Abstract
Loss of function mutations in progranulin (GRN) cause frontotemporal dementia, but how GRN haploinsufficiency causes neuronal dysfunction remains unclear. We previously showed that GRN is neurotrophic in vitro. Here, we used an in vivo axonal outgrowth system and observed a delayed recovery in GRN−/− mice after facial nerve injury. This deficit was rescued by reintroduction of human GRN and relied on its C-terminus and on neuronal GRN production. Transcriptome analysis of the facial motor nucleus post injury identified cathepsin D (CTSD) as the most upregulated gene. In aged GRN−/− cortices, CTSD was also upregulated, but the relative CTSD activity was reduced and improved upon exogenous GRN addition. Moreover, GRN and its C-terminal granulin domain granulinE (GrnE) both stimulated the proteolytic activity of CTSD in vitro. Pull-down experiments confirmed a direct interaction between GRN and CTSD. This interaction was also observed with GrnE and stabilized the CTSD enzyme at different temperatures. Investigating the importance of this interaction for axonal regeneration in vivo we found that, although individually tolerated, a combined reduction of GRN and CTSD synergistically reduced axonal outgrowth. Our data links the neurotrophic effect of GRN and GrnE with a lysosomal chaperone function on CTSD to maintain its proteolytic capacity.
Introduction
Progranulin (GRN) is a highly conserved, highly glycosylated multifunctional growth factor that is expressed in many different cell types (1). It consists of 7.5 granulin domains and upon secretion it can be cleaved by several elastases, releasing the individual granulin domains (granulin A-G and paragranulin). This cleavage can be inhibited by secretory leukocyte protease inhibitor (SLPI) and hence provides a mechanism for tight regulation (2,3). GRN can be endocytosed by binding sortilin (SORT1), a vacuolar protein sorting 10 (VPS10) protein domain receptor that mediates GRN uptake and its transport to the lysosomes (4). GRN has multiple other binding partners in different cell compartments, but it remains incompletely understood how it exerts all its biological effects (3).
In the periphery, GRN is involved mostly in cancer, inflammation, wound healing and metabolic disease (5). In the central nervous system, GRN expression is limited to neurons and microglia (6) and GRN is thought to act as a neurotrophic factor (7) and a modulator of neuroinflammation (8).
Heterozygous loss-of-function mutations in the GRN gene are a frequent cause of frontotemporal dementia (FTD), a devastating neurodegenerative disorder primarily affecting the frontal and anterior temporal lobes (9,10). Most mutations lead to a reduction of 50% of the levels of functional GRN in the blood and the cerebrospinal fluid (7,11–13) hence causing the disease through haploinsufficiency.
Homozygous loss-of-function mutations have been reported in a few patients with the lysosomal storage disorder neuronal ceroid lipofuscinosis (NCL) (14,15). Interestingly, the pathobiochemical hallmarks of this lysosomal storage disease were shown to closely resemble those seen in FTD (16), pointing towards an important role of GRN in lysosomal function. How GRN deficiency causes neuronal death is presently unknown, but exaggerated microglial activation (17–19), lysosomal dysfunction (20) and reduced neurotrophic support to neurons may be contributive factors (7,21,22). Unraveling the biological function of GRN in the central nervous system could hold far-reaching therapeutic opportunities, as GRN has also been identified as a modifier of neurodegeneration in models of Parkinson’s disease (8), Alzheimer’s disease (23), motor neuron degeneration (24,25) and stroke (26,27).
In vitro studies have shown that GRN is able to increase the survival and neurite outgrowth of both motor and cortical neurons and that these effects can be attributed to the most C-terminal granulin domain, granulin E (7,28–31). Although the 3 most C-terminal amino acids are required for SORT1 interaction (4), the stimulation of neuronal outgrowth by GRN is not mediated through its interaction with SORT1 (31,32).
In this study, we employed the in vivo paradigm of facial nerve crush injury to identify the molecular mechanisms behind the neurotrophic effects of GRN. Transcriptional changes associated with GRN-induced neuronal outgrowth in mice pointed towards the lysosomal aspartyl protease cathepsin D (CTSD) as a major player in the cascade by which GRN exerts its neurotrophic effects. In addition, we reveal a novel chaperone function of GRN on CTSD that is mediated by its C-terminal granulin domain. We show that this chaperone function is essential to stimulate axonal outgrowth after nerve crush injury and can at least in part be explained by an enhanced stabilization of the enzyme in the presence of GRN.
Results
Recovery from facial nerve crush is delayed in GRN deficient mice
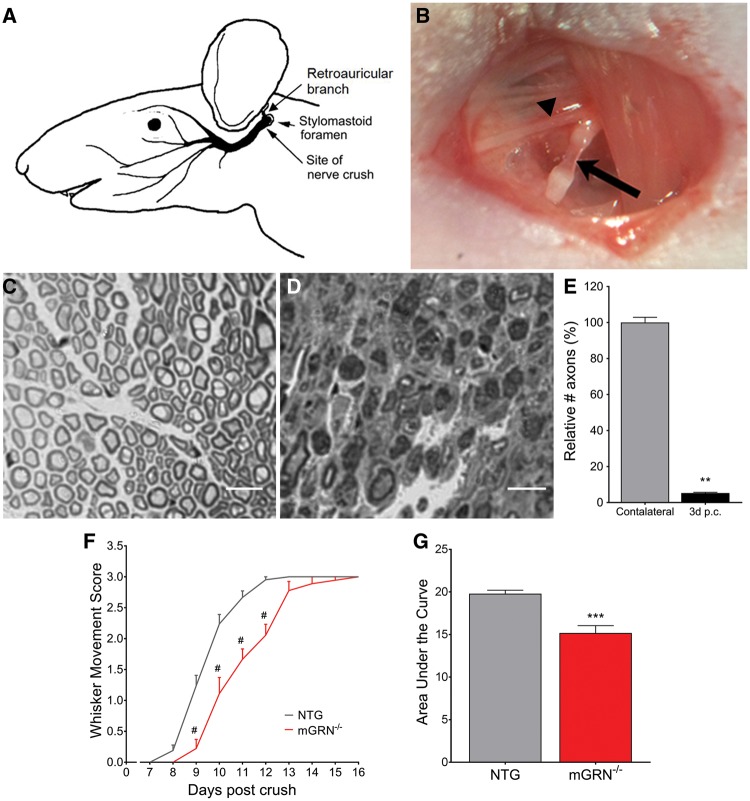
To investigate the role of GRN in nerve outgrowth in vivo, we generated GRN deficient (mGRN−/−) mice (Supplementary Material, Fig. S1) and subjected them to a nerve crush injury just distal to the retroauricular branch of the facial nerve (Fig. 1A and B), using the uninjured contralateral side as internal control. Immediately after the injury, all mice completely lost the ability to move the whiskers on the ipsilateral side. Three days after nerve crush, 95% of axons were lost in the distal segment of the facial nerve, as quantified on transverse sections (Fig. 1C–E). Functional recovery of the whisker movement was complete in non-transgenic (NTG) animals within 13 days post crush (Fig. 1F). This degeneration was not accompanied by changes in number or morphology of neuronal cell bodies in the facial motor nucleus (Supplementary Material, Fig. S2).
Figure 1.
mGRN−/− mice show delayed functional recovery after facial nerve crush injury. (A) Schematic representation of the facial nerve anatomy (adapted and adjusted from Kuzis et al. (55)). The facial nerve crush injury is performed immediately posterior from the bifurcation with the retroauricular branch (B, arrowhead), making the facial nerve completely transparent at the injury site (B, arrow). Semi-thin cross sections of contra- (C) and ipsilateral (D) facial nerve, distal from the injury site. (E) 95% of axons are lost in the distal nerve segment at 3 days post crush. **P < 0.01, Mann-Whitney test. (F) The absence of mGRN significantly delays functional recovery of the of the whisker movement. #P < 0.0001, RM Two-way ANOVA followed by Bonferroni correction. (G) Area under the curve of whisker movement recovery scores is significantly reduced in mGRN−/− mice. ***P < 0.001, Student’s t-test. Data is shown as mean ± SEM.
A comparative study between NTG and mGRN−/− mice, with daily analysis of whisker movements, showed that the functional recovery was significantly delayed in mGRN−/− mice (Fig. 1F). The curve representing the whisker movement scores of mGRN−/− mice shows an average delay of 1.5 days in recovery as calculated from the IC50 values from the interpolated standard curves (10.8 for mGRN−/− versus 9.3 for NTG mice). While this effect may seem small, it is larger than we expected, as several other studies only reported a difference of 1 day in recovery when comparing transgenic p75 or Gal1 null mutant mice with wild-type controls (33,34). As a result, it is also reflected by a significant reduction in area under the curve (AUC) of the whisker movement recovery in mGRN−/− animals (Fig. 1G). This suggests that GRN promotes, but is not indispensable for axonal regeneration in vivo.
