Abstract
Objectives
To explore the feasibility of using amide proton transfer-weighted (APTw) MRI metrics as surrogate biomarkers to identify the O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status in glioblastoma (GBM).
Methods
Eighteen newly diagnosed GBM patients, who were previously scanned at 3T and had a confirmed MGMT methylation status, were retrospectively analyzed. For each case, a histogram analysis in the tumor mass was performed to evaluate several quantitative APTw MRI metrics. The Mann-Whitney test was used to evaluate the difference in APTw parameters between MGMT methylated and unmethylated GBMs, and the receiver-operator-characteristic analysis was further used to assess the diagnostic performance.
Results
Ten GBMs were found to harbor a methylated MGMT promoter, and eight GBMs were unmethylated. The Mean, Variance, 50th percentile, 90th percentile, and Width10–90 APTw values were significantly higher in the MGMT unmethylated GBMs than in the MGMT methylated GBMs, with the areas under the receiver-operator-characteristic curves of 0.825, 0.837, 0.850, 0856, and 0.763, respectively, for the discrimination of MGMT promoter methylation status.
Conclusions
APTw signal metrics have the potential to serve as valuable imaging biomarkers for identifying MGMT methylation status in the GBM population.
Keywords: Glioblastoma, O6-methylguanine-DNA methyltransferase, Magnetic Resonance Imaging, Amide Proton Transfer-Weighted Imaging, Methylation
Introduction
Glioblastoma (GBM) is the most dismal type of primary malignant brain tumor in adults. Although tremendous effort has been made to optimize the treatment of GBM, most patients die within 15 months [1]. The miserable prognosis conferred by GBM is partly due to the tendency of GBMs to diffusely and extensively infiltrate into surrounding normal brain tissue [2]. In addition, the intrinsic or acquired resistance presents obstacles to successful chemoradiotherapy treatment [3, 4]. Recently, it has been demonstrated that some GBM cohorts, with inhibited DNA damage response mutations or pathways against chemotherapy, perceptibly modulated the tumor response [5–7]. The methylated O6-methylguanine-DNA methyltransferase (MGMT) promoter is one molecular marker that is indicative of longer survival in patients with GBM who receive alkylating agents, with both proved biochemical mechanisms and clinically verified evidence [8–10]. The postulated underlying mechanism showed that the methylated MGMT promoter could enhance chemosensitivity via silencing of the MGMT gene to eventually downregulate the expression of encoded MGMT protein, which functions as a key DNA repair enzyme that counteracts alkylate drugs [11].
Thus, the MGMT promoter methylation status test has become a routine screening test in patients with GBM as a personalized medicine strategy [12]. Unfortunately, current lab tests primarily require large tissue samples obtained from invasive surgical procedures, which are not only costly, but also pose risks. In recent years, a new hypothesis in cancer research has emerged that various genotypes or molecular alterations within GBM could lead to observable, altered imaging features [13, 14]. Hence, imaging-based methodologies have been implemented to provide insight into the MGMT methylation status in patients with GBM. Some studies have attempted to discover promising textural patterns derived from run-length textures [15] or the co-occurrence matrix [16] in MRI datasets via intricate algorithms. Several imaging textures were found to be feasible for the discrimination of GBMs with different MGMT promoter methylation statuses, despite the fact that it is time-consuming and statistic overfitting [17]. Researchers also evaluated the efficacy of applying advanced MRI techniques to determine the status of MGMT methylation in GBMs, including diffusion imaging [18], diffusion tensor imaging [19, 20], and perfusion imaging [19–21]. Positron emission tomography (PET) was also studied to discover predictive markers for MGMT-expressed glioblastoma in vitro [22]. All these studies proposed that the MGMT molecular aspects of GBMs can be characterized quite convincingly by MRI-derived tumor features or by a PET imaging agent. Yet, the results drawn from different research groups have been inconsistent, even when investigating the same advanced MRI imaging [18–21], or the sample sizes were still relatively limited with regard to compelling statistical conclusions.
Amide proton transfer (APT) imaging, the most developed branch of chemical exchange-dependent saturation transfer (CEST) imaging [23, 24], is a novel molecular MRI technique that generates contrast based on endogenous cellular proteins in vivo [25, 26]. Several studies from different labs thus far have consistently demonstrated that APT-weighted (APTw) imaging has great potential for detecting malignant brain tumors [27–30] and other cancers [31, 32]. With regard to gliomas, APTw MRI has shown its unique clinical efficiency in defining the tumor burden [33], determining tumor grade [34–36], and monitoring treatment effects [37, 38]. Notably, it has been shown that APTw signal intensities have a positive correlation with cellularity and proliferation [35, 36]. The promoter of MGMT is often hypermethylated in many cancers, suggesting the decrease of its protein expression. It may affect other protein expressions downstream of MGMT [39]. Hence, protein-based APTw MRI may provide a new approach to non-invasively investigate the MGMT methylation status in GBMs. We hypothesize that GBMs with an unmethylated MGMT promoter will present higher APTw values than the MGMT methylated GBMs. The purpose of this retrospective study is to investigate whether the MGMT methylation status in GBMs can be stratified by APTw-MRI metrics.
Materials and methods
Subjects
This retrospective study was approved by the local Institutional Review Board, and the requirement for informed consent was waived. Patients with newly diagnosed GBM (grade IV astrocytoma), treated in Zhujiang hospital hospital between July 1, 2014 and August 31, 2016, were recruited. Enrollment criteria were: age ≥ 18 years; APTw and routine MRI scanning performed within 7 days preoperatively, including T2-weighted (T2w), fluid-attenuated inversion recovery (FLAIR), T1-weighted (T1w), and gadolinium-enhanced T1-weighted (Gd-T1w) imaging; known MGMT methylation status; and no radiotherapy, chemotherapy, or surgery before imaging. Exclusion criteria included inferior image quality due to various reasons.
