Abstract
Understanding how activity patterns in specific neural circuits coordinate an animal’s behavior remains a key area of neuroscience research. Genetic tools and a brain of tractable complexity make Drosophila a premier model organism for these studies. Here, we review the wealth of reagents available to map and manipulate neuronal activity with light.
Keywords: nervous system, optogenetics, connectivity, Drosophila, functional imaging, methods, behavior, FlyBook
THE vinegar fly Drosophila melanogaster has made many contributions to our understanding of development, function, and diseases of the nervous system (Bellen et al. 2010; Hales et al. 2015). Just as its genes have shown informative homology to those of humans, the architecture and computations of its neural circuits may as well. Several relatively recent advances now permit the direct observation and manipulation of the activity of specific neurons in the brain and ventral nerve cord of the Drosophila larva and adult. “Functional imaging” allows the direct visualization of neural activity at the level of action potentials, synaptic inputs, calcium fluxes, neurotransmitter release, and intracellular signaling. “Optogenetics” enables light-controlled manipulation of specific neurons, to activate or silence them, or to drive or inhibit particular signaling pathways. Concomitant improvements in both sensors and imaging hardware have yielded improvements in sensitivity and spatiotemporal resolution. New reagent classes are being created as well, and the fly allows experiments not possible in other models due to the wealth of tools to target defined cell populations (e.g., Gal4, split-Gal4, LexA, and Q collections).
This review will focus on current methods for light-based imaging and the manipulation of neural activity in the fly nervous system. There are many experimental applications for optogenetics and functional imaging; here, we will highlight the way they have enabled the identification of neural circuits controlling fly behavior. Optogenetic control of neural activity can be used to identify neurons whose activation/silencing causes changes in behavior, while functional imaging shows which neurons have activity correlated with behavior or sensory stimulus. The two techniques are highly complementary, and can help map neurons necessary and/or sufficient for given behaviors. Often, deeper insights are possible as well, such as the activity thresholds of specific neurons and the firing sequence of distinct populations during behavior. We will describe case studies to illustrate how experiments reveal biological understanding, provide some of the technical considerations for optical imaging, and discuss the remaining challenges and prospects for future developments.
Functional Imaging: Watching Neurons in Action
In this review, we use functional imaging to refer to genetically encoded indicators of neural activity that report changes in intracellular calcium, neurotransmitter release, or voltage as changes in fluorescence. A broader definition of functional imaging might include the detection of neural activity by other means (e.g., MRI, PET, EEG, or chemical dyes). Here, we restrict discussion to protein-based reagents, mostly genetically encoded calcium indicators (GECIs) and genetically encoded voltage indicators (GEVIs), which allow us to acutely measure activity in the specific neurons in which the sensors are expressed. The archetypal GECI is GCaMP, a chimeric protein in which Green Fluorescent Protein (GFP) is circularly permuted (i.e., given new N- and C-termini) and fused to the calcium-binding protein calmodulin; in the presence of calcium, structural rearrangement of the sensor closes up the GFP barrel, dramatically increasing fluorescence output.
GECIs
GCaMP was first developed by Nakai et al. (2001). The original version has been iteratively improved (by multiple groups) through rational design and targeted mutagenesis to increase the total change in fluorescence in response to calcium (ΔF/Fo), control calcium-binding affinity, and accelerate response time—onset and offset kinetics—resulting in the recent GCaMP6 generation. All variants of GCaMP respond over relevant timescales to detect neural activity in fly neurons (although cellular calcium transients are slower than the underlying electrical activity). GECIs have been calibrated with respect to voltage- or calcium-sensitive chemical dyes (Hendel et al. 2008). Comparisons between GCaMP responses and simultaneous electrophysiological stimulation or recordings at the fly neuromuscular junction (NMJ) and in olfactory antennal lobe neurons (Jayaraman and Laurent 2007; Chen et al. 2013) demonstrate that fluorescence changes of GCaMP reliably reflect increases, and sometimes decreases, in neural activity in the populations of neurons in which it is expressed. Under some conditions and in some neurons, GCaMP6 is even capable of following single action potentials in single trials in live animals (Chen et al. 2013).
Through the use of protein-targeting sequences, GCaMP can be specifically expressed in different subcellular compartments of neurons. GCaMP is typically expressed in the neuronal cytoplasm, and changes in fluorescence can be measured in clusters of neurites forming glomeruli or in cell bodies. Synaptically targeted calcium indicators have also been made to compare activity levels of different boutons at the NMJ (Guerrero et al. 2005) and investigate sensory processing in Kenyon cell terminals in the mushroom body (Cohn et al. 2015).
Alternatives to GCaMP include ratiometric calcium sensors such as Cameleon, Camgaroo, TN-XL, or Twitch, where calcium binding induces a change in fluorescence from one wavelength to another (Fiala et al. 2002; Yu et al. 2003; Reiff et al. 2005; Mank et al. 2006, 2008; Thestrup et al. 2014). Such sensors might be a good choice when the imaging preparation is moving because sequential images can be registered by the ongoing basal fluorescence. GCaMP is also used in this way by coexpressing a standard fluorescent protein (FP) such as tdTomato in the same cells (Gruntman and Turner 2013). Alternatively, the standard FP can be directly fused to the GCaMP, for instance through the “nested doll” Green-Orange-Matryoshka-GCaMP6s fusion (Ast et al. 2017). CaMPARI (Fosque et al. 2015) is a calcium integrator, where neural activity is recorded during a user-specified time window by supply of photoconverting light, and read out later by measurement of the red-to-green ratio in whole brains. Photoconvertible calcium indicators are also available (Berlin et al. 2015); these allow the user-specified designation of specific cells of interest, for instance in areas of entangled neuron populations.
Red-shifted GECIs, including RCaMP, RGECO, and their improved variants including the jRGECO1 and jRCaMP1 series (Dana et al. 2016), have also been developed, using similar protein design strategies on red fluorescent proteins (RFPs). Red calcium indicators can be combined with Channelrhodopsin-2 (ChR2) activation in the blue–green spectrum for circuit mapping, or with green calcium indicators to visualize responses in two different neural populations at once (Sun et al. 2017) (see All Together Now section below).
GECIs do not promise “electrophysiology with microscopes,” but rather are new tools with their own applications. Experiments requiring electrophysiological precision should not realistically be attempted with GECIs. Electrophysiology remains the technique of choice to analyze the detailed electrical responses of neurons with temporal precision, and to assess the contributions of different ion channels and neurotransmitter receptors to their firing properties. Whole-cell recordings are performed at the larval NMJ and in some neurons in the adult fly brain, and multi-electrode recording of field potentials from many neurons is in progress. Paired electrophysiological recording remains the definitive measure of direct functional connectivity. While these techniques are challenging, they remain the only way to satisfactorily address some experimental questions.
GECIs enable a different type of experiment, visualizing neural activity in many identifiable neurons simultaneously. GECIs are well suited to identify brain areas with correlated activity, map regions that respond to stimuli in more intact and behaving animals, screen agnostically for connected brain regions, and visualize how activity progresses through sequential layers in a neural circuit.
Careful analysis of GECI imaging data can be challenging. Increases in cell body calcium typically track action potentials, and both presynaptic vesicle release and postsynaptic receptor activation are accompanied by calcium flux through voltage-gated calcium channels and neurotransmitter-gated ion channels, such as N-methyl-D-aspartate NMDA ionotropic glutamate and nicotinic acetylcholine receptors. This means that most aspects of neural activity can be measured to good effect with GECIs. GCaMP imaging is often described as a “proxy for neural activity,” but a nuanced view acknowledges that there are multiple sources of calcium that can produce changes in cellular levels. Since GCaMP reports calcium binding, regardless of where that calcium comes from, alternative sources should always be considered. When a neuron fires an action potential, rapid depolarization causes voltage-gated calcium channels to open, allowing an influx of extracellular calcium, localized near synapses (which then triggers synaptic vesicle release). While this source of calcium is probably the largest and fastest for most neurons, there are certainly other mechanisms that change the calcium concentration. transient receptor potential (TRP) channels and ligand-gated receptors can also admit extracellular Ca2+, and Ca2+ can be released from intracellular stores in the endoplasmic reticulum (Grienberger and Konnerth 2012; Restrepo and Basler 2016; Xu et al. 2017). Addition of a voltage-gated calcium channel blocker, or an action potential blocker, could be used to confirm that most of the fluorescence change reported by the GECI was indeed due to action potential-evoked Ca2+ influx.
While GECIs effectively report neural activity in many cells, there are regimes in which voltage indicators would be more appropriate. These include but are not limited to: (1) neurons with large amounts of calcium buffering, (2) fast-spiking neurons where the relatively slow kinetics of GECIs prohibit the measurement of spike timing or rate, (3) measurements of subthreshold events such as dendritic integration or postsynaptic potentials, and (4) measurement of inhibitory activity.
GEVIs
The first GEVI was based on the voltage paddle of the shaker potassium channel (Siegel and Isacoff 1997; Guerrero et al. 2002), and was used to assess voltage changes at the larval NMJ. ArcLight (Cao et al. 2013) is a recent variant based on a voltage-gated phosphatase rather than ion channels (and thus is potentially less disruptive to cells). ArcLight has been tested in the adult circadian circuit (Sitaraman et al. 2015), and neurons controlling courtship (Kallman et al. 2015) or sleep (Haynes et al. 2015), among others. Additional voltage sensors have been made, including those with improved kinetics [e.g., ASAP (St-Pierre et al. 2014; Yang et al. 2016; Chamberland et al. 2017)], variants with positive-going signal changes for improved signal-to-noise (Platisa et al. 2017), and red variants [FlicR1 (Abdelfattah et al. 2016)]. Some basic types of voltage sensors were reviewed in St-Pierre et al. (2015) and Vogt (2015). Other options in development have not yet been imported into flies (Inagaki et al. 2017).
Alternative Reporters of Neural Activity
Other genetically encoded reporters track neural activity by visualizing neurotransmitter release. SynaptopHluorin (Miesenbock et al. 1998) fluoresces in response to the pH change that occurs when the reporter moves from inside a synaptic vesicle to outside the cell as the vesicle fuses to the membrane. An alternative version of SynaptopHluorin, dVMAT-pHluorin, was used to track the effect of the psychostimulants methamphetamine and methylphenidate on fly synaptic activity (Freyberg et al. 2016). A similar strategy produced a connectivity tracer based on reconstitution of GFP at active synapses (Macpherson et al. 2015). Neuropeptides are released from dense core vesicles and a fusion protein, upstream activating sequence (UAS)-ANF-EMD, which combines a neuropeptide and a GFP reporter for peptide release (Rao et al. 2001; Husain and Ewer 2004). Another sensor, iGluSnFR, fluoresces upon binding the neurotransmitter glutamate (Marvin et al. 2013) and helped establish the role of fibroblast growth factor signaling in the elaboration of glial contacts with neurons and other glia in the developing fly brain (Stork et al. 2014).
There are other sensors that report neuronal activity by initiating the transcription of reporters. These are often used to identify functional connections post hoc rather than by live imaging. DopR-Tango labels neurons that respond to endogenous dopamine release (Inagaki et al. 2012) while Tango-TRACE responds to histamine (Jagadish et al. 2014). Calcium-activated transcription systems report increased neural activity, but on slower timescales than GECIs (Masuyama et al. 2012; Gao et al. 2015). Bioluminescent reporters count photon accumulation over time (Martin 2008) and have much lower background signals than fluorescent ones, at the expense of knowing when specific cells were active.
