Abstract
Safranal, a dominant component of saffron, is known to have antitumor, cytotoxic, and antibacterial properties. In this study, we examined safranal and its structural analogs— thymol, carvacrol, damascenone, cuminol, 2,6,6-Trimethyl-2-cyclohexene-1,4-dione (TMCHD), 4-Isopropylbenzyl bromide (IPBB), and 4-tert-Butylphenol (TBP) induced inhibition of Escherichia coli membrane bound F1Fo ATP synthase. Safranal and its analogs inhibited wild-type enzyme to variable degrees. While safranal caused 100% inhibition of wild-type F1Fo ATP synthase only, about 50% inhibition occurred for αR283D mutant ATP synthase. Moreover, safranal, thymol, carvacrol, damascenone, cuminol, TMCHD, IPBB, and TBP all fully abrogated the growth of wild-type E. coli cells and had partial or no effect on the growth of null and mutant E. coli strains. Therefore, the antimicrobial properties of safranal, thymol, carvacrol, damascenone, cuminol, TMCHD, IPBB, and TBP can be linked to their binding and inhibition of ATP synthase. Total loss of growth in wild-type and partial or no growth loss in null or mutant E. coli strains demonstrates that ATP synthase is a molecular target for safranal and its structural analogs. Partial inhibition of the αArg-283 mutant enzyme establishes that αArg-283 residue is required in the polyphenol binding pocket of ATP synthase for the binding of safranal. Furthermore, partial growth loss for the null and mutant strains in the presence of inhibitors also suggests the role of other targets and residues in the process of inhibition.
Keywords: E. coli ATP synthase, F1Fo-ATP synthase, ATP synthesis, safranal, thymol, cuminol, carvacrol, damascenone
Introduction
The highly conserved F1Fo ATP synthase, also known as smallest biological nanomotor, is the principal source of ATP the energy currency for all organisms [1]. ATP generation and hydrolysis occur on three catalytic sites of the water-soluble F1 sector, while protons move through the membrane-bound Fo sector [2]. In a simplified scheme, movement of protons causes the rotation of the γ-subunit, resulting in conformational changes in the α/β-subunits which in turn result in ATP synthesis or hydrolysis depending on the direction of the proton gradient. The basic reaction mechanism is ATP synthase + ADP + Pi ↔ ATP synthase + ATP [3–6].
Malfunction of ATP synthase is linked to many human disease conditions including Alzheimer’s disease, Parkinson’s disease, Batten disease, Leigh syndrome, and mitochondrial myopathies. ATP synthase has also been shown to be a useful and effective molecular drug target particularly against microbial infections and tumor progression [7]. Both F1 and Fo sectors of ATP synthase containing α3β3γδε and ab2c10–14 subunits, respectively, possess multiple inhibitor binding sites. A wide range of phytochemicals including phenolic compounds with antioxidants, chemotherapeutic, and antimicrobial properties bind and inhibit ATP synthase [7–15].
Currently, about 700,000 people die every year from microbial infections, and by 2050 antibiotic resistant microbial infections will result in ten million additional deaths worldwide per year [16]. Thus, superbugs are expected to become the top global killer, surpassing cancer. The impact of this public health crisis on the global economy is projected to have a staggering cost of $100 trillion [17]. The World Health Organization’s global report on surveillance of antimicrobial resistance estimated the yearly cost to the US health system would reach $34 billion dollars [18]. The fast encroaching antibiotic resistance by microbes in general and Escherichia coli in particular is the main reason for this alarming situation. Finding new ways to kill microbes is of paramount importance. Natural compounds from plants, vegetables, herbs, and spices that selectively bind and inhibit ATP synthase present an excellent opportunity for preventing and combating antibiotic resistant microbial infections.
ATP synthase in general and overexpression of ectopic ATP synthase in pathophysiological conditions has augmented the interest in F1Fo ATP synthase as molecular drug target. ATP synthase provides both a viable molecular drug target to counteract chronic infections sustained by therapeutically defiant bacterial strains and to cure mammalian diseases with mitochondrial dysfunctions [13]. Moreover, a wide variety of natural and synthetic chemicals have been shown to bind both F1 and Fo sectors of ATP synthase [7, 14, 19–22].
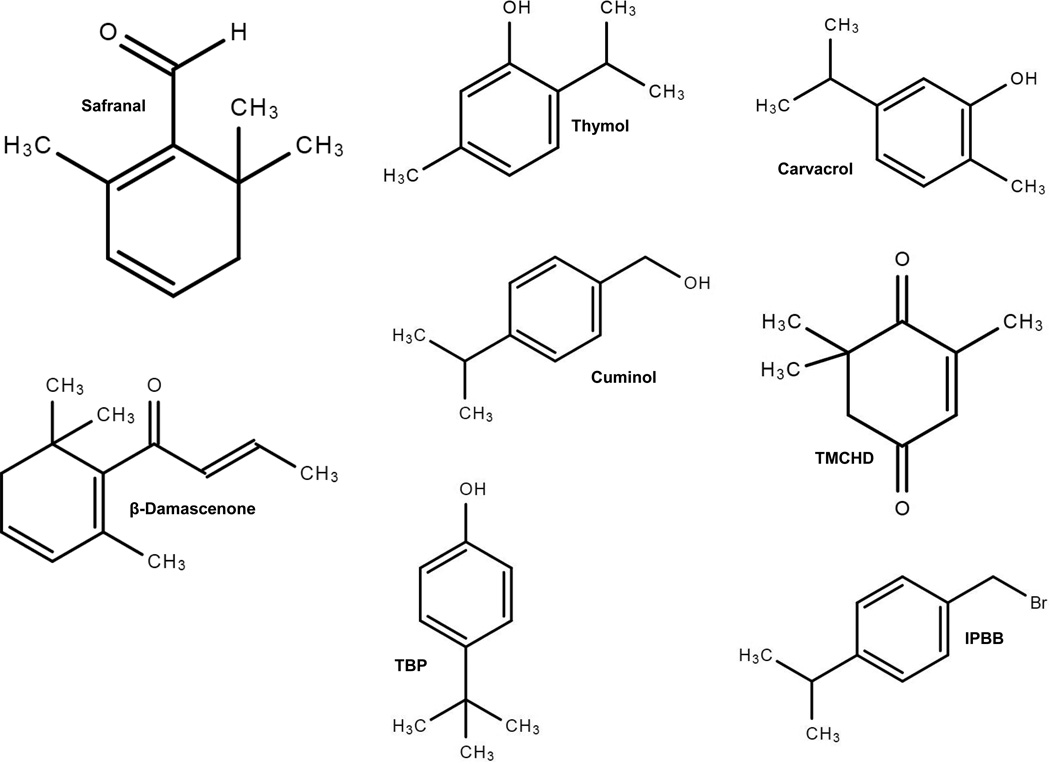
For centuries, saffron (the stigmata of Crocus sativus L. flowers) has been used as a spice, food colorant, and natural therapeutic product [23]. The structures of saffron constituent safranal and its analogs are shown in Figure 1. Throughout the world, there seems a steady increase and interest in the use of natural products as antimicrobial agents individually or in combination with other such molecules [12, 24–29]. Numerous phytochemicals have been shown to have dietary benefits and are potential antitumor or antimicrobial agents [30–33].
Fig. 1. Structures of safranal, thymol, carvacrol, damascenone, cuminol, TMCHD (2,6,6-trimethyl-2-cyclohexene-1,4-dione, IPBB (4-isopropylbenzyl bromide), and TBP (4-tertbutylphenol).
