Abstract
The formation of ribosomal subunits is a highly dynamic process that is initiated in the nucleus and involves more than 200 trans-acting factors, some of which accompany the pre-ribosomes into the cytoplasm and have to be recycled into the nucleus. The inhibitor diazaborine prevents cytoplasmic release and recycling of shuttling pre-60S maturation factors by inhibiting the AAA-ATPase Drg1. The failure to recycle these proteins results in their depletion in the nucleolus and halts the pathway at an early maturation step. Here, we made use of the fast onset of inhibition by diazaborine to chase the maturation path in real-time from 27SA2 pre-rRNA containing pre-ribosomes localized in the nucleolus up to nearly mature 60S subunits shortly after their export into the cytoplasm. This allows for the first time to put protein assembly and disassembly reactions as well as pre-rRNA processing into a chronological context unraveling temporal and functional linkages during ribosome maturation.
INTRODUCTION
Ribosomes translate the genetic information into the amino acid sequence of proteins. Eukaryotic ribosomes are composed of one small (40S) and one large (60S) subunit each containing ribosomal proteins and one (small subunit) or three (large subunit) ribosomal RNAs (rRNA). The formation of the ribosomal subunits involves more than 200 trans-acting factors and is one of the major activities in each cell (1). This process is best understood in the yeast Saccharomyces cerevisiae (2,3).
Ribosome biogenesis is initiated by transcription of rDNA repeats in the nucleolus. While the precursor of the 5S rRNA is transcribed by RNA polymerase III, the precursors of the 18S, 25S and 5.8S rRNA are present on a single RNA polymerase I transcript, the 35S pre-rRNA, which contains external (ETS) and internal (ITS) transcribed spacers (Supplementary Figure S1). After two consecutive processing reactions in the 5′ ETS of the 35S pre-rRNA, an endo-nucleolytic cleavage step in position A2 leads to a 20S and a 27SA2 pre-rRNA, the precursors of the small and large subunit rRNAs, respectively. While the 20S pre-rRNA is processed to the mature 18S rRNA of the small subunit in the cytoplasm, the 27SA2 precursor undergoes further cleavage and trimming reactions within the nucleus resulting in the 27SB pre-rRNA. This precursor is further cleaved yielding 25.5S and 7S pre-rRNA. Whereas the 25.5S pre-rRNA is converted into the mature 25S rRNA within the nucleus, the 7S pre-rRNA to 5.8S processing takes place in multiple steps first in the nucleus and finally in the cytoplasm (4).
These maturation steps are performed within pre-ribosomal particles containing ribosomal proteins and non-ribosomal factors that coordinate modification and processing reactions. Many assembly events and also the first processing steps including cleavage at site A2, occur co-transcriptionally (3,5–8). The assembly of most ribosomal proteins and transiently binding maturation factors on such pre-ribosomal particles has to occur in a defined manner to allow correct formation of ribosomal subunits (3,5,8).
Since each proliferating eukaryotic cell has to synthesize several hundred thousands of ribosomes during its cell cycle most of each cells energy spend is dedicated to ribosome biogenesis (1). Moreover, ribosome biogenesis is a highly dynamic process. Pulse analyses with radiolabeled tracers showed that the pre-rRNA processing reactions from 35S pre-rRNA to the mature 25S rRNA in yeast require <6 min (6). Investigating such a dynamic pathway using genetic manipulation of maturation factors is challenging since the onset of perturbation takes much longer than the process itself. Therefore specialized pulse chase methods involving incorporation of a non-natural amino acid and nucleotides were developed to characterize freshly synthesized pre-ribosomal particles or pre-rRNAs (9,10).
A promising strategy to investigate highly dynamic processes is the use of low molecular weight inhibitors that diffuse rapidly into the cell and block the function of their target proteins within seconds (11). We previously showed that the drug diazaborine specifically blocks 60S ribosomal subunit formation in yeast, thereby identifying the first specific inhibitor of eukaryotic ribosome biogenesis (12). Our subsequent work revealed that diazaborine targets the low abundant, cytosolic AAA-ATPase Drg1 (diazaborine resistance gene 1) and blocks ATP hydrolysis in the D2 domain of the protein (13). Drg1 binds to the pre-60S particle after nuclear export and catalyzes the release of the shuttling protein Rlp24 (13–15). This reaction initiates cytoplasmic pre-60S maturation and is a pre-requisite for all downstream processes (15,16). Inhibition of Drg1 therefore prevents release and recycling of all investigated shuttling proteins (i.e. Rlp24, Nog1, Tif6, Nmd3, Mrt4, Arx1, Alb1 and Bud20) as well as the joining of late factors (13). The failure to recycle shuttling proteins upon diazaborine treatment leads to their depletion in the nucleus, which causes early pre-60S maturation defects as obvious from the inefficient processing of 27SA2 pre-rRNA (12).
In this work we made use of the fast onset of inhibition of ribosome biogenesis by diazaborine to investigate the dynamics of the 60S subunit biogenesis pathway. The entrapment of shuttling proteins on cytoplasmic pre-60S particles after diazaborine treatment and their subsequent decline in the nucleus mimics genetic depletion. However, the onset of inhibition with chemical compounds is much faster than achievable by common technologies. This allowed us to monitor the compositional changes the pre-60S particles undergo during their transition from the nucleolus into the cytoplasm in a kinetic manner.
MATERIALS AND METHODS
Yeast strains and growth conditions
The yeast strains used in the present study are listed in Supplementary Table S1. Chromosomal gene fusions were generated by homologous recombination using polymerase chain reaction products to transform the respective yeast strain as described (17). Strains were grown either in yeast extract peptone dextrose (YPD) complex medium or in synthetic dextrose (SDC) medium supplemented with the appropriate amino acids.
Diazaborine treatment of GFP/mCherry-tagged strains
For the standard procedure strains expressing green fluorescent protein (GFP) fusions of ribosome biogenesis factors and a compartment tracker protein tagged with mCherry were grown to an OD600 0.45 in SDC-medium supplemented with all amino acids. Diazaborine was added to a final concentration of 10 μg/ml and cells from 1ml of culture were harvested after 0, 2, 5, 10, 15 and 30 min of diazaborine treatment. The cell pellet was resuspended in 5 μl SDC with inhibitor and 2 μl thereof were spotted on slides covered with 2% SD agar and containing 100 μg/ml diazaborine. The agar aimed to restrict movement of cells after spotting and to prevent cells from entering anaerobiosis during the imaging procedure. As a negative control untreated cells were spotted on SD agar coated slides without diazaborine. The cells were examined via laser scanning microscopy. In case of combined treatment of diazaborine and cycloheximide, cycloheximide was added in a concentration of 1 μg/ml.
Confocal laser scanning microscopy
Imaging was performed using a Leica SP5 confocal microscope (Leica Inc., Mannheim) with spectral detection and a HCX PL APO 63× NA 1.4 oil immersion objective. Fluorescence signal was detected using hybrid photon-detectors. GFP was excited at 488 nm, emission detected between 500 and 550 nm. mCherry was excited at 561 nm and emission detected between 570 and 650 nm. Z-stacks were recorded using 100 × 100 × 250 (x/y/z) nm sampling. Fluorescence and transmission images were acquired simultaneously.
Image processing
Image processing and quantification of fluorescent protein signal was performed using the open-source software Fiji (18). For this purpose, the nuclear membrane protein Nic96 or the nucleolar protein Nop58 were tagged with mCherry and used to identify the nucleus and the nucleolus, respectively. Initially, image noise in acquired three-dimensional (3D) fluorescence data was reduced using 3D Gaussian filtering (sigma value: 1 × 1 × 1; x/y/z).