Imaging analysis
There are several recent reviews of functional imaging (Grienberger and Konnerth 2012; Broussard et al. 2014) in Drosophila (Dipt et al. 2014), and a range of research papers have used functional imaging to determine how neural activity correlates with sensory input or behavior output. Neurons in the central complex ellipsoid body track the fly’s heading relative to landmarks in visual environments and maintain this representation in darkness by tracking self-motion (Seelig and Jayaraman 2015). Specific neurons in the subesophageal zone (SEZ) have activity patterns that correlate with different tastes (Marella et al. 2006; Harris et al. 2015). Whole-brain GCaMP imaging in the larva (Lemon et al. 2015) during crawling revealed candidate central pattern generator components (Pulver et al. 2015) and inhibitory neurons that delay activity in certain motor pools to produce behavioral sequences (Zwart et al. 2016). Whole-brain imaging of intrinsic functional activity in the adult shows correlations between connected regions (Mann et al. 2017). Lower-resolution whole-brain imaging during open field behavior in adult flies has also been attempted (Grover et al. 2016).
Most of the reporters described here require live imaging of changes in fluorescence in the nervous system. To follow the changes in fluorescence that correlate with a sensory stimulus, an optogenetic trigger or a behavioral action demands rapid imaging in the correct part of the brain. A baseline measurement of the level of fluorescence before the stimulus must be compared to the level during and after it. The GECI-response kinetics and the imaging rate must together be fast enough to capture the target calcium transients, including those resulting from action potentials and from synaptic inputs. In either case, the underlying electrical events are quite rapid, with the resulting calcium transients in the order of 100s of milliseconds for single events, and several seconds for large events. Following such events, particularly over large scan areas and even volumetric imaging, with good spatial and temporal resolution can be technically challenging.
In Drosophila, there are a variety of microscopic methods for delivering excitation light and collecting emitted photons, to focus the experiment on the region(s) of interest. Confocal microscopes (scanning or spinning disk) deliver excitation illumination broadly and collect it from planes of interest. Confocal microscopy is routine and can produce reliable results. Some concerns with the method include: optical sectioning is imperfect, so that cell bodies closely packed in the z-dimension can contribute misleading out-of-focus fluorescence; and as excitation is delivered broadly, expressed probes can photobleach during the experiment. Light sheet microscopy achieves optical sectioning at the level of excitation light delivery, and is currently applicable to Drosophila embryos and larvae (Chhetri et al. 2015). Finally, two-photon microscopy permits much tighter restriction of the excitation light and better penetration through scattering tissue, such as the fly cuticle. Downsides of two-photon microscopy include more expensive rigs and typically slower volume scan rates. GCaMP fluorescence is elicited by a fixed wavelength laser in green (488 nm) or a two-photon laser (920–1000 nm are typical wavelengths). The illumination can be wide field or targeted in the X–Y plane by galvanometer (fast) or resonant galvanometer scanning mirrors (faster). In the wide-field configuration, the resulting fluorescence can be detected by a camera (typically an electron multiplying charge coupled device (EMCCD) or Complementary metal–oxide–semiconductor (cMOS)), which detects all signals over the whole plane at once but at all depths in Z. When in the confocal or two-photon configuration, multi-alkali or gallium arsenide phosphide GaAsP photomultiplier tube detectors (PMTs) record the emitted light. The excitation and emission light signals must be separated; this is typically done with band-pass, long-pass, or dichroic filters that allow the excitation light to pass through the microscope to the sample, but only the emission light to reach the detectors. Temporal gating is also possible, where the illumination and detection are separated in time with the PMTs shuttered during illumination and then opened after illumination stops. Example microscope configurations are described in the methods sections of several recent papers [Seelig et al. 2010; Seelig and Jayaraman, 2013, 2015; Kallman et al. 2015; Green et al. 2017; Sun et al. 2017; Turner-Evans et al. 2017; for additional protocols, see also Lemon et al. (2015) and Schmied and Tomancak (2016)].
Finding the neurons that respond to a particular sensory input can be accomplished by expressing GCaMP in many neurons, applying the stimulus, and fast-scanning a focused laser beam with two-photon excitation through a volume of the brain with a scanner and a piezo motor on the objective lens to rapidly change the focal depth. Once the responding region of the brain has been identified, more detailed or rapid scans of a smaller volume (region of interest or line-scanning) can be performed.
There are tradeoffs among imaging speed, area (field of view), sensitivity, and resolution. Detection of a weak signal (sensor is dim and/or has low fluorescence change) requires longer dwell times, and reduces the speed and area that can be sampled. Imaging the whole field of view simultaneously with a camera is fast but captures unrelated light at other depths (this results in out-of-focus information, poorer spatial resolution, and lower sensitivity to activity-related fluorescence changes). It can be challenging to identify the neurons that responded if GCaMP is broadly expressed. The first problem is detecting signal from noise, given widespread low-level activity throughout the nervous system. The second problem is that even when you do detect signal, knowing precisely which neurons it came from is hard (coexpressing a photoactivatable fluorophore responding to a distinct wavelength to label the responding cells after functional imaging may solve this).
Expressing GCaMP6 in small subsets of neurons (e.g., from Gal4-UAS) allows wide-field capture but risks missing other neurons that might also be involved. The relative timing of activity can only be assessed within the expressing population. It can be hard to locate the cells of interest in the imaging preparation if GCaMP is only expressed sparsely. This can be overcome by using a GECI with higher basal fluorescence (e.g., GCaMP3 or GCaMP5), or by coexpressing a standard fluorophore (e.g., tdTomato) to target functional imaging in the preparation.
Typical functional imaging data are collected as movies or Z-stacks in software supplied by the microscope vendors. These files are often large and can be compressed, downsampled, or reduced by selecting smaller regions of interest. Popular software packages for analysis are available on the ImageJ/FIJI platform (Schindelin et al. 2012), or as custom scripts written by individual labs in MATLAB or Python, for example. Sometimes the changes in fluorescence are visible to the eye, but in other cases image processing is required to detect significant signal. Data are usually reported as the change in fluorescence divided by the baseline fluorescence (y-axis) vs. time (x-axis) with an image of the region of interest in the brain where signal is measured. A complementary presentation with single-pixel resolution is a heat map, with maximum (or average) fluorescence response for each pixel in an image shown using a color scale. Examples of these are shown in Figure 1.
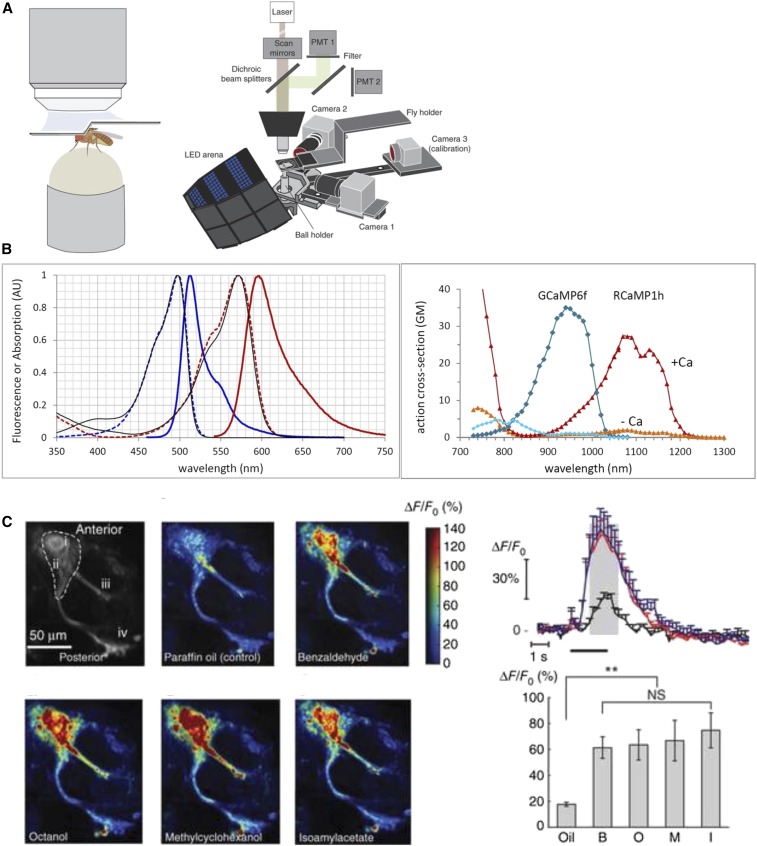
Figure 1.
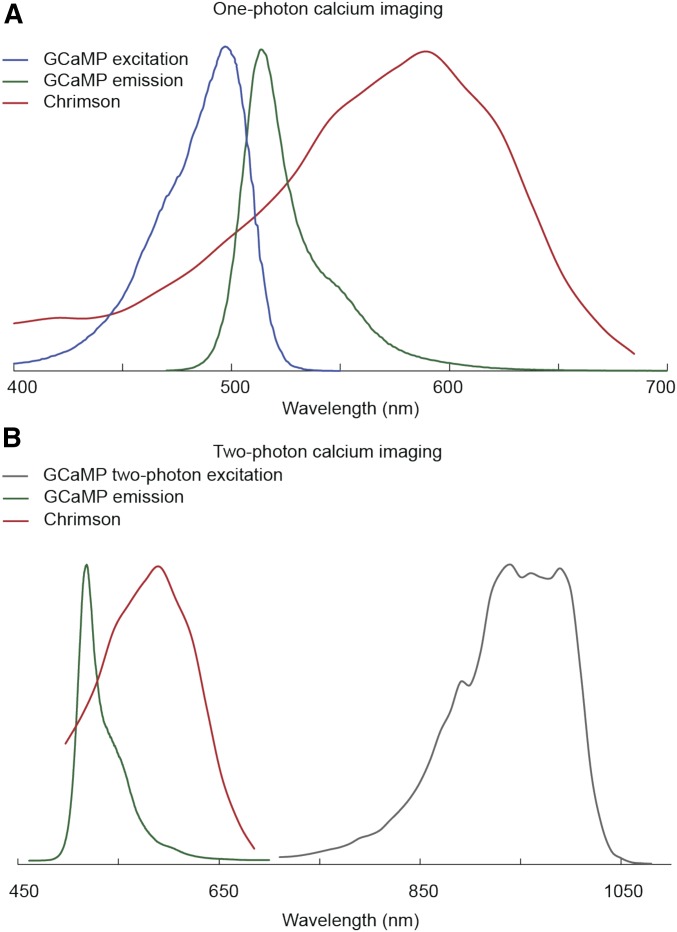
Functional imaging in the adult fly brain with GCaMP. (A) Schematic of a two-photon imaging rig, showing a tethered fly walking on a ball with a window cut in the head for visualization of GCaMP fluorescence changes in the brain. This diagram is taken from figure 1 in Seelig et al. (2010); alternative schematics are shown in Reiff et al. (2010), Maimon et al. (2010), and Mamiya and Dickinson (2015). (B) Excitation and emission spectra of GCaMP and RCaMP (Chen et al. 2013; Dana et al. 2016), taken from https://www.janelia.org/lab/harris-lab-apig/research/photophysics/two-photon-fluorescent-probes. (C) Examples of ways to show changes in GCaMP fluorescence in mushroom body neurons in response to odors [taken from figure 6 in Sejourne et al. (2011)]. AU, arbitrary units; GM, Goeppert-Mayer Units; LED, light-emitting diode; **, very significant P value between 0.001 and 0.01; NS, not significant; PMT, photomultiplier tube detector.
Examples
An elegant demonstration of the power of functional imaging comes from mapping the spatiotemporal propagation of fly taste perception from the peripheral taste cells on the proboscis to their first integration point in the SEZ (Harris et al. 2015). The study is noteworthy both for the sheer scope of the functional imaging and for the simplicity in converting functional imaging data to testable biological predictions, nicely illustrating a best-use scenario for an indicator of neural activity.
