Abstract
In order to expand the current repertoire of cancer treatments and to help circumvent limitations associated with resistance, the identification of new metallodrugs with high potency and novel mechanisms of action is of significant importance. Here we present a class of di-copper(II) complex based on the synthetic chemical nuclease [Cu(Phen)2]+ (where Phen = 1,10-phenanthroline) that is selective against solid epithelial cancer cells from the National Cancer Institute’s 60 human cell line panel (NCI-60). Two metallodrug leads are studied and in each case two [Cu(Phen)2]+ units are bridged by a dicarboxylate linker but the length and rigidity of the linkers differ distinctly. Both agents catalyze intracellular superoxide (O2•−) and singlet oxygen (1O2) formation with radical species mediating oxidative damage within nuclear DNA in the form of double strand breaks and to the mitochondria in terms of membrane depolarization. The complexes are effective DNA binders and can discriminate AT/AT from TA/TA steps of duplex DNA through induction of distinctive Z-like DNA or by intercalative interactions.
INTRODUCTION
Issues of innate or acquired drug resistance, allied with auxiliary toxicity, have limited the therapeutic application of established metallodrugs such as cisplatin. New drugs that achieve distinct mechanisms, therefore, offer an opportunity to evade resistance and deliver a broader spectrum of treatment options. The general aim here is to develop new compounds that specifically target cancer cells by exploiting alterations in biochemical function unique to cancer, while overcoming barriers to uptake and repair processes such as nucleotide or base excision repair. These targets may include a range of biomolecules, organelles and signaling pathways. Some of these avenues can target enzymatic proteins with mono- (1) and di-nuclear (2) complexes effecting DNA manipulation and proteolytic activity through their interactions with topoisomerases (3) and proteasomes (4). There is, however, intense focus—particularly in the metallodrug field—toward DNA where classical small molecule therapeutics employ several mechanisms to interact with DNA, including covalent nucleobase bonding (5,6), intercalation and groove binding (7). They may recognize particular DNA base sequences (e.g. oncogenic promoter regions (8), i-motifs (9) or G-quadruplexes (10–12)) and the binding can induce structural distortion that inhibits replication, or perturbs the stability of DNA-enzyme complexes, thereby rendering the cell incapable of mediating faithful replication.
Complexes of redox active metals, principally copper and iron, represent an important group of metallodrugs. The ligand environment in these complexes allows for the tuning of charge, redox potential, chirality and geometry to optimize DNA binding. Under aerobic conditions, or in the presence of appropriate intracellular oxidants, these compounds can undergo Fenton or Haber–Weiss chemistry to generate reactive oxygen species (ROS). If the complex is bound to DNA, then there is a high probability that ROS will oxidize DNA, leading to strand breakages. For example, the synthetic chemical nuclease [Cu(Phen)2]+ (where Phen = 1,10-phenanthroline) binds at the minor groove of DNA and mediates single-stranded breaks (SSBs) (13) through oxidation of deoxyribose, principally at thymine-adenine-thymine (TAT) rich regions (14,15). In the 35 years since its discovery, [Cu(Phen)2]+—and related complexes (14)—have found application as high-fidelity footprinting probes to map drug–DNA and protein–DNA interactions (16), as tools for recombinant genome engineering (17), and as developmental cytotoxins of human cancer. Recent advances in this last category have seen two complexes from the Casiopeińa class enter clinical trials (18) with another, structurally similar motif, displaying encouraging superoxide-mediated activity within the National Cancer Institute’s 60 human cancer cell line panel (NCI-60) (19).
As the metallodrug discovery field has matured, an increasing number of polynuclear compounds with distinctive DNA binding interactions have been reported (20–23). Sophisticated architectures complexing two or three metal ions have yielded improved oligonucleotide binding affinity and chemotherapeutic potency over mononuclear counterparts (20,24–27). Seminal examples of this approach include: development of the tri-platinum(II) complex BBR3464—an agent that facilitates long-range inter- and intrastrand platinated DNA adducts (28); the fully trans-symmetric triplatinum(II) complex class (e.g. TriplatinNC) that binds the DNA backbone through electrostatic ‘phosphate clamping’ (29,30); and, di-Cu2+ or di-Ni2+ complexes of 1,3-bis-(1,5,9-triazacyclododecyl) propane that enable transition from right-handed B-DNA to left-handed Z-DNA through electrostatic N(7) guanine interactions in synthetic polymers of guanine-cytosine(31). Accordingly, we have identified two promising dinuclear complexes [{Cu(Phen)2}2(μ-Oda)]2+ (Cu-Oda, where Oda = octanedioate) and [{Cu(Phen)2}2(μ-Terph)]2+ (Cu-Terph, where Terph = terephthalate). In each case two [Cu(Phen)2]2+ units are bridged by a dicarboxylate linker but the length and rigidity of the linkers differ distinctly (Figure 1A). Preliminary studies revealed high potency anticancer potential, including against cisplatin-resistant cell lines, along with encouraging cytoselectivity (21). Here we present the activity profile of both agents within the NCI-60 panel, their capacity to promote oxidative damage to genomic DNA and mitochondria through a combination of intracellular singlet oxygen (1O2) and superoxide (O2•−) production, and their ability to discriminate AT/AT from TA/TA steps of duplex DNA through induction of distinctive Z-like DNA or by intercalative interactions. These results, to our knowledge, are the first examples of copper complex mediated Z-DNA induction within the canonical B-DNA motif, and of non-radiative intracellular singlet oxygen production. Consequently, this work may provide important results that help expand current metallodrug treatments beyond bleomycin and cisplatin.
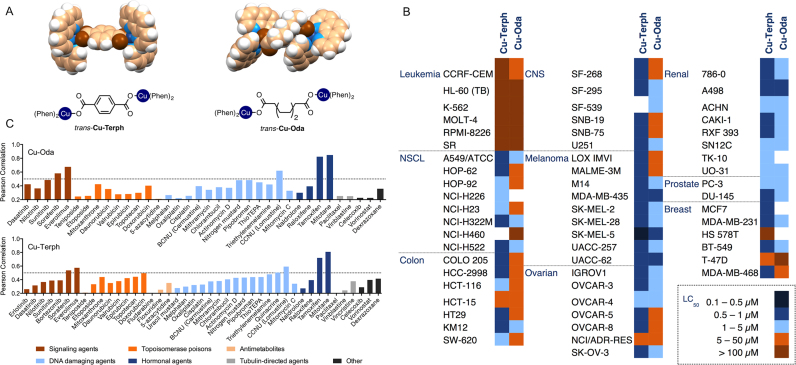
Figure 1.
(A) Crystal and molecular structures of di-nuclear copper complexes [{Cu(phen)2}2(μ-terph)]2+ (Cu-Terph) and [{Cu(phen)2}2(μ-oda)]2+ (Cu-Oda, color scheme: copper, brown; oxygen, blue; carbon, beige; hydrogen; white) (B) Heat map representation of LC50 concentrations calculated from 5-dose cytotoxicity screen across the NCI-60 human cancer cell line panel. Concentration ranges from most cytotoxic (0.1–0.5 μM shown in navy) through to least cytotoxic (>100 μM shown in brown). (C) PCCs of LC50 profiles generated from the NCI DTP COMPARE algorithm, where standard agents and market drugs are grouped according to their mode of cytotoxic action. PPCs of GI50 values are presented in Supplementary Table S2.
MATERIALS AND METHODS
NCI-60 analysis was provided by National Cancer Institute Developmental Therapeutics Program (NCI DTP) in comparison to clinical agents through the COMPARE algorithm. Cytotoxic and mechanistic properties were investigated in human ovarian cancer cells, SKOV3. Flow cytometric analysis data were acquired on a Guava EasyCyte HT flow cytometer while confocal images were obtained on a STED-Leica DMi8 super resolution confocal microscope equipped with a CCD camera. Circular dichroism studies were conducted on Applied Photophysics Circular Dichroism Spectrometer (Chirascan plus qCD). Movie clips of docking simulations are available in Supplementary Data.
Materials and reagents
All chemicals used for complex synthesis were purchased from Sigma-Aldrich without further purification. The following assays were purchased from Merck Millipore and procedures were followed as per the manufacturer protocol: Guava Nexin® Reagent (4500-0450), Guava EasyCyte™ MitoPotential Kit (4500-0250), Guava Caspase 8 6-carboxyfluorescein (FAM) and Caspase 9 SR (4500-0640) and Guava Caspase 3/7 (4500-0540). Propidium iodide (BTIU40017) was purchased from VWR. Carbonyl cyanide m-chlorophenylhydrazone (10175140), calf thymus DNA (15633-019), RNase A (12091-021), Alexa Flour 488 goat anti-mouse IgG F(ab)2 fragment (A11020), Alexa flour 488-phalloidin (A12379), 4′,6-diamidino-2-phenylindole (DAPI; D1306), NucBlue Live (R37605), singlet oxygen sensor green (S36002) and MitoTracker Deep Red (M22426) were purchased from ThermoFisher Scientific. Anti-phospho-histone H2AX (05-636) was purchased from Merck Millipore. Doxorubicin (Dox) hydrochloride (D2975000), dihydroethidium (D7008), salmon testes DNA (D1626), synthetic double stranded alternating co-polymers, poly[d(G⋅C)2] (P9389) and poly[d(A⋅T)2] (P0883) and Micrococcus Lysodeikticus (D8259) were purchased from Sigma-Aldrich. pUC19 plasmid DNA (N3041), CutSmart® buffer (B7204), 100× bovine serum albumin (BSA) (B9000) and topoisomerase I (E. coli) (M0301) were all purchased from New England Biolabs.
Preparation of complexes
[{Cu(phen)2}2(μ-octanedioate)](ClO4)2 (Cu-Oda) was prepared according to procedures reported by Devereux et al. (32). Mononuclear [Cu(μ-octanedioate)(phen)2]2+ was dissolved in an ethanolic solution of sodium perchlorate and refluxed under very mild conditions to form a light blue precipitate that was recrystallized in ethanol:water (1:1) to yield dark blue crystals (33). [{Cu(phen)2}2(μ-terephthalate)](terephthalate) (Cu-Terph) was prepared by ethanolic reflux of copper(II) terephthalate hydrate and phen in a 1:2 ratio according to reported methods by Kellett et al. (20).
NCI-60 analysis
The di-copper agents, Cu-Oda and Cu-Terph were submitted to the National Cancer Institute (NCI) Developmental Therapeutics Program (DTP) where their cytotoxicity profile was investigated across the 60 human cancer cell line panel according to sulforhodamine B protocol (34,35) (also available at https://dtp.cancer.gov/discovery_development/nci-60/default.htm). The COMPARE algorithm (https://dtp.cancer.gov/databases_tools/compare.htm) was utilized to evaluate the activity profile of Cu-Oda and Cu-Terph in comparison to known cytotoxins established in marketed drugs, mechanistic and standard agent datasets in order to correlate plausible modes of action. Criteria were set so that Pearson correlation coefficients (PCC, r) were >0.2 in a minimum of 40 common cell lines with standard deviation of 0.05.
Cell culture
SKOV3 cells were cultured in RPMI-1640 media, supplemented with 10% fetal bovine serum (FBS) and incubated at 37°C in 5% CO2 and routinely tested for mycoplasma. Cells were seeded, left to adhere and enter cell cycle (>12 h) prior to complex exposure. Cu-Oda, Cu-Terph and carbonyl cyanide m-chlorophenyl hydrazone (CCCP) stock solutions were prepared in dimethyl sulfoxide (DMSO), and Dox in 50:50 DMSO:H2O. Further dilutions were prepared in culture media daily. Stocks containing DMSO were prepared in the mM range to ensure final incubation concentrations did not exceed 0.1% v/v.
