Abstract
Members of the conserved ubiquilin (UBQLN) family of ubiquitin (Ub) chaperones harbor an antipodal UBL (Ub-like)-UBA (Ub-associated) domain arrangement and participate in proteasome and autophagosome-mediated protein degradation. Mutations in a proline-rich-repeat region (PRR) of UBQLN2 cause amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD); however, neither the normal functions of the PRR nor impacts of ALS-associated mutations within it are well understood. In this study, we show that ALS mutations perturb UBQLN2 solubility and folding in a mutation-specific manner. Biochemical impacts of ALS mutations were additive, transferable to UBQLN1, and resulted in enhanced Ub association. A Drosophila melanogaster model for UBQLN2-associated ALS revealed that both wild-type and ALS-mutant UBQLN2 alleles disrupted Ub homeostasis; however, UBQLN2ALS mutants exhibited age-dependent aggregation and caused toxicity phenotypes beyond those seen for wild-type UBQLN2. Although UBQLN2 toxicity was not correlated with aggregation in the compound eye, aggregation-prone UBQLN2 mutants elicited climbing defects and neuromuscular junctions (NMJ) abnormalities when expressed in neurons. An UBA domain mutation that abolished Ub binding also diminished UBQLN2 toxicity, implicating Ub binding in the underlying pathomechanism. We propose that ALS-associated mutations in UBQLN2 disrupt folding and that both aggregated species and soluble oligomers instigate neuron autonomous toxicity through interference with Ub homeostasis.
Introduction
The proteotoxicity hypothesis for neurodegeneration emerged from the histologic characterization of several diseases, including Alzheimer’s Disease, Parkinson’s Disease, Huntington’s Disease, amyotrophic lateral sclerosis (ALS), and frontotemporal dementia (FTD) (1). These heterogeneous conditions are collectively referred to as proteinopathies, in which pathology is characterized by aggregates of disease-specific proteins in affected tissues of the central nervous system. The potential importance of proteotoxicity mechanisms in ALS was recognized during studies of the first identified ALS gene, superoxide dismutase 1 (SOD1), which accounts for ∼20% of familial ALS (fALS) cases (2). Mutant SOD1 proteins assume toxic folds, disrupt intracellular regulation, and kill motor neurons through neuron autonomous and neuron non-autonomous mechanisms (3–5). More recently, mutations in the nuclear RNA-binding proteins, TDP-43 and FUS/TLS, were identified in ALS, with most disease mutations occurring in low-complexity regions of the proteins, resulting in a proclivity for aggregation (6–13). Furthermore, cytosolic aggregation of wild-type TDP-43 is observed in affected brain and spinal cord of >90% of sporadic ALS (sALS) cases, establishing TDP-43 inclusion pathology among the most reliable histopathologic markers of non-SOD1 ALS (14). Hexanucleotide repeat expansions in a non-coding region of C9ORF72—the most common genetic cause of ALS—may also promote neurodegeneration, in part, through production of toxic dipeptide repeat proteins [reviewed in (15)].
The toxicity of ALS disease proteins may result from burdened cellular pathways related to protein trafficking and degradation (16,17). Indeed, mutations in genes that regulate protein homeostasis have been identified in ALS and other neurodegenerative diseases, including optineurin (OPTN) and Tank-binding kinase 1 (TBK1), which participate in a common pathway to regulate autophagy (18,19); valosin-containing protein (VCP, p97 in mouse) and p62/SQSTM1, which are dual mediators of proteasomal and autophagosomal protein degradation (20–22); and additional genes involved in endosome trafficking and membrane remodeling, including CHMP2B, Alsin, and VAPB [reviewed in (23)]. Of relevance to this study, mutations in Ub chaperone ubiquilin 2 (UBQLN2) cause X-linked forms of ALS/FTD (24). UBQLN2, belongs to a family of UBQLN proteins that are thought to participate in both proteasomal and autophagosomal protein degradation (25–27). UBQLN2ALS mutants accumulate in cytosolic aggregates of degenerating neurons, suggesting that UBQLN2-associated ALS is a bona fide proteinopathy (24,28,29). Aggregation of wild-type UBQLN2 and closely related UBQLN1 is also observed in sALS, ALS-associated with C9ORF72 hexnucleotide repeat (HRE) expansions, and genetically distinct neurodegenerative diseases, implying that UBQLNs might function more generally in Ub-mediated aggregation (30,31).
UBQLNs harbor an N-terminal UBL domain, which mediates interactions with the proteasome (27,30,32) and Ub-binding endosome proteins, such as EPS15 and HRS (33), and a C-terminal UBA domain that binds to monoUb and all homotypic tetraUb chains (34,35). The central region of UBQLNs is comprised of a series of STI1 repeats and mediates protein-protein interactions and self-association (36). Through their dual UBL-UBA configuration, UBQLNs are thought to mediate delivery of Ub-modified substrates to the proteasome (27,37). UBQLNs physically and genetically interact with proteins implicated in several neurodegenerative diseases, including TDP-43 (38,39), presenilin (40,41), amyloid precursor protein (42), and polyQ expansions (43). These findings lend circumstantial support to the idea that wild-type UBQLNs are important participants in neurodegenerative processes.
Most clinically validated ALS-associated mutations in UBQLN2 occur in a functional orphan proline-rich-repeat (PRR) adjacent to the UBA domain that is not found in any other UBQLN ortholog. UBQLN2ALS mutants elicit variable phenotypes in rodents that may reflect expression-level dependent effects. Whereas mice expressing UBQLN2P497H under control of its endogenous promoter exhibited dendritic spine abnormalities and mild behavior defects (44,45), mice expressing UBQLN2ALS mutants downstream of a heterologous Thy1.2 promoter exhibited severe motor neurodegeneration and lethality (46). Viral delivery of UBQLN2P497H also elicited UBQLN2 brain pathology (47), whereas both wild-type and ALS-mutant UBQLN2 elicited neuronal death in transgenic rats (48). Most recently, Hjerpe et al. developed a knock-in mouse model of UBQLN2P506T (mUBQLN2P520T) that observed mild cognitive defects, but no motor phenotype (49). The variable phenotypes reported in the various rodent models likely reflect cell type-dependent differences in expression level. At the cellular level, it has been reported that UBQLN2ALS mutants are defective in the degradation of Ub-dependent reporter substrates (24); show reduced association with nuclear hnRNPs (50) and HSP70 (49); fail to deliver ubiquitylated substrates to the proteasome (51,52); fail to interact with the ERAD regulator UBXD8 (53); and show aberrant interactions with the autophagy regulator, OPTN (54). Which of these diverse activities are most relevant to the pathologic roles of UBQLN2 in ALS is uncertain.
Here, we show that ALS mutations in the PRR variably and additively impact UBQLN2 solubility, ubiquitylation, and Ub-binding potential. UBQLN2ALS mutants exhibited age-dependent aggregation and tissue-specific toxicities in Drosophila that required Ub-binding activity. The combined findings implicate deregulated Ub binding as a key component of UBQLN2-associated ALS and suggest that both soluble and aggregated species are responsible for UBQLN2 neurotoxicity.
Results
Differential impacts of ALS mutations on UBQLN2 solubility
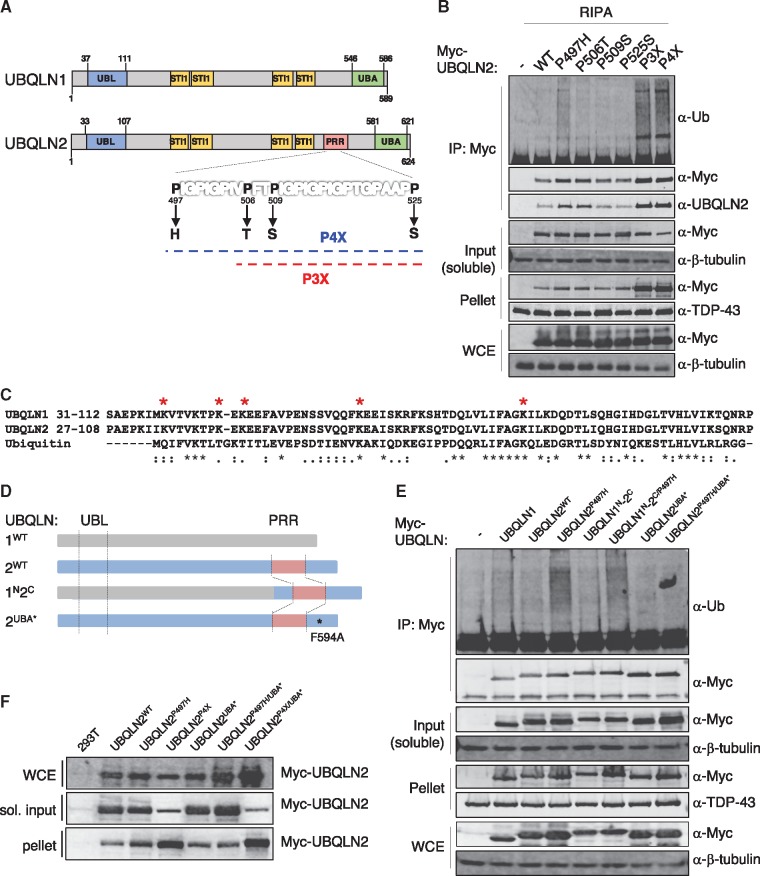
We hypothesized that ALS mutations in the UBQLN2-specific PRR (Fig. 1A) might disrupt the Ub-binding/Ub-modification cycle of UBQLN2 because of its proximity to the UBA domain. To test this, we measured Ub association of four clinical UBQLN2ALS mutants (P497H, P506T, P509S, and P525S) expressed as fusions to an amino-terminal Myc tag in HEK 293T cells. This revealed that under denaturing buffer (RIPA) conditions, UBQLN2P497H coprecipitated more Ub than either UBQLN2WT or the other three UBQLN2ALS mutants (P506T, P509S or P525S) (Fig. 1B). The bulk of this coprecipitating Ub is expected to be covalently attached to UBQLN2 as opposed to Ub that associates non-covalently through the UBA domain. We analysed Myc-UBQLN2 IPs by LC-MS/MS to confirm Ub modification of UBQLN2. DiGly-signatures in our MS dataset confirmed Ub-modified residues within UBQLN2 to several Lys (K31, K41, K43, K58, K79), all of which reside in the UBL domain (Fig. 1C). In addition to its hyperubiquitylation, a greater fraction of UBQLN2P497H was retained in the insoluble pellet following extraction in RIPA buffer, indicating this mutation diminishes UBQLN2 solubility. UBQLN2P506T and UBQLN2P525S exhibited intermediate solubility phenotypes, whereas the solubility of UBQLN2P509S resembled UBQLN2WT (Fig. 1B).
