Oxidation of a cysteine residue in translation factor EF-Tu inhibits the synthesis of proteins that are essential for the repair of photosystem II, resulting in stimulation of photoinhibition.
Abstract
The repair of photosystem II (PSII) is particularly sensitive to oxidative stress and the inhibition of repair is associated with oxidative damage to the translational elongation system in the cyanobacterium Synechocystis sp. PCC 6803. However, the molecular mechanisms underlying this inhibition are unknown. We previously demonstrated in vitro that EF-Tu, a translation factor that delivers aminoacyl-tRNA to the ribosome, is inactivated by reactive oxygen species via oxidation of the Cys residue Cys-82. In this study, we examined the physiological role of the oxidation of EF-Tu in Synechocystis. Under strong light, EF-Tu was rapidly oxidized to yield oxidized monomers in vivo. We generated a Synechocystis transformant that expressed mutated EF-Tu in which Cys-82 had been replaced with a Ser residue. Under strong light, the de novo synthesis of proteins that are required for PSII repair, such as D1, was enhanced in the transformant and photoinhibition of PSII was alleviated. However, photodamage to PSII, measured in the presence of lincomycin, was similar between the transformant and wild-type cells, suggesting that expression of mutated EF-Tu might enhance the repair of PSII. Alleviating photoinhibition through mutation of EF-Tu did not alter cell growth under strong light, perhaps due to the enhanced production of reactive oxygen species. These observations suggest that the oxidation of EF-Tu under strong light inhibits PSII repair, resulting in the stimulation of photoinhibition.
Photosystem II (PSII), a protein-pigment complex that converts light energy into chemical energy, is sensitive to light, and light-induced damage (photodamage) to PSII occurs under light at any intensity (Tyystjärvi and Aro, 1996). Damaged PSII is immediately repaired via the de novo synthesis of proteins, such as the D1 protein, that form the reaction center of PSII (Aro et al., 1993). Thus, photoinhibition of PSII becomes apparent when the rate of photodamage exceeds the rate of repair of damaged PSII under strong light (Nishiyama et al., 2006). The rate of photodamage can be monitored in the presence of an inhibitor of protein synthesis, such as lincomycin or chloramphenicol, that blocks the repair of PSII. Photoinhibition in the presence of inhibitors of protein synthesis has been examined in several photosynthetic organisms, including cyanobacteria and Arabidopsis (Arabidopsis thaliana; Wada et al., 1994; Moon et al., 1995). Exploration of photoinhibition using this strategy has revealed that reactive oxygen species (ROS) act primarily by inhibiting the repair of damaged PSII during photoinhibition (Nishiyama et al., 2001, 2004; Murata and Nishiyama, 2018).
Under strong light, ROS are produced in abundance as a result of the photosynthetic transfer of excitation energy and the transport of electrons (Asada, 1999). Polysome analysis in the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) showed that ROS inhibit the elongation step in the translation of psbA mRNA, which encodes the D1 protein (Nishiyama et al., 2001, 2004). Biochemical studies revealed that two translation factors, EF-G and EF-Tu, which are responsible for translational elongation, are the targets of ROS within the translational machinery of Synechocystis (Kojima et al., 2007; Yutthanasirikul et al., 2016). EF-G translocates peptidyl-tRNA from the A site to the P site of the ribosome, and it is inactivated by H2O2 via oxidation of Cys-105 and Cys-242, with subsequent formation of an intramolecular disulfide bond (Kojima et al., 2009). EF-Tu delivers aminoacyl-tRNA to the A site of the ribosome, and it is inactivated by H2O2 via oxidation of a single Cys residue, Cys-82, with subsequent formation of both sulfenic acid and an intermolecular disulfide bond (Yutthanasirikul et al., 2016). Oxidized EF-G and EF-Tu can be reduced and reactivated by thioredoxin, a small redox protein that regulates the activity of target proteins by reducing disulfide bonds. This observation suggests that reducing power from photosynthetic electron transport might be transmitted to EF-G and EF-Tu via thioredoxin in vivo (Kojima et al., 2009; Nishiyama et al., 2011; Yutthanasirikul et al., 2016). Interactions of thioredoxin with EF-G and EF-Tu in Synechocystis were also suggested by results of studies using thioredoxin-affinity chromatography (Lindahl and Florencio, 2003) and similar results were obtained with spinach (Spinacia olerace) chloroplasts (Balmer et al., 2003). Expression in Synechocystis of mutated EF-G, in which Cys-105 had been replaced by a Ser residue, enhanced both protein synthesis and the repair of PSII under strong light, with the resultant alleviation of photoinhibition of PSII, confirming that the oxidation of EF-G might be a critical event that stimulates the photoinhibition of PSII (Ejima et al., 2012), However, the extent of the protective effect on photoinhibition was as little as 20%, and this modest effect suggested that not only EF-G but also some other factor(s), for example EF-Tu, might be a target of oxidation that stimulates the photoinhibition of PSII (Ejima et al., 2012).
In this study, we generated a transformant of Synechocystis that expressed a mutant form of EF-Tu wherein Cys-82, the target of ROS, was replaced by a Ser residue and we examined the effects of this mutation on protein synthesis and the photoinhibition of PSII in vivo. We found that the expression of mutated EF-Tu in Synechocystis enhanced the de novo synthesis of proteins and the repair of PSII under strong light, with the resultant alleviation of photoinhibition of PSII. However, the expression of mutated EF-Tu stimulated oxidative stress by accelerating the production of ROS under strong light. Thus, we report here the importance of the redox state of EF-Tu in the photoinhibition of PSII as well as in protection from oxidative stress under strong light.
RESULTS
Oxidation of Cys-82 of EF-Tu under Strong Light
Cys-82, a single Cys residue in EF-Tu of Synechocystis, is oxidized by H2O2 to form sulfenic acid and an intermolecular disulfide bond, and the oxidized forms of EF-Tu are reduced by thioredoxin in vitro (Yutthanasirikul et al., 2016). To capture the oxidized forms of EF-Tu in vivo, we harvested cells rapidly by filtration immediately after illumination with strong light and then lysed them with glass beads. We separated the resultant cell extracts into cytosol and thylakoid membrane fractions and subjected them to a thiol-modification assay with methoxypoly(ethyleneglycol) maleimide (PEG-maleimide; average molecular mass, 5 kD), which reacts with the thiol groups of Cys residues. We also detected EF-Tu immunologically using antibodies specific for EF-Tu after separation of proteins by nonreducing SDS-PAGE.
