Significance
ATPases have diverse cellular roles, including extracting proteins from membranes to maintain organellar function. The PEX1–PEX6 heterohexameric ATPases are thought to retrotranslocate PEX5 from the peroxisome membrane, and PEX1–PEX6 dysfunction impairs peroxisome biogenesis in humans and plants. We implemented a pex6 suppressor screen in Arabidopsis and recovered a compensatory pex1 allele that rescues several pex6 defects. Preventing autophagy also improved pex6 peroxisome function, and combining the pex1 and autophagy lesions delivered synergistic benefits. Surprisingly, these different alterations ameliorated pex6 symptoms without notably restoring the sole known function of PEX6, suggesting that PEX1–PEX6 has unexplored functions. Because the pex6 mutations ameliorated by pex1 are analogous to those in human pex6 patients, this study informs research on peroxisome dysfunction in other eukaryotes.
Keywords: peroxisome, peroxin, pexophagy, AAA ATPase, oil bodies
Abstract
Peroxisomes are eukaryotic organelles critical for plant and human development because they house essential metabolic functions, such as fatty acid β-oxidation. The interacting ATPases PEX1 and PEX6 contribute to peroxisome function by recycling PEX5, a cytosolic receptor needed to import proteins targeted to the peroxisomal matrix. Arabidopsis pex6 mutants exhibit low PEX5 levels and defects in peroxisomal matrix protein import, oil body utilization, peroxisomal metabolism, and seedling growth. These defects are hypothesized to stem from impaired PEX5 retrotranslocation leading to PEX5 polyubiquitination and consequent degradation of PEX5 via the proteasome or of the entire organelle via autophagy. We recovered a pex1 missense mutation in a screen for second-site suppressors that restore growth to the pex6-1 mutant. Surprisingly, this pex1-1 mutation ameliorated the metabolic and physiological defects of pex6-1 without restoring PEX5 levels. Similarly, preventing autophagy by introducing an atg7-null allele partially rescued pex6-1 physiological defects without restoring PEX5 levels. atg7 synergistically improved matrix protein import in pex1-1 pex6-1, implying that pex1-1 improves peroxisome function in pex6-1 without impeding autophagy of peroxisomes (i.e., pexophagy). pex1-1 differentially improved peroxisome function in various pex6 alleles but worsened the physiological and molecular defects of a pex26 mutant, which is defective in the tether anchoring the PEX1–PEX6 hexamer to the peroxisome. Our results support the hypothesis that, beyond PEX5 recycling, PEX1 and PEX6 have additional functions in peroxisome homeostasis and perhaps in oil body utilization.
Plants and animals can store fixed carbon as triacylglycerol (TAG), which can then be mobilized when energy is required. Arabidopsis seedling germination and early growth are fueled by breakdown of TAG stored in oil bodies via fatty acid β-oxidation in peroxisomes, single membrane-bound organelles. During germination, fatty acids hydrolyzed from TAG are activated with CoA before import through the peroxisomal ABC transporter PXA1 (1). In the peroxisome, fatty acids undergo β-oxidation to acetyl-CoA, which ultimately can be converted to sucrose. If germinating seedlings inefficiently catabolize fatty acids, growth is impeded. However, growth can be restored by providing sucrose in the medium (reviewed in ref. 2). In addition to fatty acid β-oxidation, plant peroxisomes host various other oxidative reactions (reviewed in refs. 3 and 4), including conversion of the protoauxin indole-3-butyric acid (IBA) to the active auxin indole-3-acetic acid (IAA; reviewed in ref. 5). The IAA generated following IBA treatment inhibits root elongation and promotes lateral root formation in WT, and mutants with dysfunctional β-oxidation enzymes or impaired import of these enzymes into the organelle often display decreased IBA responsiveness (reviewed in ref. 2).
Peroxisomal enzymes are posttranslationally imported into the organelle. Peroxisomal matrix proteins are recognized in the cytosol by the receptors PEX5 and PEX7, which bind proteins harboring one of two peroxisomal targeting signals (PTSs). PEX5 recognizes a C-terminal PTS1 (6), and PEX7 recognizes a PTS2 near the N terminus (7). These receptor–cargo complexes dock with peroxisomal membrane proteins, PEX13 and PEX14 (reviewed in ref. 8), and PEX14 is thought to aid PEX5 in forming a dynamic pore through which cargo is translocated to the matrix (9). Following cargo release, yeast PEX5 in the peroxisomal membrane (10) is ubiquitinated by PEX4 (11, 12) and the RING complex peroxins (PEX2, PEX10, and PEX12) (13) and then retrotranslocated to the cytosol with the assistance of the PEX1–PEX6 ATPase complex (14), which is tethered to the peroxisome by PEX15 in yeast (15) and PEX26 in mammals (16) and plants (17). When PEX5 is inefficiently retrotranslocated (as in pex1 or pex6 mutants), PEX5 is polyubiquitinated, which can trigger degradation of PEX5 by the proteasome (18, 19) or degradation of the entire organelle via specialized autophagy (i.e., pexophagy) (20).
PEX1 and PEX6 are type II AAA proteins (ATPases Associated with various cellular Activities) with two AAA domains. These domains generally include Walker A and Walker B motifs, but the PEX6 AAA1 domain lacks a canonical Walker B motif and is unable to hydrolyze ATP (reviewed in ref. 21). Saccharomyces cerevisiae PEX1 and PEX6 form a heterohexamer (22) of alternating subunits (23–25). Biochemical experiments (26) indicate that PEX1–PEX6 can thread client proteins through a central pore revealed in structural studies (23–25, 27). Intriguingly, PEX15 is not only a tether (15), but PEX15 lacking its C-terminal transmembrane domain is an in vitro client of the ATPase (26). Human PEX1 also forms a PEX5-interacting homotrimer in the cytosol (28) that may modulate cytosolic PEX5 homooligomerization (29). Moreover, overexpressing PEX6 in yeast suppresses the import defects of a mitochondrial ATPase missense mutation (30), suggesting that PEX6 may also function beyond peroxisomes.
In humans, the generally fatal peroxisomal biogenesis disorders (PBDs) are caused by mutations in various PEX genes, most often PEX1 and PEX6 (reviewed in ref. 31). Arabidopsis PEX1 and PEX6 are 28% and 33% identical at the amino acid level with human PEX1 and PEX6, respectively, and appear to function similarly to their human counterparts (32–34). For example, expressing a human PEX6 cDNA rescues an Arabidopsis pex6 mutant (35), and like human PBD patients with dysfunctional PEX6 (36), several Arabidopsis pex6 mutants have low PEX5 levels (35, 37). Moreover, three of the six reported Arabidopsis pex1 (38) or pex6 (35, 37) missense alleles are equivalent to causal mutations in patients with PBD (www.dbpex.org/home.php) (39–41).
The peroxisomal ATPase complex is also vital for plants. Arabidopsis PEX1- (42) and PEX26- (17, 43) null alleles confer embryo lethality, and the pex1-3 missense allele is lethal when homozygous (38). pex1-3 and pex1-2 alter conserved AAA2 residues, and PEX1/pex1-3 and pex1-2 plants display β-oxidation and matrix protein import defects (38). Moreover, pex1-2 has decreased PEX1 and PEX6 levels, suggesting that this allele confers instability to the complex. The pex6-1, pex6-3, and pex6-4 missense mutations also lie in or near the AAA2 domain (Fig. 1B and Fig. S1) and result in small pale green plants with β-oxidation defects and low PEX5 levels (35, 37). In contrast, the mild pex6-2 allele lies N-terminal to the AAA1 domain (Fig. 1B), lacks notable β-oxidation defects, complements pex6-1 (44), and displays elevated PEX5 levels (37). The two viable pex26 mutants, pex26-1 (37) and aberrant peroxisome morphology9 (apem9) (17), display distinct phenotypes as well. apem9-1 is a mild missense allele that lacks notable β-oxidation defects and has matrix protein import defects only in a subset of tissues (17), whereas the pex26-1 splice-site mutation confers seedling β-oxidation defects and clustered peroxisomes (37). Lipid droplets accumulate in PEX5−/− mouse hepatocytes (45), and oil bodies persist during pex6-1, pex6-3, pex6-4, and pex26-1 seedling development after oil bodies have been consumed in WT (37), indicating that the peroxisome retrotranslocation machinery directly or indirectly promotes oil body utilization. Finally, increasing PEX5 levels in pex6-1, pex6-3, pex6-4, and pex26-1 has disparate effects. Excess PEX5 restores growth and oil body utilization in pex6-1 and pex6-3 but not in pex6-4 or pex26-1 (37), hinting that PEX5 recycling is not the sole function of the peroxisomal ATPase.
