Seasonal wood transcriptomes of Norway spruce suggest mechanisms underlying specific aspects of gymnosperm wood biology, such as latewood formation, lignin biosynthesis, and tracheid longevity.
Abstract
Seasonal cues influence several aspects of the secondary growth of tree stems, including cambial activity, wood chemistry, and transition to latewood formation. We investigated seasonal changes in cambial activity, secondary cell wall formation, and tracheid cell death in woody tissues of Norway spruce (Picea abies) throughout one seasonal cycle. RNA sequencing was performed simultaneously in both the xylem and cambium/phloem tissues of the stem. Principal component analysis revealed gradual shifts in the transcriptomes that followed a chronological order throughout the season. A notable remodeling of the transcriptome was observed in the winter, with many genes having maximal expression during the coldest months of the year. A highly coexpressed set of monolignol biosynthesis genes showed high expression during the period of secondary cell wall formation as well as a second peak in midwinter. This midwinter peak in expression did not trigger lignin deposition, as determined by pyrolysis-gas chromatography/mass spectrometry. Coexpression consensus network analyses suggested the involvement of transcription factors belonging to the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES and MYELOBLASTOSIS-HOMEOBOX families in the seasonal control of secondary cell wall formation of tracheids. Interestingly, the lifetime of the latewood tracheids stretched beyond the winter dormancy period, correlating with a lack of cell death-related gene expression. Our transcriptomic analyses combined with phylogenetic and microscopic analyses also identified the cellulose and lignin biosynthetic genes and putative regulators for latewood formation and tracheid cell death in Norway spruce, providing a toolbox for further physiological and functional assays of these important phase transitions.
Conifers are gymnosperm trees or bushes that dominate vast areas in terrestrial ecosystems, especially in the northern hemisphere, where they are widely utilized for industrial purposes. Traditionally, conifers have been used as a source of lignocellulose for the production of biomaterials and pulp, but they recently emerged as a major source of renewable raw material for the production of liquid biofuels and other value-added products (Ragauskas et al., 2014; Isikgor and Becer, 2015). Conifers represent an evolutionarily primitive branch of vascular plants and, therefore, also are an attractive target for comparative evo-devo studies of functional and genetic diversification within higher plants.
The most striking differences between the woody tissues of conifer and angiosperm tree species lie in the anatomy and chemistry of the water-conducting xylem tissues. Conifer wood consists mainly of tracheids that operate in both water transport and physical support. In the angiosperm lineage, these functions are divided between two different cell types: the vessel elements that are responsible for water conduction and the libriform fibers that give physical support to the stem. The differentiation of conifer tracheids is a relatively slow process; for instance, Norway spruce (Picea abies) latewood tracheids have been reported to die 2 to 3 months after their formation (Dieset, 2011). Therefore, the rate of conifer tracheid differentiation is more similar to the slow differentiation rate of the angiosperm fibers than to that of angiosperm vessel elements, which differentiate and die within a few days (Courtois-Moreau et al., 2009). This is not surprising, considering the common ontogeny of conifer tracheids and angiosperm fibers (Carlquist, 2001). Vessel elements appeared only after the split between the gymnosperms and the angiosperms and were shaped by the requirement for rapid and efficient water transport capacity in these elements, which subsequently became the only bulk water-transporting elements of the angiosperms (Sperry et al., 2006). In terms of the lignin composition of cell walls, conifer tracheids are more similar to angiosperm vessel elements in that they are composed mostly of guaiacyl-type lignin, whereas syringyl-enriched lignin (S-type lignin) is deposited in the cell walls of angiosperm fibers (Li et al., 2001). Therefore, it seems that guaiacyl-type lignin is preferred in water transport cell types, although a full understanding of these aspects requires further studies. For instance, several taxa that are more primitive than the conifers as well as a few conifer species (yew plum pine [Podocarpus macrophyllus] and sandarac cypress [Tetraclinis articulata]) contain S-type lignin (Weng and Chapple, 2010); therefore, the capability of the conifers to synthesize S-type lignin remains elusive.
Another interesting aspect of xylem differentiation in forest trees is variation in cambial activity throughout the seasonal cycle of tree growth. In order to ensure survival and productivity, perennials in temperate climates utilize cyclical environmental signals, such as daylength and seasonal temperature patterns. The influence of seasonality can be observed readily in the structure of wood as annual growth rings, whereby xylem tissue forms earlywood early in the growing season and latewood later in the season. Latewood tracheids have smaller lumens, smaller radial diameters, and thicker cell walls than earlywood cells (Zimmermann and Brown, 1971). In addition, latewood cells typically have a smaller cellulose microfibril angle, providing mechanical stiffness to the wood (Anagnost et al., 2002). Even though we now know a lot about the seasonal aspects of conifer tracheid differentiation and maturation, the genetic control of these processes is not known. A better understanding of gene expression profiles throughout the growing season would increase our understanding of these seasonal processes, which is important given their substantial impact on wood properties and the subsequent utilization of wood.
Until recently, genomic analyses of conifers were hindered by the large genome sizes and high heterozygosity of coniferous species. For example, Norway spruce has a genome size of 20 Gb (Nystedt et al., 2013), which is 7 times the size of the human genome. Therefore, candidate gene approaches have frequently been undertaken to correlate gene expression to processes such as wood formation (Kumar et al., 2009), natural variation (Palle et al., 2011), and association with woody traits (Beaulieu et al., 2011; Palle et al., 2013) in various conifer species. It was only recently that genome-wide analysis of gene expression became possible in conifers, first using EST sequencing (Kirst et al., 2003; Ralph et al., 2008) and later whole-genome sequencing and RNA sequencing (RNA-Seq; Qiu et al., 2013; Cronn et al., 2017; Jokipii-Lukkari et al., 2017). Today, draft genome assemblies are available for four conifer species: Norway spruce (Nystedt et al., 2013), white spruce (Picea glauca; Birol et al., 2013), loblolly pine (Pinus taeda; Neale et al., 2014), and sugar pine (Pinus lambertiana; Stevens et al., 2016). Therefore, we have an unprecedented opportunity to explore the unique processes of conifer biology utilizing genome-wide gene expression analyses.
In this work, we present data on the seasonal aspects of tracheid differentiation and maturation using comprehensive RNA-Seq combined with anatomical and chemical analyses of Norway spruce xylem tissues. We identified the suite of Norway spruce gene families involved in monolignol biosynthesis and polymerization as well as tracheid cell death and identified potential novel regulators of these processes. We also show evidence of the viability of tracheids throughout the winter period and propose some of the underlying mechanisms associated with the extended lifespan of these cells.
RESULTS
The Seasonal Cycle of Wood Formation
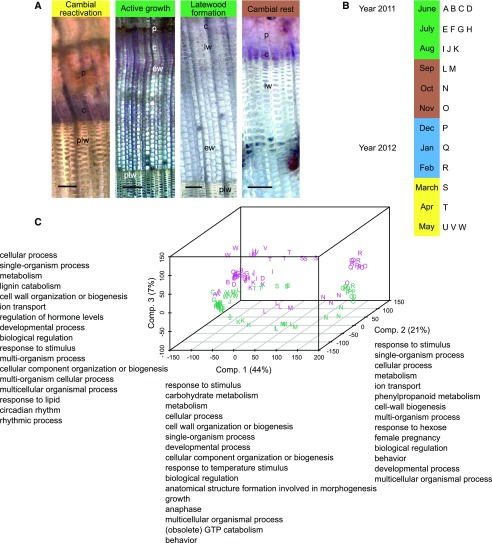
Cambial activity and wood formation of forest trees are strictly controlled by seasonal variation in temperature and daylength. In Norway spruce, the daily sum of temperature is the most important cue for cambial reactivation in the beginning of the growth season (Sarvas, 1969), whereas daylength is more important in influencing cambial dormancy at the end of the summer and autumn (Heide, 1974). To study the underlying molecular mechanisms, we collected stem samples from a clonal collection of a single Norway spruce genotype grown in northern Sweden throughout 1 year for RNA-Seq and simultaneously characterized cambial activity and wood properties in adjacent stem samples. Anatomical inspection revealed that cambial activity resumed after winter dormancy in mid-May, the formation of thick-walled latewood cells could be observed on July 11, and cambial activity declined from August 8 onward (Fig. 1A; Supplemental Table S1).
Figure 1.
Seasonal variation in cambial activity and wood formation on the anatomical and RNA-Seq levels during one year in Hissjö, Sweden. A, Light microscopy sections from four different sampling dates representing different stages of cambial activity. Viability of the cells was assessed by nitroblue tetrazolium (NBT) staining. Images were taken (from left to right) on May 11, 2012; June 13, 2011; July 11, 2011; and September 6, 2011. c, Cambium; ew, earlywood; lw, latewood; p, phloem; plw, latewood of the previous growing season. Bars = 100 µm (first, third, and fourth images) and 200 µm (second image). B, Scheme of sample collection. Green rectangle, Summer months June, July, and August; brown rectangle, autumn months September, October, and November; blue rectangle, winter months December, January, and February; and yellow rectangle, spring months March, April, and May. C, PCA plot of the sequencing reads showing tissue type separation, the clustering of biological replicates, and the effect of time in a three-dimensional space. Different sampling points are represented by letters from A to W. Cambium/phloem and xylem samples are marked with green and magenta, respectively. A semantic summary (less than log10 P < −1.3) of GO term enrichment for the genes with highest loadings is given next to each component (Comp.).
The Woody Tissues of Norway Spruce Express a Large Proportion of All Known Spruce Genes
For seasonal gene expression analysis, we collected stem samples that were divided into two fractions: a xylem sample, which consisted of the living tracheids, and a cambium/phloem sample, which consisted of the main part of the cambium as well as the functional phloem. Altogether, the transcriptomes of 127 xylem and cambium/phloem samples from 23 time points (Fig. 1B) were sequenced successfully (average number of biological replicates per time point = 2.95 [cambium/phloem] and 2.83 [xylem]). The vast majority of RNA-Seq reads were aligned uniquely to a gene model (Supplemental Fig. S1). Out of the 70,736 annotated gene models (Nystedt et al., 2013), 51.1% were represented by transcripts. We detected fewer expressed genes in the winter compared with the summer, which resulted in a higher average gene expression in the winter compared with the summer (Supplemental Fig. S2). We accounted for this in the relevant analyses by requiring a fold change above the level of this systematic effect.
A principal component analysis (PCA) plot of the whole data set revealed a sequential order of the different samples, which corresponded to the chronological order of sampling. The last sampling time of May 2012 clustered close to the first sampling point in June 2011, highlighting the cyclic nature of seasonal gene expression patterns (Fig. 1C). Replicate samples clustered together, and there was a clear separation between the samples derived from the xylem and the cambium/phloem (Fig. 1C). In total, 3.1% of the expressed genes were xylem specific (i.e. aligned reads were derived only from xylem samples), whereas 1.6% of the genes were expressed specifically in the cambium/phloem samples (Supplemental Table S2). Functional enrichment analysis of genes of high influence in the principal component loadings suggested a strong influence of stimuli, such as temperature, on the observed expression profiles, as the most enriched Gene Ontology (GO) terms included response to stimulus and response to temperature stimulus (Fig. 1C).
