Summary
Zika virus (ZIKV) remained obscure until the recent explosive outbreaks in French Polynesia (2013-2014) and South America (2015-2016). Phylogenetic studies reveal that ZIKV has evolved into African and Asian lineages. The Asian lineage of ZIKV is responsible for the recent epidemics in the Americas. However, the underlying mechanisms through which ZIKV rapidly and explosively spread from Asia to the Americas are limited. We have recently shown that nonstructural protein 1 (NS1) facilitates flavivirus acquisition by mosquitoes from an infected mammalian host and subsequently enhances viral prevalence in mosquitoes. Here, we report that the antigenemia of NS1 determines ZIKV infectivity in its mosquito vector Aedes aegypti, which acquires ZIKV via a blood meal. Clinical isolates from the most recent outbreak in the Americas were much more infectious in mosquitoes than the FSS13025 strain, which was isolated in Cambodia in 2010. Further analyses showed that these epidemic strains have more robust NS1 antigenemia than the FSS13025 strain because of an alanine-to-valine amino acid substitution at the 188th residue in NS1. ZIKV infectivity was enhanced by this residue substitution in the ZIKV FSS13025 strain in mosquitoes that acquired ZIKV from a viremic Type I and II interferon receptor-deficient (ifnagr-/-) C57BL/6 (AG6) mouse. Our results reveal that ZIKV evolved to acquire a spontaneous mutation in its NS1 protein, resulting in increased antigenemia of the protein. Enhancement of NS1 antigenemia in infected hosts promotes ZIKV infectivity and prevalence in mosquitoes, which potentially facilitates transmission during the recent ZIKV epidemics.
Zika virus (ZIKV), a mosquito-borne virus belonging to the genus Flavivirus of the family Flaviviridae, is transmitted to humans by Aedes mosquito species1,2. Several neurological complications, such as Guillain-Barré syndrome in adults3 and microcephaly in neonates4, are potentially associated with ZIKV infection. Before 2007, cases of ZIKV infection were only sporadically detected2,5. The first reported mild ZIKV outbreak occurred in 2007 on a Western Pacific Micronesian Yap Island6. In 2013, a larger ZIKV epidemic was recorded in French Polynesia and other Southern Pacific islands7,8. In 2015, ZIKV emerged in the Americas for the first time, rapidly spreading to 20 countries or territories9. Phylogenetic studies have revealed that ZIKV has evolved into African and Asian clusters2,9. Sequence analyses revealed that the ZIKV strains causing earlier outbreaks in Micronesia (2007) and French Polynesia (2013-2014) and later outbreaks in the Americas (2015-2016) all belong to the Asian clade2, suggesting that increased infectivity of the Asian ZIKV lineage might contribute to the recent worldwide epidemic.
Evolutionary enhancement of infectivity of mosquito-borne viruses within their vectors results in high epidemic potential10,11. We speculate that the adaptability and infectivity of ZIKV within its mosquito vectors might have evolved with time, contributing to the prevalence and spread of the virus from Asia to the Americas. To test this hypothesis, we compared the viral infectivity of two clinical ZIKV isolates of the Asian lineage, GZ01 isolated in 201612 and FSS13025 in 201013, in the primary urban mosquito vector, Aedes aegypti. Phylogenetic analysis shows that these 2 strains represent the clades corresponding to the epidemic strains of ZIKV that are prevalent in Americas (2015-2016) and the Southeastern Asia (2007-2012) (Extended Data Fig. 1). Thus, we exploited a "host-mosquito" acquisition model to assess the infectivity of these ZIKV strains, via mosquitoes acquiring ZIKV from a viremic AG6 mouse 14–16 (Fig. 1a). Both strains exhibited similar replication kinetics in the infected AG6 mouse blood (Fig. 1b). However, the serum level of secreted nonstructural protein 1 (NS1) in the FSS13025-infected AG6 mice was much lower (Fig. 1c). The infected mice were subjected to daily biting by the Rockefeller laboratory strain of A. aegypti from days 1 to 5 post-mouse infection (Fig. 1a). Compared to the GZ01 strain, the FSS13025 strain had a significantly lower infection prevalence in the mosquitoes that fed on the infected mice (Fig. 1d, e).
Figure 1. Comparing the infectivity of ZIKV isolates of Asian lineage in mosquitoes.
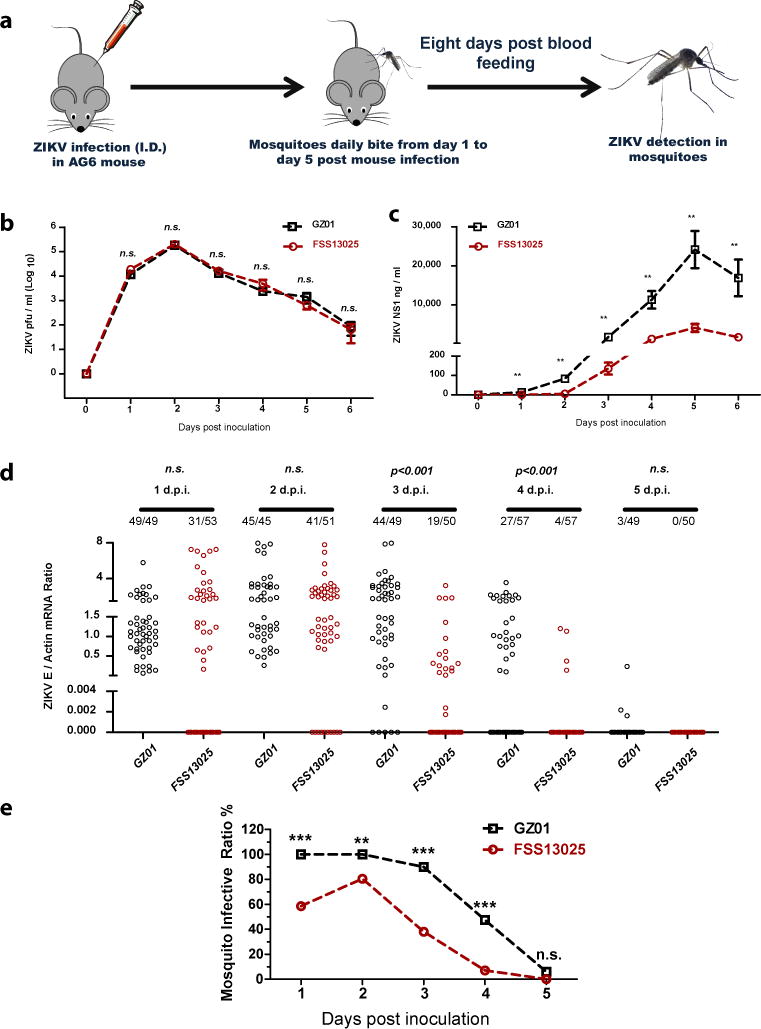
a, Schematic representation of the study design. Separated groups of AG6 mice were intradermally infected with 1×104 pfu of the GZ01 and FSS13025 strains.
b, Detection of the ZIKV load in the blood plasma by a plaque assay (n=14 mice per group pooled from 5 independent biological replicates).
c, ZIKV NS1 measurement by ELISA (n=6 mice per group pooled from 3 independent biological replicates).
d, e, Comparison of the infectivity of two ZIKV isolates in A. aegypti (n=6 mice per group pooled from 3 independent biological replicates). The number at the top of each column represents infected number/total number. Each dot represents a mosquito (d). The data are represented as the percentage of mosquito infections (e).
Data are mean ± s.e.m. (b, c). p values were determined by two-tailed Mann-Whitney test (b-d) or two-sided Fisher’s exact test (e). **p< 0.01, ***p< 0.001, n.s., not significant.
Secretion of NS1, a virus-encoded nonstructural protein, into the serum of an infected host is a common property of many flaviviruses15,17–20. Our previous study demonstrated that circulating NS1 acquired by mosquitoes together with virions when mosquitoes feed on a viremic mammalian host enhances viral infectivity15. NS1 antigenemia plays a key role in maintaining the flavivirus life cycle15. We hypothesize that NS1 antigenemia is responsible for the differential mosquito infectivity between the GZ01 and FSS13025 strains. We therefore infected Vero cells with the American isolates GZ01 and PRVABC5921, the SZ01 strain representing the French Polynesian clade22, and the FSS13025 strain. Three post-epidemic strains showed higher NS1 secretion than the FSS13025 strain (Fig. 2a–c), even when NS1 amount was normalized to viral titer (Fig. 2c).
