Abstract
Two integral outer envelope GTPases, Toc34 and Toc86, are proposed to regulate the recognition and translocation of nuclear-encoded preproteins during the early stages of protein import into chloroplasts. Defining the precise roles of Toc86 and Toc34 has been complicated by the inability to distinguish their GTPase activities. Furthermore, the assignment of Toc86 function is rendered equivocal by recent reports suggesting that the standard protocol for the isolation of chloroplasts results in significant proteolysis of Toc86 (B. Bolter, T. May, J. Soll [1998] FEBS Lett 441: 59–62; G. Schatz [1998] Nature 395: 439–440). We demonstrate that Toc86 corresponds to a native protein of 159 kD in pea (Pisum sativum), designated Toc159. We take advantage of the proteolytic sensitivity of Toc159 to selectively remove its 100-kD cytoplasmic GTPase domain and thereby distinguish its activities from other import components. Proteolysis eliminates detectable binding of preproteins at the chloroplast surface, which is consistent with the proposed role of Toc159 as a receptor component. Remarkably, preprotein translocation across the outer membrane can occur in the absence of the Toc159 cytoplasmic domain, suggesting that binding can be bypassed. Translocation remains sensitive to GTP analogs in the absence of the Toc159 GTP-binding domain, providing evidence that Toc34 plays a key role in the regulation of translocation by GTP.
Plant cells have evolved a complex targeting system to selectively import nuclear-encoded chloroplast proteins into the organelle after synthesis on cytoplasmic ribosomes. Import is mediated by interactions between the intrinsic N-terminal transit sequences of the preproteins and a common recognition and translocation machinery at the chloroplast envelope (Cline and Henry, 1996; Chen and Schnell, 1999). The import apparatus consists of translocon complexes at the outer (Toc complex) and inner (Tic complex) envelope membranes that cooperate to facilitate the direct transport of preproteins from the cytoplasm to the stromal compartment (Schnell et al., 1997).
The early stages of protein import are mediated by three core components of the Toc complex. The initial binding of preproteins to the Toc complex is detected in the absence of energy and appears to be a low-affinity interaction (Perry and Keegstra, 1994; Ma et al., 1996). Covalent cross-linking studies indicate that the Toc86 subunit forms the principal contact with preproteins during binding (Perry and Keegstra, 1994; Ma et al., 1996). These data, in conjunction with the observation that Toc86-specific antibodies inhibit the early stages of protein import (Hirsch et al., 1994), have led to the hypothesis that Toc86 acts as a primary receptor for preproteins at the Toc complex (Hirsch et al., 1994; Kessler et al., 1994).
Translocation across the outer membrane is an energy-dependent process, requiring both GTP and ATP hydrolysis at the chloroplast surface or within the intermembrane space (Olsen and Keegstra, 1992; Ma et al., 1996). Toc75 makes contact with regions of preproteins that are inserted across the outer membrane, indicating that it forms a major component of the protein-conducting channel in the outer membrane translocon (Ma et al., 1996; Kouranov and Schnell, 1997). This hypothesis is supported by the observation that recombinant Toc75 possesses ion channel activity when reconstituted in lipid bilayers (Hinnah et al., 1997). The requirement for ATP has been attributed to the activity of molecular chaperones that are postulated to bind and stabilize the vectorial insertion of preproteins across the outer membrane (Schnell et al., 1994; Chen and Schnell 1999). Upon insertion across the outer membrane, preproteins associate with components of the Tic machinery and translocation proceeds simultaneously across the outer and inner envelope membranes at the expense of stromal ATP (Pain and Blobel, 1987; Kouranov and Schnell 1997). These functional contact sites are formed by direct interactions between Toc and Tic components (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998).
Several lines of evidence suggest that GTP-binding and hydrolysis regulate the initial stages of translocation across the envelope membranes. Non-hydrolyzable analogs of GTP block preprotein translocation, but do not block initial binding at the Toc complex (Olsen and Keegstra 1992; Kessler et al., 1994). Toc86 and a third Toc subunit, Toc34, contain cytoplasmically exposed GTP-binding domains (Hirsch et al., 1994; Kessler et al., 1994; Seedorf et al., 1995), providing a putative site for the activity of GTP. Although Toc34 does not appear to interact directly with transit sequences, GTP binding at the Toc complex induces detectable changes in the interaction between Toc components and bound preproteins (Kouranov and Schnell, 1997). These observations have led to a model in which Toc86 and Toc34 cooperate to regulate the transition of preprotein from reversible binding to insertion in the protein-conducting channel of the Toc complex. In this scenario, Toc86 forms the primary receptor, and GTP binding and hydrolysis at one or both Toc GTPases acts as a molecular switch that regulates the entry of proteins into the translocation pathway (Kouranov and Schnell, 1997).
Recently, we and others showed that Toc86 results from the proteolysis of a larger native polypeptide (Bolter et al., 1998; Schatz, 1998). The appearance of a Toc86 ortholog in the Arabidopsis genomic sequence database prompted Bolter et al. (1998) to suggest by analogy that the native pea (Pisum sativum) protein is in fact 160 kD. In this report, we demonstrate that pea Toc86 corresponds to a native protein of 159 kD designated Toc159. We take advantage of the proteolytic sensitivity of the cytoplasmic GTPase domain of Toc159 to discriminate its activity from the Toc34 GTPase. Our results suggest that the initial binding of preproteins at Toc159 can be bypassed, and that translocation across the outer membrane in the absence of the Toc159 GTPase domain requires the activity of the Toc34 GTPase.
MATERIALS AND METHODS
cDNA Cloning and Northern-Blot Analysis
Poly(A+) RNA was purified from the leaves of 12-d-old pea (Pisum sativum var Green Arrow) seedlings using a mRNA purification system (MPG Direct, CPG, Lincoln Park, NJ) according to the manufacturer's recommendations. For northern-blot analysis, 1 μg of poly(A+) RNA was separated on a 1% (w/v) agarose/2.2 m formaldehyde gel and transferred to a nitrocellulose filter. The filter was hybridized by standard methods (Maniatis et al., 1982) to a [32P]cDNA probe (4 × 109 cpm/μg DNA) corresponding to nucleotides 2,277 to 4,725 of Toc159. This sequence encompassed the region encoding the C-terminal 816 amino acids of the original Toc86 deduced sequence (Kessler et al., 1994). The [32P]cDNA was radiolabeled using the random primer method (Feinberg and Vogelstein, 1983). The blots were washed and quantitated using a phosphor imager (Molecular Dynamics, Sunnyvale, CA).
