Abstract
The DNMT3A R882H mutation is frequently observed in acute myeloid leukemia (AML). It is located in the subunit and DNA binding interface of DNMT3A and has been reported to cause a reduction in activity and dominant negative effects. We investigated the mechanistic consequences of the R882H mutation on DNMT3A showing a roughly 40% reduction in overall DNA methylation activity. Biochemical assays demonstrated that R882H does not change DNA binding affinity, protein stability or subnuclear distribution of DNMT3A. Strikingly, DNA methylation experiments revealed pronounced changes in the flanking sequence preference of the DNMT3A-R882H mutant. Based on these results, different DNA substrates with selected flanking sequences were designed to be favored or disfavored by R882H. Kinetic analyses showed that the R882H favored substrate was methylated by R882H with 45% increased rate when compared with wildtype DNMT3A, while methylation of the disfavored substrate was reduced 7-fold. Our data expand the model of the potential carcinogenic effect of the R882H mutation by showing CpG site specific activity changes. This result suggests that R882 is involved in the indirect readout of flanking sequence preferences of DNMT3A and it may explain the particular enrichment of the R882H mutation in cancer patients by revealing mutation specific effects.
INTRODUCTION
Cancers acquire somatic DNA mutations as well as epigenetic changes, which ultimately confer a growth advantage to the affected cells (1–5). These two mechanisms of carcinogenesis are directly connected in cases when mutations occur in epigenetic enzymes like DNA and histone methyltransferases and demethylases. Such mutations have recently been observed in many human cancers (6–9). In 2010, first studies have reported particularly high frequencies of missense mutations in the DNMT3A DNA methyltransferase in acute myeloid leukemia (AML) (10,11). They were confirmed by several other follow up reports (reviews: (9,12)) showing that up to 25% of all AML patients contain DNMT3A mutations, and even 37% of all AML patients with a normal karyotype (10). Recurrent DNMT3A mutations have also been identified at lower frequencies in other cancers, including myelodysplastic syndrome (MDS) and T-cell lymphomas (reviews: (9,12)). DNMT3A mutations in AML usually occur heterozygously in only one allele that is accompanied by an intact wildtype allele and they show a strong enrichment of missense mutations (73% of all mutations, Supplementary Figure S1A). In particular arginine 882 in the catalytic C-terminal domain of DNMT3A is often mutated (65% of all missense mutations in DNMT3A, Supplementary Figure S1B), it is most often exchanged by histidine (66% of all R882 mutations). DNMT3A mutations are a negative prognostic marker correlated with a reduced time to treatment failure, shorter duration of complete remission and decreased disease-free survival in cytogenetically normal AML (12,13). Recent studies demonstrated that DNMT3A mutations occur early in pre-leukemic cells and they act as driver mutations of the carcinogenic process (12–15).
The structure of the C-terminal domain of DNMT3A (DNMT3AC) in complex with the C-terminal domain of its stimulator DNMT3L forms a linear heterotetramer consisting of two DNMT3L subunits (at the edges of the tetramer) and two DNMT3A molecules (in the center) (16) (Supplementary Figure S1C). The DNMT3A/3L interface is called FF interface and also used for multimerization of DNMT3A (17,18). The inner DNMT3A/3A interface, called RD interface, also forms the DNA binding site of the DNMT3A complex (16,17). This is a polar interface mediating a reversible oligomerization of DNMT3A FF-interface dimers into a mixture of dimers, tetramers and higher aggregates depending on the buffer conditions and enzyme concentrations (17–19). The R882H mutation is located in the RD interface implying potential effects on multimerization of DNMT3A and its DNA interaction (Supplementary Figure S1C).
Despite intensive work, the molecular and biochemical mechanism of leukemia initiation through the DNMT3A-R882H mutation has not yet been fully clarified (13). The high prevalence of this particular missense mutation, together with the low frequency of non-sense and frameshift mutations indicates that it must have a specific mechanistic effect at the molecular level. Different studies have determined the catalytic activity of the purified R882H mutant and often observed 50–70% residual activity of the mutant enzyme (11,20,21). Later, studies reported that the R882H mutation exhibits a dominant negative effect in cells and in vitro. Kim et al. showed that the exogenously expressed murine R878H mutant (corresponding to human R882H) interacts with the wildtype enzyme, but it was less efficiently methylating major satellite repeats in mouse ES cells (22). Russler-Germain et al. described that mixed enzyme preparations obtained after co-expression of wildtype and R882H in mammalian cells only showed a weak residual activity, suggesting that the complex formation of wildtype and R882H subunits inactivates the wildtype subunits in a dominant negative manner (23). However, it remains unclear, why the heterozygous R882H mutation is so strongly enriched and why it is not accompanied by a larger number of cases showing other forms of loss-of-function mutations like frameshifts or non-sense mutations in both alleles of the Dnmt3a gene.
MATERIALS AND METHODS
Site-directed mutagenesis, protein expression and purification
Mutagenesis was performed using the megaprimer site-directed mutagenesis method (24) and confirmed by restriction marker analysis and DNA sequencing. The C-terminal domain of human DNMT3A (Q9Y6K1.4) (amino acids 612–912) and its R882H mutant were cloned into pET28+ (Novagen) creating an N-terminal His6-tag fusion. His-tagged DNMT3AC and R882H were overexpressed in BL21 (DE3) Codon+ RIL Escherichia coli cells (Stratagene) and purified as described (25). Briefly, cells were grown in TB medium until an A600nm of 0.6 was reached and protein expression was induced for 12 h at 20°C by addition of 0.5 mM isopropyl-1-thio-β-d-galactopyranoside. The proteins were purified at high micromolar concentrations using Ni–NTA agarose and stored in 20 mM HEPES pH 7.5, 200 mM KCl, 0.2 mM DTT, 1 mM EDTA and 10% glycerol at –80°C. The concentrations of the proteins were determined by UV spectrophotometry and confirmed by densitometric analysis of Coomassie stained SDS-polyacrylamide gels.