Afterward generated 3D data were projected using the maximum-intensity method. Projected Nic96-mCherry labeled nuclear structures were automatically segmented using the Yen’s thresholding method implemented in Fiji. In a second step, still unsegmented nuclear areas were registered using the ‘fill holes’ command. This allowed definition of the nucleus occupied area in each cell. Accordingly, Nop58-mCherry labeled nucleoli were registered. Segmented nuclei and nucleoli were converted into binary masks. The raw intensity in maximum-intensity projections of acquired GFP signal within these masks was computed using the particle analyzer tool and allowed estimation of the fusion proteins distribution in the individual compartments.
Cytosolic GFP signal in maximum-intensity projections was registered using Yen’s automated thresholding method and by excluding projected masks representing nuclei or nucleoli from the segmentation process. Again, the intensity of cytosolic GFP signal was computed using the particle analyzer tool.
Affinity purification of pre-ribosomal particles using magnetic beads
Strains expressing TAP-tagged maturation factors were grown in YPD medium at 30°C to late log phase (OD600 1–1.2). Afterward diazaborine was added to a final concentration of 100 μg/ml and incubation continued for up to 60 min. Cells were harvested by centrifugation at 4500 × g for 1 min at 25°C and the cell pellets were frozen. Extracts were prepared from cell pellets corresponding to one liter of culture by breaking the cells in a bead mill (Merkenschlager) in the presence of 0.6 mm glass beads for 4 min with CO2 cooling in buffer A (20 mM HEPES-NaOH, pH 7.5, 10 mM KCl, 2.5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.5 mM PMSF, FY® protease inhibitor cocktail, Serva). After centrifugation at 40 000 × g for 30 min, supernatants were loaded on magnetic beads coupled to rabbit IgG as described (19,20). After extensive washing, 20% of the beads were used for RNA extraction. Another 20% of the beads were suspended in tobacco etch virus (TEV) protease containing cleavage buffer (buffer A, 100 mM NaCl) together with 40 U Ribolock RNAse inhibitor (Life Tech) and incubated for 1 h at 22°C as described (15). The supernatant was analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting. The residual 60% of the beads were washed twice with 1 ml AC buffer (100 mM NH4Oac, pH 7.4, 0.1 mM MgCl2). Bound proteins were eluted two times with 500 μl of freshly prepared 500 mM NH4OH solution for 20 min at RT. Eluate fractions were pooled, lyophilized and further processed for semi-quantitative mass spectrometric protein analyses using iTRAQ reagents as described (20,21). Briefly, lyophilized proteins were suspended in dissolution buffer (iTRAQ™ labeling kit, Invitrogen), reduced with Tris-(2-carboxyethyl)phosphine and cysteins blocked with 10 mM methyl-methanethiosulfonate. After trypsin digestion for 20 h at 37°C, the primary amines of the resulting peptides were labeled with different combinations of the four iTRAQ™ reagents, each containing a different isobaric tag. The labeled peptides from tandem affinity purifications (TAP) from the untreated strains were combined with differently labeled peptides from TAP purifications from a treated strain. The combined differently labeled peptides were analyzed by MALDI-TOF/TOF mass spectrometry. The peak area for iTRAQ™ reporter ions were interpreted and corrected by the GPS-Explorer software (Applied Biosystems) and Excel (Microsoft). An iTRAQ ratio average of all peptides of a given protein was calculated. For each bait protein at least two biological replicates were analyzed.
Data visualization of iTRAQ results
Peptides used for protein identification and quantitation were identified with a confidence interval of more than 95%. All observed iTRAQ ratios were normalized to the iTRAQ ratio of the respective bait protein and expressed in log2 scale. Heat map visualization was performed with Java Treeview (22).
RNA extraction and northern blotting
RNA was extracted from purified magnetic beads after their resuspension in buffer TES (10 mM Tris–HCl, pH 7.5, 10 mM ethylenediaminetetraacetic acid (EDTA), 0.5% SDS) using two subsequent extractions with phenol: chloroform: isoamyl alcohol (25:24:1) and one extraction with chloroform: isoamyl alcohol (24:1). Thereafter, RNA was precipitated from the aqueous phase by addition of 1/10 volume of 3 M sodium acetate and 2.5 volumes of ethanol and after drying dissolved in nuclease-free water. After purification, RNA was separated on 1.2% agarose gels and blotted onto nylon membranes by capillary transfer. Hybridization was performed at 42°C by overnight incubation with suspensions of the 32P 5′ radiolabeled oligonucleotides (described in Supplementary Table S2) in 0.5M Na2HPO4, pH 7.2, 7% SDS and 1 mM EDTA After washing signal were detected using X-ray films.
SDS-PAGE and western blot analysis
Protein samples were separated on precast 4–12% NuPAGE gels (Novex life technologies) and blotted on a polyvinylidene fluoride (PVDF) membrane (Carl Roth GmbH) using the tank-blot-system (Hoefer). Chemiluminescence signals were detected using the Clarity™ Western Blotting Detection Reagent (Bio-Rad) and the ChemiDoc™ Touch Imaging System (Bio-Rad). All antibodies used in this study were described previously (13,23).
RESULTS
Visualizing pre-60S particle migration from the nucleolus to the cytoplasm
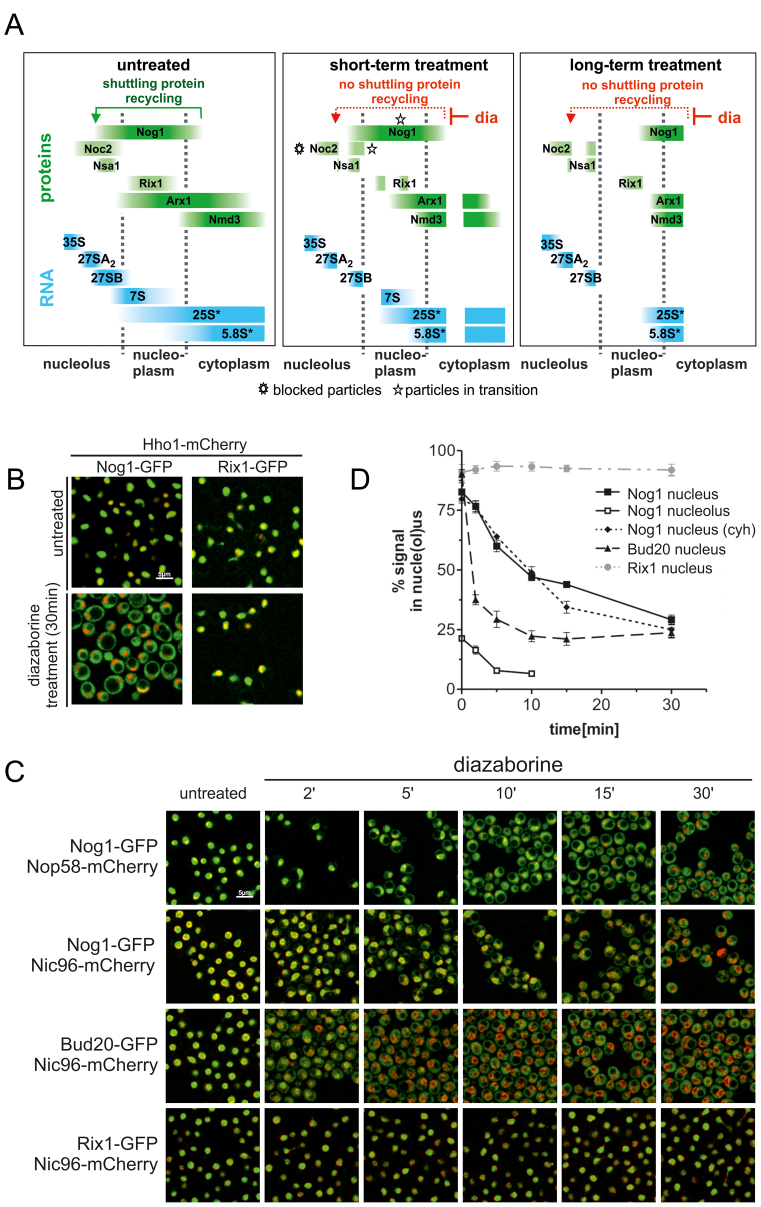
The entrapment of the shuttling proteins on cytoplasmic pre-60S particles after diazaborine treatment results in their depletion from the nucleus, causing a rapid blockage of early pre-60S assembly steps. This blockage prevents freshly formed particles from entering the maturation pathway, while pre-60S particles beyond the roadblock at the time of application of the inhibitor pass through the maturation cascade until they get trapped shortly after export in the cytoplasm due to the action of diazaborine. These facts should enable us to follow the maturation path in a timely manner (see Figure 1A for a scheme). Indeed, a GFP fusion with Nog1, which associates at an early nucleolar step with the 27SA2 pre-rRNA (24) and accompanies the particle into the cytoplasm, is progressively transferred from the nucleolus to the cytoplasm after increasing treatment periods with diazaborine (Figure 1). The distribution of Nog1-GFP within the cell after different treatment periods was estimated based on co-localization with Nop58-, Hho1- or Nic96-mCherry, which served as tracking proteins for nucleolus, nucleoplasm and nuclear membrane, respectively (Figure 1B and C). Semi-quantitative assessment of co-localization using the program Fiji showed that under steady state conditions 82.0(±5.9) % of Nog1-GFP was located inside the Nic96-mCherry rim and hence localized in the nucleus, while 21.3(±1.8) % of total Nog1-GFP co-localized with Nop58-mCherry indicating nucleolar localization (Figure 1C and D).
