Summary
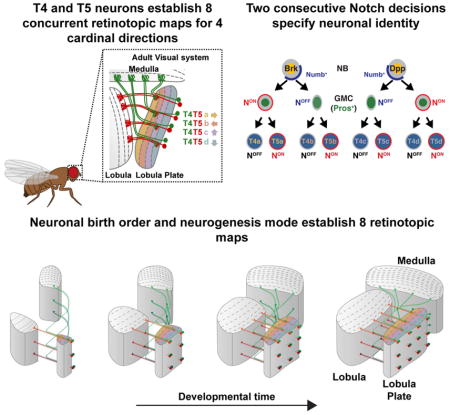
Understanding how complex brain wiring is produced during development is a daunting challenge. In Drosophila, information from 800 retinal ommatidia is processed in distinct brain neuropiles, each subdivided into 800 matching retinotopic columns. In the Lobula Plate there are four T4 and four T5 neuronal subtypes. T4 neurons respond to bright edge motion while T5 neurons to dark edge motion, and each is tuned to motion in one of the four cardinal directions, effectively establishing eight concurrent retinotopic maps to support wide-field motion. We discovered a mode of neurogenesis where two sequential Notch-dependent divisions of either a horizontal or a vertical progenitor produces matching sets of two T4 and two T5 neurons retinotopically coincident with pair-wise opposite direction selectivity. We show that retinotopy is an emergent characteristic of this neurogenic program and derives directly from neuronal birth order. Our work illustrates how simple developmental rules can implement complex neural organization.
Keywords: Drosophila, retinotopy, pattern formation, direction selective neurons, optic lobe, neural development, neural connectivity, Notch
In Brief
The circuit for motion perception emerges out of the developmental program that specifies the identity of the neurons
Introduction
A central question in neuroscience is how individual neurons are assembled into functional neural networks. In both vertebrates and invertebrates, visual input from the retina is retinotopically mapped onto the brain such that neighboring pixels are represented as neighboring regions on the retinotopic map. An example of such complex organization is found in the Drosophila optic lobe, which receives retinotopic input from photoreceptors in the eye (Fischbach and Dittrich, 1989; Morante and Desplan, 2008). The compound eye of the fly is made up of ~800 ommatidia (unit eyes). Photoreceptors from each ommatidium send their axons into the optic lobe, which is comprised of four neuropiles that each contains ~800 columns: the Lamina, Medulla, Lobula and Lobula Plate (Figure 1A–C) (Bausenwein et al., 1992; Fischbach and Dittrich, 1989; Morante and Desplan, 2008; Takemura et al., 2008). Each column processes visual information from one point in space, maintaining retinotopy (Morante and Desplan, 2004, 2008).
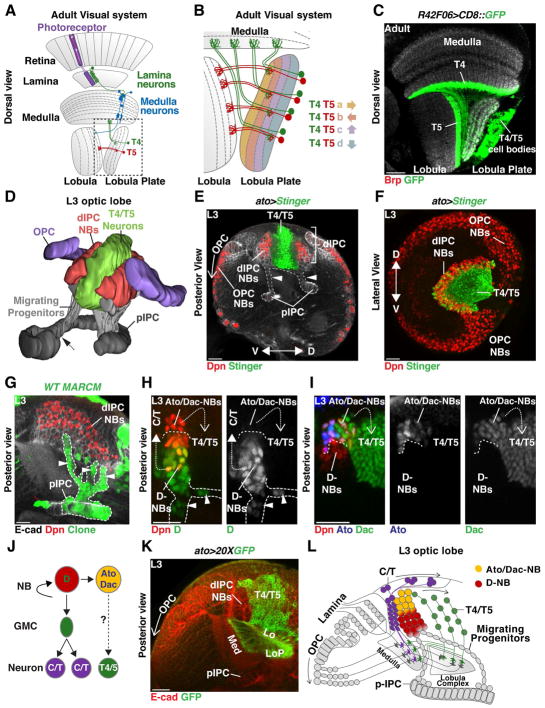
Figure 1. Development of the dIPC.
(A) Schematic of an adult optic lobe highlighting the ON (T4) and OFF (T5) pathways of the motion detection circuit. Visual input propagates retinotopically via T4 and T5 neurons to the four layers of the Lobula Plate. Dashed line shows detail in panel (B).
(B) Four subtypes of T4 and of T5 neurons, each directionally tuned to one of the four cardinal directions, converge onto layers a and b of the horizontal system, or layers c and d of the vertical system in the Lobula Plate.
(C) Dorsal cross-section of an adult brain in which R42F06-Gal4 (T4/T5-Gal4) drives membrane-GFP (green) expression in T4 and T5 neurons. T4 dendrites project to the Medulla and T5 to the Lobula (neuropiles labeled in grey by anti-Brp). (A, B, C) anterior is to the left.
(D) Posterior view of a 3-D model of the developing larval Outer (purple) and Inner (grey) Proliferation Centers (OPC and IPC). Dashed arrow marks the boundary between the proximal-IPC (pIPC) and the surface IPC. Progenitors delaminate from the pIPC (grey) to become neuroblasts (NBs) in the distal-IPC (dIPC) (red). T4 and T5 neurons (green) are generated by dIPC NBs and form inside the dIPC crescent. See also Movie 1.
(E, F) Red-Stinger driven by Ato-Gal4 labels T4/T5 neurons.
(E) Posterior cross- section of a developing larval optic lobe. The pIPC-neuroepithelium is localized between the OPC and the central brain. Cells delaminating from the pIPC (arrowheads) migrate to form the dIPC-NBs (Dpn, red) that will generate T4 and T5 neurons (green). In the OPC, a proneural wave converts the OPC-neuroepithelium into Medulla NBs (Dpn, red) from proximal to distal (open arrow).
(F) dIPC-NBs form a U-shaped cluster (Dpn, red) along the dorsal-ventral axis that surrounds T4 and T5 neurons (green). Anterior is to the left.
(G) pIPC-MARCM clone (green). Migrating cells (arrowheads) delaminate from specific neuroepithelial locations and form dIPC NBs (Dpn, red) that spread along the dorso-ventral axis (see (F) and Movie 1).
(H,I,J) dIPC-NBs transit trough two temporal stages: Migrating cells (arrowheads) and newly produced NBs express Dichaete (D) [green in (H)] that produce distal C/T neurons (closed dashed arrow); Older NBs express Ato [blue in (I)] and Dac (green) and produce Dac+ T4/T5 neurons (open dashed arrow).
(J) Schematic summarizing neurogenesis of the dIPC.
(K) T4/T5 neurons project to their target Medulla, Lobula or Lobula Plate (Ecad, red) neuropiles during development.
(L) Schematic summarizing the development and neurogenesis of the dIPC. In this and subsequent figures scale bar is 20 μm.
The retinotopic organization of the fly visual system is crucial for circuit function, as exemplified by motion detection circuits. Within the optic lobe, visual motion information is processed in two parallel pathways, the ON pathway detecting bright edge motion, and the OFF pathway that processes dark edge motion (Clark et al., 2011; Eichner et al., 2011; Joesch et al., 2010; Joesch et al., 2013). The two pathways bifurcate early since distinct lamina neurons, the first to make contact with photoreceptors, connect to different sets of Medulla neurons, which themselves then synapse onto dendrites of T4 neurons (ON) in the Medulla, or T5 neurons (OFF) in the Lobula (Figure 1A–C) (Behnia et al., 2014; Clark et al., 2011; Rister et al., 2007; Shinomiya et al., 2014; Silies et al., 2013; Takemura et al., 2013). T4 and T5 neurons are the first neurons in each pathway that are direction selective (Maisak et al., 2013). They process the visual signal originating from one main column and integrate it with ~7 neighboring columns to compute local motion (Fisher et al., 2015; Leong et al., 2016; Strother et al., 2017; Takemura et al., 2013; Takemura et al., 2017).
Both T4 and T5 neurons exist in four subtypes (termed a,b,c,d), directionally tuned to one of the four cardinal directions (front-to-back, back-to-front, upward, downward) (Figure 1B). Thus, for each column, four T4 and four T5 neurons, one of each subtype, represent eight independent motion detectors. T4 (ON) and T5 (OFF) neurons with the same directional tuning project retinotopically to one of the four layers of the Lobula Plate (Maisak et al., 2013) that is organized into two layers for horizontal motion (layer a: front-to-back; layer b: back-to-front) and two layers for vertical motion (layer c: upward; layer d: downward) (Figure 1B). Within each layer, T4 and T5 neurons synapse with the dendrites of Lobula Plate Tangential Cells (Mauss et al., 2014; Schnell et al., 2012) that integrate the retinotopic local motion signals from T4 and T5 neurons to produce direction selective wide-field motion responses. Thus, the retinotopy of the T4/T5 circuit is crucial for detecting broad field motion: at the level of T4 (Medulla) and T5 (Lobula) dendrites, where the retinotopic organization of the inputs onto T4/T5 dendrites allows direction selectivity to first emerge (Fisher et al., 2015; Strother et al., 2017; Takemura et al., 2013), and at the level of their axons, where retinotopic organization allows for efficient, selective coding of specific global motion patterns (Barnhart et al., 2018; Mauss et al., 2015). It is therefore critical that the correct number of each T4 and T5 neuronal subtype be produced so that each Medulla column is innervated by the four T4 neuronal subtypes and each Lobula column by the four T5 subtypes. Furthermore, all eight subtypes of T4 and T5 neurons must project retinotopically to individual layers of the Lobula Plate.
The four neuropiles of the optic lobes develop during the larval and early pupal stages from two crescent-shaped neuroepithelial domains: the Outer Proliferation Center (OPC), which produces neurons of the Lamina and Medulla, and the Inner Proliferation Center (IPC), which generates neurons of the Lobula and Lobula plate (Figure 1D,E,F and Movie 1) (Apitz and Salecker, 2015; Egger et al., 2007; Hofbauer and Campos-Ortega, 1990; Nassif et al., 2003; Ngo et al., 2017). The IPC crescent is localized between the OPC and the developing central brain (Figure 1D,E and Movie 1). It is divided into three domains: the surface IPC (sIPC) marked by Wingless expression (which will not be discussed further in this work) that is attached to the proximal IPC (pIPC), and a distal domain (dIPC) (Figure 1D). T4/T5 neurons are produced by progenitors that originate from the pIPC (Apitz and Salecker, 2015). Unlike the OPC neuroepithelium that is sequentially converted by a proneural wave into neuroprogenitors (neuroblasts (NBs) in the fly) (Egger et al., 2007; Egger et al., 2010; Reddy et al., 2010), the pIPC-neuroepithelium produces Dichaete+ migrating progenitors that move distally to generate the dIPC (Figures 1D,E,G,H and Movie 2) (Apitz and Salecker, 2015). Once migrating progenitors reach the dIPC, they acquire a NB identity and divide to produce neurons (Figure 1E,I,J,K,L).
dIPC NBs progress through two temporal windows (Apitz and Salecker, 2015). First, Dichaete+ NBs divide to self-renew and produce the distal C2, C3, T2 and T3 neurons (C/T neurons) to the outside of the dIPC crescent (Figure 1H, J, L) (Apitz and Salecker, 2015). In the second temporal window, these NBs express Atonal (Ato) and Dachshund (Dac), and produce T4 and T5 neurons to the inside of the crescent (Figure 1I,J,K,L) (Apitz and Salecker, 2015; Oliva et al., 2014).