Human GRN overexpression rescues nerve outgrowth defect in mGRN−/− mice via its C-terminus
Using our facial nerve crush model, we were unable to identify a dose-dependent effect of GRN as heterozygous mGRN+/- animals did not differ in recovery speed compared to NTG controls (Supplementary Material, Fig. S3A). Interestingly, mGRN+/- animals did show a significant upregulation of GRN mRNA in the facial nerve at 5 days post injury, reaching up to 3-fold of endogenous GRN mRNA levels of uninjured NTG mice (Supplementary Material, Fig. S3B). This amount of locally transcribed GRN might be sufficient to mediate its protective effects.
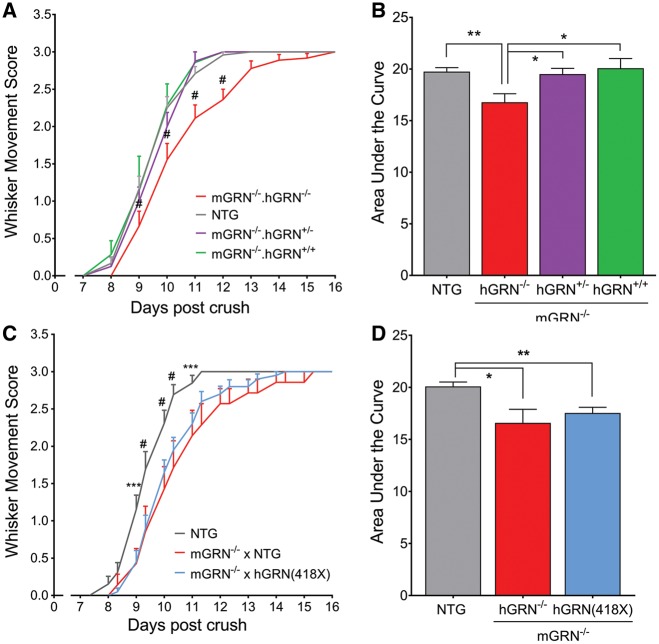
To further study dose-dependency of the GRN effect, we made use of our previously published mouse model that overexpresses human GRN (hGRN) by introduction of its cDNA in the ROSA26 locus (35). Increasing the GRN levels in NTG mice did not result in a faster recovery from facial nerve crush injury (Supplementary Material, Fig. S3C). However, the introduction of hGRN in a mGRN−/− background, by intercrossing these two transgenic lines, revealed a complete rescue of the delay in whisker movement recovery observed in mGRN−/− mice (Fig. 2A). This was true for both heterozygous and homozygous overexpression of hGRN, resulting in a significant increase in the AUC of the whisker movement recovery (Fig. 2B). These observations are a clear indication for the specificity of our effect which depends particularly on the presence of GRN. It also shows that the neurotrophic function of GRN in vivo is preserved by the hGRN protein.
Figure 2.
Re-introduction of hGRN rescues whisker movement recovery in mGRN−/− mice, but only if the C-terminal granulin domains are present. (A) mGRN−/− mice that contain one or two copies of the hGRN cDNA show a similar recovery as NTG mice, which is significantly faster than mGRN−/− mice. #P < 0.001, RM Two-way ANOVA followed by Bonferroni correction. (B) Analysis of the area under the curve of whisker movement recovery is increased to baseline levels upon introduction of hGRN in mGRN−/− mice. *P < 0.05, **P < 0.01, One-way ANOVA. (C) The introduction of one or two copies of hGRN(418X) cDNA in mGRN−/− mice did not rescue the delay in whisker movement recovery. ***P < 0.001, #P < 0.0001, RM Two-way ANOVA followed by Bonferroni correction. (D) Analysis of the are under the curve is still reduced upon introduction of hGRN(418X) in mGRN−/− mice. *P < 0.05, **P < 0.01, One-way ANOVA. Data is shown as mean ± SEM.
We previously showed that the neurotrophic properties of GRN are depending specifically on its C-terminal granulin domain, granulin E (7,31). To study this observation in our in vivo model, we created a mouse model that expresses a truncated hGRN protein, by introducing the hGRN cDNA carrying the R418X mutation in the ROSA26 locus (Supplementary Material, Fig. S4), and crossed them with mGRN−/− mice. In contrast to what we observed when expressing the full length hGRN in a mGRN−/− background, expression of the truncated hGRN protein in these mice did not rescue the delay in whisker movement recovery after facial nerve crush injury (Fig. 2C and D). These data suggest that also in vivo, the neurotrophic effect of GRN is mediated by its C-terminus.
Neuronal GRN is essential for nerve regeneration
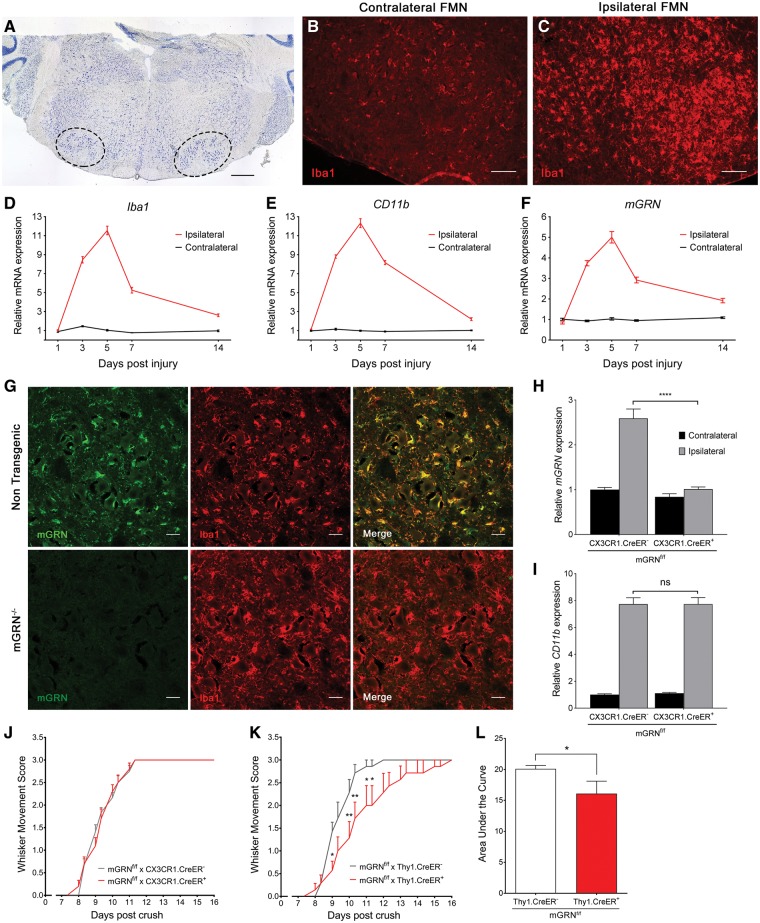
Upon facial nerve crush injury, a clear microglial response can be observed in the ipsilateral facial motor nucleus (Fig. 3A–C). A time course analysis of transcriptional changes showed that the upregulation of microglia specific genes such as Iba1 and CD11b is maximal at 5 days post crush, with an approximate 12-fold increase over the uninjured contralateral side (Fig. 3D and E). GRN mRNA expression in the facial motor nucleus followed a very similar pattern after the injury, with a maximal 5-fold increase at 5 days post crush (Fig. 3F). At this timepoint, GRN protein expression showed high co-localization with Iba1 positive microglia (Fig. 3G). Therefore, we explored the contribution of microglial GRN to in vivo neurite outgrowth using microglial specific mGRN knockout mice (mGRNf/f x CX3CR1cre/ERT2). Microglial GRN deletion abrogated the injury-induced upregulation of GRN (Fig. 3H), but the recovery of whisker movements in the mGRNf/f.CX3CR1cre+ mice occurred at exactly the same speed as in the mGRNf/f.CX3CR1cre- mice (Fig. 3J). While we observed a clear lack of mGRN upregulation during the inflammatory response due to the microglial GRN deletion, CD11b upregulation was unaltered and no reduction in GRN mRNA was seen in the uninjured contralateral facial motor nucleus (Fig. 3H and I). This strongly suggests that microglial GRN is not driving the observed protective effects. Moreover, GRN expression in basal conditions was mainly neuronal and might be essential to stimulate axonal outgrowth after nerve crush injury. We therefore created a neuronal-specific knockout of GRN (mGRNf/f x Thy1cre/ER) and observed a significant delay in axonal regrowth and functional recovery after crush (Fig. 3K and L), with a similar effect size compared to the full mGRN knockout mice. Taken together, these data suggest that neuronal production of GRN is both necessary and sufficient to stimulate nerve regeneration in vivo.
