Abstract
Loss of platelet quality during ex vivo storage is a major concern in the transfusion medicine field and it has been known that platelet mitochondrial dysfunction is associated with storage time. In the last decade, small noncoding RNAs also known as microRNAs (miRNAs) have been reported to regulate key cellular processes through their target sequence interactions with selected mRNAs. In this study, we focused on understanding the mechanisms of platelet mitochondrial dysfunction during storage through miRNA regulation of mRNAs. RNA was isolated from day 0, day 5, and day 9 of stored human leukocyte-depleted platelets and subjected to differential miRNA and mRNA profiling. The miRNA profiling identified several miRNAs at low levels including a set of 12 different miR-548 family members (miR-548a-3p, miR-548aa, miR-548x, miR-548ac, miR-548c-3p, miR-603, miR-548aj, miR-548ae, miR-548z, miR-548u, miR-548al, and miR-570-3p). The mRNA profiling identified, among many, the mitochondrial ATP synthase subunit g (ATP5L) mRNA at high levels during storage. Target Scan algorithm for potential targets of miR-570-3p also identified ATP5L as one of its targets. We further identified two target sites for miR-570-3p in the 3′ untranslated region (3′UTR) of ATP5L mRNA. While ATP5L is a subunit of F0ATPase complex, its function is not established yet. Overexpression of miR-570-3p in platelets resulted in reduced levels of ATP5L mRNA and concomitant ATP loss. These experimental results provide first-time insights into the miRNA–mRNA interactions underlying mitochondrial dysfunction in ex vivo stored platelets and warrants further investigation.
Keywords: ATP5L, microarray, microRNA, platelets, storage lesion
Introduction
Platelets collected and stored under standard blood bank conditions in a concentrated form for transfusion are known as ex vivo stored platelet concentrates (PCs) and they play an important role in transfusion medicine. They are one of the life-saving transfusion products in controlling bleeding associated with surgery, trauma, and accident victims in day-to-day life. During storage under standard blood bank conditions, platelets normally start to lose their quality with time and it has been identified that one of the reasons for this loss of quality is due to mitochondrial dysfunction [1–3]. Understanding the molecular mechanisms underlying mitochondrial dysfunction and ATP regulation in platelets during storage would facilitate developing targeted strategies toward enhancing the shelf-life of ex vivo stored platelets.
Small noncoding RNAs known as microRNAs (miRNAs) have been reported to regulate key cellular processes. In the recent past, we have shown that platelets do contain miRNAs [4,5], and others have shown that mature platelets contain active miRNA-processing proteins such as endonuclease DICER1 and members of RNA-induced silencing complex (RISC), including Argonaute RISC catalytic component 2 (Ago2) [6]. The experimental evidence of miRNA-mediated regulation of mRNA in platelets [6] and the presence of miRNA:mRNA regulatory network in platelets during storage [7–14] suggest that they might be playing an important role in platelet mitochondrial dysfunction as well.
Here we report that during platelet storage, miR-570, a member of miR-548 family [15], has demonstrated its potential to target and downregulate an mRNA encoding the mitochondrial ATP synthase subunit g (ATP5L) and promotes ATP loss in platelets.
Materials and methods
Platelet samples
Platelet samples were obtained from the National Institutes of Health (NIH) blood bank, Department of Transfusion Medicine, Clinical Center, NIH (Bethesda, MD). The IRB approval for this study was exempted under FDA-RIHSC approved protocol #03-120B. Platelets were collected from healthy donors (n = 13) on different days by apheresis and stored at 22°C under agitation. The number of platelets in each sample was quantified using CELL-DYNE 3700 (Abbott Laboratories, Abbott Park, IL, USA). Five milliliters of platelet samples were withdrawn on day 0, day 5, and day 9 from platelet bag stored at 22°C under agitation. Each sample was centrifuged at 2400 rpm for 10 minutes and pellet was suspended in 1 ml of isolation buffer 1 (Ca2+ and Mg2+ free phosphate buffered saline (PBS) supplemented with 0.1% bovine serum albumin (BSA) and 2 mM ethylenediaminetetraacetic acid (EDTA), pH 7.2). These samples were centrifuged again at 800 rpm for 5 minutes and the supernatants were transferred to a new tube. Other residual contaminating leukocytes were removed by using CD45 conjugated magnetic beads pull-down (Dynabeads, Life Technologies, Carlsbad, CA, USA) followed by centrifugation. Then 100 μl of the beads prewashed with isolation buffer 1 (supplied by the vendor) were incubated with platelets at room temperature with gentle tilting and rotation for 30 minutes. Tubes were placed on magnetic stand for 2 minutes and supernatant was transferred into another tube. Samples were centrifuged at 2400 rpm for 5 minutes, supernatant was discarded and pellets were stored at −80°C for future RNA isolation.
