Abstract
DNA topoisomerase 3B (TOP3B) is unique among all mammalian topoisomerases for its dual activities that resolve both DNA and RNA topological entanglements to facilitate transcription and translation. However, the mechanism by which TOP3B activity is regulated is still elusive. Here, we have identified arginine methylation as an important post-translational modification (PTM) for TOP3B activity. Protein arginine methyltransferase (PRMT) 1, PRMT3 and PRMT6 all methylate TOP3B in vitro at its C-terminal arginine (R) and glycine (G)-rich motif. Site-directed mutagenesis analysis identified R833 and R835 as the major methylation sites. Using a methylation-specific antibody, we confirmed that TOP3B is methylated in cells and that mutation of R833 and R835 to lysine (K) significantly reduces TOP3B methylation. The methylation-deficient TOP3B (R833/835K) is less active in resolving negatively supercoiled DNA, which consequently lead to accumulation of co-transcriptionally formed R-loops in vitro and in cells. Additionally, the methylation-deficient TOP3B (R833/835K) shows reduced stress granule localization, indicating that methylation is critical for TOP3B function in translation regulation. Mechanistically, we found that R833/835 methylation is partially involved in the interaction of TOP3B with its auxiliary factor, the Tudor domain-containing protein 3 (TDRD3). Together, our findings provide the first evidence for the regulation of TOP3B activity by PTM.
INTRODUCTION
DNA topoisomerases are a group of enzymes that resolve DNA topological tensions generated during the fundamental biological processes of transcription, replication, DNA recombination, DNA repair and chromatin remodeling (1). Based on their structures and mechanisms of unwinding, topoisomerases can be classified into type I and type II groups. Type I enzymes mediate DNA strand passage through a reversible single-strand break, whereas type II enzymes mediate transport through a double-strand DNA gate (1). Each of these groups is further divided into A and B subfamilies. In metazoans and higher eukaryotes, DNA topoisomerase III (TOP3) is the major type IA topoisomerase, which contains two isoforms named α (TOP3A) and β (TOP3B) (2). TOP3A and TOP3B have similar biochemical activities and both preferentially unwind negatively supercoiled DNA substrates (3). However, they seem to have distinct biological functions in vivo. TOP3A was found in complex with Bloom syndrome (BLM) helicase, RMI1 and RMI2, and it plays a major role in dissolving double holiday junctions, which are DNA intermediates formed during homology directed DNA repair (4). TOP3B shows both DNA and RNA topoisomerase activity in vitro and is involved in distinct biological processes in vivo. In the nucleus, TOP3B is targeted to chromatin by the epigenetic effector molecule Tudor domain-containing protein 3 (TDRD3) (5), where it unwinds negative supercoiled DNA in the wake of RNAP II (6). Defects in this activity lead to accumulation of R-loops, three-stranded nucleic acid structures consisting of a DNA–RNA hybrid and a displaced single-stranded DNA (6,7). In the cytoplasm, TOP3B interacts with TDRD3 and other proteins involved in RNA metabolism, including the fragile X mental retardation protein (FMRP) and functions as an RNA topoisomerase to facilitate translation (8,9).
TDRD3 is the major regulator of TOP3B function. It is essential for TOP3B chromatin localization (6), protein stability (6), association with polyribosomes (8,9) and more importantly, TOP3B enzymatic activity (10). A study from Hsieh’s group identified TDRD3 as a nucleic acid binding protein that preferentially interacts with single-stranded DNA and RNA (10). It is proposed that this feature of TDRD3 stabilizes TOP3B-bound nucleic acid complexes and shifts the unwinding reaction from a distributive to a processive mode (10). Additionally, TOP3B harbors a C-terminal domain containing an RG/RGG motif that mediates nucleic acid binding (11), a distinguishing feature not found in TOP3A and other mammalian topoisomerases. Deletion of the C-terminal domain significantly reduces TOP3B topoisomerase activity toward both DNA and RNA (8,12), suggesting that the RG/RGG motif is crucial for maximizing TOP3B activity.
The arginine in the RG/RGG motif is often subjected to methylation by a family of protein arginine methyltransferases (PRMTs) (13). The human genome encodes nine PRMTs (PRMT1–9), which catalyze the deposition of three types of methylated arginine residues—mono-methylated arginine (MMA), asymmetrical dimethylated arginine (ADMA) and symmetrical dimethylated arginine. Arginine methylation is abundantly found on proteins involved in regulation of transcription, replication, mRNA splicing and signaling transduction (13–15). For example, the DNA double-strand break repair protein MRE11 contains an RG/RGG motif that is methylated by PRMT1 (16,17). Defects in arginine methylation cause reduction of MRE11 exonuclease activity and impaired DNA damage checkpoint control (16). Furthermore, arginine methylation of the RG/RGG motif by PRMT1 reduces the affinity of FMRP to RNA and polyribosomes (18). Given the unique enzymatic activity of TOP3B, and its diverse biological function (in transcription and translation), we sought to explore the regulation of TOP3B activity by arginine methylation, with special focus on its RG/RGG motif-containing C-terminal domain.
Here, we identified arginine methylation of TOP3B using in vitro and in vivo assays. Two major methylation sites (R833/835) located at the RG/RGG motif-containing C-terminal domain were identified through site-directed mutagenesis. Arginine methylation deficient TOP3B (R833/835K) exhibited weaker DNA topoisomerase activity compared to the wild-type enzyme, which consequently led to accumulation of R-loops at target gene promoters and inhibition of gene transcription. Additionally, inhibition of TOP3B methylation using chemical inhibitors dampened its stress granule localization, indicating that methylation may also be critical for TOP3B function in translation regulation. Mechanistically, we identified that the Tudor domain of TDRD3 interacts with TOP3B in a methylation-dependent manner, raising the possibility that TOP3B arginine methylation may provide an additional layer of regulation with its auxiliary partner TDRD3. Altogether, we identified arginine methylation as an important post-translational modification (PTM) for modulating TOP3B function.
MATERIALS AND METHODS
Plasmids, antibodies and cell lines
The human PRMT1, PRMT3, CARM1/PRMT4, PRMT6 and PRMT8 cDNAs were cloned into a pGEX-6P-1 vector (GE Healthcare Life Sciences). The GST-Tudor (wild-type) and GST-Tudor (E691K) of TDRD3 have been described before (19). Human TOP3B cDNA and its truncated cDNAs (1-707 and 708–862) were cloned into pGEX-6P-1 and p3xFLAG-CMV vector (Sigma). All R-to-K mutants of TOP3B (708–862) were generated using site-directed mutagenesis (Agilent Technologies). The sequences of all primers are listed in Supplementary Table S3. The pUC19 plasmid used in the in vitro topoisomerase assay and the pFC53 plasmid used in the in vitro DNA/RNA immnoprecipitation (DRIP) assay have been described before (6). Rabbit monoclonal anti-MMA antibody (#8711), anti-ADMA antibody (#13522) and anti-TDRD3 antibody (#5942) were purchased from Cell Signaling Technology. Rabbit polyclonal anti-TDRD3 antibody used in the immunofluorescence were made in our laboratory (5,6) and are currently available as cat #ABE1868 from Millipore. Mouse monoclonal anti-TOP3B antibody (ab56445) and Rabbit monoclonal anti-TOP3B antibody (ab183520) were purchased from Abcam. Mouse monoclonal anti-DNA/RNA hybrid S9.6 antibody used for DRIP assay has been described before (6). Mouse monoclonal anti-Flag M2 antibody (F1804) and anti-ACTIN antibody (A5316) were purchased from Sigma. The human cervical cancer HeLa cell line was purchased from American Type Culture Collection (ATCC). The human VMRC-LCD cell, which doesn’t express TDRD3 due to gene deletion, has been described before (5).
Inhibitor and siRNA
The global methylation inhibitor adenosine dialdehyde (AdOx) (A7154) was purchased from Sigma. The type I PRMT inhibitor MS023 (18361) was purchased from Cayman Chemical. The siRNA targeting human TOP3B (J-005282-10) was purchased from Dharmacon.
