Abstract
Although the genetic contribution is under debate, biological studies in multiple mouse models have suggested that the Disrupted-in-Schizophrenia-1 (DISC1) protein may contribute to susceptibility to psychiatric disorders. In the present study, we took the advantages of the Drosophila model to dissect the molecular pathways that can be affected by DISC1 in the context of pathology-related phenotypes. We found that three pathways that include the homologs of Drosophila Dys, Trio, and Shot were downregulated by introducing a C-terminal truncated mutant DISC1. Consistently, these three molecules were downregulated in the induced pluripotent stem cell-derived forebrain neurons from the subjects carrying a frameshift deletion in DISC1 C-terminus. Importantly, the three pathways were underscored in the pathophysiology of psychiatric disorders in bioinformatics analysis. Taken together, our findings are in line with the polygenic theory of psychiatric disorders.
Introduction
Psychiatric disorders, such as schizophrenia and bipolar disorder, are heritable (1,2). Indeed, plenty of genomic loci are reported to confer risk for these disorders (3,4). Yet the functional significance of most genetic risk variants has been difficult to investigate, due to that a single risk variant may only contribute to a small fraction of the disorder (5–7).
Disrupted-in-schizophrenic-1 (DISC1) was initially discovered at the breakpoint of a balanced chromosome translocation that co-segregated with a wide range of major mental illnesses, including bipolar disorder, major depression and schizophrenia, within a large Scottish family with high statistical significance (8). Later, another rare mutation of a 4 base-pair frameshift deletion at the DISC1 carboxyl-terminus was identified in a smaller American family with a history of mental illnesses (9). Many efforts in human genetics have been made to test the general validity of the role of DISC1 in psychiatric disorders (10). Nonetheless, the genetic/genomic role for DISC1 is still under debate (11,12). In contrast, animal studies from multiple groups demonstrated that DISC1 is involved in neuronal development and synaptic functions (13–15). Furthermore, perturbation of DISC1 in rodent models leads to alteration in multiple behavioral dimensions relevant to major mental illnesses (16–19). Taken together, these data suggest the potential contribution of the DISC1 protein to the etiopathology of psychiatric disorders (10).
Given that major mental illnesses are multi-factorial, it is important to pay attention to the biological pathways that involve in the functional orchestration of multiple molecules. To dissect such pathways, Drosophila is advantageous for the manipulation of multiple genes, and for the maneuverability of neuronal endophenotypes (20). Although Drosophila lacks homologs of DISC1, the majority of the proteins that have interaction with DISC1 are conserved in fruit fly (21), suggesting that fruit fly could provide a reliable platform for dissecting the molecular framework of DISC1 functions. Additionally, devoid of interference of the endogenous protein could be advantageous in this study to differentiate the effects of different DISC1 forms. In fact, our understanding of the mechanisms of neurodegenerative diseases, including Alzheimer’s disease and Parkinson’s disease, has been advanced by ectopically expressing human disease related molecules, such as Aß42 and α-synuclein, in Drosophila system (22).
To address pathways involving DISC1 in the context of mental conditions, we aimed to overexpress the DISC1 full-length (DISC1-FL) and a DISC1 mutant (C-terminal truncated DISC1 1-597: DISC1-TR) that could elicit neurobiological and behavioral changes in previous studies (23,24), and to examine molecular and phenotypic changes in Drosophila. Moreover, to validate our findings in Drosophila research, we also aimed to analyze forebrain neurons derived from induced pluripotent stem cells (iPSCs) of patients carrying a 4 base-pair frameshift deletion in DISC1 that is known for transcriptional dysregulation of a large array of genes and for alteration of synaptic functions (25).
In the present study, we expressed DISC1-TR and DISC1-FL in Drosophila melanogaster. Intriguingly, we found that DISC1-TR exerts specific effects on neurodevelopment, neurophysiology, and cognitive functions in a gain-of-function manner. We further identified downstream molecules that are underscored in the context of mental disorders, synergistically contributing to the neural effects of DISC1-TR. Importantly, we confirmed the regulation of these molecules by DISC1-TR in a human glioblastoma cell line as well as in the iPSC-derived forebrain neurons carrying a DISC1 frameshift mutation. At last, these molecules were ranked as the top disease risk factors in bio-informatics analysis of candidate genes among general human population (26). The above findings consistently support our hypothesis that DISC1 together with other interacting risk molecules function synergistically and contribute to the pathology of major mental illnesses.
Results
Expression of DISC1-TR but not DISC1-FL results in cognitive and synaptic phenotypes in flies
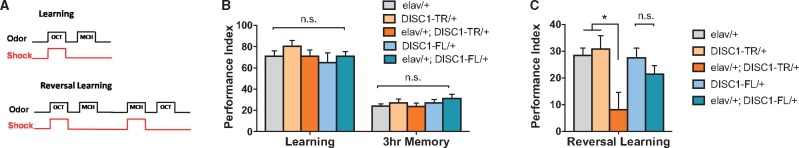
To differentiate functional effects of different forms of DISC1, we expressed transgenes of DISC1-FL and DISC1-TR respectively in a pan-neuronal pattern driven by elav-Gal4. Firstly, we assay these transgenetic flies with aversive associative olfactory learning and 3-h memory. In this task, flies were trained to associate an odor with the electric shock and would avoid the odor in the subsequent test (upper panel, Fig. 1A) (27). All genotypes concerned showed normal performance in these assays (Fig. 1B). Then cognitive flexibility was measured with the reversal-learning paradigm, in which the odor-electric shock contingency was reversed in two successive training sessions (lower panel, Fig. 1A). Reversal-learning disturbance is a reliable manifestation in many human cognitive disorders including psychiatric disorders, and thus is a useful readout for cognitive or behavior inflexibility in animal models. Pan-neuronal expression of DISC1-TR (elav/+; DISC1-TR/+), but not DISC1-FL (elav/+; DISC1-FL/+) resulted in a significant reversal learning deficit (Fig. 1C).
Figure 1.
Expression of Schizophrenia-related form of DISC1 (DISC1-TR) affects cognitive function in Drosophila. Illustration of the training paradigm for Pavlovian olfactory conditioning. In learning, flies were presented to the first odor (OCT) paired with electric shock, and then the second odor (MCH) without electric shock. In reversal learning, the odor-electric shock pair was reversed in each training session. Neuronal expression of DISC1-FL (elav/+; DISC1-FL/+) and DISC1-TR (elav/+; DISC1-TR/+) did not affect learning and 3-h memory performances in Drosophila. (ANOVA, n = 6 for learning, n = 13 for 3-h memory, n.s. P > 0.05) Reversal learning performance was imparied only when DISC1-TR (elav/+; DISC1-TR/+) was expressed pan-neuronally. (ANOVA, n = 8, P = 0.0106, 0.7387 compared to elva/+; P = 0.0035, 0.8302 compared to UAS control). All data are represented as mean ± SEM.
To identify the neuronal populations that mediate the observed behavioral phenotype, transgene expression was directed with Gal4 drivers labeling mushroom bodies (DISC1-TR/+; OK107-Gal4/+), dopaminergic neurons (DISC1-TR/+; TH-Gal4/+), GABAergic neurons (Gad1-Gal4/+; DISC1-TR/+), glutamatergic neurons (VgluT-Gal4/+; DISC1-TR/+), and rutabaga-adenylyl cyclase (rut-AC) expressing neurons (rut1047-Gal4/+; DISC1-TR/+), respectively. Only the expression of DISC1-TR in glutamatergic neurons and rut-AC expressing neurons duplicated the reversal-learning deficiency exhibited in the pan-neuronal expression (Supplementary Material, Fig. S1), suggesting possible roles for altered glutamatergic synaptic function and cAMP pathway in DISC1-TR induced behavioral inflexibility.
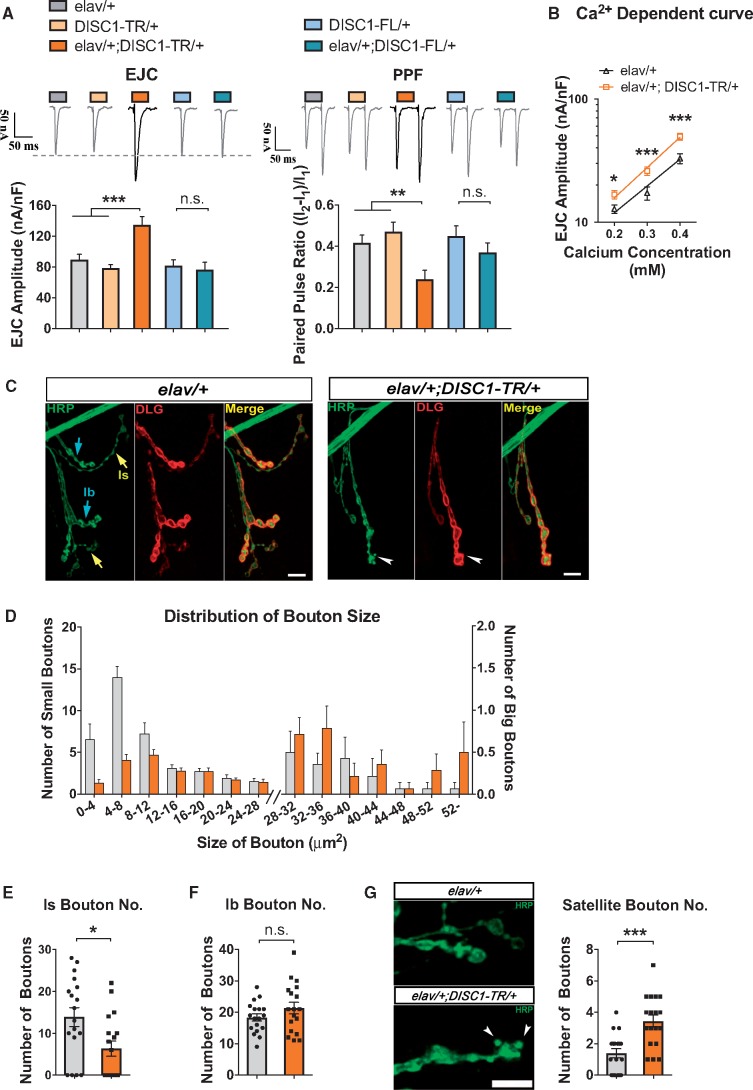
The larval neuromuscular junction (NMJ) is an ideal preparation for quantitative analysis of synaptic transmission and synapse morphology at identifiable glutamatergic synapses in Drosophila (28). Extensive studies indicate that synaptic transmission and structure of this preparation are sensitive to mutations that impair learning and memory (29) and associate with neurological or psychiatric disorders (30). Consistent with the behavioral data, both synaptic transmission and nerve terminal organization in transgenic larval NMJs were perturbed only by the expression of DISC1-TR, but not by the expression of DISC1-FL (Fig. 2).