Cellular viability
SKOV3 cells were seeded at 5 × 103 cells in a 96-well plate and exposed to drug treatment for 24 h with dinuclear complexes (0.5, 1.0 and 2.0 μM), Dox (1.0 μM) or CCCP (75 μM). For co-treatment experiments, antioxidants tiron (4, 5-Dihydroxy-1, 3-benzenedisulfonic acid disodium salt (Tiron or Tir) (36), D-mannitol (Man) (37), L-histidine (His) (38), (±)-α-lipoic acid (Lip) (39), taurine (2-aminoethanesulfonic acid, Taur) (40), L-methionine (Met) (41) and sodium pyruvate (Py) (42) were treated at 1 mM for 2 h prior to drug exposure to facilitate intracellular accumulation. After drug exposure, spent media was removed, cells were detached with trypsin:EDTA (ethylenediaminetetraacetic acid) (0.25%:0.02% in phosphate-buffered saline (PBS)) and whole samples (100 μl) were added to 100 μl Guava Viacount reagent and incubated for 10 min at room temperature. Viability samples were acquired on Guava EasyCyte HT with Viacount software.
Confocal microscopy
Cells (1.4 × 105) were seeded in 35 mm glass bottom petri dishes and exposed to drug for 24 h. Samples prepared for morphological analysis were incubated with media containing MitoTracker Deep Red (150 nM, 30 min, 37°C) and fixed with 4% paraformaldehyde (30 min, room temperature). Samples were permeabilized with 0.25% Triton X-100 (15 min). To avoid non-specific staining, cells were blocked with 1% BSA in PBS (30 min, 37°C) and subsequently stained with Alexa Flour 488-phalloidin (10U/100 μl, 30 min, 37°C), followed by DAPI (4:10 000, 10 min) and mounted in ProLong Gold. Images were acquired on a STED-Leica DMi8 super resolution confocal microscope equipped with CCD camera and 100 × oil immersion objective. DAPI was excited with a 405 nm picoquant laser unit and emission captured between 387 and 474 nm. Alexa Fluor 488 was excited at 499 nm with emission captured between 490 and 566 nm, and MitoTracker Deep Red was excited at 653 nm where emission was captured at 658–779 nm. Images were acquired whereby combinations of excitation and emission wavelengths for specific dyes were applied sequentially. Samples prepared for live cell imaging of ROS were drug treated in a similar manner to that stated above. After exposure to Cu-Terph, cells were washed with PBS and stained with Mitotracker Deep Red, followed by 5 μM dihydroethidium (DHE), MitoSOX red (15 min, 37°C) or SOSG (30 min, 37°C) and finally NucBlue Live (20 min, 37°C). Images were collected on a STED-Leica DMi8 super resolution confocal microscope equipped with CCD camera and 63 × water immersion objective. NucBlue Live was excited with a 405 nm picoquant laser unit and emission captured between 429 and 481 nm, and MitoTracker Deep Red was excited at 653 nm where emission was captured at 650–716 nm. Singlet oxygen sensor green (SOSG) was excited at 504 nm where emission was captured at 506–560 nm, while DHE and MitoSOX red was excited at 535 and emission were captured between 573 and 654 nm.
Nexin® assay
SKOV3 cells were seeded at 3 × 104 cells in 24-well plates and incubated with drug containing media. After the exposure period, spent media was removed, cells were washed with PBS and washings were kept in 1.5 ml microtubes. Cells were detached with trypsin, neutralized with fresh media, transferred to microtubes with PBS and centrifuged at 1300 rpm for 5 min. A sufficient volume of media was added to cell pellet for densities ranging from 2 × 105 to 1 × 106 cell/ml. 100 μl of sample was then transferred to 96-well round bottom plate containing 100 μl of Guava Nexin® Reagent and incubated for 20 min at room temperature. Samples were acquired on the Guava EasyCyte HT flow cytometer using Nexin software. Inherent Dox fluorescence in filter 583/26 nm was accounted for and subtracted within the relevant quadrants.
Caspase 8 FAM and 9 SR, caspase 3/7 FAM
The following fluorescent labeled inhibitors of caspase (FLICA) were used to ascertain the activation of caspase 3/7, 8 and 9 respectively; FAM-DEVD-FMK, FAM-LETD-FMK and SR-LEHD-FMK. Samples were prepared prior to staining in a similar manner as that described in Nexin Assay. Cells were transferred to 1.5 ml microtubes and resuspended in 100 μl media. 10 μl of 10X caspase 9 SR working solution and 10 μl of caspase 8 FAM were added and incubated for 1 h at 37°C. Cells were resuspended in 200 μl of caspase 7-aminoactinomycin D (7-AAD) working solution and transferred to 96-well round bottom plate and incubated for 10 min at room temperature. Samples were acquired on the Guava EasyCyte HT flow cytometer using Guava Caspase software. Compensation to correct fluorescent overlap between filters was conducted pre-acquisition and innate Dox fluorescence in filter 583/26 nm was accounted for within the relevant quadrants.
Mitochondrial membrane potential
Cells were treated as previously described in the Nexin V assay. Samples were resuspended in 600 μl of fresh media to give cell concentration 2 × 104 to 5 × 105 cells/ml from which 200 μl was transferred to a 96-well round bottom plate. The 50× staining solution (4 μl) containing JC-1 and 7-AAD was added to each sample, subsequently incubated at 37°C in darkness (30 min) and acquired on Guava EasyCyte HT using MitoPotential software. Compensation to correct fluorescent overlap between filters was conducted pre-acquisition.
Superoxide and singlet oxygen detection
SKOV3 cells (6 × 104) were seeded in 12-well plates. After drug treatment, samples were harvested, washed with PBS and incubated in 200 μl of 5.0 μM MitoSOX Red or dihydroethidium (DHE) (15 min at 37°C), or 5.0 μM singlet oxygen green sensor (SOSG) (1 h, 37°C). Samples were then repeatedly washed with PBS, transferred to 96-well round bottom plates and acquired on Guava EasyCyte flow cytometer using ExpressPro software.
DNA degradation with comet assay
Cells were seeded overnight at 1.5 × 105 cells/well in 6-well plates prior to drug addition. After 24 h of drug incubation, cells were harvested and 50 μl was resuspended in 500 μl low melting point agarose to give a final density of 1.5 × 105 cell/ml. A total of 50 μl of this was spread onto Trevigen Comet slides and allowed to solidify for 1 h at 4°C. Samples were lysed in a Coplin jar (2.5 M NaCl, 100 mM EDTA, 10 mM Tris–HCl, pH 10) overnight at 4°C (43). Slides were allowed to equilibrate in cold electrophoresis buffer (300 mM NaOH, 1.0 mM EDTA, pH 13) for 30 min at 4°C prior to electrophoresis run at 300 mA for 30 min with buffer levels adjusted to give a consistent voltage of 25 V. Slides were then washed with water, neutralized in buffer (400 mM Tris–HCl, pH 7.5), fixed with 70% ethanol (30 min) and dried for desiccated storage. Prior to scoring, slides where rehydrated for 15 min before staining with a PI solution (10 μg/ml, 10 min) and imaged through a 10 × lens and Leica DFC 500 epi-fluorescent microscope. Images were analysed with Open Comet plugin on Image J V2.0.
Computional procedures
The three exchange functionals used in this study were the three parameter hybrid functional developed by Becke (44) (B3), Handy’s Optimized Exchange Density Functional (OPTX) modification of Becke’s exchange functional (45) (O) and the exchange component of the Perdew-Wang 91 functional (PW91) (46–48). The first of these are hybrid functionals (B3 and O) used the correlation functional of Lee, Yang and Parr (LYP) designated B3LYP and OLYP, respectively (49). B3LYP has performed well for systems containing first and second row elements and mono-nuclear transition metal complexes. However B3LYP exhibits an undue preference for high spin states. OLYP is a more recent development and has been applied successfully to bioinorganic systems but it appears to favor more spin polarized descriptions. Finally the pure PW91 exchange functional was used in conjunction with Perdew and Wang’s gradient-corrected correlation functional (PW91) designated PW91PW91 (46). This combination proved problematic in its implementation for the system reported here, but has correctly described the spin state of some manganese-containing porphyrins (50,51).
The LanL2DZ basis set was chosen for all calculations (uses the Dunning Huzinaga (D95) valence double ζ description for first row elements (52) and the Los Alamos effective core potentials plus double ζ functions on elements from Na to Bi) (53–55). All calculations were modeled in water using the Polarisable Continuum Model (56), and implemented using the Gaussian 09 (Revision E.01) program suite at the Irish Centre for High End Computing. Analysis of molecular orbital compositions and spin density were performed using the AOMix package (57).
Those methods which best preserve the symmetry around the two copper atoms are assumed to have performed well. To approach these calculations, a molecular structure was generated by molecular modeling techniques. This was then refined for each selected charge and multiplicity to tight convergence criteria using a pruned (99,590) integration grid. The single determinant wavefunction was then tested for multiplicity instabilities before calculating the second order derivatives of energy with respect to nuclear coordinates. The absence of negative eigenvalues of the Hessian matrix confirmed that the molecular geometry represents a minimum on the potential energy hypersurface. In all cases, the asymmetric coordination of the carboxylate coordination to the copper atoms was observed, with this coordination mode in agreement with that obtained from single crystal X-ray diffraction (20). Consequently two isomers of the complex are possible, denoted cis and trans (Supplementary Figure S1). Calculations were performed on both isomers, however as there were no significant difference between the isomers the results from the trans-isomer are presented here; although this does not preclude the presence of the cis-isomer present in solution. The full numbering system of trans-[(Cu(Phen)2)2(μ-Terph)]2+ is presented in Supplementary Figure S2. Identical numbering conventions were used for all calculations in this study irrespective of charge or multiplicity.
Nuclease activity
DNA cleavage profiles of the copper complexes were investigated in the presence of hydrogen peroxide scavengers. To a final volume of 20 μl, 80 mM HEPES (pH = 7.2), 25 mM NaCl, 1 mM Na-L-ascorbate and 400 ng of pUC19 DNA were treated with drug concentrations of 250 nM to 2.5 μM in the presence of 10 mM hydrogen peroxide scavengers pyruvate or 1, 3-dimethylthiourea (DMTU). Reactions were incubated for 30 min at 37°C and quenched with 6 × loading buffer containing 10 mM Tris–HCl, 0.03% bromophenol blue, 0.03% xylene cyanole FF, 60% glycerol and 60 mM EDTA and loaded onto an agarose gel (1.2%) containing 8 μl EtBr. Electrophoresis was run at 70 V for 1 h in 1 × Tris-Acetate-EDTA (TAE) buffer.
Immunodection of double strand breaks (DSBs)
Samples were prepared similarly to previously reported methods (58). The 6 × 104 cells were seeded in 12-well plates and treated with complexes and Dox for 24 h. Transferred samples in 1.5 ml microtubes were fixed with 1.5% formaldehyde (15 min, room temperature) followed by ice-cold 70% ethanol and stored at −20°C. Samples were resuspended in permeabilization buffer (0.25% Triton X-100 in PBS) for 30 min on ice and blocked with 2% BSA (30 min, room temperature). The primary antibody (1:500) was incubated for 2 h at room temperature followed by secondary antibody (1:1000) for 1 h at room temperature and co-stained for 10 min with 5 μg/ml of PI. Samples were acquired using ExpressPro software on the Guava EasyCyte instrument.
