Abstract
Epidemiologic studies support that at least part of the risk of chronic diseases in childhood and even adulthood may have an in utero origin, and the placenta is a key organ that plays a pivotal role in fetal growth and development. The transcriptomes of 159 human placenta tissues were profiled by genome-wide RNA sequencing (Illumina High-Seq 2500), and linked to fetal genotypes assessed by a high density single nucleotide polymorphism (SNP) genotyping array (Illumina MegaEx). Expression quantitative trait loci (eQTLs) across all annotated transcripts were mapped and examined for enrichment for disease susceptibility loci annotated in the genome-wide association studies (GWAS) catalog. We discovered 3218 cis- and 35 trans-eQTLs at ≤10% false discovery rate in human placentas. Among the 16 439 known disease loci of genome-wide significance, 835 were placental eSNPs (enrichment fold = 1.68, P = 7.41e−42). Stronger effect sizes were observed between GWAS SNPs and gene expression in placentas than what has been reported in other tissues, such as the correlation between asthma risk allele, rs7216389-T and Gasdermin-B (GSDMB) in placenta (r2=27%) versus lung (r2=6%). Finally, our results suggest the placental eQTLs may mediate the function of GWAS loci on postnatal disease susceptibility. Results suggest that transcripts in placenta are under tight genetic control, and that placental gene networks may influence postnatal risk of multiple human diseases lending support for the Developmental Origins of Health and Disease.
Introduction
There is increasing recognition that the period of intrauterine development constitutes one of the most critical periods that can define disease risk later in life (1,2). The fetus is most susceptible to maternal or environmental influences during cellular proliferation, specification and differentiation stages, and relies closely on the placenta to assure this occurs appropriately. The placenta, situated at the maternal–fetal interface, is a key organ for fetal growth and development through a variety of functions including controlling fetal access to nutrients, hormone production and mitigation of adverse effects from the environment. Thus, the placenta plays a pivotal role in the healthy development of multiple organ systems of the fetus, which may ultimately influence postnatal disease susceptibility. Although the human fetus is well protected by the placenta, perturbation of the intrauterine environment by exogenous chemicals, maternal nutritional imbalances, stress and certainly genetic polymorphisms can interfere with regulatory pathways to disrupt placental and fetal development. Placental dysfunction has been associated with postnatal chronic diseases such as coronary heart disease, sudden cardiac death, type 2 diabetes, and cancers (2–5). To date, the placental transcriptome profile and its regulatory control, which are keys to understanding the link between placenta and adulthood diseases, have not been thoroughly studied.
Recent genome-wide association studies (GWAS) have identified loci that harbor susceptibility genes for many chronic diseases and phenotypes; however, the biologic implications of the identified variants remain unclear (6). First, many of the most significant GWAS hits are in loci with unknown function and have not previously been considered biologically plausible candidates for disease pathogenesis. Extensive linkage disequilibrium (LD) within many of these associated loci makes it difficult to identify the casual susceptibility variant, let alone which genes or proteins they influence. Second, the vast majority of GWAS hits can only explain a relatively small proportion of the variability of the phenotype in human populations (7,8). Some of the missing heritability may be owing to the limited power of GWAS to detect small-to-moderate effect sizes. Third and most importantly, genetic polymorphisms are static across all tissue types so that GWAS results alone cannot reveal the tissue/organ or stage of development in which SNPs are functional in modifying disease risk or phenotypes. Recent advances in integrative genomics, including expression quantitative trait loci (eQTL) analysis, provide tools to address these limitations by identifying true functional genes and variants and the target site/stage of relevance, with improved statistical power. eQTLs characterize the function of genetic variants at the transcriptome level by capturing the association between DNA single nucleotide polymorphisms (SNPs) and RNA transcript levels (9,10). By using gene expression as a phenotype and examining how DNA polymorphisms contribute to gene expression, true functional relationships can be discovered (9,10). The subsequent mapping of GWAS SNPs among the identified eQTLs becomes a powerful tool to empirically link functional variants and their downstream genes to disease traits. Finally, as the variability in expression owing to DNA polymorphisms can vary by tissue (11), evaluating tissue-specific eQTLs and their associations with disease traits can highlight relevant target sites of specific functional variants, further elucidating the molecular etiology of complex diseases. To date, systematic efforts, for example GTEx (12,13) and STARNET (14), have documented eQTLs of large number of adult tissues. However, eQTLs in placenta tissues are still understudied. Given the important role of the placenta in early development and growing recognition of the importance of the placenta in fetal programming, identifying placental eQTLs and their role in postnatal disease risk has the potential to provide early screening and interventional entry-points, when they may be most effective. To our knowledge, this article is the first to report large-scale placenta eQTL profiling and provides an application of leveraging the generated profile to study the potential in utero origins of post-natal diseases.
Taking advantage of an existing biorepository of placenta tissues from the Rhode Island Child Health Study (RICHS), we systematically profiled the Genome-wide transcriptome by RNAseq and genetic polymorphisms by Genome-wide SNP array in placenta tissues and characterized the placental eQTLs. Leveraging a large dataset of GWAS (see Materials and Methods) (6), we carried out enrichment analyses to explore possible biological functions or disease associations of eQTLs, GWAS-eQTL co-localization, and pathway enrichment.