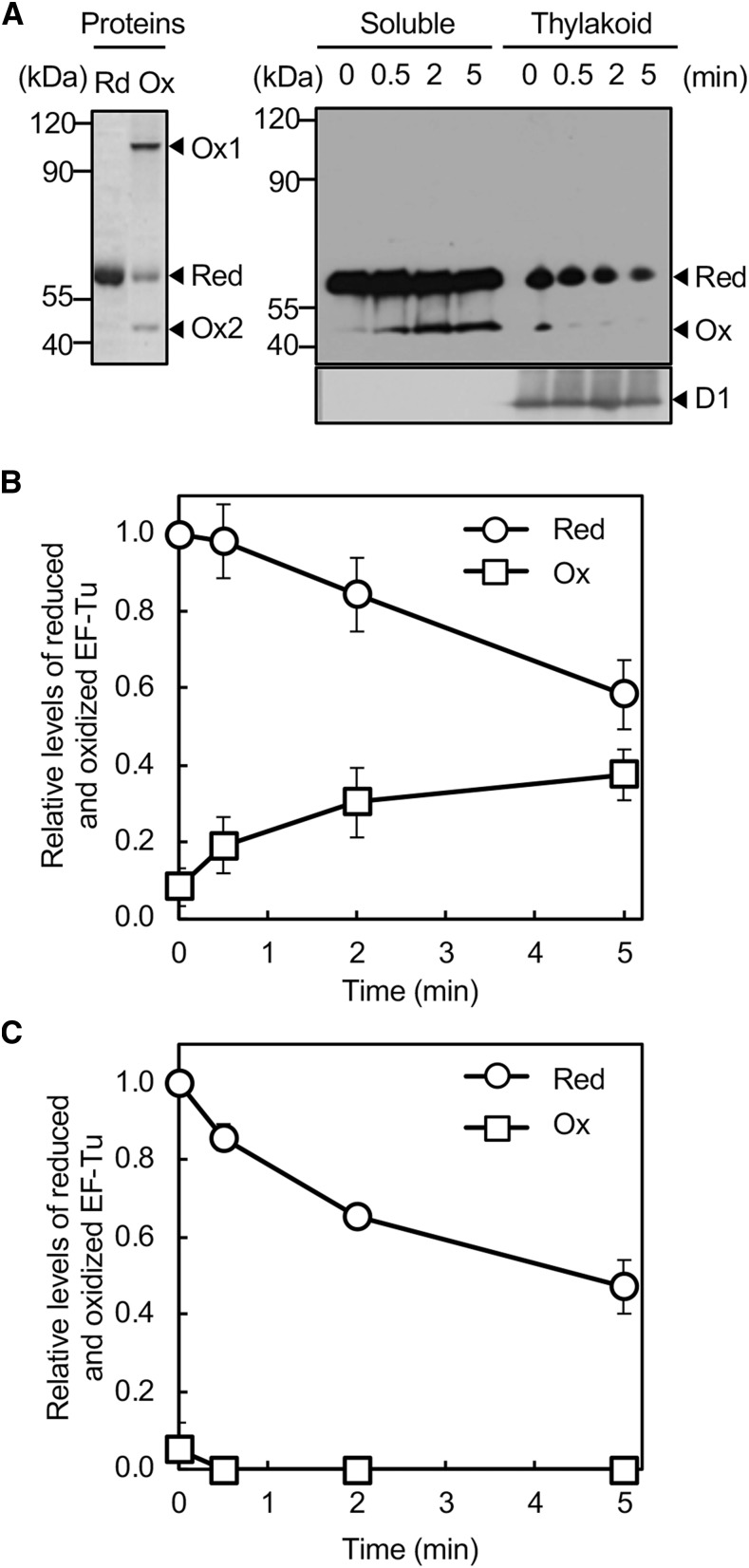
In cytosolic fractions, EF-Tu was detected in its reduced form on a nonreducing gel when we examined fractions from cells cultured under growth-light conditions (Fig. 1A). When cells were exposed to strong light at 1000 µmol photons m−2 s−1, an oxidized form of EF-Tu appeared within 30 s (Fig. 1A). The level of the oxidized form increased upon prolonged illumination, whereas the level of the reduced form decreased (Fig. 1B). A comparison with the molecular masses of recombinant His-tagged EF-Tu proteins that had been oxidized suggested that the oxidized form consisted of monomers that contained sulfenic acid (Ox2; Fig. 1A). Although oxidized dimers with an intermolecular disulfide bond appeared in vitro (Ox1; Fig. 1A), no oxidized dimers with an intermolecular disulfide bond were detected under the conditions examined. By contrast, in thylakoid membranes, EF-Tu was mostly present in the reduced form, with a corresponding minor amount in the oxidized form (Fig. 1C). The level of the reduced form decreased upon illumination with strong light and the oxidized form disappeared immediately. Because various thylakoid-localized proteins, such as the D1 protein, are synthesized on thylakoid membranes, to which the protein-synthetic machinery is bound in cyanobacteria (Nishiyama et al., 2004), it is likely that oxidized EF-Tu might not have been associated with ribosomes and, thereby, might not have been captured in thylakoid fractions.
Figure 1.
Oxidation of EF-Tu under strong light in Synechocystis. A, Wild-type cells were exposed to strong light at 1000 μmol photons m−2 s−1 without aeration. Cells were harvested by filtration and lysed with glass beads. Cell extracts were separated into soluble and thylakoid membrane fractions and subjected to the thiol-modification assay with PEG-maleimide. Proteins were separated by nonreducing SDS-PAGE and EF-Tu was detected immunologically. For comparison, His-tagged EF-Tu protein that had been treated with 5 mm DTT (Red) or 1 mm H2O2 (Ox) was subjected to the thiol-modification assay and then to nonreducing SDS-PAGE. The D1 protein in membrane fractions was also detected immunologically. B, Quantitation of relative levels of reduced and oxidized EF-Tu in soluble fractions. C, Quantitation of relative levels of reduced and oxidized EF-Tu in thylakoid membrane fractions. Levels of reduced EF-Tu at 0 time in each fraction were taken as 1.0. Values are means ± sd (bars) of results from three independent experiments. The absence of bars in all figures indicates that the sd falls within the symbol. Ox1, oxidized dimer; Ox2, oxidized monomer; Red, reduced form.
Expression of Mutated EF-Tu in Synechocystis
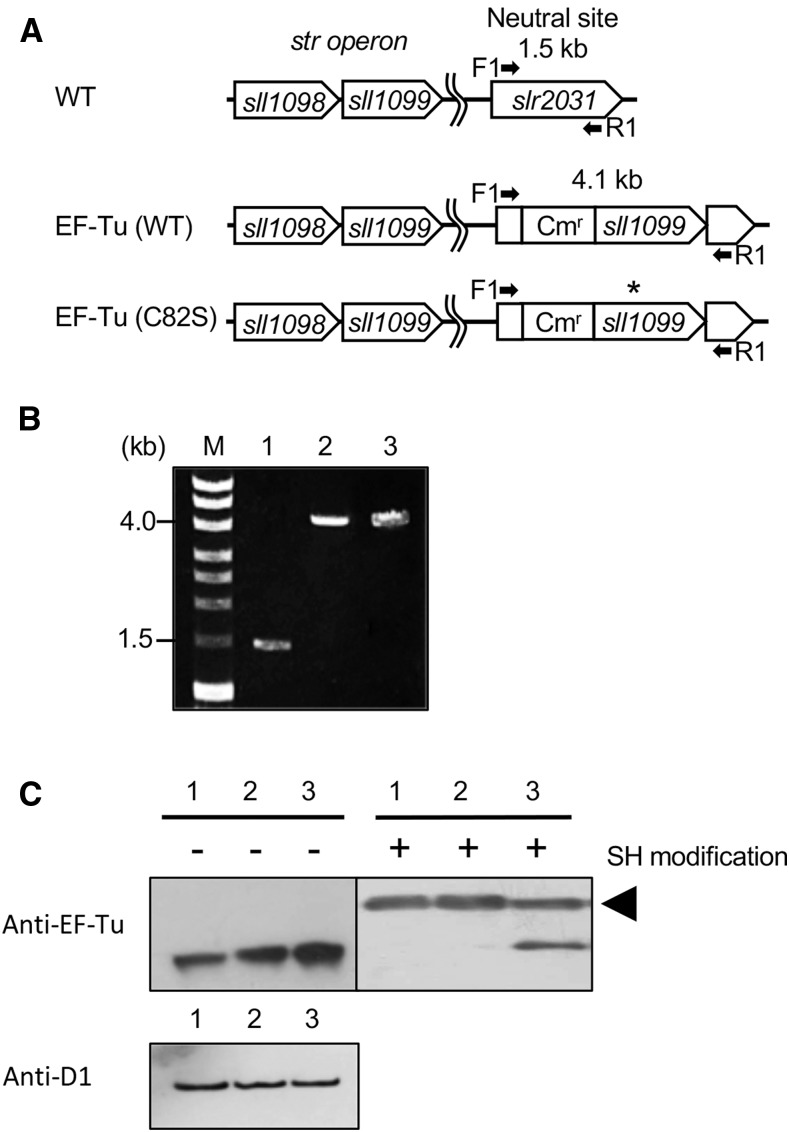
Replacement of Cys-82 by a Ser residue in the recombinant EF-Tu rendered EF-Tu resistant to oxidation by H2O2, without any adverse effects on translational activity in vitro (Yutthanasirikul et al., 2016). This observation in vitro led us to examine the effects of expression, in Synechocystis, of oxidation-insensitive EF-Tu on protein synthesis and on photosynthesis under strong light. We introduced a mutation into the sll1099 gene for EF-Tu, replacing Cys-82 by Ser-82, and inserted the mutated gene, together with the promoter region of the sll1098 gene, which consists of the str operon with the sll1099 gene, into a neutral site between slr2030 and slr2031 in the genome of Synechocystis by homologous recombination (Fig. 2A). Then we attempted to knock out the intrinsic sll1099 gene in the transformant to replace all Cys-82 with Ser-82 in EF-Tu. However, we were unable to obtain the desired transformant. Complete removal of Cys-82 from EF-Tu might be lethal, as discussed below. We also generated a control strain that expressed wild-type EF-Tu from the neutral site by inserting the sll1099 gene with the promoter of the str operon into the neutral site. The complete segregation of genomes was confirmed by PCR with the genomic DNA as template (Fig. 2B). The transformants that expressed mutated EF-Tu and wild-type EF-Tu were designated EF-Tu (C82S) and EF-Tu (wild type), respectively (Fig. 2A). We anticipated that transformants would express EF-Tu proteins from both the endogenous gene and the gene integrated at the neutral site.
