Summary
Traditional variable selection methods are compromised by overlooking useful information on covariates with similar functionality or spatial proximity, and by treating each covariate independently. Leveraging prior grouping information on covariates, we propose partition-based screening methods for ultrahigh-dimensional variables in the framework of generalized linear models. We show that partition-based screening exhibits the sure screening property with a vanishing false selection rate, and we propose a data-driven partition screening framework with unavailable or unreliable prior knowledge on covariate grouping and investigate its theoretical properties. We consider two special cases: correlation-guided partitioning and spatial location- guided partitioning. In the absence of a single partition, we propose a theoretically justified strategy for combining statistics from various partitioning methods. The utility of the proposed methods is demonstrated via simulation and analysis of functional neuroimaging data.
Keywords: Correlation-based variable screening, Partition, Spatial variable screening, Ultrahigh-dimensional variable screening
1. Introduction
Biotechnological advances have resulted in an explosion of ultrahigh-dimensional data, where the dimension of the data can be of exponential order in the sample size. Because of high computational cost and poor numerical stability, ultrahigh-dimensional data have long defied existing regularization approaches designed for high-dimensional data analysis (Tibshirani, 1996; Fan & Li, 2001; Zou & Hastie, 2005; Meinshausen & Bühlmann, 2006; Yuan & Lin, 2006; Zhao & Yu, 2006; Zou, 2006; Candès & Tao, 2007; Zhang & Lu, 2007; Huang et al., 2008; Zou & Zhang, 2009; Meinshausen & Bühlmann, 2010; Wang & Leng, 2012). An overarching goal of ultrahigh-dimensional data analytics is to effectively reduce the dimension of covariates.
Sure independence screening (Fan & Lv, 2008) has been extended to generalized linear models (Fan & Fan, 2008; Fan et al., 2009; Fan & Song, 2010), generalized additive models (Fan et al., 2012) and proportional hazards models (Zhao & Li, 2012; Gorst-Rasmussen & Scheike, 2013; Hong et al., 2016a; Li et al., 2016). By extending screening criteria that are solely based on marginal correlations between the outcome and predictors, a variety of statistics that account for dependence between predictors have been proposed to improve screening accuracy and robustness (Hall & Miller, 2009; Zhu et al., 2011; Cho & Fryzlewicz, 2012; Li et al., 2012; Cui et al., 2015). In particular, high-dimensional ordinary least squares projection (Wang & Leng, 2016), which uses the generalized inverse of the design matrix in lieu of marginal correlations, has good theoretical properties and high computational efficiency.
In many cases, scientists have knowledge about important predictors from previous research. For example, neuroimaging studies have identified voxel-level imaging predictors clustered in certain brain regions that are linked to brain functions or diseases. Genome-wide association studies have detected single nucleotide polymorphisms that are strongly associated with clinical outcomes. However, most variable screening approaches are not designed to make use of such information.
As an alternative to marginal screening approaches, conditional sure independence screening methods have been developed for generalized linear models (Barut et al., 2016) and proportional hazards models (Hong et al., 2016a). By including important predictors, conditional screening ranks the marginal utility of each variable after adjusting for variables in the conditioning set.
Partitioning biomarkers into smaller groups according to biological knowledge or other useful information may facilitate variable selection. In biological studies, leveraging information about groups of weak predictors is often useful because such predictors may have a nontrivial impact on outcomes as a group, and without considering the group structure these features might be missed. We exemplify the merit of using the grouping structure with a simple example.
Suppose that we want to identify the important associations between the outcome Y and X1,…, X1000, where Y = 0.5X1 − X2 + ε with ε ~ N(0, 1.6) and (X1,…, X1000) follows a multivariate normal distribution with mean zero, unit marginal variance and correlation corr (Xj, Xk) = 0.5 for any j ⧧ k ∈{1,…, 1000}. To screen for important variables, marginal screening would fit 1000 variate regression models, for j = 1, …, 1000, and use , the estimate of βj, as the screening statistic. Suppose that we partition these 1000 predictors into 200 groups such that the group membership index sets are . An alternative screening approach would fit 200 multivariate regression models along the group partition, , and use the corresponding as the screening statistic. We examine the performances of the two approaches based on 300 samples and 400 replicates. Figure 1 shows plots of the densities of and and the mixture density of for both approaches. Due to signal cancellation, the univariate regression introduces large biases in estimating β1 and β2, causing considerable overlap between the distribution of or and those of the estimates for the noise variables, whereas groupwise multivariate regression separates the distribution of or from the distributions of the others.
Fig. 1.
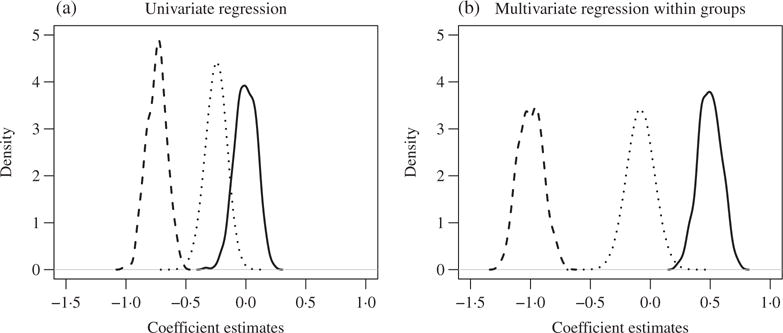
Simple example simulations: (a) summary distributions of 400 replicates of coefficient estimates in 1000 univariate regression model fits; (b) summary of 200 group-specific multivariate regression model fits. Plotted are the estimated densities of (solid) and (dashed) and the estimated mixture densities of (dotted); true coefficients are β1 = 0.5, β2 = − 1 and β3 = ⋯ = β1000 = 0.
