Summary
CRISPR-Cas immune systems integrate short segments of foreign DNA as spacers into the host CRISPR locus to provide molecular memory of infection. Cas4 proteins are widespread in CRISPR-Cas systems and are thought to participate in spacer acquisition, although their exact function remains unknown. Here we show that Bacillus halodurans type I-C Cas4 is required for efficient prespacer processing prior to Cas1-Cas2 mediated integration. Cas4 interacts tightly with the Cas1 integrase, forming a heterohexameric complex containing two Cas1 dimers and two Cas4 subunits. In the presence of Cas1 and Cas2, Cas4 processes double-stranded substrates with long 3'-overhangs through site-specific endonucleolytic cleavage. Cas4 recognizes PAM sequences within the prespacer and prevents integration of unprocessed prespacers, ensuring that only functional spacers will be integrated into the CRISPR array. Our results reveal the critical role of Cas4 in maintaining fidelity during CRISPR adaptation, providing a structural and mechanistic model for prespacer processing and integration.
eTOC Blurb
Lee et al. show that Cas4, a core family of CRISPR-associated proteins, associates with the Cas1 spacer integrase and is required for efficient prespacer processing during CRISPR-Cas adaptation. Cas4 processing prevents non-functional spacers from being integrated into the CRISPR array, ensuring the fidelity of the adaptation process.
Introduction
In bacteria and archaea, clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR associated (Cas) proteins provide an adaptive immune system against mobile genetic elements (Barrangou et al., 2007; Brouns et al., 2008; Marraffini and Sontheimer, 2008). A CRISPR array consists of a series of direct repeats that are flanked by short sequences derived from a foreign genome, called spacers (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). CRISPR-Cas immunity proceeds through three stages: adaptation, expression and interference (reviewed in (Marraffini, 2015; Mohanraju et al., 2016)). During adaptation, small fragments of foreign DNA are captured and integrated as new spacers into the CRISPR array by the Cas1-Cas2 complex (Yosef et al., 2012; Nuñez et al., 2014, 2015a; Wang et al., 2015; Jackson et al., 2017; Xiao et al., 2017). During the expression stage, the array is transcribed and processed into short CRISPR RNAs (crRNAs), which assemble with Cas proteins to form a RNA-guided surveillance complex (Brouns et al., 2008; Carte et al., 2008; Haurwitz et al., 2010; Deltcheva et al., 2011; Gesner et al., 2011; Sashital et al., 2011; Hatoum-Aslan et al., 2013; Hochstrasser and Doudna, 2015; Jackson and Wiedenheft, 2015). Finally, during the interference stage, the surveillance complex recognizes targets, often by locating a protospacer adjacent motif (PAM) that can be found immediately next to target protospacer sequence (Mojica et al., 2009; Semenova et al., 2011; Sternberg et al., 2014; Redding et al., 2015; Xue et al., 2017). Following complementary base pairing between the crRNA and protospacer, a Cas nuclease degrades the target and neutralizes the infection (Barrangou et al., 2007; Brouns et al., 2008; Marraffini and Sontheimer, 2008; Garneau et al., 2010; Westra et al., 2012).
CRISPR systems can be classified into two classes, six types (types I–VI), and many subtypes based on the architecture and composition of their cas gene loci (Koonin et al., 2017). Despite this divergence, Cas1 and Cas2 are conserved among all CRISPR systems, suggesting that spacers are acquired via a universal mechanism. Cas1 functions as an integrase while Cas2 provides a structural scaffold that enhances the integration activity of Cas1 (Yosef et al., 2012; Nuñez et al., 2014, 2015a; Wang et al., 2015; Xiao et al., 2017). The Cas1-Cas2 integrase complex captures prespacers that are flanked by 3'-hydroxyl groups on each end and catalyzes the integration reaction at the leader-proximal repeat through direct nucleophilic attack (Nuñez et al., 2015b). The A-T rich leader is found directly upstream of the repeat-spacer array and provides polarized spacer acquisition that is governed by the intrinsic sequence specificity of Cas1-Cas2 for leader-specific sites (Rollie et al., 2015; McGinn and Marraffini, 2016; Wright and Doudna, 2016; Xiao et al., 2017), and enables rapid defense against the most recent invader. The type I-E Cas1-Cas2 complex additionally relies on association with the integration host factor (IHF), which induces DNA bending within the leader and provides additional sequence specificity to Cas1-Cas2 for the leader-repeat junction (Nuñez et al., 2016; Wright et al., 2017).
Although Cas1 and Cas2 are universally required for spacer integration, other type-specific cas genes have also been implicated in adaptation (Koonin et al., 2017). In particular, Cas4 is a core family of Cas proteins present in several sub-types within type I, II and V systems (Hudaiberdiev et al., 2017). In vivo studies have shown that deletion of the cas4 gene prevents the uptake of new spacers (Li et al., 2014). Cas4 contains four conserved cysteine residues that coordinate an iron-sulfur cluster and RecB-like nuclease motifs that are required for DNA binding and cleavage activity (Lemak et al., 2013, 2014). Biochemical studies have revealed that Cas4 exhibits DNA unwinding, exonuclease and endonuclease activity, although the exact activity varies between different orthologs (Zhang et al., 2012; Lemak et al., 2013, 2014). Based on this nuclease activity, it has been hypothesized that Cas4 is involved in spacer generation for Cas1-Cas2 meditated integration, and recent evidence suggests that Cas4 nuclease activity may trim the ends of precursor integration substrates (Rollie et al., 2017).
Of the core Cas family proteins, Cas4 remains one of the few for which the mechanistic role in CRISPR-Cas immunity remains poorly understood. Here, we show that Cas4 plays an integral role in prespacer processing prior to integration by the Cas1-Cas2 complex. Cas4 processes long 3'-DNA overhangs on precursor substrates through Cas1-Cas2-dependent endonucleolytic activity, generating correctly sized substrates prior to integration. The adaptation complex selectively processes pre-spacers with correct PAM sequences, ensuring that only functional spacers are captured during acquisition. While the Cas1-Cas2 complex integrates longer precursor substrates in the absence of Cas4, the presence of Cas4 prevents premature integration and promotes preferential integration of only processed prespacers into the CRISPR locus. Combined with structural analysis of the Cas4-Cas1 complex, these biochemical results indicate that the Cas4 and Cas1 active sites compete for single-stranded overhangs, and that cleavage by Cas4 is prerequisite to integration within the full adaptation complex. Overall, these findings reveal the role of Cas4 in prespacer generation and in ensuring the fidelity of spacer length and PAM selection during spacer integration.
Results
Complex formation by type I-C Cas4, Cas1 and Cas2
Unlike the well-studied type I-E and I-F systems, other type I systems, including the widespread type I-C, contain cas4 genes (Figure 1A). We wondered whether Cas4 from the type I-C system interacts with either Cas1 or Cas2, or with the Cas1-Cas2 complex. Cas4 is found as a fusion with Cas1 in some systems (Hudaiberdiev et al., 2017), and Cas4 from type I-A has previously been shown to interact with a Cas1/2 fusion and the sub-type specific Csa1 protein (Plagens et al., 2012). However, this reconstitution was only achieved upon refolding of denatured proteins, and it is unclear whether the native proteins interact. To test potential interactions under native conditions, we co-expressed Cas1, Cas2, and Cas4 from the B. halodurans type I-C system in E. coli. Although Cas2 did not co-purify with the complex, an amino-terminal poly-histidine-tagged Cas4 co-purified with untagged Cas1 and the complex was maintained when the affinity tag was moved to the amino terminus of Cas1 (Figures S1A–B). Regardless of which subunit was tagged, the two proteins formed a stable complex that eluted as a single peak on a size exclusion column with an estimated size of ~150 kDa (Figure S1C). These results demonstrate that Cas4 directly interacts with Cas1 under native folding conditions, and that the two proteins form a stable complex.
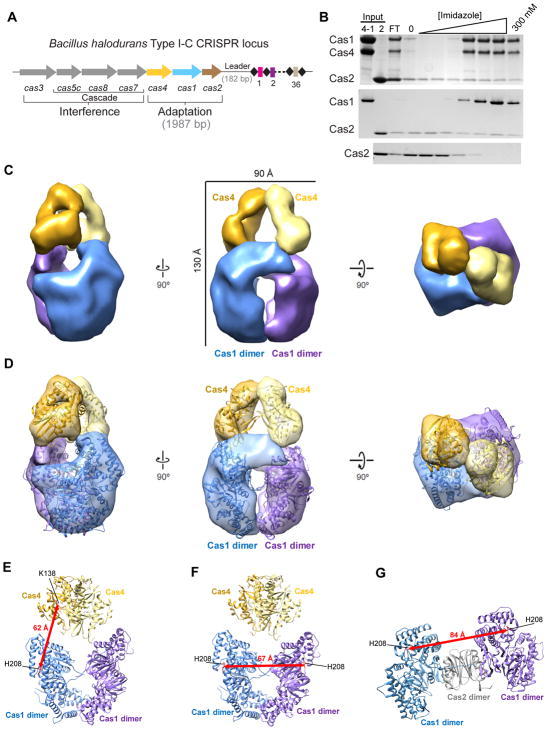
Figure 1. Structure of the B. halodurans Cas4-Cas1 complex.
