Abstract
Microbial eukaryotes, including amoeboids, display diverse and complex life cycles that may or may not involve sexual reproduction. A recent comprehensive gene inventory study concluded that the Amoebozoa are ancestrally sexual. However, the detection of sex genes in some lineages known for their potentially sexual life cycle was very low. Particularly, the genus Cochliopodium, known to undergo a process of cell fusion, karyogamy, and subsequent fission previously described as parasexual, had no meiosis genes detected. This is likely due to low data representation, given the extensive nuclear fusion observed in the genus. In this study, we generate large amounts of transcriptome data for 2 species of Cochliopodium, known for their high frequency of cellular and nuclear fusion, in order to study the genetic basis of the complex life cycle observed in the genus. We inventory 60 sex-related genes, including 11 meiosis-specific genes, and 31 genes involved in fusion and karyogamy. We find a much higher detection of sex-related genes, including 5 meiosis-specific genes not previously detected in Cochliopodium, in this large transcriptome data. The expressed genes form a near-complete recombination machinery, indicating that Cochliopodium is an actively recombining sexual lineage. We also find 9 fusion-related genes in Cochliopodium, although no conserved fusion-specific genes were detected in the transcriptomes. Cochliopodium thus likely uses lineage specific genes for the fusion and depolyploidization processes. Our results demonstrate that Cochliopodium possess the genetic toolkit for recombination, while the mechanism involving fusion and genome reduction remains to be elucidated.
Keywords: fusion, gene inventory, karyogamy, meiosis, sexual reproduction, transcriptome
Sexual reproduction, commonly defined as a process of ploidy reduction and restoration via meiosis and nuclear fusion (karyogamy) of gametes, respectively, is one of the major features of eukaryotes. Although well-documented in multicellular eukaryotes (i.e., plants and animals), sexual processes are generally poorly understood in most microbial eukaryotes including Amoebozoa (Wenrich 1954; Raikov 1982; Kondrashov 1997; Parfrey et al. 2008; Lahr et al. 2011; Tekle et al. 2014; Garg and Martin 2016). This is mainly due to diverse and complex life cycles and difficulties in cultivating and observing sexual reproduction in microorganisms. Moreover, most eukaryotic microbes were previously assumed be to asexual (Haeckel 1866; Maynard Smith 1978). More recently, however, sex or recombination has been observed in a number of microbial lineages across the eukaryotic tree of life (Erdos et al. 1973, 1975; Blanc et al. 1989; Mihake 1996; Chepurnov et al. 2004; Singh et al. 2013). In others, processes similar to sex but apparently missing one or more parts of the full sexual cycle, including meiosis, have been documented (Poxleitner et al. 2008; Carpenter et al. 2012; Tekle et al. 2014). These sex-like processes, where parts of the conventional sexual stages as seen in plants and animals are not observed, are collectively called “parasexual.” Amoebozoans display diverse parasexual life cycles, involving presumed sexual stages during the cyst (Erdos et al. 1973; Mignot and Raikov 1992; Singh et al. 2013) or vegetative stage (Tekle et al. 2014), or alternating sexual and asexual stages (Schaudinn 1899). The genetic basis of these various parasexual pathways and their evolutionary origin is poorly understood. However, genetic evidence, including gene inventories documenting meiosis and karyogamy, has been reported in various microbial eukaryotes including amoebozoans, indicating that sex is ancestral in eukaryotes (Ramesh et al. 2005; Malik et al. 2008; Chi et al. 2014a, 2014b; Patil et al. 2015; Speijer et al. 2015; Tekle et al. 2017). Genetic and cytological studies elucidating the diverse parasexual pathways observed in microbial eukaryotes will greatly enhance our understanding of the evolution and origin of sex in eukaryotes.
Cochliopodium, the focus of this study, is a genus of discoid or globose amoebae within the supergroup Amoebozoa (order Himatismenida), characterized by a dorsal covering, or tectum, composed of surface microscales. The genus consistently forms a strongly-supported monophyletic group (Kudryavtsev et al. 2005, 2011; Lahr et al. 2011; Tekle et al. 2008, 2016). The genus includes around 25 described species (Kudryavtsev et al. 2005, 2011; Anderson and Tekle 2013; Tekle et al. 2013, 2015), several of which are known to undergo a unique and unusual process of cellular fusion, karyogamy, and fission (Tekle et al. 2014). During this process, multiple cells from the same culture undergo cellular fusion resulting in giant, multinucleated cells, which then undergo karyogamy to form polyploid nuclei. These cells and their nuclei eventually fragment back into single, uninucleate cells, apparently without undergoing visible meiosis. This lack of observation of conventional meiosis, has led to the description of this process as parasexual (Tekle et al. 2014). The genetic basis of fusion and karyogamy in Cochliopodium, as well as the mechanism of the fragmentation process, is currently unknown.
