Abstract
The endoplasmic reticulum (ER) not only performs its basic function of regulating calcium homeostasis, lipid biosynthesis, folding, modifying and transporting proteins but also plays a decisive role in regulating multiple cellular processes ranging from cell growth and differentiation to apoptosis and autophagy. Disturbances in ER homeostasis initiate the unfolded protein response (UPR) implicated in the pathogenesis of many human diseases. Drugging the UPR components for therapeutic interventions has received considerable attention. The purpose of this study is to identify genes that are previously unsuspected to be regulated under ER stress. Because ER stress-inducible gene expression is majorly regulated under ERSE elements, we screened human genome by adopting an in silico approach using ERSE elements (I, II, III) as probes and identified 337 candidate genes. Having knowledge of the importance of E3 ubiquitin ligase in the ERAD machinery; we validated our preliminary search by focusing on one of the hits i.e. ASB7 gene that encodes E3 ubiquitin ligase. In HeLa cells, we found that pharmacological induction of ER stress led to an increase in the expression of ASB7 with simultaneous activation of UPR pathways. Although knockdown of ASB7 expression leads to significant reduction in GRP78 and CHOP mRNA levels, it did not protect cells from ER stress-induced cell death. Also, an up-regulation in the expression of pro-inflammatory genes like TNF-α and IL-1β in ASB7 knockdown cells was observed under ER stress. Collectively, our findings suggest that ASB7 is regulated under ER stress and this study also identifies several other genes that could apparently be regulated under ER stress.
Introduction
ER is an essential organelle involved in various cellular processes including protein folding, sorting and transportation [1, 2]. Proteins enter the ER as unfolded polypeptides, from which they change into their correct conformation; then these secreted and transmembrane proteins are transported to the desired destination [3]. Cellular disturbances, inefficient clearance of misfolded proteins or change in the Ca2+ homeostasis leads to accumulation of unfolded proteins in the ER. The ER responds by increasing its protein folding capacity through specialized signaling pathways that are collectively known as the UPR which restores the ER protein homeostasis and further regulates cell survival [4, 5]. UPR increases transcription of genes encoding enzymes and chaperones involved in protein folding, secretion and degradation of misfolded proteins, and thereby constituting a coordinated regulatory mechanism that restores protein-folding in the ER and re-establishes normal cellular function [6, 7].
The UPR pathway is a highly conserved mechanism between yeast and human. UPR is a linear signaling pathway in budding yeast controlling the expression of numerous genes in response to ER stress [7]. Meanwhile, in mammalian cells, the UPR has diversified and comprises at least three parallel signaling sensors in the membrane of ER that respond to increased levels of unfolded proteins: IRE-1α (inositol-requiring kinase-1), ATF6 (activating transcription factor 6) and PERK (RNA-dependent protein kinase-like ER kinase) [8, 9]. During unstressed conditions, the ER chaperone, GRP78 binds to the luminal domains of these key regulators keeping them inactive. Upon ER stress, GRP78 dissociates from these sensors resulting in their activation [10]. IRE-1α a type I ER transmembrane kinase undergoes auto phosphorylation, which activates its intrinsic RNase activity and leads to splicing of XBP1 mRNA to produce the active transcription factor sXBP1. ATF6 is a type II ER transmembrane transcription factor which is proteolytically cleaved upon trafficking to the Golgi apparatus to generate the soluble active product, which initiates a transcriptional program to relieve ER stress. Activated PERK a type I ER transmembrane kinase phosphorylates the eukaryotic initiation factor 2 (eIF2α) on the alpha subunit, resulting in an overall attenuation of mRNA translation. Although global protein production is reduced following UPR, the translation of certain mRNAs, including the transcription factor ATF4, is increased following PERK activation. Transcription factor C/EBP homologous protein (CHOP) can activate components of the cell death and promote apoptosis downstream of the UPR [11]. CHOP expression is low in non-stressed conditions but increases in response to ER stress, hypoxia and amino acid starvation in cells [12–14]. Although most of these molecular events are clearly established, the mechanism leading to the transcriptional regulation of specific genes under ER stress remains poorly understood.
ERSE (ER stress-response element) is short sequences of DNA located within a gene promoter region that contains binding sites for specific transcription factors which regulate ER stress [15]. However, many ER stress-responsive genes possess proximal promoter regions without an ERSE sequence, such as FKBP13 [16], asparagine synthetase [17], ATF3 [18], and RTP/NDRG [19]. ERSE possess a consensus sequence of tripartite structure CCAAT-N9-CCACG, (N-Nucleotides) which is necessary for the induction of the major ER chaperones (GRP and Calreticulin) [16]. The general transcription factor, NF-Y/CBF binds to the CCAAT motif of ERSE [20–22]. While under conditions of ER stress, ATF6 binds to the CCACG motif resulting in the transcriptional induction of ER chaperones. ERSE-II (ATTGG-N-CCACG) was first found in the promoter region of Herp gene which is involved in ER stress-associated protein degradation, and it is a direct target of lumen [23]. ERSE-II was also dependent on p50ATF6, in a manner similar to that of ERSE, despite the disparate structure. Finally, the third novel ERSE like element (CCAAT-N26-CCACG) was identified by luciferase reporter assays in the human PRNP promoter regulated in ER stress by XBP1, but not via ATF6 [24]. Both IRE-1/XBP-1 and ATF6 mediate transcriptional regulation of molecular chaperones that support folding in the ER and protein degradation mechanisms [25], metabolism [26] and apoptosis [27].
In eukaryotes, ASB (Ankyrin SOCS box) gene family is the largest family of SOCS box proteins, having 18 members (ASB1–18). Most of the ASB proteins have been defined as E3 ubiquitin ligases as they contain a SOCS box domain for substrate recognition forming a subset of Elongin-Cullin-SOCS (ECS) complex [28]. Despite having 18 members in humans, only a few ASB family proteins are examined for their E3 ligase activity and few subsets of substrates identified and explored to date. All members of ASB proteins have two functional domains: an ankyrin repeat region at its amino-terminal which is involved in specific protein-protein interactions, and a SOCS box domain at its carboxyl-terminal, which function as an adaptor providing a link for the degradation of proteins which are targeted by the ankyrin repeat region. Recent studies indicate the roles of ASBs in regulating both normal and pathological conditions in various systems in different vertebrates. Some of them have been reported in various cellular functions such as ubiquitination of different proteins via SOCS box domain, as degradation of TNFR (ASB3) [29], creatine kinase B (ASB9) [30, 31] but also in regulatory like functions as the inhibition of mitochondrial function by (ASB9) [30], spermatogenesis (ASB9) [32], alteration of myoblast differentiation (ASB15) [33] and stimulation of angiogenesis (ASB5) [34]. However, it remains to be determined whether any of the mammalian ASB genes are involved in UPR signaling. The majority of ASB family members remain poorly characterized, lacking in most cases in vitro and in vivo verification of their biological functions. Hence, a better understanding of ASB interaction partners in ER stress can potentially shed light on the inexplicable physiological actions of this family and unveil elusive aspects of the ubiquitination machinery as a whole.
Several approaches have been used to identify genes induced by ER stress including microarray technique, analysis of gene expression, networking, and representational difference analysis [35–37]. However, identification of ER stress-inducible targets by gene expression profiling is highly dependent on the expression level of the target gene, potentially overlooking genes with low expression in microarray techniques. In this case, in silico approach is an effective tool for identification and determination of novel ER stress target genes. In the present study, the aim was to identify ERSE target genes in the whole human genome using Python programming with ERSE (I, II, III) elements as probes. In our study we identified 337 candidate genes; among them, we examined the role of ASB7 gene in ER stress. We show that ER stress increases the expression of ASB7 in vitro conditions. However, ASB7 knockdown had reduced GRP78 and CHOP levels but had no effect on ER stress-mediated cell death in HeLa cells. Additionally, we also provide evidence that the induction of ASB7 expression seems to be involved in the PERK/ATF4/CHOP pathway during ER stress. Our results indicate that this in silico approach is an effective tool for identification of ERSE sequence containing genes, and help determine new targets in UPR signaling and identifies ASB7 as an ER stress-responsive gene.
Materials and methods
Bioinformatic analysis
Dataset extraction and prediction of ERSE element containing genes
To find genes in the human genome containing ERSE I-CCAAT-N9-CCACG, ERSE II-ATTGG-N-CCACG and ERSE III-CCAAT-N26-CCACG (N-nucleotides), a program was written in Python. It is a text-based programming language where the programmer type text based instructions using the language syntax to determine the function of the program and get the required results [38]. The sequences of the complete human genome were downloaded from the National Center for Biotechnology Information (NCBI) Human Assembly GRCh37.p13 (June 28, 2013) [39]. The Genome Reference Consortium (GRC) was formed in 2008 to maintain the reference assembly for the human genome. Further, the graphical sequences viewer (GSV) is used here to determine the distance between the ERSE element and their closest genes. After finding the elements by python programming, each ERSE element was subjected to the GSV in find box to pick the corresponding closest gene by using the zoom out and zoom in control (S1 Fig). By using GSV we found the gene, gene ID, gene name, ERSE location in the chromosome, gene location and distance between ERSE element and gene. Additionally, we further calculated the distance between each ERSE element and the corresponding closest gene and retrieved only the ERSE element located within 10 kb upstream and 20 kb region downstream of the gene from TSS (transcription start site).
In silico promoter analysis of ASB7 and protein-protein interactions of ASB gene family
The promoter of ASB7 was scanned further for binding of transcription factors 5 kb upstream of TSS using consensus sequence for transcription factors by using ‘MEME analysis’ tool (http://meme suite.org/tools/fime) (p-value <0.0001). Additionally, ERSE-II element in ASB7 gene sequence was checked for the evolutionary conservation in mouse, rat and zebrafish genome. Besides, this ASB7 gene and its family members were checked for protein-protein interaction network between the potential UPR components by utilizing the available physical interactions in protein databases (APID interactome, Innate DB-all, Intact, MINT) to delineate the critical regulators and the network of interactions controlling the expression of ASB genes using Cytoscape [40].