Flies expressing the sensitive, slow GECI GCaMP6s were head-fixed and lightly dissected to gain optical access to the SEZ. The entire SEZ was imaged on a spinning-disk confocal microscope, encompassing 23 separate, 250 × 250 μm planes. Specific tastants (sweet, bitter, and water) were added either individually or in mixtures to the flies’ taste cells, with volumetric imaging of the SEZ being done in the 2 sec following taste stimulation. Regions of interest around individual cells were automatically segmented, and the three-dimensional Z-stack was flattened to aid in visualization. Robust (∼20–100% ΔF/F0) GCaMP6 responses were seen following each addition of tastant, which in combination with a nuclear-localized RFP allowed clear segmentation of active cell bodies.
First, some technical notes on the study. The SEZ neurons fire many action potentials in response to taste stimulation, leading to robust Ca2+ transients in the cell bodies. This is not always the case in Drosophila; many neurons have graded potentials instead of action potentials, fire too few spikes to be detected, or fire spikes yet have only small concomitant somatic Ca2+ rises. In these latter instances, a user might prefer a voltage indicator, which could potentially pick up subthreshold and/or neurite-specific activity. However, in the current example, the choice of GCaMP6s was excellent: the sensitivity of the indicator presumably allowed the detection of all active cells; the slow decay of the signal permitted volumetric imaging without noticeable signal loss in later frames; and the concentration of the signal in the cell body permitted facile segmentation for use in delimiting taste maps. In this example, the use of a GEVI would have been problematic, as their lower signal-to-noise, response across somata and neuropils, and rapid kinetics would have led to much smaller signal changes spread across much larger areas, likely complicating the determination of active cells during volumetric scanning. Another important point is that the authors expressed their indicator in all the cells of the SEZ, allowing the same fly to be used for multiple experiments. The reversible nature of the GCaMP indicator is also critical to this; for instance, irreversible integrators like CaMPARI or immediate early gene-based readouts would be much more complicated to use.
The results from this study are as elegant as the method: the sweet, bitter, and water tastants activate nonoverlapping cells, consistent with the “labeled-line” notion of sensory processing. Furthermore, taste mixes recruited no additional cells, but rather dropped cells out, consistent with mutual inhibition (“mixture suppression”), and increased tastant concentrations recruited more cells in the SEZ. Finally, the authors traced the activity from the SEZ to higher brain centers, including the mushroom body and the pars intercerebralis, with sweet- and bitter-evoked activity remaining in labeled lines as far as the activity was traced.
Finally, the study is informative because of the performance of control experiments to aid in functional imaging interpretation. First, calcium activity in the SEZ corresponds precisely with the fly’s appropriate behavioral output, proboscis extension, even over a broad tastant concentration range, suggesting that the cells being imaged are relevant. The activation of SEZ sweet cells leads to the recruitment of directly adjacent motor neurons known to control proboscis extension (and bitter cells with retraction), strongly suggesting that the neuronal activity being imaged is not only relevant but causal. The comprehensive scope (1000s of neurons imaged near simultaneously), the easy trial repetition, the ability to image diverse stimuli in the same animal (brains are not completely stereotyped, even in the fly), the rapid experimental turnaround, and the unambiguous designation of active and inactive cells are direct products of the advanced state of functional imaging, with robust activity reporters, turnkey microscopy, and straightforward data analysis.
A second example of the power of functional imaging in the fly takes advantage of both GECIs and GEVIs (in different animals), with each providing specific insights [Yang et al. 2016; reviewed in Kaschula and Salecker (2016)]. This study uses specific Gal4 lines to target specific neuronal cell types in the adult fly visual system, from the lamina (the lamina monopolar cells L1 and L2, which collect the output of the retinal photoreceptors) to the medulla (the three transmedullary neurons Tm1, Tm2, and Tm3, and the medulla-intrinsic neuron Mi1), which then sends projections to visual output areas such as the lobula and lobular plate. Each of these neurons arborizes in multiple layers of the medulla, allowing the targeted imaging of subcellular compartments of each neuron to follow the transformation of the sensory input across multiple processing levels. Both the GECIs (GCaMP6f, fast; and GCaMP6m, medium) and the GEVI (ASAP2f) used in the study were compatible with two-photon imaging of the individual subcellular arborizations (both axons and dendrites) in defined layers of the medulla.
The authors first imaged voltage responses in the Mi1 interneuron, from its cell body to subsequent arborizations in medulla layers 1 and 5 (where input from L1, the dominant presynaptic partner, is received) and layer 10 (where Mi1 axons target the columnar interneuron T4). ASAP2f signal changes (increasing in response to hyperpolarization and decreasing following depolarization) were observable at all four imaging locations, although trial-to-trial variability was quite high, with 4–5% peak | ΔF/F0 | obtainable from 100-trial averages. ASAP2f fluorescence responses were strongest in the proximal arborization in layer 1, and fell off progressively in amplitude; responses in layer 10 were similar to those in the cell body (most electrical activity in Drosophila neurons occurs in the processes). However, the time courses of the voltage traces were largely unchanged, suggesting that depolarization propagation (the cells in the fly visual system are thought to be nonspiking) and GEVI-response kinetics are essentially identical in the different subcellular locales.
The study next examined the transformation of voltage signals across synapses, namely from laminar L2 to medullar Tm1 and Tm2, and separately from laminar L1 to medullar Tm3 and Mi1. In L2, axon terminals hyperpolarized in response to light and depolarized in the dark, and this property was preserved in postsynaptic partners Tm1 and Tm2 (imaged at their layer 2 arborizations with L2). Tm1/2 voltage signals were larger and slower than those in L2, suggesting that the dendrites integrate L2 input to ensure reliable propagation. The L1 neuron arborizes with Tm3 and Mi1 in both layers 1 and 5, and L1 ASAP2f imaging in layer 1 was similar to that of L2. However, strikingly, the ASAP2f responses of Mi1 and Tm3 showed sign inversion relative to L1, and as before were larger and slower than their presynaptic partners. Taken together, these GEVI imaging data confirm a large body of work on the fly visual system, most basically that there are parallel “OFF” (i.e., L2 and its sign-preserving, light-inhibited postsynaptic partners) and “ON” (i.e., L1 and its sign-inverting, light-activated postsynaptic partners) pathways. At a more detailed level, both the duration of Tm1/2 and Tm3/Mi1 voltage changes, as well as their relative timing, is consistent with electrical recordings from all the cells involved (Behnia et al. 2014).
The authors then imaged GCaMP6f in the same neuron combinations (in separate lines from the ASAP flies). In contrast to the voltage data, calcium imaging revealed dramatically different responses in the individual compartments that were not even rank-ordered with the voltage changes (i.e., calcium signals were highest in the proximal and distal arborizations of Tm3, and much lower in the cell body and intermediate arborization). In the Mi1 neuron, the GCaMP6m response amplitudes in the three layers rank-ordered according those of ASAP6f, but intriguingly the kinetics did not: layers 1 and 5 showed large, slow responses, whereas layer 10 showed a more rapid, biphasic response (indicating that local Ca2+ concentration dips below resting levels during the rebound phase of the response). Taken together, these data reveal that different compartments of the same neuron have diverse calcium-handling machinery, most obviously beginning with voltage-gated Ca2+ (CaV) channels. Thus, a single neuron can have multiple output mechanisms across its extent, as Ca2+ directly regulates properties such as neurotransmitter release, long-term synaptic potentiation and depression, local mRNA translation, and more.
Such an experiment is only possible with modern functional imaging and the current generation of voltage and calcium indicators. Electrophysiological recordings from Drosophila cell bodies are difficult due to their small size, and targeted recordings from axons and dendrites are likely impossible. Previous voltage indicators were incompatible with two-photon excitation (required for the subcellular resolution in this study) and/or had miniscule responses. Further experiments can determine the sources of local Ca2+ signaling, and eventually its purpose in neural computations and circuit function.
Fly line recommendations
There are numerous GECI fly lines available at the Bloomington Drosophila Stock Center (http://fly.bio.indiana.edu), including the GCaMP6 indicators under the control of UAS (Bloomington #42746–42750), LexAop (44273–44277, 44588–44590), or piggybac (52869), and synaptotagmin-fused GCaMP6 (sytGCaMP) for imaging in presynaptic terminals (64413–64416). In Drosophila, users have typically favored the GCaMP6m (medium) over the GCaMP6s (sensitive) and GCaMP6f (fast) variants, although the best choice depends on the firing rate of the neurons. For imaging activity in axonal terminals, the sytGCaMP variants are probably preferred. Many labs make and optimize genetically encoded indicators for use in flies; one site that we recommend that potential users check is the Howard Hughes Medical Institute Janelia Genetically Encoded Neuronal Indicator and Effector (GENIE) Project site (https://www.janelia.org/project-team/genie/tools).
ASAP2f fly lines are available from Bloomington as well, under UAS (65414), piggybac (65415), or LexAop (67674) control. The ArcLight GEVIs are available from Bloomington (51056–51057). A red GEVI, FlicR1, has been recently developed and fly stocks are available (63434). Another new GEVI, Marina (Platisa et al. 2017), has positive fluorescence response to depolarization and thus has greater signal-to-noise for most voltage-imaging applications. Fly lines are constantly being added to the Bloomington collection: https://bdscweb.webtest.iu.edu/stock/gfp/gfp_markers.php. Currently, ASAP2f or Marina is likely to provide the best results for GEVI imaging in the fly brain. Many groups are working to optimize GEVIs for use in all applications; there are likely to be a great many developments in this field over the next several years. Potential users should be on the lookout for new variants with improved membrane targeting, brightness, photostability, and signal change.
Optogenetics: Altering Neural Activity
Optogenetics uses genetically targeted expression of light-activated proteins, particularly ion channels and pumps, to alter neural activity. It was the Science “breakthrough of the year in 2005” and there were several reviews marking the 10-year anniversary (Boyden 2015; Deisseroth 2015). The first combination of transgenes and light to activate neurons expressed components of the Drosophila phototransduction cascade in mammalian neurons, or the ATP-gated P2X2 ion channel in Drosophila, activated by optically-uncaged ATP (Zemelman et al. 2002; Lima and Miesenbock 2005). Currently, preferred neuronal activators include ChR2, a green light (470 nm)-sensitive nonselective cation channel derived from the green alga Chlamydomonas reinhardtii, and CsChrimson, a red light (590 nm)-activated channel from the red alga C. subdivisa. Spectral profiles are shown in Figure 2A.
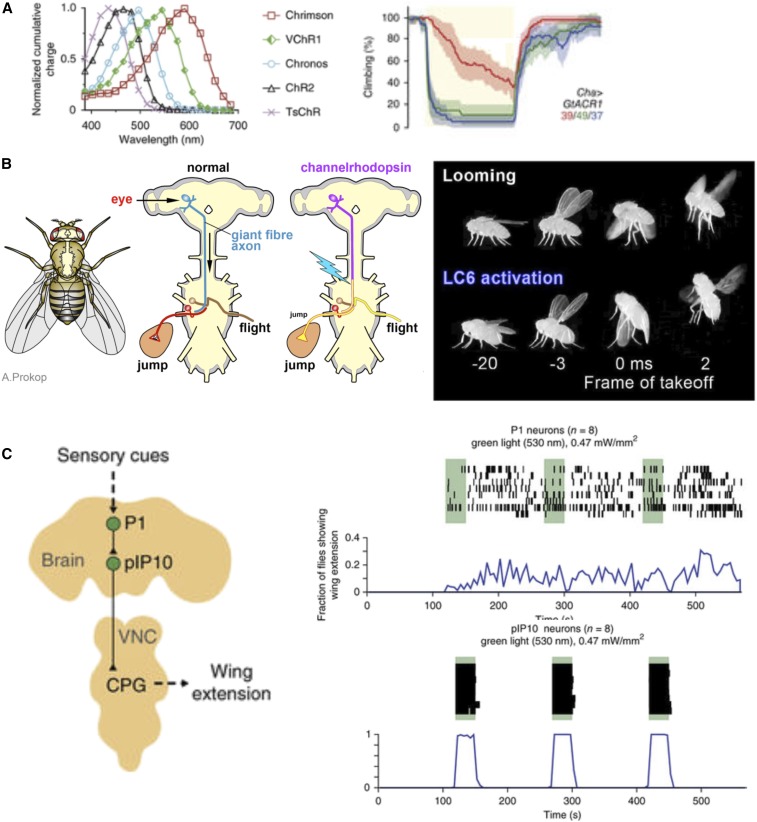
Figure 2.