Figure 2. Differences in NS1 secretion determine ZIKV infectivity in mosquitoes.
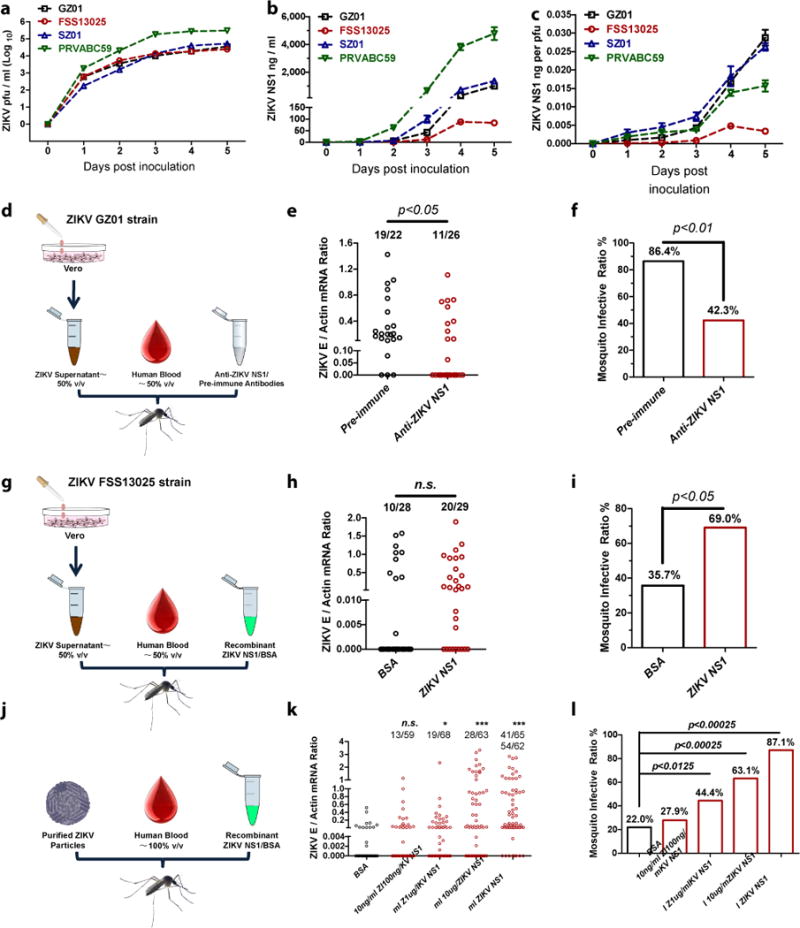
a-c, NS1 secretability in ZIKV-infected Vero cells. Vero cells were infected with 0.01 M.O.I. viruses. c, NS1 quantity normalized against average viral titer (ng per pfu). The data were pooled from 3 independent biological replicates.
d-f, Immunoblockade of NS1 in GZ01-infected supernatants reduced ZIKV acquisition. Either 10 μl of murine ZIKV NS1 antisera or pre-immune sera were mixed with supernatant from ZIKV GZ01-infected Vero cells (500 μl) and fresh human blood (500 μl) for in vitro membrane feeding of A. aegypti. The data were pooled from 2 independent biological replicates.
g-i, Addition of NS1 in FSS13025-infected supernatant increased the mosquito infection. Either 10 μg of purified NS1 or BSA was incubated with fresh human blood (500 μl) and supernatant from ZIKV FSS13025-infected Vero cells (500 μl) to feed A. aegypti. The data were pooled from 2 independent biological replicates.
j-l, Assessment of the threshold NS1 concentration that enhances ZIKV acquisition. Serial dilutions of purified ZIKV NS1 (10 μl) were mixed with purified GZ01 ZIKV virions (10 μl) and fresh human blood (1000 μl) for A. aegypti feeding. The data were pooled from 4 independent biological replicates.
A final concentration of 1×105 pfu/ml ZIKV was used for mosquito oral infection (d, g, j).
The number at the top of each column represents infected number/total number. Each dot represents a mosquito (e, h, k).
Data are mean ± s.e.m. (a-c). p values were determined by two-tailed Mann-Whitney test (e, h, k) or two-sided Fisher’s exact test (f, i, l), and adjusted using Bonferroni correction to account for multiple comparisons (k, l). The p value represents a comparison between BSA and other groups (k, l). *p< 0.0125, ***p< 0.00025, n.s., not significant.
To determine whether NS1 secretability correlates with ZIKV infectivity in mosquitoes, we generated a murine NS1 polyclonal antibody (Extended Data Fig. 2a). Neutralization of NS1 via the antibody treatment reduced the mosquito infection prevalence of the GZ01 strain (Fig. 2d–f). The murine NS1 antibodies did not neutralize ZIKV virions (Extended Data Fig. 3). We also determined whether the addition of NS1 protein could enhance ZIKV infectivity. A recombinant GZ01 NS1 protein was expressed and purified using a Drosophila S2 expression system (Extended Data Fig. 2b). The presence of NS1 increased the infectivity of the FSS13025 strain in mosquitoes (Fig. 2g–i). We next assessed the threshold concentration of NS1 that can effectively enhance ZIKV acquisition. Compared to BSA, the presence of 100 ng/ml or higher concentrations of NS1 significantly increased the ZIKV prevalence in A. aegypti (Fig. 2j–l). To determine the role of animal NS1 antigenemia in ZIKV infectivity in mosquitoes, we allowed mosquitoes to feed on viremic AG6 mice (Extended Data Fig. 4a). The NS1 antisera treatment reduced the quantity of circulating NS1 in mouse serum (Extended Data Fig. 4b) but did not influence GZ01 ZIKV replication in AG6 mice (Extended Data Fig. 4c). The infection ratios of fed A. aegypti were reduced as a result of the antisera-mediated neutralization of the NS1 in mice (Extended Data Fig. 4d, e).
Considering the differential secretability of NS1 between the GZ01 and FSS13025 strains (Fig. 1c and Fig. 2a–c), we next attempted to identify the amino acids critical for NS1 secretion. We focused on the ER transmembrane region of pre-Membrane (prM), the full length of Envelope (E), and NS1 (prM-E-NS1)23 (Fig. 3a). There are only two variable sites between the GZ01 and FSS13025 prM-E-NS1 proteins, including the 473rd residue in the E protein (Val/FSS13025 to Met/GZ01) and the 188th residue in the NS1 protein (Ala/FSS13025 to Val/GZ01) (Fig. 3a). We therefore individually subcloned the prM-E-NS1 region from the GZ01 and FSS13025 strains and subsequently transfected these segments into 293T cells. Ectopic expression of the GZ01 prM-E-NS1 protein, but not the FSS13025 protein, led to abundant NS1 secretion in the supernatants of transfected cells (Fig. 3b). Mutation from valine to methionine at the 473rd residue of the E protein in the FSS13025 prM-E-NS1 protein did not improve NS1 secretion; however, the alanine to valine substitution at the NS1 188th residue rendered NS1 secretable (Fig. 3b), demonstrating that this single mutation determines NS1 secretability. The 188th residue is located within interface between two NS1 monomers24 (Extended Data Fig. 5a). Nonetheless, the alanine-to-valine substitution at position 188 does not influence NS1 dimerization (Extended Data Fig. 5b), suggesting that the mechanism how the single mutation affects NS1 remains to be defined.
Figure 3. A single amino acid residue in NS1 determines its secretability in the Asian lineage of ZIKV.