Figure 2.
Expression of schizophrenia-related form of DISC1 (DISC1-TR) leads to aberrant glutamatergic transmission, plasticity and morphology of Drosophila larval neuromuscular junction (NMJ). Representative EJC traces and group data showed that pan-neuronal expression of DISC1-TR (elav-Gal4/+; DISC1-TR/+) but not DISC1-FL (elav-Gal4/+; DISC1-FL/+) led to increased EJC amplitude and reduced PPF ratio. The extracellular calcium concentration for recording was 0.4 mM. (ANOVA, n = 10; in DISC1-TR expressing group, P < 0.0001 for EJC, P = 0.0065 for PPF; in DISC1-FL expressing group, P = 0.5072 for EJC, P = 0.7231 for PPF). Calcium-dependent curve of EJC. Pan-neuronal expression of DISC1-TR (elav-Gal4/+; DISC1-TR/+) led to augemented EJC amplitude at 0.2, 0.3, and 0.4 mM calcium concentrations. (t-test, n = 23 and 18 for 0.2 mM; n = 10 and 12 for 0.3 mM, n = 17 and 14 for 0.4 mM, P < 0.05 for 0.2mM Ca2+; P < 0.001 for 0.3mM and 0.4mM Ca2+). The morphology of NMJ boutons in the control animals (elav-Gal4/+, left panel) and DISC1-TR larvae (elav-Gal4/+; DISC1-TR/+, right panel). Drosophila NMJs were co-immunostained with antibodies against horseradish peroxidase (HRP) (green; labeling presynaptic motor neuron membrane) and discs large (DLG) (red; labeling postsynaptic musicular membrane). Satellite boutons are indicated by white arrowheads, the type I big (Ib) boutons, the type I small (Is) boutons, and satellite boutons are indicated by blue arrow, yellow arrow, and white arrowhead respectively. Boutons from muscle 4 of the 3rd or 4th segments of larvea were statistically analyzed. The distribution of bouton size in control (elav-Gal4/+) and DISC1-TR expressing larvae (elav-Gal4/+; DISC1-TR/+). Epression of DISC1-TR led to a reduced number of small boutons (left to the double slash), and an augmented number of large boutons (right to the double slash). The number of Is boutons was decreased by the expression of DISC1-TR (elav-Gal4/+; DISC1-TR/+). (t-test, n = 14, P = 0.0128). The number of Ib buotons of DISC1-TR expressing larvae (elav-Gal4/+; DISC1-TR/+) was the same as that of the control group (elav-Gal4/+). (t-test, n = 18, P = 0.1803). The number of satellite boutons was dramatically increased in larvae expressing DISC1-TR (elav/+; DISC1-TR/+) (t-test, n = 18, P = 0.0002). White arrowheads in the left panel of 2C indicate satellite boutons. Scale bars are 5 μm. All data are represented as mean ± SEM.
Synaptic transmission was determined with the two-electrode voltage-clamp method. The amplitude of evoked excitatory junctional currents (EJCs) was significantly increased only in DISC1-TR transgenic larvae but not in the DISC1-FL larvae (left panel, Fig. 2A) at various extracellular calcium concentrations (Fig. 2B), while ion channel properties remained the same as control flies when different forms of DISC1 were expressed (Supplementary Material, Fig. S2). In addition, short-term synaptic plasticity assayed via the paired-pulse facilitation (PPF) paradigm was attenuated in DISC1-TR expressing larvae (right panel, Fig. 2A). To determine whether defective PPF resulted from an increased basal level of EJCs, we also compared PPF of the control (elav/+ at 0.4mM Ca2+) with the DISC1-TR group (elav/+; DISC1-TR/+ at 0.3mM) at comparable EJC amplitudes. The PPF was still significantly decreased (Supplementary Material, Fig. S3).
Next, nerve terminal morphology was visualized by immunohistochemical staining of Horseradish Peroxidase (HRP) and Discs large (DLG, homologous to vertebrate postsynaptic density). Only synaptic arborizations on muscle 4 at segments 3 and 4 were observed to minimize intrinsic variation. There are two subtypes of type I boutons in larval NMJ, i.e. type I small (Is, yellow arrow) and type I big (Ib, blue arrow, Fig. 2C). Ib boutons are distinguishable from Is by their visibly stronger staining of DLG. From the distribution of the bouton size, we found that the expression of DISC1-TR diminished the number of the boutons with smaller size (Fig. 2D). Indeed, the number of Is boutons innervating individual muscle fibers was significantly decreased (Fig. 2E). In fact, there was a much larger fraction of the nerve terminals on muscle 4 showing no observable Is boutons in DISC1-TR larvae than that in the control (see accumulated ‘0’ data points in the histogram of Fig. 2E). Despite the decrease of Is bouton number, the average number of Ib boutons remained unchanged in the DISC1-TR larvae (Fig. 2F). Interestingly, we found that the number of satellite boutons (arrowhead, Fig. 2C right panel, and 2G), which are tiny boutons budding off from the central Ib boutons, was greatly increased in the DISC1-TR larvae (Fig. 2G). Both Is number and satellite boutons remained unaffected in DISC1-FL expressing larvae (Supplementary Material, Fig. S4).
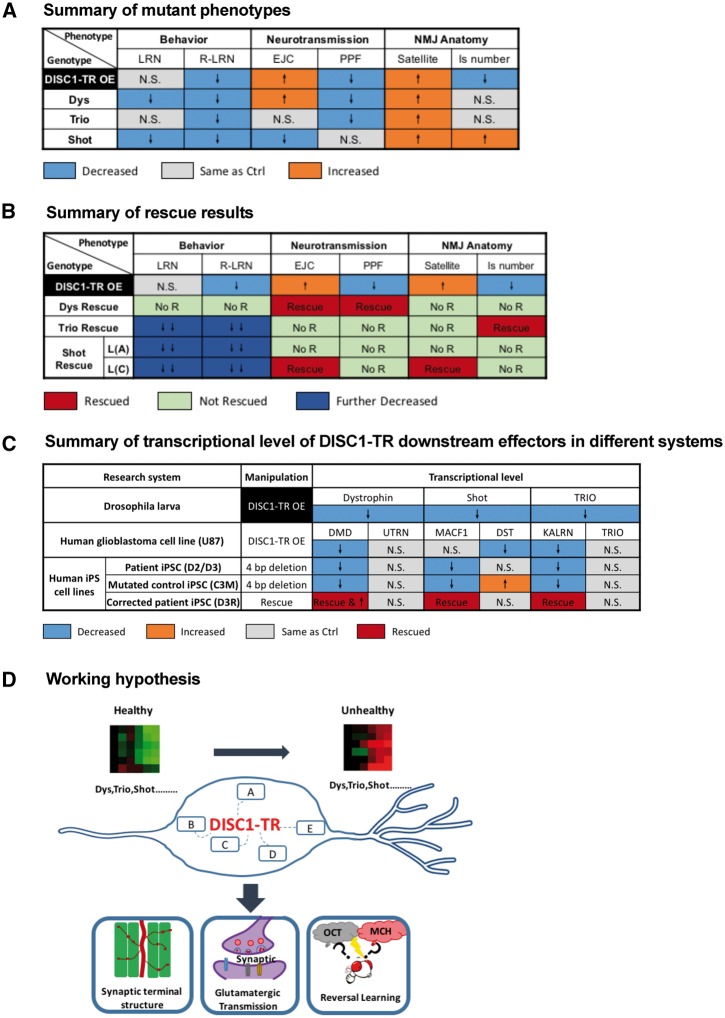
Data presented above show that the expression of DISC1-TR, but not DISC1-FL, leads to alterations in reversal learning, glutamatergic transmission, short-term synaptic plasticity, and nerve terminal organization. Although previously studies showed that the expression of DISC1-TR led to abnormal neuronal functions in mice models (17,31,32), due to the presence of the endogenous DISC1, it is largely unknown whether these abnormalities were led by the DISC1-TR independent of DISC1-FL. With the advantage of the absence of the endogenous homolog of DISC1 in Drosophila, our study demonstrated that DISC1-TR has specific gain-of-function neural effects independent of DISC1-FL in the five neuronal functional parameters, including EJC amplitude, PPF, Is number, satellite bouton, and reversal learning (see Fig. 7A for summary of the phenotypes induced by DISC1-TR).
Figure 7.
Summary of mutant phenotypes, rescue results, and transcriptional level of DISC1-TR regulated genes. (A) The neural phenotypes of DISC1-TR and mutants of its regulated genes. DISC1-TR OE (elav/+; DISC1-TR/+), Dys (DysDP186), Trio (Trio8595/+) and Shot (shot15072) are shown. In the table, grey cells indicate test groups showed the same phenotypes as the control group (elav/+). Orange and blue cells indicate that test groups had increased or decreased phenotypes compared to the control group but showed no significant difference with the DISC1-TR OE group (elav/+; DISC1-TR/+). (B) Phenotypes of flies that overexpressed DISC1-TR regulated genes in DISC1-TR expressing neurons. DISC1-TR OE (elav/+; DISC1-TR/+), Dys Rescue (elav/+; DISC1-TR/UAS-DysDp186), Trio Rescue (elav/+; DISC1-TR/UAS-Trio9513) and Shot Rescue (upper row, elav/+; DISC1-TR/UAS-Shot.L(A)29044 and lower row, elav/+; DISC1-TR/UAS-Shot.L(C)29042) are shown. Green cells indicate no rescue effect in the tested group, and red cells indicate that tested groups showed rescue effects compared to the control group. The cells in pink indicate that the phenotypes of the tested group were even severer than that of the DISC1-TR OE group. (C) The transcriptional level of DISC1-TR regulated genes in different research systems. In fly study, DISC1-TR OE is to express UAS-DISC1-TR transgene with elav-Gal4 driver in Drosophila larvae (elav/+; DISC1-TR/+). In human glioblastoma cell line study, DISC1-TR OE is to express DISC1-TR construct (PLVX-DISC1-TR-GFP) in the U87 cell line. In patient iPSC study, D2/D3 is the iPSC-differentiated neurons from patients; C3M is the iPSC-differentiated neurons with 4-bp frameshift deletion introduced to the original inside family control C3; D3R is iPSC-differentiated neurons with the 4-bp frameshift deleted mutation corrected in the original patient D3. (D) Cartoon of the working hypothesis on the function of DISC1-TR and its related genes.
