Abstract
Background
Rare multicentric lower-grade gliomas (LGGs) represent a unique opportunity to study the heterogeneity among distinct tumor foci in a single patient and to infer their origins and parallel patterns of evolution.
Methods
In this study, we integrate clinical features, histology, and immunohistochemistry for 4 patients with multicentric LGG, arising both synchronously and metachronously. For 3 patients we analyze the phylogeny of the lesions using exome sequencing, including one case with a total of 8 samples from the 2 lesions.
Results
One patient was diagnosed with multicentric isocitrate dehydrogenase 1 (IDH1) mutated diffuse astrocytomas harboring distinct IDH1 mutations, R132H and R132C; the latter mutation has been associated with Li–Fraumeni syndrome, which was subsequently confirmed in the patient’s germline DNA and shown in additional cases with The Cancer Genome Atlas data. In another patient, phylogenetic analysis of synchronously arising grade II and grade III diffuse astrocytomas demonstrated a single shared mutation, IDH1 R132H, and revealed convergent evolution via non-overlapping mutations in ATRX and TP53. In 2 cases, there was divergent evolution of IDH1-mutated and 1p/19q-codeleted oligodendroglioma and IDH1-mutated and 1p/19q-intact diffuse astrocytoma, occurring synchronously in one case and metachronously in a second.
Conclusions
Each tumor in multicentric LGG cases may arise independently or may diverge very early in their development, presenting as genetically and histologically distinct tumors. Comprehensive sampling of these lesions can therefore significantly alter diagnosis and management. Additionally, somatic IDH1 R132C mutation in either multicentric or solitary LGG identifies unsuspected germline TP53 mutation, validating the limited number of published cases.
Keywords: astrocytoma, bilateral, IDH1, multicentric, oligodendroglioma
Importance of the study
Multicentric lower-grade gliomas are characterized by spatially discrete foci, often in different lobes. These tumors present unique challenges in diagnosis and management, and their genomic alterations have not been well characterized. Previous sequencing studies of multifocal and multicentric glioblastomas have revealed a clonal process with early divergence from a common ancestor and distant migration. Here, we profile 4 patients with multicentric lower-grade gliomas using multisector exome sequencing. In 3 of 4 patients, the paired glioma foci harbored a single shared mutation at IDH1 R132H. A fourth patient with Li–Fraumeni syndrome was found to have entirely independent foci, one of which contained an IDH1 R132C mutation. We show that careful genomic characterization of each tumor focus improves diagnostic accuracy and alters patient management. To our knowledge, this is the first study of the origins and evolution of multicentric lower-grade glioma using multisector next-generation sequencing.
Molecular testing has tremendous prognostic power for gliomas and has been incorporated as an essential component of the 2016 World Health Organization (WHO) Classification of Tumors of the Central Nervous System.1 Isocitrate dehydrogenase (IDH) 1 and 2 mutation status and codeletion of chromosomes 1p and 19q define 3 categories of lower-grade gliomas (LGGs): IDH-wildtype astrocytomas, IDH-mutant and 1p/19q-intact astrocytomas, and IDH-mutant 1p/19q-codeleted oligodendrogliomas. Intriguingly, study of these conserved molecular alterations suggests that these different tumor types have fundamentally different origins and implicates distinct evolutionary steps on the path toward gliomagenesis.2–5
Molecular diagnostic tests are often performed on small representative samples of tumor, utilizing techniques such as immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), or sequencing. IDH mutation and 1p/19q codeletion appear to be ubiquitous within individual solitary tumors, suggesting that they are among the very first alterations that initiate LGG. In contrast, the intratumoral heterogeneity of numerous associated genetic alterations—for example, mutations in tumor protein 53 (TP53), alpha thalassemia/mental retardation syndrome X-linked (ATRX), far upstream element-binding protein 1, and capicua transcriptional repressor—suggests that they typically arise after IDH1/2 mutation.3,6 Mutation of IDH1 codon 132 most commonly results in an alteration from arginine (R) to histidine, although less common amino acids at the R132 locus include cysteine, serine, leucine, and glycine.7
Multiple synchronous LGGs are uncommon and present unique diagnostic and management challenges. Both multicentric and multifocal tumors refer to tumors that have discrete foci, separated by some distance within the brain, and may arise either independently or via migration and expansion of early tumor cells. In this study, we show how multisector profiling and sequencing can impact diagnosis and management of multicentric diffuse LGG. To our knowledge, this is the first study to investigate the clonality and evolution of multicentric LGG using next-generation sequencing.8,9 Although the genesis of multifocal and multicentric primary glioblastoma may be different, their origin and evolution defined by genomics have been previously studied, with most studies assuming a single clonal process.10 The genetic heterogeneity of multicentric or multifocal glioblastomas was significantly greater than that for single glioblastomas; however, all of the cases contained multiple clonal genetic alterations, suggesting early divergence and parallel evolution rather than independent gliomagenesis events.11
Materials and Methods
In Vivo Magnetic Resonance Exam
Informed written consent from patients and approval by the institutional review board at the University of California San Francisco (UCSF) and the ethics committee at the Institute of Neurology at the Medical University of Vienna were obtained for collection and research on these samples. Standard anatomic imaging included T2-weighted (fluid attenuated inversion recovery [FLAIR] and fast spin echo) as well as T1-weighted pre- and post-gadolinium contrast images obtained according to previous protocols.12
Sample Acquisition
All tumor samples were collected during surgical resection and either snap frozen in liquid nitrogen and stored at −80°C or formalin fixed and paraffin embedded. In the case of image-guided biopsy, each sample was an independent, geographically distinct piece derived from different stages of the surgery. Multicentric glioma was defined as having at least one region of tumor, either enhancing or non-enhancing, that is not contiguous with the main lesion and is outside of the region of T2 hyperintensity (edema) surrounding the main mass. Samples from different foci of each patient were obtained in separate surgeries of at least 3 days apart. For Patient 2, both lesions were present at diagnosis and the lesions were identified as independent and not connected by the anterior corpus callosum. Heterogeneity on the imaging prompted image-guided biopsy of 4 samples from each lesion, in surgeries 2 months apart. Two from the right lesion were not used due to poor quality and/or low tumor percentage. One piece of additional bulk tissue (not image-guided) from each lesion was also obtained. For Patient 3, both lesions were present at the time of diagnosis and were sampled separately, one from each lesion, 3 months apart. Patient 4 had one lesion at diagnosis and this was sampled 4 years before another lesion occurred in another lobe (one sample from each lesion). Samples from Patient 4 were not processed for sequencing, as they were obtained from a different hospital.
