Abstract
Genital malformations are among the most common human birth defects, and both genetic and environmental factors can contribute to these malformations. Development of the external genitalia in mammals relies on complex signaling networks, and disruption of these signaling pathways can lead to genital defects. Islet-1 (ISL1), a member of the LIM/Homeobox family of transcription factors, has been identified as a major susceptibility gene for classic bladder exstrophy in humans, a common form of the bladder exstrophy-epispadias complex (BEEC), and is implicated in a role in urinary tract development. We report that deletion of Isl1 from the genital mesenchyme in mice led to hypoplasia of the genital tubercle and prepuce, with an ectopic urethral opening and epispadias-like phenotype. These mice also developed hydroureter and hydronephrosis. Identification of ISL1 transcriptional targets via ChIP-Seq and expression analyses revealed that Isl1 regulates several important signaling pathways during embryonic genital development, including the BMP, WNT, and FGF cascades. An essential function of Isl1 during development of the external genitalia is to induce Bmp4-mediated apoptosis in the genital mesenchyme. Together, these studies demonstrate that Isl1 plays a critical role during development of the external genitalia and forms the basis for a greater understanding of the molecular mechanisms underlying the pathogenesis of BEEC and urinary tract defects in humans.
Introduction
Malformations of the urogenital tract are among the most common congenital anomalies in humans. Hypospadias, which is characterized by an ectopic urethral meatus on the ventral surface of the penis, is the second most common birth defect in boys and is estimated to affect 1 in every 200–300 live male births (1,2). Other rarer genital anomalies, such as those represented within the bladder exstrophy-epispadias complex (BEEC), range from 1 in 117, 000 in males and 1 in 484, 000 in females for the mildest epispadias phenotype, to 1 in 30, 000–50, 000 for the more common classic bladder exstrophy (CBE) (3–5). Surgery can help to correct these defects but is frequently unable to completely restore normal function, leading to medical and psychosocial complications (6–8). Because the underlying cause of many genital anomalies in human patients remains unknown, an increased understanding of the biological and molecular mechanisms that control urogenital development will be important for improved diagnosis and clinical management.
Recently, Islet-1 (ISL1), a member of the LIM/Homeodomain (LHX) family of transcription factors (9), was identified as a major susceptibility gene for classic bladder exstrophy in humans. Genome-wide association studies were conducted in cohorts of 110 and 268 CBE patients, revealing that ISL1 resides within the CBE locus (10,11). Furthermore, studies in animal models showed that Isl1-expressing cells contribute to several tissues in the urinary tract, and that Isl1 plays a role in its embryonic development (11). Isl1 is involved in the regulation of many cell types and organs, and its importance in development was demonstrated by early lethality (E9.5) of embryos lacking Isl1 (12). Isl1 has numerous roles that include the control of motor neuron and interneuron specification (12) as well as development of the pituitary (13), pancreas (14), heart (15), and hindlimb (16,17). In addition, removal of Isl1 from the mesenchyme surrounding the ureteric bud results in ectopic budding and abnormal formation of the bladder-ureter connection (18).
Lineage tracing using an inducible Isl1Cre allele showed that Isl1-expressing cells also contribute to the dorsal genital tubercle (GT), the anlage to the external genitalia, although in situ hybridization shows a broader pattern of expression in the embryonic GT (11,19). Expression of Isl1 in the genital mesenchyme, together with previous reports of urogenital abnormalities in Isl1 mutant mice (11,18), led us to investigate the role of Isl1 during embryonic development of the external genitalia. We found that Isl1 is required for proper embryonic GT development and present evidence that ISL1 directly regulates factors that are critical for GT formation, including Bmp4, Fgf10, and Wnt5a. Our results indicate that a critical function of Isl1 is to regulate BMP4-mediated apoptosis in the mesenchymal compartment.
Results
Isl1 shows dynamic patterns of expression in the developing external genitalia
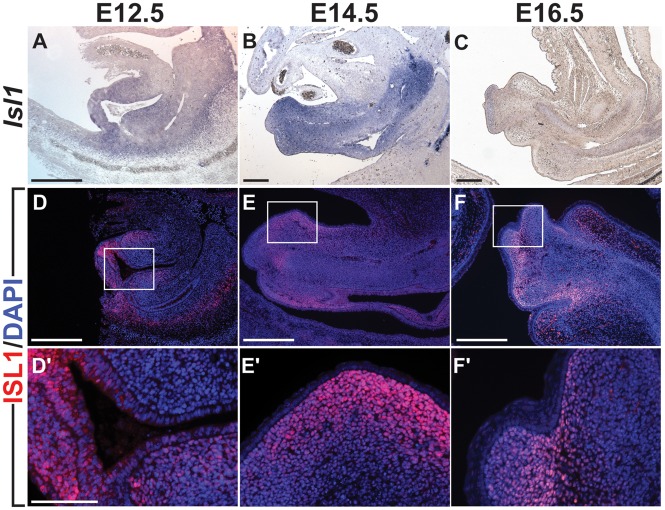
We first assessed Isl1 mRNA transcript expression and protein localization during urogenital development to identify regions of ISL1 activity. In situ hybridization (ISH) detection of Isl1 revealed that Isl1 mRNA was expressed throughout the developing GT mesenchyme at E12.5 and E14.5, but expression was significantly reduced by E16.5 (Fig. 1A–C). Isl1 transcripts were also detected in the mesenchyme surrounding the distal region of the urogenital sinus at E12.5 and E14.5. The peak of Isl1 expression appeared to be at E14.5. At E16.5, Isl1 expression was restricted to a mesenchymal domain at the distal tip of the GT, although low levels of Isl1 persisted in the mesenchyme flanking the urethra. Expression of Isl1 was also detected at low levels in the urethral plate epithelium at E12.5 and E14.5, but not at E16.5. Immunohistochemical detection of ISL1 protein revealed that its distribution in the developing genitalia closely reflected mRNA localization. At E12.5, ISL1 was found in the distal genital mesenchyme, as well as in the distal urethral plate epithelium. In the E14.5 GT, ISL1 was found throughout the distal GT mesenchyme, but was detected at substantially lower levels in the urethral plate epithelium. ISL1 protein was completely absent from the overlying ectoderm-derived GT epithelium (Fig. 1D–F′). This analysis of Isl1 expression and localization pointed to the mesenchyme as the principal region for ISL1 activity during embryonic GT development, and the strong expression of Isl1 at E12.5 and E14.5 suggests that this is the developmental window during which Isl1 acts.
Figure 1.
Isl1 expression is localized to the genital mesenchyme throughout genital development. Detection of Isl1 RNA transcripts by in situ hybridization in sagittal sections of the male GT at E12.5, E14.5, and E16.5 (A–C). Immunodetection of ISL1 in sagittal male GT sections at E12.5, E14.5, and E16.5 (D,F′). White boxes in (D–F) indicate regions of high magnification shown in D′-F′. High magnification images of ISL1 immunolocalization surrounding the urethral epithelium at E12.5 (D′), dorsal region of the distal GT at E14.5 (E′), and distal tip of the GT at E16.5 (F′). Scale bars: 300 µm (A-F); 100 µm (D′-F′).
Deletion of Isl1 from the genital mesenchyme causes abnormal development of the external genitalia
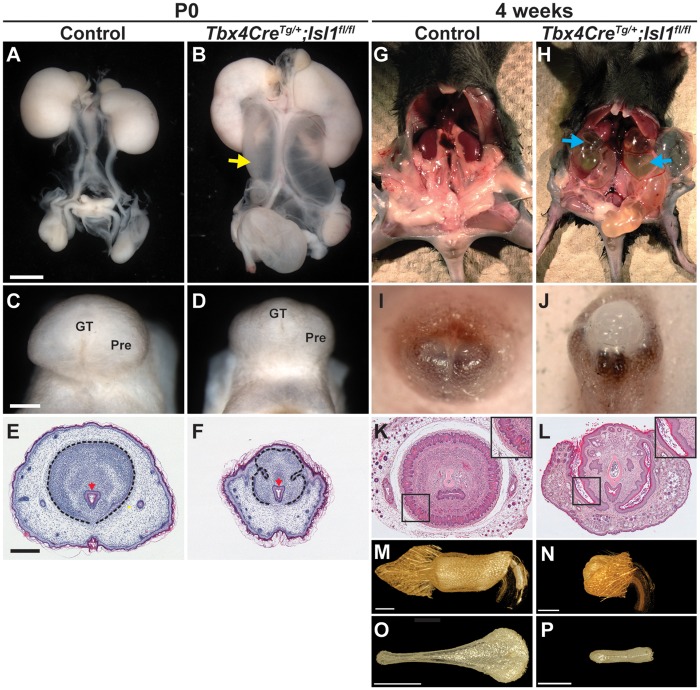
We next performed tissue-specific deletion of Isl1 to test whether its absence in the GT mesenchyme would lead to aberrant development of the external genitalia. The Tbx4CreTg allele is robustly expressed in the hindlimb and pericloacal mesenchyme beginning early in embryogenesis (20). Generation of Tbx4CreTg; Isl1fl/flmice revealed significant abnormalities in the development of the urogenital tract (Fig. 2). Tbx4CreTg; Isl1fl/fland control offspring appeared at the expected Mendelian ratio, indicating that deletion of Isl1 in the pericloacal and genital mesenchyme was not embryonic lethal (data not shown). At birth, no difference in overall weight and size was noted between control and Tbx4CreTg; Isl1fl/fl pups. However, dramatic defects were seen throughout the urogenital tract and external genitalia. In Tbx4CreTg; Isl1fl/fl pups, failure to drain urine into the bladder led to abnormally enlarged kidneys (hydronephrosis) and ureters (hydroureter) (Fig. 2A and B). Effects of Isl1 deletion on kidney and ureter development were consistent with the defects previously described when Isl1 was deleted from the pericloacal mesenchyme using the Hoxb6Cre allele (18). In addition to the defects in the urinary tract, the external genitalia of Isl1 mutants were hypoplastic (Fig. 2C and D). Histological analysis showed that the prepuce incompletely surrounded the dorsal aspect of the GT and failed to separate from the GT along the ventral midline. Moreover, the mesenchymal condensations within the GT were absent. Formation of the internal urethra in Tbx4CreTg; Isl1fl/fl mice did not appear to be adversely affected (Fig. 2E and F).