(A) Overview of the type I-C cas genes and CRISPR locus found in Bacillus halodurans type I-C system. Spacers are shown in rectangles, repeats are shown in diamonds, each cas gene is shown as arrows and gene products involved in adaptation or interference are indicated. (B) Nickel affinity pull-down of poly-histidine-tagged Cas4-Cas1 complex or His6-Cas1 and untagged Cas2. A stepwise elution using an imidazole titration (20–300 mM) was performed. FT: Flow through. Untagged Cas2 alone was used as a control (bottom panel). (C) Negative-stain reconstruction of the Cas4-Cas1 complex at ~21-Å resolution (using the gold-standard 0.143 FSC criterion) with subunits labeled and colored as follows: gold, Cas4; yellow, second Cas4; blue, Cas1 dimer; purple, second Cas1 dimer. (D) The crystal structures of the Cas4 protein Pcal_0546 from Pyrobaculum calidifontis (PDB ID: 4R5Q) and the Cas1 dimers in the Cas1-Cas2 complex from E.coli (PDB ID: 4P6I; blue, chain C and D; purple, chain E and F) are docked into the negative-stain reconstruction of the Cas4-Cas1 complex. (E) The distance between the active sites of Cas1 (His234) and Cas4 (Lys110) in the Cas4-Cas1 complex is ~62 Å (red line). (F) The distance between the active sites of two Cas1 (His234) in the Cas4-Cas1 complex is ~67 Å (red line). (G) The distance between the active sites of Cas1 (His234) in E.coli Cas1-Cas2 complex is ~84 Å (red line).
To investigate whether Cas1 or the Cas4-Cas1 complex interacts with Cas2, we incubated poly-histidine tagged Cas1 or Cas4-Cas1 complex and untagged Cas2 and performed pull-down assays using nickel affinity chromatography. The untagged Cas2 alone eluted at low imidazole concentrations, whereas some Cas2 co-eluted with Cas1 or Cas4-Cas1 at high imidazole concentrations (Figure 1B). These data suggest that both Cas1 and Cas4-Cas1 form higher-order complexes with Cas2, supporting a role for Cas4 within the adaptation machinery. However, the complexes do not appear to have the expected stoichiometry (Nuñez et al., 2014), and Cas2 partially eluted at low imidazole concentrations in the presence of His6-tagged Cas1 or Cas4-Cas1 (Figure 1B). Moreover, neither the Cas1-Cas2 nor the Cas4-Cas1-Cas2 complex could be co-purified by size exclusion chromatography. These data suggest that interactions between Cas1 and Cas2 are relatively weak for these orthologs, unlike Cas1 and Cas2 from other sub-types (Nuñez et al., 2015a; Wang et al., 2015; Fagerlund et al., 2017; Rollins et al., 2017; Xiao et al., 2017).
Molecular architecture of the Cas4-Cas1 complex
To characterize the molecular architecture of the Cas4-Cas1 complex, we performed single-particle electron microscopy (EM) of negatively stained Cas4-Cas1 complexes. Raw micrographs showed globular, monodispersed particles with internal structural features (Figure S1D). Two rounds of reference-free two-dimensional (2D) alignment and classification were performed to remove low quality particles, resulting in a total of ~13,000 particles that were used for further analysis. Distinct 2-fold symmetry was observed in many of the 2D class averages (Figure S1F). A 3D model generated by ab initio 3D reconstruction in cryoSPARC (Punjani et al., 2017) was used as an initial model for 3D classification using 3 classes in RELION (Scheres, 2012). 4,590 structurally homogenous particles were extracted from the best 3D model for iterative refinement of the model with imposed C2 symmetry, which led to a final 3D reconstruction of the Cas4-Cas1 complex at ~21 Å resolution using the gold standard 0.143 criterion (Figure S1I).
The 2-fold symmetric Cas4-Cas1 complex resembles a crab-claw and is ~130 Å in the longest dimension and ~90 Å wide, with four distinct subunits (Figure 1C). Notably, the crystal structure of the Cas4 protein Pcal_0546 from Pyrobaculum calidifontis (PDB ID: 4R5Q) (Lemak et al., 2014) fits perfectly into the two small subunits at the top of the claw, while the crystal structure of the Cas1 dimer in the Cas1-Cas2 complex from E. coli (PDB ID: 4P6I) (Nunez et al., 2014) were easily accommodated into the two larger subunits at the base of the claw in the final 3D reconstruction (Figure 1D), suggesting a stoichiometry of 4 Cas1 and 2 Cas4 proteins for the Cas4-Cas1 complex. Docking of these crystal structures into our map places the Cas4 monomer directly above the Cas1 monomer, with the active site residues of K138 and H208 for Cas4 and Cas1, respectively, roughly parallel to each other within the complex (Figure 1E).
Interestingly, although possessing similar stoichiometry to the Cas4-Cas1 complex, the Cas1-Cas2 complex has a remarkably different molecular architecture (Nuñez et al., 2014). The distances between Cas1 dimers is smaller in the Cas4-Cas1 complex as compared to the Cas1 dimers in E. coli Cas1-Cas2 complex with distances between active site residues H208 of 67 and 84- Å for each complex, respectively (Figure 1F–G). Thus, it is unlikely that two Cas2 molecules could be accommodated in the interface between Cas1 dimers without significantly altering the conformation of Cas4-Cas1. Notably, EM micrographs of Cas4-Cas1-Cas2 complex co-eluted from nickel-affinity purification (Figure 1B) showed particles of a larger size than for Cas4-Cas1 (Figure S1E). However, attempts at 3D reconstruction from these images proved unsuccessful, potentially because of sample heterogeneity due to incomplete formation of the putative Cas4-Cas1-Cas2 complex.
Cas4 enhances prespacer processing
The previously demonstrated nuclease activities of Cas4 suggest that it may be involved in prespacer processing prior to integration. To test this possibility, we tested cleavage of dsDNA substrates containing blunt ends or a 24-bp duplex flanked by 15-nt 3'- or 5'- overhangs (Figure S2A). The design of the short duplex substrates was based on previous crystal structures of E. coli Cas1-Cas2 complex bound to prespacers and the average length (34.4 bp) of the 35 spacers found in the B. halodurans CRISPR locus 4 (Nuñez et al., 2015b; Wang et al., 2015). We investigated the effect of temperature on processing activity by incubating reactions either at 37 or 65°C (Figure S2B,C). Bacillus strains are facultive alkaliphiles and specifically B. halodurans strains are polyextremophiles to temperature up to 90–100 °C and high salt concentrations (Smaali et al., 2006; Dua and Gupta, 2017). Notably, while we observed cleavage of the 3'-overhang substrates at 65ºC (Figure S2B), we did not observe cleavage of 5'-overhang or blunt end substrates under similar conditions (Figure S2C–E). Exonucleolytic cleavage of the blunt end substrate was observed using free Cas4, but only at very high concentrations (Figure S2E). We therefore proceeded with experiments testing cleavage of substrates with long 3' overhangs (Figure 2A).
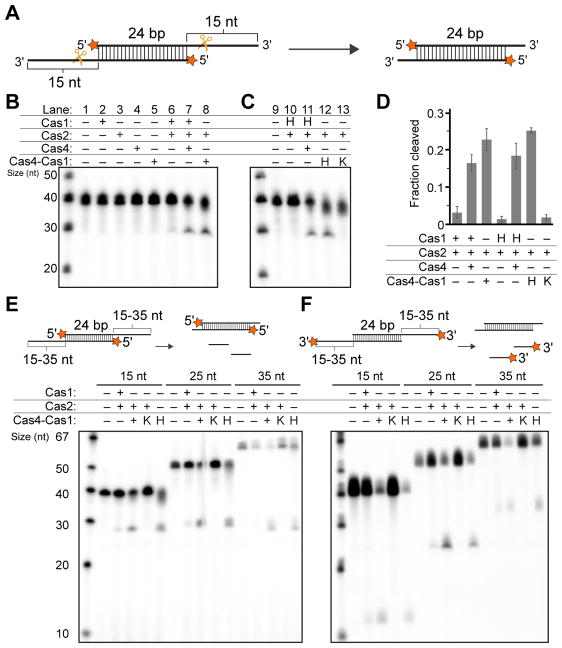
Figure 2. Prespacer processing by the Cas4-Cas1-Cas2 complex.