The full-length Toc159 cDNA was isolated by three consecutive rounds of 5′-RACE (CLONTECH, Palo Alto, CA). The template for 5′-RACE was random-primed double-stranded cDNA from pea seedlings that had been ligated to short adaptor sequences. This generated three overlapping fragments that encompassed the entire Toc159 cDNA. Each fragment was cloned and sequenced by the dideoxy method using an automated DNA sequencer (ABI 373, Perkin-Elmer-Applied Biosystems, Foster City, CA).
Chloroplast Isolation and Membrane Preparation
Intact chloroplasts were isolated from 14-d-old pea seedlings by homogenization and Percoll silica gel gradient centrifugation as previously described (Pain and Blobel, 1987) with the following modifications. A protease inhibitor cocktail (no. P9599, Sigma Chemical, St. Louis) was included at all stages of the chloroplast and membrane isolation procedures at a final concentration of 5 μL mL−1 protease inhibitor cocktail. All solutions were maintained at 0°C to 2°C, and the duration of the isolation procedure was shortened to 20 min or less. Isolated chloroplasts were resuspended in 50 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-KOH, pH 7.7, and 0.33 m sorbitol (HS buffer) containing 5 μL/mL of protease inhibitor cocktail to a concentration equivalent to 2 to 3 mg chlorophyll/mL. The preparation of chloroplast envelope membranes was performed as described previously (Keegstra and Yousif, 1986).
Preprotein Binding and Import Reactions
The precursor to the small subunit of ribulose-1,5-bisphosphate carboxylase (preSSU) was synthesized in a coupled transcription-translation system containing reticulocyte lysate according to the supplier's recommendations (Promega, Madison, WI) using T7 RNA polymerase in the presence of [35S]Met. The translation mixture was gel filtered using Sephadex G-25 (Amersham-Pharmacia Biotech, Piscataway, NJ) to remove free nucleotides before use in the import reactions.
Isolated chloroplasts were sedimented at 2,000g for 1 min and washed twice with HS buffer to remove the protease inhibitor cocktail. Thermolysin-treated chloroplasts were prepared by resuspending the chloroplasts to 1 mg chlorophyll/mL in HS buffer containing 10 μg/mL thermolysin on ice for 30 min. Intact control chloroplasts were diluted to 1 mg/mL with HS buffer containing 5 μL/mL protease inhibitor mixture, and incubated for 30 min on ice. After the incubations, both thermolysin-treated and control chloroplasts were diluted with an equal volume of HS buffer containing 10 μL/mL protease inhibitor mixture and re-isolated through 40% (v/v) Percoll silica gel in HS buffer containing 5 μL/mL protease inhibitor mixture. The chloroplasts were resuspended in 2 mg chlorophyll/mL in HS buffer containing 5 μL/mL protease inhibitor mixture. The assays of energy-independent binding, the early import intermediate, and the import of [35S]preSSU were performed as described previously (Kouranov and Schnell, 1997) by diluting chloroplasts (25 μg of chlorophyll) into 150 μL of HS buffer containing 50 mm KOAc, 4 mm MgOAc, and 400 nm nigericin (import buffer). No additional protease inhibitors were added to the binding/import reactions.
The import of the envelope-bound early import intermediate was assayed by a modification of the procedure of Young et al. (1999). Chloroplasts were re-isolated from an incubation with [35S]preSSU in the presence of 25 μm ATP by sedimentation through a 40% (v/v) Percoll silica gel. The isolated chloroplasts were resuspended in 150 μL of import buffer containing 1 mm ATP. The reaction was incubated for 10 min at 26°C, and the chloroplasts were re-isolated and analyzed by SDS-PAGE. Radioactive signals in dried gels were quantitated using a phosphor imager (Molecular Dynamics).
Preprotein Cross-Linking
Preparation of the pS-1 preprotein and its modification with 125I-APDP for label-transfer cross-linking were performed as described previously (Kouranov and Schnell, 1997). Cross-linking was performed under standard conditions in a 1-mL volume containing chloroplasts equivalent to 2 mg of chlorophyll and 200 nm 125I-pS-1. Following irradiation, the chloroplasts were fractionated, and the total envelope membrane fractions were analyzed by SDS-PAGE and phosphor imager scanning.
Antibodies and Immunoblotting
Toc34, Toc75, and Toc86 antisera were prepared as described previously (Ma et al., 1996). All antibodies were affinity-purified using the corresponding recombinant antigens coupled to Sepharose prior to use (Harlow and Lane, 1988). Immunoblotting with all antibodies was performed as described previously (Ma et al., 1996).
RESULTS
Deduced Amino Acid Sequence of Toc159
As a first step in our investigation of the role of Toc34 and Toc159, we wished to obtain the full-length sequence of the pea Toc159. We optimized the chloroplast isolation procedure to minimize proteolysis of Toc159 by testing the effects of a number of commercially available protease inhibitors with a broad range of specificities. Although many of the individual reagents partially inhibited degradation, none of them was completely effective in preventing proteolysis of Toc159 to the 86-kD fragment (data not shown). Therefore, we resorted to the use of a mixture of inhibitors during all stages of the chloroplast and envelope isolation procedures. We also shortened the length of the isolation procedure and took extra care to avoid exposure of cell and chloroplast lysates to temperatures exceeding 4°C, as suggested by Bolter et al. (1998).