Figure 1.
Differential impacts of ALS mutations on UBQLN2 ubiquitylation and solubility. (A) Schematic diagram of UBQLN1WT and UBQLN2WT, which share 74% amino acid identity and 95% similarity. ALS-causing mutations in the PRR are highlighted. Approximate positions of UBA (Ub-associated domain) and UBL (Ub-like domain) and STI1 repeats are shown. (B) HEK 293T cells transfected with indicated Myc-tagged UBQLN2 plasmids were lysed in RIPA buffer and immunoprecipitated with α-Myc antibody. Whole cell extract (WCE), input, pellet, and IP fractions were immunoblotted with α-Myc, α-Ub, α-β-tubulin and α-TDP-43 (control) antibodies. (C) Sequence alignment of the UBL domains of UBQLN1, UBQLN2, and Ub, red * marks a lysine amino acid in UBQLN2 positively identified as Ub-modified. (D) Schematic of UBQLN chimeras: grey corresponds to UBQLN1 and blue to UBQLN2. The UBA* F594A mutation disrupts Ub binding. (E) HEK 293T cells were transfected with the indicated plasmids and soluble extracts were immunoprecipitated with α-Myc antibody in RIPA buffer. WCE, input, pellet, and IP fractions were analysed by immunoblotting with α-Myc, α-Ub, α-β-tubulin and α-TDP-43 (control) antibodies. (F) HEK 293T cells were transfected with the indicated plasmids, and WCE, soluble and pellet fractions were extracted in 1% TX-100 buffer. The fractions were analysed by immunoblotting with α-Myc antibodies.
To investigate whether the biochemical impacts of ALS mutations are additive we constructed an UBQLN2 triple mutant harboring P506T, P509S, and P525S mutations (UBQLN2P3X) and an UBQLN2 quadruple mutant harboring P497H, P506T, P509S, and P525S mutations (UBQLN2P4X). Whereas individual P506T, P509S, or P525S mutations had marginal impacts, UBQLN2P3X showed dramatically reduced solubility and increased ubiquitylation. As would be expected from the inclusion of the phenotypically strong P497H mutation, UBQLN2P4X showed a further reduction of solubility as well as increased ubiquitylation (Fig. 1B). We surmise that ALS mutations impact UBQLN2 solubility and ubiquitylation to different extents and that these effects are additive.
UBA domain mutation uncouples UBQLN2P497H ubiquitylation and insolubility
UBQLN1 and UBQLN2 are highly similar with the notable exception of the UBQLN2-specific PRR (Fig. 1A). A second region of sequence divergence between UBQLN1 and UBQLN2 exists at the amino-termini of the proteins, which is the basis for commercially available antibodies that can distinguish between them. Notably, UBQLN2 harbors two conserved PXXP motifs in its first 31 aa that are absent from UBQLN1 (Supplementary Material, Fig. S1A). To investigate possible functional differences between UBQLN1 and UBQLN2, we first assessed association with endogenous Ub in co-immunoprecipitation (co-IP) experiments. We found that Myc-UBQLN1 was more enriched in the insoluble phase (pellet) than Myc-UBQLN2, indicating that despite sharing, greater than 70% amino acid identity, UBQLN1 and UBQLN2 are biochemically distinct (Fig. 1E, Supplementary Material, Fig. S1B). To determine whether ALS mutations could influence ubiquitylation or solubility of UBQLN1, we measured the ubiquitylation and solubility of a UBQLN1-UBQLN2 chimera harboring the amino terminus of UBQLN1 and the PRR/UBA domain of UBQLN2, either with or without the P497H mutation (Fig. 1D). These experiments revealed that the UBQLN1N-UBQLN2C/P497H chimera was substantially more Ub-modified than UBQLN1N-UBQLN2C/WT (Fig. 1E) and that the UBQLN1N-UBQLN2C/P497H chimera was less soluble than UBQLN1N-UBQLN2C/WT, indicating that the P497H mutation is sufficient to decrease UBQLN1 as well as UBQLN2 solubility. Next, we disrupted the UBA domains by installing a single point mutation, F594A in UBQLN2 (denoted UBA*). The UBA* mutation abolished Ub association of UBQLN2P497H (Fig. 1E) and increased its solubility slightly (Fig. 1F). Interestingly, the UBA* mutation did not rescue the insolubility of UBQLN2P4X (Fig. 1F). These findings suggest that UBQLN2 PRR mutants aggregate through Ub-binding dependent and Ub-binding independent mechanisms.
ALS mutations alter UBQLN2 folding and Ub binding
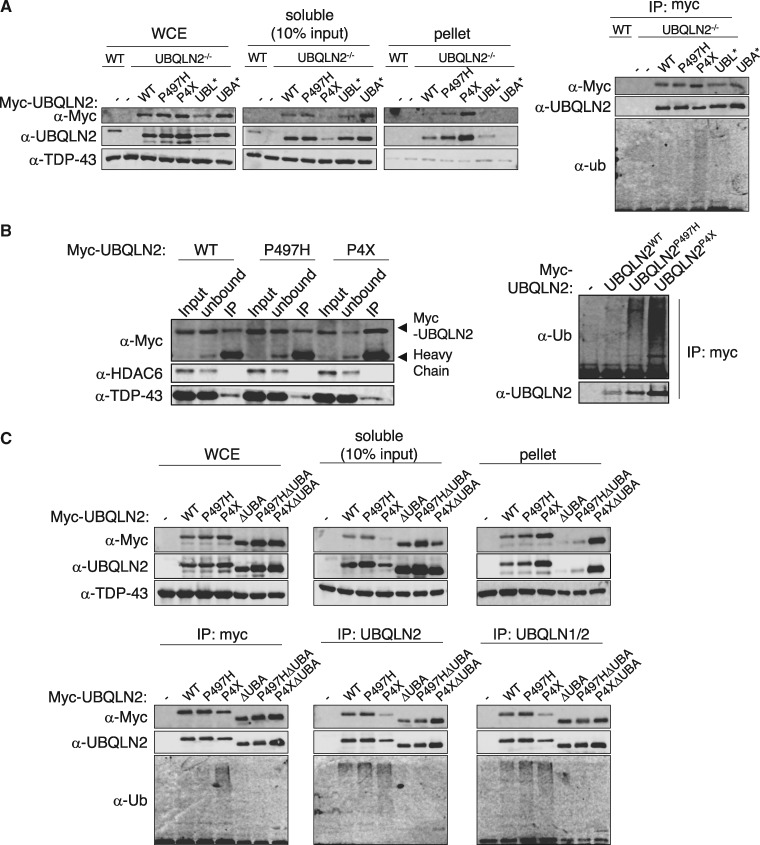
To investigate impacts of UBQLN2ALS mutants in the absence of endogenous UBQLN2, we used CRISPR-mediated gene editing to introduce a null mutation into the UBQLN2 locus of immortalized MEFs (Supplementary Material, Fig. S2). Into UBQLN2 knockout cells, we transiently introduced Myc-epitope tagged UBQLN2WT, UBQLN2P497H, UBQLN2P4X, UBQLN2UBL* and UBQLN2UBA* expression plasmids. UBL* contains an I75T mutation in a region corresponding to the hydrophobic patch of Ub (55). Consistent with findings in HEK 293T cells, UBQLN2P497H and UBQLN2P4X exhibited decreasing solubility and enhanced polyubiquitylation in UBQLN2-/- MEFs (Fig. 2A). During the course of these experiments we also observed that UBQLN2P497H and UBQLN2P4X were more efficiently immunoprecipitated than UBQLN2WT with α-Myc in either RIPA (Fig. 2A) or NP-40-containing buffers (Supplementary Material, Fig. S3). Enhanced immunoprecipitation of UBQLN2P497H and UBQLN2P4X relative to UBQLN2WT was also observed using antibodies directed against amino-terminal (α-UBQLN2, CST) or central regions of UBQLN2 (α-UBQLN1/2, Invitrogen) (Fig. 2C), indicating it is not an artifact of the Myc-epitope tag. We also measured UBQLN2 proteins in the unbound fraction to assess IP efficiency. UBQLN2WT was almost equally distributed between the bound and unbound fraction whereas UBQLN2P4X was relatively depleted from the unbound fraction, which is consistent with its enhanced IP efficiency. The recovery of UBQLN2P497H in the unbound fraction was intermediate to that of UBQLN2WT and UBQLN2P4X (Fig. 2B). Finally, neither wild-type nor ALS-mutant UBQLN2 proteins bound IgG or protein A/G agarose (data not shown). We conclude that UBQLN2P497H and UBQLN2P4X are more efficiently immunoprecipitated than UBQLN2WT and that this could partially or fully account for the apparent increase in their Ub association in co-IP experiments (Fig. 1B).
Figure 2.
ALS mutations in UBQLN2 expose antibody epitopes and alter Ub association. (A) UBQLN2-/- MEFs were transfected with the indicated Myc-UBQLN2 expression constructs. The cells were lysed with RIPA buffer, and separated into soluble and pellet fractions. The RIPA-soluble fractions were immunoprecipitated with α-Myc antibody and immunoblotted with α-Myc, α-UBQLN2 and α-Ub antibodies (right). (B) HEK 293T were transfected with the indicated Myc-UBQLN2 expression constructs. The cells were lysed with RIPA buffer, and the soluble fractions were immunoprecipitated with α-Myc antibody. Input, unbound and IP fractions were immunoblotted with α-Myc, α-UBQLN2, α-Ub, α-TDP43 (control) and α-HDAC6 (control) antibodies. (C) UBQLN2-/- MEFs were transfected with the indicated Myc-UBQLN2 constructs, lysed in RIPA buffer, and the soluble fractions immunoprecipitated with UBQLN2 antibodies targeting either the amino terminus of UBQLN2 or middle region of UBQLN1/2. Soluble, insoluble, and immunoprecipitated proteins (bottom panels) were immunoblotted with indicated UBQLN2 antibodies.
