Abstract
Using Arabidopsis, we analyzed the effect of omission of a nitrogen source and of the addition of different nitrogen-containing compounds on the extractable activity and the enzyme and mRNA accumulation of adenosine 5′-phosphosulfate reductase (APR). During 72 h without a nitrogen source, the APR activity decreased to 70% and 50% of controls in leaves and roots, respectively, while cysteine (Cys) and glutathione contents were not affected. Northern and western analysis revealed that the decrease of APR activity was correlated with decreased mRNA and enzyme levels. The reduced APR activity in roots could be fully restored within 24 h by the addition of 4 mm each of NO3−, NH4+, or glutamine (Gln), or 1 mm O-acetylserine (OAS). 35SO42− feeding showed that after addition of NH4+, Gln, or OAS to nitrogen-starved plants, incorporation of 35S into proteins significantly increased in roots; however, glutathione and Cys labeling was higher only with Gln and OAS or with OAS alone, respectively. OAS strongly increased mRNA levels of all three APR isoforms in roots and also those of sulfite reductase, Cys synthase, and serine acetyltransferase. Our data demonstrate that sulfate reduction is regulated by nitrogen nutrition at the transcriptional level and that OAS plays a major role in this regulation.
Several studies have established regulatory interactions between assimilatory sulfate and nitrate reduction in plants (Rennenberg and Bergmann, 1979; Reuveny et al., 1980; Cacco et al., 1983; Brunold and Suter, 1984; Haller et al., 1986; Takahashi and Saito, 1996). The two assimilatory pathways are very similar and well coordinated (Brunold, 1993). Deficiency for one element was shown to repress the other pathway (Reuveny et al., 1980; Neuenschwander et al., 1991). The activities of ATP sulfurylase (ATPS) and adenosine 5′-phosphosulfate (APS) reductase (APR), which has been shown to be identical to APS sulfotransferase (Suter et al., 2000), decreased under nitrogen-deficient conditions in Lemna minor L. and cultured tobacco cells (Reuveny et al., 1980; Brunold and Suter, 1984). At the same time, the addition of nitrate or ammonia to the medium quickly restored the activity of these two enzymes. Later, ATPS was shown to not always be repressed by the absence of nitrogen (Haller et al., 1986), whereas the activity of APR always rapidly decreased under these conditions (Brunold, 1990). Activity of the last enzyme of sulfate assimilation pathway, Cys synthase (CS), can also be critical in varying the flux through the sulfate reduction pathway, since the precursor of Cys, O-acetylserine (OAS), is derived from the carbon and nitrogen assimilation pathways. Accordingly, several studies showed that expression of different isoforms of CS is regulated differently under both sulfur and nitrogen starvation (Takahashi and Saito, 1996; Warrilow and Hawkesford, 1998).
ATPS and APR occupy a central control position of sulfate reduction (Brunold, 1990). Three isoforms of ATPS were cloned from Arabidopsis (Leustek et al., 1994; Klonus et al., 1995; Murillo and Leustek, 1995), all possessing a putative chloroplast-targeting peptide. Also, APR forms a small family of presumably chloroplast localized proteins. Three cDNA clones coding for APR isoforms were obtained from Arabidopsis (Gutierrez-Marcos et al., 1996; Setya et al., 1996). Other enzymes involved in sulfate reduction have already been characterized on a molecular level (Hell et al., 1994; Ruffet et al., 1995; Brühl et al., 1996). The availability of molecular probes now makes it possible to study the regulation of the whole pathway at a molecular level. Takahashi et al. (1997) demonstrated that sulfate-deficiency-induced expression of three genes of the pathway only, the sulfate transporter AST68, APR1, and SAT-1 (Ser acetyltransferase).
We report the effects of nitrogen deficiency and the subsequent addition of different nitrogen sources on APR mRNA and protein accumulation and activity in Arabidopsis shoots and roots. To address the flux through the sulfate assimilation pathway under these conditions, we also determined the incorporation of [35S]sulfate into Cys, glutathione (GSH), and proteins.
MATERIALS AND METHODS
Plant Cultivation and Treatment
Seeds of Arabidopsis var Columbia, were germinated in plastic pots (5 cm high, 5 cm in diameter) filled with small moistened balls (2–6 mm in diameter) of burned clay (Blähton, Migros, Switzerland). The pots were placed in trays containing Hentschel nutrient solution (Hentschel, 1970). The plants (three to five per pot) were grown in a day/night cycle of 10 h/14 h and a light intensity of 115 to 160 μmol s−1 m−2. Average temperature was 25°C to 27°C during the day and 23°C at night. Relative humidity ranged between 40% and 60%. All experiments were performed with 4.5-week-old plants. For establishing nitrogen deficiency, the plants were provided with modified Hentschel nutrient solution prepared by substitution of all nitrogen-containing components by the corresponding chlorides. Control plants obtained fresh nutrient solution at the beginning of the experiment. Plants were incubated for 72 h under the same conditions, then nutrient solution with 4 mm NaNO3, 4 mm ammonium tartrate, 4 mm Gln, and 0.4 or 1 mm OAS was added to the nitrogen-deficient plants and they were cultivated for an additional 24 h.