Patient-matched normal samples were obtained from peripheral blood or muscle tissue. Samples from Patients 1–3 were obtained from the Brain Tumor Center Tissue Bank at UCSF and samples from Patient 4 were obtained from the Institute of Neurology, Medical University of Vienna. All tumor samples were reviewed by a board- certified neuropathologist to classify and grade according to the 2016 WHO guidelines.1
Exome Sequencing, Mutation Identification, and Phylogenetic Tree Construction
Genomic DNA was extracted with either a Qiagen DNA extraction kit following the manufacturer’s instructions or isolated by phenol:chloroform extraction as previously described.2 Exome capture was performed using the NimbleGen (SeqCap EZ Exome v3) exome capture kit according to manufacturer’s protocol. Paired-end sequencing data from exome capture libraries were processed as previously described.2 Phylogenetic trees were built using an ordinary least squares minimum evolution13 approach from the ape R package14 as previously described.15 Only single nucleotide mutations were included for building trees due to high false positive and false negative rates in calling indels in exome data.16 Germline single nucleotide polymorphisms (SNPs) were processed with the Genome Analysis Toolkit according to the Broad Institute best practice guidelines and called using UnifiedGenotyper.17 SNPs called in genes associated with a predisposition to glioma or genes implicated in a glioma genome-wide association study (GWAS)18,19 (CCDC26, CDK4, CDKN2A, CDKN2B, NF1, NF2, PHLDB1, PIK3CA, POT1, PTEN, RTEL1, TERC, TERT, TP53, TSC1, and TSC2) were assessed using ClinVar20 and Combined Annotation Dependent Depletion (CADD) scores21 for Patients 1–3. Exome sequencing data are deposited in the European Genome-phenome Archive (EGA) as EGAS00001002495. When the results of the IHC and exome were discordant, a missense TP53 mutation was considered the final result, as frequently reported in COSMIC (Catalogue of Somatic Mutations in Cancer). 22
Copy Number and Loss of Heterozygosity Analysis from Exome Sequencing
For each window, we estimate the decrease of heterozygosity (DoH) between tumor and normal,23 which represents the difference between the tumor and normal allelic ratios across heterozygous SNPs contained in the window. Specifically, we used SAMtools mpileup24 and joint sequenza::pileup2seqz()25 to obtain tumor and normal total copy number (TCN) counts at each genomic position and DoH estimates at heterozygous SNPs (as called from normal allele counts). These estimates were then averaged in discrete 100-kb windows resulting in (TCN, DoH) mean estimates for each window. Based on these window averages, we used PSCBS::segmentedByPairedPSCBS()23 to partition the (TCN, DoH) estimates into genomic regions of [piecewise] constant parent specific copy number (PSCN) levels. For each PSCN segment (focal copy number alteration [CNA]), we obtained a genomic start and end position, TCN and DoH mean levels, and noise estimates. Shared CNAs were determined after lowering the resolution to a cytoband. Scripts for this pipeline are available at https://github.com/UCSF-Costello-Lab/Multicentric_glioma_CN, last accessed November 18, 2017.
The Cancer Genome Atlas Data
We curated grade II or III glioma samples lacking somatic TP53 mutations using cBioPortal26 and Ceccarelli et al.27TP53 and CHEK2 germline mutation data were extracted from variant call format (VCF) files for The Cancer Genome Atlas (TCGA) diffuse LGG samples downloaded from the Genomic Data Commons data portal (n = 393). VCF files were used to calculate CADD scores at http://cadd.gs.washington.edu/, last accessed September 11, 2017.21
Sanger Sequencing of the TERT Promoter
DNA samples from Patients 1–3 were amplified for the telomerase reverse transcriptase promoter (TERT-p) region and Sanger sequenced as previously described.28 For Patient 4, the relevant TERT-p sequence was amplified from genomic DNA and analyzed on a 3130 DNA sequencer. The following primers were used: TERTProm1-MTR (5ʹ-CAGGAAACAGCTATGACGCA CAGACGCCCAGG ACCG CGCT), TERTProm1-M13 (5ʹ-GTAAAACGACGGCCA GTTTCCC ACGTGCGCAGCAG GACGCA).
