Abstract
During growth under conditions of phosphate limitation, suspension-cultured cells of tomato (Lycopersicon esculentum Mill.) secrete phosphodiesterase activity in a similar fashion to phosphate starvation-inducible ribonuclease (RNase LE), a cyclizing endoribonuclease that generates 2′:3′-cyclic nucleoside monophosphates (NMP) as its major monomeric products (T. Nürnberger, S. Abel, W. Jost, K. Glund [1990] Plant Physiol 92: 970–976). Tomato extracellular phosphodiesterase was purified to homogeneity from the spent culture medium of phosphate-starved cells and was characterized as a cyclic nucleotide phosphodiesterase. The purified enzyme has a molecular mass of 70 kD, a pH optimum of 6.2, and an isoelectric point of 8.1. The phosphodiesterase preparation is free of any detectable deoxyribonuclease, ribonuclease, and nucleotidase activity. Tomato extracellular phosphodiesterase is insensitive to EDTA and hydrolyzes with no apparent base specificity 2′:3′-cyclic NMP to 3′-NMP and the 3′:5′-cyclic isomers to a mixture of 3′-NMP and 5′-NMP. Specific activities of the enzyme are 2-fold higher for 2′:3′-cyclic NMP than for 3′:5′-cyclic isomers. Analysis of monomeric products of sequential RNA hydrolysis with purified RNase LE, purified extracellular phosphodiesterase, and cleared −Pi culture medium as a source of 3′-nucleotidase activity indicates that cyclic nucleotide phosphodiesterase functions as an accessory ribonucleolytic activity that effectively hydrolyzes primary products of RNase LE to substrates for phosphate-starvation-inducible phosphomonoesterases. Biosynthetical labeling of cyclic nucleotide phopshodiesterase upon phosphate starvation suggests de novo synthesis and secretion of a set of nucleolytic enzymes for scavenging phosphate from extracellular RNA substrates.
Phosphorus is one of the most important, yet least available, mineral nutrients required by plants. The element is an essential structural component of many biomolecules and functions at the nexus of photosynthesis, energy conservation, and carbon metabolism. Consequently, assimilation, storage, and metabolism of phosphorus are highly regulated processes that immediately affect plant growth. However, low availability of soluble inorganic phosphate (Pi), the assimilated form of phosphorus, is a common phenomenon in many ecosystems and often limits plant growth (Bieleski, 1973; Schachtman et al., 1998; Raghothama, 1999). To cope with low Pi availability, plants have evolved sophisticated developmental and metabolic adaptations to enhance Pi acquisition from the rhizosphere. Such strategies include morphological changes in root architecture to accelerate soil exploration and biochemical responses to chemically increase Pi availability from insoluble salt complexes and organophosphates present in recalcitrant soil matter (Theodorou and Plaxton, 1993; Lynch, 1995; Johnson et al., 1996; Raghothama, 1999).
At the molecular level, much has been learned from the microbial response to Pi starvation. When faced with limited Pi availability, both Escherichia coli and Saccharomyces cerevisiae activate a multigene emergency rescue system to scavenge traces of usable phosphorus from the surrounding medium. Both systems are known as the pho regulon and consist of at least 30 genes that are under the same physiological and genetic control (Torriani, 1990; Lenburg and O'Shea, 1996). Phosphorus starvation leads to an increased expression of Pi mobilizing enzymes and regulatory proteins such as nucleases, phosphatases, high-affinity Pi transporters, Pi-binding proteins, and Pi sensor protein kinases that monitor extracellular Pi availability (Oshima and Halvorson, 1994; Torriani-Gorini, 1994). In higher plants, the existence of an analogous multigene Pi-starvation-inducible rescue system has been proposed (Goldstein et al., 1988a, 1988b). However, evidence for a hypothetical plant pho regulon is fragmentary. Several putative components have been described and include Pi- starvation-inducible acid phosphatases (Duff et al., 1994), phosphoenolpyruvate phosphatase (Duff et al., 1989), phosphoenolpyruvate carboxylase (Johnson et al., 1996), pyrophosphate-dependent phosphofructokinase (Theodorou et al., 1992), ribonucleases (Green, 1994; Köck et al., 1995; Dodds et al., 1996), Pi transporters (Raghothama, 1999), a β-glucosidase (Malboobi and Lefebvre, 1997), and several genes of unknown function (Liu et al., 1997; Burleigh and Harrison, 1997, 1999).
The most complete picture of Pi-starvation-inducible plant gene regulation and function in Pi-recycling from organophosphates has emerged from studies of suspension-cultured cells of black mustard (Brassica nigra) and tomato (Lycopersicon esculentum). Induction of phosphoenolpyruvate phosphatase and pyrophosphate-dependent phosphofructokinase in B. nigra cells has been proposed as a Pi-recycling system that bypasses Pi- and adenylate-requiring steps in glycolysis, thus permitting carbon metabolism to proceed in Pi-starved cells (Duff et al., 1989; Theodorou et al., 1992). Pi starvation of tomato cell cultures leads to the co-regulated induction and secretion of extracellular acid phosphatase (Goldstein et al., 1988a, 1988b) and of extracellular and intracellular ribonucleases (Nürnberger et al., 1990; Löffler et al., 1992).
Induction of ribonucleases and their coding mRNAs by Pi limitation is a rapid and reversible process that is sensitive to changes in extra- and intracellular Pi concentration (Köck et al., 1995, 1998). The combined extracellular activities of Pi-starvation-inducible ribonuclease and acid phosphatase are likely involved in degrading extracellular RNA substrates and subsequent recycling of Pi (Nürnberger et al., 1990). An analogous function has been proposed for Pi-starvation-inducible vacuolar ribonucleases. Vacuoles are equipped with an equivalent set of nucleolytic enzymes (Löffler et al., 1992) and have been shown to contain intermediates and end products of RNA degradation (Leinhos et al., 1986; Abel et al., 1990). Thus, remobilization of Pi from intra- and extracellular nucleic acid substrates to maintain constant cytoplasmic Pi concentrations is a plausible function of a subset of Pi-starvation-inducible genes.
Both extracellular and intracellular ribonucleases of cultured tomato cells have been purified and characterized in detail (Abel and Glund, 1987; Abel et al., 1989; Nürnberger et al., 1990; Jost et al., 1991; Löffler et al., 1992, 1993). All tomato ribonucleases thus far studied, extracellular RNase LE, vacuolar RNases LV-1 to LV-3, and extravacuolar RNase LX, are of the RNase I-type (EC 3.1.27.1). RNase I enzymes are endoribonucleases that generate 2′:3′-cyclic NMP as obligate monomeric products (Wilson, 1982). Further hydrolysis of 2′:3′-cyclic NMP is a side reaction of tomato extracellular and intracellular ribonucleases, which proceeds significantly slower than the generation of 2′:3′-cyclic NMP from RNA substrates (Abel et al., 1989; Nürnberger et al., 1990; Löffler et al., 1992). Therefore, we have hypothesized the existence of a cyclic nucleotide phosphodiesterase activity to provide the link between ribonuclease and phosphomonoesterase action in RNA degradation via 2′:3′-cyclic NMP intermediates (Abel et al., 1989). In this study, we describe the purification and enzymatic characterization of an extracellular cyclic nucleotide phosphodiesterase that is coordinately induced with RNase LE and acid phosphatase upon Pi starvation of suspension-cultured tomato cells. Moreover, we provide evidence that tomato cyclic nucleotide phosphodiesterase is an accessory ribonucleolytic activity required for complete degradation of extracellular RNA.