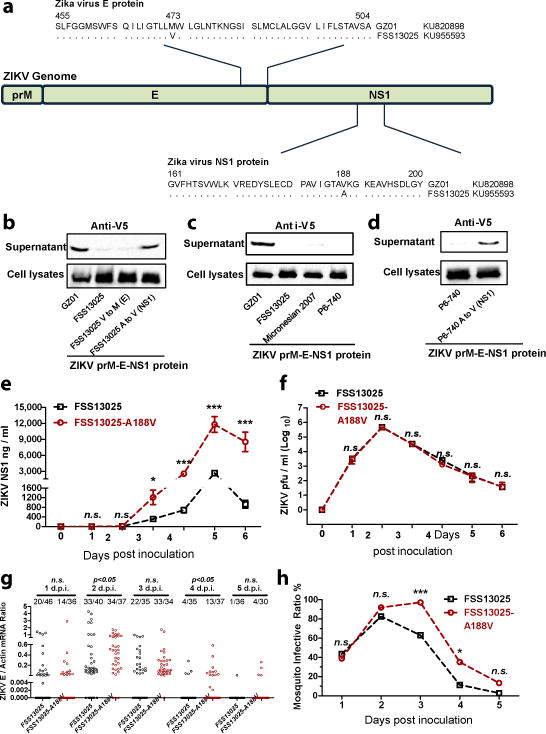
a, Comparison of the prM-E-NS1 sequence between GZ01 and FSS13025 isolates.
b, Detection of NS1 secretion in ectopically expressed prM-E-NS1 proteins of the subcloned and mutated GZ01 and FSS13025 strains.
c, Detection of NS1 secretion in ectopically expressed prM-E-NS1 proteins of ZIKV Asian strains.
d, Mutation of alanine to valine in the NS1 protein conferred NS1 secretability to the prM-E-NS1 proteins of a Malaysia P6-740 strain.
b-d, The experiments were reproduced 3 times with similar results. For gel source data, see Supplementary Fig. 1.
e, Measurement of ZIKV NS1 antigenemia by ELISA (n=10 mice per group pooled from 5 independent biological replicates).
f, Detection of the ZIKV load in the mouse blood by a plaque assay (n=8 mice per group pooled from 4 independent biological replicates).
g, h, Comparison of mosquito infectivity (g) and infective ratio (h) of ZIKV strains (n=6 mice per group pooled from 3 independent biological replicates). The number at the top of each column represents infected number/total number. Each dot represents a mosquito (g).
The AG6 mice were intradermally inoculated with 1×104 pfu of the FSS13025 and FSS13025-A188V strains, data are mean ± s.e.m. (e, f). p values were determined by two-tailed Mann-Whitney test (e-g) or two-sided Fisher’s exact test (h). *p< 0.05, ***p< 0.001, n.s., not significant.
We next performed an alignment of the NS1 proteins from the Asian clade of ZIKV. In ZIKV isolates collected before 2012, the 188th residue of the NS1 protein is an alanine. However, this residue is mutated to valine in all isolates after 2013 (Extended Data Fig. 6), suggesting that the Asian linage of ZIKV evolved to acquire NS1 secretability when it spread from Southeastern Asia to the Southern Pacific islands around 2013. Correspondingly, Singapore reported local ZIKV transmission in August 2016. The Singaporean strains stemmed from an ancestral node of the Asian lineage25. Indeed, in all ZIKV isolates from Singapore, the 188th residue of the NS1 protein is a valine (Extended Data Fig. 6), suggesting that the alanine-to-valine substitution may have contributed to the recent ZIKV epidemics. Therefore, we synthesized the prM-E-NS1 genes from a Malaysia P6-740 strain isolated in 1966 and a Micronesian 2007 strain and ectopically expressed these genes in 293T cells. Compared to the secretion of NS1 in the supernatants of GZ01 prM-E-NS1-transfected cells, the NS1 protein was hardly detectable in the supernatants of these pre-epidemic ZIKV prM-E-NS1-expressing cells (Fig. 3c). However, the alanine-to-valine mutation in the NS1 protein of the P6-740 strain conferred NS1 secretability (Fig. 3d).
Next, we created the alanine-to-valine mutation at the 188th position of the NS1 protein in an infectious clone of the FSS13025 strain (FSS13025-A188V)26. The substitution significantly increased NS1 secretion in the supernatant of infected Vero cells (Extended Data Fig. 7a–c). The FSS13025-A188V strain showed higher ZIKV infectivity in mosquitoes than wild type virus (Extended Data Fig. 7d, e). In the "mouse-mosquito" acquisition model, NS1 antigenemia was increased in the sera of FSS13025-A188V ZIKV-infected mice (Fig. 3e). Mutation of the NS1 188th position did not influence viral infectivity or dissemination kinetics in infected AG6 mouse blood (Fig. 3f). However, the substitution enhanced ZIKV transmission from mice to mosquitoes (Fig. 3g, h). Moreover, neutralization of NS1 with murine ZIKV NS1 antisera effectively reduced the prevalence of FSS13025-A188V ZIKV in mosquitoes (Extended Fig. 8a–d), highlighting that the 188th valine in NS1 has a critical role in the antigenemia, which is critical for ZIKV acquisition by mosquitoes.
We next investigated whether NS1-mediated enhancement of ZIKV infectivity influences the prevalence of ZIKV-carrying mosquitoes using a "mosquito-mouse-mosquito" transmission cycle. We thoracically inoculated an equal titer (5 pfu) of the GZ01, FSS13025 and FSS13025-A188V mutant strains, into female A. aegypti mosquitoes. Six days following virus inoculation, two infected mosquitoes were allowed to feed on AG6 mice (Fig. 4a). The mosquito-bitten animals developed high-titer viremia of all three ZIKV strains (Fig. 4b). Replication of both the GZ01 and FSS13025-A188V ZIKV strains, but not the FSS13025 strain, resulted in efficient NS1 antigenemia (Fig. 4c). Subsequently, the infected mice were subjected to daily biting by naïve A. aegypti from days 1 to 8 post-mouse infection (Fig. 4a). Through a complete transmission cycle, the FSS13025-A188V strain showed a significantly higher prevalence of mosquito infection than did the FSS13025 strain (Fig. 4d, e), demonstrating that the 188th valine is required for the generation of NS1 antigenemia which enhances ZIKV transmissibility from hosts to vectors.
Figure 4. A single amino acid substitution influences the prevalence of mosquito infection during the "mosquito-host-mosquito" life cycle.
a, Schematic representation of the study design.
b, Detection of the ZIKV viremia by a plaque assay (n=12 mice per group pooled from 4 independent biological replicates).
c, Measurements of ZIKV NS1 antigenemia by ELISA (n=12 mice per group pooled from 4 independent biological replicates).
d, e, Comparison of mosquito infectivity (d) and infective ratio (e) of ZIKV strains (n=9 mice per group pooled from 3 independent biological replicates). The number at the top of each column represents infected number/total number. Each dot represents a mosquito (d).
Data are mean ± s.e.m. (b, c). p values were determined by two-tailed Mann-Whitney test (b-d) or two-sided Fisher’s exact test (e), and adjusted using Bonferroni correction to account for multiple comparisons (b-e). The black p value represents a comparison between GZ01 and FSS13025. The blue p value represents a comparison between FSS13025-A188V and FSS13025. *p< 0.025, **p< 0.005, ***p< 0.0005, n.s., not significant.
ZIKV independently separated into African and Asian lineages. We found that the 188th residue in all ZIKV African strains is valine (Extended Data Fig. 9a). The Uganda MR766-infected Vero cells abundantly secreted NS1 (Extended Data Fig. 9b–d), implicating that infection of the African ZIKV might produce high NS1 antigenemia. ZIKV has two ecologically distinct transmission cycles: a sylvatic cycle (mosquitoes to non-human primates) and a human cycle (mosquitoes to humans)27. Knowledge of ZIKV before 2012 was almost exclusively centered on the enzootic sylvatic cycle in Africa27,28. A mosquito survey in Africa showed that ZIKV was detected in mosquitoes collected in multiple surveyed landscapes, except for indoor locations within villages28. Therefore, we hypothesize that the presence of the NS1 188th valine might bestow the African lineage of ZIKV with a highly efficient enzootic or epizootic transmission cycle27,28. Other unknown factor(s) might prevent African ZIKV from efficiently establishing a human cycle. However, the Asian strains may have evolved to target humans with high infectivity; indeed, one such strains caused a medium-scale epidemic in Micronesia in 20072,6. The alanine-to-valine substitution at position 188 further improved ZIKV transmission efficiency from humans to mosquitoes and thus increased its prevalence in mosquitoes. Our data offer a potential explanation for the recent re-emergence of ZIKV, and suggest that coevolution of arboviruses with their hosts/vectors contributes to their long-term existence in nature and occasional widespread re-emergence.