Screen for DISC1-TR regulated effectors
These findings prompted us to search for mechanisms underlying the gain-of-function of DISC1-TR. A possible mechanism is that the observed effects may be led by altered expression of multiple genes caused by the expression of DISC1-TR. To investigate this possibility, we set out to identify genes whose expression is altered by the expression of DISC1-TR.
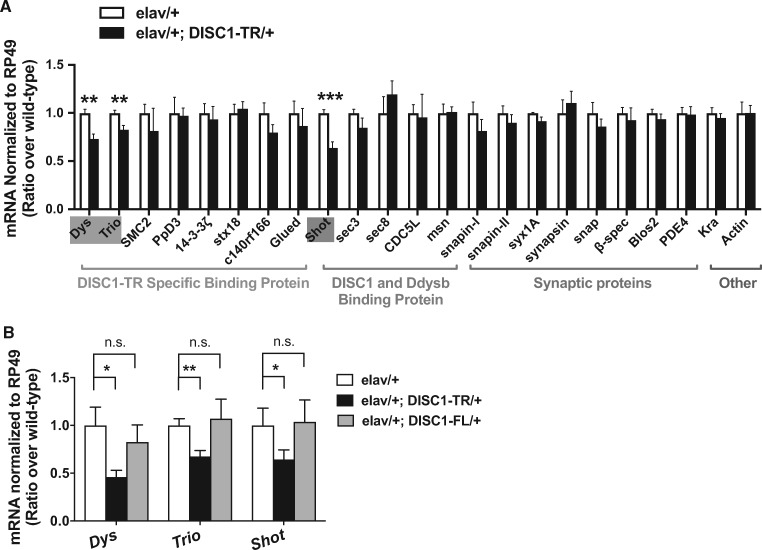
Three categories of candidate genes were chosen for a primary screening. DISC1 is a scaffold protein with its C-terminal domains mediating protein-protein interaction. In vitro biochemical and bioinformatics research showed that after losing two coiled-coil domains in the C-terminus, DISC1-TR acquires new binding partners without intersection with the full-length form (Supplementary Material, Fig. S5) (21). Therefore, the first group of genes for screening is DISC1-TR binding proteins (Fig. 3A, orange), including Dys, Trio, SMC2, PpD3, 14-3-3β, stx18, c140rf166, Glued (for the description of the genes concerned, see Supplementary Material, Table S1). In addition, it is reported that susceptibility genes associated with psychiatric disorders tend to converge on common biological pathways by sharing interconnected protein-protein interaction networks (33). Dysbindin, a susceptibility molecule for mental disorders, has been shown to have physical interaction with DISC1 (30,34,35). They were shown to interact epistatically and may share the interacting network associated with the altered neuronal functions (36,37). Moreover, our previous study showed the involvement of dysbindin in synaptic transmission and cognitive function in Drosophila (30). Thus, the second group includes genes that fall into the DISC1-Dysbindin interacting network (Fig. 3A, green) (21), including Shot, sec3, sec8, CDC5L and msn (Supplementary Material, Table S1). To exclude non-specific effects of DISC1-TR, an array of genes encoding synaptic proteins was also profiled as internal controls (Fig. 3A, purple and blue), including snapin-I, snapin-II, syx1A, synapsin, snap, PDE4, β-spec, Blos2, Kra, and Actin (Supplementary Material, Table S1).
Figure 3.
Transcriptional profiles of candidate DISC1-TR regulated effectors. (A) The primary screen of gene transcriptional level by RT-PCR. The categories for the candidate genes were indicated below the bar graph. The mRNA levels of genes Dys, Trio, and Shot in the DISC1-TR expressing larvae (elav-Gal4/+; DISC1-TR/+) were decreased. (t-test, P = 0.006 for Dys, 0.005 for trio, and 0.001 for shot, n.s., P > 0.05, n = 6-11). (B) The confirmation of the DISC1-TR regulated effectors with q-PCR. The mRNA levels of Dys, Trio, and Shot were decreased in the DISC1-TR expressing larvae (elav-Gal4/+; DISC1-TR/+), but not in the DISC1-FL expressing larvae. (ANOVA, for DISC1-TR group, P = 0.03 for Dys, 0.04 for shot, and 0.006 for shot; for DISC1-FL group, n.s., P > 0.05; n = 5-6). Statistical significance (*P < 0.05; **P < 0.01) or no significance (n.s.) is indicated. All data are represented as mean ± SEM.
In the primary screen, we measured transcriptional levels of all candidate genes in fly larvae with semi-quantitative reverse-transcript PCR (RT-PCR) and found that mRNA levels of Dys, Trio, and Shot were essentially reduced in larvae expressing DISC1-TR (Fig. 3A). We next measured the transcriptional level of the three hits from the primary screen in both DISC1-TR and DISC1-FL larvae with the quantitative real-time PCR (q-PCR), and confirmed the results from the primary screen. The mRNA level of Dys, Trio, and Shot was significantly decreased in DISC1-TR larvae but not affected in the larvae expressing DISC1-FL (Fig. 3B). Dys is the Drosophila homolog of vertebrate utrophin (UTRN) and dystrophin (DMD); Trio, a RhoGEF, is the homolog of vertebrate kalirin (KALRN) and TRIO; Shot is the homolog of dystonin (DST), and a potential functional homolog of Microtubule Actin Cross-linking Factor1 (MACF1) (38) (Supplementary Material, Table S1). Notably, although some of the vertebrate homologs of the identified proteins are thought capable of binding with both DISC1-TR and DISC1-FL, the expression was only altered in larvae expressing DISC1-TR.
Mutants of DISC1-TR regulated genes phenocopy its effects
All three identified DISC1-TR regulated genes are reported to affect some aspects of neuronal functions in Drosophila, with Dys in NMJ neurotransmission (39), Trio in dendritic morphogenesis (40) and axon growth (41), and Shot in sensory and motor axon guidance (42). Therefore, we tested the DISC1-TR-related parameters in mutants of the three genes to inspect whether they contribute to the phenotypes of DISC1-TR.
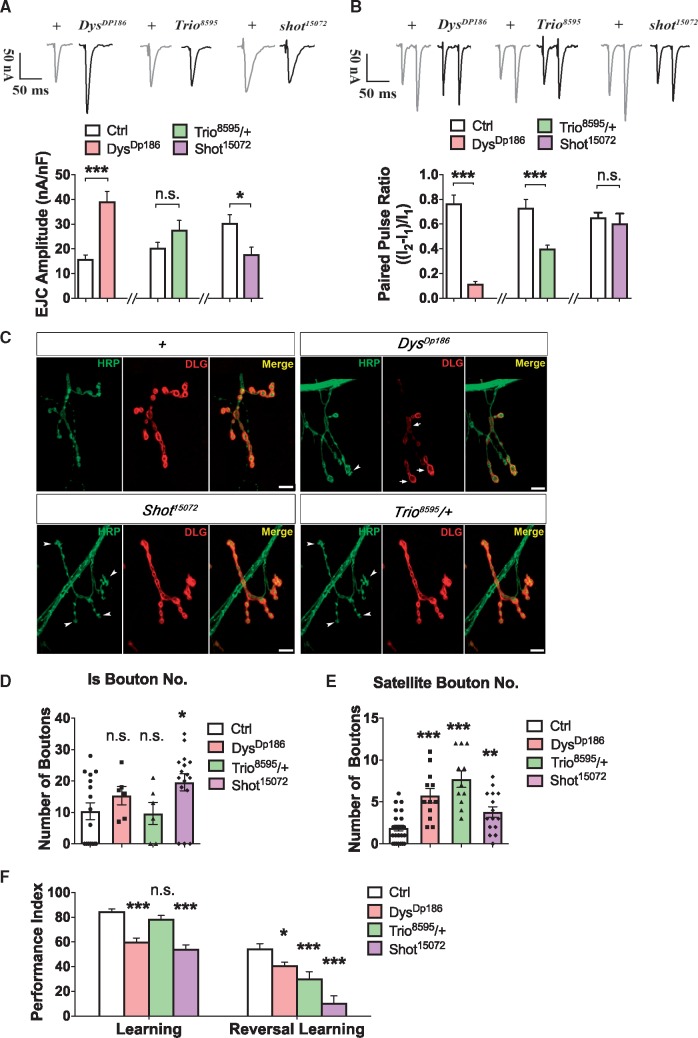
The EJC amplitude was increased (Fig. 4A) and PPF ratio was decreased (Fig. 4B) in the null mutant, DysDp186, showing a trend similar to DISC1-TR effects. For the heterozygous mutant of Trio, Trio8595/+, PPF was also reduced while EJC amplitude was not affected significantly (Fig. 4A and B). In the hypomorphic mutant Shot15072, the trend was different from the effects of DISC1-TR, with a reduced EJC amplitude and normal PPF (Fig. 4A and B).
Figure 4.