We investigated the developmental program that establishes the identity of the four T4 and four T5 neuronal subtypes and how this program leads to their eight coincident retinotopic maps. We identified a causal link between a mode of neurogenesis and retinotopy in which a single NB produces two ON and two OFF neurons with opposite motion direction selectivity (along the horizontal or the vertical axis) that innervate a single column in three neuropiles. We also show that vertical and horizontal T4/T5 motion detectors are produced by different NBs distinguished by Decapentaplegic (Dpp) activity. We conclude that retinotopy results from the features of this neurogenic program, which depends on neuronal birth order and a unique mode of NB division to pattern a complex and highly organized neural network. Thus, simple developmental rules can generate a complex neural organization across three neuropils of the optic lobes.
Results
Vertical and horizontal motion sensitive T4 and T5 neurons originate from two distinct populations of progenitors
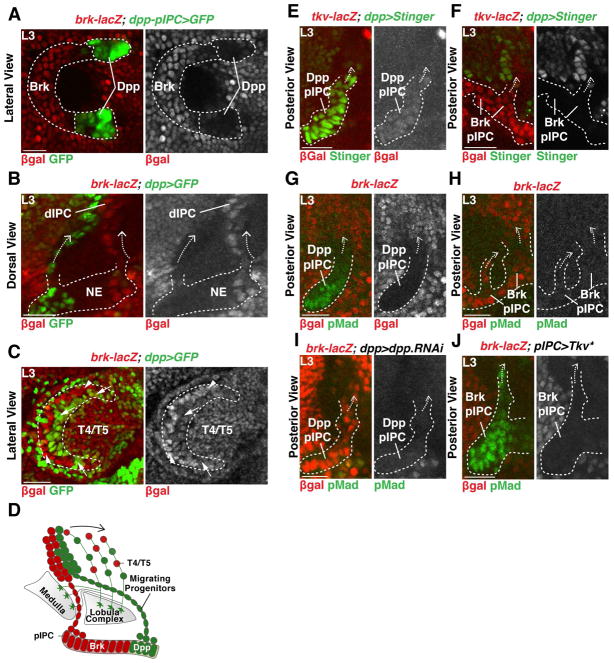
The dIPC is formed by migrating progenitors produced by distinct pIPC-neuroepithelial domains expressing either Dpp or Brinker (Brk) (Apitz and Salecker, 2015). The two Dpp-neuroepithelial domains are labeled by dpp-Gal4 (Apitz and Salecker, 2015) (Figure 2A,B) and by R45H05-Gal4 (termed Dpp-pIPC-Gal4) (Figure S1). The complementary Brk (a negative regulator of the Dpp pathway) domain sits between the two Dpp domains and is labeled by brk-lacZ and brk-Gal4, but does not express dpp-Gal4 (Figure 2A,B and Figure S1). Mosaic analysis with a repressible cellular marker (MARCM) and live imaging revealed that, even though cells delaminated from specific pIPC-neuroepitelial locations (Figure 1D,E,G and Movie 1), they spread along the entire dorso-ventral axis when reaching the dIPC such that Brk and Dpp derived NBs distributed along the entire dIPC dorsal-ventral axis (Figure 2A–D).
Figure 2. IPC-derived migrating progenitors populate the dIPC.
(A) Lateral view of the pIPC-neuroepithelial Brk domain marked by brk-lacZ (red) flanked by Dpp domains marked by dpp-pIPC>GFP (green). Anterior is to the left.
(B) Chains of migrating progenitors (arrows) emerging from the Dpp (green) or Brk domains (red).
(C) Lateral view of the dIPC. Anterior is to the left. Dpp-derived NBs (arrows, green) and Brk-derived NBs (arrowheads, red) spread equally within the dIPC. T4/T5 neurons are located in the inside of the crescent.
(D) Schematic summarizing the development of the dIPC.
(E,F) The Dpp receptor Tkv is expressed in both the Dpp (E) and Brk domains (F).
(G,H) Dpp signalling was active (pMad) in the Dpp (G), but not in the Brk domain (H).
(I) Expression of dpp-RNAi in the Dpp domains lead to the expansion of brk-lacZ expression in the Dpp domain and loss of pMad.
(J) Expression of an activated form of Tkv (Tkv*) downregulated brk-lacZ and leads to ectopic pMad in the Brk domain.
We hypothesized that Dpp signaling compartmentalizes the neuroepithelium and that the Brk and Dpp domains produce distinct subsets of T4 and T5 neurons. Upon binding Dpp, its receptor Thickveins (Tkv) phosphorylates the transcriptional effector Mad (pMad) that in turn regulates target gene expression. A lacZ reporter for Tkv was expressed within both the Dpp- and Brk-positive neuroepithelial domains (Figure 2E,F). However, pMad was only present in the Dpp domains, showing that the Dpp pathway is only active in the Dpp compartments of the neuroepithelium (Figure 2G,H). To test the role of Dpp in the specification of neuroepithelial identity, we used dpp-Gal4 to express a UAS-dpp-RNAi transgene; this led to the loss of pMad and to the expansion of the brk-lacZ reporter into the Dpp domains (Figure 2I). We also overexpressed a constitutively active form of the Tkv receptor (UAS-Tkv*) in the entire pIPC-neuroepithelium using the pIPC-specific driver line R35B01-Gal4 (termed pIPC-Gal4) (Figure S1). This resulted in the loss of brk-lacZ expression and the expansion of pMad to the Brk domain (Figure 2J).
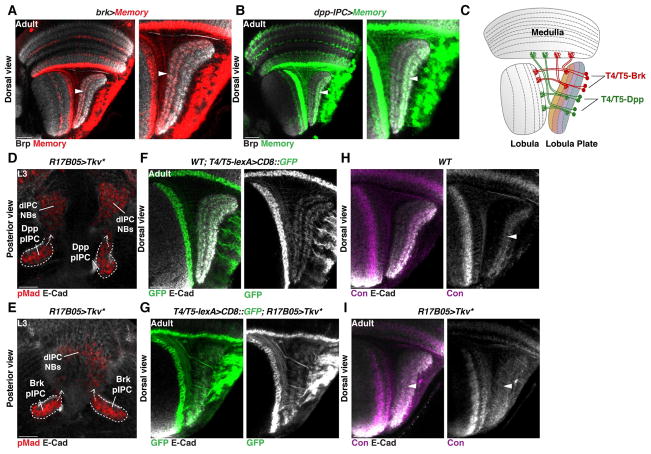
To test the contribution of the Brk- and Dpp-neuroepithelial domains to the production of T4 and T5 subtypes, we used memory cassettes (Bertet et al., 2014) in combination with the T4/T5 specific line R42F06-lexA (termed T4/T5-lexA) (Schnell et al., 2012). Activation of the memory cassette in early larvae (L2) within the Brk-neuroepithelial domain (using brk-Gal4) labeled all T4/T5 neurons of the horizontal system (subtypes a and b) (Figure 3A,C), while activation in the Dpp-neuroepithelial domains (using dpp-pIPC-Gal4) labeled all T4/T5 neurons of the vertical system (subtypes c and d) (Figure 3B,C). This demonstrates that the motion-detecting T4/T5 neurons of the horizontal and vertical systems derive from distinct neuroepithelial domains.
Figure 3. Horizontal and Vertical motion detection systems originate from distinct neuroepithelial domains.
(A,B) Memory cassette for brk-Gal4 (A) and dpp-pIPC-Gal4 (B): The horizontal system (layers a and b of the Lobula Plate, arrowhead in (A)) emerged entirely from the Brk domain, while the vertical system (layers c and d, arrowhead in (B)) was generated from the Dpp domain.
(C) Schematic of an adult brain representing horizontal, Brk-derived (red), and vertical sensing, Dpp-derived (green), T4 and T5 neurons.
(D,E) R17B05-Gal4 was used to drive the expression of an activated form of Tkv (Tkv*) in the entire pIPC neuroepithelium, resulting in the expression of pMad (Red) in the entire pIPC neuroepithelium, migrating chains and dIPC progenitors.
(F,G) Dorsal cross section of an adult wild type optic lobe (F) and an optic lobe where Tkv* was expressed in the pIPC neuroepithelium during development using R17B05-Gal4 (G). In (F) and (G) T4/T5-lexA drives the expression of membrane-GFP (green) in T4 and T5 neurons.
(H,I) Overexpression of Tkv* in the pIPC produced a Lobula Plate neuropile with only two layers that express Connectin (Con) (arrowheads; magenta) (I), which specifically labels neurons innervating layers c and d (H).
Given that Dpp signaling established neuroepithelial identity, we hypothesized that Dpp signaling specifies the identity of horizontal and vertical system neurons. To test this hypothesis we overexpressed Tkv* in the entire pIPC-neuroepithelium from late L2 until early pupae using the pIPC-specific driver line R17B05-Gal4 (Figure S1) and visualized the neurons produced using T4/T5-lexA. Expression of Tkv* resulted in pMad expression in the entire pIPC neuroepithelium and in all migrating chains (Figure 3D,E), and in the production of a Lobula Plate consisting of only c and d layers (compare Figure 3F to 3G) that are specifically labeled by the cell adhesion molecule Connectin (compare Figure 3H to 3I) (Gao et al., 2008).
Altogether, these observations show that Dpp signaling establishes the identity of the Brk- and Dpp-neuroepithelial domains, which give rise to horizontal (Brk) and vertical (Dpp) motion sensitive T4/T5 neurons, respectively.
Notch signaling specifies T4 (OFF) versus T5 (ON) neuronal identity
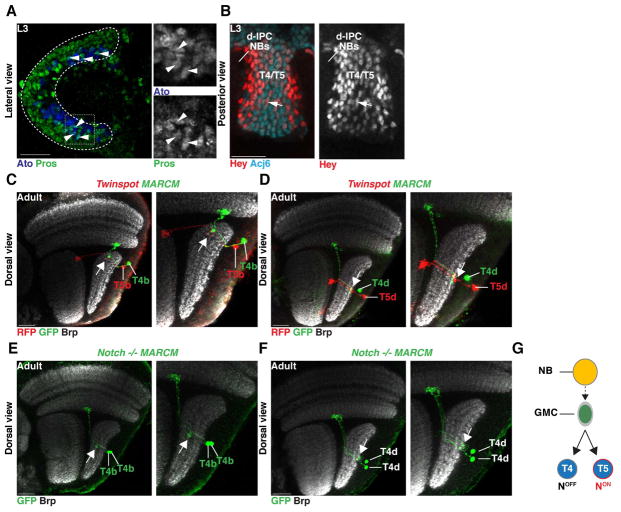
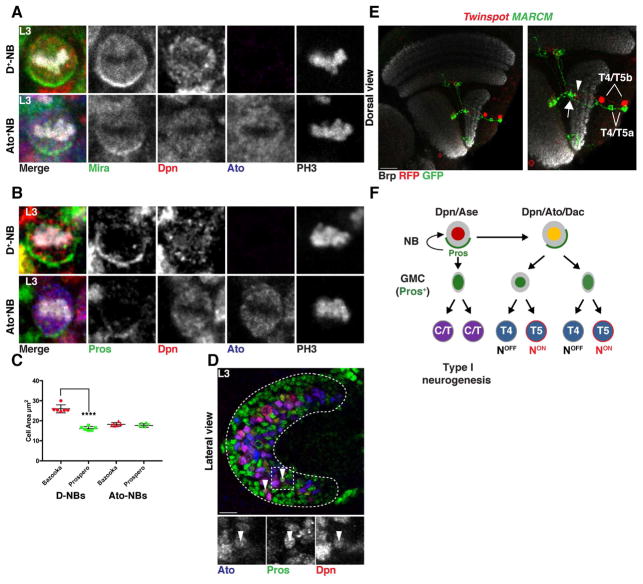
Upon delamination, migrating progenitors reach the dIPC where they assume a NB identity (Figure 1L). dIPC NBs first express Dichaete and divide to self-renew and produce Pros+ GMCs that generate neurons with C/T identity (Figure 1H,J,L) (Apitz and Salecker, 2015). These NBs then age and express Ato and Dac, and exclusively produce T4 and T5 neurons (Figure 1,H,I,J,K,L) (Apitz and Salecker, 2015; Oliva et al., 2014). We observed that newborn Ato+/Dac+/Pros+ GMCs were distributed in a salt and pepper manner surrounding T4/T5 Ato+/Pros− NBs (Figure 4A, Figure S3). The GMCs lost Ato expression by the time they divided to produce T4 and T5 neurons (Figure S3). During a typical GMC division, Notch signaling triggers a binary fate decision establishing the distinct identity of its two neuronal progeny: a NotchON and a NotchOFF neuron (Bertet et al., 2014; Buescher et al., 1998; Li et al., 2013b; Truman et al., 2010). To test the involvement of Notch in the specification of T4 versus T5 neuronal identity, we first determined the Notch status of developing neurons by monitoring the expression of the bHLH-O protein Hairy/enhancer-of-split-related with YRPW motif (Hey), which is a reliable sensor of Notch activity in optic lobe neurons (Bertet et al., 2014). We found that Hey was expressed in a salt and pepper manner in early born T4/T5 neurons (Figure 4B). This suggests that dIPC NBs that are Ato+/Dac+ produce GMCs that divide once to produce two neurons with alternative identities dictated by their Notch status. To test this hypothesis, we generated MARCM and Twin-Spot MARCM clones during the larval and early pupal stages that labeled the two progeny of Ato+/Dac+ GMCs with distinct fluorescent proteins (Yu et al., 2009). We visualized them in the adult using R42F06-Gal4 (termed T4/T5-Gal4). All 2-cell clones (N=15/15) contained one T4 and one T5 neuron of the same subtype, i.e. they projected to the same Lobula Plate layer at the same retinotopic position (Figure 4C,D and Figure S2).