Enzyme Assays
For preparing extracts, whole shoot or root systems of three to five plants were pooled and homogenized 1:10 and 1:20 (w/v), respectively, in 50 mm Na/K phosphate buffer, pH 8.0, supplemented with 30 mm Na2SO3, 0.5 mm 5′-AMP, and 10 mm dithioerythritol (DTE) (Imhof, 1994) using a glass homogenizer. The crude extracts were filtered through two layers of Miracloth (Migros, Bern, Switzerland). APR activity in extracts was measured as the production of [35S]sulfite, which was assayed as acid-volatile radioactivity formed in the presence of [35S]APS and DTE (Brunold and Suter, 1990). CS activity was determined by measuring the Cys produced from OAS and S2−, as described in Pieniazek et al. (1973). The protein concentrations in the extracts were determined according to the method of Bradford (1976) with bovine serum albumin as a standard.
Isolation of Total RNA and Northern Blotting
Plant material was pulverized with mortar and pestle in liquid nitrogen, and RNA was isolated by phenol extraction and selective precipitation with LiCl. Electrophoresis of RNA was performed on formaldehyde-agarose gels at 120 V. RNA was transferred onto Hybond-N nylon membranes (Amersham-Pharmacia Biotech, Uppsala) and hybridized with 32P-labeled cDNA probes. The membranes were washed four times at different concentrations of SSC in 0.1% (w/v) SDS for 20 min, with the final washing step being 0.5× SSC, 0.1% (w/v) SDS at 65°C, and exposed to x-ray film (medical RX, Fuji Photo Film, Tokyo) at −80°C for 3 to 8 d. The autoradiograms were quantified with a densitometer (model GS-670, Bio-Rad Laboratories, Hercules, CA) using the software Molecular Analyst (Bio-Rad). Under the hybridization and washing conditions applied there was no cross-hybridization among the APR and CS isoforms. The cDNA probes for APR1, APR2, and APR3 were obtained from Dr. T. Leustek (the Center for Agricultural Molecular Biology, Rutgers University, New Brunswick, NJ), and the cytosolic and chloroplastic CS and SAT-A cDNAs from Dr. R. Hell (Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany). The ATPS1 and SiR cDNAs corresponding to accession numbers U05218 and Z49217, respectively, were amplified by RT-PCR from Arabidopsis total RNA, and the identity of the PCR fragments was verified by sequencing. Northern analysis was performed on two independent RNA preparations with the same results.
Western-Blot Analysis
Protein extracts were prepared as described by Zavgorodnyaya et al. (1997). Aliquots representing 10 μg of protein were subjected to SDS-PAGE and electrotransferred to nitrocellulose filters (0.2-μm pore size; Schleicher & Schuell, Dassel, Germany). The blots were analyzed with antisera against APR and developed with the SuperSignal western-blotting system (Pierce Chemical, Rockford, IL). The antisera were produced in rabbits from purified APR2 protein overexpressed in Escherichia coli by the pET His-Tag system (Novagen, Madison, WI). The antisera cross-reacted with the recombinant APR1 and APR3 proteins. The analysis was performed on two independent protein preparations with the same results.
Feeding of 35SO42− and Determination of 35S in Thiols and Proteins
Four pots with Arabidopsis plants were fed with Hentschel nutrient solution containing 0.75 mm SO42− supplemented with 4 mCi of 35SO42− and the different nitrogen sources for 4 h. Shoots and roots were extracted with 0.1 m HCl containing 1 mm Na2EDTA and the extracts were centrifuged for 30 min at 4°C. The thiols in the supernatant were reduced with bis-(2-mercaptoethylsulfone) (BMS) (Bernhard et al., 1998) and labeled by monobromobimane as described by Newton et al. (1981) and as modified by Kranner and Grill (1996). A 100-μL aliquot of each sample was separated by reverse-phase HPLC, as previously described (Rüegsegger and Brunold, 1992) and fractions of 0.75 mL were collected in scintillation vials. The 35S radioactivity was determined in a liquid scintillation counter (Betamatic V, Kontron, Zurich). The radioactivity in the first five fractions of the eluate corresponded to 35SO42−. Total Cys, γ-EC, and GSH were analyzed by the HPLC system described by Schupp and Rennenberg (1988) and modified by Rüegsegger and Brunold (1992). For measurement of 35S incorporation into proteins, proteins were precipitated from 200 μL of extract with 10% (w/v) TCA, washed twice with 1% (w/v) TCA and once with 96% (v/v) ethanol, and redissolved in 400 μL of 0.2 m NaOH. Radioactivity in an aliquot of the protein solution was determined using a liquid scintillation counter.
Statistical Analysis
The Student Newmann Keuls method (SigmaStat for Windows, Version 1.0, 1992–94, SPSS, Chicago) was used to determine significant differences in the enzyme activities and the contents of labeled thiols.