Involvement of DISC1-TR regulated effectors in neuronal development and function of Drosophila NMJ. The mutant of Dys (DysDP186) exhibited dramatically increased EJC compared to the wild-type control (w1118). The heterozygous mutant of trio (Trio8595/+) showed normal basal neurotransmission. The mutant of shot (shot15072), however, showed lower EJC amplitude than the control. (t-test, n = 10 for DysDP186 and its control, n = 14 for Trio8595 and its control, n = 10 for Shot15072 and its control, P < 0.001 of DysDP186, P < 0.05 of shot15072). The PPF ratio was dramatically decreased in mutants of Dys (DysDP186) and Trio (Trio8595/+), while no significant changes of PPF ratio were observed in a mutant of Shot (shot15072) (t-test, n = 10 for DysDP186 and its control, n = 14 for Trio8595/+ and its control, n = 10 for Shot15072 and its control, P = 3.383E-07 of DysDP186, 0.0005 of Trio8595/+, 0.6029 of shot15072). Representative immunostainings of morphological changes in mutants of Dys (DysDP186), Trio (Trio8595/+) and Shot (shot15072) compared to wild-type control (W1118). Drosophila NMJs were co-immunostained with antibodies against HRP (green; motor neuron membrane) and DLG (red; postsynaptic muscle membrane). Arrowheads indicate satellite boutons and arrows indicate Ib boutons. Scale bars are 5μm. (D,E) Statistical analysis of properties of NMJ morphology, including the number of Is and satellite boutons. Is bouton number was significantly increased in a mutant of shot (shot15072) (t-test, P = 0.495, 0.300, 0.021; n = 6 for DysDP186 and Trio8595/+, n = 16 for Shot15072) and mutants of Dys (DysDP186), Trio (Trio8595/+), and Shot (shot15072) all exhibited increased number of satellite boutons (t-test, P = 0.001, <0.001, =0.008, n = 12 for DysDP186 and Trio8595/+, n = 15 for Shot15072). (F) Learning and reversal learning performance of DISC1-TR regulated effectors. Mutant Trio8595/+ had the same learning performance as controls (W1118), while the learning performance of DysDP186 and Shot15072 mutants was decreased. (t-test, P = 0.001, <0.001, =0.008; n = 8-10) Mutants of Dys (DysDP186), Trio (Trio8595/+) and Shot (Shot15072) all had impaired reversal learning performance. (t-test, P = 0.012, 0.003, 0.0001; n = 5-8.). All data are represented as mean ± SEM.
In larval NMJ morphology, satellite boutons were increased in all three mutants (white arrowhead, Fig. 4C and E), resembling the DISC1-TR transgenic larvae. However, the number of Is boutons, if not augmented, remained the same in all three mutants (Fig. 4D). Notably, the number of satellite boutons might be an essential parameter of the NMJ structure influenced by the function of DISC1, because DISC1-TR and all the mutants of the DISC1-TR effectors showed increments in satellite bouton number (Fig. 2G and 4E).
As to cognitive function, the behavioral phenotype in the Trio8595/+ mutant completely replicated the phenotype of DISC1-TR expression flies, with normal learning but impaired reversal learning (Fig. 4F). However, the DysDp186 and Shot15072 mutants exhibited decrease in both learning and reversal learning performances (Fig. 4F), which is consistent with the cognitive disruption in the DMD-related Duchenne muscular dystrophy and the DST-related sensory aberration in dystonia musculorum deformans (43). It is interesting that DISC1-TR flies exhibited normal learning ability in spite of the down-regulated expression of Dys and Shot. This could be explained by the disturbance of the expression level of Dys and Shot in the DISC1-TR flies was not as strong as that in the mutants of these genes.
Thus, every parameter that was disturbed in DISC1-TR flies (EJC amplitude, PPF, Is number, satellite bouton, and reversal learning) could be well phenocopied by attenuating the expression of one or more DISC1-TR regulated genes. As summarized in Figure 7A, the phenotypes of increased EJC and decreased PPF were mimicked in Dys mutant, the phenotypes of normal learning and impaired reversal learning were mimicked in Trio mutant, and the phenotype of increased satellite bouton number was phenocopied in all three mutants.
Rescue of DISC1-TR phenotypes via overexpression of its regulated effectors
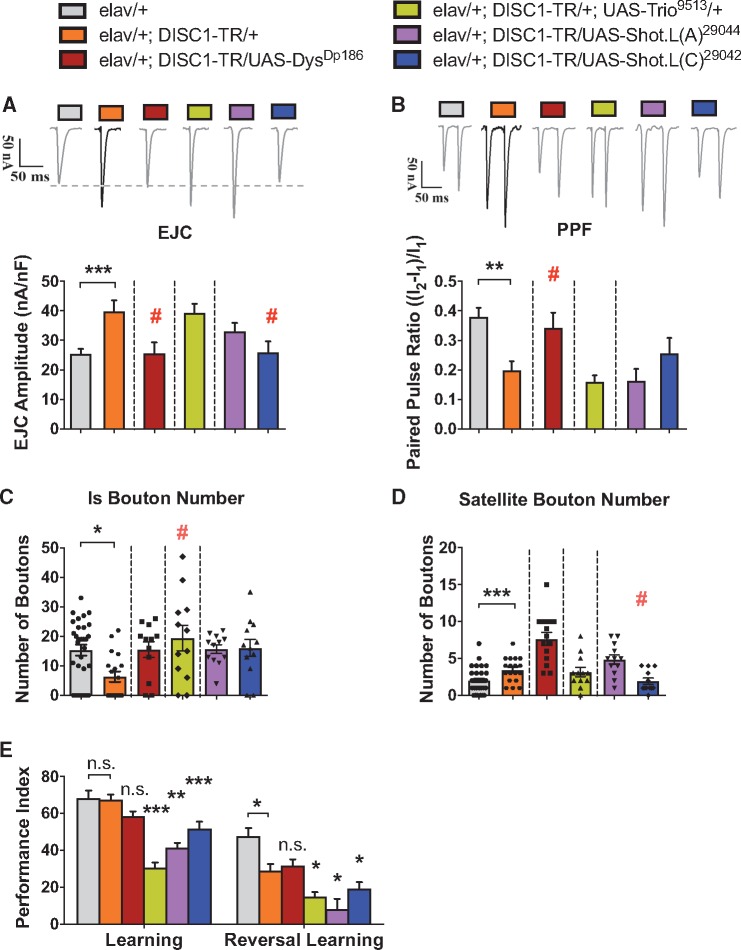
To further determine whether Dys, Trio, and Shot are bona fide functional effectors regulated by DISC1-TR, we tested whether restoring the expression of these genes rescues corresponding DISC1-TR phenotypes. Indeed, the expression of Dys (elav/+; DISC1-TR/UAS-DysDP186) overrode the increased EJC amplitude and decreased PPF phenotypes in DISC1-TR expressing flies, whereas the expression of Trio had no effects as expected (Fig. 5A and B). Additionally, the expression of a long-form C of Shot (elav/+;DISC1-TR/UAS-Shot.L(C)29042) rescued the EJC phenotype but not the PPF phenotype, while the expression of the long-form A of Shot (elav/+;DISC1-TR/UAS-Shot.L(C)29044) did not have any rescue effect on either phenotypes (Fig. 5A and B). For nerve terminal organization, we focused on the number of Is and satellite boutons. The expression of Trio (elav/+;DISC1-TR/+;UAS-Trio9513/+) rescued the DISC1-TR-induced decreased number of Is boutons (Fig. 5C), while the expression of the long-form C of Shot (elav/+;DISC1-TR/UAS-Shot.L(C)29042) rescued the increased number of satellite bouton in DISC1-TR expressing flies (Fig. 5D). The expression of Dys failed to do so even though satellite bouton number is also increased in the Dys mutant.
Figure 5.
Rescue of DISC1-TR induced neurodevelopmental and neurofunctional phenotypes via DISC1-TR regulated effector expression.(A, B) Representative traces and group data showed that expression of Dys (elav/+; DISC1-TR/UAS-DysDp186) successfully restored both the increased EJC amplitude and reduced PPF ratio induced by expression of DISC1-TR (elav/+; DISC1-TR/+) (n = 16). Expression of the L(C) isoform of Shot (elav/+; DISC1-TR/UAS-Shot.L(C)29042) only rescued increased EJC amplitude phenotype of DISC1-TR expressed flies (elav/+; DISC1-TR/+) (n = 16). The groups that showed rescue are marked with #, which indicates a statistical significance (P < 0.05, post-hoc test) with DISC1-TR expression group (elav/+; DISC1-TR/+) and statistical no significance (P > 0.05, post-hoc test) with control group (elav/+) after ANOVA. (C, D) Statistical analysis of properties of NMJ boutons morphology. Only Trio overexpression (elav/+; DISC1-TR/UAS-Trio9513) succeeded in restoring the number of Is boutons (n = 12), while larvae with overexpression of the L(C) isoform of Shot (elav/+; DISC1-TR/UAS-Shot.L(C)29042) rescued satellite bouton number (n = 14). The statistical analysis is the same as that in figure A. (E) Flies co-expressing Dys with DISC1-TR (elav/+; DISC1-TR/UAS-DysDp186) exhibited same learning performance as controls (elav/+) and the DISC1-TR group (elav/+;DISC1-TR/+). However, the expression of Trio (elav/+; DISC1-TR/UAS-Trio9513) and both isoforms of Shot (elav/+; DISC1-TR/UAS-Shot.L(A)29044 and elav/+; DISC1-TR/UAS-Shot.L(C)29042) showed decreased learning performance compare to those two groups (ANOVA, n = 6-8). The reversal learning performance was further reduced in Trio (elav/+; DISC1-TR/UAS-Trio9513) and both isoforms of Shot (elav/+; DISC1-TR/UAS-Shot.L(A)29044 and elav/+; DISC1-TR/UAS-Shot.L(C)29042) expressing flies compared to that of the DISC1-TR expressing flies, while the Dys co-expressing flies (elav/+; DISC1-TR/UAS-DysDp186) showed the same reversal learning ability as the DISC1-TR expressing flies, which was lower than that of the control (elav/+) (ANOVA, n = 6). All data are represented as mean ± SEM.
Even though the learning and reversal learning phenotypes were replicated in trio mutants, expression of Trio in the DISC1-TR expressing neurons severely impaired learning (Fig. 5E), making it impossible to test whether the reversal learning could be rescued. Similarly, the expression of other transgenes also disrupted learning (Fig. 5E), suggesting learning, or cognitive function, is more susceptible to the expression levels of these genes.