Cellular fusion is commonly observed in most eukaryotic clades including several members of Amoebozoa (Page 1971; Seravin and Goodkov 1984; Michel and Smirnov 1999; Lahr et al. 2011; Tekle et al. 2014). However, the genetic basis of cellular fusion and karyogamy in eukaryotes has been difficult to elucidate. This is mainly due to the fact that many of the involved genes are lineage-specific (Chen et al. 2007; Aguilar et al. 2013). However, a few genes involved in fusion seem to be conserved across eukaryotic species. For example, the cell fusion protein HAP2 (GCS1), an ancient derivative of the viral class II fusogens (Fedry et al. 2017; Pinello et al. 2017) has been detected across major eukaryotic clades (Wong and Johnson 2010; Speijer et al. 2015). The functional conservation of HAP2 has been confirmed in deletion and rescue assays in diverse lineages such as Chlamydomonas and Plasmodium (Hirai et al. 2008; Liu et al. 2008), Tetrahymena (Cole et al. 2014), and Dictyostelium (Okamoto et al. 2016). Even in some organisms where HAP2 does not seem to be present, such as nematodes and vertebrates (Wong and Johnson 2010; Speijer et al. 2015), other viral class II fusogens have been described which function in gamete or somatic cell fusion (Avinoam and Podbilewicz 2011; Perez-Vargas et al. 2014). Another conserved protein is GEX1 (fungal ortholog: KAR5; Ning et al. 2013), a karyogamy gene, which is even more widespread in eukaryotes than HAP2 (Speijer et al. 2015). GEX2 is known to be functionally conserved in fungi (Beh et al. 1997; Melloy et al. 2009), Chlamydomonas, Plasmodium (Ning et al. 2013), and vertebrates (Abrams et al. 2012). These genes are among the prime targets for detection in Cochliopodium, given the extensive cellular and nuclear fusion observed in the genus. The detection of these genes in Cochliopodium would indicate fusion and karyogamy in Cochliopodium are derived from evolutionarily conserved eukaryotic gamete fusion factors, supporting sexual nature of the behavior observed in this amoeba.
Meiosis, a signature of sexual reproduction, is a process by which genetic material is exchanged and halved in progeny cells. Unlike fusion, the genetic toolkit of meiosis is well-conserved across eukaryotes, enabling the formation of a core inventory of meiotic machinery, which can be tested in diverse eukaryotes (Ramesh et al. 2005; Malik et al. 2008; Schurko and Logsdon 2008). Meiotic gene inventories take a genomics-level approach to documenting meiosis in lineages where sexual reproduction has not previously been recognized. This approach has successfully been used to detect complements of meiotic genes in diverse eukaryotic microbial lineages such as Diplomonads (Ramesh et al. 2005), Parabasalia (Malik et al. 2008), Ciliates (Chi et al. 2014), Dinoflagellates (Chi et al. 2014), Diatoms (Patil et al. 2015), and Amoebozoa (Tekle et al. 2017). The presence of meiosis genes in a lineage is thought to indicate the presence of meiosis, as these genes would likely decay by genetic drift or other processes if meiosis was lost. Because of its life cycle, Cochliopodium is an excellent target for an inventory of meiosis and sex-related genes to determine whether it is (or was recently) capable of true sexual reproduction. In general, these inventories are performed on whole genomes of an organism (e.g., Ramesh et al. 2005; Malik et al. 2008; Chi et al. 2014a, 2014b; Patil et al. 2015) to ensure maximal detection of sex-related genes. However, no Cochliopodium genome has been sequenced, and sequencing such a genome is challenging at present due to the size and complexity of Amoebozoa genomes (Glöckner and Noegel 2012). Instead, transcriptome data, which is easier to collect from Amoebozoans, can be de novo assembled and inventoried. This will likely miss some genes that are lowly- or nonexpressed, however, it will also provide evidence for the activity and functionality of the genes in the organism.
A previous study found that Amoebozoa is ancestrally sexual, with sex genes well distributed across its phylogeny (Tekle et al. 2017). However, the life cycles found in the supergroup are extremely diverse and can be complex, with varying amounts of a/sexuality (Parfrey et al. 2008; Lahr et al. 2011). The genomes included in the previous study showed an abundance of sex and meiosis-specific genes. However, the wide variation in detection of the sex genes across clades and species (Tekle et al. 2017) resulted in little ability to determine possible genetic bases for the life cycles represented by the transcriptomes. One such transcriptome of a Cochliopodium species, Cochliopodium minutoidum, had a very low detection of sex genes and no meiosis-specific genes, which was surprising given the parasexual life cycle of the genus. However, the transcriptome used had very low amount of data, which may have led to an artificially low detection of sex genes (Tekle et al. 2017). Additionally, this species fuses much less frequently than some other species within the genus (Tekle et al. 2014), so genes related to its life cycle may not have been highly expressed when the data were collected.
In this study, we used next-generation sequencing methods to generate more comprehensive transcriptomes from 2 cochliopodiums, Cochliopodium pentatrifurcatum and Cochliopodium minus, known to undergo extensive cellular and nuclear fusion. Using these data we inventoried 31 genes involved in fusion and karyogamy in various species, including HAP2 and GEX1, as well as a general meiotic inventory of 11 meiosis-specific and 49 sex-related genes in Cochliopodium, to determine whether its unusual life cycle could be based on conserved sex genes. This deep sequencing enabled a much higher detection of the sex-related genes, including 5 meiosis-specific genes, than from the previous C. minutoidum study (Tekle et al. 2017). Three fungal plasmogamy and 6 fungal karyogamy genes were also detected. Although neither HAP2 nor GEX1 were detected in Cochliopodium, the much greater detection of the sex genes lends strong support to the sexual nature of the complex life cycle observed in Cochliopodium.