Methods
Ethics statement
Human blood for mosquito feeding was taken from healthy donors who provided written informed consent. The collection of human blood samples was approved by the local ethics committee at Tsinghua University.
Mice, mosquitoes, cells and viruses
AG6 mice were donated from Institute Pasteur of Shanghai, Chinese Academy of Sciences5. The mice were bred and maintained under a specific pathogen-free animal facility at Tsinghua University. Groups of age- and sex-matched AG6 mice, 6–8 weeks of age, were used for the animal studies15. All experiments were approved by and performed under the guidelines of the Experimental Animal Welfare and Ethics Committee of Tsinghua University. A. aegypti (Rockefeller strain) were reared in a low-temperature, illuminated incubator (Model 818, Thermo Electron Corporation, Waltham, MA) at 28 °C and 80% humidity according to standard rearing procedures29. The ZIKV GZ01 strain (KU820898), PRVABC59 (KU501215), SZ01 strain (KU866423), MR766 (NC_012532) and FSS13025 strain (KU955593) were grown in Vero cells for blood meals15. The Drosophila melanogaster S2 cell line was cultured in Schneider’s medium with 10% heat-inactivated fetal bovine serum and 1% antibiotic–antimycotic (15240-062, Invitrogen). The Vero cells and 293T cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% heat-inactivated fetal bovine serum (16000-044, Gibco). For ZIKV infection, the Vero cells were maintained in VP-SFM medium (11681-020, Gibco). The Vero and 293T cell lines were purchased from the ATCC (CCL-81, CRL-3216). The Drosophila S2 cell line was sourced from a Drosophila expression system of Invitrogen (R690-07). None of these cell lines were found in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample. All these cell lines were authenticated by ATCC or Invitrogen and do not have mycoplasma contamination. ZIKV was titered by a plaque formation (PFU) assay29.
Antibodies
The ZIKV NS1 genes were amplified from the cDNA of infected cells and cloned into a pET-28a (+) expression vector (69864, Millipore). The cloning primers are presented in Extended Data Table 1. Recombinant NS1 proteins were expressed in the Escherichia coli BL21 DE3 strain in an insoluble form in inclusion bodies. The proteins were then solubilized by 8 M urea and purified with a TALON Purification Kit (635515, Clontech). Murine antisera were produced via the inoculation of recombinant ZIKV NS1 with 3 boosts. Polyclonal antibodies were purified from the immunized antisera using Protein A/G agarose (20423, Thermo). The anti-V5-HRP antibody for tag detection were purchased from Invitrogen (R96125, Thermo).
Extended Data Table 1.
Primers and probes for gene cloning and qPCR.
| Primers for cloning into pET28a(+) | Upper primer | Lower primer | |
|---|---|---|---|
| ZIKV NS1 (His tag m both N and C-terminal) | TATCTAGCTAGCGATGTGGGGTGCTCG | GATTCCGCTCGAGTGCAGTCACCATTGA | |
| Primers for cloning into pMT/BiP/V5-His/A | Upper primer | Lower primer | |
| ZIKV NS1 (V5 tag in C-terminal) | TCTCGGGGTACCTGATGTGGGGTGCTCG | TATTACGCTCGAGTGCAGTCACCATTGAC | |
| Primers for cloning into pcDNA3.1/V5-His/TOPO | Upper primer | Lower primer | |
| Zika prM-E-NS1 (V5 Tag in C-terminal) | TAAGGCGGTACCGCCACCATGCAAAAAGTCATATAC | TATCCGCTCGAGCGTGCAGTCACCATTGA | |
| FSS13025 V-M mutation (E) | ATGTGGTTGGGTCTGAATACAAAGAATGGA | TCCATTCTTTGTATTCAGACCCAACCACAT | |
| FSS13025 A-V mutation (NS1) | ATTGGAACAGCCGCTAAGGGAAAGGAGGCT | AGCCTCCTTTCCCTTAGCGGCTGTTCCAAT | |
| P6-740 A-V mutation (NS1) | GTCATAGGAACAGCTGTTAAGGGAAAGGAG | CTCCTTTCCCTTAACAGCTGTTCCTATGAC | |
| The primers for Taqman RT-qPCR | Upper primer | Lower primer | Probe (for Taqman QPCR) |
| ZIKV envelope gene | CCGCTGCCCAACACAAG | CCACTAACGTTCTTTTGCAGACAT | FAM-AGCCTACCTTGACAAGCARTCAGACACTCAA-TAMRA |
| Aedes aegypti actin | GAACACCCAGTCCTGCTGACA | TGCGTCATCTTCTCACGGTTAG | FAM-AGGCCCCGCTCAACCCGAAG-TAMRA |
| Mouse actin | AGCCATGTACGTAGCCATCCA | TCTCCGGAGTCCATCACAATG | FAM-TGTCCCTGTATGCCTCTGGTCGTACCAC-TAMRA |
Expression of prM-E-NS1 protein in human cells
The prM-E-NS1 regions (820 bp-3438 bp in GZ01 KU820898; 927 bp-3544 bp in FSS13025 KU955593) were amplified from cDNA of infected cells. The prM-E-NS1 genes from the P6-740 strain (820 bp-3438 bp, HQ234499) and the Micronesian 2007 strain (820 bp-3438 bp, EU545988) were commercially synthesized (Genescript, China). All the genes were subcloned into a pcDNA3.1/V5-His/TOPO DNA vector (K4800-01, Invitrogen), and subsequently transfected by the X-treme Gene HP DNA transfection reagent (06366236001, Roche) for ectopically expression in 293T cells. The amount of NS1 was determined in the supernatants of the 293T cells at 48 hrs post-transfection. The cloning primers are shown in Extended Data Table 1.
ZIKV NS1 protein generation in a Drosophila expression system
The ZIKV NS1 gene was cloned into a pMT/BiP/V5-His A vector (V4130-20, Invitrogen) for expression in Drosophila S2 cells. The cloning primers are shown in Extended Data Table 1. The procedure for ZIKV NS1 expression and purification in a Drosophila expression system has been described in our previous studies15,29.
ZIKV NS1 detection by ELISA
Secretion of ZIKV NS1 into Vero cell supernatant and mouse serum was measured using a ZIKV NS1 Antigen ELISA Kit (ZIKV-NS1-EK, BioFront MonoTrace). The experiment was performed according to the kit’s manual. Samples were diluted to the range of quantification of the kit using the sample dilution buffer provided in the kit. Standard materials for the generation of a standard curve were also provided in the kit. The measured amount of ZIKV NS1 matched the linearity in the assay. Optical density (OD) was measured at 450 nm with an ELISA reader (Varioskan Flash Multimode Reader, Thermo Scientific). The detection limit of NS1 protein by ELISA is 0.1 ng/ml.
Thoracic inoculation of ZIKV in mosquitoes
Detailed procedures for microinjection in mosquitoes have been described by our previous studies15,30. Briefly, the female A. aegypti mosquitoes were anesthetized on a cold tray, and subsequently a certain titer of ZIKV was microinjected into the mosquito thoraxes. The infected mosquitoes were used for ZIKV transmission to AG6 mice by a blood meal.
Membrane blood feeding
Fresh human blood was placed in heparin-coated tubes (367884, BD Vacutainer) and centrifuged at 1,000×g and 4 °C for 10 min to separate the plasma from the blood cells. The plasma was collected and heat-inactivated at 55 °C for 60 min. The separated blood cells were washed 3 times with PBS to remove the anticoagulant. The cells were then resuspended in the heat-inactivated plasma. The donor's sera were negative for detection of the E and NS1 proteins of JEV, DENV and ZIKV. Either purified proteins or antibodies were mixed with the ZIKV-infected supernatant and human blood for mosquito oral feeding via a Hemotek system (6W1, Hemotek Limited, England). Engorged female mosquitoes were transferred into new containers and maintained under standard conditions for an additional 8 days. The mosquitoes were subsequently sacrificed for further analysis.