Reagents and cell culture
Human cervical carcinoma (HeLa) cells were obtained from the NCCS, India. Cells were cultured in Dulbecco’s modified Eagle’s medium containing (DMEM, HiMedia) 10% (v/v) fetal bovine serum (FBS, HiMedia), 3.7 g/L sodium bicarbonate (HiMedia), 1X glutamax (GIBCO) antibiotic and antimycotic (HiMedia) (1X).
Tunicamycin and DTT were purchased from Sigma-Aldrich. In order to block PERK pathway GSK2606414 (Medchem express) was used to pretreat cells for 1 h before cells treated with tunicamycin.
Cell viability assay (MTT assay)
Cell viability was measured using the MTT assay [3-(4, 5-dimethylthiazol-2-yl)-2, 5- diphenyltetrazolium bromide] [41]. It is a colorimetric assay which detects the reduction of MTT dependent on NAD (P) H-dependent oxidoreductase enzymes in cells. Cells were seeded in a 96-well plate at 1×104 cells/well. Treated cells were incubated with 20 μl MTT (5 mg/ml) in sterilized PBS for 3 h at 37°C in a humidified 5% CO2 atmosphere. Then, the MTT solution was removed and the formazan crystals were dissolved in 200 μL / well DMSO and absorbance was read at 570 nm using a microplate reader (Bio-Rad) and 100% viability was defined as the absorbance of the control. The experiment was triplicate-performed independently. Cell survival rates were calculated according to the following equation: survival rate = [experimental absorbance value / control absorbance value] × 100%.
Gene knockdown by RNAi transfection
Human Hela cells were reverse transfected with 0.5 μM siRNAs using Lipofectamine RNAiMax (Invitrogen) in OptiMEM medium (Invitrogen), according to the manufacturer's protocol. At 48 h after transfection, the transfection medium was replaced with the fresh culture media containing chemical inducers tunicamycin and DTT for 3, 6 and 12 h for mRNA, 6 h for protein expression and 24 h for cell viability. Human siRNAs used were purchased from Thermo scientific (Ambion, USA): ASB7 siRNA 1# Sense strand: CGGAUGUUAUAUAAUUACGTT Antisense strand: CGUAAUUAUAUAACAUCCGTG, ASB7 siRNA 2# Sense strand: CGAACACACGGAACUAUGATT, Antisense strand: UCAUAGUUCCGUGUGUUCGTG and control scramble siRNA.
Quantitative RT-PCR analysis
Total RNA was extracted using RNAiso Plus reagent (Takara) from cultured cells. Subsequently, 2 μg RNA per sample was reverse transcribed using oligo (dT) and M-MLV reverse transcriptase (RT) (Takara) according to the manufacturer’s instructions. For quantification 2 μL of the resulting RT product (1:10 dilution) was used for q-PCR using 5 μM of specific primers from Sigma in conjunction with SYBR Premix Ex Taq (Takara, Siga, Japan) and AB step one instrument (Applied Biosystems). The PCR reaction was carried out for 40 thermal cycles. Expression of the target genes was analyzed by the comparative 2−ΔΔCt method and normalized using 18S rRNA levels. The primer sequence used for qRT‐PCR analysis of transcripts was as follows (Table 1):
Table 1. Primer sequences of genes.
| S.No | Gene | Forward | Reverse |
|---|---|---|---|
| 1 | 18s | GCCGCTAGAGGTGAAATTCT | CATTCTTGGCAAATGCTTTC |
| 2 | ASB7 | TGGAGTTCAAGGCTGAGGTT | GTGTTCGTGTCTGCTCGGTA |
| 3 | GRP78 | CAACCAACTGTTACAATCAAGGC | CAAAGGTGACTTCAATCTGTGG |
| 4 | CHOP | CAGAGCTGGAACCTGAGGAG | TGGATCAGTCTGGAAAAGCA |
| 5 | PERK | CAGTGGGATTTGGATGTGG | GGAATGATCATCTTATTCCCAAA |
| 6 | IRE-1 | CCATCGAGCTGTGTGCAG | TGTTGAGGGAGTGGAGGTG |
| 7 | ATF6 | TTGGCATTTATAATACTGAACTATA | TTTGATTTGCAGGGCTCAC |
| 8 | ATF4 | GTTCTCCAGCGACAAGGCTA | ATCCTGCTTGCTGTTGTTGG, |
| 9 | sXBP-1 | CCGCAGCAGGTGCAGG | GAGTCAATACCGCCAGAATCCA |
| 10 | ASB4 | ATTTCCTTGAGGCGCTAAAGTC | GCTAGGCAACCAGTAACCTTGT |
| 11 | JUN | CCAAAGGATAGTGCGATGTTT | CTGTCCCTCTCCACTGCAAC, |
| 12 | TNF-α | AACATCCAACCTTCCCAAACG | GACCCTAAGCCCCCAATTCTC |
| 13 | IL-1β | AATCTGGTACCTGTCCTGCGTGTT | TGGGTAATTTTTGGGATCTACACT |
| 14 | XBP-1 | CGCAGCACCTCAGACTACG | ATGTTCTGGAGGGGTGACAA |
| 15 | TRB3 | ATGCGAGCCACCCCTCTAGC | CTAGCCATACAGAACCACTTC |
| 16 | DR5 | GGGAGCCGCTCATGAGGAAGTTGG | GGCAAGTCTCTCTCCCAGCGTCTC |
Western blotting
Media was aspirated and cells were washed with DPBS and lysates were obtained using RIPA buffer (20 mM Tris pH-7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% Sodium deoxycholate) supplemented with 1 mM PMSF, Protease inhibitor cocktail (1X, Sigma Aldrich) and Phosphatase inhibitor cocktail (1X, Phos STOP, Roche). Cell lysates were passed several times through an insulin syringe and incubated in ice for 30 min. The cellular extracts were centrifuged for 30 min at 13000 rpm at 4°C and the supernatant was collected. Total protein was estimated by Lowry method [42]. Total cellular extracts containing 50 μg protein, were separated by electrophoresis on 10–12% (w/v) SDS-polyacrylamide gel (SDS-PAGE) after denaturation at 95°C for 5 min in sample buffer (pH-8 in mM): 100 Tris, 100 DTT, 4% (v/v) SDS, 0.2% (w/v) bromophenol blue and 20% (v/v) glycerol. Proteins were then transferred to PVDF membranes (Trans W Pall Corporation), which were further blocked for 1 h at RT with 5% skim milk in Tris buffered saline-Tween20 (TBST) for 1 h. The Blots were blocked with specific rabbit anti human antibodies: ASB7 antibody (dilution, 1:1000, Novus), GRP78 Antibody (dilution, 1:1000, Abclonal); CHOP antibody (dilution, 1:1000, Abclonal), ATF4 (dilution, 1:1000, Elabsciences), eIF2α and phospho-eIF2α (dilution, 1:1000, Cell Signaling Technology), p38 and phospho-p38 (dilution, 1:1000, Cell Signaling Technology), IRE-1α and phospho-IRE-1α (dilution,1:1000, Cell Signaling Technology) for 3 h at room temperature. Membrane was then incubated for 1 h with anti-rabbit HRP conjugated secondary antibody (CST). Immunocomplexes were visualized using enhanced chemiluminescence (ECL) chemistry and developed on film (Fuji) [43]. Equal loading and transfer was validated by probing the membranes with β-actin antibody (dilution, 1:1000, Cell Signaling Technology).
Statistical analysis
Statistical tests were performed using Graph Pad Prism5 software. All other experiments have been done twice and performed in duplicates except for MTT cell viability assay that was performed twice in triplicates. One-way ANOVA followed by bonferroni post test was applied to compare more than two groups. Two-way ANOVA followed by bonferroni post test was applied to compare multiple groups (two parameters). Results are represented as mean ± SEM and p≤0.05 is considered as significant.
Results
Genome-wide identification of ERSE elements
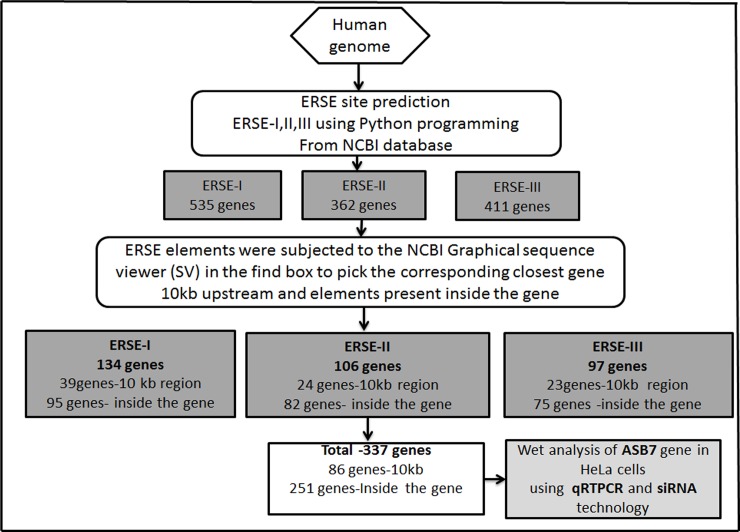
Most of the mammalian UPR-target genes contain a cis-acting ERSE element in their promoter regions. The transcription factors bind to ERSE element and transactivate UPR target genes. As a starting point to identify genes unknown to be regulated under ER stress, we used a computational approach to search for ERSE motifs within human GRC (GRCh37.p13) (Fig 1). Accordingly, Python program was written to scan the human genome using ERSE elements: ERSE-I (CCAAT-N9-CCACG), ERSE-II (ATTGG-N-CCACG) and ERSE-III (CCAAT-N26-CCACG) as probes and found 535 ERSE-I, 362 ERSE-II and 411 ERSE-III elements with their locations summarized in S1 Table. The result of the program was considered legitimate as also got known ER stress target genes like GRP94, Herp and PRNP in our list of ERSE-I, II and III containing genes. The orientation of the elements within promoters of the GRP94, Herp and PRNP are shown in S2, S3 and S4 Figs.