Figure 2.
Deletion of Isl1 in the pericloacal mesenchyme results in hydroureter, hydronephrosis, and urogenital malformations. Urogenital tract in P0 (A–F) and 4-week-old (G–P) control and Tbx4CreTg; Isl1fl/flmale mice. Hydroureter (B, yellow arrow) and GT hypoplasia (D) were observed in P0 Isl1 mutant mice. Histology of coronal sections from control external genitalia showed that the GT (black dotted line) and urethra (red arrow) are surrounded by the prepuce (E), whereas in Tbx4CreTg; Isl1fl/flmice, the GT (black dotted line) was abnormally shaped and the prepuce only covered the ventral surface of the GT (F). The urethra in Isl1 mutants was unaffected at P0 (F, red arrow). Hydronephrosis (blue arrows) in 4-week-old Tbx4CreTg; Isl1fl/flmale mice (H). Bifid prepuce in a control 4-week-old male (I), compared with a hypoplastic penile body partially surrounded by an incompletely developed prepuce in Tbx4CreTg; Isl1fl/flmale mice (J). Coronal histological sections from 4-week-old control and Tbx4CreTg; Isl1fl/flpenises show absence of surface epithelial spines in mutant mice (K, L, insets). µCT reconstruction of the penis and foreskin (M, N), and os penis (O, P) in adult control and Tbx4CreTg; Isl1fl/fl male mice. GT: genital tubercle; Pre: prepuce. Scale bars: 1 mm (A, B, I, J, M-P); 500 µm (C, D); 400 µm (E, F, K, L).
Tbx4CreTg; Isl1fl/fl mice survived until early adulthood but were consistently smaller than control littermates (not shown). Abnormal retention of urine in both the kidneys and ureters caused both structures to expand to several times their normal size (Fig. 2G and H, blue arrows). In comparison to newborn pups, the defects in the external genitalia were more pronounced in early adults. In males, a bifid prepuce normally covers the penile body, but in Tbx4CreTg; Isl1fl/fl mice, the penile body was significantly shorter, altered in structure, and only partially covered on the ventral surface by the prepuce (Fig. 2I and J). Gross examination of control and mutant external genitalia showed that a number of features found in the normal penis were absent in Tbx4CreTg; Isl1fl/fl mice. Most notably, the bifid prepuce, cartilaginous male urogenital mating protuberance (MUMP), MUMP ridge, and urethral flaps were either deformed (prepuce) or completely absent (MUMP, MUMP ridge, urethral flaps) (21–23). Histological examination of the penis at this stage revealed that in Tbx4CreTg; Isl1fl/fl mice, the penis lacked both a well-defined corpus cavernosum glandis and keratinized penile spines at the surface epithelium surrounding the glans (Fig. 2K and L, insets). In addition, the prepuce remained tethered to the ventral side of the GT in Tbx4CreTg; Isl1fl/fl mice, whereas it was completely separated in controls (Fig. 2K and L). Loss of Isl1 also resulted in pronounced defects in development of female external genitalia (Supplementary Material, Fig. S1). In adult Tbx4CreTg; Isl1fl/fl female mice, a small vaginal opening that led to a blind-ended vaginal pouch and absence of a vaginal canal were identified (Supplementary Material, Fig. S1D–F). The urethra remained patent, allowing for urine excretion and normal bladder size. However, due to vaginal atresia, uterine excretions were retained in the uterine horns, resulting in distention (hydrometrocolpos) (Supplementary Material, Fig. S1K and L).
Micro-CT was used to obtain a 3-dimensional image of the penile body, distal prepuce, and os penis in adult males (Fig. 2M–P). In early adult Tbx4CreTg; Isl1fl/fl males, the penile body was hypoplastic and the prepuce failed to cover the dorsal aspect of the distal penis, resembling an epispadias-like phenotype (Fig. 2M and N). The os penis in Tbx4CreTg; Isl1fl/fl mice was significantly shorter than controls and did not expand toward the proximal end (Fig. 2O and P). Taken together, these data demonstrate that Isl1 expression in the pericloacal and genital mesenchyme is required for proper urogenital formation.
We used scanning electron microscopy and histology to determine the stage at which the effects of Isl1 deletion on embryonic development could first be observed. At E12.5, although Tbx4CreTg; Isl1fl/fl GTs were reduced in size, the overall structure did not appear to be significantly altered compared with controls, suggesting that Isl1 is not critical for GT initiation, but may play a role in early GT outgrowth and morphogenesis (Fig. 3A and B). By E14.5, Tbx4CreTg; Isl1fl/fl GTs were noticeably hypoplastic and showed several defects, including an enlarged proximal urethral opening on the ventral GT and reduced preputial swellings (Fig. 3C and D). By E16.5, both the prepuce and GT in Isl1 mutants had developed, but these remained hypoplastic compared with controls (Fig. 3E and F). Histological analysis of mutant GTs at each stage also revealed that the volume of the dorsal mesenchyme was consistently reduced compared with controls (Fig. 3G–L; black asterisks). These data suggest that while Isl1 is not essential for initiating GT outgrowth from the paired genital swellings around E10.5, it is necessary for normal GT and preputial development shortly thereafter.
Figure 3.
Deletion of Isl1 disrupts embryonic GT development. Scanning electron micrographs of control (A, C, E) and Tbx4CreTg; Isl1fl/fl mutant (B, D, F) GTs during embryogenesis at E12.5, E14.5, and E16.5. Hypoplastic GT (yellow arrowheads), ectopic urethral opening at the base of the GT (blue arrowhead), and absent preputial swellings (red asterisks) are evident in E14.5 Tbx4CreTg; Isl1fl/fl mutant mice (D). Urethral seam is sealed in both control and mutant GTs at E16.5 (E, F, black arrowheads). Scale bar: 250 µm. Hematoxylin and eosin staining of sagittal GT sections from E12.5, E14.5, and E16.5 control (G, I, K) and Tbx4CreTg; Isl1fl/fl mutant (H, J, L) male GTs. Mesenchymal volume on the dorsal side of the distal GT is reduced at E12.5 and persists through E16.5 in Tbx4CreTg; Isl1fl/fl mutants (H, J, L, asterisks) compared with control male GTs (G, I, K). Scale bar: 250 µm. Pre: prepuce; GT: genital tubercle.
ISL1 regulates expression of Bmp4, Wnt5a and Fgf10 in the genital mesenchyme
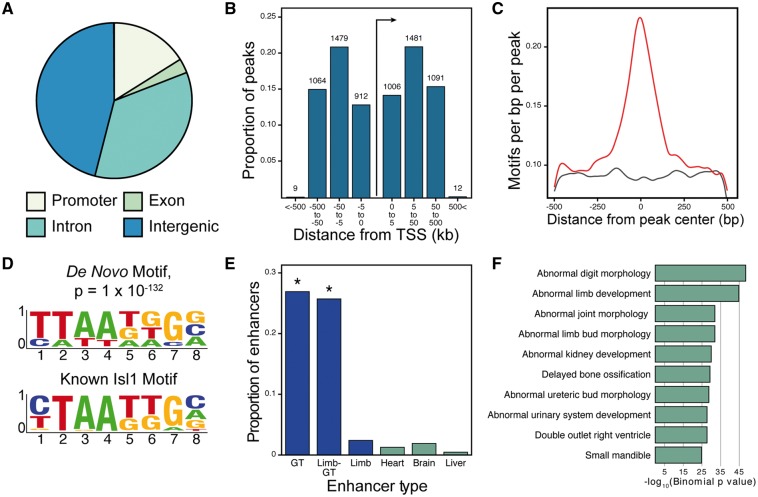
To investigate the full complement of genes that are directly under the transcriptional control of ISL1 in the embryonic GT, we performed chromatin immunoprecipitation-sequencing (ChIP-Seq) against ISL1 in the E14.5 GT. ISL1 ChIP-Seq detected 7, 054 peaks, predominantly in intergenic and intronic regions (Fig. 4A). The majority of these binding sites are located more than 5 kb from the closest annotated transcriptional start site (TSS), with nearly one third of the binding sites more than 50kb away from the nearest TSS (Fig. 4B). A de novo motif search using HOMER demonstrated that GT ISL1 ChIP-Seq peaks are enriched for a motif that matches the consensus sequence bound by ISL1 in other tissues [Fig. 4C and D; (24) and (25)].