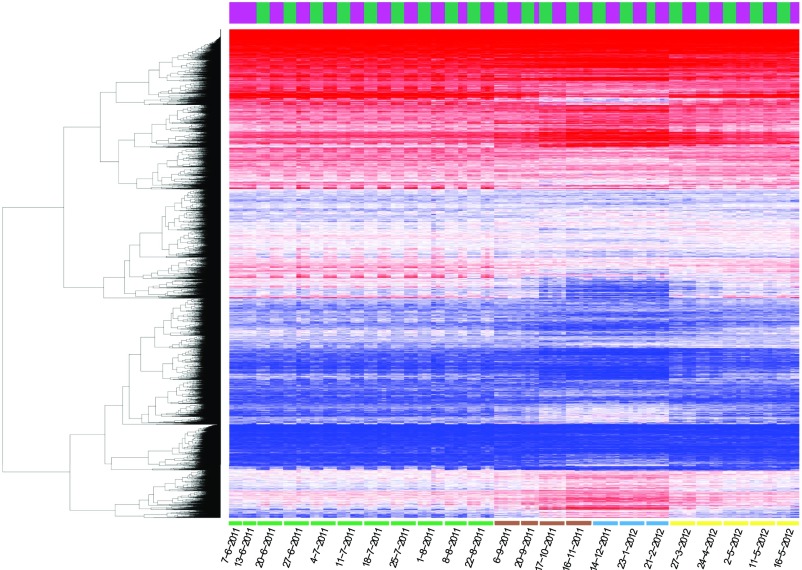
Hierarchical clustering of the genes revealed high reproducibility of the gene expression data, with replicate samples having consistent expression profiles (Fig. 2). Distinct patterns of expression were observed between the cambium/phloem and the xylem samples (Supplemental Fig. S3, A and B). Hierarchical clustering revealed clear seasonal patterns of gene expression. While gene expression during the active cambial growth (June to August) was quite consistent, a clear shift in expression occurred in September. Furthermore, a distinctive remodulation of the transcriptome could be observed in winter, with many genes showing a higher relative transcript abundance compared with other seasonal time points. According to functional enrichment analysis, the cambium/phloem winter-specific genes were especially related to UV-B response, tetrapyrrole metabolism, and carbohydrate metabolism (Supplemental Fig. S4). Hierarchical clustering of the samples and the genes confirmed the seasonal patterning of gene expression with four high-level clusters, which corresponded roughly to the active cambial growth (May to August) in the cambium/phloem, the active cambial growth in the xylem, the winter season, and a cluster containing both spring and autumn samples (Supplemental Fig. S3C). Interesting differences were observed between the cambium/phloem and the xylem samples in the beginning of the 2012 growing season in that the cambium/phloem samples of May 2 and May 16 clustered together with the samples from the active cambial growth period, while the corresponding xylem samples clustered together with the earlier time points, suggesting earlier reactivation of the phloem than the xylem (Supplemental Fig. S3C).
Figure 2.
Seasonal gene expression patterns in woody tissues of Norway spruce. Hierarchical clustering of genes is shown for all replicates across one season. Samples are ordered by sampling date. Expression values were library size corrected and variance stabilized (∼log2). The legibility of the heat map was improved by saturating the expression data between log2 values 3 and 10. The ribbon at the top of the heat map indicates the tissue type: magenta for xylem and green for cambium/phloem. Sampling months are marked by colors below the heat map: green, summer; brown, autumn; blue, winter; and yellow, spring.
Cambial Cell Expansion and Specification Are Transcriptionally Reprogrammed prior to Cell Division at the Start of the Growing Season
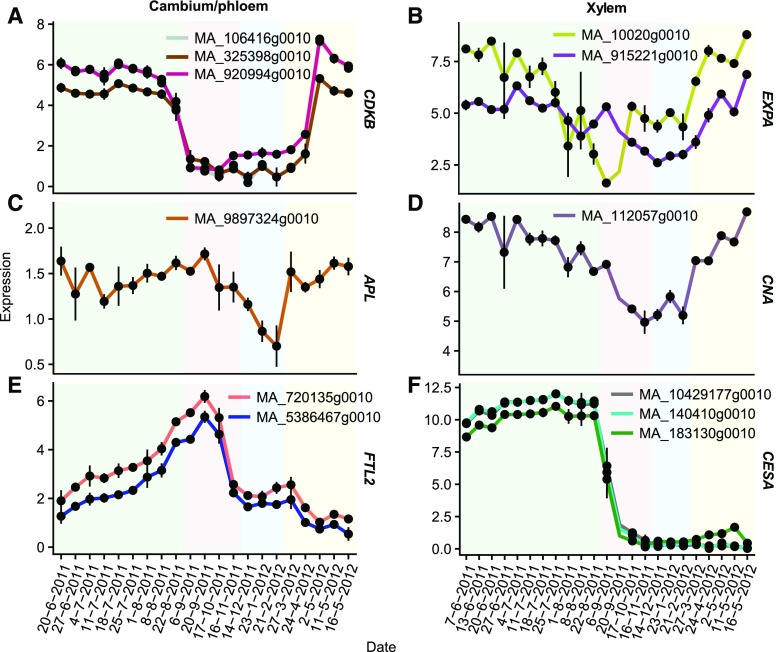
Cambial reactivation was investigated by analyzing the expression of known marker genes for cambial cell division, cell expansion, cambial derivative specification, and dormancy (for gene identities and phylogeny, see Supplemental Figs. S5–S9). The B-type cyclin kinases (CDKBs) are associated specifically with the progression of the cell cycle at the G2-to-M transition (Porceddu et al., 2001) and are known to be highly expressed in actively dividing cambial cells in Populus spp. stems (Schrader et al., 2004; Li et al., 2009). Our phylogenetic analysis identified three CDKB candidate genes in Norway spruce, which all peaked in the beginning of cambial growth in early May and declined upon the cessation of cambial growth in late August (Fig. 3A). The expression of α-expansins (EXPAs), which are marker genes for cell wall extension (Marowa et al., 2016), started to increase from basal levels after the February sampling (Fig. 3B), which was 2 months ahead of the strong increase in the expression of the CDKB genes. Similar to EXPA, a notable up-regulation of the genes marking early phloem differentiation (APL; Fig. 3C) and early xylem differentiation (CNA/ATHB15; Fig. 3D) was detected between the February and March samplings. The biosynthesis of secondary cell wall (SCW) components was examined via the expression of putative homologs of the Arabidopsis (Arabidopsis thaliana) cellulose synthase genes CESA4, CESA7, and CESA8 that have been associated specifically with the formation of the SCW (Endler and Persson, 2011). The SCW CESA genes showed high expression during the summer and were repressed in late August, when cambial activity ceased, thereafter remaining at almost undetectable levels throughout the autumn and winter (Fig. 3F). The Norway spruce FT/TFL1-LIKE2 gene (PaFTL2) is induced in needles and shoots in response to short days and has been suggested to be a repressor of growth (Klintenäs et al., 2012; Karlgren et al., 2013). Based on our in silico validation and expression data, two gene models of the Norway spruce genome assembly version 1.0 represent different regions of PaFTL2. These both had low expression in the beginning of cambial growth but increased gradually, reaching the highest expression on September 20, and then decreased rapidly to a low level during the winter and spring (Fig. 3E). Taken together, the marker gene expression data suggest that the first processes to occur in the cambial region after winter dormancy are cell expansion and specification followed by cell division.
Figure 3.
Phase transitions in cambial growth and wood formation. Gene expression is shown for putative Norway spruce marker genes of cambial activity (CDKBs), cell expansion (EXPAs), phloem (APL) and xylem (CNA) specification, dormancy (FTL2), and SCW formation (CESAs). Values are means ± se.
Expression Analysis Revealed the Core Set of Genes in Lignin Biosynthesis and Polymerization
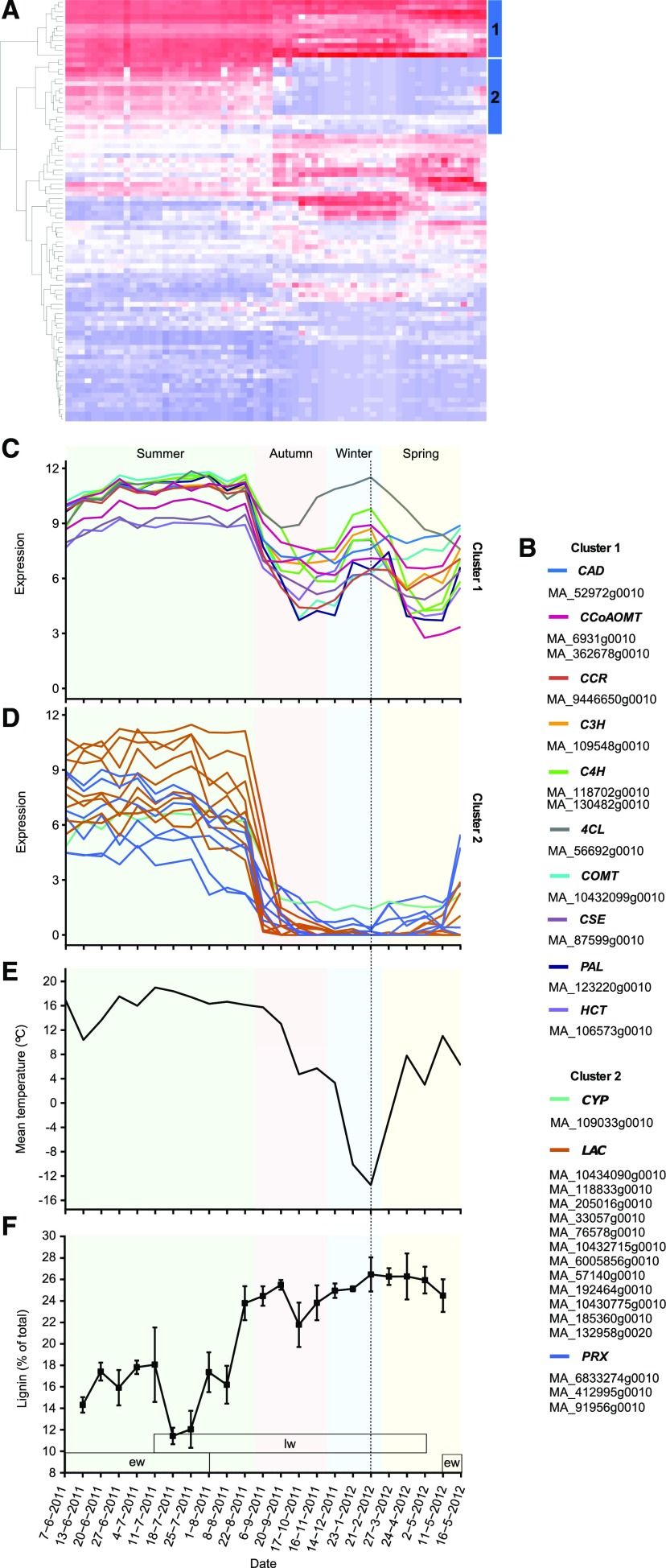
The dominating feature of wood formation is SCW formation, which involves the action of a large number of enzymes and transporters responsible for the biosynthesis and polymerization of cellulose, hemicellulose, and lignin. Here, we focused specifically on the biosynthesis of lignin, which in conifers is biochemically distinct from the angiosperm pathway. First, we identified Norway spruce genes putatively involved in monolignol biosynthesis and polymerization. Gene models assigned to monolignol biosynthesis and polymerization were derived from the ConGenIE database (Sundell et al., 2015). A large number of gene models were obtained for each associated gene family. For example, there were 215 Norway spruce gene models in the superfamily containing the Arabidopsis p-coumarate 3-hydroxylase1 gene (C3H1). After removing gene models that had low annotation quality or that were not expressed in any of the xylem samples, a set of 88 genes corresponding to the different enzymes in lignin biosynthesis and polymerization was obtained (Supplemental Fig. S10). Hierarchical clustering analysis throughout the seasonal cycle of cambial growth revealed a clear cluster of 12 genes that correspond to enzymes for all monolignol biosynthesis reactions characterized to date (cluster 1; Fig. 4, A–C; Supplemental Fig. S10), supporting the view that these genes function in the same metabolic pathway. All enzymes were represented by a single gene except for caffeoyl-CoA 3-O-methyltransferase (CCoAOMT) and cinnamate 4-hydroxylase (C4H), each of which was represented by two genes (Supplemental Figs. S11–S20). The genes in cluster 1 displayed high expression (variance stabilized transformed [VST] expression > 7) throughout the summer until the end of August, when expression declined rapidly (Fig. 4C). Interestingly, these genes showed a second, but somewhat lower, expression peak in the middle of the winter (Fig. 4C), which coincided with the lowest daily mean temperature recorded during sampling (Fig. 4E). A cluster of 16 genes (cluster 2; Fig. 4, A, B, and D) encoding mainly LACs (Supplemental Fig. S21) and PRXs (Supplemental Fig. S22) was found to be enriched for genes that were homologous to Arabidopsis lignification-related LAC11 and LAC17 (Zhao et al., 2013), supporting their role in the oxidative polymerization of lignin. The cluster 2 genes also were highly coexpressed, with a constitutively high expression throughout the period of active cambial growth and a low expression during cambial arrest (Fig. 4D). Taken together, these analyses identified a core set of genes representing candidates for the enzymes responsible for lignin biosynthesis and polymerization in Norway spruce.
Figure 4.