Figure 3.
Microgliosis in the facial motor nucleus is maximal at 5 days post injury, but only neuron specific GRN knockout mice recapitulate the delayed recovery of mGRN−/− mice. (A) Thionin stained section through the brainstem. Left and right facial motor nucleus are delineated by the dotted lines. Scale bar = 500 µm. (B–C) A strong increase in Iba1 immunoreactivity is observed in the facial motor nucleus on the ipsilateral side, at 5 days post crush. Scale bar = 100 µm. (D–F) qPCR analysis of microglial genes (Iba1, CD11b) and mGRN, in the facial motor nucleus at different days post crush. (G) mGRN staining overlaps with Iba1 positive microglia and is absent in mGRN−/− mice. Scale bar = 25 µm. (H–I) Relative mRNA expression of mGRN (H) and CD11b (I) in the ipsi- and contralateral facial motor nucleus of microglial specific mGRN knockout mice at 5 days post crush. ****P < 0.0001, Two-way ANOVA followed by Tukey’s test. (J–K) Microglial mGRN knockout mice show a normal recovery after crush (J), while neuronal GRN deletion recapitulates the delayed recovery that is observed in mGRN−/− mice (K). *P < 0.05, **P < 0.01, RM Two-way ANOVA followed by Bonferroni correction. (L) Analysis of area under the curve of whisker movement recovery in Thy1.CreER+ mGRNf/f mice is significantly reduced. *P < 0.05, Mann-Whitney test. Data is shown as mean ± SEM. FMN = facial motor nucleus.
CTSD transcription is increased in the facial motor nucleus of mGRN−/− mice after crush
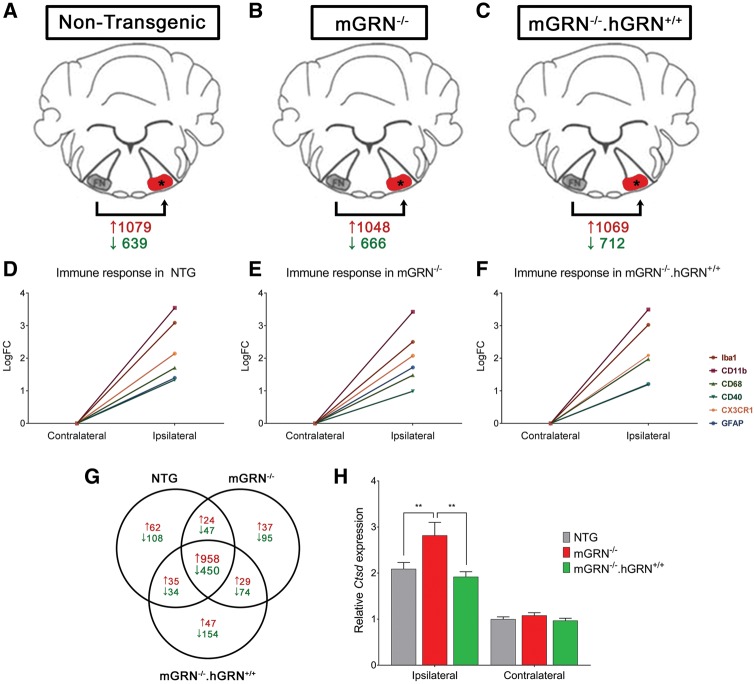
To gain more insight into the molecular mechanisms underlying the neuronal outgrowth deficit in mGRN knockout mice, we performed RNA sequencing on total RNA, extracted from the facial motor nucleus of NTG, mGRN−/− and mGRN−/−.hGRN+/+mice (Fig. 4A–C). RNA samples were collected at 5 days post crush as the transcriptional upregulation of GRN was maximal at this timepoint in NTG animals (Fig. 3F).
Figure 4.
Transcriptome analysis of facial motor nucleus RNA, 5 days post injury. (A–C) Schematic representation of the setup used for our RNA sequencing experiment. The number of significantly up- and downregulated genes (FDR < 0.05; LogFC>|1|) post injury is shown in red and green, respectively. (D–F) Graphical representation of the immune response in the three different groups. (G) Venn diagram showing the number of significantly up- and downregulated genes post crush, in red and green, respectively. (H) qPCR validation of CTSD mRNA expression after nerve crush injury confirming a significant upregulation of CTSD in the ipsilateral facial motor nucleus. **P < 0.01, Two-way ANOVA followed by Bonferroni correction. Data is shown as mean ± SEM.
Gene ontology analysis was performed on all genes with a significant differential expression pattern in the facial nuclei at the injured side of mGRN−/− versus NTG mice (P < 0.01). This analysis showed a significant overrepresentation (FDR < 0.05) of lysosome-associated genes in mGRN−/− mice (Table 1).
Table 1.
Top up- and downregulated gene ontology categories identified in mGRN−/− mice
| Upregulated GO categories | |||||
|---|---|---|---|---|---|
| GO Category | Total Genes | Overexpressed Genes | Enrichment | Log10(p) | FDR |
| Lytic vacuole | 204 | 11 | 4,897 | −4.80 | 0.015 |
| Lysosome | 204 | 11 | 4.897 | −4.80 | 0.015 |
| Vacuole | 234 | 11 | 4.269 | −4.25 | 0.040 |
| Downregulated GO categories | |||||
|---|---|---|---|---|---|
| GO Category | Total Genes | Underexpressed Genes | Enrichment | Log10(p) | FDR |
| Positive regulation of T cell activation | 70 | 6 | 10.42 | −4.63 | 0.035 |
| Positive regulation of lymphocyte activation | 104 | 7 | 8.18 | −4.63 | 0.023 |
| Antigen processing and presentation | 45 | 5 | 13.51 | −4.49 | 0.025 |
| Positive regulation of leukocyte activation | 112 | 7 | 7.60 | −4.42 | 0.020 |
| Positive regulation of cell activation | 116 | 7 | 7.34 | −4.32 | 0.025 |
| Immune system process | 778 | 18 | 2.81 | −4.20 | 0.034 |
| Hemoglobin complex | 2 | 2 | |||
| Antigen processing and presentation of peptide antigen | 31 | 4 | 15.69 | −3.94 | 0.048 |
| Cation homeostasis | 236 | 9 | 4.64 | −3.85 | 0.042 |
| Cellular metal ion homeostasis | 190 | 8 | 5.12 | −3.76 | 0.042 |
| Metal ion homeostasis | 193 | 8 | 5.04 | −3.72 | 0.042 |
| Regulation of T cell activation | 104 | 6 | 7.01 | −3.67 | 0.042 |
| Regulation of lymphocyte activation | 151 | 7 | 5.64 | −3.61 | 0.045 |
The transcriptional changes after nerve crush injury were analyzed as paired data, taking into account the expression level of each gene in the uninjured contralateral facial motor nucleus. Inflammatory responses were very similar (Fig. 4D–F), but a relatively small number of genes was different across genotypes compared to the large effect of the crush injury (Fig. 4G). A set of genes with an absolute ΔLogFC > 0.5 in mGRN−/− versus NTG mice and normal expression in mGRN−/−.hGRN+/+ mice was identified. Using this approach, cathepsin D (CTSD) ranked as the most significant upregulated gene in the facial motor nucleus of mGRN−/− mice after nerve crush injury (Table 2). CTSD expression was increased by an average ΔLogFC of +0.73, and thus the increase seen in NTG and mGRN−/−.hGRN+/+ mice was almost doubled in mGRN−/− animals. This finding was confirmed by qPCR analysis which showed an excessive upregulation of CTSD in mGRN−/− mice that was approximately 100% higher than in NTG and mGRN−/−.hGRN+/+ animals (Fig. 4H). These results show that mGRN−/− mice express an increased amount of CTSD in the facial motor nucleus after nerve crush injury, which might be necessary to mediate functional recovery.
Table 2.
Top 10 differentially regulated genes in mGRN−/− mice with normalized expression in mGRN−/−.hGRN+/+ mice at 5 days post crush, ranked according to significance
| Gene | ΔLogFC mGRN−/− vs NTG | ΔLogFC mGRN−/− vs mGRN−/− .hGRN+/+ | Average ΔLogFC | mGRN−/− FDR |
|---|---|---|---|---|
| Ctsd | 0.80 | 0.67 | 0.73 | 2.70E-26 |
| Grp | 0.50 | 0.88 | 0.69 | 6.17E-16 |
| Lrp1 | −0.64 | −0.62 | −0.63 | 1.23E-07 |
| Nefm | 0.56 | 0.51 | 0.54 | 2.40E-06 |
| Gm10800 | 2.99 | 4.17 | 3.58 | 3.54E-04 |
| Cyth4 | −1.13 | −0.63 | −0.88 | 6.38E-04 |
| Scarna17 | 1.83 | 1.38 | 1.61 | 7.66E-04 |
| Ccl12 | −0.70 | −0.77 | −0.73 | 1.30E-03 |
| 1700003F12Rik | −0.55 | −0.71 | −0.63 | 5.46E-03 |
| Lst1 | 0.88 | 0.56 | 0.72 | 1.31E-02 |
Ctsd: cathepsin D, Grp: gastrin releasing peptide, Lrp1: low density lipoprotein receptor-related protein 1, Nefm: neurofilament medium polypeptide, Gm10800: predicted gene 10800, Cyth4: cytohesin 4, Scarna17: small Cajal body-specific RNA 17, Ccl12: chemokine (C-C motif) ligand 12, 1700003F12Rik: RIKEN cDNA 1700003F12 gene, Lst1: leukocyte specific transcript 1.