Figure 4. A binary, Notch-mediated cell fate decision specifies T4 vs. T5 neurons.
(A) Ato+/Pros+ GMCs (arrowheads; blue and green) were distributed in a salt-and-pepper manner in the dIPC.
(B) T4/T5 neurons marked by Acj6 (cyan) expressed the Notch sensor Hey (arrow; red) in a salt-and-pepper manner.
(C,D) T4 and T5 neurons of the same subtype are sibling neurons and projected to the same retinotopic position in their target neuropiles (Brp, white). Twin-spot MARCM clone visualized with T4/T5-Gal4 of sibling T4 and T5 neurons of the horizontal (subtype b) [arrow in (C)], and vertical system (subtype d) [arrow in (D)]. See also Figure S2.
(E,F) Notch mutant MARCM clones visualized with T4/T5-Gal4 of a dividing GMC produced two sibling T4 neurons of the same subtypes (arrow) for both the horizontal (E) and vertical systems (F). The two T4 neurons projected to the same retinotopic position. See also Figure S2.
(G) Ato+ NBs (yellow) produce GMCs that divide once to produce sibling T4 (NotchOFF) and T5 (NotchON) neurons of the same subtype.
To further test the role of Notch in the specification of T4 vs. T5 identity, we analyzed Ato+/Dac+ GMC Notch mutant MARCM clones, which consistently showed two T4 neurons of the same subtype projecting to the same retinotopic position (N=32/32). This was true for GMCs derived from both horizontal and vertical system NBs (Figure 4E,F, S2). This shows that T4 and T5 neurons of the same subtype (a, b, c, or d) are siblings and that T4/T5 neurogenesis relies on four distinct GMC populations, each producing a T4 and a T5 neuron of a unique subtype: a or b originating from the Brk region, and c or d originating from the Dpp region (Figure 4G).
Atonal NBs undergo a terminal division to produce two GMCs that generate T4 and T5 neurons detecting motion in opposite directions
To investigate how T4/T5a neurons are distinguished from T4/T5b neurons within the horizontal system, and how T4/T5c neurons are distinguished from T4/T5d neurons within the vertical system, we characterized the mode of division of dIPC NBs. NBs can proliferate in distinct modes during development (Kohwi and Doe, 2013; Li et al., 2013a): Most divide to self-renew and to produce a smaller GMC that divides once to produce two daughter cells (“type I” NBs) (Buescher et al., 1998). Others self-renew and generate a ganglion mother cell (GMC) that does not divide, but differentiates directly into a neuron (“type 0” NBs) (Bertet et al., 2014; Karcavich and Doe, 2005; Ulvklo et al., 2012). “Type II” NBs self-renew and produce intermediate precursors that divide multiple times themselves to generate GMCs that then divide only once (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008).
To divide asymmetrically to self-renew and produce a GMC, NBs rely on the asymmetric localization of cell fate determinants that are differentially segregated between the two progeny (Homem and Knoblich, 2012). Prospero is a determinant of GMC identity that represses the expression of NB determinants (i.e. Deadpan) and of cell cycle genes (Choksi et al., 2006; Lai and Doe, 2014; Southall and Brand, 2009) to signal terminal differentiation (Doe et al., 1991). Prospero expression initiates in NBs, where it is kept inactive by the adaptor protein Miranda that tethers it to the cortex. During a NB division, Miranda and Prospero form a crescent in the basal cell cortex: In this manner, Miranda and Prospero are inherited by the GMC after cell division (Shen et al., 1997). Once in the GMC, Miranda is degraded, allowing Prospero to enter the nucleus to promote cell cycle exit and differentiation. The GMC then divides only once to produce two mitotically inactive cells (Choksi et al., 2006; Hirata et al., 1995; Ikeshima-Kataoka et al., 1997; Southall and Brand, 2009).
NB proliferation ends either when the NB undergoes apoptosis, or when the NB itself starts expressing Prospero and terminally divides. To test how Ato+ NBs end their life, we inhibited apoptosis using a mutation of the initiator caspase-9, dronc. This did not lead to NB accumulation (Figure S4), suggesting that Ato+ NBs stop proliferation by a terminal division.
To assess the mode of dIPC NB divisions, we examined the expression of the proneural factor Asense (Ase), which is expressed in “type I” NBs and their GMCs, as well as the cellular localization of both Miranda and Prospero. C/T dIPC NBs (Dichaete+) express Dpn and Ase (Figure S4), and always showed typical asymmetric cortically localized Miranda and Prospero crescents (N=15) (Figure 5A,B, Figure S4, Figure S5) (Apitz and Salecker, 2015). These NBs self-renewed and produced a smaller GMC that gave rise to C/T neurons (Figure 5C, Movie 3). In contrast, Ato+/Dac+ NBs do not express Ase but instead express another proneural protein, Ato (Figure 1I, Figure S4) (Apitz and Salecker, 2015; Oliva et al., 2014). Ato+/Dac+ NBs showed lower levels of Miranda expression as compared to Dichaete+ NBs (Figure S4). Nevertheless, they formed crescents of both Miranda and Prospero, although of weaker intensity than Dichaete+ NBs (Figure 5A,B, Figure S4, Figure S5). To unambiguously assess for Prospero crescents, we analyzed NBs at prometaphase/metaphase in both the Dichaete+ and Ato+ temporal windows, when crescents are easiest to observe (Doe et al., 1991; Shen et al., 1997). Virtually all Dichaete+ (N=15/15) and Ato+ NBs produced Prospero crescents (N=14/15) (Figure 5B). The formation of Prospero cortical crescents in Ato+ NBs was further confirmed with a Pros::GFP line (Figure S6). However, these Ato+ NBs did not self-renew: their progeny consisted of two Ato+/Dac+ cells of similar size (Figure 5C, Movie 4), a cell that inherited cortical Prospero (and Numb, see below), and a cell that transiently retained Dpn but also expressed nuclear Prospero immediately after cell division (Figure 5D), becoming the second GMC product of the Ato+ NB. This shows that Ato+ NBs divide once to effectively produce two GMCs (Figure 5F).
Figure 5. Ato+/Dac+ NBs divide symmetrically to produce two GMCs.
(A,B) First temporal window Dichaete+ and second temporal window Ato+ NBs localized Miranda (A) and Prospero (B) to the basal cortex during cell division. See also Figures S4 and S5.
(C) Quantification of the mean size of the two progeny of Dichaete+ and Ato+ NBs at the end of telophase. Dichaete+ NBs divided to self-renew (Bazooka, red) and produce a smaller GMC (Prospero, green)(see Movie 3). Ato+ NBs produced two progeny (Bazooka, red and Prospero, green) of the same size that both assumed a GMC fate (see Movie 4). Data are represented as a scattered plot ± SD. Unpaired two-tailed Student’s t test with Welch’s correction. Dichaete+ NBs progeny (t=10.79 df=7.338) and Ato+ NBs progeny (t=1.164 df=9.994), Asterisk indicates P value<0.0001.
(D) Division of an Ato+ NB produced a GMC (not shown in image) and a cell that expressed perduring Deadpan and nuclear Prospero (arrowheads; green), becoming the second GMC.
(E) A vertical system twin-spot MARCM clone of sister GMCs visualized with T4/T5-Gal4 showed two pairs of sibling T4/T5 neurons of opposite motion direction sensitivity, projecting to the same retinotopic position in the neuropile. Arrow points to layer a and arrowhead to layer b of the Lobula Plate (See also Figure S2).
(F) Schematic illustrating dIPC neurogenesis. Ato+ NBs divide to produce a GMC and a small cell that becomes a GMC.
To identify the progeny of single, terminally dividing Ato+ NBs, we analyzed MARCM and Twin-spot-MARCM clones marked by T4/T5-Gal4. In all analyzed clones (N=9/9), two pairs of T4/T5 neurons, either from the horizontal (a and b) or from the vertical (c and d) subtypes, projecting to the same retinotopic location, were observed (Figure 5E and Figure S2). This demonstrates that when horizontal (Brk) and vertical (Dpp) system dIPC NBs reach the Ato+ temporal window, they divide once to end their life, producing two sibling GMCs with different identities: a or b for Brk and c or d for Dpp (Figure 5F).
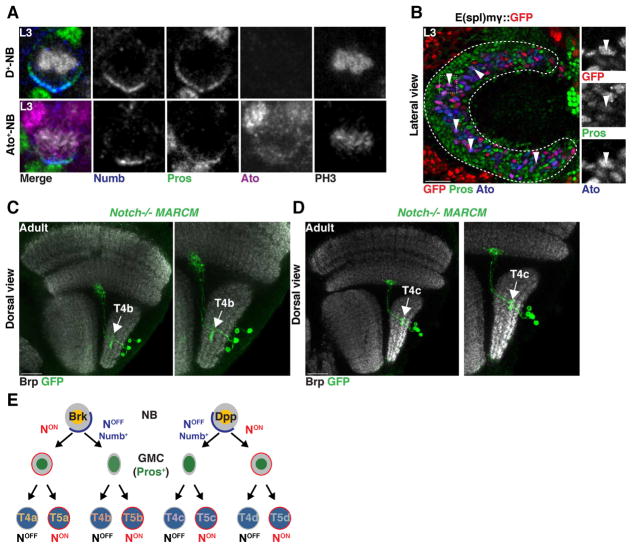
Atonal NBs divide to produce a NotchON and a NotchOFF GMC
When NBs divide, they not only segregate Prospero to the GMC, but also the Notch signaling inhibitor Numb (Knoblich et al., 1995; Rhyu et al., 1994; Wang et al., 2007)(Figure 6A). As a consequence, the NB remains NotchON while the GMC becomes NotchOFF. We hypothesized that the distinct identities of sibling T4/T5 GMCs could be the result of a Notch-mediated binary fate decision: The terminally dividing NB would produce one NotchOFF GMC and another cell that retains its NotchON identity and becomes the second GMC. This would establish the subtype identity (a vs. b or c vs. d) of the T4/T5 neurons from each GMC.. Consistent with this hypothesis, we observed that Ato+ NBs asymmetrically segregated Numb together with Prospero only one progeny cell (Figure 6A). Furthermore, we found that among the Ato+/Pros+ GMC population, we could identify GMCs that transiently expressed the Notch activity reporter E(spl)mγGFP (Almeida and Bray, 2005), corresponding to the NotchON GMC formed from the Ato+ NB (Figure 6B). To further test this hypothesis, we generated MARCM clones of Notch mutant NBs, which consistently produced clones of four T4 neurons (see above) that were all either of subtype b (from Brk+ NBs) (N=5) or of subtype c (from Dpp+ NBs) (N=9) (Figure 6C,D,E). These observations are consistent with a mode of neurogenesis in which a T4/T5 NB stops proliferation by dividing to produce two GMCs with a different Notch status: Notch first specifies the identity of the two sister GMCs (a vs. b or d vs. c), and then later the identity of sister neuronal progeny (T4 vs. T5) (Figure 6E).