In vitro methylation assay
In vitro methylation reactions were carried out in 30 μl of phosphate-buffered saline (pH = 7.4.) containing 0.5–1.0 μg of substrate, 3 μg of recombinant enzymes and 0.42 μM S-adenosyl-l- [methyl-3H]methionine (79 Ci/mmol from a 7.5 μM stock solution; PerkinElmer Life Sciences). The reaction was incubated at 30°C for 1 h and separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a Polyvinylidene difluoride (PVDF) membrane, treated with En3Hance™ (PerkinElmer Life Sciences) and exposed to film for 1 day at −80°C. After exposure, the PVDF membrane was washed with methanol and stained with coomassie to visualize total protein loaded.
Immunoprecipitation of arginine methylated proteins
To detected arginine methylated proteins, cells were either untreated or treated with methylation inhibitor AdOx (20 μM) or MS023 (10 μM) for 2 days. Cell pellets were lysed in 1× RIPA buffer (20 mM Tris HCl, (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitor) at 4°C for 1 h. The lysates were sonicated on ice and clarified by centrifugation followed by preclearing with protein G agarose. The lysates were subsequently immunoprecipitated with specific antibodies as indicated. Immunoprecipitated proteins were analyzed by western blots.
In vitro topoisomerase assay
Both wild-type and methylation-deficient TOP3B (R833/835K) were transfected into and purified from HEK293 cells using ANTI-FLAG M2 Magnetic Beads (M8823). The reaction mixture for the DNA relaxation assay consisted of 40 mM Tris–Cl, pH 7.5, 1 mM MgCl2, 5 mM dithiothreitol, 0.1 mg/ml bovine serum albumin and 400 ng of negative supercoiled pUC19 DNA in a 30 μl reaction system. The incubation was preceded at 37°C for the indicated time. The reaction was stopped with 4 μl of stop solution (1% SDS, 50% glycerol, 0.05% bromphenol blue). The sample was then loaded onto a 0.8% agarose gel. After electrophoresis, the gel was stained with ethidium bromide and photographed under UV illumination.
In vitro R-loop detection using DNA/RNA immunoprecipitation (DRIP)-qPCR
In vitro transcription of R-loop formation has been described before (6). In brief, the pFC53 R-loop prone plasmids were in vitro transcribed with T3 RNA polymerase in the presence of either wild-type or methylation-deficient TOP3B (R833/835K) enzymes at 37°C for 1 h. The reaction was then inactivated at 65°C for 10 min. Samples were equally divided and either treated with RNase A plus RNase H or only RNase A at 37°C for 30 min followed by proteinase K treatment at 37°C for 30 min. Samples were then purified using phenol/chloroform extraction. Precipitated DNA was dissolved in water.
In vitro transcription products were incubated with S9.6 antibody in binding buffer (10 mM NaPO4 pH 7.0, 140 mM NaCl, 0.05% Triton X-100) at 4°C overnight. The binding mixture was then incubated with protein A/G agarose for an additional 1 h. The bound DNA was eluted with buffer containing 50 mM Tris pH 8.0, 10 mM ethylenediaminetetraacetic acid (EDTA), 0.5% SDS and 300 μg of proteinase K at 50°C for 30 min. Samples were then purified by phenol/chloroform extraction and ethanol precipitation. Immunoprecipitated DNA was subjected to analysis by quantitative polymerase chain reaction (qPCR).
In vivo DRIP-qPCR
Total genomic DNA was extracted from cells by SDS/proteinase K treatment at 37°C overnight, followed by phenol/chloroform extraction and ethanol precipitation. DNA was fragmented using restriction enzymes HindIII, EcoRI, BsrG1, XbaI and Ssp1 and pretreated (where indicated) with RNase H overnight. After phenol/chloroform extraction and ethanol precipitation, 4 µg of each samples were subject to immunoprecipitation with 10 µg of S9.6 antibody. Immunoprecipitated DNA was analyzed by qPCR using primers listed in the Supplemental Table S2.
Reverse transcription-quantitative PCR (RT-qPCR)
Total cellular RNA was extracted by the TRIzol Reagent (Invitrogen). The RNA was analyzed for integrity using the Agilent 2100 Bioanalyzer (Agilent Technologies). The total RNA (1μg) was then used as template to synthesize cDNA using the High Capacity cDNA Archive Kit (Applied Biosystems), and qPCR was subsequently performed on the CFX96 Real-time System C1000 Touch Thermal Cycler (Bio-Rad). The RNA levels were normalized to the endogenous control gene (ACTIN). Data analysis was performed using the Bio-Rad CFX Manager 3.1. The experimental cycle threshold (Ct) was calibrated against the ACTIN control product. All amplifications were performed in triplicate. Primer sequences are listed in the Supplemental Table S1.
Immunofluorescence
The HeLa cells, either transfected with plasmids or treated with MS023, were grown on glass coverslips to the desired confluence before fixation. The cells were either left untreated or treated with 0.5 mM sodium arsenite for 1 h. The cells were rinsed with phosphate-buffered saline (PBS) and fixed with ice-cold methanol for 20 min at room temperature. After a blocking step with 20% newborn calf serum, the cells were incubated with the indicated antibodies at 4°C overnight. The cells were then stained with a fluorescence-labeled secondary antibody and stained with 4′,6-diamidino-2-phenylindole (DAPI). The coverslips were then sealed and examined using an Olympus BX50 fluorescence microscope.
GST pulldown
Glutathione-S-transferase (GST), GST-Tudor (WT) and GST-Tudor (E691K) were expressed and purified from Escherichia coli. HeLa cells (untreated or treated with MS023) were lysed in lysis buffer containing 50 mM Tris HCl, (pH 7.5), 150 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA, 5 mM EGTA, 1.5 mM MgCl2, 5% glycerol and protease inhibitors (Roche). The purified recombinant proteins were incubated with HeLa cell lysates overnight at 4°C. Glutathione Sepharose (GE Healthcare Life Sciences) beads were added to the lysate mixture and incubated for 2 h at 4°C. The eluted samples were loaded on an SDS-PAGE gel and detected by western blot using an anti-TOP3B antibody.
Co-immunoprecipitation
HeLa cells were washed with ice-cold PBS and lysed with 1 ml of co-immunoprecipitation buffer (50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40, 5 mM EDTA, 5 mM EGTA, 15 mM MgCl2) supplemented with protease inhibitor cocktail. After sonication, insoluble materials were removed by centrifugation at maximum speed for 10 min. Whole cell lysates were incubated with 2 μg of immunoprecipitation antibody overnight at 4°C. After incubation with Protein A/G agarose beads, bound proteins were eluted and analyzed using western blots.
Statistical analysis
All experiments were performed at least three times. Statistical analyses was performed by Student’s t-test. P < 0.05 was considered statistically significant. Quantification of agarose gel images, immunoblotting images and number and size of stress granules was performed using ImageJ software.