Saffron and its components were found to be promising chemopreventive agents in general and against a wide spectrum of murine tumors and human leukemia cell lines in particular [34]. Safranal was reported to inhibit 50% of the growth of HeLa tumor cells at concentrations of 0.12 mg/mL [35]. Further, the beneficial effects of safranal on neurodegenerative disorders, such as Alzheimer’s and Parkinson's diseases, are mainly due to their interactions with cholinergic, dopaminergic, and glutamatergic systems. Both animal and human studies show that saffron and its constituents, such as safranal, are effective in the treatment of mild to moderate depression, potentially through interaction with the serotonin and noradrenaline systems [36].
Saffron and its constituents were also reported to modulate obesity and associated metabolic disorders. Saffron supplementation may be responsible for lowering the risk of over snacking associated with obesity, thus promoting weight loss in overweight individuals [37]. Consumption of saffron has also been correlated with a lower risk of many other ailments, such as cardiovascular disease, gastric distress, depression, insomnia, anxiety, and premenstrual syndrome [23].
Safranal (Fig. 1) is one of the major organic compounds found in saffron. The aroma of saffron is mainly due to the safranal which accounts for about 60–70% of the volatile fraction of saffron essential oil [38]. Multiple dietary health benefits, such as protection against diabetes [39], cataracts [40], and oxidative stress in the brain, liver, and kidney [41], have been documented for saffron in general and safranal in particular. Safranal has also been shown to have antioxidant, anticancer [42], and antibacterial [35, 43] properties. Safranal and another constituent of saffron crocin were found to have a bactericidal effect on E. coli, Staphylococcus aureus, and Salmonella enterica as well as on laboratory strains, such as S. aureus 6538P and S. enterica serovar Typhimurium LT2 [35]. Recently, biological and molecular modeling studies suggested that safranal is a promising agent against Helicobacter pylori and parasitic infections caused by Plasmodium and Leishmania [44].
To our knowledge, the mode of action by which safranal and its analogs cause bactericidal effect is unknown. Data from affinity-based target deconvolution of safranal showed that the potential cellular targets of safranal include β-actin, subunit-1 of cytochrome bc1 complex, β-subunit of mitochondrial trifunctional protein, and α/β-subunits of ATP synthase in the liver [45].
The 2,6,6-trimethyl-2-cyclohexene-1,4-dione (TMCHD) is a structural analog of safranal found in kanuka honey [46]. TMCHD is also known as 4-ketoisophorone and is a major component of the aroma of saffron [38]. Carvacrol, also known as cymophenol, possesses antifungal and [47] antimicrobial properties [48]. Carvacrol was shown to be effective against antibiotic resistant microbes [49] and to inhibit E. coli cells [50]. In Pseudomonas aeruginosa, the antibacterial action of carvacrol was suggested to result from the disruption of bacterial membrane [51]. Carvacrol is found in the essential oils of oregano, thyme, pepperwort, and bergamot.
Thymol is mainly found in the oil of thyme and is an isomer of carvacrol. Thymol is known to have many health benefits: it is a potent antioxidant [52], decreases cholesterol levels [53], and stops dental biofilm formation [54]. Thymol was also shown to have antifungal [47, 55] and antimicrobial properties [48]. Moreover, thymol and carvacrol are part of naturally occurring biocides. For example, thymol and carvacrol synergistically reduce the microbial resistance to antibiotics [48].
Cuminol or 4-isopropylbenzyl alcohol is found in cumin seeds. This compound was shown to increase insulin secretion [56], has antioxidant properties [57]. β-Damascenone is another structurally related analog of safranal and is found in sherry vinegar [58]. Damascenones, in general, are structurally related chemical compounds found in variety of essential oils. The aroma of roses is mainly due to β-damascenone. It has also been shown to have antiproliferative [59] and antispasmodic activity [60].
Although it was previously documented that safranal binds to the α- and β-subunits of ATP synthase [45], its mode of action is unknown. For this reason, we studied the inhibitory effects of safranal and its analogs— thymol, carvacrol, damascenone, cuminol, TMCHD, 4-Isopropylbenzyl bromide (IPBB), and 4-tert-Butylphenol (TBP)—on F1Fo ATP synthase and the growth of E. coli cells to determine if the dietary benefits of safranal or its structural analogs are linked to the binding and inhibition of ATP synthase. E. coli mutant αR283D was used to establish the binding site for safranal and its structural analogs. The results of this work, by pointing out the effects of safranal and related compounds on the E. coli ATP synthase, elicit a molecular mechanism which may constitute the biochemical bases of the antimicrobial properties of these natural compounds.
Materials and Methods
Safranal and its analogs
Safranal with ≥88% purity (W338907-25G-K, Sigma-Aldrich), thymol with 99% purity (150335000, Acros-Organics), carvacrol with >95% purity (C0026, Tokyo Chemical Industry), damascenone with Analytical Standard purity (41163-250MG, Sigma-Aldrich), cuminol with 97% purity (187801000, Acros-Organics), TMCHD with ≥98% purity (L10199, Alfa Aesar), IPBB with 97% purity (187801000, Acros-Organics), and TBP with 97% purity (108000050, Acros-Organics) were used in this study. Many were a liquid at room temperature and were dissolved in dimethyl sulfoxide (DMSO) to obtain the desired working solution. In ATPase assays, the maximal volume of DMSO used was 3.64%. In this study and previous studies, we noted that up to 40% DMSO by itself has no effect on membrane-bound F1Fo of E. coli ATP synthase [61, 62]. All other chemicals used in this study were ultrapure, analytical grade purchased from Sigma-Aldrich Chemical Company or Fisher Scientific Company.
Construction of E. coli wild type, null, and mutant strains
The E. coli wild-type strain used in all experiments was pBWU13.4/DK8 [63]. The null strain was pUC118/DK8. E. coli αR283D mutant strain pZA84 was generated by Stratagene QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, catalog #210519-12) using CGTCCGCCAGGAGATGAAGCTTTCCCGGGCGAC primer. Bold letters show the change.
Measurement of growth in limiting glucose; preparation of E. coli membrane-bound F1Fo ATP synthase; ATPase activity assays
Oxidative and substrate-level phosphorylation were measured by growth yield on fermentable carbon source limiting glucose (3–5 mM) and non-fermentable carbon source succinate as in [64]. On succinate, the null strain shows no growth. On limiting glucose with the help of the glycolytic pathway, the null strain grows about 40–50% in comparison to the wild-type strain.
E. coli membrane-bound F1Fo ATP synthase was obtained by growing E. coli to late log phase, harvesting cells in super centrifuge, lysing cells in high pressure French Press, and separation by ultracentrifugation as in [65]. This procedure involves three washes of the initial membrane pellets. Wash one is done in a buffer containing 50 mM TES pH 7.0, 15% glycerol, 40 mM 6-aminohexanoic acid, and 5 mM p-aminobenzamidine, followed by two subsequent washes in a buffer containing 5 mM TES pH 7.0, 15% glycerol, 40 mM 6-aminohexanoic acid, 5 mM p-aminobenzamidine, 0.5 mM DTT, and 0.5 mM EDTA. Membranes are washed twice more by resuspension and ultracentrifugation in a buffer containing 50 mM TrisSO4 pH 8.0 with 2.5 mM MgSO4.
ATPase activities were measured in 1 mL ATPase cocktail containing 10 mM NaATP, 4 mM MgCl2, 50 mM TrisSO4 pH 8.5 at 37 °C. Reactions were initiated by the addition of 1 mL ATPase cocktail to membrane-bound F1Fo ATP synthase and stopped by the addition of SDS to 3.3% final concentration. Released inorganic phosphate (Pi) was measured at OD700 as in [66]. For ATPase assays, 20–30 µg wild-type and mutant proteins were used for 20–30 minute reaction time. All reactions were found to be linear with time and protein concentration. Integrity and purity of membrane-bound F1Fo ATP synthase was confirmed by 10% acrylamide SDS-gel electrophoresis and immunoblotting with rabbit polyclonal anti-F1-α and anti-F1-β antibodies [62, 67].