To determine whether the different IP efficiencies observed for UBQLN2WT, UBQLN2P497H, and UBQLN2P4X might be due to differences in intramolecular association of UBA and UBL domains, we performed IP analysis of UBQLN2WT and UBQLN2P4X in which the UBA domains were deleted. Two conclusions were drawn from these experiments. First, deletion of the UBA domain did not enhance the IP of UBQLN2WT, indicating that intramolecular UBL-UBA interaction does not account for its relatively poor IP efficiency (Fig. 2C). Second, UBA deletion reversed hyperubiquitylation of UBQLN2P4X but did not fully rescue its insolubility, indicating that ectopically expressed UBQLN2ALS mutants can aggregate independent of Ub binding when overexpressed in immortalized cells (Fig. 2C).
UBQLN2ALS mutants show enhanced Ub association in vitro
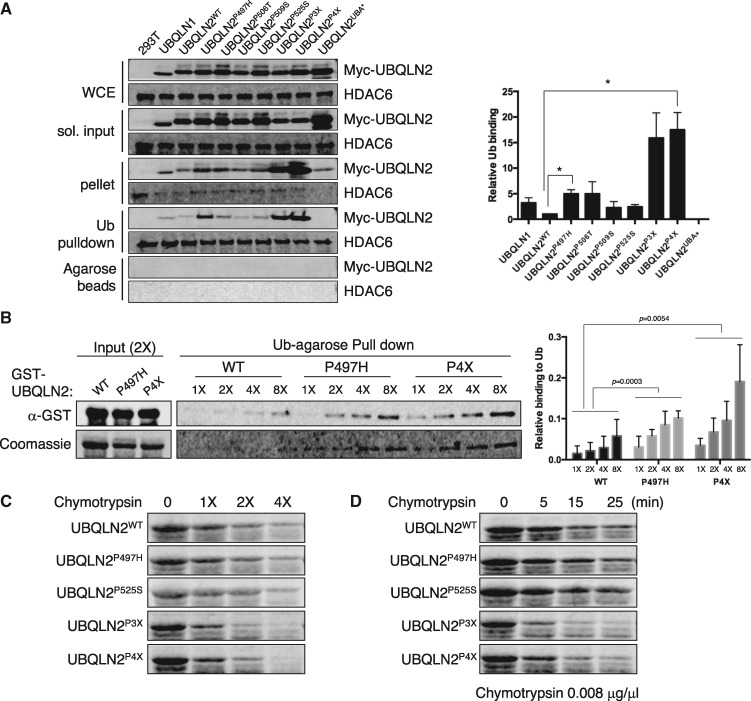
To determine whether misfolding of UBQLN2P4X and/or other ALS mutants are associated with functional changes we performed Ub-binding assays using Myc-tagged UBQLN2 proteins expressed in mammalian cells. UBQLN2P497H, UBQLN2P506T UBQLN2P3X, and UBQLN2P4X were more efficiently captured on Ub-agarose beads in comparison to UBQLN2WT, whereas UBQLN2P509S and UBQLN2P525S exhibited roughly wild-type levels of Ub-binding when accounting for input differences (Fig. 3A). Increased Ub-binding of UBQLN2P497H and UBQLN2P4X relative to UBQLN2WT was recapitulated using the respective GST-tagged proteins expressed in E. coli (Fig. 3B), whereas UBQLN2P525S exhibited wild-type levels of Ub binding in this assay (Supplementary Material, Fig. S4A). As expected, the enhanced Ub association of UBQLN2P497H and UBQLN2P4X was abolished by the UBA* mutation (Supplementary Material, Fig. S4B). Finally, given that solubility and ubiquitylation profiles of Myc-tagged UBQLN2P3X were intermediate to those of UBQLN2P497H and UBQLN2P4X, we tested its Ub association in GST pulldown assays. As shown in Supplementary Material, Figure S4A, GST-UBQLN2P3X also exhibited increased association with Ub-agarose beads. We conclude that ALS mutations affect the affinity or avidity of UBQLN2 for Ub.
Figure 3.
UBQLN2ALS mutants show enhanced Ub association. (A) Extracts from HEK 293T cells transfected with the indicated UBQLN2 plasmids were incubated with monoUb-conjugated agarose beads. Input and bound fractions were analysed by immunoblotting with α-Myc and α-HDAC6 [as a Ub-binding control (67)] antibodies. Ub-binding activity was measured by densitometric analysis (n = 3). (B) GST-tagged UBQLN2WT, UBQLN2P497H and UBQLN2P4X expressed in an E. coli were incubated with monoUb-conjugated agarose beads with either increasing amounts of GST-tagged UBQLN2 proteins. Input (2X amount of GST-tagged proteins) and bound fractions analysed by immunoblotting with α-GST or stained with Coomassie blue. Ub-binding activity was measured by densitometric analysis and normalized to soluble input (n = 3). (C, D) Purified recombinant GST-tagged UBQLN2WT, UBQLN2P497H UBQLN2P525S, UBQLN2P3X and UBQLN2P4X were incubated at room temperature with either increasing amounts of chymotrypsin for 5 min (C) or with a fixed amount of chymotrypsin for 5–30 min (D). After separation by SDS-PAGE, the proteins were visualized with Coomassie Blue.
The increased Ub association and immunoreactivity of UBQLN2P497H and UBQLN2P4X could be due to folding defects that expose antibody epitopes and/or alter the oligomerization status of UBQLN2. To investigate potential folding differences we carried out limited proteolysis assays using E. coli expressed GST-tagged UBQLN2 proteins. UBQLN2P3X and UBQLN2P4X were hypersensitive to chymotryptic cleavage relative to UBQLN2WT, UBQLN2P497H and UBQLN2P525S, which showed similar sensitivity to the enzyme (Fig. 3C and D). We infer that the folding of UBQLN2 is altered by the P3X or P4X mutations and that the P497H or P525S mutations may have more subtle impacts on folding that are not readily revealed by chymotryptic digestion.
Biochemical characterization of UBQLN2ALS mutants in Drosophila
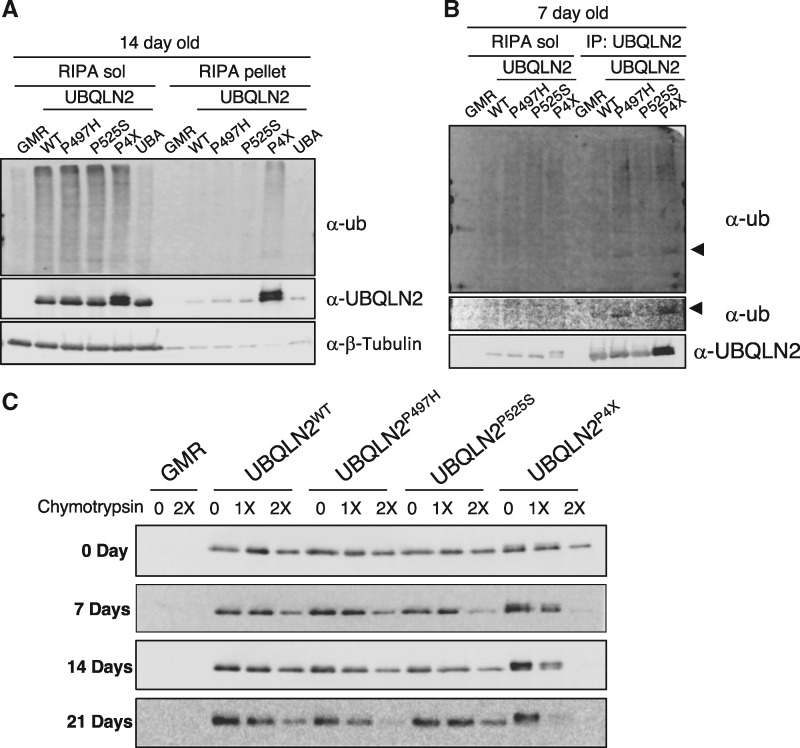
To investigate pathologic implications of UBQLN2ALS mutants we constructed isogenic Drosophila lines expressing UBQLN2WT, UBQLN2P497H, UBQLN2P525S, or the aggregation-prone UBQLN2P4X allele under control of an upstream activation sequence (UAS) conferring regulation by Gal4. To compare biochemical properties of various UBQLN2ALS mutants, we generated recombinant GMR > UBQLN2 lines expressing UBQLN2 transgenes in the eye. Western blotting of head extracts demonstrated comparable expression of the various UBQLN2 proteins and further revealed that UBQLN2WT and UBQLN2ALS mutants caused a 3–4-fold increase in soluble ubiquitylated species, whereas expression of UBQLN2UBA* failed to increase poly-Ub levels (Fig. 4A). From this, we conclude that UBA-dependent disruption of Ub homeostasis is a non-specific consequence of UBQLN2 overexpression.
Figure 4.
Biochemical characterization of UBQLN2ALS mutants in Drosophila. (A) Expression of wild-type and ALS-mutant UBQLN2 proteins leads to an increase in cellular Ub. Head extracts from GMR > UBQLN2WT, GMR > UBQLN2P497H, GMR > UBQLN2P525S, GMR > UBQLN2P4X and GMR > UBQLN2UBA* flies were separated into soluble and insoluble fractions in RIPA buffer and immunoblotted with α-UBQLN2, α-Ub, and α-β-tubulin antibodies at 14 day post-eclosion. (B) UBQLN2P497H and UBQLN2P4X are hyperubiquitylated. RIPA-soluble head lysates from 7-day-old GMR > UBQLN2WT, GMR > UBQLN2P497H, GMR > UBQLN2P525S, and GMR > UBQLN2P4X flies were immunoprecipitated with α-UBQLN2 and the input and IP fractions were immunoblotted with α-UBQLN2 and α-Ub antibodies. (C) Head extracts from GMR > UBQLN2WT, and GMR > UBQLN2P497H, GMR > UBQLN2P525S, and GMR > UBQLN2P4X flies were digested with increasing amounts of chymotrypsin for 5 min. After separation by SDS-PAGE, the proteins were immunoblotted with α-UBQLN2 antibodies.
UBQLN2WT, UBQLN2P497H, and UBQLN2P525S exhibited comparable solubility in RIPA buffer, whereas UBQLN2P4X accumulated in the RIPA-insoluble fraction and migrated as a doublet suggestive of Ub modification (Fig. 4A). To determine whether solubility of UBQLN2ALS mutants changes over time, we carried out fractionation experiments using heads from 0–21 day-old flies. Solubility of UBQLN2P497H was slightly reduced at 21-days relative to UBQLN2WT and UBQLN2P525S, whereas UBQLN2P4X was relatively insoluble at all-time points examined (Supplementary Material, Fig. S5). Consistent with cell culture findings, UBQLN2P497H and UBQLN2P4X were more efficiently immunoprecipitated than UBQLN2WT and showed a corresponding increase in coprecipitated Ub (Fig. 4B). This experiment also confirmed that UBQLN2P497H and UBQLN2P4X are Ub modified (Fig. 4B, middle).
