7 min for column reequilibration. Column temperature was maintained at 45°C. Pi-choline was identified using the retention time and the MS2 signal corresponding to the Pi group (parent ion m/z 184.1 for PCho and m/z 193.1 for PCho-d9, product ion m/z 98.9). Relative quantification of PCho in each sample was processed through the MultiQuant built-in software (AB Sciex) by comparing the peak surface areas of PCho and phosphocholine-(trimethyl-d9) internal standard.
For PEth quantification, an additional concentration step and a modified chromatography protocol were used. The chromatography mobile phases included solvent A (acetonitrile/formic acid = 100/3) and solvent B (acetonitrile/100 mm ammonium formate = 20/80). A 250-µL aliquot of the sample aqueous phase was lyophilized and then resuspended in 15 µL of the initial mobile phase (solvent A: solvent B = 8:2). HPLC was then performed using a Dionex Ultimate 3000 HPLC system equipped with an autosampler (Thermo Fisher Scientific). The HPLC system was interfaced with an EXACTIVE Plus Fourier transform mass spectrometer (Thermo Fisher Scientific) with an electrospray ionization source. An Intrada amino acid column (3 × 10 mm; Imtakt) was used. The column was developed at a flow rate of 600 μL min−1 with the following concentration gradient for solvent B: sustaining 20% B for 4 min, from 20% B to 100% B in 10 min, sustaining 100% B for 2 min, from 100% B to 20% B in 0.1 min, and finally reequilibrating with 20% B for 7 min. The electrospray ionization source was operated in positive and negative ion mode. PCho-d9 was again used as the reference compound. Data acquisition and analysis were performed using the Xcalibur software (version 2.2).
Lipid content (from the organic phase) was analyzed by an UPLC-MS/MS system (UPLC ultimate RS 3000 Dionex QTOF 5600; AB Sciex), using a Kinetex C8 2.1 × 150-mm column (Phenomenex) and a binary gradient of solution A (60v:40v water/acetonitrile) and solution B (90v:10v isopropanol/acetonitrile). Elution was achieved through a gradient of solution B from 27 to 97% as compared to solvent A within 20 min at a speed of 0.3 mL min−1 and then at 97% for 5 min. Solution B was then decreased to a 27% enrichment during 7 min for column reequilibration. Column temperature was maintained at 45°C. Lipid identification was based on retention time, mass accuracy peaks from the MS survey scan compared with theoretical masses, and fragment ions from the MS/MS scan. Relative quantification was achieved using multiquant software (AB Sciex) on the basis of intensity values after extracting masses of previously identified lipids.
Root Hair and Hypocotyl Length Measurements
Root hairs were imaged from seedlings growing on plates using a macroscope (Axio Zoom V16; Carl Zeiss) connected to an AxioCam HRm camera (Carl Zeiss). For etiolated hypocotyl measurements, hypocotyls were manually laid onto the agarose gel before image acquisition. Measures of hypocotyl and root hair length were performed in a semiautomated way using the “NeuronJ” plug-in (Meijering et al., 2004) in combination with ImageJ software (Schneider et al., 2012).
Statistics
RNA-seq data were analyzed using the Cuffdiff software from TopHat (Johns Hopkins University). RT-qPCR results were analyzed using the REST2009 relative expression software tool (Pfaffl et al., 2002). The primer efficiency factor was measured for each gene. The reference genes (SCAMP-At1g32050, ROC3-At2g16600, GAPC1-At3g04120, or GF14phi-At1g35160) used for each analysis are indicated in each figure legend. Data were analyzed after 3,000 randomizations. All other statistical analyses were performed using the GraphPad Prism6 (Graph Pad) software. Details for each test (primary and posthoc tests) are indicated in each figure legend.
Accession Numbers
Sequence data for genes from this article can be found in the GenBank/EMBL data libraries under accession numbers 838347 (AT1G17710), 843632 (AT1G73010), and 829074 (AT4G29530). The pBGWL7+ vector and map were deposited at the VIB (https://gateway.psb.ugent.be).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PromPPsPase1:GUS expression in Pi-rich plants.