Figure 2.
Expression of mutated EF-Tu in Synechocystis. A, Schematic representation of the incorporation of the wild-type sll1099 gene for EF-Tu and its mutated derivative at a neutral site between slr2030 and slr2031 in the genome of Synechocystis. The asterisk indicates the position of site-directed mutagenesis for replacement of Cys-82 by a Ser residue. Arrows indicate the positions and directions of primers (F1 and R1) for PCR. B, Confirmation by PCR of the complete incorporation of the mutated sll1099 gene at the neutral site in all the chromosomal copies of the genome. Genomes from wild-type (lane 1), EF-Tu (wild type; lane 2), and EF-Tu (C82S; lane 3) cells were extracted and used as templates for PCR. C, Expression of wild-type and mutated EF-Tu in Synechocystis cells. Proteins in cell extracts from wild-type (lane 1), EF-Tu (wild type; lane 2), and EF-Tu (C82S; lane 3) cells were separated by reducing SDS-PAGE and also by nonreducing SDS-PAGE after modification of thiol groups with PEG-maleimide, and then EF-Tu was detected immunologically. The arrowhead indicates thiol-modified EF-Tu. As a control, levels of the D1 protein were detected immunologically. Cmr, chloramphenicol-resistance gene cassette; M, molecular mass markers; SH, thiol groups; WT, wild type.
We examined the expression of EF-Tu at the protein level. Proteins in cell extracts were separated on a nonreducing gel, and EF-Tu was detected immunologically. Levels of EF-Tu in EF-Tu (C82S) and EF-Tu (wild type) cells were approximately twice those in wild-type cells (Fig. 2C). Levels of the D1 protein were the same among the three types of cells (Fig. 2C). To separate mutated EF-Tu from wild-type EF-Tu, we treated proteins in cell extracts with dithiothreitol (DTT) and then subjected proteins to the thiol-modification assay. In extracts of wild-type cells, EF-Tu appeared as a single band as a consequence of the modification of Cys-82. In extracts of EF-Tu (wild type) cells, EF-Tu appeared as a single band, as in wild-type cells, but its amount was doubled. In extracts of EF-Tu (C82S) cells, EF-Tu appeared as two bands of similar intensities, indicating that cells had expressed both wild-type and mutated EF-Tu at similar levels (Fig. 2C).
Expression of Mutated EF-Tu Enhances the De Novo Synthesis of Proteins under Strong Light
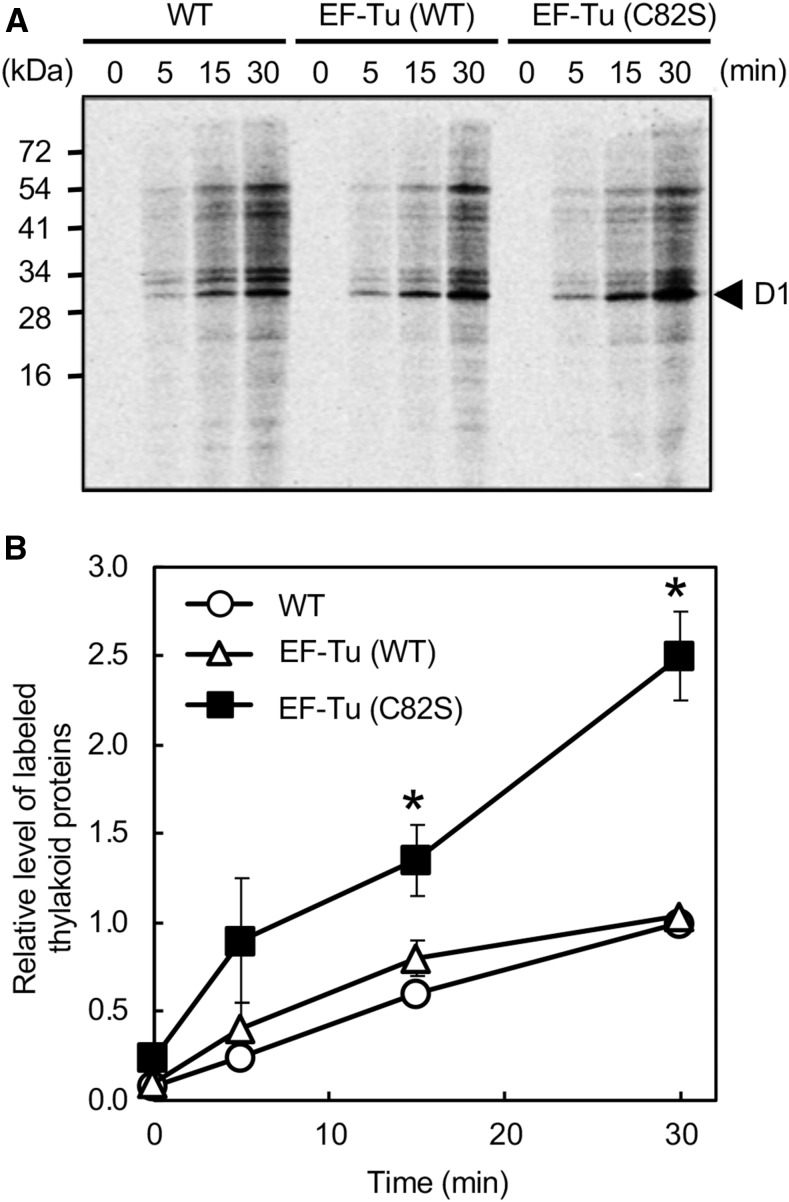
The de novo synthesis of the D1 protein plays a central role in the repair of damaged PSII (Nishiyama et al., 2011). To examine the effects of the expression of the mutated EF-Tu on the de novo synthesis of the D1 protein, we monitored the incorporation of 35S-labeled Met and Cys into proteins during the exposure of cells to strong light at 1000 μmol photons m−2 s−1. Figure 3A shows the patterns of pulse-labeled proteins from thylakoid membranes after SDS-PAGE and the time courses of the synthesis of the labeled proteins. Levels of newly synthesized D1 protein rose during exposure to strong light in all strains. The rate of synthesis of the D1 protein was similar in wild-type and EF-Tu (wild type) cells, indicating that the doubling of the amount of wild-type EF-Tu might not affect protein synthesis. By contrast, the rate of synthesis of D1 in EF-Tu (C82S) cells was approximately twice that in wild-type cells. Moreover, not only the synthesis of the D1 protein but also the synthesis of almost all proteins in thylakoid membranes was enhanced in EF-Tu (C82S) cells. Proteins on gels were also stained with Coomassie Brilliant Blue as loading controls, and the results confirmed that the basal composition of proteins did not differ among the three types of cells (Supplemental Fig. S1). The amounts of labeled proteins in thylakoid membranes were also monitored by liquid scintillation counting. The rate of de novo synthesis of proteins in EF-Tu (C82S) cells was 2.5 times higher than that in wild-type and EF-Tu (wild type) cells (Fig. 3B). These observations suggested that the expression of mutated EF-Tu might accelerate the synthesis of proteins globally under strong light.