Results
Genome-wide mapping of eQTL in human placentas
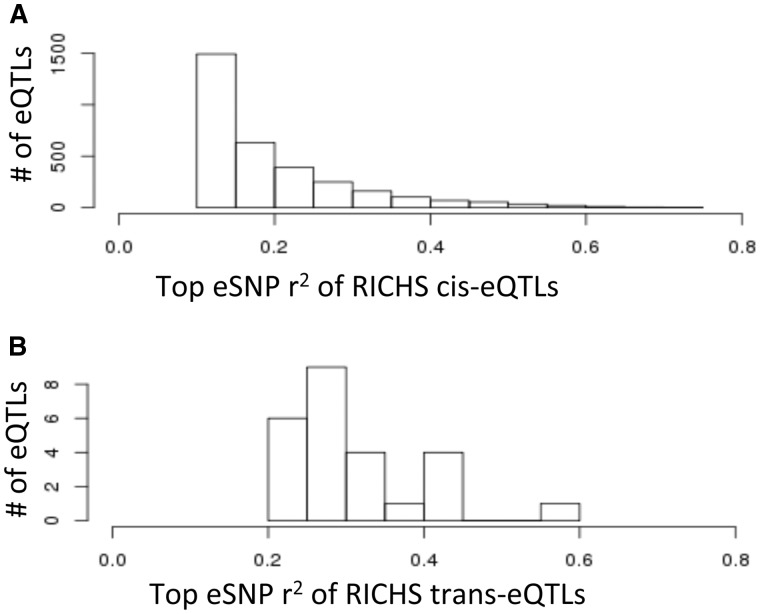
Single-end RNAseq reads were generated using the Illumina HighSeq 2500 platform. Genome-wide genotyping was performed using the Illumina MegaEx SNP array followed by genotype imputation using the Haplotype Reference Consortium (HRC) reference panel (15,16). We identified cis- and trans-acting eQTLs and quantified the false discovery rate (FDR) through permutation using established methods (9,17), considering all association signals from SNPs within 500 kb up and downstream of the transcription start site as a single cis-eQTL. For simplicity, we restricted eQTL associations so that each transcript could have no more than one cis-eQTL. Trans-eQTLs were defined as association signals from SNPs located greater than 500 kb from the transcript, or on a different chromosome. The top eSNP was defined as the SNP that was most significantly associated (i.e. having the lowest P value) with the expression levels of the corresponding gene. A summary of the eQTLs identified at a 10% FDR are reported in Supplementary Material, Table S1. Based on 159 placentas, we identified 3218 and 35 cis- and trans-eQTLs, respectively (<10% FDR). Given the modest sample size, we can detect eQTLs with moderate-to-large effect sizes, with a median r2 (gene expression variation explained by the eQTL genotypes) of 15.6 and 28.3% for cis- and trans-eQTLs, respectively (Fig. 1). Consistent to previous human eQTL studies, we capture far more cis- than trans-eQTLs because trans-eQTL detection has heavy multiple testing burden (9,18). Only strong trans-eQTLs passed the FDR threshold, thus the average r2 of trans-eQTL were substantial (Fig. 1B).
Figure 1.
Histogram of the top eSNP’s r2 values for expressional quantitative trait loci (fraction of gene expression level explained by the top eSNP) for placenta cis-eQTLs (upper) and trans-eQTLs (lower).
Placental eQTLs are enriched for disease-associated GWAS SNPs
One of the primary utilities of eQTLs is to assign functional or health relevance to GWAS findings. We surveyed the GWAS catalog (6,19,20) across all diseases and traits included in the catalog. In brief, we first retrieved the list of disease-associated SNPs (GWAS hits) from the GWAS catalog. The entries of GWAS catalog were curated using stringent thresholds (1 < e−5) based on the strict guidelines of significance levels for replication (6,19,20). Then without further filtering, we examined whether the 16 439 unique SNPs in GWAS catalog were enriched in placental eQTLs. Among 16 439 unique loci associated with any disease(s)/trait(s) in the catalog, 835 were placental eSNPs (enrichment fold = 1.68, P = 7.41e−42). In other words, a significant proportion of SNPs associated with these diseases were also functioning as eQTLs in placental tissues. Then we employed a data-driven approach to determine which disease(s)/trait(s) were most strongly enriched for placental eQTLs. This approach implicated multiple immune disorders [e.g. asthma and Crohn’s disease (CD)], growth and development (e.g. height) and lipid traits (e.g. LDL- and HDL-cholesterol and triglycerides). For example, among 137 known HDL GWAS SNPs, 17 are eSNPs found in placentae (enrichment fold = 2.68, P = 5.80e−4). Among 52 known asthma GWAS SNPs, 12 are eSNPs found in placentae (enrichment fold = 5.67, P = 8.58e−6). Finally, among 382 GWAS loci linked to inflammatory bowel disease, 34 are placental eSNPs (enrichment fold = 1.86, P = 0.00136).
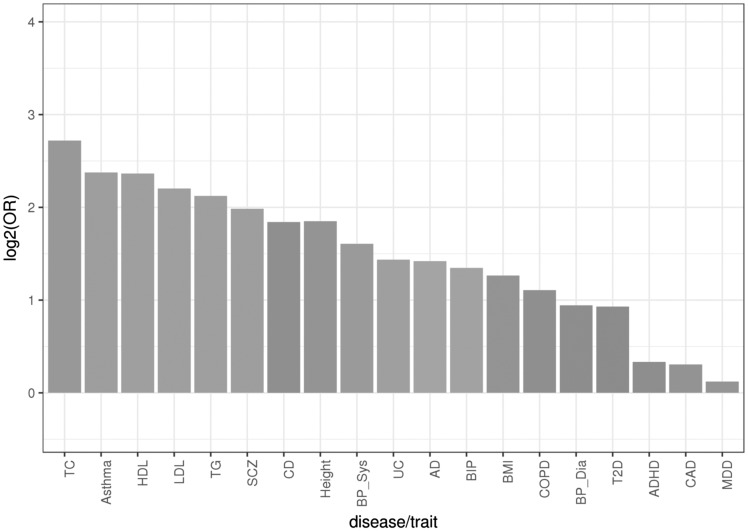
The GWAS catalog and most publications only report the top signals (e.g. P < 1e−5). While most existing GWAS only have moderate statistical power to detect moderate to small effects or suggestive P value (e.g. P < 1e−3), eQTL provides an alternative venue to identify novel susceptibility genes and pathways; however, full GWAS datasets are required instead of top hits. Herein, we leveraged large scale, full GWAS datasets on 19 traits (see Materials and Methods) for integrative analyses with the placental eQTL data (Fig. 2). The substantial overlap between GWAS signals (GWAS P ≤ 1e−3) and placental eQTLs suggests eSNPs of moderate effect size may be discovered at a less stringent P value threshold. This data-driven approach allows us to utilize both the genome-wide significant and suggestive GWAS signals to determine which disease(s)/trait(s) was most strongly enrichment for placental eQTLs. Disease-SNPs from GWAS were strongly enriched for placenta eQTLs, including psychiatric and neurological diseases, immune disease and metabolic disorders. Sixteen out of 19 GWAS results were enriched in placenta eQTLs (at Bonferroni corrected alpha level of 0.01), including total cholesterol, asthma, high-density lipoprotein, low-density lipoprotein, triglycerides, psychiatric schizophrenia, CD, body height, systolic blood pressure, ulcerative colitis, Alzheimer’s disease, psychiatric bipolar disorder, body mass index, chronic obstructive pulmonary disease, diastolic blood pressure and type 2 diabetes (Supplementary Material, Table S4). Schizophrenia, asthma, CD and lipid level are among the most enriched traits, suggesting the genetic risk loci is already active prenatally, as reflected in placenta. In contrast, the GWAS signal of coronary artery disease and major depressive disorder showed very little enrichment among placenta eQTLs (Fig. 2).