Fig 1. Schematic representation of strategy for identifying putative ERSE binding target genes in the human genome.
Putative ERSE binding target genes in the human genome were identified using an in silico approach. The ERSE elements were identified from the Human Assembly GRCh37.p13 using the algorithm which was implemented in a computer program written in the python programming language.
Further, using GSV we performed a systematic search to determine the gene, gene ID, ERSE location on the chromosome, gene location and distance between ERSE elements and the gene. Genes, having ERSE element beyond 20 kb regions to their upstream ends and elements which have none genes (marked it as zero in the S2 Table) in their vicinity up to 100 kb upstream region are also listed (S2 Table). Although genes containing ERSE elements within promoter region are certainly ER stress responsive, we hypothesized that ERSE elements present up to 10 kb region upstream of the gene and the elements present inside the gene may probably regulate the gene expression under ER stress. Applying these criteria, the list of genes narrowed down to 134 genes containing ERSE-I (39 genes having ERSE element within 10 kb region of TSS and for 95 genes the elements were present within the gene); 106 genes containing ERSE-II (24 genes having ERSE element within 10 kb region of TSS and for 82 genes the elements were present within the gene) and 97 genes containing ERSE-III (23 genes having ERSE element within 10 kb region of TSS and for 74 genes the elements were present within the gene) S3 Table. Thus in total 86 genes had ERSE element within 10 kb region of TSS and 251 genes the elements were present within the gene (highlighted in gray) (Table 2). These preliminary in silico results suggest that ER stress may regulate a wide variety of genes unknown to be involved in the ER stress response.
Table 2. ER stress target genes having ERSE elements less than -10 kb upstream and present inside the gene less than +20 kb downstream from TSS.
Genes having ERSE elements present inside the gene were marked in gray.
| SNO | Gene | SNO | Gene | SNO | Gene | SNO | Gene |
|---|---|---|---|---|---|---|---|
| ERSE-I | |||||||
| 1 | B3GALT6 | 35 | PCDHGA1 | 69 | DDX50 | 103 | GSE1 |
| 2 | TPRG1L | 36 | PCDHG@ | 70 | C10orf11 | 104 | ADORA2B |
| 3 | DARS2 | 37 | PCDHGA2 | 71 | CCDC147 | 105 | NMT1 |
| 4 | NOS1AP | 38 | PCDHGB1 | 72 | INPP5A | 106 | CCDC40 |
| 5 | DNM3 | 39 | PCDHGA3 | 73 | ART1 | 107 | DTNA |
| 6 | TDRD5 | 40 | PCDHGA4 | 74 | TRIM22 | 108 | KATNAL2 |
| 7 | IGFN1 | 41 | PCDHGB2 | 75 | TSPAN18 | 109 | KATNAL2 |
| 8 | DISP1 | 42 | CANX | 76 | ATG13 | 110 | KATNAL2 |
| 9 | SLC35F3 | 43 | BTNL9 | 77 | PPP6R3 | 111 | CTIF |
| 10 | LPIN1 | 44 | PPP1R3G | 78 | PPFIA1 | 112 | CELF5 (Partial stop) |
| 11 | NCOA1 | 45 | PGBD1 | 79 | FAT3 | 113 | KDM4B |
| 12 | KCNK3 | 46 | DNAH8 | 80 | FOXR1 | 114 | CYP4F12 |
| 13 | SLC9A2 | 47 | GPR111 | 81 | POU2F3 | 115 | UBA2 |
| 14 | STEAP3 | 48 | FAM135A | 82 | CACNA1C | 116 | CEACAM3 |
| 15 | MYO7B | 49 | ESR1 | 83 | COPS7A | 117 | ZNF580 |
| 16 | MZT2B | 50 | SDK1 | 84 | BICD1 | 118 | NLRP4 |
| 17 | BOK | 51 | RPS14P10 (pseudo) | 85 | GLIPR1L1 | 119 | ZNF17 |
| 18 | RPL23AP39 | 52 | CDK14 | 86 | HSP90B1 | 120 | PLCB4 |
| 19 | CNTN4 | 53 | FOXP2 | 87 | MIR3652 | 121 | SLC5A3 |
| 20 | GRM7 | 54 | MET | 88 | FICD | 122 | MRPS6 |
| 21 | ENPP7P4 (pseudo) | 55 | KCND2 | 89 | SMARCE1P5 (Pseudo) | 123 | DGCR5 (Partial stop) |
| 22 | LOC100128827 (pseudo) | 56 | CNTNAP2 | 90 | TEX29 | 124 | COMT |
| 23 | FRG2C | 57 | CHPF2 | 91 | TFDP1 | 125 | BCR |
| 24 | EIF2A | 58 | HSPD1P3 Pseudo | 92 | MIA2 | 126 | IGL |
| 25 | CLRN1-AS1 (pseudo) | 59 | CSPP1 | 93 | LIN52 | 127 | FUNDC2P4 (Pseudo) |
| 26 | KCNMB2 | 60 | SLC30A8 | 94 | GPATCH2L | 128 | GLRA2 |
| 27 | SORCS2 | 61 | LRRC14 | 95 | GABRG3 | 129 | KDM6A |
| 28 | SPATA18 | 62 | PIP5K1B | 96 | CHRNA7 (Partial stop) | 130 | ARAF |
| 29 | LPHN3 | 63 | PCSK5 | 97 | TCF12 | 131 | TIMP1 |
| 30 | MOB1B | 64 | TLE4 | 98 | FOXB1 | 132 | EDA |
| 31 | TRIM2 | 65 | NTRK2 | 99 | C15orf26 | 133 | PAK3 |
| 32 | ZSWIM6 | 66 | SUSD3 | 100 | SV2B | 134 | STAG2 |
| 33 | CKMT2 | 67 | C10orf68 | 101 | CCNF | ||
| 34 | CTNNA1 | 68 | WDFY4 | 102 | CDIPT-AS1 (Partial stop) | ||
| ERSE-II | |||||||
| 1 | PAPPA2 | 28 | ARIH2 | 55 | ENDOD1 | 82 | WIPF2 |
| 2 | ATP1A2 | 29 | BANK1 | 56 | RBM4 | 83 | DNAH9 |
| 3 | CAMTA1 (Partial stop) | 30 | TBC1D1 | 57 | SLC22A18 | 84 | DHX40 |
| 4 | RABGAP1L | 31 | C4orf22 | 58 | UBASH3B | 85 | ZNF473 |
| 5 | SGIP1 | 32 | TENM2 | 59 | TRIM22 | 86 | RUVBL2 |
| 6 | HTR6 | 33 | ZNF454 | 60 | VPS11 | 87 | QPCTL |
| 7 | FHAD1 | 34 | RASGRF2 | 61 | VPS11 | 88 | PIN1 (Partial stop) |
| 8 | KIF26B | 35 | SCGB3A2 | 62 | HS6ST3 | 89 | NFIC |
| 9 | PAPPA2 | 36 | RNF146 (Partial stop) | 63 | COL4A2 | 90 | RPS19 |
| 10 | PDE4B | 37 | C6orf201 | 64 | CCNK | 91 | CC2D1A |
| 11 | ST3GAL3 (Partial stop) | 38 | LY86 | 65 | NRXN3 | 92 | ZNF175 |
| 12 | FHAD1 | 39 | NRF1 | 66 | FAM177A1 | 93 | ARHGEF1 |
| 13 | SRGAP2 (Partial stop) | 40 | HECW1 | 67 | AHSA1 | 94 | FAM83D |
| 14 | ANKRD36 | 41 | POU6F2 | 68 | CALM1 | 95 | CDS2 |
| 15 | ITGA4 | 42 | LRGUK | 69 | NRXN3 | 96 | CDH4 |
| 16 | EPC2 | 43 | GTF2I | 70 | FBXO34 | 97 | CDH4 |
| 17 | FAM117B | 44 | EN2 | 71 | SNAP23 | 98 | DNAJC5 |
| 18 | ZNF804A | 45 | RDH10 | 72 | IGF1R | 99 | CTNNBL1 |
| 19 | GPD2 | 46 | COL27A1 | 73 | ASB7 | 100 | GRAP2 |
| 20 | LIMS1 | 47 | KANK1 | 74 | SV2B | 101 | CLCN4 |
| 21 | DIS3L2 (Partial stop) | 48 | PCSK5 | 75 | BANP | 102 | PCDH11X |
| 22 | PTCD3 | 49 | RUSC2 | 76 | WFDC1 | 103 | SMS |
| 23 | ANTXR1 | 50 | PALM2 | 77 | HERPUD1 | 104 | ZNF275 |
| 24 | CCRL2 | 51 | SLC18A2 | 78 | RBFOX1 | 105 | MAGEB10 |
| 25 | EEFSEC | 52 | DYDC2 (Partial stop) | 79 | PLCG2 | 106 | SURF6P1 (Pseudo) |
| 26 | LPP | 53 | ZNF365 | 80 | SCNN1B | ||
| 27 | HPS3 | 54 | FDX1 | 81 | (SEPT9) | ||
| ERSE-III | |||||||
| 1 | DFFB | 26 | PCDHB16 | 51 | ANO6 | 76 | FOXK2 |
| 2 | KAZN | 27 | PCDHB9 | 52 | TUBA1C | 77 | NDC80 |
| 3 | PADI1 | 28 | PPARD | 53 | NUP107 | 78 | SOGA2 |
| 4 | SZT2 | 29 | EYA4 | 54 | NAV3 | 79 | MALT1 |
| 5 | VPS25P1 (Pseudo) | 30 | ZNF713 | 55 | SYT1 | 80 | GADD45B |
| 6 | TUFT1 | 31 | DPP6 (Partial start) | 56 | TCP11L2 | 81 | MAST1 |
| 7 | DNM3 | 32 | DLGAP2 | 57 | BTBD11 | 82 | PLEKHG2 |
| 8 | CENPF | 33 | TGS1 | 58 | UBE3B | 83 | LIPE-AS1 (Partial stop) |
| 9 | LYPLAL1 | 34 | VPS13B | 59 | MVK | 84 | ZNF221 |
| 10 | LCLAT1 | 35 | DMRTA1 | 60 | TPCN1 | 85 | SULT2B1 |