Optogenetics. (A) Excitation spectra for ChR and Chrimson activators [figure 1E in Klapoetke et al. (2014)], and the response of flies expressing the gtAcr inhibitor to different wavelengths [figure 1A in Mohammad et al. (2017)]. (B) Schematic showing how optogenetic activation can mimic visual looming stimulus to evoke a jump-escape response. Diagrams adapted from A. Prokop: https://droso4schools.wordpress.com/about-us/. Experiment presented in Wu et al. (2016). (C) Activation of P1 neurons with ReaChR evokes a lasting courtship state, while activation of the P10 neurons elicits wing extension time-locked to stimulus (Inagaki et al. 2014). CPG, central pattern generator; VNC, ventral nerve cord.
The first reported use of ChR2 in flies activated multi-dendritic nociceptive neurons (Hwang et al. 2007), and in fact this P-element transgene insertion of UAS-ChR2 is still the most widely used stock. Protein engineering has improved the expression level and membrane trafficking of opsin constructs, and modified their electrophysiological properties. Variants exist that increase cation conductance or photocurrent amplitude (CatCh, ChIEF, ChR2-XXL, and ChR2-TC), increase onset and offset kinetics (ChETA or Chronos), and switch them between bistable open and closed states with different wavelengths of light (Step-Opsins). Chronos (Klapoetke et al. 2014) and ChR2-XXL (Dawydow et al. 2014) show increased light sensitivity in larvae and adults.
Efforts to identify or design opsins that respond to longer wavelengths resulted in a range of red-shifted variants. C1V1 and ReaChR (Inagaki et al. 2014) are chimeric opsins that respond in the red–yellow range, while CsChrimson (Klapoetke et al. 2014) is based on a naturally occurring opsin. Blue-shifted opsins have also been reported (e.g., CheRiff), although these have not yet been widely used. Currently, all opsins rely on all-trans retinal for their photoconversion steps, but semisynthetic retinals that could shift the activating wavelengths are under study (AzimiHashemi et al. 2014; Herwig et al. 2017).
ReaChR and CsChrimson have been expressed in flies. Longer wavelengths of light penetrate the cuticle better, allowing activation of deep-brain neurons in intact freely moving adult flies. Far-red wavelengths are likely out of the flies’ visual detection range, triggering fewer behavioral responses. The red light-activated reagents can be combined with ChR2 under orthogonal genetic expression control systems (Gal4, LexA, and Q) to differentially activate two distinct populations of neurons in the same preparation. Red-shifted opsins can be coupled with GCaMP imaging (see All Together Now section below).
Robustly silencing neurons with light has been more challenging. The first optogenetic silencing was achieved with a light-sensitive (570 nm) anion pump halorhodopsin (NpH 2) from the archeobacteria Natronobacterium pharaonis. Protein engineering has improved trafficking and stability, and these have been used in larvae (Inada et al. 2011), but the amount of chloride current per photon remains low, making complete silencing difficult. The next wave of silencers included Arch and Jaws (proton pumps), as well as versions of ChR2 designed to conduct chloride (ChloC). A naturally occurring light-sensitive chloride channel, gtAcr, was recently cloned from the alga Guillardia theta and has been expressed in flies (Mauss et al. 2017; Mohammad et al. 2017); this shows promise. (Figure 2A). It should be noted that chloride reversal potentials can vary dramatically both between cell types and within a single cell across developmental times, and, as such, optogenetic chloride channels and pumps might not be ideal. Similarly, proton currents are subject to rapid homeostatic compensation by neurons, meaning that proton channels and pumps might also provide unreliable silencing. An ideal optogenetic silencer would specifically conduct potassium ions, as the reversal potential is strongly hyperpolarizing in most cell types and developmental stages. Tools based on light-gated potassium channels are in their infancy (Cosentino et al. 2015) and have not yet been tested in flies.
It is possible to use light to alter some signal transduction pathways that are not specific to neurons. Cyclic AMP (cAMP) levels can be monitored with fluorescent sensors and manipulated with light using Epac1-camps, bPAC, and euPAC (Schroder-Lang et al. 2007; Shafer et al. 2008; Stierl et al. 2011); these tools have been reviewed in Leilito and Shafer (2012) and Patel and Gold (2015). Cell death can be induced by light-activated production of reactive oxygen species using reagents such as KillerRed and miniSOG, reviewed in Wojtovich and Foster (2014), or light-activated caspases (Smart et al. 2017). Some of these approaches have been employed in Drosophila neurons but are beyond the scope of this review.
Activation and Data Analysis
Optogenetic reagents are activated by light. The simplest protocol is to express UAS-ChR2 in neurons of interest with a Gal4 line, rear flies on 0.2 mM all-trans retinal (flies produce retinal, but the addition of exogenous retinal boosts performance) food in the dark, and then expose them to wide-field light and follow their behavior. Infrared illumination can be used to track the behavior of flies undergoing optogenetic stimulation with red or green light (Robie et al. 2017b). Activation light can be supplied by light-emitting diodes of specific wavelengths, either broadly or localized with fiber optics (Bath et al. 2014; Hsiao et al. 2015; Morton et al. 2016), or by lasers (Wu et al. 2014). Holographic projectors have also been used to supply custom patterns of illumination. Fixed wavelength and two-photon lasers can also be used to activate optogenetic constructs through the microscope objectives of a two-photon microscope (Rickgauer and Tank 2009; Inagaki et al. 2014), although this can be technically challenging.
There have been several recent reviews of the repertoire of genetic reagents available to manipulate neuronal activity (Venken et al. 2011; Owald et al. 2015; Hampel and Seeds 2017) Reviews and protocols specific for optogenetic manipulation of neural activity include Zhang et al. (2007), Honjo et al. (2012), Klapoetke et al. (2014), Titlow et al. (2015), and Riemensperger et al. (2016).
Earlier screens used transgenes that kill cells or interfere with neural transmission (e.g., the apoptotic gene reaper; tetanus toxin, which cleaves neural synaptobrevin; the inward-rectifying potassium channel Kir2.1; and the bacterial sodium channel NaChBac), RNA interference (RNAi) to knock down signaling molecules or receptors, or thermogenetic control of neural activity (e.g., the heat-gated calcium channel dTrpA1 or the temperature-sensitive dynamin Shibirets1 allele, which inhibits vesicle fusion) to identify neurons critical for a particular behavior. Optogenetic activation or inhibition of neuronal activity is being used in a similar manner. Most published experiments use optogenetics to test the role of candidate neurons, but large-scale unbiased screens are in progress. Optogenetic activation has mapped neurons that promote aggression (Hoopfer et al. 2015) and circuits for visual processing (Wu et al. 2016). Some experiments have taken full advantage of the precise temporal control that light and optogenetics enables; ReaChR helped identify neurons eliciting a behavioral state that promotes courtship as opposed to initiating an acute action (Inagaki et al. 2014). See Figure 2 for example uses of optogenetics. The temporally precise nature of light activation was key for determining which neurons evoke a behavior in a time-locked manner vs. which neurons initiate a reciprocal excitation loop that leads to persistence beyond the triggering stimulus in antennal grooming (Hampel et al. 2015). Numerous experiments have identified the temporal requirements for activity in specific neurons during learning, memory, and reward (Schroll et al. 2006; Claridge-Chang et al. 2009).
In electrophysiological recording, especially with extracellular electrodes, it can be difficult to identify the neurons associated with a particular activity pattern (i.e., spike sorting and then identification). Optogenetic reagents expressed in known cell types can be used after electrophysiological recording to determine which signals came from that neuron type. This use of ChR2 has been reported in mice (Lima et al. 2009) but not yet in flies.
Example
A demonstration of the power of optogenetics in reverse-engineering Drosophila neural circuits comes from larval locomotion (Fushiki et al. 2016). This study sought to determine the neurons and connections responsible for the coordinated contraction of larval body wall muscles involved in crawling. The authors first used anatomical imaging to identify specific neuronal cell types [a gamma-Aminobutyric acid (GABA) dorsolateral neuron, “GDL,” and an excitatory neuron, A27h; both present in each hemisegment of the repetitive larval body structure, and both targetable with specific Gal4 lines] deemed likely (from morphology, projection patterns, and antibody labeling) to be involved in rhythmic motor activity. This was then verified with functional imaging, by expressing GCaMP6m in GDLs and upstream motor neurons, which when imaged during fictive crawling clearly showed rhythmic activity, with GDL and motor neuron activity propagating, as expected, from the larval posterior to anterior segments. The necessity of the GDLs for rhythmic locomotion was first proven using chronic manipulations rather than acute ones such as optogenetics. Both tetanus toxin (which cleaves synaptobrevin, a key component of the SNARE vesicle machinery, thus inhibiting neurotransmission) and RNAi knockdown of the GABA biosynthetic pathway in the GDLs dramatically altered locomotion, making it less frequent and slower.
A different pipeline was used to characterize the excitatory neuron A27h, beginning with anatomy (confocal fluorescence imaging followed by sparse electron microscopy reconstruction) and antibody labeling. Conversion of photoactivatable GFP specifically in A27h identified it as being directly presynaptic (with excitatory cholinergic terminals) to the motor neuron in each segment. The conclusive demonstration that A27h activation drives motor neuron firing and contraction of a specific segment would not be possible with a gross manipulation such as thermal activation of dTRPA1, because the Gal4 expression pattern covers all larval segments, and thermal activation is not easily restricted in its scope. Activation using dTRPA1 (for instance) would lead to large-scale muscle contraction across the entire larval extent, abrogating any insights into A27h-to-motor neuron connectivity. Furthermore, the chronic nature of “thermogenetics” means that such contractions would occur over nonphysiological timescales of many minutes. Instead, the authors turned to the twin strengths of optogenetics: that light can be easily patterned over small areas (i.e., a single larval segment) and that light-gated effectors such as channelrhodopsin can be activated with milliseconds of light, and quickly return to baseline. Thus, expression of the second-generation channelrhodopsin mutant ChR2(T159C) (Berndt et al. 2011) in A27h, followed by shining light on a single segment, resulted in the contraction of that segment, and that segment alone.
Activation of ChR2(T159C) in GDL neurons led to intriguing observations about their contributions to locomotion. First, exciting all larval GDLs resulted in paralysis during light application; this result does not require the spatiotemporal specificity of optogenetics, and indeed was verified with dTRPA1 activation as well as the red light-dependent CsChrimson variant. However, the contribution of specific GDLs to wave generation and propagation again highlighted the power of optogenetics. GDL activation at the front of a wave led to muscle relaxation and wave disappearance. More anterior activation did not stop wave propagation, instead merely slowing it across those segments. Thus, the propagating wave gathers strength as it proceeds, becoming a committed movement after a few segments, with posterior GDLs being gatekeepers for wave generation. This paper nicely demonstrates the power of optogenetics relative to other neural manipulations; indeed, it uses most existing techniques in a single study.
Fly lines and recommendations
There are numerous fly lines available at the Bloomington Drosophila Stock Center (http://fly.bio.indiana.edu) for optogenetic control in flies, including ChR2 (Bloomington #52256–52260, 58373–58374, and 58400–58402), ChR2(T159C) (52256–52260 and 58373), ReaChR (53740–53749), and CsChrimson (55134–55139).