Safranal and its analogs induced inhibition of membrane-bound F1FO ATP synthase
Wild-type and mutant membrane-bound F1FO ATP synthase were preincubated with varied concentrations of safranal and other analog inhibitors for 1 hour at room temperature in 50 mM TrisSO4, pH 8.0 buffer. One mL ATPase cocktail was added to measure the ATPase activity. The reaction was stopped by the addition of 1 mL sodium dodecyl sulfate (SDS) to a final concentration of 3.3%. The addition of an equal volume of Taussky and Shorr reagent gave rise to a blue color that was assayed spectrophotometrically at OD700. Inhibitory exponential decay curves were generated using SigmaPlot 10.0. The best fit line for the curve was obtained using a single, 3-parameter model. Statistical significance of the relationship between relative ATPase activities against inhibitor concentrations was analyzed by linear regression. The range of absolute specific activity for wild-type and mutant membrane-bound F1Fo ATP synthase was 10–15 µmol/min/mg at 37 °C for the different preparations. The absolute ATPase values were used as a 100% benchmark to calculate the relative ATPase activity in the presence of inhibitors.
Growth of wild-type, mutant, and null strain in presence of safranal and its analogs
Growth of the three E. coli strains wild-type, mutant, and null was checked on Luria broth (molecular genetics grade from Fisher Scientific BP1426-2, formulation per liter was 10g trypton, 5g yeast extract, and 10g sodium chloride), limiting glucose, and succinate media in the presence and absence of inhibitors. E. coli strains were grown in a 96 well-plate for 24 hrs on AccuSkan Go Plate Reader. Data for the growth signals at OD595 were analyzed by Thermo Fisher Scientific SkanIt 4.1 Software.
Results
Safranal-induced selective inhibition of wild-type and mutant E. coli membrane-bound F1Fo ATP synthase
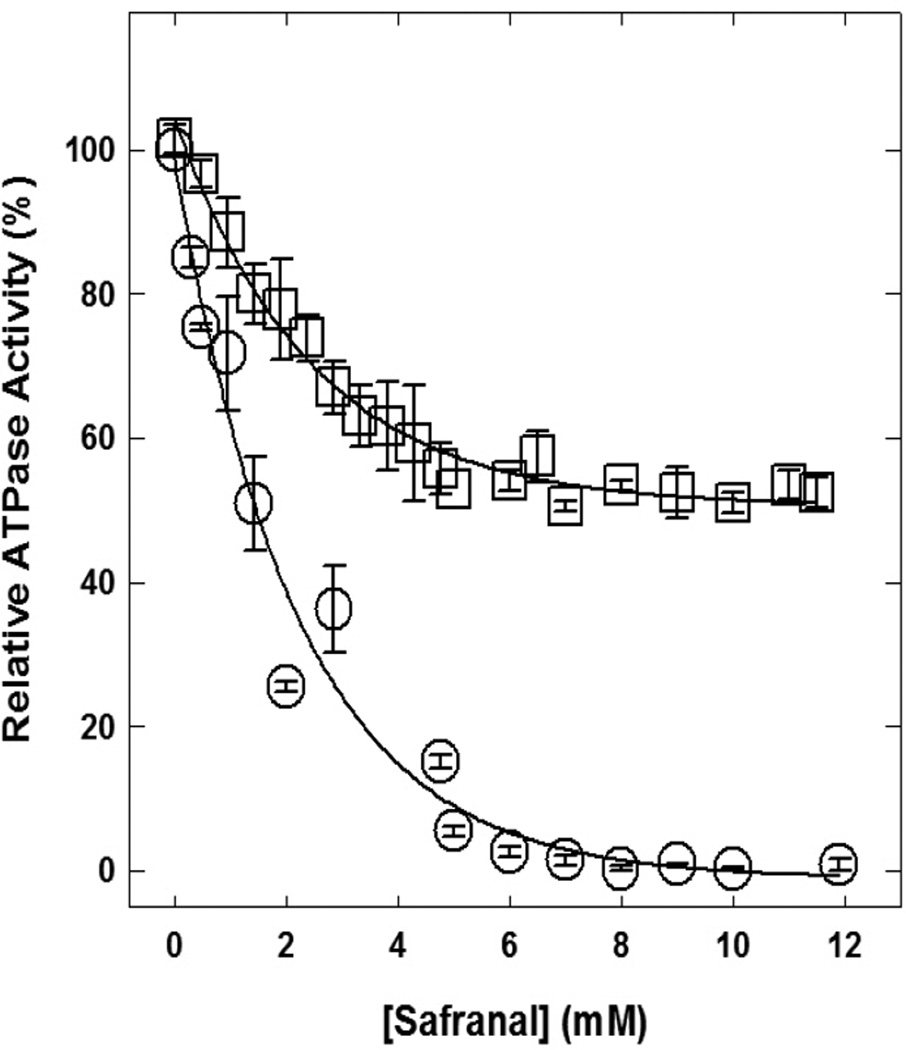
Safranal caused complete inhibition of wild-type membrane-bound F1Fo ATP synthase but caused only about 50% inhibition of the αR283D mutant enzyme (Fig. 2). Maximal inhibition of almost 100% for the wild-type enzyme occurred at about 8 mM safranal concentration. However, the mutant enzyme retained about 50% residual activity up to 12 mM safranal concentration.
Fig. 2. Safranal-induced inhibition of wild-type and mutant membrane-bound F1Fo ATP synthase.
Membranes were preincubated for 60 min at room temperature with varied concentrations of safranal and then 1 mL of ATPase cocktail was added and activity measured. Experimental details are given in the Materials and Methods section. Each data point represents the average of at least four experiments done in duplicate tubes, using 2–3 independent F1Fo membrane preparations.
Plausible safranal binding site
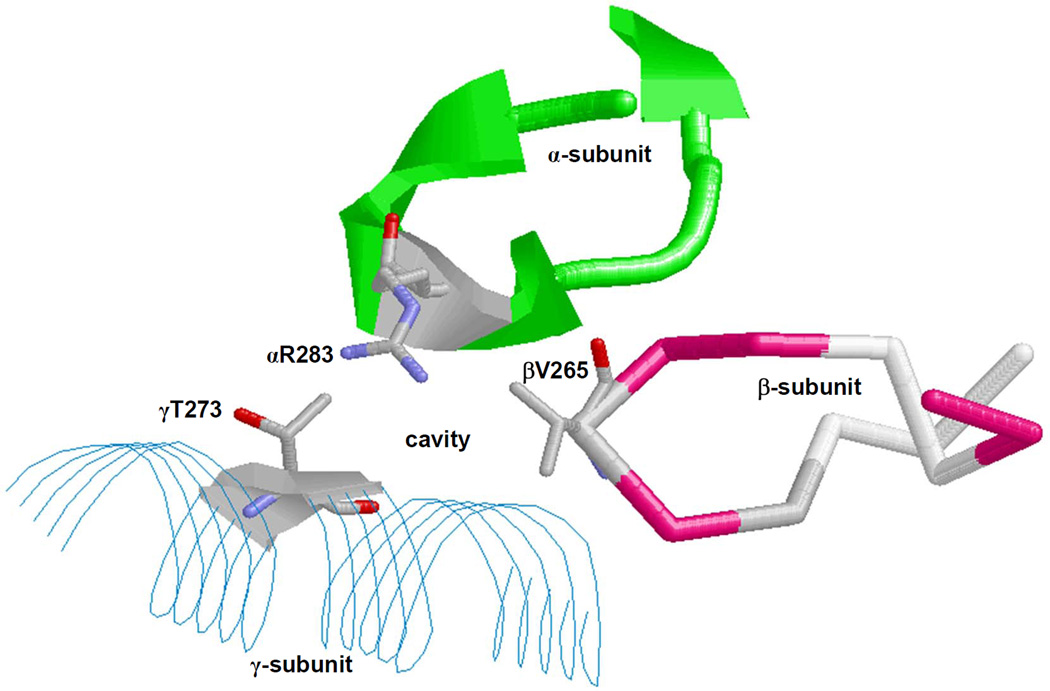
In order to investigate and identify the safranal binding site and residues, a new E. coli mutant strain pZA84 with αR283D mutation was generated. The spatial relationship between α-, β-, and γ-subunits and αArg-283 residue forming a possible safranal binding cavity is shown in Figure 3.
