Abstract
RNA–DNA hybrid primers synthesized by low fidelity DNA polymerase α to initiate eukaryotic lagging strand synthesis must be removed efficiently during Okazaki fragment (OF) maturation to complete DNA replication. In this process, each OF primer is displaced and the resulting 5′-single-stranded flap is cleaved by structure-specific 5′-nucleases, mainly Flap Endonuclease 1 (FEN1), to generate a ligatable nick. At least two models have been proposed to describe primer removal, namely short- and long-flap pathways that involve FEN1 or FEN1 along with Replication Protein A (RPA) and Dna2 helicase/nuclease, respectively. We addressed the question of pathway choice by studying the kinetic mechanism of FEN1 action on short- and long-flap DNA substrates. Using single molecule FRET and rapid quench-flow bulk cleavage assays, we showed that unlike short-flap substrates, which are bound, bent and cleaved within the first encounter between FEN1 and DNA, long-flap substrates can escape cleavage even after DNA binding and bending. Notably, FEN1 can access both substrates in the presence of RPA, but bending and cleavage of long-flap DNA is specifically inhibited. We propose that FEN1 attempts to process both short and long flaps, but occasional missed cleavage of the latter allows RPA binding and triggers the long-flap OF maturation pathway.
INTRODUCTION
All processes involving breaks in DNA, including replication, repair, recombination, transcription and related cell cycle progression, have the potential to cause genome instability and lead to disease states (1). Hence, proteins involved in these multi-step DNA metabolic pathways often function in a coordinated manner to minimize and resolve DNA breaks, and maintain genome stability (2,3). One such process is Okazaki fragment (OF) maturation, required to complete discontinuous lagging strand DNA replication (4). In eukaryotes, DNA polymerase α (Pol α) synthesizes an 8–12 nucleotide long initiator RNA (iRNA) and extends it with ∼20–30 deoxyribonucleotides (nt) to create an iRNA–DNA primer that is handed off to polymerase δ (Pol δ), which in turn extends it to an ∼200 nt OF. Approximately 10 million OFs are generated per cell cycle, all of which have to be linked efficiently during OF maturation to generate a continuous lagging strand. During this process the iRNA must be removed and the DNA fragments ligated; moreover, since Pol α lacks proofreading activity, misincorporated bases in the DNA primer need to be corrected. Thus, proper OF maturation is critical for maintaining the integrity of the genome (4,5).
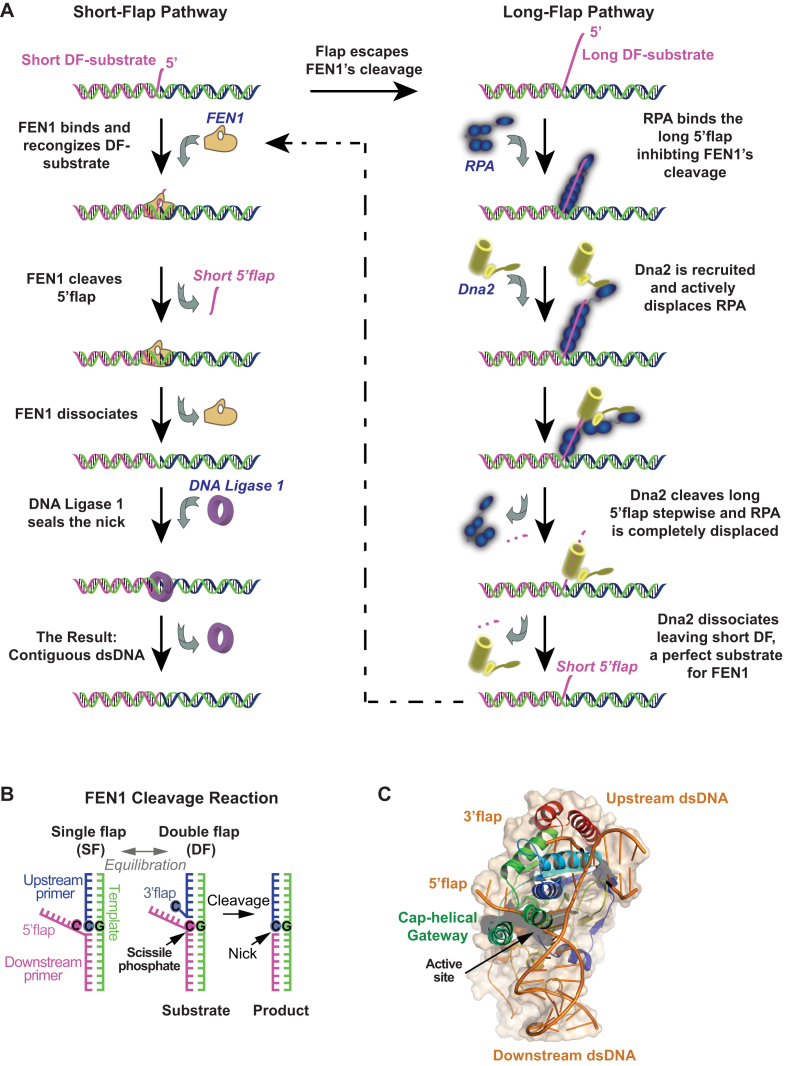
OF maturation begins when Pol δ reaches the end of a nascent OF and displaces the 5′ end of a previous fragment to generate a 5′ single-stranded iRNA/DNA flap of varying lengths protruding from a nick junction (Figure 1A and B). Complementarity between the 5′ flap and the template strand could potentially equilibrate the nick junction to form a 1-nt 3′ flap (Figure 1B) (6–8). This double flap (DF) structure is recognized by structure specific 5′ endonucleases and cleaved 1 nt into the junction to form a nick that is sealed by DNA Ligase 1 (Figure 1B). Flap Endonuclease 1 (FEN1) is primarily responsible for the flap cleavage reaction, although other pathways have been proposed depending on the flap length (for reviews, refer to (4,5,9,10)). The primary pathway involves FEN1 recognizing and cleaving short 5′ flaps while the secondary and less frequent pathway is invoked for processing longer flaps (Figure 1A). Pol δ has limited strand displacement activity, and can idle back and forth between its polymerase and exonuclease activities on the primer, thus limiting flap length. Indeed, Garg et al. (11) and Stodola and Burgers (12) have suggested that the main product of FEN1 cleavage is a monoribonucleotide resulting from an active hand-off mechanism between Pol δ and FEN1 (nick translation), in which incremental strand displacement by Pol δ and cleavage by FEN1 are tightly coupled. Nevertheless, long flaps do occur as evident from both in vitro and in vivo studies (13–17). In the secondary pathway proposed for long flaps, the iRNA/DNA strand is bound by replication protein A (RPA), FEN1 cleavage is inhibited and Dna2 helicase/nuclease is recruited to the site (18,19). Dna2 displaces RPA (20) and progressively cleaves the flap to make it shorter. At this point, Dna2 dissociates due to its lower affinity for the short flap (14) or FEN1 disengages Dna2 (21,22), and the DNA becomes a substrate for FEN1 again (Figure 1A). Notably, despite the relative infrequency of long-flap processing events, Dna2 is an essential enzyme in Saccharomyces cerevisiae while FEN1 is not (23,24). This rather counterintuitive finding can be explained by the presence of backup endonucleases such as RNaseH2 and Exo1 that can substitute for FEN1 on short flaps (25,26). On the other hand, there appears to be no equivalent substitute for Dna2 activity on long flaps, and since OF maturation defects have drastic consequences, Dna2 is essential.
Figure 1.
FEN1 actions in OF maturation. (A) Short- and long-flap pathways for processing OF. Left: schematic of the major short-flap pathway. A short 5′ flap (1–6 nt) created by limited strand displacement activity of DNA polymerase δ is recognized and cleaved by FEN1 to generate a nick that is sealed by DNA ligase 1. Right: schematic of the minor long-flap pathway. If a longer flap is formed, it is tightly bound by RPA, which inhibits FEN1 and necessitates Dna2 involvement. Dna2 displaces RPA and cleaves the long flap progressively until it is too short to maintain Dna2 binding and becomes a perfect substrate for FEN1 in the short-flap pathway. (B) Recognition and cleavage of short flaps by FEN1 in vivo involves flap equilibration. Strand complementarity between the downstream and upstream primers enables the nick junction to equilibrate. FEN1 binds this substrate and sculpts a 1 nt 3′ flap (8), resulting in a DF structure that is incised 1 nt inside the junction to generate a sealable nick. (C) Crystal structure of FEN1 in complex with a 5′ flap substrate. FEN1 makes extensive interactions with the upstream and downstream duplex regions flanking the nick junction. The DNA is bent at the junction by ∼100°, and the 5′ flap is threaded through a cap-helical gateway structure that oversees the active site. Binding of the 3′ flap transitions the gateway from a disordered to ordered form, providing a mechanism to control active site assembly (PDB accession code: 5UM9) (37).
Deletion mutations of Pif1 and the third subunit of Pol δ (Pol32) in S. cerevisiae, as well as post-translational modifications (especially acetylation) of human FEN1, Dna2 and Pol δ shed some light on how and why long flaps might occur during DNA replication. Pif1, a 5′-3′ helicase, has been shown to augment the strand displacement capacity of Schizosaccharomyces pombe Pol δ by unwinding the previous OF end (27). A similar finding has been reported for S. cerevisiae Pol δ in the presence of Pol32 as well, compared to the 2-subunit Pol δ (28). Hence, both Pol32 and Pif1 can increase the likelihood of generating longer flaps. Indeed, deletion of either protein rescues the lethality caused by deletion of Dna2 in S. cerevisiae, although ΔPol32 has a lesser effect than ΔPif1 (28), overall suggesting a lower requirement for Dna2 activity when there are fewer long flaps. Acetylation of FEN1, Dna2 and Pol δ by histone acetyltransferase p300 also provides information on the generation and processing of long flaps. While complete acetylation of FEN1 reduces its cleavage activity by about 90% (29), the same modification stimulates by many folds Dna2 helicase and nuclease activities (30) and is suggested to enhance Pol δ strand displacement activity (5,31). Thus, acetylation of these lagging strand DNA replication proteins favors the long-flap pathway. One might consider the long-flap pathway an option for removing the full error-containing iRNA/DNA primer synthesized by low fidelity Pol α. On the other hand, processing a large fraction of OFs by this multi-protein, multi-step pathway that requires significant DNA re-synthesis at each OF by Pol δ might result in untenable delays in OF maturation. It is possible that the long-flap pathway is operational in select regions of the genome (reviewed by Balakrishnan and Bambara (5)). Since p300 acetylase exhibits a preference for chromatin regions with active gene expression, acetylation of replication/repair proteins may invoke the long-flap pathway at these sites (5).
Given that longer flaps have been detected in vivo and in vitro, we set out to understand how FEN1 acts on them, and whether FEN1 actions influence the choice between short- and long-flap pathways. Specifically, we asked whether FEN1 bends long flaps and catalyzes their cleavage in a similar manner as short flaps, and whether its mechanism is affected by RPA, which can compete with FEN1 for binding long flaps.
Recent studies have provided richly detailed information on the FEN1 mechanism of action. FEN1 initially binds and bends its flap substrate to an angle of 100° at the nick junction (Figure 1C) (8,32–34). The FEN1 substrate recognition mechanism protects the template strand against inadvertent incision through a cap-helical gateway structure that oversees the active site. The cap-helical gateway selects for threading of 5′ flaps with free ends (Figure 1C). Blocking 5′ flap threading with biotin/streptavidin at the end of the 5′ flap demonstrates that threading is a prerequisite for catalysis (35). Single molecule experiments with such substrates show that FEN1 achieves a weak bent state that cannot support cleavage (8). Furthermore, the conformer of the blocked 5′ flap (33) and the degree of DNA bending (8) vary from that in the threaded complex. These single molecule findings are consistent with time-resolved crystallography data on human exonuclease 1 (Exo1), which illustrate step-wise threading of the 5′ flap through the cap-helical gateway that is coupled with progressive strengthening of the interactions with the bent DNA (36). In case of both FEN1 and Exo1, the flap strand is inverted such that the phosphates are turned away from the active site metal ions, which protects the strand from inadvertent incision during threading (36,37). Threading can also be viewed in the context of the regulation and choice between the short- and long-flap pathways; for instance, RPA-coated flaps may be blocked from threading, thus limiting catalysis. Collectively, these studies highlight the importance of 5′ flap threading in FEN1 catalytic cycle, and the possible consequences of 5′ flap length and structure on the reaction mechanism.