Methyltransferase activity assay
The methyltransferase activity of DNMT3AC and the R882H mutant was measured using 100 nM of a biotinylated 509mer containing 58 CpG sites using the following primers (Bt-AGA TTA GGG AAG GGG GTG TG and AAG ATC CTT TCA AGG CCT CAG) as described (25) (Supplementary Figure S2). To study the flanking sequence effect on the activity of DNMT3AC and R882H, DNA methylation kinetics were measured using 1 μM biotinylated double stranded 30-mer oligonucleotides containing a single CpG site, which were designed to be preferred or disfavored by R882H. In both substrates, the lower DNA strand was biotinylated. The correct annealing of the individual strands was confirmed on polyacrylamide gels stained with gel red. To study the specific methylation of one CpG site in one DNA strand, the oligonucleotide substrates were used in hemimethylated form and with the central CpG site in fully methylated form (26). The fully methylated substrate could only be methylated at non-CpG sites in the flanking regions. The rate of methylation of the upper DNA strand at the target site was calculated by subtracting the activity observed with the fully methylated substrate from the activity observed with the hemimethylated substrate. Consideration of the fully methylated control was necessary to determine methylation rates at single CpG sites, because DNMT3A shows a high activity at some non-CpG sites (27), which otherwise would obscure the results.
With both substrates, DNA methylation was measured by the incorporation of tritiated methyl groups from radioactively labeled AdoMet (Perkin Elmer) into the biotinylated substrate using an avidin–biotin methylation plate assay (28). The methylation reactions were carried out in methylation buffer (20 mM HEPES pH 7.5, 1 mM EDTA, 50 mM KCl, 0.05 mg/ml bovine serum albumin) at 37°C using 2 μM DNMT3AC wildtype or mutant enzyme. The reactions were started by adding 0.76 μM AdoMet. The initial slope of the enzymatic reaction was determined by linear regression. To determine the thermal stability of DNMT3AC and R882H, one sample mix was prepared for each enzyme without DNA and split into two reaction tubes. In one tube the DNA methylation reaction was immediately started by adding DNA and AdoMet. The second tube was kept at 22°C or 37°C for 30 min before starting the methylation reaction.
In vitro bisulfite DNA methylation studies were carried out using the 509mer DNA fragment and primers specific for the converted DNA (GTG GGA TTT GGT TTT GTT TTG TAT TTT and GAA TAT TAC TAC TAC CCT CCT CCT TCT CAA TTT AAC) covering 56 CpG sites as described (25,29) and analyzed with BISMA (30,31).
DNA binding analysis of DNMT3A
DNA binding was investigated by fluorescence depolarization using a Cy5-labeled 29mer oligonucleotide substrate (5′-ACT TGC AAC GGT CCT AAC CGT CAC CTC TT-3′) as described (25,32). The binding reactions were carried out in binding buffer containing 20 mM HEPES pH 7.5, 100 mM KCl, 1 mM EDTA, 2 nM of Cy5-labeled DNA and increasing concentration of protein after incubation at room temperature for 5 min. Each data point was measured in triplicate and the average anisotropy values were taken for the analysis. Data were least squares fitted using an equilibrium binding model to determine the dissociation constant (KD) of DNA binding. Error analysis was based on the separate analysis of experimental repeats. DNA binding of wildtype DNMT3AC and R882H was measured side by side, the results obtained with wildtype DNMT3AC have already been published (25).
Subnuclear localization of DNMT3A
The subnuclear localization of DNMT3A was analyzed by fluorescence microscopy as described using EYFP fusions of murine DNMT3A (18,32). To study the localization of R882H, the R878H mutation was introduced into the murine DNMT3A (which corresponds to R882H in the human protein).
Circular dichroism (CD) spectroscopy
Folding of purified proteins was analyzed by CD spectroscopy. CD measurements were performed using a J-815 circular dichroism spectrophotometer (JASCO Corporation, Tokyo, Japan) in 200 mM KCl, 20 mM HEPES pH 7.5, 10% glycerol using 10 μM purified His-tagged DNMT3AC or R882H. The spectra were collected at 21°C using a 0.1 mm cuvette in a wavelength range between 195 and 240 nm. For each sample, at least 30 scans were collected and averaged. CD melting experiments were performed in dialysis buffer using enzyme concentrations as indicated. The CD signal was measured at a wavelength of 222 nm in a 0.1 mm cuvette in the temperature range from 20 to 70°C applying a temperature increase of 2°C/min. The melting temperature was determined as described (33).
RESULTS
The catalytic domain of DNMT3A (DNMT3AC) is enzymatically active (34) indicating that it is a suitable model system to study the effects of DNMT3A mutations on enzymatic properties and the RD domain interface (16). To search for the enzymatic effects of DNMT3A R882H mutations, we have prepared the R882H mutant in the context of DNMT3AC and purified the proteins after recombinant expression in E. coli (Supplementary Figure S1D).