Figure 1.
Diazaborine treatment results in rapid relocalization of shuttling proteins from the nucle(ol)us into the cytoplasm. (A) Scheme demonstrating the effect of diazaborine on the pre-60S maturation pathway after short term and long term treatment. After short term treatment, cytoplasmic pre-60S particles, which are beyond the diazaborine sensitive maturation step at drug application, are able to complete maturation, while particles in maturation stages prior to the diazaborine sensitive step become entrapped, leading to two different cytoplasmic populations of pre-60S particles. This is symbolized by a gap in the cytoplasmic bars. Proteins associating with the pre-60S particle at different maturation stages which were used as bait proteins in this study (Noc2, Nsa1, Rix1) are symbolized by green bars. The individual pre-rRNAs are indicated in blue. Asterisks highlight blocked particles and particles in transition after short term treatment for the Noc2 and Nog1 bait proteins. 25S* and 5.8S* denotes the mature 25S and 5.8S rRNA including their immediate precursors, the 25.5 and 6S pre-rRNA, respectively. (B) Yeast strains expressing GFP fusion with the shuttling protein Nog1 or the nuclear resident protein Rix1 were treated with diazaborine for 30 min and inspected by fluorescence microscopy. For compartment tracking the strains also expressed a mCherry fusion with the nucleoplasmic protein histone H1 (Hho1). (C) Nog1-GFP was expressed in strains harboring mCherry fusions with the nucleolar protein Nop58 or the nuclear membrane protein Nic96 and inspected by laser scanning microscopy after different treatment periods with diazaborine. Only a section of a single scanning plane is shown. In addition, GFP-fusions with the late joining shuttling protein Bud20-GFP were expressed in the Nic96-mCherry background and treated with the inhibitor. A GFP-fusion with the nuclear resident protein Rix1 served as a control. (D) The distribution of the GFP fusions within the cell after diazaborine treatment was estimated based on co-localization of Nop58-mCherry (nucleolus) or on localization within the Nic96-mCherry ring structure (nucleus) throughout the whole stack of scans using the program Fiji (18). Nog1 nucleus (cyh): nuclear signal of Nog1-GFP after additional treatment with cycloheximide. Data points representing the average of at least three biological replicates and standard deviations are shown.
Diazaborine treatment resulted in segregation of green fluorescence and Nop58-mCherry signals indicating exit of Nog1-GFP from the nucleolus (Figure 1C). Five minutes after addition of the drug, only one-third of the initial nucleolar signal still co-localized with Nop58-mCherry (Figure 1D). Quantification of the nuclear and cytoplasmic signals using the strain co-expressing Nog1-GFP and Nic96-mCherry showed that after 15 min of treatment less than half of Nog1 was detected inside the nucleus. The decrease of Nog1-GFP in the nucleus followed a linear function during the first 10 min of the experiment but showed a significantly slower rate of decrease after longer treatment periods (Figure 1D). Since diazaborine only blocks release and recycling of shuttling proteins from cytoplasmic pre-60S particles, but not import of freshly synthesized Nog1-GFP, this slow-down in nuclear depletion could be caused by de novo synthesized protein. Indeed, simultaneous addition of cycloheximide and diazaborine resulted in lower Nog1-GFP fluorescence in the nucleus, compared to 15–30 min of diazaborine treatment but not after shorter treatment periods (Figure 1D). Thus, the bulk of Nog1 containing pre-60S particles transits from the nucleolus to the cytoplasm within 15–30 min. In contrast, Rix1 which is only a component of nuclear particles did not translocate into the cytoplasm.
To investigate the kinetics of cytoplasmic accumulation for a late-joining shuttling protein, Bud20-GFP was used. Bud20 binds to pre-60S particles late in the nucleoplasm and supports their export (25,26). The nuclear signal of Bud20-GFP was reduced from 90.4(±3)% to 37.5 (±3.6)% after 2 min of diazaborine treatment (Figure 1C and D). Thus, Bud20-GFP accumulated much faster in the cytoplasm upon diazaborine treatment than Nog1-GFP. This difference was expected, since Nog1 binds at an earlier stage than Bud20 in the nucleolus and hence requires more time to transit through the early maturation steps.