Topoisomerase inhibition
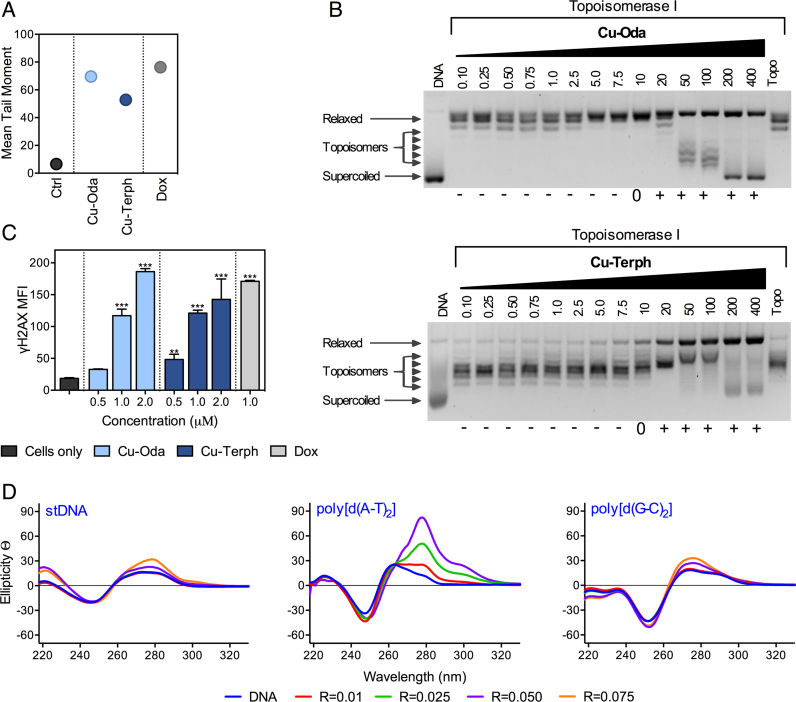
The topoisomerase I relaxation assay was carried out using a modified method to previously reported protocols (59). A total of 400 ng of pUC19 plasmid DNA was exposed to varying concentrations of drug (0.1–400 μM) for 30 min at room temperature in a final volume of 20 μl containing 80 mM HEPES buffer (pH 7.2), CutSmart® buffer and 100× BSA. Topoisomerase I (1 unit) was added to the mixture and incubated for 15 min at 37°C to ensure relaxation of plasmid DNA. The enzymatic reaction was quenched with SDS (0.25%), protein kinase (250 μg/ml) for 30 min incubation at 50°C. Samples were loaded onto 1.2% agarose gel with 6× loading buffer. Topoisomers of DNA were separated by electrophoresis in 1× Tris-Borate-EDTA (TBE) buffer at room temperature at 40 V (3 h) followed by 50 V (2.5 h). The agarose gel was post-stained using an ethidium bromide bath (25 μM) for 20 min at room temperature. Finally, the gel was soaked in deionized water for 24 h and imaged using a UV transilluminator.
Circular dichroism spectrometry
Complex–DNA interactions were analyzed within Starna quartz cuvettes using an Applied Photophysics Chirascan-Plus circular dichroism spectrometer. Solutions of oligonucleotides in H2O and 10 mM NaCl (stDNA, ϵ260 = 12824 M(bp)−1 cm−1), poly[(d(A-T)2] (ϵ260 = 13100 M(bp)−1 cm−1) and poly[(d(G-C)2] (ϵ260 = 16800 M(bp)−1 cm−1) were initially denatured by heat treatment and then allowed to slowly reanneal prior to quantification using an Agilent Cary 100 dual beam spectrophotometer to give working solutions of 2 μM. Spectra were captured in the range of 180–350 nm at 37°C with step of 1.0 nm increments, time/point of 1 s and bandwidth of 1.0 nm. DNA solutions were incubated for 30 min periods at 37°C with 2, 5 and 10 μM of complex. Studies investigating Z-DNA transitions and competitive binding interactions were conducted with 2 μM of D1 and 10 μM Cu-Oda, followed by 2.0 μM of D6. Reversion from Z to B DNA was achieved upon addition of 100 mM EDTA. Electrostatic controls used in Z-DNA experiments were NaCl (4 mM), Co(NH3)6 (10 μM) and Cu(ClO4)2 (10 and 20 μM). Cu-Terph DNA interactions were not investigated within this study due to solubility issues in CD accessible solvents. Blank water and drug samples were treated in an identical manner and subtracted from recorded data. Average traces were smoothed to four neighboring points and second order polynomial in GraphPad Prism (V4.0).
Thermal melting
Oligonucleotides were denatured by heating to 95°C at a rate of 0.1°C/s and held for 10 min. Sequences were allowed to anneal by cooling to 5°C at rate of 0.1°C/s on an Eppendorf Mastercycler Nexus prior to thermal melting experiments. Analysis was carried out on Agilent Cary 100 dual beam spectrophotometer equipped with a 6 × 6 Peltier multicell system with temperature controller. Oligonucleotides were quantified and adjusted to give 5 μM solutions with drug loading ratios of 1.0 in PBS (2.7 mM KCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4, 137 mM NaCl, pH 7.4). Melting curves were recorded at their respective λmax in the region of 260 - 280 nm upon heating from 17–95°C at rate of 0.2°C/s. Thermodynamic properties were calculated post acquisition using the built-in Cary Thermal software, hyperchomicity analysis.
Docking studies
The dockings of the dicopper ligands with chosen DNA fragments were modeled using AutoDock (Version 4.2.5). The molecular structure of Cu-Terph was obtained from single-crystal X-ray diffraction data (20) and hydrogen atoms were located at their calculated positions, while that of Cu-Oda was estimated using molecular modeling techniques. The structures of the DNA fragments were obtained from the Brookhaven Database (www.rcsb.org/pdb/home/home.do). These structures did not include hydrogen atoms, and these were added at their calculated positions using GaussView 3.0. The Gaussian 09 newzmat utility was then used to generate the .pdb input files as required by AutoDock. Analysis of the tortional freedoms in the ligands confirmed that [((Phen)2Cu)2(μ-Terph)] contains two tortional degrees of freedom while the more flexible [((Phen)2Cu)2(μ-Oda)] contains 11. In the case of [((Phen)2Cu)2(μ-Oda)], the conformation was randomized prior to docking calculations to minimize starting configuration bias. In each docking the search space encompassed the entire oligomer which was treated as a rigid structure. The docking calculations used the Lamarcian Genetic Algorithm.
Statistical analysis
Data collected from intracellular and DNA binding experiments are presented as mean ± S.D., where n = 3, and plotted in GraphPad Prism V6.0. Comparisons between the data were analyzed with two-way ANOVAs in GraphPad Prism. Differences between means were analyzed post-hoc with Dunnett’s test at 95% confidence level. Differences between groups were considered to be statistically significant if P ≤ 0.05 with *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
RESULTS AND DISCUSSION
Distinctive anticancer mechanism and broad-range chemotherapeutic response
NCI-60 analysis
Both complexes were examined within the NCI’s DTP against a variety of human cancer cells including breast, colon, central nervous system (CNS), leukaemia, non-small cell lung, melanoma, ovarian, prostate and renal. Their cytotoxic or lethal effects (50% lethal dose, LC50) were identified using a five-dose exposure level and are shown as a heat map in Figure 1B. Additional analysis to assess 50% growth inhibition (GI50), and cytostatic effects (total growth inhibition, TGI) were also undertaken (Supplementary Table S1 and Figure S3). Cu-Terph showed enhanced cytotoxic effects across the majority of cell lines within the panel (LC50 < 1 μM) with selectivity toward CNS, colon, ovarian and hypersensitivity against the melanoma line SK-MEL-5. Cu-Oda displayed a broader range of activities and was most effective against melanoma, renal and prostate cancers where LC50 concentrations typically ranged between 5.0 and 0.5 μM. Interestingly, leukemic cell lines were resistant to both di-Cu2+ complexes where lethal concentrations exceed 100 μM.
The activity profiles for both compounds were then entered into the COMPARE algorithm supplied by the NCI that associates the pattern of activity to standard clinical and developmental agents, many of which have known cytotoxic mechanism (60). Results are represented as PCCs, or r values, ranging from −1 (negative correlation) to +1 (positive correlation) where Cu-Oda or Cu-Terph samples surpassing a threshold of >0.20 were considered. From a total of 175 standard agents analyzed through COMPARE, just 43 had detectable r-values ranging up to 0.84 (Figure 1C). While medium to high PCCs were identified for a limited number of hormonal and signaling agents, the highest number of correlations appeared within the DNA damaging class followed then by topoisomerase poisons. Both copper complexes showed low correlation with cisplatin with calculated r-values of 0.26 (Cu-Oda) and 0.33 (Cu-Terph). Of further significance, clinically established metallodrugs such as bleomycin and alternative platinum chemotypes (including carboplatin and oxaliplatin) were found to be outside the measurement threshold, potentially supporting a novel mechanism of action by both di-Cu2+ complexes.
Apoptotic induction
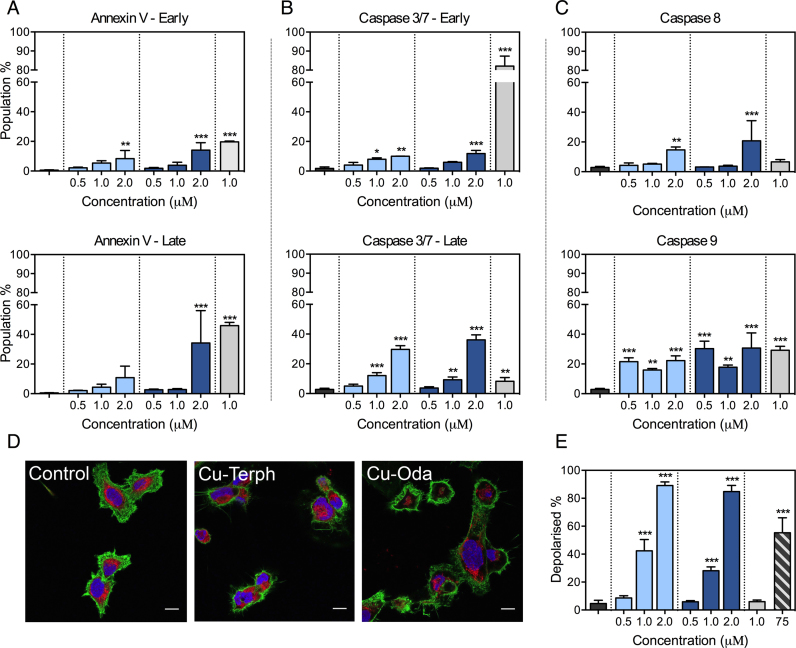
The ovarian adenocarcinoma cell line SKOV3 was selected to probe intracellular genotoxic properties of Cu-Oda and Cu-Terph as this cancer form has the leading rate of gynaecological cancer mortality, poor prognosis, and is further characterized by chemoresistance and recurrence (61). SKOV3 elicited sensitivity to both Cu-Terph and Cu-Oda treatment within the NCI-60 panel, yet distinctive GI50 activities (160 nM and 1 μM, respectively) were encountered, making this cell line an important choice to probe for differences between both the agents. SKOV3 is also innately resistant to cisplatin (62) with malfunctioning p53 (63), low expression of Ctr1 copper influx protein (64) and these are known to reduce cancer vulnerability to chemotherapeutic and radiation treatment (65). To identify mechanisms underpinning mammalian cancer cell death, morphologic changes were investigated using confocal microscopy dyes that monitor the integrity of the mitochondria, cytoskeleton and nucleus. In particular, the hallmarks of apoptosis were assessed using: cytosolic and organelle contraction; condensation and increased chromatin granularity; membrane blebbing; and the formation of apoptotic bodies (apobodies) (66). Apoptotic characteristics were evident within SKOV3 cells exposed to cytotoxic controls of carbonyl-cyanide m-chlorophenylhydrazone and Dox. Nuclear fragmentation was clearly visible here in comparison to non-treated SKOV3 cells that were typically elongated, polygonal and dome-like in morphology (Figure 2D). Cells treated with Cu-Oda and Cu-Terph also showed organelle condensation together with fragmentation of nuclear content, the remnants of apobodies and an apparent reduction in mitochondrial fluorescence. Given these observations further evidence of apoptotic induction was probed using Annexin V and caspase activation. During apoptosis, phosphatidylserine (PS) translocates from the inner to outer cellular membrane surface and Annexin V—which selectively binds to PS—can be combined with the DNA intercalator aminoactinomycin D (7-AAD) to quantify cells within early- and late-stage apoptosis by flow cytometry. At maximum exposure both complexes activated early-stage apoptosis in SKOV3 cells (Figure 2A) whereas notable late-stage apoptosis (∼34% of cells) was identified for the Cu-Terph treated population, slightly below that observed for Dox (∼45%). A selection of pathway-specific endoproteases—caspase 3/7, 8 and 9—were next examined. Late-stage activation of executioner caspase 3/7 was detected here with moderate early-stage 3/7 activity and this effect contrasted with Dox where almost exclusive early-stage caspase activation occurred (Figure 2B). In a final part of the analysis, initiator caspase 8 (extrinsic) and 9 (intrinsic) activation was monitored to identify the broad origins of apoptosis (67). In this study both di-Cu2+ complexes primarily induced apoptosis through the intrinsic caspase 9 pathway where ∼22 and ∼30% of cellular populations were responsive at an exposure level of 2.0 μM (Figure 2C). Caspase 8 was detected in a lower percentage of the cell population and this may be attributed to intercellular cross-talk from apoptotic cells (68,69).