Figure 4.
ISL1 ChIP-Seq peaks are associated with genital-specific regulatory regions. The majority of ISL1 peaks occur within intergenic regions or introns and are 5kb to 500kb from transcription start sites (A, B). The top motif in ISL1 peaks is centrally enriched and matches a known ISL1 motif (C, D). ISL1 peaks are significantly enriched on enhancers active in the developing GT compared with enhancers active in other embryonic tissues (E). The top ten gene associations from GREAT analysis indicate that ISL1 peaks are strongly enriched near genes involved in limb and urogenital system development (F).
In a recent analysis of appendage cis-regulatory activity, we identified over 1, 400 GT-specific enhancers. Sequence analyses revealed that these GT enhancers are enriched for a DNA motif that closely matches the ISL1 binding consensus (26). To directly test whether ISL1 binding events are enriched at GT enhancers, we examined the overlap between E14.5 GT ISL1 ChIP-Seq peaks and the location of GT-specific enhancers, as well as other classes of tissue-specific enhancers identified from mouse ENCODE datasets. We found that ISL1 binding sites are strongly enriched on GT enhancers (P = 7.87×10−105, Fisher’s exact test) compared with enhancers that function in other tissues (Fig. 4E). Similarly, ISL1 binding sites are also enriched on Limb-GT enhancers (P = 3.63×10−68), a class of enhancers that are active in both the genitalia and the limbs. Further analysis of the location of ISL1 peaks revealed a strong association with genes implicated in limb and urogenital development (Fig. 4F).
Given the enrichment of ISL1 peaks near urogenital genes, we examined ISL1 ChIP-Seq signal at genes that are known to play a role in GT development. This analysis revealed multiple significant ISL1 peaks in the intergenic region surrounding Bmp4, suggesting that Bmp4 is directly regulated by ISL1 (Fig. 5A). In addition, ISL1 ChIP-Seq signal was found associated with Fgf10 and Wnt5a, two genes that have previously been implicated in development of the external genitalia (27,28). ISL1 ChIP-Seq signal was observed in the intron of Fgf10, as well as in upstream and downstream regions that flank this gene. Multiple ISL1 peaks were also found in the large intergenic region located downstream of Wnt5a (Fig. 5A). Notably, all of these ISL1 ChIP-Seq peaks occur in regions that are also marked by H3K27ac, a histone modification that is enriched on active cis-regulatory elements. To determine whether ISL1 binding occurs near Bmp4, Fgf10, and Wnt5a earlier in development, we also performed ISL1 ChIP-Seq on E12.5 GTs. We found that the pattern of ISL1 binding around these loci is extremely similar at E12.5 and E14.5, with multiple significant binding events occurring at both stages of GT development (Supplementary Material, Fig. S2). Moreover, in situ hybridization of E14.5 Tbx4CreTg; Isl1fl/fl male GTs showed that expression of Bmp4, Fgf10, and Wnt5a were significantly reduced, strongly suggesting that ISL1 directly regulates expression of all three genes in the mesenchyme of the developing GT (Fig. 5B–G). Thus, our data indicate that ISL1 may influence GT development through the BMP, WNT and FGF signaling pathways.
Figure 5.
ISL1 directly binds to and regulates expression of Bmp4, Fgf10, and Wnt5a. Significant enrichment of ISL1 ChIP-Seq peaks is found near Bmp4, Fgf10, and Wnt5a loci. The majority of these ISL1 ChIP-Seq peaks overlap active enhancers that are significantly enriched for H3K27ac in E12.5 mouse GT (A). In situ hybridization of Fgf10, Wnt5a, and Bmp4 on sagittal sections of E14.5 control and Tbx4CreTg; Isl1fl/fl mutant GTs. Black boxes indicate regions of high magnification shown in insets (B–G). Scale bars: 300 µm (B–G).
BMP4-mediated apoptosis is altered in Tbx4CreTg; Isl1fl/fl GTs
The BMP signaling pathway is a critical regulator of apoptosis in the GT (29). During the earliest stages of genital development, Bmp4 expression is detected adjacent to the cloacal plate. By E11.5, Bmp4 is expressed primarily in the GT mesenchyme flanking the urethral epithelium (30). Our in situ hybridization results showed that Bmp4 expression along the dorsal GT was reduced in Tbx4CreTg; Isl1fl/fl mice at E14.5, suggesting that changes in Bmp4 expression or BMP4-mediated cell death might contribute to the observed morphological GT defects. This led us to investigate whether reduced Bmp4 expression at E14.5 altered the number of apoptotic cells or the pattern of cell death in the GT mesenchyme.
LysoTracker and TUNEL staining were used to examine whether reduced Bmp4 expression in the GT mesenchyme of Tbx4CreTg; Isl1fl/fl mice was correlated with fewer cells undergoing apoptosis. LysoTracker detection of lysosomal activity, which is closely correlated with apoptosis (31), was used to identify regions of the GT in which cells were undergoing programmed cell death. Staining of E12.5 GTs from control and mutant mice revealed a high amount of apoptotic activity in the distal tip of the GT and along the urethral seam near the base of the GT (Fig. 6A and B, yellow arrowheads). Detection of apoptotic cells by TUNEL stain on E12.5 GT sections showed that the apoptotic cells in the distal GT were largely found in the mesenchyme, whereas the apoptotic cells in the proximal urethral seam were primarily restricted to the epithelium (Fig. 6C and D, white arrows). There were no observable differences between control and Isl1 mutant GTs in the number and density of apoptotic cells at E12.5. Analysis of E14.5 mutant GTs showed that while the number of apoptotic cells at the distal GT appeared to remain constant, the localization of apoptotic cells surrounding the proximal urethral opening was altered. In control GTs at E14.5, apoptotic cells completely surrounded the opening, whereas apoptotic cells were only detected along the most proximal borders of the enlarged urethral opening in Isl1 mutant GTs (Fig. 6E and F, yellow arrowheads). Strikingly, there was also a marked reduction in apoptosis in the GT mesenchyme of Tbx4CreTg; Isl1fl/fl mice at E14.5. In normal GTs, a dense track of apoptotic cells was detected in the dorsal GT mesenchyme, extending into the bladder mesenchyme (Fig. 6G, white arrows). In addition, a cluster of apoptotic cells was found in the distal region of the dorsal GT mesenchyme (Fig. 6G, white arrows, inset). In Tbx4CreTg; Isl1fl/fl mice, however, this population of cells was almost completely absent, except for a few apoptotic cells found scattered throughout the GT mesenchyme (Fig. 6H, white arrows). Furthermore, apoptosis was undetectable in the distal GT mesenchyme of Tbx4CreTg; Isl1fl/fl mice (Fig. 6H, inset and 6K). Given the significant reduction in mesenchymal volume observed in the distal region of the dorsal GT, we expected that the number of apoptotic cells in the mesenchyme would be increased, and we explored this further by evaluating the molecular mechanisms regulating apoptosis in the GT.
Figure 6.
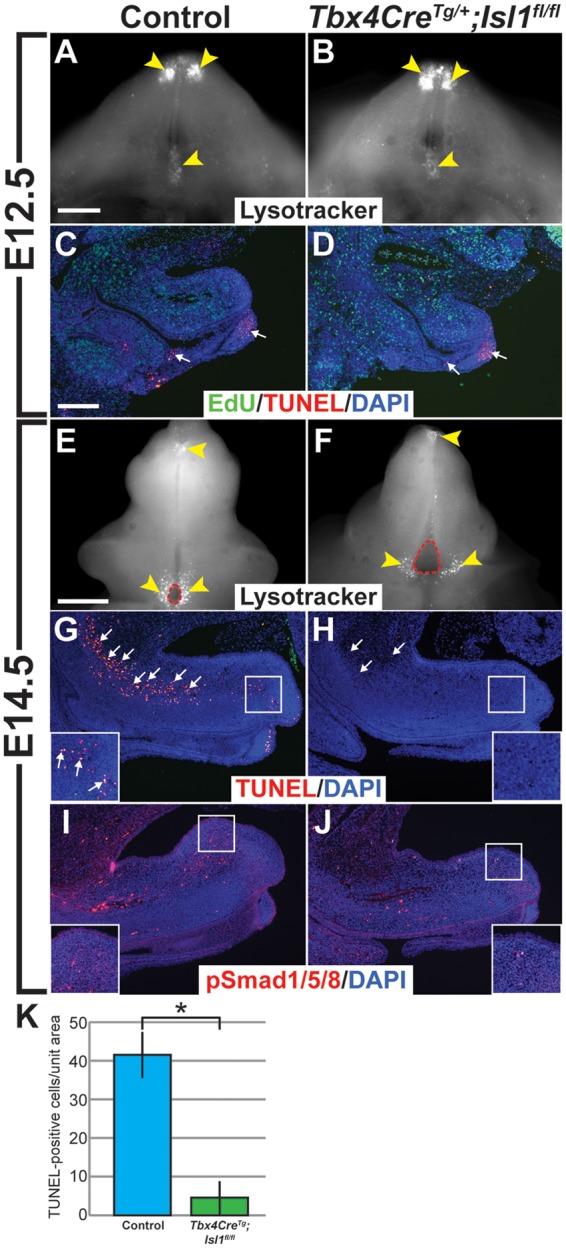
Apoptosis in the genital mesenchyme is reduced in Tbx4CreTg; Isl1fl/fl mice. Regions of cell death (yellow arrowheads) marked by LysoTracker in whole-mount E12.5 (A,B) and E14.5 (E,F) control and Tbx4CreTg; Isl1fl/fl GTs. Apoptotic cells (red, white arrows) detected by TUNEL stain in sagittal sections of E12.5 (C,D) and E14.5 (G,H) control and Tbx4CreTg; Isl1fl/fl mutant GTs. Magnified field of distal GT mesenchyme in control mice containing apoptotic cells (G, inset, white arrows), but none in Tbx4CreTg; Isl1fl/fl GTs (H, inset). Immunodetection of phospho-Smad1/Smad5/Smad8 (red) in sagittal sections of E14.5 control (I) and Tbx4CreTg; Isl1fl/fl (J) GTs. High-magnification of dorsal region of distal GT (I, J, insets). Fewer TUNEL-positive cells detected per unit area in dorsal mesenchyme of Tbx4CreTg; Isl1fl/fl GTs compared with controls (P = 0.002) (K). Nuclei (DAPI, blue) (C, D, G–J), proliferating cells (EdU, green) in E12.5 GT sections (C, D). Scale bars: 200 µm (A–D, G–J); 300 µm (E, F).