Figure 3.
Synthesis de novo of proteins in thylakoid membranes under strong light. Proteins in wild-type and transformed cells were pulse-labeled by incubation of cells at 25°C, for the indicated times, under strong light at 1000 μmol photons m−2 s−1 with standard aeration, in the presence of 35S-labeled Met and Cys. Thylakoid membranes were isolated and proteins were separated by SDS-PAGE. A, representative radiogram of pulse-labeled proteins from thylakoid membranes. B, Quantitation of the relative levels of labeled thylakoid proteins in wild-type (open circles), EF-Tu (wild type; open triangles), and EF-Tu (C82S; closed squares) cells. Values are means ± sd (bars) of results from three independent experiments. The value for wild-type cells upon incubation for 30 min was taken as 1.0. Asterisks indicate statistically significant differences (P < 0.01; Student’s t test). WT, wild type.
Expression of Mutated EF-Tu Alleviates the Photoinhibition of PSII
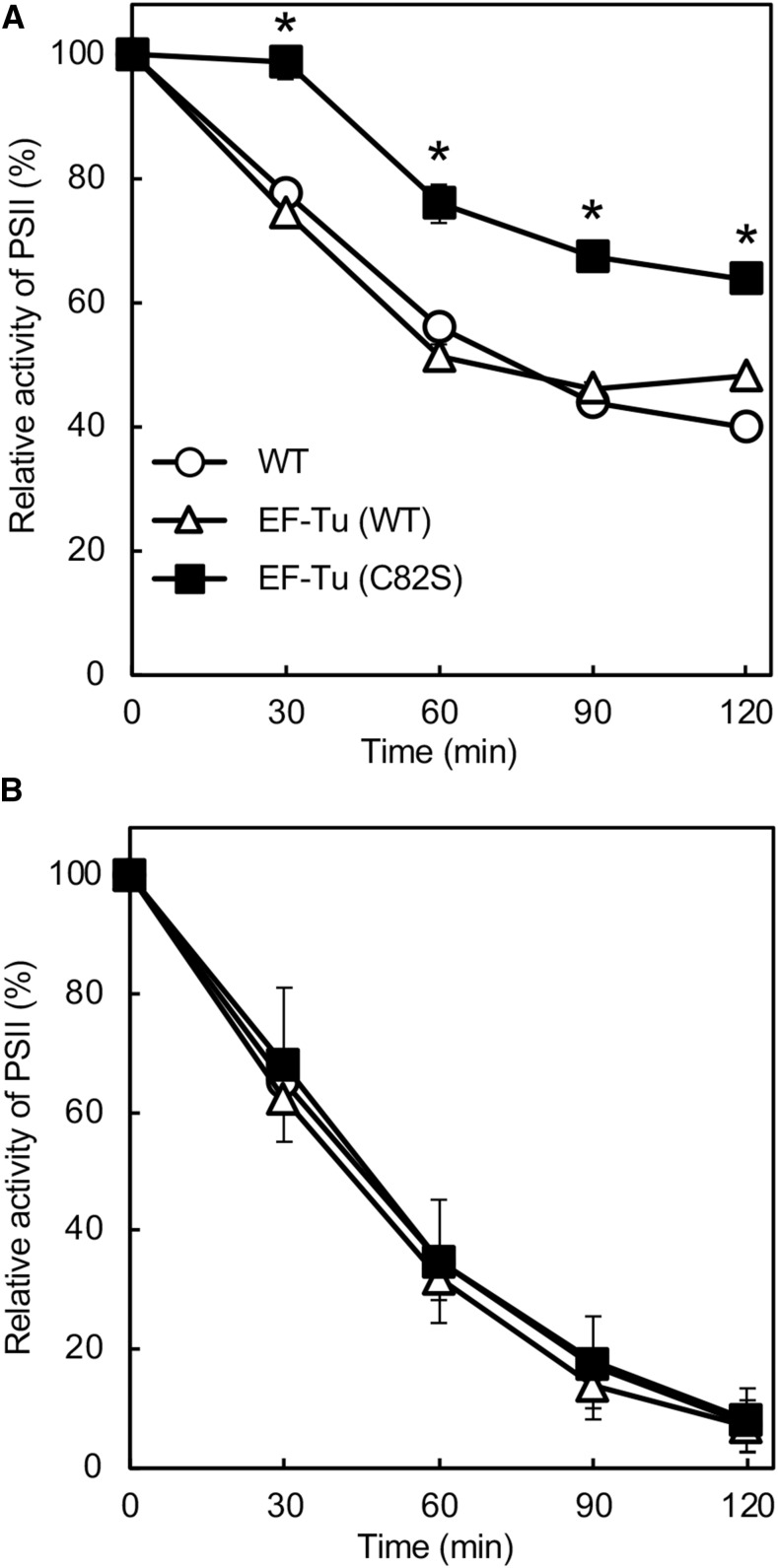
To investigate the effect of expression of mutated EF-Tu on the photoinhibition of PSII, we exposed Synechocystis cells to strong light at 1000 μmol photons m−2 s−1 at 25°C. The activity of PSII fell at similar rates in wild-type and EF-Tu (wild type) cells (Fig. 4A), indicating that doubling of the amount of wild-type EF-Tu might not affect the photoinhibition of PSII. By contrast, the activity of PSII in EF-Tu (C82S) cells remained much higher than that in wild-type cells under strong light. The activity of PSII dropped to 40% of the initial level within 120 min in wild-type cells, but it remained at 65% in EF-Tu (C82S) cells (Fig. 4A). These observations suggested that the presence of oxidation-insensitive EF-Tu might have alleviated the photoinhibition of PSII. However, when cells were exposed to strong light in the presence of lincomycin, which blocks the repair of PSII, the activity of PSII fell at the same rate in all three types of cell, indicating that the expression of mutated EF-Tu might enhance the repair of PSII without any effects on photodamage (Fig. 4B). It seems likely that the accelerated synthesis of the D1 protein in EF-Tu (C82S) cells might have enhanced the repair of PSII. To examine the neutrality of the neutral site that we used, we also generated a transformant in which the chloramphenicol resistance gene cassette had been inserted into the neutral site between slr2030 and slr2031. Neither photodamage to PSII nor repair of PSII was affected (Supplemental Fig. S2).
Figure 4.