Characterization of the properties of DNMT3AC-R882H
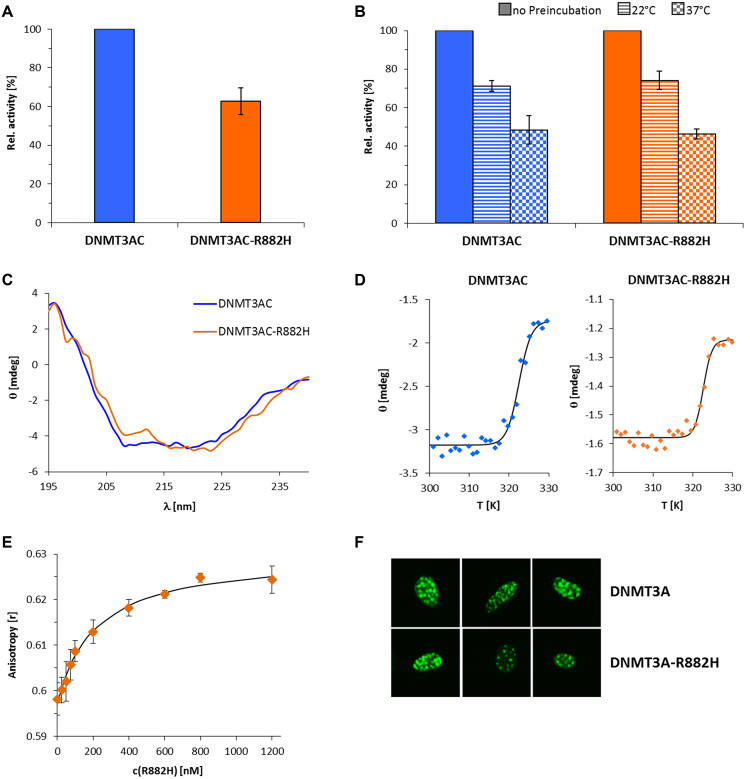
In vitro methylation experiments using identical concentrations of DNMT3AC wildtype and R882H showed that the R882H mutant has a residual activity of 62% with a 509mer DNA substrate containing 58 CpG sites, which was amplified from the CpG island upstream of the human SUHW1 gene (Figure 1A, Supplementary Figures S2 and S3). This result is comparable to residual activities of R882H observed in other studies (11,20,21). Since it represents a relatively mild reduction in activity, we were curious to study if other enzymatic properties of DNMT3A were altered by the mutation. To determine the effect of the mutation on protein stability, we pre-incubated the wildtype and mutant enzymes at 22 or 37°C before measuring the enzymatic activities. This caused a reduction of catalytic activity, which reflects the amount of enzyme denaturation during the pre-incubation interval. However, we did not observe any difference in thermal stability between wildtype and R882H in these experiments (Figure 1B), indicating that the mutation does not destabilize the enzyme folding. To directly investigate a potential effect of the R882H mutation on protein folding, far UV circular dichroism spectra of the purified DNMT3AC and R882H were determined. The CD spectra were superimposable within the level of noise indicating a similar folding of wildtype and mutant proteins (Figure 1C). To further examine the potential influence of the mutation on general protein stability, CD-melting experiments were conducted demonstrating an identical melting temperature of 49.3 ± 0.4°C and 49.5 ± 0.6°C for DNMT3AC and R882H (Figure 1D) which confirms the kinetic data shown in Figure 1B. As the R882H mutation is located in the DNA binding interface of DNMT3A, we also measured the DNA binding constant of R882H by fluorescence depolarization using a 29mer DNA containing two CpG sites. Our data show that the R882H mutant binds the substrate with a KD of 209±19 nM (Figure 1E), which is only slightly weaker than the wildtype enzyme (KD = 183±19 nM) measured under identical conditions with protein preparation that were purified in parallel (25). Previously it had been shown, that disruption of the RD interface by an R881A mutation in murine DNMT3A (the corresponding residue in human DNMT3A is R885) resulted in a loss of the normal localization of EYFP fused DNMT3A at DAPI dense heterochromatin in NIH3T3 cells (17,18). To investigate if the R882H mutation leads to a similar deterioration of the RD interface, we have prepared a murine DNMT3A R878H mutant (corresponding to the human R882H) fused to EYFP, expressed the protein in NIH3T3 cells and studied its sub-nuclear localization by fluorescence microscopy. However, there was no detectable difference in the sub-nuclear localization between the wildtype DNMT3A and R882H EYFP fusion proteins (Figure 1F).
Figure 1.
Catalytic activity and characterization of the R882H mutant. (A) DNA methylation activity of DNMT3A catalytic domain and the R882H mutant using a 509mer DNA substrate. The bars represent the average of 5 experiments, error bars indicate the standard error of the mean. See also Supplementary Figure S3. (B) Protein stability of wildtype DNMT3AC and R882H analyzed by the loss of enzymatic activity during a pre-incubation of the protein for 30 min at different temperatures. Error bars indicate the SEM based on 2 independent experiments. (C) CD spectra of wildtype DNMT3AC and R882H. The similar shape of the spectra indicates comparable folding of both proteins. (D) Thermal stability of DNMT3AC and R882H analyzed by CD melting at protein concentrations of 22 μM for DNMT3AC and 8 μM for R882H. Quantitative analysis revealed identical melting temperatures of 49.3 ± 0.4°C and 49.5 ± 0.6°C for DNMT3AC and R882H. (E) DNA binding titration of the R882H mutant to a 29mer DNA substrate. The data points show the average of two experiments and the standard error. The line shows a fit of the data with a KD of 209 nM. (F) Heterochromatic localization of EYFP fused murine DNMT3A containing the R878H mutation (corresponding to R882H in human DNMT3A).