To determine whether putative folding defects of UBQLN2ALS mutants observed in vitro are also seen in vivo, we measured chymotrypsin sensitivity of UBQLN2WT, UBQLN2P497H, UBQLN2P525S, and UBQLN2P4X in fly head extracts. Interestingly, both wild-type and ALS-mutant UBQLN2 proteins exhibited age-dependent increases in chymotryptic cleavage, and this was most pronounced for UBQLN2P497H and UBQLN2P4X (Fig. 4C). Thus, with the exception that UBQLN2P497H was not overtly insoluble when expressed from the GMR driver, the Drosophila biochemical findings are generally consistent with results employing mammalian cells. Differential solubility of UBQLN2P497H in mammalian cell culture and Drosophila may be due to lower absolute expression levels using the UAS-Gal4 system.
Eye toxicity of UBQLN2 proteins requires Ub binding and does not correlate with aggregation potential
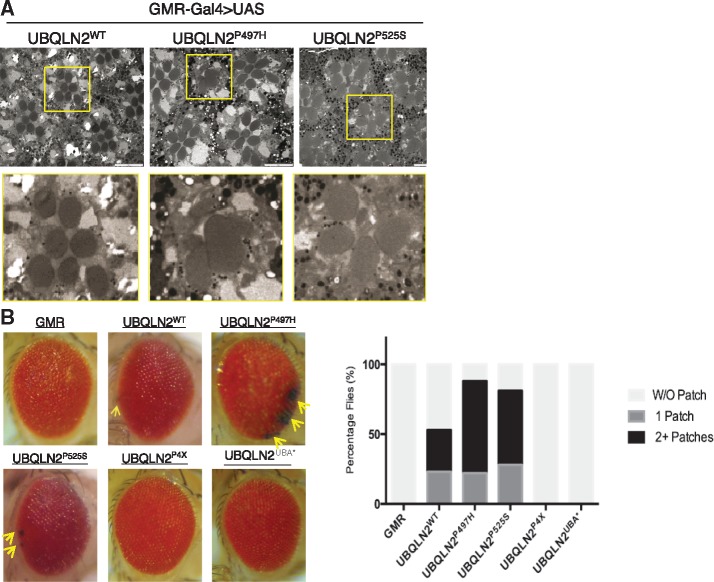
Single copy expression of wild-type or ALS-mutant UBQLN2 under control of GMR failed to elicit an external eye phenotype at either 23 or 28°C (not shown). By contrast, flies expressing two copies of UBQLN2P497H or UBQLN2P525S under control of GMR exhibited age-dependent loss of interommatidial bristles whereas UBQLN2WT expression led to mild bristle loss (Supplementary Material, Fig. S6). The external eye phenotypes of GMR > 2X-UBQLN2P497H and GMR > 2X-UBQLN2P525S flies were accompanied by photoreceptor neuron defects, including fused and missing rhabdomeres that were not observed in GMR > 2X-UBQLN2WT flies (Fig. 5A). Eye phenotypes were even more apparent in recombinant GMR > UBQLN2 lines expressing two copies of UBQLN2 and GMR driver. In addition to showing bristle loss, homozygous GMR > UBQLN2P497H and GMR > UBQLN2P525S flies exhibited hyperpigmented eye patches that were both more numerous and larger than those observed in homozygous GMR > UBQLN2WT flies and never observed in homozygous GMR-Gal4 controls, though these flies exhibited a mild rough-eye phenotype as previously described (56) (Fig. 5B). Remarkably, homozygous GMR > UBQLN2P4X flies manifested neither bristle loss nor eye patches, indicating that the eye toxicity of UBQLN2 is not directly linked to aggregation potential. The Ub-binding defective UBQLN2UBA* mutant also failed to elicit eye patches or bristle loss, indicating that eye toxicity requires Ub binding (Fig. 5B and data not shown).
Figure 5.
UBQLN2ALS mutants elicit eye degeneration. (A) Representative TEM images of rhabdomeres from GMR > UBQLN2WT, GMR > UBQLN2P497H and GMR > UBQLN2P525S flies. (B) External eye images from the indicated recombinant, homozygous GMR > UBQLN2 lines or GMR-Gal4/GMR-Gal4 controls. Hyperpigmented patches seen in UBQLN2ALS flies are denoted by arrows. Quantification of eye patch data is shown at right using a minimum of 100 flies per genotype.
Genetic interactions between UBQLN2 and other ALS genes
We crossed homozygous GMR > UBQLN2WT and GMR > UBQLN2P525S flies to flies expressing ALS-mutant alleles of TDP-43 (M337V), FUS (R518K, R521C) (57), and C9ORF72. The C9ORF72 model we used expresses 30 copies of a GGGGCC (G4C230) hexanucleotide repeat expansion (HRE) (58). UBQLN2WT exacerbated G4C230-mediated eye degeneration but had negligible effects on TDP-43 and FUS-mediated degeneration. UBQLN2P525S exhibited a similarly strong genetic interaction with G4C230, and worsened TDP-43M337V, FUSR518K, and FUSR521C-associated degeneration (Supplementary Material, Fig. S7). We conclude that wild-type and mutant UBQLN2 proteins exhibit strong genetic interactions with C9ORF72 and that UBQLN2P525S worsens eye phenotypes associated with ALS-mutant alleles of TDP-43 and FUS.
UBQLN2ALS mutants exhibit age-related protein aggregation in neurons
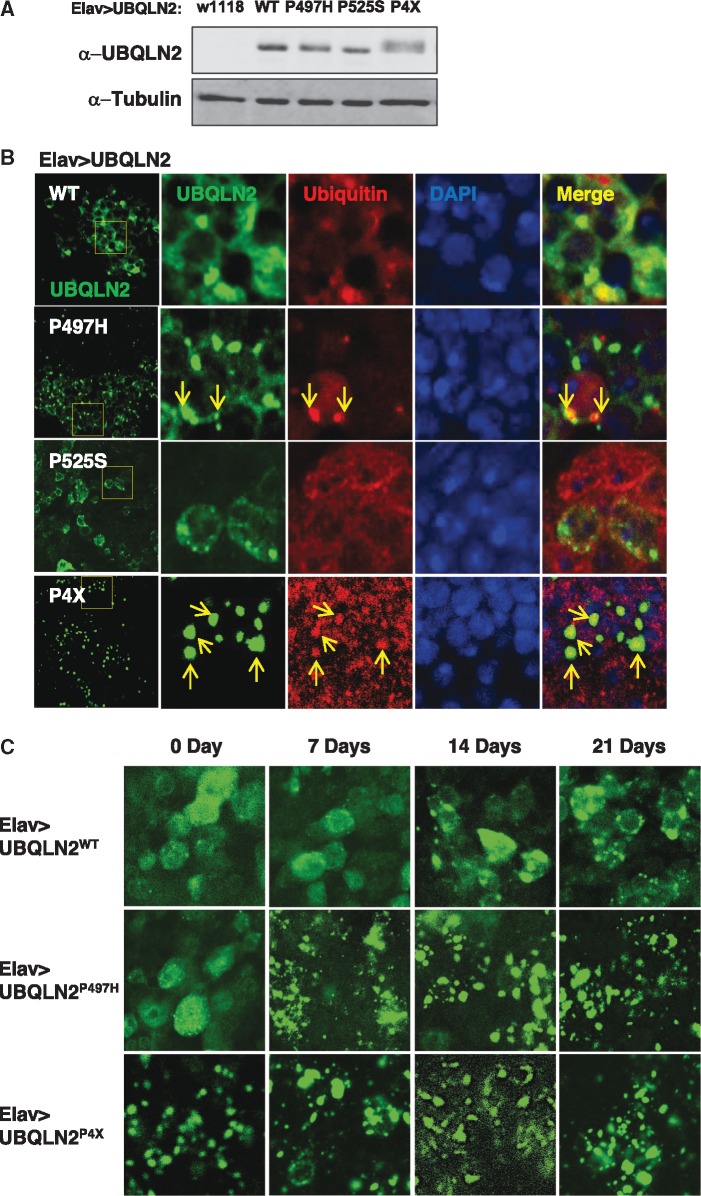
UBQLN pathology coincident with cytosolic TDP-43 pathology is a hallmark of degenerating motor neurons in sALS and is highly characteristic of C9ORF72-associated-ALS (24,31). To determine whether these pathologic features are present in UBQLN2 transgenic Drosophila, we expressed UBQLN2WT, UBQLN2P497H, UBQLN2P525S, and UBQLN2P4X under control of a neuronal Elav driver (Fig. 6A). At one-week of age UBQLN2WT was diffusely localized throughout cytoplasm of transgenic neurons, with some cells showing one to several cytosolic punctae (Fig. 6B). By contrast, UBQLN2P497H was almost exclusively localized within multiple, intense cytosolic punctae, which were partially colocalized with Ub. UBQLN2P525S exhibited an intermediate staining profile, with both diffuse staining and cytosolic punctae observed. As expected, UBQLN2P4X showed even more dramatic cytosolic punctae that were colocalized with Ub (Fig. 6B, Supplementary Material, Fig. S8).
Figure 6.
UBQLN2ALS mutants exhibit differential aggregation in Drosophila. (A) Relative expression of indicated UBQLN2 proteins expressed under control of Elav in total head extracts. (B) Localization patterns of wild-type and ALS-mutant UBQLN2 proteins. Whole brains from Elav > UBQLN2WT, Elav > UBQLN2P497H, Elav > UBQLN2P525S, and Elav > UBQLN2P4X flies were stained with α-UBQLN2 and α-Ub antibodies and imaged by confocal microscopy. Ub-positive aggregates are marked by arrows. (C) Age-dependent localization patterns of wild-type and ALS-mutant UBQLN2 proteins in fly brains. Whole brains of 0, 7, 14, 21-day-old Elav > UBQLN2WT, Elav > UBQLN2P497H and Elav > UBQLN2P4X flies were stained with α-UBQLN2 antibodies and imaged by confocal microscopy. (D) Colocalization of UBQLN2 punctae with endogenous p62. Whole brains from 14-day-old Elav > UBQLN2WT, Elav > UBQLN2P497H, and Elav > UBQLN2P4X flies were stained with α-UBQLN1/2 and α-p62 antibodies and imaged by confocal microscopy.