BMP4 signaling is mediated by phosphorylation of intracellular SMAD signal transduction proteins (32). Therefore, we assessed BMP4 signaling activity in the GT via detection of phosphorylated SMAD proteins (phospho-SMAD1/SMAD5/SMAD8; pSMAD1/5/8). Concordant with a reduction in apoptosis, we found that pSMAD1/5/8 were also reduced in the GT mesenchyme at E14.5, suggesting that decreased apoptosis observed in Tbx4CreTg; Isl1fl/fl GTs was a result of lower BMP signaling activity (Fig. 6I and J, insets). Together with our ChIP-Seq data showing direct binding of ISL1 to genomic regions surrounding Bmp4, our findings demonstrate that regulation of BMP4-mediated apoptosis within the genital mesenchyme is an important function of Isl1 in genital development.
Uptake of either EdU or BrdU was used to assay levels of cell proliferation in control and mutant GTs to determine whether changes in Bmp4, Fgf10, or Wnt5a expression altered the number of dividing cells in the GT mesenchyme. The mitotic index was determined by calculating the ratio of EdU/BrdU-positive to DAPI-positive nuclei. Interestingly, no significant changes in cell proliferation between control and mutant GTs were evident at either E12.5 or E14.5 (Supplementary Material, Fig. S3), suggesting that Isl1 primarily acts on cell death as opposed to proliferation in the GT mesenchyme.
Discussion
Through conditional deletion of Isl1 in the embryonic GT mesenchyme, we found that Isl1 is essential for normal development of the external genitalia. Isl1 is expressed in the mesenchyme of several tissues during embryogenesis, where it functions as an important transcriptional regulator of a number of signaling pathways. In humans, ISL1 is expressed in the cardiac mesenchyme of the developing outflow tract during embryogenesis, where it acts by directly binding to and driving expression of FGF10 (33). Chromatin immunoprecipitation revealed that while ISL1 binds to an enhancer in intron 1 of human FGF10 in the heart, equivalent binding was not observed in the hindlimb, suggesting that ISL1 may bind to a different FGF10 enhancer in the hindlimb, or that ISL1 acts through a different molecular mechanism in the developing hindlimb (33). In mice, it was subsequently shown that ISL1 regulates the morphoregulatory network upstream of Hand2 and Shh during establishment of the posterior hindlimb field (34). Finally, in the developing mouse kidney, Isl1 is expressed in the mesenchyme surrounding the ureteric bud. Deletion of Isl1 from the metanephric mesenchyme results in ectopic ureteric bud branching, abnormal ureterovesical junctions, and a reduction in Bmp4 expression (18). Our findings show that in the developing GT, ISL1 regulates expression of several key signaling molecules including Fgf10, Wnt5a, and Bmp4.
Interaction between Isl1 and BMP signaling has been demonstrated in the context of several other developing tissues as well. During embryonic patterning of the mouse dentition, both Isl1 and Bmp4 are expressed in the oral epithelium, where they act in a positive regulatory loop. Misexpression of Isl1 in the proximal mandible epithelium resulted in ectopic Bmp4 expression, whereas suppression of Isl1 expression using morpholino antisense oligonucleotides led to downregulation of Bmp4 expression (35). ISL1 marks progenitor cells that contribute to the majority of cells in the developing heart as well as the hindlimb. Deletion of the type 1 Bmp receptor Bmpr1a from cardiac progenitor cells using an Isl1Cre allele led to significant defects in the outflow tract and right ventricle, whereas in the hindlimb, removal of Bmpr1a produced both smaller hindlimbs and ectopic outgrowths (36). Together, these data show that Isl1-expressing cells are not only responsible for production of BMP4, but are themselves sensitive to the morphogenetic signal of BMP4. Our data showed that during GT morphogenesis, ISL1 acts in the mesenchyme by influencing expression of Bmp4, similar to its function in the metanephric mesenchyme.
Several studies have demonstrated that SHH released from the urethral plate epithelium modulates Bmp4 expression in the GT mesenchyme (30,37–39). Whereas the majority of the data suggest that Shh acts to promote transcription of Bmp4 in the GT mesenchyme, other studies have shown that Shh may act to suppress BMP signaling. Two studies showed that Bmp4 expression is downregulated in the context of deletion of Shh from the GT (30,37). In addition, Bmp4 expression was increased in the presence of exogenous SHH protein (38). However, a tamoxifen-inducible Shh conditional knockout mouse model showed that following ablation of Shh in the GT, Bmp4 expression was increased in the distal GT (39). Together, these studies underscore the importance of both temporal and spatial control of signaling pathways during embryonic GT development.
All three members of the Gli family of proteins, which act as mediators of SHH signaling, are expressed in the GT mesenchyme during embryonic development, but Gli2 was shown to be the primary downstream mediator of SHH signaling during masculinization of the GT (38,40). Whether GLI2 directly binds to and regulates expression of Bmp4 in the GT mesenchyme will be an important question to answer in order to understand the transcriptional mechanisms that contribute to normal development of the GT mesenchyme. Our ChIP-Seq data show that ISL1 is another important regulator of Bmp4, but additional work will be needed to determine the relative importance of specific ISL1 binding sites to different enhancer elements of Bmp4. In addition, further studies are needed to examine how ISL1 and SHH functions are integrated to control Bmp4 expression.
ISL1 signaling in the embryonic GT plays a role in the spatiotemporal regulation of apoptosis. We observed reduced Bmp4 expression in the GT mesenchyme in Tbx4CreTg; Isl1fl/fl male GTs, and we showed that decreased BMP signaling in the mesenchyme of Tbx4CreTg; Isl1fl/fl male GTs was associated with fewer apoptotic cells. Insights into how apoptosis shapes the overall structure of the GT during organogenesis have been provided by studies in both mice and avians. In mice, Bmp4 and Bmp7 are expressed in the GT during embryogenesis, and exposure of GT organ cultures to either protein promotes cell death, whereas exposure of the organ cultures to the BMP antagonist noggin (NOG) inhibited the number of cells undergoing apoptosis (29). Expression of Bmpr1a is detected throughout the epithelium and mesenchyme of the GT, suggesting that cells in both compartments are capable of responding to BMP4 produced in the mesenchyme (41). Deletion of Bmpr1a from the GT surface ectoderm and epithelium adjacent to the distal urethral epithelium (DUE) using a Brn4-Cre allele enhanced outgrowth of the GT. This was attributed to reduced apoptosis in the distal GT mesenchyme, which was an indirect effect of Fgf8 downregulation in the DUE (29). However, loss of BMP signaling in the DUE or the mesenchyme was not explored. In avian species, levels of Bmp4 expression and apoptosis appear to be correlated with the presence of an intromittent phallus. Avian species that have retained an intromittent phallus show reduced Bmp4 expression and reduced cell death in the distal GT, while Bmp4 expression was elevated and increased cell death was detected in a divergent clade of avian species lacking an intromittent phallus (42). These studies suggested that changes in BMP4-mediated apoptosis during evolution affected the morphology of the external genitalia in avians. Taking into consideration that similar patterns of Bmp4 expression and apoptosis have been described in the mouse GT, cell death has been presumed to perform an analogous function in the shaping of mammalian external genitalia as well (42).