MATERIALS AND METHODS
Plant Material
Cell-suspension cultures of tomato (Lycopersicon esculentum Mill. cv Lukullus) were propagated in a modified Murashige-Skoog medium (Tewes et al., 1984), and growth was monitored by cell count as previously described (Nürnberger et al., 1990). After inoculation with 2 × 105 cells mL−1, cultures grow logarithmically for about 3.5 d, thereby increasing the number of cells 10-fold (Nürnberger et al., 1990). In experiments carried out under conditions of phosphate starvation (referred to as −Pi cultures), KH2PO4 was omitted from the modified Murashige-Skoog medium. In experiments carried out under conditions of nearly constant Pi supply (referred to as +Pi cultures), the extracellular Pi concentration was determined every 12 h and adjusted to the starting concentration (2.5 mm) by adding 0.2 m KH2PO4-K2HPO4, pH 6.0. For RNA-containing medium (referred to as +RNA/−Pi), filter-sterilized, purified (desalted by gel permeation chromatography on Sephadex G-25) yeast RNA (median size of 100 nucleotides) was substituted for KH2PO4 and added to the autoclaved medium to a final concentration of 1.6 mg mL−1, equaling about 5 mm total phosphorus.
Assays
Phosphodiesterase activity was measured in a total volume of 0.02 mL containing 50 mm acetic acid-NaOH, pH 6.0, 5 mm MgCl2, 5 mm 5′dTMP p-nitrophenyl ester or bis(p-nitrophenyl) phosphate as the substrate, and appropriate amounts of enzyme. Phosphomonoesterase activity was measured with p-nitrophenyl phosphate as the substrate. Reactions were incubated in microtiter wells at 37°C, terminated by the addition of 0.1 mL of 1 m Na2CO3, and assayed spectrophotometrically using a molar extinction coefficient of 18.5 cm2 μmol−1 for p-nitrophenol at 405 nm. Ribonuclease and deoxyribonuclease activities were tested according to the method of Abel and Glund (1987). The concentration of Pi was measured as described previously (Nürnberger et al., 1990). Protein was determined according to the method of Bradford (1976) using bovine serum albumin as a standard.
Purification of Extracellular Phosphodiesterase
For purification of tomato extracellular phosphodiesterase, −Pi cultures were initiated at a high cell density (2.5 × 106 cells mL−1) with washed (−Pi medium) cells derived from mid-log phase normal cell cultures. After 4 d of cell culture in −Pi medium, the extracellular medium was filtered and diluted (1:3) into 5 mm acetic acid-NaOH, pH 5.6. All subsequent purification steps were carried out at 4°C. The diluted culture medium (7 L) was applied at a flow rate of 20 mL cm−2 h−1 to a Sephadex SP-C25 (Pharmacia Biotech, Piscataway, NJ) column (4 × 12 cm) equilibrated with 5 mm acetic acid-NaOH, pH 5.6. After washing with 10 mm Tris-HCl, pH 7.0, proteins were eluted with a linear NaCl gradient (0–1 m NaCl in 450 mL of 10 mm Tris-HCl, pH 7.0). Phosphodiesterase activity eluted between 0.3 to 0.8 m NaCl. Proteins of the combined active fractions (240 mL) were precipitated at 4°C with acetone (80%, v/v), dissolved in 10 mL of 10 mm Tris-HCl, pH 7.5, and dialyzed against 10 mm Tris-HCl, pH 7.5, 5 mm MgCl2 (1 L). The dialyzed sample was loaded onto a chromatofocusing column (1 × 30 cm; PBE 94, Pharmacia Biotech) equilibrated with 25 mm ethanolamine-acetic acid, pH 9.4. The column was developed with 250 mL of diluted (1:10) Polybuffer 96 according to the instructions of the supplier. Phosphodiesterase activity eluted as a major and minor peak at pH 8.1 and 7.5, respectively. Active fractions of the major activity peak (20 mL) were concentrated to 2 mL by ultrafiltration at 1,500g and were loaded onto a Sephadex G-100 column (0.9 cm × 110 cm) equilibrated with 20 mm Tris-HCl, pH 7.0, 5 mm MgCl2, and 500 mm NaCl. Phosphodiesterase activity eluted as a monodisperse peak. The active fractions (10 mL) were dialyzed against 10 mm Tris-HCl, pH 7.0, and 5 mm MgCl2 (1 L), concentrated by ultrafiltration, and stored at 4°C. One unit of phosphodiesterase is defined as the amount of enzyme releasing 1 nmol min−1 p-nitrophenol from bis(p-nitrophenyl) phosphate.
Preparation of RNase LE
Extracellular tomato RNase LE was purified from Pi-starved culture medium of tomato cells as previously described (Nürnberger et al., 1990). The enzyme unit is defined according to Wilson (1982) as the amount of protein causing an increase in A260 of 1.0 unit min−1 mL−1.
Protein Labeling
Cells (2 × 107) grown either under +Pi conditions for 2.5 d or under −Pi conditions for 6 to 60 h were incubated in 10 mL of +Pi and −Pi medium, respectively, with 100 μCi of a l-[U-14C] amino acid mix for 6 h. After separation of cells (1,000g, 5 min), the medium was cleared by centrifugation (10,000g, 5 min) and treated with acetone (80%, v/v, 12 h at −20°C). Precipitated material was recovered (10,000g, 15 min) and dissolved in electrophoresis sample buffer for SDS-PAGE and subsequent fluorography according to the method of Bonner (1984).
Electrophoresis
Disc gel electrophoresis in the presence of SDS was carried out using slab gels containing 12.5% (w/v) acrylamide and the discontinuous buffer system according to the method of Laemmli (1970). SDS-PAGE was carried out at 40 V for 15 h, and gels were silver-stained (Merril et al., 1983).
Enzymatic Hydrolysis of Nucleotide Substrates
Hydrolysis of 2′-NMP, 3′-NMP, 5′-NMP, 2′:3′-cyclic NMP, 3′:5′-cyclic NMP, and of diribonucleoside monophosphates (ApU, UpU, ApA, UpA, GpU) was performed in a total volume of 0.1 mL containing 50 mm acetic acid-NaOH, pH 6.0, 5 mm MgCl2, 5 mm substrate, and appropriate amounts of purified tomato extracellular phosphodiesterase (1–5 units). Control reactions received heat-inactivated enzyme. Incubations were carried out at 37°C, and reactions were terminated by injecting a 0.02-mL aliquot directly onto the HPLC column. System Ia and Ib was used for purine NMP and pyrimidine NMP hydrolysates, respectively, and system II was used for digests of diribonucleoside monophosphates (see below).