MRI data acquisition
MRI imaging was performed on a 3T MRI scanner (Achieva; Philips Medical Systems, Best, The Netherlands). A fat-suppressed, fast spin-echo pulse sequence was used to acquire APT image data, with the following parameters: radiofrequency saturation power, 2 µT; saturation time, 0.8 sec; repetition time, 3 sec; matrix, 128 ×128 (reconstructed to be 400 × 400); field of view, 240×240 mm2; slice thickness, 6 mm; sensitivity-encoding factor, 2; and turbo-spin-echo factor, 37. A single-slice, combined APTw imaging and Z-spectrum acquisition protocol was implemented on the maximum cross-sectional tumor slice (as determined by routine MRI images). As described before [33], this protocol had more acquisitions at and around ±3.5 ppm, facilitating a sufficient signal-to-noise ratio for APTw images, and higher interpolation accuracy of APTw data for B0 corrections. The Gd-T1w imaging (5 ml/s; 0.2 mL/kg body weight; Magnevist; Bayer Schering, Guangzhou, China) was acquired as the last sequence to avoid potential influence on the APTw signal intensity [40].
APTw image processing and analysis
All image data were processed using the interactive data language software (IDL, Version 7; Exelis Visual Information Solutions, Inc., Boulder, CO). To reduce possible motion artifacts during the scanning, the acquired APTw image or Z-spectrum series was registered to the saturated image at 3.5 ppm [41], which was further corrected for the B0 inhomogeneity effect on a voxel-by-voxel basis. Then, Z-spectra (Z(offset), defined as water signal intensities with and without selective radio-frequency irradiation plotted as a function of saturation frequency offset, relative to water), magnetization transfer ratio asymmetry spectra (MTRasym(offset) = Z(offset) − Z(−offset)), and MTRasym(3.5ppm) images were calculated, as previously reported [25, 26]. Because of the contributions from the nuclear Overhauser enhancement effect at −3.5 ppm [42–46], and the asymmetry of the conventional semi-solid magnetization transfer effect [47], the calculated MTRasym(3.5ppm) images are often called APTw images [48].
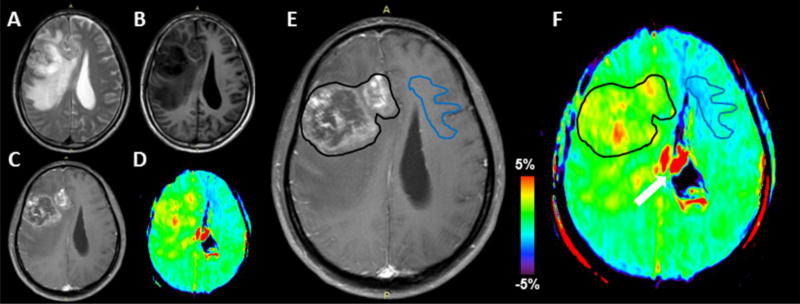
The acquired conventional MR images were co-registered to the corresponding saturated Ssat image at 3.5 ppm (which was co-registered with the APTw image) to fulfill quantitative APTw analyses [41]. Two radiologists (S.J. and X.W., with ten and seven years of experience in brain imaging, respectively) carefully drew the regions-of-interest (ROIs), in consensus. One large ROI contouring the whole area of Gd enhancement within the lesion on the Gd-T1w image was drawn and defined as the tumor mass for the histogram ROI analysis. The contralateral normal-appearing white matter (CNAWM) was also analyzed for normalization, and relative APTw values were reported (ROI APTw - CNAWM APTw). Fig. 1 shows how the ROIs were drawn.
Fig. 1.
With reference to T2w (A), T1w (B), Gd-T1w (C), and APTw (D) images, an example of the placement of ROIs on the Gd-T1w image (E) and co-registered APTw image (F). One large ROI (black lines) contouring the whole area of Gd enhancement within the lesion on the Gd-T1w image was drawn and defined as the tumor mass. In addition, one ROI was placed in the CNAWM for normalization. The white arrow indicates artifacts associated with ventricles.
For each case, the Z-spectrum data and APTw value histogram data from the large whole-tumor ROI were recorded. The histogram data were analyzed for Mean, Variance, Skewness, Kurtosis, 10th percentile, 50th percentile, 90th percentile, and Mode values, as defined previously [49, 50]. We also evaluated the Width10–90 value (90th percentile - 10th percentile) in this histogram analysis.
The qualitative APTw signal features (ring-like, nodular, or patchy) of the MGMT unmethylated and methylated GBMs were further analyzed, using standard MRI sequences as a reference by the two above-mentioned radiologists with consensus. Then, invasive lobes of each lesion were recorded. The lesions with ventricles involved were specifically noted.
Histopathological evaluation
Operative tissue samples were re-evaluated by an experienced pathologist (Y.W.), blinded to the imaging findings, using the newest 2016 WHO classification of central nervous system tumors. The MGMT methylation status was assessed with a methylation-specific polymerase chain reaction, as described in the literature [51].