GRN stabilizes CTSD, increasing its proteolytic activity to stimulate nerve regeneration
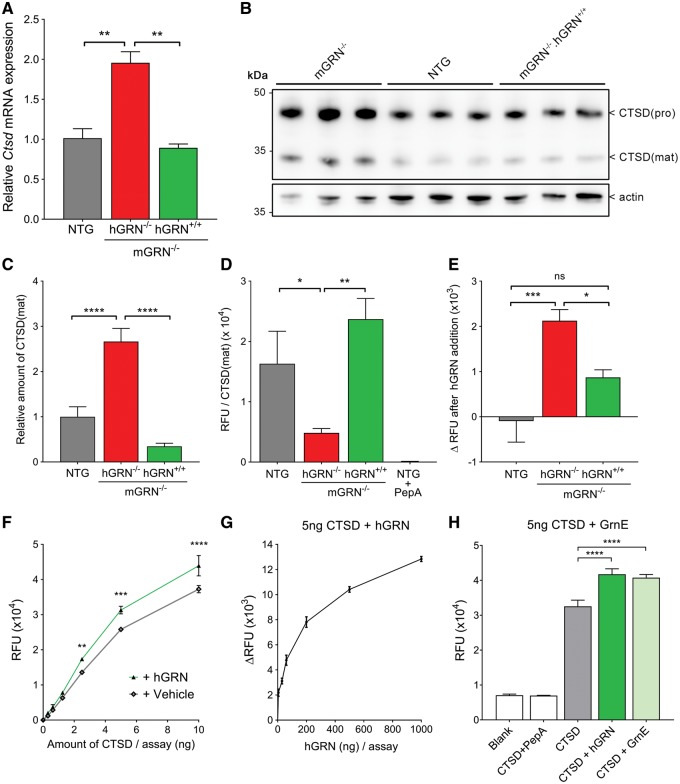
While CTSD levels were not changed in the uninjured facial motor nucleus of our young mGRN−/− mice (Fig. 4H), we did observe a significant upregulation of CTSD mRNA in brain cortices of 22–24 month old mGRN−/− mice (Fig. 5A), similar to the pattern seen in the injured facial motor nucleus and this upregulation was also present at the protein level (Fig. 5B and C). As predicted by our measurements in the facial motor nucleus, this upregulation of CTSD was completely counteracted by transgenic expression of hGRN (in mGRN−/−.hGRN+/+ mice). To further study the role of GRN in modulating CTSD activity, brain lysates were analyzed using a CTSD activity assay and normalized to the levels of mature CTSD protein in these lysates. These experiments showed that CTSD proteolytic activity is significantly decreased in old mGRN−/− mice (Fig. 5D), suggesting that the transcriptional upregulation occurs in response to decreased efficiency of the enzymatic function of CTSD. Supplementation of exogenous recombinant hGRN in this assay enhanced CTSD enzyme activity in brain lysates (Fig. 5E), and the addition of hGRN to an active recombinant CTSD showed a similar outcome (Fig. 5F). Moreover, hGRN increased CTSD activity in a dose-dependent manner (Fig. 5G). Similar to the domain specificity of the neurotrophic effect of GRN, this chaperone function was also observed for the C-terminal granulin E (GrnE) domain (Fig. 5H).
Figure 5.
GRN and GrnE increase the proteolytic activity of CTSD. (A) CTSD mRNA expression in brains of 22-24 month old mice. **P < 0.01, One-way ANOVA. (B) Western blot analysis of old mice brain lysates that were used for CTSD enzymatic assays. (C) Quantification of CTSD(mat) levels, normalized to actin. ****P < 0.0001, One-way ANOVA. (D) CTSD enzyme activity in relative fluorescent units (RFU) normalized to the levels of mature CTSD protein. 2.5 µM pepstatin A was added to NTG lysates as a specificity control. *P < 0.05, **P < 0.01, One-way ANOVA. (E) Change in CTSD enzymatic activity upon the addition of 100 ng recombinant hGRN to brain lysates. *P < 0.05, ***P < 0.001, One-way ANOVA. (F) Addition of 100 ng hGRN to an active recombinant CTSD peptide increases its proteolytic activity. **P < 0.01, ***P < 0.001, ****P < 0.0001, Two-way ANOVA followed by Bonferroni correction. (G) hGRN increases the proteolytic activity of 5 ng recombinant CTSD in a dose-dependent manner. (H) Addition of equimolar amounts GrnE and GRN increase the proteolytic activity of 5 ng CTSD to a similar extent. ****P < 0.0001, One-way ANOVA. Data are shown as mean ± SEM.
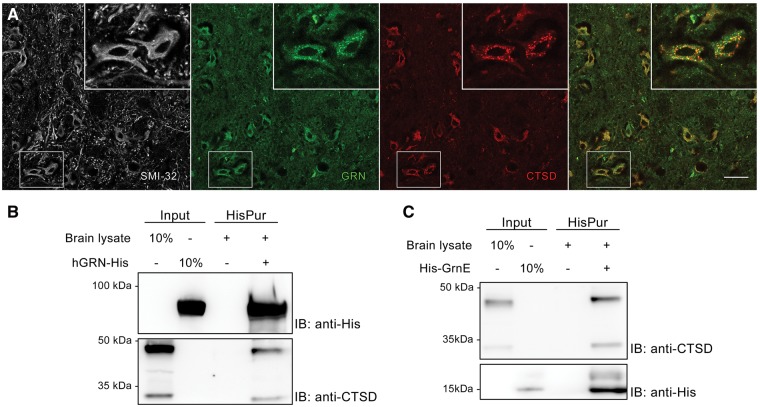
Next, we looked for a physical interaction between GRN and CTSD. GRN and CTSD are known to be localized to lysosomes, and to be two of the most abundant proteins in lysosomes (36). Also inside SMI-32 positive neurons in the facial motor nucleus, we observed significant overlap of CTSD and mGRN positive vesicles (Fig. 6A). Pull down experiments using his-tagged hGRN and GrnE, added to brain lysates, confirmed that both CTSD(pro) and CTSD(mat) co-precipitated with either the full length GRN protein as well as with the C-terminal GrnE protein (Fig. 6B and C). These experiments provide strong evidence for a direct interaction between GRN and CTSD, which is also mediated by its C-terminus.
Figure 6.
GRN and GrnE show a direct interaction with CTSD. (A) GRN and CTSD co-localize inside SMI-32 positive neurons in the facial motor nucleus. Scale bar = 50 µm. (B–C) Pull down of recombinant hGRN (B) or GrnE (C) co-precipitates both the pro- and mature form of CTSD from brain lysates.