Figure 2.
SKOV3 cells are treated over 24 h with varying concentration of Cu-Oda and Cu-Terph (0.5–2.0 μM) and control agents Dox (1.0 μM) and CCCP (75 μM). (A) Populations of early apoptotic cells exhibiting positive staining for Annexin V only with late apoptotic population detected through positive staining for both Annexin V and 7-AAD. (B) Detection of caspase 3/7 in the absences or presence of 7-AAD positive staining indicative of mid- and late-apoptotic activation. (C) Activation of caspase 8 (extrinsic pathway) and caspase 9 (intrinsic pathway). (D) 100 × magnification of SKOV3 cells treated with di-nuclear copper complexes, Cu-Oda and Cu-Terph (1.0 μM). Nuclei are stained with DAPI, F-actin with Alexa Fluor 488-Phalloidin and mitochondria with MitoTracker Deep Red. Scale bar indicates 10 μm. Individual channels are shown in Supplementary Figure S4. (E) Population of depolarized mitochondria through the detection of potential-sensitive shift in JC-1 emission. Hypsochromic shifts, evident in scatter plot, demonstrates drug-induced depolarization (Supplementary Figure S5). Non-significant (ns) P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Loss of mitochondrial membrane potential (ΔΨm)
Investigations identifying mitochondrial dysfunction were completed by examining fluorescent emission of the ΔΨm-sensitive probe JC-1. Both Cu-Oda and Cu-Terph stimulated mitochondrial depolarization in a concentration-dependent manner where >84% of cellular populations displayed a loss in membrane potential at the maximum tested dose (Figure 2E). The positive control CCCP was selected due to its proton ionophore properties and ability to readily dissipate the mitochondrial transmembrane potential. Under identical conditions a significantly higher concentration of CCCP (75 μM) was required to activate similar membrane depolarization. Drug-mediated depolarization of this magnitude, however, does not directly correlate with the comparatively modest detection of intrinsic apoptotic biomarkers (Annexin V and caspases). In this case it is likely the extent of the loss of ΔΨm results from selective mitochondrial metallodrug localization and ROS generation. It is reasonable to suggest that mitochondrial accumulation is facilitated by the delocalized cationic charge and lipophilicity of copper-coordinated 1,10-phenanthroline as this is a common feature of dyes and targeting vehicles that selectively assemble within this organelle (70).
Di-Cu2+ complexes stimulate superoxide and singlet oxygen production
Intracellular free radical production
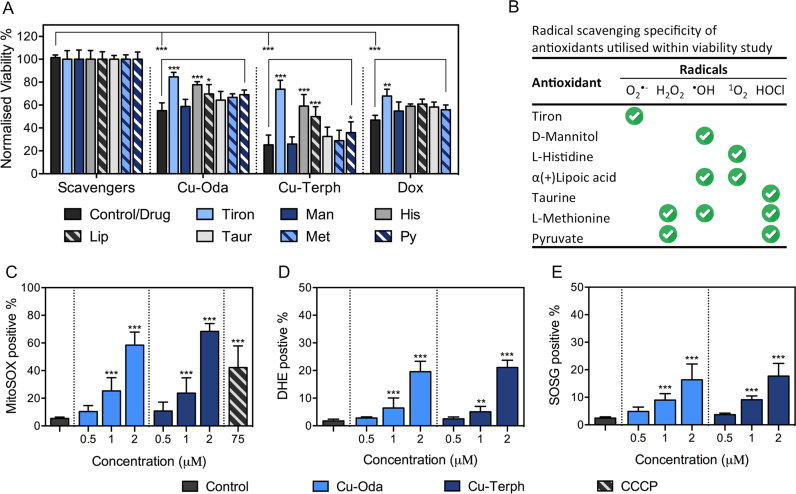
Following the observation of extensive mitochondrial depolarization, we were prompted to conduct a detailed examination of intracellular production of ROS species catalysed by these complexes. A study was designed to actively monitor cell viability upon co-treatment with the radical-specific scavengers/spin-trapping agents: tiron; mannitol; histidine; lipoic acid; taurine; methionine; and sodium pyruvate (Figure 3A and B). The extent to which cytotoxicity was averted through treatment with these ROS-selective antioxidants is remarkable with viability profiles of Cu-Terph and Cu-Oda fluctuating from 25 to 73% and 55 to 84%, respectively. The most protective scavenger identified was tiron—a known superoxide (O2•−) sequestering agent that undergoes direct oxidation to yield semiquinone radicals (36)—which increased cellular viability by ∼30% for Cu-Oda and ∼50% for Cu-Terph. Interestingly, the next most active ROS identified was singlet oxygen (1O2) that is known to readily oxidize the imidazole ring of histidine (71). Although the generation of hydrogen peroxide (H2O2) is mechanistically feasible as a precursor to deleterious species such as the hydroxyl radical, experimental observations here show considerably lower cell survival (14–16%) in the presence of the pyruvate H2O2 scavenger. The overall trend in terms of antioxidant-promoted SKOV3 survival followed: tiron >> histidine > lipoic acid > pyruvate > taurine > methionine > mannitol, with the more structurally rigid complex Cu-Terph giving higher intracellular O2•− and 1O2 production.
Figure 3.
(A) Viability results of Cu-Oda, Cu-Terph and Dox at 1.0 μM in the absence and presence of 1 mM ROS-specific scavengers listed in (B). (C) Detection of cellular population positive for MitoSOX through MitoSOX red. (D) Detection of DHE, indicative of superoxide-selective fluorescence activation within the nucleus. (E) Singlet oxygen formation through fluorescence activation of SOSG. Non-significant (ns) P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Intracellular ROS quantification
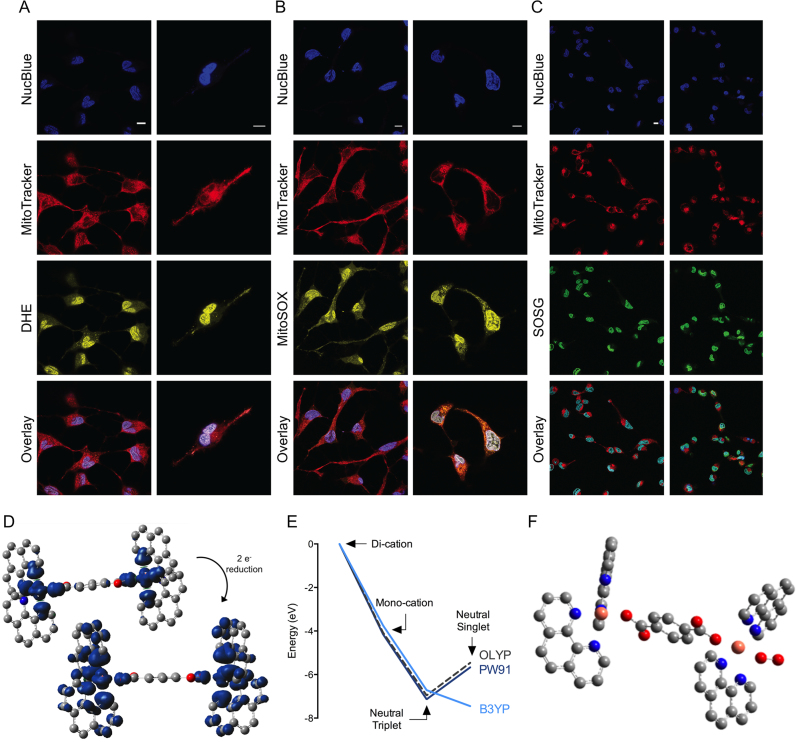
In order to probe intracellular ROS production by Cu-Oda and Cu-Terph using fluorescence, the liberation of (i) superoxide within the mitochondrial matrix was identified by MitoSOX Red, (ii) nuclear superoxide was recognized by DHE (72) and (iii) singlet oxygen was detected by SOSG (Figure 3). Results show dose-responsive mitochondrial O2•− generation by Cu-Oda and Cu-Terph complexes with ∼58 and ∼68% of the respective cellular populations containing MitoSOX Red fluorescence upon 2.0 μM drug exposure (Figure 3C). Mitochondrial generated superoxide exceeded that produced within the nucleus with DHE detected in ∼20% of SKOV3 exposed to both complexes (Figure 3D). Finally, 1O2 was detected by the photoinduced energy transfer exomarker SOSG (73) where exposure to Cu-Oda and Cu-Terph produced ∼17% positive populations for SOSG emission (Figure 3E). To corroborate ROS production, fluorogenic markers were visualized by confocal microscopy with Cu-Terph (Figure 4). Strong fluorescence emission was evident for DHE (Figure 4A) and SOSG (Figure 4C) with the dye localising primarily in nuclear and peripheral regions of SKOV3 cells. The fluorescence intensity for MitoSOX red was particularly high (Figure 4B); this increased detection of mitochondrial superoxide (MitoSOX) is in agreement with earlier flow cytometry results.
Figure 4.
Confocal images of intracellular ROS generated by Cu-Terph (0.5/1.0 μM) collected with 63 × water immersion objective. SKOV3 nuclei were stained with NucBlue Live, mitochondria with MitoTracker Deep Red and selective detection of ROS with fluorogenic probes DHE for nuclear superoxide (DHE, A), mitoSOX red for MitoSOX (B) and SOSG (C). Scale bars indicate 10 μm, D. Spin density maps upon reduction of Cu-Terph2+ to Cu-Terph0. (E) Energies of trans-Cu-Terphn+ (n = 2, 1 and 0) relative to the di-cationic complex set at 0 eV for each functional and (F) molecular structure of the oxygen adduct of triplet Cu-Terph0.
Free radical formation from doubly reduced Cu-Terph
To establish a theoretical basis for ROS generation observed in cellular systems, density functional theory (DFT) was employed. Since these calculations and description of the generalized gradient approximation (GGA) in the exchange functional can significantly distort the analysis, we used symmetry constraint—revealed by X-ray diffraction—to assess the performance of three functionals (B3LYP, OLYP and PW91PW91) at three oxidation states of the Cu-Terph complex: the di-cation singlet and triplet; the mono-cation doublet; and the neutral species—singlet and triplet. Of the three methods, B3LYP performed well at maintaining the symmetry of the two copper centres for both the singlet and triplet di-cation (Supplementary Table S3). The triplet solution had marginally lower energy and the metal-centred bond lengths were closer to the experimental values. Spin density mapping indicated the unpaired electrons of the triplet reside on the copper atoms, the coordinating oxygens, and three equatorial nitrogen atoms of the phenanthroline ligands (Figure 4D). For all functionals studied, the singly reduced form had significantly lower energy compared to the di-cation by approximately 4 eV (Supplementary Table S4). As for the biologically relevant doubly reduced species, all calculations indicated the neutral species is substantially more stable than the di-cation by between 5 and 7 eV depending on the functional used (Supplementary Table S5). The energy difference between the triplet and singlet species is dependent on the functional; the B3LYP functional suggests the singlet species had the lowest energy by some 0.8 eV while both OLYP and PW91 indicated that triplet species had lowest energy by 1.4 eV. Overall it is clear the Cu-Terph cation is a powerful oxidising agent but it is unclear as to which spin state is the most stable for the resulting neutral species (Figure 4E).
To provide a theoretical explanation for the potential biological interaction between the complex and molecular oxygen, the reaction of the doubly reduced complex with oxygen gas was investigated in DFT simulations by placing an oxygen molecule close to one copper atom within the optimized adduct structure. B3LYP and OLYP functionals were chosen for these calculations where reaction of the neutral species with triplet oxygen resulted in an oxygen adduct with either singlet or triplet multiplicity. A stable O2 adduct species was located on the triplet hyper-surface using both B3LYP and OLYP functionals and this species can be described as superoxide with a calculated O-O bond distance of 1.33 Å. The optimized molecular structure is presented in Figure 4F (also see Supplementary Figure S6 and Table S11) and switching this complex to the singlet surface resulted in a drift of the oxygen ligand away from the coordination sphere of the copper atom with release of singlet oxygen. Further attempts undertaken to locate a stable H2O2 adduct on either the singlet or triplet surfaces failed. Addition of the H2O2 into the coordination sphere of one copper atoms resulted in the dissociation of one Phen ligand from the peroxo species. Consequently, we could find no theoretical basis for the [Cu2(phen)4(μ-Terph)]° complex to act as a vector for peroxide-mediated damage. In an effort to probe the role of peroxide in the DNA damage mechanism further, the chemical nuclease activity of both complexes in the presence of pyruvate and an additional H2O2 trapping agent, DMTU, were examined (Supplementary Figure S7). Here, DNA was incubated with increasing complex concentrations (250 nM–2.5 μM) and revealed oxidation profiles that converted supercoiled (SC) plasmid DNA into open circular and linearized (L) forms. The presence of either scavengers did not significantly alter cleavage activity (in comparison to controls) to justify peroxide having a prominent mechanistic role in the DNA oxidation profile.