Our results show that BMP4-mediated apoptosis in the GT mesenchyme is partially controlled by ISL1. It should be noted that reduced cell death in Isl1 mutant GTs was only observed in the mesenchyme, while apoptosis at the distal tip of the GT and surrounding the proximal urethral opening appeared relatively unaffected. This suggests that apoptosis in the epithelial and mesenchymal compartments of the GT is independently controlled. While programmed cell death has traditionally been seen as a means to eliminate unnecessary cells (e.g. during digit individualization or degeneration of the Wolffian and Müllerian ducts) [reviewed in (43)], it can also be important in removing morphogen-producing cells that affect the patterning of the surrounding tissue. For example, during normal brain development, the anterior neural ridge (ANR) acts as a signaling center by secreting a number of morphogens including FGF8. In apoptosis-deficient Apaf-1–/– mice, these Fgf8-expressing cells are not removed and continue to release FGF8 into the surrounding telencephalon. Persistence of these cells results in neural tube closure defects as well as failure of the ventricles to expand (44). Similarly, apoptosis is responsible for silencing gene expression during mouse odontogenesis by removing the enamel knot, an important signaling center regulating tooth development (45). In Isl1 mutant GTs, it is possible that reduced apoptosis in the mesenchyme disrupts the signaling pathways necessary for normal mesenchymal expansion, thereby causing GT hypoplasia. In females, opening of the vaginal canal is an apoptosis-dependent process. Several mouse models of reduced apoptosis exhibit vaginal atresia, imperforate vagina, and hydrometrocolpos (46–48). Apoptosis in the postnatal female mouse vaginal epithelium is driven by an estrogen-dependent proteolytic cleavage of semaphorin 4D (Sema4D) (49). We found that Tbx4CreTg; Isl1fl/fl female mice phenocopied the apoptosis-deficient mouse models. These data support a role for Isl1 in regulating apoptosis not only in the developing GT mesenchyme, but within the postnatal vaginal epithelium as well. Characterization of the molecular features of these cells and the signaling molecules that induce apoptosis in them will generate valuable insight into how the overall structure of the male and female external genitalia are remodeled.
Our ChIP-Seq data show that ISL1 also directly binds to both Fgf10 and Wnt5a, two genes that are important for GT development. WNT signaling plays a prominent role in genital formation, and several Wnt genes are expressed in the endodermal, mesenchymal, and ectodermal tissues that compose the external genitalia (50). Targeted deletion of β-catenin in the mouse embryonic GT resulted in dysmorphic and hypoplastic GTs and a reduction in proliferating cells in the mutant mesenchyme (50). GT hypoplasia and reduced cell proliferation have also been reported in Wnt5a–/– mice, although variability of the genital phenotype ranges from complete genital agenesis to a mild reduction in size (29,51). Together, these data showed that WNT signaling in the GT mesenchyme is primarily responsible for regulation of cell proliferation. Loss of Fgf10 in the embryonic GT leads to defective urethral closure and GT hypoplasia (27,52). However, levels of cell proliferation were not measured in these Fgf10–/– mice, nor was there any description of overall changes in the size of the GT. Surprisingly, we did not find significant changes in the number of proliferating cells in the GT mesenchyme of Isl1 mutants. It remains a possibility that due to the abundance of proliferating cells throughout the developing embryo, subtle differences in GT cell proliferation were not captured in our analyses. Therefore, a more detailed investigation into the levels of proliferation in the GT will be needed to definitively conclude that reduced expression of both Fgf10 and Wnt5a in Tbx4CreTg; Isl1fl/fl GTs did not result in lower levels of cell proliferation.
Regulation of Wnt5a expression and WNT signaling in the GT mesenchyme has been attributed to several molecular mechanisms such as androgen receptor signaling, SHH secreted by the DUE, or endodermal WNT-β-catenin signaling (30,39,50,53,54). Downregulation of Wnt5a expression in response to exogenous BMP4 also suggested that both activating and inhibitory mechanisms are responsible for modulating levels of WNT signaling in the developing GT (29). The regulatory mechanisms that control Isl1 expression in the GT have yet to be revealed, but studies in hindlimb initiation and cardiac progenitor development showed that β-catenin can act as an upstream transcriptional regulator of Isl1 expression (16,55,56). Moreover, during hindlimb initiation, both ISL1 and β-catenin regulate proliferation of hindlimb progenitors via a feedback loop involving Fgf10 and Fgf8 (16). Our findings suggest that in the mesenchyme of the developing external genitalia, Isl1 occupies a regulatory node with high connectivity to several signaling cascades. Therefore, a focal point for future studies will be to determine whether a similar WNT-Isl1-Fgf10 regulatory mechanism is also employed during GT formation, and whether additional signaling pathways contribute to the expansion and maintenance of the GT mesenchyme. Moreover, the downstream cellular processes that are altered due to Fgf10 and Wnt5a misexpression in Isl1 mutants have yet to be fully understood.
Interestingly, despite the identification of ISL1 as a major susceptibility gene for CBE (10,11), we did not observe any evidence of bladder exstrophy in our mutants. Rather, the defects in the external genitalia that we observed in Isl1 mutant mice were consistent with hypospadias or epispadias, another component phenotype of BEEC. Various theories have been proposed to explain the pathogenesis of BEEC, including premature rupture of the cloacal membrane, lack of mesodermal ingrowth during abdominal wall development, changes in cellular functions, and abnormal cell–cell interactions (57–62). We observed defects in both the prepuce and GT of Isl1 mutants. Although epispadias has traditionally been viewed as a result of abnormal development of the cloacal membrane, our data raise the intriguing possibility that the GT mesenchyme also directly interacts with the overlying GT ectoderm to direct preputial fusion along the dorsal surface. Epithelial-mesenchymal signaling between the GT mesenchyme and urethral epithelium have been well-characterized (27,29,38,41), but further studies will be needed to determine the molecular pathways that are acting along the dorsal-ventral axis of the GT and how the dorsal GT influences the organization of the GT ectoderm and dorsal prepuce.
Our present study shows that Isl1 is an important regulator of embryonic urinary tract development. In addition to the significant association with CBE that has been previously reported, the phenotypic defects we observe in Isl1 mutant external genitalia suggest that ISL1 is a strong candidate for mutational screening in patients with BEEC and other idiopathic genital abnormalities. Surgical repair of both hypospadias and BEEC defects are often associated with complications such as urethral fistulas and wound dehiscence, which require further medical intervention. Therefore, a deeper understanding of the pathoetiology of these conditions will improve diagnosis, management, and outcomes in patients with these malformations.
Materials and Methods
Mouse maintenance and treatment
All mouse studies were carried out under approved protocols in strict accordance with the policies and procedures established by the University of California, San Francisco (UCSF) and University of Georgia (UGA) Institutional Animal Care and Use Committees (UCSF protocol AN084146; UGA protocol A2014 06–019). Mice were maintained in temperature-controlled facilities with access to food and water ad libitum. Tbx4Cre and Isl1fl/fl were previously described (20,63). Conditional knockout of Isl1 in the genital mesenchyme of the developing embryo was achieved by crossing female Isl1fl/fl mice with male Tbx4CreTg/Tg; Isl1fl/+mice. To generate embryos at specific timepoints, adult mice were mated overnight and females were checked for a vaginal plug in the morning. The presence of a vaginal plug was designated E0.5. To label proliferating cells in embryos, pregnant female mice were administered a 100 µl dose of BrdU or EdU in phosphate buffered saline (PBS) (10 mg/ml) via intraperitoneal injection. Embryos were then collected 1 h after injection.
Scanning electron microscopy
Embryonic GTs were dissected and fixed overnight in 4% PFA at 4° C. Tissue was then fixed in 0.1 M sodium cacodylate buffer, 1% osmium tetroxide in 0.1 M sodium cacodylate, and dehydrated for scanning electron microscopy (SEM). Specimens were dried in a Tousimis AutoSamdri 815 Critical Point Dryer (Tousimis, Rockville, MD) and scanning electron micrographs were obtained using a Hitachi TM-1000 scanning electron microscope (Hitachi, Schaumberg, IL). All SEM was performed at the University of California, Berkeley Electron Microscope Lab. Four embryos of each genotype at each timepoint were analysed.
Histology
At least four embryonic and adult tissue samples from each genotype were collected by dissection and fixed in 4% paraformaldehyde (PFA) overnight at 4° C. After fixation, embryos were washed in PBS, processed through an ethanol series, and dehydrated in xylene. The penis and prepuce from adult male mice were decalcified in 0.5M EDTA for 2 days at room temperature before being processed for paraffin embedding. 7 µm paraffin-embedded tissue sections were prepared using a Microm HM325 microtome and dried overnight on a slide warmer. Hematoxylin and eosin staining was performed using standard protocols. Stained slides were mounted with Permount and images were acquired in Leica Application Suite using a Leica DM5000B upright microscope.
In situ hybridization and immunohistochemistry
Three embryos of each genotype were collected and fixed in 4% paraformaldehyde overnight at 4° C, immersed in 30% sucrose in PBS overnight at 4° C, and embedded in O.C.T. compound (Sakura Finetek, Torrance, CA). 10 µm frozen sections were cut using a Microm 550 cryostat and hybridized to DIG-labeled RNA probes for in situ detection of RNA transcripts. Sections were treated with 10 µg/ml of proteinase K and acetylated prior to hybridization with probe. DIG-labeled RNA probes were synthesized from plasmids containing full-length cDNA or fragments of Isl1, Fgf10, Bmp4, and Wnt5a.