RESULTS
TOP3B is arginine methylated in vitro
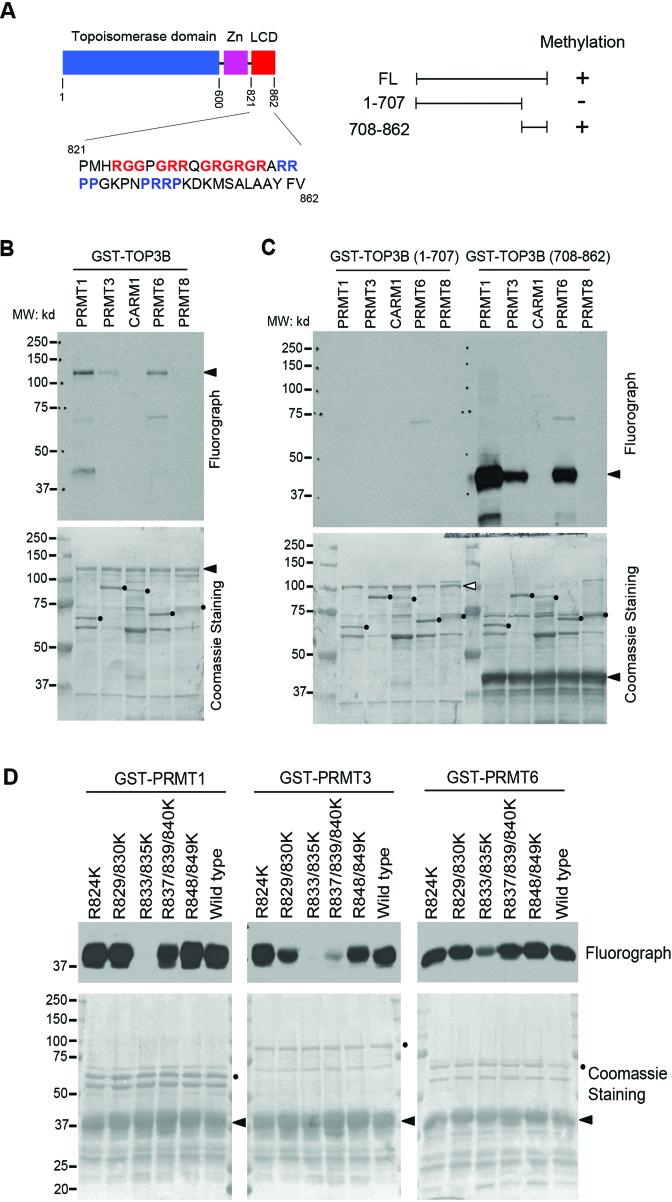
Mammalian type IA topoisomerase contains two paralogs: TOP3A and TOP3B. Although both proteins harbor a characteristic topoisomerase domain that determines their function, TOP3B, but not TOP3A, contains an array of RG/RGG and PR (P: Proline) motifs at C-terminus (Figure 1A). The arginine residues in these motifs are often subjected to methylation by PRMTs. To test if TOP3B is arginine methylated, we performed in vitro methylation assays by incubating recombinant full-length TOP3B protein with GST-tagged PRMTs (PRMT1, PRMT3, coactivator-associated arginine methyltransferase 1 (CARM1), PRMT6 and PRMT8) in the presence of 3H-labeled S-adenosyl-l-methionine ([3H]AdoMet), the universal methyl group donor. All PRMTs used in this assay are active as GST fusion proteins (13,14). We found that TOP3B can be methylated by PRMT1, PRMT6 and PRMT3, although the latter methylated TOP3B to a much lesser extent (Figure 1B). This is consistent with previous reports that these PRMTs exhibit similar substrate preference in vitro (13,14). Although equal amounts of PRMT1, PRMT3 and PRMT6 were used in the reaction (Figure 1B, lower panel), the methylation by PRMT1 generated a much stronger signal (Figure 1B, upper panel).
Figure 1.
TOP3B is methylated in vitro. (A) The locations of the TOP3B topoisomerase domain, zinc-binding domain (Zn) and C-terminal low complexity domain (LCD). Regions of arginine-glycine-rich and proline-arginine-rich motifs are highlighted in red and blue, respectively. A summary of the in vitro methylation observed in B and C is shown. (B) TOP3B is methylated in vitro. In vitro methylation assays were performed by incubating recombinant PRMTs (PRMT1, 3, CARM1, PRMT6 and PRMT8) with purified GST-TOP3B protein. (C) C-terminal domain of TOP3B is methylated. Both N-terminus and C-terminus truncations of TOP3B were subjected to in vitro methylation assay as described in (B). (D) Arginine 833 and 835 are the major methylation sites. The in vitro methylation assays were performed by incubating recombinant PRMT1, PRMT3 and PRMT6 with a series of R to K mutants of GST-TOP3B C-terminus truncation (708-862). In panels B–D, the loading of the proteins was visualized by coomassie staining the same PVDF membrane for fluorography. Arrows indicate the positions of the substrates and the solid dots indicate the positions of the recombinant enzymes.
Alternative splicing of human TOP3B gene produces three isoforms (Supplementary Figure S1). The shortest isoform (isoform 3, NP_001336780.1) lacks C-terminal RG/RGG and PR motifs (Supplementary Figure S1). Further analysis of isoform expression by RT-qPCR demonstrates that the expression of isoform 3 varies from about 1 to 4% of isoform 1 expression level across mouse tissues and human cancer cell lines (Supplementary Figure S2A and B). Western blot analysis using mouse monoclonal anti-TOP3B antibody (ab56445) detects protein bands migrate at expected molecular weight of all three isoforms. The intensity of all three bands decreases when TOP3B expression is inhibited by siRNA-mediated knockdown, indicating that the observed signals are likely from TOP3B isoforms (Supplementary Figure S2C).
To further narrow down the arginine methylation region on TOP3B, we generated two truncation proteins from the longest TOP3B isoform (isoform 1), covering amino acid 1–707 (minus C-terminal RG/RGG and PR motifs) and 708–862, respectively. In vitro methylation reactions performed using these two truncation proteins confirmed that TOP3B (708–862), but not TOP3B (1–707), was methylated by PRMT1, PRMT3 and PRMT6 (Figure 1C), which is consistent with the methylation of full-length TOP3B. The activity of recombinant PRMTs was confirmed by in vitro methylation of their respective known substrates, REF/ALY (20) and PABP1 (21) (Supplementary Figure S3A and B). We also examined the methylation of TOP3B (708–862) by PRMT5, since PRMT5 often shares similar methylation motifs with PRMT1. However, at the same experimental conditions as other PRMTs, PRMT5 does not methylate TOP3B (Supplementary Figure S3C and D). The methylation results are summarized in Figure 1A (right panel).
There are 10 arginines located within the TOP3B methylated region, six of which are clustered to form RG/RGG motifs (Figure 1A). To further map the methylation site(s), we created arginine to lysine (K) mutants that cover all 10 potential methylation sites and performed in vitro methylation assays. When comparing the methylation level among the mutants, the R833/835K mutation showed the most dramatic loss of methylation by all three PRMTs (Figure 1D). We conclude that R833 and R835 are the major arginine methylation sites of TOP3B in vitro.
TOP3B is arginine methylated in cells
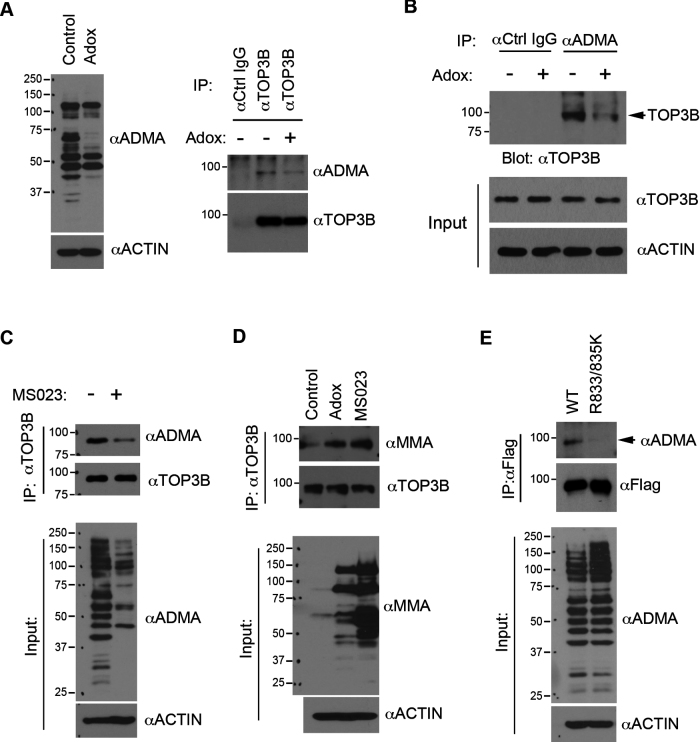
We next performed in vivo methylation assays to measure TOP3B arginine methylation in cells. Using a TOP3B antibody, we immunoprecipitated endogenous TOP3B from HeLa cells either untreated or treated with adenosine dialdehyde (AdOx), a global methylation inhibitor. The immunoprecipitated samples were detected by western blot against TOP3B methylation using a pan-ADMA antibody. A distinct band that migrated at the same molecular weight as endogenous TOP3B was detected from samples immunoprecipitated using the TOP3B antibody, but not from the samples immunoprecipitated using control IgG (Figure 2A). Importantly, the signal was reduced in the AdOx treated group (Figure 2A), suggesting that TOP3B is methylated in cells. To further confirm this, we used an ADMA antibody to perform a reciprocal immunoprecipitation from control and AdOx-treated cells. Compared to the IgG controls, TOP3B proteins were specifically detected in the ADMA antibody immunoprecipitated samples (Figure 2B). AdOx treatment reduced the detected TOP3B signal, confirming that TOP3B is arginine methylated. The immunoprecipitation was performed under denaturing conditions (0.1% SDS) to ensure that the methylation signal was not from TOP3B-interacting proteins.