Monitoring dynamic changes of pre-60S particles during the maturation path
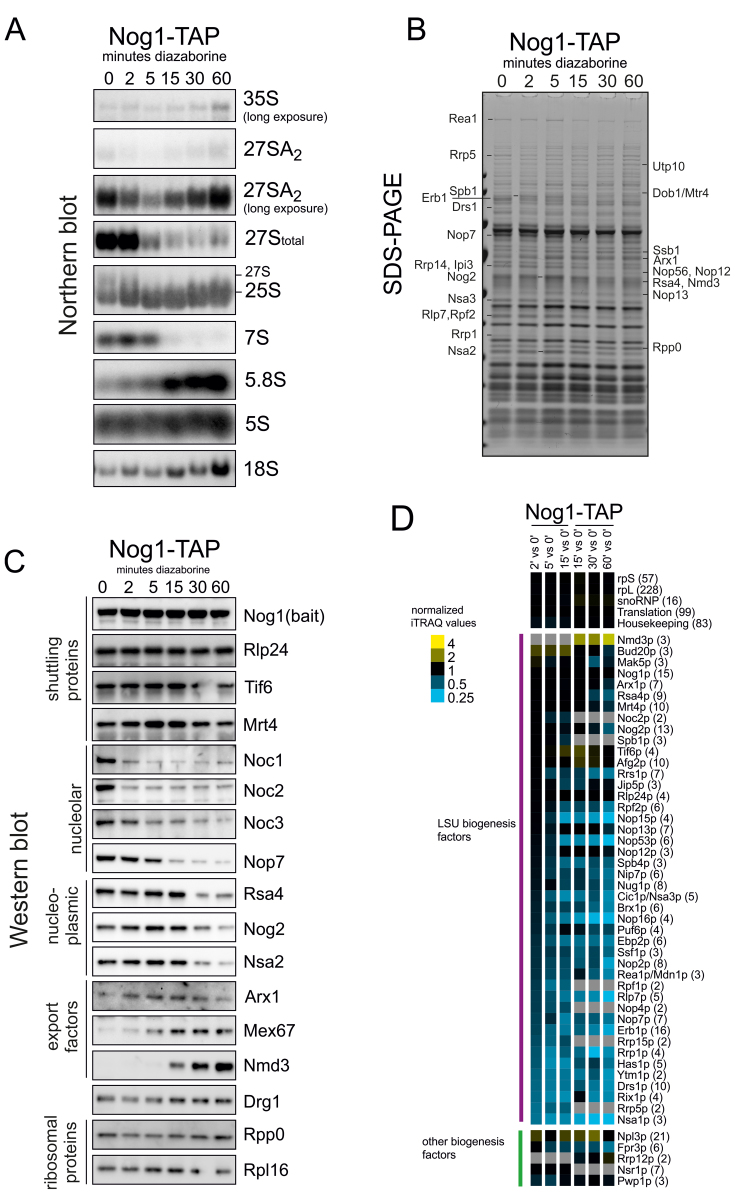
To characterize the compositional changes pre-ribosomes undergo during their passage from the nucleolus into the cytoplasm, we purified pre-60S particles using Nog1-TAP as bait protein after different times of diazaborine treatment. Co-purifying proteins and pre-rRNAs were analyzed by SDS-PAGE, western blotting, iTRAQ and northern blotting.
Analysis of the pre-rRNA composition of purified Nog1-TAP particles showed that the 5S pre-rRNA was present in all preparations, suggesting that it is incorporated into the pre-60S particle prior or concomitantly with Nog1 binding (Figure 2A). A strong signal for total 27S pre-rRNA and 7S pre-rRNA was observed in the particles from the untreated strain. The 27S pre-rRNA in these particles mainly corresponds to 27SB, since only low levels of 27SA2 pre-rRNA were present in the particles. In addition, very minor amounts of 35S pre-rRNA were detected. Signals for the mature 25S and 5.8S rRNAs, presumably also containing to some extent their immediate precursors 25.5 and 6S were also present. This pre-rRNA composition indicates that Nog1 initially associates with 27SA2 pre-rRNA and remains bound until mature 25S rRNA is formed. These results are consistent with published data of Nog1-TAP purifications (24,27).
Figure 2.
Compositional changes of Nog1-TAP containing pre-60S particles during their transition through the maturation cascade. Pre-60S particles were purified from the Nog1-TAP strain after different time periods of treatment with diazaborine. After purification, aliquots were used for protein and RNA extraction as well as iTRAQ analyses. (A) Northern blot analyses with probes hybridizing to the indicated pre-rRNAs. The blots with 27SA2 pre-rRNA are shown in normal and long exposure time to highlight changes occurring over time. 27SA2 was detected with the A2A3 probe. Total 27S pre-rRNA and 7S pre-rRNA are from the same exposure and were detected with the E-C2 probe. The other probes were specific for the mature 5.8S, 25S or 18S rRNAs (see Supplementary Table S2 for details). (B) Proteins present in the pre-60S particles were analyzed by SDS-PAGE. Proteins showing altered levels in the different preparations were identified by ESI-MS. (C) Western blotting: the Nog1-TAP preparations were analyzed by western blotting with antibodies directed to selected maturation and export factors from various steps of the pathway. (D) Proteins present in the samples were subjected to semi-quantitative iTRAQ analyses. The iTRAQ ratios for each maturation factor were normalized to the iTRAQ ratio of the bait protein and are displayed as a heat map. Mean iTRAQ ratios for small (rpS) and large (rpL) ribosomal proteins as well as translation factors (trans), snoRNPs and housekeeping proteins (housekp) are indicated. Depletion or enrichment of factors is expressed as a color gradient of blue or yellow, respectively (see color code). Numbers in brackets behind protein names indicate the average number of peptides by which the respective protein was identified. Data represent one representative biological replicate. Samples from individual treatment periods were labeled in two series with the 15 min samples present in both sets.
Diazaborine treatment for 2 min resulted in a strong reduction of 27SA2 pre-rRNA levels in Nog1-TAP particles, while the levels of total 27S pre-rRNA remained unchanged, suggesting that most of the 27SA2 pre-rRNA was processed to 27SB during this time period. After 5 min of diazaborine treatment, the 27S pre-rRNA decreased, while the level of 25S rRNA increased, demonstrating that the 27S precursor was already converted to the mature rRNA in these particles. In contrast, the 7S pre-rRNA was still present after 5 min of treatment, but had disappeared after 15 min of drug treatment. Concomitantly, the amounts of the downstream product of the 7S pre-rRNA, the mature 5.8S rRNA increased. We conclude that the major populations of the precursor RNAs present in the Nog1-containing pre-60S particles are converted from their immature forms into the mature forms within <15 min.
Diazaborine treatment periods longer than 15 min resulted in a gradual increase of 27SA2 pre-rRNA in the Nog1-TAP purification. However, the amount of 27SA2 pre-rRNA reflects only a minor portion of total 27S pre-rRNA in the particles as obvious from the total decrease of 27S signal despite the 27SA2 accumulation (Figure 2A). Since addition of cycloheximide prevented the accumulation of 27SA2 pre-rRNA after longer treatment periods with diazaborine (Supplementary Figure S2), this increase likely arises from binding of de novo synthesized Nog1-TAP to this precursor RNA.
SDS-PAGE analyses (Figure 2B) and mass spectrometric identification of selected protein bands revealed a number of proteins (e.g. Erb1, Drs1, Rrp1 Rrp5) showing reduced levels already after 2 min of treatment. In contrast, Spb1 as well as Nog2, Rea1, Nsa2, Nop13 and Arx1 showed increased levels after the same treatment period and reached highest levels after 5 min. After 15 min strongly decreased levels of Nop7 and Cic1 and a partial decrease of Nog2 and Nsa2 were observed. Longer treatment periods resulted in enrichment of Arx1 and Rpp0 in the purification.
Western blot analyses (Figure 2C) demonstrated that Nog1-TAP particles isolated from untreated cells contain very early pre-60S maturation factors, including Noc1 and Noc2. In addition, other nucleolar proteins like Nop7 or Noc3 were also present. Finally, significant amounts of the predominantly nucleoplasmic pre-60S maturation factors Nog2, Rsa4 and Nsa2 were also detected. This composition reflects the steady state localization of Nog1 in the nucleolus and in the nucleoplasm (Figure 1) and is consistent with the co-purifying pre-rRNAs (Figure 2A) and published data (24).
Treatment of the cells for 2 min with the inhibitor resulted in a pronounced decrease of very early nucleolar factors Noc1 and Noc2, while Noc3 and Nop7 could still be detected. After 5 min of treatment Noc3 levels were reduced, but Nop7 was absent only after 15 min in the presence of the inhibitor. Comparison of the Northern blot and western blot data suggests that the decrease of Noc1 and Noc2 correlates with the reduction of 27SA2 pre-rRNA in the Nog1 particle, while Noc3 decrease correlates with the decline of total 27S pre-rRNA. Since Noc1 and Noc2 are present on very early pre-60S particles and Noc3 on intermediate nucleolar pre-ribosomes these results are consistent with literature (23). In contrast, Nop7 decrease correlates with 7S pre-rRNA decrease, which is consistent with its presence on late nucleoplasmic particles and its binding to ITS2 (28,29). The nucleoplasmic factors Rsa4, Nog2 and Nsa2 reached highest levels after 5 min of treatment but decreased after 15 min of treatment, a time period when the export factors Arx1 and Mex67 reached higher levels. Interestingly, the increase of Nog2 and Nsa2 correlates with the decrease of the C2870 methyltransferase Nop2 (Supplementary Figure S3).