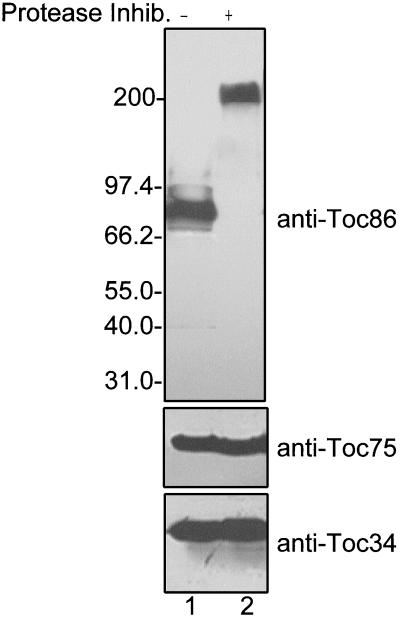
Figure 1 presents immunoblots of chloroplast envelope membrane proteins prepared by a standard isolation procedure or with the optimized procedure that includes a protease inhibitor mixture. Two components of the Toc complex, Toc34 and Toc75, were unaffected by the modified protocol (Fig. 1) indicating that they are normally resistant to proteolysis during the isolation procedure. However, the mobility of the polypeptide that reacts with anti-Toc86 serum shifted from 86 kD to approximately 200 kD on the SDS-PAGE profile (Fig. 1). These results are consistent with the observations of Bolter et al. (1998) and provide additional evidence that Toc86 corresponds to a proteolytic fragment of a larger polypeptide.
Figure 1.
Toc86 is a proteolytic fragment of a larger polypeptide. Chloroplasts were isolated from pea seedlings in the absence (−) or presence (+) of a protease inhibitor cocktail. The chloroplasts were lysed and fractionated to yield an envelope membrane fraction. Envelope membranes (25 μg of protein) were resolved by SDS-PAGE and immunoblotted with anti-Toc86 (anti-Toc86), anti-Toc75 (anti-Toc75), or anti-Toc34 (anti-Toc34) IgG. The molecular masses of known proteins are shown to the left of the figure.
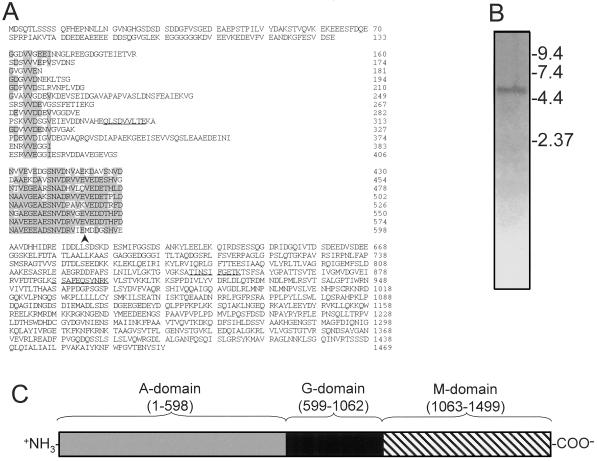
We obtained the complete cDNA sequence for Toc159 by sequencing several overlapping reverse transcriptase (RT)-PCR amplification products that encompassed the sequence corresponding to Toc86 and the previously unidentified N terminus of the polypeptide. The complete cDNA encodes a protein of 1,469 amino acids with a predicted molecular mass of 158,646 D (Fig. 2A). We refer to the polypeptide as Toc159 in accordance with the uniform nomenclature adapted for chloroplast protein import components (Schnell et al., 1997). The C-terminal 878 amino acids of the deduced sequence of Toc159, including the GTP-binding site, encompass the previously predicted 96.7-kD precursor (amino acids 591–1,469) and 86-kD mature forms (amino acids 737–1,469) of Toc86 (Fig. 2A). The sequences of three peptides obtained from the SDS-PAGE-resolved band corresponding to Toc159 are present within the cDNA, two peptides are located within the region corresponding to Toc86, and the third is located within the previously unknown N-terminal region of the Toc159 deduced sequence (Fig. 2A). These data confirm that the Toc159 cDNA corresponds to the approximately 200-kD band observed on SDS-PAGE (Fig. 1). Northern-blot analysis of pea RNA with a partial Toc159 cDNA corresponding to the sequence encoding Toc86 identifies a single mRNA of approximately 5.0 kb (Fig. 2B). This size is nearly identical to that of the 4,725-bp cDNA encoding Toc159, eliminating the remote possibility that Toc159 and Toc86 arise from different mRNAs.
Figure 2.
Deduced amino acid sequence of the pea Toc159 cDNA. A, The deduced amino acid sequence of the complete pea Toc159 cDNA. Repetitive sequences are aligned vertically and highlighted by shaded boxes. The amino acid sequences of peptides obtained from the Toc159 polypeptide are underlined. An arrowhead indicates the position of the N-terminal amino acid of the previously proposed Toc86 precursor. B, Northern-blot analysis of pea seedling poly(A+) RNA hybridized to a Toc159 cDNA 3′ fragment that corresponds to the original cDNA for Toc86. The sizes of standard RNA markers are indicated in kilobases to the right of the figure. C, Proposed tripartite domain structure of Toc159. The positions of the acidic, repetitive domain (A-domain), the GTPase domain (G-domain), and the membrane-anchor domain (M-domain) are indicated by the numbers of the corresponding amino acids in the deduced sequence in A.
Although we were able to amplify a complete 4.7-kb Toc159 cDNA from pea seedling mRNA using RT-PCR, the complete cDNA could not be stably cloned and propagated in Escherichia coli using a number of different plasmid vectors and E. coli strains (data not shown). Furthermore, three independent cDNA libraries failed to yield cDNAs that extended beyond the 5′ end of the original Toc86 cDNAs (data not shown). The cDNA instability and the fact that the site of cDNA truncation closely coincides with the site of proteolytic cleavage provide the likeliest explanation for the previous identification of Toc159 as Toc86 (Hirsch et al., 1994; Kessler et al., 1994).