Supplemental Figure S2. Expression of PPsPase1 and PECP1 is affected by the Pi concentration in the growth medium.
Supplemental Figure S3. PPsPase1 is expressed during the late stages of anther development.
Supplemental Figure S4. Characterization of the Arabidopsis PPsPase1, PECP1, and ThMPase1 knockout lines.
Supplemental Figure S5. Regulation of a set of representative genes involved in lipid recycling by Pi availability in wild-type plants.
Supplemental Movie S1. Animation showing the kinetics of luminescence signal induction in a PromPECP1:LUC+ plantlet after transfer to a –Pi medium.
Supplemental Table S1. Lipid content in wild type and ppspase1-1 pecp1-1 thmpase1-2 triple mutant.
Supplemental Table S2. Lipid content in wild type and Prom35S:PPsPase1 #A-overexpressing plants.
Supplemental Table S3. List of primers.
Acknowledgments
We acknowledge Julia Revol, Serge Chiarenza, Corinne Brouchoud, Caroline Mercier, all members of the LBDP and GRAP teams (CEA), for their technical support during the development of this project. We also thank Anne-Marie Boisson for help with the NMR sample preparation, Pierre Richaud for ICP measurements, and Brandon Loveall from Improvence for English proofreading of the manuscript. We also thank the creators of the FigureJ plug-in for their practical figure preparation tool (Mutterer and Zinck, 2013).
Footnotes
Articles can be viewed without a subscription.
References
- Alatorre-Cobos F, Cruz-Ramírez A, Hayden CA, Pérez-Torres C-A, Chauvin A-L, Ibarra-Laclette E, Alva-Cortés E, Jorgensen RA, Herrera-Estrella L (2012) Translational regulation of Arabidopsis XIPOTL1 is modulated by phosphocholine levels via the phylogenetically conserved upstream open reading frame 30. J Exp Bot 63: 5203–5221 [DOI] [PubMed] [Google Scholar]
- Angkawijaya AE, Nguyen VC, Nakamura Y (2017) Enhanced root growth in phosphate-starved Arabidopsis by stimulating de novo phospholipid biosynthesis through the overexpression of LYSOPHOSPHATIDIC ACID ACYLTRANSFERASE 2 (LPAT2). Plant Cell Environ 40: 1807–1818 [DOI] [PubMed] [Google Scholar]
- Asaoka MM, Segami S, Ferjani A, Maeshima M (2016) Contribution of PPi-hydrolyzing function of vacuolar H(+)-pyrophosphatase in vegetative growth of Arabidopsis: evidenced by expression of uncoupling mutated enzymes. Front Plant Sci 7: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi A, David P, Arrighi J-F, Chiarenza S, Thibaud M-C, Nussaume L, Marin E (2015) Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol 167: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JC, Karthikeyan AS, Cao A, Raghothama KG (2008) Biochemical and molecular analysis of LePS2;1: a phosphate starvation induced protein phosphatase gene from tomato. Planta 228: 273–280 [DOI] [PubMed] [Google Scholar]
- Baldwin JC, Karthikeyan AS, Raghothama KG (2001) LEPS2, a phosphorus starvation-induced novel acid phosphatase from tomato. Plant Physiol 125: 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzergue C, Dartevelle T, Godon C, Laugier E, Meisrimler C, Teulon J-M, Creff A, Bissler M, Brouchoud C, Hagège A, et al. (2017) Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat Commun 8: 15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R. (2014) RNA events. Cas9 targeting and the CRISPR revolution. Science 344: 707–708 [DOI] [PubMed] [Google Scholar]
- Bieleski RL. (1973) Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol 24: 225–252 [Google Scholar]
- Bieleski RLFI. (1983) Physiology and metabolism of phosphate and its compounds. In BR Läuchli, ed, Encyclopedia of Plant Physiology, Vol 15A. Springer-Verlag, Berlin, pp 422–449 [Google Scholar]
- Bligny R, Gardestrom P, Roby C, Douce R (1990) 31P NMR studies of spinach leaves and their chloroplasts. J Biol Chem 265: 1319–1326 [PubMed] [Google Scholar]
- Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L (2006) Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol 361: 1003–1034 [DOI] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrika NN, Sundaravelpandian K, Yu SM, Schmidt W (2013) ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol 198: 709–720 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zhou W, El Sheery NI, Peters C, Li M, Wang X, Huang J (2011) Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J 66: 781–795 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Cordell D, Drangert J-O, White S (2009) The story of phosphorus: Global food security and food for thought. Glob Environ Change 19: 292–305 [Google Scholar]