Enzymatic Hydrolysis of Yeast RNA
For enzymatic RNA hydrolysis, reaction mixtures (1-mL total volume) contained 50 mm acetic acid-NaOH, pH 6.0, 5 mm MgCl2, 5 mg of purified yeast RNA as the substrate, and various sources of nucleolytic enzymes. The following enzymes were used: (a) 2 Wilson units (Wilson, 1982) of purified extracellular RNase LE (Nürnberger et al., 1990); (b) 20 units of purified extracellular phosphodiesterase (this study); (c) a mixture of RNase LE (2 Wilson units) and extracellular phosphodiesterase (20 units), providing the same ratio of both enzyme activities as in 3-d-old −Pi cell culture medium; or (d) 0.2 mL of cell-free crude extracellular medium of a 3-d-old −Pi cell culture, containing 2 Wilson units RNase LE, 20 units phosphodiesterase, and 8 units phosphomonoesterase (p-nitrophenyl phosphate) activity. After incubation for 6 h at 37°C, 0.1 mL of the reaction mixture was removed and the non-hydrolyzed RNA was precipitated with 80% (v/v) ethanol. The supernatant was evaporated, the residues dissolved in 20 mm (NH4)H2PO4 (pH 6.2), and the monomeric products analyzed by HPLC using system Ic (see below). For the RNA-containing culture medium, RNA was directly ethanol precipitated, and the supernatant was processed as described above.
HPLC Analysis of Enzymatic Products
HPLC measurements were performed with a Merck-Hitachi LiChroGraph system (Darmstadt, Germany), using a L-6200 low gradient pump, a L-3000 photodiodearray detector, and an HM computing integrator. Prepacked columns (4.6 × 250 mm) were purchased from Serva Feinbiochemica (Heidelberg). Sample volumes of 0.005 to 0.020 mL were injected, eluates monitored at 254 nm, and products identified by comparing their retention times with those of authentic standards. For calibration curves, linearity was obtained in the range of 0.1 to 2.0 nmol. The following isocratic HPLC systems were used at room temperature: system Ia, reverse-phase HPLC on Butyl-Si 100 (5 μm) using 20 mm (NH4)H2PO4 (pH 6.2) as the mobile phase (Abel et al., 1989); system Ib, reverse-phase HPLC on Octyl-Si 100 (5 μm), mobile phase as in system Ia; system Ic, reverse-phase HPLC on Octadecyl-Si 100 (5 μm), mobile phase as in system Ia; and system II, boronate affinity HPLC on dihydroxyboryl-Si 100 Polyol (5 μm) with 10 mm KH2PO4, pH 6.0, as mobile phase (Abel et al., 1989).
RESULTS
Extracellular Phosphodiesterase Activity during Growth of +Pi and −Pi Tomato Cell Cultures
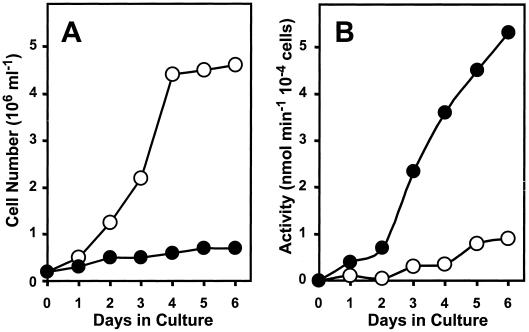
Suspension-cultured tomato cells accumulate high levels of extracellular and intracellular RNA-degrading activities during the transition from logarithmic to stationary growth or when subcultured under conditions of Pi starvation (Nürnberger et al., 1990; Löffler et al., 1992). To detect extracellular phosphodiesterase as a proposed accessory ribonucleolytic activity in tomato cell cultures and to monitor its activity during culture growth under various conditions, we initially used bis(p-nitrophenyl) phosphate as a synthetic substrate (Fig. 1). In conditions of constant Pi supply, phosphodiesterase activity is only detectable at low levels in the culture medium of logarithmically growing cells and increases about 3-fold during transition from the exponential to the stationary growth phase (4–6 d p.i.). On the contrary, in conditions of constant Pi starvation, cells secrete high phosphodiesterase activity during the 1st d of culture, exceeding about 5-fold the activity of +Pi cultures. Extracellular phosphodiesterase activity increases 12-fold during subsequent −Pi culture and accumulates to 6-fold higher levels than the extracellular activity of +Pi cultures measured at d 6 (Fig. 1). A similar activity profile during culture growth was observed with 5′dTMP p-nitrophenyl ester as the substrate. Interestingly, the profile of phosphodiesterase accumulation in +Pi and −Pi cultures closely mimics the profile of extracellular RNase LE accumulation during growth under +Pi and −Pi conditions, respectively (Nürnberger et al., 1990).
Figure 1.
Extracellular phosphodiesterase activity during growth of suspension-cultured tomato cells. Cell number (A) and extracellular phosphodiesterase activity (B) during culture growth under conditions of constant Pi supply (○) and constant Pi limitation (●).
Purification and Properties of Tomato Extracellular Phosphodiesterase
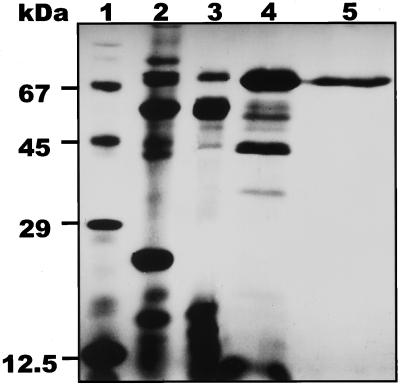
Using bis(p-nitrophenyl) phosphate as a substrate to follow purification of tomato extracellular phosphodiesterase, the enzyme was purified 15-fold from the spent medium of −Pi cultures to a specific activity of about 100 μmol min−1 mg−1 protein. The enzyme was calculated to represent approximately 6% of the total extracellular protein of Pi-starved cell cultures. All of the protein in the activity peak from the last purification step migrated as single band in SDS-PAGE (Fig. 2). The homogeneity of the enzyme preparation was further confirmed by FPLC analysis (data not shown). The purified enzyme preparation of extracellular phosphodiesterase is free of any ribonuclease, deoxyribonuclease, or nucleotidase activity.
Figure 2.
SDS-PAGE analysis of purification steps of tomato extracellular phosphodiesterase. Lane 1, Molecular mass markers; lane 2, total proteins of the spent medium of a 4-d-old −Pi culture (4 μg of protein); lane 3, pooled active fractions from Sephadex SP-C25 ion-exchange chromatography (4 μg of protein); lane 4, pooled active fractions from the chromatofocusing step eluted at pH 8.1 (3 μg of protein); lane 5, pooled active fractions from Sephadex G-100 gel permeation chromatograhy (1 μg of protein).
The molecular mass of the purified phosphodiesterase was estimated at 70 kD by SDS-PAGE (Fig. 2) and at 75 kD by gel-permeation chromatography (data not shown), indicating that tomato extracellular phosphodiesterase is a monomeric enzyme. The pI of the protein is 8.1, as inferred from the chromatofocusing purification step. Hydrolysis of bis(p-nitrophenyl) phosphate and 5′dTMP p-nitrophenyl ester by purified tomato extracellular phosphodiesterase is optimal at pH 6.2 and independent of divalent metal ions (Mg2+, Mn2+, Ca2+, and Co2+, measured at 5 mm). EDTA (5 mm) does not significantly affect enzyme activity, whereas Zn2+ and Cu2+ are strongly inhibitory (>90%) at 5 mm.