This example motivates partition-based screening, which is based on a partition of covariates using prior knowledge. Our work generalizes the univariate framework of sure independence screening (Fan & Lv, 2008) and its group version (Niu et al., 2011). Under mild conditions, partition-based screening exhibits good theoretical properties. A new functional operator, generalized linear conditional expectation, is introduced to help establish sure screening properties. When prior grouping information is available, we show that the screening accuracy of partition-based screening is superior to that of competing methods. In the absence of prior grouping information, we propose correlation-based screening and spatial partition-based screening, which make the proposed methods applicable to a wide range of problems.
2. Partition-based variable screening
Suppose that we have n independent samples D = {(Xi, Yi), i = 1, …, n}, where Yi is an outcome and Xi = (Xi,1,…, Xi,p)T is a collection of p predictors for the ith sample. Assume without loss of generality that all the covariates have been standardized so that E(Xi,j) = 0 and . We consider a class of generalized linear models by assuming that the conditional density of Yi given Xi belongs to a linear exponential family,
| (1) |
where A(·, ·) and b(·) are known functions, β = (β1,…, βp)T represent the coefficients of the predictors, and β0 is an intercept, regarded as a nuisance parameter. Let . We assume that b(·) is twice continuously differentiable, with a nonnegative second derivative b″(·). For a nonrandom function f (·) and a sequence of independent random variables ξi (i = 1,…, n), let be the empirical mean of , which are independent replicates of f (ξ). The loglikelihood function is
| (2) |
where l(θ, y) = yθ − b(θ). We assume that {Xij, Xi, Yi} are independently and identically distributed copies of {Xj, X, Y}. When p < n, the maximum likelihood estimator of β, denoted by , can be obtained by maximizing ℓ(β0, β; D). When p ⩾ n, regularization estimation is often performed under an assumption of sparsity among predictors. When p is of exponential order in n, a popular approach for reducing the dimensionality is screening.
First, we consider a simple case where the covariates can be partitioned into G disjoint groups in accordance with known information. Denote by gj the group membership of variable Xj. Let be the collection of predictors in group g, where g ∈ {1,…, G}. Additionally, let represent the corresponding coefficients and let βg,0 be the group-specific intercept in the model. Denote their estimates by and , respectively. For predictor j with gj = g, the partition-based screening statistic is defined as
We call the partition-based screening statistic. Then, for a chosen thresholding parameter γ, the set of indices selected by our proposed partition-based screening is .
When gj = j (j = 1,…, p), partition-based screening encompasses sure independence screening as a special case.
3. Sure screening properties
Let (Ω, ℱ, pr) be the probability space for all random variables considered in this paper. Let ℝd be a d-dimensional Euclidean vector space for some positive integer d. Denote by E(·), var(·) and cov(·, ·) the expectation, variance and covariance operators associated with (Ω, ℱ, pr). For any vector , let be the subvector with elements indexed by . Let be the Ld-norm for any vector a ∈ℝp, and denote the Euclidean norm by ||a|| when no confusion is likely to arise. Let λmin(M) and λmax(M) be the smallest and largest eigenvalues of the matrix M, respectively.
We start with population-level parameters for the discussion of sure screening properties. Let
| (3) |
where is the population version of . We first establish conditions to ensure that if |βj| exceeds a threshold, then will exceed a certain constant. Write and . With (2), satisfies the score equations
| (4) |
To derive the theoretical properties of the proposed methods, we introduce a functional operator on random variables.
Definition 1
For two random variables ζ : Ω → ℝ and ξ : Ω → ℝp, let h : ℝ → ℝ be a continuous link function. The generalized linear conditional expectation of ζ given ξ is
| (5) |
where (α0, αT)T is the solution to the equation E[{ζ − h(α0 + αTξ)}(1, ξT)T] = 0.
The generalized linear conditional expectation measures how ξ can explain ζ through a generalized linear model, where ζ is regarded as the outcome variable and ξ as the predictors. It can also be interpreted as the best prediction of ζ using ξ based on a generalized linear model, leading to an alternative measure of the dependence between ζ and ξ. The generalized linear conditional expectation may depend on the choice of link functions, and it is equivalent to the conditional expectation if the true conditional distribution of ζ given ξ is specified by the corresponding generalized linear model. The introduction of (5) facilitates the development of partition-based screening and its theoretical properties, and extends the linear conditional expectation proposed by Barut et al. (2016) and Hong et al. (2016a). Some basic properties are summarized below.
Lemma 1
Let ζ and ξ be random variables in (Ω, ℱ, pr).
When h(x) = 1(x) = x, Eh(ζ | ξ) is unique and has a closed-form expression. Moreover, E1(ζ | ξ) = E(ζ) + cov(ζ,ξ)var(ξ)−1{ξ −E(ξ)} and E{E1(ζ | ξ)ξ} = E(ζξ).
When the conditional distribution of ζ given ξ belongs to a linear exponential family, i.e., f(ζ | ξ) = exp{ζ(γ0 + γTξ) − b(γ0 + γTξ) + A(ζ, ξ)}, then h(x) = b'(x) and .
For any h, we have E{Eh(ζ | ξ)} = E(ζ).
These properties immediately imply the following result.
Theorem 1
Suppose that the solution to (4) is unique. For j = 1,…,p, the partition-based regression parameter equals 0 if and only if .
The sufficient part of Theorem 1 implies that if the generalized linear conditional expectation of the response given all the predictors within group gj does not involve Xj, then the regression coefficient βj will be vanishing, implying that unimportant variables would have smaller fitted coefficients.
To ensure the sure screening property at the population level, the important variables should be conditionally associated with Y given other variables within the same group . The following conditions are required.
Condition 1
For , there exist c0 > 0 and κ < 1/2 such that
Condition 2
The derivative b′(θ) satisfies a Lipschitz condition, i.e., there exists an L > 0 such that |b′(θ1) − b′(θ2)| < L|(θ1) − (θ2)|for all θ1,θ2 ∈ ℝ.
Condition 3
There exists a constant M > 0 such that for all j.
Condition 1 provides a lower bound on the generalized linear dependence between each active covariate Xj and Y conditional on other covariates within the same group, justifying the use of group partitions to retain true signals. Linear regression, logistic regression and probit regression all satisfy Condition 2.