Given that aging is a risk factor for ALS, we assessed UBQLN2 aggregation at weekly intervals for up to one month post-eclosion. UBQLN2WT and UBQLN2P497H exhibited minimal aggregation in 0-day-old flies (Fig. 6C), whereas, UBQLN2P4X was already highly aggregated at this early time point. UBQLN2P497H aggregates were observable at 7-days post-eclosion, whereas UBQLN2WT aggregates were observable beginning at day 14. However, UBQLN2WT aggregation never reached the levels seen for UBQLN2P497H and UBQLN2P4X, even in 28-day-old flies. These results suggest that intraneuronal UBQLN2 aggregation increases with age and that ALS mutations accelerate this process. Finally, to gain insights into the molecular composition of UBQLN2 aggregates we costained brains of Elav > UBQLN2 flies with α-UBQLN2 and antibodies against the Ub-binding protein p62 which plays an important role in protein degradation via proteasome and the autophagy pathways (59–61). Using 14-day-old flies that display prominent UBQLN2 pathology we found that UBQLN2P4X aggregates were intensely stained by α-p62 antibodies, suggesting that UBQLN2P4X strongly recruits p62 (Fig. 6D, Supplementary Material, Fig. S9). Although p62 pathology was less prominent in Elav > UBQLN2P497H flies, colocalization between UBQLN2P497H and p62 was still frequently observed. Rare UBQLN2WT aggregates also stained positive for p62. In sum, UBQLN2ALS mutants elicit histopathology in fly brain reminiscent of UBQLN staining pathology in ALS patient tissues, with the degree of cytoplasmic aggregation correlating roughly to the extent of insolubility seen in mammalian cells.
Neuronal expression of UBQLN2 causes climbing defects and lethality
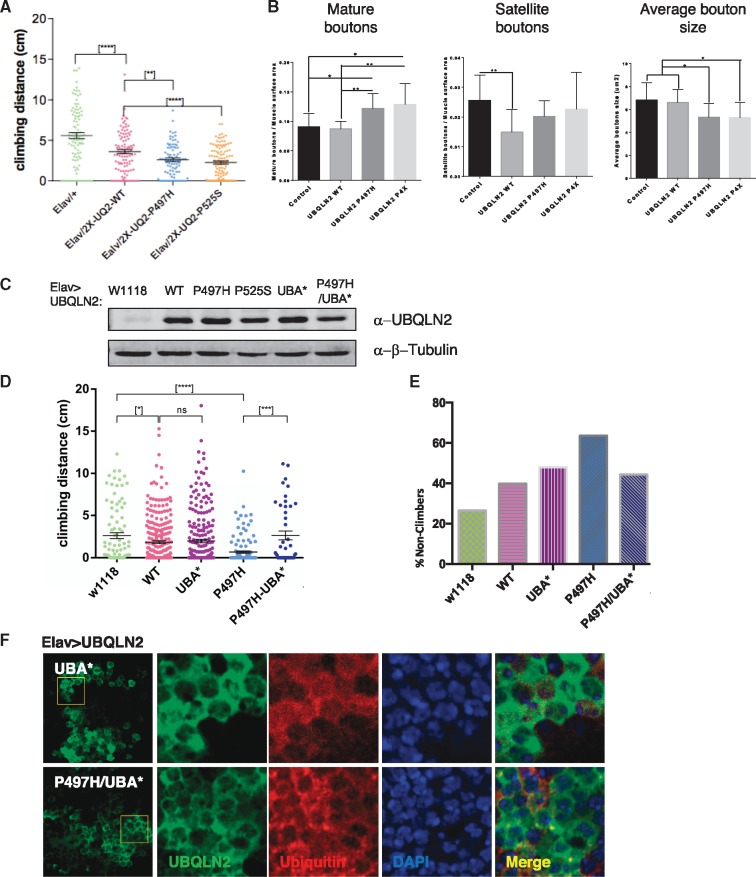
To ascertain impacts of ALS mutations on motor behavior we crossed flies homozygous for UAS-UBQLN2 transgenes on both 2nd and 3rd chromosomes (see Materials and Methods) to an Elav driver and measured climbing potential of the F1 progeny at different ages. Both UBQLN2P497H and UBQLN2P525S elicited climbing defects that were significantly worse than those elicited by expression of UBQLN2WT (Fig. 7A), supporting enhanced toxicity for these ALS mutants. Climbing defects in UBQLN2P497H flies were accompanied by alterations in neuromuscular junction (NMJ) morphology that included increased numbers of mature boutons, and features that were also seen in UBQLN2P4X flies (Fig. 7B, Supplementary Material, Fig. S10). By contrast, UBQLN2WT had no effect on the number or size of mature boutons but caused a significant decrease in satellite boutons relative to Elav controls. Despite climbing and NMJ abnormalities, lifespan was not significantly reduced in Elav > UBQLN2ALS flies relative to controls at 23°C (Supplementary Material, Fig. S11).
Figure 7.
UBQLN2ALS mutants cause climbing and NMJ defects. (A) UBQLN2ALS mutants exhibit reduced climbing ability. Climbing ability of Elav > Gal4, Elav > UBQLN2WT, Elav > UBQLN2P497H, and Elav > UBQLN2P525S flies was measured at 7-days post-eclosion as described in Materials and Methods. (*****P-value < 0.0001, **P-value < 0.01). (B) NMJ morphology analysis of Elav > Gal4, Elav > UBQLN2WT, Elav > UBQLN2P497H, and Elav > UBQLN2P4X larvae. NMJs dissected from 3rd instar larvae were stained with α-HRP and α-DLG antibodies and imaged by confocal microscopy. The number of NMJs harboring indicated phenotypes were tabulated from greater than 50 NMJs per genotype. (C) Relative expression of UBQLN2 proteins expressed under control of Elav in total head extracts. (D) Climbing ability of Elav > Gal4, Elav > UBQLN2WT, Elav > UBQLN2UBA*, Elav > UBQLN2P497H, and Elav > UBQLN2P497H/UBA* flies was measured at 10-days post-eclosion as described in Materials and Methods. A minimum of 50 flies were used for each genotype. Unpaired t-test was used for statistical analysis. (*P < 0.05, ***P < 0.0001, *****P < 0.0001). (E) Non-climbers from (D) were arbitrarily defined as flies that climbed < 1 cm. (F) Localization of UBQLN2P497H and UBQLN2P497H/UBA* proteins expressed under control of Elav.
We next crossed 2nd chromosome recombinant Elav > UBQLN2WT, Elav > UBQLN2P497H, or Elav > UBQLN2P525S lines with the corresponding 2nd chromosome UAS-UBQLN2 lines to generate flies in which both UBQLN2 transgenes were derived from the strong 51D locus. These crosses failed to yield homozygous progeny at 23°C, indicating that high-level expression of UBQLN2WT, UBQLN2P497H, and UBQLN2P525S in neurons is lethal (Table 1). The same crosses performed at 18°C yielded homozygous UBQLN2WTflies but not UBQLN2P497H progeny, indicating enhanced toxicity of the UBQLN2P497H mutant. By contrast, homozygous UBQLN2P4X progeny were observed at the expected Mendelian ratio at 23 and 18°C, but nonetheless showed severe climbing defects compare to Elav controls (Supplementary Material Fig. S12). Given roles in proteostasis, we reasoned that toxic phenotypes of UBQLN2ALS mutants might be exacerbated by thermal stress. Elav > UBQLN2P497H and Elav > UBQLN2P525S lines yielded sub-Mendelian ratios of progeny when crossed to a w1118 background at 28°C, whereas Elav > UBQLN2WT flies eclosed at the expected frequency (Table 1). These findings strongly support the idea that UBQLN2P497H and UBQLN2P525S are more toxic than UBQLN2WT when expressed in neurons, whereas UBQLN2P4X, despite being severely aggregation prone, elicits milder toxicity.
Table 1.
Viability of UBQLN2 genotypes
| Viability at indicated temperature |
||||
|---|---|---|---|---|
|
|
|
|||
| UBQLN2 | 23 °C | 18 °C | 18 °C + 28 °C HS | 28 °C |
| w1118 | Yes | Yes | n/d | n/d |
| WT | No | Yes | Yes | 49.44% |
| P497H | No | No | No | 39.86% |
| P525S | No | n/d | n/d | 37.63% |
| P4X | Yes | Yes | n/d | n/d |
HS, Heat shock.
Ub-binding contributes to the neurotoxic potential of UBQLN2
Finally, to determine whether Ub binding contributes to the neurotoxic potential of UBQLN2, we measured climbing behavior of flies expressing UBQLN2UBA* and the UBQLN2P497H/UBA* compound mutant under control of Elav (Fig. 7C). Elav > UBQLN2P497H/UBA* flies showed significantly better climbing ability than Elav > UBQLN2P497H flies, suggesting that Ub binding contributes to the neurotoxicity of this ALS mutant (Fig. 7D and E). Immunostaining studies further revealed that UBQLN2P497H/UBA* was less aggregation prone than UBQLN2P497H in Drosophila neurons (Fig. 7F). The combined findings establish enhanced toxicity of UBQLN2ALS mutants and suggest that Ub binding contributes to their neurotoxicity in vivo.
Discussion
In this study, we have characterized biochemical properties of wild-type and ALS-mutant UBQLN2 proteins and constructed Drosophila models to understand phenotypic impacts of disease-associated mutations in the UBQLN2 PRR. We have shown that ALS mutations exert non-identical effects on UBQLN2 solubility and folding and cause mutation- and tissue-specific toxicities in Drosophila. Our findings further implicate Ub binding as a central feature of UBQLN2 pathomechanisms.
The clustering of ALS mutations within or proximal to the functional-orphan PRR domain imply that such mutations interfere with cellular processes that are unique to UBQLN2 and/or initiate toxic folds that disrupt cellular regulation through GOF mechanisms. We tested five clinical mutations (P497H, P497S, P506T, P509S, P525S) and found that only P497H consistently promoted insolubility beyond wild-type levels (Fig. 1). His substitutions at disease codons Pro-506 and Pro-509 did not elicit the same changes in solubility as the P497H mutation, indicating that both the nature of the substitution (i.e. Pro to His) and its position within the PRR critically determine the extent of insolubility (data not shown). Although individual P506T, P509S, and P525S mutations had minor effects on solubility, their combination in UBQLN2P3X (P506T, P509S, and P525S) and UBQLN2P4X (P497H, P506T, P509S, and P525S) proteins led to severe reductions in solubility and patent neuronal aggregation (Figs 1B and 6B). Although results using compound mutants must be interpreted cautiously, these findings suggest that ALS mutations elicit additive or synergistic effects on UBQLN2 folding.