- Craddock CP, Adams N, Bryant FM, Kurup S, Eastmond PJ (2015) PHOSPHATIDIC ACID PHOSPHOHYDROLASE regulates phosphatidylcholine biosynthesis in Arabidopsis by phosphatidic acid-mediated activation of CTP:PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE activity. Plant Cell 27: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, López-Bucio J, Ramírez-Pimentel G, Zurita-Silva A, Sánchez-Calderon L, Ramírez-Chávez E, González-Ortega E, Herrera-Estrella L (2004) The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity. Plant Cell 16: 2020–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Ursache R, Campilho A, Miyashima S, Belevich I, O’Regan S, Mullendore DL, Yadav SR, Lanz C, Beverina L, et al. (2014) CHOLINE TRANSPORTER-LIKE1 is required for sieve plate development to mediate long-distance cell-to-cell communication. Nat Commun 5: 4276. [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Hooks MA, Williams D, Lange P, Bechtold N, Sarrobert C, Nussaume L, Graham IA (2000) Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. J Biol Chem 275: 34375–34381 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Quettier A-L, Kroon JTM, Craddock C, Adams N, Slabas AR (2010) Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22: 2796–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N (2014) Advanced methods of microscope control using μManager software. J Biol Methods 1: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79: 348–359 [DOI] [PubMed] [Google Scholar]
- Ferjani A, Segami S, Horiguchi G, Muto Y, Maeshima M, Tsukaya H (2011) Keep an eye on PPi: the vacuolar-type H+-pyrophosphatase regulates postgerminative development in Arabidopsis. Plant Cell 23: 2895–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdula DA, Sandaltzopoulos R, Tsukiyama T, Ossipow V, Wu C (1998) Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev 12: 3206–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E, Solano R, Rubio V, Leyva A, Paz-Ares J (2005) PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber BD, Giehl RFH, Friedel S, von Wirén N (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud J-B, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly J-S, Concordet J-P (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain G, Roje S, Sa N, Zallot R, Ziemak MJ, de Crécy-Lagard V, Gregory JF III, Hanson AD (2016) Bacterial and plant HAD enzymes catalyse a missing phosphatase step in thiamin diphosphate biosynthesis. Biochem J 473: 157–166 [DOI] [PubMed] [Google Scholar]
- Heinonen JK. (2001) Biological Role of Inorganic Pyrophosphate. Springer, Boston/Dordrecht/London [Google Scholar]
- Hirsch J, Misson J, Crisp PA, David P, Bayle V, Estavillo GM, Javot H, Chiarenza S, Mallory AC, Maizel A, et al. (2011) A novel fry1 allele reveals the existence of a mutant phenotype unrelated to 5′->3′ exoribonuclease (XRN) activities in Arabidopsis thaliana roots. PLoS One 6: e16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Chu C (2011) Phosphate starvation signaling in rice. Plant Signal Behav 6: 927–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Shirley N, Genc Y, Shi B, Langridge P (2011) Phosphate utilization efficiency correlates with expression of low-affinity phosphate transporters and noncoding RNA, IPS1, in barley. Plant Physiol 156: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, Baldan B, Bligny R, Joyard J, Block MA (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P (2007) Building blocks for plant gene assembly. Plant Physiol 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H (2009) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Kraner ME, Link K, Melzer M, Ekici AB, Uebe S, Tarazona P, Feussner I, Hofmann J, Sonnewald U (2017) Choline transporter-like1 (CHER1) is crucial for plasmodesmata maturation in Arabidopsis thaliana. Plant J 89: 394–406 [DOI] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu H-Y, Li S, Xing F, Chen L-L (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7: 1494–1496 [DOI] [PubMed] [Google Scholar]