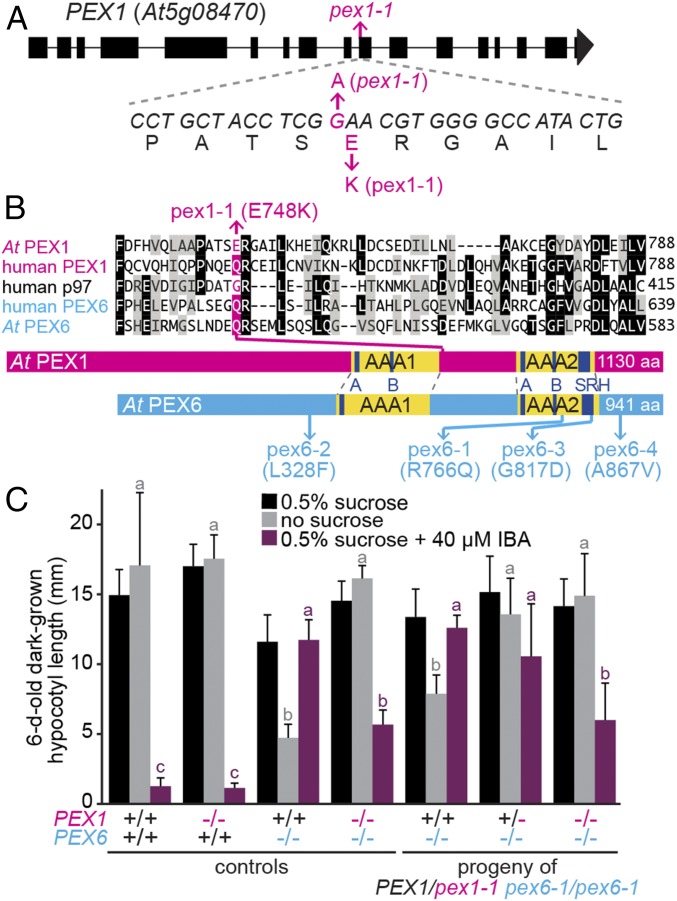
Fig. 1.
pex1-1 identification. (A) PEX1 gene diagram showing exons (rectangles), introns (lines), and the location of pex1-1, a G-to-A transition that yields a Glu748-to-Lys substitution. (B) Partial alignment of Arabidopsis and human PEX1 and PEX6 and human p97 (a related ATPase) above protein schematic illustrations depicting the locations of pex1-1 and previously described pex6 missense alleles (35, 37, 44). The complete AAA domains are in yellow, with the Walker A (A), Walker B (B), and second region of homology (SRH) domains highlighted in navy blue. (C) pex1-1 is semidominant; the PEX1/pex1-1 heterozygote partially suppresses pex6-1 sucrose dependence and IBA resistance. Seedlings were grown on the indicated media for 1 d under yellow-filtered light before moving to darkness for 5 d. Bars indicate mean hypocotyl lengths, and error bars indicate SD (n ≥ 13 except for pex6-1 PEX1 and pex6-1 pex1-1 from the segregating parent, for which n ≥ 5). Means not sharing a letter above the bar are significantly different as determined by one-way ANOVA (P < 0.001).
To learn more about peroxisomal ATPase complex functions, we employed a forward-genetic suppressor screen of Arabidopsis pex6-1 and recovered a missense mutation in PEX1 (pex1-1) that restored pex6-1 growth without an exogenous fixed carbon source. Moreover, we found that preventing autophagy also partially ameliorated pex6-1 defects. Rescue of pex6-1 by pex1-1 was more effective than preventing autophagy or reducing PEX13 expression, which also improves pex6-1 peroxisomal function (46). To explore the specificity of pex6 restoration, we characterized a suite of pex1 double mutants, and found that pex1-1 improved several physiological and molecular defects in a subset of pex6 mutants while worsening a pex26 mutant. Surprisingly, pex1-1 amelioration of pex6 physiological defects was not accompanied by restored PEX5 levels, supporting the hypothesis that PEX1 and PEX6 have roles beyond PEX5 recycling.
Results
The pex1-1 Missense Mutation Improves pex6-1 Growth.
We sought to elucidate functions of the peroxisomal ATPase complex by identifying genes that genetically interact with PEX6. Given previous demonstrations that elevating PEX5 or decreasing PEX13 expression ameliorates some physiological defects of the pex6-1 missense allele (35, 46), we screened for pex6-1 suppressors. Like other Arabidopsis pex mutants (47–53), pex6-1 displays impaired growth on medium lacking sucrose (35), providing a facile screen for suppressors with increased growth. We mutagenized pex6-1 seeds with ethyl methanesulfonate (EMS) and selected dark-grown M2 seedlings with longer hypocotyls than pex6-1 on medium lacking sucrose. One suppressor that restored pex6-1 growth and partially restored IBA responsiveness was retained for further analysis.
After backcrossing to the pex6-1 parent, we sequenced genomic DNA from the suppressor (Fig. S2) and found a mutation in PEX1 (Fig. 1A). This G4127-to-A transition in exon 10 causes a glutamic acid to lysine (E748K) change (Fig. 1A) in the region between the two AAA domains (Fig. 1B). We named this allele pex1-1. Unlike pex6-1, the pex1-1 single mutant grew similarly to WT on IBA or with and without sucrose (Fig. 1C), suggesting that pex1-1 did not markedly affect peroxisome function when PEX6 was WT. Moreover, PEX1 and PEX6 levels were similar to WT levels in the single and double mutants (Fig. 2C), indicating that pex1-1 did not notably affect stability of the ATPase hexamer components. Suppression was linked to PEX1; growth of suppressor backcross progeny on medium containing IBA or lacking sucrose correlated with pex1-1 inheritance (Fig. 1C). Moreover, pex1-1+/− pex6-1−/− seedlings displayed intermediate suppression between homozygous PEX1+/+ pex6-1−/− and pex1-1−/− pex6-1−/− phenotypes (Fig. 1C), indicating semidominance. To determine whether pex1-1 was the causal lesion suppressing pex6-1, we expressed HA-PEX1 from the constitutive 35S cauliflower mosaic virus promoter in pex1-1 pex6-1. We found that dark-grown pex1-1 pex6-1 seedlings expressing HA-PEX1 displayed reduced hypocotyl elongation without sucrose and increased IBA resistance, similar to pex6-1 (Fig. 2A).
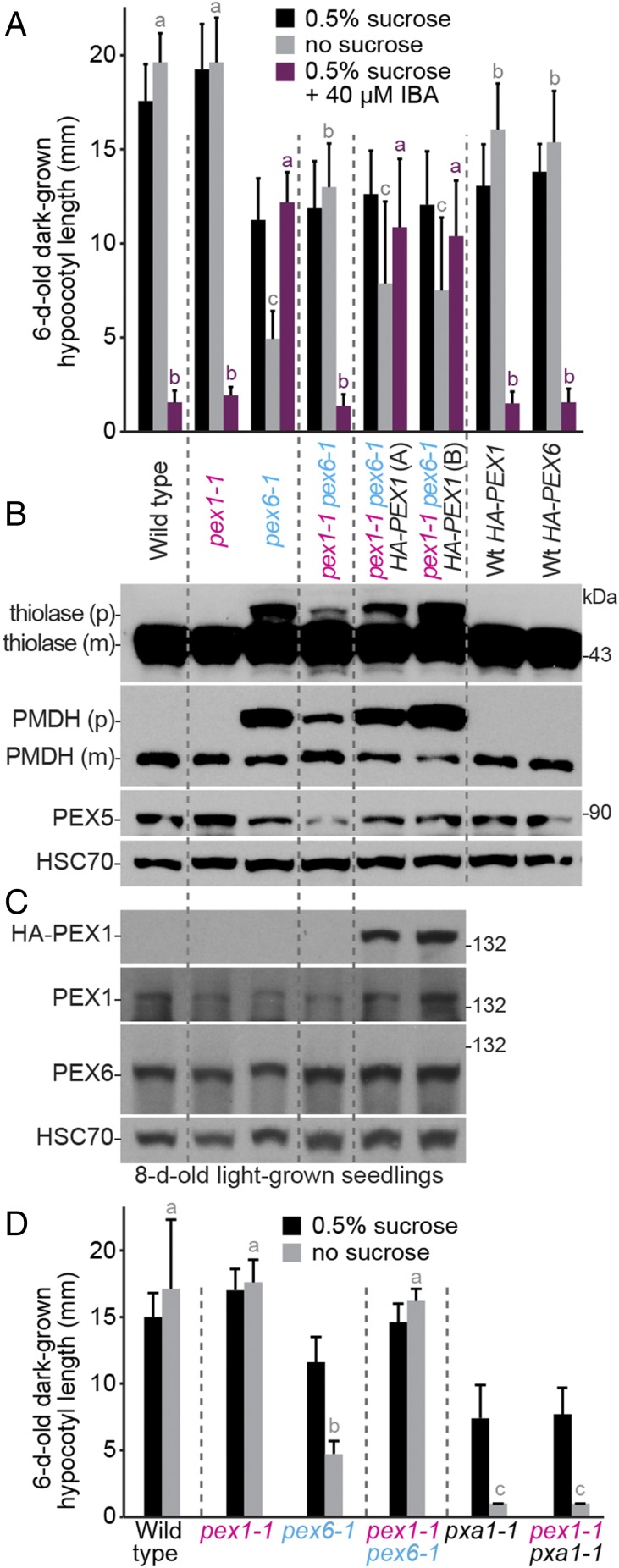
Fig. 2.
Overexpressing PEX1 in pex1-1 pex6-1 phenocopies pex6-1 defects; pex1-1 does not restore pxa1-1 growth. (A) Expressing HA-PEX1 increases resistance to the inhibitory effects of IBA and hypocotyl elongation dependence on exogenous sucrose of dark-grown pex1-1 pex6-1 seedlings. Two independent pex1-1 pex6-1 35S:HA-PEX1 lines (A and B) are shown. Seedlings were grown as in the legend to Fig. 1C. Bars indicate mean hypocotyl lengths (n ≥ 13), and error bars indicate SD. (B) PTS2-processing defects of light-grown pex1-1 pex6-1 seedlings are worsened by HA-PEX1 expression. An immunoblot of 8-d-old light-grown seedling extracts was serially probed with antibodies to the indicated proteins. For thiolase and PMDH, precursor (p) and mature (m) proteins are indicated. (C) PEX1 and PEX6 levels resemble WT levels in pex1-1. An immunoblot of 8-d-old light-grown seedling extracts was serially probed with antibodies to the indicated proteins. For B and C, the positions of molecular mass markers (in kilodaltons) are indicated on the right and HSC70 is a loading control. (D) pex1-1 does not improve growth of pxa1-1 seedlings in the absence of sucrose. Seedlings were grown as in the legend to Fig. 1C. Bars indicate mean hypocotyl lengths (n ≥ 13), and error bars indicate SD. In A and D, means not sharing a letter above the bar are significantly different as determined by one-way ANOVA (P < 0.001).
Physiological defects of pex mutants are often ascribed to matrix protein import defects (48, 50–53), and the proteolytic processing of PTS2 proteins that occurs after import (54) can be used as an import proxy. We found that the pex1-1 mutant fully processed PTS2 proteins and partially restored PTS2 processing of thiolase and PMDH in pex6-1 (Fig. 2B). Moreover, expressing HA-PEX1 decreased thiolase and PMDH processing in pex1-1 pex6-1 (Fig. 2B). Because expressing HA-PEX1 in pex1-1 pex6-1 counteracted the beneficial effects of pex1-1 (Fig. 2 A and B), we concluded that the identified pex1-1 mutation was the causal pex6-1 suppressor.
pex1-1 Effects Are Specific to the PEX1–PEX6–PEX26 Complex.