Genotoxicity causes global cellular dysfunction
Fragmentation of genomic DNA
Single-stranded breaks (SSBs) and double-stranded breaks (DSBs) along with alkali labile sites arising from genomic DNA damage were examined by the electrophoretic alkaline comet assay. Cu-Oda and Cu-Terph were found to fragment DNA with corresponding tail moments of ∼69 and ∼52 a.u., respectively (Figure 5A) with Cu-Terph exhibiting broader distribution of residual DNA damage (Supplementary Figure S8). This result was corroborated by observation of site-specific DNA damage in the topoisomerase relaxation assay (Top I) where negatively SC DNA was first unwounded due to intercalation, prior to positive supercoiling and nicking (SSBs) to the relaxed form (Figure 5B). In the COMET assay, Dox also produced extensive DNA damage with mean tail moments of ∼76 a.u. along with frequency distributions indicative of non-discriminate oxidative cleavage. Subsequent analysis using Top I unwinding showed Dox could efficiently stabilize the DNA-Topo I enzyme complex (Supplementary Figure S9).
Figure 5.
(A) Mean tail moments of untreated SKOV-3 cells and 1.0 μM Cu-Oda, Cu-Terph and Dox. (B) Topoisomerase I unwinding across concentration range 0.10–400 μM for Cu-Oda and Cu-Terph. (C). DSBs induced by Cu-Oda and Cu-Terph, detected by immunostaining of γH2AX with MFI presented for di-Cu2+ complexes and Dox. (D) CD spectra of Cu-Oda with stDNA and alternating co-polymers poly[d(A⋅T)2] and poly[d(G⋅C)2] (100 μM) at loading ratios 0.01–0.075.
The formation of DSBs was next explored through immunodetection of γH2AX. It is known that histones are indiscriminately incorporated into the nucleosome for higher order chromatin formation. The H2AX histone variant is characterized by a carboxyl tail containing a serine residue that becomes phosphorylated (γH2AX) in response to localized DSBs (74), thereby acting as signaling foci for the recruitment of DNA repair factors (75). In this study, DNA damage was estimated from the median fluorescence intensity (MFI) of γH2AX in the tested cell population (Figure 5C). Both complexes displayed concentration-dependent DSB formation with maximal MFI detection comparable to Dox exposure. It is significant to note how increased nuclearity enhances genomic DSB formation as a mononuclear analogue of these complexes—[Cu(o-phthalate)(phen)]—produced one-order of magnitude lower MFI of γH2AX under identical conditions (19).
Circular dichroism (CD) analysis with duplex polymers
Circular dichroism spectroscopy was selected to interrogate metal complex–DNA interactions. It is known that the B-form of double stranded DNA exhibits negative elliptical signals at ca. 210 and 246 nm arising from β-N-glycosidic linkages and helicity, also that positive bands at ca. 220 and 268 nm are due to H-bonding and base-pair stacking interactions. Variations in these regions arise from sequence diversity and AT content (76); however, nucleic acids with the Z-conformation display an inverse profile to B-DNA due to their left-handed helicity. Cu-Oda was examined using B-DNA polymers of salmon testes DNA (stDNA), poly[d(G-C)2], and poly[d(A-T)2] (Cu-Terph was precluded from this study due to its lack of solubility in CD accessible solvents). Concentration-dependent complex exposure gave rise to an intercalative profile in both stDNA and the alternating copolymer poly[d(G-C)2] with pronounced elliptical shifts observed at ∼275 nm (Figure 5D). This effect was enhanced within poly[d(A-T)2] where along with structural distortions identified in the form of helical unwinding (∼246 nm), a significant bathochromic shift proximate to the nitrogenous-base stacking region developed. These results suggest Cu-Oda is a high-affinity intercalator of the synthetic A-T co-polymer.
AT/AT and TA/TA steps dictate intercalative recognition
Circular dichroism (CD) analysis with short oligonucleotides
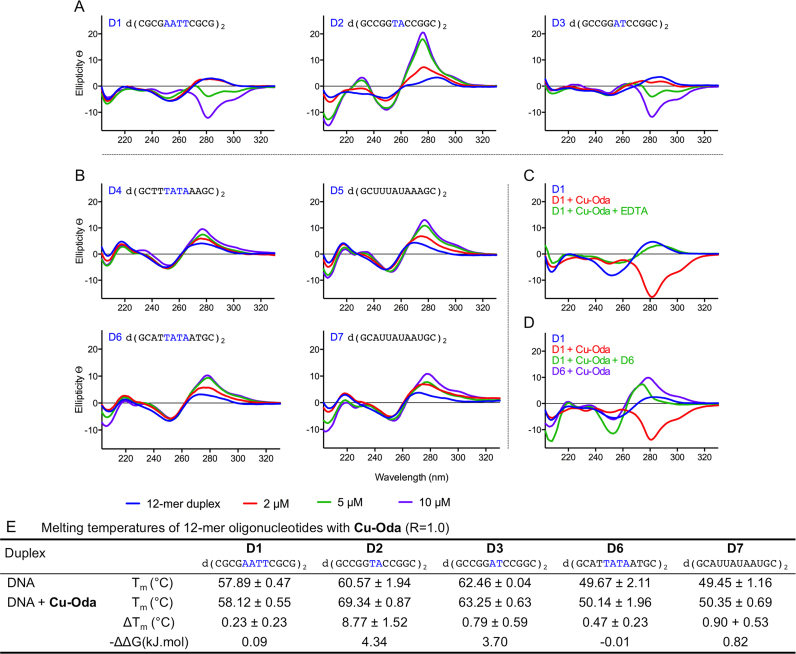
To examine the intercalation of Cu-Oda in terms of sequence specificity, a selection of palindromic dodecamers were examined with varying AT sequence content (D1-7, Figure 6). Binding of Cu-Oda to the Dickerson-Drew sequence (D1) displayed a decrease in elliptical signals mimicking a localized Z-DNA like configuration that was traced by a negative Cotton effect at 281 nm (Figure 6A). Complete conversion to Z-DNA can be disregarded here as no structural distortions in the form of DNA unwinding, helicity and left-handedness were identified at ca. 250 nm that are typically observed within the Z-DNA configuration. Furthermore, Cu-Oda was found to discriminate between TA/TA and AT/AT steps present within D2 and D3 oligomers (Figure 6A). D2 and D3 differ at this central step only and distinctly different CD structural distortions become apparent upon Cu-Oda exposure. Remarkably, positive elliptical shifts at 288 nm revealed site-specific intercalation at the TA/TA site in D2, while a strong minimum in this region indicated localized Z-like DNA conformation at the AT/AT site in D3. Binding to TATA sequences (D4 and D6) gave rise to intercalative profiles (Figure 6B) with ellipticity becoming pronounced upon substituting thymine (T) with uracil (U) in each oligonucleotide sequence (D5 and D7). Evidence here suggests at least some portion of intercalation occurs at the major groove since the absence of bulky methyl groups within T must render the major groove more accessible for the complex. Of further significance, the formation of Z-like DNA could not be replicated in the AT/AT sequence (D3) by (i) excessive salt concentration, (ii) exposure to the electrostatic binding agent [Co(NH3)6]3+ or (iii) by free Cu2+ ions alone (Supplementary Figure S10). It is also notable that Cu-Oda mediated Z-like DNA formation within the Dickerson-Drew sequence (D1) was reversed upon titrating EDTA (Figure 6C) and competitive addition of oligomer D6 (Figure 6D) with this effect further suggesting a binding preference of the complex to TA-rich sequences.
Figure 6.
(A) CD spectra of Cu-Oda (2, 5 and 10 μM) with dodecemeric duplexes (2 μM). Central AnTn steps investigated (where n = 2 in Dickerson-Drew, D1 and n = 1 in D3) and TnAn (where n = 1 in D2). (B) Differential binding mode dependent on sequence context (TA)n where n ≥ 2 for D4 and D6 and T → U equivalent sequences in D5 and D7. (C) Z → B-DNA reversion when complex is sequestered with EDTA (1 mM). (D) Localized B → Z-DNA ‘like’ transitions of D1 (2 μM) upon complexes addition (10 μM, red line) and preferential intercalation reforming right-handed DNA with D6 (2 μM, purple line). Elliptical signals-glycosidic linkages (208 nm), H-bonding (ca. 220 nm), helicity or handedness (ca. 250 nm) and base-pair stacking interactions (270–280 nm). (E) Denaturing temperatures and change in stabilization energies (ΔΔG at 37°C, where ΔΔG = ΔG(DNA+Drug) − ΔG(DNA)) of 12-mer duplexes (5 μM) in the presence of Cu-Oda (1.0 loading ratio).
Thermal melting analysis
Following CD analysis, thermal melting studies were conducted on a selection of relevant palindromic dodecamers (Figure 6E). Sequences containing AT/AT central steps (D1 and D3) were only marginally stabilized by the complex with representative ΔTm values of +0.23 and +0.79°C. These results were not surprising given the overall reduction in free energy of Z-DNA due to close-set proximity of phosphate groups in the helical backbone. Interestingly, complex binding at the TA/TA step in D2 resulted in significant stabilization of +8.77°C and enhanced duplex stability of 4.34 kJ/mol that is presumably due to intercalation. Marginal stabilization was noted for D6 (+0.47°C) and this effect was enhanced for oligomer D7 (+0.90°C) indicating the extent of duplex stability is proportional to the level of intercalation and accessibility of the major groove.
Preassociative docking studies
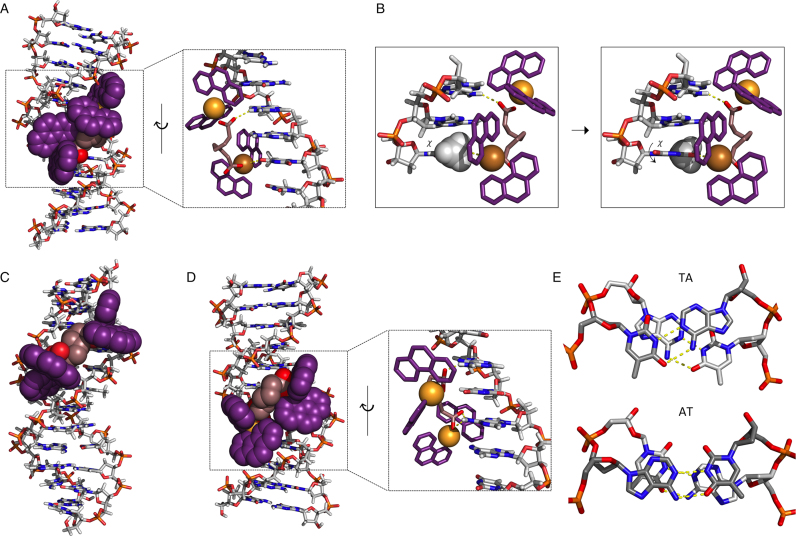
To further explore the sequence binding capabilities of this complex, molecular docking experiments were undertaken. The study treated a selection of short duplex sequences, either identical or broadly similar to those examined by circular dichroism spectroscopy, as rigid oligomers with the complex maintained in a low energy randomized configuration to constitute a non-biased docking analysis. This study might therefore give useful pre-association information of the complex–DNA interaction. Binding of Cu-Oda to the Dickerson-Drew sequence (D1, taken from PDB ID: 4C64) occurs in the major groove (Figure 7A and Supplementary Movie S1) and arises from an intrastrand electrostatic interaction between one Cu2+ ion and an uncoordinated carboxylate oxygen with A (N6) and T (O4) bases in the AAT triplet (Supplementary Movie S2). Initial pre-association shows steric hindrance between the methyl group at the C5 position of T and one phen ligand auxiliary to polar interaction Cu····T (O4) (see Supplementary Movie S3). Rotation about the glycosidic bond (χ) gives rise to a syn configuration from the sterically favored anti orientation (Figure 7B) and could explain the observed Z-like DNA configuration seen for D1 and D3 in our CD analysis. Binding to the Pribnow box sequence (TATAAT, taken from PDB ID: 5EZF) (contained within D4 and D6) can occur within both the minor and major groove, with the minor groove giving the most stable docking interaction (Figure 7C and Supplementary Movie S4). Substituting T with U within this sequence context gives rise to a favored major groove interaction (Figure 7D), with an initial electrostatic interaction (prior to intercalation) involving carboxylate oxygen atoms binding with two A bases in the AUA triplet.