Figure 2.
TOP3B is methylated in cells. (A) Endogenous TOP3B was immunoprecipitated from control and Adox-treated (20 μM, 48 h) HeLa cells under denaturing conditions. TOP3B methylation was detected using a pan-anti-ADMA antibody. Immunoprecipitated TOP3B was detected with an anti-TOP3B antibody. (B) An immunoprecipitation assay was performed using an anti-ADMA antibody to enrich arginine-methylated proteins from control and Adox treated HeLa cells. The immunoprecipitated protein complexes were detected with anti-TOP3B antibody. The expression of TOP3B was detected in input cell lysates. (C) Endogenous TOP3B was immunoprecipitated from control and MS023-treated (10 μM, 48 h) HeLa cells under denaturing conditions. TOP3B methylation was detected using an anti-ADMA antibody, as described in (A). Immunoprecipitated TOP3B was detected with an anti-TOP3B antibody. (D) Endogenous TOP3B was immunoprecipitated from control, Adox and MS023-treated HeLa cells under denaturing conditions. TOP3B methylation was detected using a pan-anti-MMA antibody. Immunoprecipitated TOP3B was detected with an anti-TOP3B antibody. (E) HeLa cells were transfected with Flag-TOP3B and Flag-TOP3B (R833/835K) constructs for 48 h. The total cell lysates were prepared under denaturing conditions and immunoprecipitated with an anti-FLAG antibody. The immunoprecipitated protein samples were detected with anti-ADMA and anti-FLAG antibodies. In panels A–E, the input samples were detected with either anti-ADMA or anti-MMA antibody to examine the global methylation level of the input cell lysates.
TOP3B is methylated by PRMT1, PRMT3 and PRMT6 in vitro (Figure 1). We next examined the impact of inhibiting these three enzymes on TOP3B methylation. HeLa cells were treated with a newly developed PRMT inhibitor, MS023, which specifically targets type I PRMTs, including PRMT1, PRMT3 and PRMT6 (22). Consistent with the results from AdOx treatment, MS023 treatment reduces TOP3B methylation (Figure 2C). Interestingly, both AdOx and MS023 treatment caused increases in global levels of arginine monomethylation and concurrent TOP3B monomethylation when detected using a pan-MMA antibody (Figure 2D). This is consistent with previous reports showing that inhibition of type I arginine methyltransferase, either by genetic depletion or chemical inhibition, caused substrate scavenging by other PRMTs (22,23). The specificity of MMA and ADMA antibodies was confirmed by Dot-blot assays using synthetic arginine-methylated peptides (Supplementary Figure S4).
To test if TOP3B is methylated at R833 and R835 in cells, we transfected cells to express either Flag-tagged wild-type or R833/835K mutant TOP3B and detected their methylation using an ADMA antibody. Compared to wild-type TOP3B, the methylation level of R833/835K mutant was significantly reduced (Figure 2E). Together with our in vitro methylation results, we demonstrated that TOP3B is methylated at R833 and R835.
Arginine methylation promotes TOP3B topoisomerase activity
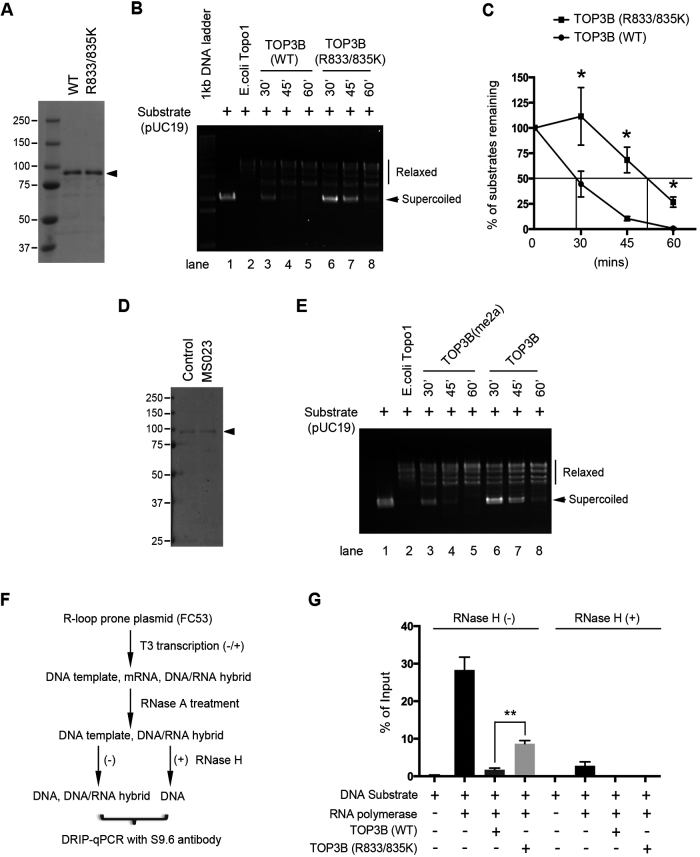
We next determined the role of arginine methylation on TOP3B topoisomerase activity using an in vitro negatively supercoiled DNA relaxation assay (24,25). Flag-tagged wild-type and methylation-deficient (R833/835K) TOP3B were purified from HEK293 cells (Figure 3A). Both enzymes were incubated with negatively supercoiled pUC19 plasmid DNA substrates at a 1:30 enzyme/DNA molar ratio for indicated time period. The methylation-deficient TOP3B (R833/835K) was significantly less active compared to the wild-type enzyme (Figure 3B). Quantitative analysis showed that wild-type TOP3B relaxed 50% of supercoiled DNA substrate in less than 30 min, whereas TOP3B (R833/835K) needed more than 45 min (Figure 3C). To further confirm that the reduced activity in TOP3B (R833/835K) is caused by lack of arginine methylation, but not due to R to K mutation, we purified Flag-tagged TOP3B from control and MS023 treated HEK293 cells (Figure 3D). MS023 treatment dramatically reduced TOP3B asymmetrical dimethylation (Figure 2C). Consistent with the results using methylation-deficient TOP3B (R833/835K), inhibiting TOP3B asymmetrical dimethylation reduced its topoisomerase activity (Figure 3E).
Figure 3.
Arginine methylation promotes TOP3B DNA topoisomerase activity. (A) A coomassie blue-stained gel image showing the purified Flag-tagged wild-type and arginine methylation-deficient (R833/835K) TOP3B from HEK293 cells. (B) Arginine methylation-deficient TOP3B (R833/835K) shows reduced activity in relaxing supercoiled plasmid DNA. Reaction mixtures containing negatively supercoiled plasmid DNA (pUC19) were incubated with either wild-type or R833/835K mutant of TOP3B protein for the indicated time points. Escherichia coli Topo1 was used as a positive control. The arrows indicate the negative supercoiled DNA substrate. (C) The topoisomerase activity was quantified by measuring the percentage of remaining substrates over the indicated time points. Statistical analysis was performed using student’s t-test from three independent experiments. *P < 0.05. (D) A coomassie blue-stained gel image showing the recombinant Flag-TOP3B purified from control and MS023-treated HEK293 cells. (E) Arginine methylation of TOP3B promotes its DNA topoisomerase activity in relaxing supercoiled plasmid DNA. The experiment was performed as described in (B), except the enzymes were purified from control and MS023-treated cells. (F) Work flow of in vitro R-loop detection using DRIP-qPCR. See details of the description in ‘Materials and Methods’ section. (G) pFC53 plasmid was subjected to in vitro transcription in the presence of either wild-type or R833/835K mutant of TOP3B as indicated. DRIP-qPCR was performed to quantify the co-transcriptional R-loop levels in individual samples. The immunoprecipitated DNA samples were analyzed by qPCR. Error bars represent standard deviation calculated from triplicate qPCR reactions of one representative experiment.