Materials and Methods
RNA Isolation and cDNA Synthesis
Cochliopodium pentatrifurcatum and C. minus (CCAP 1357/1A) were grown in a minimum of 4 plastic petri dishes with bottled natural spring water (Deer Park®; Nestlé Corp. Glendale, CA) and autoclaved grains of rice. We observed cultures to confirm that all life cycle stages were represented in large quantities before collecting RNA. After minimum 1 week incubation at room temperature, cells in culture had begun the fusion process, resulting in a mixture of the life cycle stages. Adherent amoebae were removed from dishes and transferred to 15 mL conical tubes. Total RNA was isolated from collected amoebae using a NucleoSpin® RNA kit (Macherey-Nagel, Düren, Germany) according to manufacturer’s protocol. RNA was quantified using a Qubit® RNA HS Assay Kit and Qubit® 3.0 fluorometer (Life Technologies, Carlsbad, CA). Quality of isolated RNA was measured by visualizing on an Ambion® NorthernMax® denaturing formaldehyde gel (Thermo Fisher Scientific, Waltham, MA). Small (18S) and large (28S) subunits of rRNA bands were inspected visually for intensity and smearing to assess RNA degradation. cDNA was synthesized from total RNA using a Thermo Scientific™ RevertAid™ Premium Double Stranded cDNA Synthesis Kit (Thermo Fisher Scientific). cDNA quantity was measured using a Qubit® DNA HS Assay Kit on the above Qubit® fluorometer.
To maximize our detection of the inventoried meiosis and fusion genes, data were also included from other RNA-seq experiments in both species. Additional cDNA was generated from C. pentatrifurcatum by picking 1–5 single cells from culture into 0.2 mL PCR tubes. RNA was isolated from the cells and cDNA was synthesized using a SMART-Seq® v4 Ultra® Low Input RNA Kit (Takara Bio USA, Mountain View, CA) per manufacturer’s instructions, except that one quarter of the recommended reagents were used. Additional cDNA was also generated from C. minus by isolating total RNA with the NucleoSpin® kit, then synthesizing cDNA with the SMART-Seq® kit. cDNA quantity was measured using the DNA HS Assay kit with the Qubit® flurometer, as above.
Preparation of Libraries and Sequencing
Libraries were prepared from 1 ng cDNA using a Nextera® XT DNA Library Preparation Kit (Illumina, Inc., San Diego, CA) according to manufacturer’s instructions, without the normalization step. Libraries were quantified using the DNA HS Kit with the Qubit® fluorometer, as above, and sequenced on a HiSeq 2500 in paired-end, high-output mode with 75 bp reads at Yale Center for Genomic Analysis. 10 cDNA libraries were multiplexed in one sequencing lane—4 from C. pentatrifurcatum, 2 from C. minus, and 4 from other Amoebozoa (Pellita catalonica, Mayorella cantabrigiensis, Flamella aegyptia, and Endostelium zonatum) not used in this study.
Sequence Assembly
Sequencing generated 28.8–72 million raw reads per sample (Supplementary Table S1). FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to inspect reads for quality and length. BBDuk (Joint Genome Institute, US Department of Energy, Walnut Creek, CA) was used to remove low quality bases (Phred score below 30) and Illumina adaptor sequences from the reads, discarding short reads (length below 35) after trimming (Supplementary File S1). The remaining reads from each sample were separately assembled de novo using rnaSPAdes (Bankevich et al. 2012) with default parameters (Supplementary File S1). Contigs less than 300 bp in length were removed from each assembly. Ribosomal sequences were sorted into a separate file by blasting the assemblies with blastn (Altschul et al. 1990) against a Refseq database of ribosomal sequences, with an e-value cutoff of 10−10 (Supplementary File S1). All hits below the cutoff were removed. The remaining contigs were then enriched for eukaryotic sequences using usearch, a faster alternative to blastx (Edgar 2010). Contigs were blasted with usearch against RefSeq eukaryotic and prokaryotic databases with an e-value cutoff of 10−15 (Supplementary File S1). Contigs were kept if the e-value for the best eukaryotic hit was more than 100 times smaller than the e-value for the best bacterial hit, or if there was a eukaryotic hit but no bacterial hit. The resulting eukaryotic and ribosomal contigs from each assembly were then combined into 2 files representing C. pentatrifurcatum and C. minus and duplicate contigs within each file were removed.
Gene Inventory
A total of 31 genes involved directly or indirectly in fusion or karyogamy in various organisms were chosen for inventory in Cochliopodium based on a literature review searching for factors known to be involved in fusion or karyogamy in other eukaryotic organisms—mostly fungi (Rose 1996; Melloy et al. 2009; Aguilar et al. 2013; Merlini et al. 2013). These genes were added to 11 meiosis-specific and 49 sex-related genes, chosen based on previous studies (Ramesh et al. 2005; Malik et al. 2008; Schurko and Logsdon 2008; Chi et al. 2014a, 2014b; Patil et al. 2015; Tekle et al. 2017). The new transcriptomes were inventoried for these genes following the methods of Tekle et al. (2017). Briefly, sequences for each gene were collected from orthologous groups (OGs) within the OrthoMCL database (http://www.orthomcl.org/orthomcl/) via a phylogenomic pipeline developed by Grant and Katz (2014), or directly from the database when the OG was not present in the pipeline. We used a custom Python script (Supplementary File S2) to blast each OG against each transcriptome using tblastn (Altschul et al. 1990), collecting all nonredundant hits with an e-value less than 1e-15. To ensure that no distant homologs were missed, we also used HMMer (hmmer.org, version 3.1b2) to build profiles from alignments of the OG sequences and search translated versions of the transcriptomes (Supplementary File S1). To remove potential paralogous false positives among the meiosis-specific genes with positive hits in Cochliopodium, hits were aligned with sequences previously discovered by Tekle et al. (2017) using MAFFT (Katoh and Standley 2013), columns with more than 75% missing data were removed, and gene trees were built using RAxML (Stamatakis et al. 2008) as implemented on the CIPRES portal (Miller 2010) with PROTGAMMALGF matrix and default parameters (Supplementary File S1). Similarly, to confirm hits and assess broad conservation or lineage specificity in the chosen fusion and karyogamy genes, Blast search was conducted on 6 outgroup taxa from across eukaryotic subgroups. Hits were collected and aligned with the Cochliopodium hits, and gene trees were inferred using RAxML as above (Supplementary File S1). Sequences from each cluster on the resulting trees were blasted using blastp against NCBI’s nonredundant (nr) protein database to determine cluster identity and definitively assign presence/nondetection to the new transcriptomes. When the number of returned hits for an OG was too large to infer the sequence tree in a timely manner, the alignments were manually inspected and sequences were blasted against the NCBI nr database to confirm the identity of the hits. We also re-inventoried the transcriptome of C. minutoidum as a control to ensure that genes in the new transcriptomes were not over- or under-detected compared to the previous inventory (Tekle et al. 2017).