| 11 | EIF5B | 36 | DAPK1 | 61 | SRRM4 | 86 | PRNP |
| 12 | GPR45 | 37 | ZNF618 | 62 | AKAP11 | 87 | GZF1 |
| 13 | RHBDD1 | 38 | DEC1 | 63 | CPB2-AS1 | 88 | CTNNBL1 |
| 14 | CRTAP | 39 | DBH | 64 | WDFY2 | 89 | PHACTR3 |
| 15 | RNF13 | 40 | GAD2 | 65 | RAD51B | 90 | CDH4 |
| 16 | POLR2H | 41 | INPP5A | 66 | NRXN3 | 91 | PCBP3 |
| 17 | LPP | 42 | ATHL1 | 67 | CTDSPL2 | 92 | IGL |
| 18 | SORCS2 | 43 | SMPD1 | 68 | SRRM2 | 93 | INPP5J |
| 19 | ADAD1 | 44 | GTF2H1 | 69 | RBFOX1 | 94 | TBC1D22A |
| 20 | MGST2 | 45 | GLYATL1 | 70 | EEF2K | 95 | TBC1D22A |
| 21 | PTGER4 | 46 | MS4A15 (Partial stop) | 71 | ITGAX | 96 | FRMPD4 |
| 22 | ZNF474 | 47 | FLRT1 | 72 | LLGL1 | 97 | ATG4A |
| 23 | LOC100505841 | 48 | C11orf53 | 73 | STXBP4 | ||
| 24 | PCDHB@ | 49 | UBASH3B | 74 | PITPNC1 | ||
| 25 | PCDHB8 | 50 | SSPN | 75 | RNF157-AS1 | ||
Promoter analysis of ASB7 and interaction with ER stress proteins
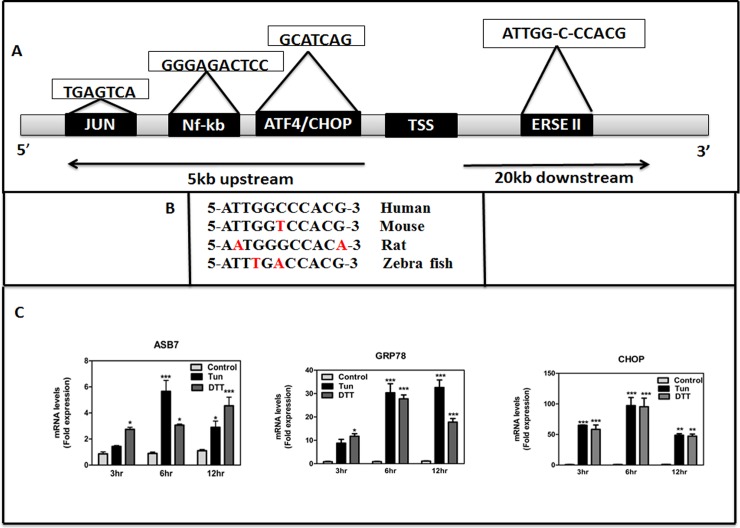
Among the ERSE containing genes identified by python programming, we were particularly interested in knowing the function of ASB7 that encodes an E3 ligase. Upregulation of E3 ubiquitin ligases is a vital mechanism through which ER stress improves degradation of misfolded ER proteins [44, 45]. Previously, ASB7 has been reported to play a crucial role in regulating spindle dynamics and genome integrity by controlling the expression of DDA3 [46]. However, the regulation of ASB7 gene in ER stress is not yet explored. Here our computational search revealed that ASB7 ERSE-II motif was in the intronic region. Looking for conservation across species is one of the best strategies for finding functional sequences in comparative genomics. Hence, we further examined whether the ERSE-II sequence is evolutionarily conserved by comparing it in the mouse, rat and zebrafish genome. As shown in Fig 1B, ERSE-II motif of mouse, rat and zebrafish ASB7 (located within the gene) were mostly homologous to that of the human ERSE motif with only 1 in mouse and 2 mismatches in rat and zebrafish, supporting the notion that this sequence is well conserved and may be functionally relevant. Additionally, we scanned the promoter region of human ASB7 for binding of transcription factors using FIMO (MEME suite) through their consensus sequences and located putative ATF/CREB binding sequence at TGAGTCA-3530 bp, NF-κB binding domain at GGGAGACTCC-2422 bp and ATF4/CHOP domain at GCATCAG-1468 bp from the TSS (Fig 2B).{Consensus sequence for ATF/CREB- TGA(C/G)TCA [47], ATF/CHOP-(GCATCAT/G) [48], NF-κB binding domain (GGGRNNYYCC)—R = purine; Y = pyrimidine [49]}.
Fig 2. In silico promoter analysis and expression of ASB7 in ER stress.
(A) Schematic representation of ASB7 gene promoter upstream (5000 bp) and downstream (20000 bp) of TSS with binding regions of ERSE-II element and transcription factor binding sequences of ATF4/CHOP, AP-1 and Nf- κB. (B) The mouse, rat and zebrafish sequence was selected as the reference species for evolutionary conservation. Numbers of mismatched nucleotides in the consensus sequence are marked in red; those conserved are marked in black. (C) HeLa cells were cultured with DMSO control (untreated cells) and concentrations of DTT (2 mM) and Tun (20 μg/ml). At 3,6 and 12 h after treatment, cells were lysed and levels of ASB7, GRP78 and CHOP mRNAs were measured by qRT-PCR and normalized relative to housekeeping gene, 18S rRNA (mean ± SEM; n = 2). “*” denotes significance with respect to control cells (DMSO treated). Error bars represent mean ± SEM. *** p≤0.001; **:p≤0.01; *: p≤0.05 calculated using two-way ANOVA with Bonferroni correction.
The function and activity of a protein are generally modulated by other proteins with which it interacts. Hence next, we wanted to investigate protein-protein interactions of ASB7 and its family members to determine whether they can interact with any of the ER stress proteins (eg. CHOP, GRP78 etc) using cytoscape open source software platform. Interestingly, from the interacting protein-protein databases we found only ASB7 in the whole ASB family to have a direct protein-protein interaction with ATF4 an ER stress transcription factor and JUN proto-oncogene (AP-1 transcription factor subunit) S4 Table. ATF4 and JUN belong to a family of DNA-binding proteins that includes the AP-1 family of transcription factors, cAMP-response element binding proteins (CREBs) and CREB-like proteins [50]. Altogether, aforementioned computational results suggest that ERSE-II present within ASB7 seem to be evolutionarily conserved which might be under the regulation of CHOP and NF-κB and ASB7 potentially interacts with transcription factors like JUN and ATF4.
ER stress increases the expression of ASB7 at the transcriptional level
To further confirm whether ERSE-II containing ASB7 gene is expressed during ER stress, we experimentally investigated its expression using chemical ER stress inducers like dithiothreitol (DTT; reduces the disulfide bridges of proteins) and tunicamycin (Tun; N-glycosylation inhibitor) in HeLa cells, a well-characterized cell line where the three major UPR pathways are active and responsive to ER stress. The expression of ASB7 was significantly elevated by both Tun (5 fold) and DTT (4 fold). The increase in ASB7 mRNA levels following stimulation with ER stress agents was evident within 6 h for Tun and 12 h for DTT after treatment (Fig 2C). To ascertain whether the treatments resulted in ER stress we checked the expression of CHOP and GRP78 mRNA (Fig 2C). These data strongly suggest that ASB7 gene expression is up regulated by ER stress in HeLa cells.
Effect of ASB7 knockdown on the expression of UPR markers and ER stress pathways
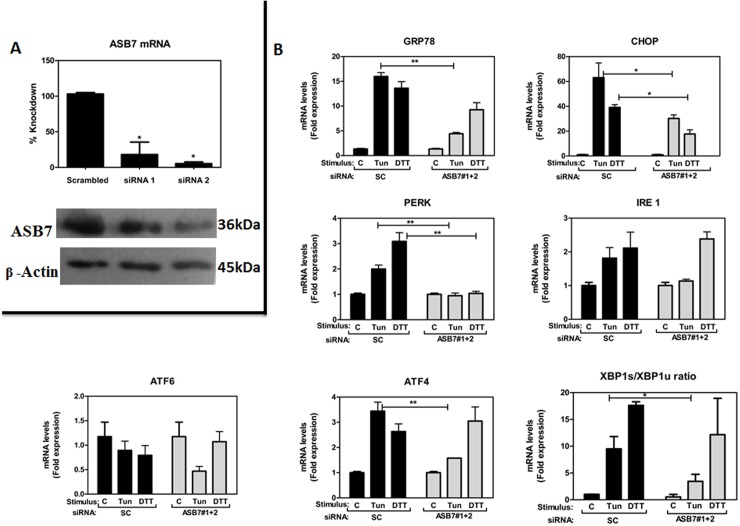
The experiments described above established that ASB7 mRNA is regulated in response to ER stress. Based on these findings, first, we examined the effect of ASB7 knockdown on upstream (GRP78) and downstream UPR signaling component (CHOP). We compared the signaling events in which ASB7 was knocked down by transfecting pooled ASB7 siRNAs to a nonspecific scramble siRNA (control) in HeLa cells. Compared to control siRNA, ASB7 knockdown reduced the expression of CHOP and GRP78 mRNA in HeLa cells treated with DTT (2 mM) and Tun (20 μg/ml). Additionally, CHOP and GRP78 protein levels were measured by western blot and GRP78 levels were dramatically reduced in cells treated with Tun (20 μg/ml) compared to CHOP which showed no significant changes.