Recently, using DF substrates containing short 5′ flaps, we showed that the DNA exists in an extended conformer that is actively bent by FEN1 in diffusion-limited kinetics (8). This bending appears to induce ordering and assembly of the FEN1 active site associated with binding of the 1-nt 3′ flap in the 3′ flap-binding pocket (Figure 1C) (8,32,38,39). This DNA–protein induced fit mechanism leads to a catalytically competent state in the first encounter between FEN1 and a cognate DF substrate. However, in the case of a non-cognate substrate, the probability of forming the catalytically competent state is lower, which favors DNA dissociation prior to flap cleavage (8). In this study, we examined equilibrated (EQ) DF substrates of different 5′ flap lengths by single molecule F?rster Resonance Energy Transfer (smFRET) and rapid quench-flow bulk cleavage assays to determine whether and how the mechanism of action of human FEN1 might vary with flap length. We found that the efficiency with which FEN1 binds and bends a longer flap containing DNA is not significantly affected, and its single turnover cleavage rate is slightly decreased. Importantly, as flap length increases, the probability of flap cleavage within the first DNA binding/bending encounter decreases. In other words, FEN1 has a higher chance of dissociating from the substrate without cleaving as flap length increases. These missed cleavage events create an opportunity for RPA to bind the flap and inhibit FEN1, thus necessitating involvement of Dna2 and initiation of the long-flap pathway.
MATERIALS AND METHODS
Proteins expression and purification
Human FEN1 (amino acids: 2−380) was cloned and expressed as described previously for single molecule (8) and ensemble experiments (40). Human RPA clone (pET11d-tRPA) was a generous gift of Professor Marc S. Wold, and the protein was purified as described (41). Briefly, the plasmid was expressed in BL21 (DE3) Escherichia coli cells and RPA was purified on a HiTrap blue HP column, followed by desalting and concentrating on a hydroxyapatite column, and finally ion exchange chromatography on a MonoQ column. Protein fractions were pooled, flash frozen and stored at −80°C.
DNA substrates
DNA oligos were synthesized and HPLC purified by Integrated DNA technologies (IDT). Substrates were annealed by mixing template: 5′ flap: 3′ flap strands in 1: 3: 5 molar ratio in TE-100 buffer (50 mM Tris–HCl pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA) pH 8.0, 100 mM NaCl), and heating at 95°C for 5 min followed by slow cooling to room temperature. For longer flaps used in cleavage assays, 10 mM MgCl2 was added to the annealing buffer. Substrates were purified to >80% purity by non-denaturing polyacrylamide gel electrophoresis (PAGE) and eluted using the crush and soak method in TE-100 buffer, shaking at 16°C for 30 min. Eluted substrates were passed through 0.2 μm filters, aliquoted and stored at −20°C. A list of these substrates is illustrated in Supplementary Figure S5A.
TIRF-based smFRET
Glass coverslips were functionalized and passivated by 1:100 molar ratio of biotinylated polyethylene glycol (Biotin-PEG-SVA MW 5,000) and polyethylene glycol (mPEG-SVA MW 5000) (Laysan Bio Inc.). These functionalized coverslips, along with a quartz slide, were used to construct an airtight microfluidic flow cell with a 3 mm-wide channel of polyethylene double-sided tape SA-S-1L (100 μm thick) (Secure-Seal, Grace BioLabs) sandwiched in between. This flow cell had one inlet and one outlet tubing attached for exchange of buffer.
For immobilization of the double-labeled biotinlated DNA substrates, just prior to any experiment, the flow cell was incubated with 0.2 mg/ml NeutrAvidin for 10–15 min. This treatment was followed by excessive washing with reaction buffer to remove excess NeutrAvidin and to block extra binding sites. The reaction buffer contained 50 mM HEPES-KOH pH 7.5, 5% (v/v) glycerol, 1 mM Dithiothreitol (DTT), 0.1 mg/ml bovine serum albumin (BSA), 100 mM KCl and 10 mM salt containing divalent cations (CaCl2 or MgCl2 to assess bending or cleavage efficiency, respectively). The DNA substrates were then immobilized on the surface (∼100–200 pM) until optimal coverage was reached, followed by excessive washing. Flowing DNA substrates and any subsequent steps were performed under imaging buffer (reaction buffer + oxygen scavenging solution).
To enhance the photostability of the fluorophores under our imaging conditions, we used an oxygen scavenging solution as described earlier (42), which relies on the enzymatic elimination of oxygen through a 6 mM proto-catechuic acid (Sigma-Aldrich, P5630) and 60 nM protocatechuate-3,4-dioxygenase system. To reduce the photo-blinking effect, we included 2 mM of the triplet-state quencher, Trolox (Sigma-Aldrich), in our imaging buffer.
All single molecule experiments were performed using a custom-built TIRF-FRET setup (43). Several movies on different fields of view were recorded for the DNA substrates and different protein additions using two-color alternating excitation (2c-ALEX) between green and red laser with a 160 ms time resolution and/or continuous excitation through green laser (CW) with a 50 or 100 ms time resolution. For each experiment, a transformation matrix file was constructed by imaging a diluted sample of fluorescent beads (FluoSpheres, F8810 Invitrogen) in TIRF mode, then linking each particle in the green channel to its corresponding pair in the red channel. This transformation matrix was then used through twotone software (44) to map the donor and acceptor positions. In this process, certain restrictions concerning the brightness of the donor and acceptor, the distance between the centers of two adjacent particles, as well as the clustering distance are applied to ensure the particles are not too dim or too bright and are well separated. The software then extracts donor and acceptor intensities by measuring the photon counts from the two-dimensionally Gaussian-fitted point spread functions in both channels. These intensities are used by the software to calculate the apparent FRET.
Ca2+-based DNA bending assays
These experiments were performed under the conditions described above in the presence of CaCl2. For each condition, DNA-only and subsequent protein titrations, at least three 2c-ALEX and three CW movies were recorded. The histograms of apparent FRET efficiencies were constructed based on 2c-ALEX movies as described (45). These histograms were analyzed and fitted using OriginPro software to determine the center of FRET peaks and to integrate the percentage of each peak. The percentages of the bent-state peak versus FEN1 concentration were plotted and fitted to a one-site binding model using GraphPad Prism software, and Kd-bending was estimated using the constraint Bmax ≤ 100. For dwell time analysis of the time traces, CW movies were analyzed using the vbFRET package implemented in Matlab (46), where the time traces were idealized and fitted to two FRET states (bent and unbent). vbFRET is based on Hidden Markov Modeling where the most likely FRET values are determined based on probability alone. The dwell times spent in each state were plotted in histograms and fitted with a single exponential decay yielding kbending (1/τbending) and kunbending (1/τunbending). kbending was plotted versus different FEN1 concentrations to obtain kon-bending from the slope of the linear fit. koff-bending was obtained from the average of kunbending at different FEN1 concentrations.
Single molecule cleavage assays
smFRET cleavage assays were performed using two labeling schemes, flap and internal. In both cases, DNA substrate immobilization and all subsequent steps were performed with imaging buffer containing 10 mM MgCl2. For experiments with RPA, the DNA-immobilized surface was pre-incubated with sufficient RPA before co-injection of RPA and FEN1; 250 nM FEN1 was injected into the chamber with or without RPA. In all experiments, recording movies with single excitation by green laser at 50 ms temporal resolution was started before the proteins reached the microfluidic flow cell. In both labeling schemes, time traces were manually screened for cleavage events.
For the flap-labeling scheme, we followed the protocol described in our previous study (8). Briefly, a cleavage event was identified as a transition from the bent to unbent FRET state (with clear anti-correlation between the donor and acceptor intensities), followed by a single-step loss of signal of both donor and acceptor. For the internal-labeling scheme, a cleavage event was identified by following transition from the unbent substrate FRET to that of the bent substrate and then the unbent nicked product. The unbent substrate and product FRET states showed a difference of ∼0.05 as evident in the DNA-only histograms of the DF substrates and nicked duplex in CaCl2 experiments. The dwell times of the substrate spent in bent state before loss of signal (in the flap-labeling scheme), and the dwell times spent in bent state before transitioning to the FRET state of the nick (in the internal-labeling scheme) were calculated by manually counting the frames in the bent state. We opted for manual counting of frames rather than automated fitting as these dwell times were too short to be picked accurately by most available tools. In both cases, distributions of the dwell times were plotted and fitted by gamma distribution using Matlab dfittool and the mean and the standard error of the mean were reported.
The internal-labeling scheme was also used to assess missed cleavage events. An event where bending occurred but the FRET state shifted back to that of the unbent DF substrate rather than the unbent nicked duplex was considered a missed event. Dwell times of the missed events were calculated as described above for cleavage events. The percentage of missed cleavage was calculated as the percentage of particles (rather than events) that showed at least one missed event.
PIFE bulk and single molecule experiments
For bulk time-resolved fluorescence lifetime measurements, a QuantaMaster 800 spectrofluorometer (Photon Technology International Inc.), coupled with a supercontinuum fiber laser source, was used. Cy3 fluorescence lifetime measurements of non-EQ DF-6,1PIFE in the absence and presence of 1 μM FEN1 were performed at room temperature in time-correlated single-photon counting mode. Cy3 was excited at 532 nm and emission was collected at 568 nm with 5 nm-wide slits for both excitation and emission. To reduce collection of scattered light, a longpass filter of 550 nm was placed on the emission side. A suspension of colloidal silica was utilized to estimate the instrument response function (IRF). In all measurements, 10 000 counts were acquired. Cy3 fluorescence lifetime in both cases was then determined by a two-exponential decay fit using FluoFit software package (PicoQuant) that implements the IRF reconvolution. The best fit was achieved with a reduced chi-square and randomness of the residuals.
Single molecule protein-induced fluorescence enhancement (smPIFE) experiments followed the standard conditions used for smFRET cleavage reactions with non-EQ DF-6,1PIFE as the substrate. The data was analyzed in a similar fashion and the number of frames a molecule spent in the enhanced-fluorescence state were counted manually and fitted to a gamma distribution.
Ensemble single turnover and steady state cleavage assays
FEN1 cleavage activity on EQ DF substrates of varying lengths (DF-6,1, DF-30,1, DF-50,1, DF-60,1) was measured by bulk single turnover experiments using a rapid quench-flow apparatus (RQF-3; KinTek Corp.). The 5′ end of the 5′ flap strand was modified with Fluorescein (FAM) dye, and substrates were prepared by mixing 5′ flap: template: 3′ flap strands in 1: 1.5: 2.5 ratio, heating the mixture at 80°C for 10 min followed by spinning down and cooling O/N to 25°C for a yield of 75–85% (annealing buffer: 50 mM HEPES-KOH, pH 7.5, 100 mM KCl, 10 mM MgCl2). A list of the substrates is shown in Supplementary Figure S5B.
In each experiment, 15 μl of FEN1 was mixed with 15 μl of DNA, incubated at 37°C for varying times and then quenched with 76 μl of 200 mM EDTA (final reaction conditions: 1.2 μM FEN1, 0.035 μM DNA in buffer containing 50 mM HEPES-KOH, pH 7.5, 40 or 100 mM KCl, 10 mM MgCl2, 0.1 mg/ml BSA, 5% glycerol, 1 mM DTT). The quenched reactions were placed on ice until analysis by denaturing PAGE. A 25 μl aliquot of each reaction was mixed with an equal volume of denaturing dye (0.3% bromophenol blue, 12 mM EDTA in formamide), heated at 95°C for 1 min and run on an 18% denaturing urea polyacrylamide gel (3.25 × 3 × 0.15 cm) for 40 min at 12 W. The substrate and 5′-flap product were quantified on a Typhoon scanner (λEx = 488 nm). The fraction of product formed was plotted versus time, and the data were fit to a single exponential equation by GraphPad Prism to obtain the cleavage rate (kSTO).