RNA extraction
Total RNA was extracted from the platelet pellets using TRIZOL method as per manufacturer’s instruction (Life Technologies). Briefly, 1 ml of TRIZOL was added to platelet pellet and homogenized using syringe and needle. Homogenized sample was incubated at room temperature for 5 minutes and 200 μl chloroform was added. After mixing for 15 seconds, the sample was kept at room temperature for additional 2–3 minutes and centrifuged at 12 000 rpm for 15 minutes at 4°C. The top aqueous phase was transferred to another tube and the RNA was precipitated using 0.5 ml of 100% isopropanol and 2 μl glycoblue followed by overnight incubation at −20°C. Next day, samples were centrifuged at 13 000 rpm for 30 minutes and the RNA pellet was washed with 75% ethanol, air dried for 10 minutes, and resuspended in RNase-free water. RNA was quantified using NanoVue GE (GE, Pittsburgh, PA, USA) and the RNA integrity and presence of small RNAs (low molecular weight) was determined by gel-on-chip analysis using Agilent bioanalyzer.
MicroRNA arrays
Affymetrix Gene chip miRNA 3.0 arrays were used for profiling the RNA from four donors, at three different storage time points, day 0, day 5, and day 9 (12 samples). After quantifying the RNA, 300 ng of total RNA was used from each time point sample for microarray profiling. Poly-A tailing and biotin labeling of the RNA samples were performed using the FlashTag Biotin HSR RNA Labeling Kit (Affymetrix, Santa Clara, CA, USA) as per the manufacturer’s protocol. An enzyme-linked oligosorbent assays (ELOSA) was performed to confirm the labeling of the RNA. Biotin-labeled RNA samples were hybridized at 48°C for 16 hours with 60 rpm rotation in the Affymetrix Hybridization Oven 645 (Affymetrix). After hybridization, washing and staining of arrays were performed in Fluidics Station 450. The microarrays were scanned using GeneChip Scanner 3000 7G and success of the labeling and array processing was evaluated by using Expression Console software (Affymetrix). Data analysis was performed using Partek Genomic Suite software, which applies robust multichip averaging (RMA), background correction, and quantile normalization. The quantile normalization is a nonlinear normalization method based on the confirmation that there is a common distribution of intensities of all miRNAs within and across miRNA arrays (quality control aspect in array printing). Differentially expressed miRNAs were identified using analysis of variance (ANOVA) as per Partek Genomics Suite workflow recommended by Affymetrix. We also used JMP 9.0 for analysis, and performed principal component analysis and hierarchical clustering using R software.
Microarray data validation by RT-qPCR analysis
RT-qPCR validation was performed on all 13 donor samples at 3 different storage time points (39 samples), according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA, USA). Briefly, cDNA was generated from 10 ng of total RNA using specific miRNA primers from TaqMan MicroRNA Reverse Transcription Kit followed by RT-qPCR using TaqMan MicroRNA Assay with the TaqMan Universal PCR Master Mix. Only for validation (not for miRNA profiling) study, RNU6B (U6snRNA) was used as an endogenous control (used 50 ng total RNA) and fold changes were calculated using 2^ddCt method [16].
mRNA microarray
To examine the change in messenger RNA (mRNA) expression pattern during storage, total RNA extracted from two different donors samples at three different time points was used for mRNA microarray profiling using HumanRef-8 Expression BeadChip (Illumina, Inc., San Diego, CA, USA). Biotinylated cRNA was prepared using Illumina RNA Amplification Kit as per manufacturer’s instructions. Hybridization, washing, and scanning were done as per Illumina Beadstation 5006 protocol and data analysis was done using Illumina Bead Studio software. Significant genes were selected based on p-value <0.05, false discovery rate (fdr) <0.05, and fold change 1.5 (for upregulated genes) or −1.5 (for downregulated genes).
Ingenuity pathway analysis
The genes upregulated in an mRNA microarray performed in our laboratory (unpublished data) were compared with Target Scan–predicted targets of two selected miRNAs. A total of 30 genes were identified. These potential mRNA targets were analyzed using ingenuity pathway analysis (IPA). We applied IPA “Core analysis” function to determine the functional role of these potential targets. The analysis generated data showing role of these potential target genes in different biological processes, pathways, and networks. The networks were generated based on the already known functions and associations of these selected genes.
miRNA overexpression, quantification, and selected mRNA estimation in platelets
Precursor miRNA, control, and pre-miR-548u were obtained from Life Technologies. About 2 × 109 platelets per well in a 6-well plate were incubated with precursor miRNAs (100 nM concentration) at 22°C under shaking. After 48 hours of incubation, total RNA was extracted using Trizol and quantified using the RNA 6000 Nano Kit. The level of miRNAs and mRNAs were assessed using miRNA and mRNA RT-qPCR as described earlier [16]. Briefly, cDNA was generated from 10 ng of total RNA using specific miRNA primers from TaqMan® MicroRNA Assays and reagents from TaqMan® MicroRNA Reverse Transcription Kit followed by qRT-PCR using TaqMan® MicroRNA Assay with the TaqMan® Universal PCR Master Mix. RNU6B (U6snRNA) was used as an endogenous control (used 50 ng total RNA) and fold changes were calculated using 2^ddCt method. For selected mRNA, RT-PCR was performed using SuperScript® III Reverse Transcriptase kit (Thermo Fisher, Waltham, MA, USA). Briefly, 250 ng of total RNA was used to generate cDNA using SuperScript® III Reverse Transcriptase. The SYBR Green real-time PCR master mix (Thermo Fisher) and QuantStudio™ 6 (Thermo Fisher) were used for detecting real-time PCR products. The primers were designed to cross intron–exon boundaries to distinguish PCR products generated from genomic versus cDNA template. The 2^ddCt method was used to determine gene expression in each sample relative to the value observed in the control miRNA versus test miRNA expression, using GAPDH as normalization controls.