Figure 2.
We retrieved full GWAS test statistics from public data bases (Materials and Methods). For each disease/trait, we filtered out the SNPs with P > 1e−3. The enrichment of GWAS SNPs in placenta eQTLs was quantified with odds ratio (OR). Positive Log2OR indicates the GWAS SNPs were enriched for placenta eQTLs, and negative Log2OR indicates GWAS SNPs were depleted for placenta expressional quantitative trait loci. The GWAS information is detailed in Supplementary Material, Table S5. TC, total cholesterol; Asthma; HDL, high-density lipoprotein; LDL, low-density lipoprotein; TG, triglycerides; SCZ, psychiatric schizophrenia; CD, Crohn’s disease; Height, body height; BP_Sys, systolic blood pressure; UC, ulcerative colitis; AD, Alzheimer’s disease; BIP, psychiatric bipolar disorder; BMI, body mass index; COPD, chronic obstructive pulmonary disease; BP_Dia diastolic blood pressure; T2D, type 2 diabetes; ADHD, psychiatric attention-deficit hyperactivity disorder; CAD, coronary artery disease; MDD, psychiatric major depressive disorder.
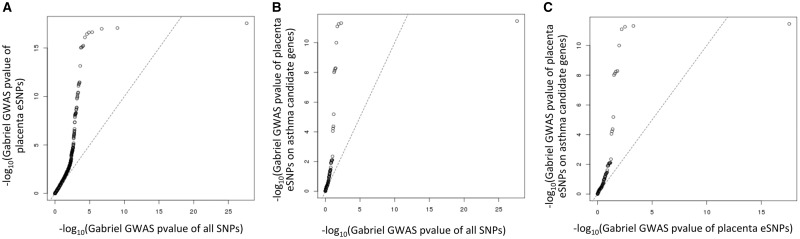
To demonstrate the validity of placental eQTLs, we focused our investigation on one disease, asthma, to investigate whether the known asthma candidate genes were controlled by placenta eQTLs. Among the 567 589 SNPs investigated in the asthma meta-GWAS (GABRIEL study) (7), 13 435 were identified as eSNPs with <10% FDR in placenta. It should be noted that we did not restrict the analysis to the top eSNP for each eQTL, but rather included all eSNPs corresponding to any given eQTL as long as their P values survived the 10% FDR threshold. The 13 435 eSNPs were significantly enriched in the GWAS hits identified in the GABRIEL study (P < 2.2e−16; Fig. 3A). This is consistent with a previous report showing that SNPs associated with complex traits are more likely to be eQTLs (18). Furthermore, we interrogated well documented asthma candidate genes (21) and found that airway eSNPs in these candidate genes were substantially enriched for small GWAS P values (Fig. 3B and C). In other words, known asthma candidate genes were more likely to be influenced by placenta eQTLs than random chance.
Figure 3.
Among all SNPs surveyed by the GABRIEL asthma meta-GWAS, 13 435 were eSNPs in the placenta at 10% FDR. (A) GABRIEL meta-GWAS P values of the 13 435 eSNPs plotted against P values for all SNPs investigated in the GABRIEL study, (B) GABRIEL meta-GWAS P values for the eSNPs in asthma candidate genes plotted against meta-GWAS P values for all SNPs; (C) GABRIEL meta-GWAS P values for the eSNPs in asthma candidate genes plotted against GABRIEL meta-GWAS P values for the 13 435 eSNPs.