Thus, the DISC1-TR-induced cellular and behavioral phenotypes could be largely mimicked through attenuating expression of identified genes (Fig. 7A). Meanwhile, a majority of observed phenotypes could also be rescued through restoring the expression of these genes (Fig. 7B). Taken together, the DISC1-TR regulated effectors work as a synergistic functional network mediating different aspects of the neural effects associated with DISC1-TR in Drosophila.
Dysregulation of DISC1-TR regulated genes in prefrontal neurons carrying DISC1 mutation
It is evident from the above data that DISC1-TR is capable of attenuating expression of multiple molecules, which contribute to its cellular and behavioral phenotypes in Drosophila. To further support our findings in Drosophila and to evaluate the relevance of such findings to mental disorders associated with DISC1 mutants, we performed the following two experiments in cultured human cells.
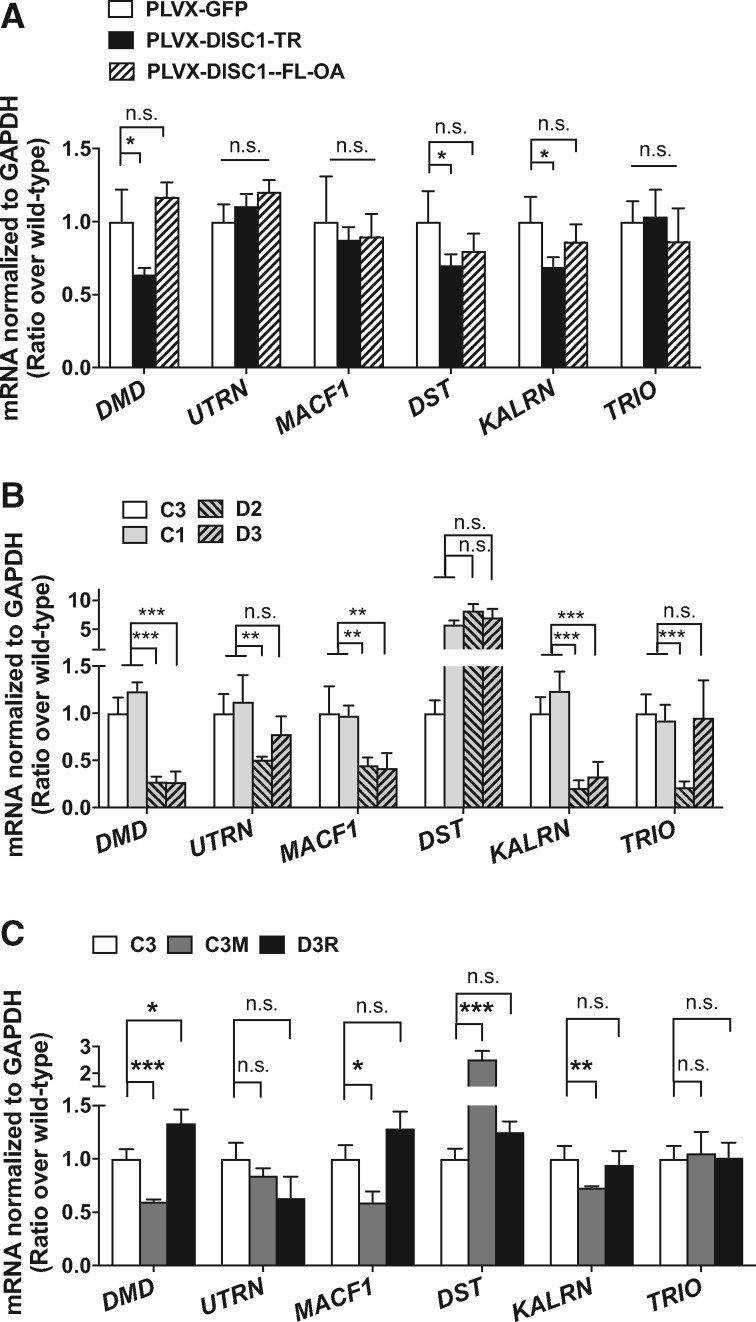
First, we assayed how DISC1-TR and DISC1-FL affect the expression of the human homologs of the identified effectors in the U87 human glioblastoma cell line. Lenti-virus (PLVX-GFP, PLVX-DISC1-TR-GFP, and PLVX-DISC1-FL-OA-GFP) was generated to overexpress two forms of DISC1 in the cell line. Through q-PCR analysis, we tested the RNA level of the effectors of DISC1-TR in 90% GFP positive U87 cells. The genes tested include DMD/UTRN (human homologs of Dys), MACF1/DST (human homologs of Shot), and KALRN/TRIO (human homologs of Trio). Compared to the control group (PLVX-GFP), the transcriptional levels of DMD, DST and KALRN, were downregulated in U87 cells expressing DISC1-TR (PLVX-DISC1-TR-GFP) but not DISC1-FL (PLVX-DISC1-FL-OA-GFP) (Fig. 6A). The profile is consistent with the results observed in Drosophila.
Figure 6.
Dysregulation of DISC1-TR regulated genes in human neurons carrying DISC1 mutations. (A) Validation of differential mRNA level of DISC1-TR regulated genes in human U87 cell lines expressed GFP, DISC1-TR (t-test, P < 0.0001, =0.234, 0.175, 0.001, 0.0003, 0.887) and DISC1-FL (ANOVA, P = 0.081, 0.023, 0.552, 0.112, 0.263, 0.655). n = 5–8. Values were normalized to those of neurons expressing PLVX-GFP. (B) Validation of differential mRNA expression of DISC1-TR regulated genes in 4-weeks-old forebrain neurons from C3, C1, D2 and D3 iPS cell lines. D2 (schizophrenia) and D3 (major depression) are two patients with the frameshift DISC1 mutation from pedigree H, C3 is one unaffected family members without the mutation, and C1 is an unrelated healthy individual as an additional outside family control. (ANOVA, **P < 0.01; ***P < 0.001; n.s., P > 0.05. n = 5-14. Values were normalized to those of C3 neurons). (C) mRNA levels of DISC1-TR regulated genes in forebrain neurons differentiated from C3, C3M (knock-in the 4-bp mutation of the control line C3) and D3R (correction of the same mutation line D3) isogenic iPS cell lines. (ANOVA, *P < 0.05; **P < 0.01; ***P < 0.001; n.s., P > 0.05. n = 6–7 for DMD, MACF1, and KALRN; n = 4-5 for UTRN, DST, and TRIO. Values were normalized to those of C3 neurons.). All data are represented as mean ± SEM.
Second, we tested how a mutation in DISC1 affects the expression of the identified effectors in the neurons differentiated from iPSC lines from a family with psychiatric disorders. A rare 4-bp frameshift deletion mutation in the C-terminus of DISC1, resulting a 1-809 C-terminal truncated DISC1, was detected in an American family (pedigree H) with schizophrenia and schizoaffective disorder (9). We utilized iPSC lines from members of this family, including two affected members and two control individuals (25). The affected members are patients D2 (schizophrenia) and D3 (major depression) with the frameshift deletion DISC1 mutation from pedigree H. The control individuals include an inside family control C3 (an unaffected family member without the mutation) and an outside family control C1 (an unrelated healthy individual). We performed q-PCR analysis on the human homologs of the DISC1-TR regulated genes in 4-week-old forebrain neurons that were differentiated from pedigree H iPSCs with about 90% glutamatergic neurons and a few GABAergic and dopaminergic neurons. To reveal the different gene expression in healthy population and patients, we compared D2 and D3 to C1 and C3. Only the genes whose expression was changed in both D2 and D3, but remained the same in both C1 and C3 were considered affected by the DISC1 mutation. Among the six human homolog genes (DMD/UTRN of fly Dys, MACF1/DST of fly Shot, KALRN/TRIO of fly Trio), the mRNA levels of DMD, MACF1 and KALRN were downregulated in the neurons carrying the DISC1 mutation (D2 and D3), but remained unchanged in the neurons from the control group (C1 and C3, Fig. 6B). There was no difference on the mRNA levels of UTRN and TRIO in the neurons from the control group and the patients (Fig. 6B). However, the mRNA level of DST was significantly higher in the neurons from the outside family control (C1) and the patients (D2 and D3) (Fig. 6B), which suggest the expression level of DST may be variable and sensitive to the genetic background in the healthy population. Notably, DMD was also identified as one of the top candidates that were essentially down-regulated by the 4-bp frameshift deletion in the previous RNAseq analysis (25). Furthermore, to exclude potential effects of other genetic components, we tested the RNA level of these three genes in neurons differentiated from two engineered isogenic iPS cell lines. One is the mutant DISC1 iPSC line that has the 4-bp frameshift deletion corrected (D3R), and the other is the control iPSC line that is introduced with the 4-bp frameshift deletion (C3M) (25). As expected, the expression levels of DMD/MACF1/KALRN were reduced in C3M neurons to a level comparable to that in DISC1 mutant neurons (D2 and D3) (Fig. 6C). Moreover, in D3R neurons, the decreased RNA levels of these genes were rescued to a level comparable to the control neurons (C3), if not even higher (Fig. 6C). Consistent with the observation in patients’ neurons, the mRNA levels of UTRN and TRIO remained the same when the 4-bp frameshift deletion was either introduced in C3M or corrected in D3R (Fig. 6C). Interestingly, the mRNA level of DST was essentially increased in C3M (Fig. 6C), similar to, but not as much as, the increment in C1, D2 and D3 (Fig. 6B). What’s more, the increase of DST in D3 was rescued in D3R (Fig. 6C). However, as stated above, the expression level of DST was variable even in the healthy controls (C1 and C3 in Fig. 6B), so the change of DST should be interpreted cautiously. These results suggest that the disease-related DISC1 mutation is necessary and sufficient for the dysregulated expression of the DISC1-TR regulated genes.
Taken together, data presented above showed that both the expression of DISC1-TR in human glioblastoma cell line and the mutation in C-terminal of DISC1 in neurons derived from patient iPSC led to the decreased transcription of the human homologs of Dys, Trio, and Shot, which support the conservation of the DISC1-TR regulated genes we identified in Drosophila (Fig. 7C).