- Li M, Qin C, Welti R, Wang X (2006) Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 140: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Ülker B, Somssich IE (2006) An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BA, Tolbert NE (1983) Factors which affect the amount of inorganic phosphate, phosphorylcholine, and phosphorylethanolamine in xylem exudate of tomato plants. Plant Physiol 73: 464–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A, Berger S, Hertel T, Köck M (2011) The Arabidopsis thaliana phosphate starvation responsive gene AtPPsPase1 encodes a novel type of inorganic pyrophosphatase. Biochim Biophys Acta 1810: 178–185 [DOI] [PubMed] [Google Scholar]
- May A, Spinka M, Köck M (2012) Arabidopsis thaliana PECP1: enzymatic characterization and structural organization of the first plant phosphoethanolamine/phosphocholine phosphatase. Biochim Biophys Acta 1824: 319–325 [DOI] [PubMed] [Google Scholar]
- McNeil SD, Nuccio ML, Ziemak MJ, Hanson AD (2001) Enhanced synthesis of choline and glycine betaine in transgenic tobacco plants that overexpress phosphoethanolamine N-methyltransferase. Proc Natl Acad Sci USA 98: 10001–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meï CE, Cussac M, Haslam RP, Beaudoin F, Wong YS, Maréchal E, Rébeillé F (2017) C1 metabolism inhibition and nitrogen deprivation trigger triacylglycerol accumulation in Arabidopsis thaliana cell cultures and highlight a role of NPC in phosphatidylcholine-to-triacylglycerol pathway. Front Plant Sci 7: 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JCF, Steiner P, Hirling H, Unser M (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58: 167–176 [DOI] [PubMed] [Google Scholar]
- Mimura M, Zallot R, Niehaus TD, Hasnain G, Gidda SK, Nguyen TND, Anderson EM, Mullen RT, Brown G, Yakunin AF, et al. (2016) Arabidopsis TH2 encodes the orphan enzyme thiamin monophosphate phosphatase. Plant Cell 28: 2683–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Thibaud M-C, Bechtold N, Raghothama K, Nussaume L (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Mongélard G, Seemann M, Boisson A-M, Rohmer M, Bligny R, Rivasseau C (2011) Measurement of carbon flux through the MEP pathway for isoprenoid synthesis by (31)P-NMR spectroscopy after specific inhibition of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate reductase. Effect of light and temperature. Plant Cell Environ 34: 1241–1247 [DOI] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143: 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutterer J, Zinck E (2013) Quick-and-clean article figures with FigureJ. J Microsc 252: 89–91 [DOI] [PubMed] [Google Scholar]
- Nakamura Y. (2013) Phosphate starvation and membrane lipid remodeling in seed plants. Prog Lipid Res 52: 43–50 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Andrés F, Kanehara K, Liu YC, Dörmann P, Coupland G (2014) Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat Commun 5: 3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K, Ohta H (2005) A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Chem 280: 7469–7476 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, Ohta H (2009) Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci USA 106: 20978–20983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud M-C (2011) Phosphate import in plants: Focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T (2003) Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Pant BD, Burgos A, Pant P, Cuadros-Inostroza A, Willmitzer L, Scheible WR (2015) The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J Exp Bot 66: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T (2011) Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci 16: 442–450 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzio GA, Paez-Valencia J, Khadilkar AS, Regmi K, Patron-Soberano A, Zhang S, Sanchez-Lares J, Furstenau T, Li J, Sanchez-Gomez C, et al. (2015) Arabidopsis type I proton-pumping pyrophosphatase expresses strongly in phloem, where it is required for pyrophosphate metabolism and photosynthate partitioning. Plant Physiol 167: 