Toc159 has a striking tripartite domain structure (Fig. 2C). The N-terminal 62-kD region corresponding to amino acids 1 to 598 is very acidic, with a calculated pI of 3.6; we refer to this region as the A-domain. This region was lacking from the original Toc86 cDNAs and contains two highly repetitive motifs (Fig. 2A). The first motif (amino acids 134–406) consists of 13 imperfect repeats with a consensus sequence of G(D/E) XVV(D/E)(D/E) X(V/I). The second motif (amino acids 407–598) consists of eight conserved, tandem repeats with a consensus sequence of NA(V/A) EG(E/D) A(E/D) SNVDRV(V/L/I)(E/D) D(E/D)(S/T) H(V/F/L) D. The A-domain exhibits no significant homology with proteins of known function in the databases. It contains no predicted membrane-spanning domains, suggesting that it extends into the cytoplasm from the surface of the outer envelope membrane. This prediction is consistent with the observation that thermolysin degrades Toc159 to a 52-kD C-terminal fragment (Fig. 4A). The second domain, designated the G-domain, includes amino acids 599 to 1,062 and encompasses the consensus motifs for the GTP-binding site. Amino acids 1,063 to 1,469 correspond to the protease-resistant membrane anchor of the protein (Hirsch et al., 1994; Kessler et al., 1994) (Fig. 3A); we designate this region the M-domain.
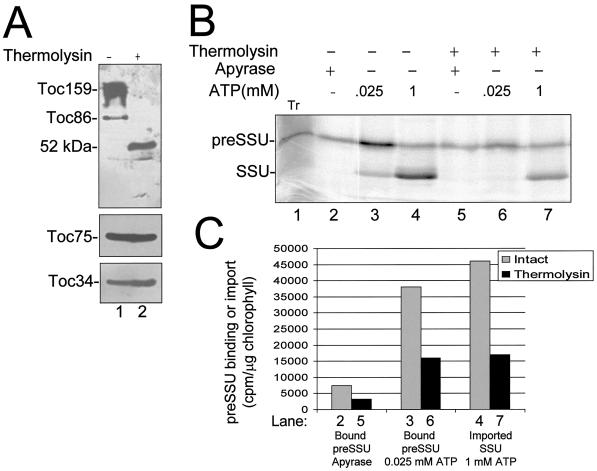
Figure 4.
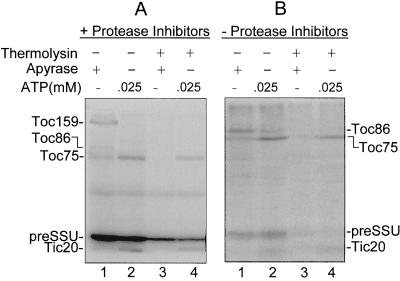
Effects of Toc159 proteolysis on preprotein binding and import. Intact chloroplasts or chloroplasts treated with 10 μg/mL thermolysin were incubated with in vitro-synthesized [35S]preSSU in a standard import reaction containing 25 μm ATP or 1 mm ATP as indicated. The chloroplasts were re-isolated and analyzed by SDS-PAGE. A, Immunoblots of envelope membrane fractions from thermolysin-treated (+) or control (−) chloroplasts. Membrane proteins (25 μg of chlorophyll) were resolved by SDS-PAGE and immunoblotted with affinity-purified antibodies to Toc86, Toc75, and Toc34. The positions of the known Toc components and the 52-kD membrane-protected fragment of Toc159 (52 kD) are indicated to the left. Tr, 10% of the [35S]preSSU translation product added to each reaction. B, Phosphor imager analysis of [35S]preSSU import. C, Quantitative analysis of the data presented in B. The numbers of the lanes in B that were used for quantitation are indicated at the bottom of the graph in C.
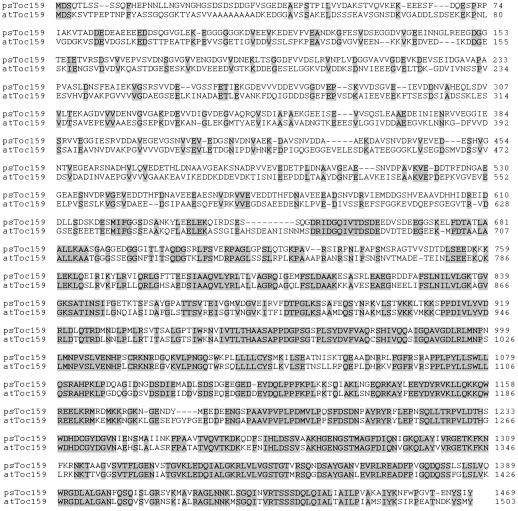
Figure 3.
Comparison of the amino acid sequences of pea and Arabidopsis Toc159. The complete deduced amino acid sequences of the pea Toc159 (psToc159) cDNA and a predicted gene encoding a related protein in Arabidopsis (atToc159) (GenBank accession no. AF069298) are aligned to maximize the occurrence of identical residues. Identical amino acids are highlighted by shaded boxes.
Overall, Toc159 exhibits 48% identity (Fig. 3) to the deduced amino acid sequence of a related Arabidopsis genomic sequence (GenBank accession no. AF069298). The highest degree of identity is found within their C-terminal 820 amino acids (72% identity), which includes the G- and M-domains, as noted by Bolter et al. (1998). However, the previously unknown N-terminal A-domain of the pea protein exhibits only 20% identity with the predicted Arabidopsis sequence. Although the A-domains of both proteins are acidic, the repetitive motifs of the predicted Arabidopsis sequence are less pronounced than those of pea Toc159. The Arabidopsis protein has a predicted size of 161 kD. Bolter et al. (1998) predicted that the intact pea Toc86 might correspond in size to the predicted Arabidopsis sequence, and proposed to change the designation of Toc86 to Toc160 on this basis. However, our pea cDNA sequence clearly encodes a protein of 159 kD, and therefore we prefer to retain the designation Toc159, which is in accordance with the adopted nomenclature for chloroplast protein import components (Schnell et al., 1997).
Effects of Toc159 Proteolysis on Protein Import
The hypersensitivity of Toc159 to proteolysis presented an opportunity to distinguish its activities from other Toc components. However, the variability in the degree of Toc159 proteolysis in standard chloroplast isolation procedures made comparisons with chloroplasts prepared with the optimized protocol unreliable. Therefore, we sought to carry out the degradation of Toc159 under controlled conditions. Chloroplasts were purified using the modified isolation procedure to minimize proteolysis of Toc159. Under these conditions, 70% to 80% of Toc159 existed in its full-length form, with the remaining protein migrating at the position of Toc86 (Fig. 4A, lane 1). We defined conditions to selectively remove the A- and G-domains of Toc159 by treating intact chloroplasts with low concentrations of the outer-membrane-impermeable protease thermolysin (Cline et al., 1984).