To determine if the pex1-1 lesion improved peroxisome function in general, we crossed pex1-1 to pxa1-1, which is impaired in the peroxisomal transporter that moves β-oxidation substrates into the peroxisome (55). pex1-1 did not significantly restore pxa1-1 growth on medium without sucrose (Fig. 2D), indicating that pex1-1 did not improve pex6-1 seedling growth by bypassing the requirement for peroxisomal fatty acid β-oxidation.
To examine the specificity of the pex1-1 suppression among pex6 alleles, we compared peroxisome function of pex1-1 pex6-1 to double mutants of pex1-1 with pex6-2 (44), pex6-3 (37), and pex6-4 (37). Intriguingly, pex1-1 did not similarly suppress all pex6 mutants. In addition to improving growth without sucrose, pex1-1 increased sensitivity of pex6-1 dark-grown hypocotyls to IBA (Fig. 3A) and slightly increased pex6-1 hypocotyl sensitivity to the IBA analog 2,4-dichlorophenoxybutyric acid (2,4-DB) (Fig. 3B). Similarly, pex1-1 rescued pex6-3 physiological defects, largely restoring IBA and 2,4-DB responsiveness and the ability to grow without sucrose in the dark (Fig. 3 A and B). Although pex1-1 did not significantly improve IBA-induced lateral rooting in pex6-1, pex1-1 largely restored the ability of pex6-3 to respond to IBA in this assay (Fig. 3C). Aside from a partial restoration of growth without sucrose in pex1-1 pex6-4 (Fig. 3A), pex1-1 did not significantly alter hypocotyl IBA (Fig. 3A) or 2,4-DB (Fig. 3B) responsiveness or IBA-responsive lateral rooting (Fig. 3C) in the weak pex6-2 allele or the strong pex6-4 allele.
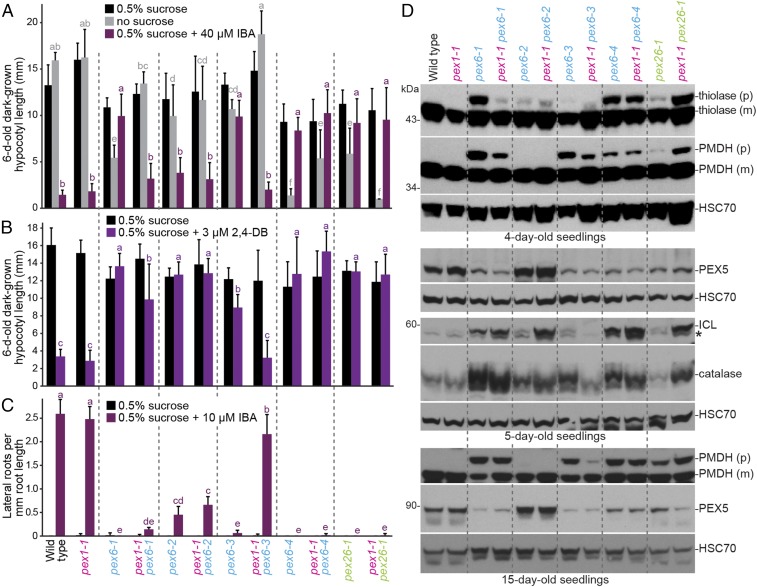
Fig. 3.
pex1-1 ameliorates pex6-1 and pex6-3 and worsens pex26-1 defects without restoring PEX5 levels. (A) pex1-1 increases the sensitivity of dark-grown pex6-1 and pex6-3 seedlings to the inhibitory effects of IBA and reduces dependence on exogenous sucrose for hypocotyl elongation but increases pex26-1 sucrose dependence. Seedlings were grown as in the legend to Fig. 1C. Bars indicate mean hypocotyl lengths (n ≥ 13), and error bars indicate SD. (B) pex1-1 increases sensitivity of dark-grown pex6-1 and pex6-3 seedlings to the inhibitory effects of 2,4-DB on hypocotyl elongation. Seedlings were grown as in the legend to Fig. 1C. Bars indicate mean hypocotyl lengths (n ≥ 12), and error bars indicate SD. (C) pex1-1 increases sensitivity of light-grown pex6-3 seedlings to the stimulatory effects of IBA on lateral root production. Seedlings were grown on control medium for 4 d followed by 4 d on medium with or without 10 μM IBA under constant yellow-filtered light. Bars indicate mean lateral root densities (n ≥ 8), and error bars indicate SD. In A–C, means not sharing a letter above the bar are significantly different as determined by one-way ANOVA (P < 0.001). (D) pex1-1 differentially impacts PTS2 processing and peroxisomal protein levels in pex6 and pex26-1 seedlings. Immunoblots of 4- (Top), 5- (Middle), and 15-d-old (Bottom) light-grown seedling extracts were serially probed with antibodies to the indicated proteins. The positions of molecular mass markers (in kilodaltons) are indicated on the left. For thiolase and PMDH, precursor (p) and mature (m) proteins are indicated. HSC70 is a loading control. An asterisk indicates a cross-reacting band in the ICL panels.
We also examined the impact of combining pex1-1 with pex26-1 (37), which is defective in the tether that recruits the PEX1–PEX6 hexamer to the peroxisome (17). In contrast to the restorative or neutral effects on pex6 mutants, we found detrimental effects when combining pex1-1 with pex26-1 (Fig. 3). The pex1-1 pex26-1 double mutant scarcely grew in the absence of sucrose (Fig. 3A) and remained fully resistant to IBA (Fig. 3 A and C) and 2,4-DB (Fig. 3B), demonstrating that the pex1-1 mutation was not beneficial to all peroxisomal ATPase complex mutants.
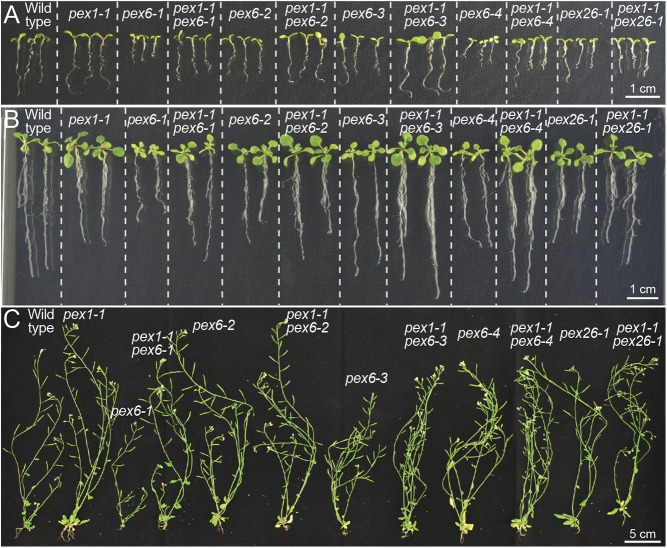
Monitoring overall growth of the suite of single and double mutants revealed that pex1-1 resembled WT throughout development (Fig. 4). As previously documented (37), pex6-2 displayed minimal growth defects, whereas pex6-1, pex6-3, pex6-4, and pex26-1 seedlings were small and pale (Fig. 4 A and B). We found that pex1-1 generally increased root growth (Fig. 4 A and B) and rosette size (Fig. 4B) of pex6 seedlings. Unlike the worsened seedling growth without sucrose (Fig. 3A), pex1-1 did not notably alter pex26-1 growth when sucrose-supplemented (Fig. 4 A and B) or following transfer to soil (Fig. 4C). Whereas mature pex6-2, pex6-4, and pex26-1 plants resembled WT with or without pex1-1, the pex1-1mutation appeared to alleviate the smaller adult plant size of pex6-1 and pex6-3 mutants (Fig. 4C).
Fig. 4.
pex1-1 lessens pex6 growth defects. (A and B) pex1-1 resembles WT and restores pex6 mutant defects in size and pigmentation. Seedlings at 7 d old (A) and 14 d old (B) were grown on sucrose-containing medium before photography (Scale bar: 1 cm.) (C) pex1-1 resembles WT and rescues pex6-1 and pex6-3 defects in adult size. Plants were transferred to soil after 14 d on sucrose-containing medium and photographed after an additional 4 wk in light. (Scale bar: 5 cm.)
To examine the molecular consequences of the pex1-1 lesion, we compared PTS2 processing in seedlings. As with pex6-1, pex1-1 slightly improved PTS2 processing of PMDH in pex6-3 (Fig. 3D). In contrast, pex1-1 did not notably alter PTS2 processing in pex6-2 or pex6-4 (Fig. 3D), and PTS2 processing of thiolase and PMDH in pex26-1 was worsened by pex1-1 (Fig. 3D). Thus, the effects of pex1-1 on PTS2 processing mirrored the suppression seen in β-oxidation phenotypes.
We also monitored levels of several peroxisomal matrix proteins in the double mutants. Catalase levels are elevated in certain peroxisome-defective mutants (56), perhaps because catalase undergoes less oxidative damage and degradation when β-oxidation is impaired. Consistent with the changes in β-oxidation phenotypes (Fig. 3 A–C), pex1-1 lessened catalase accumulation in the pex6-3 mutant and further elevated catalase levels in pex26-1 (Fig. 3D). pex1-1 did not dramatically alter catalase levels in pex6-1, pex6-2, or pex6-4 (Fig. 3D), adding to the evidence that pex1-1 does not uniformly restore all pex6 mutants’ defects.
The glyoxylate cycle enzyme isocitrate lyase (ICL) is degraded during early seedling development (57) as the glyoxylate cycle becomes obsolete, and pex6 mutants degrade ICL more slowly than WT (44, 57). We found that changes in ICL levels in 5-d-old seedlings mirrored catalase changes; pex1-1 decreased ICL levels in pex6-3 and further increased ICL levels in pex26-1 but did not decrease ICL levels in pex6-1, pex6-2, or pex6-4 (Fig. 3D).
pex1-1 Improves Oil Body Utilization in pex6-1 and pex6-3.