Figure 6. Ato+/Dac+ neuroblasts terminally divide to produce two sister GMCs with different Notch identities.
(A) First temporal window Dichaete+ and second temporal window Ato+ NBs localized the Notch inhibitor Numb to the basal cortex during cell division.
(B) Newborn Ato+/Pros+ GMCs that did not inherit Numb transiently expressed the Notch reporter mγGFP (arrowheads).
(C,D) Notch mutant MARCM clones visualized with T4/T5-Gal4 produced four T4 neurons projecting to the same retinotopic location, both of subtype b in the horizontal system (C), or subtype d for the vertical system (D) (see also Figure S2).
(E) Schematic illustrating Ato+/Dac+ neurogenesis. Two Notch binary fate decisions specify neuronal identity.
Neuronal birth location and mode of neurogenesis establish a precise retinotopic map
During development, each T4 or T5 neuron must find its correct target location along the dorsal-ventral and anterior-posterior axes of its target neuropiles: the Medulla and Lobula Plate for T4 neurons, the Lobula and Lobula Plate for T5 neurons. In the medulla, all uni-columnar neurons from a single column, such as the Medulla intrinsic 1 (Mi1) neuron, are produced by a single NB located on top of this developing column (Erclik et al., 2017; Li et al., 2013b). This facilitates establishment of retinotopy as neurons are born within the column in which they will project. Indeed, the proneural wave that converts the OPC neuroepithelium into NBs is tightly synchronized with the morphogenetic furrow in the eye disc such that all NBs along one dorsal-ventral position are produced at the same time, matching the development of the eye (Egger et al., 2007; Egger et al., 2010; Ngo et al., 2010; Sato et al., 2016).
How T4 and T5 neurons project retinotopically to their targets is more difficult to understand. In contrast to OPC-derived neurons, T4 and T5 neurons are born far away from their first target neuropile without an obvious wave that could be synchronized with the eye morphogenetic furrow or OPC proneural wave: NBs delaminate from the pIPC, either from the Dpp or from the Brk domain, and migrate to establish the dIPC-NB crescent along the dorsal-ventral axis (Figure 2A–D). Furthermore, T4 and T5 axons have to cross the Lobula Plate, then project to the Medulla (T4) or Lobula (T5), and then come back to the Lobula Plate, terminating at the correct retinotopic location.
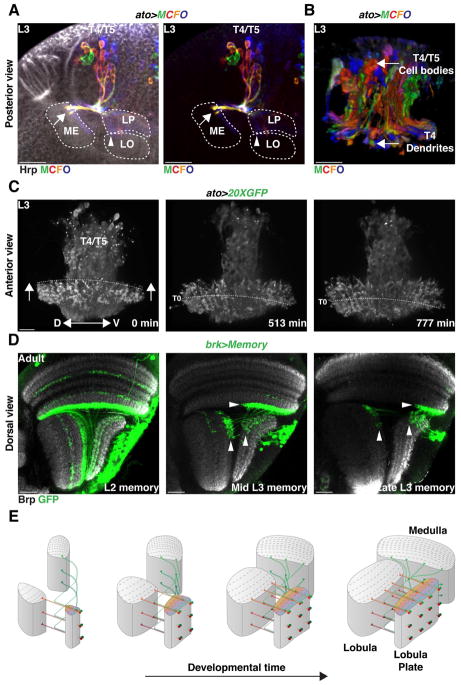
To address how retinotopy is established by the eight T4 and T5 neuronal subtypes, we investigated how they synchronize their targeting with target neuropile formation. We hypothesized that the mode of neurogenesis of Ato+ NBs, which we showed synchronously produces the full complement of T4 and T5 neurons with opposite motion direction selectivity, allows T4/T5 neurons to establish retinotopic projections. We observed that all four T4/T5 neurons produced by one NB projected to the same retinotopic position in their target neuropiles (Figure 5E Figure S2). To understand how a complex system of eight coincident retinotopic maps (one for each T4 and each T5 of the a, b, c, or d subtypes) comprising a total of 6400 neurons between three neuropiles is established, we visualized how T4/T5 neurons project along the dorsal-ventral and antero-posterior axes of the developing target neuropiles. We stochastically labeled developing T4/T5 neurons with different fluorophore combinations using Multicolor FlpOut (MCFO) (Nern et al., 2015) in combination with ato-Gal4 that labels T4/T5 neurons during development. This revealed that newborn T4/T5 neurons project to newly forming columns of their target neuropiles according to their relative birth position along the dorsal-ventral axis (Figure 7A,B and Movie 5). Based on these observations, we used ato>GFP to perform live imaging of developing T4/T5 neurons and visualized how innervation along the dorsal-ventral and antero-posterior axes of the neuropiles is coordinated. We focused on T4 neuronal projections and found that newborn T4 neurons targeted newly formed Medulla columns along the entire dorsal-ventral axis, a process that is reiterated along the anterior-posterior axis of the developing Medulla (Figure 7C and Movie 6).
Figure 7. Neurogenesis mode and neuronal birth location establish retinotopy.
(A,B) In a developing L3 larval brain, T4/T5 neurons projected to the neuropiles along the dorso-ventral axis according to their birth location.
(A) Posterior view of MCFO labeled T4/T5 neurons. Ventral is to the left.
(B) Anterior 3-D view of MCFO labeled T4/T5 neurons. Proximal is to the bottom. Neurons projected in the neuropiles along the dorsal-ventral axis according to their birth position. Ventrally born T4/T5 neurons projected ventrally in the neuropiles [arrow in (A)]. See also Movie 5.
(C) Still pictures of a time-lapse movie highlighting T4 projections in the Medulla. Anterior view with proximal to the bottom. Neurons innervating columns along the same dorso-ventral plane projected at the same time in the neuropiles. Dashed line follows the position of neurons that were targeting the medulla at T0 (see Movie 6). Arrows indicate the development along the anterior-posterior axis of the Medulla neuropile.
(D) Activation of a memory cassette in the Brk-pIPC domain at late L2, mid-L3 and Late L3 and visualized in the Adult with T4/T5-Gal4 labeled neurons of the horizontal system along the antero-posterior axis (Arrowheads) depending on their time of birth (see also Figure S7).
(E) 3-D Schematic illustrating the establishment of the retinotopy of T4 and T5 neurons.
Consistent with these observations, activation of a memory cassette in the pIPC neuroepithelium using brk-Gal4 or dpp-pIPC-Gal4 in early L2 marked all T4/T5 neurons, while activation at progressively later development stages marked neurons at more anterior parts of the visual field (Figure 7D and S7). Thus, NB time of birth establishes a temporal axis: NBs born early in development produce neurons targeting columns corresponding to the posterior eye (older columns), while later born NBs produce neurons that process information from more anterior parts of the eye (younger columns) (Figure 7E). We note that dIPC NBs do not exhibit spatial organization along the temporal axis since dIPC NBs are continuously replaced by new NBs originating from the pIPC. In contrast, the proneural wave in the OPC converts neuroepithelium crescent to NBs from its outer edge towards its inner edge, building the Medulla along a temporal axis (Figure 1K,L and Figure 7E). Because the dIPC is located distally to the developing Medulla, newborn T4 neurons find the youngest Medulla column that was just formed. Therefore, the time of birth of a dIPC-NB establishes a temporal axis that is synchronous with the conversion of the OPC neuroepithelium. Because each T4/T5 NB produces the full complement of T4/T5 neurons for either the horizontal or vertical system, this ensures that as new target columns are added through time they are supplied with the correct number of each T4/T5 neuronal subtype. Our results are consistent with a model in which matching of neuronal birth location and a specific mode of neurogenesis establishes a retinotopic, coherent connection across all optic lobe neuropiles.
Discussion
As neurons are produced and their identities are specified, they must be precisely incorporated into neuronal circuits. Understanding how neurons are specified, how the developing brain orchestrates the correct targeting of a myriad of individual neurons, and in which way these two developmental processes are related, are difficult problems to solve. We addressed these by studying how each of the eight T4/T5 neuronal subtypes is specified and how their eight retinotopic maps are precisely established.
Typically, NBs change their transcription factor identity at each division. Neuronal progeny inherit this identity through an intermediate GMC to dictate their fate (Doe, 2017; Rossi et al., 2017). Here we identified a mode of neurogenesis that relies on two consecutive Notch binary fate decisions to produce four distinct T4/T5 neurons from a single NB temporal window. Because T4/T5 neurons with opposite motion direction selectivity for one retinotopic position are produced by a single NB at the same time, these four neurons innervate their target neuropiles synchronously, connecting with the same, newly-produced target column to establish retinotopy. If each of the four T4 and T5 neurons were produced independently, synchronization of their projection patterns between three neuropiles could be much more difficult to achieve. This would require the establishment of a deterministic spatiotemporal molecular code, such that each column would use a unique molecular code recognized by all the neurons that are supposed to target it. The stepwise, synchronous production of sibling retinotopic neurons described here reduces the target possibilities at each time point since the progeny of one NB always find the newest column produced in the Medulla or Lobula. Our results illustrate how the developmental program that specifies T4/T5 fate meets the functional requirements of the motion circuit by establishing coherent retinotopic maps within horizontal and vertical systems.
Such successive divisions that rely on the reutilization of the Notch pathway are reminiscent of the divisions of Drosophila Sensory Organ Precursors (SOPs). Although these cells are not bona fide NBs, SOPs divide in a Notch-dependent manner multiple times to first produce two distinct cells (pIIa and pIIb) that divide once (pIIa) or twice (pIIb) more to give rise to the full complement of cells that form the sensory organ, only one of which is a neuron (Schweisguth, 2015). In olfactory sensilla, a similar precursor also appears to divide several times in a Notch-dependent fashion to produce up to four olfactory neurons, as well as sensilla cells. In this case, some of the four progeny die, producing 1, 2, 3 or in some rare cases four neurons per sensillum (Endo et al., 2007).
In the case of T4/T5 neurogenesis, we demonstrated that Notch signaling is used in two consecutive divisions: After the final NB division, the Notch target in one of the two GMCs is E(spl)my (but not Hey), while in the GMC division, Hey (but not E(spl)my) marks only one of the early born neurons (T5). How Notch differs in these distinct contexts, and how such precise temporal control is established is not known. However, the observation that Notch signalling activates different reporters in different contexts and cell types (Zacharioudaki and Bray, 2014) supports the notion that differential transcriptional programs are activated in different cell types. Furthermore, Notch activity is rather transient, which helps explain how Notch signalling instructs different gene expression programs at each round of division.