ATP assay
Luminescence-based ATP assay was performed as published earlier [17]. Briefly, 1 × 108 platelets were suspended in 100 μl PBS and mixed with equal volume of 6% Trichloroacetic acid (TCA). The lysates were diluted 1:50 with Tris-acetate buffer (pH 7.75) and 10 μl transferred to 96-well plate. ATP levels were determined in triplicate using ENLITEN ATP Assay System (Promega, Madison, WI, USA) and a luminometer (Perkin Elmer, Waltham, MA, USA).
Luciferase assay
Cultured HeLa cells in combination with a standard dual reporter expression plasmid system (Genecopoeia, Inc., Rockville, MD, USA) containing Gaussia Luciferase (GLuc) reporter gene and secreted alkaline phosphatase (SEAP) with a copy of ATP5L 3′ untranslated region (3′UTR) sequence was used to evaluate the interaction between miR-570 and its target site present in the ATP5L 3′UTR. In this system, if miR-570 binds to its target site in the 3′UTR of ATP5L mRNA, then such interaction down-regulates GLuc reporter protein expression, which can be assayed by measuring the luciferase activity. The miR-570 binding is specific to the target site and this specificity can be verified by using a miR that has a scrambled nucleotide sequence which cannot bind to miR-570 target site in the ATP5L 3′UTR and hence do not affect the reporter gene activity. This scrambled-miR is a standard negative control that has been well established in the field of miR–mRNA interaction assays [10]. Similarly if the miR-570 target site containing ATP5L 3′UTR sequence is not present in the GLuc mRNA generated by the plasmid, then again miR-570 cannot target GLuc mRNA 3′UTR and hence does not affect the reporter luciferase activity. This reporter plasmid serves as a negative control plasmid for the binding site (GLuc without ATP5L 3′UTR). In our experiments, the above described two standard negative controls were used to demonstrate the direct interaction of miR-570 with ATP5L 3′UTR.
The experiment was set up as follows: in one set, using Lipofectamine 2000 (Thermo Fisher), HeLa cells were transfected with luciferase reporter plasmid containing 3′UTR of ATP5L along with either precursor miR-570 or miRNA-control. In another set, the same reporter plasmid lacking the ATP5L 3′ UTR was used along with either precursor miR-570 or miRNA-control. After 24 hours of transfection, GLuc and SEAP luciferase activities were measured by using Secrete-Pair Dual Luminescence Assay Kit according to the manufacturer’s instruction (Genecopoeia, Inc.). Luciferase activity was measured using a luminometer (Perkin Elmer).
Statistical analysis
In this study the miRNA microarray data analysis was performed using Partek Genomic Suite software which applies RMA, background correction, and quantile normalization. The quantile normalization is a nonlinear normalization method based on the confirmation that there is a common distribution of intensities of all miRNAs within and across miRNA arrays (quality control aspect in array printing). ANOVA was applied to determine whether differentially expressed miRNAs during platelet storage are statistically significant based on p-values (p < 0.05).
The mRNA microarray data were analyzed using Illumina Bead Studio software. Significant genes were selected based on p-value <0.05 and false discovery rate (fdr) <0.05.
For qPCR, ATP assay and luciferase all experiments were performed at least in three independent sets and statistical significance was tested using t-test, and a p-value of <0.05 was considered significant.
Results
miRNA microarray profiling reveals downregulation of miR-548 family members in stored platelets
For assessing the changes in miRNAs expression in stored platelets, RNA extracted from platelet samples on day 0, day 5, and day 9 were subjected to miRNA microarrays. To identify significantly differentially expressed miRNAs during storage, miRNA microarray data were analyzed using Partek, JMP9.0, and R softwares. Figure 1 represents the methods used for profiling and analysis of miRNAs in platelets during storage. Our analysis identified differential expression of 302 platelet miRNAs on day 5 and day 9 compared with day 0 showing more than 1.5-fold changes in expression with significant p-value <0.05. Out of 302 differentially expressed miRNAs, 82 were downregulated and 220 were upregulated (Figure 1).
Figure 1.
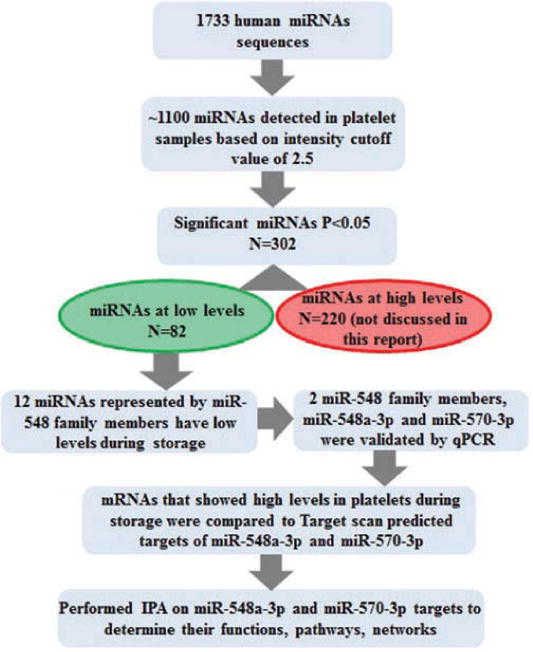
Schematic diagram showing methods applied for data analysis.