Immunohistochemistry
IHC for Patients 1–3 was performed in the Department of Pathology at UCSF using the following antibodies: IDH1-R132H mutant protein (clone H09, Dianova, 1:500 dilution), ATRX (HPA001906, Sigma, 1:100 dilution), MIB1 (CONFIRM anti–Ki-67 [30-9] rabbit monoclonal primary antibody; Ventana), and p53 (clone DO-7, Dako, 1:100 dilution). All staining was performed in Ventana or Leica Bond automated staining processors. The percentage of cells staining for Ki-67 (number of MIB1-positive tumor cells divided by the total number of tumor cells) was defined by a neuropathologist. Quantification was performed by manual counting under a light microscope. Hematoxylin and eosin (H&E) staining was performed according to standard protocols. IHC for Patient 4 was performed at the Medical University of Vienna using the following antibodies: IDH1-R132H (antigen retrieval, 1:60, Dianova #DIA-H09) and ATRX (antigen retrieval, 1:300, Sigma-Aldrich #HPA001906), both on a Ventana Benchmark automated staining system. Anti-p53 (1:50, Dako #M7001) staining was performed using a Dako Autostainer PlusLink automated staining system. In cases of discordance between ATRX results of the IHC and exome sequencing, loss of ATRX on the IHC was considered the final result, as per clinical practice.
Fluorescence In Situ Hybridization
FISH for chromosomes 1 and 19 (1p/19q) was performed on paraffin-embedded sections using Vysis LSI dual-color probes for 1p36/1q25 and 19q13/19p13 (#04N60-020, Abbott Laboratories) according to manufacturer’s instructions and imaged using a Carl Zeiss fluorescence microscope. Signal ratios were assessed across 200 nuclei for chromosomes 1 and 19. More than 30% of nuclei with impaired control-to-target signal ratios were required to identify deletions.
Results
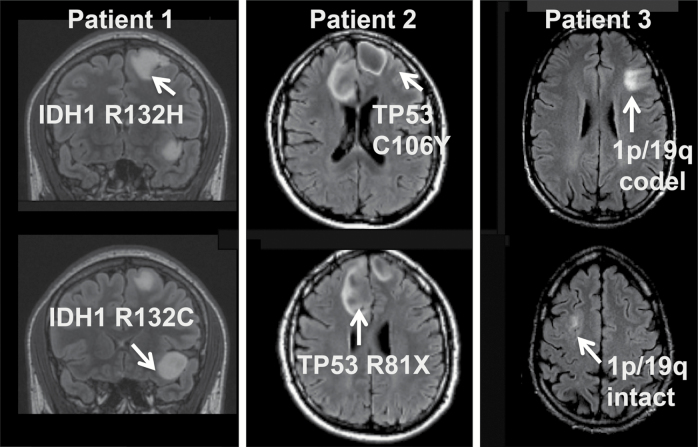
Comprehensive genomic analyses of 12 samples in total from 3 patients, and clinical genomic analysis of another patient, highlight heterogeneity of the earliest mutations in gliomagenesis (Fig. 1).
Fig. 1.
Multifocal tumors exhibit heterogeneity of core mutations required for gliomagenesis. Summary of diverging early mutations and MRI of Patients 1–3.
Patient 1: Germline TP53 Mutation and Distinct Somatic IDH1 Mutations
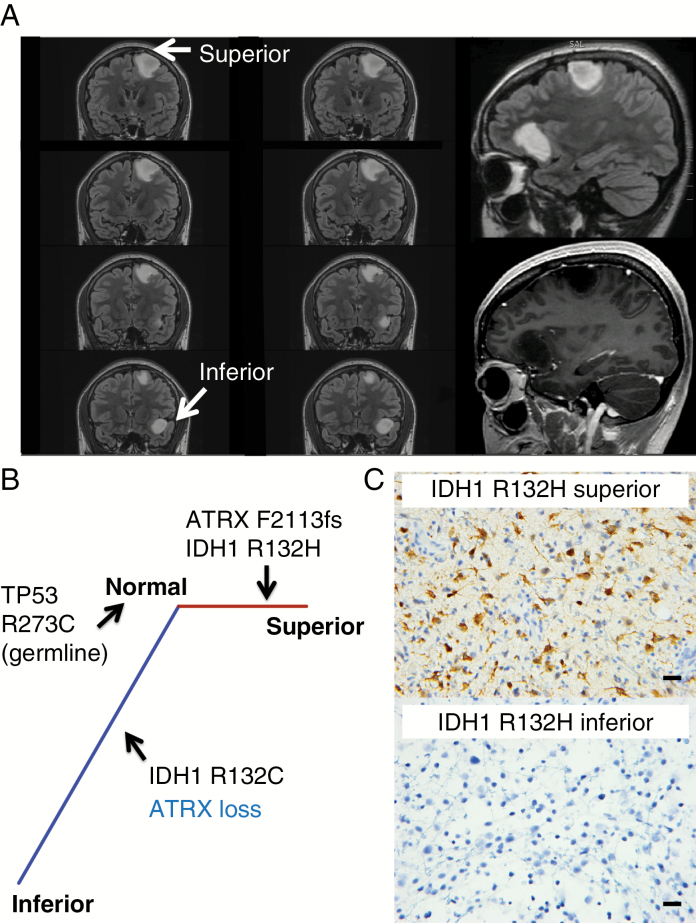
Patient 1 was a 23-year-old female at diagnosis, without significant family history of cancer, who presented with seizures and was found to have 2 distinct non-enhancing, T2 hyperintense tumors in the left frontal lobe separated by normal-appearing brain parenchyma (Fig. 2A). Craniotomy and biopsy of the superior tumor revealed a diffuse glioma with astrocytic morphology and WHO grade II histologic features. The tumor cells were positive for IDH1 R132H-mutant protein and showed strong nuclear p53 positivity by IHC. ATRX was retained on IHC (Supplementary Figure S1). The Ki-67 labeling index was approximately 2%. FISH revealed intact 1p/19q. The patient then underwent staged awake craniotomies with cortical mapping for near-total resection of both tumors. Both tumors were diffuse gliomas with astrocytic morphology and WHO grade II histologic features. The inferior tumor was negative for IDH1 R132H-mutant protein by IHC, had strong nuclear p53 immunostaining, and was 1p/19q intact by FISH with a Ki-67 labeling index of approximately 4% (images not shown). ATRX was lost on IHC (Supplementary Figure S1) As a result of the discrepancy in the IDH1 mutation status (Fig. 2C), direct sequencing of IDH1 exon 4 was performed, confirming the IDH1 R132H mutation in the left superior frontal lesion and demonstrating a disparate IDH1 R132C mutation in the left inferior frontal tumor. As this latter R132C mutation has been associated with Li–Fraumeni syndrome29 and given the presence of multifocal tumors, commercial germline analysis on a peripheral blood sample was performed (Invitae). This revealed a pathogenic R273C mutation in the TP53 gene, confirming a diagnosis of Li–Fraumeni syndrome in this patient. There were no damaging germline mutations in 15 other genes associated with glioma predisposition or implicated in glioma GWAS in this patient. Following resection of both lesions, the patient was observed with serial MRI without additional adjuvant therapy based upon her age and minimal extent of residual disease.