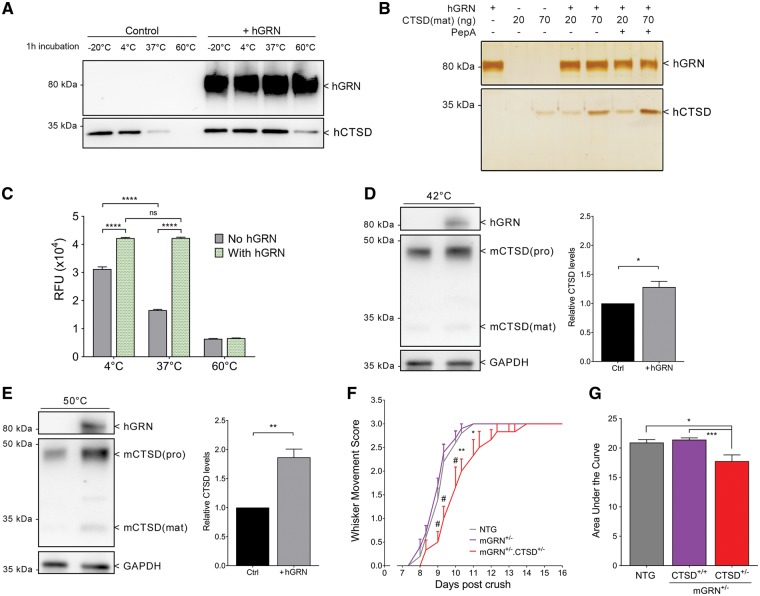
As shown by previous studies, CTSD activity is highly dependent on temperature. Even short incubations at increased temperatures have been shown to inactivate its enzymatic activity (37,38). Also in our experiments, incubating 5 ng of recombinant CTSD protein for 1 h at 37 °C led to a highly reduced detection of the protein on western blot (Fig. 7A). Interestingly, the addition of hGRN to the CTSD protein at 37 °C drastically improved the stability of CTSD in these conditions (Fig. 7A). In the presence of hGRN, a substantial amount of CTSD protein remained even after 1 h of incubation at 60 °C, while no CTSD protein could be detected in the same conditions without hGRN (Fig. 7A). This finding was highly reproducible, using different concentrations of CTSD, and was independent of its activity, as shown by co-incubation with pepstatin A (Fig. 7B). While 1 h pre-incubation of CTSD at 37 °C led to a significant reduction in CTSD proteolytic activity by approximately 50%, this function was completely preserved in the presence of hGRN (Fig. 7C). Based on these findings, we hypothesized that the rate of CTSD turnover might be higher in the absence of GRN. Therefore, we created mGRN−/− mouse embryonic fibroblasts (MEFs) to study heat inactivation of CTSD at different temperatures. As different delivery routes for extracellular GRN to the lysosome exist (39), we added hGRN to the medium of these cells. Serum-free medium was used to increase the rate of endocytosis. We could clearly detect endocytosis of hGRN in the cell lysates and found that the addition of hGRN to mGRN−/− MEFs protected against heat-induced breakdown of CTSD (Fig. 7D and E).
Figure 7.
GRN stabilizes CTSD, which is necessary to mediate axonal outgrowth in vivo. (A) Heat inactivation of CTSD is prevented by the addition of hGRN. (B) Silver staining showing that the inhibition of CTSD activity by PepA has no effect on its heat inactivation. (C) Pre-incubation of CTSD for 1 h at different temperatures leads to a significant reduction in its proteolytic capacity, which is prevented by the addition of hGRN. ****P < 0.0001, Two-way ANOVA followed by Bonferroni correction. (D,E) Heat inactivation of CTSD in mGRN−/− MEFs for 1 h at 42 °C (D) and 15’ (after 3h pre-incubation with hGRN) at 50 °C (E) is prevented by the supplementation of hGRN, in a temperature dependent manner. *P < 0.05, **P < 0.01, Student’s t-test. (F) A combined heterozygous reduction of mGRN and CTSD significantly delays whisker movement recovery. *P < 0.05, **P < 0.01, #P < 0.0001, RM Two-way ANOVA followed by Bonferroni correction. (G) Analysis of the area under the curve of whisker movement recovery. *P < 0.05, ***P < 0.001, Kruskal Wallis test followed by Dunn’s test. Data is shown as mean ± SEM. PepA = pepstatin A.
Hypothesizing that the observed effect of GRN on CTSD function and stability would be important for the functional recovery after nerve crush, we explored this interaction in the in vivo paradigm of facial nerve crush injury. Comparable to heterozygous mGRN knockout mice, heterozygous CTSD knockout mice show a normal recovery after nerve crush (Supplementary Material, Fig. S3 D). Interestingly, the combined mGRN+/-. CTSD+/- heterozygous knockout mice showed a significantly delayed recovery (Fig. 7F and G). Hence, the synergistic effect of reducing GRN and CTSD levels on neurite outgrowth indicates that the chaperone function of GRN on CTSD is necessary in vivo to stimulate nerve outgrowth and functional recovery after nerve crush injury.
Discussion
GRN is a pleiotropic growth factor of which the expression in the nervous system is largely limited to neurons and microglia (6). Currently, three main hypothesis exist that could explain the disease mechanisms in illnesses caused by GRN deficiency. First of all, GRN has been implicated in neuroinflammation, as an anti-inflammatory protein via its inhibitory effect on TNF signaling (40). It is however still a debate whether this interaction is truly important for the biology of GRN in the central nervous system (41). In our acute neurite outgrowth model, we could not find evidence for beneficial effects of microglial GRN. While tnfrsf1b, the gene encoding tumor necrosis factor receptor type 2, was one of the genes in the transcriptome analysis of which the expression was significantly increased after nerve crush injury, this response was similar in the absence or presence of GRN. Furthermore, we observed a comparable inflammatory response between the mGRN−/− mice and the NTG animals. However, these acute injury experiments do not exclude important roles of GRN in microglia and neuroinflammation.
Second, GRN acts as a neurotrophic factor to increase neuronal survival and neurite outgrowth (7,28,29,32). Two recent papers identified Notch signaling (42) and EPHA2 (43) as functional receptors for GRN. However, we did not observe transcriptional changes related to Notch receptors or Notch response genes after nerve crush injury. RNA expression of a small subset of ephrin signaling genes (EPHA4, Efna3, Efnb3) was changed after crush, but none of these changes were GRN-dependent.
A third hypothesis, that GRN is necessary to maintain proper lysosomal function, has been put forward since the discovery of homozygous GRN mutations in a few patients with NCL (14,15). However, the putative lysosomal function of GRN remains unknown and how GRN deficiency might cause neurodegeneration through lysosomal dysfunction is still largely unexplored. There is some evidence that GRN is part of a larger network of lysosomal genes that is upregulated in response to the nuclear translocation of transcription factor EB (TFEB) and that this network is over-activated in GRN knockout mice after traumatic brain injury (44). Interestingly, CTSD is also a member of this network activated by TFEB (45). It is possible that the activation of this network is necessary to generate sufficient amounts of GRN and CTSD which are both necessary for lysosomal function. This would also explain why heterozygous deletion of GRN does not lead to a delayed recovery from facial nerve injury, as the reported increase in GRN production from the remaining allele (Supplementary Material, Fig. S3B) might be sufficient to fulfill its role in our acute model.
In this study, we provide evidence for a direct link between the neurotrophic effects of both GRN and GrnE with CTSD as chaperones to stabilize the enzyme, hence increasing its proteolytic activity. GRN and GrnE co-immunoprecipitated with CTSD and GRN also dose-dependently increased the enzymatic activity of CTSD. Interestingly, homozygous loss-of-function mutations in GRN and CTSD cause similar clinical phenotypes, i.e. the lysosomal storage disease, NCL (46). This type of lysosomal storage disease is mostly observed in patients with homozygous mutations, as small amounts of lysosomal proteins can be sufficient for a normal function. Indeed, heterozygous knockout mice for GRN or CTSD do not show any pathology associated with neurodegeneration and also showed normal recovery after crush. However, a combined 50% reduction in their levels seems to be detrimental for normal functional recovery after crush. Our experiments indicate that in the absence of GRN, a less efficient and less stable CTSD enzyme is being formed. This could explain why different scenario’s leading to an activation of the lysosomal stress response results in a seemingly similar compensatory upregulation of CTSD (e.g. an acute nerve crush injury, or significant ageing of mGRN−/− mice). Although we did not observe differences in lysosomal CTSD localization or in the relative amount of mature over pro-CTSD, GRN is present in all of the cellular compartments where CTSD is found and hence could also influence different steps of its processing or trafficking to the lysosomes. Interestingly, one of the other genes that was differentially regulated in mGRN−/− mice upon nerve injury was the low-density lipoprotein receptor-related protein 1 (LRP1), a sorting receptor which is also implicated in the delivery of GRN to the lysosomes (39). Future studies will be necessary to determine if LRP1 dependent trafficking of GRN plays a role in neurite outgrowth and its chaperone function on CTSD.
In summary, our current study provides new insights into the biological functions of GRN, more specifically on its in vivo neurotrophic function and how this might be directly linked with a chaperone function of GRN, enhancing lysosomal CTSD activity and stability.