Theorem 2
If Conditions 1–3 hold, then there exists c2 > 0 such that .
To establish sure screening properties, we need regularity conditions (Fan & Song, 2010; Barut et al., 2016); see Conditions A.1–A.6 in the Supplementary Material.
Theorem 3
Let be the size of group g. Assume that Conditions A.1–A.6 in the Supplementary Material hold and that as n → ∞ for all g = 1,…, G.
- With c2 as in Theorem 2, there exists a positive constant c3 such that
where . - If Conditions 1–3 hold, then with γ = c4n−κ and c4 ≤ c2/2, we have
where .
For logistic regression, the Lipschitz constant rg,n is bounded. Therefore, the optimal rate for Rn is of order n(1−2κ)/(2+α), ensuring that Qg,n is of the same order as . This also implies that the partition-based screening method can handle group sizes of order log Sg = o(n(1−2κ)α/(α+2)) (g = 1,…, G). The same optimal rate and a similar order of dimensionality can be achieved for logistic regression by sure independence screening (Fan & Song, 2010) and conditional sure independence screening (Barut et al., 2016).
To provide an upper bound on the number of selected variables, we need the following additional conditions:
Condition 4
and b″(θ) are both bounded for all θ and β.
Condition 5
Let g = gj and with
where . Then there exists a K1 > 0 such that for all j = 1,…, p.
Condition 6
Assume that ║U║2 = o(V) where U = (U1,…, Up)T with
and . Here (β0, βT)T are the parameters that generate the data.
For a linear model, Condition 5 becomes for all g, which is a mild condition. In Condition 6, Uj = 0 for the linear model when cov(X) has a block-diagonal structure over a group partition, i.e., for g ⧧ g′, because and .
Condition 6, which requires ||U|| to be bounded, may be restrictive. This condition holds if the number of nonzero coefficients is finite or if the correlations among different partitions shrink as p → ∞. Condition 6 can be viewed as rigid, even though the group structure is natural in many biomedical applications. To overcome this difficulty, one could first perform a principal component analysis on X, and then apply the proposed procedures to the residuals of X after projecting them to a set of variables with the largest loadings on the leading eigenvectors (Hong et al., 2016a). See the Supplementary Material for more details.
Theorem 4
With γ, c3 and r2 as in Theorem 3, if Conditions 4–6 and A. 1–A.6 hold, then as n →∞,
4. Extensions of partition-based screening
4.1. Goodness-of-fit adjustment
One difficulty in the proposed partition-based screening is that the coefficient estimates from group-specific models may not be comparable because different models may have various degrees of goodness-of-fit. Under the generalized linear model framework, we propose to adjust for the goodness-of-fit by weighting the screening statistics using the deviance ratio Ψg ∈ (0,1) (Friedman et al., 2010), which is the fraction of null deviance explained by the covariates in group g and is equal to R2 in the linear model. In other words, we weight the partition-based screening statistic as for predictor j and redefine the selected index set as . However, the performance of such a procedure may be sensitive to grouping. For instance, in the example in § 1, goodness-of-fit adjustment can improve the model selection accuracy when all the true predictors are in the same group, as in Fig. 2(a). In contrast, when the true predictors are in separate groups, there is no improvement; see Fig. 2(b) and the Supplementary Material.
Fig. 2.
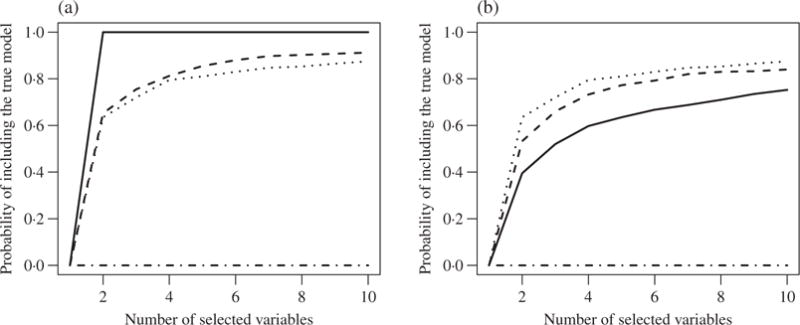
Variable screening accuracy in the example in § 1 by sure independence screening (dot-dash), high-dimensional ordinary least squares projection (dotted), partition-based screening without goodness-of-fit adjustment (dashed) and partition-based screening with goodness-of-fit adjustment (solid). Panel (a) shows results with the group partitions defined in § 1 where group 1 includes the two true predictors; panel (b) displays results with a random group partition where the two true predictors are not in the same group.
4.2. Data-driven partition
When prior partitioning information is unavailable, we may use information from the data, including correlations between predictors and the spatial locations. Let us denote the data-driven partition by . We derive two procedures for determining based on such information.
The data-driven partition can be determined by the covariance or correlation structure of the design matrix. We propose a simple correlation-guided partition procedure. We use the correlation between covariates to define a p × p distance matrix, denoted by Δ = (dj,k), where dj,k = 1 − |corr(Xk, Xj)| (1 ≤ k,j ≤ p). We apply the nearest-neighbour chain algorithm (Murtagh, 1983), a standard hierarchical clustering algorithm, to Δ to obtain an estimate of , where the number of groups, , can be determined by controlling the corresponding maximum group size for the generalized linear model fitting. Based on our experience, choosing can lead to good performance. When the correlation matrix has a block-diagonal structure and the correlations within each block are high, this procedure can correctly identify the block structure. Covariate-assisted variable screening (Ke et al., 2014) and graphlet screening (Jin et al., 2014) are also based on the covariance structure of covariates, but these procedures focus on linear regression.