Fig. 2.
Protein expression and phylogenetic analyses among the tumors in Patient 1 revealed mutational heterogeneity in IDH1 and ATRX. (A) This patient had 2 non-enhancing, T2 bright left frontal tumors. The right bottom image shows the resection cavities of the lesions. (B) Phylogenetic tree derived from somatic mutations from exome sequencing. Branch length is proportional to the number of coding single nucleotide somatic mutations. Aberrations identified by IHC are labeled in blue. (C) IHC using IDH1 R132H antibody. Scale bar, 20 μm.
Exome sequencing of both tumors demonstrated no shared somatic mutations or CNA transition points between the 2 tumors, suggesting 2 distinct gliomagenesis events initiated by disparate mutations in IDH1 (Fig. 2B, Supplementary Tables S1, S2, S3). Although both tumors harbored loss of heterozygosity on 17p (encompassing the TP53 locus), these alterations had distinct breakpoints, further suggesting that the 2 tumors arose through independent gliomagenesis events (Supplementary Figure S1).
The patient developed progressive generalized seizures and was found to have recurrent disease 5.7 years after her initial resection. Imaging demonstrated recurrent enhancing disease arising from the superior resection cavity; the inferior resection cavity remained stable. A second resection demonstrated glioblastoma, harboring the canonical IDH1 R132H mutation by IHC. At last follow-up, she remained alive and was undergoing chemoradiotherapy with concurrent temozolomide.
Lack of Somatic TP53 Mutations and Presence of IDH1 R132C Mutations in Solitary Glioma
The discovery of a TP53 germline mutation in a patient with an otherwise classic IDH-mutant diffuse astrocytoma with strong p53 positivity by IHC but lacking a somatic TP53 point mutation prompted us to assess whether other glioma patients may have similar germline TP53 mutations suggestive of potentially undiagnosed Li–Fraumeni syndrome. Analysis of 393 grade II and III gliomas from TCGA diffuse LGG cohort of astrocytoma and oligodendroglioma27 identified 8 patients (2%) whose tumors had ATRX mutations and intact 1p/19q but lacked somatic point mutations in TP53. Assessment of the germline TP53 locus of these 8 patients showed that 2 had damaging germline TP53 missense mutations consistent with Li–Fraumeni syndrome (R248W and C277F; Supplementary Table S4). Both mutations had CADD scores >30, suggesting that they are among the top 0.1% of deleterious variants in the human genome.21 The case TCGA-S9-A6U1 had a chr17:7577539 G>A mutation (CADD score 34) and TCGA-VM-A8CH had a chr17:7577108 C>A mutation (CADD score 34) (Supplementary Table S4). Like the inferior tumor in Patient 1, both TCGA tumors harbored an IDH1 R132C mutation, rather than the canonical R132H mutation. The TP53 R248W variant is one of the most commonly identified mutations in patients with Li–Fraumeni syndrome, and the 248 residue is a mutation hotspot in both Li–Fraumeni germlines and sporadic tumors.30 Although the TP53 C277F mutation has not previously been reported as a cause of Li–Fraumeni syndrome, the amino acid residue is highly conserved31 and a C277Y mutation at this position has previously been reported in Li–Fraumeni families in ClinVar and the TP53 database of the International Agency for Research on Cancer,32 suggesting that the mutation we identified is likely pathogenic. No germline CHEK2 mutations were identified in the remaining 6 TCGA glioma patients lacking a Li–Fraumeni associated TP53 mutation.