Mosquito feeding on infected mice
Eight to ten female A. aegypti mosquitoes were separated into a netting-covered container for blood feeding. The mosquitoes were starved for 24 hr before engorgement. ZIKV-infected AG6 mice were anesthetized and placed on top of the container. The mosquitoes were allowed to feed on the mice for 30 min in the dark. After anesthetization using ice, the engorged mosquitoes were transferred to new containers and maintained under standard conditions for an additional 8 days. The mosquitoes were subsequently sacrificed for further analysis.
Purification of infectious ZIKV virions
The procedure of purification of flaviviral infectious virions has been described in our previous study15. Briefly, supernatant from GZ01-infected Vero cells was collected 5 days after infection. Cell fragments were removed by centrifugation at 25,000×g and 4 °C for 20 min. Subsequently, the supernatant was carefully transferred into a clean centrifuge tube and centrifuged at 25,000×g and 4 °C for an additional 6 h to pellet the virions. The precipitated virions were washed twice and then solubilized in VP-SFM medium (11681-020, Gibco). Insoluble material was removed by an extra centrifugation step at 12,000×g and 4 °C for 2 min. The virions in the VP-SFM medium were aliquoted and stored at -80 °C.
Viral genome quantitation by TaqMan qPCR
Total RNA was isolated from homogenized mosquitoes using an RNeasy Mini Kit (74106, Qiagen) and reverse-transcribed into cDNA using an iScript cDNA synthesis kit (170-8890, Bio-Rad). Viral genomes were quantified via TaqMan qPCR amplification of ZIKV genes, normalized against A. aegypti actin (AAEL011197). The detection limit of ZIKV E/Actin mRNA ratio is 0.001. The primers and probes used for this analysis are shown in Extended Data Table 1.
Determination of the virus titer in infected mice by plaque assay
Blood samples were collected from the tail veins of infected mice in 0.4% sodium citrate and were centrifuged for 5 min at 6,000×g and 4 °C for plasma isolation. The presence of infectious viral particles in the plasma was determined using a plaque assay15. The detection limit in the plaque assay is 10 pfu/ml in cell supernatant, and 100 pfu/ml in serum.
RNA transcription, transfection and virus recovery
The pFLZIKV-FSS13025 and pFLZIKV-FSS13025-A188V26 plasmids were amplified in E. coli Top10 (C404010, Invitrogen) and purified using MaxiPrep PLUS (12965, QIAGEN). For in vitro transcription, 10 μg of plasmid was linearized with the restriction enzyme ClaI. A mMESSAGE mMACHINE T7 kit (AM1344, Ambion) was used for in vitro transcription of RNA in a 20 μl reaction with an additional 1 μl of 30 mM GTP solution. The RNA was precipitated with lithium chloride and quantified via spectrophotometry. Subsequently, the RNA was transfected into Vero cells seeded in a T175 flask with DMEM medium and incubated at 37°C and 5% CO2 using Lipofectamine 3000 reagent (L3000015, Invitrogen). Recombinant viruses in cell culture media were harvested 4 days post transfection, stored in aliquots at -80°C, and subjected to further investigation.
Statistics
Animals were randomly allocated into different groups. Mosquitoes that died before measurement were excluded from analysis. The investigators were not blinded to the allocation during the experiments or to the outcome assessment. No statistical methods were used to predetermine the sample size. Descriptive statistics have been provided in the figure legends. Kruskal-Wallis analysis of variance was conducted to detect any significant variation among replicates. If no significant variation was detected, the results were pooled for further comparison. Given the nature of the experiments and the type of samples, differences in continuous variables were assessed with a non-parametric Mann-Whitney test. Differences in mosquito infective rates were analyzed using Fisher’s exact test. p values were adjusted using Bonferroni correction to account for multiple comparisons. All analyses were performed using GraphPad Prism statistical software.
Data availability
Source data for the main and Extended Data Figures are provided in the online version of this paper. All other data are available from the corresponding authors upon reasonable request.
Extended Data
Extended Data Figure 1. Phylogenetic analysis of ZIKV isolates in the Asian lineage.
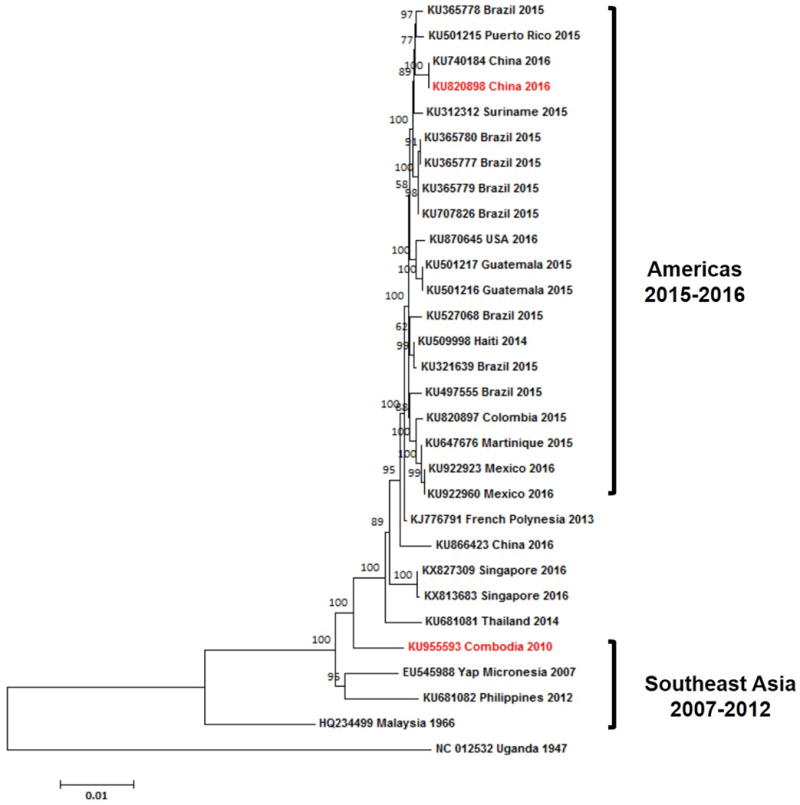
The tree was constructed using the neighbor-joining (NJ) method based on the alignment of ZIKV sequences in the Asian lineage. The bootstrap values of 2000 replicates are indicated on the branch nodes.
Extended Data Figure 2. Generation of murine anti-ZIKV NS1 polyclonal antibody and purification of the GZ01 ZIKV NS1 protein from Drosophila S2 cells.
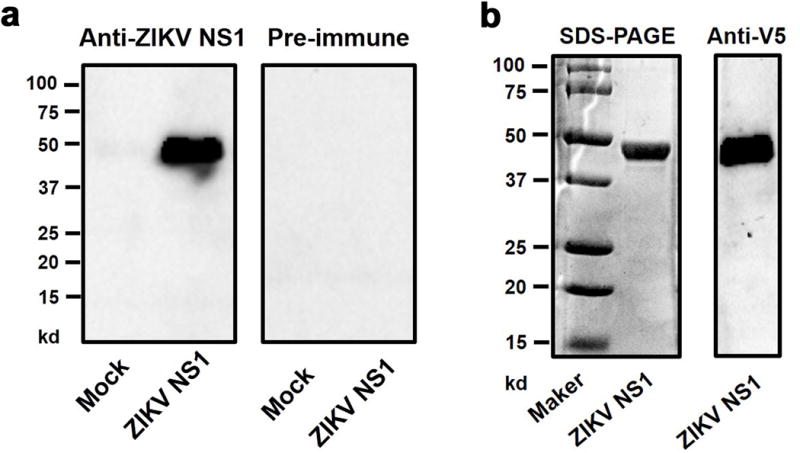
a, Generation of murine anti-ZIKV NS1 polyclonal antibody. A murine ZIKV NS1 polyclonal antibody was generated by immunization of the purified GZ01 ZIKV NS1 recombinant protein expressed in E. coli. The antibody was validated by probing an S2-expressed ZIKV NS1 recombinant protein. The same samples probed by murine pre-immune antibody served as a negative control.
b, Purification of the GZ01 ZIKV NS1 protein from Drosophila S2 cells. Full-length NS1 was cloned into the pMT/BiP/V5-His A expression vector. Recombinant ZIKV NS1 protein was expressed in Drosophila S2 cells and purified using a cobalt-His column (Left Panel). Protein quality was evaluated by Western-blotting with an anti-V5 antibody (Right Panel).
a, b, The experiments were reproduced 3 times with similar results. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 3. Anti-ZIKV NS1 antibodies did not neutralize ZIKV virions.