Results
Transcriptome Data
Extensive sequencing was used to generate transcriptomes for 2 species of Cochliopodium, C. minus and C. pentatrifurcatum. The amount of data for each of these transcriptomes is much larger than the previous transcriptome of C. minutoidum; the C. minus transcriptome contains 11078 nonredundant eukaryotic contigs in 17.5 MB of data, while the C. pentatrifurcatum transcriptome contains 9430 contigs in 11.8 MB of data (Supplementary Table S1). For comparison, the previous C. minutoidum transcriptome (Cavalier-Smith et al. 2015) had 4013 contigs in only 1.4 MB of data.
Meiosis Specific and Sex-Related Genes
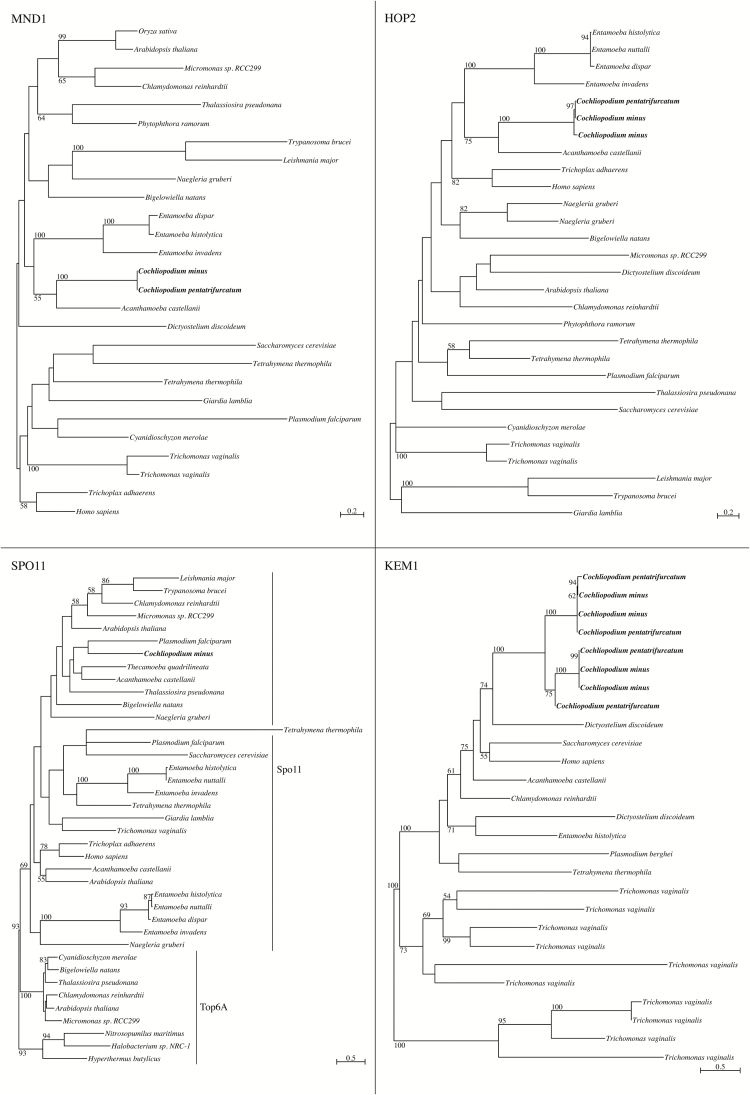
A total of 60 sex-related genes, including 11 meiosis-specific genes, were inventoried in the transcriptomes of Cochliopodium. Of the 11 meiosis-specific genes inventoried, 5 were detected in Cochliopodium (Table 1; Figure 1). Four of these were detected in both Cochliopodium species (DMC1, MND1, HOP2, and MSH4), while one, SPO11, was only detected in C. minus (Table 1; Figure 1). Of the genes not detected in Cochliopodium, 4 (HOP1, RED1, ZIP1, and REC8) are also absent in the rest of Amoebozoa and are thus unlikely to be present in Cochliopodium, while the remaining 2 (MER3 and MSH5) are detected to varying degrees in the rest of Amoebozoa (Tekle et al. 2017). None of these genes were detected in C. minutoidum (Table 1; Figure 1), congruent with previous study (Tekle et al. 2017). A total of 49 nonmeiosis-specific sex-related genes were also inventoried in Cochliopodium. Of these, 35 were detected in both C. pentatrifurcatum and C. minus, and 7 of these 35 were also detected in C. minutoidum (Table 1). An additional 4 were detected in 1 of the 2 species; 3 in C. minus and 1 in C. pentatrifurcatum (Table 1).