A recent preprint on a similar topic (Apitz and Salecker, 2018) is in line with our findings on the role of Dpp and Notch in the specification of the eight T4/T5 subtypes and shows that both Dac and Ato are required for the transition between neuroblasts competence states in the dIPC and for the switch to T4/T5 neuron formation. Another upcoming report (Mora et al., 2018) addresses the role of the temporal transition from Ase+ to Ato+ in dIPC neuroblasts and shows how Ato expression is required for subsequent neuronal differentiation of T4/T5 neurons. It further suggests that Ato+ neuroblasts divide symmetrically to self-amplify before producing the T4/T5 progeny. However, the data reported above, including our precise lineage analysis, do not support such an amplification step that would disrupt the stoichiometry of production of the C/T neurons and eight T4/T5 subtypes.
The lineage of the T4/T5 direction-selective neurons suggests how motion circuitry and the optic lobe neuropiles themselves might have evolved. We found that horizontal and vertical motion selective neurons originate from two distinct pIPC-neuroepithelial domains whose identity is established by Dpp signaling. In the absence of Dpp signaling, Brk expression was expanded to the Dpp domains, suggesting that the default status of the neuroepithelium is to express Brk. Horizontal and vertical motion-selective neurons were produced by distinct progenitor pools and both rely on the special type of neurogenesis described above to produce their complement of T4/T5 neurons. The most parsimonious evolutionary history for this developmental program is that the Notch-mediated binary fate decisions that specify layers of the Lobula Plate with opposite tuning, as well as T4 (moving bright edges) vs. T5 (moving dark edges) fate, was implemented before the specification of horizontal and vertical motion-selective subtype identity. The ancestor might have only responded to horizontal motion (Brk) before splitting of the neuroepithelium occurred, allowing the acquisition of vertical motion vision (Dpp), perhaps when the animals developed the capacity for flight.
T4 and T5 neurons share morphological and functional similarities (Haag et al., 2017), but also important differences, such as the organization of their dendritic processes in the Medulla (T4) vs. Lobula (T5) where each subtype (a,b,c, and d) must be oriented according to its local motion direction preference (Fisher et al., 2015; Strother et al., 2017). Dpp signaling and the two Notch binary fate decisions establish the specification of the four T4 and four T5 subtypes. Future studies will be required to understand how the dendrites of each subtype are properly organized.
Sensory maps and neural circuits are largely genetically “hard-wired” in Drosophila and are usually activity-independent (Hassan and Hiesinger, 2015; Hiesinger et al., 2006; Jefferis et al., 2001; Kolodkin and Hiesinger, 2017; Langen et al., 2015). Despite this developmental rigidity, we have a very limited understanding of how genetic programs drive developmental processes that are able to establish precise neural circuits. This study shows that the neurogenic program that specifies the identity of the eight T4/T5 neuron subtypes is also sufficient to establish the coherent retinotopy that supports global motion perception in the fly. It provides an example of how the establishment of connectivity within a neural circuit can only be fully understood in its developmental context. The existence of a causal link between the genetic program that specifies cell fate and the circuit these cells build provides an example of how a complex hard-wired neuronal circuit can be built from simple developmental rules.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Claude Desplan (cd38@nyu.edu).
Experimental Model and Subject Details
Fly Stocks
Flies were raised at 25°C and 60% humidity on standard cornmeal agar medium at a 12h light/dark cycle. Male and Female L3 larvae and 2–5 day old adult flies were analysed. No differences were observed between sexes. We used the following transgenic flies in this study (See Table S1 for more details); {} enclose individual genotypes, separated by commas: {yw;;}, {;UAS-CD8::GFP; T4/T5-Gal4 (R42F06)}, {;Ato-Gal4;} a gift from Bassem Hassan; {;Ato-Gal4; UAS-redStinger}, {;Ato-Gal4; 20XUAS-6XGFP}, {; UAS-redStinger; pIPC-Gal (R35B01)}, {; UAS-nls-GFP; pIPC-Gal (R35B01)}, {brkx47; UAS-redStinger; DPP-Gal4}) (brkx47 is a gift from Andrew Tomlinson), {brkx4;UAS-nlsGFP; DPP-pIPC-Gal4 (R45H05-Gal4)},{ Brk-Gal4(brk3sB); UAS-redStinger;}, {;tkvk16713; UAS-redStinger; DPP-Gal4}, {brkx4, UAS-DCR2; UAS-dppRNAi}, {brkx4;Ca-T2A-Uas-TKV*;Ca-8B3-Uas-TKV*} (TKV* lines are a gift from Julian Ng), {;Tubgal80ts;DPP-Gal4}, {;Tubgal80ts; pIPC-Gal4 (R35B01)}, {UAS-Flp;Tubgal80ts; act > y[+] > LHV2-86Fb,13XlexAop2-myr::GFP/TM6B (Flexamp)}; {;UAS-redStinger; Dpp-pIPC-Gal4 (R45H05)}; {Brk-Gal4(brk3sB); T4/T5-Lexa (R42F06);), {UAS-Flp; Tubgal80ts; LexAop<>CD8::GFP}, {yw,hsFlp122;Twinspot (BL56184); T4/T5-Gal4 (R42F06)}, {hsFlpPEST2; Twinspot (BL56184); T4/T5-Gal4 (R42F06)}, {;Twinspot (BL56185); T4/T5-Gal4 (R42F06)}, {hsFlp122, Tubgal80 Frt19; Uas-CD8::GFP; T4/T5-Gal4 (R42F06)}, {Frt19;;}, {Notch (N55e11) FRT19A;;}, {hsFlp122, Tubgal80 Frt19; UAS-lacZ, Tubg-Gal4}, {MCFO-1}, {;LexaopCD8::GFP;T4/T5-Lexa (R42F06); R17B05-gal4}, {Tubgal80ts;;Ca-8B3-Uas-TKV*}, {;E(spl)mγ-GFP;} (a gift from Sarah Bray), {;Pros::GFP;}, {GFPbaz-CC01941;;}, {;UAS-redStinger; R17B05-gal4}
Method Details
Antibodies
The following antibodies were used for this study: Guinea pig anti-Bruchpilot (Desplan Lab) (1:300), Guinea pig anti-Dichaete (1:50) (a gift from J. Nambu), Guinea pig anti-DPN (1:500) (Desplan Lab), Rabbit anti-DPN (1:500) (from Y.-N. Jan), Sheep anti-Ato (1:1000) is a gift from Bassem Hassan (Originally from by A. Jarman), Guinea pig anti-Ato (1:1000) (a gift from Daniel Marenda), Guinea pig anti-Hey (1:1000) (a gift from C. Delidakis), Rabbit anti-Ase (1:5000) (a gift from Y. Jan), Rat anti-Miranda (1/1000) and Mouse anti-Prospero (1/1000) (gifts from C. Doe); Guinea pig anti-pMad (a gift from Jessel Lab). Commercial antibodies used were: Goat anti-Numb (1:50) (Santa Cruz Biotechnology), Chicken anti-β-gal (1:2500) (Abcam), Rabbit anti-GFP (1:300) (Life technologies), Sheep anti-GFP (1:500) (Bio-Rad), Mouse anti-RFP 1/200 (MBL international), AlexaFluor405 conjugated Goat Anti-HRP (1:100; Jackson Immunochemicals). The following antibodies were obtained from the Developmental Studies Hybridoma Bank: Mouse anti-Dachshund (1:20), Mouse anti-Abnormal chemosensory jump 6 (1:20), Rat anti-DE-Cadherin (1:20), Mouse anti-Connectin (DSHB).
Secondary antibodies used at 1:200/1:500. Please view the Key resource table for additional information and providers.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea pig anti-Bruchpilot | Desplan Lab | N/A |
| Guinea Pig anti-Dichaete | J. Nambu | N/A |
| Guinea pig anti-DPN | Desplan Lab | N/A |
| Rabbit anti-DPN | Y.-N. Jan | N/A |
| Sheep anti-Ato | B. Hassan | N/A |
| Guinea pig anti-Ato | D. Marenda | N/A |
| Guinea pig anti-Dichaete | J. Nambu | N/A |
| Guinea pig anti-Hey | C. Delidakis | N/A |
| Rabbit anti-Ase | Y. Jan | N/A |
| Rabbit anti-Miranda | C. Doe | N/A |
| Mouse anti-Prospero | C. Doe | N/A |
| Guinea pig anti-PMad | T. Jessel | N/A |
| Chicken Anti-β-gal | Abcam | Code #9361 |
| Rabbit Anti GFP | Life Technologies | Catalog #11122 |
| Sheep Anti GFP | Bio-Rad | Code #4745-1051 |
| AlexaFluor405 conjugated Goat Anti-HRP | Jackson Immunochemicals | Code #123-475-021 |
| Mouse Anti-Dachshund | DSHB | Name: mAbdac2-3 |
| Mouse Anti Abnormal chemosensory jump 6 | DSHB | Name: Anti-Acj6 |
| Rat Anti DE-Cadherin | DSHB | Name: DCAD2 |
| Mouse Anti Connectin | DSHB | Name: Connectin C1.427 |
| Goat anti-Numb | Santa Cruz Biotechnology | Catalog #B0504 |
| Donkey Anti Sheep A488 | Jackson Immunochemicals | Code #713-545-147 |
| Donkey Anti Chicken A488 | Jackson Immunochemicals | Code #703-545-155 |
| Donkey Anti Rabbit A488 | Jackson Immunochemicals | Code #711-545-152 |
| Donkey Anti Mouse A488 | Jackson Immunochemicals | Code #715-545-151 |
| Dichaete Anti Guinea pig 488 | Jackson Immunochemicals | Code #706-545-148 |
| Donkey Anti Mouse A555 | Thermo Scientific | Catalog #A-31570 |
| Mouse Anti RFP | MBL | Code #M155-3 |
| Rabbit Anti RFP | MBL | Code #PM005 |
| Donkey Anti Rabbit A555 | Thermo Scientific | Catalog #A-31572 |
| Donkey Anti Guinea pig Cy3 | Jackson Immunochemicals | Code #706-165-148 |
| Donkey Anti Rat Cy3 | Jackson Immunochemicals | Code #712-165-153 |
| Donkey Anti Guinea pig A647 | Jackson Immunochemicals | Code #706-605-148 |
| Donkey Anti Rabbit A647 | Jackson Immunochemicals | Code #711-605-152 |
| Donkey Anti Mouse A647 | Jackson Immunochemicals | Code #715-605-151 |
| Donkey Anti Rat A647 | Jackson Immunochemicals | Code #712-605-153 |
| Donkey Anti Rabbit 405 | Jackson Immunochemicals | Code #711-475-152 |
| Donkey Anti Mouse 405 | Jackson Immunochemicals | Code #711-475-150 |
| Experimental Models: Organisms/Strains | ||
| Fly: D. melanogaster: T4/T5-Gal4 (R42F06) | BDSC | Stock #41253 |
| Fly: D. melanogaster: Ato-Gal4 | B. Hassan | N/A |
| Fly: D. melanogaster: 20XUAS-6XGFP | BDSC | Stock #52262 |
| Fly: D. melanogaster: pIPC-Gal (R35B01) | BDSC | Stock #49898 |
| Fly: D. melanogaster: brkx47 | A. Tomlinson | N/A |
| Fly: D. melanogaster: DPP-pIPC-Gal4 (R45H05) | BDSC | Stock #46529 |
| Fly: D. melanogaster: Brk-Gal4(brk3sB) | BDSC | Stock #53707 |
| Fly: D. melanogaster: tkvk16713 | BDSC | Stock #11191 |
| Fly: D. melanogaster: UAS-Dcr-2 | BDSC | Stock #24650 |
| Fly: D. melanogaster: UAS-dppRNAi | BDSC | Stock #36779 |
| Fly: D. melanogaster: DPP-Gal4 | BDSC | Stock #1553 |
| Fly: D. melanogaster: Ca-T2A-Uas-TKV* | Julian Ng | N/A |
| Fly: D. melanogaster: Ca-8B3-Uas-TKV* | Julian Ng | N/A |
| Fly: D. melanogaster: Flexamp Cassette | Desplan Lab | N/A |
| Fly: D. melanogaster: UAS-Flp | BDSC | Stock #8208 |
| Fly: D. melanogaster: T4/T5-Lexa (R42F06-Lexa) | BDSC | Stock #54203 |