About 15% of the downregulated miRNAs (12 out of total 82) belongs to members of miR-548 family and 11 of them were clustered together. Of the total 12 miR-548 family members, 6 showed significant downregulation on both day 5 and day 9, and 6 were significantly downregulated only on day 9. All miRNAs showed progressive downregulation (Table I). Downregulation of two selected miRNAs, miR-548a-3p and miR-570-3p, was further confirmed by RT-qPCR in the same samples that were used for microarray and this result was validated further in an independent set of RNA samples from nine different donors. qPCR data showed significant downregulation of both miR-548a-3p and miR-570-3p on day 5 and day 9 validating the microarray data (Figure 2). Overall, our data showed that miR-548 family members are significantly downregulated in platelets during storage.
Table I.
List of miR-548 family members showing decreased expression in platelets samples during storage.
| miRNA |
p-Value
|
Fold change
|
p-Value (Day 9 vs. Day 0) | Fold change (Day 9 vs. Day 0) |
|---|---|---|---|---|
| (Day 5 vs. Day 0) | (Day 5 vs. Day 0) | |||
| miR-548a-3p | 0.001 | −1.5 | <0.001 | −2.6 |
| miR-548aa | 0.016 | −1.6 | <0.001 | −2.6 |
| miR-548x | 0.043 | −1.4 | 0.001 | −2.2 |
| miR-548ac | 0.042 | −1.6 | 0.001 | −3.1 |
| miR-548c-3p | 0.830 | −1.0 | 0.002 | −2.0 |
| miR-603 | 0.036 | −1.8 | 0.004 | −2.7 |
| miR-548aj | 0.390 | −1.2 | 0.008 | −2.3 |
| miR-548ae | 0.052 | −1.8 | 0.009 | −2.5 |
| miR-548z | 0.810 | −1.1 | 0.014 | −2.6 |
| miR-570 | 0.140 | −1.6 | 0.016 | −2.6 |
| miR-548u | 0.090 | −2.3 | 0.031 | −3.1 |
| miR-548al | 0.043 | −2.1 | 0.042 | −2.1 |
Bold, significant at Day 5 versus Day 0 and Day 9 versus Day 0; italic, significant at Day 9 versus Day 0.
Figure 2.
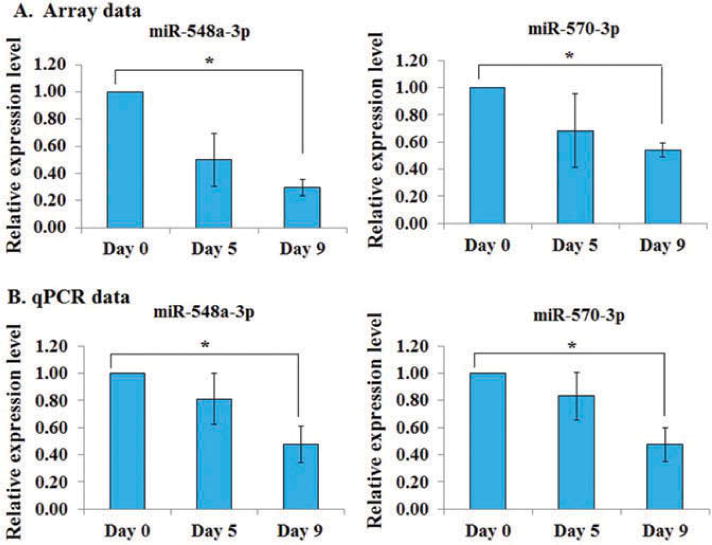
RT-qPCR validation of miR-548a-3p and miR-570. Validation in the same samples (n = 3 donors) collected on day 0, day 5, and day 9 that were used for microarray profiling and in a separate set of samples (n = 9 donors). (A) Microarray data, and (B) qPCR validation data.
In silico analysis to determine potential role of miR-548 members in platelets
In order to understand the physiological role of miR-548 family members in platelets storage, we performed in silico analysis to identify potential miR-548a and miR-570-3p targets by using mRNA microarray and predicted by Target Scan prediction algorithm. Comparison of mRNAs upregulated during storage with Target Scan–predicted targets of selected miRNAs, miR-548a-3p and miR-570-3p, identified 30 potential mRNA targets (Table II). We have utilized networking analysis tool of IPA to get further insight on the functional role of miR-548 and miR-570 in platelets during storage. Networking analysis of potential mRNA targets of miR-548a-3p and miR-570 which were found upregulated in platelets during storage identified 10 target genes involved in the network “Cellular Development, Hematological System Development and Function and Hematopoiesis” (Figure 3). These target genes are part of the pathways already known to have important functions in platelet physiology. The IPA tool also identified, mitochondrial dysfunction among the top canonical pathways (Figure 4). The mitochondrial dysfunction also topped the list in IPA-toxicity analysis of members of miR-548 targets. Platelet mRNA microarray profiling showed upregulation of mRNAs coding for CPT1A, ATP5L and COX7B as targets of miR-548, miR-570 and miR-548/miR-570 respectively, which are all involved in mitochondrial dysfunction.