Statistical analysis
Mann-Whitney U-tests were used to analyze the statistical differences between quantitative APTw parameters for methylated and unmethylated GBMs after normality testing. Receiver operating characteristic (ROC) curves for the significant different APTw parameters were used to assess the diagnostic performance. Statistical analyses were performed using statistical software (SPSS, Version 23; Chicago, IL). P values < 0.05 were considered statistically significant.
Results
Patient characteristics
Eighteen patients (aged 20–67 years old), who fulfilled the eligibility criteria, based on their medical records, were retrospectively analyzed. Tumor tissue samples were available for MGMT analysis from all subjects by gross total resection (n = 15) or subtotal resection (n = 3). A methylated MGMT promoter was found in 10 cases (55.6%, 47.3 ± 14.3 years), and an unmethylated MGMT promoter was proven in eight cases (44.4%, 51.1 ± 12.4 years). In this retrospective study, three of the subjects were included in a previous paper [29].
MTRasym spectra for MGMT unmethylated and methylated GBMs
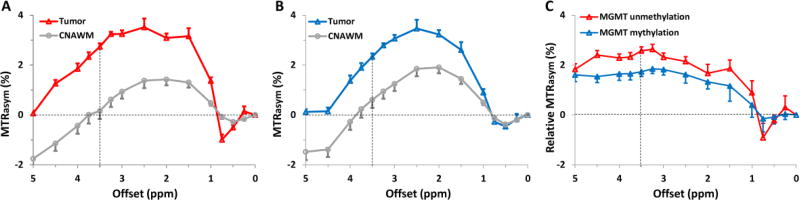
The average MTRasym spectra for the two GBM groups—with a methylated MGMT promoter and an unmethylated MGMT promoter—were compared to explore the specific characteristics of the APT effect at an offset of ~3.5 ppm downfield from water (Fig. 2). In the offset range of 1–4.5 ppm, both GBM groups demonstrated stronger protein-based APT (at 3.5 ppm) and other CEST effects, compared to the CNAWM. For human studies at 3T, no obvious APT effect can be clearly observed at 3.5 ppm offset, due to the larger voxel size, the larger B0 inhomogeneity, and the smaller absolute offset range. Notably, the CEST signal intensities in the offset range of 1–4.5 ppm were relatively higher in MGMT unmethylated GBMs than in MGMT methylated GBMs.
Fig. 2.
A,B Comparison of the average MTRasym spectra of the tumor mass and CNAWM for GBMs with an unmethylated MGMT promoter (n = 8, A) and a methylated MGMT promoter (n = 10, B). The CEST effects were clearly observed at multiple frequencies in the 1–4.5 ppm frequency offset range. C The average relative MTRasym spectra of the tumor mass (MTRasym spectra from tumor mass - MTRasym spectra from CNAWM). The error bars in the figures are shown as standard errors.
APTw image features for MGMT unmethylated and methylated GBMs
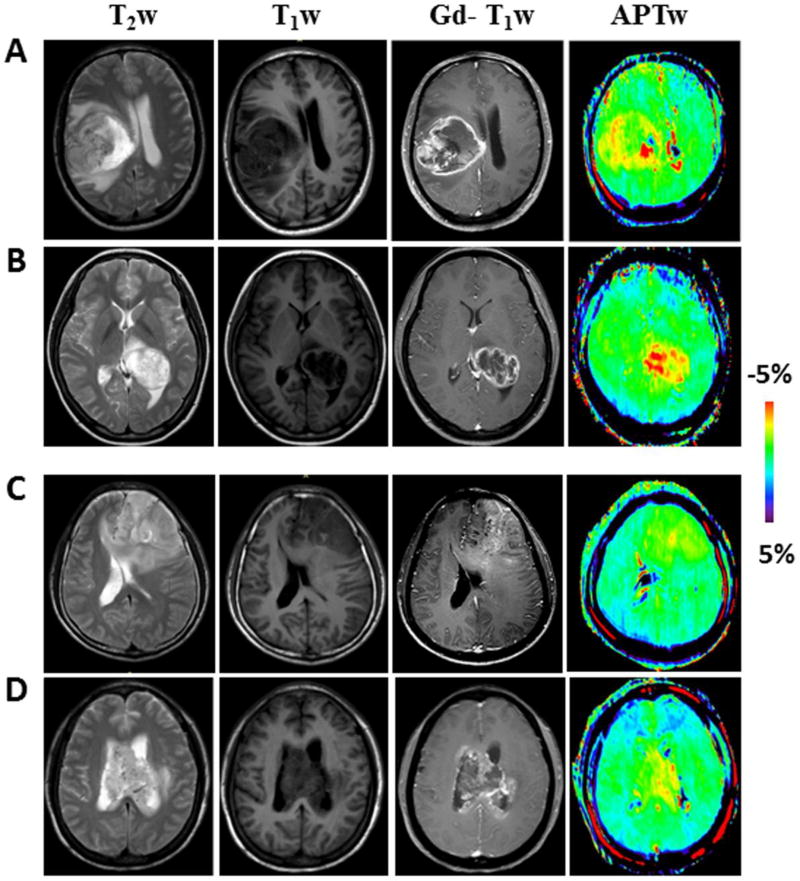
Of eight MGMT unmethylated GBMs, three lesions were limited to one single lobe, three lesions had infiltrated two lobes, and two lesions were mainly involved in the third ventricle. The APTw images often demonstrated these MGMT unmethylated GBMs as highly heterogeneous, ring-like or single nodular, hyperintense lesions, compared to the CNAWM. Figure 3A and B shows two typical examples of standard and APTw MR images for two MGMT unmethylated GBMs.
Fig. 3.