| Fly: D. melanogaster: Lexaop<>CD8::GFP | BDSC | Stock #57588 |
| Fly: D. melanogaster: Twinspot | BDSC | Stock #56184 |
| Fly: D. melanogaster: Twinspot | BDSC | Stock #56185 |
| Fly: D. melanogaster: hsFlpPEST2 | A.Nern | N/A |
| Fly: D. melanogaster: {Notch (N55e11) FRT19A | BDSC | Stock #28813 |
| Fly: D. melanogaster: hsFlp,Tubgal80 Frt19a;; | BDSC | Stock #55132 |
| Fly: D. melanogaster: Frt19a;; | BDSC | Stock #1709 |
| Fly: D. melanogaster: MCFO-1 | A.Nern | Stock #64085 |
| Fly: D. melanogaster: GFPbaz-CC01941 | BDSC | Stock #51572 |
| Fly: D. melanogaster: Canton S | BDSC | Stock #64349 |
| Fly: D. melanogaster: {;E(spl)mγ-GFP;} | S. Bray | N/A |
| Fly: D. melanogaster: Pros::GFP | BDSC | Stock #66463 |
| Fly: D. melanogaster: Tubgal80ts (x) | BDSC | Stock #7016 |
| Fly: D. melanogaster: UAS-CD8::GFP (II) | BDSC | Stock #32186 |
| Fly: D. melanogaster: Tubgal80ts (II) | BDSC | Stock #7108 |
| Fly: D. melanogaster: UAS-RedStinger (III) | BDSC | Stock #8547 |
| Fly: D. melanogaster: UAS-RedStinger (II) | BDSC | Stock #8546 |
| Fly: D. melanogaster: lexaop-CD8::GFP (x) | BDSC | Stock #32204 |
| Fly: D. melanogaster: UAS-nlsGFP | BDSC | Stock #4775 |
| Fly: D. melanogaster: UAS-lacZ | BDSC | Stock #3955 |
| Fly: D. melanogaster: hsFlp | BDSC | Stock #1929 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SlowFade Gold | Thermo Scientific | Catalog #S36936 |
| Low Melting Agarose | Sigma | SKU #39346-81-1 |
| Schneider’s Medium | Sigma | SKU #S0146 |
| Fetal Bovine Serum | Sigma | SKU #12003C |
| Penicillin-Streptomycin | Sigma | SKU #P4333 |
| Insulin | Sigma | SKU #I9278 |
| Software and Algorithms | ||
| Fluorender | (Wan et al., 2017) | http://www.sci.utah.edu/software/fluorender.html |
| Fiji | (Schindelin et al., 2012) | https://fiji.sc |
| Prism 7 GraphPad | N/A | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Cell Culture Petri Dish | Thermo Scientific | Catalog #171099 |
Immunostaining Protocol
Larval or adult brains were dissected in 1XPBS and fixed in 4% Formaldehyde at room temperature for 20 min. Brains were then rinsed 3X times in PBX (1% PBS, 0.3% triton) and left to wash for at least 2 hours and then incubated in primary antibody solution overnight at 4°C. Brains were then rinsed 3X times and left to wash for at least 2 hours and then incubated in secondary antibody solution in PBX 2 hours (with added 2% horse serum) at room temperature. They were finally rinsed 3X and left to wash overnight in PBX and mounted in Slowfade.
For the staining of Miranda, Prospero and Numb crescents, an optimized fixation protocol and immunostaining was used:
Larval brains were fixed in 4% formaldehyde with 0.1 M PIPES (pH = 6.9), 0.3% Triton X-100, 20 mM EGTA, and 1 mM MgSO4 for 20 min at room temperature. Brains were then rinsed 3X times in PBX (1% PBS, 0.3% triton) and incubated in primary antibody solution in PBX (with added 2% horse serum) for 48 hours at 4°C. Brains were then rinsed 3X and left to wash for at least 8 hours and incubated in secondary antibody solution in PBX (with added 2% horse serum) overnight at 4°C. Brains where finally rinsed 3X wi th PBX and left to wash overnight at 4°C before mounting in Slowfade.
Multicolor Stochastic Labelling (MCFO)
“Multicolor FlpOut” (MCFO) labelling was carried out as described before (Nern et al., 2015). Briefly, FLP recombinase expression was used to excise FRT-flanked transcriptional terminators from UAS reporter constructs carrying HA, V5 and FLAG epitope tags. L3 Larvae carrying Ato-Gal4 and MCFO-1 were raised at 25°C and heat shocked at 37°C for 15min, 8–12 hours before dissection and staining.
Live Imaging
Live imaging was performed in Eye-brain complexes from late L3 {;Ato-Gal4>20XUAS-6XGFP} or {;UAS-nls-GFP; pIPC-Gal (R35B01, BL49898)}. Larvae were dissected in imaging medium (Schneider’s medium supplemented with 10% Fetal Bovine Serum, 1% Penicillin-Streptomycin and 0.25% Insulin) on ice. Eye-brain complexes were transferred to a cell culture petri dish, embedded in 0.4% low temperature gelling agarose. Once agarose solidified, eye-brain complexes were incubated in imaging medium for the duration of image acquisition. Image acquisition was done using an Olympus FV1000MPE multi-photon laser-scanning microscope (at 920nm).
Quantification and Statistical Analysis
Sample sizes were based on the standard of the field. Control or experimental specimens of the correct stage and genotype were randomly and independently selected from a pool of animals. Unless otherwise noted, we examined at least 10 brains for every genotype. In the main text “N” represents the number of samples analysed. Phenotypes were fully penetrant at all conditions. Data acquisition and analysis were not performed blinded as samples were identified based on their genotypes that were not limited in repeatability. When statistical analyses were performed the experimenter was blind to the genotypes analysed. The calculation of 95% confidence interval error bars and unpaired two-tailed Student’s t test P values (Welch-corrected, not assuming equal s.d.) was performed using Prism 7.00 for Mac, GraphPad Software, La Jolla California USA. Data met the assumption of normality (D’Agostino-Pearson omnibus normality test).
Image Acquisition and Processing
Images were acquired using a Leica SP5 confocal using a 20X, 40X or 60X objective. When acquired, Z-stacks sections were of 0.6–2 μm intervals. Images were processed with Fiji (Schindelin et al., 2012) software package. Image processing, when applied, was limited to correct global images brightness, global background subtraction and Despeckle. To allow the full visualization of the cell bodies and neuronal projections of adult MARCM and TWINSPOT T4/T5 Clones in a 2D image, stacks are shown as maximum intensity Z-projections. Z-projections were processed with the Fiji software package (Schindelin et al., 2012). Images were compiled in Illustrator (CC2014).
The Supplementary Movie 2 depicting the development of the IPC was processed in Fiji (Schindelin et al., 2012) by selecting the z-slice of interest for each time point and saving as an AVI file. The Supplementary Movies 3 and 4 depicting the division of Dichaete and Ato Neuroblasts, respectively, were processed in Fiji (Schindelin et al., 2012). For each movie the Z-slices of interest were concatenated as a stack and saved as an AVI file. The Supplementary Movie 5 depicting the 3-D view of MCFO labeled T4/T5 neurons was processed using Fiji 3D viewer (Schindelin et al., 2012). The Supplementary Movie 6 of a L3 larva optic lobe highlighting T4 projections in the developing Medulla was processed with Fluorender (Wan et al., 2017) and Fiji (Schindelin et al., 2012) software packages. The Z-stack was initially opened in Fluorender to create a 4D view of the Data. The resulting TIFF File was opened in Fiji to add labels and saved as AVI file.
Generation of Three-dimensional Model of Developing Optic Lobe
Ato>Stinger optic lobes of third instar larvae were dissected and labeled with Dpn and E-cad. The optic lobe used for generating the three-dimensional model (Figure 1d) was mounted in a Posterior orientation. Sections at 0.6-μm intervals were collected with a Leica Sp5 Confocal. Isosurfaces were produced with Fiji/TrakEM2 by manually outlining the regions of interest throughout the stack: Opc neuroepithelium, IPC, progenitors cell streams, dIPC neuroblasts and T4/T5 neurons cell bodies. Segmented objects were visualized into a model using the Fiji 3D viewer (Schindelin et al., 2012).
Supplementary Material
pIPC is compartmentalized into Brk and Dpp domains
The compartmentalization of the pIPC has been described previously (Aptiz, 2015). The Dpp chains correspond to the most posterior chains, two dorsal and two ventral, while all chains anterior to these correspond to Brk derived chains.
(A) dpp-pIPC-Gal4 labels the Dpp-pIPC neuroepithelium, but not the Brk-pIPC- neuroepithelium (B).
(C) brk-Gal4 labels the Brk-pIPC neuroepithelium, but not the Dpp-pIPC neuroepithelium (D).
(E,F) pIPC-Gal4 labels both the Dpp-pIPC (E) and Brk-pIPC (F) neuroepithelium migrating chains and dIPC progenitors.
(G,H) R17B05-Gal4 labels all the pIPC neuroepithelium, migrating chains and dIPC progenitors. In all panels, dashed lines delineate the pIPC.
A three-dimensional model of the developing Larval Optic Lobe
Three dimensional model built based on the expression of Ato>Red-Singer optic lobes of third instar larvae labeled with Dpn and E-cad. For labels see Figure 1D.
Time lapse of a L3 larval developing optic lobe
The pIPC is labeled with pIPC-Gal>UAS-nls-GFP to visualize the migration of pIPC derived migrating progenitors to the dIPC. pIPC-Gal4 is described in Sup. Figure 1.
Dichaete+ NBs divide asymmetrically in a Type I manner
Z-stack highlighting the division of a Dichaete+ NB in Telophase. The dividing NB highlighted with a dashed square in the top left corner of the image is depicted in the bottom of the image. Dichaete+ NBs divided asymmetrically to self renew (Arrow, Red, Bazooka+ cell) and produce a smaller that acquires a GMC fate (Arrowhead, Green).
Ato+ NBs division produces two GMCs.
Z-stack highlighting the division of a Ato+ NB in Telophase. The dividing NB highlighted with a dashed square in top left corner of the image is depicted in the bottom of the image. Ato+ NBs divided asymmetrically to produce two similarly sized cells that acquired a GMC fate (Arrowheads).
T4 and T5 neurons of the same subtype are sibling neurons and project to the same retinotopic position in their target neuropiles
(A–D) WT-MARCM clone of sibling T4 and T5 neurons.
(E–H) A Notch binary fate decision specifies T4 vs. T5 fate. Notch Mutant MARCM clones of a dividing Notch−/− GMC produced two sibling T4 retinotopic neurons of the same subtype.
(I) WT-MARCM clone of a horizontal system NB showed two pairs of sibling T4/T5 neurons of opposite motion direction sensitivity projecting to the same retinotopic position in the neuropiles. Arrow points to “layer a” projections and arrowhead to “layer b” projections.