Spatial regression models have often been used in environmental health and neuroimaging studies, where a spatial location is attached to each covariate. For example, in the scalar-on-image regression problem for brain imaging, where the spatial location of each voxel is in a standard three-dimensional brain template, the imaging intensities at different voxels are usually considered as potential predictors for clinical outcomes. It is generally believed that spatially close predictors tend to have stronger correlations and may have more similar effects on the outcome (Wang et al., 2017). Therefore spatial location information can be useful in determining the partition for variable screening. Specifically, model-based clustering (Fraley & Raftery, 2002) and k-means clustering (Jain, 2010) can be used to assign each predictor to a fixed number of clusters or spatial locations, typically determined by controlling the corresponding maximum group size in a similar fashion to correlation-guided partitioning.
Using a partition determined by data, we can also establish the following theoretical results for data-driven partition-based screening, which can be proved by conditioning on the event and using Theorems 1–4.
Theorem 5
Suppose that is a consistent estimator of satisfying Conditions 1–6 and A.1–A.6; that is, . With the same γ, c3, r2 and r3 as in Theorem 3, as n → ∞ we have
4.3. Combined partition-based screening
Several partitioning rules for covariates may exist, but none is clearly superior. For example, in neuroimaging, brain atlases, such as Talairach–Tournoux, Harvard–Oxford, Eickoff–Zilles and automatic anatomical labelling, have variable partitioning of brain regions. In genome-wide association studies, different sources of information, including the locations of genes that harbour single nucleotide polymorphisms and linkage disequilibrium between single nucleotide polymorphisms, can be integrated to determine the groups. Using multiple sources of partitioning, we propose a strategy for combining screening statistics from different partitions and establish its theoretical properties.
Definition 2
Suppose that we have K < ∞ partitions and that partition k has G(k) groups and group indices . Let and let be the screening statistics for partition k. The combined partition-based screening statistic is with . Given a thresholding parameter γ, the selected index set is , which is referred to as combined partition-based screening selection.
Theorem 6
Suppose that Conditions 4–6 and A.1–A.6 hold for all partitions k (k = 1, …, K), and take γ = c5n−κ for some constant c5.
If there exists an l ∈ {1, …, K} such that satisfies Conditions 1–3, then .
- Let V(k) be the V term in Condition 6 for . Then combined partition-based screening controls the false positive rates; that is,
Theorem 6 suggests that combined partition-based screening has sure screening properties, even when some partition-based screening procedures do not satisfy Conditions 1–3, which are in general difficult to verify. Moreover, Conditions 4–6 and A.1–A.6 are true for many generalized linear models. Thus, combined partition-based screening can extract useful prior knowledge about partitions and maintain good theoretical properties.
4.4. Choice of thresholding parameters
The thresholding parameter γ is critical to the performance of the variable screening procedure. Overestimating γ will inflate false positive rates and underestimating γ will hinder sure screening. We define the expected false positive rate , where . To control EFPRγ, we resort to higher-criticism t statistics (Zhao & Li, 2012; Barut et al., 2016; Hong et al., 2016a). We introduce , where is the element that corresponds to βj in the information matrix . The key idea is to select γ such that of the same size as , where τ is chosen to control the expected false positive rate. Under Conditions 4 and 5 and Conditions A.1–A.6 and B in the Supplementary Material, we have the following theorem.
Theorem 7
For a given false positive number q, take τ = Φ−1 {1 − q/(2p)}, where Φ denotes the standard normal distribution function. Set , where and {(1), …, (p)} is a permutation of {1, …, p} such that . Then there exist N7 > 0 and c7 > 0 such that for any n > N7, EFPRγ ≤ q/p + c7n−1/2.
For , the set of indices selected by combined partition-based screening in § 4.3, we choose with s = min1≤k≤K s(k). Here s(k) is the size of the higher-criticism t-tests for partition k.
5. Simulation studies
We conducted simulation studies to compare the model selection accuracy of partition-based screening with that of existing variable screening methods. We generated the covariates X from multivariate normal distributions and specified the true coefficient β with four different settings.
Setting 1: (n, p) = (200, 5000) and . Thus . The covariance structure of X is compound symmetric with unit variance and correlation 0.5, i.e., .
Setting 2: (n, p) = (200, 5000) and βj = 3(−1)jI (j ≤ 10) for j ∈ {1, …, p}. Thus . The covariance structure of X is that of a block first-order autoregression model with unit variance and correlation 0.9, i.e., cov(Xj, Xj′) = 0.9|j−j′| for any , where for k = 1,…, 50, and cov(Xj, Xj′) otherwise.
Setting 3: (n, p) = (200, 5000) and βj = (−1)jI (j ≤ 10) for j ∈ {1,…, p}. Thus . The covariance structure of X is block compound symmetric with unit variance and correlation 0.9, i.e., cov(Xj, Xj′) = 0.9 for any , where for k = 1,…, 50, and cov(Xj, Xj′) = 0 otherwise.
Setting 4: (n, p) = (500, 10 000). We first define as a collection of 100 × 100 equally spaced grid points on [0, 1]2. Specifically, set with sj = 0.01 (l, k), j = (100 − 0.5l)(l − 1) + k − l for 1 ≤ l, k ≤ 100, and , where for any g ⧧ g′ ∈ {1,…, G}. All the sj were clustered into 100 exclusive spatially contiguous regions using a k-means clustering algorithm. Set βj = 3(−1)j if and βj = 0 otherwise. The covariance structure of X is exponentially decaying over space, cov(Xj, Xj′) = exp(−10‖sj − sj′‖2) for any . For example, cov(Xj, Xj′) = 0.9 when ‖sj − sj′‖2 = 0.01 and cov(Xj, Xj′) < 0.05 when ‖sj − sj′‖2 > 0.3. This configuration was designed to mimic the spatial data with an active set .
Given X and β generated from each of the above settings, we generated Y from a linear regression model and a logistic regression model. For the linear regression model, we set the variance of random errors so that the theoretical R2 is equal to 0.9. We replicated our simulation 200 times and evaluated the performance using the following criteria: probability of including the true model, minimum model size, and true positive rate.
Definition 3
Every partition belongs to at least one of the following types:
a size-reduced partition if there exists a group that contains all active covariates, which means ;
an optimal partition if there exists a group that is a collection of all active covariates, which means ;
a misspecified partition if there does not exist a group such that .