Overall, the lack of changes in DNA binding, enzyme localization and stability indicates that the DNMT3A/3A RD interface is not massively deranged by the R882H mutation. This conclusion is in agreement with the moderate reduction in catalytic activity of the R882H mutant, because disruption of the RD interface by a R881A mutation in murine DNMT3AC (corresponding to human R885) completely inactivated the enzyme (17).
Altered flanking sequence preferences of DNMT3A R882H
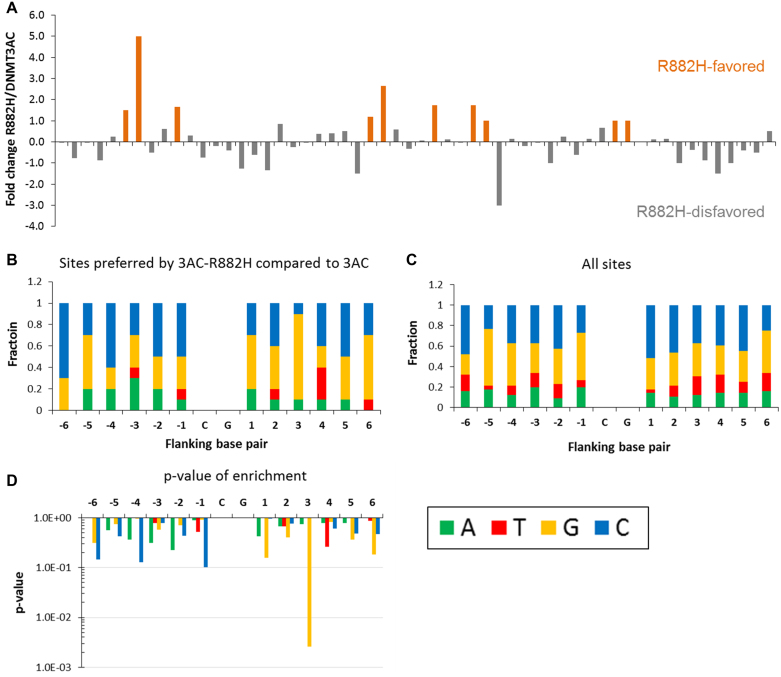
Based on the position of the R882H mutation in the DNA binding interface of DNMT3A, we next investigated if this mutation influences the specificity of the DNA interaction of DNMT3A. It has been well documented that DNMT3A shows pronounced differences in the methylation activity of CpG sites depending on their flanking sequence (26,35–37), and CpG sites have been identified which could not be methylated by DNMT3A at all (26). Moreover, the R882A mutation had already been shown to alter the relative preferences of the methylation of three DNA substrates with different flanking sequences (38). We used the purified R882H mutant protein for our experiments, because it is not possible to prepare DNMT3AC with a mixed wildtype/R882H RD interface. We conducted methylation kinetics with a fragment of the 509mer substrate used in the radioactive methylation assays as DNA substrate, which allows to study the methylation of 56 CpG sites in different flanking contexts, and measured DNA methylation at all CpG sites by wildtype DNMT3AC and R882H by bisulfite conversion and sequencing. Wildtype DNMT3A showed strong differences in the methylation of different sites in agreement with previous studies (25,26). In agreement with the general reduction of R882H activity, most CpG sites exhibited lower methylation levels by R882H. However, a comparison of the relative methylation levels of different CpG sites between wildtype and the R882H revealed a strong change of the flanking sequence preferences of the mutant (Figure 2A). Because of this, some sites showed a much more pronounced reduction of methylation activity than the 38% average reduction in activity reported above. Even more strikingly, some sites were methylated up to five times faster by R882H than by the wildtype enzyme. This was mainly observed at sites which were weakly methylated by wildtype DNMT3AC, but still methylated with good yields by R882H. We extracted the –6 to +6 flanks of the 10 sites most preferred by R882H when compared to wildtype and observed a strong overrepresentation of G at the +3 position (Figure 2B and C). Less pronounced enrichments at other positions were detected after a statistical analysis comparing the distribution of bases at each flanking position in the preferred sites with the overall distribution in all sites (Figure 2D). These results clearly indicate that the flanking sequences of R882H differ substantially from wildtype DNMT3AC.
Figure 2.
Flanking sequence analysis of R882H. (A) Bisulfite analysis of the methylation of 56 CpG sites of the 509mer PCR fragment. The figure shows for each site (1–56), the ratio of the relative methylation levels observed in the wildtype (wt) and R882H samples. In case of zero methylation by the wildtype, ratios were set to 5.0 (for details see Supplementary Table S1). (B) Distribution of bases at the flanking sequence positions in the 10 sites preferred by R882H. (C) Distribution of bases at the flanking sequence positions of all sites on the DNA substrate. (D) P-values of enrichment of bases at all flanking sites in the set of preferred sites (for details see Supplementary Table S2).