RESULTS
Effect of Nitrogen Deficiency on the Sulfate Assimilation Pathway
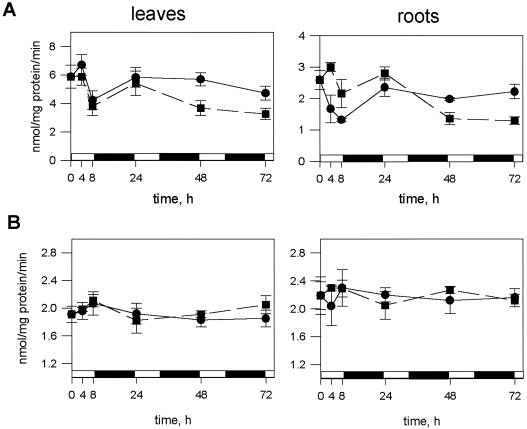
As evident from Figure 1A, APR activity was significantly decreased in both roots and leaves of Arabidopsis after 72 h of nitrogen deficiency. Significant (P < 0.05) differences in activity were detectable in leaves after 48 h. Afterward, the enzyme activity further decreased and after 72 h the extractable APR activity was reduced to about 70% of that in control plants. In roots, however, the APR activity was first increased and only after 24 h started to decrease rapidly, so that after 72 h it had decreased to 50% of the activity in control plants. The variation in activity within the first 8 h of treatment in both control and treated plants was due to diurnal changes (Kopriva et al., 1999). These changes were characterized by a rapid increase in activity, protein level, and mRNA during the morning, a decrease during the afternoon, and a slow increase during the night both in roots and shoots, and had to be taken into account in the analysis of the results described here. CS, the final enzyme in the pathway, which we measured for comparison, exhibited no diurnal changes. As shown in Figure 1B, omission of nitrogen from the nutrient solution over 3 d did not affect this enzymatic activity in roots or in shoots. Plant growth measured on a fresh weight basis and extractable protein were also not significantly affected by the omission of a nitrogen source (data not shown). The plants could only be kept on nitrogen-deficient nutrient solution for 4 d, however, since after this time the first symptoms of senescence were detectable.
Figure 1.
APR and CS activity during nitrogen deficiency. Arabidopsis plants (4.5 weeks old) were transferred onto nutrient solution without a nitrogen source. APR (A) and CS (B) activities were measured at the time points indicated in extracts from leaves and roots of nitrogen-deficient plants (▪) and controls cultivated with 4 mm NO3− (●). Mean values ± sd from five to nine measurements are shown. The differences in APR activity between nitrogen-deficient and control plants are significant (P < 0.05) from 48 h.
Figure 2 shows the changes in mRNA and protein accumulation of APR and CS during 3 d of nitrogen deficiency. The levels of mRNA for all three APR isoforms in control plants increased slightly during the course of the experiment (Fig. 2A), probably due to exchange of the nutrient solution. The APR mRNA levels in leaves and roots decreased to about 60% and 50%, respectively, of those in control plants (Fig. 2, A and B). The effect of nitrogen deficiency was thus more pronounced in roots than in leaves, which is in agreement with the changes in APR activity (Fig. 1). In leaves levels of mRNA for APR1 and APR2 isoforms decreased more rapidly in response to nitrogen starvation than APR3, whereas in roots the APR3 isoform was more affected than APR1 and APR2. Despite there being no changes in enzyme activity, the mRNA level of the chloroplastic isoform of CS decreased in leaves and roots (Fig. 2, A and B). The level of mRNA for the cytoplasmic isoform of CS in leaves was not significantly changed, but in roots a decrease of the mRNA level for this isoform was detected. The discrepancy between total CS activity and expression of the chloroplastic and cytosolic CS isoforms might be best explained by assuming a higher expression of the mitochondrial CS isoform, which would complement the decrease in expression of the two other isoforms. Indeed, this expression pattern was described for nitrogen deficiency in spinach (Takahashi and Saito, 1996).
Figure 2.
Expression analysis of APR and CS isoforms during nitrogen deficiency. A, Total RNA (10 μg) isolated from leaves or roots of plants cultivated without a nitrogen source up to 72 h and controls was analyzed with cDNA probes for APR1, APR2, and APR3, and chloroplastic (chlCS) and cytosolic (cytCS) CS. Ethidium-bromide-stained RNA is shown as a control of loading and RNA intactness. B, Relative mRNA levels of APR1 (▪), APR2 (□), and APR3 (▴) (top panels), and chloroplastic (▪) and cytosolic (□) CS (bottom panels) in leaves (left panels) and roots (right panels) during nitrogen deficiency. One-hundred percent represents the mRNA level in control plants at the selected time point. C, Protein extracts (10 μg of protein) were analyzed by western blotting with antisera against recombinant APR2. The upper panel shows the protein accumulation in control plants (C), the lower panel in plants cultivated without a nitrogen source up to 72 h (−N).
Western-blot analysis revealed a decrease in the APR protein level during nitrogen deficiency (Fig. 2C) in both leaves and roots. In contrast, there was no significant effect on the protein accumulation of both chloroplastic and cytoplasmic isoforms of CS (data not shown).