Each partition is either a size-reduced partition or a misspecified partition. An optimal partition must be a size-reduced partition. Neither the misspecified partition, the size-reduced partition, nor the optimal partition, is unique in general. For each non-optimal reduced partition, there exists at least a group containing that can be further reduced in size while containing .
We assessed the performance of the proposed methods under various partition types. In Setting 1, the size-reduced partition was specified with 357 groups and each group had 14 members except for group 357 which had 16 members; the group size was approximately n1/2. For covariate j ∈ {1, …, 4998}, its group label was assigned to be , and for j = 4999 or 5000, , where for each setting. The misspecified partitions and were respectively sampled from and . In this setting, variable X6 is marginally unimportant but conditionally important. Some studies (Barut et al., 2016; Hong et al., 2016a,b) have shown that conditional sure independence screening performs much better than sure independence screening in terms of retaining X6. In Settings 2 and 3, where the correlation matrix for covariates is block diagonal, we focused primarily on the performance of partition-based screening under reduced partitions and correlation-guided partition-based screening with the same specifications as in Setting 1. The partition determined by the estimated correlation structure is denoted by . In Setting 4, the optimal partition was designed as follows. For each covariate j, , where . To generate , we combined groups 1 and 2 while keeping other groups intact, i.e., for each covariate j, , where . To form the misspecified partitions in Setting 4, we split into two adjacent but mutually exclusive subregions and such that . We considered two different misspecified partitions, denoted by and , where and . Figure 3 is a graphical representation of Setting 4.
Fig. 3.
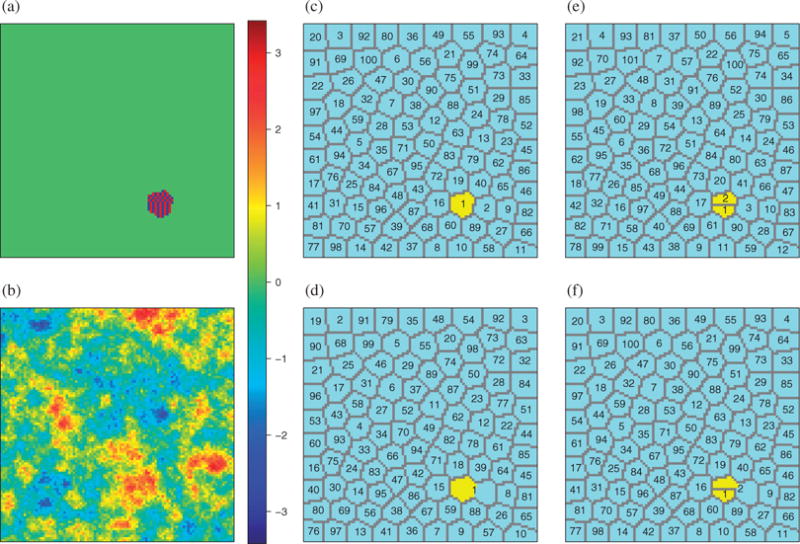
Model designs and configurations of different partitions on , the 100 × 100 equally spaced grid points in two-dimensional space [0, 1]2, in Setting 4 of the simulation study for spatial variable screening: (a) the true coefficient βj, which takes only three possible values, −3 in blue, 3 in red and 0 in green; (b) one set of simulated predictors Xj over space from a Gaussian random field on with covariance kernel exp(−10||s − s′||2) for s, s′ ∈ ; (c) the partition in Setting 4 to define the true nonzero coefficients, with βj ⧧ 0 if and only if , in yellow. Panels (c), (d), (e) and (f) respectively represent the optimal partition , the size-reduced partition , and two misspecified partitions, and , for the corresponding partition-based screening for selecting variables in Setting 4; in panels (c)–(f), is coloured yellow.
For further comparison, we investigated the performance of sure independence screening (Fan & Lv, 2008) and high-dimensional ordinary least squares projection (Wang & Leng, 2016) in Settings 1–4. In addition, conditional sure independence screening (Barut et al., 2016) with a conditioning variable X1 was considered for Settings 1–3. To evaluate the performance of high-dimensional ordinary least squares projection for logistic regression, we modified it to accommodate generalized linear models with a ridge penalty by specifying the tuning parameter to be 1. Sure and conditional sure independence screening results were obtained using the R (R Development Core Team, 2017) package SIS, while high-dimensional ordinary least squares projection was implemented using the R package screening. To make different methods comparable, we chose γ so that the number of selected indices was equal to the sample size and computed the true positive rate and the probability that the selected indices include the true model, following Fan & Lv (2008).
Table 1 summarizes the simulation results. In Settings 1–3, partition-based screening performs best for linear and logistic regression. In Setting 4, the performance of spatial-oriented reduced partition screening is almost the same as spatial-oriented optimal partition screening in linear regression, and is close to spatial-oriented optimal partition screening in logistic regression. This indicates that even when an optimal partition is not available, a size-reduced partition is a good alternative. In Settings 2 and 3, correlation-guided partition screening produces better selection accuracy than all three existing methods, and is comparable to partition-based screening. Thus, when there is insufficient prior knowledge to determine a size-reduced partition but the covariate variables have a block-diagonal correlation structure up to permutations, data-driven partition-based screening can yield improved selection accuracy. In Setting 1, where the covariance structure of covariates is compound symmetric, correlation-guided partition screening does not yield more accurate selection than high-dimensional ordinary least squares projection and conditional sure independence screening. In this case we examined the performance of partition-based screening with randomly generated misspecified partitions, as well as combined partition-based screening that combines five or ten different random partition-based screening statistics. The results indicate that combined partition-based screening with ten random is slightly better than combined partition-based screening with five random and outperforms partition-based screening with only or with only. Therefore, in the absence of an optimal partition, combining multiple partitions for variable screening fares better than relying on a single partition. Moreover, it produces better results than high-dimensional ordinary least squares projection and sure independence screening in Setting 1. The advantages of combined partition-based screening are obvious in Setting 4, where the accuracy of spatial-oriented partition screening with can be improved by combining it with two misspecified partition-based screening statistics. Thus, combining various screening statistics from multiple sources of partitions, even though they may have been misspecified, appears to be a useful strategy.