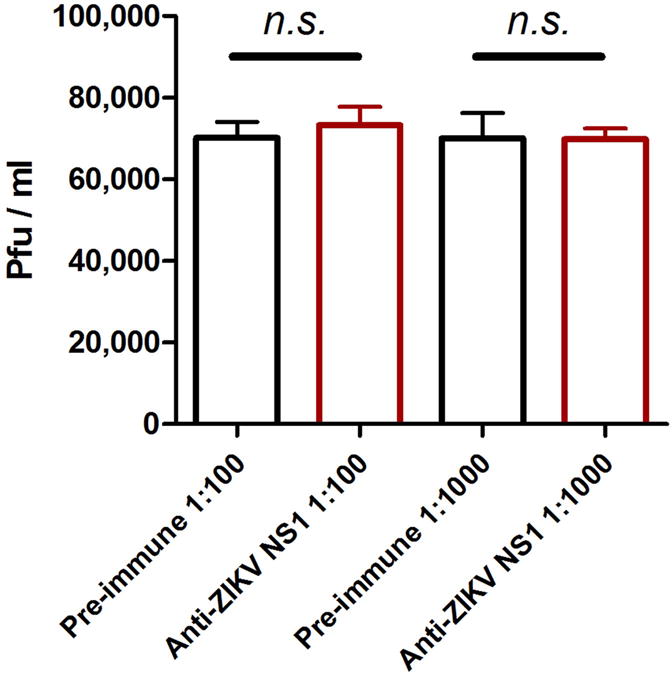
We premixed two dilutions of murine ZIKV NS1 polyclonal antisera (1:100 and 1:1000) with purified ZIKV GZ01 infectious virions. The same amount of pre-immune sera served as a negative control. After a 30 min incubation, the infectivity of the "antibody-ZIKV virion" mixture was determined by plaque assay on Vero cells. The values in the graph represent the mean ± s.e.m. p values were determined by two-tailed Mann-Whitney test. The data were combined from 3 biological repeats. n.s., not significant.
Extended Data Figure 4. Passive transfer of ZIKV NS1 antibodies into infected AG6 mice prevented ZIKV acquisition by A. aegypti.
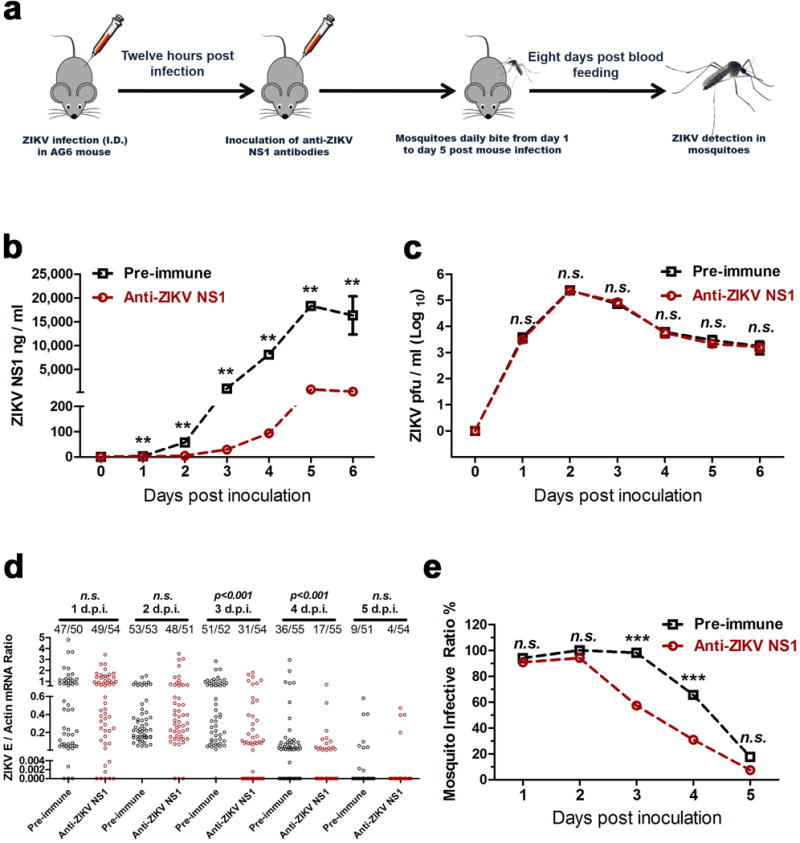
a, Schematic representation of the study design. AG6 mice were intradermally infected with 1×104 pfu of the ZIKV GZ01 strain. Subsequently, 100 μl of a murine ZIKV NS1 antibody was intraperitoneally inoculated into the mice 12 hr post-infection. The same amount of pre-immune serum was inoculated as a mock control. After 12 hr of antibody dissemination, the infected mice were subjected to daily biting by female A. aegypti from day 1 to day 5 post-mouse infection. The mouse blood-fed mosquitoes were reared for an additional 8 days for ZIKV detection.
b, ZIKV NS1 measurement by ELISA (n=6 mice per group pooled from 3 independent biological replicates). Mouse sera were collected to quantify the amounts of ZIKV NS1 protein from days 0 and 6 post-mouse infection.
c, Detection of the ZIKV load in the blood of the infected mice (n=8 mice per group pooled from 4 independent biological replicates). The presence of infectious ZIKV particles in blood plasma was determined by a plaque assay from days 0 to 6 post-mouse infection.
d, e, Immuno-blockade of ZIKV NS1 in infected AG6 mice reduced the infection of fed A. aegypti (n=6 mice per group pooled from 3 independent biological replicates). The number of infected mosquitoes relative to the total number of mosquitoes is shown at the top of each column. Each dot represents a mosquito (d). The data are represented as the percentage of mosquito infection (e).
Data are mean ± s.e.m. (b, c). p values were determined by two-tailed Mann-Whitney test (b-d) or two-sided Fisher’s exact test (e). **p< 0.01, ***p< 0.001, n.s., not significant.
Extended Data Figure 5. Modeling the location of the 188th residue of ZIKV NS1 and assessing NS1 dimer formation in cell lysates.
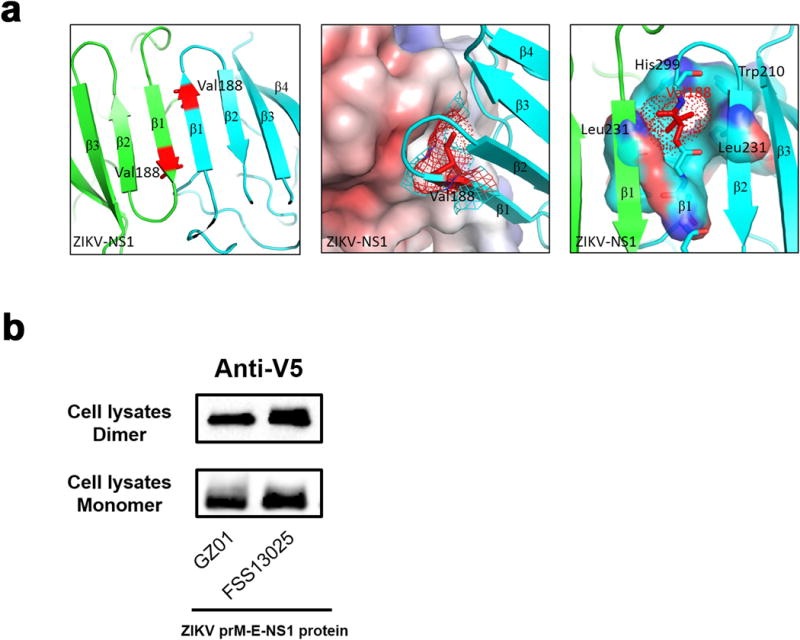
a, Modeling the location of the 188th residue of ZIKV NS1. Modeling was based on the ZIKV NS1 structure (PDB: 5K6K)30. The 188th position is located within the interface of a NS1 dimer (contoured by red), which is formed by hydrophobic interactions. The 188th amino acid is deep within the hydrophobic pocket formed by Trp-210, Leu-23, His-299 and Leu-231.
b, Assessing NS1 dimer formation in the lysates of cells transfected with GZ01 or FSS13025 prM-E-NS1 recombinant plasmids. The cell lysates, with or without heating treatment, were used for assessing NS1 monomer and dimer by Western blotting with an anti-V5 antibody, respectively. The experiment was reproduced 3 times with similar results. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 6.