In steady state experiments, 1 nM FEN1 was mixed with 800 nM DNA substrate (10×KM; (47) at 37°C, and 15 μl aliquots of the reaction were mixed with 4 μl of 100 mM EDTA at varying times to quench the reaction and determine initial velocity. The substrates and products were resolved and quantified as described above, and the data fit to a linear equation yielded the kcat (slope/[FEN1]).
Steady state bulk cleavage in the presence of RPA
EQ DF substrates of varying flap lengths (DF-2,1, DF-6,1 and DF-30,1) with Atto647 dye at the 3′ end of the 5′ flap oligo (substrates shown in Supplementary Figure S5C) were used to assess FEN1 cleavage efficiency in the absence and presence of RPA. For direct comparison, the assay was performed as described previously for yeast proteins (48). Briefly, 0.1 nM FEN1 and increasing RPA (0, 0.75, 1.25, 2.5 or 5 nM) were pre-mixed in buffer containing 30 mM HEPES-KOH, 0.5% inositol, 40 mM KCl, 4 mM MgCl2, 0.01% Nonidet P-40, 0.1 mg/ml BSA, 1 mM DTT and 5% glycerol. DNA substrates were added at 0.25 nM concentration to initiate the reaction. The 20 μl reactions were incubated at 37°C for 10 min before quenching with an equal volume of 2× buffer (90% deionized formamide, 100 mM EDTA). The samples were then heated at 95°C for 10 min, cooled immediately on ice and the products were resolved by 20% denaturing urea PAGE and quantified on a Typhoon TRIO Variable Mode Imager (GE Healthcare, Life Sciences).
RESULTS
In this study, we set out to investigate whether 5′ flap length affects FEN1 substrate recognition and catalytic efficiency, and whether any variation in FEN1 activity might influence the choice between short- versus long-flap pathways for OF maturation. We addressed these questions by measuring transient events in the reaction using smFRET complemented with bulk cleavage experiments and determining the FEN1 kinetic mechanism on short and long 5′ flaps, both in the absence and presence of RPA.
Effect of 5′ flap length on the DNA bending activity of FEN1
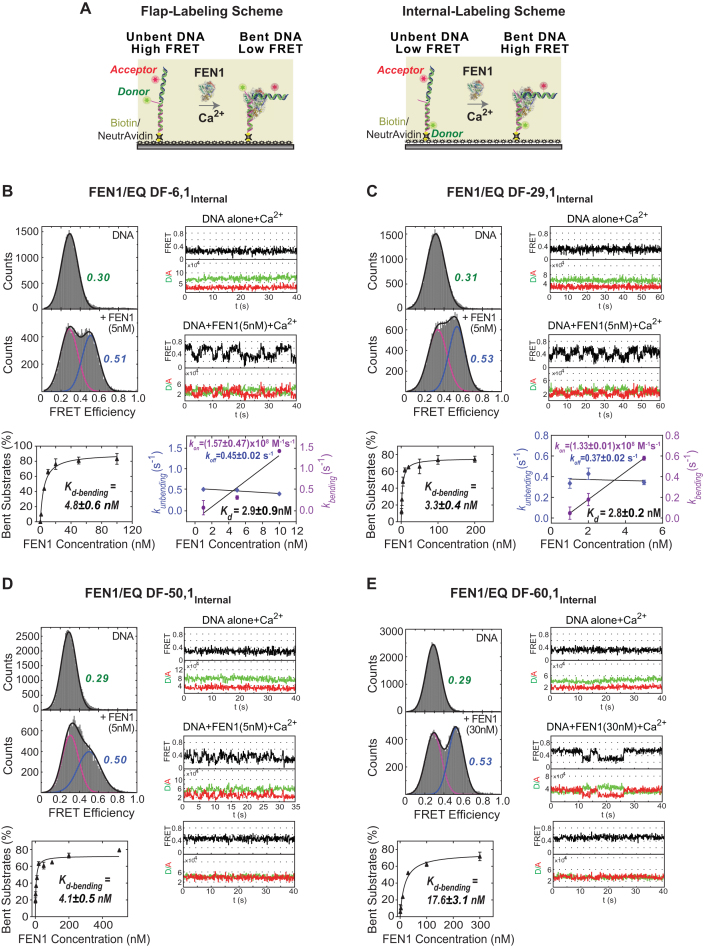
All DF substrates used in this study are capable of flap equilibration, i.e. the template base at the nick junction is complemented by both upstream and downstream strands to generate either a single flap or DF substrate (Figure 1B). EQ DF substrates best reflect the dynamicity of flap substrates encountered by FEN1 in vivo; although, it has been shown in vitro that FEN1 cleaves non-EQ DF substrates with similar single turnover rate (kSTO) as EQ DF (8,49). We used two DNA labeling schemes for the smFRET assays, namely ‘flap-labeling’ and ‘internal-labeling’. In the flap-labeling scheme (Figure 2A), the DF substrate has a Cy3 donor at the 5′-end of the flap and an Alexa Fluor 647 acceptor at nucleotide position 12 upstream of the nick junction. In the internal-labeling scheme, the dyes are located on either side of the nick junction, with Cy3 at position 15 downstream and Alexa Fluor 647 at position 12 upstream of the nick (Figure 2A). These two labeling schemes provide complementary views of DNA binding, bending, cleavage and product release steps in the reaction, as described below.
Figure 2.
Effect of 5′ flap length on FEN1 DNA bending activity. (A) Schematic of smFRET assay with two labeling schemes used to study FEN1 bending. The left panel shows the flap-labeling scheme with Cy3 donor at the end of 5′ flap and Alexa Fluor 647 acceptor in the upstream primer of the DF substrate; the FRET ratio decreases upon FEN1 binding and bending. The right panel shows the internal-labeling scheme where both donor and acceptor are in the template strand; the FRET ratio increases upon FEN1 binding and bending. (B) FEN1 bending efficiency of internally labeled EQ DF-6,1 (EQ DF-6,1Internal). Top left panel shows smFRET histograms of EQ DF-6,1Internal alone and upon addition of 5 nM FEN1. The histograms were fitted by one or two Gaussian distributions for DNA-only and DNA + FEN1, respectively. In the DNA + FEN1 histogram, the fitted unbent peak (shown in magenta) has same FRET center as DNA-only, and the fitted bent peak is shown in blue. Top right panel shows representative single molecule time traces of EQ DF-6,1Internal alone and upon addition of 5 nM FEN1. Bottom left panel is an isotherm of percentage of bent substrate (%) versus FEN1 concentration (nM). The percentage of bent substrates was estimated by the Gaussian-fitted bent peak for three replicates at each FEN1 concentration. The isotherm was fitted to a one-site binding model with Bmax ≤ 100 and yielded the DNA bending dissociation constant (Kd-bending). Error bars reflect variation of the % bent substrate from the three replicates and the reported error is the error of the fit. Bottom right panel shows a graph of kbending (s−1) and kunbending (s−1) versus FEN1 concentrations (nM). At each FEN1 concentration, time traces were idealized and fit by vbFRET to calculate the dwell times τbending and τunbending spent in the bent and unbent states, respectively. The histograms from the population of the dwell times were fit to exponential functions yielding kbending (1/τbending) and kunbending (1/τunbending). The association rate constant (kon-bending) was calculated from the slope of the linear regression fit of kbending versus FEN1 concentration. The dissociation rate constant (koff-unbending) was calculated as the mean of kunbending. The error bars correspond to the standard error of the exponential fit of kbending and kunbending and the errors reported for the association/dissociation constants correspond to the error of the fit and the standard error of the mean, respectively. Kd-bending = koff-unbending/kon-bending. (C) FEN1 bending efficiency of EQ DF-29,1Internal as described in B. (D) FEN1 bending efficiency of EQ DF-50,1Internal as described in B. (E) FEN1 bending efficiency of EQ DF-60,1Internal as described in B.
First, we assessed the ability of FEN1 to bind and bend DNA substrates with varying flap lengths. FEN1 was titrated with internally labeled EQ DF substrates with 6, 29, 50 or 60 nt long 5′ flaps under non-catalytic conditions in the presence of Ca2+ ions. Histograms were constructed for each DF substrate as described in the ‘Materials and Methods’ section. The FRET peaks were centered around 0.30 for all unbent DF substrates and around 0.52 for the FEN1-bent substrates (Figure 2B–E). This result indicates that the flap length does not affect the basic backbone structure of the duplex portion of the substrate, whether free or bound to FEN1. The percentage of bent substrate at each FEN1 concentration was determined by the integrated area of the Gaussian-fitted bent peak, and plotted versus FEN1 concentration to generate the isotherms shown in Figure 2B–E. The data yielded the following DNA bending dissociation constants (Kd-bending): EQ DF-6,1Internal = 4.8 ± 0.6 nM; EQ DF-29,1Internal = 3.3 ± 0.4 nM; EQ DF-50,1Internal = 4.1 ± 0.5 nM; and EQ DF-60,1Internal = 17.6 ± 3.1 nM (Figure 2B–E). The time traces showed that in all cases, the DNA alone has a single FRET conformer that is actively bent upon addition of FEN1, consistent with our previous results (8). While EQ DF-6,1Internal and EQ DF-29,1Internal substrates transitioned between bent and unbent states in the presence of FEN1, EQ DF-50,1Internal and EQ DF-60,1Internal displayed higher stability in the FEN1-bound bent state, including a significant portion of bent particles that did not undergo any transition (Figure 2D and E). This higher stability observed with EQ DF-50,1Internal and EQ DF-60,1Internal implies that FEN1 has a lower rate of binding/bending given that the Kd-bending does not change in case of EQ DF-50,1Internal and increases slightly in case of EQ DF-60,1Internal. EQ DF-6,1Internal and EQ DF-29,1Internal displayed diffusion-limited association rates; (1.57 ± 0.47) × 108 and (1.33 ± 0.01) × 108 M−1s−1, respectively, with similar dissociation rates.