Photoinhibition of PSII under strong light. Wild-type and transformed cells were incubated at 25°C under strong light at 1000 μmol photons m−2 s−1, with standard aeration, in the absence of lincomycin (A) and in its presence (B). The activity of PSII in wild-type, EF-Tu (wild type), and EF-Tu (C82S) cells was measured at 25°C in terms of the evolution of oxygen in the presence of 1 mm 1,4-benzoquinone and 1 mm K3Fe(CN)6. The activities taken as 100% for wild-type, EF-Tu (wild type), and EF-Tu (C82S) cells were 618 ± 35, 612 ± 26, and 593 ± 14 μmol O2 mg−1 Chl h−1, respectively. Values are means ± sd (bars) of results from three independent experiments. Asterisks indicate statistically significant differences (P < 0.01; Student’s t test). WT, wild type.
Expression of Mutated EF-Tu Stimulates the Production of ROS
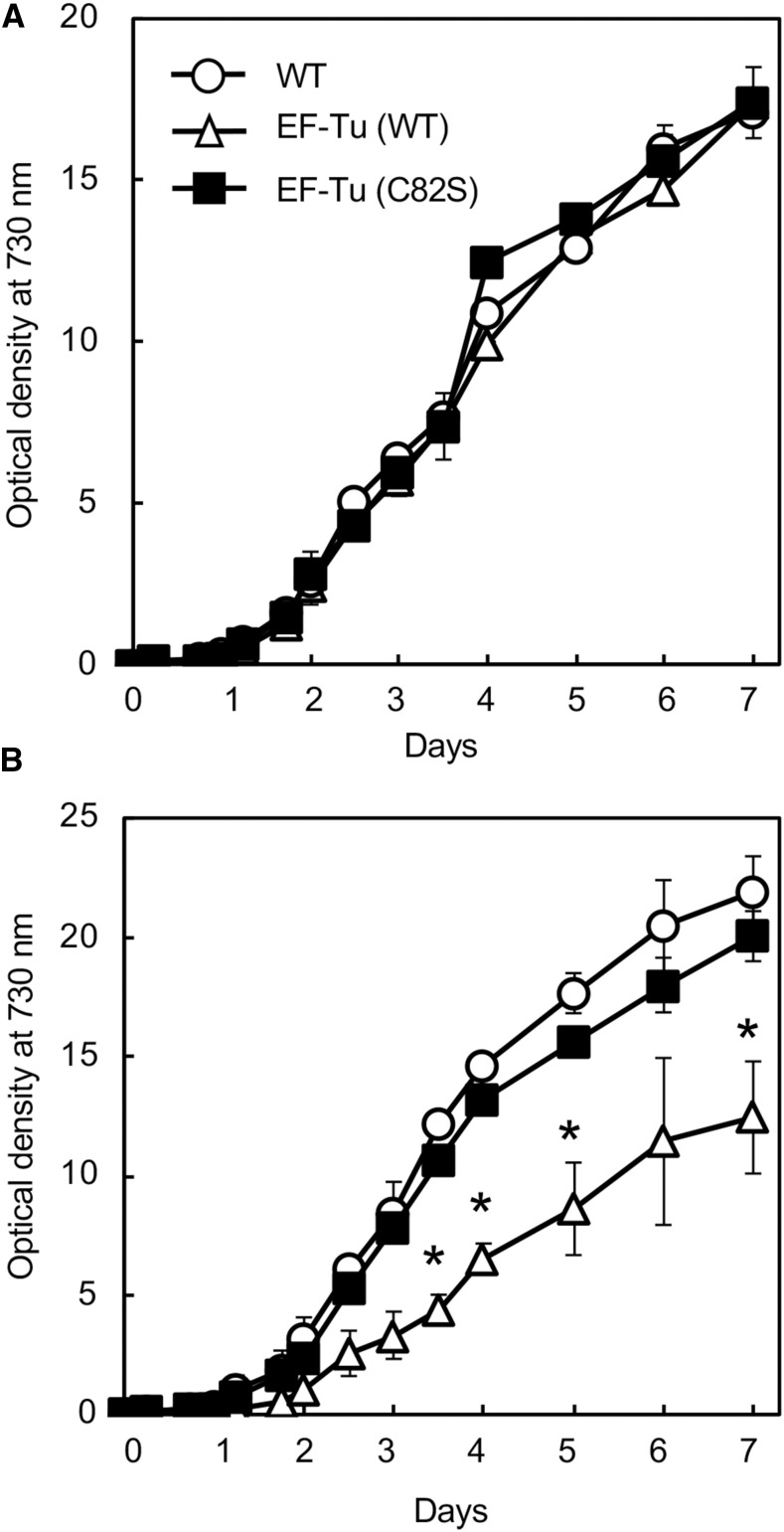
Photoinhibition of PSII is a limiting factor for growth in photosynthetic organisms (Powles, 1984; Liu and Last, 2017). To examine the effect of mutated EF-Tu on the growth of Synechocystis, we monitored photoautotrophic growth under weak light at 70 μmol photons m−2 s−1 and under strong light at 200 μmol photons m−2 s−1 at 32°C. The three types of cells grew at the same rate under weak light (Fig. 5A), suggesting that neither increased amounts of EF-Tu nor the presence of mutated EF-Tu affected growth under weak light. Under strong light, EF-Tu (C82S) cells grew at a rate similar to that of wild-type cells, whereas EF-Tu (wild type) cells grew more slowly than wild-type cells (Fig. 5B). Thus, the expression of mutated EF-Tu did not affect photoautotrophic growth under either weak or strong light. The reason for the hindered growth of EF-Tu (wild type) cells under strong light remains to be determined. However, it is possible that increased amounts of oxidized EF-Tu in EF-Tu (wild type) cells might have partially interfered with the effects of functional EF-Tu, acting as a competitive inhibitor.
Figure 5.
Photoautotrophic growth under weak and strong light. Wild-type, EF-Tu (wild type), and EF-Tu (C82S) cells were grown under weak light at 70 μmol photons m−2 s−1 (A) and strong light at 200 μmol photons m−2 s−1 (B). Cell growth was monitored as optical density at 730 nm. Values are means ± sd (bars) of results from three independent experiments. Asterisks indicate statistically significant differences (P < 0.01; Student’s t test). WT, wild type.
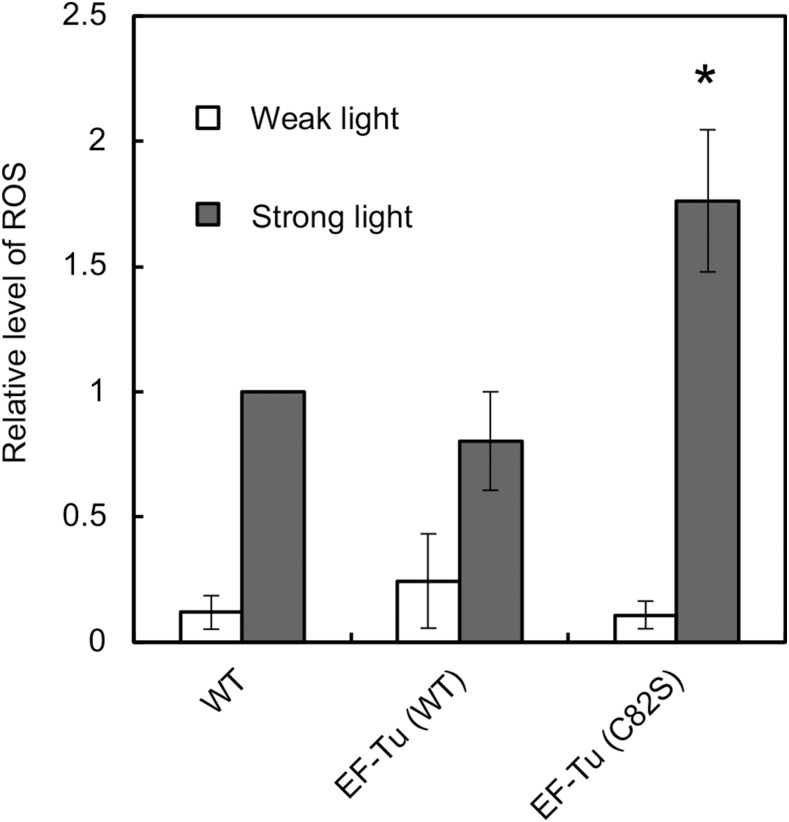
The absence of any effect of mutated EF-Tu on growth under strong light suggested that the presence of mutated EF-Tu might have had some adverse effects. We postulated that the expression of mutated EF-Tu might have stimulated oxidative stress, because the enhanced repair of PSII would maintain the activity of PSII and the subsequent transport of electrons would be more active under strong light, with resultant production of large amounts of ROS. We analyzed levels of ROS under strong light using 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA), a membrane-permeable indicator of ROS that reacts nonspecifically with ROS (Hakkila et al., 2014, Sae-Tang et al., 2016). Under weak light at 70 μmol photons m−2 s−1, the three types of cell produced similar amounts of ROS (Fig. 6). By contrast, under strong light at 1000 μmol photons m−2 s−1, EF-Tu (C82S) cells produced 1.7-fold more ROS than wild-type and EF-Tu (wild type) cells (Fig. 6). These results indicate that the expression of mutated EF-Tu increased the production of ROS under strong light. The resultant stimulation of oxidative stress might explain why cell growth was not altered, even though PSII became more tolerant to photoinhibition.