Further to investigate the impact of ASB7 knockdown on ER stress pathway, we studied the expression level of PERK, IRE-1α or ATF6, in HeLa cells upon ER stress induction (Fig 3B). These three ER stress response branches control the expression of specific transcription factors and signaling events that modulate a variety of downstream ER stress responses, which orchestrate adaptation to ER stress. Interestingly; we found reduced PERK mRNA levels compared to IRE-1α and ATF6 mRNA levels which were not much altered with respective scramble control. Indeed, ASB7 knockdown led to a decrease in Tun induced phosphorylated eIF2α levels; at 6 h post induction (Fig 4). ATF4 which gets translated under stress conditions via PERK pathway was found to show a significant reduction in their mRNA levels but was found to be expressed more at protein levels even in only ASB7 siRNA control condition also indicating its interaction with ASB7. ATF4 is one of the several key UPR transcriptional activators which have interaction with ASB7, suggesting it may play an important role in ASB7 regulation. Next, we checked IRE-1α phosphorylation protein levels which were not much altered on ASB7 knockdown. Further, the efficiency of XBP-1 splicing in these cells was also examined. The spliced XBP-1 ratio mRNA also showed decreased levels in ASB7 knockdown cells. P38 MAPK is a key kinase activated by many cellular stress conditions including ER stress. ASB7 knockdown cells significantly displayed reduced p-p38MAPK levels (Fig 4). Taken together these findings suggest that the phosphorylation of eIF2α and p38, ATF4 and GRP78 expression is mostly influenced by the ASB7 knockdown in ER stress. The reduction in the mRNA levels of PERK, ATF4 and sXBP-1 ratio and phosphorylation of eIF2α protein level may be due to a partial shutdown of the PERK-ATF4-CHOP pathway. Overall, these results establish ASB7 might act as a mediator of ER stress signaling in vitro via PERK/ATF4/CHOP pathway and suggest a possible role for ASB7 downstream of PERK signaling pathway. qRT-PCR and western blot analysis confirmed that ASB7 protein and mRNA expression level was decreased by ASB7 siRNAs compared to control siRNA (Fig 3A).
Fig 3. Effect of ASB7 knockdown on UPR markers and pathways under ER stress.
(A) HeLa cells were transiently transfected with 2 independent ASB7 siRNA and a scrambled control (SC). After 2 days, levels of endogenous ASB7 mRNA and protein levels were measured by qRT‐PCR and immunoblotting and normalized relative to the housekeeping gene, 18S rRNA and β-actin (mean ± SEM, n = 2). (B) HeLa cells were transiently transfected pooling 2 independent ASB7 siRNAs (gray bars) and compared with scrambled siRNA as control (SC) (black bars). After 2 days, cells were cultured with DTT (2 mM) and Tun (20 μg/ml) for 3 h then total RNA was extracted. Relative levels of mRNAs encoding CHOP, GRP78, PERK, IRE-1α, ATF6 and spliced XBP-1 ratio were compared by qRT-PCR and displayed as ratios relative to a housekeeping gene (18S rRNA). “*” denotes significance with respect to scramble control. Error bars represent mean ± SEM. *** p≤0.001; **: p≤0.01; *: p≤0.05 calculated using one-way ANOVA.
Fig 4. Effect of ASB7 knockdown on protein levels of UPR markers under ER stress.
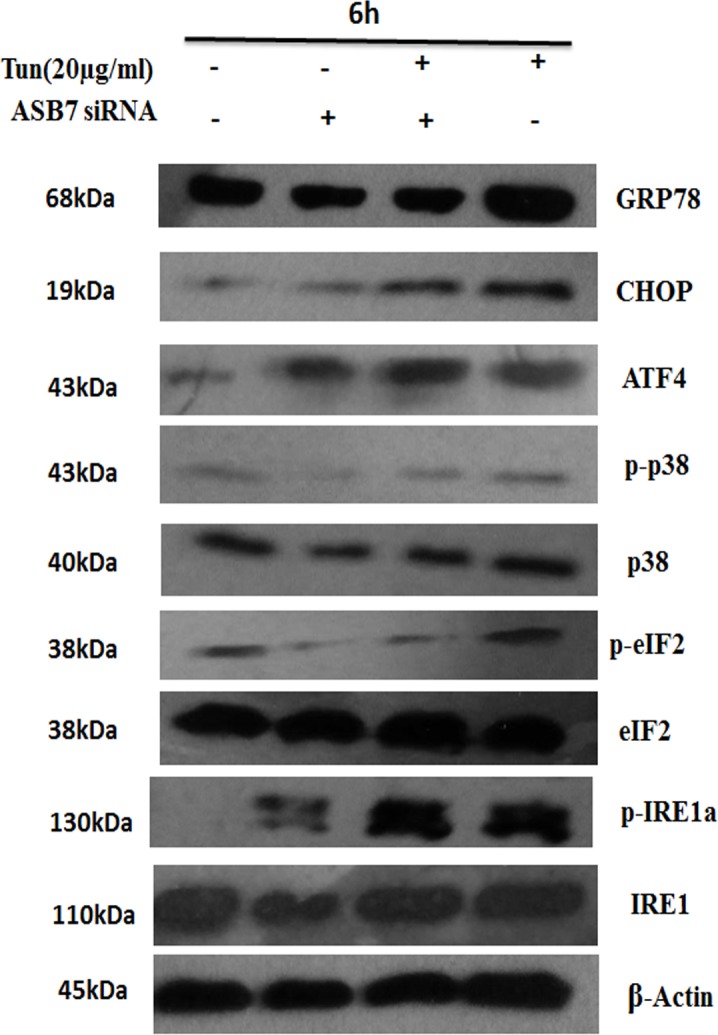
HeLa cells were transiently transfected pooling 2 independent ASB7 siRNAs, only ASB7 siRNA and compared with scrambled control. After 2 days, cells were cultured with Tun (20 μg/ml) for 6 h then total protein was extracted. Cell lysates were prepared, normalized for total protein and analyzed for immunoblotting for indicated proteins GRP78, CHOP, ATF4, IRE-1α, p-IRE-1α, p-eIF2α, eIF2α, p-p38 and p38 and normalized relative to the housekeeping gene β-Actin. Data representative of 2 experiments.
ASB7 plays an important role in the PERK/ATF4/CHOP pathway in ER stress
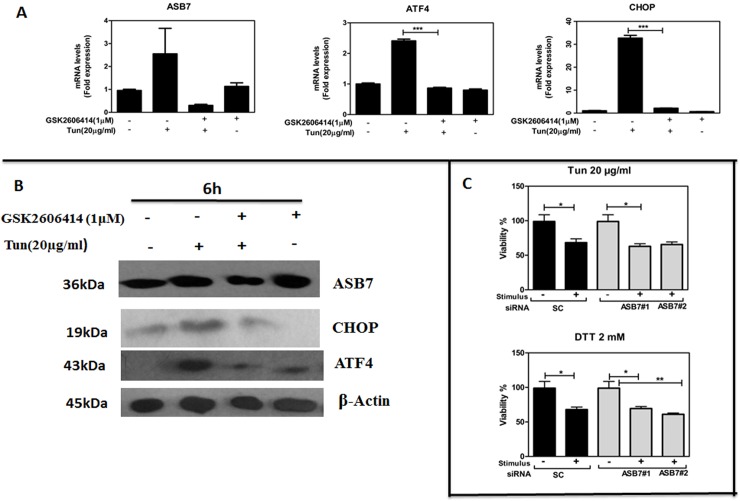
Since PERK mRNA expression and eIf2α phosphorylation was decreased on ASB7 knockdown compared to IRE-1α and ATF6 mRNA levels, we tried to investigate whether PERK pathway had its roles in the ASB7 induction. HeLa cells were treated with the ATP-competitive PERK kinase inhibitor GSK2606414 (1 μM), followed by treatment with Tun for 6 h. PERK inhibition reduced ATF4 and CHOP mRNA and protein levels, confirming PERK inhibition, and slightly attenuated ASB7 mRNA and protein expression, indicating PERK pathway can play an essential for ER stress-induced ASB7 activation (Fig 5A and 5B). Moreover, we also measured the mRNA expression levels of ATF4/CHOP signaling targeted genes, such as TRB3 and DR5 in ASB7 knockdown cells which displayed a slight but non-significant decrease in the expression of TRB3 and DR5 suggesting defective PERK/ATF4/CHOP signaling (S5 Fig). Taken together these findings suggest that, the downstream targets of PERK/ATF4/CHOP pathway are influenced by the ASB7 knockdown in ER stress. Overall, these results establish ASB7 might act as a mediator of ER stress signaling in vitro via PERK/ATF4/CHOP pathway and suggest a possible role for ASB7 downstream of PERK signaling pathway.
Fig 5. ASB7 may act downstream of PERK pathway and knockdown of ASB7 did not influence ER stress-induced cell death in HeLa cells.
(A) and (B) HeLa cells were treated with Tun (20 μg/ml, 6 h) in the absence or presence of PERK kinase inhibitor GSK2606414 (1 μM) with DMSO as the control. Total RNA and cell lysates were prepared, normalized for total protein and mRNA expression and immunoblotting of indicated proteins ASB7, CHOP and ATF4 for 6 h in HeLa cells and displayed as rations of the housekeeping gene (18s rRNA and β-actin). Data representative of 2 experiments. (C) HeLa cells were transfected with two different siRNAs targeting ASB7 and with scrambled (SC) siRNA control. After 2 days of transfection, cells were cultured with DTT (2 mM) and Tun (20 μg/ml) for 24 h. Cell viability was estimated by measuring MTT assay, expressing data as % of control untreated DMSO cells (mean ± SEM, n = 2). “ * ” denotes significance with respect to scramble and DMSO control. Error bars represent mean ± SEM. *** p≤0.001; **: p≤0.01; *: p≤0.05 calculated using one-way ANOVA.