Induction of Synthesis by Phosphate Starvation
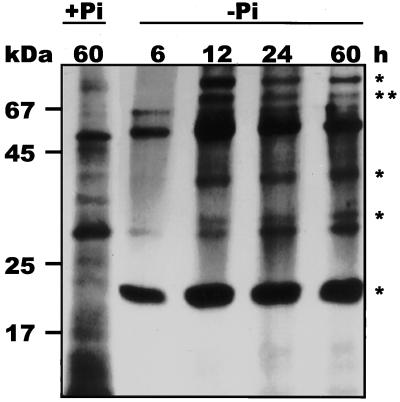
Next, we wanted to determine if accumulation of extracellular phosphodiesterase activity during growth in −Pi conditions is a consequence of de novo protein synthesis. Cell cultures grown in +Pi and −Pi conditions for 2.5 d and for 6 to 60 h, respectively, were incubated for 6 h with a mix of radioactively labeled amino acids. Biosynthetically labeled extracellular proteins were separated by SDS-PAGE and visualized by fluorography. The data in Figure 3 indicate that Pi limitation induces the synthesis of at least five proteins (molecular mass of 80, 70, 40, 35, and 23 kD) that are not biosynthetically labeled under +Pi conditions. Synthesis of Pi-starvation-inducible proteins is most active from 12 to 18 h in −Pi conditions. The biosynthetically labeled 23-kD protein was previously identified as RNase LE (Nürnberger et al., 1990), and the 70-kD protein corresponds to the purified extracellular phosphodiesterase. The addition of cycloheximide and actinomycin D prior to (1 h) or after (6 h) transfer of cells from +Pi to −Pi culture effectively inhibits accumulation of extracellular phosphodiesterase activity for at least 24 h in −Pi culture, indicating the requirement of de novo protein and RNA synthesis, respectively, for induction of phosphodiesterase activity (data not shown).
Figure 3.
De novo synthesis of extracellular proteins during −Pi culture of tomato cells. Cells (2 × 107) of a 60-h-old +Pi culture and of a −Pi culture grown for 6, 12, 24, and 60 h were incubated with 100 μCi of a l-[U-14C]amino acid mix for 6 h. Biosynthetically labeled extracellular proteins were separated by SDS-PAGE and visualized by fluorography (50,000 dpm per lane). Asterisks indicate proteins that are apparently not synthesized under +Pi conditions, including a protein of about 70 kD (two asterisks).
Characterization as Cyclic Nucleotide Phosphodiesterase
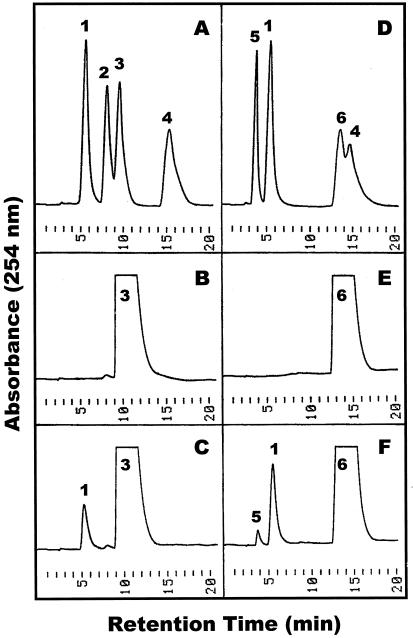
To analyze the substrate specificity of the purified tomato extracellular phosphodiesterase, we have studied the hydrolysis of 2′:3′-cyclic and 3′:5′-cyclic NMP isomers. As shown for cAMP substrates in Figure 4, the enzyme catalyzes the exclusive formation of 3′-AMP from 2′:3′-cAMP (Fig. 4, A–C), while hydrolysis of 3′:5′-cAMP (Fig. 4, D–F) results in a mixture of 3′-AMP and 5′-AMP in a ratio of 6 to 1. The same substrate-product relationship is observed for cyclic GMP, cyclic UMP, and cyclic CMP isomers. Importantly, the resulting 3′-NMP and 5′-NMP products are not further hydrolyzed to nucleosides and Pi (Fig. 4C, F). Moreover, when used directly as substrates, 2′-NMP, 3′-NMP, and 5′-(d) NMP are not hydrolyzed by tomato phosphodiesterase, indicating that the purified enzyme does not exhibit any nucleotidase activity.
Figure 4.
Hydrolysis of cAMP isomers by purified tomato extracellular phosphodiesterase. Enzymatic hydrolysis of 2′:3′-cAMP (A–C) and 3′:5′-cAMP (D–F) was performed as described in “Materials and Methods.” Standard compounds (A and D), control incubations with heat-inactivated enzyme (B and E), and enzymatic digests for 30 min (C and F) were separated by reverse-phase HPLC on Butyl-Si 100. Peak identities: 1, 3′-AMP; 2, 2′-AMP; 3, 2′:3′-cAMP; 4, adenosine; 5, 5′-AMP; 6, 3′:5′-AMP.
Table I presents the rates of cleavage of various natural and artificial substrates by purified tomato extracellular phosphodiesterase. Interestingly, the enzyme hydrolyzes both 2′:3′-cyclic NMP and 3′:5′-cyclic NMP isomers with no apparent base specificity. Specific activities of tomato extracellular phosphodiesterase are about 2-fold higher for 2′:3′-cyclic NMP than for 3′:5′-cyclic NMP substrates. Of the synthetic phosphodiesterase substrates tested, p-nitrophenyl phosphate, bis(p-nitrophenyl) phosphate, 5′dTMP p-nitrophenyl ester, 5′dTMP α-naphthylester, and various diribonucleoside monophosphates, only the p-nitrophenyl esters are hydrolyzed (see Table I). Based on its substrate specificity and on its preference for 2′:3′-cyclic NMP substrates, the tomato extracellular enzyme may be classified as a 2′:3′-cyclic nucleotide-2′-phosphodiesterase (EC 3.1.4.16).
Table I.
Substrate specificity of tomato extracellular cyclic nucleotide phosphodiesterase
| Substrate | Specific Enzyme Activity | Relative Rate |
|---|---|---|
| nmol min−1 mg−1 | % | |
| 2′:3′-Cyclic AMP | 102,375 (5,050) | 100 |
| 2′:3′-Cyclic UMP | 102,375 (6,125) | 100 |
| 2′:3′-Cyclic GMP | 54,180 (3,875) | 53 |
| 2′:3′-Cyclic CMP | 55,125 (3,625) | 54 |
| 3′:5′-Cyclic AMP | 47,565 (2,150) | 46 |
| 3′:5′-Cyclic UMP | 51,975 (2,375) | 51 |
| 3′:5′-Cyclic GMP | 18,900 (1,025) | 18 |
| 3′:5′-Cyclic CMP | 39,375 (1,975) | 38 |
| 2′-NMP | NDa | ND |
| 3′-NMP | ND | ND |
| 5′-NMP/5′-dNMP | ND | ND |
| Diribonucleoside monophosphates | ND | ND |
| bis(p-Nitrophenyl)phosphate | 153,659 (8,025) | 150 |
| 5′-dTMP p-Nitrophenyl ester | 25,659 (1,550) | 25 |
| 5′-dTMP α-naphthyl ester | ND | ND |
| p-Nitrophenyl phosphate | 10,602 (475) | 10 |
Enzymatic hydrolysis of the listed substrates was performed as described in “Materials and Methods.” Rates of hydrolysis are given as the mean (±se) and relative to the rate of 2′:3′-cyclic AMP cleavage taken as 100% (n = 3).