Norway spruce monolignol genes showed expression peaks both in the summer and in the winter. A, Hierarchical clustering of all putative monolignol, laccase (LAC), and peroxidase (PRX) genes identified based on the phylogenetic analyses and expression in xylem samples. The candidate genes for lignin biosynthesis and polymerization were identified from clusters 1 and 2, respectively. B, Names of the genes in clusters 1 and 2. C and D, Expression profiles of the genes in clusters 1 and 2. E, Daily mean temperatures of sampling dates in Umeå. F, Proportions of lignin ± se in the part of the year ring that contained living tracheids. The periods of earlywood (ew) and latewood (lw) formation are indicated. Dashed lines show the date for the coldest sampling time point during the study period.
The expression pattern of the core set of lignin biosynthetic genes suggests that lignin polymerization is restricted to the active growth period, while monolignol biosynthesis also can proceed during the period of cambial dormancy. To analyze how this affects the accumulation of lignin, a chemical pyrolysis-gas chromatography/mass spectrometry (Py-GC/MS) analysis was performed in both the living part of the year ring and the part of the year ring with dead tracheids, whenever it was present. In the living tracheids, the relative proportion of lignin was somewhat stable during earlywood formation until the initiation of latewood formation, when the relative proportion of lignin dropped rapidly, followed by a gradual increase during continued latewood maturation (Fig. 4F). The proportion of lignin did not correlate with sampling time during cambial dormancy (2012), suggesting that the observed increases in the expression of lignin biosynthetic genes were not associated with lignin accumulation.
Transcriptional Regulators of the Seasonal Control of Lignification
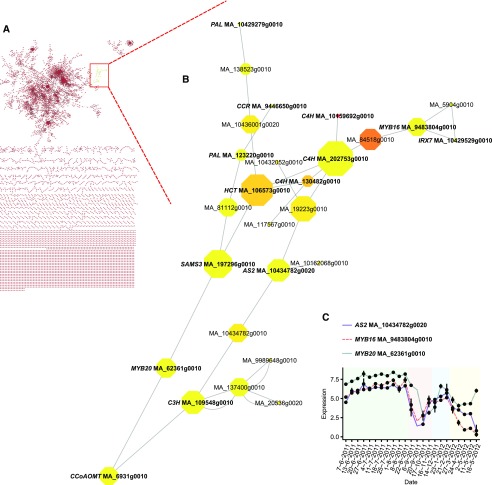
The strict transcriptional coregulation of lignin biosynthetic genes during the seasonal cycle of cambial activity implies a common transcriptional regulation of gene expression. Transcriptional control of lignin biosynthesis is well established in the developmental context (Vanholme et al., 2012), but very little information exists on the transcriptional control of lignin biosynthesis genes in response to seasonal changes. In order to identify factors involved in such control, we employed nine independent coexpression gene network inference methods and aggregated them to form a consensus network. The resulting network was subsequently filtered to retain the most supported edges, connecting approximately 7,500 genes through approximately 10,000 links. In the coexpression network, a subnet of 27 highly connected genes was found to contain six out of the 12 cluster 1 monolignol biosynthetic genes (Figs. 5C and 6B), one additional monolignol gene with a low expression level, two partial lignin biosynthetic genes, and S-ADENOSYLMETHIONINE SYNTHETASE3 and IRREGULAR XYLEM7 genes, which are known to be involved in SCW formation (Shen et al., 2002; Brown et al., 2005). In addition, gene annotation enrichment analysis showed that GO terms such as lignin metabolic process and lignin biosynthetic process were statistically enriched (P < 0.05) for the genes in the subnet. Based on a domain pattern analysis conducted using the PlantTFcat tool (Dai et al., 2013), we identified three genes encoding transcription factors (TFs). These genes represented the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) and MYELOBLASTOSIS HOMEOBOX (MYB-HB) families (Fig. 5, B and C) and had high sequence similarity with Arabidopsis AS2, ATMYB16, and ATMYB20 (Supplemental Figs. S23 and S24). They had relatively high betweenness centrality values (the number of shortest paths passing through a gene, where the shortest path is the smallest number of edges connecting a pair of genes), indicating high centrality in a network. The AS2 factor, MA_10434782g0020, was expressed exclusively in xylem samples (Supplemental Table S2), further supporting a specific role for this TF in the seasonal control of lignin biosynthesis. Most importantly, all three TF genes exhibited similar annual expression patterns with the identified monolignol genes (Figs. 4C and 5C).
Figure 5.
A subnet in the coexpression network connects several gene models related to cell wall formation and especially the monolignol pathway. A, Location of the subnet in the coexpression network. B, Gene cluster containing several cluster 1 lignin biosynthetic genes. The size of the octagons represents the betweenness centrality value, while the color denotes neighborhood connectivity (ascending from yellow to red). Double links indicate bidirectional edges. C, Expression profiles of the TF genes implicated in the seasonal control of lignification. Values are means ± se.
Figure 6.
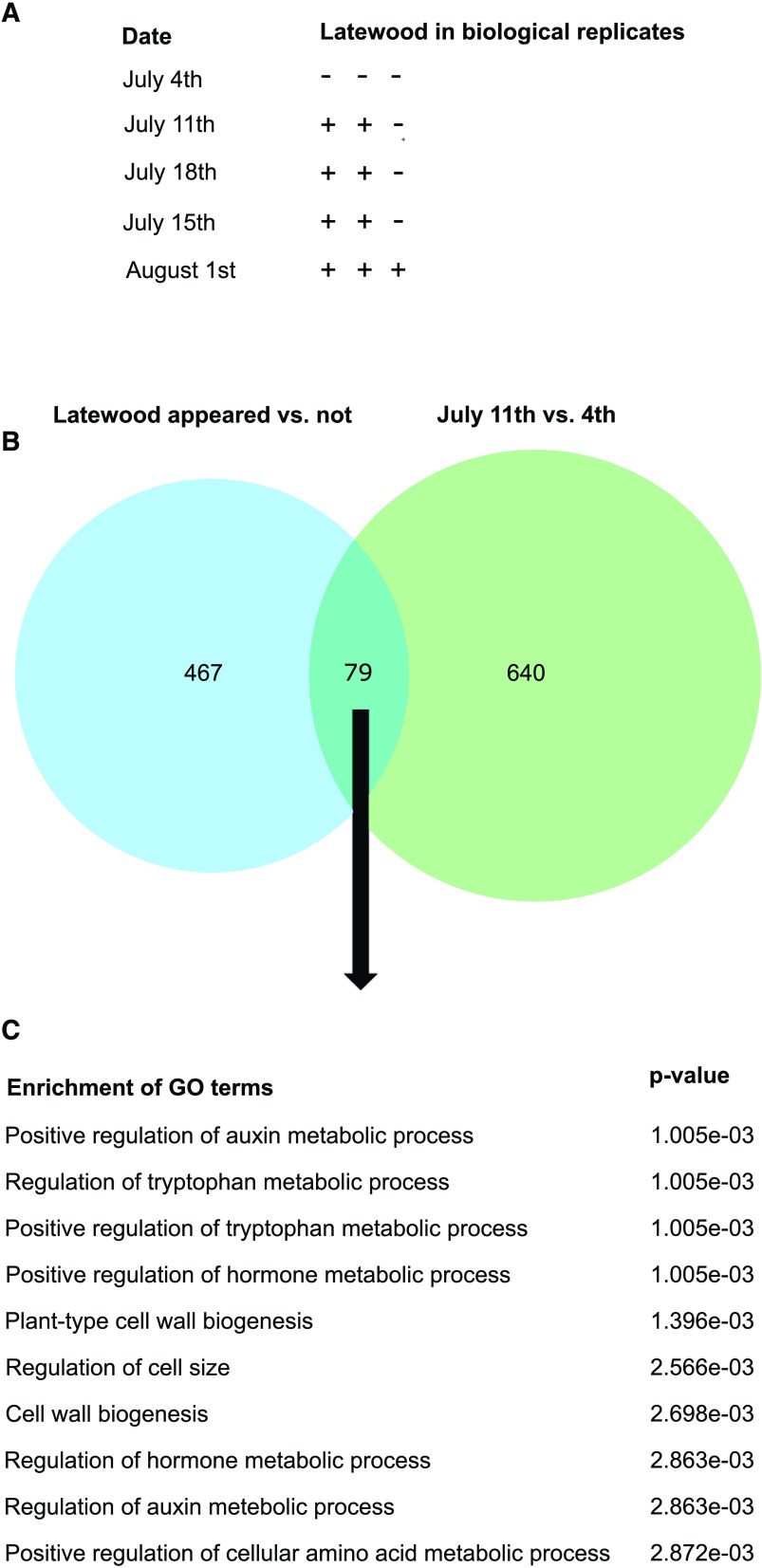
Onset of latewood formation in Norway spruce. A, Date of latewood onset and variation among the three replicate trees for each time point. B, Venn diagram showing the overlap between the results of two differential expression analyses (adjusted P < 0.01; 79 genes): the first time point when latewood cells were observed versus the last sampling before latewood detection (640 genes) and samples containing latewood versus those without over the course of latewood formation (467 genes; three samplings in July). C, GO term enrichment analysis of the common genes between comparisons.
The Formation of Latewood Tracheids
One of the most striking changes during the seasonal cycle of secondary growth is the transition from earlywood to latewood formation. In our seasonal series, the transition to latewood formation varied between the different replicate trees. Thick-walled latewood cells were observed in NBT-stained light microscopy sections from July 11 onward but were observed only in samples from all replicate trees from August 1 (Fig. 6A). When data from July 11 were compared with the previous sampling date (June 4), 719 genes were found differentially expressed (adjusted P < 0.01) in xylem samples (Fig. 6B). In turn, 546 genes showed differential expression (adjusted P < 0.01) when samples containing latewood were compared with samples without latewood between July 11 and July 25 (Fig. 6B). The overlap between the two comparisons contained 79 genes (i.e. genes that were differentially expressed with the same direction in expression change in both comparisons; Supplemental Table S3). These genes contained one A-type (MA_6619g0010) and one B-type (MA_19215g0010) cyclin, which were both suppressed during latewood initiation, and, therefore, could be related to the suppression of cambial cell division. Another interesting gene was an RNA-dependent RNA polymerase (RDR; MA_10436273g0010), which is involved in posttranscriptional gene silencing, based on its closest homolog in Arabidopsis (AT4G11130; RDR2). RDR may contribute to large-scale transcriptome reprogramming that is needed when cambial growth shifts from earlywood to latewood formation. Also, several cell wall biosynthetic and auxin-related genes were associated with latewood formation. This was also apparent from the GO term analysis of the overlapping set of genes (Fig. 6C).