Cell proliferation was assessed using immunohistochemical detection of BrdU on paraffin sections using a rat monoclonal antibody specific for BrdU (Abcam, Cambridge, MA; ab6326, 1: 1000). Slides were treated with 0.2N HCl in water prior to applying antibody, and positive cells were visualized by diaminobenzidine (DAB) staining after incubation with an HRP-conjugated secondary antibody. For double labeling of apoptosis and proliferation, the Click-iT EdU Alexa Fluor 488 Imaging Kit (ThermoFisher Scientific, Grand Island, NY) was used following the manufacturer protocol. The proportion of proliferating cells in the GT mesenchyme was determined by dividing the image into uniform grids and manually counting BrdU/EdU-positive and BrdU/EdU-negative cells in mesenchymal regions using Fiji imaging software (https://fiji.sc; date last accessed November 4, 2017) (64). A minimum of two sections from at least three independent tissue samples at each timepoint were analysed to determine the proportion of proliferating cells. ISL1 (Developmental Studies Hybridoma Bank, Iowa City, IA; 39.4D5, 1: 500) and phosphorylated Smad1/5/8 (EMD Millipore, Billerica, MA; AB3848, 1: 500) were detected by immunohistochemistry using standard protocols.
3-D reconstruction of genitalia using micro-CT (µCT)
3-D reconstructions of the genitalia were generated using µCT. The penis and prepuce were removed from 6-week old male mice and fixed overnight in 4% PFA. Following fixation, tissue was dehydrated through an ethanol series and stored in 70% ethanol. Tissues were then soaked in iodine solution (1%) overnight to differentially stain soft tissues for µCT visualization (65). Samples were scanned using a micro-focused X-ray tomographic system (MicroXCT-200, Zeiss, Pleasanton, CA), at 60 kV and 133 µA. 1200 projection images at an exposure time of 2 s with a linear magnification of 4X were taken. The final pixel size was 4.4 µm. The volume was reconstructed using a back projection filtered algorithm (Zeiss, Pleasanton, CA). Following reconstruction, tissues were manually segmented and rendered as 3-D surfaces using Drishti V2 Volume Exploration software (http://sf.anu.edu.au/Vizlab/drishti; date last accessed November 4, 2017).
ISL1 chromatin immunoprecipitation-sequencing (ChIP-seq)
Tissues for chromatin immunoprecipitation-sequencing (ChIP-Seq) were collected from timed matings of CD1 mice (Charles River). GTs were isolated from E14.5 and E12.5 embryos and cross-linked at room temperature in 1% formaldehyde in PBS for 20 min. After crosslinking, tissues were rinsed and treated with trypsin for 5 min to generate a single cell suspension. Samples were then gently sonicated with a Branson 450 Sonifier (at low amplitude for 30 s, 100% duty cycle) to create a uniform homogenate. The homogenates were sheared in a Bioruptor set to high for 15 cycles (30 s on, 30 s rest) to generate a chromatin size range of 150–400bp. PureProteomeTM Protein G Magnetic Beads (Millipore) were pre-incubated with 5 μg anti-ISL1 rabbit monoclonal antibody (Abcam #EP4182), and the beads were incubated overnight with 500 μg of the sheared GT chromatin. After washing, immune complexes were eluted from the beads, and protein-DNA crosslinks were reversed by incubating at 65 °C overnight. After treatment with RNase followed by Proteinase K, samples were purified over MicroChIP DiaPure columns (Diagenode, Inc.). Independent biological replicates were used to generate two Illumina ChIP-Seq and two control libraries. All ChIP and input chromatin control libraries were produced using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, #E7645S) as directed by the manufacturer. Libraries were sequenced at the Georgia Genomics Facility on an Illumina NextSeq 500 to produce 75bp SE reads. Sequencing reads were processed to remove adapters using Trimmomatic (v0.35) (66) with the settings “ILLUMINACLIP: TruSeq3-SE.fa: 2: 30: 12: 1 MAXINFO: 50: 0.95 LEADING: 3 TRAILING: 3 MINLEN: 50”, and then trimmed from the low quality end to a uniform length of 50bp using fastx_trimmer from the FASTX-Toolkit (v0.0.14) (http://hannonlab.cshl.edu/fastx_toolkit/; date last accessed November 4, 2017). Read quality was assessed using FastQC (v0.11.3) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; date last accessed November 4, 2017). Reads were aligned to the mouse genome (mm10) using bowtie2 (v2.2.6) (67) with the options “–end-to-end –very-sensitive”. Aligned reads were then used to identify enriched regions (peaks) by comparing the signal in the ChIP library to the input chromatin control library for each replicate using MACS2 (v2.1.0.20150731) (68) with a q-value cutoff of 0.01. To generate a consensus peak list, peaks identified in each replicate were merged using BEDTOOLS (v2.24.0) and only enriched regions present in both samples were kept. De novo enriched motifs were identified in each replicate and in the merged dataset using the HOMER command findMotifsGenome.pl with the option “-size 50” to identify centrally enriched motifs. Regions in the merged dataset were associated with gene annotations using GREAT (69). Regions were annotated further in R (https://www.r-project.org; date last accessed November 4, 2017) using the rtracklayer (70) and GenomicRanges (71) packages and the Ensembl mm10 gene annotation (v82) (72). ISL1 ChIP-Seq data has been deposited at the Gene Expression Omnibus (GSE91082).
Mouse H3K27ac ChIP-Seq alignments for two replicates each of 16 adult and embryonic tissues were downloaded from the ENCODE project website (73). Regions of significant enrichment were determined using MACS2 with the default parameters. For each replicate, significantly enriched regions within 1 kb were merged into a single region using BEDTools. Enriched regions from each tissue replicate were then combined by merging regions with a minimum overlap of 1bp. Putative enhancers were identified from the merged tissue replicates by excluding enriched regions that overlapped promoters and exons based on the UCSC genome browser Known Gene dataset (74).
Putative enhancer regions from all 16 mouse tissues were combined by merging regions with a minimum overlap of 1bp. The R package Rsubread was used to count the number of aligned reads from both replicates that overlapped regions in the combined enhancer list (75). Reads per kilobase of transcript per million mapped reads (RPKM) values were calculated from the read counts in R using the edgeR package (76). These RPKM values were then normalized based on the multi-IP normalization output across all tissue datasets calculated by CHANCE (77). These values were then transformed into a matrix with rows as putative enhancer regions and columns as normalized RPKM for each tissue type. For each row, the data were standardized further by subtracting the mean and then dividing by the standard deviation for each value. Putative enhancer regions were then grouped based on similar H3K27ac signal profiles using k-means clustering into tissue-specific categories. Coordinates for putative enhancers from embryonic heart, embryonic brain, and embryonic liver were converted to genome version mm10 using the UCSC liftOver tool (74).
Detection of programmed cell death
To label populations of cells undergoing programmed cell death, six E12.5 and E14.5 embryos of each genotype were incubated in 5 mM LysoTracker Red DND-99 (Thermo Fisher Scientific, Waltham, MA) diluted in PBS for 45 min at 37° C, then fixed overnight in 4% PFA at 4° C. Fixed embryos stained with LysoTracker were then processed through a series of methanol washes and stored in 100% methanol. Whole-mount imaging of embryos stained with LysoTracker was performed on a Leica MZ16F dissecting microscope.
To identify cells undergoing apoptosis in tissue sections, TUNEL (Terminal deoxynucleotidyl transferase dUTP nick end labeling) staining was performed on 7 µm paraffin sections of four embryos of each genotype using the in situ Cell Death Detection Kit (Roche, Indianapolis, IN). Enzymatic labeling solution containing TMR-dUTP was applied to prepared tissue sections encircled with a PAP Pen (Ted Pella, Inc., Redding, CA) and incubated at 37° C for 60 min. Sections were then counterstained with DAPI and mounted with ProLong Gold Antifade Mountant (Thermo Fisher Scientific, Waltham, MA). Imaging of TUNEL-stained tissue sections was performed on a Leica DM5000B upright microscope. Quantification of TUNEL-positive cells was determined by cell-counting in defined regions of the dorsal GT mesenchyme using Fiji.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Sarah Alto and Rebecca d’Urso for assistance with the mouse colony, Larry Baskin and Gerald Cunha for helpful discussions, and Mark Lewandoski for providing Tbx4CreTg/Tg mice.
Data Availability
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (78) and are accessible through GEO Series accession number GSE GSE91082 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE91082; date last accessed November 4, 2017).
Conflict of Interest statement. None declared.
Funding
National Institutes of Health [R01DK095002, R35DE026602 to O.D.K., R01HD081034 to D.B.M., and T32DK779010 to S.C.] and Georgia Advanced Computing Resource Center.