Materials and Methods
Animals
Generation
The conditional mGRN knockout mice were generated by homologous recombination in embryonic stem (ES) cells using a vector harboring three adjacent fragments from the mGRN locus isolated from a mouse C57Bl/6J BAC clone (#RP23-226P4, Children's Hospital Oakland Research Institute (CHORI)) and cloned into the pDELBOY conditional targeting vector (47). The targeting vector contained from 5’ to 3’: an FRT-flanked neomycin resistance cassette; a 3.9 kb mGRN fragment containing exon 1 together with ca 1.9 kb upstream sequence and ca 2 kb of intron 1; a loxP flanked MID fragment of 2.38 kb containing the remainder of intron 1 through ca 0.29 kb of intron 8; a 2.93 kb downstream adjacent mGRN fragment comprising exons 9–13; and a thymidine kinase expression cassette outside of the genomic homology regions for selection against random integrants. The targeting vector was linearized and electroporated into G4 ES cells (48) (kindly provided by J.J. Haigh, Ghent, Belgium). Targeted ES cells were subjected to positive/negative selection with G418/gancyclovir, and correctly targeted ES cell clones with absence of random integrants were identified by appropriate Southern blot analysis. Recombinant ES cells were transiently electroporated with the Flp recombinase-expressing vector pOG-Flpe6 (49) (kindly made available by A.F Stewart, Dresden, Germany) to remove the FRT-flanked neomycin cassette. Correctly excised clones were identified by PCR and were used for morula aggregation and generation of chimeric mice. These mice were bred with C57Bl/6J mice to generate germline mGRNflox mice. The resulting heterozygous mGRNflox mice were backcrossed with C57Bl/6J mice for at least 10 generations. Mice were then crossed with PGK-Cre mice (50) to obtain heterozygous and homozygous mGRN deficient mice. Human GRN (hGRN) overexpressing mice were generated as described previously (35) and mice with overexpression of a truncated GRN protein comprising only the first 417 amino acids (hGRN(418X) mice) were created in an identical way after the introduction of an R418X mutation in the hGRN cDNA using the QuikChange Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA, USA). CTSD deficient mice were created as described before (51).
Maintenance
All animals, except for the hGRN(418X) mice, were born and initially housed in type II filtertop cages in a specific pathogen free area. All animals used for experiments were transferred to a conventional animalium, where hGRN(418X) mice were born and housed in individually ventilated cages. All animals had access to food and water ad libitum with light-dark cycles of 12 h. All operations were performed under aseptic conditions and all procedures were approved by the Institutional Animal Care and Ethical Research Advisory Committee of the University of Leuven, Belgium (P132/2012 and P015/2015).
Facial nerve crush injury
Surgical procedure
At the time of nerve crush injury, animals used to study functional recovery were 8 to 12 weeks old, while animals used for cell body counts in the facial motor nucleus were 7 weeks old. After checking the mice for normal eye blinking and whisker movement, they were anesthetized with 3% inhaled isoflurane (Eurovet NV, Heusden-Zolder, Belgium) which was maintained throughout the surgical procedure. Mice were placed on a 37 °C hot plate to prevent hypothermia during the surgery. Hair was removed and an incision was made in close proximity to the stylomastoid foramen. The facial nerve was uncovered from surrounding tissues by blunt dissection after which it was crushed using an ultra-fine hemostat with smooth tips (Fine science tools, Heidelberg, Germany). The nerve crush injury was performed for a duration of 20 s, as close as possible but proximal to the bifurcation of the retroauricular branch of the facial nerve (Fig. 1A and B). Next, the skin was sutured with 6–0 absorbable sutures (Ethicon, Somerville, New Jersey, USA). Mice were then returned to their cages and recovered from the surgery under a heating lamp, preventing hypothermia. When fully recovered, the absence of whisker movement was confirmed on the ipsilateral side of the nerve crush injury to verify that the procedure was carried out successfully.
Functional analysis of the recovery
To evaluate regeneration of facial nerve axons, all mice were examined daily for whisker function by an investigator blinded for the genotypes. Whisker movement scores were given according to the following scale: a score of 0 was given for no detectable movement, 1 for detectable motion of individual whiskers, 2 for significant (but asymmetric) voluntary motion, and 3 for symmetric voluntary motion. To increase the power of our observations, behavioral assessments were made twice daily (9–10 am and 6–7 pm) in later experiments. The contralateral whiskers were used as a reference for complete functional recovery.
Immunofluorescence and histology
Facial motor neuron analysis
All animals used to study facial motor neuron vulnerability were subjected to a facial nerve crush injury at 7 weeks of age. 28 days after the injury, they were euthanized by CO2 asphyxiation, brains were rapidly removed, embedded in Tissue-Tek OCT compound (Sakura Finetek USA Inc., Torrance, CA, USA) and frozen on dry ice before being stored at -80 °C until further processing. 25 µm cryosections were made through the facial motor nucleus and stained with a 2X thionin solution to visualize the facial motor nucleus cell bodies (52). Briefly, after a 1 min fixation in 100% ethanol in the cryochamber, slides were washed twice for 15 s in dH2O, stained for 45 s in 2X thionin, again washed in dH20 two times and dehydrated in a three step ethanol series (70, 90 and 100%) for 30 s each. Slides were cleared with Histoclear solution for 1 min, mounted with PerTex mounting medium (Histolab, Göteborg, Germany) and images were acquired with a Zeiss Imager.M1 using a 10X objective. Facial motor neuron cell bodies were counted and analyzed for area, circular morphology and staining intensity, using ImageJ (v.1.49, NIH, USA). All parameters were normalized to those of the non-injured contralateral side.
Immunostaining
All facial motor nucleus stainings were performed at 5 days post injury. To this end, mice were anesthetized with an overdose of pentobarbital (120 mg/kg bodyweight, I.P, NEMBUTAL®, Ceva Santé Animale, Brussels, Belgium) and perfused with PBS and 4% PFA (Sigma-Aldrich, St. Louis, MO, USA). Whole brains were post-fixed overnight in PFA and cryoprotected by 48 h incubation in 30% sucrose (Sigma-Aldrich). 25 µm cryosections were blocked with 3% BSA (Enzo Life Sciences BVBA, Brussels, Belgium) in 0.1% Triton X-100/PBS and incubated overnight at 4 °C with primary antibodies against SMI-32 (1:200, BioLegend, San Diego, CA, USA), Iba1 (1:200, Wako Chemicals GmbH, Neuss, Germany), Cathepsin D (53) and mouse GRN (1:200, R&D systems Inc., Minneapolis, MN, USA), diluted in 0.1% Triton X-100/PBS containing 1% BSA. Alexa-conjugated secondary antibodies (Invitrogen, Carlsbad, CA, USA) were diluted in 0.1% Triton X-100/PBS and incubated for 1 h. Sections were mounted with ProLong® Gold antifade reagent (Life Technologies, Carlsbad, CA, USA). Images were acquired using the Leica SP8x confocal microscope.
Transcriptional analysis after facial nerve crush injury
For RNA extraction from the facial motor nucleus, mice were euthanized by CO2 asphyxiation and after rapid dissection, brains were frozen in Tissue-Tek OCT compound on dry ice. Sections of 100 µm thickness through the facial motor nucleus were collected in the cryochamber, picked up on RNaseZAP (Life Technologies) treated superfrost microscope slides and stained with a 2X thionin solution (as described in section 3.1 using ultrapure diethylpyrocarbonate-treated water instead of dH2O). After careful microscopic dissection, pooling the ipsi- and contralateral facial motor nucleus in separate tubes, the collected tissue was immediately submerged in RNA extraction buffer of the PicoPure® RNA isolation kit (Thermo Fisher Scientific Inc., Pittsburgh, PA, USA) and further processed according to the manufacturer’s protocol. For qRT-PCR analysis, first-strand cDNA was synthesized using SuperScript III (Invitrogen). PCR reactions were performed using TaqMan assays (Applied Biosystems, Foster City, CA, USA) for Iba1, CD11b and mGRN or SYBR Green reagents (Thermo Fisher Scientific) with the following primers: cathepsin D (CTSD) (forward, 5’-GCTTCCGGTCTTTGACAACCT-3'; reverse, 5’-CACCAAGCATTAGTTCTCCTCC-3') and normalized to three reference genes: adaptor-related protein complex 3, delta 1 subunit (Ap3d1) (forward, 5’-CAAGGGCAGTATCGACCGC-3'; reverse, 5’-GATCTCGTCAATGCACTGGGA-3'), MON2 homolog (Mon2) (forward, 5’-CTACAGTCCGACAGGTCGTGA-3'; reverse, 5’-CGGCACTGGAGGTTCTATATCTC-3') and F-box protein 38 (Fbxo38) (forward, 5’-ATGGGACCACGAAAGAAAAGTG-3'; reverse, 5’-TAGCTTCCGAGAGAGGCATTC-3'). Expression levels were normalized and analyzed using qBase+ (v.3.0, Biogazelle, Zwijnaarde, Belgium). For next-generation sequencing, samples were sent to VIB’s nucleomics core (Leuven, Belgium) and analyzed for RNA integrity (≥ 7) by running on a Bioanalyzer (Agilent) before Trueseq total stranded RNA library preparation and sequencing on a NextSeq 500 instrument (Illumina Inc., San Diego, CA, USA).