Table II.
List of potential miR-548a-3p and miR-570-3p targets upregulated in platelets during storage.
| Symbol | Entrez Gene Name | miRNA |
|---|---|---|
| CCDC53 | Coiled-coil domain containing 53 | miR-548a-3p |
| CPT1A | Carnitine palmitoyltransferase 1A (liver) | miR-548a-3p |
| IL7 | Interleukin 7 | miR-548a-3p |
| LRRC15 | Leucine-rich repeat containing 15 | miR-548a-3p |
| MYL12A | Myosin, light chain 12A, regulatory, nonsarcomeric | miR-548a-3p |
| USP33 | Ubiquitin specific peptidase 33 | miR-548a-3p |
| CISD2 | CDGSH iron sulfur domain 2 | miR-548a-3p/miR-570 |
| COX7B | Cytochrome c oxidase subunit VIIb | miR-548a-3p/miR-570 |
| DNA2 | DNA replication helicase/nuclease 2 | miR-548a-3p/miR-570 |
| FAM13A | Family with sequence similarity 13, member A | miR-548a-3p/miR-570 |
| FNIP2 | Folliculin interacting protein 2 | miR-548a-3p/miR-570 |
| FZD7 | Frizzled class receptor 7 | miR-548a-3p/miR-570 |
| LCLAT1 | Lysocardiolipin acyltransferase 1 | miR-548a-3p/miR-570 |
| LIMS1 | LIM and senescent cell antigen-like domains 1 | miR-548a-3p/miR-570 |
| LYVE1 | Lymphatic vessel endothelial hyaluronan receptor 1 | miR-548a-3p/miR-570 |
| PRKACB | Protein kinase, cAMP-dependent, catalytic, beta | miR-548a-3p/miR-570 |
| ROCK1 | Rho-associated, coiled-coil containing protein kinase 1 | miR-548a-3p/miR-570 |
| ROMO1 | Reactive oxygen species modulator 1 | miR-548a-3p/miR-570 |
| TMEM55A | Transmembrane protein 55A | miR-548a-3p/miR-570 |
| UBA52 | Ubiquitin A-52 residue ribosomal protein fusion product 1 | miR-548a-3p/miR-570 |
| AKAP7 | A kinase (PRKA) anchor protein 7 | miR-570 |
| ATP5L | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G | miR-570 |
| COL8A2 | Collagen, type VIII, alpha 2 | miR-570 |
| LARP1B | La ribonucleoprotein domain family, member 1B | miR-570 |
| PPBP | Pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | miR-570 |
| PSMA6 | Proteasome (prosome, macropain) subunit, alpha type, 6 | miR-570 |
| SIPA1L3 | Signal-induced proliferation-associated 1 like 3 | miR-570 |
| SLC25A45 | Solute carrier family 25, member 45 | miR-570 |
| TRIO | Trio Rho guanine nucleotide exchange factor | miR-570 |
| ZFAND1 | Zinc finger, AN1-type domain 1 | miR-570 |
These mRNAs are also predicted as potential targets of miR-548a-3p and miR-570-3p by Target Scan algorithms.
Figure 3.
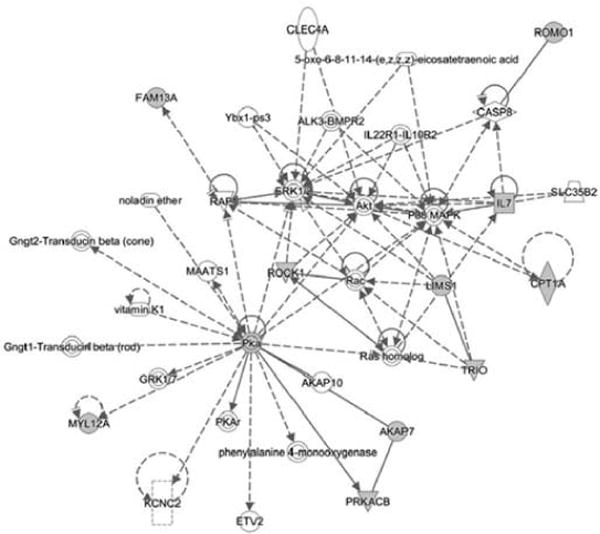
IPA-network analysis. Potential miR-548-3p and miR-570 targets revealed 10 different genes with known functions in Cellular Development, Hematological System Development and Function and Hematopoiesis.
Figure 4.