Fig. 3. X-ray crystallographic structure of the polyphenol binding pocket of F1Fo ATP synthase.
α-, β-, and γ-subunits forming an inhibitor binding cavity are shown. Some residues, including αArg-283, are identified. The figure was generated by PDB file 2JJ1 [11] using RasMol software. Residue numbers are based on E. coli numbering.
Thymol, carvacrol, damascenone, cuminol, TMCHD, IPBB, and TBP induced inhibition of E. coli membrane-bound F1Fo ATP synthase
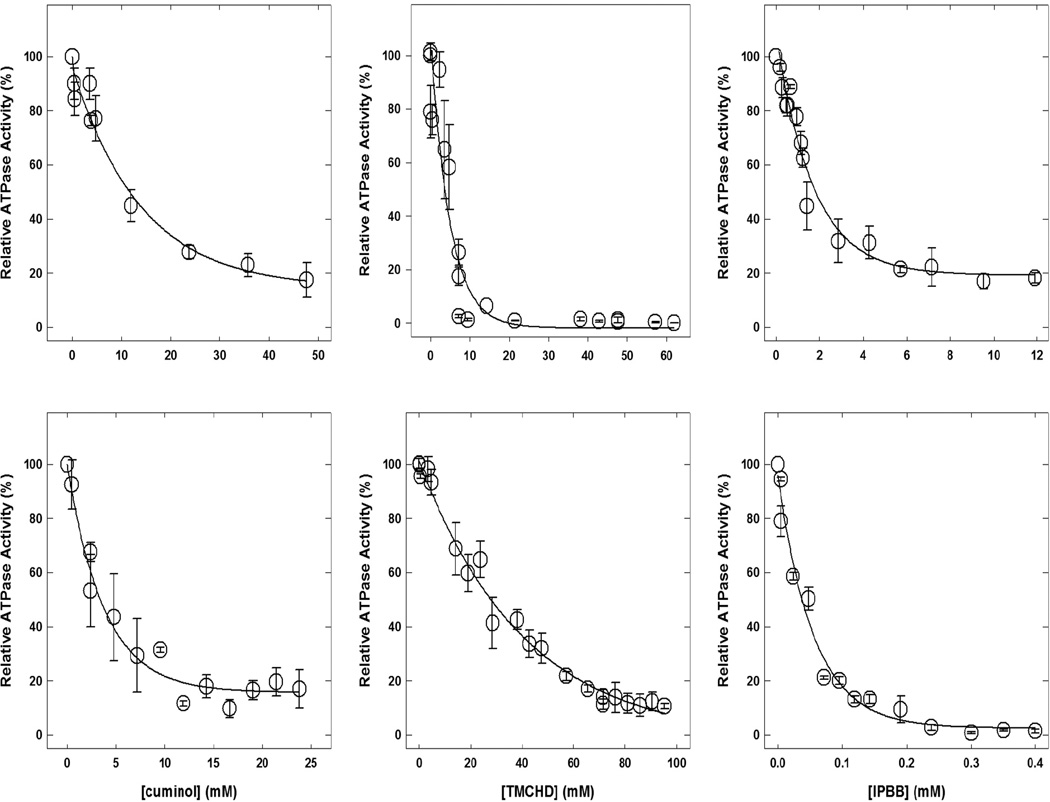
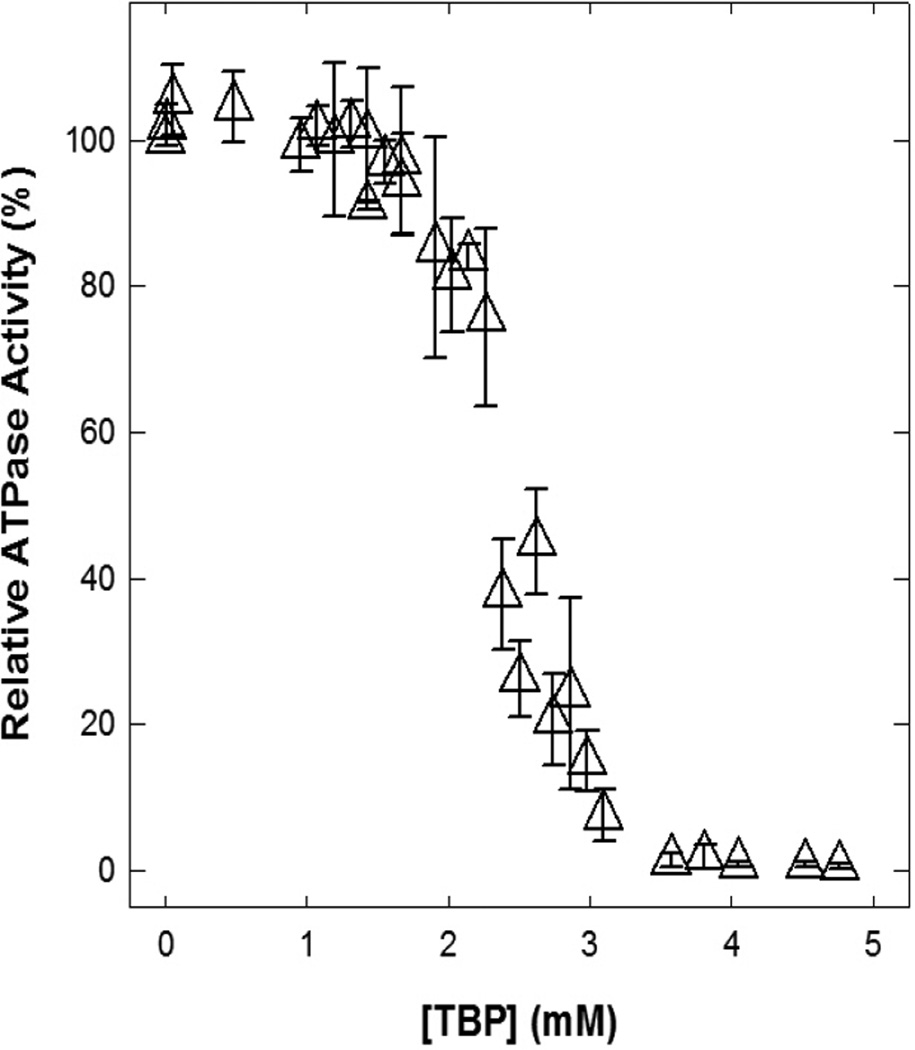
Figure 4 shows the thymol, carvacrol, damascenone, cuminol, TMCHD, and IPBB induced inhibitory profiles of wild-type E. coli membrane-bound F1Fo ATP synthase. The maximal inhibition in the presence of thymol (up to 46 mM) was about 87%, carvacrol was almost 100% at 10 mM, damascenone was about 85% at 7 mM, cuminol was about 93% at 16 mM, TMCHD was about 90% at 95 mM, and IPBB was almost 100% at 0.3 mM. TBP also induced almost 100% inhibition at 2.9 mM concentration (Fig. 5).