The Cys and GSH levels in leaves and roots did not change significantly during 72 h of nitrogen deficiency (data not shown).
Effect of Addition of Different Nitrogen Sources on APR and CS Activities
The ability of different nitrogen sources to restore the normal APR activity after 72 h of nitrogen deficiency is demonstrated in Figure 3. As in Figure 1A, the variation in APR activity in control and treated plants within the course of the experiment was due to diurnal rhythms. The addition of 4 mm nitrate and ammonia restored normal APR activity in roots within 4 h. During further treatment with nitrate, APR activity increased to about 120% of the controls. In leaves the effects of nitrate and ammonia only became significant after 24 h, and APR activity never reached that of controls. Gln only partially restored APR activity in both roots and leaves (Fig. 3). The addition of 1 mm OAS caused an increase in APR activity in roots after 4 h, to 250% of those of nitrogen-deficient plants. After 8 h, APR activity was four times as high as in the nitrogen-deficient plants and twice that of controls; however, after 24 h, it dropped to a level lower then the controls. In leaves no effect of OAS addition on the APR activity was detectable. No significant effect of the various nitrogen sources was detected on CS activity (data not shown).
Figure 3.
APR activity after addition of different nitrogen compounds. Arabidopsis plants were cultivated for 72 h without a nitrogen source, fresh media containing different nitrogen sources were added, and plants were cultivated for additional 24 h. APR activity was measured in leaf and root extracts 4, 8, and 24 h after treatment with 4 mm NO3− (●), 4 mm NH4+ (▪), 4 mm Gln (▴), or 1 mm OAS (▾), as well as in controls (○) and plants cultivated without nitrogen (□). Mean values ± sd from five to nine measurements are shown. Time point 0 corresponds to 72 h without nitrogen.
The levels of the three APR transcripts in leaves were not affected by the addition of the different nitrogen sources up to 8 h (Fig. 4). After 24 h with nitrate, however, a strong increase in the APR3 mRNA level (+200%) and a weaker increase in the APR1 and APR2 levels (+20% and +80%, respectively) were detected. The addition of NH4+ or Gln had no effects on APR1 and APR2 mRNA during the experimental period, but the APR3 mRNA level was increased at this time point by both compounds (Fig. 4, A and B). OAS had no effect on APR mRNA levels in leaves at either 0.4 or 1 mm. The decrease in APR2 mRNA levels at 4 and 8 h and the increase after 24 h in both control and treated plants is consistent with previous experiments and is controlled by natural diurnal rhythms (Kopriva et al., 1999). In roots, nitrate, NH4+, and Gln did not affect the levels of APR1 and APR2 mRNA, but those of APR3 were slightly increased 8 h after the addition of NH4+ and Gln (Fig. 4B). OAS rapidly induced the mRNA expression of all APR isoforms. Four hours of treatment with 0.4 mm OAS resulted in three times higher APR1 and APR3 mRNA levels, which dropped to the control levels after total 8 h of treatment and increased again to 150% of the controls after 24 h (Fig. 4B). On the other hand, APR2 transcript levels only increased about 20% and the increased level remained stable. The addition of higher concentrations of OAS (1 mm) had a more prolonged and slightly stronger effect on the expression of all three APRs than that caused by a 0.4 mm concentration, but, again, the APR1 and APR3 mRNA levels increased much more than the APR2 level.
Figure 4.
Northern analysis of APR after addition of different nitrogen sources. A, Total RNA (10 μg) isolated from leaves (left) or roots (right) 4, 8, and 24 h after beginning of treatment was hybridized with cDNA probes for the three APR isoforms. Ethidium-bromide-stained RNA is shown as a control of loading and RNA intactness (RNA). Time point 0 corresponds to 72 h without nitrogen. B, Relative mRNA levels of the APR isoforms after addition of different nitrogen sources. The autoradiograms (A) were quantified by densitometry and the changes in mRNA levels of the APR isoforms after further incubation without nitrogen (▪) and after additions of nitrate (□), NH4+ (●), Gln (○), 0.4 mm OAS (▴), or 1 mm OAS (▵) are presented. One-hundred percent represents the mRNA level in the nitrogen-deficient plants at time 0, corresponding to 72 h without nitrogen.
Western analysis revealed that the addition of all nitrogen-containing compounds increased APR protein accumulation in both leaves and roots (Fig. 5). APR protein gradually increased after the addition of nitrate, NH4+, and Gln; the addition of 1 mm OAS increased APR protein rapidly and transiently. Nitrate was also the most efficient in the long-term induction of APR protein accumulation in roots, whereas Gln and OAS induced the fastest response.
Figure 5.
Western-blot analysis of APR after the addition of different nitrogen sources. Protein extracts were prepared from leaves and roots at the time points indicated and analyzed with antisera against recombinant APR2, as described in experimental procedures. Time point 0 corresponds to 72 h without nitrogen.