Western blot analysis
Proteins were extracted from brain cortex samples using RIPA buffer (Sigma-Aldrich) supplemented with CompleteTM, EDTA-free protease inhibitor cocktail (Sigma-Aldrich), or with CD cell lysis buffer of the Cathepsin D activity assay kit (Abcam, Cambridge, UK) where CTSD activity was normalized to protein levels. Protein concentrations were determined using the microBCA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Reducing sample buffer (Thermo Fisher Scientific) was added to samples containing equal amounts of protein and heated for 5 min at 95 °C before separation on a sodium dodecyl sulfate–polyacrylamide electrophoresis gel. After electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane (Merck Millipore, Darmstadt, Germany). Non-specific binding was blocked using 5% blotting-grade blocker (Bio-rad, Hercules, CA, USA), diluted in Tris-Buffered Saline Tween (50 mM TRIS, 150 mM NaCl, 0.1% Tween-20; Applichem, Darmstadt, Germany) for 1 h at room temperature before. Primary antibodies were incubated overnight at 4 °C, diluted in blocking-grade buffer and directed against human progranulin (1:200, R&D Systems), mouse cathepsin D (53), 6X-His (1:1000, Abcam), beta-actin (1:5000, Sigma-Aldrich), alpha-tubulin (1:5000, Sigma-Aldrich), human cathepsin D (1:1000, Abcam) and GAPDH (1:2000, Thermo Fisher Scientific). HRP-coupled secondary antibodies (1:5000, Dako, Agilent), diluted in TBS-T, were incubated for 1 h at room temperature. Blots were visualized using the enhanced chemiluminescent substrate (Thermo Fisher Scientific) and imaged with an ImageQuant LAS 4000 system (GE Healthcare, Uppsala, Sweden). Blots were quantified in ImageQuant TL (v. 7.0).
Cathepsin D activity assay
Brain cortex samples were lysed in CD cell lysis buffer of the cathepsin D activity assay kit (Abcam). An amount of 100 ng total protein was analyzed according to the manufacturer’s instructions. Individual GrnE with its 5’ and 3’ linker domains was produced by VIB’s protein service facility (PSF, Ghent, Belgium). To this end, the cDNA encoding amino acids 497 to 593 of GRN, with an N-terminal 6X His-tag, was synthetized and cloned into a pcDNA3.1(+) expression vector (Geneart, Thermo Fisher Scientific).
Statistical analysis
All statistical analyses were performed using GraphPad Prism (v 7.01, GraphPad Software, Inc, San Diego, CA, USA). Gene ontology analysis was performed using the High-Throughput GoMiner webinterface with standard parameters (54).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors would like to thank Ir. Jurgen Haustraete and Dr. Philippe De Groote of the VIB Protein Service Facility (UGent, Belgium) for their excellent assistance in the design of the vector and the subsequent production of the recombinant GrnE. The Leica SP8x confocal microscope was provided by InfraMouse (KU Leuven-VIB) through a Hercules type 3 project (ZW09-03).
Conflict of Interest statement. None declared.
Funding
This work was supported by grants from Opening the Future Fund (KU Leuven), the Fund for Scientific Research Flanders (FWO-Flanders), the Interuniversity Attraction Poles (IUAP) program P7/16 of the Belgian Federal Science Policy Office, the ALS liga Belgium, the Alzheimer Research Foundation (SAO-FRA), the Flemish Government initiated Flanders Impulse Program on Networks for Dementia Research (VIND), the European Union Joint Programme-Neurodegenerative Disease Research (JPND) project STRENGTH, and RiMod-FTD, the European E-Rare-2 project PYRAMID and the Deutsche Forschungsgemeinschaft [SFB877]. PVD holds a senior clinical investigatorship of FWO-Vlaanderen. WR is supported through the E. von Behring Chair for Neuromuscular and Neurodegenerative Disorders and an ERC grant agreement [340429]. Funding to pay the Open Access publication charges for this article was provided by KU Leuven.
References
- 1. Bateman A., Bennett H.P. (1998) Granulins: the structure and function of an emerging family of growth factors. J. Endocrinol., 158, 145–151. [DOI] [PubMed] [Google Scholar]
- 2. Zhu J., Nathan C., Jin W., Sim D., Ashcroft G.S., Wahl S.M., Lacomis L., Erdjument-Bromage H., Tempst P., Wright C.D.. et al. (2002) Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell, 111, 867–878. [DOI] [PubMed] [Google Scholar]
- 3. De Muynck L., Van Damme P. (2011) Cellular effects of progranulin in health and disease. J. Mol. Neurosci., 45, 549–560. [DOI] [PubMed] [Google Scholar]
- 4. Zheng Y., Brady O.A., Meng P.S., Mao Y., Hu F. (2011) C-terminus of progranulin interacts with the beta-propeller region of sortilin to regulate progranulin trafficking. PLoS One, 6, e21023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petkau T.L., Leavitt B.R. (2014) Progranulin in neurodegenerative disease. Trends Neurosci., 37, 388–398. [DOI] [PubMed] [Google Scholar]
- 6. Petkau T.L., Neal S.J., Orban P.C., MacDonald J.L., Hill A.M., Lu G., Feldman H.H., Mackenzie I.R., Leavitt B.R. (2010) Progranulin expression in the developing and adult murine brain. J. Comp. Neurol., 518, 3931–3947. [DOI] [PubMed] [Google Scholar]
- 7. Van Damme P., Van Hoecke A., Lambrechts D., Vanacker P., Bogaert E., van Swieten J., Carmeliet P., Van Den Bosch L., Robberecht W. (2008) Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J. Cell Biol., 181, 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martens L.H., Zhang J., Barmada S.J., Zhou P., Kamiya S., Sun B., Min S.W., Gan L., Finkbeiner S., Huang E.J.. et al. (2012) Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J. Clin. Invest., 122, 3955–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S.. et al. (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature, 442, 916–919. [DOI] [PubMed] [Google Scholar]
- 10. Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J.J.. et al. (2006) Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature., 442, 920–924. [DOI] [PubMed] [Google Scholar]
- 11. Finch N., Baker M., Crook R., Swanson K., Kuntz K., Surtees R., Bisceglio G., Rovelet-Lecrux A., Boeve B., Petersen R.C.. et al. (2009) Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain, 132, 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghidoni R., Benussi L., Glionna M., Franzoni M., Binetti G. (2008) Low plasma progranulin levels predict progranulin mutations in frontotemporal lobar degeneration. Neurology, 71, 1235–1239. [DOI] [PubMed] [Google Scholar]
- 13. Sleegers K., Brouwers N., Van Damme P., Engelborghs S., Gijselinck I., van der Zee J., Peeters K., Mattheijssens M., Cruts M., Vandenberghe R.. et al. (2009) Serum biomarker for progranulin-associated frontotemporal lobar degeneration. Ann. Neurol., 65, 603–609. [DOI] [PubMed] [Google Scholar]
- 14. Almeida M.R., Macario M.C., Ramos L., Baldeiras I., Ribeiro M.H., Santana I. (2016) Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol. Aging, 41, 200 e201–205. [DOI] [PubMed] [Google Scholar]
- 15. Smith K.R., Damiano J., Franceschetti S., Carpenter S., Canafoglia L., Morbin M., Rossi G., Pareyson D., Mole S.E., Staropoli J.F.. et al. (2012) Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet., 90, 1102–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gotzl J.K., Mori K., Damme M., Fellerer K., Tahirovic S., Kleinberger G., Janssens J., van der Zee J., Lang C.M., Kremmer E.. et al. (2014) Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol., 127, 845–860. [DOI] [PubMed] [Google Scholar]
- 17. Yin F., Dumont M., Banerjee R., Ma Y., Li H., Lin M.T., Beal M.F., Nathan C., Thomas B., Ding A. (2010) Behavioral deficits and progressive neuropathology in progranulin-deficient mice: a mouse model of frontotemporal dementia. faseb J., 24, 4639–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Philips T., De Muynck L., Thu H.N., Weynants B., Vanacker P., Dhondt J., Sleegers K., Schelhaas H.J., Verbeek M., Vandenberghe R.. et al. (2010) Microglial upregulation of progranulin as a marker of motor neuron degeneration. J. Neuropathol. Exp. Neurol., 69, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 19. Lui H., Zhang J., Makinson S.R., Cahill M.K., Kelley K.W., Huang H.Y., Shang Y., Oldham M.C., Martens L.H., Gao F.. et al. (2016) Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell, 165, 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanaka Y., Chambers J.K., Matsuwaki T., Yamanouchi K., Nishihara M. (2014) Possible involvement of lysosomal dysfunction in pathological changes of the brain in aged progranulin-deficient mice. Acta Neuropathol. Commun., 2, 78.