Fig. 4. Thymol-, carvacrol-, damascenone-, cuminol-, TMCHD (2,6,6-trimethyl-2-cyclohexene-1,4-dione)-, and IPBB (4-isopropylbenzyl bromide)-induced inhibition of wild-type membrane-bound F1Fo ATP synthase.
E. coli F1Fo membranes were preincubated for 60 min at room temperature with varied concentrations of inhibitors and then 1 mL of ATPase cocktail was added and activity measured. Experimental details are given in the Materials and Methods section. Each data point represents the average of at least four experiments done in duplicate tubes, using 2–3 independent F1Fo membrane preparations.
Fig. 5. TBP (4-tert-butylphenol)-induced inhibition of membrane-bound F1Fo ATP synthase.
Membranes were preincubated for 60 min at room temperature with varied concentrations of TBP and then 1 mL of ATPase cocktail was added and activity measured as in Fig. 2 and Fig. 4. Each data point is the average of at least four experiments done in duplicate tubes, using 2–3 independent F1Fo membrane preparations. Results agreed within ± 8%.
Inhibition of E. coli cell growth on limiting glucose and succinate in presence of safranal and its analogs
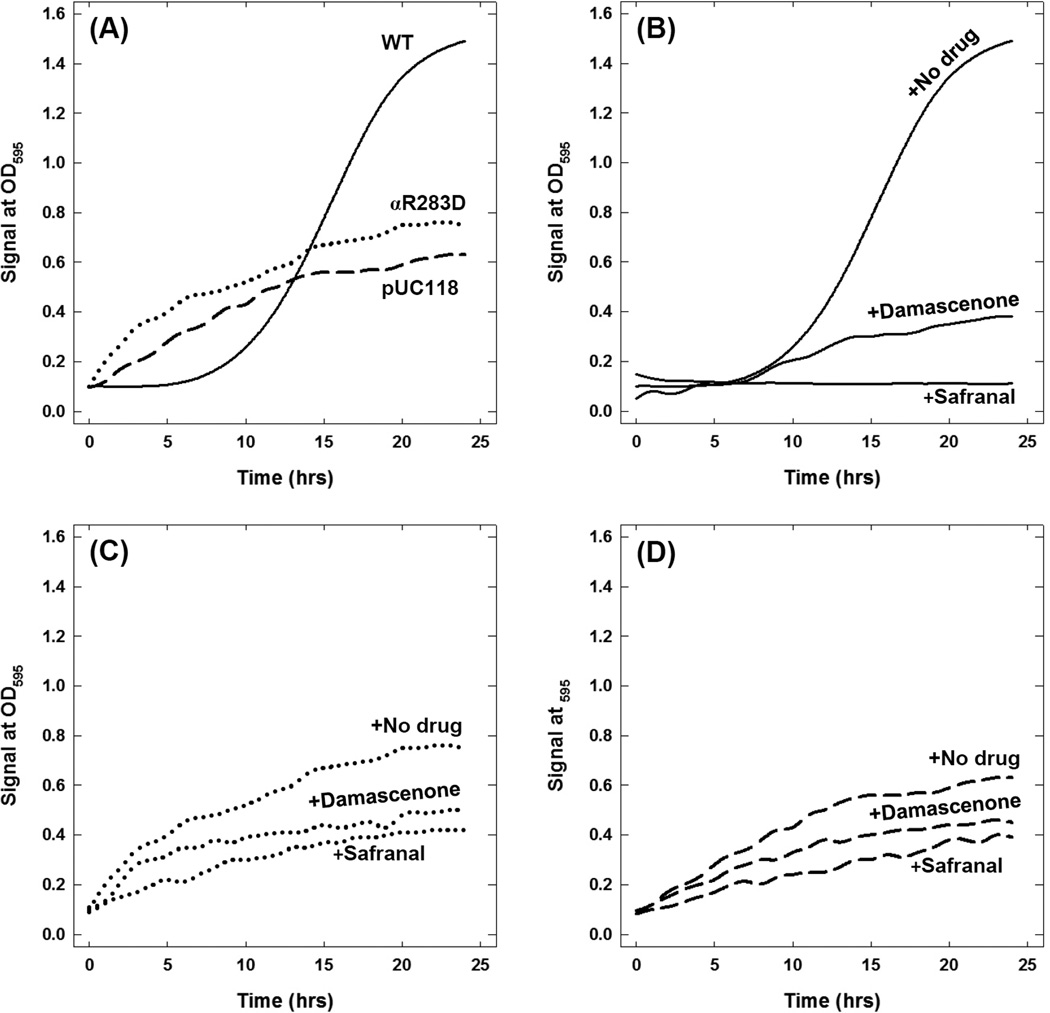
Wild-type, mutant, and null E. coli strains were grown on limiting glucose (containing Ile and Val) and succinate (non-fermentable carbon source) in the presence and absence of safranal and its analog inhibitors (Table 1). Variable reduction in wild-type growth was observed in the presence of inhibitors. Partial to no growth inhibition was observed for mutant and null strains. Figure 6 shows the growth signals in limiting glucose for wild-type, mutant, and null E. coli strains in presence and absence of safranal and damascenone. Figure 6A shows the maximal normal comparative growth where null and mutant strains grew about 40–50% in comparison to the wild-type. Figure 6B shows the complete growth inhibition in presence of safranal and partial growth inhibition in presence of damascenone. Figure 6C and 6D show the effect of safranal and damascenone on mutant and null strains respectively. Both mutant and null strains show partial growth inhibition in presence of safranal and damascenone.
Table 1.
Effect of safranal and its structural analogs on the growth of Escherichia coli cells
| Presence/absence of inhibitors |
1Growth on limiting glucose (%) |
2Growth on succinate (%) |
|---|---|---|
| 3WT (pBWU13.4) | 100 | 100 |
| 4Null (pUC118) | 48±4 | 4±3 |
| Mutant (αR283D) | 53±5 | 49±4 |
| WT + Safranal | 1±3 | 2±2 |
| Null + Safranal | 29±5 | 0 |
| Mutant + Safranal | 30±2 | 29±3 |
| WT + Thymol | 3±2 | 4±2 |
| Null + Thymol | 32±5 | 0 |
| Mutant + Thymol | 20±4 | 22±5 |
| WT + Carvacrol | 2±2 | 2±2 |
| Null + Carvacrol | 48±2 | 0 |
| Mutant + Carvacrol | 42±3 | 45±3 |
| WT+ Damascenone | 7±2 | 6±2 |
| Null + Damascenone | 34±2 | 0 |
| Mutant + Damascenone | 35±3 | 29±2 |
| WT + Cuminol | 5±2 | 6±3 |
| Null + Cuminol | 48±5 | 0 |
| Mutant + Cuminol | 40±2 | 37±3 |
| WT + TMCHD | 7±3 | 4±3 |
| Null + TMCHD | 45±3 | 0 |
| Mutant + TMCHD | 40±4 | 39±5 |
| WT + IPBB | 0 | 0 |
| Null + IPBB | 40±3 | 0 |
| Mutant + IPBB | 22±2 | 20±4 |
| WT + TBP | 2±1 | 2±2 |
| Null + TBP | 23±3 | 0 |
| Mutant + TBP | 16±2 | 14±2 |
Growth yield on limiting glucose (fermentable carbon source) was measured as OD595 with hourly reading for 25 hours at 37 °C in AccuSKan Go Plate Reader.