Regulation of Other Genes Involved in Sulfate Assimilation by OAS in the Roots
To evaluate the effect of nitrogen-deficient conditions on other enzymes of the sulfate assimilation pathway, we performed a northern analysis with probes against sulfite reductase, SAT-A, a chloroplastic isoform of ATPS (ATPS1), and chloroplastic and cytosolic CS. No significant effects on steady-state levels of these mRNAs were observed in leaves and roots during 3 d of nitrogen deficiency (data not shown). However, feeding 0.4 mm OAS to the nitrogen-deficient plants led to an increase of mRNA levels of sulfite reductase, both CS isoforms, and SAT-A in roots (Fig. 6). Only the ATPS1 mRNA was unaffected by OAS treatment. These data demonstrate that OAS has a major role in the regulation of sulfate assimilation.
Figure 6.
Effect of OAS on mRNA levels of genes involved in sulfate assimilation. Total RNA (10 μg) isolated from roots of plants kept on nitrogen-deficient nutrition (−N) and treated with 0.4 mm OAS at 4, 8, and 24 h after beginning of treatment was hybridized with cDNA probes for ATPS1, sulfite reductase, chloroplastic (chl.) and cytosolic (cyt.) CS, and the SAT-A isoform. Ethidium-bromide-stained RNA is shown as a control of loading and RNA intactness (RNA). Time point 0 corresponds to 72 h without nitrogen.
In Vivo Flux through the Sulfate Assimilation Pathway
For analyzing in vivo sulfate assimilation, plants were fed with 35SO42− in the nutrient solution for 4 h after 3 d of nitrogen deficiency and after the addition of various nitrogen sources to the culture medium of nitrogen-starved plants. The amount of radioactivity incorporated into thiols and proteins was measured. The addition of nitrate did not change the 35S uptake into leaves or its incorporation into GSH and proteins; however, it caused a significant decrease in labeling of Cys in both leaves and roots (Fig. 7). OAS increased the 35SO42− labeling of the leaves 2-fold, caused a similar increase of 35S incorporation into proteins, and caused three times higher labeling of Cys and GSH than in nitrogen-deficient controls. Although having a significant effect on APR activity and expression of most genes of the pathway, as in leaves, nitrate did not significantly alter the amount of 35SO42− incorporated into proteins but diminished the accumulation of radioactive Cys to 50% in roots. NH4+ and Gln significantly increased protein labeling; Gln had a stronger effect and also increased the amounts of labeled GSH. Most interestingly, feeding with 1 mm OAS led to a 30-fold increase in 35S-labeled Cys, a 20-fold increase in γ-EC (not shown), and six and four times higher labeling of proteins and GSH, respectively, compared with nitrogen-deficient controls.
Figure 7.
Incorporation of 35S from [35S]sulfate into thiols and proteins. Arabidopsis plants (4.5 weeks old) were fed with 35SO42− in the nutrient solution for 4 h together with different nitrogen compounds as in Figure 3. Radioactive sulfur in SO42−, Cys, GSH, and protein in leaves and roots was measured. Mean values ± sd from four measurements are presented. Values indicated by different letters are different at P ≤ 0.05.
The data demonstrate a strong positive effect of nitrogen-containing compounds on the flux through the sulfate assimilation pathway. This effect was increased the closer the nitrogen source was metabolically related to OAS, and the acceptor of sulfur was reduced to the thiol level.
DISCUSSION
Until now, our knowledge of the regulation of sulfate assimilation, and particularly APR, by nitrogen nutrition could be addressed only by activity measurements. Due to the availability of cDNA probes and antisera, the regulation of APR mRNA and protein could also be analyzed for the first time to our knowledge. During nitrogen deficiency, APR mRNA and protein accumulation decreased in correlation with APR activity. It has to be stressed that the plant growth and protein concentration in extracts did not change during nitrogen deficiency, indicating that the Arabidopsis plants had sufficient nitrogen stored to cope with this situation. This was corroborated by the finding that the Cys and GSH concentrations did not change.
Cys synthesis from OAS and sulfide by CS links assimilatory sulfate reduction with carbohydrate and nitrogen metabolism (Brunold, 1993). The availability of both substrates of CS is strongly controlled. Cys inhibits SAT at low concentrations (Brunold and Suter, 1982; Noji et al., 1998) and sulfate uptake and reduction is inhibited at high Cys concentrations (Herschbach and Rennenberg, 1994). Therefore, when these two internal control systems are circumvented by supplying plants with either OAS or H2S, Cys synthesis is enhanced and more Cys is synthesized than is needed for protein synthesis (Neuenschwander et al., 1991; Buwalda et al., 1993; Saito et al., 1994). The surplus of Cys synthesized under these conditions is used for GSH synthesis and, correspondingly, GSH levels in plants fed with OAS or H2S were shown to be highly increased (Neuenschwander et al., 1991; Buwalda et al., 1993). These findings were corroborated in our experiments using OAS. In addition, they showed that OAS not only increased APR activity, but also led to a strong increase in APR mRNA levels and protein accumulation. The 35SO42− feeding revealed that in roots OAS is rapidly metabolized to Cys and used further for synthesis of proteins and GSH. In leaves, however, APR activity and the synthesis of 35S-labeled compounds were much less enhanced by OAS compared with roots, indicating that OAS was mostly metabolized already in roots and transported to the shoots only in minute amounts. The increased 35S labeling of Cys and GSH in leaves of OAS-treated plants compared with those cultivated with the other nitrogen compounds may be based on the higher specific activity of 35SO42− (Fig. 7) transported to the leaves and/or on enhanced, OAS-driven sulfate reduction in leaf cells.