Figure 7.
Molecular docking simulations. (A) Cu-Oda binds with DDD (PDB ID: 4C64) in major groove interactions (Supplementary Movie S1). This pre-associative binding is due to electrostatic interactions N7 on A and O6 on T (yellow-dashed line, also see Supplementary Movie S2). (B) AAT triplet with rotation around glycosidic linkage (χ) of thymine (Supplementary Movie S3). (C) Minor groove binding to PDB ID: 5EZF (d(CGCTATAATGCG)) (Supplementary Movie S4) and (D) modified T→U (d(CGCUAUAAUGCG)) resulting in alternative interactions in the major groove at AUA triplet. (E) Top down view of stacking interactions in TA and AT steps from PDB ID: 167D (d(CCATTAATGG)) in which only the P and sugar O atoms have been fitted. Hydrogen bonds are shown as dashed yellow lines; the angle between the vectors representing the two N3(T)→N1(A) hydrogen bonds is 118° for the TA step and 160° for the AT step.
The observed difference between AT and TA steps in respect of intercalation might be expected to have a structural basis. Figure 7E shows a qualitative comparison of typical AT/AT and TA/TA steps (taken from PDB ID: 167D); a qualitative comparison of the two shows that the position of the backbone structure remains similar; however the relative orientations of the base pairs is very different. In the AT step the bases are clearly overlapped and the dihedral angle between the two NH····N hydrogen bonds is 160°; in the TA step the base pairs are significantly rotated, with the NH····N dihedral angle of 118° and reduced opportunity for effective π–π stacking. A likely basis for discrimination between AT and TA steps is that the more extensive overlap of the bases in the AT step allows a more extensive stacking interaction and therefore more resistance to intercalation than at the poorly-overlapped TA step. Although this simplistic view does not address the detailed nature of the stacking interactions, the conclusion is borne out by other experimental studies. Breslauer et al. analyzed the melting behavior of a series of DNA oligomers to extract thermodynamic data for each of the 10 unique nearest neighbor steps (77). Their thermodynamic data for helix-coil transition (i.e. stacking plus H-bonding) gave relative stabilities (ΔG°) of 1.5 and 0.9 kcal/mol for AT and TA steps, respectively. Subsequently Alexandrov et al. combined melting data and Monte Carlo simulations to separate H-bonding and stacking contributions (78). They extracted stacking force constants of 0.023 and 0.019 eV/Å2 for AT and TA steps, respectively. In both cases the values obtained could be used additively to predict, for example, melting temperatures for given oligomeric duplexes, amongst various other thermodynamic parameters. Recently Kilchherr et al. used single molecule atomic force microscopy (AFM) studies to measure stacking forces between all possible sets of base pairs; again the AT step was more stable than TA (free energies of stacking −2.35 and −1.02 kcal/mol with dissociation rates 3.22 × 103 and 3.11 × 104 s−1, respectively) (79). In each of these studies the results were strongly dependent on the experimental conditions, particularly the salt concentration. Cardin and co-workers have observed similar sequence bias where Λ–[Ru(phen)2dppz]2+ intercalated (via dppz) at the TA step in d(CCGGTACCGG)2, but not at the AT step of d(CCGGATCCGG)2; in this case the authors ascribed the TA specificity to the packing of the phen co-ligands against the adenosine residue in the minor groove (80).
CONCLUSION
There are several clinically established metallodrugs capable of targeting DNA, principally platinum drugs and metallo-activated bleomycin. Here, cytotoxicity arises from either platinum(II) intrastrand DNA cross-linking (81), or by single and double strand breaks following oxidative DNA lesions arising from coordination of bleomycin to first row transition metals such as iron.(82) The current study sought to identify the broad anticancer application of bioactive di-copper2+ complexes incorporating intercalating 1,10-phenanthroline (phen) ligands and to identify specific molecular targeting effects in human cancer cells. In this context, both agents (Cu-Terph and Cu-Oda) displayed selective antiproliferative effects toward solid epithelial cancers within the NCI panel that did not overlap with the cytotoxic mechanism of either platinum drugs or bleomycin when analysed by the COMPARE algorithm. Although incorporation of multiple copper centres into inorganic structures can in principle enhance cytotoxicity and free radical generation (83,84), direct comparisons are possible in this instance with a mononuclear chemotype [Cu(o-phthalate)(phen)] which mediates cell death through superoxide generation only (19). In the current work both Cu-Terph and Cu-Oda complexes yield ca. 10- to 100-fold in vitro cytotoxic increases against the same NCI-60 panel when compared with this mononuclear complex. In comparative terms, it is evident that Cu-Terph—containing a shorter and more rigid bridging dicarboxylate linker than Cu-Oda—was more effective in the majority of cell lines screened. There is some precedence for this effect as it was previously shown that ligating rigidity within pyridyl-alkylamine multinuclear Cu2+ systems altered deoxyribose oxidation, and thus DNA strand breakages (84). Morphological changes along with Annexin V and caspase activation associated with apoptosis were then identified using an NCI-60 ovarian cancer cell line (SKOV3) that is innately cisplatin resistant. The resulting spectrum of activity points to a unique cell death mechanism distinct also from the pro-apoptotic DNA damaging and topoisomerase poison Dox, which was examined in parallel as a clinical control agent. Although there is overlap between both copper complexes and Dox by way of their NCI-60 activity profile (COMPARE r-values of 0.403 (Cu-Oda) and 0.496 (Cu-Terph)), differences with Dox arise between early- and late-stage caspase activation pointing to kinetically distinct apoptotic profiles.
To probe cellular processes identified by the COMPARE algorithm findings, the next phase of our analysis revealed that both complexes can target and extensively fragment genomic DNA in SKOV3 cells through DSB formation quantified by immunodetection of γH2AX. A selection of radical-specific antioxidant traps and fluorescent exomarkers were used to identify intracellular formation of superoxide (O2•−) and singlet oxygen (1O2). These results are distinctly different from alternative Cu2+ systems such as the Casiopeina family where the most cytoactive members (CasIIgly, CasIIIEa and CasIIia) exhibit apoptotic cell death that is reliant on the mitochondrial pathway from ROS-mediated dysfunction (85). The pro-oxidant behavior of the Casiopeina complexes seems dependent on Fenton-like and Haber–Weiss type processes (86) with one key mechanistic feature: there is a requirement for redox cycling between H2O2 and GSH (87) which produces deleterious ROS species capable of generating 8-oxo-dG (88). However, for the di-nuclear agents reported here, it is likely that both O2•− and 1O2 radical species cause irreparable oxidative damage initially to the mitochondria through membrane depolarization, followed thereafter by genome oxidation in the form of DSBs. Interestingly, alongside the progression of new ROS-active drugs toward the clinic, mitochondrial targeted therapies have generated considerable recent interest in chemotherapeutic drug design (89,90). Since increased nuclearity is a defining feature of this class, alteration of the di-carboxylate bridging ligand might be expected to modify biological activity. In current examples, the more cytotoxic Cu-Terph complex contains a rigid terephthalate linker having fixed intermetal Cu-Cu distance of 11.014 Å with fixed relative orientation of the phenanthroline ligands. The more flexible Cu-Oda, with a Cu-Cu distance of 9.741 Å in the solid state, has free rotation around the octanedioate and flexible orientation of intercalating moieties. Accordingly, a theoretical basis for stability and free radical chemistry of the Cu-Terph cation was established through DFT methods which are capable of providing high-accuracy quantum chemical studies on transition–metal complexes in biological systems. These results revealed that complex stability increases going from di-Cu2+ to a singly reduced Cu2+/Cu+ state (∼4 eV), and then to the biologically relevant doubly reduced di-Cu+ adduct (∼5–7 eV). In strong agreement with our experimental observations, the role of H2O2 as a precursor in the oxidative mechanism is abrogated while the doubly reduced triplet state (Cu+/Cu+) Cu-Terph–dioxygen complex yielded a stable superoxide adduct. Switching this complex to the singlet surface resulted in release of singlet oxygen.
Given the observed intracellular DNA damaging effects by this complex class, the final part of our analysis focused on uncovering the DNA binding mode and recognition properties of Cu-Oda. Using circular dichroism analysis with long duplex polymers revealed Cu-Oda as a high-affinity intercalator of the synthetic co-polymer d(A-T)2. Probing this interaction further in terms of sequence selectively, a selection of palindromic dodecamers of varying AT content were examined. Remarkably, Cu-Oda was found to intercalate at the TA/TA step, while generating Z-like DNA at the AT/AT step. To our knowledge this is the first example of copper complex-mediated Z-like DNA formation in the Dickerson Drew dodecamer and of a probe that renders opposing elliptical CD base stacking effects upon TA and AT base discrimination. To provide a theoretical basis for this interaction, and to give reasons why substituting thymine with uracil in TATA sequence contexts enhances DNA intercalation, pre-associative docking studies were performed. Here, steric hindrance between the methyl group at the C5 position of thymine and one of the Phen ligands was calculated. Flipping this base to the syn orientation about the glycosidic bond alleviates this hindrance and provides a possible theoretical explanation for Z-like DNA formation within AT/AT containing oligomers. It is known that transitions from B to Z-DNA can be induced from high salt concentrations or the presence of multivalent and cationic metal complexes, typically in GC rich DNA sequences and/or those with modified methyl cytosines. Such metals include Mg, Co, Cu, Ni and Pt, where the majority of complex-mediated conversions still require substantial salt addition. Furthermore, polycationic Pt2+ agents facilitate Z-DNA by charge and electrostatic interactions with DNA from terminal amines (91). Few examples have previously demonstrated such conversion in AT rich sequences, however porphyrins and Ru2+ complexes have shown Z-DNA formation with complete inversion of the elliptical profile when analyzed by circular dichroism. The complex [Ru(dip)2(dppz)]2+ (where dip = 4, 7-diphenyl-1, 10-phenanthroline and dppz = dipyridophenazine) can induce B to Z-DNA transitions in poly-d(AT) tracts, promoter regions and TATA box sequences, plasmid and genomic DNA at low salt concentrations, specifically noted by a negative Cotton effect at 283 nm in CD analysis (92). In that study, conformational distortions in the helical structure was assigned by 2D NOESY where A and G bases adopt a syn rather than anti orientation. Furthermore, chiral stereospecificity of [Ru(dip)3]2+ can selectively distinguish between B and Z-DNA, where right (Δ) or left (Λ) handed enantiomers bind to DNA with the same stereochemistry, in term of handedness (93). In the context of the current study, the application of this valuable tool was precluded due to paramagnetism of the Cu2+ centre and so X-ray crystallographic analysis of complex-bound oligomers could provide a useful avenue for future research.
DATA AVAILABILITY
NCI-60 Human Tumour Cell Lines Screen is available through the NCI DTP (https://dtp.cancer.gov/discovery_development/nci-60/default.htm). The COMPARE pattern recognition algorithm associates the pattern of activity to the NCI database (https://dtp.cancer.gov/databases_tools/compare.htm).