A, B Conventional and APTw MR images of two typical GBMs with an unmethylated MGMT promoter, illustrating the heterogeneous ring-enhancement characteristic of the disease on Gd-T1w image. APTw image demonstrated the lesion with ring-like, strong hyperintensity. C, D Conventional and APTw MR images of two typical MGMT promoter-methylated GBMs. Gd-T1w image demonstrated a patchy Gd-enhancing tumor mass. APTw image showed the masses as a hyperintense nodule (C) or scattered patch (D).
Of 10 patients with MGMT methylated GBM, seven cases were located in one single lobe, one case involved the right parieto-occipital lobes, and two involved the third ventricle. These MGMT methylated GBMs showed ring-like, scattered patchy, or nodular heterogeneous APTw hyperintensity, compared to the CNAWM. Figure 3C and D shows one example of a patient with an MGMT methylated GBM.
Quantitative APTw analyses
Based on the quantitative analysis and comparison, the APTw value metrics (Mean, Variance, 50th percentile, 90th percentile, and Width10–90) were significantly different between the two groups (p < 0.05), and lesions that harbored an unmethylated MGMT promoter showed higher values than did MGMT methylated lesions, as listed in Table 1.
Table 1.
Quantitative APTw intensity values and the corresponding diagnostic performance for MGMT unmethylated and methylated GBMs.
| Unmethylated | Methylated | P value |
AUC (95% Confidence interval) |
Cut-off value |
Sensitivity | Specificity | Accuracy | |
|---|---|---|---|---|---|---|---|---|
| Mean | 2.54±0.41 | 2.01±0.42 | 0.022* | 0.825 (0.626–1.000) | 2.26 | 87.5% | 80% | 83.3% |
| Variance | 1.01±0.34 | 0.59±0.24 | 0.01 1* | 0.837 (0.649–1.000) | 0.94 | 62.5% | 90% | 77.8% |
| Skewness | 0.04±0.52 | 0.06±0.87 | 0.963 | |||||
| Kurtosis | 4.67±1.93 | 4.80±3.48 | 0.934 | |||||
| 10th percentile | 1.40±0.53 | 1.06±0.45 | 0.186 | |||||
| 50th percentile | 2.54±0.36 | 1.99±0.41 | 0.012* | 0.850 (0.672–1.000) | 2.25 | 75% | 80% | 77.8% |
| 90th percentile | 3.71±0.45 | 2.93±0.53 | 0.006* | 0.856 (0.674–1.000) | 3.25 | 87.5% | 70% | 77.8% |
| Width10–90 | 2.31±0.42 | 1.87±0.41 | 0.049* | 0.763 (0.537–0.988) | 2.15 | 62.5% | 80% | 72.2% |
| Mode | 2.45±0.38 | 2.05±0.47 | 0.086 |
Note: APTw values were recorded as average ± standard deviation (% of the water signal intensity). AUC is the area under the ROC curve. Significant difference (p < 0.05) was marked by a *.
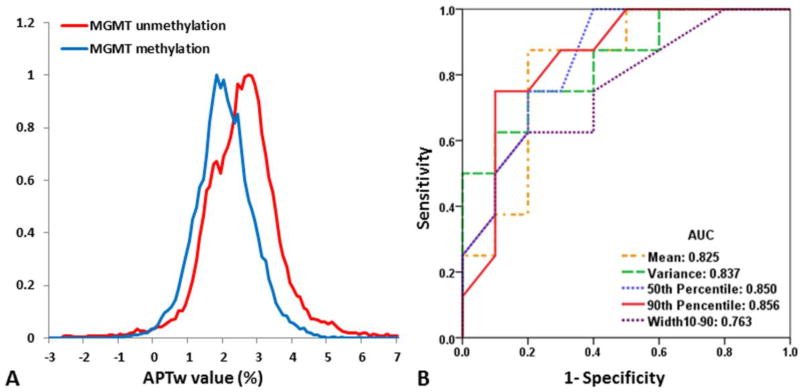
Figure 4A shows the whole-tumor APTw histograms obtained for GBMs with unmethylated and methylated MGMT promoters. The GBMs with an unmethylated MGMT promoter encompassed more voxels with APTw hyperintensity, compared to methylated GBMs. Histogram-based APTw metrics for two cohorts of GBMs are summarized in Table 1. With regards to the differences between the histogram-based APTw value metrics, the MGMT unmethylated GBMs had significantly higher Mean (2.54 ± 0.41 vs. 2.01 ± 0.42; P = 0.022), Variance (1.01 ± 0.34 vs. 0.59 ± 0.24; P = 0.011), 50th percentile (2.54 ± 0.36 vs. 1.99 ± 0.41; P = 0.012), 90th percentile (3.71 ± 0.45 vs. 2.93 ± 0.53; P = 0.006), and Width10–90 (2.31 ± 0.42 vs. 1.87 ± 0.41; P = 0.049) values than the MGMT methylated GBMs. The10th percentile and Mode values showed a higher trend in the MGMT unmethylated group, compared to the MGMT methylated (P = 0.186, 0.086, respectively). Skewness and Kurtosis showed no difference between the two groups (P = 0.963, 0.934, respectively), which indicates that the shapes of the histograms from the two groups are almost identical.
Fig. 4.
A Comparison of average tumor mass APTw histograms for all GBMs with an unmethylated (n = 8) and a methylated (n = 10) MGMT promoter. The GBMs with an unmethylated MGMT promoter show more voxels with APTw hyperintensity. B ROC analysis of APTw metrics as imaging biomarkers with which to distinguish GBMs with an unmethylated MGMT promoter from GBMs with a methylated MGMT promoter. The 90th Percentile APTw value demonstrated the highest AUC, at 0.856. AUC: area under the curve.