The Lifetime of the Tracheids Varies According to the Season
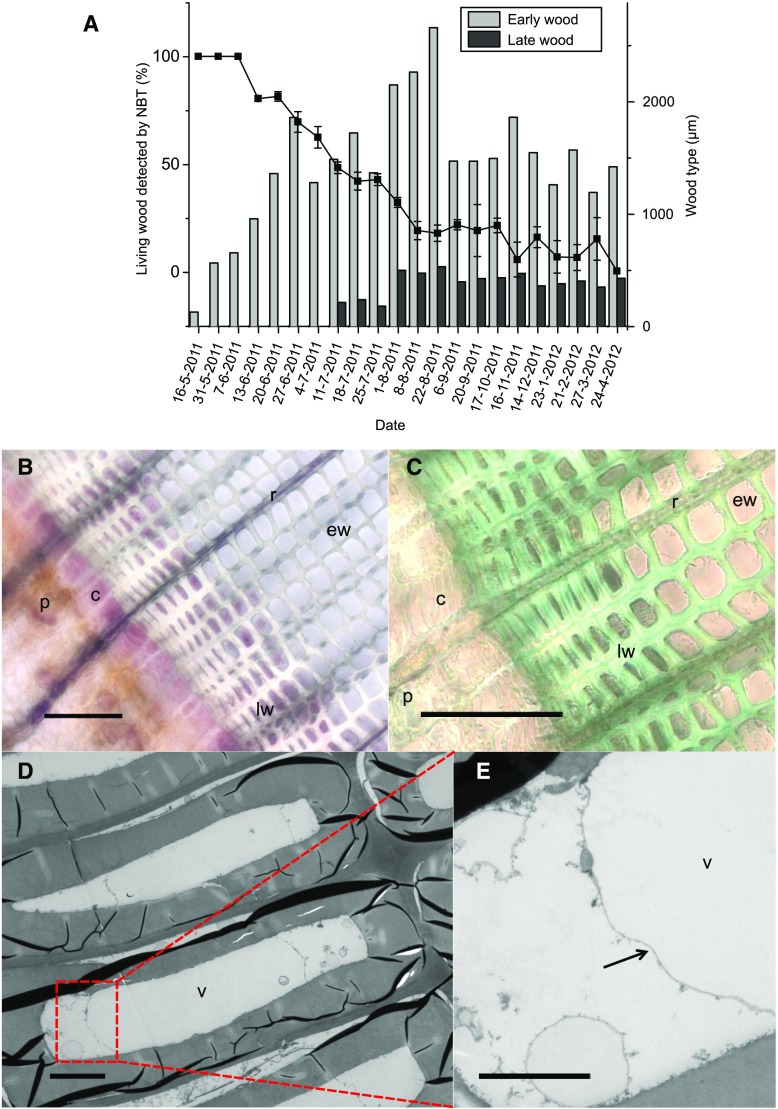
The last phase to occur during the seasonal cycle of cambial growth is cell death of the tracheids. We used NBT viability staining to define the occurrence of tracheid cell death throughout the seasonal cycle. The first dead earlywood tracheids were detected on June 13 (Fig. 7A), and all earlywood tracheids were determined to be dead on August 8. The latewood tracheids did not die at the same speed as the earlywood tracheids but remained alive even during the winter period (Fig. 7, A and B). In February and March 2012, two and one out of three replicate trees still had enzymatically active latewood tracheids, respectively. Toluidine Blue staining and electron microscopy (EM) analysis confirmed these results (Fig. 7, C–E). The latewood tracheids near the cambial area, cambial cells, as well as ray cells of the annual ring formed in the previous summer clearly showed cell contents in samples collected between January and May 2012 (Fig. 7). The existence of membranous structures in latewood tracheids of the contemporary annual ring also was observed in the winters of 2013 and 2014 (data not shown). On May 2, 2012, most of the latewood tracheids appeared empty, but it was still possible to identify a few latewood tracheids with intact tonoplasts (Fig. 7, D and E).
Figure 7.
The long lifespan of latewood tracheids. A, Average amount (%) of viable xylem tissue ± se, defined based on NBT viability staining, and average width (µm) of earlywood and latewood in sampled trees. B, Light microscopy image of a transverse section collected on March 27, 2012, and stained with NBT. The viability is shown by the presence of a pink/lilac precipitate that is formed by succinate dehydrogenase activity of living cells in the presence of NBT. Enzymatic activity was observed in several cell layers of the latewood. c, Cambium; ew, earlywood; lw, latewood; p, phloem; r, ray. C, A Toluidine Blue-stained section from March 27, 2012. D and E, Electron micrographs showing latewood tracheids with intact vacuoles in a sample collected May 2, 2012. Tonoplast is marked by the arrow. v, Vacuole. Bars = 100 µm (B and C), 5 µm (D), and 2 µm (E).
To analyze the tracheid cell death process at the molecular level, we identified Norway spruce gene models for the homologs of endopeptidases, metacaspases, papain-like Cys proteases, and ribonucleases, which all have been implicated previously in programmed cell death (PCD) of other plant species. From the investigated 19 Arabidopsis genes (Supplemental Table S4), we found clear homologs in Norway spruce for all genes except XYLEM SERINE PEPTIDASE1 (XSP1; Supplemental Fig. S25).
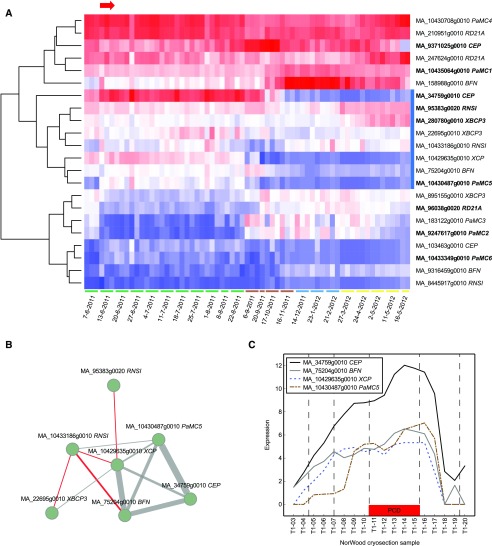
When we analyzed gene expression in the period of the first visual appearance of dead tracheids during earlywood formation in June, 3,964 genes underwent a significant expression change (adjusted P < 0.01; Supplemental Table S5). Nine putative PCD genes (Supplemental Figs. S26–S29) belonged to this group of differentially expressed genes (Fig. 8A), encoding type I and type II metacaspases, RESPONSIVE TO DEHYDRATION21A proteinase (RD21A), CYSTEINE ENDOPEPTIDASES (CEPs), XYLEM BARK CYSTEINE PEPTIDASE3 (XBCP3), and class I RNASE (RNSI). The strongest up-regulation was observed in the expression of the AtMC9 homolog MA_10430487g0010. The corresponding transcript (comp95097_c0_seq3) was annotated previously as PaMC5 in the bioinformatic analyses of Minina et al. (2013). We also cloned the coding region of PaMC5. It was 1,023 bp long, which is 93 bp more than the gene model MA_10430487g0010. The deduced amino acid sequence of PaMC5 differed by one amino acid from the MA_10430487g0010 gene model. We also found another putative AtMC9 homolog, MA_10433349g0010, by genome mining and named it PaMC6. The deduced amino acid sequence of MA_10433349g0010 has 77% identity with that of PaMC5. However, the transcript of PaMC6 was not amplified successfully from spruce wood cDNA, which may be due to the rather low expression level in xylem tissue (ConGenIE.org). When we analyzed the expression of the putative PCD genes by hierarchical clustering and used the NorWood data resource (http://NorWood.ConGenIE.org; Jokipii-Lukkari et al., 2017) based on the spatial wood section series of Norway spruce, PaMC5 (MA_10430487g0010) clustered tightly together with three other genes: putative XYLEM CYSTEINE PROTEINASE (XCP), ENDONUCLEASE1 (ENDO1, also known as BIFUNCTIONAL NUCLEASE1 [BFN1]), and CEP homologs (Fig. 8). These genes exhibited high expression, especially in the PCD zone of the NorWood wood series (Fig. 8C), strongly indicating their importance in tracheid cell death.
Figure 8.
Norway spruce PaMC5, BFN, XCP, and CEP are strong candidate genes for executors of tracheid cell death. A, Hierarchical clustering of genes putatively related to PCD selected based on phylogenetic analyses. The genes that were differentially expressed (adjusted P < 0.01) during the appearance of dead earlywood cells (time point marked with the red arrow) are written in boldface. The cluster containing homologs for Arabidopsis MC9, BFN1, and XCP genes is marked with the blue rectangle next to the heat map. Sampling months are marked by colors below the heat map; green, summer; brown, autumn; blue, winter; and yellow, spring. B, Seven out of eight genes in the identified cluster were connected in the coexpression network of the spatial wood section series of the NorWood Web resource (threshold 3; http://NorWood.ConGenIE.org). PaMC5, BFN, XCP, and CEP showed the strongest correlation in their expression. The gray and red lines indicate positive and negative correlations, respectively. The width of the line is relative to the strength of the correlation. C, PaMC5, BFN, XCP, and CEP exhibited high expression in the cell death zone (red rectangle) of the tangential wood sections of NorWood.
DISCUSSION
Temperature and daylength are the two main factors that influence seasonal patterns of secondary growth in tree stems. Our transcriptome data set that spanned the yearly cycle of secondary growth supported the importance of stimuli, such as temperature, in the seasonal control of secondary growth (Figs. 1C and 2). These data also support the fact that these external cues influencing secondary growth are sensed directly in the cambial tissues. A recent transcriptomic study identified photoperiod as the dominant driver of seasonal gene expression variation in needles of Douglas fir (Pseudotsuga menziesii; Cronn et al., 2017). Therefore, it is possible that, even though both temperature and photoperiod are important for all aspects of seasonal growth control, cambial growth is affected to a greater extent by changes in the ambient temperature than in the photoperiod, while the opposite is true for the growth of the shoots. This is supported by our data on transcriptome reprogramming in the woody tissues, which was not always gradual across the seasons, but often abrupt, and therefore apparently more related to changes in the temperature than in the photoperiod (Fig. 2). Temperature has been recognized previously as a key trigger during cambial reactivation (Sarvas, 1969; Oribe et al., 2001) and latewood formation (Begum et al., 2012) of conifer trees. Interestingly, both in our data and in the Douglas fir needles (Cronn et al., 2017), a large group of genes showed their annual expression maxima during the winter. A similar phenomenon also was observed with some of the genes that are important for heartwood formation in Scots pine (Pinus sylvestris; Lim et al., 2016). This increase in the winter-time expression of genes could be due to enhanced gene expression during ambient temperatures that allow transcription to occur, but it is also possible that posttranscriptional regulation, such as changes in RNA stability, affects transcript abundance during the winter. The winter-specific genes of this study showed functional enrichment of GO terms related to response to UV-B radiation, a condition likely to occur during late winter/spring in the northern hemisphere due to ozone depletion (Sinnhuber et al., 2011). The winter-specific transcriptome also indicated significant alterations in metabolic processes, including the biosynthesis of raffinose and starch degradation (Supplemental Table S6), that are known to prevail during winter.
The Unique Features of Lignin Biosynthesis in Norway Spruce
The transcriptome data and phylogenetic analyses enabled us to identify a core set of monolignol biosynthetic genes in Norway spruce (Fig. 4). These genes are likely to correspond to enzymes having a major role in the lignin biosynthesis of Norway spruce, but we cannot exclude the presence of additional lignin biosynthetic genes that were not identified here due to expression in specific cell types. The Norway spruce genome is still somewhat fragmented, and additional genes in lignin biosynthesis might appear along with improved genome assembly. We also identified three TFs, one AS2/LOB and two MYB factors, that, on the basis of the coexpression pattern with the core set of monolignol biosynthetic genes, seem to be involved in the seasonal control of lignification (Fig. 5). The AS2/LOB factor has high sequence similarity with Arabidopsis AS2, which forms a protein complex with AS1 to mediate the long-term repression of BREVIPEDICELLUS/KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1, which, in turn, regulates lignin deposition in the stem (Mele et al., 2003; Lodha et al., 2013). AS2 and AS1 act partially redundantly, and the dsl1-D mutant of AS1 shows irregular lignification in stems (Chalfun-Junior et al., 2005). One of the MYB factors is a close homolog of ATMYB20, which has been reported to be regulated by SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1, which is one of the master regulators of SCW formation and lignification in Arabidopsis (Zhong et al., 2008). In line with our study, P. taeda PtMYB1, a homolog of ATMYB20, has been suggested to regulate lignin biosynthesis based on its strong expression in differentiating xylem and its ability to activate the transcription of the Phe ammonia-lyase (PAL) promoter (Patzlaff et al., 2003). The second MYB factor localizing with the core set of monolignol genes in the coexpression network is homologous to ATMYB16, which was not connected earlier to the regulation of lignin biosynthesis but, instead, to the regulation of cell shape (Baumann et al., 2007) and cuticle formation (Oshima and Mitsuda, 2013). Therefore, we identified two Norway spruce TFs that were linked earlier in herbaceous species to lignification and one that is a novel TF in this process.
A striking feature in the expression of the monolignol biosynthetic genes was the second peak in midwinter, while the expression of genes putatively related to lignin polymerization or secondary cellulose biosynthesis remained at very low levels after the growing season. The phenylpropanoid pathway is known to be stimulated by low temperatures, although variation occurs according to genes, species, and tissues (Cabane et al., 2012). Low temperatures can cause excessive production of harmful reactive oxygen species, and hydroxycinnamic acids and esters are recognized as potent antioxidants (Chen and Ho, 1997; Janas et al., 2000). Phenolic compounds also have been suggested to function as UV screens in land plants (Cheynier et al., 2013). Indeed, UV-B-induced synthesis of soluble flavonoids has been documented in Scots pine needles (Schnitzler et al., 1997). Microscopy analyses in Scots and loblolly pine also have indicated increased production of cell wall-bound phenolic compounds, such as 4-coumaric acid or lignin, upon elevated UV-B radiance (Laakso et al., 2000). In this study, however, only subtle changes in the abundance of lignin were observed during the winter months, when the most prominent increase occurred in the expression of monolignol genes. Therefore, it seems possible that the winter-time induction of phenylpropanoid pathway genes contributes to protection against freezing or UV-B radiance-induced stress by the production of precursors of lignin.