The increased immunoreactivity of UBQLN2P497H, UBQLN2P3X, and UBQLN2P4X with antibodies directed against amino-terminal epitope tags and endogenous UBQLN2 peptides supports the notion that localized mutations in the PRR lead to global changes in UBQLN2 folding (Fig. 2A–C), which is further bolstered by the enhanced chymotryptic sensitivity of UBQLN2P3X and UBQLN2P4X in cell culture (Fig. 3C and D) and fly heads (Fig. 4C). Interestingly, we observed age-dependent increases in chymotryptic cleavage of UBQLN2 proteins expressed in fly heads, and aging dependent increases in the aggregation of UBQLN2WT and UBQLN2P497H in fly neurons (Fig. 6C) suggesting that UBQLN2 folding is sensitive to the aging cellular environment. We speculate that ALS mutations disrupt intramolecular folding between the PRR and amino-terminal domains and/or inhibit functional intermolecular oligomerization, which is thought to be mediated by centrally located STI1 repeats (36). Structural studies of wild-type and ALS-mutant UBQLN2 proteins should further inform these possibilities.
The Drosophila findings revealed significantly stronger degenerative phenotypes in flies expressing UBQLN2ALS alleles versus UBQLN2WT and support the idea that Ub-binding figures prominently in UBQLN2-mediated toxicity. Both wild-type and ALS-mutant UBQLN2 proteins altered Ub homeostasis in an UBA domain-dependent manner, likely contributing to non-specific impacts on eye structure, survival, and climbing behavior observed here and other studies (Figs 4A and 5B, Supplementary Material, Fig. S5) (41,48). Nevertheless, enhanced toxicity of UBQLN2ALS mutants was seen across a host of assays. Neuronally expressed UBQLN2P497H reduced viability, conferred NMJ abnormalities, and diminished climbing to a greater extent than UBQLN2WT (Table 1 and Fig. 7A and B); whereas eye-directed expression of UBQLN2P497H and UBQLN2P525S elicited severe bristle loss and hyperpigmented eye patches (Fig. 5, Supplementary Material, Fig. S6). Enhanced phenotypes in UBQLN2ALS flies may enable identification of disease-specific modifier genes.
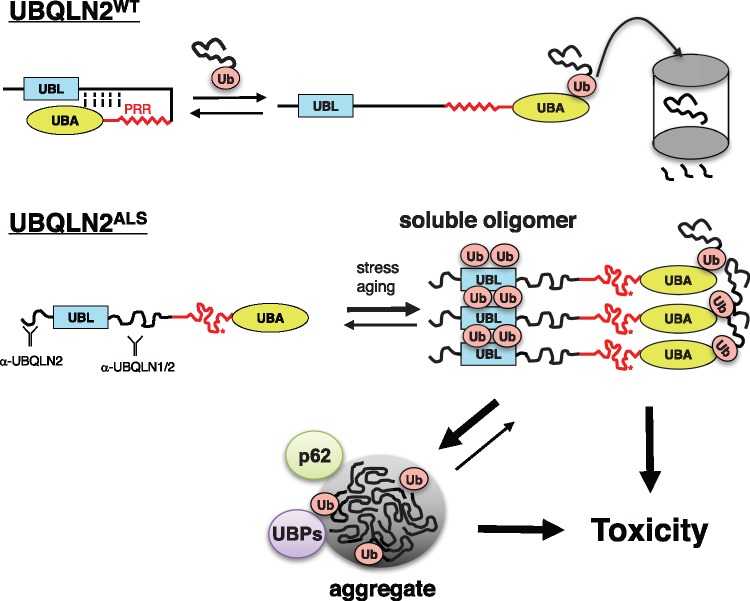
Somewhat unexpectedly, UBQLN2P4X was less toxic than UBQLN2WT, UBQLN2P497H and UBQLN2P525S in the eye, and was homozygous viable when expressed in neurons at room temperature (Fig. 5B and Table 1), indicating that cellular toxicity of UBQLN2 is not directly correlated to its aggregation potential. Nevertheless, Elav > UBQLN2P4X/UAS-UBQLN2P4X flies exhibited severe climbing defects (Supplementary Material, Fig. S12) and UBQLN2P4X conferred NMJ abnormalities that were comparable to those seen for UBQLN2P497H (Fig. 7B, Supplementary Material, Fig. S10). From the combined findings, we propose that soluble, partially misfolded UBQLN2 oligomers and/or microaggregates are the primary toxic species as has been proposed for neurodegeneration-associated mutants of SOD1 and Huntingtin (62–65). UBQLN2 macroaggregates, amplified by the artificial P4X mutation, also contribute to neurodegeneration, perhaps through pathways that are distinct from those initiated by soluble proteins (Fig. 8). Critical pathways mediating each mode of UBQLN2 toxicity should be illuminated through genetic modifier screens using the suite of Drosophila UBQLN2ALS models described here.
Figure 8.
Model for coupled misfolding, Ub-binding and ubiquitylation of UBQLN2ALS mutants. UBQLN2WT cycles between folded and open, Ub bound conformations during its activities as a Ub chaperone. PRR mutations result in misfolding of UBQLN2, leading to exposure of epitopes, increased oligomerization, and increased Ub engagement. UBQLN2 oligomers may become insoluble or engage additional Ub moieties via enhanced Ub-binding potential. Both soluble and aggregated UBQLN2 proteins are proposed to instigate toxicity. UBP, Ub-binding protein.
Materials and Methods
Plasmids and antibodies
A list of plasmids used in this study is provided in Table 2. PCR-generated hUBQLN1 and hUBQLN2 cDNAs were subcloned into the BglII-KpnI sites and EcoRI-XhoI sites of pCMV-Myc, respectively, and expressed as C-terminal fusions to an N-terminal Myc tag. The UBQLN1-UBQLN2 chimera was generated by amplifying a cDNA fragment from nt 1396 (relative to start ATG) to the 3’ end of the UBQLN2 ORF using a forward primer (5’-CCACTGAAGCCCCGGGCCTGATTCCGAGC-3’) containing a silent SmaI site. The PCR fragment was subcloned into the SmaI-KpnI sites of pCMV-Myc: UBQLN1. The UBQLN1-UBQLN2 fusion was made within a region of extended identity (Ala-Pro-Gly), just upstream of the UBQLN2 PRR. UBA* (F594A) and UBL* (I75T) mutants contain inactivating mutations in the UBA and UBL domains, respectively. All mutants were generated using a QuickChange mutagenesis strategy.
Table 2.
List of constructs
| Construct | Mutations |
|---|---|
| pCMV-myc-UBQLN1 | – |
| pCMV-myc-UBQLN2 | – |
| pCMV-myc-UBQLN2UBA* | F594A |
| pCMV-myc-UBQLN2UBL* | I75T |
| pCMV-myc-UBQLN2P497H | P497H |
| pCMV-myc-UBQLN2P506T | P506T |
| pCMV-myc-UBQLN2P509S | P509S |
| pCMV-myc-UBQLN2P525S | P525S |
| pCMV-myc-UBQLN2P3X | P506T, P509S, P525S |
| pCMV-myc-UBQLN2P4X | P497H, P506T, P509S, P525S |
| pCMV-myc-UBQLN2P497H/UBA* | P497H, F594A |
| pCMV-myc-UBQLN2P4X/UBA* | P497H, P506T, P509S, P525S, F594A |
| pCMV-myc-UBQLN2ΔUBA | R581* |
| pCMV-myc-UBQLN2P4X/ΔUBA | P497H, P506T, P509S, P525S, R581* |
Commercial antibodies used in this study listed by vendor include: Enzo Life Sciences: Ubiquitin FK2 (BML-PW8810–0500); Invitrogen: UBQLN1/2 (37–7700); Cell Signaling Technologies: UBQLN2 (85509); Proteintech Group: TDP-43 (10782–2-AP); Santa Cruz Biotechnology: Myc (sc-40), Ubiquitin P4D1 (sc-8017), HDAC6 (sc-11420); Millipore: β-Tubulin (05–661); p62 antibody was a kind gift of Dr. Gábor Juhász, Eötvös Loránd University, Budapest, Hungary.
Cell culture and transfection
HEK 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin in a humidified 5% CO2 incubator. HEK 293T cells or MEFs were transfected by Lipofectamine 3000 (Invitrogen). MEFs were isolated from 14.5-day C57BL/6J embryo and immortalized by transfection with a plasmid encoding with SV40 large T antigen. UBQLN2-/- MEFs were generated using a CRISPR-Cas9 vector pX459 [Addgene: pSpCas9(BB)-2A-Puro (PX459) V2.0] including sgRNA targeting UBQLN2 (Supplementary Material, Fig. S2). Transfected cells were briefly selected in Puro-containing media and individual clones expanded and selected for sequencing.
Cell fractionation and immunoprecipitation
HEK 293T or MEFs were harvested and lysed with lysis buffer containing 20 mM Tris-HCl (pH 8.0), 138 mM NaCl, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA and either: 1% Triton-X 100 v/v (TX buffer); 0.2% NP-40 v/v (NP-40 buffer); or 1% NP-40 v/v, 0.5% sodium deoxycholate w/v, and 0.1% SDS w/v (RIPA buffer). All lysis buffers were supplemented with protease inhibitor cocktail (Sigma, P8340), 10 mM NaF, and 1 mM DTT. Following high-speed centrifugation at 21, 000 x g for 15 min, the insoluble pellet was washed twice with PBS, then suspended and boiled in Laemmli buffer. For IP, the cell lysates were incubated with antibodies and A/G agarose. IPs were washed with the same lysis buffer for three times. After the final wash, Laemmli loading dye was added and the beads incubated at 95 °C for 5 min.