(A) Schematic view of prespacer processing assay. Radiolabel is indicated with a star. (B) Prespacer processing assay using 15 nt 3'-overhang DNA. (C) Prespacer processing assay as in (B) but with catalytic mutants in Cas1 or Cas4 active sites. H indicates H234A Cas1 catalytic mutant; K indicates K110A Cas4 catalytic mutant. (D) Fraction cleaved for prespacer processing. The average of three triplicates is shown, and error bars represent standard deviation. (E–F) Prespacer processing assay with 15 nt, 25 nt, and 35 nt 3'overhang DNA with radiolabel on (E) 5' or (F) 3' end.
Incubation of Cas1, Cas2 and an unprocessed prespacer bearing 15-nt 3' overhangs generated a small amount of cleaved products consistent with the length of a processed prespacer with short overhangs on the 3' ends (Figures 2B, lane 6). Strikingly, the processing activity was enhanced substantially in the presence of Cas4 (Figure 2B, lanes 7–8, Figure 2D), suggesting that Cas4 may be directly involved in prespacer cleavage. No cleaved products were observed when Cas1, Cas2, Cas4 or the Cas4-Cas1 complex were incubated individually with DNA (Figure 2B, lanes 2–5), indicating that optimal processing activity requires all three adaptation proteins. In addition, overhang and duplex length had little effect on overall processing activity both in the absence and presence of Cas4 (Figure S4).
To determine the extent to which each subunit of the Cas4-Cas1 complex contributes to the catalytic activity of prespacer processing, we introduced mutations in the active sites of each subunit (Figure 2C). Based on sequence alignments, Cas4 Lys110 is located in one of the conserved RecB nuclease motifs (motif III) (Figure S3A) and Cas1 His234 is found in the predicted active site as reported in E. coli and S. pyogenes (Nuñez et al., 2015b; Wright and Doudna, 2016) (Figure S3B). While the Cas1 active site mutant (H234A) ablated Cas1-Cas2 processing activity (Figure 2C, lane 10), addition of individually purified Cas4 or Cas4-Cas1 complex containing H234A Cas1 restored cleavage to wild-type levels (Figure 2C, lanes 11–12, Figure 2D). In contrast, the Cas4-Cas1 complex containing K110A Cas4 showed no detectable products (Figure 2C, lane 13, Figure 2D). Together these data reveal that while the Cas1 active site can catalyze low levels of prespacer processing in the absence of Cas4, the Cas4 catalytic site is both necessary and sufficient for processing when all three adaptation proteins are present.
Because Cas4 has exonuclease activity (Figure S2E), it is possible that the increased processing activity in the presence of Cas4 may be based on exonucleolytic degradation by free Cas4 or Cas4-Cas1. Alternatively, if Cas1-Cas2 engages DNA with overhangs positioned in the Cas4 and/or Cas1 active sites, the cleavage activity may be expected to proceed endonucleolytically. To test these two possibilities, we used substrates with 3' overhangs of different lengths labeled on either the 5' or 3' ends. Interestingly, while all products were the same length for the 5'-end labelled substrates (Figure 2E), we detected products corresponding to the length of the overhang for the 3'-end labelled substrates (Figure 2F), indicating that Cas4 processes prespacers endonucleolytically. Overall, our results suggest that Cas4 is a Cas1-Cas2 dependent endonuclease that processes prespacers at a precise site within the 3' single-stranded overhang.
PAM-dependent prespacer processing
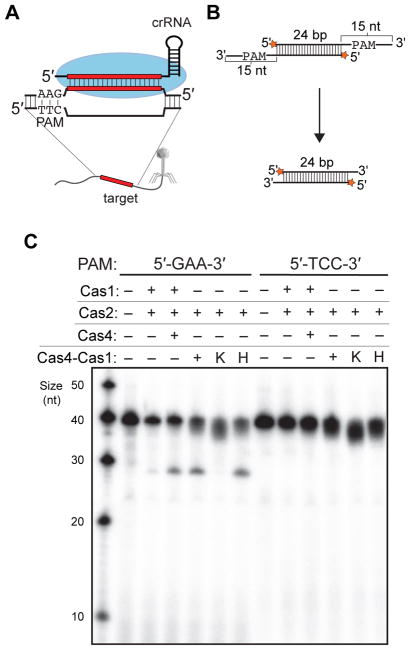
Given the importance of PAM sequences for targeting by the surveillance complex during CRISPR interference, it is critical that the adaptation complex select and integrate prespacers from sites with correct PAM sequences. In the B. halodurans type I-C system, the PAM has been characterized as 5'-GAA-3' on the target strand (Leenay et al., 2016; Rao et al., 2017) (Figure 3A). The adaptation complex is expected to recognize the PAM sequence within the 3'-overhang of the prespacer, by analogy to the E. coli Cas1-Cas2 complex (Wang et al., 2015). To determine whether Cas4-dependent prespacer processing is sequence specific, we tested the cleavage of two different prespacer substrates containing either 5'-GAA-3' (perfect) or 5'-TTC-3' (reverse) PAM on the 3' overhangs (Figure 3B). Interestingly, while the presence of Cas4 enhanced the processing of prespacers with a GAA PAM, no detectable cleavage was observed for prespacers with a TTC PAM (Figure 3C). These data suggest that the adaptation complex can specifically capture prespacers with correct PAM sequences, and that Cas4-dependent cleavage is also PAM dependent.
Figure 3. PAM-dependent cleavage by Cas4 in the presence of Cas1-Cas2.
(A) Schematic view of PAM sequence (5'-GAA-3' on the target strand) in the B. halodurans type I-C system. (B) Schematic view of PAM-dependent processing assay. The prespacer contains a PAM site within the 3' overhang. (C) PAM-dependent processing assay using either 5'-GAA-3' (perfect) or 5'-TTC-3' (reverse) PAM on the 3' overhangs.
Cas4 ensures the integration of processed prespacers
To determine whether Cas4 in the presence of Cas1-Cas2 integrates processed spacers, we designed minimal CRISPRs with varied leader sequence lengths from 10 to 50 bp, the full 32-bp repeat, and a 5-bp spacer (Figure S5A). Each minimal CRISPR substrate was labeled at the 5′ end of the plus strand resulting in leader-length products upon successful integration (Figure S5A). Leader-length products were observed for all minimal CRISPRs (Figure S5B), indicating that, like the type II-A system, integration by the type I-C adaptation complex relies on intrinsic sequence specificity rather than additional structural motifs or factors, such as IHF (Rollie et al., 2015; Wright and Doudna, 2016; Xiao et al., 2017). Surprisingly, we observe a slight increase in the amount of leader-length product in the presence of Cas4, and especially for the Cas4-Cas1 complex. However, the formation of leader-length products is dependent on Cas1 catalytic activity, as no product is observed with the catalytically dead Cas1 mutant in the absence or presence of Cas4. We also observed the formation of small amounts of leader-length product in the absence of prespacers (Figure S5C). These data suggest that Cas1 can perform site-specific cleavage at the leader end, as has been observed previously in the type II-A system (Wright and Doudna, 2016).
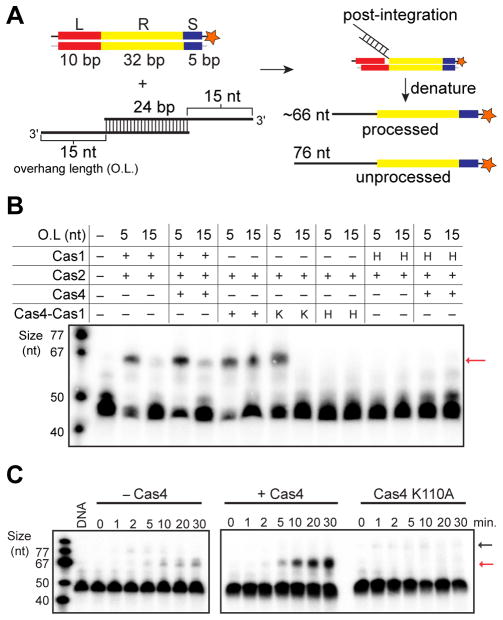
Because it is not possible to distinguish between leader-length integration or cleavage products, we also tested integration of preprocessed (5-nt 3' overhangs) or unprocessed prespacers with varied 3'-overhang lengths in the absence or presence of Cas4. This experimental design ensures the direct detection of integration products, and also enables detection of products containing processed versus unprocessed prespacers (Figure 4A). Using this experimental design, preprocessed prespacers were integrated with similar efficiency in both the absence and presence of Cas4. These results suggest that Cas4 does not improve the integration efficiency by Cas1 but may contribute to the putative leader cleavage activity observed in the absence of prespacers (Figure S5C). In contrast, when the unprocessed substrate was used for integration, production of the correct length integration product was substantially enhanced in the presence of Cas4 (Figure 4B, S5D–E), consistent with the enhanced processing activity by Cas4. Cas1 active site mutants ablated integration activity, whereas Cas4 active site mutants produced integration products only with the preprocessed prespacer, consistent with the lack of processing activity for this mutant (Figure 4B). Notably, in time-course assays, Cas1-Cas2 integrated unprocessed prespacer but quickly catalyzed the disintegration in favor of integrating processed prespacers (Figure 4C). However, we were unable to detect integration of unprocessed prespacers in the presence of WT Cas4, although we saw low levels of unprocessed integration for the K110A mutant (Figure 4C). Together these results indicate that in the presence of Cas4, the Cas1-Cas2 adaptation complex preferentially integrates processed prespacers based on the enhanced processing activity provided by Cas4.