Patient 2: Bilateral Tumor Foci with Shared IDH1 Mutations and Heterogeneity of ATRX and TP53 Mutations
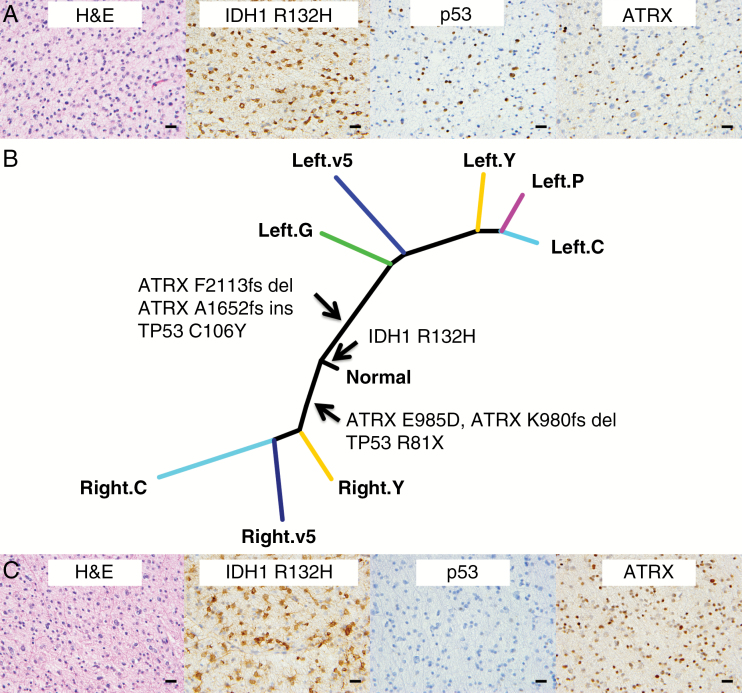
Patient 2 was a 21-year-old male at diagnosis who presented with new onset seizures. MRI of the brain demonstrated 2 distinct T2 hyperintense, non-enhancing tumors, one in each of the frontal lobes, without apparent involvement of the corpus callosum. The tumors were initially observed and his seizures were treated with anti-epileptic medications. Five years after diagnosis, the patient presented with progressive headaches, and review of the interval serial imaging demonstrated slow growth of both tumors. Both tumors remained non-enhancing and hyperintense on T2 FLAIR sequences. He underwent subtotal resection of the right frontal tumor, demonstrating a diffuse astrocytoma with WHO grade II histologic features that was positive for IDH1 R132H-mutant protein by IHC, had loss of ATRX expression, was mostly negative for p53 immunostaining, 1p/19q intact, and had a Ki-67 labeling index of approximately 2%. Staged gross total resection of the left-side tumor 6 weeks later demonstrated diffuse astrocytoma with WHO grade III histologic features that was positive for IDH1 R132H-mutant protein by IHC, had loss of ATRX expression, positive for p53 immunostaining, 1p/19q intact, and had a Ki-67 labeling index of approximately 4% (Fig. 3, Supplementary Fig. S2). Following the initial resection, strong consideration was given to treatment with temozolomide alone due to the potential morbidity of bifrontal radiotherapy in a young patient. Due to the presence of grade III disease identified in the second resection specimen, the patient received adjuvant radiotherapy with concurrent temozolomide, followed by 6 months of adjuvant temozolomide. At last follow-up, he remained free of recurrent disease 2.5 years from his initial surgery.
Fig. 3.
Protein expression and genetic differences among the image-guided biopsies of the bilateral tumors from Patient 2. (A) H&E and IHC for IDH1 R132H, p53, and ATRX on the left-side tumor. (B) Phylogenetic tree derived from somatic mutations from exome sequencing. Genes frequently mutated in glioma inferred from exome sequencing data are highlighted. (C) H&E and IHC for IDH1 R132H, p53, and ATRX on the right-side tumor (IHC staining from a separate piece to the image-guided biopsies). Scale bar, 20 μm.
Six image-guided biopsy specimens, 2 from the right and 4 from the left, plus 1 bulk tumor piece for each lesion, were suitable for exome sequencing. The image-guided specimens were mapped to preoperative multiparametric MRI and MR spectroscopy, which allowed us to correlate high-resolution imaging features with genomic information. Exome sequencing revealed considerable intratumoral heterogeneity: The left- and right-side specimens shared only a single point mutation at the IDH1 R132 locus. Both tumors harbored mutations in TP53 and ATRX, which were conserved among samples from the same tumor but distinct between the 2 tumors, suggesting a high degree of convergent evolution (Fig. 3B, Supplementary Table S1). There were CNAs that were shared among the samples from both lesions on 13 autosomes, at the resolution of a cytoband (approximately 1 Mb) (Supplementary Tables S2, S3). Scoring of the proliferative index of the image-guided biopsies showed discrete regions in both sides with increased Ki-67 staining: right-C (5.8%) and left-Y (4.3%) (Supplementary Table S5). Multiparametric MRI with diffusion and perfusion sequences demonstrated increased diffusivity and reduced fractional anisotropy in the higher-grade left-side tumor, consistent with invasive tumor and loss of normal tissue architecture (Supplementary Figure S3, Supplementary Table S6). There were no damaging germline mutations in 16 genes associated with glioma predisposition or implicated in glioma GWAS in this patient.
Patients 3 and 4: Oligodendroglioma and Diffuse Astrocytoma in Spatially Distinct Foci
These 2 patients were found to have both oligodendroglioma, IDH mutant and 1p/19q codeleted, and diffuse astrocytoma, IDH mutant and 1p/19q intact, occurring in spatially distinct lesions.