DISC1-TR regulated effectors ranked as top disease-risk factors
Last, we evaluated the rankings of disease risk of the DISC1-TR regulated genes in human genetic studies. Extensively mining the online resources and literature, we collected three independent gene sets from human genetic studies. These genes are (a) DISC1-Interacting (154 genes): genes involved in DISC1 related pathways or selected as the candidate genes for schizophrenia (21), (b) HuGE-SZ (1499 genes): candidate genes associated with schizophrenia from HuGE Navigator (https://www.cdc.gov/genomics/hugenet/hugenavigator.htm; date last accessed April 30, 2017), and (c) Polygenic-SZ (1796 genes): genes associated with schizophrenia based on human variant data from whole exome sequencing (44). As shown in Table 1, most of the human homologs of candidate DISC1-TR regulated effectors tested in our screening in flies are present in the three gene sets. We adopted a gene-based scoring tool, Residual Variation Intolerance Score (RVIS) (26), to quantitatively measure the intolerance of genes to functional mutations. The intolerance score is designed to rank genes in terms of whether they have more or less common functional genetic variation relative to the genome-wide expectation given the amount of apparently neutral variation the gene has (26). Genes with less common functional variation have low RVIS scores; functional mutants of these genes are more likely to be involved in the development of diseases.
Table 1.
Disease risk assessment of DISC1-TR downstream candidate genes in genome wide analysis. Gene-based scoring analysis of the downstream candidate genes of DISC1-TR. Residual Variation Intolerance Score (RVIS) is a gene-based scoring tool, which systematically quantified the amount of functional mutations in each gene based on NHLBI GO Exome Sequencing Project (ESP) 6500 and established a genome-wide scoring system to prioritize genes based on their intolerance to standing functional mutations in the human population. The RVIS score is color-coded (red to dark grey, ranging from −4 to 4). The intolerance of functional mutation is positively correlated with the RVIS score. The genes that we did the transcriptional screen with were ranked by RVIS score in three gene collections: DISC1-Interacting (154 genes); HuGE-SZ (1499 genes); Polygenic-SZ (1796 genes). Within each collection, the relative RVIS score is color-coded (orange to white, ranging from the highest to the lowest RVIS score in the collection). %set is the percentile rank of each gene in each collection.
| Gene in fly | Gene in human | RVIS (EA_0.1%) | DISC1-Interacting (n = 154) | HuGE-SZ (n = 1499) | Polygenic-SZ (n = 1796) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Score | % | Rank | % Set | Rank | % Set | Rank | % Set | ||
| Trio | TRIO | −3.68 | 0.2 | 1 | 0.6 | NA | NA | 16 | 0.9 |
| Shot | MACF1 | −3.29 | 0.3 | 2 | 1.3 | 12 | 0.8 | 30 | 1.7 |
| Trio | KALRN | −2.76 | 0.6 | 4 | 2.6 | 24 | 1.6 | 63 | 3.5 |
| beta-spec | SPTBN1 | −2.5 | 0.9 | 8 | 5.2 | NA | NA | 89 | 5.0 |
| Shot | DST | −1.51 | 3.4 | 21 | 13.6 | 107 | 7.1 | 239 | 13.3 |
| Dys/dystrophin | DMD | −1.39 | 4.2 | 23 | 14.9 | 127 | 8.5 | 281 | 15.6 |
| msn/misshapen | TNIK | −1.27 | 5.4 | 26 | 16.9 | 152 | 10.1 | 333 | 18.5 |
| CDC5L | CDC5L | −1.23 | 5.8 | 27 | 17.5 | NA | NA | NA | NA |
| Dys/dystrophin | UTRN | −1.19 | 6.2 | 28 | 18.2 | 166 | 11.1 | 357 | 19.9 |
| Gl/Glued | DCTN1 | −0.75 | 13.9 | 39 | 25.3 | NA | NA | 589 | 32.8 |
| Actin | ACTB | −0.52 | 22.0 | NA | NA | NA | NA | 764 | 42.5 |
| PpD3 | PPP5C | −0.48 | 24.0 | 54 | 35.1 | NA | NA | NA | NA |
| syx1A | STX1A | −0.46 | 25.0 | NA | NA | 504 | 33.6 | 819 | 45.6 |
| SMC2 | SMC2 | −0.4 | 27.3 | NA | NA | NA | NA | NA | NA |
| Krasviazs/Kra | BZW1 | −0.16 | 43.8 | NA | NA | NA | NA | NA | NA |
| snapin-I | SNAPIN | −0.02 | 53.6 | NA | NA | NA | NA | NA | NA |
| snapin-II | SNAPIN | −0.02 | 53.6 | NA | NA | NA | NA | NA | NA |
| snap | NAPA | −0.01 | 54.5 | NA | NA | NA | NA | 1245 | 69.3 |
| c14orf166 | C14orf166 | 0.06 | 58.8 | 91 | 59.1 | NA | NA | NA | NA |
| Syntaxin18 | STX18 | 0.07 | 59.3 | 92 | 59.7 | NA | NA | NA | NA |
| 14-3-3zeta | YWHAZ | 0.08 | 60.1 | 93 | 60.4 | 919 | 61.3 | 1318 | 73.4 |
| blos2, PED4 | BLOC1S2 | 0.22 | 68.2 | NA | NA | NA | NA | NA | NA |
| ref. | −4 | 0.0 | 1 | 0.0 | 1 | 0.0 | 1 | 0.0 | |
| 4 | 100.0 | 154 | 100.0 | 1499 | 100.0 | 1796 | 100.0 | ||
We downloaded the RVIS scores (EA_0.1%) from the server (http://genic-intolerance.org/; date last accessed April 30, 2017) and ranked all candidate effectors based on their RVIS percentile (Table 1). The TRIO (Trio human homolog) has the lowest RVIS score of -3.68 and percentile of 0.17%, and MACF1 (Shot human homolog) has the second lowest RVIS score of -3.29 and percentile of 0.31% among all candidates. We also ranked DISC1 interacting partners and psychiatric disorders candidate genes based on their RVIS percentiles, and the TRIO and MACF1 are among 2% of the most intolerant genes in each gene sets. KALRN and DST, the other human homologs of trio and shot, are also in the top intolerant genes group. In addition, DMD (human homolog of Dys) has a considerable intolerance to mutation with an RVIS score of -1.39 and a percentile of 4.24%, also in the foremost intolerance group.
Among all the DISC1 interacting partners and the schizophrenia-related factors, the DISC1-TR regulated genes we identified are in the top ranks of factors potentially associated with human illnesses.
Discussion
The current study tests the hypothesis that perturbation of DISC1 protein may be involved in the pathology of mental disorders through attenuating the expression of multiple disease-risk factors, each of which has smaller effects on particular subsets of phenotypes (Fig. 7D). Results obtained from a range of experiments in flies and in human cell lines reported above are consistent with the proposed hypothesis. The three major discoveries in this study are outlined below.
First, the expression of DISC1-TR yields a range of pathology-related cellular and behavioral endophenotypes that differ significantly from the phenotypes of DISC1-FL expression. With the neuronal expression of DISC1-TR, flies had reduced reversal-learning performance, augmented glutamatergic transmission, decreased short-term synaptic plasticity, and abnormal morphological development of larval NMJ (Figs 1 and 2). Additional relevant endophenotypes have been reported in mouse studies. Three transgenic mouse models were generated by expressing human or mouse C-terminal truncated DISC1, either constitutively (24), or an inducible manner under the controls of the CAMKII promoter (19) or the endogenous mouse DISC1 promoter (45). These mouse models exhibited overlapping abnormal phenotypes in sensorimotor gating (pre-pulse inhibition), working memory and latent inhibition, partially emulating certain symptoms of psychiatric disorders (10,46). Thus, the effects of DISC1-TR and its interacting network are conserved evolutionarily from Drosophila, which has no DISC1 homologous gene reported, to mammals.
Although the expression of DISC1-FL did not show detectable phenotypes in the physiological and behavioral assays tested above (Figs 1 and 2 and Supplementary Material, S4), we found that both DISC1-FL and DISC1-TR expression reduced lifespan in two environmental stress tests (Supplementary Material, Fig. S6). Notably, DISC1-FL expression caused aberrant sleep which was not perturbed by neuronal DISC1-TR expression in Drosophila (47). The involvement of DISC1-FL in sleep was further confirmed recently in mouse model (48). Interestingly, the subcellular distribution of DISC1 protein was significantly changed when the truncated DISC1 was expressed in cultured cells (23,49). More investigations are required to determine whether the very different intracellular distribution of DISC1-TR and DISC1-FL may be involved in producing the neural effects (Supplementary Material, Fig. S7).
It is noteworthy that despite above discussion on the gain-of-function of truncated DISC1, the evidence supporting the DISC1 loss-of-function hypothesis still stands. DISC1 protein level was found to be decreased in the lymphocytes and the brain tissue of schizophrenia patients (50,51). Furthermore, animal models, including the spontaneous occurring point mutation or deletion in the endogenous mouse DISC1 and the knockdown of mouse endogenous DISC1 with RNAi (these models are extensively reviewed in 10, 46), revealed that the loss-of-function of endogenous DISC1 led to abnormalities in the neurogenesis, neurodevelopment, and behaviors (13,14,52). Therefore, above evidence indicates that both truncated form of DISC1 and the loss of functional endogenous DISC1 protein contribute to the disease mechanism.
Second, a functional network of molecules underlying the neural effects of DISC1-TR, including Dys, Trio, and Shot was identified. We found that mutants of these DISC1-TR regulated genes showed different phenotypes in behavior, neurotransmission, and neurodevelopment, which mimicked the abnormal phenotypes of DISC1-TR flies (Figs 4 and 7A). Further, co-expression of these genes with DISC1-TR rescued different aspects of the cellular phenotypes of DISC1-TR flies (Figs 5 and 7B). Consequently, instead of a single signaling pathway, the DISC1-TR regulated effectors are more likely to act as a synergistic network in translating DISC1-TR-dependent neural effects. In this network, each gene plays distinct roles in different aspects of the DISC1-TR neural effects (Fig. 7D). Presumably, there are more genes involved in this network to be discovered. Such DISC1-TR regulated genes are again conserved across species, since the expression of the homologs of these genes are also perturbed in U87 human glioblastoma cell lines overexpressing truncated DISC1 (Figs. 6A and 7C).