A kinetic mechanism for short 5′ flap recognition and cleavage by FEN1
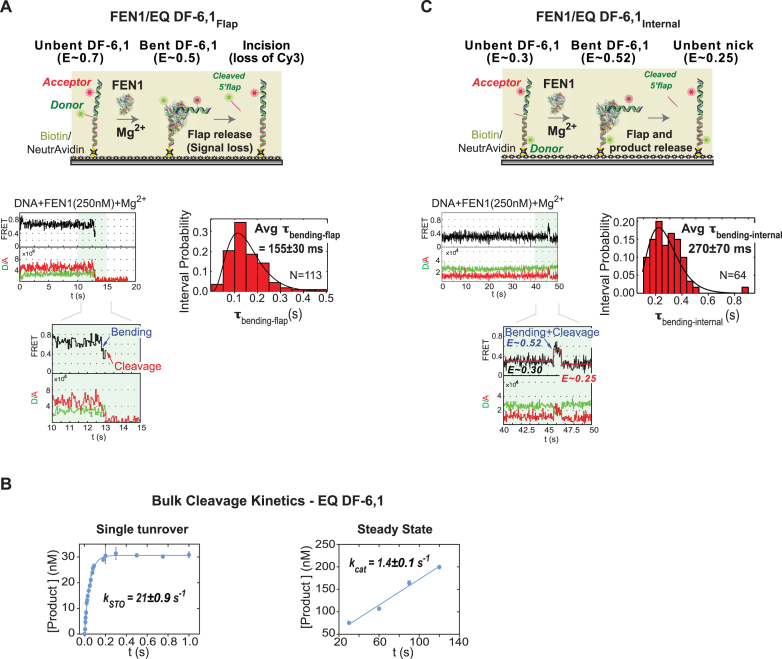
Our next goal was to assess the catalytic efficiency of FEN1 on substrates with varying flap lengths. We first investigated the kinetic mechanism of FEN1 on a short flap substrate to establish a basis for detailed comparison of the different DNAs. Addition of FEN1 to EQ DF-6,1Flap (Cy3 donor-labeled 5′ flap) in the presence of catalytically competent Mg2+ ions resulted in FEN1 bending the substrate, followed by a brief period with the complex in bent conformation and then flap cleavage (Figure 3A), as reported previously (8). We previously showed that FEN1 always cleaves the 5′ flap in DF-6,1 within the first DNA bending event, without a missed opportunity (8). The diffusion-limited association rate (Figure 2B) (8) and the productive catalysis from the first bending event (8) demonstrate that FEN1 cleavage is not limited by substrate binding/bending. Since the steady state cleavage kinetics (kcat) is significantly slower than the single turnover kinetics (kSTO) (47,49), FEN1 is widely accepted to be product-inhibited. The reaction generates two products, a 5′ flap and a nicked DNA duplex, and based on 5′flap and nicked product competition studies (8,47), only the latter was a competitor. Therefore, it has been proposed that 5′ flap release is fast following cleavage whereas nicked duplex release limits the kcat. The cleavage kinetics were determined by monitoring the bending step just before loss of the donor signal. Thus, the experiment with donor-labeled 5′ flap DNA reports the kSTO rate, which includes DNA bending, protein ordering for active site assembly, chemistry and flap release (8). By fitting the dwell time spent in the bent state (τbending-flap) for each cleavage event with a gamma distribution, we obtained an average τbending-flap of 155±30 ms and calculated the kSTO (1/τbending-flap) to be 6.5±1.2 s−1 for the EQ DF-6,1Flap substrate (Figure 3A); this result is consistent with our previous single molecule measurements (8). Rapid quench-flow bulk cleavage experiments at 35:1 ratio of FEN1:DNA determined the kSTO as 21 ± 0.9 s−1 (Figure 3B), which is consistent with previous reports (47,49). We attributed the ∼3-fold slower kSTO in single molecule experiments to the difference in temperature; 22°C in single molecule versus 37°C in bulk.
Figure 3.
Single molecule and bulk cleavage kinetics of FEN1 on short DF-substrates. (A) Flap-labeling smFRET cleavage assay. Top: schematic of the assay. EQ DF-6,1Flap is labeled as described in Figure 2A. In the presence of Mg2+, FEN1 binds and bends EQ DF-6,1Flap, decreasing FRET from ∼0.7 to ∼0.5. Upon cleavage, the Cy3-labeled flap is released and the signal is lost. The assay follows the time spent by DNA in bent state before loss of signal. Bottom left: a representative single molecule time trace showing FEN1 bending and cleaving the substrate before the 5′ flap is released; the inset zooms in on the cleavage event preceded by a brief bending step showing clear anti-correlation between donor and acceptor intensities. Bottom right: distribution of the dwell times spent in bent state (τbending-flap) for N = 113 cleavage events fitted to a gamma distribution. The average τbending-flap is reported with the standard error of the mean. The cleavage reaction was performed at 50 ms temporal resolution. More representative traces are shown in Supplementary Figure S1B. (B) Ensemble cleavage kinetics of FEN1 on EQ-DF6,1. Left: single turnover cleavage was measured on a rapid quench-flow instrument at a FEN1:DNA ratio of 35:1. The amount of 5′ flap product formed was analyzed by denaturing PAGE. Average product concentration from two replicates was plotted versus time and fitted to a single exponential equation to determine the cleavage rate (kSTO). Right: steady state cleavage was measured with FEN1:DNA at a ratio of 1:800. Average data from two replicates fitted to a linear regression yielded kcat (slope/[FEN1]). Error bars correspond to the variation of the two replicates, and the error of the fit is reported. (C) Internal-labeling smFRET cleavage assay. Top: schematic of the assay. EQ DF-6,1Internal is labeled as described in Figure 2A. In the presence of Mg2+, FEN1 binds and bends EQ DF-6,1Internal, increasing FRET from 0.3 to 0.52. Upon cleavage, the 5′ flap is released and a nicked duplex is generated, which has a FRET of 0.25 when unbent (Supplementary Figure S1A). The assay follows the time spent by DNA in bent state (0.52) before the product achieves unbent state. Bottom left: a representative single molecule time trace showing FEN1 bending and cleaving the substrate before FRET drops to 0.25; the inset zooms in on a vbFRET-fitted version of the cleavage event showing a three-state fit (0.3, 0.52 and 0.25) corresponding to the three DNA conformers, unbent EQ DF-6,1Internal, bent EQ DF-6,1Internal and unbent nicked product, respectively. Bottom right: distribution of the dwell times spent in bent state (τbending-internal) for N = 64 cleavage events fitted to a gamma distribution. The average τbending-internal is reported with the standard error of the mean. The cleavage reaction was performed at 50 ms temporal resolution. More representative traces are shown in Supplementary Figure S1C.
The flap-labeling scheme follows the cleavage reaction up to the 5′ flap release step but offers no information about the nicked DNA product; therefore, we used a complementary assay based on the internal-labeling scheme to monitor steps subsequent to flap release. This assay detects the time spent by the substrate in bent state at a high FRET of E ∼0.52 before the signal decreases to E ∼0.25, which we interpret as the nicked product in an unbent state (Figure 3C). This interpretation is supported by the following information. The unbent EQ DF-6,1Internal substrate exhibits a difference of E ∼0.05 from the unbent nicked product, as evident from the DNA-only histograms of DF-6,1 and nicked DNA (Figure 2B and Supplementary Figure S1A). Thus, if DNA bending is not followed by 5′ flap cleavage, the FRET should change from 0.3 (unbent substrate) to 0.52 (bent substrate) and back to 0.3 (unbent substrate), whereas in the case of DNA bending followed by 5′ flap cleavage, the FRET should change from 0.3 (unbent substrate) to 0.52 (bent substrate) and back to 0.25 (unbent nicked product). Importantly, the experiment was designed to minimize FEN1 rebinding and bending the nicked product while allowing substrate binding/bending. Specifically, the cleavage reaction was performed at 250 nM FEN1, which is ∼50-fold higher than Kd-bending of DF-6,1Internal (4.8 nM; Figure 2B) but well below Kd-bending of the nicked product (lower estimate of 580 nM; Supplementary Figure S1A). Furthermore, since FEN1 always cleaves DF-6,1 after the first encounter (8), this FEN1 concentration ensures a high fraction of cleavage within a single turnover, and no signal from nicked product rebinding and bending after it is released.
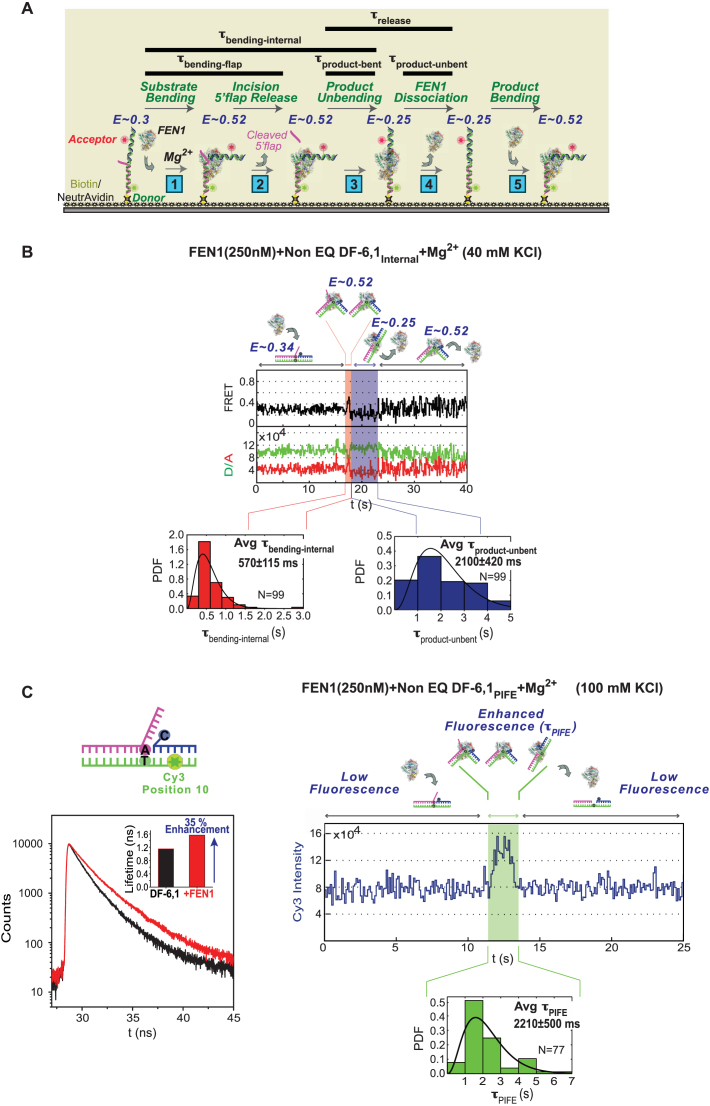
The data show that the time spent by the internal-labeled substrate in bent state (τbending-internal) is 270 ± 70 ms (Figure 3C), while that of the flap-labeled substrate (τbending-flap) is 155±30 ms (Figure 3A). The difference between τbending-flap and τbending-internal, which reports the dwell time of the nicked product in bent state (τproduct-bent), is only 115 ± 75 ms. This value yields an apparent rate of 8.7 ± 5.7 s−1 (1/τproduct-bent) for nicked product release if we assume that FEN1 dissociation is coupled with unbending of the DNA. This rate is comparable to the kSTO of 6.5 ± 1.2 s−1 (Figure 3A), and is much higher than the kcat measured in bulk under similar reaction conditions (1.4 ± 0.1 s−1; Figure 3B), which would imply that another step after nicked DNA release limits steady state turnover. However, as noted above, a product inhibition study predicted that release of nicked DNA is the rate-limiting step (47). In order to reconcile these contradictory findings, we considered the possibility that FEN1 remains bound to the unbent nicked product for some time before dissociating into solution. In order to test this hypothesis, we performed the same experiment under conditions that favor FEN1 binding to nicked DNA. The prediction was that if after cleavage FEN1 remains bound to the unbent nicked product for some time, we would observe a lag phase at low FRET of E ∼0.25, which reports the dwell time of this complex before it dissociates and another FEN1 rapidly binds and bends the released product to increase the signal to E ∼0.52 (as illustrated in Figure 4A). Lowering KCl concentration to 40 mM from 100 mM increases the affinity of FEN1 for the DNA products of both EQ DF-6,1 and Non-EQ DF-6,1 (Kd = 62 and 12 nM, respectively; Supplementary Figures S2A and B). A control experiment with flap-labeled EQ DF-6,1 showed that kSTO is not affected by lower KCl (τbending-flap is 155 ± 30 ms at 100 mM KCl and 180 ± 40 ms at 40 mM KCl; Figure 3A and Supplementary Figure S2C, respectively). Since FEN1 bends (Figure 2B, bottom right) and cleaves EQ and Non-EQ substrates with similar kSTO (8,49), Non-EQ DF-6,1Internal was used as the substrate in this experiment to ensure rapid and high affinity rebinding of FEN1 to the nicked product. The results show an increase in τbending-internal from 270 ± 70 ms at 100 mM KCl (Figure 3C) to 570 ± 115 ms at 40 mM KCl (Figure 4B), indicating that the dwell time of bent product, τproduct-bent, has been extended from 115 ± 75 ms to 390 ± 120 ms with lower KCl. Notably, we observed an extended phase with unbent DNA at E ∼0.25 that lasted for 2100 ± 420 ms (τproduct-unbent), before FEN1 dissociation followed by fast rebending re-established the bent state at E ∼0.52 (Figure 4B). These results could be interpreted such that product release by FEN1 occurs in two steps: τproduct-bent wherein FEN1 briefly holds the product in bent state for 390 ± 120 ms after 5′ flap departure and τproduct-unbent wherein FEN1 remains bound to the unbent product for 2100 ± 420 ms before dissociating into solution. Therefore, the actual τrelease could be the sum of the two dwell times, which yields a krelease of 0.40 ± 0.07 s−1 for the nicked product. The resulting kcat of 0.37 ± 0.06 s−1 (1/(τbending-flap + τrelease)) is in line with rates determined by bulk experiments at 1:800 of FEN1:DNA (Supplementary Figures S2D and 3B at 40 and 100 mM KCl, respectively) and previous reports (47,49). Taken together, these results suggest that FEN1 turnover is limited by release of the nicked DNA product, mainly from an unbent state.