Treatment of chloroplasts with 10 μg/mL thermolysin for 30 min on ice resulted in the complete degradation of Toc159 to a 52-kD fragment (Fig. 4A, lane 2). The 52-kD fragment was previously shown to correspond to the C-terminal M-domain (residues 1063–1469 of Toc159) (Hirsch et al., 1994). Intact Toc159 was undetectable over a broad range of chloroplast protein concentrations after thermolysin treatment, indicating that the proteolysis of the protein was complete (data not shown). Two additional components of the outer membrane import complex, Toc34 and Toc75, were resistant to digestion at the low levels of thermolysin used in the experiment (Fig. 4A), indicating that digestion does not result in complete degradation of the Toc complex. Thermolysin treatment did not affect known components of the inner membrane import apparatus (data not shown).
We compared the effects of proteolysis on translocation across the outer and inner membranes using in vitro import assays consisting of isolated chloroplasts and in vitro-synthesized preSSU (Kouranov and Schnell, 1997). Three steps in the in vitro import of preSSU were assayed. Energy-independent binding of the preprotein was assayed using ATP-depleted chloroplasts in the presence of the nucleoside-triphosphate-hydrolyzing enzyme apyrase. In previous studies, preSSU binding under these conditions occurred primarily through contacts with Toc86 (Perry and Keegstra, 1994; Ma et al., 1996), and was shown to be readily reversible (Friedman and Keegstra, 1989). Translocation across the outer membrane was assayed in the presence of low concentrations of ATP and was monitored by the appearance of an envelope-bound early import intermediate. Preproteins trapped at this stage are partially inserted across the outer membrane and are in contact with components of the Tic machinery at the inner envelope membrane (Ma et al., 1996; Kouranov and Schnell, 1997). An optimal ATP concentration of 25 μm was determined for maximal formation of the preSSU early import intermediate in our studies (data not shown).
Finally, translocation across the inner membrane was performed in the presence of 1 mm ATP, and was assayed by the appearance of mature SSU that had been fully imported to the stroma and processed by the stromal processing peptidase. Figure 4B shows that thermolysin treatment decreased the levels of all stages of import. Binding, outer membrane translocation, and import decreased by 57%, 59%, and 69%, respectively (Fig. 4C). Thus, proteolytic treatments of the Toc complex that remove the N-terminal region of Toc159 significantly decrease the efficiency of import.
Toc86 is proposed to mediate the initial energy-independent binding of preproteins at the chloroplast surface (Perry and Keegstra, 1994; Ma et al., 1996). Therefore, the effect of proteolysis on import could result from a selective defect in the initial energy-independent binding of preproteins at the Toc complex. To directly assess the effect of proteolysis on this step in import, we mapped the association of preSSU with Toc components by trapping the interaction with covalent cross-linking. We used a label-transfer cross-linking method that results in the transfer of a radiolabel from the transit sequence of a modified version of preSSU, pS-1, to neighboring proteins upon photoactivation and cleavage of the cross-linking agent (Ma et al., 1996). This approach was shown previously to stabilize binding (Perry and Keegstra, 1994; Ma et al., 1996) and was used to map the interactions of preproteins with import components at each stage in import (Kouranov and Schnell, 1997).
Toc159 is the major target of preSSU cross-linking during energy-independent binding (Fig. 5A, lane 1). A minor fragment corresponding to the Toc86 fragment is also observed (Fig. 5A, lane 1). In addition, Toc75 cross-links at the energy-independent binding stage are consistent with the proposal that Toc159 and Toc75 cooperate in transit sequence recognition (Ma et al., 1996). Thermolysin treatment of the chloroplasts abolishes detectable cross-linking to envelope proteins under binding conditions (Fig. 5A, lane 3), indicating that the recognition site for preproteins at the chloroplast surface has been eliminated. Although we cannot rule out the possibility that proteolysis digests unknown factors that participate in binding, the fact that Toc159 is the major cross-linking target suggests that its cytoplasmic domain is essential for the interaction of preproteins with the Toc complex at this stage in import.
Figure 5.
Label-transfer cross-linking of pre-SSU to intact and protease-treated chloroplasts. Intact chloroplasts (−) or chloroplasts treated with 10 μg/mL thermolysin (+) were incubated with 200 nm 125I-pS-1 in a standard import reaction containing apyrase or 25 μm ATP as indicated. The chloroplasts were irradiated with UV light, re-isolated, and fractionated to yield a total envelope membrane fraction. The envelope membrane proteins were analyzed by SDS-PAGE and phosphor imager analysis. A, 125I-pS-1 cross-linking to intact chloroplasts isolated in the presence of protease inhibitors. The protein samples were resolved on 8% (w/v) polyacrylamide gels. B, 125I-pS-1 cross-linking to chloroplasts isolated in the absence of protease inhibitors. The protein samples were resolved on 12% (w/v) polyacrylamide gels. The positions of known Toc and Tic components and 125I-pS-1 (pS-1) are indicated to the left of A and the right of B.
Although the cytoplasmic domain of Toc159 appears to be crucial for binding in the absence of ATP, the pattern of cross-linking to the Toc complex in intact chloroplasts (Fig. 5A) is comparable to that observed with chloroplasts containing predominantly Toc86 (Fig. 5B). Toc159 is preferentially cross-linked approximately 2:1 over Toc75 in the absence of energy (Fig. 5A, lane 1). These cross-linking ratios are nearly identical to those observed in chloroplasts prepared in the absence of protease inhibitors, with the exception that Toc159, rather than Toc86, is the major cross-linked target (Fig. 5B, lane 1). Therefore, it appears that the 86-kD fragment of Toc159 is sufficient to interact with preproteins in vitro.