The export factor Nmd3 was detectable in Nog1-TAP purifications only after 15 min of drug treatment and showed a strong increase at longer treatment periods. Considering that more than half of Nog1-GFP was localized to the cytoplasm after 15 min of treatment (Figure 1), this result suggests that Nmd3 binds to the pre-60S particle immediately before export. The levels of the shuttling proteins Rlp24, Tif6 and Mrt4 behaved similar to the bait protein Nog1, which was expected since they join the particles at a similar time as the bait protein and leave it concomitantly (Rlp24) or later (Tif6 and Mrt4) (13,15,16,24,27).
To obtain a more global understanding of the changes taking place on the particles during their transition from the nucleolus through the nucleoplasm into the cytoplasm we performed iTRAQ analyses. As shown in Figure 2D, a pronounced reduction of the levels of Nsa1 occurred within 2 min of treatment. We conclude that Nsa1 is released from the pre-60S particle shortly after Nog1 binding. This view is in accordance with the strong depletion of Nog1 on the Nsa1-TAP particle after short treatment periods (Figure 4A and B). Incubation with the drug for 5 min resulted in additional reduction of Rrp1, Erb1, Ytm1, Rrp5, Rrp15, Rpf1, Ebp2, Nop16 and Nop2. The earlier leave of Ytm1 and Erb1 compared to Nop7 was confirmed by western blotting (Supplementary Figure S3). This indicates that although the proteins Ytm1, Nop7 and Erb1 form a trimeric complex (30,31), they leave the pre-60S particle at different maturation stages. This view is consistent with the detection of Nop7 but not Ytm1 and Erb1 on nucleoplasmic pre-60S particles (28,29). Moreover, the coordinate leave of Ebp2, Rpf1 and Nop16 might be an indication for their involvement in the same process, which is in line with their previously demonstrated genetic linkage (32).
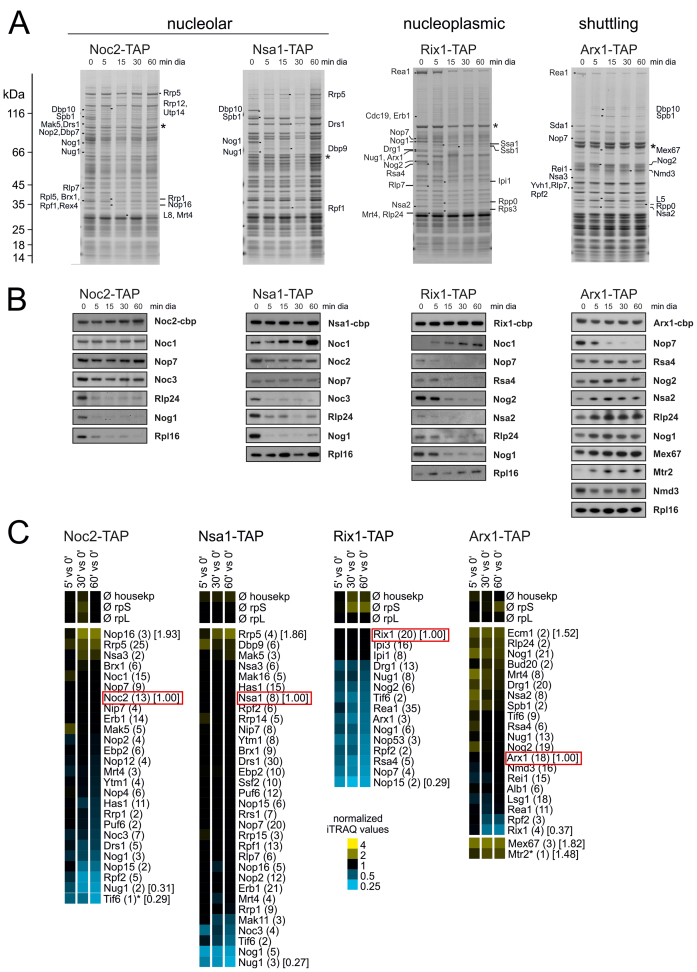
Figure 4.
Impact of blocked shuttling protein recycling on composition of nucleolar pre-60S particles. Particles were purified using IgG coated magnetic beads with TAP-tagged nucleolar (Noc1, Nsa1), nucleoplasmic (Rix1) and shuttling proteins (Arx1) as bait proteins and analyzed by SDS-PAGE, western blotting and iTRAQ. (A) SDS-PAGE of TAP purifications after treatment of the yeast strains for 0–60 min. Protein bands differing between treated and untreated samples in the SDS-PAGE were identified by ESI-MS. Asterisks denote bait proteins. (B) Western blot analyses with selected maturation and export factors are shown. (C) Proteomic analysis of purified pre-60S particles by semi-quantitative mass spectrometry using iTRAQ reagents. Mean iTRAQ ratios for small (rpS) and large (rpL) ribosomal proteins as well as housekeeping proteins (housekp) are indicated. The iTRAQ ratios for each maturation factor after 5, 30 and 60 min of drug treatment were normalized to the iTRAQ ratio of the respective bait proteins and displayed as a heat map (see color code). Numbers in brackets behind protein names indicate the average number of peptides by which the respective protein was identified; numbers in squared brackets display the iTRAQ ratios for selected proteins. Samples were derived from the same purification and biological replicate as in Figure 3 and panels (A and B).
After 15 min of diazaborine treatment, Nop7, Nsa3, Nop15 and Nop53 were largely gone from Nog1-TAP particles. These factors bind to the ITS2-containing foot structure of the pre-60S particle ((28), Figure 5 and Supplementary Table S3). It is therefore feasible that their decrease is coordinated and correlates with the processing of 7S pre-rRNA (Figure 2A). In contrast, proteins Rpf2 and Rrs1, which are associated with 5S pre-rRNA, become depleted significantly after and to a lesser extent than the ITS2 binding factors. Finally, Rsa4 and Nog2 only decreased upon longer treatment periods, which is in agreement with the western blot data (Figure 2C and D). Concomitantly, Tif6 and Drg1 together with export factor Nmd3 showed a gradual increase for up to 15–30 min. These changes likely reflect the predominantly cytoplasmic localization of the Nog1 bait protein after this treatment period (Figure 1). Treatment periods from 30 min to 1 h also resulted in increased levels of the early factor Rrp12 co-purifying with the Nog1 bait protein (Figure 2D and Supplementary Figure S2). This increase correlates with the observed increase in 27SA2 and 35S pre-rRNA using Northern blotting (Figure 2A) and can be suppressed by addition of cycloheximide (Supplementary Figure S2). It therefore likely reflects the binding of de novo synthesized Nog1-TAP to early precursor rRNA.
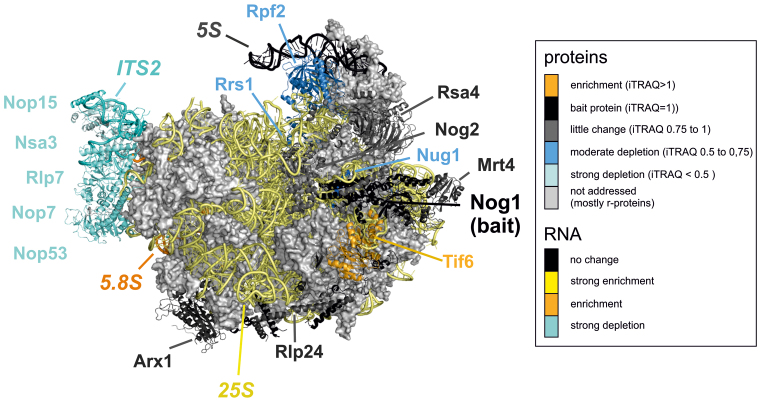
Figure 5.