The DNA topoisomerase activity of TOP3B is crucial to suppress the formation of co-transcriptional R-loops, which are three-stranded nucleic acid structures containing a DNA/RNA hybrid and a single-stranded DNA (6,7). We hypothesized that reduced topoisomerase activity observed in hypomethylated TOP3B would increase the formation of co-transcriptional R-loops. To test this, we performed in vitro transcription of an R-loop prone plasmid (FC53) (26) in the presence of either methylation-proficient (wild-type) and -deficient (R833/835K) TOP3B (Figure 3A). The in vitro transcribed product mixture was subjected to sequential treatment with RNASE A to remove RNA in the product and/or RNASE H to remove DNA/RNA hybrids (serving as negative controls). The amount of R-loops was quantified by performing a DNA/RNA immunoprecipitation (DRIP)-qPCR using an S9.6 antibody that specifically recognizes DNA/RNA hybrids (Figure 3F). Adding wild-type TOP3B to the in vitro transcription system dramatically reduced R-loops in the final product (Figure 3G, compare the second and third column). The methylation-deficient TOP3B (R833/835K) was less efficient in suppressing the formation of co-transcriptional R-loops, leading to ∼4 times more R-loops compared to the wild-type TOP3B (Figure 3G, compare the third and fourth column). RNASE H treatment abolished the DRIP-qPCR signal, confirming the specificity of the S9.6 antibody (Figure 3G, RNase H+ group). Altogether, we demonstrated that arginine methylation of TOP3B at R833 and R835 enhances TOP3B topoisomerase activity and suppresses formation of co-transcriptional R-loops in vitro.
Arginine methylation promotes TOP3B function in transcription regulation
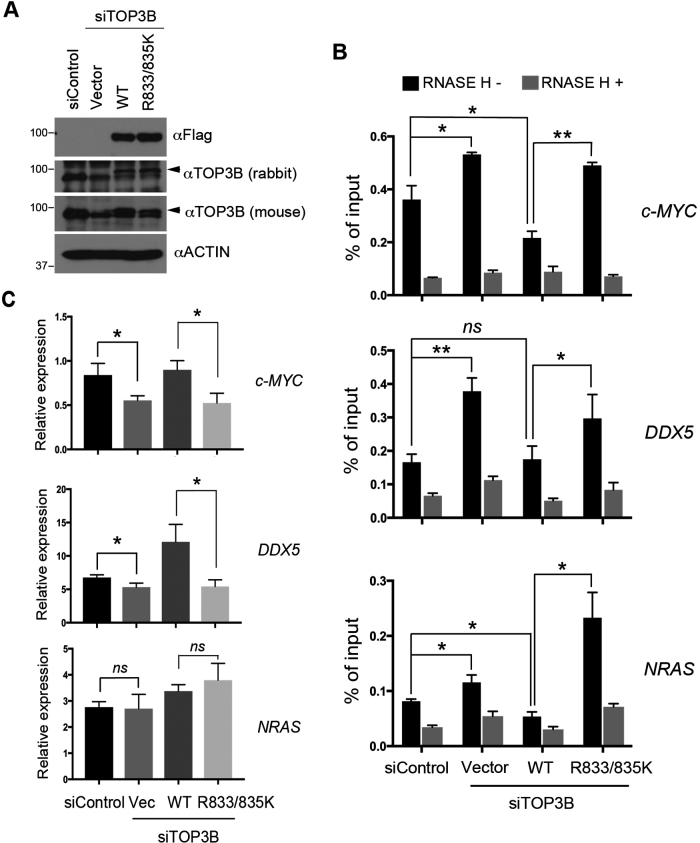
In the nucleus, TOP3B is targeted to transcriptional start site and the elongating RNAP II by the methylarginine effector molecule TDRD3 (5,6). Through relaxing negatively supercoiled DNA and suppressing R-loop formation, TOP3B facilitates transcription and maintains genome integrity (6). To determine the role of arginine methylation in regulating TOP3B function in vivo, we compared the activity of wild-type and methylation-deficient TOP3B (R833/835K) in suppressing the formation of co-transcriptional R-loops in cells. HeLa cells were transfected with control siRNA (small interfering RNA) and siRNA that specifically targets 3′-UTR (untranslated region) of TOP3B to knockdown endogenous TOP3B expression. Both wild-type and methylation-deficient TOP3B (R833/835K) were re-expressed in the knockdown cells by transient transfection to establish the rescue-expressing cells. Protein expression was confirmed by Western blot (Figure 4A). The genomic DNA from these cells were extracted and subjected to DRIP-qPCR following a previously established protocol (6,26). We examined the formation of R-loops at three previously identified TDRD3/TOP3B targeted loci, namely c-MYC, DDX5 and NRAS (6). Knockdown of TOP3B caused accumulation of R-loops at all three tested loci (Figure 4B). Importantly, re-expressing wild-type TOP3B, but not the methylation-deficient TOP3B (R833/835K), reduced R-loop accumulation at all three loci, when compared to TOP3B knockdown cells (Figure 4B).
Figure 4.
Arginine methylation promotes TOP3B DNA topoisomerase activity in cells. (A) HeLa cells were transfected with scramble siRNA or siRNA that targets TOP3B (siTOP3B). After 24 h, the siTOP3B-transfected cells were transfected with either empty vector, wild-type or R833/835K mutant TOP3B for an additional 48 h. Total cell lysates were prepared, and protein expression was examined by western blot using indicated antibodies. (B) DRIP-qPCR was performed using genomic DNA extracted from the cells described in (A). The levels of R loops at three TDRD3-targeted genomic loci were examined by qPCR. *P < 0.05, **P < 0.01. (C) RT-qPCR was performed using the RNA extracted from the cells described in (A) to examine the expression of three TDRD3 target genes. *P < 0.05, ns: not significant.
We next examined the impact of wild-type and methylation-deficient TOP3B (R833/835K) on gene expression using RT-qPCR. Knockdown of TOP3B reduced the mRNA level of c-MYC and DDX5 (Figure 4C). Re-expressing wild-type TOP3B, but not the methylation-deficient TOP3B (R833/835K), rescued the expression of c-MYC and DDX5 (Figure 4C). Interestingly, the mRNA expression of NRAS was not regulated by TOP3B (Figure 4C), suggesting that uncharacterized mechanisms may counteract the impact of R-loops for NRAS transcription regulation in HeLa cells. Altogether, these results demonstrated that arginine methylation of TOP3B is essential for suppressing the formation of co-transcriptional R-loops, likely by promoting TOP3B topoisomerase activity.
TOP3B arginine methylation regulates its stress granule localization
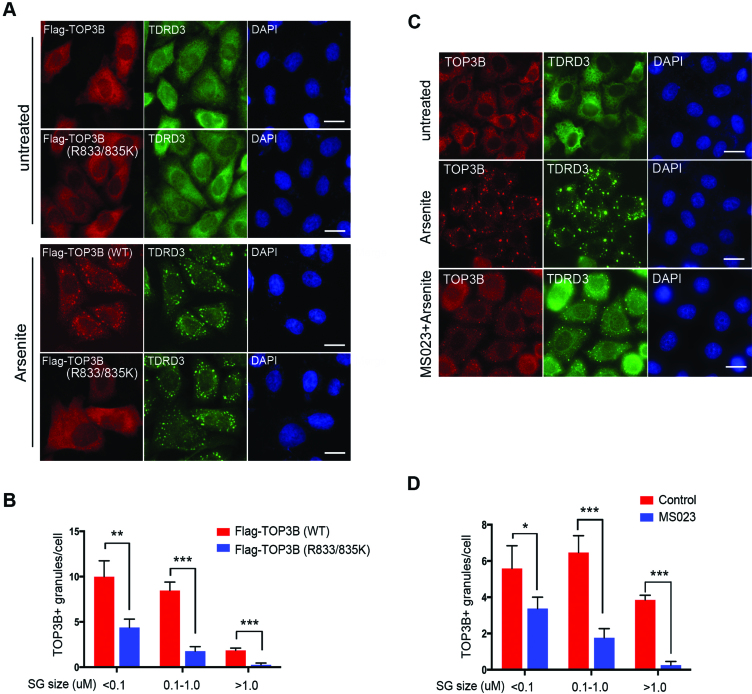
In the cytoplasm, TOP3B associates with polyribosomes and localizes to stress granules in response to oxidative stress induced by arsenite (8,9). We next determined the role of arginine methylation on TOP3B cytoplasmic function, particularly stress granule localization. HeLa cells were transfected with either Flag-tagged wild-type or methylation-deficient TOP3B (R833/835K). Immunofluorescence assays were performed using an anti-Flag antibody. Similar to endogenous TOP3B, the localization of Flag-tagged TOP3B was in the cytoplasm, co-localizing with its binding partner TDRD3 (Figure 5A, untreated). No obvious subcellular localization change was observed, comparing the wild-type to the methylation-deficient TOP3B (R833/835K) (Figure 5A, untreated). When the cells were treated with arsenite, wild-type TOP3B formed large, distinct cytoplasmic foci that co-localized with TDRD3 (Figure 5A, arsenite treated). Compared to the wild-type TOP3B, the methylation-deficient TOP3B (R833/835K) formed much fewer and smaller foci, whereas the majority of the mutant protein remained dispersed in the cytoplasm (Figure 5A, arsenite treated). This is likely not caused by disruption of the overall stress granule formation, because the localization of TDRD3 was not affected. Quantitative analysis of stress granules formed in the wild-type and methylation-deficient TOP3B (R833/835K) expressing cells confirmed that the size and the number of TOP3B positive (TOP3B+) stress granules were diminished when TOP3B methylation was blocked (Figure 5B).