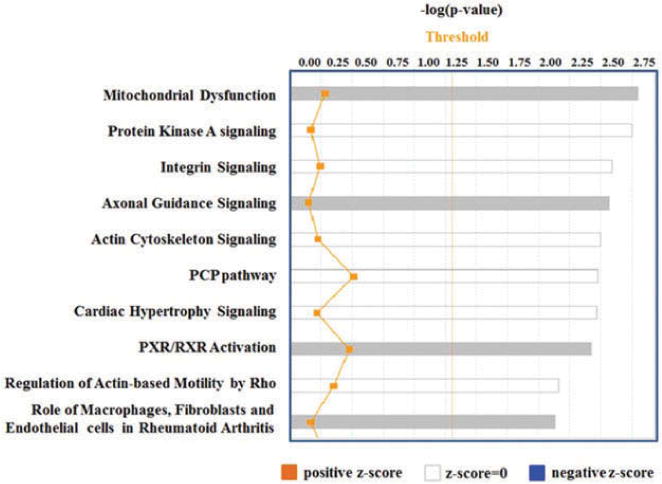
IPA-based Canonical Pathway Analysis. Note that Mitochondrial Dysfunction is at the top of 10 pathways identified.
Overexpression of miR-570-3p downregulates ATP5L mRNA and ATP levels in platelets
We have identified that during platelet storage (a) miR-548 family members stay at low levels, (b) ATP5L mRNA stays at high levels, and (c) ATP5L mRNA has two target sites for miR-570-3p (Figure 5). From these observations, we have generated a testable hypothesis that overexpression of miR-570 in platelets (i.e., perturbing the inverse relationship between the levels of miR-570 being low and ATP5L mRNA being high) should negatively affect ATP levels in platelets through miR-570 targeted regulation of ATP5L mRNA. When miR-570 was overexpressed by tranfecting platelet with precursor miR-570 (Figure 6A), we observed a reduction in ATP5L mRNA levels (~75% reduction, p < 0.05) (Figure 6B), and the same sample also demonstrated a modest reduction in platelet ATP levels (Figure 6C).
Figure 5.
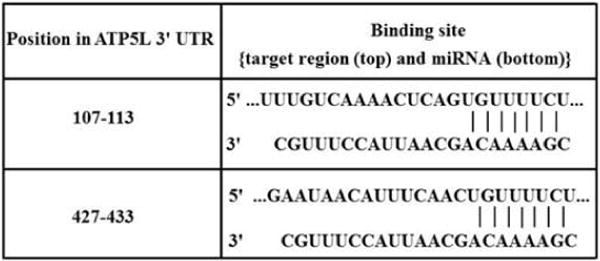
Binding sites of miR-570 in 3′UTR of ATP5L. Two target sites of miR-570 in the 3′UTR region of ATP5L mRNA as predicted by Target Scan software.
Figure 6.
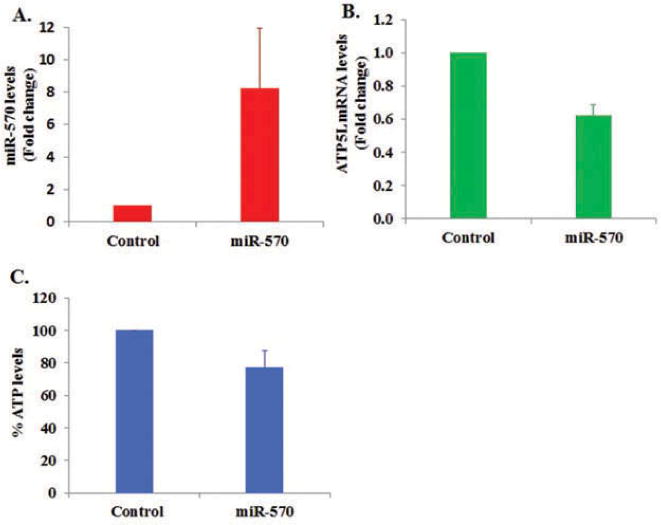
Overexpression of miR-570 in platelets and its effect on ATP5L mRNA and ATP levels in platelets. (A) Levels of miR-570 in platelets transfected with precursor miR-570 compared with nonspecific control miRNA. (B) Levels of ATP5L mRNA in platelets transfected with precursor miR-570 compared with nonspecific control miRNA. (C) Effect of miR-570 overexpression on ATP levels in platelets compared with nonspecific control miRNA.
miR-570 targets the ATP5L 3′UTR in a luciferase reporter system and downregulates the luciferase activity
To further validate that miR-570 is in fact able to target the 3′UTR of ATP5L at its target site (please refer to Figure 5) and downregulate a reporter gene such as luciferase, we co-transfected miR-570 and luciferase reporter plasmid carrying the 3′UTR of ATP5L mRNA, and after 24 hours luciferase activity was evaluated in HeLa cells. The luciferase assay results identified that there was approximately 17% reduction in luciferase activity (p < 0.05) after 24 hours of transfection. In the same set of experiments, miR-570 did not affect the luciferase activity of a negative control vector (Figure 7). The decrease in luciferase activity in miR-570 transfected cells, compared with that of control miRNA tranfected cells, suggests direct downregulation of ATP5L by miR-570. There was also no significant reduction in luciferase activity in cells transfected with control plasmid and miR-570 or control miRNA. Our data support that ATP5L is the direct target of miR-570.
Figure 7.