Our analysis of the Norway spruce genome resulted in the unexpected finding of genes associated with S-type lignin biogenesis. Although some specific gymnosperm lineages produce significant amounts S-type lignin, only traces of S units have been detected in common softwood lignin (Baucher et al., 1998). Therefore, it was previously considered unlikely that conifers possess ferulate 5-hydroxylase1 (F5H) and caffeic acid O-methyltransferase (COMT), which are required for the production of sinapyl alcohol. However, we detected a COMT-like gene model (Supplemental Fig. S18) that showed a highly similar expression pattern with the majority of the identified monolignol genes. COMT-like sequences also have been found previously in Picea sitchensis (Friedmann et al., 2007) and shown to be up-regulated during lignin-forming conditions in Norway spruce cell culture (Laitinen et al., 2017). Furthermore, a loblolly pine O-methyltransferase that was capable of methylating both caffeic acid and 5-hydroxyferulic acid and the corresponding CoA esters was similar in sequence to angiosperm COMTs but not CCoAOMTs (Li et al., 1997). This led to the proposal that dual methylation pathways, operating both at the acid and CoA ester levels, exist in loblolly pine xylem (Li et al., 1997). It was also obvious from the metabolic engineering of S-type lignin biosynthesis in radiata pine (Pinus radiata; Wagner et al., 2015) that transformation with the angiosperm F5H was sufficient for the synthesis of S-type lignin in the transgenic pine tracheary elements (TEs) and, therefore, that endogenous COMT activities must exist in pine that are adequate to allow the synthesis of S-type lignin. Therefore, the earlier work combined with our transcriptomic and phylogenetic analyses support the capacity of at least some of the conifer species to synthesize S-type lignin.
The Long Life of Spruce Latewood Tracheids
Earlywood and latewood tracheids both undergo PCD at the end of their maturation. Previously, Norway spruce tracheids were reported to die 1 to 3 months after their formation (Dieset, 2011; Bollhöner et al., 2012), and the last-formed one or two rows of latewood tracheids of another conifer, Pinus strobus, were shown to occasionally possess cellular contents even in January in Wisconsin (Murmanis and Sachs, 1969). We observed that spruce latewood cells remained alive substantially longer than this on the basis of both enzymatic activity assays and cellular morphology. Some latewood cells adjacent to the cambium showed intact vacuoles even until the beginning of the next growing season in May. Therefore, the tracheids were not only arrested in cellular autolysis but apparently still alive and metabolically active. Previously, the aspect of overwintering xylem derivatives was studied only with vessel elements of deciduous species. The earlywood vessels of species such as Ulmus davidiana (Imagawa and Ishida, 1972) and Aesculus hippocastanum (Barnett, 1992) were found to overwinter in an undifferentiated state and continue development in the spring without undergoing further division.
What might be the purpose of the long lifespan of the latewood tracheids? One possibility lies in the poor water transport capacity of the latewood tracheids. It has been recorded that the earlywood of Douglas fir has about 11 times the water conductivity of the latewood and that 90% of the total water flow occurs through earlywood (Domec and Gartner, 2002). Therefore, it seems that latewood tracheids do not need to properly control their demise and maybe even actively suppress this process, which demands a lot of energy and strict transcriptional and translational control. Another possibility is that it is important to keep living cells next to the vascular cambium during the period of cambial arrest. It is well established that several angiosperm species, including Salix and Populus spp., form a so-called terminal parenchyma layer in the secondary xylem before resuming cambial arrest (Carlquist, 2001). The purpose of this cell layer is not clear but could be related to cross talk between cambial and xylem cells during cambial reactivation. It is possible that spruce and maybe other conifers retain the viability of at least parts of the latewood tracheids for the same purposes.
Regulation of Tracheid PCD
Metacaspases are ancestral relatives of caspases, the Cys-dependent Asp proteases that are central players in the metazoan apoptotic pathway. Several Arabidopsis metacaspases have been shown to be involved in stress and hypersensitive response-related cell death (Watanabe and Lam, 2005; van Baarlen et al., 2007; He et al., 2008; Coll et al., 2010). AtMC9 has been connected specifically to cell death in various developmental processes, including TE differentiation (Bollhöner et al., 2013; Olvera-Carrillo et al., 2015).
Our combined phylogenetic and expression analyses strongly indicate a role for PaMC5, the Norway spruce homolog of AtMC9, in the PCD of tracheids. Upon initiation of the anatomically determined death of the earlywood tracheids, PaMC5 showed the strongest up-regulation among the identified putative PCD genes. We also successfully cloned the complete coding sequence of PaMC5, facilitating future functional assays of this gene. Previously, a spruce metacaspase gene, mcIIPa (later named PaMC4; Minina et al., 2013), was shown to be required for differentiation and cell death in embryo suspensor (Suarez et al., 2004). In this study, PaMC4 showed high and nearly constant expression in xylem over the course of the year, suggesting that this gene does not encode a protein that would be involved in tracheid PCD or that its possible function in this process is not controlled at the transcriptional level.
The expression of PaMC5 showed a strong positive correlation with the Norway spruce genes ENDO1, XCP, and CEP (Fig. 7). So far, functional evidence for the control of TE PCD has been obtained only for AtMC9, Zinnia elegans ZEN1 (the homolog of ENDO1), and Arabidopsis XCP1 and XCP2. AtMC9 is involved in degrading the vessel cell content postmortem (i.e. after the rupture of the tonoplast; Bollhöner et al., 2013), whereas the ZEN1 nuclease is required for the postmortem degradation of DNA in TEs (Ito and Fukuda, 2002). Arabidopsis XCP1 and XCP2 function both in microautolysis and megaautolysis of vessel elements (Avci et al., 2008). Also, AtMC9 and the Arabidopsis XCPs and ENDO1 are highly coregulated (Olvera-Carrillo et al., 2015). The observed coexpression neighborhood and, more importantly, the expression peaking in the PCD zone of the NorWood wood section series emphasize CEPs as novel candidates for executors of PCD in xylem cells. Thus far, CEP1 has been found to be crucial for the degeneration of tapetal cells and functional pollen formation (Zhang et al., 2014).
CONCLUSION
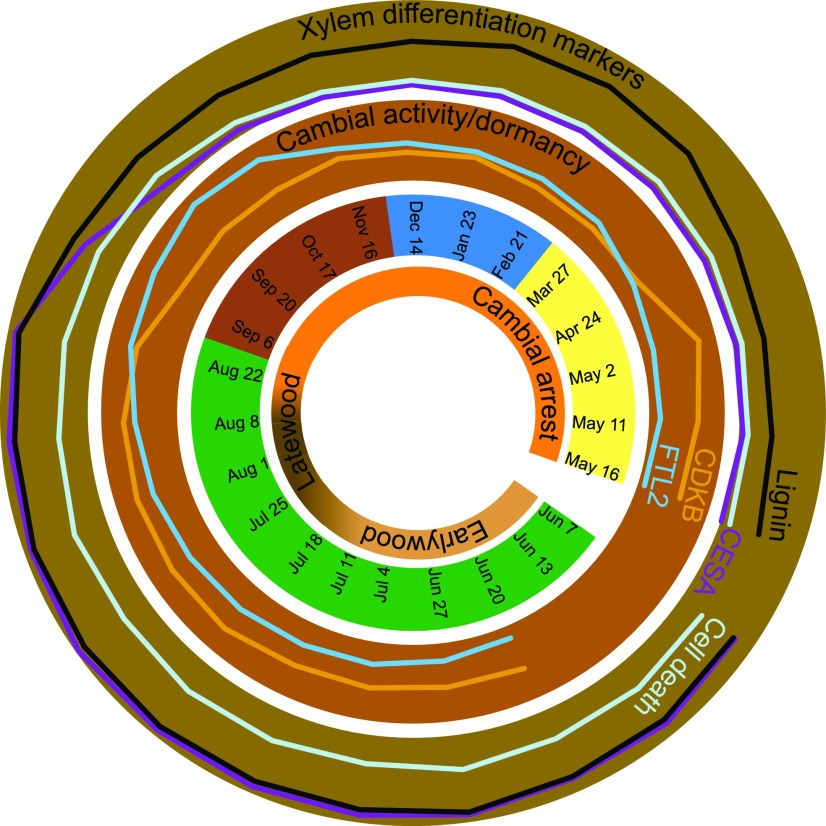
Our analyses provide new information about several aspects of seasonality in the cambial growth of Norway spruce. In the beginning of the growing season, cambial cell expansion and cell specification preceded cambial reactivation based on the transcriptome changes first on the phloem side. Cambial reactivation was followed by activation of the complete program of xylem differentiation, as shown by the simultaneous induction of genes encoding cellulose and lignin biosynthesis as well as PCD genes (Fig. 9). Transcriptomic changes were only subtle during the periods of earlywood and latewood formation. A rather sudden transcriptomic reprogramming coincided with the loss of cambial activity at the end of the growing season, when the expression of both cell division and xylem differentiation markers decreased rapidly. Surprisingly, the expression of the lignin biosynthetic genes remained at a relatively high level during the period of cambial arrest and showed even increased expression during the coldest months of the winter. Also, a large number of other genes showed high transcript abundance at the same time. Another surprising finding during the cambial arrest was the long lifetime of the latewood tracheids: at least parts of the latewood retained viability throughout the winter season, and living tracheids were observed until cambial reactivation in the spring.
Figure 9.
Marker genes define the main phase transitions during the annual cycle of cambial growth in Norway spruce. The outermost and second outermost rings indicate the transcript abundances in xylem and cambium/phloem samples, respectively. The profiles represent the means of the specific gene groups.
Transcriptomic changes in the described phase transitions of cambial growth provide information about the underlying molecular mechanisms and point to interesting candidate genes for future work. For instance, the genes that were linked to the earlywood-to-latewood transition as well as the seasonality of lignin biosynthesis need further functional validation. The extent and significance of the winter-time gene expression as well as the factors controlling latewood tracheid viability are two other intriguing findings that need further exploration.
MATERIALS AND METHODS
Plant Material
Samples were collected from clonally replicated 48-year-old Norway spruce (Picea abies) trees of the genotype Z4006, which were used for Norway spruce genome sequencing (Nystedt et al., 2013). The trees grew among other genotypes randomly spread within a spruce plantation in Hissjö, Sweden (63°55′50′′N, 20°08′47′′E). The stem pieces were collected from three replicate trees between spring 2011 and spring 2012 (Supplemental Table S7). During the growing season, the sampling was conducted on a weekly basis, and it continued at least once per month outside of the active growing season (Fig. 1B). The samples for EM were collected between January and May 2012, in January and July 2013, as well as in January 2014.
Sampling of the Stem Material
A stem piece was collected from breast height of the stem in an area devoid of damage or traumatic resin ducts (Supplemental Fig. S30). Before sampling, the shearing forces of the stem were released around the stem piece by using a carpet knife to cut deep into the wood around the sampling area. Then, the stem pieces, including bark, phloem, cambium, developing xylem, and two to three year rings of the previous years, were collected by placing a chisel at a 90° angle above the pieces and hammering tangentially downward until the piece was released from the stem. Separate samples were taken for RNA extraction and microscopy next to each other. The pieces used for RNA-Seq were immediately flash frozen in liquid N2 and then kept on dry ice until storage at −80°C. The stem pieces used for light microscopy and EM were kept on ice until fixation for microscopy in the laboratory.
Light Microscopy
Cell viability was determined by NBT (Berlyn and Miksche, 1976; Gahan, 1984) staining. Cross sections of 20 to 30 µm were cut with a cylinder hand microtome and incubated for 1.5 h in 50 mm sodium phosphate buffer (pH 7.6) containing 50 mm sodium succinate and 500 mg L−1 NBT under strong light conditions. On March 27, fresh tissue samples also were stained in 0.05% Toluidine Blue. All the sections were examined with an Axioplan 2 microscope (Zeiss), and micrographs were captured with an AcioCam HRC camera. The width of the living xylem (determined by the presence of blue precipitates) of each tree was measured in at least three different loci of the xylem using AxioVision LE software.