Figure 6.
Intracellular levels of ROS under strong light. Wild-type, EF-Tu (wild type), and EF-Tu (C82S) cells were exposed to weak light at 70 μmol photons m−2 s−1 (white bars) or strong light at 1000 μmol photons m−2 s−1 (gray bars) for 30 min in the presence of the fluorescent indicator carboxy-H2DCFDA. Fluorescence was normalized by reference to Chl fluorescence at 680 nm. The level of ROS in wild-type cells that had been exposed to light was taken as 1.0. Asterisk indicates statistically significant differences (P < 0.01; Student’s t test). WT, wild type.
DISCUSSION
Oxidation of EF-Tu under Strong Light
Recent studies in vitro revealed that EF-Tu of Synechocystis is inactivated via oxidation of Cys-82, a single Cys residue, with formation of sulfenic acid and an intermolecular disulfide bond (Yutthanasirikul et al., 2016). In this study, we found that Cys-82 of EF-Tu was oxidized under strong light in vivo in Synechocystis (Fig. 1). The oxidation of Cys-82 occurred immediately after exposure of cells to strong light, yielding an oxidized form of EF-Tu. The oxidized form is likely to be EF-Tu with sulfenic acid at Cys-82, as observed in vitro, in view of its apparent molecular mass. However, it is also possible that the oxidized form might bind a small biomolecule, such as glutathione, at Cys-82 via a disulfide bond.
Relatively minimal levels of oxidized EF-Tu were present in the cytosol under weak light, and reduced forms were more abundant than oxidized forms even under strong light (Fig. 1). These observations might reflect the incomplete fixation of oxidized forms in vivo in our assay. Oxidized forms of EF-Tu are immediately reduced by m-type thioredoxin in vitro (Yutthanasirikul et al., 2016). If oxidized forms were more abundantly generated under strong light, some might have been reduced by endogenous thioredoxins immediately after the light was turned off. The rapid harvesting of cells by filtration and their subsequent rapid disruption allowed us to capture oxidized forms of EF-Tu, but capture was necessarily incomplete. Therefore, we cannot exclude the possibility that oxidized dimers with an intermolecular disulfide bond might also have been formed, as observed in vitro, even though they were not detected under the conditions of this study.
Redox analysis in vivo also revealed that levels of reduced forms of EF-Tu that had been present in thylakoid membranes fell during exposure of cells to strong light (Fig. 1). Under strong light, ROS are produced in abundance from the photosynthetic machinery that is embedded in thylakoid membranes (Asada, 1999). In cyanobacteria, various proteins that are localized in thylakoid membranes, including the D1 protein, are initially synthesized in the cytosol. After nascent polypeptides reach a certain length, they are inserted into thylakoid membranes and elongation continues (Tyystjärvi et al., 2001). Thus, cotranslational insertion allows the polypeptide-ribosome complexes to be transiently attached on thylakoid membranes. It seems that EF-Tu that was associated with the polypeptide-ribosome complexes might have been captured in the thylakoid membrane fraction. In fact, thioredoxin-targeted proteomic analysis of thylakoid membranes from Synechocystis showed that EF-Tu was present together with several ribosomal proteins, such as S2, S3, and L3 (Mata-Cabana et al., 2007), indicating that polypeptide-ribosome complexes that include EF-Tu are bound to thylakoid membranes. Immunogold analysis in corn (Zea mays) revealed that EF-Tu was localized in both the stroma and the grana membranes of chloroplasts (Momcilovic and Ristic, 2004), suggesting that some EF-Tu proteins are associated with thylakoid membranes via polypeptide-ribosome complexes. Such polypeptide-ribosome complexes are likely to be susceptible to oxidation by ROS, and EF-Tu proteins that are approaching these complexes near thylakoid membranes might also be a target of oxidation. It seems that once EF-Tu is oxidized, it is no longer associated with polypeptide-ribosome complexes and remains dissociated from them until it has been rereduced by thioredoxin. Hence, oxidized EF-Tu appears to be present only in the cytosol under strong light (Fig. 1).
Oxidation of EF-Tu Inhibits the Repair of PSII
In this study, we demonstrated that expression of mutated EF-Tu, in which Cys-82 had been replaced by a Ser residue, enhanced the de novo synthesis of proteins, including the D1 protein, under strong light and activated the repair of PSII, resulting in alleviation of photoinhibition of PSII (Figs. 3 and 4). These observations indicate that oxidation of Cys-82 in EF-Tu under strong light might depress the de novo synthesis of proteins and inhibit the repair of PSII, resulting in the stimulation of photoinhibition. In previous studies, we observed that increases in intracellular levels of ROS, such as H2O2 and singlet oxygen, depressed the synthesis of the D1 protein at the elongation step of translation, resulting in inhibition of the repair of PSII in Synechocystis (Nishiyama et al., 2001, 2004). This study confirmed that it is translational elongation that is primarily inhibited by ROS and, in addition, it allowed us to identify EF-Tu as a critical target of ROS among the various macromolecules involved in translational elongation. EF-G was initially identified as a target of ROS within the translational machinery of Synechocystis (Kojima et al., 2007, 2009). Expression in Synechocystis of mutated EF-G with a target Cys residue replaced by Ser enhanced the de novo synthesis of proteins and protected the repair of PSII under strong light (Ejima et al., 2012). However, the protective effect of mutated EF-G was only approximately 20%. Compared to EF-G, mutated EF-Tu appears to be more effective in terms of the protection of PSII from photoinhibition; the protective effect of mutated EF-Tu was approximately 60% (Fig. 4A). Even though the sensitivity to ROS of EF-G and EF-Tu cannot be compared directly, it is likely that both elongation factors might be critical targets of ROS that are responsible for the inhibition of the repair of PSII under photooxidative stress.