Prediction of MGMT promoter methylation status with APTw metrics
Mean, Variance, 50th percentile, 90th percentile, and Width10–90 APTw values differed significantly between the MGMT unmethylated and MGMT methylated groups. Based on ROC curve analyses (Fig. 4B), the 90th percentile values showed the highest area under the ROC curve (AUC = 0.856), and the Mean showed the highest accuracy (83.3%) in predicting the MGMT methylation status (Table 1). MGMT methylation status was, thus, predictable with APTw imaging, non-invasively.
Discussion
Recently, increasing evidence suggests that MGMT methylation status could be a strong predictive and prognostic factor in the GBM patient cohort who are undergoing chemotherapy with alkylating agents, both in newly diagnosed [8, 52–54] and recurrent [55, 56] patients. Thus, MGMT methylation status has been served as a stratification marker in randomized clinical trials. Predicting the MGMT methylation status before treatment, with methods such as MRI, is of paramount importance. Nevertheless, the early results published, to date, have presented some controversial conclusions with regard to the correlation between MRI parameters and MGMT methylation status [19, 57, 58]. When applying machine-learning and texture analysis, the texture features originating from T2w images were claimed to reach an AUC of 0.85 [16], or an accuracy of 71% [15]. The texture analysis reveals subtleties not seen by an observer to aid in determining the MGMT status, but at the cost of a huge amount of computational power and time-consuming registration.
Our study shows that GBMs with an unmethylated MGMT promoter are typically associated with relatively higher APTw signal intensity values, compared to methylated GBMs. Multiple APTw-MRI metrics are potentially capable of identifying the MGMT methylation status in GBMs. To the best of our knowledge, this is the first presentation to evaluate correlations between APTw imaging features and MGMT promoter methylation status in patients with GBM. It was anticipated that the potential to determine MGMT status noninvasively could improve treatment efficacy in patients with recurrent GBM, because the repeated surgery only for MGMT status determination is not the optimized regimen in clinical practice.
In this study, we implemented the whole-tumor-ROI-based histogram analysis for APTw value assessment. Strong and significant higher histogram-based Mean, Variance, 50th percentile, 90th percentile, and Width10–90 APTw values were identified in MGMT unmethylated GBMs, compared with methylated GBMs (Table 1). The corresponding AUCs were 0.813, 0.825, 0.837, 0.850, 0.856, and 0.763, respectively, for the determination of the MGMT methylation status (Fig. 4B). Among the aforementioned APTw parameters for the feasibility of the discrimination of the MGMT genotype of GBMs, the Mean APTw value showed the highest diagnostic accuracy (83.3%). Therefore, the APTw signals could be valuable imaging biomarkers with which to predict MGMT methylation status in GBMs.
Among the histogram parameters that were utilized in this study, the Skewness and Kurtosis, which depict the lack of symmetry and normal distribution of the data shape, as well as the Mode, which represents the value with highest frequency to be sampled, showed no difference (Fig. 4A). The most significantly different portion was found at the right tail in the histogram, where the 90th percentile was assigned. Thus, it could be reasonably extrapolated that the GBMs with an unmethylated MGMT promoter usually present a larger proportion of voxels with hyperintensity on APTw images than the MGMT methylated GBMs.
Generally, the stratification accuracy of our proposed APTw-MRI approach is a little higher than other methods reported, with quantitative imaging biomarkers extracted from some advanced MRI sequences, and is comparable with the texture analysis when investigating big MRI datasets. The MGMT promoter methylation in cancer suggests a decrease in protein expression [59]. This may affect other protein expressions downstream of MGMT. Therefore, protein-based APTw-MRI is potentially a novel imaging biomarker for the prediction of MGMT methylation status. It should be kept in mind that the APTw-MRI signal is associated with a large group of cellular (mainly cytosolic) proteins, each contributing multiple amide groups. The exact quantitative explanation demands a further proteomics study, as shown in the previous work [60].
There are a few limitations that relate to our conclusions, which merit discussion. The first limitation was the relatively small sample size used in our study, necessitating a future large-scale study to obtain more conclusive results. The second was the semi-quantitative nature of the current APTw signal intensity, due to the contamination by the upfield nuclear Overhauser enhancement and other effects [43, 48]. Fortunately, several improved APTw imaging analysis or acquisition approaches [61–66] have been proposed to achieve a more pure APT effect. Notably, evidence has also shown that the APT effect would be the major contributor to the APTw image contrast obtained in our experimental setting [43, 67, 68]. Finally, APTw MRI is very sensitive to motion. Artifacts associated with the intraventricular CSF pulsation can often be found in ventricles on APTw images that could disturb the clinical diagnosis.
In conclusion, our early results show that multiple APTw-MRI metrics provide valuable information for the noninvasive discrimination of MGMT promoter methylation status in GBMs. The findings are of paramount importance for the management of alkylating agent chemotherapy and of particular benefit for ambiguous recurrent cases where biopsy or reoperation could potentially be avoided.
Key Points.
APTw-MRI is applied to predict MGMT promoter methylation status in GBMs.
GBMs with unmethylated MGMT promoter present higher APTw-MRI than methylated GBMs.
Multiple APTw histogram metrics can identify MGMT methylation status.
Mean APTw values showed the highest diagnostic accuracy (AUC = 0.825).
Acknowledgments
The authors thank Ms. Mary McAllister for editorial assistance.