Third, the expression of human homologs of the DISC1-TR regulated genes, DMD, MACF1, and KALRN, is also downregulated in neurons from subjects carrying another DISC1 mutation (Figs 6B and C, 7C). Furthermore, the downregulated genes are characterized as top-ranked human disease-risk factors in bioinformatics analysis (Table 1). All these data support the hypothesis that the interaction between DISC1 and its effectors we found in Drosophila applies to human conditions.
Based on above discussion, we further address the implications and insight we gain from our findings in the following sections.
DISC1 interacting network converges at cytoskeleton organization
DISC1 has been considered as a scaffold protein and possesses a highly intertwined molecular network including many risk factors possibly for psychiatric disorders (21). We point out that although these molecules interact with both forms of DISC1, the expression of all three regulated genes is attenuated significantly with the expression of a mutant DISC1-TR. The function analysis of these molecules from previous studies helps us to uncover the fundamental basis of the multiple disease-risk pathways perturbed by DISC1-TR. DMD/UTRN, as membrane cytoskeletal associated glycoproteins, connect the cytoskeleton to the cell membrane through binding with actin (39,53) and play a central role in the molecular pathogenesis of several muscular dystrophies (54). TRIO/KALRN are both Guanine-nucleotide Exchange Factors of GTPases Rac1/RhoG and RhoA (RhoGEF), essential members in signaling pathways critical in the regulation of dynamics of cytoskeleton organization (55,56). What’s more, KALRN is a genetic risk factor for Alzheimer’s disease, attention deficit hyperactivity disorder, and schizophrenia (57,58). The human homologs of Shot include cytoskeleton linker proteins DST and MACF1 (59), both associated with multiple disorders (43). Variations of MACF1 were reported in schizophrenia patients (60). Taken together, their functions converge at the regulation of cytoskeleton organization and their mutations are critically involved in human diseases.
More important, when we applied DAVID (http://david.abcc.ncifcrf.gov/; date last accessed April 30, 2017) (61) for Gene Ontology (GO) enrichment analysis of the DISC1/psychiatric disorders related genes, We identified cytoskeleton-related GO terms, such as "cytoskeleton", "actin cytoskeleton", "microtubule cytoskeleton" and "cytoskeletal protein binding", overrepresented in the DISC1-TR regulated effectors, as well as in DISC1-Interacting and Polygenic-SZ gene sets (Supplementary Material, Table S3). For example, 12 DISC1-TR downstream effectors (55%), 41 DISC1-Interacting partners (34%) and 303 schizophrenia candidate genes (17%) are enriched for "cytoskeleton". More interestingly, 6 DISC1-TR downstream effectors (27%) and 90 schizophrenia candidate genes (17%) are enriched for "actin cytoskeleton", and KALRN (Trio), MACF1/DST (Shot), UTRN (Dys) are involved in the GO category. All these results suggest that psychiatric disorders risk pathways, especially the DISC1 interacting network, converge at cytoskeleton organization.
Implications for disease
A key challenge and opportunity for Drosophila disease modeling, or any other animal modeling studies, is to extend the findings to human patients. In the current research, we validate our findings from Drosophila research in human cell line study and bioinformatics analysis with human genetic data. The dysregulation of the human homologs of the DISC1-TR regulated molecules, (DMD/MACF1/KALRN) in human glioblastoma cells expressing DISC1-TR as well as in human forebrain neurons derived from human iPSCs carrying another mutant DISC1, suggest that the study of DISC1 in Drosophila could be relevant to the molecular and cellular effects in human conditions (Fig. 7C). The top ranking of the DISC1-TR regulated molecules in RVIS analysis further confirmed the mutations of these genes are likely to be involved in the development of diseases. Taken together, the Drosophila model, with powerful genetic accessibility and robust disease-related phenotypes from cellular to behavioral level, provides a feasible strategy to generally evaluate the function-related significance of risk factors for diseases.
Materials and Methods
Fly stocks and detailed procedures for the immunostaining, confocal microscopy, electrophysiology, behavior, longevity assay, differentiation of iPSCs, RT-PCR, q-PCR, and bioinformatics analysis are described in Supplemental Text.
Statistical Analysis
Unless stated otherwise, the data are shown as means ± SEM and analyzed by Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test using GraphPad Prism 7.0 (GraphPad Software, CA) statistical software. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., no significance (P > 0.05).
Supplementary Material
Acknowledgements
We thank Bloomington Stock Center and Professor Furukubo-Tokunaga for fly stocks. We are grateful to Ha Nam Nguyen, Wei-kai Wang and Lianzhang Wang for technical support, and Dr. Clement Kent, Dr. Hsiang-Ting Lei, Dr. Mark Eddison, Mr. Weiwei Ma for discussion and critical reading of the manuscript. Y.Z. was supported by grants from the National Science Foundation of China, and the National Basic Research Project (973 program) of the Ministry of Science and Technology of China, the Beijing Municipal Science & Technology Commission. H.S. was supported by NIH, G.L.M was supported by MSCRF, A.S. was supported by NIH, NARSAD, Stanley, RUSK and S-R foundations.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
National Science Foundation of China (91332207 and 91632301), National Basic Research Project (973 program) of the Ministry of Science and Technology of China (2013cb835100), Beijing Municipal Science & Technology Commission (Z161100002616010), NIH [NS047344, NS048271, MH105128] and MSCRF, MH-084018, MH-094268, MH-069853, MH-085226, MH-088753, and MH-092443, Silvo O. Conte Center, DA-040127, NARSAD, Stanley, RUSK, and S-R Foundations.
References
- 1. Gottesman I.I., Shields J. (1967) A polygenic theory of schizophrenia. Proc. Natl Acad. Sci. U.S.A., 58, 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sullivan P.F., Daly M.J., O'Donovan M. (2012) Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet., 13, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M.. et al. (2014) De novo mutations in schizophrenia implicate synaptic networks. Nature, 506, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gulsuner S., Walsh T., Watts A.C., Lee M.K., Thornton A.M., Casadei S., Rippey C., Shahin H., Consortium on the Genetics of S., Group P.S.. et al. (2013) Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell, 154, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schizophrenia Psychiatric Genome-Wide Association Study, C. (2011) Genome-wide association study identifies five new schizophrenia loci. Nat. Genet., 43, 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gratten J., Wray N.R., Keller M.C., Visscher P.M. (2014) Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat. Neurosci., 17, 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McClellan J.M., Susser E., King M.C. (2007) Schizophrenia: a common disease caused by multiple rare alleles. Br. J. Psychiatry, 190, 194–199. [DOI] [PubMed] [Google Scholar]
- 8. Millar J.K., Christie S., Anderson S., Lawson D., Hsiao-Wei Loh D., Devon R.S., Arveiler B., Muir W.J., Blackwood D.H., Porteous D.J. (2001) Genomic structure and localisation within a linkage hotspot of disrupted in schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol. Psychiatry, 6, 173–178. [DOI] [PubMed] [Google Scholar]
- 9. Sachs N.A., Sawa A., Holmes S.E., Ross C.A., DeLisi L.E., Margolis R.L. (2005) A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol. Psychiatry, 10, 758–764. [DOI] [PubMed] [Google Scholar]
- 10. Brandon N.J., Sawa A. (2011) Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat. Rev. Neurosci., 12, 707–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Porteous D.J., Thomson P.A., Millar J.K., Evans K.L., Hennah W., Soares D.C., McCarthy S., McCombie W.R., Clapcote S.J., Korth C.. et al. (2014) DISC1 as a genetic risk factor for schizophrenia and related major mental illness: response to Sullivan. Mol. Psychiatry, 19, 141–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sullivan P.F. (2013) Questions about DISC1 as a genetic risk factor for schizophrenia. Mol. Psychiatry, 18, 1050–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duan X., Chang J.H., Ge S., Faulkner R.L., Kim J.Y., Kitabatake Y., Liu X.B., Yang C.H., Jordan J.D., Ma D.K.. et al. (2007) Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell, 130, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mao Y., Ge X., Frank C.L., Madison J.M., Koehler A.N., Doud M.K., Tassa C., Berry E.M., Soda T., Singh K.K.. et al. (2009) Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell, 136, 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishizuka K., Kamiya A., Oh E.C., Kanki H., Seshadri S., Robinson J.F., Murdoch H., Dunlop A.J., Kubo K., Furukori K.. et al. (2011) DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature, 473, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niwa M., Jaaro-Peled H., Tankou S., Seshadri S., Hikida T., Matsumoto Y., Cascella N.G., Kano S., Ozaki N., Nabeshima T.. et al. (2013) Adolescent stress-induced epigenetic control of dopaminergic neurons via glucocorticoids. Science, 339, 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ayhan Y., Abazyan B., Nomura J., Kim R., Ladenheim B., Krasnova I.N., Sawa A., Margolis R.L., Cadet J.L., Mori S.. et al. (2011) Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence for neurodevelopmental origin of major psychiatric disorders. Mol. Psychiatry, 16, 293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnstone M., Thomson P.A., Hall J., McIntosh A.M., Lawrie S.M., Porteous D.J. (2011) DISC1 in schizophrenia: genetic mouse models and human genomic imaging. Schizophrenia Bulletin, 37, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pletnikov M.V., Ayhan Y., Nikolskaia O., Xu Y., Ovanesov M.V., Huang H., Mori S., Moran T.H., Ross C.A. (2008) Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol. Psychiatry, 13, 173–186. 115. [DOI] [PubMed] [Google Scholar]