References
- 1. Paulozzi L.J., Erickson J.D., Jackson R.J. (1997) Hypospadias trends in two US surveillance systems. Pediatrics, 100, 831–834. 10.1542/peds.100.5.831 [DOI] [PubMed] [Google Scholar]
- 2. Nordenvall A.S., Frisén L., Nordenström A., Lichtenstein P., Nordenskjöld A. (2014) Population based nationwide study of hypospadias in Sweden, 1973 to 2009: incidence and risk factors. J. Urol., 191, 783–789. [DOI] [PubMed] [Google Scholar]
- 3. Gearhart J.P. (2002) Exstrophy, epispadias, and other bladder anomalies. Campbells. Urol., 3, 2136–2196. [Google Scholar]
- 4. Caton A.R., Bloom A., Druschel C.M., Kirby R.S. (2007) Epidemiology of bladder and cloacal exstrophies in New York State, 1983-1999. Birt. Defects Res. A. Clin. Mol. Teratol., 79, 781–787. [DOI] [PubMed] [Google Scholar]
- 5. Siffel C., Correa A., Amar E., Bakker M.K., Bermejo-Sánchez E., Bianca S., Castilla E.E., Clementi M., Cocchi G., Csáky-Szunyogh M.. et al. (2011) Bladder exstrophy: an epidemiologic study from the International Clearinghouse for Birth Defects Surveillance and Research, and an overview of the literature. Am. J. Med. Genet. C Semin. Med. Genet., 157, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tourchi A., Hoebeke P. (2013) Long-term outcome of male genital reconstruction in childhood. J. Pediatr. Urol., 9, 980–989. [DOI] [PubMed] [Google Scholar]
- 7. Park W., Zwink N., Rösch W.H., Schmiedeke E., Stein R., Schmidt D., Noeker M., Jenetzky E., Reutter H., Ebert A.-K. (2015) Sexual function in adult patients with classic bladder exstrophy: A multicenter study. J. Pediatr. Urol., 11, 125.e1–125.e6. [DOI] [PubMed] [Google Scholar]
- 8. Reddy S.S., Inouye B.M., Anele U.A., Abdelwahab M., Le B., Gearhart J.P., Rao P.K. (2015) Sexual health outcomes in adults with complete male epispadias. J. Urol., 194, 1091–1095. [DOI] [PubMed] [Google Scholar]
- 9. Karlsson O., Thor S., Norberg T., Ohlsson H., Edlund T. (1990) Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature, 344, 879–882. [DOI] [PubMed] [Google Scholar]
- 10. Draaken M., Knapp M., Pennimpede T., Schmidt J.M., Ebert A.-K., Rösch W., Stein R., Utsch B., Hirsch K., Boemers T.M.. et al. (2015) Genome-wide association study and meta-analysis identify ISL1 as genome-wide significant susceptibility gene for bladder exstrophy. PLoS Genet., 11, e1005024.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang R., Knapp M., Suzuki K., Kajioka D., Schmidt J.M., Winkler J., Yilmaz Ö., Pleschka M., Cao J., Kockum C.C.. et al. (2017) ISL1 is a major susceptibility gene for classic bladder exstrophy and a regulator of urinary tract development. Sci. Rep., 7, 42170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pfaff S.L., Mendelsohn M., Stewart C.L., Edlund T., Jessell T.M. (1996) Requirement for LIM Homeobox Gene Isl1 in motor neuron generation reveals a motor neuron– dependent step in interneuron differentiation. Cell, 84, 309–320. [DOI] [PubMed] [Google Scholar]
- 13. Ericson J., Norlin S., Jessell T.M., Edlund T. (1998) Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Dev. Camb. Engl., 125, 1005–1015. [DOI] [PubMed] [Google Scholar]
- 14. Hunter C.S., Dixit S., Cohen T., Ediger B., Wilcox C., Ferreira M., Westphal H., Stein R., May C.L. (2013) Islet α-, β-, and δ-cell development is controlled by the Ldb1 coregulator, acting primarily with the islet-1 transcription factor. Diabetes, 62, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Witzel H.R., Jungblut B., Choe C.P., Crump J.G., Braun T., Dobreva G. (2012) The LIM protein Ajuba restricts the second heart field progenitor pool by regulating Isl1 activity. Dev. Cell, 23, 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawakami Y., Marti M., Kawakami H., Itou J., Quach T., Johnson A., Sahara S., O’Leary D.D.M., Nakagawa Y., Lewandoski M.. et al. (2011) Islet1-mediated activation of the β-catenin pathway is necessary for hindlimb initiation in mice. Dev. Camb. Engl., 138, 4465–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Narkis G., Tzchori I., Cohen T., Holtz A., Wier E., Westphal H. (2012) Isl1 and Ldb co-regulators of transcription are essential early determinants of mouse limb development. Dev. Dyn., 241, 787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaku Y., Ohmori T., Kudo K., Fujimura S., Suzuki K., Evans S.M., Kawakami Y., Nishinakamura R. (2013) Islet1 deletion causes kidney agenesis and hydroureter resembling CAKUT. J. Am. Soc. Nephrol., 24, 1242–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suzuki K., Adachi Y., Numata T., Nakada S., Yanagita M., Nakagata N., Evans S.M., Graf D., Economides A., Haraguchi R.. et al. (2012) Reduced BMP signaling results in hindlimb fusion with lethal pelvic/urogenital organ aplasia: a new mouse model of sirenomelia. PLoS One, 7, e43453.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luria V., Krawchuk D., Jessell T.M., Laufer E., Kania A. (2008) Specification of motor axon trajectory by ephrin-B: EphB signaling: symmetrical control of axonal patterning in the developing limb. Neuron, 60, 1039–1053. [DOI] [PubMed] [Google Scholar]
- 21. Yang J.H., Menshenina J., Cunha G.R., Place N., Baskin L.S. (2010) Morphology of mouse external genitalia: implications for a role of estrogen in sexual dimorphism of the mouse genital tubercle. J. Urol., 184, 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodriguez E., Weiss D.A., Yang J.H., Menshenina J., Ferretti M., Cunha T.J., Barcellos D., Chan L.Y., Risbridger G., Cunha G.R.. et al. (2011) New insights on the morphology of adult mouse penis. Biol. Reprod., 85, 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Phillips T.R., Wright D.K., Gradie P.E., Johnston L.A., Pask A.J. (2015) A Comprehensive atlas of the adult mouse penis. Sex. Dev., 9, 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cho H.-H., Cargnin F., Kim Y., Lee B., Kwon R.-J., Nam H., Shen R., Barnes A.P., Lee J.W., Lee S.. et al. (2014) Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet., 10, e1004280.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liang X., Zhang Q., Cattaneo P., Zhuang S., Gong X., Spann N.J., Jiang C., Cao X., Zhao X., Zhang X.. et al. (2015) Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Invest., 125, 3256–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Infante C.R., Mihala A.G., Park S., Wang J.S., Johnson K.K., Lauderdale J.D., Menke D.B. (2015) Shared enhancer activity in the limbs and phallus and functional divergence of a limb-genital cis-regulatory element in snakes. Dev. Cell, 35, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haraguchi R., Suzuki K., Murakami R., Sakai M., Kamikawa M., Kengaku M., Sekine K., Kawano H., Kato S., Ueno N.. et al. (2000) Molecular analysis of external genitalia formation: the role of fibroblast growth factor (Fgf) genes during genital tubercle formation. Development, 127, 2471–2479. [DOI] [PubMed] [Google Scholar]
- 28. Yamaguchi T.P., Bradley A., McMahon A.P., Jones S. (1999) A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development, 126, 1211–1223. [DOI] [PubMed] [Google Scholar]
- 29. Suzuki K., Bachiller D., Chen Y.P., Kamikawa M., Ogi H., Haraguchi R., Ogino Y., Minami Y., Mishina Y., Ahn K.. et al. (2003) Regulation of outgrowth and apoptosis for the terminal appendage: external genitalia development by concerted actions of BMP signaling [corrected]. Development, 130, 6209–6220. 10.1242/dev.00846 [DOI] [PubMed] [Google Scholar]
- 30. Perriton C.L., Powles N., Chiang C., Maconochie M.K., Cohn M.J. (2002) Sonic hedgehog signaling from the urethral epithelium controls external genital development. Dev. Biol., 247, 26–46. [DOI] [PubMed] [Google Scholar]
- 31. Fogel J.L., Thein T.Z.T., Mariani F.V. (2012) Use of lysotracker to detect programmed cell death in embryos and differentiating embryonic stem cells. J. Vis. Exp., 10.3791/4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larsson J., Karlsson S. (2005) The role of Smad signaling in hematopoiesis. Oncogene, 24, 5676–5692. 10.1038/sj.onc.1208920 [DOI] [PubMed] [Google Scholar]
- 33. Golzio C., Havis E., Daubas P., Nuel G., Babarit C., Munnich A., Vekemans M., Zaffran S., Lyonnet S., Etchevers H.C., Schubert M. (2012) ISL1 directly regulates FGF10 transcription during human cardiac outflow formation. PLoS One, 7, e30677.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Itou J., Kawakami H., Quach T., Osterwalder M., Evans S.M., Zeller R., Kawakami Y. (2012) Islet1 regulates establishment of the posterior hindlimb field upstream of the Hand2-Shh morphoregulatory gene network in mouse embryos. Development, 139, 1620–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitsiadis T.A., Angeli I., James C., Lendahl U., Sharpe P.T. (2003) Role of Islet1 in the patterning of murine dentition. Development, 130, 4451–4460. [DOI] [PubMed] [Google Scholar]