Funding:
This study was partially supported by grants from National Natural Science Foundation of China (81171322), Guangdong Provincial Natural Science Foundation (2014A030313271, S2012010009114), Guangdong Provincial Science and Technology Project (2014A020212726), Southern Medical University clinical research project (LC2016ZD028), and the National Institutes of Health (R01EB009731, R01CA166171).
Abbreviations and acronyms
- APTw
amide proton transfer-weighted
- MRI
magnetic resonance imaging
- ROC
receiver operator characteristic curve
- AUC
area under the curve
- MGMT
O6-methylguanine-DNA methyltransferase
- GBM
glioblastoma
- CEST
chemical exchange-dependent saturation transfer
- T2w
T2-weighted
- T1w
T1-weighted
- FLAIR
fluid-attenuated inversion recovery
- Gd-T1w
gadolinium-enhanced T1-weighted
- ADC
apparent diffusion coefficient
Footnotes
Compliance with ethical standards:
Ethical approval:
Institutional Review Board approval was obtained.
Informed consent:
Written informed consent was waived by the Institutional Review Board.
Guarantor:
The scientific guarantor of this publication is Zhibo Wen, MD, PhD.
Conflict of interest:
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry:
One of the authors (Dr. Fangyao Chen) has significant statistical expertise. No complex statistical methods were necessary for this paper.
Study subjects or cohorts overlap:
Three study subjects have been previously reported in one of our previous papers, in which we evaluated the diagnostic values of APTw imaging in differentiate PCNSL and malignant gliomas, see Ref. [29].
Methodology: retrospective, diagnostic or prognostic study, performed at one institution.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Claes A, Idema AJ, Wesseling P. Diffuse glioma growth: a guerilla war. Acta Neuropathol. 2007;114:443–458. doi: 10.1007/s00401-007-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Stevens MF, Laughton CA, Madhusudan S, Bradshaw TD. Acquired resistance to temozolomide in glioma cell lines: molecular mechanisms and potential translational applications. Oncology. 2010;78:103–114. doi: 10.1159/000306139. [DOI] [PubMed] [Google Scholar]
- 4.Ramirez YP, Weatherbee JL, Wheelhouse RT, Ross AH. Glioblastoma multiforme therapy and mechanisms of resistance. Pharmaceuticals (Basel) 2013;6:1475–1506. doi: 10.3390/ph6121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woods D, Turchi JJ. Chemotherapy induced DNA damage response: convergence of drugs and pathways. Cancer Biol Ther. 2013;14:379–389. doi: 10.4161/cbt.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi C, Ganji S, Hulsey K, et al. A comparative study of short- and long-TE (1)H MRS at 3 T for in vivo detection of 2-hydroxyglutarate in brain tumors. NMR Biomed. 2013;26:1242–1250. doi: 10.1002/nbm.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed SU, Carruthers R, Gilmour L, Yildirim S, Watts C, Chalmers AJ. Selective Inhibition of Parallel DNA Damage Response Pathways Optimizes Radiosensitization of Glioblastoma Stem-like Cells. Cancer Res. 2015;75:4416–4428. doi: 10.1158/0008-5472.CAN-14-3790. [DOI] [PubMed] [Google Scholar]
- 8.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 9.Zhao F, Li M, Kong L, Zhang G, Yu J. Delineation of radiation therapy target volumes for patients with postoperative glioblastoma: a review. Onco Targets Ther. 2016;9:3197–3204. doi: 10.2147/OTT.S104241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 11.Sarkaria JN, Kitange GJ, James CD, et al. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–2908. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 13.Pope WB, Chen JH, Dong J, et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology. 2008;249:268–277. doi: 10.1148/radiol.2491072000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellingson BM. Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr Neurol Neurosci Rep. 2015;15:506. doi: 10.1007/s11910-014-0506-0. [DOI] [PubMed] [Google Scholar]
- 15.Drabycz S, Roldan G, de Robles P, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage. 2010;49:1398–1405. doi: 10.1016/j.neuroimage.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 16.Korfiatis P, Kline TL, Coufalova L, et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys. 2016;43:2835–2844. doi: 10.1118/1.4948668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassner A, Thornhill RE. Texture analysis: a review of neurologic MR imaging applications. AJNR Am J Neuroradiol. 2010;31:809–816. doi: 10.3174/ajnr.A2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris RJ, Cloughesy TF, Liau LM, et al. pH-weighted molecular imaging of gliomas using amine chemical exchange saturation transfer MRI. Neuro-Oncol. 2015;17:1514–1524. doi: 10.1093/neuonc/nov106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon WJ, Choi JW, Roh HG, Lim SD, Koh YC. Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology. 2012;54:555–563. doi: 10.1007/s00234-011-0947-y. [DOI] [PubMed] [Google Scholar]
- 20.Corrigan F, Mander KA, Leonard AV, Vink R. Neurogenic inflammation after traumatic brain injury and its potentiation of classical inflammation. J Neuroinflammation. 2016;13:264. doi: 10.1186/s12974-016-0738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta A, Omuro AM, Shah AD, et al. Continuing the search for MR imaging biomarkers for MGMT promoter methylation status: conventional and perfusion MRI revisited. Neuroradiology. 2012;54:641–643. doi: 10.1007/s00234-011-0970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koyama H, Ikenuma H, Toda H, et al. Synthesis of PET probe O6-[(3-[11C]methyl)benzyl]guanine by Pd0-mediated rapid C-[11C]methylation toward imaging DNA repair protein O6-methylguanine-DNA methyltransferase in glioblastoma. Bioorg Med Chem Lett. 2017;27:1892–1896. doi: 10.1016/j.bmcl.2017.03.045. [DOI] [PubMed] [Google Scholar]