Figure 4. Prespacer processing by Cas4 enhances integration.
(A) Schematic view of integration assay. Red, leader; yellow, repeat; blue, spacer; star, radiolabel. The lengths of substrates and expected products are indicated. (B) Integration assay with Cas1 (1 μM), Cas2 (1 μM) and Cas4 (2 μM) individually or Cas4-Cas1 (1 μM) complex containing wild-type (WT) subunits, or Cas1 (H, H234A) or Cas4 active site mutants (K, K110A). Red arrow indicates the integrated products of processed prespacer. O.L is overhang length. (C) Time-course integration assay of WT Cas1 + Cas2, WT Cas4-Cas1 + Cas2, or Cas4-Cas1 + Cas2 with Cas4 active site mutant (K110A) using 15-nt 3'overhang prespacers. Red arrow indicates the integrated products of processed prespacers while black arrow indicates the integrated products of unprocessed prespacers.
Sequence-specific integration and asymmetric prespacer processing by the adaptation complex
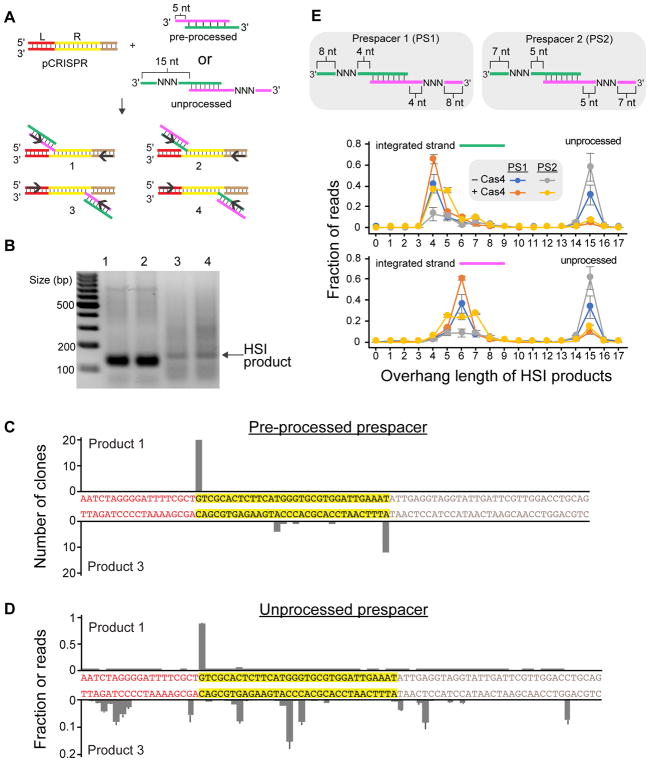
To determine the processing and integration site for HSI products formed following prespacer processing, we developed an integration assay using a plasmid bearing a portion of the native B. halodurans CRISPR locus (pCRISPR). Prespacer integration converts a negatively supercoiled plasmid into different plasmid species, such as relaxed or linear forms of the plasmid for successful integration events or plasmid topoisomers for integration followed by disintegration events (Nuñez et al., 2015b; Wright and Doudna, 2016) (Figure S6A). We detected relaxed or linearized plasmid species when Cas1, Cas2 or Cas4 were incubated alone with pCRISPR, consistent with native integrase activity of Cas1 or nuclease activities of Cas2 and Cas4 (Nuñez et al., 2015b; Nam et al., 2012; Zhang et al., 2012; Lemak et al., 2013, Figure S2E). When both Cas1 and Cas2 were added to the integration reaction, we observed robust integration activity where supercoiled plasmids were converted to a linear form at 37°C in both the presence and absence of Cas4 (Figure S6B). However, at 65°C we observed multiple plasmid topoisomers under both conditions, which were visible as a ladder of slow migrating DNA bands on a post-stained agarose gel, suggesting disintegration is favored at higher temperature.
Using this plasmid integration assay, we sought to determine the integration and processing sites for prespacers. Integration events are expected to occur at either the leader-repeat junction of the plus strand or at the first repeat-spacer junction of the minus strand within the CRISPR (Figure 5A). To determine whether the prespacers were correctly integrated into either the leader-repeat or repeat-spacer junction, we PCR amplified the half-site integrated (HSI) products of Cas1-Cas2 in the presence of Cas4 using a preprocessed prespacer (Figure 5B). Plus-strand PCR amplicons ran as a single band, while amplification reactions against the minus strand resulted in less specific bands (Figure 5B), suggesting that plus-strand integration is more specific. We cloned the amplicons into pRSF and sequenced 20 clones for each integration site. All integration events on the plus strand of the CRISPR occurred at the leader-repeat junction, while only 60% (12 out of 20) of integration events on the minus strand occurred precisely at the repeat-spacer junction (Figure 5C). These results indicate that while the majority of integration events occur at the correct site, minus-strand integration is less specific and may be specified following plus-strand integration at the leader-repeat site. Moreover, non-specific minus-strand HSI products may be subject to disintegration, resulting in the formation of topoisomer products at 65°C (Figure S7B).
Figure 5. Sequencing half-site products reveal integration and processing sites.
(A) Schematic view of half-site integration events. Four different events occurred due to two orientations of prespacers and two different integration sites. Substrates are either preprocessed prespacers or prespacers containing 15 nt 3' overhangs with degenerate sequences. (B) PCR products for the half-site integrated (HSI) products of Cas1-Cas2 in the presence of Cas4 using a preprocessed prespacer. The numbers indicate the four different events that are depicted in (A). (C) Integration sites for HSI products of Cas1-Cas2 in the presence of Cas4 using the preprocessed spacer. The regions of the CRISPR are colored as in (A). (D) Integration sites for HSI products of Cas1-Cas2 using unprocessed prespacer. The average fraction of read counts at each start site from three separate replicates are plotted, with error bars representing standard deviation. Other conditions are shown in Figure S7. (E) Processing sites for two prespacers for either the top (green) or bottom (magenta) strand in the absence or presence of Cas4. The average fraction of read counts for three separate replicates are plotted, with error bars representing standard deviation.
We next developed a high throughput sequencing assay with unprocessed prespacers containing degenerate sequences on the 3'overhangs (Figure 5A). The degenerate sequences mimic the effects of varied sequences that would be present in prespacers encountered in endogenous situations. These prespacers were used for integration assays into pCRISPR in the presence or absence of Cas4. To limit disintegration of unprocessed HSI products, experiments were performed at 37ºC (Figure S6B). PCR amplification reactions were performed (Figure S6C) and the products were sequenced by Illumina MiSeq to determine the integration sites for unprocessed prespacers. Similar to the preprocessed prespacer, the vast majority of HSI products were integrated precisely at the leader-repeat junction in both the presence and absence of Cas4 (Figure 5D, S7). However, only a small fraction of prespacers were integrated at the expected repeat-spacer junction site on the minus strand (Figure 5D, S7). The lower specificity of minus strand HSI products for unprocessed prespacers may be due to decreased processing activity observed at 37ºC (Figure S2B), resulting in HSI products where the non-integrated overhang remains unprocessed and unsuitable for full-site integration. Overall, our results suggest that spacer acquisition proceeds through initial integration at the leader-repeat site, and that correct integration at the repeat-spacer site is partially dependent on complete prespacer processing.
Sequencing of HSI products also revealed the extent and site of processing for the prespacer substrates. Consistent with our processing and integration assays (Figures 2 and 4), the presence of Cas4 greatly enhanced the integration of processed prespacers, although low levels of unprocessed HSI products were detected, potentially because Cas4 was added in trans rather than as part of the Cas4-Cas1 complex (Figure 5E). Intriguingly, when the degenerate sequence was placed at positions 5–7 of the overhang (Prespacer 1, Figure 5E), the HSI products displayed a marked asymmetry for the processing sites for each of the prespacer strands. While the “top” strand (green strand, Figure 5E) was mainly processed following position 4 of the overhang, the “bottom” strand (magenta strand, Figure 5E) was processed following position 6. For Prespacer 2, in which the degenerate sequence was placed at positions 6–8 of the overhang, the asymmetrical processing sites were also observed, although the processing sites were more variable for this substrate. The variability in processing position for the two different prespacers suggests that sequence specificity plays a role in processing site selection.