RNA Extraction and Sequencing
For RNA extraction from phloem, cambium, and the living part of the wood, bark was removed from the stem pieces by peeling. A sample consisting of the vascular cambium and the living part of the phloem was scraped from the inner surface of the bark. This sample is hereafter referred to as the cambium/phloem sample. Subsequently, the exposed surface of the wood was collected by scraping until the appearance of dead wood. This sample is hereafter referred to as the xylem sample. The scraped material was homogenized to powder and stored at −80°C. Total RNA was extracted with the cetyl-trimethyl-ammonium bromide-based method (Chang et al.,1993) and purified with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. The RNA concentration and purity were measured by a NanoDrop 2000 spectrophotometer (NanoDrop Technologies). The integrity of the RNA was analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies).
RNA library preparation and subsequent sequencing were performed at the SciLifeLab. Strand-specific RNA libraries for sequencing were prepared with the TruSeq Stranded mRNA Sample prep kit of 96 dual indexes (Illumina) according to the manufacturer’s instructions, except for the following changes. The protocols were automated in Agilent NGS workstation (Agilent) using purification steps as described by Lundin et al. (2010) and Borgström et al. (2011).
Clonal clusters were generated using cBot (Illumina) and sequenced with a 2x101 setup on HiSeq2500 (Illumina) according to the manufacturer’s instructions. Bcl-to-Fastq conversion was performed with bcl2Fastq version 1.8.3 from the CASAVA software suite. The quality scale was Sanger/phred33/Illumina 1.9.
Preprocessing of RNA-Seq Data and Differential Expression Analyses
The data preprocessing was performed as described at http://www.epigenesys.eu/en/protocols/bio-informatics/1283-guidelines-for-rna-seq-data-analysis. Briefly, the quality of the raw sequence data was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The residual rRNA contamination was assessed and filtered using SortMeRNA (version 1.9; Kopylova et al., 2012), with settings -n 6 -a 8 -v, using the rRNA sequences provided with SortMeRNA (rfam-5s-database-id98.fasta, rfam-5.8s-database-id98.fasta, silva-bac-16s-database-id85.fasta, silva-euk-18s-database-id95.fasta, silva-bac-23s-database-id98.fasta, and silva-euk-28s-database-id98.fasta). Data were then filtered to remove adapters and trimmed for quality using Trimmomatic (version 0.32; Bolger et al., 2014), with settings TruSeq3-PE-2.fa:2:30:10 LEADING:3 SLIDINGWINDOW:5:20 MINLEN:50. After both filtering steps, FastQC was run again to ensure that no technical artifacts were introduced. Filtered reads were aligned to version 1.0 of the Norway spruce genome (retrieved from the ConGenIE resource; Sundell et al., 2015) using STAR (version 2.3.1e; Dobin et al., 2013), with nondefault settings –outReadsUnmapped Fastx–alignIntronMax 11000. The annotations obtained from the Norway spruce version 1.0 GFF file contain only one transcript per gene model. This GFF file and the STAR read alignments were used as input to the HTSeq (Anders et al., 2015) htseq-count python utility to calculate exon-based read count values. The htseq-count utility takes only unique mapping reads into account. Statistical analysis of single-gene differential expression (DE) between conditions was performed in R (version 3.2.3; R Core Team, 2015) using the Bioconductor (version 3.3; Gentleman et al., 2004) DESeq2 package (version 1.10.1; Love et al., 2014). False discovery rate-adjusted P values were used to assess significance; a common threshold of 1% was used throughout. For the data quality assessment and visualization, the read counts were normalized using a variance-stabilizing transformation as implemented in DESeq2. This was run in a blind manner for the quality assessment but otherwise used the model ∼ sampling + tissue, where sampling represents the date of sampling and tissue represents either cambium/phloem or xylem. The biological relevance of the data (e.g. biological replicate similarity) was assessed by PCA and other visualizations (e.g. heat maps) using custom R scripts. For all subsequent expression analyses, which also were performed in R, the normalized read counts obtained from DESeq2 were used. The DE analysis for the death of the early wood was performed for the xylem between the corresponding two time points (three replicates each), modeling for the time of sampling. The DE analyses for the late wood formation were performed for the xylem samples, first by modeling for the time of sampling, comparing July 11 versus July 4 (three replicates each), and second by modeling for the presence versus the absence of latewood in samples collected from July 11 to July 25 (three sampling times, three replicates each); six versus three samples, respectively. The R scripts to reproduce the analyses are available from https://github.com/UPSCb/UPSCb/tree/master/manuscripts/Jokipii-Lukkari2017. An overview of the data, including raw and post-quality control read counts and alignment rates, is given in Supplemental Table S8. For the heat map and the gene network inference, the data were filtered to remove lowly expressed genes, provided that they did not have 10 reads in at least two replicates of any sampling. This was done independently by tissue, resulting in 32,830 common genes and 1,136 and 2,177 genes specific to cambium/phloem and xylem, respectively.
Gene Network Inference
Nine gene network inference methods were run: Anova (Küffner et al., 2012), Aracne2 (Margolin et al., 2006), CLR (Faith et al., 2007), GeneNet (Opgen-Rhein and Strimmer, 2007), GENIE3 (Huynh-Thu et al., 2010), NARROMI (Zhang et al., 2013), Pearson, Spearman, and a modified implementation of TIGRESS (Haury et al., 2012); their results were aggregated into a consensus network using the Top1 method (Hase et al., 2013). An assessment of the scale-free property of the consensus network, fitting a heavy-tailed distribution using a log-log linear model, was performed at several candidate thresholds in the range of 0.9999 to 1 with a step size of 1e-5. At the same steps, the network transitivity or average cluster criterion was calculated. The best threshold was chosen at 0.99997 with an R2 scale-free fit of 0.95 and an average cluster criterion of 0.4. The obtained network was further visualized and processed using the Cytoscape (Shannon et al., 2003) tool. The network was further partitioned using Infomap (Rosvall and Bergstrom, 2008), with the parameters -z -i link-list–markov-time 0.01. These methods were combined into a software utility suite named seidr (Schiffthaler et al., 2018).
PCA Loading Analysis
Using the modeled vst expression data, a PCA was performed using the R prcomp function. The rotations (or loadings) from the first three components were selected. All followed an acceptable Gaussian distribution. The genes having rotations more extreme than the mean ± 3 sd of the corresponding component were extracted. These three lists of genes were then analyzed for GO enrichment using ConGenIE.org (Sundell et al., 2015), and the enrichment results were summarized and visualized using REVIGO (http://revigo.irb.hr; Supek et al., 2011) websites.
Analysis of Winter Genes
The filtered, modeled vst data were split by tissue and subjected to hierarchical clustering. A manual observation of the obtained dendrogram revealed five clusters that we could assign as season specific for both tissues and that we designated as Summer, Summer-early-late, Autumn, Winter, and Spring and as Summer-early, Summer-late, Autumn, Winter, and Spring for cambium/phloem and xylem, respectively. The season specificity of every gene was calculated using the tissue specificity score, which ranged from 0 to 1, indicating ubiquitous to specific expression, respectively (Yanai et al., 2005). A total of 9,462 cambium/phloem and 9,546 xylem genes had their highest abundance in the winter. We observed a systematic pattern of fewer low-abundance genes and a set of consistently higher abundance genes during the winter compared with other periods (Supplemental Fig. S2). To account for this, we only considered genes with a change greater than the observed effect (log2 fold change > 2) as significantly different, resulting in 207 and 217 genes with a season expression specificity score > 0.6 in cambium/phloem and xylem, respectively (Supplemental Table S6), of which 117 were in common. GO enrichment analysis was performed as described above. The scripts for this analysis are available in the above-mentioned Git repository as manuscripts/Jokipii-Lukkari2017/src/R/seasonalAbundanceEffectAssessment.html and manuscripts/Jokipii-Lukkari2017/src/R/winterGenes.html.
FTL2 Gene in Silico Validation
Approximately 30% of annotated genes were predicted to be fragmented across two or more scaffolds in the current spruce genome assembly (Nystedt et al., 2013). A tBLASTn search against the Norway spruce genome assembly (performed at ConGenIE; Sundell et al., 2015) using the Norway spruce FLT2 protein sequence (Nystedt et al., 2013) identified two genes: MA_720135g0010 and MA_5386467g0010. Both genes are classified as medium confidence, a classification that indicates that their respective sequence covers between 30% and 70% of an existing protein in the National Center for Biotechnology Information nonredundant database (Nystedt et al., 2013), suggesting fragmentation of the FLT2 gene. Using paired-end read alignments, where the forward and reverse reads of single paired-end reads align to different scaffolds (reported as chimeric alignments by STAR), there was evidence supporting a connection between the two identified scaffolds in the genome assembly. This was further supported by the high correlation of expression (Pearson’s correlation of 0.97) between the two gene models.
Phylogenetic and Expression Analyses of Norway Spruce Genes Putatively Related to Cell Division, Secondary Cell Wall Formation, and PCD of Tracheids
tBLASTn searches using the protein sequences of genes known or suggested to be involved in cell division (Porceddu et al., 2001), phloem and xylem differentiation (Miyashima et al., 2013), cell expansion (Marowa et al., 2016), cellulose biosynthesis of SCWs (Endler and Persson, 2011), monolignol biosynthesis (Vanholme et al., 2013; Wang et al., 2014), polymerization (Gabaldón et al., 2005; Liang et al., 2006; Berthet et al., 2011; Fernández-Pérez et al., 2015), and PCD (Ito and Fukuda, 2002; Turner et al., 2007; Avci et al., 2008; Helm et al., 2008; Ondzighi et al., 2008; Ohashi-Ito et al., 2010; Bollhöner et al., 2013) were performed against the v1.0 Gene Prediction and contaminant-free Trinity Transcript Assembly databases at ConGenIE.org. Gene family members of the identified gene models were extracted from the Gene Family tab of a representative gene model per family in ConGenIE.org. Thereafter, the nucleotide sequences of gene models and transcripts were aligned with corresponding Arabidopsis (Arabidopsis thaliana), Populus trichocarpa, and Selaginella moellendorffii nucleotide sequences in Clustal Omega (Sievers et al., 2011), and any aberrant sequences (e.g. truncated or those missing conserved amino acids; Zhao et al., 2000; Vercammen et al., 2004; Triques et al., 2007; Hillwig et al., 2010; Richau et al., 2012; Supplemental Table S4) were manually identified and subsequently discarded from further analyses.
The phylogenetic trees were created in Galaxy (Goecks et al., 2010), which provides access to the genomes and associated annotations hosted at PlantGenIE.org. The PlantGenIE Galaxy workflow was constructed using the OSIRIS suite (Oakley et al., 2014), allowing users to create phylogenetic trees with given input sequences. The workflow utilized MUSCLE version 3.8.31 (maximum number of iterations, 16) for multiple alignment and PhyML 3.1 (substitution model, WAG; aLRT test, SH-like; tree topology search operation, Nearest Neighbor Interchange) and Tree Vector for building and drawing phylogenetic trees, respectively. In the case of lignin genes, the mapping of sequencing reads derived from xylem samples was examined with Integrated Genomics Viewer (https://www.broadinstitute.org/igv/), and only the genes showing expression in xylem were selected for further analyses. Subsequently, coregulation and expression profiles of the putative Norway spruce orthologs were studied with heat.map2 and the ggplot2 function in R, respectively.
Cloning of the AtMC9 Homolog, PaMC5
A sample representing the date when the first dead tracheids were observed in maturing xylem was used for cloning. Total RNA was amplified with the MessageAmp II Amplification Kit (Ambion) and transcribed into cDNA with SuperScript II Reverse Transcriptase (Invitrogen) from random hexamers. The primers used to amplify the AtMC9 homolog were designed based on the transcript comp95097_c0_seq3, showing the highest sequence similarity from the Norway spruce sequences. The transcript was associated, along with two other mRNA sequences, to PaMC5 in the phylogenetic analyses of Minina et al. (2013). The coding sequence of PaMC5 was amplified with primer pair 5′-CACCATGGGCAACAACGACAACAAGAAATAC-3′ (forward, underlined bases added for directional cloning) and 5′-TCAGTGCTTCGCGGAGTCCTCCTC-3′ (reverse) using Phusion High-Fidelity DNA polymerase in a two-step PCR with an extension temperature of 72°C. The resultant PCR product was gel purified, cloned into the pENTR/D-TOPO vector, and sequenced at Eurofins Genomics.