ASB7 knockdown does not influence ER stress-induced cell death in HeLa cells
As ASB7 was up regulated in ER stress and ASB7 knockdown reduced UPR markers we investigated the ability of ASB7 to protect cells from ER stress-induced cell death in HeLa cells, we transfected HeLa cells with two different ASB7 siRNAs and a non-specific scramble siRNA (control) exposed to the ER stressors Tun (20 μg/ml) and DTT (2 mM) 48 h post transfection. MTT assay was performed after 24 h to confirm the effect of ASB7 knockdown on cell viability. We found ablation of ASB7 confers no protection in ER stress-induced cell death in HeLa cells (Fig 5C). These results suggested that ASB7 knockdown cells retain their sensitivity to ER stress-induced cytotoxicity despite the up-regulation of ASB7 in response to ER stress.
ASB7 knockdown alters pro-inflammatory markers in ER Stress
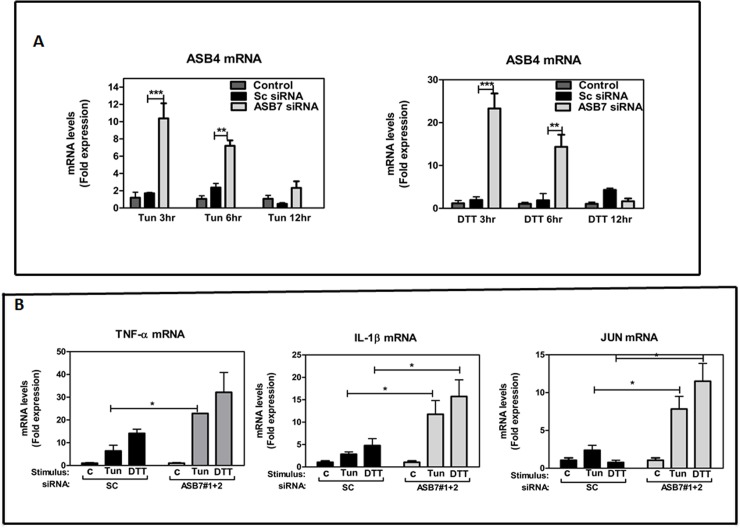
Recent research reveals connections between UPR and inflammation at multiple levels [51]. Finally, we wanted to investigate the effect of ASB7 knockdown on the regulation of ER stress-induced inflammation and the expression of proinflammatory markers. At first, we found that knockdown of ASB7 showed significant upregulation of ASB4 mRNA with respective scramble control (Fig 6A). Recent studies, identified dimers of ASB7 and ASB4, but no evidence of target-induced dimerization are reported, as the putative interactions identified do not contain overlapping proteins [52]. Previously, ASB4 has also been reported to act downstream of NF-κB in the TNF-α signaling cascade which can aid in the potential regulation of TNF-α’s numerous functions linked with inflammation, angiogenesis, and apoptosis [53]. We were particularly interested in understanding whether ASB4 upregulation influences the pro-inflammatory cytokines expression such as TNF-α, IL-1β, and IL-6 in ER stress. As shown in Fig 6B; TNF-αand IL-1β mRNA levels were strongly upregulated in the ASB7 knockdown cells, compared to the scramble control. Additionally, we checked JUN proto-oncogene AP-1 transcription factor subunit which was also found to be upregulated in ASB7 knockdown cells. Current studies indicate that there may be interactions between the AP-1 activating pathways and NF-κB pathways, in which TNF receptor associated factor (TRAF-2) (activated by TNF-α) and TRAF-6 (activated by IL-1β) may both activate NF-κB inducing kinase (NIK) and subsequently IκB kinases (IKK) [54]. Collectively, above result suggests that knockdown of ASB7 and upregulation of ASB4 in HeLa cells can be associated with alterations of immune/inflammatory players, such as pro-inflammatory cytokines, and JUN under ER stress conditions showing its signaling link between AP-1 and NF-κB signaling pathway.
Fig 6. Effect of ASB7 knockdown on pro-inflammatory markers in ER stress.
(A) HeLa cells were transiently transfected pooling 2 independent ASB7 siRNA (gray bars) and compared with scrambled siRNA control (black bars) (SC). Cells were cultured with Tun (20 μg/ml) and DTT (2 mM) and total RNA was extracted. Levels of endogenous ASB4 mRNA levels were measured by qRT‐PCR for different time points 3, 6, 12 h and normalized relative to housekeeping gene, 18S rRNA (mean ± SEM, n = 2). “*” denotes significance with respect to scramble control. Error bars represent mean ± SEM. ***: p≤0.001; **: p≤0.01; *: p≤0.05 calculated using two-way ANOVA with Bonferroni correction. (B) HeLa cells were transiently transfected pooling 2 independent ASB7 siRNA (gray bars) and compared with scrambled siRNA control (black bars) (SC). After 2 days, total RNA was extracted for 6 h. Relative levels of mRNAs encoding TNF-α (left), IL-1β (center), and JUN (right) were compared by qRT-PCR and displayed as ratios relative to a housekeeping gene (18S rRNA). Data are mean ± SEM, n = 2. “*” denotes significance with respect to scramble control. Error bars represent mean ± SEM. *** p≤0.001; **: p≤0.01; *: p≤0.05 calculated using one-way ANOVA with Bonferroni correction.
Discussion
ER stress is implicated in numerous diseases [55–57]. In spite of our current understanding of ER stress and related pathways mechanisms underlying cell decision to undergo adaptation versus death programs has been poorly understood. In this study, we performed a genome-wide search using computational programming approach to predict ER stress target genes regulated under ERSEs (I, II, II). Our results show that putative ERSE are present, on average, on every chromosome in the human genome, although it is likely that most of the genes found may or not be regulated under ER stress, at least by transactivation. In this regard, we screened genes having ERSE in the 10 kb region upstream of the promoter and inside the gene, out of which we found 337 candidate genes might be ER stress targeted genes. Most of these genes had not been identified previously as ERSE containing or ER stress-responsive genes. Nevertheless, the data obtained in the present study should be useful not only for identifying ER stress target genes but also for identifying transcriptional factors that act cooperatively and regulate their expression. Among the found candidate genes, we were interested in ASB7 gene, which belongs to ASB family, composed of 18 members. At present, knowledge on the function of the ASB members as a substrate-recognition component of the ECS complex is limited. Most of these proteins have interaction with Cul5 and Rbx2 which act as components of ubiquitin ligase complexes [58]. Ubiquitin ligases can regulate factors that improve the magnitude and duration of the UPR and thus impact overall cellular stress outcome.
In our findings, we found ERSE-II located within ASB7 in the intronic region. Previous studies have shown intron sequences to be involved in transcriptional regulation of many other genes. First known intronic enhancement of gene expression was recognized in Human alpha-1 collagen gene [59] and was further found in other genes including the CD21 [60], ALAS2 [61], delta1-crystallin [62], CRP1 [63] and Opalin gene. Mostly, intronic control of gene transcription usually involves the binding of transcription factors to the DNA sequence [64, 65]. Additionally, the sequence surrounding the ERSE in the ASB7 seems to be conserved across species. Other species have limited number of ASB proteins; in spite of this, they show very high conservation with human ASBs. Moreover, we identified several putative cis-acting elements for ATF4/CHOP, NF-κB, and AP-1, within 5 kb upstream of ASB7. Moreover, all UPR branches can be modulated at the level of ER sensors and transcription factors, through the binding of co-factors and post-translational modifications. It is thus possible that this consensus sequences close to promoter may play a role in recruiting this transcriptional co-activators to activate ASB7 gene expression.
Gene expression in response to numerous stimuli is regulated by conserved cis-elements found in the promoter region of the gene. Most UPR-target genes, such as GRP78, GRP94, and calreticulin, are fully activated by multiple copies of ERSE (CCAAT-N9-CCACG/A). We found that expression of the ASB7 gene is directly up-regulated by ER stress, which confirmed an association between ERSE and ASB7 and helps to elucidate one aspect of the biological significance of elevated ASB7 expression. During ER stress GRP78 dissociate from PERK, ATF6 and IRE-1α, and leads to downstream UPR signaling [10]. The regulatory interplay between ER stress and other pathway operates at multiple levels, suggesting that fine-tuning the balance is of importance to cellular physiology and metabolism [66]. In this study, we suggest ASB7 might collaborate with signaling enzymes or transcription factors such as ATF4 and CHOP that can regulate the PERK pathway. With regards to ER stress, ASB7 potentially could operate at several levels to impact PERK signaling. Two aspects of our findings in this report suggest that ASB7 may play a major role in PERK signaling. First, we found ASB7 has interaction with ATF4 and regulates ATF4 protein and PERK mRNA levels, GRP78 and CHOP levels, eIF2α phosphorylation and decreases induction of PERK/ATF4/CHOP target genes like TRB3 and DR5 mRNA under ER stress. Second, we found inhibition of PERK pathway using PERK inhibitor slightly reduced the levels of ASB7 protein levels in Tun treated cells. However, it could not be elucidated whether ASB7 promoted ATF4 expression directly or indirectly in this study. Comprehending detailed mechanism of ATF4 regulation by ASB7 will help us elucidate its role in ER stress. However, knockdown of ASB7 in HeLa cells did not influence cell survival in ER stress-induced cell death. Although these observations could suggest ASB7 is not required for ER stress induced cell death in HeLa cells as CHOP protein levels were not much regulated in ASB7 knockdown. All three UPR signaling events finally converge into the activation of CHOP a pro-apoptotic transcription factor that induces apoptosis by modulating the expression of several genes. The other reason may be only the inhibition of ASB7 gene alone would likely not be sufficient to protect cells during ER stress in HeLa cells.