Most enzymes of the sulfate assimilation pathway are present in multiple isoforms in plants. In Arabidopsis three organelle-specific isoforms of SAT and CS occur, and ATPS and APR also form small families of three isoenzymes (however, these are all localized in the chloroplast) (Murillo and Leustek, 1995; Gutierrez-Marcos et al., 1996; Setya et al., 1996). Our results clearly showed that the APR mRNAs were differently regulated by nitrogen compounds and differences were also observed between roots and leaves (Fig. 4). APR1 and APR2 decreased in a similar manner during nitrogen deficiency, while APR1 and APR3 mRNAs responded very similarly to the different nitrogen sources. Also, the diurnal changes in APR1 and APR3 mRNA levels were not as pronounced as those of APR2. Interestingly, the amino acid sequence of APR1 is more related to APR3, being 86.7% and 91.9% identical to APR2 and APR3, respectively (Gutierrez-Marcos et al., 1996). In addition, both APR1 and APR3 are located on chromosome IV of Arabidopsis, whereas APR2 resides on chromosome I. Nevertheless, the special roles of the different APR isoforms remain to be elucidated.
Although regulatory interactions between nitrate and sulfate assimilation have already been described, the mechanism is not yet clear. Previous reports (Reuveny et al., 1980; Brunold and Suter, 1984; Haller et al., 1986) documented a strong decrease in APR and ATPS activities upon nitrogen deficiency. These experiments were, however, performed with cell cultures or L. minor, thus circumventing the roots and root/shoot interactions. It is therefore significant that by using whole Arabidopsis plants, the same effects of nitrogen deficiency on APR activity were detected. This enzyme activity could be quickly restored in roots, where reduced nitrogen compounds were more effective than nitrate. In leaves, only nitrate was able to fully restore APR activity, probably because of limited transport of the other nitrogen compounds applied. In L. minor both nitrate and NH4+ were very efficient in restoring APR activity, which achieved normal values 4 h after beginning feeding (Brunold and Suter, 1984).
Our feeding experiments demonstrated that the closer the nitrogen sources were metabolically related to OAS, the higher was their impact on 35SO42− incorporation (Fig. 7). The lower 35S labeling of Cys in roots after the addition of NO3−, NH4+, or Gln compared with nitrogen-deficient controls indicates that the newly produced radioactive Cys is used for protein synthesis to a high degree, and therefore does not accumulate. In addition, northern analysis revealed that OAS feeding increased mRNA levels of a wide range of genes involved in sulfate reduction (Figs. 4 and 6). Our data thus extend the results of Smith et al. (1997), who showed that in barley roots OAS induced not only sulfate uptake but also the mRNA level of a sulfate transporter. However, the experiments with OAS were performed on plants deficient in nitrogen and, therefore, in free amino acids, including OAS. Cys synthesis may be thus limited by low concentrations of OAS. The addition of this compound might thus remove this limitation, leading to a rapid depletion of sulfide and other intermediates of the sulfate assimilation pathway. This might result in sulfur deficiency, which is known to induce expression of several genes of the sulfate assimilation pathway (Takahashi et al., 1997). In contrast to sulfur deficiency, however, feeding with OAS led to a high increase of mRNA levels for sulfite reductase, chloroplastic and cytosolic CS, and SAT-A (Fig. 6). It seems therefore likely that OAS is a direct transcriptional regulator of genes involved in sulfate assimilation. This view is supported by the fact that in bacteria, an isomer of OAS, N-acetylserine, induces transcription of several genes in the Cys regulon (Kredich, 1993).
In conclusion, the data presented here show that APR activity in plants is controlled by nitrogen availability on a transcriptional level, and that the three APR isoforms are differently regulated. Furthermore, our results lead to the conclusion that OAS plays a central role in coordinating sulfate with nitrate assimilation and in the regulation of the whole assimilatory sulfate reduction pathway.
ACKNOWLEDGMENTS
We thank Dr. T. Leustek and Dr. R. Hell for cDNA probes of APR, CS, and SAT, respectively. We would also like to acknowledge Dr. M. Hawkesford for helpful discussions.
Footnotes
This work was supported by the Swiss National Foundation (grant no. 3149246–96 to C.B.).