Chemical Analyses
After scraping the material for RNA sequencing, the dead earlywood of the contemporary year ring was scraped for Py-GC/MS analysis, freeze dried, and ball milled with an MM400 mixer mill (Retch; 2 min, 30 Hz). Aliquots of RNA sequencing samples (that represented the living part of the xylem) also were included into the analyses. A total of 50 ± 10 µg of ball-milled wood powder was applied to a pyrolyzer equipped with an autosampler (PY-2020iD and AS-1020E; Frontier Laboratories) connected to a GC/MS device (7890A/5975C; Agilent Technologies). The pyrolysate was separated and analyzed according to Gerber et al. (2012). The statistical analysis for the correlation between sampling time and relative lignin content was analyzed by linear regression using the lm function.
Transmission EM
The stem pieces for EM were fixed in 2.5% (w/v) glutaraldehyde in 0.1 m cacodylate buffer overnight at 4°C. After washing three times for 10 min in the buffer, the specimens were postfixed in 1% (w/v) osmium tetroxide in water for 2 h. The samples were dehydrated with 50%, 70%, 80%, 90%, and 100% ethanol and infiltrated and embedded in Spurr resin. The ultrathin sections of 80 nm, prepared as described by Bollhöner et al. (2013), were examined in a JEM 1230 (JEOL) transmission electron microscope. Micrographs were captured with an MSC 600 CW (Gatan) camera.
Accession Numbers
The obtained RNA sequencing data were deposited to the European Nucleotide Archive and are accessible under accession number PRJEB9578 (ERP010702). The nucleotide sequence of the PaMC5 coding region was deposited into the National Center for Biotechnology Information database under accession number KP058500.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment state of preprocessed reads for all the libraries.
Supplemental Figure S2. Seasonal expression variability of mRNA.
Supplemental Figure S3. Companion heat maps to Figure 2.
Supplemental Figure S4. REVIGO Treemap of the cambium/phloem winter gene GO enrichment.
Supplemental Figure S5. Phylogeny of CDKB genes.
Supplemental Figure S6. Phylogeny of EXPA genes.
Supplemental Figure S7. Phylogeny of the APL gene.
Supplemental Figure S8. Phylogeny of the CNA gene.
Supplemental Figure S9. Phylogeny of SCW CESA genes.
Supplemental Figure S10. Hierarchical clustering of genes putatively related to lignification based on their expression in xylem samples.
Supplemental Figure S11. Phylogeny of cinnamyl alcohol dehydrogenase genes.
Supplemental Figure S12. Phylogeny of CCoAOMT genes.
Supplemental Figure S13. Phylogeny of cinnamoyl-CoA reductase and CCR-like genes.
Supplemental Figure S14. Phylogeny of C3H1 and F5H genes.
Supplemental Figure S15. Phylogeny of the C4H gene.
Supplemental Figure S16. Phylogeny of 4CL and 4CL-like genes.
Supplemental Figure S17. Phylogeny of COMT and COMT-like genes.
Supplemental Figure S18. Phylogeny of the caffeoyl shikimic acid esterase gene.
Supplemental Figure S19. Phylogeny of the hydroxycinnamoyl-CoA shikimate hydroxycinnamoyl transferase gene.
Supplemental Figure S20. Phylogeny of PAL genes.
Supplemental Figure S21. Phylogeny of LAC genes.
Supplemental Figure S22. Phylogeny of PRX genes.
Supplemental Figure S23. Phylogeny of the AS2 gene.
Supplemental Figure S24. Phylogeny of MYB genes.
Supplemental Figure S25. Phylogeny of the XSP1 gene.
Supplemental Figure S26. Phylogeny of type I and type II metacaspase genes.
Supplemental Figure S27. Phylogeny of papain-like Cys protease genes CEP, RD21A, XCP, and XBCP3.
Supplemental Figure S28. Phylogeny of RNSI genes.
Supplemental Figure S29. Phylogeny of ENDO genes.
Supplemental Figure S30. Sampling of the trees.
Supplemental Table S1. Onset of earlywood cell death and latewood formation.
Supplemental Table S2. Genes expressed only in specific tissues.
Supplemental Table S3. Genes putatively related to latewood formation.
Supplemental Table S4. PCD-related candidate genes used in this study.
Supplemental Table S5. Genes showing differential expression (adjusted P < 0.01) during the visual appearance of dead earlywood cells.
Supplemental Table S6. List of annotated winter genes.
Supplemental Table S7. Sampling dates.
Supplemental Table S8. RNA-Seq reads preprocessing summary.
Acknowledgments
We thank the National Genomics Infrastructure, funded by the Swedish Research Council, and the Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. We also thank the plant cell wall and carbohydrate analytical facility at Umeå Plant Science Centre/Sveriges Lantbruksuniversitet, supported by Bio4Energy and the TC4F project, for the Py-GC/MS analysis.
Footnotes
This work was supported by funds from the Knut and Alice Wallenberg Foundation (KAW 2013.0305), the Kempe Foundation (SMK-1340), the Swedish Research Council VR (621-2013-4949), the Swedish Governmental Agency for Innovation Systems Vinnova (2015-02290), and the Foundation for Forest Tree Breeding (Finland).
References
- Anagnost SE, Mark RE, Hanna RB (2002) Variation of microfibril angle within individual tracheids. Wood Fiber Sci 34: 337–349 [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avci U, Petzold HE, Ismail IO, Beers EP, Haigler CH (2008) Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J 56: 303–315 [DOI] [PubMed] [Google Scholar]
- Barnett JR. (1992) Reactivation of the cambium in Aesculus hippocastanum L.: a transmission electron microscope study. Ann Bot 70: 169–177 [Google Scholar]
- Baucher M, Monties B, Van Montagu M, Boerjan W (1998) Biosynthesis and genetic engineering of lignin. Crit Rev Plant Sci 17: 125–197 [Google Scholar]
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C (2007) Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134: 1691–1701 [DOI] [PubMed] [Google Scholar]
- Beaulieu J, Doerksen T, Boyle B, Clément S, Deslauriers M, Beauseigle S, Blais S, Poulin PL, Lenz P, Caron S, et al. (2011) Association genetics of wood physical traits in the conifer white spruce and relationships with gene expression. Genetics 188: 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum S, Nakaba S, Yamagishi Y, Yamane K, Islam MA, Oribe Y, Ko JH, Jin HO, Funada R (2012) A rapid decrease in temperature induces latewood formation in artificially reactivated cambium of conifer stems. Ann Bot 110: 875–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn GP, Miksche JP (1976) Botanical Microtechnique and Cytochemistry. Iowa State University Press, Ames [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol I, Raymond A, Jackman SD, Pleasance S, Coope R, Taylor GA, Yuen MM, Keeling CI, Brand D, Vandervalk BP, et al. (2013) Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29: 1492–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollhöner B, Prestele J, Tuominen H (2012) Xylem cell death: emerging understanding of regulation and function. J Exp Bot 63: 1081–1094 [DOI] [PubMed] [Google Scholar]
- Bollhöner B, Zhang B, Stael S, Denancé N, Overmyer K, Goffner D, Van Breusegem F, Tuominen H (2013) Post mortem function of AtMC9 in xylem vessel elements. New Phytol 200: 498–510 [DOI] [PubMed] [Google Scholar]
- Borgström E, Lundin S, Lundeberg J (2011) Large scale library generation for high throughput sequencing. PLoS ONE 6: e19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabane M, Afif D, Hawkins S (2012) Lignins and abiotic stresses. Adv Bot Res 61: 220–262 [Google Scholar]
- Carlquist S. (2001) Comparative Wood Anatomy: Systematic, Ecological, and Evolutionary Aspects of Dicotyledon Wood. Springer-Verlag, Berlin [Google Scholar]
- Chalfun-Junior A, Franken J, Mes JJ, Marsch-Martinez N, Pereira A, Angenent GC (2005) ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol Biol 57: 559–575 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Chen JH, Ho CT (1997) Antioxidant activities of caffeic acid and its related hydroxycinnamic acid compounds. J Agric Food Chem 45: 2374–2378 [Google Scholar]
- Cheynier V, Comte G, Davies KM, Lattanzio V, Martens S (2013) Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol Biochem 72: 1–20 [DOI] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P (2010) Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- Courtois-Moreau CL, Pesquet E, Sjödin A, Muñiz L, Bollhöner B, Kaneda M, Samuels L, Jansson S, Tuominen H (2009) A unique program for cell death in xylem fibers of Populus stem. Plant J 58: 260–274 [DOI] [PubMed] [Google Scholar]
- Cronn R, Dolan PC, Jogdeo S, Wegrzyn JL, Neale DB, St Clair JB, Denver DR (2017) Transcription through the eye of a needle: daily and annual cyclic gene expression variation in Douglas-fir needles. BMC Genomics 18: 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Sinharoy S, Udvardi M, Zhao PX (2013) PlantTFcat: an online plant transcription factor and transcriptional regulator categorization and analysis tool. BMC Bioinformatics 14: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieset A. (2011) Genetic variation of xylem formation in Norway spruce (Picea abies (L.) Karst.) clones with contrasting growth rhythm. Master thesis. Norwegian University of Life Sciences, As, Norway [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domec JC, Gartner BL (2002) How do water transport and water storage differ in coniferous earlywood and latewood? J Exp Bot 53: 2369–2379 [DOI] [PubMed] [Google Scholar]
- Endler A, Persson S (2011) Cellulose synthases and synthesis in Arabidopsis. Mol Plant 4: 199–211 [DOI] [PubMed] [Google Scholar]
- Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS (2007) Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol 5: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Pérez F, Vivar T, Pomar F, Pedreño MA, Novo-Uzal E (2015) Peroxidase 4 is involved in syringyl lignin formation in Arabidopsis thaliana. J Plant Physiol 175: 86–94 [DOI] [PubMed] [Google Scholar]
- Friedmann M, Ralph SG, Aeschliman D, Zhuang J, Ritland K, Ellis BE, Bohlmann J, Douglas CJ (2007) Microarray gene expression profiling of developmental transitions in Sitka spruce (Picea sitchensis) apical shoots. J Exp Bot 58: 593–614 [DOI] [PubMed] [Google Scholar]
- Gabaldón C, López-Serrano M, Pedreño MA, Barceló AR (2005) Cloning and molecular characterization of the basic peroxidase isoenzyme from Zinnia elegans, an enzyme involved in lignin biosynthesis. Plant Physiol 139: 1138–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan PB. (1984) Plant Histochemistry and Cytochemistry. Academic Press, London [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber L, Eliasson M, Trygg J, Moritz T, Sundberg B (2012) Multivariate curve resolution provides a high-throughput data processing pipeline for pyrolysis-gas chromatography/mass spectrometry. J Anal Appl Pyrolysis 95: 95–100 [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase T, Ghosh S, Yamanaka R, Kitano H (2013) Harnessing diversity towards the reconstructing of large scale gene regulatory networks. PLOS Comput Biol 9: e1003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haury AC, Mordelet F, Vera-Licona P, Vert JP (2012) TIGRESS: Trustful Inference of Gene REgulation using Stability Selection. BMC Syst Biol 6: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T, Woltering EJ, Gallois P (2008) Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J Biol Chem 283: 774–783 [DOI] [PubMed] [Google Scholar]
- Heide OM. (1974) Growth and dormancy in Norway spruce ecotypes (Picea abies). I. Interaction of photoperiod and temperature. Physiol Plant 30: 1–12 [Google Scholar]
- Helm M, Schmid M, Hierl G, Terneus K, Tan L, Lottspeich F, Kieliszewski MJ, Gietl C (2008) KDEL-tailed cysteine endopeptidases involved in programmed cell death, intercalation of new cells, and dismantling of extensin scaffolds. Am J Bot 95: 1049–1062 [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Liu X, Liu G, Thornburg RW, Macintosh GC (2010) Petunia nectar proteins have ribonuclease activity. J Exp Bot 61: 2951–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P (2010) Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 5: e12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa H, Ishida S (1972) Study on the wood formation in trees. Report III. Occurrence of the overwintering cells in cambial zone in several ring-porous trees. Res Bull Coll Exp For Hokkaido Univ 27: 373–393 [Google Scholar]
- Isikgor FH, Becer CR (2015) Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers. Polym Chem 6: 4497–4559 [Google Scholar]
- Ito J, Fukuda H (2002) ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 14: 3201–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas KM, Cvikrova M, Palagiewicz A, Eder J (2000) Alterations in phenylpropanoid content in soybean roots during low temperature acclimation. Plant Physiol Biochem 68: 1674–1682 [Google Scholar]