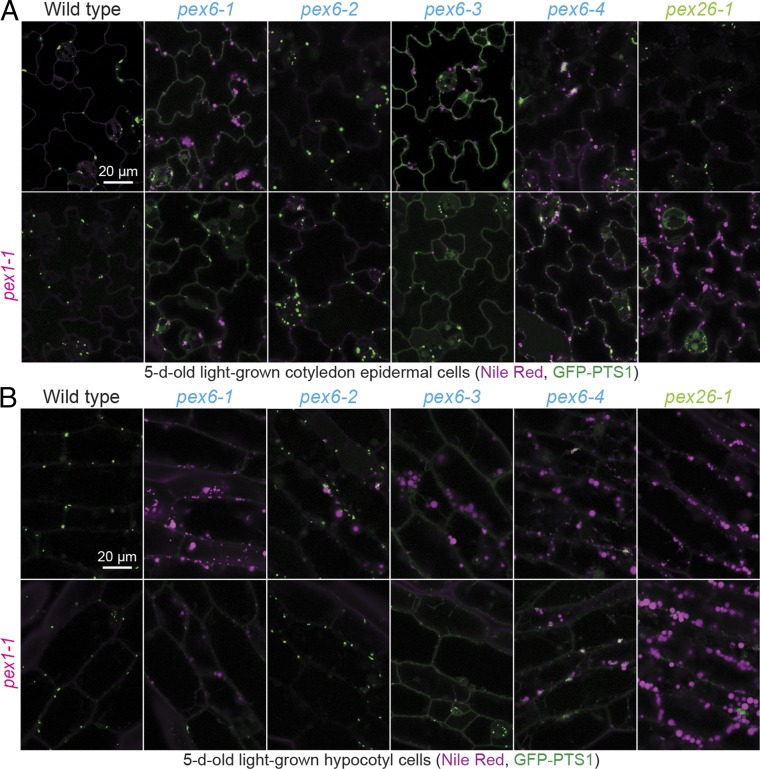
Because pex6 and pex26 mutants inefficiently utilize cotyledon oil bodies (37), we examined oil bodies by staining neutral lipids with Nile red. Confocal microscopy revealed persisting oil bodies in 5-d-old pex6-1, pex6-3, pex6-4, and pex26-1 pavement and hypocotyl cells that were not observed in WT or pex1-1 (Fig. 5). pex1-1 alleviated oil body persistence in pex6-1 and pex6-3 and partially reduced oil body persistence in pex6-4, but failed to alleviate oil body persistence in pex26-1 (Fig. 5). The improvement of oil body utilization conferred by pex1-1 mirrored and presumably explains the improved growth of these lines without supplemental sucrose (Fig. 3A).
Fig. 5.
pex1-1 slightly improves pex6-1 and pex6-3 GFP-PTS1 import and improves oil body utilization in pex6 mutants but does not improve oil body utilization in pex26-1. Confocal images of cotyledon epidermal cells (A) and hypocotyl cells (B) of 5-d-old seedlings carrying 35S:GFP-PTS1 (green) stained with Nile red (magenta). Emissions were collected at 490–519 nm for GFP and 587–643 nm for Nile red. Seedlings were screened for germination 2 d after plating, and individuals that had germinated by day 2 were imaged on day 5. (Scale bar: 20 μm.)
pex1-1 Slightly Improves GFP-PTS1 Import in pex6-1 and pex6-3.
The minor improvement of PTS2 processing observed when pex1-1 was combined with pex6-1 or pex6-3 (Fig. 3D) suggested that pex1-1 might be improving matrix protein import. To directly assess import, we examined the localization of GFP-PTS1 in the pex1-1 double mutants by using confocal microscopy. WT, pex1-1, pex6-2, and pex1-1 pex6-2 displayed similar GFP-PTS1 puncta, and GFP-PTS1 remained largely cytosolic in the pex1-1 pex6-4 double mutant (Fig. 5). In contrast, the predominantly cytosolic GFP-PTS1 in pex6-1 and pex6-3 mutants was slightly improved by pex1-1, as evidenced by the appearance of some GFP-PTS1 puncta in the pex1-1 pex6-1 and pex1-1 pex6-3 double mutants (Fig. 5).
pex1-1 Does Not Restore PEX5 Levels in pex6 Mutants.
Most Arabidopsis pex6 mutants (35, 37) and human pex6 patients (36) have low PEX5 levels, presumably because PEX5 is polyubiquitinated and degraded in the absence of efficient recycling. In contrast, immunoblotting revealed that PEX5 levels were not decreased in pex1-1 (Figs. 2B and 3D). Surprisingly, PEX5 levels remained low in seedlings when pex1-1 was combined with pex6-1, pex6-3, pex6-4, or pex26-1 (Fig. 3D). Moreover, PEX5 levels remained elevated when pex1-1 was combined with pex6-2 (Fig. 3D). As expected, PEX5 levels remained low in pex1-1 pex6-1 35S:HA-PEX1 (Fig. 2B).
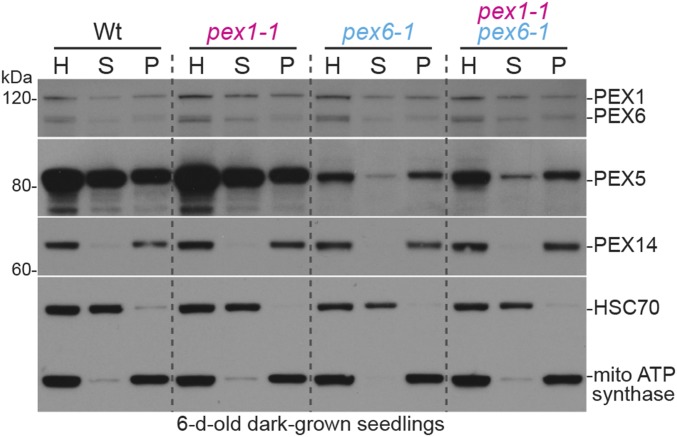
Because pex1-1 did not restore PEX5 levels in pex6 mutants, we interrogated PEX5 localization by using centrifugation to separate cytosolic and membrane-associated proteins. PEX5 is excessively membrane associated in pex6-1 (46) and pex6-3 (37). We found that PEX5 distribution between the supernatant and pellet in pex1-1 resembled WT and that pex1-1 did not dramatically alter the excessive association of PEX5 with the membrane fraction in pex6-1 (Fig. 6) or pex6-3 (Fig. S3).
Fig. 6.
PEX5 remains excessively membrane-associated in pex1-1 pex6-1. Homogenates (H) prepared from 6-d-old dark-grown WT, pex1-1, pex6-1, and pex1-1 pex6-1 seedlings were separated by centrifugation to isolate cytosolic supernatant (S) and an organellar pellet, which was resuspended and recentrifuged to provide a final organellar pellet (P) fraction. Fractions were subjected to immunoblotting with the indicated antibodies. HSC70 is cytosolic, and mitochondrial (mito) ATP synthase subunit α and PEX14 localize in the organelle fraction. The positions of molecular mass markers (in kilodaltons) are indicated on the left.
We also examined PEX1 and PEX6 localization in these fractionation experiments. Like PEX5, PEX1 and PEX6 were approximately evenly distributed between the supernatant and pellet fractions in WT (Fig. 6 and Fig. S3). This distribution did not markedly change in the pex1-1 or pex6 single or double mutants (Fig. 6 and Fig. S3), suggesting that the pex6-1 and pex6-3 mutations do not impair peroxisome association of the pex6 protein and that altering pex6 localization is not the basis of pex1-1 suppression.
Overexpressing PEX5 in pex1-1 pex6-1 Further Rescues pex6-1 Deficiencies.
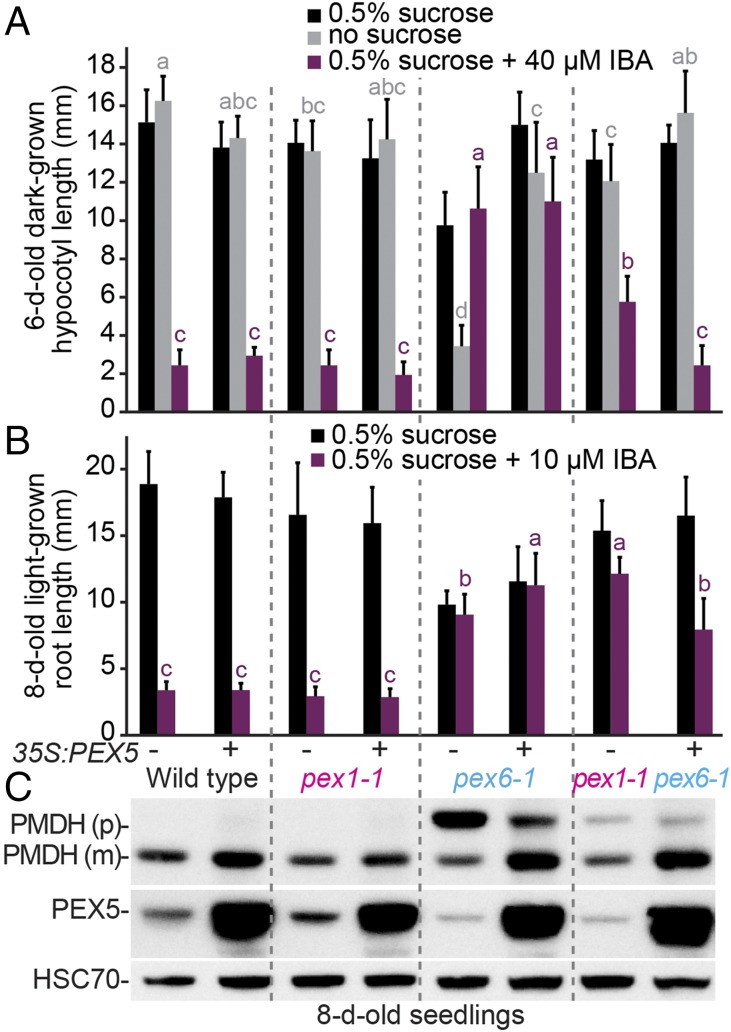
Because PEX5 levels remained low in pex1-1 pex6-1 (Fig. 3D), we tested whether overexpressing PEX5 could further restore peroxisome function. We found no discernible effect of excess PEX5 on pex1-1 or WT growth, IBA sensitivity, or PTS2 processing (Fig. 7). As previously reported (35, 44), excess PEX5 improved pex6-1 growth without sucrose (Fig. 7A) and partially restored PTS2 processing (Fig. 7C). We found that overexpressing PEX5 in pex1-1 pex6-1 further improved growth without sucrose (Fig. 7A) and IBA responsiveness (Fig. 7 A and B), although a slight defect in PTS2 processing remained (Fig. 7C). This additive pex6-1 suppression by pex1-1 and 35S:PEX5 suggests that pex1-1 pex6-1 remains limited by decreased PEX5 recycling.