Lines should be chosen so that the activation light (action spectrum) of the opsin does not interfere with the imaging channel used or evoke undesirable behavioral responses in the fly. Light-gated opsins are almost always used in fusion to an FP, which both facilitates the imaging of expression patterns and improves membrane trafficking. If possible, the fusion proteins for the opsin should be optimized so as not to interfere with any imaging channels as well. Due to the good penetration of red light through the fly cuticle, ReaChR and CsChrimson have become the light-gated activators of first use in Drosophila. When the blue light-activated ChR2 is preferred, the Thr159Cys (“TC”) mutant is typically selected. For a discussion of considerations for choosing an excitatory opsin (particularly between ReaChR and CsChrimson), and the challenges of using inhibitory opsins, see Kim et al. (2015).
All Together Now
Functional imaging enables the measurement of activity in many neurons simultaneously and in behaving flies. Optogenetic manipulation of neural activity identifies critical circuit components governing behavior. The combination of optogenetic activation in presynaptic neurons and functional imaging in candidate postsynaptic neurons allows us to test whether they are functionally connected. This strategy was used to map circuits governing antennal grooming behavior (Hampel et al. 2015), courtship (Zhou et al. 2015), and aggression (Hoopfer et al. 2015), among others.
For example, UAS-ChR2 (blue–green activation)/LexAop-RCaMP (yellow excitation) or LexAop-CsChrimson (orange–red activation)/UAS-GCaMP (cyan excitation) can be expressed in potentially connected populations. It is important to configure the imaging setup to minimize spectral overlaps in both excitation and emission, and to use transgenes controlled by independent gene expression systems (Gal4, LexA, and Q). The reporters should be inserted into different chromosomal locations to avoid transvection, that is, interactions between two transgenes inserted at the same locus (attP site) on homologous chromosomes (Mellert and Truman 2012). These experiments are compatible with split-Gal4 or other strategies to target restricted neural populations (Venken et al. 2011; Diao et al. 2015; Riabinina and Potter 2016; Fisher et al. 2017). Recommended combinations include codon-optimized GCamP6s and Chrimson-tdTomato (A. Wong, personal communication) (Shirangi et al. 2016; Strother et al. 2017). Examples are shown in Figures 2, 3, and 4.
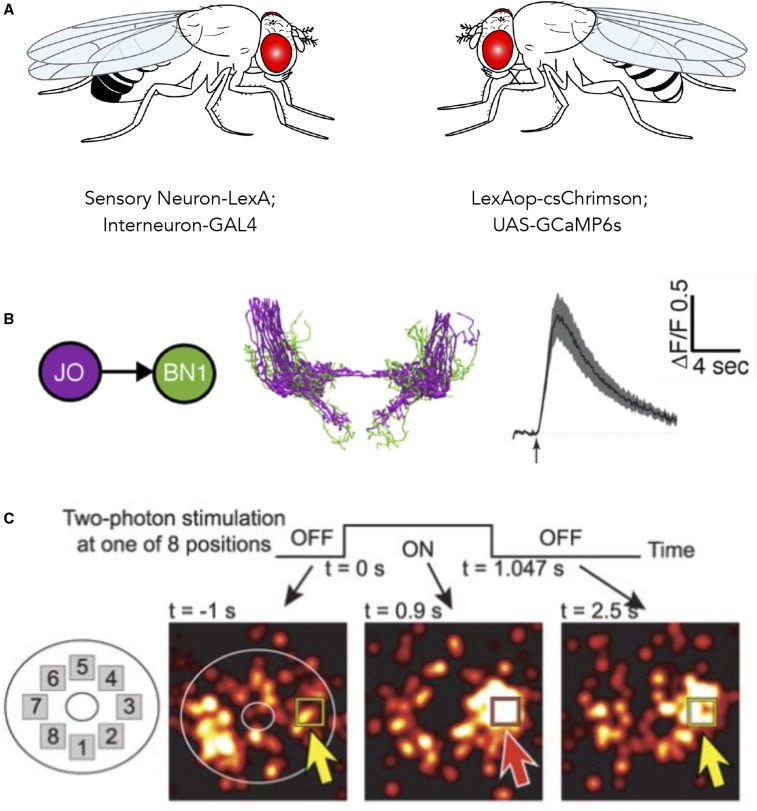
Figure 3.
Using optogenetics and functional imaging together. (A) Example cross scheme for functional connectivity mapping, showing independent transcriptional control of optogenetic activator and genetically encoded calcium reporter. (B) Excitation of CsChrimson (590 nm light-emitting diode through objective) in Johnston's Organ (JO) sensory neurons evokes changes in GCaMP6 fluorescence (Bruker two-photon 920 nm) in candidate postsynaptic interneurons Brain Neurons (BN1) in the antennal grooming circuit (Hampel et al. 2015). (C) Optogenetic activation of specific ellipsoid body neurons coexpressing CsChrimson and GCaMP6f shifts the location of the bump of population activity, suggesting mutual suppression consistent with ring attractor models of circuits that maintain a unique representation of an animal’s heading [figure 2C in Kim et al. (2017)].
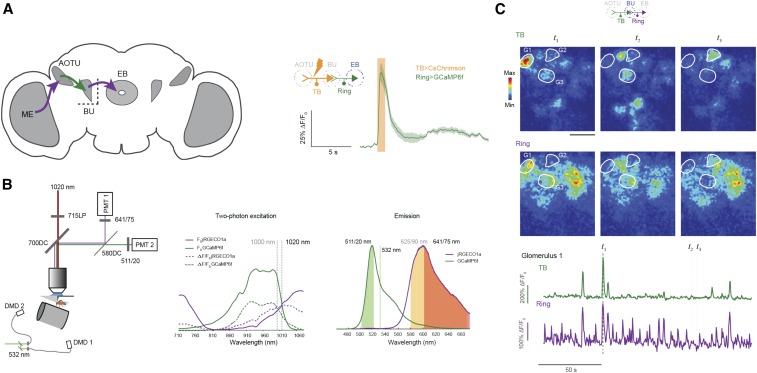
Figure 4.
Mapping how neural activity is transformed through a circuit. (A) Example configuration for functional connectivity. In this experiment, a visual pathway was mapped by establishing functional connectivity between TB neurons, which project from the anterior optical tubercle (AOTU) to the bulb (BU), and the downstream ring neurons that convey information from the BU to the ellipsoid body (EB). The optogenetic activator, CsChrimson, was expressed in tubercle-bulb (TB) neurons using the LexA system, while the GCaMP6f calcium indicator was expressed in ring neurons using GAL4 (13XLexAop2-CsChrimson-tdTomato@su(Hw)attP5/R76B06-LexAp65@attP40; 20xUAS-IVS-GCaMP6f@VK0005/R56H10-GAL4@VK00027, VT002072-GAL4@attP2). CsChrimson was activated with wide-field illumination through the microscope objective 40×) using a 590 nm light-emitting diode (50 μW/mm2) with 2 msec pulses at 30 Hz for 20 sec. GCaMP was imaged using a two-photon laser-scanning microscope with 920 nm excitation and a 520/35 nm band-pass filter. Regions of interest for imaging were spatially separated from the processes of optogenetically activated neurons to avoid unintended two-photon excitation of CsChrimson. Adapted from figure 1 in Sun et al. (2017). (B) Example configuration for dual-color functional imaging. To track the transformation of sensory information as it passes through this visual pathway, functional imaging was performed in two different neuron populations (TB, green, GCaMP6f and ring, magenta, jRGECO1a) using genetically encoded calcium indicators with different fluorescence spectra. GCaMP6f and jRGECO1a were expressed in different neural populations using the GAL4 and LexA transcription systems (R76B06-LexAp65@attP40/13xLexAop2-IVS-GCaMP6f-10@su(Hw)attP5; R56H10-GAL4@VK00027, VT002072-GAL4@attP2/20xUAS-IVS-NES-jRGECO1a-p10@VK0005). A visual stimulus was delivered to the fly at 532 nm. A narrow band-pass green channel filter (511/20) was used to isolate the GCaMP signal from contamination by the 532 nm visual stimulation light. A 641/75 filter was chosen for the red channel, with a long-pass 715 nm filter to eliminate additional light contamination from the laser delivering the two-photon excitation light. The emission spectra of GCaMP6f and jRGECO1a overlap, and the GCaMP signal can enter the red imaging channel unless proper precautions are taken, something that can be assessed by examining the red channel signal when only GCaMP is expressed in the sample. Here, shifting the two-photon excitation wavelength to 1020 nm (to enhance/balance the weaker jRGECO1a excitation) and including stringent band-pass filters in two detection streams was critical to achieving effective spectral separation [Figure S3 in Sun et al. (2017), adapted from Dana et al. (2016)]. PMT, photomultiplier tube detector.
Alternative methods of circuit connectivity mapping include optogenetic activation of presynaptic neurons coupled with electrophysiological recording in candidate postsynaptic neurons (Chang et al. 2016), and presynaptic expression of P2X2 activated by uncaged ATP with either GCaMP or ePAC1-camps [a cAMP sensor (Yao et al. 2012)] as the postsynaptic reporter. Closed-loop experiments to test how specific patterns of olfactory sensory neuron activity (selected based on GCaMP functional imaging and electrophysiology in separate experiments) affect behavioral decisions in larval navigation were accomplished by temporally precise optogenetic activation (Schulze et al. 2015).
Example
The simplest demonstration of the power of combined neuronal activation and imaging is “functional connectomics,” i.e., the activation of one cell and the subsequent visualization of a response in a second, in effect an all-optical approximation of a paired-patch experiment. In addition to the fundamental demonstration that one cell lies downstream of another, the size and shape of the functional response can, coupled with other experiments, suggest whether such connections are direct or multi-synaptic, and excitatory, inhibitory, or mixed. For example, a recent study (Kim et al. 2017; Turner-Evans et al. 2017) elucidated how a circuit in the adult fly’s central complex uses angular velocity integration to update a representation of head direction [a mechanism independently identified in a parallel study (Green et al. 2017)]. The authors first identified two neuronal populations of interest from anatomy and functional imaging, denoted “E-PG” (meaning that their postsynaptic processes lie within the Ellipsoid body, whereas their axons project to the Protocerebral bridge and the Gall) and “P-EN” (postsynaptic processes in the Protocerebral bridge, and axons project to the Ellipsoid body and Noduli). After an elegant set of functional imaging and electrophysiological experiments, the authors proposed that the E-PG and P-EN cells together comprise a recurrent excitatory loop, with the former receiving inputs from the latter in the bridge and projecting onto the latter in the ellipsoid body, and vice versa. Synaptotagmin (a presynaptic molecule) localization was consistent with the proposed connectivity, but further experiments were needed to prove functional synaptic connectivity.
Thus, the authors expressed the red-shifted CsChrimson in either E-PG or P-EN neurons, and GCaMP6m or GCaMP6f in the other population. Optogenetic activation of P-EN neurons robustly elicited calcium activity in E-PG neurons, with the kinetics suggesting a direct connection. Strongly activating E-PG neurons led to biphasic (two peaks) responses in P-EN, consistent with both direct and indirect excitatory connections. Furthermore, weakly activating E-PG neurons led to complicated, peak-then-dip responses in P-EN, again suggesting that both excitatory and inhibitory connections may exist. Such data-rich functional connectomics experiments not only confirm synaptic connectivity, but also propose hypotheses about wiring diagrams and involved transmitter systems. No other experimental design allows such rapid testing and characterization of connectivity, which can be enhanced with pharmacology and RNAi, etc.
Challenges
Interpreting global activity maps:
Broad expression of GECIs allows us to image the whole brain or large areas simultaneously, generating a global view of activity patterns in different contexts. These data sets can be challenging to interpret. Repeating a specific sensory trigger, imaging the response to controlled activation of command-like neurons, and correlating activity with behavioral responses has revealed variability in large-scale activity patterns. These complex responses are likely to be a real biological feature of flexible neural networks, but this makes analysis and interpretation complicated. Examples from similar data sets in leech, fish, and worms provide some guidance (Briggman et al. 2005; Gordus et al. 2015; Kato et al. 2015; Keller and Ahrens 2015), and strategies for interpreting large-scale electrophysiological recording or MRI/PET data sets may present a guide. Even static measures correlating expression to phenotype are complex (Vogelstein et al. 2014; Robie et al. 2017a), but both present opportunities to better understand how the brain processes sensory information and controls behavior.