ND, Not detectable.
Function in Extracellular RNA Degradation
To further illustrate the substrate specificity of tomato extracellular cyclic nucleotide phosphodiesterase and to demonstrate its function as an accessory activity in extracellular RNA degradation, yeast RNA was sequentially incubated with purified extracellular RNase LE, purified extracellular cyclic nucleotide phosphodiesterase, and with cell-free medium of a −Pi cell culture as a source of 3′-nucleotidase activity. The RNA hydrolysates were subsequently analyzed by HPLC for enzymatic NMP products. Hydrolysis of yeast RNA by purified RNase LE results in the formation of 2′:3′-cyclic NMP as primary monomeric products and of the corresponding 3′-NMP to lesser amounts (Fig. 5B). However, when RNA is exposed to the combined action of purified RNase LE and purified extracellular cyclic nucleotide phosphodiesterase (using the same ratio of enzyme activities as in 3-d-old −Pi culture medium, see “Materials and Methods”), the 2′:3′-cyclic NMP products of RNase LE action are completely converted to 3′-NMP by cyclic nucleotide phosphodiesterase, which alone does not release any NMP products from the RNA substrate (Fig. 5C). Finally, the addition of cleared −Pi culture medium to the RNA digest with RNase LE and extracellular phosphodiesterase leads to the formation of ribonucleosides and, implicitly, of Pi (data not shown; Fig. 6).
Figure 5.
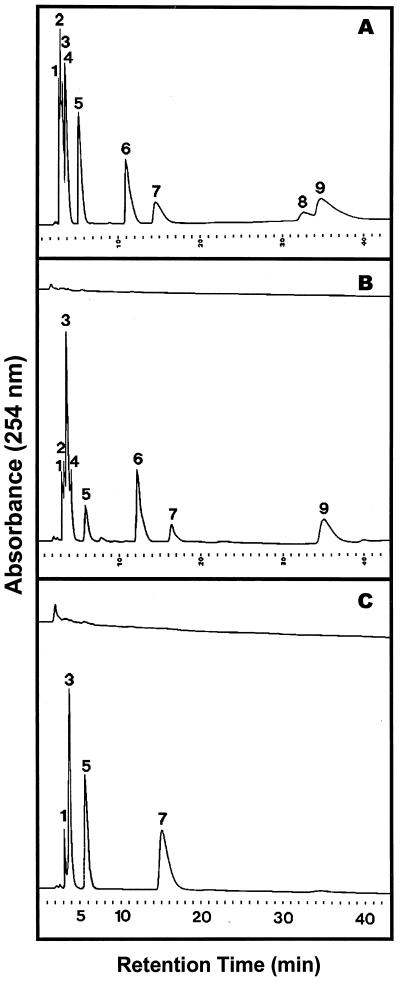
Elution profile of ribonucleoside monophosphates after hydrolysis of yeast RNA with purified RNase LE and purified tomato extracellular phosphodiesterase. Enzymatic hydrolysis of RNA and HPLC separation of monomeric products on Octadecyl-Si 100 were performed as described in “Materials and Methods.” Shown are elution profiles of standard compounds (A), monomeric products of RNA hydrolysis with RNase LE for the zero-time control (B, upper tracing) and after 6 h of reaction (B, lower tracing), and monomeric products of RNA hydrolysis with extracellular phosphodiesterase for 6 h (C, upper tracing) and with RNase LE for 6 h followed with extracellular phosphodiesterase for 3 h (C, lower tracing). Peak identities: 1, 3′-CMP/2′-CMP; 2, 2′:3′-cyclic CMP; 3, 3′-UMP/2′-UMP; 4, 2′:3′-cyclic UMP; 5, 3′-GMP/2′-GMP; 6, 2′:3′-cyclic GMP; 7, 3′-AMP; 8, 2′-AMP; 9, 2′:3′-cAMP.
Figure 6.
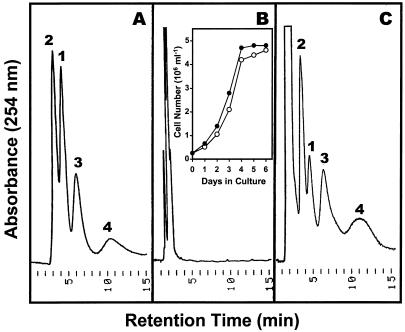
Growth of suspension-cultured tomato cells in +RNA/−Pi medium and analysis of monomeric products of RNA hydrolysis during culture growth. Yeast RNA (1.6 mg mL−1) was substituted for KH2PO4 as a source of phosphorus, and growth of +Pi (○) and +RNA/−Pi (●) cultures was monitored (inset in B). HPLC separation of ribonucleosides on dihydroxyboryl-Si 100 were performed as described in “Materials and Methods.” Shown are elution profiles of standard ribonucleosides (A) and of monomeric RNA degradation products at 0 h (B) and 3 d (C) after cell transfer to +RNA/−Pi medium. Peak identities: 1, Uridine; 2, cytidine; 3, guanosine; 4, adenosine.
To determine if extracellular RNA hydrolysis is indeed relevant and sufficient to sustain cell growth under conditions in which RNA is the only source of phosphorus, we followed tomato cell culture growth in +Pi and +RNA/−Pi medium (Fig. 6). Interestingly, when purified yeast RNA was substituted for KH2PO4 in the modified Murashige-Skoog medium, cells grew without any detectable lag phase at growth rates comparable to +Pi cell cultures (Fig. 6B, inset). This implies degradation of RNA to ribonucleosides and Pi to maintain cell growth. As expected, when analyzed after 3 d of culture in +RNA/−Pi medium, ribonucleosides were the prominent monomeric products of RNA hydrolysis in the cell culture medium (Fig. 6C), demonstrating the significance of secreted nucleolytic enzymes for cell survival during Pi limitation.