Figure 4.
Kinetic scheme for short 5′ flap recognition and cleavage by FEN1. (A) Schematic of the multi-step FEN1 reaction as revealed by the internal-labeling scheme: (1) Substrate bending: the extended DF substrate exhibits FRET of 0.3, which increases to 0.52 upon FEN1 binding and bending. (2) Incision/5′ flap release: FEN1 incises the 5′ flap 1 nt inside the junction, and the flap is instantaneously released (8); at this stage the FRET is 0.52. The time spent by the substrate in bent state just before 5′ flap release is accessed by the flap-labeling cleavage assay (τbending-flap). (3) Product unbending: the nicked duplex product remains bound and bent by FEN1 at a FRET of 0.52 (similar to that of bent substrate) before unbending to FRET ∼0.25. The time spent by the product in bent state (τproduct-bent) is the difference between the bent state dwell times in the internal-labeling (τbending-internal) and flap-labeling (τbending-flap) assays. (4) FEN1 dissociation: FEN1 remains bound to the unbent product (E ∼0.25) for some time (τproduct-unbent) before dissociating into solution. Thus, product release occurs in two steps and τrelease is the sum of τproduct-bent and τproduct-unbent. (5) Product bending: FEN1 can rebind/rebend the product again; hence, FRET fluctuates between 0.25 (unbent product) and 0.52 (bent product) at the end of the reaction. The rebinding step is detected by lowering KCl from 100 to 40 mM to increase FEN1 affinity for nicked DNA (Supplementary Figure S2B). (B) smFRET cleavage of non-EQ DF-6,1Internal. Top: representative single molecule time trace showing cleavage of DF-6,1Internal and exhibiting the substrate and product dynamics described in (A). The FRET state and the substrate/product conformer in each step is illustrated. τbending-internal is highlighted in red and τproduct-unbent is highlighted in blue on the time trace. The distributions of τbending-internal (bottom left) and τproduct-unbent (bottom right) for N = 99 cleavage events were fitted to gamma distributions, and the means with standard errors are reported. The cleavage reaction was performed at 100 ms temporal resolution. More representative traces are shown in Supplementary Figure S2E. (C) Bulk and single molecule PIFE experiments. Left: bulk time-resolved fluorescence lifetime measurements of non-EQ DF-6,1PIFE in the absence (black curve) and presence (red curve) of 1 μM FEN1. Inset shows the quantification of fluorescence lifetime in the absence and presence of FEN1. As described in ‘Materials and Methods’ section, the lifetimes are determined using a 2-exponential decay fit and show 35% fluorescence enhancement with FEN1. Right: representative time trace showing a smPIFE cleavage experiment with non-EQ DF-6,1PIFE. The substrate/product conformer in each state is illustrated. The time spent in the enhanced-fluorescence state τPIFE is highlighted in green. The distribution of τPIFE for N = 77 cleavage events was fitted to gamma distribution, with the mean and standard error of the mean reported. The cleavage reaction was performed at 100 ms temporal resolution. Additional traces are shown in Supplementary Figure S2F.
However, the smFRET data described above does not provide direct evidence that FEN1 is still bound to the unbent product during the lag phase. For more direct evidence, we employed PIFE, in which the fluorescence of certain fluorophores (mainly cyanine dyes) is enhanced upon protein binding rather than induced conformational changes. We placed Cy3 at position 10 on the downstream duplex DNA (Figure 4C) and observed 35% enhancement of Cy3 fluorescence with non-EQ DF-6,1PIFE in the presence of FEN1 (Figure 4C). smPIFE experiments were performed under standard smFRET cleavage conditions (100 mM KCl, 250 nM FEN1). In this assay, the time spent in the enhanced-fluorescence state (τPIFE) is interpreted as time spent by FEN1 binding to the substrate, cleaving the flap, and any subsequent binding to the nicked product. By comparing the kinetics from the smFRET and smPIFE cleavage reaction, we hypothesize that if FEN1 releases the product from a bent state right after cleavage, τPIFE should be relatively short while a longer-lived enhanced-fluorescence state would imply that FEN1 remains bound to the product after unbending occurs. The smPIFE assay was performed with DF-6,1, which is cleaved by FEN1 almost always in the first encounter (8), and at 100 mM KCl and 250 nM FEN1 where no product rebending is observed, as shown in Figure 3C and discussed above. These conditions support our interpretation that the observed enhanced-fluorescence state stems from a productive binding event that leads to cleavage. Fitting the τPIFE for N = 77 cleavage events with a gamma distribution yields a lengthy average τPIFE of 2210 ± 500 ms (Figure 4C) which translates to a rate of 0.45 ± 0.10 s−1. This rate is in line with the kcat we observed in bulk as well as the suggested kcat in smFRET experiments. Taken together, smFRET and smPIFE assays potentially explain FEN1 product release mechanism with two steps, a fast unbending step and a relatively slow release of product after it achieves an unbent state.
Finally, based on our interpretation of the smFRET data from experiments with both flap- and internal-labeled DNAs, we introduced a comprehensive reaction mechanism of FEN1 action on short flaps (Figure 5). To summarize, FEN1 binds and actively bends DF-6,1 by diffusion-limited kinetics, cleaves the 5′ flap rapidly after protein ordering and active site assembly (8), followed by slow product release that occurs in two steps, highlighting the relatively high affinity and stability of the FEN1-nicked product complex.
Figure 5.
Kinetic scheme for the FEN1 reaction. FEN1 binds and bends a short DF substrate at diffusion-limited kinetics and commits to cleavage within the first encounter. After cleavage, the 5′ flap is released instantaneously while nicked product release occurs in two steps. FEN1 can rebind/rebend the nicked product at diffusion-limited kinetics, albeit with a lower on-rate than the substrate. With longer flaps, FEN1 can miss cleavage and dissociate from the bent substrate, requiring more than one attempt at cleavage. kbending-internal and product release rates are shown for non-EQ DF-6,1Internal at 40 mM KCl in presence of Mg2+. kSTO at 40 mM KCl is shown for EQ DF-6,1Flap in Supplementary Figure S2C; kSTO is independent of KCl concentration and it has been shown to be similar for both EQ DF-6,1Flap and non-EQ DF-6,1Flap (8). The substrate and product on/off rates are determined in presence of Ca2+; substrate EQ DF-6,1Internal at 100 mM KCl (Figure 2B) and product at 40 mM KCl (Supplementary Figure S2B). Based on data with the product (Supplementary Figure S2B), we do not anticipate a significant change in kon-bending of the substrate with lower KCl concentration.
Effect of 5′ flap length on the FEN1 kinetic mechanism
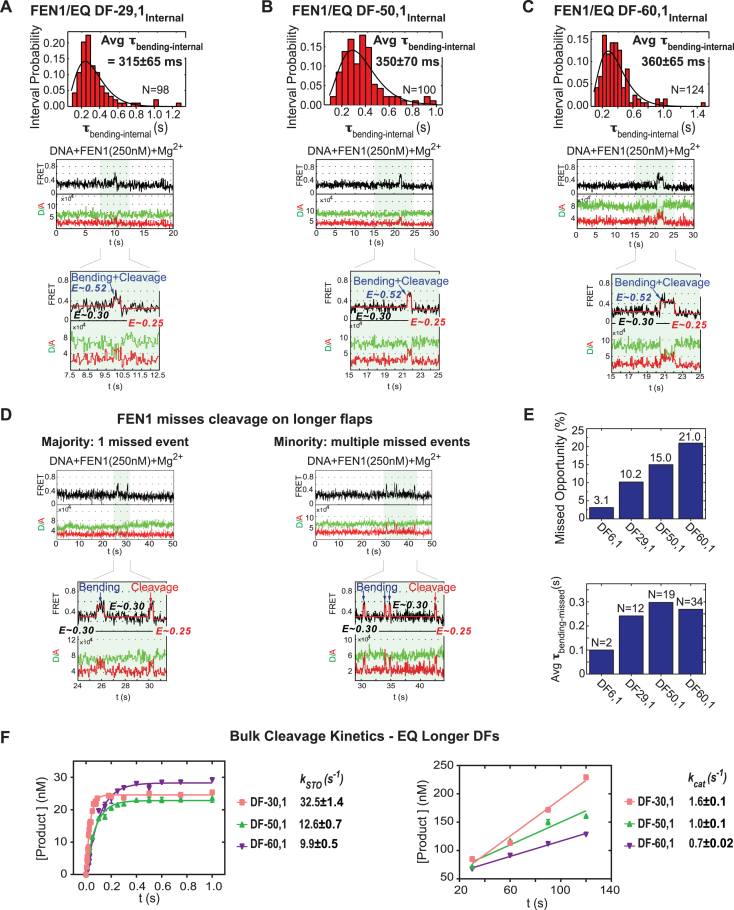
After establishing the FEN1 kinetic mechanism on a short 6 nt flap substrate, we tested its activity on longer flaps (29, 50 and 60 nt). In this case, we performed experiments mainly with internal-labeled substrates since the higher flexibility of long flaps increased the noise level with flap-labeled substrates, complicating identification of real bending and cleavage events (data not shown). smFRET cleavage experiments were performed with 250 nM FEN1, which is ∼15-fold higher than the weakest Kd-bending measured for the DF substrates (17.6 nM for DF-60,1Internal; Figure 2C–E), but lower than the Kd-bending of nicked DNA (lower estimate of 580 nM; Supplementary Figure S1A). As with DF-6,1Internal, we were able to identify cleavage events due to the slight difference in FRET efficiency between unbent DF substrates (E∼0.3) and the unbent nicked product (E∼0.25), and thus obtain the average τbending-internal for each DF substrate as shown in Figure 6A–C. Interestingly, there were no significant differences in τbending-internal values across the flap lengths (315 ± 65 ms for DF-29,1, 350 ± 70 ms for DF-50,1 and 360 ± 65 ms for DF-60,1), and even compared to DF-6,1 (270 ± 70 ms) (Figure 3C). Given that krelease for the common nicked product is not expected to vary with 5′ flap length, similar τbending-internal values mean that kSTO is similar across the flap lengths as well.
Figure 6.