Fig. 7.
pex1-1 and overexpressing PEX5 additively restore pex6-1 defects. (A and B) Overexpressing PEX5 improves pex1-1 pex6-1 dark-grown (A) and light-grown (B) physiology. Plant lines with (+) and without (−) 35S:PEX5 were grown on the indicated media for 1 d under yellow-filtered light before moving to darkness for 5 d (A) or were grown for 8 d under continuous yellow-filtered light (B). Bars indicate mean hypocotyl lengths (A; n ≥ 14) or root lengths (B; n ≥ 13), and error bars indicate SD. Means not sharing a letter above the bar are significantly different as determined by one-way ANOVA (P < 0.001). (C) Overexpressing PEX5 improves PTS2 processing in pex1-1 pex6-1 seedlings. An immunoblot of 8-d-old light-grown seedling extracts was serially probed with antibodies to the indicated proteins. Precursor (p) and mature (m) PMDH are indicated. HSC70 is a loading control.
Preventing Autophagy Also Restores Peroxisome Function in pex6-1.
To illuminate the mechanism of pex6-1 suppression, we compared the suppression effects of pex1-1 vs. a previously characterized pex6 suppressor, pex13-1, a partial loss-of-function allele that may ameliorate pex6-1 defects by decreasing PEX5 docking at the peroxisome (46). We found that pex1-1 was more effective than pex13-1 in restoring growth without sucrose, IBA responsiveness, and PTS2 processing to pex6-1 (Fig. 8 A and B). The pex1-1 pex6-1 pex13-1 triple mutant resembled the pex1-1 pex6-1 double mutant in these assays (Fig. 8 A and B), indicating that decreasing PEX13 levels did not provide additional benefit to pex1-1 pex6-1.
Fig. 8.
pex1-1 augments benefits provided to pex6-1 by preventing autophagy without increasing PEX5 levels. (A) pex1-1, atg7-3, and pex13-1 improve pex6-1 IBA responsiveness and growth without sucrose, and pex1-1 further increases atg7-3 benefits. Seedlings were grown as in the legend to Fig. 1C. Bars indicate mean hypocotyl lengths (n = 16), and error bars indicate SD. Means not sharing a letter above the bar are significantly different as determined by one-way ANOVA (P < 0.001). (B) Preventing autophagy increases PTS2 processing in pex6-1, and this increase is further improved by pex1-1. An immunoblot of 8-d-old light-grown seedling extracts was serially probed with antibodies to the indicated proteins. Precursor (p) and mature (m) PMDH are indicated. HSC70 is a loading control. (C) Preventing autophagy and pex1-1 synergistically improve GFP-PTS1 import in pex6-1. Confocal images of cotyledon epidermal and hypocotyl cells were collected after staining 5-d-old seedlings carrying 35S:GFP-PTS1 (green) with Nile red (magenta). Emissions were collected at 490–526 nm for GFP and 586–643 nm for Nile red.
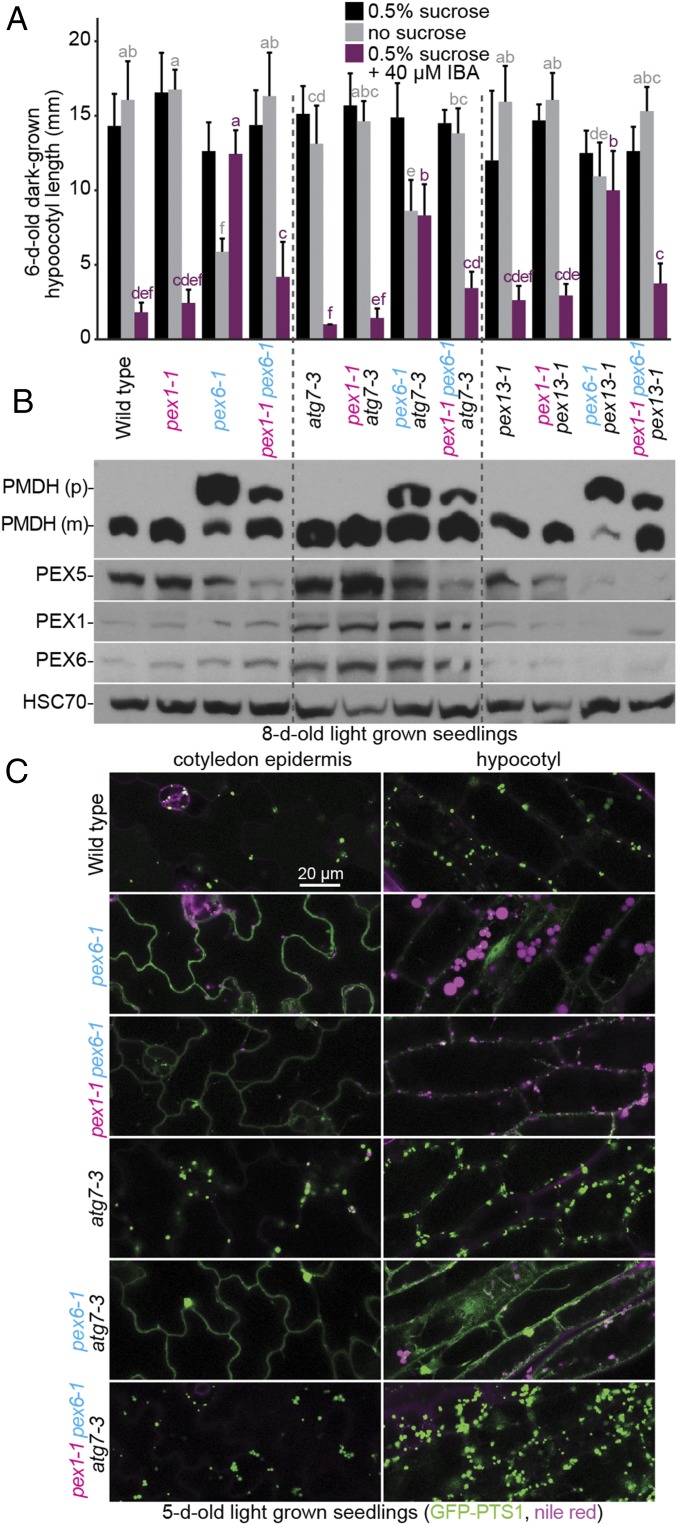
Autophagy is a process by which eukaryotes degrade organelles (reviewed in ref. 58), including peroxisomes (reviewed in ref. 59). Because preventing autophagy ameliorates some pex1-3 defects (38), we examined the contributions of autophagy to pex6-1 phenotypes by crossing to the atg7-3–null allele (60, 61). atg7-3 improved pex6-1 growth without sucrose, IBA responsiveness, and PTS2 processing, although not as markedly as pex1-1 (Fig. 8 A and B). Introducing pex1-1 to pex6-1 atg7-3 further improved PTS2 processing (Fig. 8B). PEX5 levels remained low in the pex1-1 pex6-1 atg7-3 and pex1-1 pex6-1 pex13-1 triple mutants (Fig. 8B), indicating that the physiological rescue (Fig. 8A) was accomplished without restoring PEX5 levels. We also examined PEX1 and PEX6 levels in these mutants and found that both peroxins were elevated in all genotypes that included the atg7-3 mutation, suggesting that PEX1 and PEX6 levels can be modulated by autophagy.
Because atg7-3 improved PTS2 processing in pex6-1, we also monitored GFP-PTS1 localization and oil body persistence in these mutants. Like WT, 5-d-old atg7-3 seedlings lacked oil bodies and displayed GFP-PTS1 puncta (Fig. 8C). Consistent with the restoration of growth without sucrose (Fig. 8A), atg7-3 restored oil body utilization in pex6-1 (Fig. 8C). However, GFP-PTS1 remained largely cytosolic in pex6-1 atg7-3 (Fig. 8C). In contrast, GFP-PTS1 appeared largely punctate in the pex1-1 pex6-1 atg7-3 triple mutant (Fig. 8C), indicating that pex1-1 and atg7-3 synergistically improved pex6-1 matrix protein import without increasing PTS1 receptor levels.
Discussion
The peroxins in the ATPase complex (PEX1, PEX6, and PEX26) are implicated in removing PEX5 from the peroxisomal membrane after cargo delivery (14), allowing recycled PEX5 to promote another round of matrix protein import. Analyses of Arabidopsis pex1 (38), pex6 (35, 37, 44), and pex26 (17, 37) loss-of-function mutants have begun to elucidate the roles of the ATPase complex peroxins in PEX5 management (reviewed in ref. 3). For example, four Arabidopsis pex6 missense mutations confer disparate phenotypes that suggest impairment of different aspects of PEX6 function (35, 37, 44). In an effort to expand our understanding of PEX6, we conducted a screen for pex6-1 suppressors and characterized the pex1-1 missense allele that emerged from this screen (Fig. 1).