- 36. Yang L., Cai C.-L., Lin L., Qyang Y., Chung C., Monteiro R.M., Mummery C.L., Fishman G.I., Cogen A., Evans S. (2006) Isl1Cre reveals a common Bmp pathway in heart and limb development. Dev. Camb. Engl., 133, 1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seifert A.W., Zheng Z., Ormerod B.K., Cohn M.J. (2010) Sonic hedgehog controls growth of external genitalia by regulating cell cycle kinetics. Nat. Commun., 1, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haraguchi R., Mo R., Hui C., Motoyama J., Makino S., Shiroishi T., Gaffield W., Yamada G. (2001) Unique functions of Sonic hedgehog signaling during external genitalia development. Development, 128, 4241–4250. [DOI] [PubMed] [Google Scholar]
- 39. Lin C., Yin Y., Veith G.M., Fisher A.V., Long F., Ma L. (2009) Temporal and spatial dissection of Shh signaling in genital tubercle development. Development, 136, 3959–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miyagawa S., Matsumaru D., Murashima A., Omori A., Satoh Y., Haraguchi R., Motoyama J., Iguchi T., Nakagata N., Hui C-c., Yamada G. (2011) The role of sonic hedgehog-Gli2 pathway in the masculinization of external genitalia. Endocrinology, 152, 2894–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morgan E.A., Nguyen S.B., Scott V., Stadler H.S. (2003) Loss of Bmp7 and Fgf8 signaling in Hoxa13-mutant mice causes hypospadia. Development, 130, 3095–3109. [DOI] [PubMed] [Google Scholar]
- 42. Herrera A.M., Shuster S.G., Perriton C.L., Cohn M.J. (2013) Developmental basis of phallus reduction during bird evolution. Curr. Biol., 23, 1065–1074. [DOI] [PubMed] [Google Scholar]
- 43. Meier P., Finch A., Evan G. (2000) Apoptosis in development. Nature, 407, 796–801. 10.1038/35037734 [DOI] [PubMed] [Google Scholar]
- 44. Nonomura K., Yamaguchi Y., Hamachi M., Koike M., Uchiyama Y., Nakazato K., Mochizuki A., Sakaue-Sawano A., Miyawaki A., Yoshida H. (2013) Local apoptosis modulates early mammalian brain development through the elimination of morphogen-producing cells. Dev. Cell, 27, 621–634. [DOI] [PubMed] [Google Scholar]
- 45. Vaahtokari A., Aberg T., Thesleff I. (1996) Apoptosis in the developing tooth: association with an embryonic signaling center and suppression by EGF and FGF-4. Dev. Camb. Engl., 122, 121–129. [DOI] [PubMed] [Google Scholar]
- 46. Rodriguez I., Araki K., Khatib K., Martinou J.C., Vassalli P. (1997) Mouse vaginal opening is an apoptosis-dependent process which can be prevented by the overexpression of Bcl2. Dev. Biol., 184, 115–121. [DOI] [PubMed] [Google Scholar]
- 47. Lindsten T., Ross A.J., King A., Zong W.-X., Rathmell J.C., Shiels H.A., Ulrich E., Waymire K.G., Mahar P., Frauwirth K.. et al. (2000) The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol. Cell, 6, 1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hübner A., Cavanagh-Kyros J., Rincon M., Flavell R.A., Davis R.J. (2010) Functional cooperation of the proapoptotic Bcl2 family proteins Bmf and Bim in vivo. Mol. Cell. Biol., 30, 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ito T., Bai T., Tanaka T., Yoshida K., Ueyama T., Miyajima M., Negishi T., Kawasaki T., Takamatsu H., Kikutani H.. et al. (2015) Semaphorin 4D induces vaginal epithelial cell apoptosis to control mouse postnatal vaginal tissue remodeling. Mol. Med. Rep., 11, 829–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin C., Yin Y., Long F., Ma L. (2008) Tissue-specific requirements of beta-catenin in external genitalia development. Dev. Camb. Engl., 135, 2815–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seifert A.W., Yamaguchi T., Cohn M.J. (2009) Functional and phylogenetic analysis shows that Fgf8 is a marker of genital induction in mammals but is not required for external genital development. Development, 136, 2643–2651. 10.1242/dev.036830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Satoh Y., Haraguchi R., Wright T.J., Mansour S.L., Partanen J., Hajihosseini M.K., Eswarakumar V.P., Lonai P., Yamada G. (2004) Regulation of external genitalia development by concerted actions of FGF ligands and FGF receptors. Anat. Embryol. (Berl.), 208, 479–486. [DOI] [PubMed] [Google Scholar]
- 53. Miyagawa S., Satoh Y., Haraguchi R., Suzuki K., Iguchi T., Taketo M.M., Nakagata N., Matsumoto T., Takeyama K., Kato S.. et al. (2009) Genetic interactions of the androgen and Wnt/beta-catenin pathways for the masculinization of external genitalia. Mol. Endocrinol., 23, 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miyagawa S., Moon A., Haraguchi R., Inoue C., Harada M., Nakahara C., Suzuki K., Matsumaru D., Kaneko T., Matsuo I.. et al. (2009) Dosage-dependent hedgehog signals integrated with Wnt/beta-catenin signaling regulate external genitalia formation as an appendicular program. Dev. Camb. Engl., 136, 3969–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin L., Cui L., Zhou W., Dufort D., Zhang X., Cai C.-L., Bu L., Yang L., Martin J., Kemler R.. et al. (2007) Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc. Natl. Acad. Sci. U. S. A., 104, 9313–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu H., Li Y., Wang Y., Liu Y., Wang W., Jia Z., Chen P., Ma K., Zhou C. (2014) Wnt-promoted Isl1 expression through a novel TCF/LEF1 binding site and H3K9 acetylation in early stages of cardiomyocyte differentiation of P19CL6 cells. Mol. Cell. Biochem., 391, 183–192. [DOI] [PubMed] [Google Scholar]
- 57. Muecke E.C. (1964) The role of the cloacal membrane in exstrophy: the first successful experimental study. J. Urol., 92, 659–667. [DOI] [PubMed] [Google Scholar]
- 58. Marshall V.F., Muecke E.C. (1968) Congenital Abnormalities of the Bladder In Malformations, Handbuch der Urologie/Encyclopedia of Urology/Encyclopédie d’Urologie. Springer Berlin Heidelberg, pp. 165–223. [Google Scholar]
- 59. Thomalla J.V., Rudolph R.A., Rink R.C., Mitchell M.E. (1985) Induction of cloacal exstrophy in the chick embryo using the CO2 laser. J. Urol., 134, 991–995. [DOI] [PubMed] [Google Scholar]
- 60. Wei X., Sulik K.K. (1993) Pathogenesis of craniofacial and body wall malformations induced by ochratoxin A in mice. Am. J. Med. Genet., 47, 862–871. [DOI] [PubMed] [Google Scholar]
- 61. Cheng W., Jacobs W.B., Zhang J.J.R., Moro A., Park J.-H., Kushida M., Qiu W., Mills A.A., Kim P.C.W. (2006) DeltaNp63 plays an anti-apoptotic role in ventral bladder development. Dev. Camb. Engl., 133, 4783–4792. [DOI] [PubMed] [Google Scholar]
- 62. Mahfuz I., Darling T., Wilkins S., White S., Cheng W. (2013) New insights into the pathogenesis of bladder exstrophy–epispadias complex. J. Pediatr. Urol., 9, 996–1005. [DOI] [PubMed] [Google Scholar]
- 63. Pan L., Deng M., Xie X., Gan L. (2008) ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Dev. Camb. Engl., 135, 1981–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.. et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Metscher B.D. (2009) MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev. Dyn., 238, 632–640. [DOI] [PubMed] [Google Scholar]
- 66. Bolger A.M., Lohse M., Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl., 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.. et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol., 9, R137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol, 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lawrence M., Gentleman R., Carey V. (2009) rtracklayer: an R package for interfacing with genome browsers. Bioinformatics, 25, 1841–1842. 10.1093/bioinformatics/btp328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J., Prlic A. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol., 9, e1003118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L.. et al. (2016) Ensembl 2016. Nucleic Acids Res., 44, D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V.. et al. (2012) A map of the cis-regulatory sequences in the mouse genome. Nature, 488, 116–120. 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M.. et al. (2015) The UCSC Genome Browser database: 2015 update. Nucleic Acids Res., 43, D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liao Y., Smyth G.K., Shi W. (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res., 41, e108.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Robinson M.D., McCarthy D.J., Smyth G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl., 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Diaz A., Nellore A., Song J.S. (2012) CHANCE: comprehensive software for quality control and validation of ChIP-seq data. Genome Biol., 13, R98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Edgar R., Domrachev M., Lash A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (78) and are accessible through GEO Series accession number GSE GSE91082 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE91082; date last accessed November 4, 2017).
Conflict of Interest statement. None declared.