Experimental evidence has demonstrated that synthesis of many pro-inflammatory cytokines takes place during ER stress-induced UPR signaling. It has been shown that activation of UPR itself induces inflammation through c-Jun N-terminal kinase (JNK)-activator protein-1 (AP-1) [67] and inhibitor of κB (IκB) kinase (IKK)-nuclear factor-κ B (NF-κB) pathways [68]. ASB7 can play a pro-inflammatory role comes from our following observation: first, ASB7 knockdown upregulated pro-inflammatory genes as TNF-α, IL-1β and protoncogene JUN. Studies have revealed AP-1 as a transcription factor can activate an inflammatory gene program in ER stress [69]. Also the co-activation of NF-κB and AP-1 can synergistically and effectively enhance the transcription of genes in response to a variety of stimuli [70]. Second, we also found an NF-κB binding site in the promoter region of ASB7. Binding of NF-κB to the promoter region of ASB7 can affect transcriptional regulation of several cellular and viral genes expressed in ER stress. These observations could suggest a potential role for ASB7 in inflammatory processes; although no known association of ASB7 in inflammatory response has been reported earlier. These data propose a link between ER stress and inflammation. The mechanism behind the action of ASB7 and ASB4 under ER stress on pro-inflammation is still unclear. Dimers of ASB4/ASB7 has been described, but no evidence of target-induced dimerization, as the putative interactions identified do not contain overlapping proteins, suggesting that if they bind to the same targets they will do it with different affinities. However, if similar regulation of ASB7 is demonstrated in other cell lines, it could suggest that ASB7 is potentially instrumental in ER stress-induced inflammation and UPR signaling.
In summary, we have identified 337 ERSE containing genes by in silico computational approach using python programming. Our results indicate that in silico approach is a commanding method for identification of ERSE containing genes and could serve to facilitate analysis of the function of them in UPR signaling. In particular, we have shed light on the regulation of ASB7 in vitro and highlight the complexity of the regulatory pathways involved in modulating the effects of this protein in ER stress. The involvement of ASB7 in ER stress signaling is particularly exciting as the data suggests a potential role for ASB7 in UPR and inflammatory responses which are involved in numerous disease conditions. ASB7 being an E3 ligase, it is also critical to determine the experimental substrate, cellular localization of ASB7 under ER stress. Further studies are required to determine the tissue specific functions of ASB7 in different cell lines and detailed mechanisms of how ASB7 contributes to ER stress. Our study identifies genes previously unsuspected to be involved in ER stress; however their experimental validation is demanded. Although ASB7 did not regulate ER stress induced cell death, it decreased UPR signaling events indicating its involvement in other cellular signaling pathways. We believe that our finding contributes to the existing knowledge on ER stress target genes and may open new therapeutics perspectives by providing a suitable starting point for further study of UPR target genes.
Supporting information
(DOCX)
(DOCX)
(DOC)
(DOCX)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Acknowledgments
The authors wish to greatly acknowledge VIT University for providing research facilities to carry out this work. We would also like to acknowledge Namrata Chaudhari for reviewing the manuscript. Additionally, PT and PR wish to acknowledge Department of Science and Technology (DST, Govt. of India) Young Scientist Scheme grants (YSS/2014/000915 and SB/FT/LS-419/2012 respectively), for supporting this work.
Abbreviations
- ER
Endoplasmic reticulum
- ERAD
Endoplasmic reticulum associated protein degradation
- ERSE
Endoplasmic reticulum stress element
- UPR
Unfolded protein response
- ASB7
Ankyrin repeat SOCS box protein 7
- TNF-α
Tumor necrosis factor alpha
- TNFR
Tumor necrosis factor receptor
- IL-1β
Interleukin 1 beta
- CHOP
C/EBP Homologous Protein
- GRP78
Glucose-regulated protein 78
- XBP1
X-Box binding protein 1
- ATF4
Activating transcription factor 4
- FKBP13
FK506 Binding Protein 1A
- ATF3
Activating transcription factor 3
- RTP/NDRG
Receptor transporter protein/ N-Myc Downstream Regulated
- HERP
Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member protein
- PRNP
Prion protein
- SOCS
Suppressor of cytokine signaling
- DDA3
Differential Display and Activated By P53
- TRB3
Tribbles Pseudo kinase 3
- DR5
Death receptor 5
- Cul5
Cullin 5 protein
- Rbx2
Ring box protein 2
- CD21
Complement C3d Receptor 2
- ALAS2
Aminolevulinate, Delta Synthase 2
- CRP1
Cruciform DNA-Recognizing Protein
- AP-1
Activator protein 1
- TSS
Transcription start site
- TRAF2
TNF receptor associated factor 2
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- PMSF
Phenyl methane sulfonyl fluoride or phenylmethylsulfonyl fluoride
- PVDF
Polyvinylidene difluoride
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
PT and PR wish to acknowledge Department of Science and Technology (DST, Govt. of India) Young Scientist Scheme grants (YSS/2014/000915 and SB/FT/LS-419/2012 respectively) for supporting this work.
References
- 1.Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harbor perspectives in biology. 2013;5(5):a013201 doi: 10.1101/cshperspect.a013201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleizen B, Braakman I. Protein folding and quality control in the endoplasmic reticulum. Current opinion in cell biology. 2004;16(4):343–9. doi: 10.1016/j.ceb.2004.06.012 [DOI] [PubMed] [Google Scholar]
- 3.Helenius A, Marquardt T, Braakman I. The endoplasmic reticulum as a protein-folding compartment. Trends in cell biology. 1992;2(8):227–31. [DOI] [PubMed] [Google Scholar]
- 4.Malhotra JD, Kaufman RJ, editors. The endoplasmic reticulum and the unfolded protein response. Seminars in cell & developmental biology; 2007: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang K, Kaufman RJ. The unfolded protein response A stress signaling pathway critical for health and disease. Neurology. 2006;66(1 suppl 1):S102–S9. [DOI] [PubMed] [Google Scholar]
- 6.Zhang K, Kaufman RJ. Signaling the unfolded protein response from the endoplasmic reticulum. Journal of Biological Chemistry. 2004;279(25):25935–8. doi: 10.1074/jbc.R400008200 [DOI] [PubMed] [Google Scholar]
- 7.Patil C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Current opinion in cell biology. 2001;13(3):349–55. [DOI] [PubMed] [Google Scholar]
- 8.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134 [DOI] [PubMed] [Google Scholar]
- 9.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the mammalian unfolded protein response. Annual review of cell and developmental biology. 2002;18(1):575–99. [DOI] [PubMed] [Google Scholar]
- 10.Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35(4):373–81. doi: 10.1016/j.ymeth.2004.10.010 [DOI] [PubMed] [Google Scholar]
- 11.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death & Differentiation. 2004;11(4):381–9. [DOI] [PubMed] [Google Scholar]
- 12.Zheng X, Wang X, Ma Z, Sunkari VG, Botusan I, Takeda T, et al. Acute hypoxia induces apoptosis of pancreatic β-cell by activation of the unfolded protein response and upregulation of CHOP. Cell death & disease. 2012;3(6):e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jousse C, Bruhat A, Harding HP, Ferrara M, Ron D, Fafournoux P. Amino acid limitation regulates CHOP expression through a specific pathway independent of the unfolded protein response. FEBS letters. 1999;448(2–3):211–6. [DOI] [PubMed] [Google Scholar]
- 14.Nishitoh H. CHOP is a multifunctional transcription factor in the ER stress response. Journal of biochemistry. 2012;151(3):217–9. doi: 10.1093/jb/mvr143 [DOI] [PubMed] [Google Scholar]
- 15.Roy B, Lee AS. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic acids research. 1999;27(6):1437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins Involvement of basic leucine zipper transcription factors. Journal of Biological Chemistry. 1998;273(50):33741–9. [DOI] [PubMed] [Google Scholar]
- 17.Barbosa-Tessmann IP, Chen C, Zhong C, Siu F, Schuster SM, Nick HS, et al. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. Journal of Biological Chemistry. 2000;275(35):26976–85. doi: 10.1074/jbc.M000004200 [DOI] [PubMed] [Google Scholar]
- 18.Cai Y, Zhang C, Nawa T, Aso T, Tanaka M, Oshiro S, et al. Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH2-terminal kinase and promoter response element. Blood. 2000;96(6):2140–8. [PubMed] [Google Scholar]
- 19.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes & development. 1999;13(10):1211–33. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Molecular and cellular biology. 2000;20(18):6755–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy B, Lee AS. Transduction of calcium stress through interaction of the human transcription factor CBF with the proximal CCAAT regulatory element of the grp78/BiP promoter. Molecular and cellular biology. 1995;15(4):2263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy B, Li WW, Lee AS. Calcium-sensitive transcriptional activation of the proximal CCAAT regulatory element of the grp78/BiP promoter by the human nuclear factor CBF/NF-Y. Journal of Biological Chemistry. 1996;271(46):28995–9002. [DOI] [PubMed] [Google Scholar]
- 23.Kokame K, Kato H, Miyata T. Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. Journal of Biological Chemistry. 2001;276(12):9199–205. doi: 10.1074/jbc.M010486200 [DOI] [PubMed] [Google Scholar]