DISCUSSION
In this communication we report the purification and characterization of a Pi starvation-inducible, secretory phosphodiesterase from cultured tomato cells, and provide evidence for its participation in extracellular RNA degradation. Based on enzymatic properties of Pi-starvation-inducible extracellular RNase LE (Nürnberger et al., 1990) and vacuolar RNase LV-3 from tomato (Abel and Glund, 1987; Abel et al., 1989; Löffler et al., 1992), we hypothesized the existence of a Pi-starvation-inducible 2′:3′-cyclic nucleotide-2′-phosphodiesterase as an auxiliary ribonucleolytic activity that is required for complete RNA degradation. RNase LE and RNase LV-3 are non-specific, EDTA-insensitive endoribonucleases that hydrolyze single-stranded RNA substrates preferentially adjacent to purine residues by a phosphotransferase reaction, thereby generating 2′:3′-cyclic NMP as obligate primary monomeric products. Consistent with their classification as RNase I (Wilson, 1982), tomato extracellular and vacuolar RNases hydrolyze 2′:3′-cyclic NMP to 3′-NMP in a secondary reaction. However, the subsequent decyclization step proceeds 200- to 1,000-fold slower than the generation of respective 2′:3′-cyclic NMP from diribonucleoside monophosphate substrates (Abel et al., 1989; Nürnberger et al., 1990). Consequently, RNA degradation by RNase I activity may lead to accumulation of 2′:3′-cyclic NMP intermediates, which are not immediate substrates of phosphomonoesterases. As predicted, even extended incubation of yeast RNA with purified tomato secretory RNases demonstrates preferential formation and accumulation of 2′:3′-cyclic NMP products (Abel et al., 1989; Löffler et al., 1992). On the other hand, analysis of the culture medium of Pi-starved tomato cells indicates secretion of high levels of phosphodiesterase activity, and the ability of the cell culture medium to completely hydrolyze extracellular RNA to nucleosides and Pi.
Purification of tomato extracellular phosphodiesterase to apparent homogeneity and subsequent characterization of its substrate specificity classify the tomato enzyme as cyclic nucleotide phosphodiesterase. The purified enzyme hydrolyzes both 2′:3′-cyclic NMP and 3′:5′-cyclic isomers with no apparent base specificity. The tomato enzyme evidently prefers 2′:3′-cyclic NMP as substrates, which are hydrolyzed to 3′-NMP, whereas hydrolysis of 3′:5′-cyclic NMP results in a mixture of 3′-NMP and 5′-NMP. Calculations of specific activities for 2′:3′-cyclic NMP hydrolysis by tomato secretory RNases (5,000–10,000 units mg−1; Abel et al., 1989; Nürnberger et al., 1990) and by tomato extracellular phosphodiesterase (50,000–100,000 units mg−1; Table I) indicate a 10-fold higher specific activity for the latter enzyme. Thus, the nucleobase-nonspecific cyclic nucleotide phosphodiesterase of tomato may efficiently convert 2′:3′-cyclic NMP products of RNase action to 3′-NMP substrates for hydrolysis by 3′-nucleotidase. Indeed, analysis of sequential hydrolysis of yeast RNA by purified RNase LE, purified cyclic nucleotide phosphodiesterase, and cell-free culture medium as a source of 3′-nucleotidase activity provides compelling evidence for a function of tomato cyclic nucleotide phosphodiesterase in extracellular RNA degradation.
Two classes of cyclic nucleotide phosphodiesterase activities have been reported in plants. Phosphodiesterases from wheat germ (Tyc et al., 1987) and Arabidopsis (Genschik et al., 1997) have been characterized as 2′:3′-cyclic nucleotide-3′-phosphodiesterase activities (EC. 3.1.4.37), forming 2′-NMP products, and may function in tRNA splicing (Culver et al., 1994), or in unknown biological processes. Physical and catalytic properties of the tomato extracellular phosphodiesterase are markedly different from members of this class, but are strikingly similar to enzymes that have been studied during efforts to identify 3′:5′-cAMP phosphodiesterase activity as indirect evidence for the presence and function of 3′:5′-cAMP in plants (Lin and Varner, 1972; Vandepeute et al., 1973; Ashton and Polya, 1975; Brown et al., 1977; Junker et al., 1977, 1979, 1980; Zan-Kowalczewska et al., 1984; Dupon et al., 1987; Chiatante et al., 1988; Gangwani et al., 1994). However, properties of most plant cyclic nucleotide phosphodiesterases characterized during those investigations have been found to differ significantly from animal and bacterial 3′:5′-cyclic nucleotide-3′-phosphodiesterases. Intriguingly, characteristics of the second class of plant cyclic nucleotide phosphodiesterases suggest a function in RNA degradation, which was first proposed by Lin and Varner (1972). Therefore, it is not surprising that tomato extracellular cyclic nucleotide phosphodiesterase shares several key characteristics with preparations of phosphodiesterases from pea (Lin and Varner, 1972), barley (Vandepeute et al., 1973), potato (Ashton and Polya, 1975; Zan-Kowalczewska et al., 1984), sunflower (Junker et al., 1977), spinach (Brown et al., 1980), lettuce (Chiatante et al., 1988), or Lemna (Gangwani et al., 1994), such as molecular mass of the monomer unit (65–80 kD), an acidic pH optimum (pH 5–7), lack of requirement for bivalent metal ions, and broad substrate specificity. In general, higher plant cyclic nucleotide phosphodiesterases of this class hydrolyze, irrespective of the nucleobase present, 2′:3′-cyclic NMP to 3′-NMP at a higher rate than 3′:5′-cyclic NMP to a mixture of 3′-NMP and 5′-NMP, typically in a ratio of 7 to 1 (Lin and Varner, 1972), which is similar to the ratio observed for the tomato enzyme (6:1). The susceptibility of p-nitrophenyl phosphate may indicate that tomato extracellular phosphodiesterase co-purifies with a minor acid phosphatase. However, absence of any nucleotidase activity suggests that hydrolysis of p-nitrophenyl phosphate is a catalytic capability of the tomato enzyme, which has been proposed for cyclic nucleotide phosphodiesterase from potato (Ashton and Polya, 1975; Zan-Kowalczewska et al., 1984). In summary, catalytic properties of tomato extracellular phosphodiesterase validate that the enzyme belongs to a class of common plant cyclic nucleotide phosphodiesterases that likely play a role in RNA degradation.
A function of tomato extracellular cyclic nucleotide phosphodiesterase in RNA hydrolysis is also implied by its induction kinetics during cell culture growth, which are mirrored by induction kinetics of extracellular RNase LE (Nürnberger et al., 1990; Köck et al., 1995, 1998) and extracellular acid phosphatase (Goldstein et al., 1988a, 1988b). The data suggest co-regulated de novo synthesis and secretion of a set of Pi-starvation-inducible nucleolytic enzymes to rapidly mobilize Pi from RNA substrates under Pi-limiting conditions, which is observed for tomato cell cultures grown in +RNA/−Pi medium. Reports on the expression of Pi-starvation-inducible gene products in roots and their localization to the rhizosphere provide evidence that this model is also valid in the context of whole plants. For example, Pi-starvation-inducible acid phosphatase is excreted from tomato roots (Goldstein et al., 1988a), and activities of several phosphohydrolases (including acid phosphatase and ribonuclease) are significantly increased in roots of Pi-deficient tomato seedlings (Bosse and Köck, 1998). Furthermore, tissue-specific expression of Pi-starvation-inducible, high-affinity Pi transporters in tomato epidermal root cells points to a significant role of inducible transporters in Pi acquisition under natural conditions (Liu et al., 1998; Muchhal and Raghothama, 1999). Consistently, overexpression of a high-affinity Pi transporter from Arabidopsis in cultured tobacco cells results in increased biomass production under Pi-limiting conditions (Mitsukawa et al., 1997), pointing to an essential role of enhanced Pi uptake in overcoming adverse effects of Pi starvation on plant growth. Thus, the collective evidence suggests that, upon Pi starvation, plants coordinately induce a set of genes that are members of the proposed plant pho regulon, and whose encoded proteins ensure complete degradation of extracellular nucleic acid substrates and efficient uptake of the recycled Pi (see Fig. 7).