Methylation kinetics with designed oligonucleotide substrates
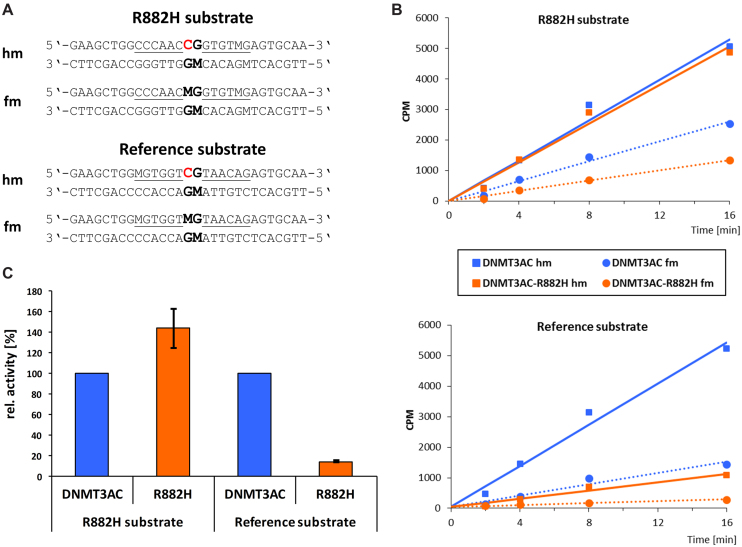
Based on the analysis described in the last section, we designed model substrates containing one central CpG site with flanks optimized for methylation of the upper DNA strand by R882H (‘R882H substrate’) and flanks not containing features preferred by R882H (‘reference substrate’) (Figure 3A). To study only the specific methylation of the central CpG in the upper strand, we used the designed substrates in hemimethylated form. Moreover, it has been repeatedly observed, that DNMT3A also methylates other cytosine residues in oligonucleotide substrates, which are not situated in a CpG context (26,27,39,40). This additional activity could obscure the comparison of the methylation rates of the two substrates. Therefore, both designed substrates were used in an additional control form, where the central CpG site is methylated in both strands (fully methylated). Hence, these control substrates could only be methylated at other cytosine residues. To determine the methylation activity only for the central CpG in the upper strand, the activity observed with the fully methylated substrates was subtracted from the activity observed with the corresponding hemimethylated substrates (Figure 3B).
Figure 3.
Design of a preferred R882H substrate. (A) Based on the sequences of the 10 most preferred methylation sites by R882H, a substrate with R882H optimized flanks was designed together with a reference substrate not containing R882H preferred residues. For each substrate, a hemimethylated version (hm) and a fully methylated (fm) version was used to allow measurement of the specific methylation activity in the upper DNA strand of the central CpG site. ‘M’ denotes 5-methyldeoxycytosine, the target C is highlighted in red. (B) Example kinetic data of the methylation of the hm (squares, continuous lines) and fm (circles, dotted lines) substrates by DNMT3AC (blue) and R882H (orange). (C) Average target site methylation rates of the R882H substrate and reference substrate by wildtype DNMT3AC and DNMT3AC-R882H. To determine the specific methylation of the central CpG site in the upper DNA strand, the methylation of the fm substrate is subtracted from the methylation of the hm substrate. Error bars indicate the SEM based on four independent experiments.
Methylation experiments of the two substrates in their hemi- and fully methylated forms with wildtype DNMT3AC and R882H revealed that methylation of the R882H substrate by the R882H mutant was 45% faster than by wildtype DNMT3AC (Figure 3C). In contrast, the residual activity of R882H on the reference substrate was <15% when compared to the wildtype enzyme, indicating that the lack of sequence features preferred by R882H has led to a very low activity. Strikingly, the overall difference of the catalytic activities of wildtype and R882H on these two substrates was ∼10-fold. These results confirm the conclusions of the flanking sequence analysis of methylated sites by demonstrating that it is possible to design an R882H preferred and a disfavored substrate on the basis of the flanking sequence preferences.
Inhibition of DNMT3A R882H by small compounds
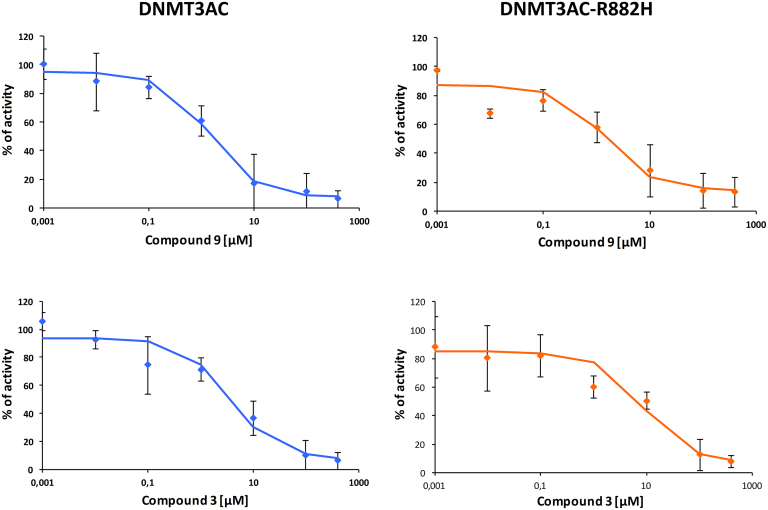
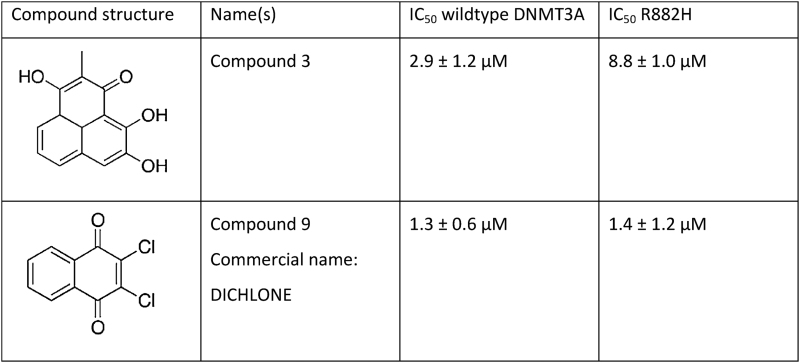
Our data indicate that the R882H mutation causes a defined hyperactivity of DNMT3A at a subset of CpG sites. Hence, an inhibition of the mutated DNMT3A might be a therapeutic option. Initial studies indeed indicated that DNMT3A mutant cancer patients respond better to treatment with nucleosidic hypomethylating agents (41,42). To investigate the possibility of a direct inhibition of DNMT3A R882H, we tested two previously validated DNMT3A inhibitors (43) for their effects on the activity of the R882H mutant. As shown in Table 1 and Figure 4, one of them (compound 9) showed equal inhibition of both wildtype and mutant DNMT3AC, while the other (compound 3) showed a threefold reduced inhibition of the R882H mutant. Thus, these inhibitors, in particular compound 9, are still active against the mutant DNMT3A.