Ub-agarose binding assay
Ub-agarose beads (Boston Biochem, Cambridge, MA) were incubated with lysates from either HEK 293T cells expressing WT or ALS mutations of Myc-UBQLN2, or Escherichia coli (Origami) expressing WT or ALS mutations of GST-UBQLN2. After overnight incubation in 1% TX-100 IP buffer, Ub-agarose beads were washed with 1% TX-100 buffer three times. The washed beads were boiled in SDS-loading buffer for 5 min. Ub-agarose beads binding proteins were analysed by Western blotting with α-Myc or α-GST antibodies, or staining with Coomassie blue.
Limited proteolysis
GST-tagged UBQLN2 proteins were purified from E. coli extracts using GST affinity chromatography. A total of 50 ng of each protein in 10 μl was digested with increasing amounts of chymotrypsin (0.008 μg/ml to 0.032 μg/ml, final volume, Sigma) for 5 min or 0.008 μg/ml chymotrypsin for the indicated length of time at room temperature. Limited proteolysis of UBQLN2 proteins expressed under control of GMR was performed by incubating 1: 100 dilution of detergent extracts from 10 fly heads in total volume of 20 μl in the presence of 0.001 μg/ml or 0.002 μg/ml chymotrypsin for 5 min. Digestion was terminated by addition of Laemmli loading dye and boiling at 95 °C for 5 min. The digested proteins were analysed by Western blotting or staining with Coomassie blue.
Generation of transgenic flies
The w1118 strain was used as wild-type control and host for transgenesis. For transgene insertion at specific genomic sites, we used fly strains in which the phiC31 integrase is inserted on the X chromosome and attP landing sites is located in 51D (2nd chromosome) or 86Fb (3rd chromosome). Drosophila Kozak sequence (caacATG) was designed in the UBQLN2 forward primer to facilitate expression. UBQLN2 open-reading frames were subcloned into the pUASTattB vector using EcoRI and XhoI. White gene is used to screen for positive transformants, which shows red eyes in w1118 background. The WT, P497H, and P525S lines were injected by Rainbow Transgenic Flies, Inc.; the P4X, UBA*, and P497H/UBA* lines were injected and balanced by BestGene. UBQLN2 (2X) lines were made through genetic crossing from the two original lines. To drive transgene expression in tissue-specific manner, we used the pan-neuronal diver Gal4-Elav/CyO (Bloomingston Stock FBst0008765) and the eye specific Gal4-ninaE.GMR (Bloomington Stock FBst0001104). Second chromosome recombinant GMR > UBQLN2 and Elav > UBQLN2 fly lines were made using standard methods.
Drosophila climbing
Flies were maintained with the standard cornmeal-yeast medium (Nutri-Fly (BF), Genesee scientific) supplemented with propionic acid and all crosses were performed at 23 °C. Climbing assay was modified from methods described previously (66). Climbing ability was measured by tapping ∼10 flies to the bottom of a graduated testing vial and taking a time-delayed photograph taken 5 or 10 s later. Position of individual flies was determined using the PointPicker plugin in Image J. Data showing velocity of each individual fly were graphed as scattered plots with mean climbing distance and SEM. Unpaired t-test with Welch’s correction were used for statistical analysis for different groups of flies. Non-climbers were arbitrarily defined as flies that climbed < 1 cm and plotted separately.
NMJ assay
Third-instar, wandering larvae from the F1 generation were rinsed in ice-cold PBS (Lonza, 17–512F) and dissected along the dorsal midline. All tissues except the brain and nerves were removed to expose the muscles and NMJs. The dissected larval pelt was fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. The larval pelts were given a wash with PBS followed by blocking with 5% Normal Goat Serum (NGS) (Abcam ab7481) in 0.1% PBST (0.1% Triton-X-100 in PBS). Following blocking, the larval pelts were probed with primary antibodies overnight at 4 °C. They were then washed several times with 0.1% PBST followed by incubation with secondary antibodies for 2 h at room temperature, subsequently followed by washes with 0.1% PBST. Larvae were then mounted onto slides using Prolong Gold (Invitrogen, P36930) mounting media. Confocal images were acquired using Zeiss LSM 710 confocal microscope and a 60x oil objective was used to image the NMJs. Both primary and secondary antibody solutions were prepared in 5% NGS in 0.1% PBST. For primary antibodies, the following dilutions were used: 1: 100 Cy3-conjugated goat anti-HRP (Jackson ImmunoResearch, 123–165-021); 1: 100 mouse anti-DLG 4F3 (DSHB). For secondary antibodies, the following antibody dilutions were used: 1: 250 Alexa Fluor 647-conjugated anti-Phalloidin (Invitrogen, A22287); 1: 500 goat anti-mouse Alexa Fluor 488 (Invitrogen, A-11029). For the analyses, NMJs innervating muscle 4 on segments A2-A3 were imaged and analysed for synaptic bouton quantification. Mature boutons are defined as boutons that are included in a chain of two or more boutons. Satellite boutons are defined as a single bouton that is not included in a chain of boutons, and instead, sprout off of a mature bouton or branch. The groups were compared using Unpaired Student t-test on GraphPad Prism software. P-value less than 0.05 was considered statistically significant.
UBQLN2 solubility and ubiquitylation assay
Fly lysates were prepared by homogenizing 10 fly heads (5 from each gender) in 90 μl of 4X sample loading buffer containing 250 mM Tris-HCl (pH 6.8), 5% SDS, 10% glycerol and 5% 2-mercaptoethanol by pestle. 10% of protein lysate were resolved in 10% SDS-PAGE gel and detected via standard Western blotting procedure. For soluble/insoluble extraction, fly heads were homogenized in RIPA buffer at volume of 10 μl per head. Soluble fraction was taken after spinning in a microcentrifuge at 14 000 rpm for 10 min. The insoluble pellet was washed twice before re-suspension in buffer and boiled for 20 min. To perform IP analysis, RIPA-soluble fractions were incubated with UBQLN2 antibody and protein A/G agarose beads. After washing with 0.2% TX-100/PBS for three times, the IP samples were analysed with Western blot.
Microscopy
The protocol for fly immunohistochemistry was adapted from previously published protocol (66). Adult fly brains were dissected using a pair of fine forceps in PBS (or 0.3% TX-100 in PBS), fixed, blocked with NGS, and stained with primary antibodies at 1: 500 dilution for two overnights at 4 °C. After subsequent secondary antibody staining, DAPI was added for the nuclear staining. Images were acquired using a Nikon A1 confocal microscope using either a 10X lens or a 60X oil lens. Eye pictures were acquired using Leica S9 i Stereomicroscope.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors would like to thank Peyton Uhl and Lance Rodenkirch (UW-Madison Optical Imaging Core) for expert assistance with confocal microscopy, David A. Wassarman and Rebeccah J. Katzenberger (UW-Madison, Department of Medical Genetics) for providing reagents for Drosophila experiments, Arash Bashirullah (UW-Madison, School of Pharmacy) for access to microscopy, Qijing Xie for generation of UBQLN2 transgenic flies, and Samuel Smukowski for UBQLN2 protein purification.
Conflict of Interest statement. None declared.
Funding
National Cancer Institute [R01CA180765-01 to RST], National Institute for Neurological Disorders and Stroke [1R21NS090313-01A1, 1R21NS101661-01-A1 to RST], ALS Association [Proteostatic regulation by Ubiquilins in ALS, and Drosophila models for ubiquilin-associated ALS (18-IIA-414) to RST], NIH [NS094921, R01NS081303, R21NS100055, R21NS098379 to UBP], the Robert Packard Center for ALS at Johns Hopkins to UBP, the Muscular Dystrophy Association to UBP.