- 23.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143:79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, van Zijl PC. Chemical exchange saturation transfer imaging and spectroscopy. Progr NMR Spectr. 2006;48:109–136. [Google Scholar]
- 25.Zhou J, Payen J, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature Med. 2003;9:1085–1090. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 26.Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PCM. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med. 2003;50:1120–1126. doi: 10.1002/mrm.10651. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Zhu H, Lim M, et al. Three-dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement. J Magn Reson Imaging. 2013;38:1119–1128. doi: 10.1002/jmri.24067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Togao O, Yoshiura T, Keupp J, et al. Amide proton transfer imaging of adult diffuse gliomas: correlation with histopathological grades. Neuro-Oncology. 2014;16:441–448. doi: 10.1093/neuonc/not158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang S, Yu H, Wang X, et al. Molecular MRI differentiation between primary central nervous system lymphomas and high-grade gliomas using endogenous protein-based amide proton transfer MR imaging at 3 Tesla. Eur Radiol. 2016;26:64–71. doi: 10.1007/s00330-015-3805-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu H, Lou H, Zou T, et al. Applying protein-based amide proton transfer MR imaging to distinguish solitary brain metastases from glioblastoma. Eur Radiol. 2017 doi: 10.1007/s00330-00017-04867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia G, Abaza R, Williams JD, et al. Amide proton transfer MR imaging of prostate cancer: A preliminary study. J Magn Reson Imaging. 2011;33:647–654. doi: 10.1002/jmri.22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan J, Chen S, King AD, et al. Amide proton transfer-weighted imaging of the head and neck at 3 T: a feasibility study on healthy human subjects and patients with head and neck cancer. NMR Biomed. 2014;27:1239–1247. doi: 10.1002/nbm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen Z, Hu S, Huang F, et al. MR imaging of high-grade brain tumors using endogenous protein and peptide-based contrast. Neuroimage. 2010;51:616–622. doi: 10.1016/j.neuroimage.2010.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi YS, Ahn SS, Lee SK, et al. Amide proton transfer imaging to discriminate between low- and high-grade gliomas: added value to apparent diffusion coefficient and relative cerebral blood volume. Eur Radiol. 2017 doi: 10.1007/s00330-00017-04732-00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Togao O, Hiwatashi A, Yamashita K, et al. Grading diffuse gliomas without intense contrast enhancement by amide proton transfer MR imaging: comparisons with diffusion- and perfusion-weighted imaging. Eur Radiol. 2017;27:578–588. doi: 10.1007/s00330-016-4328-0. [DOI] [PubMed] [Google Scholar]
- 36.Jiang S, Eberhart CG, Zhang Y, et al. Amide proton transfer-weighted MR image-guided stereotactic biopsy in patients with newly diagnosed gliomas. Eur J Cancer. 2017;83:9–18. doi: 10.1016/j.ejca.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma B, Blakeley JO, Hong X, et al. Applying amide proton transfer-weighted MRI to distinguish pseudoprogression from true progression in malignant gliomas. J Magn Reson Imaging. 2016;44:456–462. doi: 10.1002/jmri.25159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park KJ, Kim HS, Park JE, Shim WH, Kim SJ, Smith SA. Added value of amide proton transfer imaging to conventional and perfusion MR imaging for evaluating the treatment response of newly diagnosed glioblastoma. Eur Radiol. 2016;26:4390–4403. doi: 10.1007/s00330-016-4261-2. [DOI] [PubMed] [Google Scholar]
- 39.Oue N, Shigeishi H, Kuniyasu H, et al. Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int J Cancer. 2001;93:805–809. doi: 10.1002/ijc.1403. [DOI] [PubMed] [Google Scholar]
- 40.Tee YK, Donahue MJ, Harston GW, Payne SJ, Chappell MA. Quantification of amide proton transfer effect pre- and post-gadolinium contrast agent administration. J Magn Reson Imaging. 2014;40:832–838. doi: 10.1002/jmri.24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Heo HY, Lee DH, et al. Selecting the reference image for registration of CEST series. J Magn Reson Imaging. 2016;43:756–761. doi: 10.1002/jmri.25027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST) Proc Natl Acad Sci (USA) 2008;105:2266–2270. doi: 10.1073/pnas.0707666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, Hong X, Zhao X, Gao J-H, Yuan J. APT-weighted and NOE-weighted image contrasts in glioma with different RF saturation powers based on magnetization transfer ratio asymmetry analyses. Magn Reson Med. 2013;70:320–327. doi: 10.1002/mrm.24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones CK, Huang A, Xu J, et al. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7T. Neuroimage. 2013;77:114–124. doi: 10.1016/j.neuroimage.2013.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heo H-Y, Zhang Y, Lee D-H, Hong X, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear Overhauser enhancement (NOE) imaging with extrapolated semi-solid magnetization transfer reference (EMR) signals: Application to a rat glioma model at 4.7 T. Magn Reson Med. 2016;75:137–138. doi: 10.1002/mrm.25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paech D, Zaiss M, Meissner JE, et al. Nuclear overhauser enhancement mediated chemical exchange saturation transfer imaging at 7 Tesla in glioblastoma patients. PLoS One. 2014;9:e104181. doi: 10.1371/journal.pone.0104181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hua J, Jones CK, Blakeley J, Smith SA, van Zijl PCM, Zhou J. Quantitative description of the asymmetry in magnetization transfer effects around the water resonance in the human brain. Magn Reson Med. 2007;58:786–793. doi: 10.1002/mrm.