Co-localization of GWAS lead SNPs and placenta eSNPs
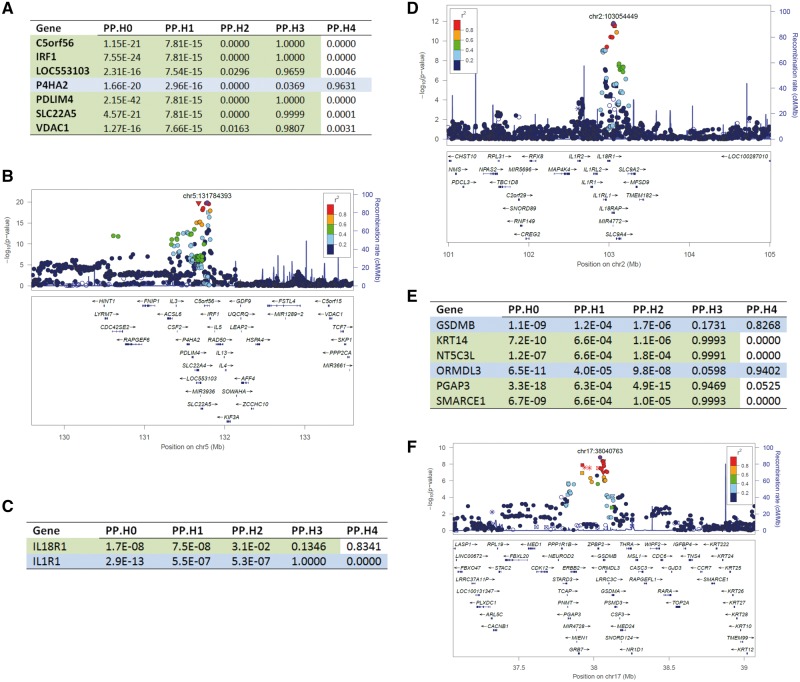
As shown in Figure 2, several disease GWAS SNPs (e.g. CD and asthma) strongly overlapped placenta eQTLs. That is the same SNP is associated with disease risk (e.g. GWAS P < 5e−8) and placenta gene expression (e.g. eQTL FDR ≤10%). However, given that neighboring SNPs were often in tight LD, the overlap of GWAS signals and eSNPs do not guarantee that the disease risk and placenta gene expression variability is caused by the same variant. Recently developed methods allow more advanced integration of GWAS and eQTL results to co-localize GWAS and eQTL signals and identify functional genes and SNPs underlying biologic traits and diseases (22). Herein, we used CD and asthma GWAS loci (11,23) as examples to demonstrate the co-localization of disease risk and placenta eQTLs. For each locus, the co-localization methods (22) evaluated five hypotheses (see Materials and Methods), where we were interested in Hypothesis 3 (H3: disease risk and gene expression alternation were caused by two independent SNPs in the locus), and Hypothesis 4 (H4: disease risk and gene expression alternation were caused by the same SNP in the locus). For example, among the 42 genes where we observed an influence on expression by CD-associated eSNPs (P ≤ 5e−8) (23), we were able to resolve the co-localization of the SNPs that control gene expression and the SNPs that control disease risk. The expression levels of nine genes (ERAP2, P4HA2, ORMDL3, SCAMP3, ARHGEF2, IL18R1, GSDMB, ZNF300P1 and YY1AP1) were controlled by SNPs that were also functional to CD susceptibility (Supplementary Material, Table S2). In contrast, the empirical data suggested variability in the expression level of 33 genes were caused by different SNPs that reside in the same locus as the CD functional SNPs (Supplementary Material, Table S3). Leveraging placental eQTLs, we show that in this locus only P4HA2 is influenced by the CD functional variant (Fig. 4A), and many nearby candidate genes (Fig. 4B) were also influenced by genetic variants in this locus but different from the CD functional variant (Fig. 4A and B). Additionally, we were able to resolve whether CD risk and gene expression variability were driven by the same functional variants for the IL1R1-IL18R1 locus and the GSDM-ORMDL3 locus. The placenta eQTLs indicated that IL18R1 expression level was controlled by the same variant that controlled CD risk, but not ILR1 (Fig. 4C and D). Further, both GSDM and ORMDL3 expression levels were driven by the same functional variant that influenced CD risk (Fig. 4E and F). Leveraging RICHS placenta data, we found their expression level were also driven by the asthma functional variants (Fig. 4G and H), while the expression of KRT14, NT5C3L, SMARCE1 and PGAP3 were not directly influenced by asthma functional variants.
Figure 4.
Co-localization of GWAS and eQTL. The Co-localization algorithms evaluate five hypotheses: PP.H0: No association with either disease risk (i.e. trait 1) or placenta gene expression (i.e. trait 2); PP.H1: Association with trait 1, not with trait 2; PP.H2: Association with trait 2, not with trait 1; PP.H3: Association with trait 1 and trait 2, two independent SNPs; PP.H4: Association with trait 1 and trait 2, one shared SNP. (A) Crohn’s Disease (CD) and placenta eQTL COLOC posterior probability of P4HA2 locus. P4HA2 gene showed large H4 posterior probability, indicating the expression level of P4HA2 was driven by the same genetic variants that caused CD risk. Other genes in this locus showed large H3 posterior probability indicating these genes’ expression levels were influenced by variants in this locus but not that caused CD risk. (B) –log10(P) of the CD GWAS at P4HA2 locus. (C) CD and placenta eQTL COLOC posterior probability of IL18R1 locus. (D) –log10(P) of the CD GWAS at IL18R1 locus. (E) CD and placenta eQTL COLOC posterior probability of GSDMB locus. (F) –log10(P) of the CD GWAS at GSDMB locus. (G) Asthma GWAS and placenta eQTL COLOC posterior probability of GSDMB locus. (H) –log10(P) of the asthma GWAS at GSDMB locus.
Pathway enrichment analysis for biological functions using eQTL
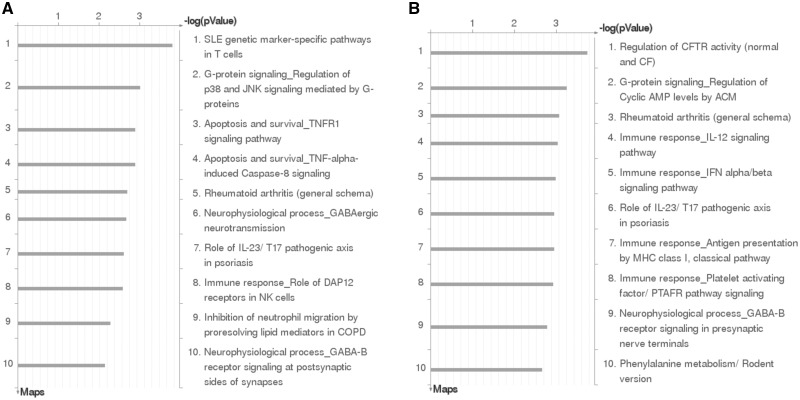
Complex diseases are believed to be polygenic, meaning the genetic susceptibility is attributable to many genetic loci and each locus may only have a moderate effect size. However, as the genetic loci likely act on several common pathways that collectively contribute to disease etiology, the application of pathway and network analyses are powerful approaches that can reveal the common biological processes underlying complex diseases. Typically, such analyses are limited in scope to either focus on gene transcripts associated with disease or GWAS-identified disease-associated SNPs. By applying pathway enrichment analysis on the identified placental eQTLs, we are able to empirically bridge the link between GWAS loci, the associated variability in genes, perturbed tissue-specific biologic processes and ultimate disease. Herein, we showcase this added utility of eQTL enrichment analyses using Asthma and CD disease endpoints as examples. Herein, eQTLs serve as an empirical ‘bridge’ between GWAS SNPs and gene/transcripts under influence and disease, and enable us to empirically link GWAS loci to genes and carry out pathway enrichment test (see Materials and Methods). We intersected each SNP set with the placental eQTLs to identify genes influenced by the SNP set in placenta. In detail, 117 genes were affected by asthma-associated SNPs (GWAS P value ≤0.01), and 237 genes were affect by CD-associated SNPs (GWAS P value ≤0.01). Afterwards, the resulting genes were tested for enrichment in pathways and biological processes. Pathways related to immune function were enriched in both asthma and CD gene sets (Fig. 5). For example, asthma gene set were enriched for SLE genetic marker-specific pathways in T-cell and G-protein mediated P38 and JNK signaling (Fig. 5A); and CD gene set was enriched for IL-12 signaling pathway (Fig. 5B).