Visualizing inhibition:
GCaMP has been used to demonstrate an inhibitory connection by visualizing a decrease in basal fluorescence, but this only works if the neuron is normally active and then shows a reduction. Sometimes this is artificially enhanced by activating the neuron, increasing the GCaMP signal, and then determining whether an inhibitory input can decrease this fluorescence. Negative-responding GECIs such as Inverse Pericam or CaMPARI (which is a negative calcium indicator in addition to being a positive calcium integrator), and improved versions thereof, could greatly improve the signal-to-noise ratio of measurements of inhibition, since this would lead to fluorescent increases on a dim background, rather than decreases on a bright background. Voltage sensors can be used in situations where resting calcium is insufficiently high to facilitate robust imaging of small downward deflections, and likely offer a better long-term prospect for imaging inhibition overall. Chloride sensors exist (Kuner and Augustine 2000) and have been successfully used in flies (Haynes et al. 2015).
Generating realistic neural activation patterns:
One promise of activating or inhibiting neurons with light is the temporal precision to mimic or disrupt normal electrical activity and action potentials, i.e., to “speak the language” of neurons. But we do not always know what range of stimuli to try. Most neurons in the brain or ventral nerve cord have not been recorded electrophysiologically, and the range of spike frequencies that a given neuron can produce under different circumstances can only be estimated. Sustained light exposure could result in patterned neuronal outputs due to the refractory properties of the neuron, or feedback inhibition or excitation from circuit connectivity. Research groups conducting electrophysiological recordings (Gruntman and Turner 2013; Murthy and Turner 2013; von Reyn et al. 2014; Hsu and Bhandawat 2016; Nagel and Wilson 2016; Tuthill and Wilson 2016; Schnell et al. 2017) expand our repertoire of realistic activity patterns that we could attempt to replay with optogenetic reagents, as was done in Schulze et al. (2015).
The exact electrical consequences of expressing of ChR2 or CsChrimson and applying a given light stimulus in a specific neuron can only be inferred based on behavioral outcome (Pauls et al. 2015), or comparison to the few places where the activity of that neuron has actually been measured by GCaMP coexpression or electrophysiological recording, and this level of correlation or validation is rare. The larval NMJ, where an optogenetic reagent can be expressed in the motor neurons and the degree of electrical activation in response to light measured by recording from the muscle, is a common test-bed (Hornstein et al. 2009; Pulver et al. 2009). One alternative way that the output from optogenetic activation has been measured is cyclic voltammetry to measure the release of biogenic amines such as dopamine (Xiao et al. 2014; Privman and Venton 2015). Another method combining optogenetics and functional imaging involves coexpression of both CsChrimson and GCaMP in the same neurons to evaluate how effectively different red-light stimulation paradigms activate a particular neural population.
Inferring direct synaptic connectivity:
Because of the difficulty of activating one set of neurons and then rapidly imaging from another set, as well as the delay imposed by the response and decay of GECI fluorescence, inferring whether a connection between two neurons is direct or indirect remains challenging. Including an action potential blocker can show that a connection is direct or monosynaptic, since optogenetic activation still occurs in the presence of the blocker because it uses a light-gated channel to admit depolarizing ions. Correlated noise or subthreshold depolarizations have also been used to argue for monosynaptic connectivity. Dual-patch electrophysiology or electron microscopy still provides the highest proof of connectivity, but showing the behavioral or biological function of those connections is also required.
Light can influence behavior
One challenge for optogenetics and functional imaging is that light itself can affect the experiment. Flies detect light using their eyes, ocelli, Bolwig’s organ, larval epidermis, and even deep-brain nuclei. Very small amounts of light are sufficient to reset circadian rhythms, and high-intensity light can cause heat or pain, so responses to light itself should always be considered as a potential cause of a behavioral phenotype or change in neural activity when we do optogenetic activation or one-photon functional imaging. The choice of far-red excitation minimizes this, as does mounting flies with the eyes shielded below the area of illumination. Using blind flies is sometimes an alternative (Schulze et al. 2015). In some cases, a light-evoked behavior can even be suppressed by preexposure or habituation (Zhou et al. 2015). Appropriate behavioral controls—comparing light responses in flies that do not express channelrhodopsins or were not fed retinal—allow the effects of optogenetic activation to be isolated (note that many fly food recipes contain chemical precursors to retinal so using a truly retinal-free food requires care). So, while the fly’s response to light itself must be considered in experimental design and interpretation, it can usually be managed with appropriate conditions and controls. In cases where visual sensory processing or a visually evoked behavior is the subject of study, temperature-sensitive effectors (Venken et al. 2011; Owald et al. 2015; Hampel and Seeds 2017) might be a more appropriate alternative way to manipulate neural activity. These may also be preferable in experiments at early developmental stages, where prefeeding retinal can be challenging. One additional caution is that the improved cuticle penetration of the red-shifted opsins and the high photo-sensitivity of both CsChrimson and GtACR can cause unwanted activation by ambient light. This makes it advisable to rear flies in the dark before optogenetic activation and behavioral testing.
Technical hurdles:
How do you evoke neural activity with light at one wavelength in one location and then image calcium-induced fluorescent changes with another wavelength in another location fast enough to capture synaptic transmission and trains of action potentials? On a two-photon microscope, both regions must be in the same field of view if both the excitation and imaging lasers pass through the same lens. The optogenetic stimulation can be delivered with a fixed-wavelength laser targeted with one set of galvos while light to the PMT detectors is shuttered or filtered. The two-photon laser is then targeted to a region of interest for a fast, volumetric scan using independent galvos to drive the scan in XY and a piezo on the lens to capture a Z-stack. Optogenetic activation and functional imaging are synchronized with a camera that records the fly’s behavioral responses See Figure 5 for one solution.
Figure 5.
Two-photon excitation of GCaMP can minimize spectral overlap with Chrimson. (A) One-photon action spectrum of Chrimson (red), as well as one-photon excitation (blue) and emission (green) spectra of GCaMP. Extensive overlap between Chrimson action spectrum and GCaMP excitation spectrum indicates potential spectral contamination between Chrimson stimulation and wide-field calcium imaging with GCaMP. (B) One-photon action spectrum of Chrimson (red) as well as two-photon excitation spectrum (gray) of GCaMP. Using one-photon optogenetic stimulation with Chrimson and two-photon calcium imaging with GCaMP improves spectral separation (figure: Y. Sun personal communication from Yi Sun, modified from Sun et al 2017.).
Future Developments
The future is quite bright for both functional imaging and optogenetics in the fly. New variants of both green and red calcium indicators are being released at a brisk pace, with each new generation offering improvements in sensitivity, brightness, photostability, and kinetics. Specialized GECIs are being developed for Drosophila, such as those with very low-affinity and fast kinetics, which could potentially give better resolution of action potential number and timing for fast-spiking neurons. Voltage indicators are also being rapidly improved upon. More red-shifted versions of both sensor types would facilitate better cuticle penetration, and lower background and phototoxicity. Genetically encoded indicators are being developed for the entire complement of neurotransmitters, neuromodulators, and secondary messengers, potentially allowing the full interrogation of molecular signaling within a neural circuit. Improved imaging methods (Tao et al. 2017) will facilitate easier access to multiple, deep-brain regions with better light collection and decreased out-of-focus excitation, photobleaching, and phototoxicity. Light-gated effectors beyond ion channels are also being engineered, permitting targeted control of specific pathways. And, as always, concomitant additions to the complement of available expression lines (e.g., sparse or split Gal4 lines) will improve the specificity with which monitoring and manipulation can occur. The parallel solution of the connectivity diagram (“connectome”) of the larva and adult through electron microscopy drives hypothesis generation for neural circuit structure, which can then be validated by functional imaging and/or optogenetics for every behavior of interest.
Conclusions
The combination of optogenetics and functional imaging is a powerful strategy for investigating neural function (Emiliani et al. 2015). We are just beginning to explore the power of light to interrogate (image and manipulate) the brain; the fly is a technically and biologically advantageous place to discover general rules for how specific circuit motifs perform the calculations that integrate sensory and state cues into appropriate behavioral responses. These new tools open the door to answering new, harder questions. With the ready availability of well-characterized and sparse reporter, effector, and driver lines, the commoditization of imaging/recording/optogenetic rigs, and the low barrier-to-entry into fly work, times have never been better for rapid, effective circuit mapping in any model organism. We hope that this review provides ideas and technical suggestions that open some doors for you.
Footnotes
Communicating editor: N. Perrimon
Available freely online through the author-supported open access option.