Table 1. Inhibition of wildtype and R882H mutant by DNMT3A inhibitors (43). IC50 values were derived from 2 independent experiments, in which inhibitor concentrations were titrated between 1 nM and 400 μM. Error margins denote the SD of the fit of the IC50 values. The titration curves are shown in Figure 4.
|
Figure 4.
Inhibition of wildtype DNMT3AC and R882H by compound 3 and compound 9. Averages and standard errors from two independent experiments are shown. IC50-values are listed in Table 1.
DISCUSSION
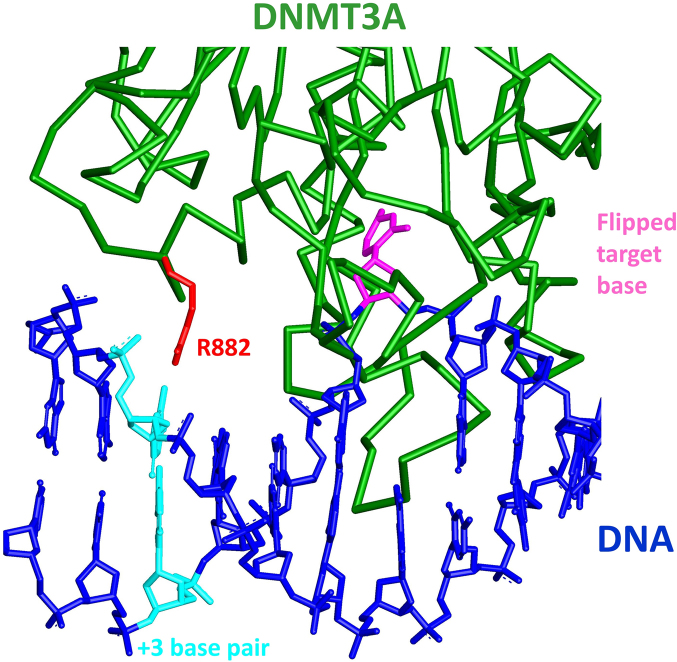
Despite the fact that the R882H mutation in the DNA binding interface of DNMT3A is highly prevalent in AML, its pathogenic mechanism has not been finally clarified at the molecular level (13). Previous data showed a moderately reduced activity of the mutant and a dominant negative effect of the R882H mutation (23). Here, we show that the R882H mutation leads to strong changes in the flanking sequence preferences of DNMT3A most prominently at the +3 position but also at other sites. DNMT3A has been shown previously to exhibit pronounced flanking sequence preferences in the methylation of different CpG sites (26,35–37), which could be due to flanking sequence effects on the DNA binding or conformational changes of the DNA during complex formation (like DNA bending or unwinding), but also effects of the flanking sequence on dynamic steps during catalysis, like target base flipping. Our findings suggests that the R882H exchange directly affects the DNA interaction of DNMT3A. To further explore this point, we prepared a model of the DNMT3A-DNA complex structure by using the DNA from the M.HhaI-DNA complex (44) after superposition of the conserved catalytic methyltransferase residues of DNMT3A and M.HhaI (Figure 5). Strikingly, in this model the R882 side chain is positioned directly adjacent to the +3 base pair in an ideal position to contact the DNA backbone at this site. Thereby, R882 could probe sequence dependent conformational preferences of the DNA substrate in a process reminiscent of the classic indirect readout of DNA sequence by proteins (45,46). This leads to a complex readout of the DNA sequence dependent bending and twisting properties of the DNA, mediated by contacts of the protein to the phosphodiester backbone of the DNA.
Figure 5.
Details of a model of the DNMT3A-DNA complex structure. The Cα-backbone of DNMT3A (pdb: 2QRV) is shown in green, R882 is in red. DNA (shown in blue) was modelled into the DNMT3A structure using the DNA molecule from the M.HhaI-DNA complex structure (pdb: 1MHT) by superposition of the conserved catalytic methyltransferase residues of M.HhaI and DNMT3A. The flipped target cytosine is shown in pink and the +3 base pair in cyan.