References
- 1. Douglas P.M., Dillin A. (2010) Protein homeostasis and aging in neurodegeneration. The J. Cell Biol., 190, 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J.P., Deng H.-X.. et al. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature, 362, 59–62. [DOI] [PubMed] [Google Scholar]
- 3. Ilieva H., Polymenidou M., Cleveland D.W. (2009) Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. 187, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boillée S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., Cleveland D.W. (2006) Onset and progression in inherited ALS determined by motor neurons and microglia. Science, 312, 1389–1392. [DOI] [PubMed] [Google Scholar]
- 5. Cleveland D.W., Rothstein J.D. (2001) From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci., 2, 806–819. [DOI] [PubMed] [Google Scholar]
- 6. Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M.. et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science, 314, 130–133. 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- 7. Johnson B.S., Snead D., Lee J.J., Mccaffery J.M., Shorter J., Gitler A.D. (2009) TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem., 284, 20329–20339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwiatkowski T.J., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T.. et al. (2009) Mutations in the FUS/TLS Gene on Chromosome 16 cause familial amyotrophic lateral sclerosis. Science, 323, 1205–1208. 10.1126/science.1166066 [DOI] [PubMed] [Google Scholar]
- 9. Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P.. et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science, 323, 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gitcho M.A., Baloh R.H., Chakraverty S., Mayo K., Norton J.B., Levitch D., Hatanpaa K.J., White C.L., Bigio E.H., Caselli R.. et al. (2008) TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol., 63, 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Vande Velde C., Bouchard J.-P., Lacomblez L., Pochigaeva K., Salachas F.. et al. (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet., 40, 572–574. [DOI] [PubMed] [Google Scholar]
- 12. Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E.. et al. (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science, 319, 1668–1672. 10.1126/science.1154584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yokoseki A., Shiga A., Tan C.-F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A.. et al. (2008) TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann. Neurol., 63, 538–542. [DOI] [PubMed] [Google Scholar]
- 14. Mackenzie I.R.A., Bigio E.H., Ince P.G., Geser F., Neumann M., Cairns N.J., Kwong L.K., Forman M.S., Ravits J., Stewart H.. et al. (2007) Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann. Neurol., 61, 427–434. [DOI] [PubMed] [Google Scholar]
- 15. Haeusler A.R., Donnelly C.J., Rothstein J.D. (2016) The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease. Nat. Rev. Neurosci., 17, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morimoto R.I. (2008) Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev., 22, 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taylor J.P., Brown R.H., Cleveland D.W. (2016) Decoding ALS: from genes to mechanism. Nature, 539, 197–206. 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maruyama H., Morino H., Ito H., Izumi Y., Kato H., Watanabe Y., Kinoshita Y., Kamada M., Nodera H., Suzuki H.. et al. (2010) Mutations of optineurin in amyotrophic lateral sclerosis. Nature, 465, 223–226. 10.1038/nature08971 [DOI] [PubMed] [Google Scholar]
- 19. Cirulli E.T., Lasseigne B.N., Petrovski S., Sapp P.C., Dion P.A., Leblond C.S., Couthouis J., Lu Y.-F., Wang Q., Krueger B.J.. et al. (2015) Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science, 347, 1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J.. et al. (2010) Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron, 68, 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tresse E., Salomons F.A., Vesa J., Bott L.C., Kimonis V., Yao T.-P., Dantuma N.P., Taylor J.P. (2010) VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy, 6, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fecto F., Yan J., Vemula S.P., Liu E., Yang Y., Chen W., Zheng J.G., Shi Y., Siddique N., Arrat H.. et al. (2011) SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol., 68, 1440–1446. [DOI] [PubMed] [Google Scholar]
- 23. Renton A.E., Chiò A., Traynor B.J. (2013) State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci., 17, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deng H.-X., Chen W., Hong S.-T., Boycott K.M., Gorrie G.H., Siddique N., Yang Y., Fecto F., Shi Y., Zhai H.. et al. (2011) Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature, 477, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee D.Y., Arnott D., Brown E.J. (2013) Ubiquilin4 is an adaptor protein that recruits Ubiquilin1 to the autophagy machinery. EMBO Rep., 14, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. N'Diaye E.-N., Kajihara K.K., Hsieh I., Morisaki H., Debnath J., Brown E.J. (2009) PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep., 10, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ko H.S., Uehara T., Tsuruma K., Nomura Y. (2004) Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett., 566, 110–114. [DOI] [PubMed] [Google Scholar]
- 28. Rothenberg C., Srinivasan D., Mah L., Kaushik S., Peterhoff C.M., Ugolino J., Fang S., Cuervo A.M., Nixon R.A., Monteiro M.J. (2010) Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet., 19, 3219–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mah A.L., Perry G., Smith M.A., Monteiro M.J. (2000) Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J.Cell Biol., 151, 847–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walters K.J., Kleijnen M.F., Goh A.M., Wagner G., Howley P.M. (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry, 41, 1767–1777. [DOI] [PubMed] [Google Scholar]
- 31. Brettschneider J., Van Deerlin V.M., Robinson J.L., Kwong L., Lee E.B., Ali Y.O., Safren N., Monteiro M.J., Toledo J.B., Elman L.. et al. (2012) Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol., 123, 825–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X., Randles L., Shi K., Tarasov S.G., Aihara H., Walters K.J. (2016) Structures of Rpn1 T1: Rad23 and hRpn13: hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure. 24, 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Regan-Klapisz E., Sorokina I., Voortman J., de Keizer P., Roovers R.C., Verheesen P., Urbé S., Fallon L., Fon E.A., Verkleij A.. et al. (2005) Ubiquilin recruits Eps15 into ubiquitin-rich cytoplasmic aggregates via a UIM-UBL interaction. J. Cell. Sci., 118, 4437–4450. [DOI] [PubMed] [Google Scholar]
- 34. Raasi S., Varadan R., Fushman D., Pickart C.M. (2005) Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol., 12, 708–714. [DOI] [PubMed] [Google Scholar]
- 35. Kristariyanto Y.A., Abdul Rehman S.A., Campbell D.G., Morrice N.A., Johnson C., Toth R., Kulathu Y. (2015) K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin. Mol. Cell, 58, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ford D.L., Monteiro M.J. (2006) Dimerization of ubiquilin is dependent upon the central region of the protein: evidence that the monomer, but not the dimer, is involved in binding presenilins. Biochem. J., 399, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kleijnen M.F., Shih A.H., Zhou P., Kumar S., Soccio R.E., Kedersha N.L., Gill G., Howley P.M. (2000) The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell, 6, 409–419. [DOI] [PubMed] [Google Scholar]
- 38. Kim S.H., Shi Y., Hanson K.A., Williams L.M., Sakasai R., Bowler M.J., Tibbetts R.S. (2009) Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, ubiquilin 1. J. Biol. Chem., 284, 8083–8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hanson K.A., Kim S.H., Wassarman D.A., Tibbetts R.S. (2010) Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS). J. Biol. Chem., 285, 11068–11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Massey L.K., Mah A.L., Ford D.L., Miller J., Liang J., Doong H., Monteiro M.J. (2004) Overexpression of ubiquilin decreases ubiquitination and degradation of presenilin proteins. J. Alzheimers Dis., 6, 79–92. [DOI] [PubMed] [Google Scholar]
- 41. Ganguly A., Feldman R.M.R., Guo M. (2008) ubiquilin antagonizes presenilin and promotes neurodegeneration in Drosophila. Hum. Mol. Genet., 17, 293–302. [DOI] [PubMed] [Google Scholar]
- 42. Ayadi E.,A., Stieren E.S., Barral J.M., Boehning D. (2012) Ubiquilin-1 regulates amyloid precursor protein maturation and degradation by stimulating K63-linked polyubiquitination of lysine 688. Proc. Natl Acad. Sci., 109, 13416–13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang H., Lim P.J., Yin C., Rieckher M., Vogel B.E., Monteiro M.J. (2006) Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum. Mol. Genet., 15, 1025–1041. [DOI] [PubMed] [Google Scholar]
- 44. Gorrie G.H., Fecto F., Radzicki D., Weiss C., Shi Y., Dong H., Zhai H., Fu R., Liu E., Li S.. et al. (2014) Dendritic spinopathy in transgenic mice expressing ALS/dementia-linked mutant UBQLN2. Proc. Natl Acad. Sci., 111, 14524–14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Radzicki D., Liu E., Deng H.-X., Siddique T., Martina M. (2016) Early impairment of synaptic and intrinsic excitability in mice expressing ALS/dementia-linked mutant UBQLN2. Front. Cell Neurosci., 10, 216.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Le N.T.T., Chang L., Kovlyagina I., Georgiou P., Safren N., Braunstein K.E., Kvarta M.D., Van Dyke A.M., LeGates T.A., Philips T.. et al. (2016) Motor neuron disease, TDP-43 pathology, and memory deficits in mice expressing ALS-FTD-linked UBQLN2 mutations. Proc. Natl Acad. Sci., 113, E7580–E7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ceballos-Diaz C., Rosario A.M., Park H.-J., Chakrabarty P., Sacino A., Cruz P.E., Siemienski Z., Lara N., Moran C., Ravelo N.. et al. (2015) Viral expression of ALS-linked ubiquilin-2 mutants causes inclusion pathology and behavioral deficits in mice. Mol. Neurodegener., 10, 25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang B., Wu Q., Zhou H., Huang C., Xia X.-G. (2016) Increased Ubqln2 expression causes neuron death in transgenic rats. J. Neurochem., 139, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hjerpe R., Bett J.S., Keuss M.J., Solovyova A., McWilliams T.G., Johnson C., Sahu I., Varghese J., Wood N., Wightman M.. et al. (2016) UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell, 166, 935–949. 10.1016/j.cell.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gilpin K.M., Chang L., Monteiro M.J. (2015) ALS-linked mutations in ubiquilin-2 or hnRNPA1 reduce interaction between ubiquilin-2 and hnRNPA1. Hum. Mol. Genet., 24, 2565–2577. [DOI] [PubMed] [Google Scholar]
- 51. Itakura E., Zavodszky E., Shao S., Wohlever M.L., Keenan R.J., Hegde R.S. (2016) Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol. Cell, 63, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang L., Monteiro M.J., Pandey U. (2015) Defective proteasome delivery of polyubiquitinated proteins by ubiquilin-2 proteins containing ALS mutations. PLoS ONE, 10, e0130162.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xia Y., Yan L.H., Huang B., Liu M., Liu X., Huang C. (2014) Pathogenic mutation of UBQLN2 impairs its interaction with UBXD8 and disrupts endoplasmic reticulum-associated protein degradation. J. Neurochem., 129, 99–106. [DOI] [PubMed] [Google Scholar]
- 54. Osaka M., Ito D., Yagi T., Nihei Y., Suzuki N. (2015) Evidence of a link between ubiquilin 2 and optineurin in amyotrophic lateral sclerosis. Hum. Mol. Genet., 24, 1617–1629. [DOI] [PubMed] [Google Scholar]
- 55. Beal R., Deveraux Q., Xia G., Rechsteiner M., Pickart C. (1996) Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl. Acad. Sci. U.S.A, 93, 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kramer J.M., Staveley B.E. (2003) GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet. Mol. Res., 2, 43–47. [PubMed] [Google Scholar]
- 57. Lanson N.A., Maltare A., King H., Smith R., Kim J.H., Taylor J.P., Lloyd T.E., Pandey U.B. (2011) A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum. Mol. Genet., 20, 2510–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu Z., Poidevin M., Li X., Li Y., Shu L., Nelson D.L., Li H., Hales C.M., Gearing M., Wingo T.S., Jin P. (2013) Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl Acad. Sci., 110, 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.-A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem., 282, 24131–24145. [DOI] [PubMed] [Google Scholar]
- 60. Bjørkøy G., Lamark T., Johansen T. (2006) p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy, 2, 138–139. [DOI] [PubMed] [Google Scholar]
- 61. Seibenhener M.L., Babu J.R., Geetha T., Wong H.C., Krishna N.R., Wooten M.W. (2004) Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol., 24, 8055–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schaffar G., Breuer P., Boteva R., Behrends C., Tzvetkov N., Strippel N., Sakahira H., Siegers K., Hayer-Hartl M., Hartl F.U. (2004) Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell, 15, 95–105. [DOI] [PubMed] [Google Scholar]
- 63. Takahashi T., Kikuchi S., Katada S., Nagai Y., Nishizawa M., Onodera O. (2008) Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum. Mol. Genet., 17, 345–356. [DOI] [PubMed] [Google Scholar]
- 64. Brotherton T.E., Li Y., Glass J.D. (2013) Cellular toxicity of mutant SOD1 protein is linked to an easily soluble, non-aggregated form in vitro. Neurobiol. Dis., 49, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Todd T.W., Lim J. (2013) Aggregation formation in the polyglutamine diseases: protection at a cost? Mol. Cells, 36, 185–194. 10.1007/s10059-013-0167-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhan L., Hanson K.A., Kim S.H., Tare A., Tibbetts R.S. (2013) Identification of genetic modifiers of TDP-43 neurotoxicity in Drosophila. PLoS ONE, 8, e57214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Boyault C., Gilquin B., Zhang Y., Rybin V., Garman E., Meyer-Klaucke W., Matthias P., Müller C.W., Khochbin S. (2006) HDAC6-p97/VCP controlled polyubiquitin chain turnover. embo J., 25, 3357–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.