(J) WT-MARCM clone of a vertical system NB showed two pairs of sibling T4/T5 neurons of opposite motion direction sensitivity projecting to the same retinotopic position in the neuropiles. Arrow points to “layer c” projections and arrowhead to “layer d” projections.
Ato+/Dac+ NBs produce Ato+/Dac+ GMCs, which divide to exclusively produce Dac+ T4 and T5 neurons. (A) Newborn Dac+ GMCs (open arrowheads) inherited atonal from the NB. As GMCs matured and moved towards the inside of the dIPC crescent they lost Atonal expression (arrows) such that dividing Dac+ GMCs did not express Atonal (closed arrowheads). (B) E(spl)mγ::GFP was only transiently expressed in newborn Ato+/Dac+ GMCs (open arrowheads). Dac+ dividing GMCs did not express E(spl)mγ::GFP (closed arrowheads) (N=15).
Dichaete+ and Ato+ NBs localize Miranda and Prospero during cell division
(A) Cross sections of a posterior view of the dIPC-NB population of a WT and dronc mutant optic lobe. (B) Quantification of the number of NBs in cross sections of WT and dronc Mutants. WT and dronc mutants showed equal number of dIPC-neuroblasts. Data are represented as scattered plot with mean ± SD. Two-tailed unpaired Student’s t test with Welch’s correction. P= 0,3579; t=0,9493; df=14,59.(C) dIPC Dichaete+ (D) NBs express Asense (Ase) (Green). In the second temporal window, dIPC-NBs expressed Dac (blue) and Ato (see also Figure 1I). (D,E) Overall levels of Miranda were reduced in Ato+ NBs. (E,F) Miranda (E) and Prospero (F) crescents were weaker in Ato+ (open arrowheads) than in Dichaete+ NBs (closed arrowheads). In (D) Fire-Lut represents Miranda signal intensity between the two temporal stages.
dIPC-NBs form Miranda and Prospero crescents
(A–D) Lateral views of developing optic lobes highlighting the formation of Miranda and Prospero crescents in Dichaete+ (A,B) and Ato+ NBs (C,D). The NBs highlighted in each panel with a dashed square are depicted in Figure 5.
Pros::GFP labels a Pros crescent in Ato+ NB
(A,B) Pros::GFP co-localizes with Prospero (A) and showed a higher signal to noise ratio than antibody staining, allowing the unambiguous identification of Prospero crescents in Ato+ NBs. Arrowheads identify crescents of Prospero. (B). Pros:GFP did not label the neuronal tracks identified with Asterisks in (A).
Neurons project along the antero-posterior axis depending on their time of birth.
Activation of the FLEXAMP cassette in the Dpp-pIPC domain at L2, early-L3 and Late L3 labeled neurons of the vertical system along the A-P axis (Arrowheads) depending on their time of birth.
3-D view of MCFO labeled T4/T5 neurons
Anterior 3-D view of MCFO labeled T4/T5 neurons. Proximal is to the bottom. Neurons projected in the neuropiles along the dorsal-ventral axis according to their time of birth.
Time-lapse movie of a L3 larva optic lobe highlighting T4 projections in the developing Medulla
Time-lapse movie highlighting T4 projections in the Medulla. Ato-Gal4 (described in Figure 1) was used to drive GFP in T4/T5 neurons. View is anterior and proximal is to the bottom. Neurons innervating columns along the same dorso-ventral plane projected at the same time in the neuropiles. Arrows indicate the development along the Medulla neuropile anterior-posterior axis. Dashed line follows the position of dendrites that were targeting the medulla at T0.
Highlights.
A neuroblast produces 4 neurons detecting 2 directions of bright or dark edge motion
Distinct progenitors produce neurons processing vertical vs. horizontal motion.
Notch specifies neurons responding to bright or dark edge motion.
This mode of neurogenesis establishes retinotopy in the fly global motion system.
Acknowledgments
We would like to dedicate this manuscript to the memory of Jean-Philippe Grossier with whom we had extensive discussions on the establishment of retinotopy. We would like to acknowledge Laura Quintana-Rio for helping building the 3D model of the developing optic lobe; Chris Doe and Cheng-Yu Lee for sharing reagents and suggestions for improving the antibody staining protocol, and Sarah Bennert for help with the elaboration of the illustrations. We also thank the fly community, and especially the Doe lab and Janelia for gifts of antibodies and fly stocks, and the Desplan laboratory members for essential discussion and support.
This work was supported by the Center for Genomics and Systems Biology of NYU Abu Dhabi and NIH grant R01 EY017916 to C.D. F. P-T., N.N. and A. D-V-R. were supported by the CGSB at NYUAD; C.B. by fellowships from EMBO (ALTF 680-2009) and HFSPO (LT000077/2010-L).
Footnotes
Author Contributions
F.P-T. and C.D. conceived the project, analyzed the data and wrote the manuscript. F.P-T. contributed to all the described experiments. C.K. performed many of the MARCM, MCFO and IPC experiments. A.M.R. characterized the Miranda and Prospero crescents. N.N and C.B performed early IPC characterization. X.L. performed preliminary FlexAmp analysis. A.D-V-R. performed early Twin-Spot MARCM analysis.
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almeida MS, Bray SJ. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech Dev. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Apitz H, Salecker I. A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system. Nature neuroscience. 2015;18:46–55. doi: 10.1038/nn.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apitz H, Salecker I. Spatio-temporal relays control layer specificity of motion-direction sensitive neurons in Drosophila. bioRxiv. 2018 doi: 10.1038/s41467-018-04592-z. https://doi.org/10.1101/262451. [DOI] [PMC free article] [PubMed]
- Barnhart EL, Wang IE, Wei H, Desplan C, Clandinin TR. Multilayered non-linear integration of local motion cues underpins efficient coding of global motion patterns in Drosophila. 2018 Submitted. [Google Scholar]
- Bausenwein B, Dittrich AP, Fischbach KF. The optic lobe of Drosophila melanogaster. II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res. 1992;267:17–28. doi: 10.1007/BF00318687. [DOI] [PubMed] [Google Scholar]
- Behnia R, Clark DA, Carter AG, Clandinin TR, Desplan C. Processing properties of ON and OFF pathways for Drosophila motion detection. Nature. 2014;512:427–430. doi: 10.1038/nature13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural development. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell. 2014;158:1173–1186. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Developmental neurobiology. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Developmental cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher M, Yeo SL, Udolph G, Zavortink M, Yang X, Tear G, Chia W. Binary sibling neuronal cell fate decisions in the Drosophila embryonic central nervous system are nonstochastic and require inscuteable-mediated asymmetry of ganglion mother cells. Genes & development. 1998;12:1858–1870. doi: 10.1101/gad.12.12.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Developmental cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Clark DA, Bursztyn L, Horowitz MA, Schnitzer MJ, Clandinin TR. Defining the computational structure of the motion detector in Drosophila. Neuron. 2011;70:1165–1177. doi: 10.1016/j.neuron.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Temporal Patterning in the Drosophila CNS. Annual review of cell and developmental biology. 2017;33:219–240. doi: 10.1146/annurev-cellbio-111315-125210. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural development. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development (Cambridge, England) 2010;137:2981–2987. doi: 10.1242/dev.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner H, Joesch M, Schnell B, Reiff DF, Borst A. Internal structure of the fly elementary motion detector. Neuron. 2011;70:1155–1164. doi: 10.1016/j.neuron.2011.03.028. [DOI] [PubMed] [Google Scholar]
- Endo K, Aoki T, Yoda Y, Kimura K, Hama C. Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nature neuroscience. 2007;10:153–160. doi: 10.1038/nn1832. [DOI] [PubMed] [Google Scholar]
- Erclik T, Li X, Courgeon M, Bertet C, Chen Z, Baumert R, Ng J, Koo C, Arain U, Behnia R, et al. Integration of temporal and spatial patterning generates neural diversity. Nature. 2017;541:365–370. doi: 10.1038/nature20794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach K-F, Dittrich A. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989:441–475. [Google Scholar]
- Fisher YE, Silies M, Clandinin TR. Orientation Selectivity Sharpens Motion Detection in Drosophila. Neuron. 2015;88:390–402. doi: 10.1016/j.neuron.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Takemura SY, Ting CY, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong ST, et al. The neural substrate of spectral preference in Drosophila. Neuron. 2008;60:328–342. doi: 10.1016/j.neuron.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J, Mishra A, Borst A. A common directional tuning mechanism of Drosophila motion-sensing neurons in the ON and in the OFF pathway. Elife. 2017:6. doi: 10.7554/eLife.29044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan BA, Hiesinger PR. Beyond Molecular Codes: Simple Rules to Wire Complex Brains. Cell. 2015;163:285–291. doi: 10.1016/j.cell.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesinger PR, Zhai RG, Zhou Y, Koh TW, Mehta SQ, Schulze KL, Cao Y, Verstreken P, Clandinin TR, Fischbach KF, et al. Activity-independent prespecification of synaptic partners in the visual map of Drosophila. Current biology : CB. 2006;16:1835–1843. doi: 10.1016/j.cub.2006.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- Hofbauer A, Campos-Ortega JA. Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Roux’s archives of developmental biology : the official organ of the EDBO. 1990;198:264–274. doi: 10.1007/BF00377393. [DOI] [PubMed] [Google Scholar]
- Homem CC, Knoblich JA. Drosophila neuroblasts: a model for stem cell biology. Development (Cambridge, England) 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- Joesch M, Schnell B, Raghu SV, Reiff DF, Borst A. ON and OFF pathways in Drosophila motion vision. Nature. 2010;468:300–304. doi: 10.1038/nature09545. [DOI] [PubMed] [Google Scholar]
- Joesch M, Weber F, Eichner H, Borst A. Functional specialization of parallel motion detection circuits in the fly. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:902–905. doi: 10.1523/JNEUROSCI.3374-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcavich R, Doe CQ. Drosophila neuroblast 7–3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. The Journal of comparative neurology. 2005;481:240–251. doi: 10.1002/cne.20371. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nature reviews Neuroscience. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin AL, Hiesinger PR. Wiring visual systems: common and divergent mechanisms and principles. Current opinion in neurobiology. 2017;42:128–135. doi: 10.1016/j.conb.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Doe CQ. Transient nuclear Prospero induces neural progenitor quiescence. Elife. 2014:3. doi: 10.7554/eLife.03363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen M, Agi E, Altschuler DJ, Wu LF, Altschuler SJ, Hiesinger PR. The Developmental Rules of Neural Superposition in Drosophila. Cell. 2015;162:120–133. doi: 10.1016/j.cell.2015.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong JC, Esch JJ, Poole B, Ganguli S, Clandinin TR. Direction Selectivity in Drosophila Emerges from Preferred-Direction Enhancement and Null-Direction Suppression. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36:8078–8092. doi: 10.1523/JNEUROSCI.1272-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen Z, Desplan C. Temporal patterning of neural progenitors in Drosophila. Current topics in developmental biology. 2013a;105:69–96. doi: 10.1016/B978-0-12-396968-2.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013b;498:456–462. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisak MS, Haag J, Ammer G, Serbe E, Meier M, Leonhardt A, Schilling T, Bahl A, Rubin GM, Nern A, et al. A directional tuning map of Drosophila elementary motion detectors. Nature. 2013;500:212–216. doi: 10.1038/nature12320. [DOI] [PubMed] [Google Scholar]