The extent of depletion of individual pre-60S maturation factors after diazaborine treatment is linked to their position on the pre-60S particle. Projection of the results of the iTRAQ analyses on the structure of a late nuclear pre-60S particle. The structure of the recently described Nog2 containing particle (28) was used to color the maturation factors according to the extent of their depletion after 15 min of diazaborine treatment. A similar color scheme as for the iTRAQ analysis was used with factors depleted in different shades of blue and factors enriched in yellow. Factors behaving similar as the bait protein Nog1 are colored in dark gray. Data are based on iTRAQ values representing the mean of two independent biological replicates. Only factors with standard deviations of <30% were considered (Supplementary Table S3). Factors not addressed (ribosomal proteins) or not detected in at least three iTRAQ analyses are shown in light gray.
Taken together, these results demonstrate that during treatment with diazaborine, maturation factors show a distinct pattern of association and dissociation from the Nog1-containing pre-60S particle, which indicates functional linkages and reflects their chronological sequence during the maturation cascade.
Dissecting early pre-60S maturation steps
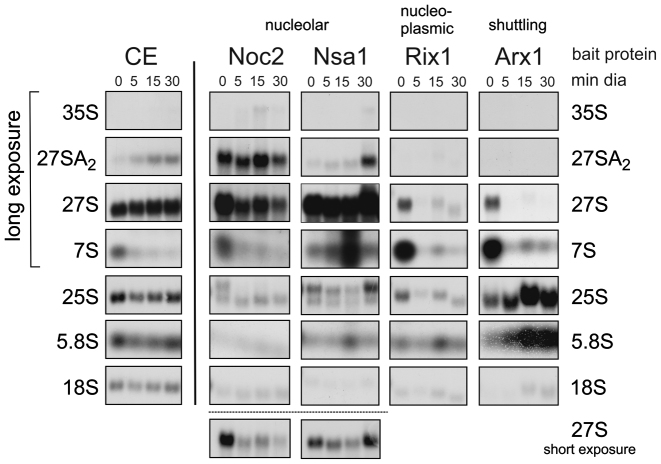
The depletion of shuttling proteins and the resulting rapid block of early pre-60S maturation also allows dynamic investigations of early assembly steps. For this purpose we isolated pre-60S particles from early nucleolar and late nucleolar stages of maturation after different treatment periods using Noc2 and Nsa1 as bait proteins and analyzed their protein and pre-rRNA composition (Figures 3 and 4). To facilitate interpretation, we analyzed the pre-60S particles after short and long-term treatment with diazaborine. Short-term treatment is expected to lead to isolation of blocked particles together with particles that are still transiting through the maturation path. In contrast, after longer treatment, the particles in transition are expected to vanish due to ongoing downstream maturation allowing exclusive isolation of blocked particles (see Figure 1A for a scheme). Consistent with this assumption, 27SA2 pre-rRNA is retained in the purifications using Noc2 as bait protein, while the total 27S pre-rRNA strongly decreases with longer treatment periods (Figure 3).
Figure 3.
Impact of blocking shuttling protein recycling on pre-rRNA composition of nucleolar and nucleoplasmic pre-60S particles. RNA was extracted from an aliquot of the purifications shown in this figure, separated by agarose gel electrophoresis and analyzed by northern blotting using radiolabeled oligonucleotides directed to the indicated precursor rRNAs. For comparison, northern blot analysis of total RNA from the crude extract (CE) is also shown.
As shown in Figure 4, shuttling proteins Nog1 and Rlp24 are completely depleted from Noc2 and Nsa1 particles after 5 min, confirming the fast onset of blockage upon diazaborine treatment. Nop16, Rrp5, Cic1 and Brx1 showed increased co-purification with Noc2-TAP after long term diazaborine treatment, while Mak5, Nop4 and Rrp1 showed a transient behavior, reaching highest levels after 5 min but decreasing after longer treatment (Figure 4C). This result shows that Nop16, Rrp5 and Cic1 associate already with 27SA2 pre-rRNA containing Noc2-TAP particles and accumulate with the blocked particle, while Mak5, Nop4 and Rrp1 are present on particles in transition and are reduced after longer treatment. Nop7, Erb1, Ytm1 and Nop12 showed only minor changes. In contrast, Spb1, Dbp10 (Figure 4A), Drs1, Has1 and Nop15 as well as Nug1 and Rpf2 (Figure 4C) decreased strongly from the Noc2 particle after diazaborine treatment and hence likely bind after the drug induced blockage or require one of the depleted shuttling proteins for their loading. For Spb1 and Dbp10 this view is consistent with current literature because it was shown previously that Nug1 is required for loading of these proteins (33).
Late nucleolar pre-60S particles purified via Nsa1 as bait protein co-purified 27S pre-rRNA (most likely 27SB) with only a minor content of 27SA2. Treatment with the inhibitor for 5 min had almost no effect on total 27S pre-rRNA levels and resulted only in a minor accumulation of 27SA2. Similar as observed for the Noc2 particle, Nog1, Noc3, Dbp10, Spb1 and Nug1 were absent from late nucleolar Nsa1-TAP particles already after 5 min of diazaborine treatment (Figure 4A). However, the levels of Drs1, Rpf2, Rrs1 and Nop2, which decreased on the Noc2 particle after diazaborine treatment, were unaltered in the Nsa1-TAP purifications. Therefore these proteins might associate with the pre-60S particle prior to the Nsa1-bait protein. In contrast, Dpb9, Rrp14, Rrp15 and Ebp2 showed a transient accumulation after 5 min of treatment (Figure 4C). Since these proteins leave the particle after Nsa1 (Figure 2D), their accumulation cannot be explained by effects on their release. We therefore speculate that they associate with the pre-ribosome after the Nsa1 bait protein. Incubation with the inhibitor for 30 min resulted in increased levels of 27SA2 pre-rRNA co-purifying with the Nsa1-bait protein, which was accompanied with increased levels of the very early factors Rrp5, Mak5 and Cic1. Taken together, these data provide interesting insights into the assembly line of early pre-60S particles.
Response of nucleoplasmic maturation steps to the nucleolar roadblock
To determine the response of nucleoplasmic components of the pathway, we purified pre-60S particles via Rix1 and Arx1 as bait proteins. Rix1 is a central component of late nucleoplasmic pre-60S particles and is required for their restructuring prior to nuclear export (34). Arx1 is a shuttling protein that associates with pre-60S particles containing 27SB pre-rRNA and remains bound until the cytoplasm, where it is released and recycled (35–37).
In contrast to the early pre-60S particles purified with nucleolar bait proteins, the shuttling proteins Nog1 and Rlp24 disappeared from nucleoplasmic particles purified via Rix1-TAP as bait protein only after longer drug treatment (Figure 4B). This finding shows that shuttling proteins require longer time periods to be depleted from late particles, which can be explained by the progression of the particles through the maturation cascade. The ITS2 binding proteins Nop15, Nop7, Rlp7 and Nop53 showed strong depletion from the Rix1-containing particle already after 5 min of drug treatment suggesting that they are released shortly after Rix1 binding (Figure 4A–C). Fifteen minutes of treatment also resulted in decline of the nucleoplasmic proteins Nsa2, Nog2 and Rsa4 from Rix1-TAP particles (Figure 4B). In addition, the Rix1-TAP purification from the treated strain exhibited decreased levels of the AAA-ATPase Rea1 (Figure 4A). iTRAQ analyses showed that with the exception of Ipi1 and Ipi3, which are known to form a low molecular weight complex with Rix1 (30,38), all co-purifying proteins undergo a marked depletion from the particle after treatment (Figure 4C). This finding suggests that a significant portion of the bait protein is not associated with pre-ribosomes. In line with this suggestion, all pre-rRNA species are reduced in the Rix1-TAP purification upon diazaborine treatment (Figure 3).