The change in flanking sequence preferences of R882H reported here uncovers an additional dimension in the effects of the R882H mutation on the methylation activity of DNMT3A by showing that the influence of the mutation is CpG site specific. In addition to the moderate global reduction of activity, our data show that specific sites are affected much more strongly and strong hypomethylation as well as hypermethylation can occur depending on the target site. Unfortunately, these changes in flanking sequence preferences observed in vitro cannot be directly compared with patient data, because in cells DNMT3A R882H can methylate the upper or lower DNA strand creating a hemimethylated CpG site. Afterwards, DNMT1 will methylate the second strand due to its high activity on hemimethylated CpG sites. Hence, in cellular methylation data sets, one cannot determine which strand was initially methylated by DNMT3A. As the flanking preferences are not palindromic, in cellular data all flanking signatures are mixed resulting in very poor statistics.
A gain-of-function effect of the R882H mutation potentially related to hyperactivity at particular sites is in agreement with its positive correlation with other mutations blocking DNA demethylation, which would enhance these effects. This refers to the co-occurrence of R882H with TET2 inactivating mutations in peripheral T-cell lymphomas (12,47), which were experimentally shown to cooperate in the induction of lymphoid malignancies in mice (47). Along the same line, IDH mutations (which indirectly reduce the activity of TET enzymes) are also positively correlated with DNMT3A R882H in AML and myeloproliferative neoplasm (3,12,48). Finally, some studies indicated that DNMT3A mutant patients responded better to treatment with hypomethlyating agents (41,42), which could be explained by the site specific gain-of-function model of R882H. Future work in hematopoietic stem cells and AML precursor cells, will be needed to identify the relevant methylation sites which are hyper- and hypomethylated by R882H, connect them with corresponding genes, and find out, if inhibition of hypermethylation of CpG sites by DNMT3A-R882H in AML precursor cells may delay the onset of the disease.
DATA AVAILABILITY
All data are included in the manuscript and its supplemental material.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks are due to Drs Christine Maulay-Bailly and Séverine Amand for the initial synthesis of compounds 3 and 9.
Author contributions: A.J., M.E. and A.R. devised the project. M.E. and A.R. conducted the biochemical assays with contributions from S.K. R.R. conducted DNA sequencing for the bisulfite methylation analysis. P.B.A. provided the DNMT3A inhibitors. R.Z.J. and A.J. supervised research. A.J. wrote the manuscript draft. All authors contributed to data interpretation and discussion, read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft Priority Program SPP1463 [JE 252/20-1]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Baylin S.B., Jones P.A.. A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer. 2011; 11:726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. You J.S., Jones P.A.. Cancer genetics and epigenetics: two sides of the same coin. Cancer Cell. 2012; 22:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L. et al. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013; 502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suva M.L., Riggi N., Bernstein B.E.. Epigenetic reprogramming in cancer. Science. 2013; 339:1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shih A.H., Abdel-Wahab O., Patel J.P., Levine R.L.. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 2012; 12:599–612. [DOI] [PubMed] [Google Scholar]
- 7. Aumann S., Abdel-Wahab O.. Somatic alterations and dysregulation of epigenetic modifiers in cancers. Biochem. Biophys. Res. Commun. 2014; 455:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kudithipudi S., Jeltsch A.. Role of somatic cancer mutations in human protein lysine methyltransferases. Biochim. Biophys. Acta. 2014; 1846:366–379. [DOI] [PubMed] [Google Scholar]
- 9. Hamidi T., Singh A.K., Chen T.. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015; 7:247–265. [DOI] [PubMed] [Google Scholar]
- 10. Ley T.J., Ding L., Walter M.J., McLellan M.D., Lamprecht T., Larson D.E., Kandoth C., Payton J.E., Baty J., Welch J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010; 363:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamashita Y., Yuan J., Suetake I., Suzuki H., Ishikawa Y., Choi Y.L., Ueno T., Soda M., Hamada T., Haruta H. et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010; 29:3723–3731. [DOI] [PubMed] [Google Scholar]
- 12. Yang L., Rau R., Goodell M.A.. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015; 15:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roy D.M., Walsh L.A., Chan T.A.. Driver mutations of cancer epigenomes. Protein Cell. 2014; 5:265–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu J., Wang Y.Y., Dai Y.J., Zhang W., Zhang W.N., Xiong S.M., Gu Z.H., Wang K.K., Zeng R., Chen Z. et al. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:2620–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shlush L.I., Zandi S., Mitchell A., Chen W.C., Brandwein J.M., Gupta V., Kennedy J.A., Schimmer A.D., Schuh A.C., Yee K.W. et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014; 506:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X.. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007; 449:248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jurkowska R.Z., Anspach N., Urbanke C., Jia D., Reinhardt R., Nellen W., Cheng X., Jeltsch A.. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 2008; 36:6656–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jurkowska R.Z., Rajavelu A., Anspach N., Urbanke C., Jankevicius G., Ragozin S., Nellen W., Jeltsch A.. Oligomerization and binding of the Dnmt3a DNA methyltransferase to parallel DNA molecules: heterochromatic localization and role of Dnmt3L. J. Biol. Chem. 2011; 286:24200–24207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kareta M.S., Botello Z.M., Ennis J.J., Chou C., Chedin F.. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 2006; 281:25893–25902. [DOI] [PubMed] [Google Scholar]