Table 1.
Meiosis gene inventory in 3 Cochliopodium species
| Gene | OG | Cochliopodium minus | Cochliopodium pentatrifurcatum | Cochliopodium minutoidum |
|---|---|---|---|---|
| Bouquet formation | ||||
| SAD1 | OG5_129586 | + | + | nd |
| Cell cycle regulation | ||||
| CDC20 | OG5_126765 | + | + | nd |
| Crossover regulation | ||||
| DMC1 | OG5_126834 | + | + | nd |
| HOP1 | OG5_128667 | nd | nd | nd |
| HOP2 | OG5_128568 | + | + | nd |
| MER3 | OG5_129931 | nd | nd | nd |
| MND1 | OG5_127882 | + | + | nd |
| MSH4 | OG5_130077 | + | + | nd |
| MSH5 | OG5_129379 | nd | nd | nd |
| RED1 | OG5_180525 | nd | nd | nd |
| ZIP1 | OG5_171209 | nd | nd | nd |
| DNA damage sensing/response | ||||
| MEC1/ATR | OG5_128386 | + | + | + |
| MRE11 | OG5_127969 | + | + | nd |
| RAD1/MEI9 | OG5_130438 | + | + | nd |
| RAD17 | OG5_127538 | nd | + | nd |
| RAD23 | OG5_130351 | + | + | nd |
| RAD24 | OG5_126706 | + | + | + |
| RAD50 | OG5_127792 | + | + | + |
| TEL1/ATM | OG5_128955 | + | + | nd |
| XRS2 | OG5_180328 | nd | nd | nd |
| Double-strand break formation | ||||
| SPO11 | OG5_127274 | + | nd | nd |
| REC114 | OG5_142109 | nd | nd | nd |
| Double-strand break repair (nonhomologous end joining) | ||||
| KU70 | OG5_129086 | + | + | nd |
| KU80 | OG5_129372 | + | + | nd |
| LIG4/DNL1 | OG5_130132 | + | + | nd |
| XRCC4/LIF1 | OG5_135131 | nd | nd | nd |
| Recombinational repair | ||||
| BRCA1 | OG5_159932 | + | + | nd |
| BRCA2 | OG5_131863 | + | nd | nd |
| DNA2 | OG5_129631 | + | + | nd |
| EXO1 | OG5_127511 | + | + | nd |
| FEN1 | OG5_127472 | + | + | nd |
| MLH1 | OG5_127201 | + | + | nd |
| MLH2 | OG5_202562 | nd | nd | nd |
| MLH3 | OG5_130552 | + | + | nd |
| MMS4/EME1 | OG5_135664 | nd | nd | nd |
| MPH1/FANCM | OG5_128649 | + | + | nd |
| MSH2 | OG5_127538 | + | + | nd |
| MSH3 | OG5_130351 | + | + | nd |
| MSH6 | OG5_126895 | + | + | nd |
| MUS81 | OG5_129162 | + | nd | nd |
| PMS1 | OG5_128001 | + | + | nd |
| RAD51 | OG5_126834 | + | + | nd |
| RAD52 | OG5_130806 | nd | nd | nd |
| RAD54 | OG5_127098 | + | + | nd |
| RTEL1 | OG5_127294 | + | + | nd |
| SAE2 | OG5_138817 | nd | nd | nd |
| SGS1 | OG5_126644 | + | + | nd |
| SLX1 | OG5_128732 | nd | nd | nd |
| SLX4 | OG5_136855 | nd | nd | nd |
| SMC5 | OG5_128615 | + | + | nd |
| SMC6 | OG5_127751 | + | + | + |
| YEN1 | OG5_132593 | nd | nd | nd |
| Sister chromatid cohesion | ||||
| PDS5 | OG5_128901 | + | nd | nd |
| RAD21 | OG5_129513 | + | + | nd |
| REC8 | OG5_150817 | nd | nd | nd |
| SCC3 | OG5_127983 | + | + | nd |
| SMC1 | OG5_127449 | + | + | + |
| SMC2 | OG5_127360 | + | + | + |
| SMC3 | OG5_127789 | + | + | nd |
| SMC4 | OG5_127440 | + | + | + |
| Total detected | 42 | 39 | 7 | |
Meiosis-specific genes are highlighted in bold. "+" = detected, "nd" = not detected.
Figure 1.
Maximum-likelihood phylogenetic trees for the meiosis-specific genes MND1, HOP2, and SPO11 and the fusion-related gene KEM1. Trees inferred with RAxML on the Cipres Science Gateway using GAMMALGF model of evolution. Bootstrap support values above 50% are shown above or below their respective branches. MND1, HOP2, and KEM1 trees rooted at midpoint; SPO11 tree rooted based on prokaryotic outgroup position.