Surprisingly, also Drg1 was detected in the Rix1-TAP preparation from untreated cells. However, this co-purification occurred only when the bait protein was pulled down using IgG coupled magnetic beads and not in parallel purifications with IgG coupled agarose beads (data not shown). Moreover, Drg1 was sometimes also found in earlier pre-60S particles. Possibly, Drg1 associates with these particles after cell lysis due to its affinity for Rlp24. The significance of its presence on nucle(ol)ar particles is therefore not yet clear.
When pre-60S particles were purified via the shuttling factor Arx1 as bait protein, steadily increasing levels of other shuttling proteins and of the export factors Mex67 and Mtr2 were observed with increasing treatment periods with diazaborine. The nucleoplasmic factors Rsa4, Nog2 and Nsa2 showed transient accumulation after 15 min of treatment and decreased again after longer treatment periods, correlating well with the Nog1-TAP data (Figure 2C and D). As in case of Nog1-TAP, the proteins Spb1 and Dbp10, which were depleted from early nucleolar pre-60S particles also showed a transient behavior upon diazaborine treatment, reaching maximal levels on Arx1-TAP particles after 15 min. We conclude that these proteins stay attached to the particles for a longer period of time and are released shortly before export. In contrast, the nucleolar factors Cic1 and Nop7 showed a rapid decrease from Arx1-particles and were completely lost after 15 min of treatment (Figure 4A and B). The significant presence of the late cytoplasmic factor Rei1 in the TAP purification from the untreated strain together with relatively high levels of Mex67 and Nmd3 compared to the Nog1-TAP purifications (compare Figures 2C and 4B) demonstrates that Arx1 exhibits a pronounced cytoplasmic phase. Consistently, the levels of Rei1 and Lsg1, which join the particle after Rlp24 release in the cytoplasm, decreased rapidly after diazaborine treatment (Figure 4C).
Northern blot analyses of the Arx1 containing pre-ribosomes demonstrated that the co-purifying 7S and 27S pre-rRNAs were converted into mature 5.8S and 25S rRNAs or their immediate precursors with increasing treatment periods (Figure 3). This finding confirms the results of the Nog1-TAP purification experiments and shows that particles, which were assembled with shuttling proteins in the nucleus prior to application of diazaborine, are fully capable to transit through the maturation cascade until they become trapped shortly after export.
DISCUSSION
During the last decade the ribosome biogenesis research field has witnessed a boost of exciting developments that were facilitated by technological improvements in mass spectrometry, affinity purifications and structural methods including cryo-EM. These developments resulted in a detailed characterization of a number of pre-ribosome intermediates from different maturation steps. However, all these particles represent static snapshots that lack information about temporal and functional linkages of the individual maturation steps. However, such information is required to fully understand the pathway and coordination of the restructuring and processing reactions.
In this work, we challenged the ribosome biogenesis pathway with a low molecular weight inhibitor to monitor the dynamic changes pre-60S particles undergo during maturation on their way from the nucleolus to the cytoplasm. The main advantage of our strategy is the fast onset of inhibition, which blocks the maturation pathway immediately. Given that the entire process of ribosome biogenesis is finished within about 15 min, such short response times are essential to correctly reflect the dynamics of the process and to dissect individual maturation steps. The utilization of a low molecular weight inhibitor allowed to accurately resolve this timeframe and enabled us to follow the undisturbed maturation of the particles from the very early, 27SA2 containing pre-ribosomes to almost mature particles shortly after export without using loss of function alleles, or changes in growth condition or temperature. The facts that pre-rRNAs are successfully converted into their mature forms within the time course of our experiment and that a major population of the particles is capable of binding export factors proof that they obtain their correct conformation during their transit through the maturation cascade. This confirms that the outcome of our experiments correctly reflects the maturation process. Moreover, the conversion of 27SA2 pre-rRNA to mature 25S pre-rRNA in our experiment took about 5 min. This period is in reasonable agreement with previous estimates derived from a dedicated pulse assay with radioactive metabolites (6). Depending on whether A2 cleavage occurs co- or post-transcriptionally, a 260–340 s period from the 35S pre-rRNA to 25S rRNA was measured (6). Consistently, the total 27S pre-rRNA largely disappeared from the investigated particles after 5 min of drug treatment in our experiments. The 7S pre-rRNA was still detected after 5 min of treatment, but was not detected after 15 min of treatment.
Comparison of the northern blot, iTRAQ and western blot data suggest a clear correlation between progression of the particle through the maturation cascade and the coordinated release of maturation factors (summarized in Supplementary Figure S4). The kinetics of decrease from pre-60S particles are frequently similar for factors positioned in close vicinity to each other on the particle and therefore not only reflect temporal but also functional linkages (Figure 5). This is particularly obvious for the ITS2 binding factors Cic1, Nop7, Nop15, Rlp7 and Nop53 that disappear once 7S pre-rRNA processing is completed. This tight temporal correlation of ITS2 binding factor release reflects their functional coordination in ITS2 processing and can experimentally clearly be separated from other events like the decrease of Rrs1/Rpf2 or Rsa4, Nog2 and Nsa2 (Figure 5).
Moreover, our data revealed an inverse correlation between the levels of Nop2 and Nog2 in the Nog1-TAP purification. Indeed, the residue modified by Nop2 is positioned in a cavity formed by Nog1, Nsa2 and Nog2 with the modified C5 of nucleotide C2870 positioned in a pocket of Nog2 (Supplementary Figure S3). Therefore Nop2 likely has to perform its activity and must leave the pre-60S particle prior to Nog2 binding. Together with the previously described requirement of Nop2 for Nog2 loading (27), our results suggest that modification of C2870 might facilitate Nog2 association with the pre-60S particle. We conclude that our methodology was successful to also resolve functional linkages between different maturation steps within the pathway in a temporal manner.
We also performed a detailed characterization of the effect of shuttling protein depletion on a set of overlapping pre-60S particles from early to late nucleolar maturation steps. We observed a rapid blockage of 27SA2 processing in the Noc2-TAP particle (Figure 3) and the persistence of total 27S (likely 27SB pre-rRNA) in the late nucleolar Nsa1-TAP particle. Together with the 35S pre-rRNA increase and 7S pre-rRNA decrease observed in the crude extract upon short term diazaborine treatment, these effects point to a dual effect of shuttling protein depletion on pre-rRNA processing affecting both 27SA2 and 27SB processing. These effects could arise from the simultaneous depletion of Nog1, Tif6 and Nug1 (Figure 4C), which join the maturation path at different stages (24,39) and are trapped in the cytoplasm in the dominant negative drg1 mutant which phenocopies the diazaborine effects (13,25). Depletion of these essential factors in the nucleus therefore could result in a blockage at different stages of the pathway.
Due to the overlap of particles purified with Noc2 and Nsa1 as bait proteins it is not surprising that certain late joining proteins (Dbp10, Spb1 and Noc3) are depleted in both particles. In contrast, Nop15 and Rpf2 show only a decrease in the earlier Noc2-TAP particle (Figure 4C) which suggests that those proteins bind after shuttling protein joining but prior to Nsa1.