Table 1.
Model selection accuracy of variable screening methods for linear and logistic regression in Settings 1–4
| Setting 1 | Linear regression | Logistic regression | ||||
|---|---|---|---|---|---|---|
| PIT (%) | MMS | TPR (%) | PIT (%) | MMS | TPR (%) | |
| SIS | 10 | 2273 | 84 | 12 | 2590 | 80 |
| HOLP | 91 | 24 | 98 | 53 | 173 | 90 |
| CSIS | 97 | 11 | 100 | 73 | 62 | 95 |
| PartS ( ) | 100 | 7 | 100 | 100 | 6 | 100 |
| PartS ( ) | 84 | 36 | 97 | 46 | 226 | 88 |
| PartS ( ) | 87 | 34 | 98 | 60 | 145 | 91 |
| CombPartS (5 ) | 93 | 25 | 99 | 64 | 115 | 93 |
| CombPartS (l0 ) | 96 | 22 | 99 | 74 | 105 | 96 |
| CorrPartS ( ) | 78 | 64 | 96 | 47 | 219 | 88 |
| Setting 2 | Linear regression | Logistic regression | ||||
| PIT (%) | MMS | TPR (%) | PIT (%) | MMS | TPR (%) | |
| SIS | 0 | 3982 | 56 | 0 | 4243 | 41 |
| HOLP | 0 | 3721 | 59 | 0 | 4231 | 43 |
| CSIS | 69 | 52 | 96 | 0 | 311 | 90 |
| PartS ( ) | 100 | 10 | 100 | 100 | 10 | 100 |
| CorrPartS ( ) | 91 | 10 | 98 | 88 | 10 | 98 |
| Setting 3 | Linear regression | Logistic regression | ||||
| PIT (%) | MMS | TPR (%) | PIT (%) | MMS | TPR (%) | |
| SIS | 0 | 3671 | 20 | 100 | 1367 | 60 |
| HOLP | 65 | 110 | 100 | 16 | 560 | 90 |
| CSIS | 6 | 3033 | 70 | 6 | 2265 | 70 |
| PartS ( ) | 100 | 11 | 100 | 100 | 10 | 100 |
| CorrPartS ( ) | 62 | 101 | 100 | 18 | 426 | 80 |
| Setting 4 | Linear regression | Logistic regression | ||||
| PIT (%) | MMS | TPR (%) | PIT (%) | MMS | TPR (%) | |
| SIS | 0 | 9890 | 17 | 0 | 9860 | 26 |
| HOLP | 0 | 9886 | 18 | 0 | 9876 | 27 |
| SpatPartS ( ) | 100 | 100 | 100 | 100 | 100 | 100 |
| SpatPartS ( ) | 100 | 100 | 100 | 73 | 174 | 100 |
| SpatPartS ( ) | 0 | 8232 | 65 | 0 | 9033 | 65 |
| SpatPartS ( ) | 0 | 9190 | 55 | 0 | 9269 | 74 |
| CombPartS ( , , ) | 100 | 100 | 100 | 80 | 174 | 100 |
SIS, sure independence screening; HOLP, high-dimensional ordinary least squares projection; CSIS, conditional sure independence screening; PartS, partition-based screening; CombPartS, combined partition-based screening; CorrPartS, correlation-guided partition screening; SpatPartS, spatial-oriented partition screening; MMS, median minimum size of the selected models that are required to have a sure screening; TPR, average true positive rate; PIT, estimated probability of including all true predictors in the top n selected predictors.
6. Application
We applied the proposed methods to analyse resting-state functional magnetic resonance imaging data from the Autism Brain Imaging Data Exchange study (Di Martino et al., 2014). The primary goal of this study was to understand how brain activity is associated with autism spectrum disorder, a disease with substantial heterogeneities among children. Functional magnetic resonance imaging measures blood oxygen levels linked to neural activity, and resting-state functional magnetic resonance imaging measures brain activity only when the brain is not performing any tasks. This study aggregated 20 resting-state functional magnetic resonance imaging datasets from 17 experiment sites. For each subject, the resting-state functional magnetic resonance imaging signal was recorded for each voxel in the brain over multiple time-points. Standard imaging pre-processing steps (Di Martino et al., 2014) included motion correction, slice-timing correction, and spatial smoothing. The entire brain was registered into the 3 mm standard Montreal Neurological Institute space, which consists of 38 547 voxels in 90 brain regions defined by the automated anatomical labelling system (Hervé et al., 2012). After removal of missing values, the complete dataset included 819 subjects, consisting of 378 patients and 441 age-matched controls. To select imaging biomarkers for autism spectrum disorder risk prediction, we considered the fractional amplitude of low-frequency fluctuations (Zou et al., 2008), defined as the ratio of the power spectrum for frequencies 0.01–0.08 Hz to the entire frequency range. This measure has been widely used as a voxel-wise measure of the intrinsic functional brain architecture derived from resting-state functional magnetic resonance imaging data (Zuo et al., 2010).
We constructed a spatial logistic regression model that has clinical diagnosis of autism spectrum disorder as the outcome and the voxel-wise fractional amplitudes of frequency fluctuations as imaging predictors, adjusting for age at scan, sex and intelligence quotient. Because imaging predictors on the risk of autism spectrum disorder are spatially clustered and sparse (Liu & Calhoun, 2014), the primary aim of this study is to identify imaging biomarkers among 38 547 voxel-level fractional amplitudes of low-frequency fluctuations that predict the autism spectrum disorder risk. We applied partition-based screening by using anatomical information, correlation among imaging predictors and spatial information. Specifically, we considered the following methods: brain region partition-based screening on 90 brain regions; correlation-guided partition screening, which partitions the 38 547 voxels into G groups using the clustering algorithm introduced in § 4–2, where G is taken to be 256, 128, 64, 32, 16 or 8; spatial-oriented partition screening, which partitions the 38 547 voxels into 1024 equal-sized regions where the voxels are spatially contiguous within each region; and combined partition-based screening that combines all of the above. We also applied high-dimensional ordinary least squares projection for logistic regression with the ridge penalty, as implemented in the R package screening.