Atomic coordinates and structure factors for molecular docking studies were obtained from the Protein Data bank under accession numbers PDB 4C64, 5EZF and 167D.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to acknowledge the DJEI/DES/SFI/HEA Irish Centre for High-End Computing (ICHEC) for the provision of computational facilities and support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Science Foundation Ireland [15/CDA/3648]; Irish Research Council [GOIPG/2014/1182]; Dublin City University (Faculty of Science and Health Distinguished Studentship); Confocal microscopy and circular dichroism was carried out at the Nano Research Facility in Dublin City University which was funded under the Programme for Research in Third Level Institutions (PRTLI) Cycle 5. The PRTLI is co-funded through the European Regional Development Fund (ERDF), part of the European Union Structural Funds Programme 2011–2015; Marie Skłodowska-Curie Innovative Training Network (ITN) ClickGene [H2020-MSCA-ITN-2014-642023 to A.K., T.B.]. Funding for open access charge: Marie Skłodowska-Curie Innovative Training Network (ITN) ClickGene [H2020-MSCA-ITN-2014-642023].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen Z.-F., Tan M.-X., Liu L.-M., Liu Y.-C., Wang H.-S., Yang B., Peng Y., Liu H.-G., Liang H., Orvig C.. Cytotoxicity of the traditional chinese medicine (TCM) plumbagin in its copper chemistry. Dalton Trans. 2009; 28:10824–10833. [DOI] [PubMed] [Google Scholar]
- 2. Tabassum S., Al-Asbahy W.M., Afzal M., Arjmand F., Bagchi V.. Molecular drug design, synthesis and structure elucidation of a new specific target peptide based metallo drug for cancer chemotherapy as topoisomerase I inhibitor. Dalton Trans. 2012; 41:4955–4964. [DOI] [PubMed] [Google Scholar]
- 3. Santini C., Pellei M., Gandin V., Porchia M., Tisato F., Marzano C.. Advances in copper complexes as anticancer agents. Chem. Rev. 2014; 114:815–862. [DOI] [PubMed] [Google Scholar]
- 4. Frezza M., Hindo S., Chen D., Davenport A., Schmitt S., Tomco D., Dou Q.P.. Novel metals and metal complexes as platforms for cancer therapy. Curr. Pharm. Des. 2010; 16:1813–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Todd R.C., Lippard S.J.. Inhibition of transcription by platinum antitumor compounds. Metallomics. 2009; 1:280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang D., Lippard S.J.. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005; 4:307–320. [DOI] [PubMed] [Google Scholar]
- 7. Komor A.C., Barton J.K.. The path for metal complexes to a DNA target. Chem. Commun. 2013; 49:3617–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Domarco O., Lötsch D., Schreiber J., Dinhof C., Schoonhoven S.V., García M.D., Peinador C., Keppler B.K., Berger W., Terenzi A.. Self-assembled Pt2L2 boxes strongly bind G-quadruplex DNA and influence gene expression in cancer cells. Dalton Trans. 2017; 46:329–332. [DOI] [PubMed] [Google Scholar]
- 9. Yu H., Wang X., Fu M., Ren J., Qu X.. Chiral metallo-supramolecular complexes selectively recognize human telomeric G-quadruplex DNA. Nucleic Acids Res. 2008; 36:5695–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma D.-L., Che C.-M., Yan S.-C.. Platinum(II) complexes with dipyridophenazine ligands as human telomerase inhibitors and luminescent probes for G-quadruplex DNA. J. Am. Chem. Soc. 2009; 131:1835–1846. [DOI] [PubMed] [Google Scholar]
- 11. Cao Q., Li Y., Freisinger E., Qin Z.P., Sigel R.K.O., Mao Z.-W.. G-quadruplex DNA targeted metal complexes acting as potential anticancer drugs. Inorg. Chem. Front. 2017; 4:10–32. [Google Scholar]
- 12. Yu Z., Han M., Cowan J.A.. Toward the design of a catalytic metallodrug: selective cleavage of G-quadruplex telomeric DNA by an anticancer copper–acridine–ATCUN complex. Angew. Chem. Int. Ed. 2015; 54:1901–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pitié M., Pratviel G.. Activation of DNA carbon-hydrogen bonds by metal complexes. Chem. Rev. 2010; 110:1018–1059. [DOI] [PubMed] [Google Scholar]
- 14. Sigman D.S., Mazumder A., Perrin D.M.. Chemical nucleases. Chem. Rev. 1993; 93:2295–2316. [Google Scholar]
- 15. Veal J.M., Rill R.L.. Sequence specificity of DNA cleavage by Bis (1, 10)-phenanthroline copper (I): effects of single base pair transitions on the cleavage of preferred pyrimidine-purine-Pyrimidine triplets. Biochemistry. 1989; 28:3243–3250. [DOI] [PubMed] [Google Scholar]
- 16. Basak S., Nagaraja V.. A versatile in vivo footprinting technique using 1, 10-phenanthroline-copper complex to study important cellular processes. Nucleic Acids Res. 2001; 29:E105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larragy R., Fitzgerald J., Prisecaru A., McKee V., Leonard P., Kellett A.. Protein engineering with artificial chemical nucleases. Chem. Commun. 2015; 51:12908–12911. [DOI] [PubMed] [Google Scholar]
- 18. Serment-Guerrero J., Cano-Sanchez P., Reyes-Perez E., Velazquez-Garcia F., Bravo-Gomez M.E., Ruiz-Azuara L.. Genotoxicity of the copper antineoplastic coordination complexes casiopeinas®. Toxicol. In Vitro. 2011; 25:1376–1384. [DOI] [PubMed] [Google Scholar]
- 19. Slator C., Barron N., Howe O., Kellett A.. [Cu(o-phthalate)(phenanthroline)] exhibits unique superoxide-mediated NCI-60 chemotherapeutic action through genomic DNA damage and mitochondrial dysfunction. ACS Chem. Biol. 2016; 11:159–171. [DOI] [PubMed] [Google Scholar]
- 20. Kellett A., O’Connor M., McCann M., McNamara M., Lynch P., Rosair G., McKee V., Creaven B., Walsh M., McClean S. et al. . Bis-phenanthroline copper(II) phthalate complexes are potent in vitro antitumour agents with ‘self-activating’ metallo-nuclease and DNA binding properties. Dalton Trans. 2011; 40:1024–1027. [DOI] [PubMed] [Google Scholar]
- 21. Kellett A., O’Connor M., McCann M., Howe O., Casey A., McCarron P., Kavanagh K., McNamara M., Kennedy S., May D.D. et al. . Water-soluble bis(1, 10-phenanthroline) octanedioate Cu2+ and Mn2+ complexes with unprecedented nano and picomolar in vitro cytotoxicity: promising leads for chemotherapeutic drug development. Medchemcomm. 2011; 2:579–584. [Google Scholar]
- 22. Suntharalingam K., Hunt D.J., Duarte A.A., White A.J.P., Mann D.J., Vilar R.. A tri-copper(II) complex displaying DNA-cleaving properties and antiproliferative activity against cancer cells. Chem. Eur. J. 2012; 18:15133–15141. [DOI] [PubMed] [Google Scholar]
- 23. Prisecaru A., Devereux M., Barron N., McCann M., Colleran J., Casey A., McKee V., Kellett A.. Potent oxidative DNA cleavage by the di-copper cytotoxin: [Cu2(μ-terephthalate)(1, 10-phen)4]2+. Chem. Commun. 2012; 48:6906–6908. [DOI] [PubMed] [Google Scholar]
- 24. Molphy Z., Prisecaru A., Slator C., Barron N., McCann M., Colleran J., Chandran D., Gathergood N., Kellett A.. Copper phenanthrene oxidative chemical nucleases. Inorg. Chem. 2014; 53:5392–5404. [DOI] [PubMed] [Google Scholar]
- 25. Kellett A., Howe O., O’Connor M., McCann M., Creaven B.S., McClean S., Kia A.F.-A., Casey A., Devereux M.. Radical-induced DNA damage by cytotoxic square-planar copper(II) complexes incorporating o-phthalate and 1, 10-phenanthroline or 2, 2′-dipyridyl. Free Radic. Biol. Med. 2012; 53:564–576. [DOI] [PubMed] [Google Scholar]
- 26. Prisecaru A., McKee V., Howe O., Rochford G., McCann M., Colleran J., Pour M., Barron N., Gathergood N., Kellett A.. Regulating bioactivity of Cu2+ bis-1, 10-phenanthroline artificial metallonucleases with sterically functionalized pendant carboxylates. J. Med. Chem. 2013; 56:8599–8615. [DOI] [PubMed] [Google Scholar]
- 27. Galindo-Murillo R., García-Ramos J.C., Ruiz-Azuara L., Cheatham T.E., Cortés-Guzmán F.. Intercalation processes of copper complexes in DNA. Nucleic Acids Res. 2015; 43:5364–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hegmans A., Berners-Price S.J., Davies M.S., Thomas D.S., Humphreys A.S., Farrell N.P.. Long Range 1, 4 and 1, 6-Interstrand Cross-Links Formed by a Trinuclear Platinum Complex. Minor Groove Preassociation Affects Kinetics and Mechanism of Cross-Link Formation as Well as Adduct Structure. J. Am. Chem. Soc. 2004; 126:2166–2180. [DOI] [PubMed] [Google Scholar]
- 29. Komeda S., Moulaei T., Woods K.K., Chikuma M., Farrell N.P., Williams L.D.. A third mode of DNA binding: phosphate clamps by a polynuclear platinum complex. J. Am. Chem. Soc. 2006; 128:16092–16103. [DOI] [PubMed] [Google Scholar]
- 30. Prisecaru A., Molphy Z., Kipping R.G., Peterson E.J., Qu Y., Kellett A., Farrell N.P.. The phosphate clamp: sequence selective nucleic acid binding profiles and conformational induction of endonuclease inhibition by cationic Triplatin complexes. Nucleic Acids Res. 2014; 42:13474–13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Medina-Molner A., Spingler B.. When two metal centres are needed instead of one: exclusive induction of Z-DNA by dinuclear metal complexes. Chem. Commun. 2012; 48:1961–1963. [DOI] [PubMed] [Google Scholar]
- 32. Devereux M., McCann M., Cronin J.F., Ferguson G.. Binuclear and polymeric copper(II) dicarboxylate complexes: syntheses and crystal structures of [Cu2(pda)(Phen)4](ClO4)2⋅5H2O⋅C2H5OH, [Cu2(oda)(Phen)4](ClO4)2⋅2.67H2O⋅C2H5OH and [Cu2(pda)2(NH3)4(H2O)2]⋅4H2On. Polyhedron. 1999; 18:2141–2148. [Google Scholar]
- 33. McCann M., Cronin J.F., Devereux M., Ferguson G.. Copper(II) complexes of heptanedioic acid (hdaH2) and octanedioic acid (odaH2): X-ray crystal structures of [Cu(η2-hda)(phen)2]⋅11.73H2O and [Cu(η2-oda)(phen)2]⋅12H2O (phen = 1, 10-Phenanthroline). Polyhedron. 1995; 14:2379–2387. [Google Scholar]
- 34. Holbeck S.L., Collins J.M., Doroshow J.H.. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol. Cancer Ther. 2010; 9:1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Monks A., Scudiero D., Skehan P., Shoemaker R., Paull K., Vistica D., Hose C., Langley J., Cronise P., Vaigro-Wolff A.. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J. Natl. Cancer Inst. 1991; 83:757–766. [DOI] [PubMed] [Google Scholar]
- 36. Taiwo F.A. Mechanism of tiron as scavenger of superoxide ions and free electrons. J. Spectrosc. 2008; 22:491–498. [Google Scholar]
- 37. Goldstein S., Czapski G.. Mannitol as an OH. scavenger in aqueous solutions and in biological systems. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1984; 46:725–729. [DOI] [PubMed] [Google Scholar]
- 38. Matheson I.B.C., Lee J.. Chemical reaction rates of amino acids with singlet oxygen. Photochem. Photobiol. 1979; 29:879–881. [Google Scholar]
- 39. Packer L., Witt E.H., Tritschler H.J.. α-Lipoic acid as a biological antioxidant. Free Radic. Biol. Med. 1995; 19:227–250. [DOI] [PubMed] [Google Scholar]
- 40. Weiss S.J., Klein R., Slivka A., Wei M.. Chlorination of taurine by human neutrophils. J. Clin. Invest. 1982; 70:598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vogt W. Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Radic. Biol. Med. 1995; 18:93–105. [DOI] [PubMed] [Google Scholar]
- 42. Giandomenico A.R., Cerniglia G.E., Biaglow J.E., Stevens C.W., Koch C.J.. The importance of sodium pyruvate in assessing damage produced by hydrogen peroxide. Free Radic. Biol. Med. 1997; 23:426–434. [DOI] [PubMed] [Google Scholar]