- Mauss AS, Meier M, Serbe E, Borst A. Optogenetic and pharmacologic dissection of feedforward inhibition in Drosophila motion vision. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:2254–2263. doi: 10.1523/JNEUROSCI.3938-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauss AS, Pankova K, Arenz A, Nern A, Rubin GM, Borst A. Neural Circuit to Integrate Opposing Motions in the Visual Field. Cell. 2015;162:351–362. doi: 10.1016/j.cell.2015.06.035. [DOI] [PubMed] [Google Scholar]
- Mora N, Oliva C, Fiers M, Ejsmont R, Soldano A, Zhang T-T, Yan J, Claeys A, De Geest N, CBA A Temporal transcriptional switch governs stem cell division, neuronal numbers and maintenance of differentiation. Developmental Cell. 2018 doi: 10.1016/j.devcel.2018.02.023. In Press. [DOI] [PubMed] [Google Scholar]
- Morante J, Desplan C. Building a projection map for photoreceptor neurons in the Drosophila optic lobes. Seminars in cell & developmental biology. 2004;15:137–143. doi: 10.1016/j.semcdb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Morante J, Desplan C. The color-vision circuit in the medulla of Drosophila. Current biology : CB. 2008;18:553–565. doi: 10.1016/j.cub.2008.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Early development of the Drosophila brain: III. The pattern of neuropile founder tracts during the larval period. The Journal of comparative neurology. 2003;455:417–434. doi: 10.1002/cne.10482. [DOI] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, Rubin GM. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E2967–2976. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo KT, Andrade I, Hartenstein V. Spatio-temporal pattern of neuronal differentiation in the Drosophila visual system: A user’s guide to the dynamic morphology of the developing optic lobe. Developmental biology. 2017;428:1–24. doi: 10.1016/j.ydbio.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo KT, Wang J, Junker M, Kriz S, Vo G, Asem B, Olson JM, Banerjee U, Hartenstein V. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Developmental biology. 2010;346:284–295. doi: 10.1016/j.ydbio.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva C, Choi CM, Nicolai LJ, Mora N, De Geest N, Hassan BA. Proper connectivity of Drosophila motion detector neurons requires Atonal function in progenitor cells. Neural development. 2014;9:4. doi: 10.1186/1749-8104-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BV, Rauskolb C, Irvine KD. Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development (Cambridge, England) 2010;137:2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Rister J, Pauls D, Schnell B, Ting CY, Lee CH, Sinakevitch I, Morante J, Strausfeld NJ, Ito K, Heisenberg M. Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron. 2007;56:155–170. doi: 10.1016/j.neuron.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Rossi AM, Fernandes VM, Desplan C. Timing temporal transitions during brain development. Current opinion in neurobiology. 2017;42:84–92. doi: 10.1016/j.conb.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Yasugi T, Minami Y, Miura T, Nagayama M. Notch-mediated lateral inhibition regulates proneural wave propagation when combined with EGF-mediated reaction diffusion. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E5153–5162. doi: 10.1073/pnas.1602739113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell B, Raghu SV, Nern A, Borst A. Columnar cells necessary for motion responses of wide-field visual interneurons in Drosophila. Journal of comparative physiology A, Neuroethology, sensory, neural, and behavioral physiology. 2012;198:389–395. doi: 10.1007/s00359-012-0716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F. Asymmetric cell division in the Drosophila bristle lineage: from the polarization of sensory organ precursor cells to Notch-mediated binary fate decision. Wiley interdisciplinary reviews Developmental biology. 2015;4:299–309. doi: 10.1002/wdev.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- Shinomiya K, Karuppudurai T, Lin TY, Lu Z, Lee CH, Meinertzhagen IA. Candidate neural substrates for off-edge motion detection in Drosophila. Current biology : CB. 2014;24:1062–1070. doi: 10.1016/j.cub.2014.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silies M, Gohl DM, Fisher YE, Freifeld L, Clark DA, Clandinin TR. Modular use of peripheral input channels tunes motion-detecting circuitry. Neuron. 2013;79:111–127. doi: 10.1016/j.neuron.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall TD, Brand AH. Neural stem cell transcriptional networks highlight genes essential for nervous system development. The EMBO journal. 2009;28:3799–3807. doi: 10.1038/emboj.2009.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strother JA, Wu ST, Wong AM, Nern A, Rogers EM, Le JQ, Rubin GM, Reiser MB. The Emergence of Directional Selectivity in the Visual Motion Pathway of Drosophila. Neuron. 2017;94:168–182. e110. doi: 10.1016/j.neuron.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Lu Z, Meinertzhagen IA. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. The Journal of comparative neurology. 2008;509:493–513. doi: 10.1002/cne.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Nern A, Chklovskii DB, Scheffer LK, Rubin GM, Meinertzhagen IA. The comprehensive connectome of a neural substrate for ‘ON’ motion detection in Drosophila. Elife. 2017:6. doi: 10.7554/eLife.24394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development (Cambridge, England) 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvklo C, MacDonald R, Bivik C, Baumgardt M, Karlsson D, Thor S. Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression. Development (Cambridge, England) 2012;139:678–689. doi: 10.1242/dev.074500. [DOI] [PubMed] [Google Scholar]
- Wan Y, Otsuna H, Holman HA, Bagley B, Ito M, Lewis AK, Colasanto M, Kardon G, Ito K, Hansen C. FluoRender: joint freehand segmentation and visualization for many-channel fluorescence data analysis. BMC Bioinformatics. 2017;18:280. doi: 10.1186/s12859-017-1694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Chen CH, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nature neuroscience. 2009;12:947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharioudaki E, Bray SJ. Tools and methods for studying Notch signaling in Drosophila melanogaster. Methods. 2014;68:173–182. doi: 10.1016/j.ymeth.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
pIPC is compartmentalized into Brk and Dpp domains
The compartmentalization of the pIPC has been described previously (Aptiz, 2015). The Dpp chains correspond to the most posterior chains, two dorsal and two ventral, while all chains anterior to these correspond to Brk derived chains.
(A) dpp-pIPC-Gal4 labels the Dpp-pIPC neuroepithelium, but not the Brk-pIPC- neuroepithelium (B).
(C) brk-Gal4 labels the Brk-pIPC neuroepithelium, but not the Dpp-pIPC neuroepithelium (D).
(E,F) pIPC-Gal4 labels both the Dpp-pIPC (E) and Brk-pIPC (F) neuroepithelium migrating chains and dIPC progenitors.
(G,H) R17B05-Gal4 labels all the pIPC neuroepithelium, migrating chains and dIPC progenitors. In all panels, dashed lines delineate the pIPC.
A three-dimensional model of the developing Larval Optic Lobe
Three dimensional model built based on the expression of Ato>Red-Singer optic lobes of third instar larvae labeled with Dpn and E-cad. For labels see Figure 1D.
Time lapse of a L3 larval developing optic lobe
The pIPC is labeled with pIPC-Gal>UAS-nls-GFP to visualize the migration of pIPC derived migrating progenitors to the dIPC. pIPC-Gal4 is described in Sup. Figure 1.
Dichaete+ NBs divide asymmetrically in a Type I manner
Z-stack highlighting the division of a Dichaete+ NB in Telophase. The dividing NB highlighted with a dashed square in the top left corner of the image is depicted in the bottom of the image. Dichaete+ NBs divided asymmetrically to self renew (Arrow, Red, Bazooka+ cell) and produce a smaller that acquires a GMC fate (Arrowhead, Green).
Ato+ NBs division produces two GMCs.
Z-stack highlighting the division of a Ato+ NB in Telophase. The dividing NB highlighted with a dashed square in top left corner of the image is depicted in the bottom of the image. Ato+ NBs divided asymmetrically to produce two similarly sized cells that acquired a GMC fate (Arrowheads).
T4 and T5 neurons of the same subtype are sibling neurons and project to the same retinotopic position in their target neuropiles
(A–D) WT-MARCM clone of sibling T4 and T5 neurons.
(E–H) A Notch binary fate decision specifies T4 vs. T5 fate. Notch Mutant MARCM clones of a dividing Notch−/− GMC produced two sibling T4 retinotopic neurons of the same subtype.
(I) WT-MARCM clone of a horizontal system NB showed two pairs of sibling T4/T5 neurons of opposite motion direction sensitivity projecting to the same retinotopic position in the neuropiles. Arrow points to “layer a” projections and arrowhead to “layer b” projections.
(J) WT-MARCM clone of a vertical system NB showed two pairs of sibling T4/T5 neurons of opposite motion direction sensitivity projecting to the same retinotopic position in the neuropiles. Arrow points to “layer c” projections and arrowhead to “layer d” projections.
Ato+/Dac+ NBs produce Ato+/Dac+ GMCs, which divide to exclusively produce Dac+ T4 and T5 neurons. (A) Newborn Dac+ GMCs (open arrowheads) inherited atonal from the NB. As GMCs matured and moved towards the inside of the dIPC crescent they lost Atonal expression (arrows) such that dividing Dac+ GMCs did not express Atonal (closed arrowheads). (B) E(spl)mγ::GFP was only transiently expressed in newborn Ato+/Dac+ GMCs (open arrowheads). Dac+ dividing GMCs did not express E(spl)mγ::GFP (closed arrowheads) (N=15).
Dichaete+ and Ato+ NBs localize Miranda and Prospero during cell division
(A) Cross sections of a posterior view of the dIPC-NB population of a WT and dronc mutant optic lobe. (B) Quantification of the number of NBs in cross sections of WT and dronc Mutants. WT and dronc mutants showed equal number of dIPC-neuroblasts. Data are represented as scattered plot with mean ± SD. Two-tailed unpaired Student’s t test with Welch’s correction. P= 0,3579; t=0,9493; df=14,59.(C) dIPC Dichaete+ (D) NBs express Asense (Ase) (Green). In the second temporal window, dIPC-NBs expressed Dac (blue) and Ato (see also Figure 1I). (D,E) Overall levels of Miranda were reduced in Ato+ NBs. (E,F) Miranda (E) and Prospero (F) crescents were weaker in Ato+ (open arrowheads) than in Dichaete+ NBs (closed arrowheads). In (D) Fire-Lut represents Miranda signal intensity between the two temporal stages.
dIPC-NBs form Miranda and Prospero crescents
(A–D) Lateral views of developing optic lobes highlighting the formation of Miranda and Prospero crescents in Dichaete+ (A,B) and Ato+ NBs (C,D). The NBs highlighted in each panel with a dashed square are depicted in Figure 5.
Pros::GFP labels a Pros crescent in Ato+ NB
(A,B) Pros::GFP co-localizes with Prospero (A) and showed a higher signal to noise ratio than antibody staining, allowing the unambiguous identification of Prospero crescents in Ato+ NBs. Arrowheads identify crescents of Prospero. (B). Pros:GFP did not label the neuronal tracks identified with Asterisks in (A).
Neurons project along the antero-posterior axis depending on their time of birth.
Activation of the FLEXAMP cassette in the Dpp-pIPC domain at L2, early-L3 and Late L3 labeled neurons of the vertical system along the A-P axis (Arrowheads) depending on their time of birth.
3-D view of MCFO labeled T4/T5 neurons
Anterior 3-D view of MCFO labeled T4/T5 neurons. Proximal is to the bottom. Neurons projected in the neuropiles along the dorsal-ventral axis according to their time of birth.
Time-lapse movie of a L3 larva optic lobe highlighting T4 projections in the developing Medulla
Time-lapse movie highlighting T4 projections in the Medulla. Ato-Gal4 (described in Figure 1) was used to drive GFP in T4/T5 neurons. View is anterior and proximal is to the bottom. Neurons innervating columns along the same dorso-ventral plane projected at the same time in the neuropiles. Arrows indicate the development along the Medulla neuropile anterior-posterior axis. Dashed line follows the position of dendrites that were targeting the medulla at T0.