To assess the performance of the different methods, we used ten-fold crossvalidation, randomly splitting the data into ten equal-sized subsets. We applied the variable screening methods to the training dataset and obtained a set of selected voxels, based on which we made a prediction about the disease status in the testing dataset using logistic regression with the elastic net penalty, as implemented in the R package glmnet. We repeated this ten times and computed the crossvalidation accuracy. Among all the methods, combined partition-based screening achieved the smallest crossvalidation prediction error, 37%, and high-dimensional ordinary least squares projection had the largest crossvalidation prediction error, 48%. All the other partition-based screening methods achieved a prediction error of approximately 40%. More details and the receiver operating characteristic curves are given in the Supplementary Material.
Next, we applied combined partition-based screening to the entire dataset, using the method in § 4–4 to determine the threshold by taking an upper bound on the expected false positive rate to be 0.20. A total of 6142 important voxels were selected. Eight regions with more than 60 selected voxels are reported in Table 2, along with the median rank of voxel-specific screening statistics within each region. These regions are known to be involved in specific brain functions related to autism (Friederici et al., 2003; Japee et al., 2015).
Table 2.
Eight automated anatomical labelling regions with more than 60 voxels that are selected by combined partition-based screening
| Selected region | Voxel counts | Median rank | Selected region | Voxel counts | Median rank |
|---|---|---|---|---|---|
| Frontal_Mid_R | 95 | 5 | Frontal_Sup_L | 71 | 14 |
| Temporal_Mid_R | 78 | 27 | Frontal_Mid_L | 71 | 47 |
| Temporal_Mid_L | 75 | 154 | Frontal_Sup_R | 66 | 35 |
| Precuneus_L | 73 | 56 | Precuneus_R | 65 | 41 |
7. Discussion
The method proposed in this paper can be improved. First, our framework requires that the size of each partition group be less than the sample size to make (3) sensible. If this condition is not met, penalized likelihood methods such as the lasso can be applied, though these approaches may involve the selection of tuning parameters and the correction of biases due to penalization. A simple but efficient remedy would be to further refine the groups randomly. This refining procedure can be performed multiple times, and the resulting screening statistics can be combined using the rule in § 4.3. Second, although this paper has focused on non-overlapping partitions for ease of theoretical development, our screening framework can accommodate overlapping partitions. According to the combination rule in § 4.3, for each predictor that is covered by more than one partition, we can simply choose the screening statistic with larger value. Third, the time complexity of correlation-guided partition screening is O(p2), mainly due to the need to compute the correlation matrix and clustering predictors. To compute the correlations among ultrahigh-dimensional predictors more efficiently, we suggest adopting parallel computing techniques. To speed up the clustering of predictors, we propose to threshold the correlation matrix and generate a binary matrix, regarded as the adjacency matrix of an undirected graph. The connected components corresponding to group partitions can be obtained by using the breadth-first or depth-first search algorithms with time complexity between O(p) and O(p2).
Supplementary Material
Acknowledgments
The authors are grateful to the editor, the associate editor and the referees for their constructive comments and suggestions, which have substantially improved the manuscript. Kang was supported in part by the U.S. National Institutes of Health. Hong was supported in part by the U.S. National Security Agency. Li was supported in part by the U.S. National Institutes of Health and the National Natural Science Foundation of China.
Footnotes
Supplementary material available at Biometrika online includes theoretical results and further results and figures for the simulation study.
Contributor Information
JIAN KANG, Department of Biostatistics, University of Michigan, 1415 Washington Heights, Ann Arbor, Michigan 48109, U.S.A.
HYOKYOUNG G. HONG, Department of Statistics and Probability, Michigan State University, 619 Red Cedar Rd, East Lansing, Michigan 48823, U.S.A
YI LI, Department of Biostatistics, University of Michigan, 1415 Washington Heights, Ann Arbor, Michigan 48109, U.S.A.