- 20. Ugur B., Chen K., Bellen H.J. (2016) Drosophila tools and assays for the study of human diseases. Dis. Model. Mech., 9, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Camargo L.M., Collura V., Rain J.C., Mizuguchi K., Hermjakob H., Kerrien S., Bonnert T.P., Whiting P.J., Brandon N.J. (2007) Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry, 12, 74–86. [DOI] [PubMed] [Google Scholar]
- 22. Lu B., Vogel H. (2009) Drosophila models of neurodegenerative diseases. Ann. Rev. Pathol., 4, 315–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M.. et al. (2005) A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol., 7, 1167–1178. [DOI] [PubMed] [Google Scholar]
- 24. Hikida T., Jaaro-Peled H., Seshadri S., Oishi K., Hookway C., Kong S., Wu D., Xue R., Andrade M., Tankou S.. et al. (2007) Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc. Natl Acad. Sci. U S A, 104, 14501–14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wen Z., Nguyen H.N., Guo Z., Lalli M.A., Wang X., Su Y., Kim N.S., Yoon K.J., Shin J., Zhang C.. et al. (2014) Synaptic dysregulation in a human iPS cell model of mental disorders. Nature, 515, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. (2013) Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet., 9, e1003709.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tully T., Quinn W.G. (1985) Classical conditioning and retention in normal and mutant Drosophila melanogaster. J. Comp. Physiol. A., 157, 263–277. [DOI] [PubMed] [Google Scholar]
- 28. Jan L.Y., Jan Y.N. (1976) Properties of the larval neuromuscular junction in Drosophila melanogaster. J. Physiol., 262, 189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhong Y., Wu C.F. (1991) Altered synaptic plasticity in Drosophila memory mutants with a defective cyclic AMP cascade. Science, 251, 198–201. [DOI] [PubMed] [Google Scholar]
- 30. Shao L., Shuai Y., Wang J., Feng S., Lu B., Li Z., Zhao Y., Wang L., Zhong Y. (2011) Schizophrenia susceptibility gene dysbindin regulates glutamatergic and dopaminergic functions via distinctive mechanisms in Drosophila. Proc. Natl Acad. Sci. U S A, 108, 18831–18836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li W., Zhou Y., Jentsch J.D., Brown R.A., Tian X., Ehninger D., Hennah W., Peltonen L., Lonnqvist J., Huttunen M.O.. et al. (2007) Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc. Natl Acad. Sci. U S A, 104, 18280–18285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abazyan B., Nomura J., Kannan G., Ishizuka K., Tamashiro K.L., Nucifora F., Pogorelov V., Ladenheim B., Yang C., Krasnova I.N.. et al. (2010) Prenatal Interaction of Mutant DISC1 and Immune Activation Produces Adult Psychopathology. Biol. Psychiatry, 68, 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo X., Huang L., Jia P., Li M., Su B., Zhao Z., Gan L. (2014) Protein-protein interaction and pathway analyses of top schizophrenia genes reveal schizophrenia susceptibility genes converge on common molecular networks and enrichment of nucleosome (chromatin) assembly genes in schizophrenia susceptibility loci. Schizophr. Bull., 40, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee S.A., Kim S.M., Suh B.K., Sun H.Y., Park Y.U., Hong J.H., Park C., Nguyen M.D., Nagata K., Yoo J.Y.. et al. (2015) Disrupted-in-schizophrenia 1 (DISC1) regulates dysbindin function by enhancing its stability. J. Biol. Chem., 290, 7087–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ottis P., Bader V., Trossbach S.V., Kretzschmar H., Michel M., Leliveld S.R., Korth C. (2011) Convergence of two independent mental disease genes on the protein level: recruitment of dysbindin to cell-invasive disrupted-in-schizophrenia 1 aggresomes. Biol. Psychiatry, 70, 604–610. [DOI] [PubMed] [Google Scholar]
- 36. Nicodemus K.K., Kolachana B.S., Vakkalanka R., Straub R.E., Giegling I., Egan M.F., Rujescu D., Weinberger D.R. (2007) Evidence for statistical epistasis between catechol-O-methyltransferase (COMT) and polymorphisms in RGS4, G72 (DAOA), GRM3, and DISC1: influence on risk of schizophrenia. Hum. Genet., 120, 889–906. [DOI] [PubMed] [Google Scholar]
- 37. Papaleo F., Burdick M.C., Callicott J.H., Weinberger D.R. (2014) COMT-Dysbindin epistatic interaction. Mol. Psychiatry, 19, 273.. [DOI] [PubMed] [Google Scholar]
- 38. Leung C.L., Sun D., Zheng M., Knowles D.R., Liem R.K. (1999) Microtubule actin cross-linking factor (MACF) a Hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J. Cell Biol., 147, 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fradkin L.G., Baines R.A., van der Plas M.C., Noordermeer J.N. (2008) The dystrophin Dp186 isoform regulates neurotransmitter release at a central synapse in Drosophila. J. Neurosci., 28, 5105–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shivalkar M., Giniger E. (2012) Control of dendritic morphogenesis by Trio in Drosophila melanogaster. PloS One, 7, e33737.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Awasaki T., Saito M., Sone M., Suzuki E., Sakai R., Ito K., Hama C. (2000) The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron, 26, 119–131. [DOI] [PubMed] [Google Scholar]
- 42. Lee S., Nahm M., Lee M., Kwon M., Kim E., Zadeh A.D., Cao H., Kim H.J., Lee Z.H., Oh S.B.. et al. (2007) The F-actin-microtubule crosslinker Shot is a platform for Krasavietz-mediated translational regulation of midline axon repulsion. Development, 134, 1767–1777. [DOI] [PubMed] [Google Scholar]
- 43. Ferrier A., Sato T., De Repentigny Y., Gibeault S., Bhanot K., O'Meara R.W., Lynch-Godrei A., Kornfeld S.F., Young K.G., Kothary R. (2014) Transgenic expression of neuronal dystonin isoform 2 partially rescues the disease phenotype of the dystonia musculorum mouse model of hereditary sensory autonomic neuropathy VI. Hum. Mol. Genet., 23, 2694–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Purcell S.M., Moran J.L., Fromer M., Ruderfer D., Solovieff N., Roussos P., O'Dushlaine C., Chambert K., Bergen S.E., Kahler A.. et al. (2014) A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 506, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shen S., Lang B., Nakamoto C., Zhang F., Pu J., Kuan S.L., Chatzi C., He S., Mackie I., Brandon N.J.. et al. (2008) Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J. Neurosci., 28, 10893–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomson P.A., Malavasi E.L., Grunewald E., Soares D.C., Borkowska M., Millar J.K. (2013) DISC1 genetics, biology and psychiatric illness. Front. Biol., 8, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sawamura N., Ando T., Maruyama Y., Fujimuro M., Mochizuki H., Honjo K., Shimoda M., Toda H., Sawamura-Yamamoto T., Makuch L.A.. et al. (2008) Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol. Psychiatry, 13, 1138–1148. 1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jaaro-Peled H., Altimus C., LeGates T., Cash-Padgett T., Zoubovsky S., Hikida T., Ishizuka K., Hattar S., Mongrain V., Sawa A. (2016) Abnormal wake/sleep pattern in a novel gain-of-function model of DISC1. Neurosci. Res., 112, 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eykelenboom J.E., Briggs G.J., Bradshaw N.J., Soares D.C., Ogawa F., Christie S., Malavasi E.L., Makedonopoulou P., Mackie S., Malloy M.P.. et al. (2012) A t(1;11) translocation linked to schizophrenia and affective disorders gives rise to aberrant chimeric DISC1 transcripts that encode structurally altered, deleterious mitochondrial proteins. Hum. Mol. Genet., 21, 3374–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trossbach S.V., Fehsel K., Henning U., Winterer G., Luckhaus C., Schable S., Silva M.A., Korth C. (2014) Peripheral DISC1 protein levels as a trait marker for schizophrenia and modulating effects of nicotine. Behav. Brain Res., 275, 176–182. [DOI] [PubMed] [Google Scholar]
- 51. Ratta-Apha W., Hishimoto A., Mouri K., Shiroiwa K., Sasada T., Yoshida M., Supriyanto I., Ueno Y., Asano M., Shirakawa O.. et al. (2013) Association analysis of the DISC1 gene with schizophrenia in the Japanese population and DISC1 immunoreactivity in the postmortem brain. Neurosci. Res., 77, 222–227. [DOI] [PubMed] [Google Scholar]
- 52. Kim J.Y., Liu C.Y., Zhang F., Duan X., Wen Z., Song J., Feighery E., Lu B., Rujescu D., St Clair D.. et al. (2012) Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell, 148, 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van der Plas M.C., Pilgram G.S., Plomp J.J., de Jong A., Fradkin L.G., Noordermeer J.N. (2006) Dystrophin is required for appropriate retrograde control of neurotransmitter release at the Drosophila neuromuscular junction. J. Neurosci., 26, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Culligan K.G., Mackey A.J., Finn D.M., Maguire P.B., Ohlendieck K. (1998) Role of dystrophin isoforms and associated proteins in muscular dystrophy (review). Intl J. Mol. Med., 2, 639–648. [DOI] [PubMed] [Google Scholar]
- 55. Bateman J., Van Vactor D. (2001) The Trio family of guanine-nucleotide-exchange factors: regulators of axon guidance. J. Cell Sci., 114, 1973–1980. [DOI] [PubMed] [Google Scholar]
- 56. O'Brien S.P., Seipel K., Medley Q.G., Bronson R., Segal R., Streuli M. (2000) Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc. Natl Acad. Sci. U.S.A., 97, 12074–12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Youn H., Ji I., Ji H.P., Markesbery W.R., Ji T.H. (2007) Under-expression of Kalirin-7 Increases iNOS activity in cultured cells and correlates to elevated iNOS activity in Alzheimer's disease hippocampus. J. Alzheimers Dis., 12, 271–281. [DOI] [PubMed] [Google Scholar]
- 58. Kushima I., Nakamura Y., Aleksic B., Ikeda M., Ito Y., Shiino T., Okochi T., Fukuo Y., Ujike H., Suzuki M.. et al. (2012) Resequencing and association analysis of the KALRN and EPHB1 genes and their contribution to schizophrenia susceptibility. Schizophr. Bull., 38, 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brown A., Dalpe G., Mathieu M., Kothary R. (1995) Cloning and characterization of the neural isoforms of human dystonin. Genomics, 29, 777–780. [DOI] [PubMed] [Google Scholar]
- 60. Costas J., Suarez-Rama J.J., Carrera N., Paz E., Paramo M., Agra S., Brenlla J., Ramos-Rios R., Arrojo M. (2013) Role of DISC1 interacting proteins in schizophrenia risk from genome-wide analysis of missense SNPs. Annals of Human Genetics, 77,504–512. [DOI] [PubMed] [Google Scholar]
- 61. Huang da W., Sherman B.T., Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocol., 4, 44–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.