Discussion
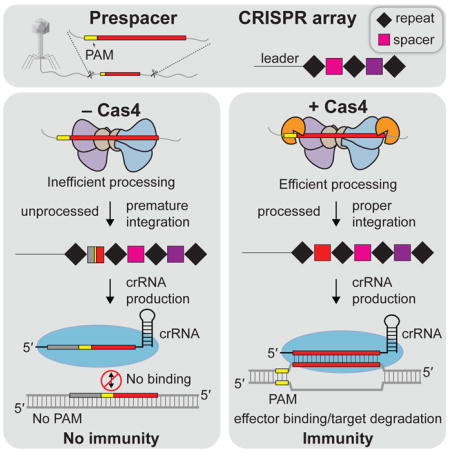
Despite the recognition of Cas4 as a core family of adaptation proteins, it has remained unclear whether it is directly involved in spacer acquisition. Our results reveal that Cas4 is a key factor in ensuring PAM-dependent prespacer processing prior to integration by the Cas1-Cas2 complex. Cas4 is required for efficient processing of precursor prespacers with 3' overhangs, which may be generated from RecBCD or Cas3 activities (Levy et al., 2015; Kunne et al., 2016; Staals et al., 2016). Previous biochemical studies found that some Cas4 variants exhibit bidirectional exonuclease activity (Lemak et al., 2013), suggesting that Cas4 5'-3' exonucleolytic activity against blunt DNA ends may also generate precursors with 3' overhangs. A recent study also showed that Cas4 can trim the ends of precursor substrates at multiple sites that are not protected by Cas1-Cas2 to generate the final length of prespacers prior to integration (Rollie et al., 2017). Our results show that Cas4 cuts precursors at specific locations through endonucleolytic cleavage, suggesting that Cas4 associates with Cas1-Cas2 and the complex positions the 3' overhangs in the Cas4 active site to dictate the exact cut sites.
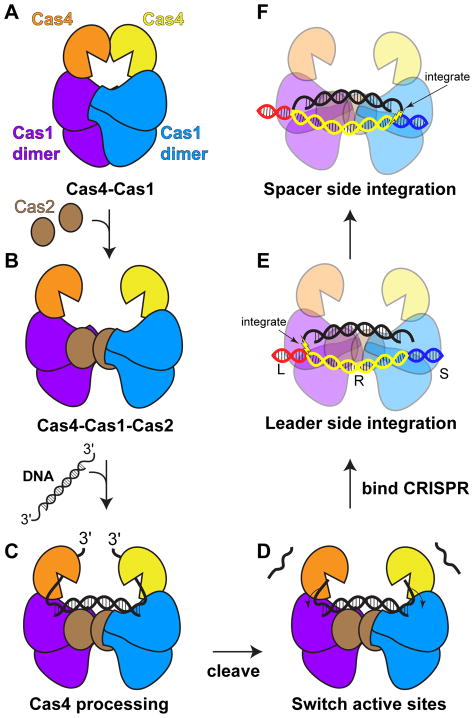
Our structural studies of the Cas4-Cas1 complex reveal a surprising architecture that may be mutually exclusive with formation of the Cas1-Cas2 complex structure observed in other sub-types. It is possible that type I-C Cas1-Cas2 adopts a different overall conformation. Alternatively, it is possible that the Cas4-Cas1 complex sequesters Cas1, preventing it from forming a productive Cas1-Cas2 complex for integrating dsDNA substrates (Figure 6A–B). Thus, these competing structures could provide a regulatory mechanism for the adaptation stage of CRISPR immunity. Future structural work will be required to determine how the Cas4-Cas1 structure transitions to the overall adaptation complex.
Figure 6. Model of processing and integration by Cas4-Cas1-Cas2.
(A–B) Transition of (A) Cas4-Cas1 to (B) a putative Cas4-Cas1-Cas2 complex. The two structures may be mutually exclusive. (C) Upon binding of precursor DNA substrates with long 3' overhangs, Cas4 subunits within the putative Cas4-Cas1-Cas2 complex binds the overhangs in their active sites. (D) Following cleavage by Cas4, the shortened 3' overhangs are transferred to the adjacent Cas1 active sites. (E) Following binding of the complex at the CRISPR locus, Cas1 integrates the substrate at the leader-spacer junction. (F) Integration at the repeat-spacer junction is dictated by the length of the substrate and only proceeds following leader-side integration and complete substrate processing.
Our structural studies also indicate that the Cas4 and Cas1 active sites are relatively distal from one another within the Cas4-Cas1 complex (Figure 1E). This arrangement of the two active sites disfavors a model in which both proteins simultaneously contribute to cleavage. Consistently, our mutagenesis studies reveal that the Cas4 active site is necessary and sufficient for efficient prespacer processing, while the Cas1 active site can catalyze low levels of cleavage in the absence of Cas4. Together, these results suggest that long single-stranded 3' overhangs may shuttle between the Cas4 and Cas1 active sites, and may be preferentially bound by Cas4 until cleavage occurs, preventing either cleavage or integration by the Cas1 active site (Figure 6C–D). Consistently, we mainly observe integration following prespacer processing in the presence of Cas4, while integration of unprocessed prespacers was much more prevalent in the absence of Cas4. In addition, we observe low specificity for minus-strand integration when using unprocessed prespacers, suggesting that specific integration at the repeat-spacer junction only occurs during full-site integration and requires processing of both ends of the substrate (Figure 6E–F). Integration at the correct repeat-spacer junction is also dictated by specific sequences within the repeat, which provides an additional ruler mechanism to dictate the site of integration (Goren et al., 2016; Wang et al., 2016).
In order to generate functional spacers, prespacers with correct PAMs must be captured by the adaptation complex and processed just upstream of the PAM. Previous studies of the type I-E Cas1-Cas2 complex revealed that Cas1 can recognize and cleave PAM sequences within the 3'-overhangs of prespacers (Wang et al., 2015), while studies of type I-A adaptation indicated that Cas4 trims prespacers in a PAM-dependent manner (Rollie et al., 2017). Similarly, our results show that the type I-C adaptation complex processes prespacers in a PAM-specific manner based on either Cas1-dependent cleavage in the absence of Cas4, or Cas4-dependent cleavage in the presence of all three adaptation proteins. In addition to enhancing prespacer cleavage, Cas4 also provides an important fidelity check to ensure that prespacers are only integrated following removal of the PAM sequence. Integration prior to processing is likely to result in a spacer targeting a non-PAM region. Thus, our results suggest that Cas4-dependent processing is critical for maintaining a fully functional CRISPR array, without the addition of defective spacers through aberrant integration prior to prespacer processing.
Functional spacers also require that the PAM-proximal end of the prespacer be integrated at the repeat-spacer junction, although it remains unclear how the orientation of spacer integration is achieved. Our results reveal that prespacer processing occurs asymmetrically, with different length overhangs produced following processing. It is possible that asymmetrical overhang length may help to dictate spacer orientation during half-site integration. However, it remains unclear what factors dictate asymmetrical processing. Notably, outside of the degenerate sequence, the overhang sequences were identical for both strands of the prespacers used in our experiments, suggesting that the duplex sequence may affect processing asymmetry rather than the overhang sequence. Further experiments will be needed to determine the exact mechanism for spacer orientation by the adaptation complex.
Our analysis of cleavage site selection also suggests that prespacer length may vary based on “slipping” within the active site during prespacer processing, as processing sites varied for some prespacers tested. These results are consistent with in vivo analysis of type I-C spacer acquisition, which has shown that PAM slipping can cause aberrant spacer lengths (Rao et al., 2017). Similarly, the lengths of the existing spacers in the B. halodurans CRISPR array varies between 33–36 bp. Together, these data suggest that selection of functional spacers in vivo may play a key role in determining the spacer size.
Overall, our data support a model in which a putative Cas4-Cas1-Cas2 complex controls processing and integration of prespacers (Figure 6). Cas4 cleavage of precursor prespacers establishes spacer length and PAM site, while active site switching ensures that only cleaved ends enter the Cas1 active site and that integration only occurs following processing at both ends of the precursor (Figure 6C–D). Precise integration at the leader-repeat junction establishes the location for spacer insertion (Figure 6E), while integration at the repeat-spacer junction is dictated by the length of the prespacer substrate (Figure 6F). Our work establishes the core role for Cas4 in type I-C adaptation and suggests a similar integral function in other Cas4-containing systems.