Effect of 5′ flap length on FEN1 cleavage activity. (A) smFRET cleavage of EQ DF-29,1Internal. Top: distribution of the dwell times spent in bent state (τbending-internal) for N = 98 cleavage events fitted to a gamma distribution. Average τbending-internal is reported with the standard error of the mean. Bottom: a representative single molecule time trace showing FEN1 bending and cleaving the substrate before FRET drops to 0.25; the inset zooms in on a vbFRET-fitted version of the cleavage event showing a three-state fit (0.3, 0.52 and 0.25) corresponding to the three DNA conformers, unbent DF-29,1Internal, bent DF-29,1Internal and unbent nicked product. The cleavage reaction was performed at 50 ms temporal resolution. More representative traces are shown in Supplementary Figure S3A. (B) smFRET cleavage of EQ DF-50,1Internal as described in (A). More representative traces are shown in Supplementary Figure S3B. (C) smFRET cleavage of EQ DF-60,1Internal as described in (A). More representative traces are shown in Supplementary Figure S3C. (D) FEN1 misses cleavage on longer DF substrates. FEN1 almost always cleaves a short-flap substrate (EQ DF-6,1Internal) in the first bending event (8). With longer flaps, FEN1 exhibits missed opportunities at cleavage (defined as an unproductive bending event wherein FRET drops to unbent substrate state (0.3) rather than unbent product state (0.25)). Representative single molecule time traces show a majority of these events are single missed opportunities (left) and a minority are multiple missed opportunities (right). Insets zoom in on vbFRET-fitted versions of the missed events as well as the cleavage events with three-state fits. The fits show FRET returning to the substrate unbent state (0.3) in missed events and dropping to the product unbent state (0.25) in cleavage events. More representative traces are shown in Supplementary Figure S3D. (E) Quantification of missed cleavage events. Top: a bar chart shows that the percentage of particles exhibiting cleavage with missed opportunities increases with flap length from 3.1% (DF-6,1) to 21.0% (DF-60,1). Bottom: a bar chart shows the average time spent in bent state during missed events (τbending-missed) by the different DF substrates. The reported N is the number of missed events, not number of particles, accounting for particles with multiple missed events. (F) Ensemble cleavage kinetics of FEN1 on longer flap EQ DF substrates. Left: single turnover cleavage of DF-30,1, DF-50,1 and DF-60,1. Right: steady state cleavage of DF-30,1, DF-50,1 and DF-60,1. The rates were measured and reported as described in Figure 3B.
Interestingly, a closer look at the time traces revealed some particles undergoing cleavage after missed opportunities in which FEN1 bends the DF substrate but does not cleave the flap (Figure 6D). These missed events resulted in a return to the unbent substrate with FRET efficiency ∼0.3 rather than the unbent nicked product (E ∼0.25), followed by one or more additional binding and bending events until cleavage occurred. The assignment of a bending event as either a missed cleavage or cleavage event was further confirmed by constructing FRET histograms from the traces that showed multiple bending events in DF-50,1. As shown in Supplementary Figure S3E, the FRET values of the section of traces before any bending occurs averaged around 0.29 (highlighted in green), those of sections with periods separating any two bending events also averaged around 0.29 (highlighted in blue), but the FRET values of the section after the last bending event averaged around 0.23 (highlighted in red). This pattern confers confidence in the assignment of the first bending event(s) as missed cleavage event(s) and the last one as a cleavage event. Note that FEN1 concentration is below Kd-bending of nicked DNA and the experiments are performed at 100 mM KCl; therefore, FRET increases subsequent to the first one do not represent FEN1 binding to nicked product. On some of the particles, FEN1 went through multiple missed tries at cleavage, but the majority were cleaved after one miss. Interestingly, the percentage of particles with at least one missed event increased significantly from 3.1% in the case of DF-6,1 to 21% in the case of DF-60,1 (Figure 6E). We also found that the fraction of particles with more than one missed event increased with flap length. Analysis of the dwell time spent in the bent state during missed events (τbending-missed) showed that τbending-missed did not differ significantly across the flap lengths (note: in case of DF-6,1 the sample size of 3.1% was too small to draw any conclusion). Moreover, the τbending-missed (∼270 ms) was in the same range as τbending-internal (Figure 6E and A–C, respectively). This result indicates that during the missed opportunities FEN1 can access and bend all the DF substrates with similar efficiency but still cannot assemble a catalytically competent active site. We also note that the 5′ flap might be getting threaded into the cap-helical gateway in these missed-cleavage bending events since a DF substrate in which the 5′ flap is blocked from threading dissociates ∼13-fold faster at 47.2 s−1 (8) than the ∼3.7 s−1 rate (1/τbending-missed) measured here (Figure 6E). It is also possible that these events result from trapping partially threaded complexes in a fashion similar to that observed with Exo1 (36).
We also noticed that in contrast to DF-6,1, cleavage of DF-50,1 and DF-60,1 occurs in an asynchronous manner after FEN1 enters the flow cell, even under single turnover conditions. This behavior was clear when we plotted the distribution of the initial time point in each cleavage event for DF-6,1, DF-50,1 and DF-60,1. The distributions were broader for the 50 and 60 nt flaps as compared to the 6 nt flap, suggesting that a decrease in the DNA binding and bending rate with increasing flap length also influences cleavage (Supplementary Figure S3F). We speculate that this reduction reflects challenges faced by FEN1 in binding the nick junction in the context of long 5′ flaps rather than flap threading, since significant DNA bending occurs even without threading of the 5′ flap into the cap-helical gateway (8).
Finally, we conducted complementary rapid quench-flow bulk cleavage experiments with the same EQ DF substrates to validate the finding that kSTO and kcat do not vary significantly with increasing 5′ flap length. As described previously for DF-6,1, kSTO was determined at 35:1 ratio of FEN1:DNA, while kcat was determined at 1:800 ratio of FEN1:DNA. The kSTO was slightly higher for 30 nt (32.5 ± 1.4 s−1; Figure 6F) compared to 6 nt (21 ± 0.9 s−1; Figure 3B) and slightly lower for 50 nt (12.6 ± 0.7 s−1) and 60 nt flaps (9.9 ± 0.5 s−1). The kcat also followed the same trend as kSTO, showing a small change with flap length relative to DF-6,1 (2-fold maximum; Figure 6F), which is not surprising since the rate-limiting product release step should be comparable for all the substrates. While this finding is consistent with our smFRET results, it contrasts with a recent report suggesting that the catalytic efficiency of FEN1 is significantly compromised with 5′ flaps longer than 45 nt (∼10-fold decrease in kSTO) (49). The authors compared kSTO to kcat and suggested that beyond 45 nt flaps, the rate limiting step in the reaction switches from product release to 5′ flap threading. We note, however, that while we utilized poly(T) flaps in this study, Tarantino et al., (49) had mixed sequence flaps, and an analysis of the sequences by IDT OligoAnalyzer 3.1 indicates a propensity for secondary structure, especially in their DF-60,1M substrate (hairpin of Tm = 51.9°C). The kSTO of FEN1 is significantly reduced on DF substrates containing 5′ flap hairpins (7,47); hence, it is possible that secondary structures formed due to the mixed sequence rather than flap length account for the reported impairment in FEN1 activity on long flap substrates. To summarize our results thus far, both single molecule and bulk cleavage analyses demonstrate that 5′ flap length has a minor inhibitory effect on the FEN1 reaction mechanism. The missed cleavage behavior on long flap substrates indicates that the most striking difference occurs at a step after DNA binding and bending but before, chemistry and product release (Figure 5). However, the steady state rate-limiting step in the reaction remains nicked product release. Accordingly, the overall effect of increasing flap length on FEN1 catalytic efficiency is relatively small.
To date, most in vitro characterizations of FEN1 substrate recognition and catalytic efficiency have involved substrates with DNA flaps. Since RNA flaps arise in vivo and could be substrates for FEN1 cleavage, we investigated the effect of replacing a DNA flap with RNA in EQ DF-6,1 and EQ DF-29,1 substrates. Single molecule bending experiments in the presence of Ca2+ showed that FEN1 is only modestly (∼3-fold) defective in accessing both RNA-flap substrates. However, the stability of the bent complexes varies with flap length, such that FEN1 shows a modest, ∼3-fold reduction in stability on EQ DF-6,1-RNA but ∼10-fold reduction on EQ DF-29,1-RNA when compared with the corresponding DNA substrates (Supplementary Table S1). Nonetheless, single molecule cleavage assays with internal-labeled substrates showed slightly faster cleavage kinetics with RNA-flap substrates as compared with corresponding DNA substrates. These findings are in line with a previous report on single flap RNA substrates (50). Importantly, the missed cleavage behavior of FEN1 remains the same with an RNA flap as with DNA. Overall, we conclude that although FEN1 exhibits reduced stability on RNA-flap substrates, its catalytic efficiency is not limited significantly by the higher dissociation rate.
Coordination between RPA and FEN1 on short- and long-flap substrates
We have shown that the probability of FEN1 missing cleavage within its first encounter with a DNA substrate increases with 5′ flap length. While this probability seems low, the outcomes of any missed cleavage event can be significant. If left unprocessed, long flaps can form secondary structures that may impede DNA replication and repair, undergo recombination at ectopic sites, and result in duplication of sequences, among other deleterious effects (5); hence, they are not generally tolerated by the cell. As noted earlier, deletion of Dna2, which is required to process long flaps along with RPA (Figure 1), is lethal in S. cerevisiae whereas deletion of the FEN1 ortholog (Rad27) is tolerated, albeit with a severe mutator phenotype (23,24). We posit that the availability of an alternate pathway for processing long flaps is very important, given our discovery that FEN1 occasionally misses cleaving such DNA substrates. To test this idea, we asked whether FEN1 activity has any impact on how long flaps are diverted to the secondary long-flap pathway.
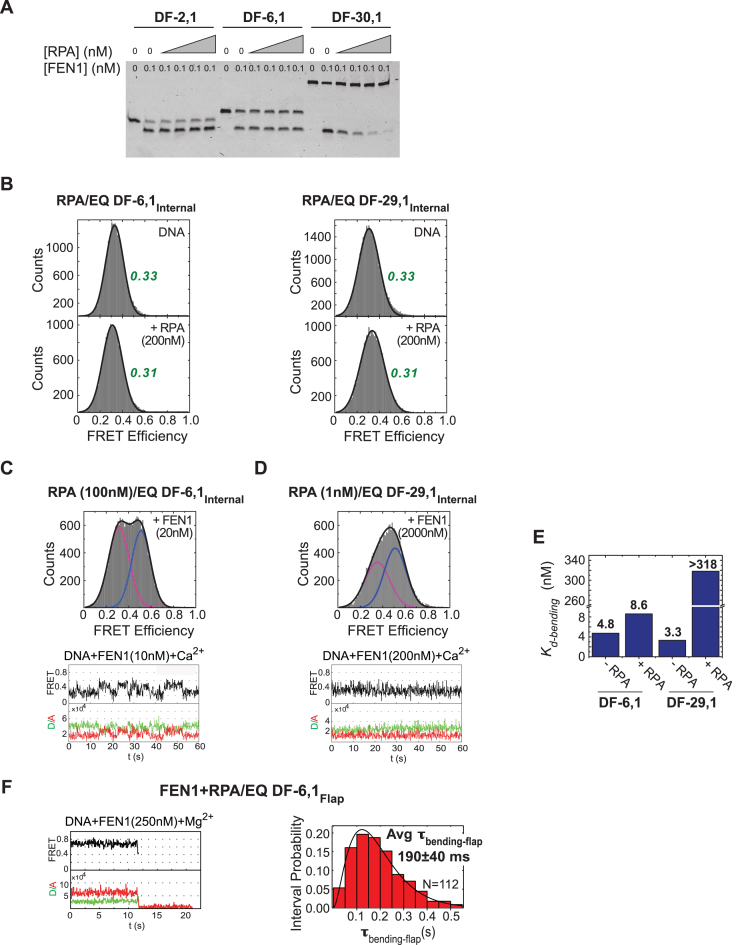
One way to address this question was to monitor the actions of RPA and FEN1 with respect to each other on short- as well as long-flap substrates. A gel-based assay showed that FEN1 cleavage activity on DF substrates with varying flap lengths (EQ DF-2,1, EQ DF-6,1 and EQ DF-30,1) remains unchanged on EQ DF-2,1 and EQ DF-6,1 but is inhibited on EQ DF-30,1 with increasing RPA (Figure 7A). This result can be explained by the dual single stranded DNA-binding modes of RPA: a weak, transient 8-nucleotide mode (Kd-binding ∼50 nM) and a stronger, more stable 30-nt mode (Kd-binding ∼0.5 nM) (51), indicating that RPA forms a stable complex with the 30 nt flap. The next series of experiments measured DNA binding/bending and cleavage by FEN1 using the smFRET assays described above. First, as a control, we tested the effect of RPA binding to two internal-labeled short- and long-flap DF substrates (EQ DF-6,1Internal and EQ DF-29,1Internal) in the presence of Ca2+. For both substrates, FRET efficiency histograms with DNA alone or DNA with increasing concentrations of RPA showed a single peak centered at ∼0.33 and ∼0.31, respectively (Figure 7B); thus, RPA does not appear to distort these DF substrates, which simplified analysis of FEN1 activity in the presence of RPA. Subsequent experiments were performed with 100 nM RPA for EQ DF-6,1 and 1 nM RPA for EQ DF-29,1, which is two fold higher than Kd-binding of both binding modes. As shown in Figure 7C, FEN1 binds and bends EQ DF-6,1Internal similarly in the absence or presence of 100 nM RPA. The histograms in presence of both proteins showed two separable peaks centered around the same FRET efficiencies of E∼0.3 and E∼0.52 (Figure 7C) as with FEN1 alone (Figure 2B). The time traces also showed similar transitions between bent and unbent states as seen with FEN1 alone. Moreover, the bending efficiency of FEN1 in presence of RPA was comparable to that of FEN1 alone, as evident from similar Kd-bending constants (4.8 ± 0.6 and 8.6 ± 1.2 nM in the absence and presence of RPA, respectively; Figure 7E). In sharp contrast, with EQ DF-29,1Internal, the presence of 1 nM RPA caused the FRET histogram peaks to merge with the centers shifted (Figure 7D) when compared with FEN1 alone (Figure 2C); note: the time traces showed that the bent and unbent state transitions were faster than our 100-ms temporal resolution, resulting in an averaging effect and merged peaks in the FRET histograms. With DF-29,1, unlike DF-6,1, RPA had an inhibitory effect on FEN1 binding and bending efficiency. The averaging effect complicated data fitting and determination of Kd-bending for FEN1 in the presence of RPA, nevertheless, a lower estimate of 318.0 ± 30.7 nM (Figure 7E) was ∼100-fold higher than that in the absence of RPA (3.3 ± 0.4 nM; Figure 2C). This result indicates that RPA significantly lowers FEN1 affinity for a long-flap substrate, but FEN1 can still access the DNA and bend it (Figure 7D). In other words, the inhibitory effect of RPA does not stem from complete blockage of FEN1 from the substrate, but is more likely due to the inability of FEN1 to form a stable bent conformer as a result of blocked 5′ flap threading by bound RPA. By analogy with 5′ flaps bound and blocked from threading by biotin/streptavidin, the lifetime of the bent conformer with an RPA block is significantly less than that required for FEN1 to catalyze cleavage (8).