Figure 5.
Pathway enrichment of genes transcriptionally influenced by GWAS loci in placenta. (A) A total of 117 genes were influenced by asthma-associating SNPs; (B) 237 genes were influenced by Crohn’s disease-associating SNPs. The GWAS loci for each disease are SNPs retrieved from NHGRI catalog (6) combined with full GWAS results (Materials and Methods) filtered by P value cut-off.
Discussion
The intrauterine developmental period represents a critical window of susceptibility to a myriad of environmental exposures and conditions with potentially lifelong impacts on health and disease. The placenta is the first complex fetal organ to form during development. Once developed it serves as the source of fetal nutrients, water, gas exchange, excretion and immune regulation. These effects are modulated by simultaneous production of many pregnancy-related hormones, proteins and growth factors thereby fulfilling a critical role in proper intrauterine development. In order to perform this multitude of functions, the placenta must regulate a complex series of molecular pathways. These genes and pathways can be perturbed by environmental insults or genetic polymorphisms and such alterations can lead to important effects on fetal growth and health outcome.
In this study, we took a systematic approach to conduct a large scale population-based study to assess the genetically influenced placental gene expression profile in tissues collected from clinically normal pregnancies free of pathological complications. The large number of placental expressional QTLs discovered in placenta tissue (Supplementary Material, Table S1, 3218 cis- and 35 trans-eQTLs at 10% FDR) was comparable to liver and adipose tissue sets of the same sample size (14) demonstrating that the placenta processes a very active transcriptome under strong genetic influence. In detail, we down-sampled STARNET data (14) to a similar sample size as our placenta set, and found 4467 and 3641 cis-eQTLs in liver and adipose tissue, respectively. It should be noted the moderately more eQTLs in liver and adipose could also owing to difference in experiment protocols, where STARNET data have deeper RNA seq and longer reads (100 bp) than RICHS placenta data (50 bp reads). We assessed the overlap between the placental eQTLs identified in our study and the eQTLs identified by the Genotype-Tissue Expression (GTEx) project (13), which queried eQTLs across various human post-mortem tissues. We observed significant overlap between the placental eQTLs identified in our study and GTEx-identified eQTLs across multiple tissues/organs, with the strongest overlaps between placenta and fibroblast and the lowest overlap between placenta and testis (Supplementary Material, Fig. S1), indicating many eQTLs were shared among multiple tissues/organs including placenta. In addition, leveraging large GWAS databases, we queried the identified placental eQTLs against a global survey of human disease risk loci, and we provide evidence that GWAS loci associated with multiple diseases/traits actively influence placental gene transcription.
The enriched phenotypes among our placental eQTLs included growth/metabolism traits loci as well as immune-related disorders, including asthma and CD (Figs 2–4). In fact, regulating the maternal immune system to tolerate the genetically distinct fetus is a key function of the placenta to ensure a healthy pregnancy. These findings suggest that the underlying biologic pathways for these diseases are already active in placentas and lend support to placenta eQTLs as a valuable resource not only for fetal development research but also to study potential placental origins of postnatal diseases.
Finding eSNPs in placenta that share the same functional genetic variants with diseases can provide insights into the disease origins and mechanisms as showcased by our in-depth analyses of asthma and CD as disease endpoints. For example, we showed that the gene expression level of ORMDL3 and GSDMB were driven by the asthma functional variants. ORMDL3 and GSDMB have previously been suggested as the genes mediating rs7216389-T’s association with asthma (11,18), based on blood and lung tissue gene expression studies. Collectively, these results suggest that the asthma-associated SNP at 17q21 is already active in early development, i.e. prenatally. Consistent with previous findings in lung tissues, the rs7216389-T allele up-regulates the ORMDL3 and GSDMB genes in placentas. Interestingly, the effect size (r2, variance explained) of the rs7216389 eQTL is much higher in placenta compared with lung tissue (18). Specifically, rs7216389 explains 22.2 and 19.8% of expression variance of GSDMB and ORMDL3 in placenta, respectively. In contrast, contribution of rs7216389 eQTLs to GSDMB variations were intensively studied in lungs in multiple cohorts (Laval, Groningen, UBC) (18), and the top GSDMB eSNPs only had moderate r2 in the three cohorts (4.4, 9.2 and 4.6%) (18). Furthermore, GSDMB was not influenced by any eQTL in airway epithelium even at 20% FDR (24). Verlaan et al. (3) showed that SNPs in 17q21 demonstrated domain-wide cis-regulatory effects, suggesting long-range chromatin interactions. It was also shown that asthma risk alleles on 17q21 elevate IL-17 secretion in cord blood mononuclear cells (25). Clearly, the mechanism by which 17q21 SNPs modify asthma risk requires additional research, including multi-tissue analyses and evaluating different developmental stages (i.e. pre- and post-natal). For another example in CD study of PDLIM4-P4HA2 locus. It is noteworthy that the locus has been reported to be significantly associated with CD risk (P < 5e−8) (23) and several candidate genes in this locus were influenced by eQTLs and proposed to underlie CD etiology, including PDLIM4, SLC22A4, SLC22A5 and IRF1 (26) (Fig. 4A and B). For GSDM-ORMDL3 locus, previous reports also showed GSDM-ORMDL3 locus significantly associated with asthma risk (24), and our results showed both genes driven by the same eQTLs, suggesting both genes underlie CD molecular etiology.