- Jokipii-Lukkari S, Sundell D, Nilsson O, Hvidsten TR, Street NR, Tuominen H (2017) NorWood: a gene expression resource for evo-devo studies of conifer wood development. New Phytol 216: 482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlgren A, Gyllenstrand N, Clapham D, Lagercrantz U (2013) FLOWERING LOCUS T/TERMINAL FLOWER1-like genes affect growth rhythm and bud set in Norway spruce. Plant Physiol 163: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst M, Johnson AF, Baucom C, Ulrich E, Hubbard K, Staggs R, Paule C, Retzel E, Whetten R, Sederoff R (2003) Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda L.) with Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 7383–7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klintenäs M, Pin PA, Benlloch R, Ingvarsson PK, Nilsson O (2012) Analysis of conifer FLOWERING LOCUS T/TERMINAL FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytol 196: 1260–1273 [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H (2012) SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28: 3211–3217 [DOI] [PubMed] [Google Scholar]
- Küffner R, Petri T, Tavakkolkhah P, Windhager L, Zimmer R (2012) Inferring gene regulatory networks by ANOVA. Bioinformatics 28: 1376–1382 [DOI] [PubMed] [Google Scholar]
- Kumar M, Saranpää P, Barnett JR, Wilkinson MJ (2009) Juvenile-mature wood transition in pine: correlation between wood properties and candidate gene expression profiles. Euphytica 166: 341–355 [Google Scholar]
- Laakso K, Sullivan JH, Huttunen S (2000) The effects of UV-B radiation on epidermal anatomy in loblolly pine (Pinus taeda L.) and Scots pine (Pinus sylvestris L.). Plant Cell Environ 23: 461–472 [Google Scholar]
- Laitinen T, Morreel K, Delhomme N, Gauthier A, Schiffthaler B, Nickolov K, Brader G, Lim KJ, Teeri TH, Street NR, et al. (2017) A key role for apoplastic H2O2 in Norway spruce phenolic metabolism. Plant Physiol 174: 1449–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cheng XF, Leshkevich J, Umezawa T, Harding SA, Chiang VL (2001) The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13: 1567–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Popko JL, Zhang XH, Osakabe K, Tsai CJ, Joshi CP, Chiang VL (1997) A novel multifunctional O-methyltransferase implicated in a dual methylation pathway associated with lignin biosynthesis in loblolly pine. Proc Natl Acad Sci USA 94: 5461–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WF, Ding Q, Chen JJ, Cui KM, He XQ (2009) Induction of PtoCDKB and PtoCYCB transcription by temperature during cambium reactivation in Populus tomentosa Carr. J Exp Bot 60: 2621–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Davis E, Gardner D, Cai X, Wu Y (2006) Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Lim KJ, Paasela T, Harju A, Venäläinen M, Paulin L, Auvinen P, Kärkkäinen K, Teeri TH (2016) Developmental changes in Scots pine transcriptome during heartwood formation. Plant Physiol 172: 1403–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodha M, Marco CF, Timmermans MCP (2013) The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev 27: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin S, Stranneheim H, Pettersson E, Klevebring D, Lundeberg J (2010) Increased throughput by parallelization of library preparation for massive sequencing. PLoS ONE 5: e10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A (2006) ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics (Suppl 1) 7: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marowa P, Ding A, Kong Y (2016) Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep 35: 949–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S (2003) The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina EA, Filonova LH, Fukada K, Savenkov EI, Gogvadze V, Clapham D, Sanchez-Vera V, Suarez MF, Zhivotovsky B, Daniel G, et al. (2013) Autophagy and metacaspase determine the mode of cell death in plants. J Cell Biol 203: 917–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Sebastian J, Lee JY, Helariutta Y (2013) Stem cell function during plant vascular development. EMBO J 32: 178–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmanis L, Sachs IB (1969) Seasonal development of secondary xylem in Pinus Strobus L. Wood Sci Technol 3: 177–193 [Google Scholar]
- Neale DB, Wegrzyn JL, Stevens KA, Zimin AV, Puiu D, Crepeau MW, Cardeno C, Koriabine M, Holtz-Morris AE, Liechty JD, et al. (2014) Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol 15: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Oakley TH, Alexandrou MA, Ngo R, Pankey MS, Churchill CKC, Chen W, Lopker KB (2014) Osiris: accessible and reproducible phylogenetic and phylogenomic analyses within the Galaxy workflow management system. BMC Bioinformatics 15: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Oda Y, Fukuda H (2010) Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22: 3461–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Van Bel M, Van Hautegem T, Fendrych M, Huysmans M, Simaskova M, van Durme M, Buscaill P, Rivas S, Coll NS, et al. (2015) A conserved core of programmed cell death indicator genes discriminates developmentally and environmentally induced programmed cell death in plants. Plant Physiol 169: 2684–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondzighi CA, Christopher DA, Cho EJ, Chang SC, Staehelin LA (2008) Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 20: 2205–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opgen-Rhein R, Strimmer K (2007) From correlation to causation networks: a simple approximate learning algorithm and its application to high-dimensional plant gene expression data. BMC Syst Biol 1: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oribe Y, Funada R, Shibagaki M, Kubo T (2001) Cambial reactivation in locally heated stems of the evergreen conifer Abies sachalinensis (Schmidt) masters. Planta 212: 684–691 [DOI] [PubMed] [Google Scholar]
- Oshima Y, Mitsuda N (2013) The MIXTA-like transcription factor MYB16 is a major regulator of cuticle formation in vegetative organs. Plant Signal Behav 8: e26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palle SR, Seeve CM, Eckert AJ, Cumbie WP, Goldfard B, Loopstra CA (2011) Natural variation in expression of genes involved in xylem development in loblolly pine (Pinus taeda L.). Tree Genet Genomes 7: 193–206 [Google Scholar]
- Palle SR, Seeve CM, Eckert AJ, Wegrzyn JL, Neale DB, Loopstra CA (2013) Association of loblolly pine xylem development gene expression with single-nucleotide polymorphisms. Tree Physiol 33: 763–774 [DOI] [PubMed] [Google Scholar]
- Patzlaff A, Newman LJ, Dubos C, Whetten RW, Smith C, McInnis S, Bevan MW, Sederoff RR, Campbell MM (2003) Characterisation of Pt MYB1, an R2R3-MYB from pine xylem. Plant Mol Biol 53: 597–608 [DOI] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheld JP, Segers G, De Veylder L, Barroco RP, Casteels P, Van Montagu M, Inzé D, Mironov V (2001) A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J Biol Chem 276: 36354–36360 [DOI] [PubMed] [Google Scholar]
- Qiu Z, Wan L, Chen T, Wan Y, He X, Lu S, Wang Y, Lin J (2013) The regulation of cambial activity in Chinese fir (Cunninghamia lanceolata) involves extensive transcriptome remodeling. New Phytol 199: 708–719 [DOI] [PubMed] [Google Scholar]
- Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, et al. (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344: 1246843. [DOI] [PubMed] [Google Scholar]
- Ralph SG, Chun HJE, Kolosova N, Cooper D, Oddy C, Ritland CE, Kirkpatrick R, Moore R, Barber S, Holt RA, et al. (2008) A conifer genomics resource of 200,000 spruce (Picea spp.) ESTs and 6,464 high-quality, sequence-finished full-length cDNAs for Sitka spruce (Picea sitchensis). BMC Genomics 9: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
- Richau KH, Kaschani F, Verdoes M, Pansuriya TC, Niessen S, Stüber K, Colby T, Overkleeft HS, Bogyo M, Van der Hoorn RAL (2012) Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol 158: 1583–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall M, Bergstrom CT (2008) Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci USA 105: 1118–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvas R. (1969) Genetical adaptation of forest trees to the heat factor of the climate. In Second World Consultation on Forest Tree Breeding, Washington, DC, USA, 7–16 August 1969. FAO, Rome: pp. 187–202 [Google Scholar]
- Schiffthaler B, Serrano A, Delhomme N, Street NR (2018) Seidr: a toolkit for calculation of crowd networks. bioRxiv [Google Scholar]
- Schnitzler JP, Jungblut TP, Feicht C, Köfferlein M, Langebartels C, Heller W, Sandermann H Jr (1997) UV-B induction of flavonoid biosynthesis in Scots pine (Pinus sylvestris L.) seedlings. Trees (Berl) 11: 162–168 [Google Scholar]
- Schrader J, Moyle R, Bhalerao R, Hertzberg M, Lundeberg J, Nilsson P, Bhalerao RP (2004) Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J 40: 173–187 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Li C, Tarczynski MC (2002) High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-L-methionine synthetase 3 gene. Plant J 29: 371–380 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnhuber BM, Stiller G, Ruhnke R, von Clarmann T, Kellmann S, Aschmann J (2011) Arctic winter 2010/2011 at the brink of an ozone hole. Geophys Res Lett 38: L24814 [Google Scholar]
- Sperry JS, Hacke UG, Pittermann J (2006) Size and function in conifer tracheids and angiosperm vessels. Am J Bot 93: 1490–1500 [DOI] [PubMed] [Google Scholar]
- Stevens KA, Wegrzyn JL, Zimin A, Puiu D, Crepeau M, Cardeno C, Paul R, Gonzalez-Ibeas D, Koriabine M, Holtz-Morris AE, et al. (2016) Sequence of the sugar pine megagenome. Genetics 204: 1613–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez MF, Filonova LH, Smertenko A, Savenkov EI, Clapham DH, von Arnold S, Zhivotovsky B, Bozhkov PV (2004) Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr Biol 14: R339–R340 [DOI] [PubMed] [Google Scholar]
- Sundell D, Mannapperuma C, Netotea S, Delhomme N, Lin YC, Sjödin A, Van de Peer Y, Jansson S, Hvidsten TR, Street NR (2015) The Plant Genome Integrative Explorer Resource: PlantGenIE.org. New Phytol 208: 1149–1156 [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triques K, Sturbois B, Gallais S, Dalmais M, Chauvin S, Clepet C, Aubourg S, Rameau C, Caboche M, Bendahmane A (2007) Characterization of Arabidopsis thaliana mismatch specific endonucleases: application to mutation discovery by TILLING in pea. Plant J 51: 1116–1125 [DOI] [PubMed] [Google Scholar]
- Turner S, Gallois P, Brown D (2007) Tracheary element differentiation. Annu Rev Plant Biol 58: 407–433 [DOI] [PubMed] [Google Scholar]
- van Baarlen P, Woltering EJ, Staats M, van Kan JAL (2007) Histochemical and genetic analysis of host and non-host interactions of Arabidopsis with three Botrytis species: an important role for cell death control. Mol Plant Pathol 8: 41–54 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inzé D, Van Breusegem F (2004) Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem 279: 45329–45336 [DOI] [PubMed] [Google Scholar]
- Wagner A, Tobimatsu Y, Phillips L, Flint H, Geddes B, Lu F, Ralph J (2015) Syringyl lignin production in conifers: proof of concept in a pine tracheary element system. Proc Natl Acad Sci USA 112: 6218–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, Naik PP, Chen HC, Shi R, Lin CY, Liu J, Shuford CM, Li Q, Sun YH, Tunlaya-Anukit S, et al. (2014) Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26: 894–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Lam E (2005) Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J Biol Chem 280: 14691–14699 [DOI] [PubMed] [Google Scholar]
- Weng JK, Chapple C (2010) The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, et al. (2005) Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21: 650–659 [DOI] [PubMed] [Google Scholar]
- Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z, Li F, Lu H (2014) The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26: 2939–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu K, Liu ZP, Duval B, Richer JM, Zhao XM, Hao JK, Chen L (2013) NARROMI: a noise and redundancy reduction technique improves accuracy of gene regulatory network inference. Bioinformatics 29: 106–113 [DOI] [PubMed] [Google Scholar]
- Zhao C, Johnson BJ, Kositsup B, Beers EP (2000) Exploiting secondary growth in Arabidopsis: construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol 123: 1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA (2013) Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann MH, Brown CL (1971) Trees: Structure and Function. Springer-Verlag, New York [Google Scholar]