Figure 7.
Coordination between FEN1 and RPA actions on short- and long-flap substrates. (A) Steady state FEN1 cleavage activity in the presence of RPA. Gel showing short- and long-flap substrates cleaved by FEN1 in the presence of increasing RPA (0–5 nM). While RPA has no effect in the case of short flaps (DF-2,1 and DF-6,1), it inhibits FEN1 activity on DF-30,1 in a concentration-dependent manner. (B) RPA shows no effect on the structure, and thus the FRET, of EQ DF-6,1Internal and EQ DF-29,1Internal. Left: EQ DF-6,1Internal DNA-only histogram (top) with FRET centered around 0.33, and upon addition of 200 nM RPA (bottom) with FRET centered around 0.31. Right: corresponding histograms for EQ DF-29,1Internal substrate. (C) smFRET bending of EQ DF-6,1Internal by FEN1 in the presence of 100 nM RPA. Top: histogram showing distribution of FRET states upon addition of 20 nM FEN1 (unbent peak shown in magenta, bent peak shown in blue). The peaks are well separated and centered around the same FRET values as in the absence of RPA (Figure 2B). Bottom: a representative single molecule time trace showing similar transitioning rates between bent and unbent states as seen without RPA. (D) smFRET bending of EQ DF-29,1Internal by FEN1 in the presence of 1 nM RPA. Top: histogram showing distribution of FRET states upon addition of 2000 nM FEN1. The peaks (unbent in magenta, bent in blue) are merged and the centers are shifted from those seen in the absence of RPA (Figure 2C). Bottom: a representative single molecule time trace showing fast transitions between bent and unbent states that cannot be resolved within the temporal resolution of acquisition (100 ms). With such fast transitions, the FRET state captured within each frame (100 ms period) is an average and not a true FRET state. This averaging explains why the traces do not show distinct FRET states, and why the full bending (0.5) state is not reached. This effect appears as merging of the peaks in the histograms. Therefore, at any particular concentration, the percentage of the bent peak is underestimated, and consequently the Kd-bending as well. (E) A bar chart illustrating RPA effect on FEN1 Kd-bending. RPA has no effect on the Kd-bending of FEN1 for DF-6,1, but increases Kd-bending by >100-fold for DF-29,1 (note that this value is a lower estimate, due to the averaging effect noted above, given that the bent state does not saturate even at 2000 nM FEN1; panel D). (F) smFRET cleavage in the presence of RPA. FEN1 cleavage efficiency on EQ DF-6,1Flap was assayed in the presence of 100 nM RPA and at 50 ms temporal resolution. Left: single molecule time trace showing cleavage wherein a brief bending event is followed by loss of signal due to flap release. Right: the distribution of τbending-flap for N = 112 cleavage events in the presence of RPA was fitted with a gamma distribution and the mean with standard error is reported. More representative traces are shown in Supplementary Figure S4A.
Finally, the effect of RPA on FEN1 cleavage activity was measured under single turnover conditions using flap-labeled EQ DF-6,1Flap and EQ DF-29,1Flap substrates. The DNAs were pre-incubated with 100 or 1 nM RPA, respectively, prior to co-injection of the same concentration of RPA and 250 nM FEN1. FEN1 cleaved EQ DF-6,1Flap with comparable efficiency in the presence (τbending-flap = 190 ± 40 ms; Figure 7F) or absence of RPA (τbending-flap = 155 ± 30 ms; Figure 3A). We do not anticipate any effect of RPA on kcat with DF-6,1 since Kd-bending of the product was not affected by RPA (Supplementary Figure S4B). Together, these results show that RPA neither stimulates nor inhibits FEN1 binding, bending, cleavage or multiple turnover kinetics on short-flap substrates. Moreover, as predicted by RPA-induced inhibition of a stable DF-29,1 bent conformer bound to FEN1 (Figure 7D), cleavage of this substrate was severely inhibited by RPA (data not shown, since cleavage events were scarce). It should be noted that a previous study suggests that S. cerevisiae RPA stimulates FEN1 activity on short flaps and inhibits it on long flaps (48). According to the results of our study, human RPA does not affect FEN1 activity on short flaps but inhibits it on long flaps. S. cerevisiae and human RPA have been found to have differential effects on another endonuclease, EXO1, as well (52). While the molecular basis of the variation is unknown at this time, it may reflect a subtle difference between the structure and function of RPA in these two organisms.
Based on the findings of this study, we propose that while FEN1 is capable of processing a long flap by itself, it misses cleavage every so often, allowing the abundant RPA at the replication fork to compete effectively for binding the flap. The resulting inhibition of cleavage requires Dna2 to displace RPA and shorten the flap as part of the long-flap processing pathway.
DISCUSSION
OF maturation involves removal of RNA/DNA primers in the form of 5′ single-stranded flaps to precisely create nicks that are ligated in order to complete lagging strand DNA synthesis. Defective or incomplete processing of 5′ flaps can interfere with DNA replication and promote sequence expansions, especially of repeat sequences, among other outcomes that have detrimental impacts on genome integrity and stability (37,53,54). The critical importance of accurate and efficient OF maturation is highlighted by the fact that deletion mutations of enzymes primarily responsible for this process are linked to cancer predisposition and neurodegenerative diseases (3,55–57).
It has been proposed that when Pol δ generates a short 5′ flap at a downstream OF, there is tight coupling and highly efficient hand-off of the DNA to FEN1 for cleavage (11,12). However, there is also evidence that longer 5′ flaps are formed. For example, in wild-type S. pombe cells, 5′ flaps visualized by electron microscopy have a mean length of 51 nt, with some exceeding 100 nt (13); deletion of FEN1 in S. cerevisiae results in duplications that indicate flap lengths as long as 100 nt as well (15,58). In this case, the evidence indicates that RPA and Dna2 helicase/nuclease are involved in flap removal in addition to FEN1 (18,19,59,60). Processing of long flaps has the benefit of removing both the iRNA and the error-containing DNA portion of the primer generated by low fidelity Pol α, but that comes with the cost of significant DNA re-synthesis and potential delays in OF maturation given the larger number of proteins required to perform the task. Not surprisingly, the long-flap pathway is considered a secondary or backup option to the more predominant short-flap pathway (48,61). The choice between these pathways can have important consequences, and therefore likely involves coordination between the proteins involved and may be subject to regulation as well (5,29,30,62).
In this study, we examined FEN1 activity on short and long flaps in order to elucidate the events leading to OF maturation by the short- versus long-flap pathways. Interestingly, we found that in bulk experiments, FEN1 exhibits only 2-fold lower single turnover and steady state cleavage rates on 60 nt flaps compared with 6 nt flaps (Figures 3B and 6F, respectively), which suggests that it is capable of acting on longer flaps by itself during OF maturation. However, single molecule analysis of FEN1 activity revealed key transient events where its actions differ on short- versus long-flap DNA substrates. First, in smFRET DNA bending experiments we found that substrate recognition, in particular bending efficiency, is not affected by flap length (Figure 2B–E). This finding is supported by the FEN1-flap DNA crystal structure, which shows that most of the interactions of FEN1 are with the duplex portion of DNA, not the flap (32,37). In smFRET single turnover cleavage experiments, again we found little difference in the rates of multiple steps in the reaction with increasing flap length (Figures 3C and 6A–C). Therefore, we concluded that flap length has no significant impact on bending of the DNA substrate, cleavage chemistry, 5′ flap release and unbending of the nicked DNA product by FEN1 during the reaction. Importantly, we did find that while FEN1 always cleaves a short flap within the first encounter with the substrate, it increasingly misses cleavage as the flap length increases (Figure 6E). As noted above, FEN1 binds and bends both short and long flaps with similar efficiency. In addition, the average lifetime of the bent FEN1–DNA conformer is the same whether FEN1 misses or completes flap cleavage (Figures 3C and 6A–C versus 6E). Hence, the missed cleavage opportunities indicate that longer flaps pose a challenge for conversion of the bent FEN1–DNA complex into a catalytically active state. Threading a long 5′ flap through the cap-helical gateway to position the scissile phosphate in the active site may pose difficulties that increase the odds of FEN1 dissociating from DNA rather than cleaving the flap.
To determine the fate of longer flaps that can escape FEN1 cleavage, we examined the interplay between FEN1 and RPA on substrates with varying flap lengths. We found that RPA strongly inhibits cleavage of long-flap substrates, but it has no effect on short-flap substrates at the bending, cleavage or product release steps in the reaction (Figure 7 and Supplementary Figure S4). Notably, while the presence of RPA weakens FEN1 affinity for a long-flap substrate, the enzyme can still access the DNA and bend it. Cleavage appears to be inhibited mainly because the FEN1–DNA complex cannot achieve a stable, catalytically active conformation with RPA bound to the flap (Figure 7D). Thus, when FEN1 misses cleaving a long flap, RPA has the opportunity to bind it and block subsequent attempts at cleavage even if FEN1 rebinds the DNA. Given the high affinity and stability of RPA interaction with ssDNA, we expect that this protein–DNA complex will remain in pause mode until RPA is actively displaced by Dna2 helicase/nuclease. In short, the competition between FEN1 and RPA determines the choice between the short- and long-flap pathways. The ability of FEN1 to bind and bend an RPA-bound long-flap substrate could facilitate substrate transfer to FEN1 as Dna2 shortens the 5′ flap and displaces RPA. Moreover, our finding that the nicked product is released in two steps indicates a product hand-off mechanism. Since FEN1 holds on to the DNA in unbent form, we speculate that perhaps it contacts part of the duplex while allowing DNA ligase to access the newly formed nick and complete OF maturation.