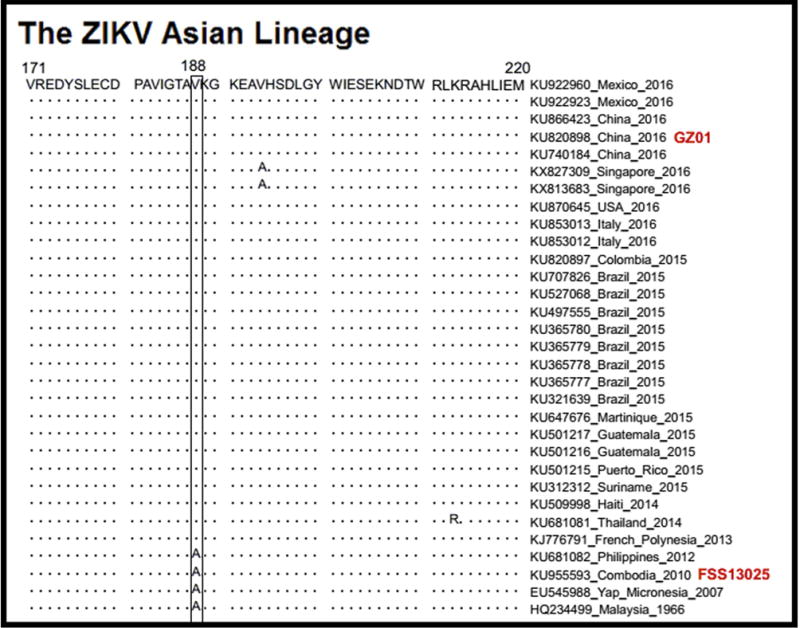
Sequence alignment of NS1 sequences in ZIKV isolates of the Asian lineage.
Extended Data Figure 7. A single amino acid substitution enhanced NS1 secretability and ZIKV infectivity in mosquitoes.
a–c, NS1 secretability in ZIKV-infected Vero cells. We mutated the single amino acid from alanine to valine at the 188th position of NS1 protein on an infectious clone of ZIKV Cambodian FSS13025 strain (FSS13025-A188V strain). FSS13025 or FSS13025-A188V strain (0.01 M.O.I.) was used to infect Vero cells. Supernatants from the infected Vero cells were collected from 0 to 5 days post-infection. a, The amount of ZIKV NS1 protein was determined by ELISA. b, ZIKV titer was determined by a plaque assay. c, NS1 quantity normalized against average viral titer (ng per pfu). The data were pooled from 3 independent biological replicates.
d–e, NS1 secretability determined ZIKV infectivity in mosquitoes. The supernatant, isolated from the FSS13025-infected or FSS13025-A188V-infected Vero cells, was premixed with fresh human blood at a 1:1 ratio for an in vitro membrane feeding experiment (final titer is 5×104 pfu/ml), respectively. The fed mosquitoes were scarified on 8 days after a blood meal to determine the ZIKV load by qPCR. The number of infected mosquitoes relative to the total number of mosquitoes (infected number/total number) is shown at the top of each column. Each dot represents a mosquito (d). The data are represented as the percentage of mosquito infections (e). The data were pooled from 3 independent biological replicates.
Data are mean ± s.e.m. (a-c). p values were determined by two-tailed Mann-Whitney test (a-d) or two-sided Fisher’s exact test (e). *p< 0.05, **p< 0.01, n.s., not significant.
Extended Data Figure 8. Neutralization of NS1 by murine ZIKV NS1 antisera effectively reduced the prevalence of FSS13025-A188V ZIKV in A. aegypti.
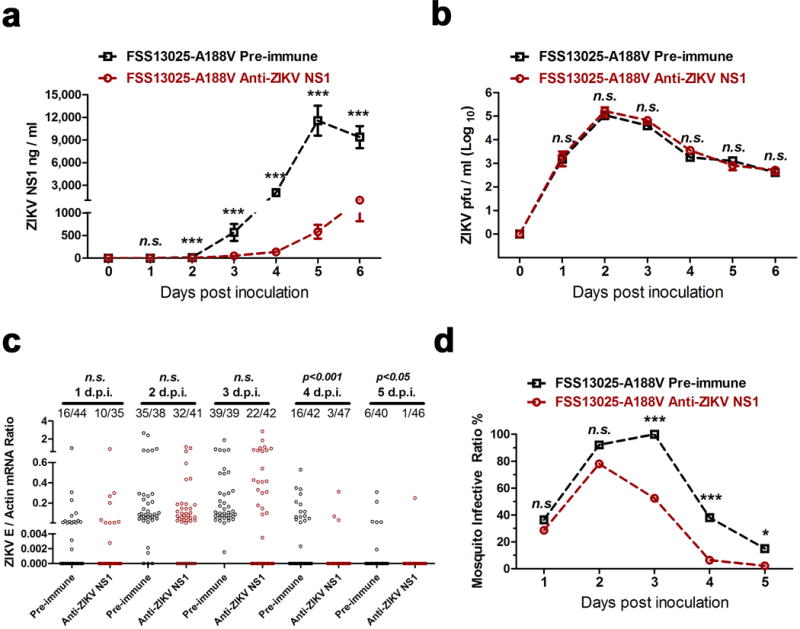
a, b, Infection by FSS13025-A188V ZIKV in AG6 mice. The AG6 mice were intradermally inoculated with these viruses (1×104 pfu). ZIKV NS1 measurement by ELISA. Mouse sera were collected to qualify the amounts of ZIKV NS1 protein from days 0 to 6 post-mouse infection (a). Detection of the ZIKV load in the blood of infected AG6 mice. The presence of infectious ZIKV particles in blood plasma was determined by a plaque assay (b). n=10 mice per group pooled from 4 independent biological replicates.
c, d, Immuno-blockade of NS1 in the FSS13025-A188V ZIKV infected AG6 mice reduced the acquisition of fed A. aegypti (n=7 mice per group pooled from 3 independent biological replicates). The number of infected mosquitoes relative to the total number of mosquitoes is shown at the top of each column. Each dot represents a mosquito (c). The data are represented as the percentage of mosquito infection (d).
Data are mean ± s.e.m. (a, b). p values were determined by two-tailed Mann-Whitney test (a-c) or two-sided Fisher’s exact test (d). *p< 0.05, ***p< 0.001, n.s., not significant.
Extended Data Figure 9. Sequence alignment of NS1 sequences in African ZIKV isolates, and NS1 secretability in African ZIKV-infected Vero cells.
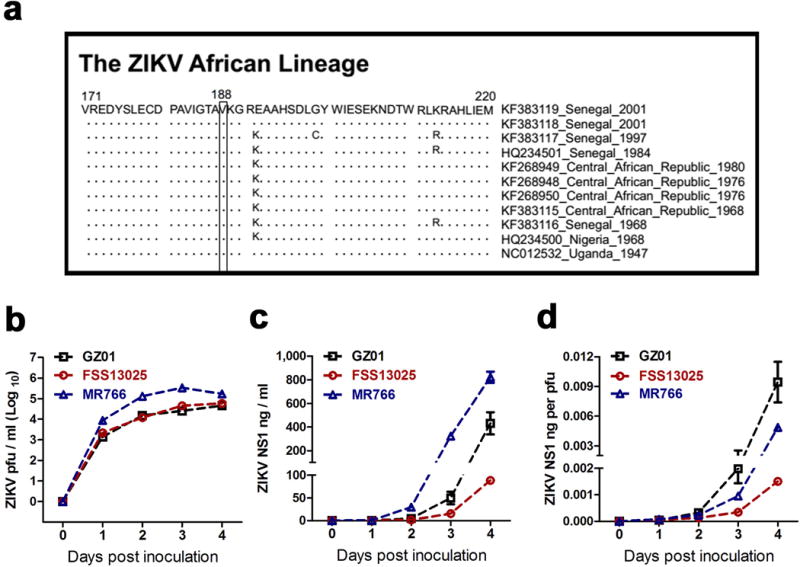
a, Sequence alignment of NS1 sequences in African ZIKV isolates.
b-d, NS1 secretability in African ZIKV-infected Vero cells. Vero cells were infected at an M.O.I. of 0.01 with the GZ01, FSS13025 and MR766 strains. Supernatants from the infected Vero cells were collected from 0 to 4 days post-infection. b, ZIKV titer was determined by a plaque assay. c, The amount of ZIKV NS1 protein was determined by ELISA. d, NS1 quantity normalized against average viral titer (ng per pfu). The data were pooled from 3 independent biological replicates.