Literature Cited
- Abdelfattah A. S., Farhi S. L., Zhao Y., Brinks D., Zou P., et al. , 2016. A bright and fast red fluorescent protein voltage indicator that reports neuronal activity in organotypic brain slices. J. Neurosci. 36: 2458–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast C., Foret J., Oltrogge L. M., De Michele R., Kleist T. J., et al. , 2017. Ratiometric Matryoshka biosensors from a nested cassette of green- and orange-emitting fluorescent proteins. Nat. Commun. 8: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AzimiHashemi N., Erbguth K., Vogt A., Riemensperger T., Rauch E., et al. , 2014. Synthetic retinal analogues modify the spectral and kinetic characteristics of microbial rhodopsin optogenetic tools. Nat. Commun. 5: 5810. [DOI] [PubMed] [Google Scholar]
- Bath D. E., Stowers J. R., Hormann D., Poehlmann A., Dickson B. J., et al. , 2014. FlyMAD: rapid thermogenetic control of neuronal activity in freely walking Drosophila. Nat. Methods 11: 756–762. [DOI] [PubMed] [Google Scholar]
- Behnia R., Clark D. A., Carter A. G., Clandinin T. R., Desplan C., 2014. Processing properties of ON and OFF pathways for Drosophila motion detection. Nature 512: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Tong C., Tsuda H., 2010. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat. Rev. Neurosci. 11: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S., Carroll E. C., Newman Z. L., Okada H. O., Quinn C. M., et al. , 2015. Photoactivatable genetically encoded calcium indicators for targeted neuronal imaging. Nat. Methods 12: 852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A., Schoenenberger P., Mattis J., Tye K. M., Deisseroth K., et al. , 2011. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. USA 108: 7595–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden E. S., 2015. Optogenetics and the future of neuroscience. Nat. Neurosci. 18: 1200–1201. [DOI] [PubMed] [Google Scholar]
- Briggman K. L., Abarbanel H. D., Kristan W. B., Jr., 2005. Optical imaging of neuronal populations during decision-making. Science 307: 896–901. [DOI] [PubMed] [Google Scholar]
- Broussard G. J., Liang R., Tian L., 2014. Monitoring activity in neural circuits with genetically encoded indicators. Front. Mol. Neurosci. 7: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Platisa J., Pieribone V. A., Raccuglia D., Kunst M., et al. , 2013. Genetically targeted optical electrophysiology in intact neural circuits. Cell 154: 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberland S., Yang H. H., Pan M. M., Evans S. W., Guan S., et al. , 2017. Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. Elife 6: e25690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. E., Vaughan A. G., Wilson R. I., 2016. A mechanosensory circuit that mixes opponent channels to produce selectivity for complex stimulus features. Neuron 92: 888–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. W., Wardill T. J., Sun Y., Pulver S. R., Renninger S. L., et al. , 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhetri R. K., Amat F., Wan Y., Hockendorf B., Lemon W. C., et al. , 2015. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat. Methods 12: 1171–1178. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A., Roorda R. D., Vrontou E., Sjulson L., Li H., et al. , 2009. Writing memories with light-addressable reinforcement circuitry. Cell 139: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn R., Morantte I., Ruta V., 2015. Coordinated and compartmentalized neuromodulation shapes sensory processing in Drosophila. Cell 163: 1742–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino C., Alberio L., Gazzarrini S., Aquila M., Romano E., et al. , 2015. Optogenetics. Engineering of a light-gated potassium channel. Science 348: 707–710. [DOI] [PubMed] [Google Scholar]
- Dana H., Mohar B., Sun Y., Narayan S., Gordus A., et al. , 2016. Sensitive red protein calcium indicators for imaging neural activity. Elife 5: e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawydow A., Gueta R., Ljaschenko D., Ullrich S., Hermann M., et al. , 2014. Channelrhodopsin-2-XXL, a powerful optogenetic tool for low-light applications. Proc. Natl. Acad. Sci. USA 111: 13972–13977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K., 2015. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F., Ironfield H., Luan H., Diao F., Shropshire W. C., et al. , 2015. Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep. 10: 1410–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipt S., Riemensperger T., Fiala A., 2014. Optical calcium imaging using DNA-encoded fluorescence sensors in transgenic fruit flies, Drosophila melanogaster. Methods Mol. Biol. 1071: 195–206. [DOI] [PubMed] [Google Scholar]
- Emiliani V., Cohen A. E., Deisseroth K., Hausser M., 2015. All-optical interrogation of neural circuits. J. Neurosci. 35: 13917–13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala A., Spall T., Diegelmann S., Eisermann B., Sachse S., et al. , 2002. Genetically expressed Cameleon in Drosophila melanogaster is used to visualize olfactory information in projection neurons. Curr. Biol. 12: 1877–1884. [DOI] [PubMed] [Google Scholar]
- Fisher Y. E., Yang H. H., Isaacman-Beck J., Xie M., Gohl D. M., et al. , 2017. FlpStop, a tool for conditional gene control in Drosophila. Elife 6: e22279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosque B. F., Sun Y., Dana H., Yang C. T., Ohyama T., et al. , 2015. Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science 347: 755–760. [DOI] [PubMed] [Google Scholar]
- Freyberg Z., Sonders M. S., Aguilar J. I., Hiranita T., Karam C. S., et al. , 2016. Mechanisms of amphetamine action illuminated through optical monitoring of dopamine synaptic vesicles in Drosophila brain. Nat. Commun. 7: 10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushiki A., Zwart M. F., Kohsaka H., Fetter R. D., Cardona A., et al. , 2016. A circuit mechanism for the propagation of waves of muscle contraction in Drosophila. Elife 5: e13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. J., Riabinina O., Li J., Potter C. J., Clandinin T. R., et al. , 2015. A transcriptional reporter of intracellular Ca(2+) in Drosophila. Nat. Neurosci. 18: 917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordus A., Pokala N., Levy S., Flavell S. W., Bargmann C. I., 2015. Feedback from network states generates variability in a probabilistic olfactory circuit. Cell 161: 215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Adachi A., Shah K. K., Hirokawa J. D., Magani P. S., et al. , 2017. A neural circuit architecture for angular integration in Drosophila. Nature 546: 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C., Konnerth A., 2012. Imaging calcium in neurons. Neuron 73: 862–885. [DOI] [PubMed] [Google Scholar]
- Grover D., Katsuki T., Greenspan R. J., 2016. Flyception: imaging brain activity in freely walking fruit flies. Nat. Methods 13: 569–572. [DOI] [PubMed] [Google Scholar]
- Gruntman E., Turner G. C., 2013. Integration of the olfactory code across dendritic claws of single mushroom body neurons. Nat. Neurosci. 16: 1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero G., Siegel M. S., Roska B., Loots E., Isacoff E. Y., 2002. Tuning FlaSh: redesign of the dynamics, voltage range, and color of the genetically encoded optical sensor of membrane potential. Biophys. J. 83: 3607–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero G., Reiff D. F., Agarwal G., Ball R. W., Borst A., et al. , 2005. Heterogeneity in synaptic transmission along a Drosophila larval motor axon. Nat. Neurosci. 8: 1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales K. G., Korey C. A., Larracuente A. M., Roberts D. M., 2015. Genetics on the fly: a primer on the Drosophila model system. Genetics 201: 815–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel, S., and A. M. Seeds, 2017 Targeted manipulation of neural activity in behaving adult flies, in Decoding Neural Circuit Structure and Function, edited by A. Celik and M. F. Wernet. Springer-Verlag, Berlin, Germany. [Google Scholar]
- Hampel S., Franconville R., Simpson J. H., Seeds A. M., 2015. A neural command circuit for grooming movement control. Elife 4: e08758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D. T., Kallman B. R., Mullaney B. C., Scott K., 2015. Representations of taste modality in the Drosophila brain. Neuron 86: 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes P. R., Christmann B. L., Griffith L. C., 2015. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife 4: e03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel T., Mank M., Schnell B., Griesbeck O., Borst A., et al. , 2008. Fluorescence changes of genetic calcium indicators and OGB-1 correlated with neural activity and calcium in vivo and in vitro. J. Neurosci. 28: 7399–7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwig L., Rice A. J., Bedbrook C. N., Zhang R. K., Lignell A., et al. , 2017. Directed evolution of a bright near-infrared fluorescent rhodopsin using a synthetic chromophore. Cell Chem. Biol. 24: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo K., Hwang R. Y., Tracey W. D., Jr., 2012. Optogenetic manipulation of neural circuits and behavior in Drosophila larvae. Nat. Protoc. 7: 1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopfer E. D., Jung Y., Inagaki H. K., Rubin G. M., Anderson D. J., 2015. P1 interneurons promote a persistent internal state that enhances inter-male aggression in Drosophila. Elife 4: e11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein N. J., Pulver S. R., Griffith L. C., 2009. Channelrhodopsin2 mediated stimulation of synaptic potentials at Drosophila neuromuscular junctions. J. Vis. Exp. DOI: 10.3791/1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao P. Y., Tsai C. L., Chen M. C., Lin Y. Y., Yang S. D., et al. , 2015. Non-invasive manipulation of Drosophila behavior by two-photon excited red-activatable channelrhodopsin. Biomed. Opt. Express 6: 4344–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. T., Bhandawat V., 2016. Organization of descending neurons in Drosophila melanogaster. Sci. Rep. 6: 20259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain Q. M., Ewer J., 2004. Use of targetable gfp-tagged neuropeptide for visualizing neuropeptide release following execution of a behavior. J. Neurobiol. 59: 181–191. [DOI] [PubMed] [Google Scholar]
- Hwang R. Y., Zhong L., Xu Y., Johnson T., Zhang F., et al. , 2007. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol. 17: 2105–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada K., Kohsaka H., Takasu E., Matsunaga T., Nose A., 2011. Optical dissection of neural circuits responsible for Drosophila larval locomotion with halorhodopsin. PLoS One 6: e29019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki H. K., Ben-Tabou de-Leon S., Wong A. M., Jagadish S., Ishimoto H., et al. , 2012. Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell 148: 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki H. K., Jung Y., Hoopfer E. D., Wong A. M., Mishra N., et al. , 2014. Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat. Methods 11: 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S., Tsutsui H., Suzuki K., Agetsuma M., Arai Y., et al. , 2017. Genetically encoded bioluminescent voltage indicator for multi-purpose use in wide range of bioimaging. Sci. Rep. 7: 42398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadish S., Barnea G., Clandinin T. R., Axel R., 2014. Identifying functional connections of the inner photoreceptors in Drosophila using Tango-Trace. Neuron 83: 630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman V., Laurent G., 2007. Evaluating a genetically encoded optical sensor of neural activity using electrophysiology in intact adult fruit flies. Front. Neural Circuits 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallman B. R., Kim H., Scott K., 2015. Excitation and inhibition onto central courtship neurons biases Drosophila mate choice. Elife 4: e11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschula R., Salecker I., 2016. Neuronal computations made visible with subcellular resolution. Cell 166: 18–20. [DOI] [PubMed] [Google Scholar]
- Kato S., Kaplan H. S., Schrodel T., Skora S., Lindsay T. H., et al. , 2015. Global brain dynamics embed the motor command sequence of Caenorhabditis elegans. Cell 163: 656–669. [DOI] [PubMed] [Google Scholar]
- Keller P. J., Ahrens M. B., 2015. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 85: 462–483. [DOI] [PubMed] [Google Scholar]
- Kim S. S., Franconville R., Turner-Evans D., Jayaraman V., 2015. Optogenetics in Drosophila melanogaster, pp. 147–176 in New Techniques in Systems Neuroscience, edited by Douglass A. D. Springer-Verlag, Berlin. [Google Scholar]
- Kim S. S., Rouault H., Druckmann S., Jayaraman V., 2017. Ring attractor dynamics in the Drosophila central brain. Science 356: 849–853. [DOI] [PubMed] [Google Scholar]
- Klapoetke N. C., Murata Y., Kim S. S., Pulver S. R., Birdsey-Benson A., et al. , 2014. Independent optical excitation of distinct neural populations. Nat. Methods 11: 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner T., Augustine G. J., 2000. A genetically encoded ratiometric indicator for chloride: capturing chloride transients in cultured hippocampal neurons. Neuron 27: 447–459. [DOI] [PubMed] [Google Scholar]
- Leilito K. R., Shafer O. T., 2012. Imaging cAMP dynamics in the Drosophila brain with the genetically encoded sensor Epac1-camps, in Genetically Encoded Functional Indicators, edited by Martin J. R. Humana Press, Totowa, NJ. [Google Scholar]
- Lemon W. C., Pulver S. R., Hockendorf B., McDole K., Branson K., et al. , 2015. Whole-central nervous system functional imaging in larval Drosophila. Nat. Commun. 6: 7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima S. Q., Miesenbock G., 2005. Remote control of behavior through genetically targeted photostimulation of neurons. Cell 121: 141–152. [DOI] [PubMed] [Google Scholar]
- Lima S. Q., Hromadka T., Znamenskiy P., Zador A. M., 2009. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS One 4: e6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson L. J., Zaharieva E. E., Kearney P. J., Alpert M. H., Lin T. Y., et al. , 2015. Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat. Commun. 6: 10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon G., Straw A. D., Dickinson M. H., 2010. Active flight increases the gain of visual motion processing in Drosophila, Nat. Neurosci. 13: 393–399. [DOI] [PubMed] [Google Scholar]