Physiological Implications of the Role of Cys-82 in EF-Tu
The alleviation of photoinhibition of PSII did not affect cell growth under strong light. There was no significant difference between wild-type cells and transformed cells that expressed mutated EF-Tu (Fig. 5). In the transformant, ROS were produced in greater abundance than in wild-type cells under strong light (Fig. 6). The elevated oxidative stress appeared to offset any advantage due to alleviated photoinhibition. What might be responsible for such elevated oxidative stress? The presence of mutated EF-Tu might facilitate the rapid synthesis of proteins and the efficient repair of PSII under strong light, which would maintain PSII in an active state. The consequent activation of electron transport would produce excess ROS on the acceptor side of photosystem I under strong light, stimulating oxidative stress. In this context, the oxidation of Cys-82 in EF-Tu might play a role in suppressing the repair of PSII to avoid further oxidative stress. In this study, we were unable to replace all Cys-82 in EF-Tu by inactivating the intrinsic gene for EF-Tu in the transformed cells that expressed mutated EF-Tu. If all Cys-82 residues were replaced, it is likely that the activation of PSII would no longer be regulated under strong light. It is even possible that Cys-82 in EF-Tu might act as a regulator to suppress the photosynthetic machinery under strong light or fluctuating light. Cys-82 is strongly conserved in EF-Tu from cyanobacteria to higher plants (De Laurentiis et al., 2011). As suggested by Sonoike and colleagues, it seems that inactivation of PSII is necessary under strong light to protect PSI from photoinhibition, which cannot be efficiently reversed (Sonoike, 1996, 2011). Stimulation of the photoinhibition of PSII via the oxidation of EF-Tu appears to be a plausible strategy that allows Synechocystis to survive under changes in ambient light conditions and, in particular, under strong light.
CONCLUSION
Translation factor EF-Tu of Synechocystis includes a single Cys residue at position 82 that is strongly conserved among photosynthetic organisms. Under strong light, Cys-82 in EF-Tu is rapidly oxidized to yield oxidized monomers, and the presence of these oxidized monomers depresses the de novo synthesis of proteins, such as the D1 protein. As a result, the repair of PSII is inhibited and photoinhibition of PSII is stimulated. The sensitivity of EF-Tu to oxidation might act as a regulator for transient depression of the synthesis of proteins required for the repair of PSII to prevent excessive oxidative stress that would be caused by photosynthesis under strong light.
MATERIALS AND METHODS
Strains and Culture Conditions
Wild-type and transformed cells of Synechocystis sp. PCC 6803 were grown photoautotrophically at 32°C in liquid BG-11 medium under light at 70 μmol photons m−2 s−1 and aeration by sterile air that contained 1% (v/v) CO2 (Kusama et al., 2015). Cultures with an optical density at 730 nm of 1.0 ± 0.1, in which the concentration of chlorophyll a (Chl a) was approximately 4.0 μg mL−1, were used for assays.
Detection of the Redox State of EF-Tu In Vivo
Cells in culture at an optical density at 730 nm of 1.0 ± 0.1 were exposed to light at 1000 μmol photons m−2 s−1 at 25°C. Aliquots of 10 mL each were withdrawn at designated times for analysis. Cells were collected rapidly by vacuum filtration on a membrane filter with a pore size of 1.0 μm (Omnipore; Merck Millipore). Redox reactions were terminated in vivo by addition of 30% (v/v) methanol to the membrane filter and harvested cells were suspended in 30% methanol. Cells were lysed with glass beads, and the soluble and membrane fractions were separated by centrifugation, as described in Nishiyama et al. (2004). To separate oxidized EF-Tu from reduced EF-Tu, all Cys residues were subjected to the modification of thiol groups with 2 mm PEG-maleimide reagent, which has an average molecular mass of 5 kD (Nihon Yushi). Proteins (5 μg in soluble fractions) and those that corresponded to 0.5 μg Chl in membrane fractions of cells were separated by nonreducing SDS-PAGE on a 12.5% polyacrylamide gel, and EF-Tu was detected immunologically with specific antibodies against EF-Tu, as described in Yutthanasirikul et al. (2016). For comparison, His-tagged EF-Tu that had been treated with 5 mm DTT or 1 mm H2O2 was subjected to the thiol-modification assay and then to nonreducing SDS-PAGE, as described in Yutthanasirikul et al. (2016). The D1 protein in membrane fractions of cells was also detected immunologically with specific antibodies against the D1 protein of Synechocystis (provided by Dr. Hajime Wada, the University of Tokyo).
Site-Directed Mutagenesis
The sll1099 gene that encodes EF-Tu, together with 36 bp of the sd region, was amplified from the genomic DNA of Synechocystis with the forward and reverse primers 5′-GGATCCCAATCTGTGTAGAAACAATCA-3′ and 5′-CATATGGGCTGGCCCAGTCAGA-3′, respectively. To obtain the promoter region from the str operon, which consists of the sll1098 and sll1099 genes (Krásný et al., 2000), a DNA region of 156 bp upstream of the sll1098 gene was amplified with the forward and reverse primers 5′-GACGTCGGCAAGGCAACATTGATAC-3′ and 5′-GGATCCAAAACGGGAGGCCATT-3′, respectively. The amplified DNA fragments were cloned into the pGEM-T vector (Promega). The plasmid DNA containing the promoter region was digested with NdeI and BamHI, and the resultant DNA fragment was inserted between the NdeI and BamHI sites of the pGEM-T easy vector that harbored the sll1099 gene for EF-Tu. To replace Cys-82 with a Ser residue in EF-Tu, site-directed mutagenesis was performed with a KOD-Plus Mutagenesis Kit (Toyobo) by PCR with the forward and reverse primers 5′-CTCCTGGTCACGCTGACTATG-3′ and 5′-AGTCTACGTGGGCATAGTGACG-3′, respectively. The plasmid DNA with the appropriate mutation in sll1099 was digested with NdeI and AatII. The DNA fragment that contained the mutated sll1099 gene and the promoter region was inserted between the NdeI and AatII sites of the pTCP2031 vector, which had been designed to incorporate a gene of interest at a neutral site flanked by slr2030 and slr2031 in the genome of Synechocystis (Yamauchi et al., 2011). This site is originally disrupted in Glc-tolerant Synechocystis sp. PCC 6803 (Kanesaki et al., 2012). The resultant plasmid DNA was used to transform wild-type cells of Synechocystis by homologous recombination. For the insertional control, we generated a mutant in which the chloramphenicol resistance gene cassette had been inserted into the neutral site.
Expression of Mutated EF-Tu
The complete incorporation of the mutated sll1099 gene at the neutral site in all the chromosomal copies of the genome was examined by PCR with the forward and reverse primers 5′-CCGAGTTGTAGTCGGCAGTC-3′ and 5′-CCGCAGAACGACCAATGTC-3′, respectively. The appropriate site-directed mutagenesis of the genome was confirmed by sequencing the amplified DNA fragment. For examination of the expression of wild-type and mutated EF-Tu, cell extracts were treated with PEG-maleimide, fractionated by reducing SDS-PAGE on a 12.5% polyacrylamide gel, and subjected to immunological detection of EF-Tu with EF-Tu-specific antibodies.
Labeling of Proteins In Vivo
For pulse labeling of proteins, 25 mL of cell culture were supplemented with 240 kBq mL 35S-labeled Met and Cys (Easy Tag EXPRE35S; PerkinElmer) and incubated at 25°C under light at 1000 μmol photons m−2 s−1, as described previously in Kusama et al. (2015). Aliquots of 7 mL each were withdrawn at designated times for analysis of proteins. Labeling was terminated by the addition of nonradioactive Met and Cys to a final concentration of 2 mm each, with immediate cooling of samples on ice. Thylakoid membranes were isolated from cells and then membrane proteins were separated by SDS-PAGE on a 12.5% polyacrylamide gel that contained 6 m urea, as described in Nishiyama et al. (2004). Labeled proteins on the gel were visualized with an imaging analyzer (FLA-7000; Fujifilm) and levels of the D1 protein were determined densitometrically, as described in Nishiyama et al. (2004). Levels of labeled proteins in thylakoid membranes were also quantitated by liquid scintillation counting, as described in Kojima et al. (2007).