References
- Barut E, Fan J, Verhasselt A. Conditional sure independence screening. J Am Statist Assoc. 2016;111:1266–77. doi: 10.1080/01621459.2015.1092974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candès E, Tao T. The Dantzig selector: Statistical estimation when p is much larger than n. Ann Statist. 2007;35:2313–51. [Google Scholar]
- Cho H, Fryzlewicz P. High dimensional variable selection via tilting. J R Statist Soc B. 2012;74:593–622. [Google Scholar]
- Cui H, Li R, Zhong W. Model-free feature screening for ultrahigh dimensional discriminant analysis. J Am Statist Assoc. 2015;110:630–41. doi: 10.1080/01621459.2014.920256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M, et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Molec Psychiat. 2014;19:659–67. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Fan Y. High dimensional classification using features annealed independence rules. Ann Statist. 2008;36:2605–37. doi: 10.1214/07-AOS504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Feng Y, Song R. Nonparametric independence screening in sparse ultra-high dimensional additive models. J Am Statist Assoc. 2012;106:544–57. doi: 10.1198/jasa.2011.tm09779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. J Am Statist Assoc. 2001;96:1348–60. [Google Scholar]
- Fan J, Lv J. Sure independence screening for ultrahigh dimensional feature space. J R Statist Soc B. 2008;70:849–911. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Samworth R, Wu Y. Ultrahigh dimensional feature selection: Beyond the linear model. J Mach Learn Res. 2009;10:2013–38. [PMC free article] [PubMed] [Google Scholar]
- Fan J, Song R. Sure independence screening in generalized linear models with NP-dimensionality. Ann Statist. 2010;38:3567–604. [Google Scholar]
- Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. J Am Statist Assoc. 2002;97:611–31. [Google Scholar]
- Friederici AD, Rüschemeyer SA, Hahne A, Fiebach CJ. The role of left inferior frontal and superior temporal cortex in sentence comprehension: Localizing syntactic and semantic processes. Cereb Cortex. 2003;13:170–7. doi: 10.1093/cercor/13.2.170. [DOI] [PubMed] [Google Scholar]
- Friedman JH, Hastie TJ, Tibshirani RJ. Regularization paths for generalized linear models via coordinate descent. J Statist Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Gorst-Rasmussen A, Scheike T. Independent screening for single-index hazard rate models with ultrahigh dimensional features. J R Statist Soc B. 2013;75:217–45. [Google Scholar]
- Hall P, Miller H. Using generalized correlation to effect variable selection in very high dimensional problems. J Comp Graph Statist. 2009;18:533–50. [Google Scholar]
- Hervé PY, Razafimandimby A, Vigneau M, Mazoyer B, Tzourio-Mazoyer N. Disentangling the brain networks supporting affective speech comprehension. NeuroImage. 2012;61:1255–67. doi: 10.1016/j.neuroimage.2012.03.073. [DOI] [PubMed] [Google Scholar]
- Hong HG, Kang J, Li Y. Conditional screening for ultra-high dimensional covariates with survival outcomes. Lifetime Data Anal. 2016a doi: 10.1007/s10985-016-9387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong HG, Wang L, He X. A data-driven approach to conditional screening of high-dimensional variables. Stat. 2016b;5:200–12. [Google Scholar]
- Huang J, Horowitz JL, Ma S. Asymptotic properties of bridge estimators in sparse high-dimensional regression models. Ann Statist. 2008;36:587–613. [Google Scholar]
- Jain AK. Data clustering: 50 years beyond k-means. Pat Recog Lett. 2010;31:651–66. [Google Scholar]
- Japee S, Holiday K, Satyshur MD, Mukai I, Ungerleider LG. A role of right middle frontal gyrus in reorienting of attention: A case study. Front Syst Neurosci. 2015;9 doi: 10.3389/fnsys.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Zhang CH, Zhang Q. Optimality of graphlet screening in high dimensional variable selection. J Mach Learn Res. 2014;15:2723–72. [Google Scholar]
- Ke T, Jin J, Fan J. Covariance assisted screening and estimation. Ann Statist. 2014;42:2202–42. doi: 10.1214/14-AOS1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Peng H, Zhang J, Zhu L. Robust rank correlation based screening. Ann Statist. 2012;40:1846–77. [Google Scholar]
- Li J, Zheng Q, Peng L, Huang Z. Survival impact index and ultrahigh-dimensional model-free screening with survival outcomes. Biometrics. 2016;72:1145–54. doi: 10.1111/biom.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Calhoun VD. A review of multivariate analyses in imaging genetics. Front Neuroinfo. 2014;8:1–11. doi: 10.3389/fninf.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinshausen N, BÜhlmann P. High-dimensional graphs and variable selection with the lasso. Ann Statist. 2006;34:1436–62. [Google Scholar]
- Meinshausen N, BÜhlmann P. Stability selection (with Discussion) J R Statist Soc B. 2010;72:417–73. [Google Scholar]
- Murtagh F. A survey of recent advances in hierarchical clustering algorithms. Comp J. 1983;26:354–9. [Google Scholar]
- Niu Y, Hao N, An L. Detection of rare functional variants using group ISIS. BMC Proc. 2011;5:S108. doi: 10.1186/1753-6561-5-S9-S108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. http://www.R-project.org. [Google Scholar]
- Tibshirani RJ. Regression shrinkage and selection via the Lasso. J R Statist Soc B. 1996;58:267–88. [Google Scholar]
- Wang H, Leng C. Unified LASSO estimation by least squares approximation. J Am Statist Assoc. 2012;102:1039–48. [Google Scholar]
- Wang X, Leng C. High dimensional ordinary least squares projection for screening variables. J R Statist Soc B. 2016;78:589–611. [Google Scholar]
- Wang X, Zhu H, for the Alzheimer’s Disease Neuroimaging Initiative Generalized scalar-on-image regression models via total variation. J Am Statist Assoc. 2017 doi: 10.1080/01621459.2016.1194846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J R Statist Soc B. 2006;68:49–67. [Google Scholar]
- Zhang HH, Lu W. Adaptive lasso for Cox’s proportional hazards model. Biometrika. 2007;94:691–703. [Google Scholar]
- Zhao P, Yu B. On model selection consistency of Lasso. J Mach Learn Res. 2006;7:2541–63. [Google Scholar]
- Zhao SD, Li Y. Principled sure independence screening for Cox models with ultra-high-dimensional covariates. J Mult Anal. 2012;105:397–411. doi: 10.1016/j.jmva.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LP, Li L, Li R, Zhu LX. Model-free feature screening for ultrahigh-dimensional data. J Am Statist Assoc. 2011;696:1464–75. doi: 10.1198/jasa.2011.tm10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. J Am Statist Assoc. 2006;101:1418–29. [Google Scholar]
- Zou H, Hastie TJ. Regularization and variable selection via the elastic net. J R Statist Soc B. 2005;67:301–20. [Google Scholar]
- Zou H, Zhang HH. On the adaptive elastic-net with a diverging number of parameters. Ann Statist. 2009;37:1733–51. doi: 10.1214/08-AOS625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q-H, Zhu CZ, Yang Y, Zuo XN, Long XY, Cao QJ, Wang YF, Zang YF. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: Fractional ALFF. J Neurosci Meth. 2008;172:137–41. doi: 10.1016/j.jneumeth.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Di Martino A, Kelly C, Shehzad ZE, Gee DG, Klein DF, Castellanos FX, Biswal BB, Milham MP. The oscillating brain: Complex and reliable. NeuroImage. 2010;49:1432–45. doi: 10.1016/j.neuroimage.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