Cross-linking to Toc159 is not detected in intact chloroplasts in the presence of 25 μm ATP, whereas the levels of Toc75 cross-linking increase approximately 5-fold under these conditions (Fig. 5A, lane 2). These results are consistent with the previous conclusion that the transit sequence is inserted across the outer membrane to form an early import intermediate under these conditions (Ma et al., 1996). Cross-linking to Toc75 is detected in both intact (Fig. 5, lane 2) and protease-treated (Fig. 5, lane 4) chloroplasts. In addition, bands corresponding to the inner membrane component Tic20 are observed with both chloroplast preparations (Fig. 5, lanes 2 and 4), which is consistent with the previous observation that preproteins are in contact with the Tic machinery at this stage in import. The cross-linking efficiencies of Toc75 and Tic20 in proteolyzed chloroplasts are approximately 40% of those observed in intact chloroplasts, which is consistent with the 50% to 60% drop in the efficiency of outer membrane insertion observed in the import assays (Fig. 4). On the basis of these observations, we conclude that proteolysis has removed the binding site for energy-independent association of the preprotein with the Toc complex without destroying the protein-conducting channel of the outer membrane or blocking the ability of Toc and Tic components to form functional contact sites.
Low levels of cross-linking to Toc86 are detectable in the presence of 25 μm ATP using chloroplasts isolated by standard methods (Fig. 5B, lane 2), suggesting that degradation of the N-terminal domain might affect energy-driven dissociation of bound preprotein from Toc86.
Control of Translocation by GTP
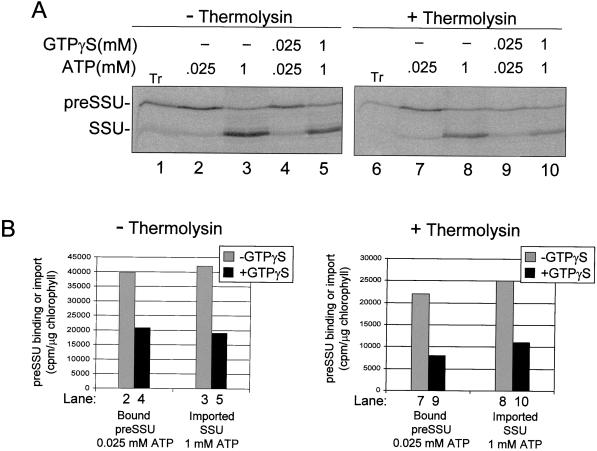
The hydrolysis of GTP is a prerequisite for envelope translocation and consequently, the formation of early import intermediates is inhibited by the presence of non-hydrolyzable GTP analogs (Olsen and Keegstra, 1992; Kessler et al., 1994). The ability to selectively remove the GTP-binding domain of Toc159 while leaving Toc34 intact allowed us to investigate the relative contributions of the Toc GTPases to the regulation of translocation. As expected, the GTP analog GTPγS inhibited the formation of the preSSU early import intermediate by 48% (Fig. 6, A, lanes 2 and 4, and B) and preSSU import by 55% (Fig. 6, A, lanes 3 and 5, and B) in intact chloroplasts. GTPγS had a similar effect on translocation in proteolyzed chloroplasts. Formation of the early import intermediate (Fig. 6, A, lanes 7 and 9, and B) and import across the envelope (Fig. 6A, lanes 8 and 10) were inhibited by 66% and 50%, respectively (Fig. 6B). Therefore, GTP is able to regulate import independent of Toc159 GTPase activity.
Figure 6.
Effect of a GTP analog on the binding and import of preSSU in intact or protease-treated chloroplasts. Intact chloroplasts or chloroplasts treated with 10 μg/mL thermolysin were incubated with in vitro synthesized [35S]preSSU in a standard import reaction containing 25 μm ATP, or 1 mm ATP as indicated. GTPγS was included in the binding or import reactions at the concentrations indicated. The chloroplasts were re-isolated and analyzed by SDS-PAGE. A, Phosphor imager analysis of SDS-PAGE-resolved chloroplast proteins. Tr, 10% of the [35S]preSSU translation product added to each reaction. B, Quantitative analysis of the data presented in A. The numbers of the lanes in A that were used for quantitation are indicated at the bottom of the graph in B.
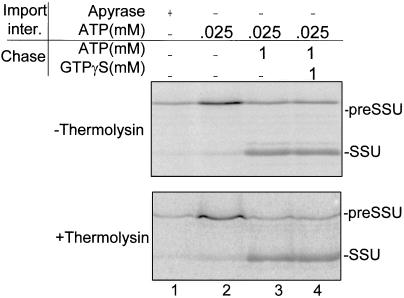
To determine whether the action of GTP is specific for translocation across the outer membrane, we tested the effect of GTPγS on the import of preSSU from the early import intermediate stage (Young et al., 1999). The early intermediate was trapped by incubation of preSSU with chloroplasts in the presence of 25 μm ATP. The chloroplasts were re-isolated and incubated with 1 mm ATP to promote complete import of the intermediate. The role of GTP was assessed by including equimolar concentrations of GTPγS in the import reaction. The analog had no effect on the import of the early import intermediate in intact or proteolyzed chloroplasts (Fig. 7, lanes 3 and 4). Therefore, the inhibition by GTPγS is specific for translocation across the outer membrane. Furthermore, the efficiency of import of the early import intermediate was 80% to 85% in intact and proteolyzed chloroplasts (Fig. 7, lanes 2 and 3), indicating that proteolysis does not affect steps in import subsequent to outer membrane translocation.
Figure 7.
Effect of a GTP analog on the import of a preSSU early import intermediate in intact or protease-treated chloroplasts. Intact chloroplasts or chloroplasts treated with 10 μg/mL thermolysin were incubated with in vitro-synthesized [35S]preSSU in a standard import reaction containing apyrase or 25 μm ATP as indicated (Import inter.). The chloroplasts were re-isolated and resuspended in import buffer containing 1 mm ATP and 1 mm GTPγS as indicated and incubated at 26°C for 10 min (Chase). The chloroplasts were re-isolated and analyzed by SDS-PAGE and phosphor imager analysis.