LITERATURE CITED
- Bernhard MC, Junker E, Hettinger A, Lauterburg BH. Time course of total cysteine, glutathione and homocysteine in plasma of patients with chronic hepatitis C treated with interferon-a with and without supplementation of N-acetylcysteine. J Hepatol. 1998;28:751–755. doi: 10.1016/s0168-8278(98)80223-7. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brühl A, Haverkamp T, Gisselmann G, Schwenn JD. A cDNA clone from Arabidopsis thalianaencoding plastidic ferredoxin: sulfite reductase. Biochem Biophys Acta. 1996;1295:119–124. doi: 10.1016/0167-4838(96)00066-0. [DOI] [PubMed] [Google Scholar]
- Brunold C. Reduction of sulfate to sulfide. In: Rennenberg H, Brunold C, De Kok LJ, Stulen I, editors. Sulfur Nutrition and Sulfur Assimilation in Higher Plants. The Hague, The Netherlands: SPB Academic Publishing; 1990. pp. 13–31. [Google Scholar]
- Brunold C. Regulatory interactions between sulfate and nitrate assimilation. In: De Kok LJ, Stulen I, Rennenberg H, Brunold C, Rauser WE, editors. Sulfur Nutrition and Sulfur Assimilation in Higher Plants. The Hague, The Netherlands: SPB Academic Publishing; 1993. pp. 61–75. [Google Scholar]
- Brunold C, Suter M. Intracellular localization of serine acetyltransferase in spinach leaves. Planta. 1982;155:321–327. doi: 10.1007/BF00429459. [DOI] [PubMed] [Google Scholar]
- Brunold C, Suter M. Regulation of sulfate assimilation by nitrogen nutrition in the duckweed Lemna minorL. Plant Physiol. 1984;76:579–583. doi: 10.1104/pp.76.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunold C, Suter M. Adenosine 5′-phosphosulfate sulfotransferase. In: Lea P, editor. Methods in Plant Biochemistry. Vol. 3. London: Academic Press; 1990. pp. 339–343. [Google Scholar]
- Buwalda F, De Kok LJ, Stulen I. Effects of atmospheric H2S on thiol composition of crop plants. J Plant Physiol. 1993;142:281–285. [Google Scholar]
- Cacco G, Saccomani M, Ferrari G. Changes in the uptake and assimilation efficiency for sulfate and nitrate in maize hybrids selected during the period of 1930 through 1975. Physiol Plant. 1983;58:171–174. [Google Scholar]
- Gutierrez-Marcos JF, Roberts MA, Campbell EI, Wray JL. Three members of a novel smallgene-family from Arabidopsis thaliana able to complement functionally an Escherichia colimutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc Natl Acad Sci USA. 1996;93:13377–13382. doi: 10.1073/pnas.93.23.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller E, Suter M, Brunold C. Regulation of ATP-sulfurylase and adenosine 5′-phosphosulfate sulfotransferase by the sulfur and the nitrogen source in heterotrophic cell suspension cultures of Paul's scarlet rose. J Plant Physiol. 1986;125:275–286. [Google Scholar]
- Hell R, Bork C, Bogdanova N, Frolov I, Hauschild R. Isolation and characterization of two cDNAs encoding for compartment specific isoforms of O-acetylserine (thiol) lyase from Arabidopsis thaliana. FEBS Lett. 1994;351:257–262. doi: 10.1016/0014-5793(94)00872-8. [DOI] [PubMed] [Google Scholar]
- Hentschel G. Untersuchungen über die Aufnahme von 15N-markiertem Harnstoff bei Phaseolus vulgaris L. PhD thesis. Germany: University of Hohenheim/Stuttgart; 1970. [Google Scholar]
- Herschbach C, Rennenberg H. Influence of glutathione (GSH) on net uptake of sulphate and sulphate transport in tobacco plants. J Exp Bot. 1994;45:1069–1076. [Google Scholar]
- Imhof M. Die Rolle von Glutathion bei der Resistenz von Arabidopsis thaliana gegen den pathogenen Pilz Pythium paroecandrum. Diploma work. Switzerland: Institute for Plant Physiology of the University of Berne; 1994. [Google Scholar]
- Klonus D, Riesmeier JW, Willmitzer L. A cDNA clone for an ATP-sulfurylase from Arabidopsis thaliana. Plant Physiol. 1995;107:653–654. doi: 10.1104/pp.107.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Muheim R, Koprivova A, Trachsel N, Catalano C, Suter M, Brunold C. Light regulation of assimilatory sulfate reduction in Arabidopsis thaliana. Plant J. 1999;20:37–44. doi: 10.1046/j.1365-313x.1999.00573.x. [DOI] [PubMed] [Google Scholar]
- Kranner I, Grill D. Determination of glutathione and glutathione disulfide in lichens: a comparison of frequently used methods. Phytochem Anal. 1996;7:24–28. [Google Scholar]
- Kredich NM. Gene regulation of sulfur assimilation. In: De Kok LJ, Stulen I, Rennenberg H, Brunold C, Rauser WE, editors. Sulfur Nutrition and Sulfur Assimilation in Higher Plants. The Hague, The Netherlands: SPB Academic Publishing; 1993. pp. 37–47. [Google Scholar]