Figure 5.
TOP3B arginine methylation is important for its stress granule localization. (A) HeLa cells cultured on glass cover slips were transfected with Flag-tagged wild-type or R833/835K mutant TOP3B. After 24 h of transfection, cells were treated with 0.5 mM sodium arsenite (Arsenite) for 1 h. The cells were fixed and immunostained with anti-FLAG and anti-TDRD3 to detect the protein localization. DAPI was used to stain the nuclear DNA. Scale bar = 20 µM (B) At least 200 of both wild-type and R833/835K mutant TOP3B-transfected cells were imaged and analyzed for comparison. The number and size of the stress granules were analyzed with ImageJ software. (C) HeLa cells cultured on glass cover slides were untreated or treated with MS023 for 2 days before the cells were treated with 0.5 mM Arsenite for 1 h. The samples were processed as described in (A), except that anti-TOP3B and anti-TDRD3 antibodies were used to examine the endogenous protein localization. Scale bar = 20 µM (D) The number and size of stress granules in control and MS023-treated cells were analyzed as described in (B).
To further confirm that the reduced stress granule localization of methylation-deficient TOP3B (R833/835K) is due to lack of arginine methylation, but not by R to K mutation, we treated cells with the type I PRMT inhibitor MS023 and examined endogenous TOP3B stress granule localization after arsenite stress. Similar to the results observed with methylation-deficient TOP3B (R833/835K), inhibition of arginine methylation led to smaller and fewer TOP3B positive stress granules (Figure 5C and D; Supplementary Figure S5), supporting the conclusion that arginine methylation is important for TOP3B stress granule localization. This is not surprising, because many stress granule proteins have been reported to be arginine methylated and arginine methylation is essential for their stress granule localization (27–29). Inhibition of PRMT activity is likely to cause broader impacts on stress granule formation because localization of TDRD3 to stress granules is also decreased (Figure 5C). Altogether, we demonstrated that arginine methylation is important for TOP3B stress granule localization.
Arginine methylation enhances TOP3B interaction with TDRD3
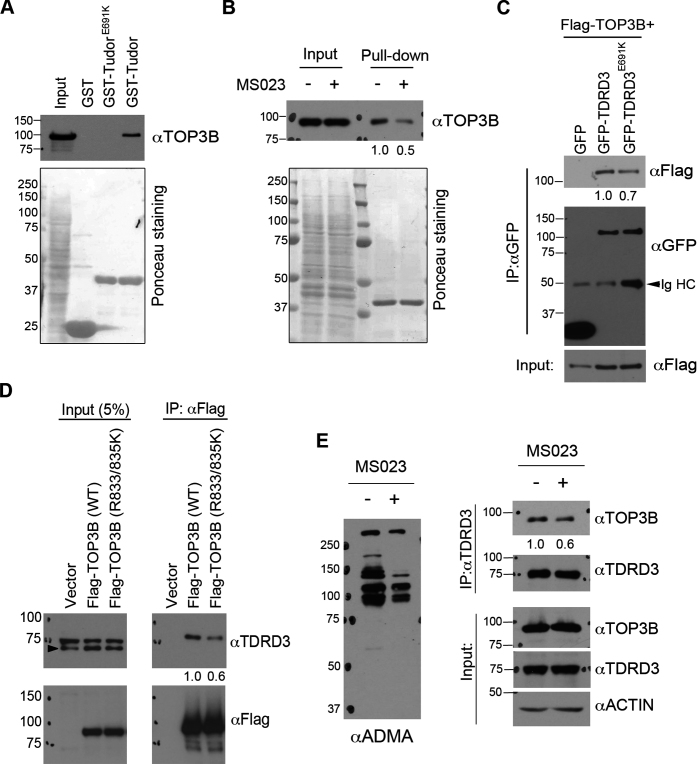
TOP3B forms a stable protein complex with TDRD3 (6,8,9). The N-terminus oligosaccharide/nucleotide-binding (OB)-fold of TDRD3 interacts with the topoisomerase enzymatic domain of TOP3B (6,8). This protein–protein interaction is important for TOP3B stability (6) and its enzymatic activity toward both DNA and RNA (10). The Tudor domain of TDRD3 prefers to interact with methylated arginine motifs on both histone and non-histone proteins (30–32). To test if arginine methylation enhances TOP3B interaction with TDRD3, we performed a GST pulldown assay to determine if the Tudor domain of TDRD3 interacts with TOP3B. Recombinant proteins of GST, GST-Tudor and mutant GST-Tudor (E691K, which disrupts Tudor-methylarginine interaction (6,31), Supplementary Figure S6A) were incubated with HeLa cell total lysate, and pulldown samples were detected by western blot using an anti-TOP3B antibody. Wild-type Tudor domain, but not the methylarginine-binding deficient Tudor (E691K), interacted with endogenous TOP3B (Figure 6A). To test if this interaction is dependent upon TOP3B arginine methylation, we performed a GST pull-down assay by incubating recombinant GST-Tudor protein with HeLa cell lysates pre-treated with or without PRMT inhibitor MS023. The interaction of the Tudor domain with TOP3B was reduced when the arginine methylation was inhibited (Figure 6B), suggesting that the Tudor–TOP3B interaction is methylation dependent.
Figure 6.
The Tudor domain of TDRD3 interacts with TOP3B in a methylation-dependent manner. (A) GST-pulldown was performed by incubating recombinant wild-type and methylarginine-binding deficient (E691K) Tudor domain of TDRD3 with HeLa total cell lysate. The pulldown samples were detected using an anti-TOP3B antibody by western blots. The membrane was stained with Ponceau S to visualize the recombinant proteins used. (B) GST-pulldown was performed by incubating recombinant TDRD3 Tudor domain with HeLa cell lysate either untreated or pretreated with MS023 for 2 days. The samples were processed as described in (A). (C) Co-immunoprecipitation experiments were performed by transfecting HeLa cells with Flag-TOP3B and GFP-tagged variants of either wild-type or Tudor domain mutant (E691K) TDRD3. Anti-GFP antibody was used for immunoprecipitation. The samples were detected by western blots using antibodies as indicated. IgHC: IgG heavy chain. (D) Co-immunoprecipitation experiment was performed by transfecting HeLa cells with Flag-tagged variants of either wild-type or methylation-deficient (R833/835K) TOP3B. Anti-Flag antibody was used for immunoprecipitation. (E) Inhibition of arginine methylation reduces TOP3B and TDRD3 interaction. Co-immunoprecipitation experiment was performed in control and MS023-treated HeLa cells. Anti-TDRD3 antibody was used for immunoprecipitation. Quantification of the immunoblotting images was performed using ImageJ software. The relative ratio of IP signals for MS023 treated (B and E), Tudor mutant (E691K) (C) and methylation-deficient TOP3B (R833/835K) (D) were calculated using the signals of the wild-type or untreated protein as the standard (set as 1.0) and were listed below each image.