Growth on succinate medium (non-fermentable carbon source) was measured as OD595 with hourly reading for 25 hours at 37 °C in AccuSKan Go Plate Reader.
Wild-type (pBWU13.4/DK8) contains UNC+ gene encoding ATP synthase
Null, (pUC118/DK8) is UNC− gene encoding ATP synthase is removed.
The absolute wild-type OD values were used as 100% bench mark to calculate the relative OD values for null and mutant. Data points are average of at least three sample assays.
Fig.6. Wild-type, mutant, and null E. coli growth signals at OD595 in the presence and absence of safranal and damascenone.
200 µl E. coli culture was grown in limiting glucose and succinate with maximal inhibitory concentrations of safranal (10 mM) and damascenone (8 mM) in 96-well plates. Optical density at λ595 nM was set to be measured every 10 min for 25 hours with shaking. Experimental details are given in the Materials and Methods section. Each data point is the average of at least three sample readings.
Discussion
The fast encroaching antibiotic resistance by microbes in general and E. coli in particular warrants finding alternative ways to combat the microbial infections. The goal of this study was to determine if the antimicrobial or anticancer properties of safranal and its structural analogs may be associated with the inhibition of ATP synthase. Additionally, we investigated whether safranal binds at the polyphenol binding pocket of ATP synthase.
Safranal fully inhibited wild-type membrane-bound F1Fo ATP synthase and caused about 50% inhibition to the αR283D mutant enzyme (Fig. 2). About 50% inhibition of mutant enzyme by safranal agrees with our assumption that safranal may also bind at the polyphenol binding pocket of ATP synthase. Further, αArg-283 is one of the key residues required for the binding of phenolic compounds at this site (Fig. 3). Recently, thymoquinone which is a structurally comparable compound to safranal with three methyl groups was also found to completely inhibit ATP synthase and was suggested to bind at polyphenol binding pocket of ATP synthase [62].
Previously, resveratrol-, piceatannol-, and quercetin-bound ATP synthase x-ray crystal structures showed that the polyphenol binding pocket for resveratrol, piceatannol, and quercetin was contributed to by residues from the α-, β-, and γ-subunits [11]. Although in earlier studies, several other polyphenolic compounds structurally related to safranal were shown to bind to the polyphenol binding pocket [61, 62, 68, 69]. In those studies, no site-directed mutations were performed to specify the key residues. Therefore, we mutated αArg-283 to αAsp-283 to confirm its role in safranal binding.
As shown in Figure 3, αArg-283 protrudes into the cavity where safranal is expected to bind. The αR283D mutant ATP synthase retains about 50% residual activity in comparison to the wild-type enzyme with the substitution of Arg →Asp (Fig. 2). Safranal-induced inhibition of wild-type and mutant enzymes suggests and establishes that the guanidino group of αArg-283 is required for proper binding of safranal at the polyphenol binding pocket of E. coli ATP synthase. Moreover, the -CH3 groups of safranal may be involved in hydrophobic, nonpolar interactions with other polyphenol binding pocket residues, such as γGln274, γThr-277, βAla-264, βVal-265, γAla-270, γThr-273, γGlu-278, αGly-282, or αGlu-284 residues.
Carvacrol and IPBB induced complete inhibition of F1Fo membranes, while thymol-, damascenone-, cuminol-, and TMCHD-induced inhibition resulted in about 10–15% residual activity. TBP a prooxidant, clastogenic, vitiligo causing compound [70] also fully inhibits membrane-bound F1Fo ATP synthase. The toxic compound TBP was used to check if ATP synthase is its possible target. TBP induced complete inhibition of ATP synthase along with total abrogation of wild-type E. coli and significant growth inhibition of mutant and null E. coli strains (Fig. 5 and Table 1). The nature and spatial orientation of functional groups of inhibitors seems to plays important role in inducing complete or partial inhibition [69]. Variable maximal inhibition of ATP synthase has been observed in several previous studies where wild-type or mutant enzymes were partially or incompletely inhibited by phytochemicals, peptides, NBD-Cl, NaN3, AlCl3, or ScCl3 [61, 67, 71].
To confirm that the wild-type and mutant enzymes were maximally inhibited by safranal and its structural analogs, wild-type and mutant membrane-bound F1Fo preparations were incubated with maximal inhibitory concentrations of each inhibitor for 1 hour as in Figure 2, Figure 4, and Figure 5. This inhibition was followed by an additional pulse of the same inhibitory concentration of inhibitors, and then incubation was continued for an additional hour before ATPase assay. Very little or no additional inhibition was observed. These results established that the inhibitors fully reacted at the binding site, which resulted in maximal inhibition, and yet residual activity could be retained by the wild-type and mutant enzymes in the presence of certain inhibitors.
According to the International Union of Pure and Applied Chemistry, in order for a compound to be considered pharmacophoric, the compound must have a group of steric and electronic features that ensure its optimal supramolecular interactions with a specific biological target to trigger or block the biological response [72]. The presence of an unsaturated nucleus within a compound is one such pharmacophoric property. Safranal and all its analogs (thymol, carvacrol, damascenone, cuminol, TMCHD, IPBB, and TBP) possess an unsaturated nucleus (Fig. 1). Safranal and its structural analogs induced inhibition profiles of membrane-bound F1Fo ATP synthase (Fig. 2, Fig. 4, and Fig. 5) provide experimental evidence for both ATP synthase as potent molecular target and safranal as a lead molecule for the development of selective antimicrobial agents.
Table 1 shows the growth properties of wild-type, mutant, and null E. coli strains in the presence and absence of safranal and its analogs. Safranal, thymol, carvacrol, damascenone, cuminol, TMCHD, IPBB, and TBP all fully abrogated the wild-type (pBWU13.4/DK8) E. coli cell growth. On limiting glucose, the null strain (pUC118/DK8) typically grew about 40–50% in comparison to the wild-type. In the absence of ATP synthase, the null strain growth depends on glycolysis to generate ATP, whereas the wild-type strain uses all glycolysis, tricarboxylic acid cycle, and oxidative phosphorylation pathways. Mutant αR283D also showed about 50% growth both on limiting glucose and succinate. Near complete inhibition of wild-type E. coli growth in comparison to about 85–90% inhibition of membrane-bound F1Fo by thymol, damascenone, cuminol, or TMCHD could be attributed to additional targets or damage to bacterial membranes. Table 1 also shows the partial growth loss for null and mutant strains, which may also be attributed to additional targets or damage to bacterial membranes.
Figure 6 presents the growth signals for wild-type, mutant, and null E. coli strains in the presence or absence of safranal and damascenone. Growth retention in both mutant and null cells can be credited to ATP production through the glycolytic pathway. A complete loss of growth in the wild-type in the presence of safranal results from the loss of oxidative phosphorylation through inhibition of ATP synthesis. Moreover, inhibition of wild-type growth in succinate as the sole carbon source in the presence of safranal establishes the inhibition of F1-ATPase activity (Table 1). These results demonstrate that safranal-induced abrogation of microbial growth is through the inhibition of ATP synthase.
High concentrations of safranal and its analogs caused the inhibition of ATP synthase. It has been documented that very high concentrations of saffron and its constituents are required to cause the pharmacological effects and these concentrations are non-toxic in animal studies and human trials [73]. In mice models peritoneum administered lethal dose (LD50) values for saffron stigma and petal were found to be 1.6 and 6 g/kg, respectively [74]. Moreover, in both mice and rats models safranal was found to be low-toxic in acute intraperitoneal route and practically nontoxic in acute oral administration [75]. The intraperitoneal LD50 values of safranal were 1.48 mL/kg in male mice, 1.88 mL/kg in female mice, and 1.50 mL/kg in male rats. Oral LD50 values were 21.42 mL/kg in male mice, 11.42 mL/kg in female mice, and 5.53 mL/kg in male rats [75]. In human studies ingestion of up to 1.5 g of saffron was found to be non-toxic while ingestion of more than 5 g doses is considered toxic and could be lethal if taken about 20 g/day ([73] and reference therein). Once administered inside the body safranal and its analogs may or may not undergo any metabolic transformation. So far we have not knowledge for this effect.