Fusion- and Karyogamy-Related Gene Inventory
Of the 31 fusion- and karyogamy-related proteins inventoried in this study, 9 were detected in Cochliopodium (Table 2; Figure 1). Two of these genes—BNI1 and MYO2—are actin-associated proteins involved in plasmogamy in fungi; one, KEX2, is a protein processing enzyme acting upstream of plasmogamy factors; and the remaining 6—CDC28, KEM1, KAR3, CIN4, CDC4, and KAR2—are fungal karyogamy factors. Notably, neither HAP2 nor GEX1 were detected in Cochliopodium (Table 2). Homologs of all of the above proteins except CDC4, as well as 4 other fusion and karyogamy genes (CIN2, KAR4, JEM1, and SEC63), were found across eukaryotic supergroups (Table 2; Figure 1). CDC4 and 2 other genes (RVS161 and CDC34) were detected only in Amorphea (Opisthokonts + Amoebozoa) (Table 2). The remaining genes were specific to Opisthokonts (Table 2), many of which were only found in the group they were originally described in, either fungi (Saccharomyces) or metazoa (Homo) (data not shown).
Table 2.
Inventory of genes involved in plasmogamy (including cellular congression) and karyogamy (including nuclear congression) in Cochliopodium and in genomes from across the major eukaryotic supergroups
| Gene | OG(s) | Cochliopodium | Other Amoebozoa | Opisthokonts | Excavates | Plants | SAR |
|---|---|---|---|---|---|---|---|
| Cellular congression | |||||||
| BNI1 | OG5_127406 | + | + | + | n/d | + | + |
| TPM1 | OG5_127228 | n/d | n/d | + | n/d | n/d | n/d |
| Plasmogamy | |||||||
| CD9 | OG5_136376 | n/d | n/d | + | n/d | n/d | n/d |
| FIG1a | OG5_138630 | n/d | n/d | + | n/d | n/d | n/d |
| FUS1a | OG5_156961 | n/d | n/d | + | n/d | n/d | n/d |
| FUS2a | OG5_171667, OG5_216847 | n/d | n/d | + | n/d | n/d | n/d |
| HAP2 (GCS1) | OG5_135474, OG5_143221 | n/d | + | n/d | n/d | + | + |
| IZUMO1a | OG5_151190 | n/d | n/d | + | n/d | n/d | n/d |
| KEX2 | OG5_127788 | + | + | + | + | n/d | + |
| MYO2 | OG5_126577 | + | + | + | + | + | + |
| PRM1a | OG5_136560 | n/d | n/d | + | n/d | n/d | n/d |
| RVS161 | OG5_129890 | n/d | + | + | n/d | n/d | n/d |
| Nuclear congression | |||||||
| BIK1a | OG5_139423 | n/d | n/d | + | n/d | n/d | n/d |
| CDC4 | OG5_129129 | + | + | + | n/d | n/d | n/d |
| CDC28 | OG5_126712 | + | + | + | + | + | + |
| CDC34 | OG5_129084 | n/d | + | + | n/d | n/d | n/d |
| CDC37a | OG5_133705, OG5_135666 | n/d | n/d | + | n/d | n/d | n/d |
| CIK1a | OG5_147436 | n/d | n/d | + | n/d | n/d | n/d |
| CIN1a | OG5_156139 | n/d | n/d | + | n/d | n/d | n/d |
| CIN2a | OG5_132672, OG5_180432 | n/d | + | + | + | + | + |
| CIN4 | OG5_128319 | + | + | + | + | + | + |
| KAR3 | OG5_126975, OG5_136029 | + | + | + | + | + | + |
| KAR4 | OG5_130096 | n/d | n/d | + | n/d | + | + |
| KAR9a | OG5_156957 | n/d | n/d | + | n/d | n/d | n/d |
| KEM1 | OG5_126774 | + | + | + | + | + | + |
| Karyogamy | |||||||
| JEM1a | OG5_163504 | n/d | + | + | + | n/d | n/d |
| KAR2 | OG5_126588 | + | + | + | + | + | + |
| GEX1/KAR5 | OG5_142622, OG5_144651, OG5_156781, OG5_168665 | n/d | + | + | + | + | + |
| SEC63 | OG5_128413 | n/d | + | + | n/d | + | + |
| SEC66a | OG5_135424 | n/d | n/d | + | n/d | n/d | n/d |
| SEC72a | OG5_136566 | n/d | n/d | + | n/d | n/d | n/d |
Taxa included in Amoebozoa were Acanthamoeba castellanii, Dictyostelium discoideum, and Entamoeba histolytica; in Opisthokonts, Homo sapiens and Saccharomyces cerevisiae; In Excavates, Trichomonas vaginalis; in Plants, Arabidopsis thaliana and Chlamydomonas reinhardtii; and in SAR, Plasmodium berghei and Tetrahymena thermophila. “+” means gene was detected in the group; “n/d” means the gene was not detected but may still be present in other members of the group not included here.
aBased on initial blastp results only; not confirmed by alignment inspection or tree inference.