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raitano S., Ordovas L., De Muynck L., Guo W., Espuny-Camacho I., Geraerts M., Khurana S., Vanuytsel K., Toth B.I., Voets T.. et al. (2015) Restoration of progranulin expression rescues cortical neuron generation in an induced pluripotent stem cell model of frontotemporal dementia. Stem Cell Rep., 4, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Almeida S., Zhang Z., Coppola G., Mao W., Futai K., Karydas A., Geschwind M.D., Tartaglia M.C., Gao F., Gianni D.. et al. (2012) Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep., 2, 789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Minami S.S., Min S.W., Krabbe G., Wang C., Zhou Y., Asgarov R., Li Y., Martens L.H., Elia L.P., Ward M.E.. et al. (2014) Progranulin protects against amyloid beta deposition and toxicity in Alzheimer's disease mouse models. Nat. Med., 20, 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laird A.S., Van Hoecke A., De Muynck L., Timmers M., Van den Bosch L., Van Damme P., Robberecht W. (2010) Progranulin is neurotrophic in vivo and protects against a mutant TDP-43 induced axonopathy. PLoS One, 5, e13368.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chitramuthu B.P., Baranowski D.C., Kay D.G., Bateman A., Bennett H.P. (2010) Progranulin modulates zebrafish motoneuron development in vivo and rescues truncation defects associated with knockdown of Survival motor neuron 1. Mol. Neurodegener., 5, 41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jackman K., Kahles T., Lane D., Garcia-Bonilla L., Abe T., Capone C., Hochrainer K., Voss H., Zhou P., Ding A.. et al. (2013) Progranulin deficiency promotes post-ischemic blood-brain barrier disruption. J. Neurosci., 33, 19579–19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanazawa M., Kawamura K., Takahashi T., Miura M., Tanaka Y., Koyama M., Toriyabe M., Igarashi H., Nakada T., Nishihara M.. et al. (2015) Multiple therapeutic effects of progranulin on experimental acute ischaemic stroke. Brain, 138, 1932–1948. [DOI] [PubMed] [Google Scholar]
- 28. Gao X., Joselin A.P., Wang L., Kar A., Ray P., Bateman A., Goate A.M., Wu J.Y. (2010) Progranulin promotes neurite outgrowth and neuronal differentiation by regulating GSK-3beta. Protein Cell, 1, 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ryan C.L., Baranowski D.C., Chitramuthu B.P., Malik S., Li Z., Cao M., Minotti S., Durham H.D., Kay D.G., Shaw C.A.. et al. (2009) Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci., 10, 130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu J., Xilouri M., Bruban J., Shioi J., Shao Z., Papazoglou I., Vekrellis K., Robakis N.K. (2011) Extracellular progranulin protects cortical neurons from toxic insults by activating survival signaling. Neurobiol. Aging, 32, 2326 e2325–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De Muynck L., Herdewyn S., Beel S., Scheveneels W., Van Den Bosch L., Robberecht W., Van Damme P. (2013) The neurotrophic properties of progranulin depend on the granulin E domain but do not require sortilin binding. Neurobiol. Aging, 34, 2541–2547. [DOI] [PubMed] [Google Scholar]
- 32. Gass J., Lee W.C., Cook C., Finch N., Stetler C., Jansen-West K., Lewis J., Link C.D., Rademakers R., Nykjaer A.. et al. (2012) Progranulin regulates neuronal outgrowth independent of sortilin. Mol. Neurodegener., 7, 33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferri C.C., Moore F.A., Bisby M.A. (1998) Effects of facial nerve injury on mouse motoneurons lacking the p75 low-affinity neurotrophin receptor. J. Neurobiol., 34, 1–9. [PubMed] [Google Scholar]
- 34. McGraw J., McPhail L.T., Oschipok L.W., Horie H., Poirier F., Steeves J.D., Ramer M.S., Tetzlaff W. (2004) Galectin-1 in regenerating motoneurons. Eur. J. Neurosci., 20, 2872–2880. [DOI] [PubMed] [Google Scholar]
- 35. Herdewyn S., De Muynck L., Van Den Bosch L., Robberecht W., Van Damme P. (2013) Progranulin does not affect motor neuron degeneration in mutant SOD1 mice and rats. Neurobiol. Aging, 34, 2302–2303. [DOI] [PubMed] [Google Scholar]
- 36. Markmann S., Krambeck S., Hughes C.J., Mirzaian M., Aerts J.M., Saftig P., Schweizer M., Vissers J.P., Braulke T., Damme M. (2017) Quantitative Proteome Analysis of Mouse Liver Lysosomes Provides Evidence for Mannose 6-phosphate-independent Targeting Mechanisms of Acid Hydrolases in Mucolipidosis II. Mol. Cell Proteomics, 16, 438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karwowska A., Roszkowska-Jakimiec W., Dabrowska E., Gacko M., Chlabicz M. (2006) The effect of temperature, acidification, and alkalization changes as well as ethanol on salivary cathepsin D activity. Adv. Med. Sci., 51 Suppl 1, 59–61. [PubMed] [Google Scholar]
- 38. Hayes M.G., Hurley M.J., Larsen L.B., Heegaard C.W., Magboul A.A., Oliveira J.C., McSweeney P.L., Kelly A.L. (2001) Thermal inactivation kinetics of bovine cathepsin D. J. Dairy Res., 68, 267–276. [DOI] [PubMed] [Google Scholar]
- 39. Zhou X., Sun L., Bastos de Oliveira F., Qi X., Brown W.J., Smolka M.B., Sun Y., Hu F. (2015) Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J. Cell Biol., 210, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang W., Lu Y., Tian Q.Y., Zhang Y., Guo F.J., Liu G.Y., Syed N.M., Lai Y., Lin E.A., Kong L.. et al. (2011) The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science, 332, 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen X., Chang J., Deng Q., Xu J., Nguyen T.A., Martens L.H., Cenik B., Taylor G., Hudson K.F., Chung J.. et al. (2013) Progranulin does not bind tumor necrosis factor (TNF) receptors and is not a direct regulator of TNF-dependent signaling or bioactivity in immune or neuronal cells. J. Neurosci., 33, 9202–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Altmann C., Vasic V., Hardt S., Heidler J., Haussler A., Wittig I., Schmidt M.H., Tegeder I. (2016) Progranulin promotes peripheral nerve regeneration and reinnervation: role of notch signaling. Mol. Neurodegener., 11, 69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neill T., Buraschi S., Goyal A., Sharpe C., Natkanski E., Schaefer L., Morrione A., Iozzo R.V. (2016) EphA2 is a functional receptor for the growth factor progranulin. J. Cell Biol., 215, 687–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tanaka Y., Matsuwaki T., Yamanouchi K., Nishihara M. (2013) Increased lysosomal biogenesis in activated microglia and exacerbated neuronal damage after traumatic brain injury in progranulin-deficient mice. Neuroscience, 250, 8–19. [DOI] [PubMed] [Google Scholar]
- 45. Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S.. et al. (2009) A gene network regulating lysosomal biogenesis and function. Science, 325, 473–477. [DOI] [PubMed] [Google Scholar]
- 46. Siintola E., Partanen S., Stromme P., Haapanen A., Haltia M., Maehlen J., Lehesjoki A.E., Tyynela J. (2006) Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain, 129, 1438–1445. [DOI] [PubMed] [Google Scholar]
- 47. Rossi D.J., Londesborough A., Korsisaari N., Pihlak A., Lehtonen E., Henkemeyer M., Makela T.P. (2001) Inability to enter S phase and defective RNA polymerase II CTD phosphorylation in mice lacking Mat1. embo J., 20, 2844–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. George S.H., Gertsenstein M., Vintersten K., Korets-Smith E., Murphy J., Stevens M.E., Haigh J.J., Nagy A. (2007) Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc. Natl Acad. Sci. U S A, 104, 4455–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Buchholz F., Angrand P.O., Stewart A.F. (1998) Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol., 16, 657–662. [DOI] [PubMed] [Google Scholar]
- 50. Lallemand Y., Luria V., Haffner-Krausz R., Lonai P. (1998) Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res., 7, 105–112. [DOI] [PubMed] [Google Scholar]
- 51. Saftig P., Hetman M., Schmahl W., Weber K., Heine L., Mossmann H., Koster A., Hess B., Evers M., von Figura K.. et al. (1995) Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. embo J., 14, 3599–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mesnard N.A., Sanders V.M., Jones K.J. (2011) Differential gene expression in the axotomized facial motor nucleus of presymptomatic SOD1 mice. J. Comp. Neurol., 519, 3488–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Claussen M., Kubler B., Wendland M., Neifer K., Schmidt B., Zapf J., Braulke T. (1997) Proteolysis of insulin-like growth factors (IGF) and IGF binding proteins by cathepsin D. Endocrinology, 138, 3797–3803. [DOI] [PubMed] [Google Scholar]
- 54. Zeeberg B.R., Qin H., Narasimhan S., Sunshine M., Cao H., Kane D.W., Reimers M., Stephens R.M., Bryant D., Burt S.K.. et al. (2005) High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID). BMC Bioinformatics, 6, 168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuzis K., Coffin J.D., Eckenstein F.P. (1999) Time course and age dependence of motor neuron death following facial nerve crush injury: role of fibroblast growth factor. Exp. Neurol., 157, 77–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.