- 43. Speit G., Rothfuss A.. Bjergbæk L. The comet assay: a sensitive genotoxicity test for the detection of DNA damage and repair. DNA Repair Protocols. 2012; 920:Totowa: Humana Press; 79–90. [DOI] [PubMed] [Google Scholar]
- 44. Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1998; 98:5648–5652. [Google Scholar]
- 45. Handy N.C., Cohen A.J.. Left-right correlation energy. Mol. Phys. 2001; 99:403–412. [Google Scholar]
- 46. Perdew J.P., Burke K., Wang Y.. Generalized gradient approximation for the exchange-correlation hole of a many-electron system. Phys. Rev. B. 1996; 54:16533–16539. [DOI] [PubMed] [Google Scholar]
- 47. Perdew J.P., Chevary J.A., Vosko S.H., Jackson K.A., Pederson M.R., Singh D.J., Fiolhais C.. Atoms, molecules, solids, and surfaces—applications of the generalised gradient approximation for exchange and correlation. Phys. Rev. B. 1993; 48:4978–4978. [DOI] [PubMed] [Google Scholar]
- 48. Perdew J.P., Chevary J.A., Vosko S.H., Jackson K.A., Pederson M.R., Singh D.J., Fiolhais C.. Atoms, molecules, solids, and surfaces: Applications of the generalized gradient approximation for exchange and correlation. Phys. Rev. B. 1992; 46:6671–6687. [DOI] [PubMed] [Google Scholar]
- 49. Lee C., Yang W., Parr R.G.. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988; 37:785–789. [DOI] [PubMed] [Google Scholar]
- 50. De Angelis F., Jin N., Car R., Groves J.T.. Electronic structure and reactivity of isomeric Oxo-Mn(V) porphyrins: effects of spin-state crossing and pKa modulation. Inorg. Chem. 2006; 45:4268–4276. [DOI] [PubMed] [Google Scholar]
- 51. De Angelis F., Car R., Jin N., Groves J.T.. Reduction of oxomanganese (V) porphyrins by bromide ions: A DFT study. Abstr. Pap. Am. Chem. Soc. 2003; 226:U436. [Google Scholar]
- 52. Dunning T.H., Hay P.J.. Schaefer HF. Gaussian basis sets for molecular calculations. Methods of Electronic Structure Theory: Modern Theoretical Chemistry. 1977; 3:Boston: Springer; 1–27. [Google Scholar]
- 53. Wadt W.R., Hay P.J.. Ab initio effective core potentials for molecular calculations. Potentials for main group elements Na to Bi. J. Chem. Phys. 1985; 82:284–298. [Google Scholar]
- 54. Hay P.J., Wadt W.R.. Ab initioeffective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985; 82:270–283. [Google Scholar]
- 55. Hay P.J., Wadt W.R.. Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitals. J. Chem. Phys. 1985; 82:299–310. [Google Scholar]
- 56. Tomasi J., Mennucci B., Cammi R.. Quantum mechanical continuum solvation models. Chem. Rev. 2005; 105:2999–3093. [DOI] [PubMed] [Google Scholar]
- 57. Gorelsky S. I., Ghosh S. A., Solomon E. I.. Mechanism of N2O reduction by the μ4-S tetranuclear CuZ cluster of nitrous oxide reductase. J. Am. Chem. Soc. 2006; 128:278–290. [DOI] [PubMed] [Google Scholar]
- 58. Brzozowska K., Pinkawa M., Eble M.J., Müller W.-U., Wojcik A., Kriehuber R., Schmitz S.. In vivo versus in vitro individual radiosensitivity analysed in healthy donors and in prostate cancer patients with and without severe side effects after radiotherapy. Int. J. Radiat. Biol. 2012; 88:405–413. [DOI] [PubMed] [Google Scholar]
- 59. Peixoto P., Bailly C., David-Cordonnier M.H.. Fox KR. Topoisomerase I-mediated DNA relaxation as a tool to study intercalation of small molecules into supercoiled DNA. Drug-DNA Interaction Protocols. 2010; 613:Totowa: Humana Press; 235. [DOI] [PubMed] [Google Scholar]
- 60. Paull K.D., Shoemaker R.H., Hodes L., Monks A., Scudiero D.A., Rubinstein L., Plowman J., Boyd M.R.. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 1989; 81:1088–1092. [DOI] [PubMed] [Google Scholar]
- 61. Shaw T.J., Senterman M.K., Dawson K., Crane C.A., Vanderhyden B.C.. Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2004; 10:1032–1042. [DOI] [PubMed] [Google Scholar]
- 62. Hills C.A., Kelland L., Abel G., Siracky J., Wilson A.P., Harrap K.R.. Biological properties of ten human ovarian carcinoma cell lines: calibration in vitro against four platinum complexes. Br. J. Cancer. 1989; 59:527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yaginuma Y., Westphal H.. Abnormal structure and expression of the p53 gene in human ovarian carcinoma cell lines. Cancer Res. 1992; 52:4196–4199. [PubMed] [Google Scholar]
- 64. Liang Z.D., Long Y., Tsai W.-B., Fu S., Kurzrock R., Gagea-Iurascu M., Zhang F., Chen H.H.W., Hennessy B.T., Mills G.B. et al. . Mechanistic basis for overcoming platinum resistance using copper chelating agents. Mol. Cancer Ther. 2012; 11:2483–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lowe S.W., Ruley H.E., Jacks T., Housman D.E.. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993; 74:957–967. [DOI] [PubMed] [Google Scholar]
- 66. Kerr J.F., Wyllie A.H., Currie A.R.. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972; 26:239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fulda S., Debatin K.-M.. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006; 25:4798–4811. [DOI] [PubMed] [Google Scholar]
- 68. Kang K.S., Wang P., Yamabe N., Fukui M., Jay T., Zhu B.T.. Docosahexaenoic acid induces apoptosis in MCF-7 cells in vitro and in vivo via reactive oxygen species formation and caspase 8 activation. PLoS One. 2010; 5:e10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Inoue A., Muranaka S., Fujita H., Kanno T., Tamai H., Utsumi K.. Molecular mechanism of diclofenac-induced apoptosis of promyelocytic leukemia: dependency on reactive oxygen species, Akt, Bid, cytochrome and caspase pathway. Free Radic. Biol. Med. 2004; 37:1290–1299. [DOI] [PubMed] [Google Scholar]
- 70. Horobin R.W., Trapp S., Weissig V.. Mitochondriotropics: a review of their mode of action, and their applications for drug and DNA delivery to mammalian mitochondria. J. Control. Release. 2007; 121:125–136. [DOI] [PubMed] [Google Scholar]
- 71. Méndez-Hurtado J., López R., Suárez D., Menéndez M.I.. Theoretical study of the oxidation of histidine by singlet oxygen. Chem. Eur. J. 2012; 18:8437–8447. [DOI] [PubMed] [Google Scholar]
- 72. Robinson K.M., Janes M.S., Pehar M., Monette J.S., Ross M.F., Hagen T.M., Murphy M.P., Beckman J.S.. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:15038–15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kim S., Fujitsuka M., Majima T.. Photochemistry of singlet oxygen sensor green. J. Phys. Chem. B. 2013; 117:13985–13992. [DOI] [PubMed] [Google Scholar]
- 74. Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M.. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998; 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 75. Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M.. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000; 10:886–895. [DOI] [PubMed] [Google Scholar]
- 76. Chang Y.-M., Chen C.K.M., Hou M.-H.. Conformational changes in DNA upon ligand binding monitored by circular dichroism. Int. J. Mol. Sci. 2012; 13:3394–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Breslauer K.J., Frank R., Blöcker H., Marky L.A.. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Alexandrov B.S., Gelev V., Monisova Y., Alexandrov L.B., Bishop A.R., Rasmussen K.Ø., Usheva A.. A nonlinear dynamic model of DNA with a sequence-dependent stacking term. Nucleic Acids Res. 2009; 37:2405–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kilchherr F., Wachauf C., Pelz B., Rief M., Zacharias M., Dietz H.. Single-molecule dissection of stacking forces in DNA. Science. 2016; 353:aaf5508-9. [DOI] [PubMed] [Google Scholar]
- 80. Niyazi H., Hall J.P., O’Sullivan K., Winter G., Sorensen T., Kelly J.M., Cardin C.J.. Crystal structures of Λ-[Ru(phen)2dppz]2+ with oligonucleotides containing TA/TA and AT/AT steps show two intercalation modes. Nat. Chem. 2012; 4:621–628. [DOI] [PubMed] [Google Scholar]
- 81. Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer. 2007; 7:573–584. [DOI] [PubMed] [Google Scholar]
- 82. Chen J., Stubbe J.. Bleomycins: towards better therapeutics. Nat. Rev. Cancer. 2005; 5:102–112. [DOI] [PubMed] [Google Scholar]
- 83. Humphreys K.J., Karlin K.D., Rokita S.E.. Targeted strand scission of DNA substrates by a tricopper(II) coordination complex. J. Am. Chem. Soc. 2002; 124:8055–8066. [DOI] [PubMed] [Google Scholar]
- 84. Li L., Karlin K.D., Rokita S.E.. Changing selectivity of DNA oxidation from deoxyribose to guanine by ligand design and a new binuclear copper complex. J. Am. Chem. Soc. 2005; 127:520–521. [DOI] [PubMed] [Google Scholar]
- 85. Ruiz-Azuara L., Bravo-Gómez M.E.. Copper compounds in cancer chemotherapy. Curr. Med. Chem. 2010; 17:3606–3615. [DOI] [PubMed] [Google Scholar]
- 86. García-Ramos J.C., Gutiérrez A.G., Vázquez-Aguirre A., Toledano-Magaña Y., Alonso-Sáenz A.L., Gómez-Vidales V., Flores-Alamo M., Mejía C., Ruiz-Azuara L.. The mitochondrial apoptotic pathway is induced by Cu(II) antineoplastic compounds (Casiopeínas®) in SK-N-SH neuroblastoma cells after short exposure times. Biometals. 2017; 30:43–58. [DOI] [PubMed] [Google Scholar]
- 87. Kachadourian R., Brechbuhl H.M., Ruiz-Azuara L., Gracia-Mora I., Day B.J.. Casiopeína IIgly-induced oxidative stress and mitochondrial dysfunction in human lung cancer A549 and H157 cells. Toxicology. 2010; 268:176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rivero-Müller A., De Vizcaya-Ruiz A., Plant N., Ruiz L., Dobrota M.. Mixed chelate copper complex, Casiopeina IIgly®, binds and degrades nucleic acids: A mechanism of cytotoxicity. Chem. Biol. Interact. 2007; 165:189–199. [DOI] [PubMed] [Google Scholar]
- 89. Trachootham D., Alexandre J., Huang P.. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach. Nat. Mol. Cell Biol. 2009; 8:579–591. [DOI] [PubMed] [Google Scholar]
- 90. Fulda S., Galluzzi L., Kroemer G.. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 2010; 9:447–464. [DOI] [PubMed] [Google Scholar]
- 91. Johnson A., Qu Y., Van Houten B., Farrell N.. B—Z DNA conformational changes induced by a family of dinuclear bis(platinum) complexes. Nucleic Acids Res. 1992; 20:1697–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wu Z., Tian T., Yu J., Weng X., Liu Y., Zhou X.. Formation of sequence-independent Z-DNA induced by a ruthenium complex at low salt concentrations. Angew. Chem. Int. Ed. 2011; 50:11962–11967. [DOI] [PubMed] [Google Scholar]
- 93. Barton J.K., Basile L.A., Danishefsky A., Alexandrescu A.. Chiral probes for the handedness of DNA helices: enantiomers of tris(4, 7-diphenylphenanthroline)ruthenium(II). Proc. Natl. Acad. Sci. U.S.A. 1984; 81:1961–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NCI-60 Human Tumour Cell Lines Screen is available through the NCI DTP (https://dtp.cancer.gov/discovery_development/nci-60/default.htm). The COMPARE pattern recognition algorithm associates the pattern of activity to the NCI database (https://dtp.cancer.gov/databases_tools/compare.htm).
Atomic coordinates and structure factors for molecular docking studies were obtained from the Protein Data bank under accession numbers PDB 4C64, 5EZF and 167D.