1541–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT (2011) Metabolic adaptations of phosphate-starved plants. Plant Physiol 156: 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Bucher M (2002) Phosphate transport and homeostasis in Arabidopsis. Arabidopsis Book 1: e0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt J, Boisson AM, Gout E, Bligny R, Douce R, Aubert S (2009) Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells: an in vivo 31P-nuclear magnetic resonance study using methylphosphonate as a Pi analog. Plant Physiol 151: 1646–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, et al. (2014) SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA 111: 14947–14952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Rojas-Triana M, de Lorenzo L, Leyva A, Rubio V, Paz-Ares J (2017) Novel signals in the regulation of Pi starvation responses in plants: facts and promises. Curr Opin Plant Biol 39: 40–49 [DOI] [PubMed] [Google Scholar]
- Qin C, Wang X (2002) The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLD zeta 1 with distinct regulatory domains. Plant Physiol 128: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Raghothama KG, Karthikeyan AS (2005) Phosphate acquisition. Plant Soil 274: 37–49 [Google Scholar]
- Roberts SJ, Stewart AJ, Schmid R, Blindauer CA, Bond SR, Sadler PJ, Farquharson C (2005) Probing the substrate specificities of human PHOSPHO1 and PHOSPHO2. Biochim Biophys Acta 1752: 73–82 [DOI] [PubMed] [Google Scholar]
- Rouached H, Arpat AB, Poirier Y (2010) Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3: 288–299 [DOI] [PubMed] [Google Scholar]
- Rowland O, Lee R, Franke R, Schreiber L, Kunst L (2007) The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett 581: 3538–3544 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Wang C, Shou H, Schultz MD, Chiarenza S, Nussaume L, Ecker JR, Whelan J, Lister R (2015) Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4: e09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifried A, Schultz J, Gohla A (2013) Human HAD phosphatases: structure, mechanism, and roles in health and disease. FEBS J 280: 549–571 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Siebers M, Dörmann P, Hölzl G (2015) Membrane remodelling in phosphorus-deficient plants. In Annual Plant Reviews, Plaxton WC, Lambers H, eds, Vol 48. John Wiley & Sons Hoboken, NJ, pp 237–263 [Google Scholar]
- Stewart AJ, Schmid R, Blindauer CA, Paisey SJ, Farquharson C (2003) Comparative modelling of human PHOSPHO1 reveals a new group of phosphatases within the haloacid dehalogenase superfamily. Protein Eng 16: 889–895 [DOI] [PubMed] [Google Scholar]
- Sun L, Song L, Zhang Y, Zheng Z, Liu D (2016) Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170: 499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Tannert M, May A, Ditfe D, Berger S, Balcke GU, Tissier A, Köck M (2018) Pi starvation-dependent regulation of ethanolamine metabolism by phosphoethanolamine phosphatase PECP1 in Arabidopsis roots. J Exp Bot 69: 467–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Abel S (2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang H, Li K, Wu P, Shou H (2009) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wang L, Li Z, Qian W, Guo W, Gao X, Huang L, Wang H, Zhu H, Wu J-W, Wang D, Liu D (2011) The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157: 1283–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasaki J, Yonetani R, Kuroda S, Shinano T, Yazaki J, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T, et al. (2003) Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ 26: 1515–1523 [Google Scholar]
- Wild R, Gerasimaite R, Jung JY, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M, Mayer A (2016) Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352: 986–990 [DOI] [PubMed] [Google Scholar]
- Xie K, Zhang J, Yang Y (2014) Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant 7: 923–926 [DOI] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Lu S, Liu D (2014) A major root-associated acid phosphatase in Arabidopsis, AtPAP10, is regulated by both local and systemic signals under phosphate starvation. J Exp Bot 65: 6577–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]