To further determine if the Tudor domain contributes to TDRD3–TOP3B interaction in vivo, we performed a co-immunoprecipitation assay by transfecting HeLa cells with Flag-tagged TOP3B together with GFP-tagged variants of either wild-type or methylarginine-binding deficient TDRD3 (E691K) (Supplementary Figure S6B) (6). Compared to wild-type TDRD3, less GFP-TDRD3 (E691K) was immunoprecipitated (Figure 6C), suggesting that functional integrity of the Tudor domain may help maximize the TDRD3–TOP3B interaction. To further confirm the role of arginine methylation in enhancing TOP3B–TDRD3 interaction, we performed a co-immunoprecipitation assay to compare the interaction of wild-type TOP3B and the methylation-deficient TOP3B (R833/835K) with TDRD3. Although equal amount of proteins were immunoprecipitated, less TDRD3 co-immunoprecipitated with TOP3B (R833/835K) compared to wild-type TOP3B (Figure 6D). To further assess the contribution of C-terminal arginine methylation to TOP3B–TDRD3 interaction, we treated cells with PRMT inhibitor MS023 and performed co-immunoprecipitation using anti-TDRD3 antibody. As shown in Figure 6E, MS023 treatment reduced TOP3B–TDRD3 interaction, confirming that arginine methylation of the RG/RGG motif enhances TOP3B interaction with TDRD3 through the Tudor domain.
DISCUSSION
TOP3B is unique among all mammalian topoisomerases because it has activity toward both DNA and RNA substrates (33). Additionally, TOP3B contains a C-terminal RG/RGG motif unlike TOP3A, the only other topoisomerase in the type IA subfamily. The goal of this study was to reveal the molecular mechanisms that regulate TOP3B function. We identified arginine methylation as a novel PTM that regulates TOP3B topoisomerase activity. Methylation of the C-terminus RG/RGG motifs is essential for maximizing TOP3B activity toward transcriptional and translational regulation. Understanding how methylation regulates TOP3B will help to further elucidate normal TOP3B biological function and its dysregulation, which has been linked to both cancer and neurological diseases (8,9,34).
The C-terminal domain regulates TOP3B function
RG/RGG motifs, which include RG and RGG repeats of varied lengths interspersed with spacers of different amino acids (35), are often presented within low complexity, unstructured regions of protein. Although a clear structure has not been defined, RG/RGG motif plays important roles in mediating nucleic acid binding, protein–protein interactions and protein subcellular localization (11). For example, RG/RGG motifs from nucleolin and FMRP can bind and stabilize nucleic acid secondary structures known as G-quadruplexes (G4s), which are structures formed by guanine tetrads arranged in a planar conformation (36–38). The RG/RGG motif located at the C-terminus of TOP3B was recently reported to mediate DNA and RNA binding (8). Deletion of the RG/RGG motif significantly reduced TOP3B topoisomerase activity (8), suggesting that DNA and RNA-binding activity of the RG/RGG motif is crucial for TOP3B function. Interestingly, alternative splicing of the human TOP3B gene locus produces three protein isoforms with distinct C-terminal domains (Supplementary Figure S1). One of the isoforms lacks the RG/RGG motif. It is not uncommon for the presence or absence of certain RG/RGG motifs to be regulated by alternative splicing (39–41). For example, a splice variant of FMR1 is missing exon 12 and 14, which introduces a frameshift resulting in truncation and the loss of the RGG motif. This isoform showed reduced localization to dendritic RNA granules (39). However, the tissue-specific expression and the biological significance of TOP3B isoforms without an RG/RGG motif are still not clear.
In addition to the RG/RGG motif, the TOP3B C-terminus contains two PR motifs (Figure 1A). PR motifs are preferred methylation substrates for CARM1/PRMT4 (14). In vitro methylation assay did not detect TOP3B methylation signal when reacted with CARM1/PRMT4 (Figure 1B and C). However, PR motifs can also mediate protein–protein interaction with WW domains and Src homology 3 (SH3) domains (42–44). These domains are often presented in signaling transduction molecules, such as proto-oncogene tyrosine-protein kinase Fyn and formin-binding protein 30 (FBP30) (42,43), raising the possibility that PR motifs may mediate protein-protein interaction and allow the regulation of TOP3B function by cellular signaling.
Arginine methylation regulates TOP3B DNA topoisomerase activity and stress granule localization
RG/RGG motifs are preferred substrates for PRMTs (11,13). Many of the RG/RGG motif functions are regulated, at least in part, by arginine methylation. Although methylation does not neutralize the cationic charge of an arginine residue (45), each addition of a methyl group to an arginine residue removes a potential hydrogen bond donor and changes its structure. Thus, arginine dimethylation imparts bulkiness and hydrophobicity to a protein and can affect protein–protein and protein–nucleic acid interactions. For example, methylation of FMRP at its RGG motif results in reduced binding to a subset of mRNAs (18), likely due to the reduced hydrogen bonding between the RGG motif and RNA. TOP3B is asymmetrically dimethylated at its C-terminus RG/RGG motif by type I PRMTs—PRMT1, 3, 6 (Figures 1 and 2). Arginine methylation deficient TOP3B exhibited reduced DNA topoisomerase activity and impeded stress granule localization (Figures 3–5), supporting the importance of arginine methylation in regulating TOP3B function. To test if TOP3B RG/RGG motif methylation affects its interaction with nucleic acids, we performed pulldown experiments using recombinant proteins and biotinylated-DNA oligonucleotides (data not shown). We did not observe a significant difference in binding affinity between the methylation-proficient and -deficient TOP3B (R833/835K). However, we cannot exclude the possibility that pulldown and immunoblotting-based approaches are not ideal for detecting such a difference.
Many RG/RGG motif-containing proteins translocate to SGs after cellular stress, possibly through their interaction with mRNA. Although the underlying molecular mechanism is still unclear, arginine methylation of the RG/RGG motifs was reported to modulate this process both positively and negatively (29). We found that inhibition of arginine methylation using chemical inhibitor (MS023) imparts TOP3B localization to SGs (Figure 5C and D). Although it is possible that globally reduced arginine methylation affects the general mechanism of SG formation, our complementary experiment using methylation-deficient TOP3B mutant (R833/835K) confirmed that methylation is important for efficient TOP3B SG localization (Figure 5A and B). Similar regulatory mechanisms have also been reported on other RG/RGG motif-containing proteins, such as FMRP (29), hnRNP A1 (28) and SERBP1 (27).
The Tudor domain of TDRD3 is involved in the interaction with TOP3B
Methylated arginine creates a docking site for protein-protein interaction with the Tudor domain-containing proteins (30). We found that the Tudor domain of TDRD3 interacts with TOP3B in a methylation-dependent manner and that methylation enhances TDRD3–TOP3B interaction (Figure 6). TDRD3 is by far the most significant regulator of TOP3B function. It regulates TOP3B protein stability (6), chromatin localization (6), association with polyribosomes (8,9) and topoisomerase activity (10). Although previous studies demonstrated that the catalytic domain (amino acid 125–625) of TOP3B is sufficient for TDRD3 interaction (6,8), the extent to which this interaction contributes to TOP3B stress granule localization has not been determined. We examined the stress granule localizations of three GFP-fusion TOP3B truncations that were previously used for mapping TDRD3 interaction (6). Surprisingly, the TDRD3 interacting catalytic domain of TOP3B does not exhibit obvious stress granule localization. Instead, the GFP-fusion TOP3B C-terminal domain (amino acid 600–862) does (Supplementary Figure S7), arguing that the C-terminal domain is important for TOP3B stress granule localization. Further experiments are needed to determine the extent to which Tudor domain-mediated interaction with arginine methylated C-terminal domain contributes to TOP3B stress granule localization. Nevertheless, RG/RGG motif methylation-mediated interactions with the Tudor domain may provide additional regulatory flexibility for this protein complex.