Because PEX1 and PEX6 are present in a 1:1 ratio in a heterohexamer, the pex1-1 semidominant suppression (Fig. 1C) likely reflects the multiple possible combinations of pex1-1 and PEX1 with pex6-1. For example, only 12.5% of heterohexamers would be expected to carry three WT PEX1 subunits in a PEX1/pex1-1 heterozygote. Indeed, the Arabidopsis pex1-3 missense allele confers peroxisome-related defects when heterozygous and lethality when homozygous (38), highlighting the sensitivity of this hexamer to perturbation even when some WT PEX1 remains.
pex1-1 Ameliorates Defects in a Subset of pex6 Alleles Without Restoring PEX5 Levels.
pex1-1 did not similarly rescue all pex6 mutants (Table S1). pex1-1 had little or negative effects on pex6-2 physiological and molecular defects (Fig. 3). pex1-1 slightly improved pex6-4 oil body utilization (Fig. 5) and growth in the absence of sucrose (Fig. 3A) without notably improving IBA responsiveness or PTS2 processing (Fig. 3 A, C, and D). In contrast, pex1-1 improved pex6-1 and pex6-3 IBA responsiveness (Fig. 3 A and C), growth on medium lacking sucrose (Fig. 3A), PTS2 processing (Fig. 3D), GFP-PTS1 import (Fig. 5), and oil body utilization (Fig. 5). Interestingly, pex1-1 improved pex6-3 more fully than the pex6-1 allele used in our original screen (Fig. 3). As implied by the linear model shown in Fig. 1B, examination of analogous residues in the crystal structure of the related human p97 protein (62, 63) reveals that pex1-1 alters a residue adjacent to the AAA1 domain (Fig. S1), which is separated by a linker from the AAA2 domain where the lesions in pex6-1, pex6-3, and pex6-4 mutants are found (37). The pex1-1 analogous residue in human p97 (Gly376) is at least 25 Å from the residues corresponding to the pex6-1, pex6-3, and pex6-4 mutations in the neighboring subunit (Fig. S1), indicating that suppression is unlikely to arise from direct interaction of the mutated residues. In contrast, the analogous pex1-1 residue is only ∼7 Å from the ATP bound by the AAA1 domain and ∼11 Å from the linker that connects the AAA1 and AAA2 domains, hinting that pex1-1 might influence AAA1 ATP binding and/or communication between the AAA1 and AAA2 domains (Fig. S1).
Adding to the evidence of pex1-1 rescue specificity is the observation that the other two reported PEX1 missense alleles, the mild pex1-2 allele and the more severe pex1-3 allele, confer lethality in combination with pex6-1 (38), suggesting that simply decreasing PEX1 function does not improve activity of complexes carrying mutated PEX6. Moreover, pex1-1 worsened rather than improved pex26-1 growth defects (Fig. 3A), PTS2 processing (Fig. 3D), catalase and ICL levels (Fig. 3D), and oil body utilization (Fig. 5). This exacerbation suggests that pex1-1 further impedes ATPase function in pex26-1 even though pex1-1 does not notably impair peroxisome function as a single mutant.
Interestingly, the pattern of pex1-1 suppression of pex6 and pex26 alleles mirrored the range of restorative or exacerbative effects of overexpressing PEX5 in these mutants. PEX5 overexpression worsens the peroxisome-related defects of pex26-1 (37), fails to rescue pex6-2 (44), slightly rescues a subset of pex6-4 defects (37), substantially rescues pex6-1 (35, 37), and most fully rescues pex6-3 (37). PEX5 accumulation in the peroxisomal membrane is detrimental to peroxisome function (46, 64), and the parallel suppression patterns conferred by PEX5 overexpression or pex1-1 reinforces the hypothesis that these pex6 and pex26-1 mutations differentially disrupt retrotranslocation of monoubiquitinated PEX5 for recycling vs. retrotranslocation of polyubiquitinated PEX5 for proteasomal degradation (37). Analysis of PEX5 ubiquitination states in these mutants would be useful to test this hypothesis.
We initially expected that pex1-1 improvement of pex6 physiology and matrix protein import might be caused by improved PEX5 recycling. The pex6-1, pex6-3, pex6-4, and pex26-1 mutants have low PEX5 levels (35, 37) that can be increased by proteasomal inhibition (37, 52), suggesting that PEX5 is ubiquitinated and degraded when the ATPase complex is dysfunctional. Contrary to our expectations, PEX5 levels remained low in all pex1-1 combinations with pex6 and pex26 mutant except pex6-2 (Fig. 3D), which has elevated rather than reduced PEX5 levels (37). Moreover, examination of PEX5 membrane association to determine whether pex1-1 might improve removal of PEX5 from the membrane without decreasing PEX5 degradation revealed that PEX5 remained excessively membrane-associated in pex1-1 pex6-1 and pex1-1 pex6-3 (Fig. 6 and Fig. S3). Although a slight recycling improvement would be difficult to discern in this assay, these results hint that dysregulation of PEX6 functions in addition to PEX5 retrotranslocation may contribute to pex6 phenotypes, as has been suggested in plants (44, 57) and yeast (65). These hypothetical ubiquitinated PEX1–PEX6 clients might be generated by the PEX2–PEX10–PEX12 complex (66) or other peroxisome-associated ubiquitin–protein ligases (67).
Diverse pex6-1 Suppression Mechanisms Suggest Complexity of PEX6 Functions.
Decreasing PEX13 expression via the pex13-1 allele partially suppresses pex6-1 physiological defects without notably improving PTS2 processing or PEX5 levels (Fig. 8 A and B) (46). These findings suggest that pex6-1 defects stem not only from reduced matrix protein import but also from excessive PEX5 buildup in the membrane and that pex13-1 relieves this detrimental accumulation by decreasing PEX5 peroxisomal docking. Our observation that pex1-1 slightly improves pex6-1 and pex6-3 PTS2 processing (Fig. 3D) and matrix protein import (Fig. 5A) implies that pex1-1 does not act by reducing PEX5 docking at the peroxisome.
We found that the autophagy-defective atg7-3–null allele also partially ameliorated physiological and molecular defects of pex6-1 (Fig. 8), suggesting that decreasing constitutive pexophagy is beneficial or that pex6-1 suffers from heightened pexophagy. Similarly, preventing autophagy lessens the physiological defects of yeast pex1 and pex6 mutants (65), mammalian PEX1 and PEX26 knockdown cells (20), and the Arabidopsis PEX1/pex1-3 heterozygote (38). This suppression implies that, when the retrotranslocation machinery is disabled, polyubiquitinated PEX5 (or other ATPase clients) marooned on the peroxisome membrane triggers not only proteasomal degradation of PEX5 (37, 52), but also pexophagy of the entire organelle. Alternatively or in addition, the elevated PEX1–PEX6 levels that accompany autophagy prevention (Fig. 8B) could provide additional ATPase function that slightly improves peroxisome function.
Preventing autophagy also improves peroxisome function in the Arabidopsis lon2 mutant. LON2 is a peroxisomal protease that promotes sustained matrix protein import (68) by preventing premature autophagy of peroxisomes (69). Unlike the apparently complete suppression of multiple lon2-2 phenotypes when autophagy is precluded (69, 70), preventing autophagy by introducing atg7-3 only slightly ameliorated pex6-1 (Fig. 8) or PEX1/pex1-3 (38) defects, confirming that the PEX1–PEX6 hexamer contributes to peroxisome functions beyond the prevention of pexophagy.
Because PEX5 levels remained low in pex1-1 pex6-1 (Fig. 3D), we hypothesized that pex1-1 might increase removal and proteasomal degradation of polyubiquitinated PEX5 in the peroxisome membrane, thus reducing PEX5-triggered pexophagy and increasing pex6-1 peroxisome function without restoring PEX5 levels. However, we found that combining pex1-1 with pex6-1 atg7-3 further improved peroxisome function; matrix protein import resembled WT in pex1-1 pex6-1 atg7-3 (Fig. 8C) despite low PEX5 levels (Fig. 8B). The observation that pex1-1 further improved pex6-1 import even in the complete absence of autophagy (Fig. 8C) indicates that pex1-1 does not primarily suppress pex6-1 by preventing pexophagy. Because preventing autophagy in pex6-1 was insufficient to restore import, it follows that pex1-1 partially increases pex6-1 function. Perhaps preventing autophagy allows more time for the pex1-1 pex6-1 hexamer to extract PEX5 (for recycling or degradation), resulting in improved peroxisome import and function.
In addition to decreased PEX5 levels (35), PEX5 is excessively membrane-associated in pex6-1 (46); neither defect was relieved by pex1-1 (Figs. 3D and 6). Moreover, increasing PEX5 levels via transgenics (35) or by combination with a ubiquitination machinery mutant (71) partially suppresses pex6-1 defects. Overexpressing PEX5 further ameliorated defects remaining in the pex1-1 pex6-1 double mutant (Fig. 7). This additive rescue suggests that pex1-1 and PEX5 overexpression may restore different facets of pex6 dysfunction.
Our observations that pex6 symptoms can be relieved through multiple avenues (Table S1) without restoring PEX5 recycling, the sole known function of PEX6, suggest that PEX1–PEX6 has additional functions. For example, our results suggest that PEX1–PEX6 removes a pexophagy-promoting signal from the peroxisome membrane. Moreover, several Arabidopsis pex6 mutants retain seedling oil bodies after they have been fully utilized in WT seedlings (37). Similarly, mice lacking PEX5 accumulate lipid droplets in liver cells (45). Regulation of TAG mobilization is complex; overexpressing PEX5 (37), preventing autophagy (Fig. 8C), or introducing pex1-1 (Figs. 5 and 8C) each restore oil body utilization in Arabidopsis pex6-1. Our characterization of this pex1 allele that selectively restores a subset of pex6 mutant defects expands our understanding of the peroxisomal ATPase complex and provides tools for the future investigation of the roles of this essential ATPase complex in recycling PEX5, preventing pexophagy, and promoting oil body utilization.
Materials and Methods
Plant Materials and Growth Conditions.