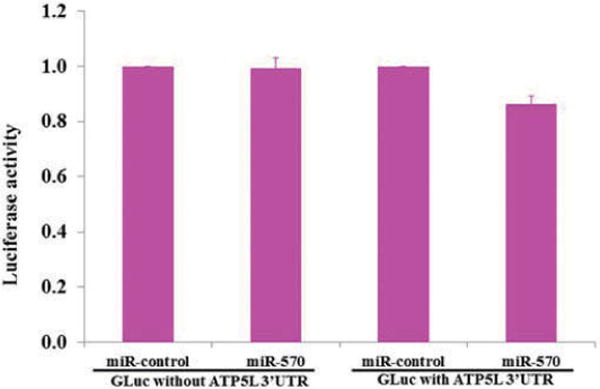
Demonstration of miR-570 targeting of ATP5L 3′UTR in a luciferase reporter system. Note that the luciferase activity was downregulated only in cells cotransfected with GLuc plasmid carrying ATP5L 3′UTR and pre-miR-570, but not in cells cotransfected with either control plasmid lacking ATP5L 3′UTR and miRNA-control or, with control plasmid lacking ATP5L 3′UTR and Pre-miR-570; miRNA-control, scrambled sequence containing miRNA.
Discussion
Early studies on platelet storage have suggested that ATP and ADP levels in platelets fall rapidly during storage [18,19]. Subsequent studies on platelet storage have identified that mitochondrial bioenergetics plays important roles in platelet activation and in the development of platelet storage lesion (PSL) [20]. Increased platelet storage time was shown to associate with mitochondrial dysfunction and impaired platelet function [1]. However, to date, molecular mechanisms underlying mitochondrial dysfunction and ATP regulation in platelets during storage have been poorly understood.
During last decade, small 22–29 nucleotide-noncoding RNAs aka miRNAs have been shown to play important post-transcriptional regulatory role in cells and such a regulatory mechanism is more apt for enucleated platelets. Therefore, in this report, we attempted to unravel the mechanisms of mitochondrial dysfunction during platelet storage by studying platelet miRNA and mRNAs during storage for up to 9 days by focusing on selected miRNAs and mRNAs that were identified in our bioinformatics analyses.
Our miRNA profiling analysis identified low levels of 12 different miR-548 family members (miR-548a-3p, miR-548aa, miR-548x, miR-548ac, miR-548c-3p, miR-603, miR-548aj, miR-548ae, miR-548z, miR-548u, miR-548al, and miR-570) beside other miRNAs (not part of this report), suggesting existence of a unique miRNA signature in platelets during storage, a feature that might serve as a biomarker of platelet quality. Downregulation of miR-548 family members has been reported earlier in dilated cardiomyopathy patients, and miR-548c was identified as a potential marker for the dilated cardiomyopathy patients [21].
The mRNA profiling and subsequent bioinformatics analysis identified among many, the ATP5L encoding mRNA as the potential target for miR-570-3p that demonstrated high levels during platelet storage. From these observations, we have generated a testable hypothesis that overexpression of miR-570 in platelets, that is, by perturbing the inverse relationship between the levels of miR-570 and ATP5L mRNA will negatively affect ATP levels in platelets through miR-570 targeted downregulation of ATP5L mRNA. We have experimentally tested this hypothesis by demonstrating that levels of platelet ATP5L mRNA and ATP go down relative to the controls, as we expressed precursor miR-570-3p in platelets. The results of luciferase assay further confirm that ATP5L is a direct target of miR-570.
In conclusion, these studies provide first insight into the miRNA–mRNA interactions underlying mitochondrial dysfunction in ex vivo stored platelets, which needs further investigation. Members of miR-548, especially miR-570, appear to regulate platelet functions, and hence miR-548 family members may serve as biomarkers of stored platelets quality. Further studies in this direction are warranted to validate these observations.
Acknowledgments
We thank Dr. Valerie W. Hu from the Department of Biochemistry and Molecular Biology, The George Washington University School of Medicine and Health Sciences, for allowing us to use the Ingenuity Pathway Analysis software.
Funding
Chintamani D. Atreya received funding for this study from the Center for Biologics Evaluation and Research (CBER), U.S. Food and Drug Administration. Neetu Dahiya and Tewarit Sarachana are recipients of a postdoctoral fellowship at the Center for Biologics Evaluation and Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration. The collaborative work performed by the coauthors Kevin G. Becker, William H. Wood, and Yongqing Zhang at the National Institute on Aging, NIH, was supported by their Intramural Research Program.
Footnotes
Neetu Dahiya conceived and designed the study, performed the experiments, did IPA analysis, and wrote the manuscript. Tewarit Sarachana assisted Neetu Dahiya with the microRNA microarray experiments described here. Chintamani D. Atreya provided training to Neetu Dahiya and Tewarit Sarachana, and also participated in designing the study and writing and editing the manuscript. Illumina microarrays were conducted in the laboratory of Kevin G. Becker, where William H. Wood and Yongqing Zhang participated in microarray experiments and data analysis, respectively. Bi-Dar Wang performed bioinformatics analysis on microRNA microarrays. Sandhya Kulkarni assisted Neetu Dahiya in obtaining samples from NIH Blood Bank and ordering laboratory supplies and reagents needed for the experiments described in this report.