Analysis of the Photoinhibition of PSII
Cells in culture at an optical density at 730 nm of 1.0 ± 0.1 were exposed to light at 1000 μmol photons m−2 s−1 at 25°C for designated times to induce the photoinhibition of PSII, as described in Kusama et al. (2015). For assays of photodamage, lincomycin at a final concentration of 200 μg mL−1 was added to the suspension of cells just before the onset of illumination. The activity of PSII was measured at 25°C in terms of the evolution of oxygen in the presence of 1 mm 1,4-benzoquinone and 1 mm K3Fe(CN)6 with a Clark-type oxygen electrode (Hansatech Instruments). The concentration of Chl was determined by measuring A663 in 80% acetone.
Detection of Intracellular Levels of ROS
To detect the production of ROS under strong light, carboxy-H2DCFDA, a membrane-permeable fluorescent indicator, was used as described in Sae-Tang et al. (2016). Cells in aliquots of 1 mL were incubated with 15 μm carboxy-H2DCFDA (Thermo Fisher Scientific) in darkness for 1 h for activation of the reagent by intracellular esterase. After the cells had been washed with fresh BG11 medium, cells in suspension were exposed to strong light at 1000 μmol photons m−2 s−1 or to weak light at 70 μmol photons m−2 s−1 at 32°C for 30 min. The fluorescence emitted from the reaction product upon excitation at 485 nm was detected at 520 nm with a FLUOstar OPTIMA system (BMG Labtech). Fluorescence from Chl a at 680 nm was used for normalization of all data.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effects of the expression of EF-Tu (C82S) on the relative levels of proteins in thylakoid membranes in Synechocystis.
Supplemental Figure S2. Neutrality of the neutral site that was used in this study.
Acknowledgments
The authors thank Mr. Tomohisa Niimi (Saitama University) for the radioisotopic analysis of proteins and Dr. Hajime Wada (University of Tokyo) for providing D1-specific antibodies.
References
- Aro EM, McCaffery S, Anderson JM (1993) Photoinhibition and D1 protein degradation in peas acclimated to different growth irradiances. Plant Physiol 103: 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schürmann P, Buchanan BB (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 100: 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurentiis EI, Mo F, Wieden HJ (2011) Construction of a fully active Cys-less elongation factor Tu: functional role of conserved cysteine 81. Biochim Biophys Acta 1814: 684–692 [DOI] [PubMed] [Google Scholar]
- Ejima K, Kawaharada T, Inoue S, Kojima K, Nishiyama Y (2012) A change in the sensitivity of elongation factor G to oxidation protects photosystem II from photoinhibition in Synechocystis sp. PCC 6803. FEBS Lett 586: 778–783 [DOI] [PubMed] [Google Scholar]
- Hakkila K, Antal T, Rehman AU, Kurkela J, Wada H, Vass I, Tyystjärvi E, Tyystjärvi T (2014) Oxidative stress and photoinhibition can be separated in the cyanobacterium Synechocystis sp. PCC 6803. Biochim Biophys Acta 1837: 217–225 [DOI] [PubMed] [Google Scholar]
- Kanesaki Y, Shiwa Y, Tajima N, Suzuki M, Watanabe S, Sato N, Ikeuchi M, Yoshikawa H (2012) Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res 19: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Motohashi K, Morota T, Oshita M, Hisabori T, Hayashi H, Nishiyama Y (2009) Regulation of translation by the redox state of elongation factor G in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 284: 18685–18691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Oshita M, Nanjo Y, Kasai K, Tozawa Y, Hayashi H, Nishiyama Y (2007) Oxidation of elongation factor G inhibits the synthesis of the D1 protein of photosystem II. Mol Microbiol 65: 936–947 [DOI] [PubMed] [Google Scholar]
- Krásný L, Vacík T, Fucík V, Jonák J (2000) Cloning and characterization of the str operon and elongation factor Tu expression in Bacillus stearothermophilus. J Bacteriol 182: 6114–6122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusama Y, Inoue S, Jimbo H, Takaichi S, Sonoike K, Hihara Y, Nishiyama Y (2015) Zeaxanthin and echinenone protect the repair of photosystem II from inhibition by singlet oxygen in Synechocystis sp. PCC 6803. Plant Cell Physiol 56: 906–916 [DOI] [PubMed] [Google Scholar]
- Lindahl M, Florencio FJ (2003) Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc Natl Acad Sci USA 100: 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Last RL (2017) A chloroplast thylakoid lumen protein is required for proper photosynthetic acclimation of plants under fluctuating light environments. Proc Natl Acad Sci USA 114: E8110–E8117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Cabana A, Florencio FJ, Lindahl M (2007) Membrane proteins from the cyanobacterium Synechocystis sp. PCC 6803 interacting with thioredoxin. Proteomics 7: 3953–3963 [DOI] [PubMed] [Google Scholar]
- Momcilovic I, Ristic Z (2004) Localization and abundance of chloroplast protein synthesis elongation factor (EF-Tu) and heat stability of chloroplast stromal proteins in maize. Plant Sci 166: 81–88 [Google Scholar]
- Moon BY, Higashi S, Gombos Z, Murata N (1995) Unsaturation of the membrane lipids of chloroplasts stabilizes the photosynthetic machinery against low-temperature photoinhibition in transgenic tobacco plants. Proc Natl Acad Sci USA 92: 6219–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N, Nishiyama Y (2018) ATP is a driving force in the repair of photosystem II during photoinhibition. Plant Cell Environ 41: 285–299 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Allakhverdiev SI, Murata N (2006) A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim Biophys Acta 1757: 742–749 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Allakhverdiev SI, Murata N (2011) Protein synthesis is the primary target of reactive oxygen species in the photoinhibition of photosystem II. Physiol Plant 142: 35–46 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Allakhverdiev SI, Yamamoto H, Hayashi H, Murata N (2004) Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemistry 43: 11321–11330 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N (2001) Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J 20: 5587–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles SB. (1984) Photoinhibition of photosynthesis induced by visible light. Annu Rev Plant Physiol 35: 15–44 [Google Scholar]
- Sae-Tang P, Hihara Y, Yumoto I, Orikasa Y, Okuyama H, Nishiyama Y (2016) Overexpressed superoxide dismutase and catalase act synergistically to protect the repair of PSII during photoinhibition in Synechococcus elongatus PCC 7942. Plant Cell Physiol 57: 1899–1907 [DOI] [PubMed] [Google Scholar]
- Sonoike K. (2011) Photoinhibition of photosystem I. Physiol Plant 142: 56–64 [DOI] [PubMed] [Google Scholar]
- Sonoike K. (1996) Photoinhibition of photosystem I: its physiological significance in the chilling sensitivity of plants. Plant Cell Physiol 37: 239–247 [Google Scholar]
- Tyystjärvi E, Aro EM (1996) The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc Natl Acad Sci USA 93: 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjärvi T, Herranen M, Aro EM (2001) Regulation of translation elongation in cyanobacteria: membrane targeting of the ribosome nascent-chain complexes controls the synthesis of D1 protein. Mol Microbiol 40: 476–484 [DOI] [PubMed] [Google Scholar]
- Wada H, Gombos Z, Murata N (1994) Contribution of membrane lipids to the ability of the photosynthetic machinery to tolerate temperature stress. Proc Natl Acad Sci USA 91: 4273–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Kaniya Y, Kaneko Y, Hihara Y (2011) Physiological roles of the cyAbrB transcriptional regulator pair Sll0822 and Sll0359 in Synechocystis sp. strain PCC 6803. J Bacteriol 193: 3702–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutthanasirikul R, Nagano T, Jimbo H, Hihara Y, Kanamori T, Ueda T, Haruyama T, Konno H, Yoshida K, Hisabori T, Nishiyama Y (2016) Oxidation of a cysteine residue in elongation factor EF-Tu reversibly inhibits translation in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 291: 5860–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]