The Columbia-0 (Col-0) accession of Arabidopsis thaliana was the WT control, and all mutants were in the Col-0 background. atg7-3 (SAIL_11_H07; 60, 61), pex6-1 (35), pex6-2 (44), pex6-3 (37), pex6-4 (37), pex13-1 (SALK_006744; 46), pex13-1 pex6-1 (46), pex26-1 (37), and pxa1-1 (55) were previously described. pex1-1 and pex1-1 pex6-1 mutants were backcrossed at least once before phenotypic assays. Genotypes were determined by using PCR-based markers (Table S2). Seedlings were moved from plates to soil (SunGro MetroMix 366) after 1–2 wk and were grown under constant white light at 22 °C.
We crossed previously described WT 35S:PEX5 and pex6-1 35S:PEX5 expressing a PEX5 cDNA (35) to pex1-1 pex6-1 to isolate pex1-1 and pex1-1 pex6-1 lines overexpressing PEX5, respectively. Transgenic plants were selected by using Basta resistance and by PCR-amplifying PEX5 using intron-spanning primers (Table S2).
To isolate peroxisome-targeted GFP in pex1-1 and double/triple mutants, we crossed pex1-1 to WT, pex6-2, pex6-3, pex6-4, or pex26-1 containing 35S:GFP-PTS1 (35, 37, 44), crossed pex1-1 pex6-1 to pex6-1 35S:GFP-PTS1 (35), and crossed pex1-1 pex6-1 35S:GFP-PTS1 to atg7-3 35S:GFP-PTS1. The transgene was tracked by using PCR amplification with primers annealing to the promoter and GFP (Table S2).
Seedlings were grown on plant nutrient (PN) medium (72) solidified with 0.6% (wt/vol) agar and supplemented with 0.5% (wt/vol) sucrose (PNS medium) and IBA from an ethanol-dissolved stock solution as indicated. Physiological assays monitoring seedling growth and IBA responsiveness were conducted as described previously (37) and were repeated at least twice with similar results.
Mutant Isolation.
pex6-1 seeds were soaked in 0.12% EMS for 16 h in the dark, washed extensively, and allowed to germinate in liquid 1/6× PN medium supplemented with 0.5% sucrose to promote germination. Seedlings were transferred to soil to allow M2 seed set. pex1-1 was isolated by screening pex6-1 M2 seedlings grown on PN for individuals with elongated dark-grown hypocotyls. Putative suppressors were transferred to PNS in the light to recover before transferring to soil, and progeny were retested for the ability to elongate hypocotyls in the dark without sucrose.
Whole-Genome Sequencing.
Approximately 2,000 backcrossed seedlings were grown under continuous white light for 14 d on sterile filter paper on PNS. Seedlings from three lines were pooled to reduce the appearance of homozygous noncausal background mutations, and genomic DNA was prepared as described previously (73). DNA was sequenced at the Genome Technology Access Center at Washington University in St. Louis by using an Illumina HiSEq 2000 sequencer and was analyzed as described previously (69, 74) to identify EMS-consistent lesions in predicted coding sequences, introns, and UTRs that were absent in our laboratory version of Col-0. More than 80% of the genome had 10-fold sequence coverage.
DNA Constructs and Plant Transformation.
35S:HA-PEX6 was previously described (37). A PEX1 cDNA in pENTR/SD-DTOPO was transferred into the pEG201 destination vector (75) using LR clonase II (Invitrogen) to give 35S:HA-PEX1, which was transformed into Agrobacterium tumefaciens GV3101 (pMP90) (76) and used to transform pex1-1 pex6-1 and WT using the floral dip method (77). Transformants were selected on PNS plates containing 8–10 μg/mL Basta; homozygous lines were identified by monitoring Basta resistance in the T3 generation.
Immunoblot Analysis.
Seedling tissue was prepared for immunoblotting as described previously (37). Primary antibodies were incubated with membranes overnight as follows: rabbit anti-PEX1 (1:200; ref. 38), anti-PEX5 (1:100; ref. 35), anti-PEX6 (1:800–1,000; ref. 46), anti-PEX14 (1:10,000; Agrisera AS08), anti-catalase (1:20,000; ref. 78), anti-ICL (1:1,000; ref. 79), anti-PMDH2 (1:2,000; ref. 80), anti-thiolase (1:2,500–1:5,000; ref. 57), mouse anti-mitochondrial ATP synthase subunit α (1:2,000; MitoScience MS507) and anti-HSC70 (1:50,000–1:100,000; SPA-817; StressGen Biotechnologies), and rat anti-HA (1:100–1:500; clone 3F10; Roche). Membranes were incubated for 3–6 h with HRP-conjugated secondary antibodies (1:5,000) before washing and imaging using WesternBright ECL substrate (Advansta). Membranes were sequentially probed with various antibodies without stripping. Films were scanned by using a flatbed scanner. Immunoblotting experiments were performed at least twice with similar results.
Confocal Fluorescence Microscopy.
Seedlings were grown under continuous light on PNS, and germination was scored 2 or 3 d after plating. Cotyledons and hypocotyls from individuals with emerged radicles at germination scoring were stained with 5 μg/mL Nile red and imaged on day 5. A Carl Zeiss 710 confocal microscope with a 63× oil-immersion objective and a Meta detector was used to capture fluorescence. A 488-nm Argon laser was used to image 0.8-μm optical sections. Each image is an average of four images with 12-bit depth. Confocal microscopy experiments were repeated at least four times with similar results.
Cell Fractionation.
An approximately equal mass (±0.03 g) of seedlings was collected from each genotype. Tissue was processed as described previously (64). Fractionation experiments were repeated twice with similar results.
Statistical Analysis.
One-way ANOVA with Duncan’s test was used to assess statistical significance (SPSS Statistics, version 24; IBM). Different letters above bars in the figures denote significant differences (P < 0.001).
Accession Numbers.
Sequence data can be found in the Arabidopsis Genome Initiative under the accession numbers At5g08470 (PEX1) and At1g03000 (PEX6). Other proteins aligned in Fig. 1B were human PEX1 (NP_000457), PEX6 (NP_000278.3), and p97 (NP_009057).
Supplementary Material
Acknowledgments
We thank Shino Goto and Mikio Nishimura for the PEX1 cDNA; Masayoshi Maeshima, Steven Smith, and Richard Trelease antibodies recognizing ICL, PMDH, and catalase, respectively; and Yun-Ting Kao, Kathryn Smith, and Zachary Wright for critical comments on the manuscript. This research was supported by National Institutes of Health (NIH) Grant R01GM079177, National Science Foundation (NSF) Grant MCB-1516966, Robert A. Welch Foundation Grant C-1309, NSF Graduate Research Fellowship DGE-0940902 (to K.L.G.), and an NSF training program EHR-0966303 (to K.L.G.). Genomic sequencing at the Genome Technology Access Center at Washington University School of Medicine was supported by NIH Grants P30CA91842 and UL1RR024992. Confocal microscopy performed in this study used equipment obtained through NIH Shared Instrumentation Grant S10RR026399.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721279115/-/DCSupplemental.
References
- 1.Nyathi Y, et al. The Arabidopsis peroxisomal ABC transporter, comatose, complements the Saccharomyces cerevisiae pxa1 pxa2Δ mutant for metabolism of long-chain fatty acids and exhibits fatty acyl-CoA-stimulated ATPase activity. J Biol Chem. 2010;285:29892–29902. doi: 10.1074/jbc.M110.151225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel B, Burkhart SE, Fleming WA. Protein transport in and out of plant peroxisomes. In: Brocard C, Hartig A, editors. Molecular Machines Involved in Peroxisome Biogenesis and Maintenance. Springer; Vienna: 2014. pp. 325–345. [Google Scholar]
- 3.Kao YT, Gonzalez KL, Bartel B. Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol. 2018;176:162–177. doi: 10.1104/pp.17.01050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu J, et al. Plant peroxisomes: biogenesis and function. Plant Cell. 2012;24:2279–2303. doi: 10.1105/tpc.112.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strader LC, Bartel B. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant. 2011;4:477–486. doi: 10.1093/mp/ssr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azevedo JE, Schliebs W. Pex14p, more than just a docking protein. Biochim Biophys Acta. 2006;1763:1574–1584. doi: 10.1016/j.bbamcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Meinecke M, et al. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol. 2010;12:273–277. doi: 10.1038/ncb2027. [DOI] [PubMed] [Google Scholar]
- 10.Kerssen D, et al. Membrane association of the cycling peroxisome import receptor Pex5p. J Biol Chem. 2006;281:27003–27015. doi: 10.1074/jbc.M509257200. [DOI] [PubMed] [Google Scholar]
- 11.Williams C, van den Berg M, Sprenger RR, Distel B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem. 2007;282:22534–22543. doi: 10.1074/jbc.M702038200. [DOI] [PubMed] [Google Scholar]
- 12.Platta HW, et al. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol. 2007;177:197–204. doi: 10.1083/jcb.200611012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platta HW, et al. Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol. 2009;29:5505–5516. doi: 10.1128/MCB.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platta HW, Grunau S, Rosenkranz K, Girzalsky W, Erdmann R. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat Cell Biol. 2005;7:817–822. doi: 10.1038/ncb1281. [DOI] [PubMed] [Google Scholar]
- 15.Birschmann I, et al. Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6p to peroxisomal membranes. Mol Biol Cell. 2003;14:2226–2236. doi: 10.1091/mbc.E02-11-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto N, Tamura S, Fujiki Y. The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat Cell Biol. 2003;5:454–460. doi: 10.1038/ncb982. [DOI] [PubMed] [Google Scholar]
- 17.Goto S, Mano S, Nakamori C, Nishimura M. Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell. 2011;23:1573–1587. doi: 10.1105/tpc.110.080770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Platta HW, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem J. 2004;384:37–45. doi: 10.1042/BJ20040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiel JA, Emmrich K, Meyer HE, Kunau WH. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem. 2005;280:1921–1930. doi: 10.1074/jbc.M403632200. [DOI] [PubMed] [Google Scholar]