- 20. Yan X.J., Xu J., Gu Z.H., Pan C.M., Lu G., Shen Y., Shi J.Y., Zhu Y.M., Tang L., Zhang X.W. et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 2011; 43:309–315. [DOI] [PubMed] [Google Scholar]
- 21. Holz-Schietinger C., Matje D.M., Reich N.O.. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J. Biol. Chem. 2012; 287:30941–30951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S.J., Zhao H., Hardikar S., Singh A.K., Goodell M.A., Chen T.. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013; 122:4086–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Russler-Germain D.A., Spencer D.H., Young M.A., Lamprecht T.L., Miller C.A., Fulton R., Meyer M.R., Erdmann-Gilmore P., Townsend R.R., Wilson R.K. et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014; 25:442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeltsch A., Lanio T.. Site-directed mutagenesis by polymerase chain reaction. Methods Mol. Biol. 2002; 182:85–94. [DOI] [PubMed] [Google Scholar]
- 25. Emperle M., Rajavelu A., Reinhardt R., Jurkowska R.Z., Jeltsch A.. Cooperative DNA binding and protein/DNA fiber formation increases the activity of the Dnmt3a DNA methyltransferase. J. Biol. Chem. 2014; 289:29602–29613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jurkowska R.Z., Siddique A.N., Jurkowski T.P., Jeltsch A.. Approaches to enzyme and substrate design of the murine Dnmt3a DNA methyltransferase. Chembiochem. 2011; 12:1589–1594. [DOI] [PubMed] [Google Scholar]
- 27. Gowher H., Jeltsch A.. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J. Mol. Biol. 2001; 309:1201–1208. [DOI] [PubMed] [Google Scholar]
- 28. Roth M., Jeltsch A.. Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol. Chem. 2000; 381:269–272. [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y., Rohde C., Tierling S., Stamerjohanns H., Reinhardt R., Walter J., Jeltsch A.. DNA methylation analysis by bisulfite conversion, cloning, and sequencing of individual clones. DNA Methylation: Methods Protoc. 2009; 177–187. [DOI] [PubMed] [Google Scholar]
- 30. Rohde C., Zhang Y., Reinhardt R., Jeltsch A.. BISMA - fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010; 11:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rohde C., Zhang Y., Jurkowski T.P., Stamerjohanns H., Reinhardt R., Jeltsch A.. Bisulfite sequencing Data Presentation and Compilation (BDPC) web server—a useful tool for DNA methylation analysis. Nucleic Acids Res. 2008; 36:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rajavelu A., Jurkowska R.Z., Fritz J., Jeltsch A.. Function and disruption of DNA methyltransferase 3a cooperative DNA binding and nucleoprotein filament formation. Nucleic Acids Res. 2012; 40:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Greenfield N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006; 1:2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gowher H., Jeltsch A.. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 2002; 277:20409–20414. [DOI] [PubMed] [Google Scholar]
- 35. Lin I.G., Han L., Taghva A., O’Brien L.E., Hsieh C.L.. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell. Biol. 2002; 22:704–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Handa V., Jeltsch A.. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005; 348:1103–1112. [DOI] [PubMed] [Google Scholar]
- 37. Wienholz B.L., Kareta M.S., Moarefi A.H., Gordon C.A., Ginno P.A., Chedin F.. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet. 2010; 6:e1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gowher H., Loutchanwoot P., Vorobjeva O., Handa V., Jurkowska R.Z., Jurkowski T.P., Jeltsch A.. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J. Mol. Biol. 2006; 357:928–941. [DOI] [PubMed] [Google Scholar]
- 39. Ramsahoye B.H., Biniszkiewicz D., Lyko F., Clark V., Bird A.P., Jaenisch R.. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arand J., Spieler D., Karius T., Branco M.R., Meilinger D., Meissner A., Jenuwein T., Xu G., Leonhardt H., Wolf V. et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012; 8:e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Traina F., Visconte V., Elson P., Tabarroki A., Jankowska A.M., Hasrouni E., Sugimoto Y., Szpurka H., Makishima H., O’Keefe C.L. et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014; 28:78–87. [DOI] [PubMed] [Google Scholar]
- 42. Metzeler K.H., Walker A., Geyer S., Garzon R., Klisovic R.B., Bloomfield C.D., Blum W., Marcucci G.. DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia. 2012; 26:1106–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ceccaldi A., Rajavelu A., Ragozin S., Senamaud-Beaufort C., Bashtrykov P., Testa N., Dali-Ali H., Maulay-Bailly C., Amand S., Guianvarc’h D. et al. Identification of novel inhibitors of DNA methylation by screening of a chemical library. ACS Chem. Biol. 2013; 8:543–548. [DOI] [PubMed] [Google Scholar]
- 44. Klimasauskas S., Kumar S., Roberts R.J., Cheng X.. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994; 76:357–369. [DOI] [PubMed] [Google Scholar]
- 45. Garvie C.W., Wolberger C.. Recognition of specific DNA sequences. Mol. Cell. 2001; 8:937–946. [DOI] [PubMed] [Google Scholar]
- 46. Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S.. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010; 79:233–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scourzic L., Couronne L., Pedersen M.T., Della Valle V., Diop M., Mylonas E., Calvo J., Mouly E., Lopez C.K., Martin N. et al. DNMT3A(R882H) mutant and Tet2 inactivation cooperate in the deregulation of DNA methylation control to induce lymphoid malignancies in mice. Leukemia. 2016; 30:1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shen Y., Zhu Y.M., Fan X., Shi J.Y., Wang Q.R., Yan X.J., Gu Z.H., Wang Y.Y., Chen B., Jiang C.L. et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011; 118:5593–5603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and its supplemental material.