Finally, inhibition of microbial cell growth in the presence of phytochemicals from previous studies [10, 61, 62, 68] and safranal, thymol, carvacrol, damascenone, cuminol, TMCHD, IPBB, or TBP from this study suggests that ATP synthase can be used as a potential molecular drug target to combat microbial infections. In conclusion, safranal induced abrogation of ATPase activity and inhibition of E. coli cell growth shows that the antimicrobial properties of safranal can be linked to its inhibitory effects on ATP synthase.
Acknowledgments
This work was supported by the National Institutes of Health Grant GM085771 to ZA and A.T. Still University-Kirksville College of Osteopathic Medicine Biomedical Science Graduate Program Funding #850-611 to ZA and ML. ZA is grateful to Dr. Margaret Wilson, dean of Kirksville College of Osteopathic Medicine for providing funding to purchase the AccuSkan Go Plate reader. We are also thankful to Deborah Goggin, scientific writer, research support, A.T. Still University for reviewing the manuscript.
Abbreviations used
- TMCHD
2,6,6-Trimethyl-2-cyclohexene-1,4-dione
- IPBB
4-Isopropylbenzyl bromide
- TBP
4-tert-Butylphenol
- NBD-Cl
7-chloro-4-nitrobenzo-2-oxa-1, 3-diazole
- TES
(2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid)
- Membrane
membrane-bound F1Fo ATP synthase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Senior AE. J Biol Chem. 2012;287:30049–30062. doi: 10.1074/jbc.X112.402313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senior AE, Nadanaciva S, Weber J. Biochim Biophys Acta. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 3.Weber J, Senior AE. FEBS Lett. 2003;545:61–70. doi: 10.1016/s0014-5793(03)00394-6. [DOI] [PubMed] [Google Scholar]
- 4.Noji H, Yoshida M. J Biol Chem. 2001;276:1665–1668. doi: 10.1074/jbc.R000021200. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad Z, Cox JL. The Scientific World Journal. 2014;2014:10. [Google Scholar]
- 6.Ahmad Z, Okafor F, Laughlin TF. J Amino Acids. 2011;2011:785741. doi: 10.4061/2011/785741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong S, Pedersen PL. Microbiol Mol Biol Rev. 2008;72:590–641. doi: 10.1128/MMBR.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng J, Ramirez VD. Br J Pharmacol. 2000;130:1115–1123. doi: 10.1038/sj.bjp.0703397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piotto S, Concilio S, Sessa L, Porta A, Calabrese EC, Zanfardino A, Varcamonti M, Iannelli P. Eur J Med Chem. 2013;68:178–184. doi: 10.1016/j.ejmech.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Vik SB, Tu Y. J Nutr Biochem. 2012;23:953–960. doi: 10.1016/j.jnutbio.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. Proc Natl Acad Sci U S A. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cushnie TP, Lamb AJ. Int J Antimicrob Agents. 2011;38:99–107. doi: 10.1016/j.ijantimicag.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Pagliarani A, Nesci S, Ventrella V. Mini Rev Med Chem. 2016;16:815–824. doi: 10.2174/1389557516666160211120955. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad Z, Laughlin TF. Curr Med Chem. 2010;17:2822–2836. doi: 10.2174/092986710791859270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad Z, Okafor F, Azim S, Laughlin TF. Curr Med Chem. 2013;20:1956–1973. doi: 10.2174/0929867311320150003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Neill J. Review on antimicrobial resistance. 2016:1–20. [Google Scholar]
- 17.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. Nature. 2015 [Google Scholar]
- 18.World Health Organization. Antimicrobial resistance : global report on surveillance [Google Scholar]
- 19.Nakanishi-Matsui M, Sekiya M, Futai M. Biochim Biophys Acta. 2016;1857:129–140. doi: 10.1016/j.bbabio.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Nesci S, Ventrella V, Trombetti F, Pirini M, Pagliarani A. Med Hypotheses. 2014;83:160–165. doi: 10.1016/j.mehy.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Nesci S, Ventrella V, Trombetti F, Pirini M, Pagliarani A. Biochim Biophys Acta. 2014;1840:1882–1891. doi: 10.1016/j.bbagen.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Nesci S, Trombetti F, Pirini M, Ventrella V, Pagliarani A. Chem Biol Interact. 2016;260:42–49. doi: 10.1016/j.cbi.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Melnyk JP, Wang S, Marcone MF. Food Research International. 2010;43:1981–1989. [Google Scholar]
- 24.Ahmad A, Khan A, Yousuf S, Khan LA, Manzoor N. Fitoterapia. 2010;81:1157–1162. doi: 10.1016/j.fitote.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 25.Caselli A, Cirri P, Santi A, Paoli P. Curr Med Chem. 2016;23:774–791. doi: 10.2174/0929867323666160106150821. [DOI] [PubMed] [Google Scholar]
- 26.Karygianni L, Al-Ahmad A, Argyropoulou A, Hellwig E, Anderson AC, Skaltsounis AL. Front Microbiol. 2015;6:1529. doi: 10.3389/fmicb.2015.01529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandoval-Acuna C, Ferreira J, Speisky H. Arch Biochem Biophys. 2014;559:75–90. doi: 10.1016/j.abb.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Madrigal-Perez LA, Ramos-Gomez M. Int J Mol Sci. 2016;17:368. doi: 10.3390/ijms17030368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nesci S, Trombetti F, Ventrella V, Pagliarani A. J Membr Biol. 2016;249:11–21. doi: 10.1007/s00232-015-9860-3. [DOI] [PubMed] [Google Scholar]
- 30.Ahmad I, Muneer KM, Tamimi IA, Chang ME, Ata MO, Yusuf N. Toxicol Appl Pharmacol. 2013;270:70–76. doi: 10.1016/j.taap.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 31.Chaieb K, Kouidhi B, Jrah H, Mahdouani K, Bakhrouf A. BMC Complement Altern Med. 2011;11:29. doi: 10.1186/1472-6882-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azevedo C, Correia-Branco A, Araujo JR, Guimaraes JT, Keating E, Martel F. Nutr Cancer. 2015;67:504–513. doi: 10.1080/01635581.2015.1002625. [DOI] [PubMed] [Google Scholar]
- 33.Hassan SM, Youakim MF, Rizk AA, Thomann C, Ahmad Z. Neurourol Urodyn. 2016 doi: 10.1002/nau.23109. [DOI] [PubMed] [Google Scholar]
- 34.Abdullaev FI. Exp Biol Med (Maywood) 2002;227:20–25. doi: 10.1177/153537020222700104. [DOI] [PubMed] [Google Scholar]
- 35.Pintado C, Miguel AD, Acevedo O, Nozal L, Novella JS, Rotger R. Food Control. 2011;22:638–642. [Google Scholar]