- 20.Law KB, et al. The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy. 2017;13:868–884. doi: 10.1080/15548627.2017.1291470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimm I, Erdmann R, Girzalsky W. Role of AAA(+)-proteins in peroxisome biogenesis and function. Biochim Biophys Acta. 2016;1863:828–837. doi: 10.1016/j.bbamcr.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Saffian D, Grimm I, Girzalsky W, Erdmann R. ATP-dependent assembly of the heteromeric Pex1p-Pex6p-complex of the peroxisomal matrix protein import machinery. J Struct Biol. 2012;179:126–132. doi: 10.1016/j.jsb.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Gardner BM, Chowdhury S, Lander GC, Martin A. The Pex1/Pex6 complex is a heterohexameric AAA+ motor with alternating and highly coordinated subunits. J Mol Biol. 2015;427:1375–1388. doi: 10.1016/j.jmb.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciniawsky S, et al. Molecular snapshots of the Pex1/6 AAA+ complex in action. Nat Commun. 2015;6:7331. doi: 10.1038/ncomms8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blok NB, et al. Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc Natl Acad Sci USA. 2015;112:E4017–E4025. doi: 10.1073/pnas.1500257112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner BM, et al. The peroxisomal AAA-ATPase Pex1/Pex6 unfolds substrates by processive threading. Nat Commun. 2018;9:135. doi: 10.1038/s41467-017-02474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saffert P, Enenkel C, Wendler P. Structure and function of p97 and Pex1/6 type II AAA+ complexes. Front Mol Biosci. 2017;4:33. doi: 10.3389/fmolb.2017.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura S, Yasutake S, Matsumoto N, Fujiki Y. Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J Biol Chem. 2006;281:27693–27704. doi: 10.1074/jbc.M605159200. [DOI] [PubMed] [Google Scholar]
- 29.Tamura S, Matsumoto N, Takeba R, Fujiki Y. AAA peroxins and their recruiter Pex26p modulate the interactions of peroxins involved in peroxisomal protein import. J Biol Chem. 2014;289:24336–24346. doi: 10.1074/jbc.M114.588038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo JG, Lai CY, Miceli MV, Jazwinski SM. A novel role of peroxin PEX6: suppression of aging defects in mitochondria. Aging Cell. 2007;6:405–413. doi: 10.1111/j.1474-9726.2007.00291.x. [DOI] [PubMed] [Google Scholar]
- 31.Braverman NE, et al. Peroxisome biogenesis disorders in the Zellweger spectrum: An overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab. 2016;117:313–321. doi: 10.1016/j.ymgme.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M. Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 2007;48:763–774. doi: 10.1093/pcp/pcm053. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan CP, Thomas JE, Charlton WL, Baker A. Identification and characterisation of PEX6 orthologues from plants. Biochim Biophys Acta. 2001;1539:173–180. doi: 10.1016/s0167-4889(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A. Stress induces peroxisome biogenesis genes. EMBO J. 2000;19:6770–6777. doi: 10.1093/emboj/19.24.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zolman BK, Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA. 2004;101:1786–1791. doi: 10.1073/pnas.0304368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dodt G, Gould SJ. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol. 1996;135:1763–1774. doi: 10.1083/jcb.135.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez KL, et al. Disparate peroxisome-related defects in Arabidopsis pex6 and pex26 mutants link peroxisomal retrotranslocation and oil body utilization. Plant J. 2017;92:110–128. doi: 10.1111/tpj.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinaldi MA, et al. The PEX1 ATPase stabilizes PEX6 and plays essential roles in peroxisome biology. Plant Physiol. 2017;174:2231–2247. doi: 10.1104/pp.17.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith CE, et al. Spectrum of PEX1 and PEX6 variants in Heimler syndrome. Eur J Hum Genet. 2016;24:1565–1571. doi: 10.1038/ejhg.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maxwell MA, Leane PB, Paton BC, Crane DI. Novel PEX1 coding mutations and 5′ UTR regulatory polymorphisms. Hum Mutat. 2005;26:279. doi: 10.1002/humu.9356. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, et al. Genomic structure and identification of 11 novel mutations of the PEX6 (peroxisome assembly factor-2) gene in patients with peroxisome biogenesis disorders. Hum Mutat. 1999;13:487–496. doi: 10.1002/(SICI)1098-1004(1999)13:6<487::AID-HUMU9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 42.Muralla R, Lloyd J, Meinke D. Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS One. 2011;6:e28398. doi: 10.1371/journal.pone.0028398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li XR, et al. Arabidopsis DAYU/ABERRANT PEROXISOME MORPHOLOGY9 is a key regulator of peroxisome biogenesis and plays critical roles during pollen maturation and germination in planta. Plant Cell. 2014;26:619–635. doi: 10.1105/tpc.113.121087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burkhart SE, Lingard MJ, Bartel B. Genetic dissection of peroxisome-associated matrix protein degradation in Arabidopsis thaliana. Genetics. 2013;193:125–141. doi: 10.1534/genetics.112.146100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dirkx R, et al. Absence of peroxisomes in mouse hepatocytes causes mitochondrial and ER abnormalities. Hepatology. 2005;41:868–878. doi: 10.1002/hep.20628. [DOI] [PubMed] [Google Scholar]
- 46.Ratzel SE, Lingard MJ, Woodward AW, Bartel B. Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic. 2011;12:121–134. doi: 10.1111/j.1600-0854.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodward AW, Bartel B. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell. 2005;16:573–583. doi: 10.1091/mbc.E04-05-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17:3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monroe-Augustus M, et al. Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol Biol. 2011;77:1–15. doi: 10.1007/s11103-011-9782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodward AW, et al. A viable Arabidopsis pex13 missense allele confers severe peroxisomal defects and decreases PEX5 association with peroxisomes. Plant Mol Biol. 2014;86:201–214. doi: 10.1007/s11103-014-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kao YT, Fleming WA, Ventura MJ, Bartel B. Genetic interactions between PEROXIN12 and other peroxisome-associated ubiquitination components. Plant Physiol. 2016;172:1643–1656. doi: 10.1104/pp.16.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayashi M, et al. AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 2000;19:5701–5710. doi: 10.1093/emboj/19.21.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helm M, et al. Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA. 2007;104:11501–11506. doi: 10.1073/pnas.0704733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zolman BK, Silva ID, Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 2001;127:1266–1278. [PMC free article] [PubMed] [Google Scholar]
- 56.Kao YT. 2017. Insights from forward- and chemical-genetic studies on peroxisome functions, peroxisome-environment interactions, and the peroxisome-associated ubiquitination. PhD thesis (Rice University, Houston)
- 57.Lingard MJ, Monroe-Augustus M, Bartel B. Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:4561–4566. doi: 10.1073/pnas.0811329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17:526–537. doi: 10.1016/j.tplants.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 59.Young PG, Bartel B. Pexophagy and peroxisomal protein turnover in plants. Biochim Biophys Acta. 2016;1863:999–1005. doi: 10.1016/j.bbamcr.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai Z, Wang F, Zheng Z, Fan B, Chen Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011;66:953–968. doi: 10.1111/j.1365-313X.2011.04553.x. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Nishimura MT, Zhao T, Tang D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011;68:74–87. doi: 10.1111/j.1365-313X.2011.04669.x. [DOI] [PubMed] [Google Scholar]
- 62.Hänzelmann P, Schindelin H. The interplay of cofactor interactions and post-translational modifications in the regulation of the AAA+ ATPase p97. Front Mol Biosci. 2017;4:21. doi: 10.3389/fmolb.2017.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 64.Kao YT, Bartel B. Elevated growth temperature decreases levels of the PEX5 peroxisome-targeting signal receptor and ameliorates defects of Arabidopsis mutants with an impaired PEX4 ubiquitin-conjugating enzyme. BMC Plant Biol. 2015;15:224. doi: 10.1186/s12870-015-0605-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nuttall JM, Motley AM, Hettema EH. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy. 2014;10:835–845. doi: 10.4161/auto.28259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaur N, Zhao Q, Xie Q, Hu J. Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins(F) J Integr Plant Biol. 2013;55:108–120. doi: 10.1111/jipb.12014. [DOI] [PubMed] [Google Scholar]
- 67.Pan R, Satkovich J, Hu J. E3 ubiquitin ligase SP1 regulates peroxisome biogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2016;113:E7307–E7316. doi: 10.1073/pnas.1613530113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lingard MJ, Bartel B. Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 2009;151:1354–1365. doi: 10.1104/pp.109.142505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farmer LM, et al. Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell. 2013;25:4085–4100. doi: 10.1105/tpc.113.113407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goto-Yamada S, et al. Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol. 2014;55:482–496. doi: 10.1093/pcp/pcu017. [DOI] [PubMed] [Google Scholar]
- 71.Burkhart SE, Kao YT, Bartel B. Peroxisomal ubiquitin-protein ligases peroxin2 and peroxin10 have distinct but synergistic roles in matrix protein import and peroxin5 retrotranslocation in Arabidopsis. Plant Physiol. 2014;166:1329–1344. doi: 10.1104/pp.114.247148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- 73.Thole JM, Beisner ER, Liu J, Venkova SV, Strader LC. Abscisic acid regulates root elongation through the activities of auxin and ethylene in Arabidopsis thaliana. G3 (Bethesda) 2014;4:1259–1274. doi: 10.1534/g3.114.011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rinaldi MA, et al. The roles of β-oxidation and cofactor homeostasis in peroxisome distribution and function in Arabidopsis thaliana. Genetics. 2016;204:1089–1115. doi: 10.1534/genetics.116.193169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 76.Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 77.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 78.Kunce CM, Trelease RN, Turley RB. Purification and biosynthesis of cottonseed (Gossypium hirsutum L.) catalase. Biochem J. 1988;251:147–155. doi: 10.1042/bj2510147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maeshima M, Yokoi H, Asahi T. Evidence for no proteolytic processing during transport of isocitrate lyase into glyoxysomes in castor bean endosperm. Plant Cell Physiol. 1988;29:381–384. [Google Scholar]
- 80.Pracharoenwattana I, Cornah JE, Smith SM. Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J. 2007;50:381–390. doi: 10.1111/j.1365-313X.2007.03055.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.