The values in the graph represent the mean ± s.e.m. (b-d).
Supplementary Material
Acknowledgments
This work was funded by grants from the National Key Research and Development Plan of China (2016YFD0500400, 2016YFC1201000, 2016ZX10004001-008, 2016YFD0500300), the National Natural Science Foundation of China (81422028, 81571975, 81522025, 31300600), the National Key Basic Research Program of China 2013CB911500, SZSTI-JCYJ20160331115853521, the National Institutes of Health of the United States (R01AI087856, AI103807), and Shenzhen San-Ming Project for prevention and research on vector-borne diseases. G.C. is a Newton Advanced Fellow awarded by the Academy of Medical Sciences and the Newton Fund. G.C. is also a Janssen Investigator at Tsinghua University. P.Y.S. was supported by University of Texas Medical Branch (UTMB) startup award and University of Texas STARs Award. We thank the core facilities of the Center for Life Sciences and Center of Biomedical Analysis for technical assistance (Tsinghua University).
Footnotes
Author Contributions
G.C. and P.Y.S. designed the experiments and wrote the manuscript; Y.L., J.L., S.D. and C.S. performed the majority of the experiments and analyzed the data; K.N. and R.Z. assisted in RNA isolation and qPCR detection; C.S., X.F.L., C.F.Q., and P.Y.S. provided the ZIKV strains; X.F.L., R.Z., T.W., C.F.Q. and P.W. contributed experimental suggestions and strengthened the writing of the manuscript. All authors reviewed, critiqued and provided comments on the text.
Author information
The authors declare that they have no competing financial interests.
References
- 1.Marchette NJ, Garcia R, Rudnick A. Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am J Trop Med Hyg. 1969;18:411–415. doi: 10.4269/ajtmh.1969.18.411. [DOI] [PubMed] [Google Scholar]
- 2.Musso D, Gubler DJ. Zika Virus. Clin Microbiol Rev. 2016;29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oehler E, et al. Zika virus infection complicated by Guillain-Barre syndrome-case report, French Polynesia, December 2013. Eurosurveillance. 2014;19:4–6. doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- 4.Cordeiro MT, Pena LJ, Brito CA, Gil LH, Marques ET. Positive IgM for Zika virus in the cerebrospinal fluid of 30 neonates with microcephaly in Brazil. Lancet. 2016;387:1811–1812. doi: 10.1016/S0140-6736(16)30253-7. [DOI] [PubMed] [Google Scholar]
- 5.Faye O, et al. Molecular evolution of Zika virus during its emergence in the 20th century. Plos Neglect Trop D. 2014;8:2–3. doi: 10.1371/journal.pntd.0002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffy MR, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. New Engl J Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 7.Caolormeau VM. Zika virus, French Polynesia, South Pacific, 2013. Emerg Infect Dis. 2014;20:1085–1086. doi: 10.3201/eid2006.140138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musso D, Nilles EJ, Cao-Lormeau VM. Rapid spread of emerging Zika virus in the Pacific area. Clin Microbiol Infec. 2014;20:O595–O596. doi: 10.1111/1469-0691.12707. [DOI] [PubMed] [Google Scholar]
- 9.Enfissi A, Codrington J, Roosblad J, Kazanji M, Rousset D. Zika virus genome from the Americas. Lancet. 2016;387:227–228. doi: 10.1016/S0140-6736(16)00003-9. [DOI] [PubMed] [Google Scholar]
- 10.Brault AC, et al. Venezuelan equine encephalitis emergence: enhanced vector infection from a single amino acid substitution in the envelope glycoprotein. P Natl Acad Sci USA. 2004;101:11344–11349. doi: 10.1073/pnas.0402905101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. Plos Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang FC, Li XF, Deng YQ, Tong YG, Qin CF. Excretion of infectious Zika virus in urine. Lancet Infect Dis. 2016;16:641–642. doi: 10.1016/S1473-3099(16)30070-6. [DOI] [PubMed] [Google Scholar]
- 13.Heang V, et al. Zika virus infection, Cambodia, 2010. Emerg Infect Dis. 2012;18:349–351. doi: 10.3201/eid1802.111224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng G, Liu Y, Wang P, Xiao X. Mosquito defense strategies against viral infection. Trends Parasitol. 2016;32:177–186. doi: 10.1016/j.pt.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, et al. Flavivirus NS1 protein in infected host sera enhances viral acquisition by mosquitoes. Nat Microbiol. 2016;1:16087. doi: 10.1038/nmicrobiol.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazear HM, et al. A mouse model of Zika virus pathogenesis. Cell Host Microbe. 2016;19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller DA, Young PR. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir Res. 2013;98:192–208. doi: 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Young PR, Hilditch PA, Bletchly C, Halloran W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J Clin Microbiol. 2000;38:1053–1057. doi: 10.1128/jcm.38.3.1053-1057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macdonald J, et al. NS1 protein secretion during the acute phase of West Nile virus infection. J Virol. 2005;79:13924–13933. doi: 10.1128/JVI.79.22.13924-13933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takamatsu Y, et al. NS1’ protein expression facilitates production of Japanese encephalitis virus in avian cells and embryonated chicken eggs. J Gen Virol. 2014;95:373–383. doi: 10.1099/vir.0.057968-0. [DOI] [PubMed] [Google Scholar]
- 21.Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, Signor LDCC. Phylogeny of Zika Virus in Western Hemisphere, 2015. Emerg Infect Dis. 2016;22:933–935. doi: 10.3201/eid2205.160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng YQ, et al. Isolation, identification and genomic characterization of the Asian lineage Zika virus imported to China. Sci China Life Sci. 2016;59:428–430. doi: 10.1007/s11427-016-5043-4. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Structures of immature flavivirus particles. The EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song H, Qi J, Haywood J, Shi Y, Gao GF. Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat struct Mol biol. 2016;23:456–458. doi: 10.1038/nsmb.3213. [DOI] [PubMed] [Google Scholar]
- 25.Maurer-Stroh S, et al. South-east Asian Zika virus strain linked to cluster of cases in Singapore, August 2016. Eurosurveillance. 2016;21 doi: 10.2807/1560-7917.ES.2016.21.38.30347. Article 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shan C, et al. An Infectious cDNA Clone of Zika Virus to Study Viral Virulence, Mosquito Transmission, and Antiviral Inhibitors. Cell Host Microbe. 2016;19:891–900. doi: 10.1016/j.chom.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buechler CR, et al. Prevalence of Zika virus infection in wild African primates. bioRxiv. 2016;077628 [Google Scholar]
- 28.Diallo D, et al. Zika virus emergence in mosquitoes in southeastern Senegal, 2011. Plos one. 2014;9:e109442. doi: 10.1371/journal.pone.0109442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, et al. Transmission-blocking antibodies against mosquito C-type lectins for dengue prevention. Plos Pathog. 2014;10:e1003931. doi: 10.1371/journal.ppat.1003931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown WC, et al. Extended surface for membrane association in Zika virus NS1 structure. Nat Struct Mol Biol. 2016;23:865–867. doi: 10.1038/nsmb.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for the main and Extended Data Figures are provided in the online version of this paper. All other data are available from the corresponding authors upon reasonable request.