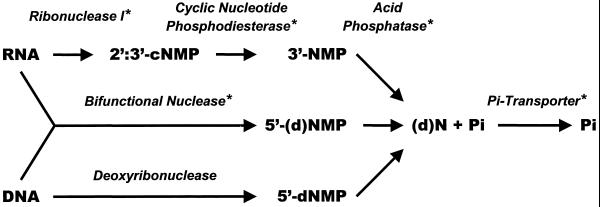
Figure 7.
Model of extracellular nucleic acid degradation and Pi recycling by secretory nucleolytic enzymes. Asterisks indicate proteins known to be inducible by Pi starvation.
The biological significance of the proposed pho regulon for plant phosphorus nutrition is further supported by the high fraction of organic phosphorus present in the organic soil matter, which ranges from 30% to 95% in most agricultural and forest soils (Marschner, 1995). Although data on total nucleic acid content of soils are not available, extraction of ribosomal RNA and genomic DNA from intact indigenous bacterial soil communities suggests that the occurrence of DNA and ribosomal RNA amounts to at least 30 μg g−1 soil and 2 μg g−1 soil, respectively (Duarte et al., 1998). It has been suggested that, due to their compact secondary structure, both free DNA and RNA might become stabilized in soil following the release from lysing cells (Nannipieri et al., 1986). In view of the high turnover of organic phosphorus in the rhizosphere (Helal and Dressler, 1989), the importance of secretory nucleolytic enzymes for effective phosphorus acquisition of plants is evident. This is further demonstrated by normal growth and development of Arabidopsis plants on synthetic media containing purified nucleic acids as the only source of phosphorus (S. Abel, unpublished data).
ACKNOWLEDGMENTS
The authors thank Donna Chen for preparation of figures and Kristin Morgan for critically reading and editing of the manuscript.
LITERATURE CITED
- Abel S, Blume B, Glund K. Evidence for RNA-oligo-nucleotides in plant vacuoles isolated from cultured tomato cells. Plant Physiol. 1990;94:1163–1171. doi: 10.1104/pp.94.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Glund K. Ribonuclease in plant vacuoles: purification and molecular properties of the enzyme from cultured tomato cells. Planta. 1987;172:71–78. doi: 10.1007/BF00403030. [DOI] [PubMed] [Google Scholar]
- Abel S, Krauss G-J, Glund K. Ribonuclease in tomato vacuoles: high-performance liquid chromatographic analysis of ribonucleolytic activities and base specificity. Biochim Biophys Acta. 1989;998:145–150. [Google Scholar]
- Ashton AR, Polya GM. Higher plant cyclic nucleotide phosphodiesterases. Biochem J. 1975;149:329–339. doi: 10.1042/bj1490329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski RL. Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol. 1973;24:225–252. [Google Scholar]
- Bonner WM. Fluorography for the detection of radioactivity in gels. Methods Enzymol. 1984;104:460–465. doi: 10.1016/s0076-6879(84)04115-x. [DOI] [PubMed] [Google Scholar]
- Bosse D, Köck M. Influence of phosphate starvation on phosphohydrolases during development of tomato seedlings. Plant Cell Environ. 1998;21:325–332. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown EG, Al-Najafi T, Newton RP. Cyclic nucleotide phosphodiesterase activity in Phaseolus vulgaris. Phytochemistry. 1977;16:1333–1337. [Google Scholar]
- Brown EG, Edwards MJ, Newton RP, Smith CJ. Plurality of cyclic nucleotide phosphodiesterase in Spinacea oleracea: subcellular distribution, partial purification, and properties. Phytochemistry. 1979;18:1943–1948. [Google Scholar]
- Brown EG, Edwards MJ, Newton RP, Smith CJ. The cyclic nucleotide phosphodiesterases of spinach chloroplasts and microsomes. Phytochemistry. 1980;19:23–30. [Google Scholar]
- Burleigh SH, Harrison MJ. A novel gene whose expression in Medicago trancatula roots is suppressed in response to colonization by vesicular-arbuscular mucorrhizal (VAM) fungi and to phosphate nutrition. Plant Mol Biol. 1997;34:199–208. doi: 10.1023/a:1005841119665. [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 1999;119:241–248. doi: 10.1104/pp.119.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiatante D, Balconi C, Newton RP, Brown EG. Immunoaffinity purification of cyclic nucleotide phosphodiesterase from Lactuca cotyledons. Phytochemistry. 1988;27:2477–2483. [Google Scholar]
- Culver GM, Consaul SA, Tycowski KT, Filipowicz W, Phizicky EM. tRNA splicing in yeast and wheat germ. J Biol Chem. 1994;269:24928–24934. [PubMed] [Google Scholar]
- Dodds P, Clarke A, Newbigin E. Molecular characterization of an S-like Rnase of Nicotiana alata that is induced by phosphate starvation. Plant Mol Biol. 1996;31:227–238. doi: 10.1007/BF00021786. [DOI] [PubMed] [Google Scholar]
- Duarte FD, Rosado AS, Seldin L, Keijzer-Wolters AC, van Elsas JD. Extraction of ribosomal RNA and genomic DNA from soil for studying the diversity of the indeginous bacterial community. J Microbiol Methods. 1998;32:21–29. [Google Scholar]
- Duff SMG, Moorhead GBG, Lefebvre DD, Plaxton WC. Phosphate starvation inducible bypasses of adenylate and phosphate-dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol. 1989;90:1275–1278. doi: 10.1104/pp.90.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC. The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant. 1994;90:791–800. [Google Scholar]
- Dupon M, Van Onckelen HA, De Greef JA. Characterization of cyclic nucleotide phosphodiesterase activity in Phaseolus vulgaris. Physiol Plant. 1987;69:361–365. [Google Scholar]
- Gangwani L, Khurana JP, Maheshwari SC. Cyclic nucleotide phosphodiesterase from Lemna paucicostata: effect of calmodulin and theophyllin. Phytochemistry. 1994;35:857–861. [Google Scholar]
- Genschik P, Hall J, Filipowicz W. Cloning and characterization of the Arabidopsis cyclic phosphodiesterase which hydrolyzes ADP-ribose 1“,2”-cyclic phosphate and nucleoside 2′,3′-cyclic phosphates. J Biol Chem. 1997;272:13211–13219. doi: 10.1074/jbc.272.20.13211. [DOI] [PubMed] [Google Scholar]
- Goldstein AH, Beartlein DA, McDaniel RG. Phosphate starvation inducible metabolism in Lycopersicon esculentum: I. Excretion of acid phosphatase by tomato plants and suspension cultured cells. Plant Physiol. 1988a;87:711–715. doi: 10.1104/pp.87.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AH, Danon A, Beartlein DA, McDaniel RG. Phosphate starvation inducible metabolism in Lycopersicon esculentum: II. Characterization of the phosphate starvation-inducible-excreted acid phosphatase. Plant Physiol. 1988b;87:716–720. doi: 10.1104/pp.87.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ. The ribonucleases of higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:421–445. [Google Scholar]