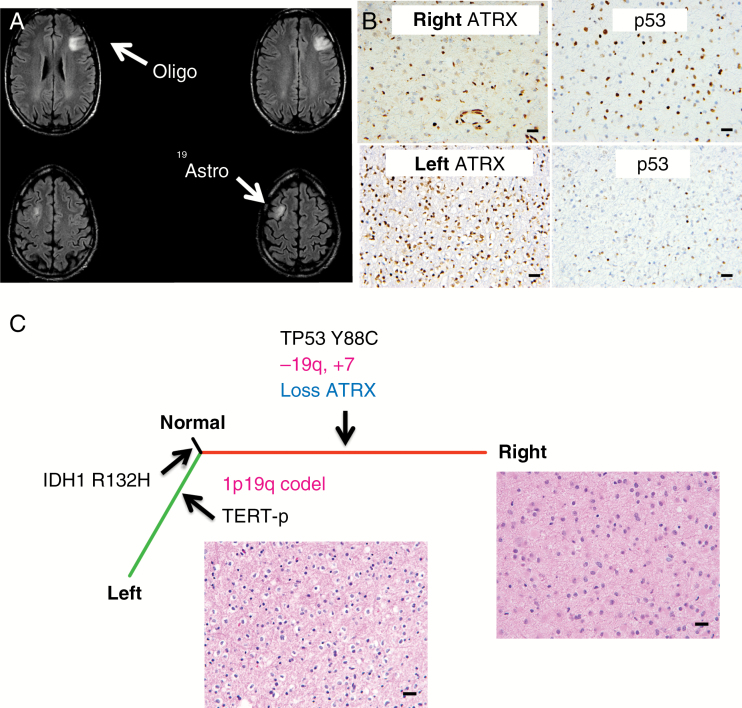
Patient 3 was a 35-year-old male at diagnosis who was found to have bifrontal non-enhancing, T2 hyperintense tumors consistent with LGGs (Fig. 4A). He was observed for several years with stable disease. Six years after diagnosis, the patient developed new absence seizures, and MRI revealed progression in the right frontal tumor. Craniotomy and biopsy of the right frontal tumor revealed a diffuse astrocytoma with WHO grade II histologic features, 1p/19q intact by FISH and exome sequencing (Fig. 4, Supplementary Figures S4, S5), IDH1 R132H mutant, p53 immunopositive, and ATRX loss of expression by IHC. The Ki-67 labeling index was approximately 2%. He underwent awake craniotomy with language mapping for gross total resection of the slowly progressive left frontal tumor. Pathology from the left-side tumor revealed an oligodendroglioma with WHO grade II histologic features, 1p/19q codeleted by FISH and exome sequencing, IDH1 R132H mutant, with intact ATRX expression and no significant p53 staining by IHC (Fig. 4, Supplementary Figures S4, S5, Supplementary Table S1). The Ki-67 labeling index was approximately 1%. There were no shared CNAs between the 2 tumors (Supplementary Tables S2, S3) and there were no damaging germline mutations in 16 genes associated with glioma predisposition or implicated in glioma GWAS.
Fig. 4.
Protein expression and genetic differences of the bilateral tumors of Patient 3. (A) Patient 3 presented with bilateral tumors. The left side has a 1p/19q codeletion and the right side has 1p/19q intact with 19q loss (different breakpoints than the 1p/19q codeletion; Supplementary Figure S4). (B) P53 and ATRX IHC on the left- and right-side tumors. Scale bar, 20 μm. (C) A phylogenetic tree derived from somatic mutations from exome sequencing. Genes frequently mutated in glioma, copy number changes (pink) inferred from exome sequencing, and protein expression (blue) are highlighted.
PCR amplification and Sanger sequencing revealed a TERT-p g.1,295,250 G>A mutation (Supplementary Figure S4). Three months later, Patient 3 underwent staged gross total resection of the right-side tumor.
Due to the complete resection of both tumors, the patient was observed without immediate adjuvant therapy. The right-side tumor had progressed on 9-month follow-up imaging, and he was treated with concurrent radiation and temozolomide, followed by adjuvant temozolomide. Radiation was directed to the right-side lesion only. At last follow-up he was without evidence of recurrent disease and continued on adjuvant temozolomide.
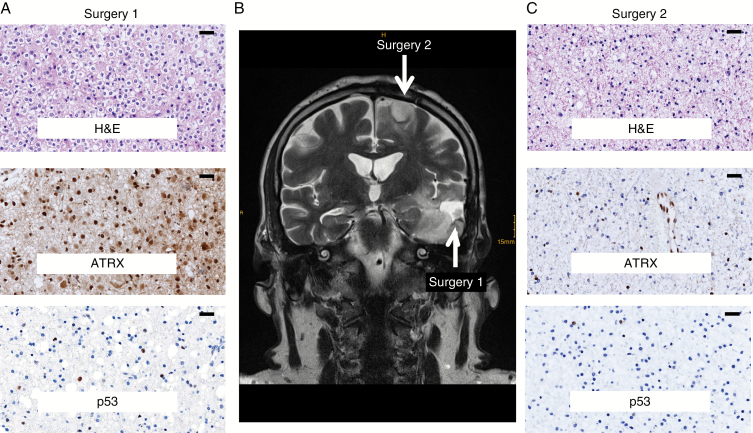
Patient 4 was a 44-year-old male at diagnosis who presented with a left temporal mass (Fig. 5). He underwent resection, which demonstrated a WHO grade II oligodendroglioma, IDH1 R132H mutant by IHC, 1p/19q codeleted by FISH. On IHC, the tumor was minimally positive for TP53, and ATRX was retained. The patient was monitored without intervention for 4 years, after which MRI showed a separate tumor in the ipsilateral frontal lobe and he underwent resection for that lesion. This demonstrated a WHO grade II diffuse astrocytoma, IDH1 R132H mutated by IHC, 1p/19q intact by FISH and loss of ATRX expression. A minor fraction of tumor cells were immunopositive for TP53. He was monitored for another 6 years before he developed recurrent disease in the left temporal lobe.