The emergence of the Developmental Origins of Health and Disease (DOHaD) hypothesis proposes that the early environment, from the periconceptional period until early childhood, can predispose an individual to disease later in life (1,27). This hypothesis is supported by animal studies, and recently human data in cardiovascular disease, obesity, type 2 diabetes, osteoporosis, chronic obstructive pulmonary disease and depression (3–5,25,27). While the DOHaD hypothesis emphasizes the role of the in utero environment on shaping the developmental trajectory of the fetus and, thereby, subsequent health outcomes across the lifespan, our findings showcasing the influence of genetically controlled in utero expression patterns linked to later life health effects also support the hypothesis. By considering genetic association findings as providing insight into the effects of naturally occurring genetic perturbation on gene transcription, translation and downstream phenotypes (28), we were able to investigate the effect of many genes/loci genome-wide in vivo, which is often prohibitively time-consuming using other experimental approaches (e.g. RNAi). As such, our results infer the potential consequences of introducing variability in important genes/transcripts, where such variability could be caused by genetic variants and importantly also environmental exposure (e.g. nutrition or stress pregnancy). Therefore, the eQTL findings presented in this article provide valuable guidance to developmental origins research in terms of potential post-natal disease risk caused by in utero transcriptome disruption.
The power of the eQTL analysis is moderate in rejecting a particular null hypothesis (i.e. detecting a particular eQTL), given that the typical sample size of eQTL are in the hundreds. In contrast, a powerful GWAS meta-analysis employs tens or hundreds of thousands of individual to ensure the statistical power to identify a particular genetic susceptibility locus. Based on our power estimation, at α = 5e−4 (about the 10% FDR threshold for cis-eQTLs), we have 80% power to detect an eQTLs of r2 = 0.11 (11% of the expression level variance explained by eSNP), and only 25% power to detect an eQTLs of r2=0.05. While we discovered 3218 cis- and 35 trans-eQTLs at ≤10% FDR in this study, it is likely that we failed to capture many placenta eQTLs, especially the eQTLs with moderate effect size (e.g. eQTLs of r2 ≤ 0.05), which could be captured with larger studies or eQTL meta-analysis.
We acknowledge several limitations in our study. The placenta is a heterogeneous organ, consisting of various cell sub-types of both the fetal and maternal origin. To address this issue, placental specimens were exclusively biopsied from four quadrants on the fetal membrane side, free from maternal decidua, and within 2 cm from the cord insertion site, a region identified as the least variable in the placenta (29). This strategy ensured representative sampling of the fetal membrane side of the placenta (29). Also this strategy results in very low levels of maternally derived cells/tissues in our specimens. We are aware of additional limitations associated with this study. There is limited follow-up of the study subjects, therefore little postnatal outcome information were available to directly link placenta eQTLs and postnatal outcome in the same study population. Alternatively, we applied the latest statistical approaches and demonstrated such associations (i.e. genetic variants → placenta transcriptome changes → postnatal disease risk) in other independent studies, such as Gabriel Asthma GWAS. Nevertheless, future studies with postnatal following-up should carefully monitor post-natal trait outcomes. To fully define the causal roles, the next steps would be to consider modification of these genes and pathways in an experimental setting, and our results helps to spur this line of research.
In summary, this report is the first attempt to construct the architecture of genetic control of gene expression in human placenta. We conducted a systematic RNA sequencing study on the placenta whole-transcriptome and show that the placenta has an actively transcribed genome, which is profoundly influenced by genetic polymorphisms. Many placenta eQTLs had very large effect sizes, for example, the rs7216389 genotype explains 27.0% of the placental gene expression variance of GSDMB, which is strongly associated with asthma and CD. Immune and developmental disease genetic loci showed particularly high overlap and co-localization with placenta eQTLs, suggesting that placental gene expression and genetic control may play a role in the fetal origins of these traits/disorders. The functional inference that the genetic regulation of the placental transcriptome contributes post-natal disease risk will need to be confirmed in additional studies.
Materials and Methods
Datasets
Placenta tissues were collected as part of the Rhode Island Child Health Study (RICHS) (30). This population consists of singleton, term infants (≥37 weeks gestation) born without serious pregnancy complications or congenital or chromosomal abnormalities. Given an a priori interest to study fetal growth, the RICHS population was oversampled for both large for gestational age (LGA, >90% 2013 Fenton Growth Curve) and small for gestational age (SGA, <10% 2013 Fenton Growth Curve) infants. Written informed consent was obtained from all participants (n = 841). The study protocol was reviewed and approved by the Office of Human Research Protections registered Institutional Review Boards. RNAseq (Illumina HiSeq) and genotyping (Illumina MegaEX array) was performed on a subset of RICHS samples (n = 200) selected to be representative of the total cohort. Forty-one DNA samples failed genotype QC (sample call rate <90%), and the remaining N = 159 samples with high quality genotype and RNAseq data were subsequently used in eQTL computation. The demographic characteristics of the study samples were summarized in Table 1.
Table 1.
Demographic characteristics of the study population (n = 159)
| Variables | Mean | (Min, max) |
|---|---|---|
| Birth weight (g) | 3543 | (2030, 4940) |
| Gestational age (weeks) | 39.1 | (37.0, 41.0) |
| Maternal age (years) | 30.9 | (18.0, 40.0) |
| Maternal BMI (kg/m2) | 26.6 | (16.0, 46.7) |
| N | (%) | |
| Maternal ethnicity (self-report) | ||
| White | 123 | (77.4) |
| Black | 10 | (6.3) |
| Other | 23 | (16.3) |
| Unknown | 3 | (1.9) |
| Infant Ethnicity (genotype-inferred) | ||
| White | 142 | (89.3) |
| Black | 11 | (6.9) |
| Mixed | 6 | (3.8) |
| Infant gender | ||
| Female | 79 | (49.7) |
| Male | 80 | (50.3) |
Placental specimens were collected from four quadrants exclusively on the fetal membrane side and within 2 cm from the cord insertion site, a region identified as the least variable in the placenta (29). Approximately 10 g of villous tissue was obtained for nucleic acid extraction, immediately rinsed and stored in RNALater (Life Technologies, AM7024) and stored at 4 °C. Within 72 h, samples were snap-frozen in liquid nitrogen, homogenized and stored at -80 °C. Nucleic acids (e.g. genomic DNA and total RNA) were extracted from the samples stored in RNALater using the QIAmp DNA mini kit (Qiagen, #51306) and RNeasy mini kit (Qiagen, #74106), respectively, according to the manufacturer’s instruction (Qiagen, 80204). Genomic DNA and total RNA samples were quantified using a Nanodrop ND-1000 instrument (Thermo Fisher, CA, USA). RNA sequencing library prepared using RiboZero kit (Illumina, San Diego, CA, USA), followed with sequencing on Illumina High-Seq 2500 platform. On each sample, we generated about 20 million single-end RNAseq reads.