To summarize, in this study, we defined the kinetic mechanism of FEN1 in greater detail, especially regarding recognition and cleavage of long 5′ flap-containing DNA substrates. We found that as the flap gets longer, the probability of escaping cleavage increases, giving RPA the chance to get involved and trigger the long-flap pathway. What might be the structure/dynamics basis for FEN1 dissociation from a long-flap substrate, and how Dna2 acts on a DNA substrate possibly crowded with RPA and FEN1, are intriguing follow-up questions to be addressed in future studies of this system.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Vlad-Stefan Raducanu and Susan E. Tsutakawa for helpful discussions. We thank Prof. Marc S. Wold for the generous gift of RPA expression plasmid.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
King Abdullah University of Science and Technology Core Funding (to S.M.H.); Competitive Research Award (CRG3) (to S.M.H.); National Institutes of Health [R15 GM114743 to M.M.H.]. Funding for open access charge: Global Collaborative Research, King Abdullah University of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Tubbs A., Nussenzweig A.. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017; 168:644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Branzei D., Foiani M.. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Bology. 2010; 11:208–219. [DOI] [PubMed] [Google Scholar]
- 3. Zheng D.Q., Zhang K., Wu X.C., Mieczkowski P.A., Petes T.D.. Global analysis of genomic instability caused by DNA replication stress in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E8114–E8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng L., Shen B.. Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell Biol. 2011; 3:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balakrishnan L., Bambara R.A.. Okazaki fragment metabolism. Cold Spring Harb. Perspect. Biol. 2013; 5:a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kao H.I., Henricksen L.A., Liu Y., Bambara R.A.. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002; 277:14379–14389. [DOI] [PubMed] [Google Scholar]
- 7. Henricksen L.A., Tom S., Liu Y., Bambara R.A.. Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J. Biol. Chem. 2000; 275:16420–16427. [DOI] [PubMed] [Google Scholar]
- 8. Rashid F., Harris P.D., Zaher M.S., Sobhy M.A., Joudeh L.I., Yan C., Piwonski H., Tsutakawa S.E., Ivanov I., Tainer J. A, Habuchi S., Hamdan S. M.. Single-molecule FRET unveils induced-fit mechanism for substrate selectivity in flap endonuclease 1. Elife. 2017; 6:e21884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Y., Kao H.I., Bambara R.A.. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004; 73:589–615. [DOI] [PubMed] [Google Scholar]
- 10. Burgers P.M. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 2009; 284:4041–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garg P., Stith C.M., Sabouri N., Johansson E., Burgers P.M.. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004; 18:2764–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stodola J.L., Burgers P.M.. Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale. Nat. Struct. Mol. Biol. 2016; 23:402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu B., Hu J., Wang J., Kong D.. Direct visualization of RNA-DNA primer removal from Okazaki fragments provides support for flap cleavage and exonucleolytic pathways in eukaryotic cells. J. Biol. Chem. 2017; 292:4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gloor J.W., Balakrishnan L., Campbell J.L., Bambara R.A.. Biochemical analyses indicate that binding and cleavage specificities define the ordered processing of human Okazaki fragments by Dna2 and FEN1. Nucleic Acids Res. 2012; 40:6774–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tishkoff D.X., Filosi N., Gaida G.M., Kolodner R.D.. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997; 88:253–263. [DOI] [PubMed] [Google Scholar]
- 16. Maga G., Villani G., Tillement V., Stucki M., Locatelli G.A., Frouin I., Spadari S., Hubscher U.. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:14298–14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Masuda-Sasa T., Imamura O., Campbell J.L.. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006; 34:1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bae S.H., Bae K.H., Kim J.A., Seo Y.S.. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001; 412:456–461. [DOI] [PubMed] [Google Scholar]
- 19. Bae S.H., Seo Y.S.. Characterization of the enzymatic properties of the yeast dna2 Helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 2000; 275:38022–38031. [DOI] [PubMed] [Google Scholar]
- 20. Zhou C., Pourmal S., Pavletich N.P.. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife. 2015; 4:e09832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stewart J.A., Campbell J.L., Bambara R.A.. Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 2009; 284:8283–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stewart J.A., Campbell J.L., Bambara R.A.. Flap endonuclease disengages Dna2 helicase/nuclease from Okazaki fragment flaps. J. Biol. Chem. 2006; 281:38565–38572. [DOI] [PubMed] [Google Scholar]
- 23. Budd M.E., Choe W., Campbell J.L.. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 2000; 275:16518–16529. [DOI] [PubMed] [Google Scholar]
- 24. Lee K.H., Kim D.W., Bae S.H., Kim J.A., Ryu G.H., Kwon Y.N., Kim K.A., Koo H.S., Seo Y.S.. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 2000; 28:2873–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tishkoff D.X., Boerger A.L., Bertrand P., Filosi N., Gaida G.M., Kane M.F., Kolodner R.D.. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen J.Z., Qiu J., Shen B., Holmquist G.P.. Mutational spectrum analysis of RNase H(35) deficient Saccharomyces cerevisiae using fluorescence-based directed termination PCR. Nucleic Acids Res. 2000; 28:3649–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pike J.E., Burgers P.M., Campbell J.L., Bambara R.A.. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J. Biol. Chem. 2009; 284:25170–25180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stith C.M., Sterling J., Resnick M.A., Gordenin D.A., Burgers P.M.. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 2008; 283:34129–34140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hasan S., Stucki M., Hassa P.O., Imhof R., Gehrig P., Hunziker P., Hubscher U., Hottiger M.O.. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell. 2001; 7:1221–1231. [DOI] [PubMed] [Google Scholar]
- 30. Balakrishnan L., Stewart J., Polaczek P., Campbell J.L., Bambara R.A.. Acetylation of Dna2 endonuclease/helicase and flap endonuclease 1 by p300 promotes DNA stability by creating long flap intermediates. J. Biol. Chem. 2010; 285:4398–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M.. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science (New York, N.Y.). 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- 32. Tsutakawa S.E., Classen S., Chapados B.R., Arvai A.S., Finger L.D., Guenther G., Tomlinson C.G., Thompson P., Sarker A. H., Shen B. et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011; 145:198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sobhy M.A., Joudeh L.I., Huang X., Takahashi M., Hamdan S.M.. Sequential and multistep substrate interrogation provides the scaffold for specificity in human flap endonuclease 1. Cell Rep. 2013; 3:1785–1794. [DOI] [PubMed] [Google Scholar]
- 34. Craggs T.D., Hutton R.D., Brenlla A., White M.F., Penedo J.C.. Single-molecule characterization of Fen1 and Fen1/PCNA complexes acting on flap substrates. Nucleic Acids Res. 2014; 42:1857–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patel N., Atack J.M., Finger L.D., Exell J.C., Thompson P., Tsutakawa S., Tainer J.A., Williams D.M., Grasby J.A.. Flap endonucleases pass 5′-flaps through a flexible arch using a disorder-thread-order mechanism to confer specificity for free 5′-ends. Nucleic Acids Res. 2012; 40:4507–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi Y., Hellinga H.W., Beese L.S.. Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:6010–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsutakawa S.E., Thompson M.J., Arvai A.S., Neil A.J., Shaw S.J., Algasaier S.I., Kim J.C., Finger L.D., Jardine E., Gotham V.J.B. et al. Phosphate steering by Flap Endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability. Nat. Commun. 2017; 8:15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chapados B.R., Hosfield D.J., Han S., Qiu J., Yelent B., Shen B., Tainer J.A.. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004; 116:39–50. [DOI] [PubMed] [Google Scholar]
- 39. Sakurai S., Kitano K., Yamaguchi H., Hamada K., Okada K., Fukuda K., Uchida M., Ohtsuka E., Morioka H., Hakoshima T.. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005; 24:683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee B.I., Wilson D.M. 3rd. The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J. Biol. Chem. 1999; 274:37763–37769. [DOI] [PubMed] [Google Scholar]
- 41. Henricksen L.A., Umbricht C.B., Wold M.S.. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994; 269:11121–11132. [PubMed] [Google Scholar]
- 42. Aitken C.E., Marshall R.A., Puglisi J.D.. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008; 94:1826–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sobhy M.A., Elshenawy M.M., Takahashi M., Whitman B.H., Walter N.G., Hamdan S.M.. Versatile single-molecule multi-color excitation and detection fluorescence setup for studying biomolecular dynamics. Rev. Sci. Instrum. 2011; 82:113702. [DOI] [PubMed] [Google Scholar]
- 44. Holden S.J., Uphoff S., Hohlbein J., Yadin D., Le Reste L., Britton O.J., Kapanidis A.N.. Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys. J. 2010; 99:3102–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kapanidis A.N., Laurence T.A., Lee N.K., Margeat E., Kong X., Weiss S.. Alternating-laser excitation of single molecules. Acc. Chem. Res. 2005; 38:523–533. [DOI] [PubMed] [Google Scholar]
- 46. Bronson J.E., Fei J., Hofman J.M., Gonzalez R.L. Jr, Wiggins C.H.. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 2009; 97:3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Finger L.D., Blanchard M.S., Theimer C.A., Sengerova B., Singh P., Chavez V., Liu F., Grasby J.A., Shen B.. The 3′-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J. Biol. Chem. 2009; 284:22184–22194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Henry R.A., Balakrishnan L., Ying-Lin S.T., Campbell J.L., Bambara R.A.. Components of the secondary pathway stimulate the primary pathway of eukaryotic Okazaki fragment processing. J. Biol. Chem. 2010; 285:28496–28505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tarantino M.E., Bilotti K., Huang J., Delaney S.. Rate-determining step of flap endonuclease 1 (FEN1) reflects a kinetic bias against long flaps and trinucleotide repeat sequences. J. Biol. Chem. 2015; 290:21154–21162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qian L., Yuan F., Rodriguez-Tello P., Padgaonkar S., Zhang Y.. Human Fanconi anemia complementation group a protein stimulates the 5′ flap endonuclease activity of FEN1. PLoS One. 2013; 8:e82666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fan J., Pavletich N.P.. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012; 26:2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Myler L.R., Gallardo I.F., Zhou Y., Gong F., Yang S.H., Wold M.S., Miller K.M., Paull T.T., Finkelstein I.J.. Single-molecule imaging reveals the mechanism of Exo1 regulation by single-stranded DNA binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1170–E1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu Y., Zhang H., Veeraraghavan J., Bambara R.A., Freudenreich C.H.. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol. Cell. Biol. 2004; 24:4049–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim J.C., Mirkin S.M.. The balancing act of DNA repeat expansions. Curr. Opin. Genet. Dev. 2013; 23:280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kucherlapati M., Yang K., Kuraguchi M., Zhao J., Lia M., Heyer J., Kane M.F., Fan K., Russell R., Brown A.M. et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:9924–9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bentley D.J., Harrison C., Ketchen A.M., Redhead N.J., Samuel K., Waterfall M., Ansell J.D., Melton D.W.. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J. Cell Sci. 2002; 115:1551–1561. [DOI] [PubMed] [Google Scholar]
- 57. Larsen E., Gran C., Saether B.E., Seeberg E., Klungland A.. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell. Biol. 2003; 23:5346–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gordenin D.A., Kunkel T.A., Resnick M.A.. Repeat expansion–all in a flap. Nat. Genet. 1997; 16:116–118. [DOI] [PubMed] [Google Scholar]
- 59. Ayyagari R., Gomes X.V., Gordenin D.A., Burgers P.M.. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem. 2003; 278:1618–1625. [DOI] [PubMed] [Google Scholar]
- 60. Kao H.I., Veeraraghavan J., Polaczek P., Campbell J.L., Bambara R.A.. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 2004; 279:15014–15024. [DOI] [PubMed] [Google Scholar]
- 61. Rossi M.L., Bambara R.A.. Reconstituted Okazaki fragment processing indicates two pathways of primer removal. J. Biol. Chem. 2006; 281:26051–26061. [DOI] [PubMed] [Google Scholar]
- 62. Kao H.I., Bambara R.A.. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit. Rev. Biochem. Mol. Biol. 2003; 38:433–452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.