Fig. 5.
Protein expression and genetic characteristic exhibited by the multicentric lesions of Patient 4. (A) H&E and IHC for p53 and ATRX on the first surgery. (B) MRI scan of the lesions of the first and second surgeries of Patient 4. (C) H&E and IHC for p53 and ATRX on the second surgery. Scale bar, 20 μm.
Discussion
Our molecular analysis of the intrapatient and intralesion heterogeneity shows that multicentric IDH-mutant WHO grade II and III diffuse gliomas diverge at an early point in tumor evolution. In one case (Patient 1), a clearly independent origin of the 2 lesions was established. The remaining 3 cases shared just one mutation between each separate lesion, which could also represent independent origins, or alternatively, early divergence as we described previously in a patient with a solitary IDH1 R132H-mutant LGG.2 These findings and the concomitant alterations made to treatment protocols indicate the need for molecular characterization of each lesion in patients with multicentric glioma.
The interesting identification of 2 patients with spatially separated lesions composed of oligodendroglioma, IDH mutant and 1p/19q codeleted, in one lesion and diffuse astrocytoma, IDH mutant, in the other, has not been described previously to our knowledge. Six cases of solitary glioma have been previously documented with regional heterogeneity of oligodendroglioma and astrocytoma cells, and our findings add weight to the hypothesis that rare “dual-genotype” oligodendroglioma and astrocytoma may exist.33–36 Neuropathologists may incorrectly discount classification as diffuse astrocytoma due to the finding of a 1p/19q codeletion, which has ramifications on prognosis and patient therapy.
We discovered a pathogenic missense mutation of TP53 in the germline of Patient 1, which had previously been observed in Li–Fraumeni families, thus establishing Li–Fraumeni syndrome. Subsequent screening with whole body MRI revealed an additional ductal carcinoma in situ, which was excised. Although germline TP53 mutations are rare in patients with glioma, their incidence is increased in patients with multifocal and multicentric presentation, as well as history of extracranial malignancies.37 Somatic IDH1 R132C has previously been reported in 5 cases of Li–Fraumeni associated gliomas.38 The 3 additional cases provided here (1 from UCSF and 2 in the cohort from TCGA) support the hypothesis that presentation of IDH1 R132C-mutant glioma is an indication for testing for Li–Fraumeni syndrome. This further highlights the importance of follow-up sequencing for less common IDH1 or IDH2 mutations in diffuse LGG that are negative for IDH1 R132H-mutant protein by IHC. Additionally, this may be the reason for previous reports that multicentric glioma is not IDH mutated (since these cases were assessed by IHC only).39,40
For each of these multicentric cases, sampling and molecular analysis of both tumor foci were critical for establishing an optimal treatment strategy. In the first case, recognition of the IDH1 R132C mutation in the second resection specimen led to diagnosis of Li–Fraumeni syndrome, which had long-term implications for adjuvant glioma-directed therapy, as well as routine screening for extracranial malignancies. In the second case, identification of a high-grade component in the second resected tumor specimen prompted an escalation of therapy from the initial plan for treatment with chemotherapy alone on an institutional phase II clinical trial for grade II gliomas to treatment with radiation and temozolomide, which has resulted in 2.5 years of disease stability at last follow-up. In the third and fourth cases, sampling of both tumors resulted in identification of distinct tumor types, with prognosis and adjuvant therapy implications.
In summary, genomic characterization of 4 rare, multicentric IDH-mutant diffuse LGGs indicates independent evolution and early divergence of the separate lesions, high heterogeneity of early drivers in gliomagenesis, and a high likelihood of underlying pathogenic germline mutations. These findings, when extrapolated to solitary gliomas, shed light on the “gaps and overlaps” identified in the classification of diffuse LGG.41
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This project was supported by the National Institutes of Health (R01CA169316 to J.F.C.; P01CA118816-06 to J.F.C., J.J.P., and S.M.C.; P50CA097257 to J.J.P., S.M.C., and J.F.C.; 5T32CA151022-07 to L.E.J.; DP5 OD021403 to D.A.S.), the National Cancer Institute Cancer Center (5P30CA082103 to H.B.), and the Austrian Science Fund (KLI 394 to A.W.).
Acknowledgments
The authors thank the UCSF Brain Tumor Tissue Bank and the UCSF Department of Pathology for timely and significant contributions of key samples.
Conflict of interest statement. J.F.C. is a cofounder of Telo Therapeutics, Inc.