In parallel, we carried out genome wide SNP genotyping with Illumina MegaEX chip; 1 730 225 SNP was successfully genotyped and passed QC. The QC steps excluded SNPs with low call rate (<0.9), SNPs deviating from Hardy-Weinberg equilibrium (P < 1e−6). Also, rare SNPs (minor allele was observed less than 5 times in dataset) were excluded from the eQTL analysis.
The GWAS catalog table (version 1.0.1) was downloaded from NHGRI website (20), then without further filtering, we examined whether the 16 439 unique SNPs in GWAS catalog were enriched in placental eQTLs.
Data quality control and eQTLs construction
Recorded gender was also confirmed with computational inferred gender, based on expression of the Y linked gene RPSY41, and ChrX SNP heterozygosity rate. A total of 1 730 225 SNPs passed genotype QC. This dataset was then leveraged for genotype imputation based on the HRC reference (15,16). The reference panel contains individuals with predominantly European ancestry and the 1000 Genomes Project data. In total, 5 748 854 SNPs of high imputation quality and minor allele count (MAC) no <5 were utilized for the eQTL computation. Gene expression values were adjusted for gender and the top 10 transcriptome-derived principle components (derived from the gene expression data itself to control for experimental artifacts) and top 3 genotype-derived principal components in a robust linear model to accommodate potential outliers for expression level. The linear model residuals underwent inverse normal transformation before entering eQTL construction. We applied a previously described (9) method to identify cis- and trans-eQTLs. SNPs within 500 kb up and downstream of the transcription start site as a single cis-eQTL. For simplicity, we restricted eQTL associations so that each transcript could have no more than one cis-eQTL. Trans-eQTLs were defined as association signals from SNPs located greater than 500 kb from the transcript, or on a different chromosome. The top eSNP was defined as the SNP that was most significantly associated (i.e. having the lowest P value) with the expression levels of the corresponding gene.
Testing GWAS SNPs enrichment among placenta eQTLs
The GWAS catalog (6) was downloaded. SNPs reported in multiple studies for related traits were pooled by selecting the study reporting the smallest association P value. The SNPs were intersected with placenta eQTLs to identify the disease-associating SNPs that also influence gene expression in placenta (Supplementary Material, Table S1). Enrichment fold between eSNPs and GWAS SNPs were then computed with logistic regression, in which the outcome variable was the 10% FDR eQTL significance status of the SNP, and the regressors were indicator variables for each of the diseases/traits.
To measure enrichment for diseases/traits among placenta eQTLs, we also leveraged GWAS whose full test statistics are available. Specifically, we retrieved the statistics of the following studies, total cholesterol (10), asthma (GABRIEL consortium) (7), high-density lipoprotein (10), low-density lipoprotein (10), triglycerides (10), psychiatric schizophrenia (31), CD (23), body height (32), systolic blood pressure (33), ulcerative colitis (23), Alzheimer’s disease (34), psychiatric bipolar disorder (35), body mass index (36), chronic obstructive pulmonary disease (37), diastolic blood pressure (33), type 2 diabetes (38), psychiatric attention-deficit hyperactivity disorder (39), coronary artery disease (40) and psychiatric major depressive disorder (41). We filtered the full GWAS summary statistics with P value ≤1e−3 threshold, overlap with RICHS placenta eQTLs (≤10% FDR), and computed overlap odds ratio and enrichment P values (hyper-geometric test). The studies sample size and endpoints are summarized in Supplementary Material, Table S5.
Co-localization of GWAS top SNPs and placenta eSNPs
CD (23) and asthma (7) GWAS results were used in co-localization analysis, which is performed using COLOC version 2.3-6 in R (22). This method assesses whether two association signals, GWAS summary statistics and eQTL statistics, are consistent with shared functional variant(s) (22). Default priors of the software were used. We also filtered the data requiring the GWAS P ≤ 5e−8 and eSNP FDR ≤10% before running COLOC. In total, five hypotheses were evaluated. H0: No association with either disease risk (i.e. trait 1) or placenta gene expression (i.e. trait 2); H1: Association with trait 1, not with trait 2; H2: Association with trait 2, not with trait 1; H3: Association with trait 1 and trait 2, two independent SNPs; H4: Association with trait 1 and trait 2, one shared SNP. Genes that demonstrated a high posterior probability of hypothesis 4 (PP.H4 >75%) indicate the disease risk and placenta gene expression were controlled by the same genetic variant; and genes that demonstrated a high posterior probability of hypothesis 3 (PP.H3 >75%) indicate the disease risk and placenta gene expression were controlled by distinct genetic variant at the locus.
Pathway enrichment analyses
To further characterize the regulatory and pathway nature, enrichment analysis of the genes influenced by GWAS SNPs through eQTLs were performed for pathways and biological processes using the METACORE integrated software suite (https://portal.genego.com/; date last accessed July 13, 2017).
Supplementary Material
Supplementary Material is available here.
Conflict of Interest statement. None declared.