Declaration of interest
The authors report no conflicts of interest.
References
- 1.Perales Villarroel JP, Figueredo R, Guan Y, Tomaiuolo M, Karamercan MA, Welsh J, Selak MA, Becker LB, Sims C. Increased platelet storage time is associated with mitochondrial dysfunction and impaired platelet function. J Surg Res. 2013;184:422–429. doi: 10.1016/j.jss.2013.05.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skripchenko A, Myrup A, Awatefe H, Thompson-Montgomery D, Wagner SJ. A rest period before agitation may improve some in vitro apheresis platelet parameters during storage. Transfusion. 2012;52:1433–1438. doi: 10.1111/j.1537-2995.2011.03493.x. [DOI] [PubMed] [Google Scholar]
- 3.Sandgren P, Stjepanovic A. High-yield platelet units revealed immediate pH decline and delayed mitochondrial dysfunction during storage in 100% plasma as compared with storage in SSP+ Vox Sang. 2012;103:55–63. doi: 10.1111/j.1423-0410.2011.01581.x. [DOI] [PubMed] [Google Scholar]
- 4.Kannan M, Mohan KV, Kulkarni S, Atreya C. Membrane array-based differential profiling of platelets during storage for 52 miRNAs associated with apoptosis. Transfusion. 2009;49:1443–1450. doi: 10.1111/j.1537-2995.2009.02140.x. [DOI] [PubMed] [Google Scholar]
- 5.Dahiya N, Sarachana T, Vu L, Becker KG, Wood WH, 3rd, Zhang Y, Atreya CD. Platelet MicroRNAs: An Overview. Transfus Med Rev. 2015;29:215–219. doi: 10.1016/j.tmrv.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16:961–966. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stakos DA, Gatsiou A, Stamatelopoulos K, Tselepis AD, Stellos K. Platelet microRNAs: From platelet biology to possible disease biomarkers and therapeutic targets. Platelets. 2013;24:579–589. doi: 10.3109/09537104.2012.724483. [DOI] [PubMed] [Google Scholar]
- 8.Osman A, Hitzler WE, Meyer CU, Landry P, Corduan A, Laffont B, Boilard E, Hellstern P, Vamvakas EC, Provost P. Effects of pathogen reduction systems on platelet microRNAs, mRNAs, activation, and function. Platelets. 2014:1–10. doi: 10.3109/09537104.2014.898178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edelstein LC, McKenzie SE, Shaw C, Holinstat MA, Kunapuli SP, Bray PF. MicroRNAs in platelet production and activation. J Thromb Haemost. 2013;11:340–350. doi: 10.1111/jth.12214. [DOI] [PubMed] [Google Scholar]
- 10.Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, Chen J, McKnight GS, López JA, Yang L, et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117:5189–5197. doi: 10.1182/blood-2010-09-299719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray PF, McKenzie SE, Edelstein LC, Nagalla S, Delgrosso K, Ertel A, Kupper J, Jing Y, Londin E, Loher P, et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics. 2013;14:1–15. doi: 10.1186/1471-2164-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu S, Deng G, Qian D, Xie Z, Sun H, Huang D, Li Q. Detection of apoptosis-associated microRNA in human apheresis platelets during storage by quantitative real-time polymerase chain reaction analysis. Blood Transfus. 2014;5:1–7. doi: 10.2450/2014.0291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osman A, Fälker K. Characterization of human platelet microRNA by quantitative PCR coupled with an annotation network for predicted target genes. Platelets. 2011;22:433–441. doi: 10.3109/09537104.2011.560305. [DOI] [PubMed] [Google Scholar]
- 14.Plé H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7:e50746. doi: 10.1371/journal.pone.0050746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang T, Guo L, Liu C. Genome-Wide Analysis of mir-548 Gene Family Reveals Evolutionary and Functional Implications. J Biomed Biotechnol. 2012;2012:679563. doi: 10.1155/2012/679563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;254:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Sarachana T, Kulkarni S, Atreya CD. Evaluation of small non-coding RNAs in ex vivo stored human mature red blood cells: Changes in noncoding RNA levels correlate with storage lesion events. Transfusion. 2015;55:2672–2683. doi: 10.1111/trf.13235. [DOI] [PubMed] [Google Scholar]
- 18.Rao AK, Niewiarowski S, Murphy S. Acquired granular pool defect in stored platelets. Blood. 1981;57:203–208. [PubMed] [Google Scholar]
- 19.Edenbrandt CM, Murphy S. Adenine and guanine nucleotide metabolism during platelet storage at 22 degrees C. Blood. 1990;76:1884–1892. [PubMed] [Google Scholar]
- 20.Saillant NN, Sims CA. Platelet dysfunction in injured patients. Mol Cell Ther. 2014;2:37. doi: 10.1186/s40591-014-0037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta MK, Halley C, Duan ZH, Lappe J, Viterna J, Jana S, Augoff K, Mohan ML, Vasudevan NT, Na J, et al. miRNA-548c: A specific signature in circulating PBMCs from dilated cardiomyopathy patients. J Mol Cell Cardiol. 2013;62:131–141. doi: 10.1016/j.yjmcc.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