- 24.Misiewicz M, Déry M-A, Foveau B, Jodoin J, Ruths D, LeBlanc AC. Identification of a novel endoplasmic reticulum stress response element regulated by XBP1. Journal of Biological Chemistry. 2013;288(28):20378–91. doi: 10.1074/jbc.M113.457242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9(6):861–70. doi: 10.1111/j.1600-0854.2008.00729.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1 a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. The Journal of cell biology. 2004;167(1):35–41. doi: 10.1083/jcb.200406136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. Journal of molecular biology. 2002;318(5):1351–65. [DOI] [PubMed] [Google Scholar]
- 28.Heuzé ML, Guibal FC, Banks CA, Conaway JW, Conaway RC, Cayre YE, et al. ASB2 is an Elongin BC-interacting protein that can assemble with Cullin 5 and Rbx1 to reconstitute an E3 ubiquitin ligase complex. Journal of Biological Chemistry. 2005;280(7):5468–74. doi: 10.1074/jbc.M413040200 [DOI] [PubMed] [Google Scholar]
- 29.Chung AS, Guan Y-j, Yuan Z-L, Albina JE, Chin YE. Ankyrin repeat and SOCS box 3 (ASB3) mediates ubiquitination and degradation of tumor necrosis factor receptor II. Molecular and cellular biology. 2005;25(11):4716–26. doi: 10.1128/MCB.25.11.4716-4726.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon S, Kim D, Rhee JW, Park J-A, Kim D-W, Kim D-S, et al. ASB9 interacts with ubiquitous mitochondrial creatine kinase and inhibits mitochondrial function. BMC biology. 2010;8(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Debrincat MA, Zhang J-G, Willson TA, Silke J, Connolly LM, Simpson RJ, et al. Ankyrin repeat and suppressors of cytokine signaling box protein asb-9 targets creatine kinase B for degradation. Journal of Biological Chemistry. 2007;282(7):4728–37. doi: 10.1074/jbc.M609164200 [DOI] [PubMed] [Google Scholar]
- 32.Lee MR, Kim SK, Kim JS, Rhim SY, Kim K-S. Expression of murine Asb-9 during mouse spermatogenesis. Molecules & Cells (Springer Science & Business Media BV). 2008;26(6). [PubMed] [Google Scholar]
- 33.McDaneld T, Spurlock D. Ankyrin repeat and suppressor of cytokine signaling (SOCS) box-containing protein (ASB) 15 alters differentiation of mouse C C myoblasts and phosphorylation of mitogen-activated protein kinase and Akt. Journal of animal science. 2008;86(11):2897–902. doi: 10.2527/jas.2008-1076 [DOI] [PubMed] [Google Scholar]
- 34.Boengler K, Pipp F, Fernandez B, Richter A, Schaper W, Deindl E. The ankyrin repeat containing SOCS box protein 5: a novel protein associated with arteriogenesis. Biochemical and biophysical research communications. 2003;302(1):17–22. [DOI] [PubMed] [Google Scholar]
- 35.Lee A-H, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Molecular and cellular biology. 2003;23(21):7448–59. doi: 10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nayak RR, Bernal WE, Lee JW, Kearns MJ, Cheung VG. Stress-induced changes in gene interactions in human cells. Nucleic acids research. 2014;42(3):1757–71. doi: 10.1093/nar/gkt999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Déry M-A, Jodoin J, Ursini-Siegel J, Aleynikova O, Ferrario C, Hassan S, et al. Endoplasmic reticulum stress induces PRNP prion protein gene expression in breast cancer. Breast cancer research. 2013;15(2):R22 doi: 10.1186/bcr3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassi S. A primer on python for life science researchers. PLoS Comput Biol. 2007;3(11):e199 doi: 10.1371/journal.pcbi.0030199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9(7):e1001091 doi: 10.1371/journal.pbio.1001091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrari M, Fornasiero MC, Isetta AM. MTT colorimetric assay for testing macrophage cytotoxic activity in vitro. Journal of immunological methods. 1990;131(2):165–72. [DOI] [PubMed] [Google Scholar]
- 42.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–75. [PubMed] [Google Scholar]
- 43.Mruk DD, Cheng CY. Enhanced chemiluminescence (ECL) for routine immunoblotting: An inexpensive alternative to commercially available kits. Spermatogenesis. 2011;1(2):121–2. doi: 10.4161/spmg.1.2.16606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101(3):249–58. [DOI] [PubMed] [Google Scholar]
- 45.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nature cell biology. 2000;2(7):379–84. doi: 10.1038/35017001 [DOI] [PubMed] [Google Scholar]
- 46.Uematsu K, Okumura F, Tonogai S, Joo-Okumura A, Alemayehu DH, Nishikimi A, et al. ASB7 regulates spindle dynamics and genome integrity by targeting DDA3 for proteasomal degradation. J Cell Biol. 2016;215(1):95–106. doi: 10.1083/jcb.201603062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulpaep EL, Boron WF, Caplan MJ, Cantley L, Igarashi P, Aronson PS, et al. Medical Physiology a Cellular and Molecular Approach. Signal Transduct. 2009;48:27. [Google Scholar]
- 48.Han J, Back SH, Hur J, Lin Y-H, Gildersleeve R, Shan J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature cell biology. 2013;15(5):481–90. doi: 10.1038/ncb2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baeuerle PA. The inducible transcription activator NF-κB: regulation by distinct protein subunits. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 1991;1072(1):63–80. [DOI] [PubMed] [Google Scholar]
- 50.Wingender E, Schoeps T, Dönitz J. TFClass: an expandable hierarchical classification of human transcription factors. Nucleic acids research. 2013;41(D1):D165–D70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaudhari N, Talwar P, Parimisetty A, Lefebvre d’Hellencourt C, Ravanan P. A molecular web: endoplasmic reticulum stress, inflammation, and oxidative stress. Frontiers in cellular neuroscience. 2014;8:213 doi: 10.3389/fncel.2014.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andresen CA, Smedegaard S, Sylvestersen KB, Svensson C, Iglesias-Gato D, Cazzamali G, et al. Protein interaction screening for the ankyrin repeats and suppressor of cytokine signaling (SOCS) Box (ASB) family identify Asb11 as a novel endoplasmic reticulum resident ubiquitin ligase. Journal of Biological Chemistry. 2014;289(4):2043–54. doi: 10.1074/jbc.M113.534602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bode M, Wu Y, Pi X, Lockyer P, Dechyapirom W, Portbury AL, et al. Regulation of ankyrin repeat and suppressor of cytokine signalling box protein 4 expression in the immortalized murine endothelial cell lines MS1 and SVR: a role for tumour necrosis factor alpha and oxygen. Cell biochemistry and function. 2011;29(4):334–41. doi: 10.1002/cbf.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eder J. Tumour necrosis factor α and interleukin 1 signalling: do MAPKK kinases connect it all? Trends in pharmacological sciences. 1997;18(4):319–22. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida H. ER stress and diseases. FEBS journal. 2007;274(3):630–58. doi: 10.1111/j.1742-4658.2007.05639.x [DOI] [PubMed] [Google Scholar]
- 56.Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death & Differentiation. 2006;13(3):385–92. [DOI] [PubMed] [Google Scholar]
- 57.Lee AS, Hendershot LM. ER stress and cancer. Cancer biology & therapy. 2006;5(7):721–2. [DOI] [PubMed] [Google Scholar]
- 58.Kohroki J, Nishiyama T, Nakamura T, Masuho Y. ASB proteins interact with Cullin5 and Rbx2 to form E3 ubiquitin ligase complexes. FEBS letters. 2005;579(30):6796–802. doi: 10.1016/j.febslet.2005.11.016 [DOI] [PubMed] [Google Scholar]
- 59.Bornstein P, McKay J, Morishima JK, Devarayalu S, Gelinas RE. Regulatory elements in the first intron contribute to transcriptional control of the human alpha 1 (I) collagen gene. Proceedings of the National Academy of Sciences. 1987;84(24):8869–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zabel MD, Weis JH. Cell-specific regulation of the CD21 gene. International immunopharmacology. 2001;1(3):483–93. [DOI] [PubMed] [Google Scholar]
- 61.Surinya KH, Cox TC, May BK. Identification and characterization of a conserved erythroid-specific enhancer located in intron 8 of the human 5-aminolevulinate synthase 2 gene. Journal of Biological Chemistry. 1998;273(27):16798–809. [DOI] [PubMed] [Google Scholar]
- 62.Kondoh H, Uchikawa M, Kamachi Y. Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation. International Journal of Developmental Biology. 2004;48(8–9):819–27. doi: 10.1387/ijdb.041868hk [DOI] [PubMed] [Google Scholar]
- 63.Najwer I, Lilly B. Ca2+/calmodulin-dependent protein kinase IV activates cysteine-rich protein 1 through adjacent CRE and CArG elements. American Journal of Physiology-Cell Physiology. 2005;289(4):C785–C93. doi: 10.1152/ajpcell.00098.2005 [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. Journal of Biological Chemistry. 2000;275(35):27013–20. doi: 10.1074/jbc.M003322200 [DOI] [PubMed] [Google Scholar]
- 65.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of thecis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. Journal of Biological Chemistry. 1999;274(4):2592–. [DOI] [PubMed] [Google Scholar]
- 66.Hasnain SZ, Lourie R, Das I, Chen AC, McGuckin MA. The interplay between endoplasmic reticulum stress and inflammation. Immunology and cell biology. 2012;90(3):260–70. doi: 10.1038/icb.2011.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–6. [DOI] [PubMed] [Google Scholar]
- 68.Tam AB, Mercado EL, Hoffmann A, Niwa M. ER stress activates NF-κB by integrating functions of basal IKK activity, IRE1 and PERK. PLoS One. 2012;7(10):e45078 doi: 10.1371/journal.pone.0045078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schonthaler HB, Guinea-Viniegra J, Wagner EF. Targeting inflammation by modulating the Jun/AP-1 pathway. Annals of the rheumatic diseases. 2011;70(Suppl 1):i109–i12. [DOI] [PubMed] [Google Scholar]
- 70.Fujioka S, Niu J, Schmidt C, Sclabas GM, Peng B, Uwagawa T, et al. NF-κB and AP-1 connection: mechanism of NF-κB-dependent regulation of AP-1 activity. Molecular and cellular biology. 2004;24(17):7806–19. doi: 10.1128/MCB.24.17.7806-7819.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOC)
(DOCX)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.