References
- 1. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK.. WHO Classification of Tumours of the Central Nervous System. 4 ed Lyon: International Agency for Research on Cancer; 2016. [Google Scholar]
- 2. Johnson BE, Mazor T, Hong C et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suzuki H, Aoki K, Chiba K et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–468. [DOI] [PubMed] [Google Scholar]
- 4. Louis DN, Perry A, Reifenberger G et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 5. Eckel-Passow JE, Lachance DH, Molinaro AM et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watanabe K, Sato K, Biernat W et al. Incidence and timing of p53 mutations during astrocytoma progression in patients with multiple biopsies. Clin Cancer Res. 1997;3(4):523–530. [PubMed] [Google Scholar]
- 7. Yan H, Parsons DW, Jin G et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Terakawa Y, Yordanova YN, Tate MC, Duffau H. Surgical management of multicentric diffuse low-grade gliomas: functional and oncological outcomes: clinical article. J Neurosurg. 2013;118(6):1169–1175. [DOI] [PubMed] [Google Scholar]
- 9. Sridharan V, Urbanski LM, Bi WL et al. Multicentric low-grade gliomas. World Neurosurg. 2017;84(4):1045–1050. [DOI] [PubMed] [Google Scholar]
- 10. Liu Q, Liu Y, Li W et al. Genetic, epigenetic, and molecular landscapes of multifocal and multicentric glioblastoma. Acta Neuropathol. 2015;130(4):587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee J-K, Wang J, Sa JK et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;54:3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jalbert LE, Neill E, Phillips JJ et al. Magnetic resonance analysis of malignant transformation in recurrent glioma. Neuro Oncol. 2016;18(8):1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol. 2002;9(5):687–705. [DOI] [PubMed] [Google Scholar]
- 14. Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20(2):289–290. [DOI] [PubMed] [Google Scholar]
- 15. Mazor T, Pankov A, Johnson BE et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28(3):307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fang H, Wu Y, Narzisi G et al. Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med. 2014;6(10):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Van der Auwera GA, Carneiro MO, Hartl C et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kyritsis AP, Bondy ML, Rao JS, Sioka C. Inherited predisposition to glioma. Neuro Oncol. 2010;12(1):104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rice T, Lachance DH, Molinaro AM et al. Understanding inherited genetic risk of adult glioma—a review. Neurooncol Pract. 2016;3(1): 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Landrum MJ, Lee JM, Riley GR et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Forbes SA, Beare D, Boutselakis H et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olshen AB, Bengtsson H, Neuvial P, Spellman PT, Olshen RA, Seshan VE. Parent-specific copy number in paired tumor-normal studies using circular binary segmentation. Bioinformatics. 2011;27(15):2038–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, Handsaker B, Wysoker A et al. ; 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Favero F, Joshi T, Marquard AM et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26(1):64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cerami E, Gao J, Dogrusoz U et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ceccarelli M, Barthel FP, Malta TM et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bell RJ, Rube HT, Kreig A et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348(6238):1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Varley JM. Germline TP53 mutations and Li-Fraumeni syndrome. Hum Mutat. 2003;21(3):313–320. [DOI] [PubMed] [Google Scholar]
- 31. Gagnebin J, Kovar H, Kajava AV et al. Use of transcription reporters with novel p53 binding sites to target tumour cells expressing endogenous or virally transduced p53 mutants with altered sequence-specificity. Oncogene. 1998;16(5):685–690. [DOI] [PubMed] [Google Scholar]
- 32. Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19(6):607–614. [DOI] [PubMed] [Google Scholar]
- 33. Barresi V, Lionti S, Valori L, Gallina G, Caffo M, Rossi S. Dual-genotype diffuse low-grade glioma: is it really time to abandon oligoastrocytoma as a distinct entity?J Neuropathol Exp Neurol. 2017;76(5):342–346. [DOI] [PubMed] [Google Scholar]
- 34. Huse JT, Diamond EL, Wang L, Rosenblum MK. Mixed glioma with molecular features of composite oligodendroglioma and astrocytoma: a true “oligoastrocytoma”?Acta Neuropathol. 2015;129(1):151–153. [DOI] [PubMed] [Google Scholar]
- 35. Wilcox P, Li CC, Lee M et al. Oligoastrocytomas: throwing the baby out with the bathwater?Acta Neuropathol. 2015;129(1):147–149. [DOI] [PubMed] [Google Scholar]
- 36. Qu M, Olofsson T, Sigurdardottir S et al. Genetically distinct astrocytic and oligodendroglial components in oligoastrocytomas. Acta Neuropathol. 2007;113(2):129–136. [DOI] [PubMed] [Google Scholar]
- 37. Kyritsis AP, Bondy ML, Xiao M et al. Germline p53 gene mutations in subsets of glioma patients. J Natl Cancer Inst. 1994;86(5):344–349. [DOI] [PubMed] [Google Scholar]
- 38. Watanabe T, Vital A, Nobusawa S, Kleihues P, Ohgaki H. Selective acquisition of IDH1 R132C mutations in astrocytomas associated with Li-Fraumeni syndrome. Acta Neuropathol. 2009;117(6):653–656. [DOI] [PubMed] [Google Scholar]
- 39. Karlowee V, Amatya VJ, Hirano H et al. Multicentric glioma develops via a mutant IDH1-independent pathway: immunohistochemical study of multicentric glioma. Pathobiology. 2017;84(2):99–107. [DOI] [PubMed] [Google Scholar]
- 40. Inoue A, Ohnishi T, Kohno S et al. A case of multicentric gliomas in both supra- and infratentorial regions with different histology: a case report. World J Surg Oncol. 2016;14(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. The Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.