Taken together, our results provide a first glimpse of the dynamics of the pre-60S maturation pathway by directly relating pre-rRNA conversions with assembly and disassembly of trans-acting factors in real time. Our investigations thus provide a major step toward a dynamic understanding of the process and open up the possibility to follow the maturation process with very high temporal resolution in the 1–2 min range. This should enable us to further break down the individual maturation steps into a detailed sequence of biochemical reactions and set them into a temporal context with pre-rRNA processing and assembly or disassembly reactions of trans-acting factors. Such information will provide new insights into functional linkages of the individual processes and might finally enable a detailed in silico modeling of the pathway.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are indebted to Micheline Fromont-Racine, Melanie Oakes, Ed Hurt, Dieter Kressler, Arlen Johnson, Mercedes Dosil, Sabine Rospert, John Woolford and Jesus de la Cruz for their generous gift of antibodies, strains and plasmids.
Author contributions: P.M. and H.B. planned the experiments; G.Z., U.O., C.M., V.M., I.K., H.W., G.N.R., M.P. and P.M. performed the experiments. All authors discussed the results. U.O., P.M., C.M. and H.B. performed data analysis. M.P. and H.B. wrote the manuscript. V.M., H.W., B.P. and P.M. commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Austrian Science Fund FWF [P26136, P29451 to H.B.]; Deutsche Forschungsgemeinschaft (DFG) [Grant SFB960 to P.M.]. Funding for open access charge: FWF P29451.
Conflict of interest statement. None declared.
REFERENCES
- 1. Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999; 24:437–440. [DOI] [PubMed] [Google Scholar]
- 2. Fernández-Pevida A., Kressler D., de la Cruz J.. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip. Rev. RNA. 2015; 6:191–209. [DOI] [PubMed] [Google Scholar]
- 3. Woolford J.L., Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomson E., Tollervey D.. The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol. Cell. Biol. 2010; 30:976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaker-Margot M., Hunziker M., Barandun J., Dill B.D., Klinge S.. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat. Struct. Mol. Biol. 2015; 22:920–923. [DOI] [PubMed] [Google Scholar]
- 6. Kos M., Tollervey D.. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010; 37:809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turowski T.W., Tollervey D.. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip. Rev. RNA. 2015; 6:129–139. [DOI] [PubMed] [Google Scholar]
- 8. Zhang L., Wu C., Cai G., Chen S., Ye K.. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev. 2016; 30:718–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stelter P., Kunze R., Radwan M., Thomson E., Thierbach K., Thoms M., Hurt E.. Monitoring spatiotemporal biogenesis of macromolecular assemblies by pulse-chase epitope labeling. Mol. Cell. 2012; 47:788–796. [DOI] [PubMed] [Google Scholar]
- 10. Swiatkowska A., Wlotzka W., Tuck A., Barrass J.D., Beggs J.D., Tollervey D.. Kinetic analysis of pre-ribosome structure in vivo. RNA. 2012; 18:2187–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stokes J.M., Brown E.D.. Chemical modulators of ribosome biogenesis as biological probes. Nat. Chem. Biol. 2015; 11:924–932. [DOI] [PubMed] [Google Scholar]
- 12. Pertschy B., Zisser G., Schein H., Köffel R., Rauch G., Grillitsch K., Morgenstern C., Durchschlag M., Högenauer G., Bergler H.. Diazaborine treatment of yeast cells inhibits maturation of the 60S ribosomal subunit. Mol. Cell. Biol. 2004; 24:6476–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loibl M., Klein I., Prattes M., Schmidt C., Kappel L., Zisser G., Gungl A., Krieger E., Pertschy B., Bergler H.. The drug diazaborine blocks ribosome biogenesis by inhibiting the AAA-ATPase Drg1. J. Biol. Chem. 2014; 289:3913–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., Liebminger E., Nobis B., Kappel L., van der Klei I. et al. . Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 2007; 27:6581–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kappel L., Loibl M., Zisser G., Klein I., Fruhmann G., Gruber C., Unterweger S., Rechberger G., Pertschy B., Bergler H.. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J. Cell Biol. 2012; 199:771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lo K.-Y., Li Z., Bussiere C., Bresson S., Marcotte E.M., Johnson A.W.. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 2010; 39:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 18. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. . Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oeffinger M., Wei K.E., Rogers R., DeGrasse J.A., Chait B.T., Aitchison J.D., Rout M.P.. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods. 2007; 4:951–956. [DOI] [PubMed] [Google Scholar]
- 20. Ohmayer U., Gamalinda M., Sauert M., Ossowski J., Pöll G., Linnemann J., Hierlmeier T., Perez-Fernandez J., Kumcuoglu B., Leger-Silvestre I. et al. . Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS One. 2013; 8:e68412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ross P.L., Huang Y.N., Marchese J.N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S. et al. . Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004; 3:1154–1169. [DOI] [PubMed] [Google Scholar]
- 22. Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 23. Milkereit P., Strauss D., Bassler J., Gadal O., Kühn H., Schütz S., Gas N., Lechner J., Hurt E., Tschochner H.. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 2003; 278:4072–4081. [DOI] [PubMed] [Google Scholar]
- 24. Saveanu C., Namane A., Gleizes P.-E., Lebreton A., Rousselle J.-C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M.. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003; 23:4449–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altvater M., Chang Y., Melnik A., Occhipinti L., Schütz S., Rothenbusch U., Picotti P., Panse V.G.. Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export. Mol. Syst. Biol. 2012; 8:628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bassler J., Klein I., Schmidt C., Kallas M., Thomson E., Wagner M.A., Bradatsch B., Rechberger G., Strohmaier H., Hurt E. et al. . The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol. Cell. Biol. 2012; 32:4898–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Talkish J., Zhang J., Jakovljevic J., Horsey E.W., Woolford J.L.. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 2012; 40:8646–8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu S., Tutuncuoglu B., Yan K., Brown H., Zhang Y., Tan D., Gamalinda M., Yuan Y., Li Z., Jakovljevic J. et al. . Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016; 534:133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu S., Tan D., Woolford J.L., Dong M.-Q., Gao N.. Atomic modeling of the ITS2 ribosome assembly subcomplex from cryo-EM together with mass spectrometry-identified protein-protein crosslinks. Protein Sci. Publ. Protein Soc. 2017; 26:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krogan N.J., Peng W.-T., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P. et al. . High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004; 13:225–239. [DOI] [PubMed] [Google Scholar]
- 31. Miles T.D., Jakovljevic J., Horsey E.W., Harnpicharnchai P., Tang L., Woolford J.L.. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 2005; 25:10419–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pratte D., Singh U., Murat G., Kressler D.. Mak5 and Ebp2 act together on early pre-60S particles and their reduced functionality bypasses the requirement for the essential pre-60S factor Nsa1. PLoS One. 2013; 8:e82741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manikas R.-G., Thomson E., Thoms M., Hurt E.. The K+-dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res. 2016; 44:1800–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barrio-Garcia C., Thoms M., Flemming D., Kater L., Berninghausen O., Baßler J., Beckmann R., Hurt E.. Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat. Struct. Mol. Biol. 2016; 23:37–44. [DOI] [PubMed] [Google Scholar]
- 35. Hung N.-J., Johnson A.W.. Nuclear Recycling of the Pre-60S Ribosomal Subunit-Associated Factor Arx1 Depends on Rei1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006; 26:3718–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lebreton A., Saveanu C., Decourty L., Rain J.-C., Jacquier A., Fromont-Racine M.. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 2006; 173:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nissan T.A., Bassler J., Petfalski E., Tollervey D., Hurt E.. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002; 21:5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nissan T.A., Galani K., Maco B., Tollervey D., Aebi U., Hurt E.. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol. Cell. 2004; 15:295–301. [DOI] [PubMed] [Google Scholar]
- 39. Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E.. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001; 8:517–529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.