STAR Methods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli BL21 Star (DE3) | Thermo Fisher Scientific | C6010-03 |
| Escherichia coli BL21 (DE3) | New England Biolabs | C2527I |
| Escherichia coli TOP10 | Thermo Fisher Scientific | C4040-03 |
| Biological Samples | ||
| Bacillus halodurans JM 9153 genomic DNA | ATCC | BAA-125D-5 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Q5 High Fidelity DNA Polymerase | New England Biolabs | #M0491L |
| T4 DNA Ligase | New England Biolabs | #M0202L |
| BamHI-HF | New England Biolabs | #R3136L |
| XhoI-HF | New England Biolabs | #R0146L |
| EcoRI-HF | New England Biolabs | #R3101L |
| T4 Polynucleotide Kinase | New England Biolabs | #M0201L |
| GoTaq DNA polymerase | Promega | #M3008 |
| DpnI | New England Biolabs | #R0176L |
| Agar | AMRESCO | #J637-1kg |
| Carbenicillin disodium salt | RPI | #C46000-25.0 |
| Kanamycin monosulfate | RPI | #K22000-25.0 |
| Ampicillin | RPI | #A40040-100.0 |
| Tetracycline HCl | RPI | #T17000-25.0 |
| LB Broth (Miller) | Thermo Fisher Scientific | #BP1426-2 |
| IPTG | RPI | #I56000-100.0 |
| DTT | RPI | #D11000-100.0 |
| Tryptone | RPI | #T60060-5000.0 |
| Sodium chloride | AMRESCO | #7647-14.5 |
| Yeast extract | Thermo Fisher Scientific | #BP1422-2 |
| Agarose | Thermo Fisher Scientific | #BP160-500 |
| Invitrogen SYBR Safe DNA Gel Stain | Thermo Fisher Scientific | #S33102 |
| HEPES | Thermo Fisher Scientific | #BP310-1 |
| Sodium phosphate dibasic heptahydrate | Thermo Fisher Scientific | #S373-3 |
| Glycerol | VWR analytical BDH | #BDH1172-4LP |
| Imidazole | Thermo Fisher Scientific | #O31960599 |
| PMSF | RPI | #P20270-25.0 |
| Ferrous sulfate | Thermo Fisher Scientific | #I146-500 |
| Ferric sulfate | Sigma | #F3388-250G |
| L-Cysteine free base | MP Biomedicals | #194646 |
| TEMED | RPI | #T18000-0.25 |
| Manganese chloride tetrahydrate | Thermo Fisher Scientific | #M87-100 |
| Potassium chloride | RPI | #D41000-2500.0 |
| Brilliant blue R-250 | RPI | #B43000-50.0 |
| Ammonium persulfate | RPI | #A20500-100.0 |
| 40% Acrylamide/Bis solution, 19:1 | Thermo Fisher Scientific | #BP1406-1 |
| Urea | RPI | #U20200-25000.0 |
| Boric acid | RPI | #B32050-5000.0 |
| Tris | RPI | #T60040-5000.0 |
| EDTA | Thermo Fisher Scientific | #BP120-1 |
| 2X RNA loading dye | New England Biolabs | #B0363A |
| Invitrogen GlycoBlue | Thermo Fisher Scientific | #00548854 |
| NEB Buffer2 | New England Biolabs | #B7002S |
| DNA polymerase I, Large (Klenow) fragment | New England Biolabs | #M0210S |
| dNTP solution set | New England Biolabs | #N0446S |
| Magnesium chloride | AMRESCO | #J364-500 |
| [α-32P]-dATP | Perkin Elmer | #BLU512H250UC |
| [γ32P]-ATP | Perkin Elmer | #BLU502A250UC |
| Phenol:Chloroform:Isoamyl Alcohol (25:24:1) | Thermo Fisher Scientific | #15593049 |
| Terminal Transferase | New England Biolabs | #M0315S |
| Critical Commercial Assays | ||
| QIAprep Spin Miniprep Kit | Qiagen | #27106 |
| Wizard SV Gel and PCR Clean-up system | Promega | #A9282 |
| Wizard Plus SV minipreps DNA purification system | Promega | #A1460 |
| HisPur Ni-NTA Spin columns | Thermo Fisher Scientific | #88224 |
| HisPur Ni-NTA resin | Thermo Fisher Scientific | #88223 |
| HiTrap SP HP | GE Healthcare | #7115201 |
| HiTrap Heparin HP | GE Healthcare | #17-0407-03 |
| HiLoad 16/600 Superdex 200 | GE Healthcare | #28989335 |
| HiLoad 16/600 Superdex 75 | GE Healthcare | #28989333 |
| TapeStation 2200 High Sensitivity D1000 kit | Agilent Technologies | 5067-5584 |
| TruSeq DNA Nano Library Preparation | Illumina | 20015964 |
| Deposited Data | ||
| Pyrobaculum calidifontis Cas4 | Lemak et al, 2014 | PDB: 4R5Q |
| Archaeoglobus fulgidus Cas1 | Kim et al, 2013 | PDB: 4N06 |
| Escherichia coli Cas1 | Nuñez et al, 2014 | PDB: 4P6I |
| Bacillus halodurans Cas4-Cas1 | This paper | EMDB-7485 |
| Gel images | This paper | http://dx.doi.org/doi:10.17632/w2gbyz6228.1 |
| Oligonucleotides | ||
| See Table S1 and S2 for sequences of oligonucleotides used in this study. | ||
| Recombinant DNA | ||
| pET52b | EMD Millipore | 72554 |
| pET52b/ His6 Cas4 | This paper | N/A |
| pET52b/ His6 Cas4-Cas1 | This paper | N/A |
| pET52b/ His6 Cas4-Cas1 H234A | This paper | N/A |
| pET52b/ His6 Cas4-Cas1 K110A | This paper | N/A |
| pSV272 | James Berger Lab | N/A |
| pSV272/ His6-MBP-TEV Cas1 | This paper | N/A |
| pSV272/ His6-MBP-TEV Cas1 H234A | This paper | N/A |
| pSV272/ His6-MBP-TEV Cas2 | This paper | N/A |
| pUC19 | New England Biolabs | N3041S |
| pUC19/Repeat-spacer-repeat CRISPR array | This Paper | N/A |
| pRSF-1b | EMD Millipore | 71363 |
| pRSF/Cas4 | This paper | N/A |
| pRKSUF017 | Takahashi et al, 2002 | N/A |
| Software and Algorithms | ||
| Appion | Lander et al, 2009 | nramm.nysbc.org/software |
| cryoSPARC | Punjani et al, 2017 | cryosparc.com |
| RELION | Scheres, 2012 | mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| Chimera | Pettersen et al, 20014 | cgl.ucsf.edu/chimera/ |
| Segger | Pintilie et al, 2010 | cryoem.bcm.edu/cryoem/downloads/segger |
| GSNAP-GMAP | Wu and Watanbe, 2005 | Research-pub.gene.com/gmap/ |
| Other | ||
| Formvar/Carbon 400 mesh, Copper approx. grid hole size: 42μm | Ted Pella, Inc. | 01754-F |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dipali Sashital (sashital@iastate.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Escherichia coli BL21 (DE3)
E. coli BL21 (DE3) cells were used for protein production of Cas4-Cas1, Cas1 and Cas2 for in vitro experiments. Cells were grown at 16°C in LB medium supplemented with 100 ug/mL ampicillin for Cas4-Cas1 and 50 μg/mL kanamycin for Cas1 or Cas2.
Escherichia coli BL21 Star (DE3)
E. coli BL21 Star (DE3) cells were used for protein production of Cas4 with pRKSUF017 for in vitro experiments. Cells were grown at 18°C in 2xYT medium supplemented with 25 μg/mL carbenicillin and 2.5 μg/mL tetracycline.
Escherichia coli TOP10
E. coli TOP10 cells were used for cloning of plasmids. Cells were grown at 37°C in LB medium supplemented with 50 μg/mL kanamycin or 100 ug/mL ampicillin.
Method Details
Cloning
Genomic DNA from Bacillus halodurans was obtained from ATCC. The cas4, cas1, and cas2 genes were PCR amplified from the genomic DNA using the indicated primers in Table S1 and cloned into pET52b for cas4 and pSV272 for cas2 and cas1 for individual expression. For co-expression with N-terminal His6-tagged cas4, cas4 and cas1 or all three genes were PCR amplified as a single operon and cloned into pET52b. For co-expression with N-terminal His6-tagged cas1 (Figure S1A), cas1 was cloned into pET52b and untagged cas4 was cloned into pRSF. The pET52b expression vector encodes an N-terminal His6-tag and pSV272 expression vector encodes an N-terminal His6-MBP (maltose binding protein) tag followed by a tobacco etch virus (TEV) protease recognition site. pCRISPR was generated by PCR amplification of the CRISPR array and leader sequence from the genomic DNA using the indicated primers in Table S1 and ligation into BamHI- and EcoRI-digested pUC19. All sequences were verified by Sanger sequencing (Eurofins Genomics).