Discussion
Extensive Transcriptome Sequencing Reveals Meiosis Genes in Cochliopodium
A recent study of sex gene inventory in Amoebozoa based on genome and transcriptome data showed that most amoebae possess and express several sex-related and meiosis-specific genes (Tekle et al. 2017). This study concluded that Amoebozoa is ancestrally sexual. However, the detection of sex genes in Amoebozoans was not uniform. Many of the transcriptomes showed a lower detection of sex genes than the genomes, and this detection varied with the amount of data; that is, amoebae represented by large transcriptomes had more sex-related/meiosis genes detected than those represented by smaller amounts of transcriptome data (Tekle et al. 2017). This, combined with the diversity and complexity of Amoebozoan parasexual cycles, limited the ability to determine the genetic basis for any particular Amoebozoan life cycle. In C. minutoidum, none of the meiosis exclusive genes were detected (Tekle et al. 2017). This result was surprising given the reported parasexual life cycle of Cochliopodium involving extensive cellular and nuclear fusion (Tekle et al. 2014). Cochliopodium minutoidum is among species that is represented by a very low amount of transcriptome data, therefore, it is likely that the lower detection of sex genes might be due to the amount of data analyzed. It is also possible that available data reflected the physiological state of the amoeba at the time the transcriptome data were collected, or might correspond to the lower rate of fusion observed in this amoeba (Tekle et al. 2014). In this study, we collected much larger amounts of transcriptome data, representing the different phases of the life cycle, of 2 cochliopodiums that are known for their high frequency of cellular and nuclear fusion (Tekle et al. 2014). The newly sequenced transcriptomes of C. pentatrifurcatum and C. minus both have a much higher representation of data than the previously sequenced transcriptome of C. minutoidum. These newly generated data not only show a much higher detection of sex-related genes in general but also detection of meiosis specific genes in both cochliopodiums (Table 1; Figure 1)—in fact, the detection of sex genes in this study is comparable to that of sequenced genomes in the previous study (Tekle et al. 2017). This finding lends support to the sexual nature of the life cycle of Cochliopodium involving extensive cellular and nuclear fusion followed by fission.
The 5 meiosis-specific genes detected in Cochliopodium (Table 1; Figure 1) are all directly involved in the recombination process. SPO11 initiates recombination by causing double-stranded breaks (Keeney et al. 1997). HOP2 and MND1 form a heterodimer, which interacts with DMC1 and its nonmeiosis-specific homolog, RAD51, to promote homologous recombination (Chen et al. 2004; Bugreev et al. 2011). Finally, MSH5 acts to resolve crossover intermediates (Hollingsworth et al. 1995), usually as a heterodimer with MSH4 (Novak et al. 2001; Nishant et al. 2010). Thus, genes for the full primary pathway of recombination are present in Cochliopodium. Furthermore, the functions performed by the meiotic genes not detected in Cochliopodium could likely be replaced using nonmeiosis-specific genes. For example, REC8, which is involved in establishing the cohesin complex that holds sister chromatids together during meiosis (Watanabe and Nurse 1999), has a nonmeiosis-specific homolog, RAD21, which performs the same function in mitosis (Michaelis et al. 1997). RAD21 is present in both cochliopodiums, and the functions of RAD21 and REC8 appear to be interchangeable when only one is present in an organism (Xu et al. 2004; Howard-Till et al. 2013; Patil et al. 2015). As in the rest of Amoebozoa, the elements forming the synaptonemal complex (SC) are also not detected in Cochliopodium (Table 1; Tekle et al. 2017). However, alternate pathways to meiosis not involving the synaptonemal complex do exist and likely replace the conventional pathway in Cochliopodium (Lukaszewicz et al. 2013; Chi et al. 2014; Shodhan et al. 2014). This is also congruent with the lack of observation of structures such as meiotic spindle fibers and SC during the Cochliopodium life cycle (Tekle et al. 2014). Taken together, this evidence indicates that a functional recombination pathway exists in Cochliopodium, most likely facilitating genetic exchange in the giant, polyploid nuclei during karyogamy (Tekle et al. 2014).
Despite the larger transcriptome sampling, some notable nondetections among the meiosis-specific genes may indicate that the new transcriptomes still do not capture all of the genes present in Cochliopodium. For example, MSH4 is not detected in either of the new transcriptomes, while its interaction partner, MSH5, is detected in both transcriptomes (Table 1). The 2 genes are almost always either both present or both absent in other inventoried genomes (e.g., Malik et al. 2008; Schurko and Logsdon 2008), including other amoebozoans (Tekle et al. 2017), due to their shared function during meiosis (Nishant et al. 2010). Thus, MSH4 is likely present in Cochliopodium, but not detected in our transcriptomes, probably due to low or no expression when the transcriptomes were collected. Additionally, SPO11, the near-universally conserved initiator of meiosis, was detected in C. minus but not in C. pentatrifurcatum (Table 1; Figure 1). It is likely that, similar to MSH4, our C. pentatrifurcatum samples were not at the right life stage to express SPO11. Alternatively, SPO11 may have been so lowly expressed as to be missed in our transcriptome data, even with our extensive sequencing of the transcriptome. While transcriptome data are easy to generate and can ascertain the functionality of detected genes in the life cycle of an organism, lack of detection has no conclusive biological meaning. To fully understand the exact mechanism and sexual nature of Cochliopodium, data from the whole genome of a Cochliopodium is required.
Exploring the Genetic Basis of Cellular and Nuclear Fusion in Cochliopodium
The genetic basis of the life cycle of Cochliopodium, involving plasmogamy, karyogamy, and fission, is not well understood. The lack of detection of the conserved gamete fusion factors HAP2 and GEX1 in Cochliopodium deepens this mystery. It is possible that, as with the meiosis genes, these fusion factors are present in the Cochliopodium genome, but were not found in the transcriptome due to low or no expression in the cells when our samples were collected. Alternately, our homology methods may not be sensitive enough to detect Cochliopodium HAP2 and GEX1 orthologs. Unlike the well-conserved meiosis genes, HAP2, GEX1 and other fusion genes are highly divergent in different species (as low as 20% protein sequence identity in GEX1 orthologs, for example). This is evident given their spread across multiple OGs in the OrthoMCL database (Table 2). BLAST and HMMer based search tools were unable to detect even known orthologs in the wider eukaryote inventory if the full complement of OGs for each gene were not included (data not shown). If Cochliopodium sequences are similarly divergent from other eukaryote sequences, an alternate, more sensitive method of detection will be needed to detect them.