- Helal HM, Dressler A. Mobilization and turnover of soil phosphorus in the Rhizosphere. Z Pflanzenernähr Bodenk. 1989;152:175–180. [Google Scholar]
- Johnson JF, Vance CP, Allan DL. Phosphorus deficiency in Lupinus albus. Plant Physiol. 1996;112:31–41. doi: 10.1104/pp.112.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost W, Bak H, Glund K, Terpstra P, Beintema JJ. Amino acid sequence of an extracellular, phosphate-starvation induced ribonuclease from cultured tomato (Lycopersicon esculentum) cells. Eur J Biochem. 1991;198:1–6. doi: 10.1111/j.1432-1033.1991.tb15978.x. [DOI] [PubMed] [Google Scholar]
- Junker S, Verbeek-Wyndaele R, Truelsen TA. Characterization of cyclic nucleotide phosphodiesterase activity in sunflower callus. Physiol Plant. 1977;39:45–50. [Google Scholar]
- Köck M, Löffler A, Abel S, Glund K. cDNA structure and regulatory properties of a family of starvation-induced ribonucleases from tomato. Plant Mol Biol. 1995;27:477–485. doi: 10.1007/BF00019315. [DOI] [PubMed] [Google Scholar]
- Köck M, Theierl K, Stenzel I, Glund K. Extracellular administration of phosphate-sequestering metabolites induces ribonucleases in cultured tomato cells. Planta. 1998;204:404–407. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leinhos V, Krauss GJ, Glund K. Evidence that a part of cellular uridine of a tomato cell suspension culture is located in the vacuoles. Plant Sci. 1986;47:15–20. [Google Scholar]
- Lenburg ME, O'Shea EK. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- Lin PP-C, Varner JE. Cyclic nucleotide phosphodiesterase in pea seedlings. Biochim Biophys Acta. 1972;276:454–474. doi: 10.1016/0005-2744(72)91007-8. [DOI] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Raghothama KG. Differential expression of TPS11, a phosphate starvation-induced gene in tomato. Plant Mol Biol. 1997;33:867–874. doi: 10.1023/a:1005729309569. [DOI] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Uthappa M, Kononowicz, Raghothama KG. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998;116:91–99. doi: 10.1104/pp.116.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler A, Abel S, Jost W, Beintema JJ, Glund K. Phosphate-regulated induction of intracellular ribonucleases in cultured tomato (Lycopersicon esculentum) cells. Plant Physiol. 1992;98:1472–1478. doi: 10.1104/pp.98.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler A, Glund K, Irie M. Amino acid sequence of an intracellular, phosphate-starvation-induced ribonuclease from cultured tomato (Lycopersicon esculentum) cells. Eur J Biochem. 1993;214:627–633. doi: 10.1111/j.1432-1033.1993.tb17962.x. [DOI] [PubMed] [Google Scholar]
- Lynch J. Root architecture and plant productivity. Plant Physiol. 1995;109:7–13. doi: 10.1104/pp.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malboobi MA, Lefebvre DD. A phosphate-starvation inducible β-glucosidase gene isolated from Arabidopsis thaliana is a member of a distinct subfamily of the BGA family. Plant Mol Biol. 1997;34:57–68. doi: 10.1023/a:1005865406382. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. London: Academic Press; 1995. [Google Scholar]
- Merril CR, Goldman D, Van Keulen ML. Silver staining methods for acrylamide gel electrophoresis. Methods Enzymol. 1983;96:230–239. doi: 10.1016/s0076-6879(83)96021-4. [DOI] [PubMed] [Google Scholar]
- Mitsukawa N, Okumura S, Shirano Y, Sato S, Kato T, Harashima S, Shibata D. Overexpresssion of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances growth under phosphate-limited conditions. Proc Natl Acad Sci USA. 1997;94:7098–7102. doi: 10.1073/pnas.94.13.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Raghothama KG. Transcriptional regulation of plant phosphate transporters. Proc Natl Acad Sci USA. 1999;96:5868–5872. doi: 10.1073/pnas.96.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannipieri P, Ciardi C, Badalucco L, Casella S. A method to determine soil DNA and RNA. Soil Biol Biochem. 1986;18:275–281. [Google Scholar]
- Nürnberger T, Abel S, Jost W, Glund K. Induction of an extracellular ribonuclease in cultured tomato cells upon phosphate starvation. Plant Physiol. 1990;92:970–976. doi: 10.1104/pp.92.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Halvorson H. Regulation of phosphate metabolism in Saccharomyces cerevisiae. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in Microorganisms: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1994. [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM. Phosphorus uptake by plants: from soil to cell. Plant Physiol. 1998;116:447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewes A, Glund K, Walther R, Reinbothe H. High yield isolation and rapid recovery of protoplasts from suspension culture of tomato (Lycopersicon esculentum) Z Pflanzenphysiol. 1984;113:141–150. [Google Scholar]
- Theodorou ME, Cornel FA, Duff SMG, Plaxton WC. Phosphate starvation-inducible synthesis of the α-subunit of the pyrophosphate-dependent phosphofructokinase in black mustard suspension cells. J Biol Chem. 1992;267:21901–21905. [PubMed] [Google Scholar]
- Theodorou ME, Plaxton WC. Metabolic adaptations of plant respiration to nutritional phosphate deprivation. Plant Physiol. 1993;101:339–344. doi: 10.1104/pp.101.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani A. From cell membrane to nucleotides: the phosphate regulon in Escherichia coli. Bioessays. 1990;12:371–376. doi: 10.1002/bies.950120804. [DOI] [PubMed] [Google Scholar]
- Torriani-Gorini A. The Pho regulon of Escherichia coli. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in Microorganisms: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1994. pp. 1–4. [Google Scholar]
- Tyc K, Kellenberger C, Filipowicz W. Purification and characterization of wheat germ 2′,3′-cyclic nucleotide 3′-phosphodiesterase. J Biol Chem. 1987;262:12994–13000. [PubMed] [Google Scholar]
- Vandepeute J, Huffaker RC, Alvarez R. Cyclic nucleotide phosphodiesterase acticvity in barley seeds. Plant Physiol. 1973;52:278–282. doi: 10.1104/pp.52.3.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CM. Plant nucleases: biochemistry and development of multiple molecular forms. Isozymes Curr Top Biol Med Res. 1982;6:33–54. [PubMed] [Google Scholar]
- Zan-Kowalczewska M, Bartkiewicz M, Sierakowska H, Shugar D. Purification and resolution of potato tuber cyclic nucleotide phosphodiesterase from nucleotide pyrophosphatase. Biochim Biophys Acta. 1984;788:62–73. [Google Scholar]