DISCUSSION
We have shown that the Toc86, the 86-kD subunit of the outer membrane protein translocon of chloroplasts, is a proteolytic fragment of a native 159-kD polypeptide designated Toc159. The original identification of the polypeptide as Toc86 (Hirsch et al., 1994; Schnell et al., 1994) can be attributed to the hypersensitivity of the cytoplasmic A-domain of Toc159 (Fig. 2A) to proteolysis during standard chloroplast isolation procedures. Without adequate precautions, proteolysis results in the quantitative conversion of Toc159 to the 86-kD fragment. By unusual coincidence, the cDNA sequences corresponding to the A-domain of Toc159 cannot be stably cloned and propagated in E. coli, precluding the isolation of full-length cDNA clones that include the coding region for the A-domain. We were able to obtain the full-length cDNA sequence for Toc159 by sequencing several overlapping RT-PCR products that encompassed the 5′ half of the complete cDNA.
The coding region of the original Toc86 cDNAs encoded a polypeptide of 96.7 kD (Hirsch et al., 1994; Schnell et al., 1994). Studies of the targeting of the 96.7-kD fragment in in vitro import assays led to the conclusion that the 1.7-kD N-terminal amino acid sequence of the polypeptide participated in targeting and integration of Toc86 into the outer membrane (Hirsch et al., 1994; Muckel and Soll, 1996). In these studies, the N terminus was shown to be proteolytically removed during the import of the 96.7-kD in vitro translation product into isolated chloroplasts, yielding Toc86 (Muckel and Soll, 1996). Our results indicate that the proteolytic processing observed in the Toc86 import studies was due to non-specific proteolysis of the truncated 96.7-kD translation product during the import reaction. As a consequence, the factors responsible for selective targeting of Toc159 to the outer membrane remain to be established in assays using the full-length polypeptide.
Previous antibody inhibition (Hirsch et al., 1994) and covalent cross-linking studies (Ma et al., 1996; Kouranov and Schnell, 1997) provided evidence that Toc86 participates in the recognition and binding of preproteins at the outer membrane translocon. Our data support this conclusion by demonstrating that intact Toc159 is the major cross-linking target for the preSSU transit sequence during energy-independent binding (Fig. 5). Furthermore, complete degradation of the cytoplasmic region of Toc159 encompassing the A- and G-domains eliminates detectable binding under these conditions. Although we cannot rule out the possibility that unknown components of the import apparatus are affected by proteolysis, the fact that Toc159 is the major target of cross-linking to the preSSU transit sequence strongly implies that the cytoplasmic domains of Toc159 are essential for the initial binding of preproteins at the chloroplast surface. Removal of this reversible binding step is likely to account for the decrease in overall import capacity that is observed with proteolyzed chloroplasts (Fig. 4). In our previous studies, we demonstrated that Toc86 and Toc75 both interact with transit sequences in the absence of energy (Ma et al., 1996; Kouranov and Schnell, 1997), raising the possibility that Toc159 and Toc75 act cooperatively during binding. In vivo, the cooperative binding activity of Toc159 and Toc75 could increase the efficiency of preprotein targeting to the sites of outer membrane translocation and therefore increase the efficiency of subsequent translocation. Conversely, elimination of the Toc159 cytoplasmic domain would eliminate the cooperative interaction and reduce the overall efficiency of translocation.
The ability of proteolyzed chloroplasts to import proteins indicates that the cytoplasmic domain of Toc159 is not required for translocation across the outer membrane. This remarkable finding suggests that the binding and translocation stages of import are not a priori linked. This scenario is reminiscent of the relationship between the protein import receptors and the general import pore in the outer membrane of mitochondria. Yeast cells lacking the Tom70, Tom20 (Hines and Schatz, 1993; Ramage et al., 1993; Lithgow et al., 1994), and Tom37 (Gratzer et al., 1995) mitochondrial preprotein receptors are viable and are therefore able to import proteins in the absence of the receptor system. These results suggest that the receptor components in both mitochondria and chloroplasts evolved during endosymbiosis to increase the efficiency of preprotein targeting to translocation channels.
The ability of preproteins to bypass binding indicates that the outer membrane translocon possesses a distinct transit sequence recognition site that allows preproteins to engage the Toc complex. Hinnah et al. (1997) previously reported that the ion channel activity of isolated, recombinant Toc75 is altered by the presence of preSSU but not mature SSU. Therefore, Toc75 alone possesses an ability to transiently interact with transit sequences that could account for the preprotein recognition in the absence of intact Toc159. The ability to detect cross-linking of preSSU to Toc75 in the presence of energy is consistent with this hypothesis (Fig. 5). We do not detect energy-independent binding of preSSU to Toc75 in the absence of the cytoplasmic domain of Toc159 (Fig. 5), as predicted by the ability of preSSU to alter the channel properties of isolated Toc75 (Hinnah et al., 1997). However, this is not surprising given the relative insensitivity of the cross-linking assay compared with the electrophysiological measurements.
The elimination of the Toc159 cytoplasmic domain allowed us to discriminate between the roles of the Toc159 and Toc34 GTPase domains. The role of Toc34 in import was unclear, but our observation that GTPγS inhibits import in the absence of the Toc159 GTPase strongly implies that GTP hydrolysis at Toc34 is a prerequisite for translocation at the outer membrane (Fig. 6). Therefore, the Toc34 GTPase plays a direct role in regulating translocation. The role of GTP in regulating import appears to be specific for the initial insertion of the preprotein into the outer membrane translocon because translocation of an early import intermediate that is partially inserted across the outer membrane is not affected by the GTP analog (Fig. 7).
Previous cross-linking studies indicated that Toc34 is in close proximity to preproteins that are bound in the absence of energy (Kouranov and Schnell, 1997). Toc34 does not appear to form part of the transit sequence receptor site, because its interactions are not specific to this region of the preprotein. However, binding of a GTP analog to the Toc complex alters the cross-linking pattern of the preprotein to Toc34 (Kouranov and Schnell, 1997). On the basis of this observation, we proposed that GTP binding induced a conformational shift in the Toc complex that was a prerequisite for subsequent insertion into the protein-conducting channel. Our current findings support this conclusion and provide additional evidence that Toc34 functions as a regulatory subunit of the Toc complex. Our results are consistent with a model in which Toc159 participates in the formation of a primary receptor site for the binding of nuclear-encoded preproteins to the Toc complex at the outer chloroplast membrane. A cycle of GTP binding and hydrolysis at Toc34 would promote productively bound preprotein into the protein-conducting channel, thereby committing the preprotein to translocation.