- 36.Khazdair MR, Boskabady MH, Hosseini M, Rezaee R, A MT. Avicenna J Phytomed. 2015;5:376–391. [PMC free article] [PubMed] [Google Scholar]
- 37.Mashmoul M, Azlan A, Khaza'ai H, Yusof BN, Noor SM. Antioxidants (Basel) 2013;2:293–308. doi: 10.3390/antiox2040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amanpour A, Sonmezdag AS, Kelebek H, Selli S. Food Chem. 2015;182:251–256. doi: 10.1016/j.foodchem.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Samarghandian S, Azimi-Nezhad M, Samini F. Biomed Res Int. 2014;2014:920857. doi: 10.1155/2014/920857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makri OE, Ferlemi AV, Lamari FN, Georgakopoulos CD. Mol Vis. 2013;19:1188–1197. [PMC free article] [PubMed] [Google Scholar]
- 41.Bandegi AR, Rashidy-Pour A, Vafaei AA, Ghadrdoost B. Adv Pharm Bull. 2014;4:493–499. doi: 10.5681/apb.2014.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samarghandian S, Borji A. Pharmacognosy Res. 2014;6:99–107. doi: 10.4103/0974-8490.128963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cosano I, Pintado C, Acevedo O, Novella JL, Alonso GL, Carmona M, de la Rosa C, Rotger R. J Food Prot. 2009;72:2217–2220. doi: 10.4315/0362-028x-72.10.2217. [DOI] [PubMed] [Google Scholar]
- 44.De Monte C, Bizzarri B, Gidaro MC, Carradori S, Mollica A, Luisi G, Granese A, Alcaro S, Costa G, Basilico N, Parapini S, Scaltrito MM, Masia C, Sisto F. J Enzyme Inhib Med Chem. 2015;30:1027–1033. doi: 10.3109/14756366.2014.1001755. [DOI] [PubMed] [Google Scholar]
- 45.Hosseinzadeh H, Mehri S, Abolhassani MM, Ramezani M, Sahebkar A, Abnous K. Daru. 2013;21:25. doi: 10.1186/2008-2231-21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beitlich N, Koelling-Speer I, Oelschlaegel S, Speer K. J Agric Food Chem. 2014;62:6435–6444. doi: 10.1021/jf501818f. [DOI] [PubMed] [Google Scholar]
- 47.Abbaszadeh S, Sharifzadeh A, Shokri H, Khosravi AR, Abbaszadeh A. J Mycol Med. 2014;24:e51–e56. doi: 10.1016/j.mycmed.2014.01.063. [DOI] [PubMed] [Google Scholar]
- 48.Palaniappan K, Holley RA. Int J Food Microbiol. 2010;140:164–168. doi: 10.1016/j.ijfoodmicro.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Ait Said L, Zahlane K, Ghalbane I, El Messoussi S, Romane A, Cavaleiro C, Salgueiro L. Nat Prod Res. 2015;29:582–585. doi: 10.1080/14786419.2014.954246. [DOI] [PubMed] [Google Scholar]
- 50.Du WX, Olsen CW, Avena-Bustillos RJ, McHugh TH, Levin CE, Friedman M. J Agric Food Chem. 2008;56:3082–3088. doi: 10.1021/jf703629s. [DOI] [PubMed] [Google Scholar]
- 51.Di Pasqua R, Betts G, Hoskins N, Edwards M, Ercolini D, Mauriello G. J Agric Food Chem. 2007;55:4863–4870. doi: 10.1021/jf0636465. [DOI] [PubMed] [Google Scholar]
- 52.Undeger U, Basaran A, Degen GH, Basaran N. Food Chem Toxicol. 2009;47:2037–2043. doi: 10.1016/j.fct.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 53.Abdel-Wareth AA. Br Poult Sci. 2016;57:114–122. doi: 10.1080/00071668.2015.1123219. [DOI] [PubMed] [Google Scholar]
- 54.Abdul Rahim ZH, Shaikh S, Hasnor Wan Ismail WN, Wan Harun WH, Razak FA. J Coll Physicians Surg Pak. 2014;24:796–801. [PubMed] [Google Scholar]
- 55.de Castro RD, de Souza TM, Bezerra LM, Ferreira GL, Costa EM, Cavalcanti AL. BMC Complement Altern Med. 2015;15:417. doi: 10.1186/s12906-015-0947-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patil SB, Takalikar SS, Joglekar MM, Haldavnekar VS, Arvindekar AU. Br J Nutr. 2013;110:1434–1443. doi: 10.1017/S0007114513000627. [DOI] [PubMed] [Google Scholar]
- 57.Teissedre PL, Waterhouse AL. J Agric Food Chem. 2000;48:3801–3805. doi: 10.1021/jf990921x. [DOI] [PubMed] [Google Scholar]
- 58.Acena L, Vera L, Guasch J, Busto O, Mestres M. J Agric Food Chem. 2011;59:4062–4070. doi: 10.1021/jf104763u. [DOI] [PubMed] [Google Scholar]
- 59.Nazaruk J, Karna E, Kalemba D. Nat Prod Commun. 2012;7:269–272. [PubMed] [Google Scholar]
- 60.Pongprayoon U, Baeckstrom P, Jacobsson U, Lindstrom M, Bohlin L. Planta Med. 1992;58:19–21. doi: 10.1055/s-2006-961381. [DOI] [PubMed] [Google Scholar]
- 61.Chinnam N, Dadi PK, Sabri SA, Ahmad M, Kabir MA, Ahmad Z. Int J Biol Macromol. 2010;46:478–486. doi: 10.1016/j.ijbiomac.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmad Z, Laughlin TF, Kady IO. PLoS One. 2015;10:e0127802. doi: 10.1371/journal.pone.0127802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ketchum CJ, Al-Shawi MK, Nakamoto RK. Biochem J. 1998;330(Pt 2):707–712. doi: 10.1042/bj3300707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Senior AE, Latchney LR, Ferguson AM, Wise JG. Arch Biochem Biophys. 1984;228:49–53. doi: 10.1016/0003-9861(84)90045-6. [DOI] [PubMed] [Google Scholar]
- 65.Senior AE, Langman L, Cox GB, Gibson F. Biochem J. 1983;210:395–403. doi: 10.1042/bj2100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taussky HH, Shorr E. J Biol Chem. 1953;202:675–685. [PubMed] [Google Scholar]
- 67.Zhao C, Syed H, Hassan SS, Singh VK, Ahmad Z. Arch Biochem Biophys. 2016;592:27–37. doi: 10.1016/j.abb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dadi PK, Ahmad M, Ahmad Z. Int J Biol Macromol. 2009;45:72–79. doi: 10.1016/j.ijbiomac.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 69.Ahmad Z, Ahmad M, Okafor F, Jones J, Abunameh A, Cheniya RP, Kady IO. Int J Biol Macromol. 2012;50:476–486. doi: 10.1016/j.ijbiomac.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Babu S, Uppu S, Claville MO, Uppu RM. Toxicol Mech Methods. 2013;23:273–280. doi: 10.3109/15376516.2012.753969. [DOI] [PubMed] [Google Scholar]
- 71.Azim S, McDowell D, Cartagena A, Rodriguez R, Laughlin TF, Ahmad Z. Int J Biol Macromol. 2016;87:246–251. doi: 10.1016/j.ijbiomac.2016.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wermuth CG, Ganellin CR, Lindberg P, Mitscher LA. Pure and Applied Chemistry. 2009;70:1129–1143. [Google Scholar]
- 73.Moshiri M, Vahabzadeh M, Hosseinzadeh H. Drug Res (Stuttg) 2015;65:287–295. doi: 10.1055/s-0034-1375681. [DOI] [PubMed] [Google Scholar]
- 74.Modaghegh MH, Shahabian M, Esmaeili HA, Rajbai O, Hosseinzadeh H. Phytomedicine. 2008;15:1032–1037. doi: 10.1016/j.phymed.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 75.Hosseinzadeh H, Sadeghi Shakib S, Khadem Sameni A, Taghiabadi E. Iran J Pharm Res. 2013;12:93–99. [PMC free article] [PubMed] [Google Scholar]