Supplementary Material
Funding
This work is supported by NIH-NIMH R01MH094609, NIH-NIEHS R01ES022223, NIH-NIDA 1R41DA042464-01 and NIH-NIEHS R01ES022223-03S1. Dr Ke Hao is partially supported by the National Natural Science Foundation of China (Grant No. 21477087, 91643201) and by the Ministry of Science and Technology of China (Grant No. 2016YFC0206507).
References
- 1. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc., 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li J., Liu S., Li S., Feng R., Na L., Chu X., Wu X., Niu Y., Sun Z., Han T.. et al. (2017) Prenatal exposure to famine and the development of hyperglycemia and type 2 diabetes in adulthood across consecutive generations: a population-based cohort study of families in Suihua, China. Am. J. Clin. Nutr., 105, 221–227. [DOI] [PubMed] [Google Scholar]
- 3. Barker D.J., Larsen G., Osmond C., Thornburg K.L., Kajantie E., Eriksson J.G. (2012) The placental origins of sudden cardiac death. Int. J. Epidemiol., 41, 1394–1399. [DOI] [PubMed] [Google Scholar]
- 4. Barker D.J., Thornburg K.L. (2013) Placental programming of chronic diseases, cancer and lifespan: a review. Placenta, 34, 841–845. [DOI] [PubMed] [Google Scholar]
- 5. Barker D.J., Thornburg K.L. (2013) The obstetric origins of health for a lifetime. Clin. Obstet. Gynecol., 56, 511–519. [DOI] [PubMed] [Google Scholar]
- 6. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L.. et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res., 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moffatt M.F., Gut I.G., Demenais F., Strachan D.P., Bouzigon E., Heath S., von Mutius E., Farrall M., Lathrop M., Cookson W.O. (2010) A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med., 363, 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A.. et al. (2009) Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S.. et al. (2008) Genetics of gene expression and its effect on disease. Nature, 452, 423–428. [DOI] [PubMed] [Google Scholar]
- 10. Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J.. et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E.. et al. (2007) Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature, 448, 470–473. [DOI] [PubMed] [Google Scholar]
- 12. Bahcall O.G. (2015) Human genetics: GTEx pilot quantifies eQTL variation across tissues and individuals. Nat. Rev. Genet., 16, 375.. [DOI] [PubMed] [Google Scholar]
- 13.(2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Franzen O., Ermel R., Cohain A., Akers N.K., Di Narzo A., Talukdar H.A., Foroughi-Asl H., Giambartolomei C., Fullard J.F., Sukhavasi K.. et al. (2016) Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science, 353, 827–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das S., Forer L., Schonherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M.. et al. (2016) Next-generation genotype imputation service and methods. Nat. Genet., 48, 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K.. et al. (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet., 48, 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Di Narzo A., Cheng H., Lu J., Hao K. (2014) Meta-eQTL: a tool set for flexible eQTL meta-analysis. BMC Bioinformatics, 15, 392.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hao K., Bosse Y., Nickle D.C., Pare P.D., Postma D.S., Laviolette M., Sandford A., Hackett T.L., Daley D., Hogg J.C.. et al. (2012) Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet., 8, e1003029.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A., 106, 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., Boehm F., Caporaso N.E., Cornelis M.C., Edenberg H.J.. et al. (2010) Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol., 34, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bosse Y., Hudson T.J. (2007) Toward a comprehensive set of asthma susceptibility genes. Annu. Rev. Med., 58, 171–184. [DOI] [PubMed] [Google Scholar]
- 22. Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet., 10, e1004383.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A.. et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature, 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Myatt L. (2006) Placental adaptive responses and fetal programming. J. Physiol., 572, 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lluis A., Schedel M., Liu J., Illi S., Depner M., von Mutius E., Kabesch M., Schaub B. (2011) Asthma-associated polymorphisms in 17q21 influence cord blood ORMDL3 and GSDMA gene expression and IL-17 secretion. J. Allergy Clin. Immunol., 127, 1587–1594.e6. [DOI] [PubMed] [Google Scholar]
- 26. Ning K., Gettler K., Zhang W., Ng S.M., Bowen B.M., Hyams J., Stephens M.C., Kugathasan S., Denson L.A., Schadt E.E.. et al. (2015) Improved integrative framework combining association data with gene expression features to prioritize Crohn's disease genes. Hum. Mol. Genet., 24, 4147–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verlaan D.J., Berlivet S., Hunninghake G.M., Madore A.M., Lariviere M., Moussette S., Grundberg E., Kwan T., Ouimet M., Ge B.. et al. (2009) Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am. J. Hum. Genet., 85, 377–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang L., Kim S. (2014) Learning gene networks under SNP perturbations using eQTL datasets. PLoS Comput. Biol., 10, e1003420.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wyatt S.M., Kraus F.T., Roh C.R., Elchalal U., Nelson D.M., Sadovsky Y. (2005) The correlation between sampling site and gene expression in the term human placenta. Placenta, 26, 372–379. [DOI] [PubMed] [Google Scholar]
- 30. Paquette A.G., Lester B.M., Koestler D.C., Lesseur C., Armstrong D.A., Marsit C.J. (2014) Placental FKBP5 genetic and epigenetic variation is associated with infant neurobehavioral outcomes in the RICHS cohort. PLoS One, 9, e104913.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z.. et al. (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet., 46, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J.. et al. (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature, 478, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B.. et al. (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet., 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.(2011) Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet., 43, 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J.. et al. (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature, 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cho M.H., McDonald M.L., Zhou X., Mattheisen M., Castaldi P.J., Hersh C.P., Demeo D.L., Sylvia J.S., Ziniti J., Laird N.M.. et al. (2014) Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir. Med., 2, 214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A.. et al. (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet., 44, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neale B.M., Medland S.E., Ripke S., Asherson P., Franke B., Lesch K.P., Faraone S.V., Nguyen T.T., Schafer H., Holmans P.. et al. (2010) Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J. Am. Acad. Child Adolesc. Psychiatry, 49, 884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C.. et al. (2011) Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Na. Genet., 43, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ripke S., Wray N.R., Lewis C.M., Hamilton S.P., Weissman M.M., Breen G., Byrne E.M., Blackwood D.H., Boomsma D.I., Cichon S.. et al. (2013) A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry, 18, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.