Interestingly, chloroplasts containing intact Toc159 did not have a significantly higher binding efficiency than those containing the Toc86 fragment, as measured by the label-transfer cross-linking assay (Fig. 5). Thus, it appears that the GTPase domain (G-domain) and the membrane anchor domain (M-domain) corresponding to the Toc86 fragment of Toc159 are sufficient to form a transit sequence binding site in vitro. Our previous observation that cross-linking of the transit sequence of preSSU during energy-independent binding mapped to the M-domain of Toc86 (Kouranov and Schnell, 1997) is consistent with this conclusion. This leaves open the question of the function of the N-terminal A-domain. Although unrelated in primary structure to known proteins, the highly repetitive nature of this domain is reminiscent of structured regions involved in protein-to-protein interactions. For example, the A-domain might act as a binding site for an unknown factor(s) that participates in the targeting of preproteins to the Toc complex or a factor that regulates the GTPase activity of Toc159 in vivo. The activity of the Toc159 GTPase domain also remains unclear. The GTPase might act in cooperation with Toc34 to regulate insertion of preproteins into the translocon. Alternatively, Toc159 GTPase activity might act at a previous step to regulate the initial binding of the preprotein at the Toc complex. The functions of the A- and G-domains in import have not been apparent in in vitro assays because of the proteolytic sensitivity of Toc159. However, the availability of a potential homolog of Toc159 in Arabidopsis provides the potential to investigate the in vivo role of these domains in chloroplast biogenesis through molecular genetic approaches.
Footnotes
This work was supported by grants from the National Science Foundation (MCB–9722914 and BIR–94131980) and a Charles and Johanna Busch Memorial Fund Award.
LITERATURE CITED
- Akita M, Nielsen E, Keegstra K. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J Cell Biol. 1997;136:983–994. doi: 10.1083/jcb.136.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolter B, May T, Soll J. A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide. FEBS Lett. 1998;441:59–62. doi: 10.1016/s0014-5793(98)01525-7. [DOI] [PubMed] [Google Scholar]
- Chen X, Schnell DJ. Protein import into chloroplasts. Trends Cell Biol. 1999;9:222–227. doi: 10.1016/s0962-8924(99)01554-8. [DOI] [PubMed] [Google Scholar]
- Cline K, Henry R. Import and routing of nucleus-encoded chloroplast proteins. Annu Rev Cell Dev Biol. 1996;12:1–26. doi: 10.1146/annurev.cellbio.12.1.1. [DOI] [PubMed] [Google Scholar]
- Cline K, Werner-Washburne M, Andrews J, Keegstra K. Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 1984;74:675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Friedman AL, Keegstra K. Chloroplast protein import: quantitative analysis of precursor binding. Plant Physiol. 1989;89:993–999. doi: 10.1104/pp.89.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratzer S, Lithtgow T, Bauer RE, Lamping E, Paltauf F, Kohlwein SD, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hines V, Schatz G. Precursor binding to yeast mitochondria: a general role for the outer membrane protein Mas70p. J Biol Chem. 1993;268:449–454. [PubMed] [Google Scholar]
- Hinnah SC, Hill K, Wagner R, Schlicher T, Soll J. Reconstitution of a chloroplast protein import channel. EMBO J. 1997;16:7351–7360. doi: 10.1093/emboj/16.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Muckel E, Heemeyer F, von Heijne G, Soll J. A receptor component of the chloroplast protein translocation machinery. Science. 1994;266:1989–1992. doi: 10.1126/science.7801125. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Yousif AE. Isolation and characterization of chloroplast envelope membranes. Methods Enzymol. 1986;118:316–325. [Google Scholar]
- Kessler F, Blobel G, Patel HA, Schnell DJ. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Schnell DJ. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J Cell Biol. 1997;139:1677–1685. doi: 10.1083/jcb.139.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow T, Junne T, Wachter C, Schatz G. Yeast mitochondria lacking the tow import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J Biol Chem. 1994;269:15325–15330. [PubMed] [Google Scholar]
- Ma Y, Kouranov A, LaSala S, Schnell DJ. Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J Cell Biol. 1996;134:1–13. doi: 10.1083/jcb.134.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Muckel E, Soll J. A protein import receptor of chloroplasts is inserted into the outer envelope membrane by a novel pathway. J Biol Chem. 1996;371:23846–23852. doi: 10.1074/jbc.271.39.23846. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. Stable association of chloroplastic precursors with protein-translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen LJ, Keegstra K. The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J Biol Chem. 1992;267:433–439. [PubMed] [Google Scholar]
- Pain D, Blobel G. Protein import in chloroplasts requires a chloroplast ATPase. Proc Natl Acad Sci USA. 1987;84:3288–3292. doi: 10.1073/pnas.84.10.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Keegstra K. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 1994;6:93–105. doi: 10.1105/tpc.6.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage L, Junne T, Hahne K, Lithgow T, Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz G. Protein transport: the door to organelles. Nature. 1998;395:439–440. doi: 10.1038/26620. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G, Keegstra K, Kessler F, Ko K, Soll J. A nomenclature for the protein import components of the chloroplast envelope. Trends Cell Biol. 1997;7:303–304. doi: 10.1016/S0962-8924(97)01111-2. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Waegemann K, Soll J. A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 1995;7:401–411. doi: 10.1046/j.1365-313x.1995.7030401.x. [DOI] [PubMed] [Google Scholar]
- Young ME, Keegstra K, Froehlich JE. GTP promotes the formation of early-import intermediates but is not required during the translocation step of protein import into chloroplasts. Plant Physiol. 1999;121:237–244. doi: 10.1104/pp.121.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]