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J, Blakeley JO, Hua J, et al. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med. 2008;60:842–849. doi: 10.1002/mrm.21712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang HY, Huang YQ, Yang ZX, Ying D, Zeng MS, Rao SX. Potential of MR histogram analyses for prediction of response to chemotherapy in patients with colorectal hepatic metastases. Eur Radiol. 2016;26:2009–2018. doi: 10.1007/s00330-015-4043-2. [DOI] [PubMed] [Google Scholar]
- 50.Wang HY, Su ZH, Xu X, et al. Dynamic contrast-enhanced MR imaging in renal cell carcinoma: Reproducibility of histogram analysis on pharmacokinetic parameters. Sci Rep. 2016;6:29146. doi: 10.1038/srep29146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paz MF, Yaya-Tur R, Rojas-Marcos I, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–4938. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 52.Ahluwalia MS. American Society of Clinical Oncology 2011 CNS tumors update. Expert Rev Anticancer Ther. 2011;11:1495–1497. doi: 10.1586/era.11.151. [DOI] [PubMed] [Google Scholar]
- 53.Reifenberger G, Hentschel B, Felsberg J, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2012;131:1342–1350. doi: 10.1002/ijc.27385. [DOI] [PubMed] [Google Scholar]
- 54.Muldoon LL, Gahramanov S, Li X, Marshall DJ, Kraemer DF, Neuwelt EA. Dynamic magnetic resonance imaging assessment of vascular targeting agent effects in rat intracerebral tumor models. Neuro Oncol. 2011;13:51–60. doi: 10.1093/neuonc/noq150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baur M, Preusser M, Piribauer M, et al. Frequent MGMT (0(6)-methylguanine-DNA methyltransferase) hypermethylation in long-term survivors of glioblastoma: a single institution experience. Radiol Oncol. 2010;44:113–120. doi: 10.2478/v10019-010-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirk P, He T, Anderson LJ, et al. International reproducibility of single breathhold T2* MR for cardiac and liver iron assessment among five thalassemia centers. J Magn Reson Imaging. 2010;32:315–319. doi: 10.1002/jmri.22245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pope WB, Lai A, Mehta R, et al. Apparent diffusion coefficient histogram analysis stratifies progression-free survival in newly diagnosed bevacizumab-treated glioblastoma. AJNR Am J Neuroradiol. 2011;32:882–889. doi: 10.3174/ajnr.A2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romano A, Calabria LF, Tavanti F, et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. Eur Radiol. 2013;23:513–520. doi: 10.1007/s00330-012-2601-4. [DOI] [PubMed] [Google Scholar]
- 59.Oue N, Shigeishi H, Kuniyasu H, et al. Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int J Cancer. 2001;93:805–809. doi: 10.1002/ijc.1403. [DOI] [PubMed] [Google Scholar]
- 60.Yan K, Fu Z, Yang C, et al. Assessing amide proton transfer (APT) MRI contrast origins in 9L gliosarcoma in the rat brain using proteomic analysis. Mol Imaging Biol. 2015;17:479–487. doi: 10.1007/s11307-015-0828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaiss M, Schmitt B, Bachert P. Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of z-spectra. J Magn Reson. 2011;211:149–155. doi: 10.1016/j.jmr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 62.Jin T, Wang P, Zong X, Kim S-G. MR imaging of the amide-proton transfer effect and the pH-insensitive nuclear overhauser effect at 9.4 T. Magn Reson Med. 2013;69:760–770. doi: 10.1002/mrm.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zu Z, Janve VA, Xu J, Does MD, Gore JC, Gochberg DF. A new method for detecting exchanging amide protons using chemical exchange rotation transfer. Magn Reson Med. 2013;69:637–647. doi: 10.1002/mrm.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JS, Xia D, Ge Y, Jerschow A, Regatte RR. Concurrent saturation transfer contrast in in vivo brain by a uniform magnetization transfer MRI. Neuroimage. 2014;95:22–28. doi: 10.1016/j.neuroimage.2014.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zaiss M, Windschuh J, Paech D, et al. Relaxation-compensated CEST-MRI of the human brain at 7 T: Unbiased insight into NOE and amide signal changes in human glioblastoma. Neuroimage. 2015;112:180–188. doi: 10.1016/j.neuroimage.2015.02.040. [DOI] [PubMed] [Google Scholar]
- 66.Zaiss M, Windschuh J, Goerke S, et al. Downfield-NOE-suppressed amide-CEST-MRI at 7 Tesla provides a unique contrast in human glioblastoma. Magn Reson Med. 2017;77:196–208. doi: 10.1002/mrm.26100. [DOI] [PubMed] [Google Scholar]
- 67.Heo HY, Zhang Y, Jiang S, Lee DH, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semisolid magnetization transfer reference (EMR) signals: II. Comparison of three EMR models and application to human brain glioma at 3 Tesla. Magn Reson Med. 2016;75:1630–1639. doi: 10.1002/mrm.25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee DH, Heo HY, Zhang K, et al. Quantitative assessment of the effects of water proton concentration and water T1 changes on amide proton transfer (APT) and nuclear overhauser enhancement (NOE) MRI: The origin of the APT imaging signal in brain tumor. Magn Reson Med. 2017;77:855–863. doi: 10.1002/mrm.26131. [DOI] [PMC free article] [PubMed] [Google Scholar]