- Leustek T, Murillo M, Cervantes M. Cloning of a cDNA encoding ATP sulfurylase from Arabidopsis thaliana by functional expression in Saccharomyces cerevisiae. Plant Physiol. 1994;105:897–902. doi: 10.1104/pp.105.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo M, Leustek T. Adenosine-5′-triphosphate-sulfurylase from Arabidopsis thaliana and Escherichia coli are functionally equivalent but structurally and kinetically divergent: nucleotide sequence of two adenosine-5′-triphosphate-sulfurylase cDNAs from Arabidopsis thalianaand analysis of recombinant enzyme. Arch Biochem Biophys. 1995;323:195–204. doi: 10.1006/abbi.1995.0026. [DOI] [PubMed] [Google Scholar]
- Neuenschwander U, Suter M, Brunold C. Regulation of sulfate assimilation by light and O-acetyl-l-serine in Lemna minorL. Plant Physiol. 1991;97:253–258. doi: 10.1104/pp.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Dorian R, Fahey RC. Analysis of biological thiols: derivatisation with monobromobimane and separation by reversed-phase high performance liquid chromatography. Anal Biochem. 1981;114:383–387. doi: 10.1016/0003-2697(81)90498-x. [DOI] [PubMed] [Google Scholar]
- Noji M, Inoue K, Kimura N, Gouda A, Saito K. Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J Biol Chem. 1998;273:32739–32745. doi: 10.1074/jbc.273.49.32739. [DOI] [PubMed] [Google Scholar]
- Pieniazek N, Stepien PP, Paszewski A. An Aspergillus nidulans mutant lacking cystathionine-synthase: identity of l-serine sulfhydrylase with cystathionine-synthase and its distinctness from O-acetyl-l-serine sulfhydrylase. Biochim Biophys Acta. 1973;297:37–47. doi: 10.1016/0304-4165(73)90047-0. [DOI] [PubMed] [Google Scholar]
- Rennenberg H, Bergmann L. Influences of ammonium and sulfate on the production of glutathione in suspension cultures of Nicotiana tabacum. Z Pflanzenphysiol. 1979;92:133–142. [Google Scholar]
- Reuveny Z, Dougall DK, Trinity PM. Regulatory coupling of nitrate and sulfate assimilation pathways in cultured tobacco cells. Proc Natl Acad Sci USA. 1980;77:6670–6672. doi: 10.1073/pnas.77.11.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger A, Brunold C. Effect of cadmium on γ-glutamylcysteine synthesis in maize seedlings. Plant Physiol. 1992;99:428–433. doi: 10.1104/pp.99.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffet M-L, Lebrun M, Droux M, Douce R. Subcellular distribution of serine acetyltransferase from Pisum sativum and characterization of an Arabidopsis thalianaputative cytosolic isoform. Eur J Biochem. 1995;227:500–509. doi: 10.1111/j.1432-1033.1995.tb20416.x. [DOI] [PubMed] [Google Scholar]
- Saito K, Kurosawa M, Tatsuguchi K, Takagi Y, Murakoshi I. Modulation of cysteine biosynthesis in chloroplasts of transgenic tobacco overexpressing cysteine synthase [O-acetylserine(thiol)-lyase] Plant Physiol. 1994;106:887–895. doi: 10.1104/pp.106.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp R, Rennenberg H. Diurnal changes in glutathione content of spruce needles (Picea abiesL.) Plant Sci. 1988;57:113–117. [Google Scholar]
- Setya A, Murillo M, Leustek T. Sulfate reduction in higher plants: molecular evidence for a novel 5′-adenylsulfate reductase. Proc Natl Acad Sci USA. 1996;93:13383–13388. doi: 10.1073/pnas.93.23.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Hawkesford MJ, Ealing PM, Clarkson DT, van den Berg PJ, Belcher AR, Warrilow AG. Regulation of expression of a cDNA from barley roots encoding a high affinity sulphate transporter. Plant J. 1997;12:875–884. doi: 10.1046/j.1365-313x.1997.12040875.x. [DOI] [PubMed] [Google Scholar]
- Suter M, von Ballmoos P, Kopriva S, Op den Camp R, Schaller J, Kuhlemeier C, Schürmann P, Brunold C. Adenosine 5′-phosphosulfate sulfotransferase and adenosine 5′-phosphosulfate reductase are identical enzymes. J Biol Chem. 2000;275:930–936. doi: 10.1074/jbc.275.2.930. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Saito K. Subcellular localization of spinach cysteine synthase isoforms and regulation of their gene expression by nitrogen and sulfur. Plant Physiol. 1996;112:273–280. doi: 10.1104/pp.112.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, de Almeida Engler J, Engler G, Van Montagu M, Saito K. Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:11102–11107. doi: 10.1073/pnas.94.20.11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow AGS, Hawkesford MJ. Separation, subcellular location and influence of sulfur nutrition on isoforms of cysteine synthase in spinach. J Exp Bot. 1998;49:1625–1636. [Google Scholar]
- Zavgorodnyaya A, Papenbrock J, Grimm B. Yeast 5-aminolevulinate synthase provides additional chlorophyll precursor in transgenic tobacco. Plant J. 1997;12:169–178. doi: 10.1046/j.1365-313x.1997.12010169.x. [DOI] [PubMed] [Google Scholar]