Recently, TDRD3 was identified as a nucleic acid binding protein that has a strong preference for single-stranded DNA and/or RNA structures (10). This activity of TDRD3 significantly stimulates the strand-annealing activity of TOP3B. Although a possible mechanism was proposed involving the TDRD3 OB-fold mediated interaction with both TOP3B and nucleic acids, further biochemical studies are required to determine if the Tudor domain mediated interaction with the methylated C-terminal RG/RGG motif may also contribute to this process.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Xiaochun Yu and members of the Yang laboratory for their helpful discussions on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
V Scholar Award from the V Foundation for Cancer Research (to Y.Y.); City of Hope Start-up Fund (to Y.Y.); Core facility support grant from National Cancer Institute of the National Institutes of Health [P30CA33572]. Funding for open access charge: City of Hope Start-up Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen S.H., Chan N.L., Hsieh T.S.. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 2013; 82:139–170. [DOI] [PubMed] [Google Scholar]
- 2. Pommier Y., Sun Y., Huang S.N., Nitiss J.L.. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016; 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker N.M., Rajan R., Mondragon A.. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009; 37:693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachrati C.Z., Hickson I.D.. Dissolution of double Holliday junctions by the concerted action of BLM and topoisomerase IIIalpha. Methods Mol. Biol. 2009; 582:91–102. [DOI] [PubMed] [Google Scholar]
- 5. Yang Y., Lu Y., Espejo A., Wu J., Xu W., Liang S., Bedford M.T.. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell. 2010; 40:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang Y., McBride K.M., Hensley S., Lu Y., Chedin F., Bedford M.T.. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell. 2014; 53:484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 8. Xu D., Shen W., Guo R., Xue Y., Peng W., Sima J., Yang J., Sharov A., Srikantan S., Yang J. et al. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013; 16:1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stoll G., Pietilainen O.P., Linder B., Suvisaari J., Brosi C., Hennah W., Leppa V., Torniainen M., Ripatti S., Ala-Mello S. et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013; 16:1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siaw G.E., Liu I.F., Lin P.Y., Been M.D., Hsieh T.S.. DNA and RNA topoisomerase activities of Top3beta are promoted by mediator protein Tudor domain-containing protein 3. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E5544–E5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thandapani P., O’Connor T.R., Bailey T.L., Richard S.. Defining the RGG/RG motif. Mol. Cell. 2013; 50:613–623. [DOI] [PubMed] [Google Scholar]
- 12. Ahmad M., Shen W., Li W., Xue Y., Zou S., Xu D., Wang W.. Topoisomerase 3beta is the major topoisomerase for mRNAs and linked to neurodevelopment and mental dysfunction. Nucleic Acids Res. 2017; 45:2704–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bedford M.T., Clarke S.G.. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009; 33:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang Y., Bedford M.T.. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013; 13:37–50. [DOI] [PubMed] [Google Scholar]
- 15. Yang Y., Hadjikyriacou A., Xia Z., Gayatri S., Kim D., Zurita-Lopez C., Kelly R., Guo A., Li W., Clarke S.G. et al. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 2015; 6:6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boisvert F.M., Dery U., Masson J.Y., Richard S.. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005; 19:671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boisvert F.M., Hendzel M.J., Masson J.Y., Richard S.. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle. 2005; 4:981–989. [DOI] [PubMed] [Google Scholar]
- 18. Blackwell E., Zhang X., Ceman S.. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 2010; 19:1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Narayanan N., Wang Z., Li L., Yang Y.. Arginine methylation of USP9X promotes its interaction with TDRD3 and its anti-apoptotic activities in breast cancer cells. Cell Discov. 2017; 3:16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hung M.L., Hautbergue G.M., Snijders A.P., Dickman M.J., Wilson S.A.. Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1. Nucleic Acids Res. 2010; 38:3351–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee J., Bedford M.T.. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002; 3:268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eram M.S., Shen Y., Szewczyk M., Wu H., Senisterra G., Li F., Butler K.V., Kaniskan H.U., Speed B.A., Dela Sena C. et al. A potent, selective, and cell-active inhibitor of human Type I protein arginine methyltransferases. ACS Chem. Biol. 2016; 11:772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dhar S., Vemulapalli V., Patananan A.N., Huang G.L., Di Lorenzo A., Richard S., Comb M.J., Guo A., Clarke S.G., Bedford M.T.. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 2013; 3:1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seki T., Seki M., Onodera R., Katada T., Enomoto T.. Cloning of cDNA encoding a novel mouse DNA topoisomerase III (Topo IIIbeta) possessing negatively supercoiled DNA relaxing activity, whose message is highly expressed in the testis. J. Biol. Chem. 1998; 273:28553–28556. [DOI] [PubMed] [Google Scholar]
- 25. Wilson T.M., Chen A.D., Hsieh T.. Cloning and characterization of Drosophila topoisomerase IIIbeta. Relaxation of hypernegatively supercoiled DNA. J. Biol. Chem. 2000; 275:1533–1540. [DOI] [PubMed] [Google Scholar]
- 26. Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chedin F.. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012; 45:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee Y.J., Wei H.M., Chen L.Y., Li C.. Localization of SERBP1 in stress granules and nucleoli. FEBS J. 2014; 281:352–364. [DOI] [PubMed] [Google Scholar]
- 28. Wall M.L., Lewis S.M.. Methylarginines within the RGG-motif region of hnRNP A1 affect its IRES trans-acting factor activity and are required for hnRNP A1 stress granule localization and formation. J. Mol. Biol. 2017; 429:295–307. [DOI] [PubMed] [Google Scholar]
- 29. Xie W., Denman R.B.. Protein methylation and stress granules: posttranslational remodeler or innocent bystander. Mol. Biol. Int. 2011; 2011:137459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen C., Nott T.J., Jin J., Pawson T.. Deciphering arginine methylation: tudor tells the tale. Nat. Rev. Mol. Cell Biol. 2011; 12:629–642. [DOI] [PubMed] [Google Scholar]
- 31. Cote J., Richard S.. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005; 280:28476–28483. [DOI] [PubMed] [Google Scholar]
- 32. Lasko P. Tudor domain. Curr. Biol. 2010; 20:R666–R667. [DOI] [PubMed] [Google Scholar]
- 33. Ahmad M., Xue Y., Lee S.K., Martindale J.L., Shen W., Li W., Zou S., Ciaramella M., Debat H., Nadal M. et al. RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res. 2016; 44:6335–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saviozzi S., Ceppi P., Novello S., Ghio P., Lo Iacono M., Borasio P., Cambieri A., Volante M., Papotti M., Calogero R.A. et al. Non-small cell lung cancer exhibits transcript overexpression of genes associated with homologous recombination and DNA replication pathways. Cancer Res. 2009; 69:3390–3396. [DOI] [PubMed] [Google Scholar]
- 35. Corley S.M., Gready J.E.. Identification of the RGG box motif in Shadoo: RNA-binding and signaling roles. Bioinform. Biol. Insights. 2008; 2:383–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanakahi L.A., Sun H., Maizels N.. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999; 274:15908–15912. [DOI] [PubMed] [Google Scholar]
- 37. Zanotti K.J., Lackey P.E., Evans G.L., Mihailescu M.R.. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA. Biochemistry. 2006; 45:8319–8330. [DOI] [PubMed] [Google Scholar]
- 38. Vasilyev N., Polonskaia A., Darnell J.C., Darnell R.B., Patel D.J., Serganov A.. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5391–E5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blackwell E., Ceman S.. A new regulatory function of the region proximal to the RGG box in the fragile X mental retardation protein. J. Cell Sci. 2011; 124:3060–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ina S., Tsunekawa N., Nakamura A., Noce T.. Expression of the mouse Aven gene during spermatogenesis, analyzed by subtraction screening using Mvh-knockout mice. Gene Expr. Patterns. 2003; 3:635–638. [DOI] [PubMed] [Google Scholar]
- 41. Zullo A.J., Michaud M., Zhang W., Grusby M.J.. Identification of the small protein rich in arginine and glycine (SRAG): a newly identified nucleolar protein that can regulate cell proliferation. J. Biol. Chem. 2009; 284:12504–12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bedford M.T., Sarbassova D., Xu J., Leder P., Yaffe M.B.. A novel pro-Arg motif recognized by WW domains. J. Biol. Chem. 2000; 275:10359–10369. [DOI] [PubMed] [Google Scholar]
- 43. Bedford M.T., Frankel A., Yaffe M.B., Clarke S., Leder P., Richard S.. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 2000; 275:16030–16036. [DOI] [PubMed] [Google Scholar]
- 44. Kowanetz K., Szymkiewicz I., Haglund K., Kowanetz M., Husnjak K., Taylor J.D., Soubeyran P., Engstrom U., Ladbury J.E., Dikic I.. Identification of a novel proline-arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors. J. Biol. Chem. 2003; 278:39735–39746. [DOI] [PubMed] [Google Scholar]
- 45. Tripsianes K., Madl T., Machyna M., Fessas D., Englbrecht C., Fischer U., Neugebauer K.M., Sattler M.. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat. Struct. Mol. Biol. 2011; 18:1414–1420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.