Protein purification
Cas1, Cas2 and Cas4-Cas1 were overexpressed in BL21(DE3) and grown to 0.6 OD600 in LB media, followed by overnight induction at 16°C with 0.5 mM IPTG. The cells were harvested and lysed using a homogenizer (Avestin, Inc.). All proteins were initially purified using HisPur Ni-NTA affinity resin in recommended buffers (Thermo Fisher Scientific). His6-MBP-Cas1 and His6-MBP-Cas2 were cleaved using TEV protease overnight at 4°C to remove His6-MBP tag. The cleaved Cas1 and Cas2 were flowed through a Ni-NTA column and further purified using a Superdex 200 16/60 or Superdex 75 16/60 GL column (GE Healthcare), respectively, in a size exclusion buffer (20 mM HEPES (pH 7.5), 100mM KCl, 5% glycerol and 2mM DTT).
Cas4 and pRKSUF017 (carrying sufABCDSE (Takahashi and Tokumoto, 2002)) were co-expressed in BL21(DE3) star cells and grown to 0.7–0.8 OD600 in 2xYT (pH 7.0) media with 100 mg of ferric citrate (Sigma), ferrous sulfate (Fisher), and L-cysteine (MP biomedicals), followed by overnight induction at 18 °C with 1 mM IPTG. Cas4 was purified as described above through a Ni-NTA column and further purified by using Superdex 200 16/600 in size exclusion buffer. All final stocks were concentrated, aliquoted, flash frozen in liquid nitrogen, and stored at −80°C.
Pull-down assays
32 μM His6-Cas4-Cas1 or 32 μM His6-Cas1 was incubated with Ni-NTA SpinTrap columns (Thermo) at 4 °C for 15min in size exclusion buffer. 27 uM or 12.8 μM untagged Cas2 was added and incubated at 4 °C for 15 min. As a negative control, 27 μM untagged Cas2 was incubated in SpinTrap columns at 4ºC for 15 min in the absence of His6-Cas4-Cas1. The columns were washed with size exclusion buffer supplemented with 20 mM, 40mM, 80 mM, 150 mM, 200 mM, 250 mM, and 300 mM imidazole, and flow through for each wash was collected for analysis. Samples were run on 4–20% SDS-PAGE (NEB) and the gels were stained by Coomassie blue.
Negative-stain electron microscopy
4 μL Cas4-Cas1 complex (~100 nM) was applied to a glow-discharged copper 400-mesh continuous carbon grid. After one-minute adsorption, the grid was blotted on a filter paper to remove the majority of the protein buffer and immediately stained with 2% (w/v) uranyl acetate solution on the continuous carbon side. The grid was then blotted on a filter paper to remove residual stain and air-dried in a fume hood. The grid was observed with a JEOL 2010F transmission electron microscope operated at 200 keV with a nominal magnification of x60,000 (3.6 Å at the specimen level). Each image was acquired using a 1 s exposure time with a total dose of ~30–35 e-Å-2 and a defocus between –1 and –2 μm. A total of 50 micrographs were manually recorded on a Gatan OneView camera.
Single-particle pre-processing
The image processing and two-dimensional (2D) classification were performed in Appion (Lander et al., 2009). A total of ~21,300 particles were picked from 50 micrographs using a template generated from a 2D class average of another random protein complex with a similar size as the Cas4-Cas1 complex. Particles were extracted using a 64 x 64-pixel box size. Reference-free 2D class averages were generated using a total of 146 classes. 13,855 particles were left after removal of junk 2D classes in Appion.
Three-dimensional reconstruction and analysis
The 13,855 good particles left after removing those contributing to junk class averages in Appion were first used for ab initio three-dimensional (3D) reconstruction in cryoSPARC (Punjani et al., 2017). These particles were further subjected to reference-free 2D classification with 100 classes in RELION (Scheres, 2012). After further removal of bad 2D classes, 12,967 particles were left for the 3D classification in RELION using the 3D model generated by cryoSPARC as a starting model. The best class (clearest features and with the largest number of particles) was refined using Autorefine within RELION. The reference-free 2D class averages showed excellent agreement with the reprojections of the final 3D model (Figure S1G). The Euler angle plot created in RELION showed a good distribution of Euler angles, despite some preferred orientations (Figure S1H). The final 3D reconstruction, which showed structural features to ~21 Å resolution based on the 0.143 gold standard FSC criterion (Figure S1H), was segmented using Segger (Pintilie et al., 2010) in Chimera (Pettersen et al., 2004). The crystal structures of the Cas4 protein Pcal_0546 from Pyrobaculum calidifontis (PDB ID: 4R5Q) (Lemak et al., 2014), Cas1 from Archaeoglobus fulgidus (PDB ID: 4N06) (Kim et al., 2013), and the Cas1 dimers in the Cas1-Cas2 complex from E. coli (PDB ID: 4P6I; blue, chain C and D; purple, chain E and F) (Nuñez et al., 2014) were docked into the final reconstruction of the Cas4-Cas1 complex. We chose to use the E. coli crystal structure for the model shown in Figure 1 because the Cas1 core fits unambiguously into the map. While the A. fulgidus variant is a closer homolog to B. halodurans Cas1, a large portion of the crystal structure lies outside of the electron density when fit to the map (Figure S1J). It appears that the conformation of the crystal structure is not the same as in our map and would require us to flexibly fit a large alpha-helical domain into the density which would be over-interpreting our structure at the current resolution.
DNA substrate preparation
All oligonucleotides were synthesized by Integrated DNA Technologies. Sequences of all DNA substrates are shown in Table S2. Prespacers and minimal dsDNA CRISPR arrays were hybridized by heating to 95 °C for 5 minutes and slow cooling to room temperature in oligo annealing buffer (20 mM HEPES (pH 7.5), 25 mM KCl, 10 mM MgCl2) and purified on 8% native PAGE. Prespacers were labeled with [γ-32P]-ATP (PerkinElmer) and T4 polynucleotide kinase (NEB) for 5'-end labelling or with [α-32P]-dATP (PerkinElmer) and Terminal Transferase (NEB) for 3'-end labelling. The double-stranded minimal CRISPRs were labeled with [α-32P]-dATP (PerkinElmer) and Klenow-fragment (NEB) for 3'-end labelling.
Integration assays
For plasmid integration assays, Cas4-Cas1 complex was formed by incubating 200 nM Cas1 and 500 nM Cas4 at 37 °C in integration buffer (20 mM HEPES (pH 7.5), 100 mM KCl, 5% glycerol, 2 mM MnCl2, and 2 mM DTT) for 10 min. 200 nM Cas2 was added and the complex was incubated on ice for 10 min. For Cas1-Cas2, 200 nM Cas1 was incubated with 200 nM Cas2 on ice for 10 min. Both Cas1-Cas2 and Cas4-Cas1-Cas2 were incubated with 500 nM of prespacer for 5 min at room temperature. 7.5 nM pCRISPR were added and incubated for one hour either at 37 °C or 65 °C, as indicated. Reactions were quenched by addition of phenol-chloroform-isoamyl alcohol. Samples were analyzed on 1 % unstained agarose gel at 18 V overnight and post-stained with SYBR Safe (Invitrogen) for 30 min.
Integration assays with 5'- or 3'-radiolabelled minimal CRISPRs were carried out with 2 μM Cas4, 1 μM Cas1, 1 μM Cas4-Cas1, and 1 μM Cas2 in integration buffer. Cas1-Cas2 or Cas4-Cas1-Cas2 complexes were formed from individual protein components through incubation steps described above. The co-purified Cas4-Cas1 complex was incubated with Cas2 at 4 ºC for 10 min to form Cas4-Cas1-Cas2. Complexes were incubated with 1 μM prespacer and minimal CRISPRs at 65 °C for 30 minutes. Reactions were quenched by the addition of phenol-chloroform-isoamyl alcohol. Samples were extracted and run on 8% urea-PAGE. The gels were dried and imaged using phosphor screens on a Typhoon imager (GE Life Sciences).
Prespacer processing assays
Prespacer processing assays were performed using 5'or 3'-radiolabelled prespacer with 500 nM Cas4, 200 nM Cas1, 200 nM Cas4-Cas1 and 50 nM Cas2 in integration buffer. The complexes were incubated as described above and all reactions were performed at 65 °C for 20 min. Reactions were quenched with phenol-chloroform-isoamyl alcohol. Samples were extracted and were run on 12% urea-PAGE. The gels were dried and imaged using phosphor screens.