- Mamiya A., Dickinson M. H., 2015. Antennal mechanosensory neurons mediate wing motor reflexes in flying Drosophila, J. Neurosci. 35: 7977–7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank M., Reiff D. F., Heim N., Friedrich M. W., Borst A., et al. , 2006. A FRET-based calcium biosensor with fast signal kinetics and high fluorescence change. Biophys. J. 90: 1790–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank M., Santos A. F., Direnberger S., Mrsic-Flogel T. D., Hofer S. B., et al. , 2008. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat. Methods 5: 805–811. [DOI] [PubMed] [Google Scholar]
- Mann K., Gallen C. L., Clandinin T. R., 2017. Whole-brain calcium imaging reveals an intrinsic functional network in Drosophila. Curr. Biol. 27: 2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella S., Fischler W., Kong P., Asgarian S., Rueckert E., et al. , 2006. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron 49: 285–295. [DOI] [PubMed] [Google Scholar]
- Martin J. R., 2008. In vivo brain imaging: fluorescence or bioluminescence, which to choose? J. Neurogenet. 22: 285–307. [DOI] [PubMed] [Google Scholar]
- Marvin J. S., Borghuis B. G., Tian L., Cichon J., Harnett M. T., et al. , 2013. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10: 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama K., Zhang Y., Rao Y., Wang J. W., 2012. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J. Neurogenet. 26: 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauss A. S., Busch C., Borst A., 2017. Optogenetic neuronal silencing in Drosophila during visual processing. Sci. Rep. 7: 13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert D. J., Truman J. W., 2012. Transvection is common throughout the Drosophila genome. Genetics 191: 1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenbock G., De Angelis D. A., Rothman J. E., 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394: 192–195. [DOI] [PubMed] [Google Scholar]
- Mohammad F., Stewart J. C., Ott S., Chlebikova K., Chua J. Y., et al. , 2017. Optogenetic inhibition of behavior with anion channelrhodopsins. Nat. Methods 14: 271–274. [DOI] [PubMed] [Google Scholar]
- Morton A., Murawski C., Pulver S. R., Gather M. C., 2016. High-brightness organic light-emitting diodes for optogenetic control of Drosophila locomotor behaviour. Sci. Rep. 6: 31117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy M., Turner G., 2013. Whole-cell in vivo patch-clamp recordings in the Drosophila brain. Cold Spring Harb. Protoc. 2013: 140–148. [DOI] [PubMed] [Google Scholar]
- Nagel K. I., Wilson R. I., 2016. Mechanisms underlying population response dynamics in inhibitory interneurons of the Drosophila antennal lobe. J. Neurosci. 36: 4325–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J., Ohkura M., Imoto K., 2001. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 19: 137–141. [DOI] [PubMed] [Google Scholar]
- Owald D., Lin S., Waddell S., 2015. Light, heat, action: neural control of fruit fly behaviour. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370: 20140211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N., Gold M. G., 2015. The genetically encoded tool set for investigating cAMP: more than the sum of its parts. Front. Pharmacol. 6: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls D., von Essen A., Lyutova R., van Giesen L., Rosner R., et al. , 2015. Potency of transgenic effectors for neurogenetic manipulation in Drosophila larvae. Genetics 199: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platisa J., Vasan G., Yang A., Pieribone V. A., 2017. Directed evolution of key residues in fluorescent protein inverses the polarity of voltage sensitivity in the genetically encoded indicator ArcLight. ACS Chem. Neurosci. 8: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privman E., Venton B. J., 2015. Comparison of dopamine kinetics in the larval Drosophila ventral nerve cord and protocerebrum with improved optogenetic stimulation. J. Neurochem. 135: 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver S. R., Pashkovski S. L., Hornstein N. J., Garrity P. A., Griffith L. C., 2009. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J. Neurophysiol. 101: 3075–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver S. R., Bayley T. G., Taylor A. L., Berni J., Bate M., et al. , 2015. Imaging fictive locomotor patterns in larval Drosophila. J. Neurophysiol. 114: 2564–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S., Lang C., Levitan E. S., Deitcher D. L., 2001. Visualization of neuropeptide expression, transport, and exocytosis in Drosophila melanogaster. J. Neurobiol. 49: 159–172. [DOI] [PubMed] [Google Scholar]
- Reiff D. F., Ihring A., Guerrero G., Isacoff E. Y., Joesch M., et al. , 2005. In vivo performance of genetically encoded indicators of neural activity in flies. J. Neurosci. 25: 4766–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiff D. F., Plett J., Mank M., Griesbeck O., Borst A., 2010. Visualizing retinotopic half-wave rectified input to the motion detection circuitry of Drosophila, Nat. Neurosci. 13: 973–978. [DOI] [PubMed] [Google Scholar]
- Restrepo S., Basler K., 2016. Drosophila wing imaginal discs respond to mechanical injury via slow InsP3R-mediated intercellular calcium waves. Nat. Commun. 7: 12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O., Potter C. J., 2016. The Q-system: a versatile expression system for Drosophila. Methods Mol. Biol. 1478: 53–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer J. P., Tank D. W., 2009. Two-photon excitation of channelrhodopsin-2 at saturation. Proc. Natl. Acad. Sci. USA 106: 15025–15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemensperger T., Kittel R. J., Fiala A., 2016. Optogenetics in Drosophila neuroscience. Methods Mol. Biol. 1408: 167–175. [DOI] [PubMed] [Google Scholar]
- Robie A. A., Hirokawa J., Edwards A. W., Umayam L. A., Lee A., et al. , 2017a Mapping the neural substrates of behavior. Cell 170: 393–406.e28. [DOI] [PubMed] [Google Scholar]
- Robie A. A., Seagraves K. M., Egnor S. E., Branson K., 2017b Machine vision methods for analyzing social interactions. J. Exp. Biol. 220: 25–34. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmied C., Tomancak P., 2016. Sample preparation and mounting of Drosophila embryos for multiview light sheet microscopy. Methods Mol. Biol. 1478: 189–202. [DOI] [PubMed] [Google Scholar]
- Schnell B., Ros I. G., Dickinson M. H., 2017. A descending neuron correlated with the rapid steering maneuvers of flying Drosophila. Curr. Biol. 27: 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder-Lang S., Schwarzel M., Seifert R., Strunker T., Kateriya S., et al. , 2007. Fast manipulation of cellular cAMP level by light in vivo. Nat. Methods 4: 39–42. [DOI] [PubMed] [Google Scholar]
- Schroll C., Riemensperger T., Bucher D., Ehmer J., Voller T., et al. , 2006. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr. Biol. 16: 1741–1747. [DOI] [PubMed] [Google Scholar]
- Schulze A., Gomez-Marin A., Rajendran V. G., Lott G., Musy M., et al. , 2015. Dynamical feature extraction at the sensory periphery guides chemotaxis. Elife 4: e06694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig J. D., Jayaraman V., 2013. Feature detection and orientation tuning in the Drosophila central complex. Nature 503: 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig J. D., Jayaraman V., 2015. Neural dynamics for landmark orientation and angular path integration. Nature 521: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig J. D., Chiappe M. E., Lott G. K., Dutta A., Osborne J. E., et al. , 2010. Two-photon calcium imaging from head-fixed Drosophila during optomotor walking behavior. Nat. Methods 7: 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejourne J., Placais P. Y., Aso Y., Siwanowicz I., Trannoy S., et al. , 2011. Mushroom body efferent neurons responsible for aversive olfactory memory retrieval in Drosophila, Nat. Neurosci. 14: 903–910. [DOI] [PubMed] [Google Scholar]
- Shafer O. T., Kim D. J., Dunbar-Yaffe R., Nikolaev V. O., Lohse M. J., et al. , 2008. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron 58: 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi T. R., Wong A. M., Truman J. W., Stern D. L., 2016. Doublesex regulates the connectivity of a neural circuit controlling Drosophila male courtship song. Dev. Cell 37: 533–544. [DOI] [PubMed] [Google Scholar]
- Siegel M. S., Isacoff E. Y., 1997. A genetically encoded optical probe of membrane voltage. Neuron 19: 735–741. [DOI] [PubMed] [Google Scholar]
- Sitaraman D., Aso Y., Jin X., Chen N., Felix M., et al. , 2015. Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr. Biol. 25: 2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart A. D., Pache R. A., Thomsen N. D., Kortemme T., Davis G. W., et al. , 2017. Engineering a light-activated caspase-3 for precise ablation of neurons in vivo. Proc. Natl. Acad. Sci. USA 114: E8174–E8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierl M., Stumpf P., Udwari D., Gueta R., Hagedorn R., et al. , 2011. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem. 286: 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T., Sheehan A., Tasdemir-Yilmaz O. E., Freeman M. R., 2014. Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron 83: 388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F., Marshall J. D., Yang Y., Gong Y., Schnitzer M. J., et al. , 2014. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 17: 884–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F., Chavarha M., Lin M. Z., 2015. Designs and sensing mechanisms of genetically encoded fluorescent voltage indicators. Curr. Opin. Chem. Biol. 27: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strother J. A., Wu S. T., Wong A. M., Nern A., Rogers E. M., et al. , 2017. The emergence of directional selectivity in the visual motion pathway of Drosophila. Neuron 94: 168–182.e10. [DOI] [PubMed] [Google Scholar]
- Sun Y., Nern A., Franconville R., Dana H., Schreiter E. R., et al. , 2017. Neural signatures of dynamic stimulus selection in Drosophila. Nat. Neurosci. 20: 1104–1113. [DOI] [PubMed] [Google Scholar]
- Tao X., Lin H. H., Lam T., Rodriguez R., Wang J. W., et al. , 2017. Transcutical imaging with cellular and subcellular resolution. Biomed. Opt. Express 8: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thestrup T., Litzlbauer J., Bartholomaus I., Mues M., Russo L., et al. , 2014. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat. Methods 11: 175–182. [DOI] [PubMed] [Google Scholar]
- Titlow J. S., Johnson B. R., Pulver S. R., 2015. Light activated escape circuits: a behavior and neurophysiology lab module using Drosophila optogenetics. J. Undergrad. Neurosci. Educ. 13: A166–A173. [PMC free article] [PubMed] [Google Scholar]
- Turner-Evans D., Wegener S., Rouault H., Franconville R., Wolff T., et al. , 2017. Angular velocity integration in a fly heading circuit. Elife 6: e23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill J. C., Wilson R. I., 2016. Parallel transformation of tactile signals in central circuits of Drosophila. Cell 164: 1046–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., Simpson J. H., Bellen H. J., 2011. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 72: 202–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein J. T., Park Y., Ohyama T., Kerr R. A., Truman J. W., et al. , 2014. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science 344: 386–392. [DOI] [PubMed] [Google Scholar]
- Vogt N., 2015. Voltage sensors: challenging, but with potential. Nat. Methods 12: 921–924. [DOI] [PubMed] [Google Scholar]
- von Reyn C. R., Breads P., Peek M. Y., Zheng G. Z., Williamson W. R., et al. , 2014. A spike-timing mechanism for action selection. Nat. Neurosci. 17: 962–970. [DOI] [PubMed] [Google Scholar]
- Wojtovich A. P., Foster T. H., 2014. Optogenetic control of ROS production. Redox Biol. 2: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Nern A., Williamson W. R., Morimoto M. M., Reiser M. B., et al. , 2016. Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. Elife 5: e21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. C., Chu L. A., Hsiao P. Y., Lin Y. Y., Chi C. C., et al. , 2014. Optogenetic control of selective neural activity in multiple freely moving Drosophila adults. Proc. Natl. Acad. Sci. USA 111: 5367–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N., Privman E., Venton B. J., 2014. Optogenetic control of serotonin and dopamine release in Drosophila larvae. ACS Chem. Neurosci. 5: 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Luo J., He L., Montell C., Perrimon N., 2017. Oxidative stress induces stem cell proliferation via TRPA1/RyR-mediated Ca(2+) signaling in the Drosophila midgut. Elife 6: e22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. H., St-Pierre F., Sun X., Ding X., Lin M. Z., et al. , 2016. Subcellular imaging of voltage and calcium signals reveals neural processing in vivo. Cell 166: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Macara A. M., Lelito K. R., Minosyan T. Y., Shafer O. T., 2012. Analysis of functional neuronal connectivity in the Drosophila brain, J. Neurophysiol. 108: 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Baird G. S., Tsien R. Y., Davis R. L., 2003. Detection of calcium transients in Drosophila mushroom body neurons with camgaroo reporters. J. Neurosci. 23: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemelman B. V., Lee G. A., Ng M., Miesenbock G., 2002. Selective photostimulation of genetically chARGed neurons. Neuron 33: 15–22. [DOI] [PubMed] [Google Scholar]
- Zhang W., Ge W., Wang Z., 2007. A toolbox for light control of Drosophila behaviors through Channelrhodopsin 2-mediated photoactivation of targeted neurons. Eur. J. Neurosci. 26: 2405–2416. [DOI] [PubMed] [Google Scholar]
- Zhou C., Franconville R., Vaughan A. G., Robinett C. C., Jayaraman V., et al. , 2015. Central neural circuitry mediating courtship song perception in male Drosophila. Elife 4: e08477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart M. F., Pulver S. R., Truman J. W., Fushiki A., Fetter R. D., et al. , 2016. Selective inhibition mediates the sequential recruitment of motor pools. Neuron 91: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]