Low- and high-throughput sequencing of HSI products
HSI products were amplified from plasmid integration assay products generated as described above, with 2 μM Cas4, 1 μM Cas1 and Cas2, 1 μM of prespacer 1 or 2, and 7.5 nM pCRISPR at 37 ºC. For low-throughput sequencing, samples were purified with Wizard SV Gel and PCR Clean-Up kit (Promega) and amplified using primers (Table S1) containing BamHI or XhoI sites by PCR using GoTaq polymerase (Promega). The amplicons were digested, ligated into BamHI- and XhoI- digested pRSF, and plasmids extracted from 20 transformants for each sample were analyzed by Sanger sequencing. For high-throughput sequencing, three separate samples were prepared for each condition and treated as separate replicates. Samples were purified with PCR clean-up kit before amplification. The samples were amplified by PCR using GoTaq polymerase (Promega) with barcoded primers. Amplification products were analyzed on 2% SYBR Safe stained agarose gels and quantified using densitometry. Samples were mixed in equal quantities and were run on 2% agarose gel. The band was excised and DNA was purified using a Wizard SV Gel and PCR Clean-Up kit (Promega). The DNA was analyzed on a TapeStation 2200 High Sensitivity D1000 kit (Agilent Technologies), libraries were prepared using a TruSeq DNA Nano Library Preparation (Illumina), and libraries were sequenced (2 x 150 paired-end reads) on an Illumina MiSeq by Admera Health, LLC (New Jersey, USA).
HSI Data processing and analysis
Because the vast majority of products were less than 150 bp in length, only the R1 reads were analyzed from the paired-end read output. Sequences were demultiplexed and sorted into separate files for each sample condition and replicate based on the presence of specific pairs of barcodes at both ends of the read using a bash script. To determine the site of integration, the reads were matched to the pCRISPR sequence using GMAP (Wu and Watanabe, 2005). The site of integration was considered to be the position at which the match between the read and pCRISPR began, with the exception noted below. An output file was generated for each condition and replicate containing the number of counts at each start site position. The plots in Figures 5D and S7 show the average number reads at each start site for the three replicates, with standard deviation represented as error bars. For plus strand HSI products, many prespacer cleavage products ended in T, and were assumed to be integrated at the end of the leader rather than at the -1 position within the leader, which is also a T. Because minus strand integration was less specific, it was not possible to determine both the precise site of integration and the processing site, therefore this assumption was not applied to these products.
To determine the processing site of the positive strand HSI products, the length of sequence between the end of the duplex sequence within the prespacer and the beginning of the repeat sequence within pCRISPR was determined for all reads for amplification products 1 and 2 for all conditions (Figure 5A). The average number of counts for products of each length for the three replicates is plotted in Figure 5D, with error bars representing standard deviation.
QUANTIFICATION AND STATISTICAL ANALYSIS
All in vitro experiments were repeated three times, and representative gel images were shown. Quantification of data shown in Figure 2D was performed using ImageJ. All plotted data are the average of three replicates with error bars representing standard deviation.
DATA AND SOFTWARE AVAILABILITY
The accession number for the Cas4-Cas1 EM density reported in this paper is EMDB-7485. All gel images are available on Mendeley Data at http://dx.doi.org/doi:10.17632/w2gbyz6228.1.
Supplementary Material
Highlights.
Cas4 forms a tight heterohexameric complex with the Cas1 spacer integrase
Cas4 is a Cas1-Cas2-dependent endonuclease that cleaves 3' overhangs of prespacers
Cas4 cleavage is sequence and site specific and depends on the presence of a PAM
Cas4 blocks premature integration of uncleaved prespacers, ensuring spacer fidelity
Acknowledgments
We thank Raimund Nagel and Reuben Peters for providing pRKSUF017 plasmid and members of the Taylor and Sashital laboratories for helpful discussion. This work was supported by NIH R01 GM115874 (to D.G.S.) and Welch Foundation Grant F-1938 (to D.W.T.). D.W.T is a CPRIT Scholar supported by the Cancer Prevention and Research Institute of Texas (RR160088).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Author Contributions
H.L. performed all biochemical and sequencing experiments. Y.Z. performed single particle electron microscopy and structure determination. H.L., Y.Z., D.W.T., and D.G.S. analyzed and interpreted the results. H.L. and D.G.S. wrote the manuscript with Y.Z. and D.W.T. contributing. D.W.T and D.G.S. supervised research and secured funding for the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bolotin A, et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brouns SJJ, et al. Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes Stan. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, et al. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund RD, et al. Spacer capture and integration by a type I-F Cas1–Cas2-3 CRISPR adaptation complex. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1618421114. 201618421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gesner EM, et al. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- Goren MG, et al. Repeat Size Determination by Two Molecular Rulers in the Type I-E CRISPR Array. Cell Rep. 2016;16:2811–2818. doi: 10.1016/j.celrep.2016.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, et al. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem. 2013;288:27888–27897. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, et al. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Hudaiberdiev S, et al. Phylogenomics of Cas4 family nucleases. BMC Evol Biol. 2017;17:232. doi: 10.1186/s12862-017-1081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Wiedenheft B. A Conserved Structural Chassis for Mounting Versatile CRISPR RNA-Guided Immune Responses. Mol Cell. 2015;58:722–728. doi: 10.1016/j.molcel.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, et al. CRISPR-Cas: Adapting to change. Science. 2017;356 doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- Kim TY, et al. Crystal structure of Cas1 from Archaeoglobus fulgidus and characterization of its nucleolytic activity. Biochem Biophys Res Commun. 2013;441:720–725. doi: 10.1016/j.bbrc.2013.10.122. [DOI] [PubMed] [Google Scholar]
- Koonin EV, et al. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunne T, et al. Cas3-Derived Target DNA Degradation Fragments Fuel Primed CRISPR Adaptation Article Cas3-Derived Target DNA Degradation Fragments Fuel Primed CRISPR Adaptation. 2016:1–13. doi: 10.1016/j.molcel.2016.07.011. [DOI] [PubMed] [Google Scholar]
- Lander GC, et al. Image Processing. Access. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenay RT, et al. Identifying and Visualizing Functional PAM Diversity across CRISPR-Cas Systems. Mol Cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak S, et al. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from sulfolobus solfataricus. J Am Chem Soc. 2013;135:17476–17487. doi: 10.1021/ja408729b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak S, et al. The CRISPR-associated Cas4 protein Pcal 0546 from Pyrobaculum calidifontis contains a [ 2Fe-2S ] cluster3: crystal structure and nuclease activity. 2014;42:11144–11155. doi: 10.1093/nar/gku797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, et al. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn J, Marraffini LA. CRISPR-Cas Systems Optimize Their Immune Response by Specifying the Site of Spacer Integration. Mol Cell. 2016;64:616–623. doi: 10.1016/j.molcel.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraju P, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Nuñez JK, et al. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez JK, et al. Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature. 2015a;527:535–538. doi: 10.1038/nature15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez JK, et al. Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature. 2015b;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez JK, et al. CRISPR Immunological Memory Requires a Host Factor for Specificity. Mol Cell. 2016;62:824–833. doi: 10.1016/j.molcel.2016.04.027. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, et al. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie GD, et al. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, et al. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, et al. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Punjani A, et al. CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rao C, et al. Priming in a permissive type I-C CRISPR-Cas system reveals distinct dynamics of spacer acquisition and loss. RNA. 2017;23:1525–1538. doi: 10.1261/rna.062083.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding S, et al. Surveillance and Processing of Foreign DNA by the Escherichia coli CRISPR-Cas System. Cell. 2015 doi: 10.1016/j.cell.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollie C, et al. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. Elife. 2015;4 doi: 10.7554/eLife.08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollie C, et al. Prespacer processing and specific integration in a Type I-A CRISPR system. Nucleic Acids Res. 2017:1–14. doi: 10.1093/nar/gkx1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MF, et al. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc Natl Acad Sci. 2017;1:201616395. doi: 10.1073/pnas.1616395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, et al. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RHJ, et al. Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR–Cas system. Nat Commun. 2016;7:1–13. doi: 10.1038/ncomms12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, et al. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems. Cell. 2015;163:840–853. doi: 10.1016/j.cell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Wang R, et al. DNA motifs determining the accuracy of repeat duplication during CRISPR adaptation in Haloarcula hispanica. Nucleic Acids Res. 2016;44:4266–4277. doi: 10.1093/nar/gkw260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, et al. CRISPR Immunity Relies on the Consecutive Binding and Degradation of Negatively Supercoiled Invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, et al. Structures of the CRISPR genome integration complex. Science. 2017;357:1113–1118. doi: 10.1126/science.aao0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Doudna JA. Protecting genome integrity during CRISPR immune adaptation. Nat Struct Mol Biol. 2016;23:876–883. doi: 10.1038/nsmb.3289. [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Xiao Y, et al. How type II CRISPR–Cas establish immunity through Cas1–Cas2-mediated spacer integration. Nature. 2017;550:137–141. doi: 10.1038/nature24020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, et al. Real-Time Observation of Target Search by the CRISPR Surveillance Complex Cascade. Cell Rep. 2017;21 doi: 10.1016/j.celrep.2017.11.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, et al. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. The CRISPR Associated Protein Cas4 Is a 5’ to 3’ DNA Exonuclease with an Iron-Sulfur Cluster. PLoS One. 2012;7 doi: 10.1371/journal.pone.0047232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.