Some other fusion and karyogamy factors are detected in Cochliopodium (Table 2; Figure 1). However, despite these genes being required for fusion and karyogamy in fungi, they may not be similarly required for Cochliopodium fusion. None of the detected fusion factors are specific to fusion. Some are involved in more general cellular processes related to fusion, such as regulation of the cytoskeleton (Stearns et al. 1990; Sagot et al. 2002) or the cell cycle (Mendenhall and Hodge 1998; Goh and Surana 1999). Others act upstream of fusion as gene regulators (Fuller et al. 1989; Rose et al. 1989; Mendenhall and Hodge 1998; Kim et al. 2004) or as transporters (Johnston et al. 1991; Endow et al. 1994). The inventoried factors directly involved in the fusion process were mostly lineage-specific in our wider eukaryote inventory (Table 2), and it is likely that the same is true for direct fusion factors in Cochliopodium. Finding these lineage-specific genes will require an alternate approach, such as whole genome data coupled with differential gene expression analyses and cytological studies across the different stages of the Cochliopodium life cycle.
Cochliopodium Is an Unusual Sexual Lineage
The detection of many of the sex-related genes, including several meiosis-specific genes, in Cochliopodium transcriptomes (Table 1; Figure 1) lends evidence to the hypothesis that the genus is actively sexual. It is likely that recombination takes place in the large, polyploid nuclei produced after karyogamy. Despite the presence and expression of the meiosis genes, conventional meiosis has never been observed during the polyploid stage of the life cycle (Tekle et al. 2014). Instead, nuclear fission (karyotomy) appears to involve stretching and deforming of the nucleus, a likely process of depolyplodizion in a cell simultaneously undergoing cellular fission. Cells undergoing fission and nuclear fragmentation have no apparent chromosomal condensation, mitotic/meiotic spindle fibers, or conventional signatures of chromosome segregation (Tekle et al. 2014). Adaption of the meiosis toolkit for recombination without classical or observed accompanying meiosis also occurs in several other microbial lineages such as Giardia (Poxleitner et al. 2008; Carpenter et al. 2012), Candida (Forche et al. 2008), and Entamoeba (Singh et al. 2013). It is possible that the meiosis genes detected here are also primarily used for recombination in Cochliopodium, while another unconventional process performs the actual segregation of the chromosomes. Such a process may be similar to or derived from unusual forms of mitosis documented in several eukaryotic lineages including other Amoebozoa (Raikov 1994). In particular, genome reduction by a process resembling closed intranuclear pleuromitosis (Raikov 1994) could account for the lack of observation of mitotic- or meiotic-like structures in dividing polyploid nuclei. This also occurs in endopolyploid mammalian cells to produce diploid or near-diploid proliferative cells, possibly contributing to genomic instability associated with cancer (Walen 2010). Interestingly, meiosis-specific genes such as SPO11, DMC1, and REC8 are activated in polyploid tumor cells undergoing meiotic-like reductional divisions, which may be a contributing factor to treatment resistance (Erenpreisa et al. 2009; Ianzini et al. 2009). Because of the similarity between these processes, Cochliopodium may provide a good model for studying these ploidy cycling processes.
Several other microbial eukaryotes including amoebae are assumed to undergo meiosis during dormant cyst stages, which creates problems for live experimentation (Erdos et al. 1973; Goodfellow et al. 1974; Mignot and Raikov 1992). By contrast, Cochliopodium genome fragmentation occurs during the vegetative stages of its life cycle (Tekle et al. 2014). Cochliopodium is thus an ideal organism for the study of genome reduction or depolyploidization processes. Further genome-wide exploration coupled with live experimentation in Cochliopodium will unravel the mysteries surrounding the mechanism of genome reduction in eukaryotes with similar life cycles.
Supplementary Material
Supplementary data are available at Journal of Heredity online.
Funding
This work was supported by the National Institutes of Health (1R15GM116103-01 to Y.I.T.).
Data Availability
Transcriptomes used in this study have been deposited at the National Center for Biotechnology Information (NCBI) under BioProject 394890.
Supplementary Material
Acknowledgments
We would like to thank Laura Katz, Xyrus X. Maurer-Alcalá, Mario Alberto Cerón Romero, and Jessica Grant for their assistance and sharing scripts for data analysis. The authors thank the 3 anonymous reviewers for their constructive comments.
References
- Abrams EW, Zhang H, Marlow FL, Kapp L, Lu S, Mullins MC. 2012. Dynamic assembly of brambleberry mediates nuclear envelope fusion during early development. Cell. 150:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar PS, Baylies MK, Fleissner A, Helming L, Inoue N, Podbilewicz B, Wang H, Wong M. 2013. Genetic basis of cell–cell fusion mechanisms. Trends Genet. 29:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Anderson OR, Tekle YI. 2013. A description of Cochliopodium megatetrastylus n. sp. isolated from a freshwater habitat. Acta Protozool. 52:55–64. [Google Scholar]
- Avinoam O, Podbilewicz B. 2011. Eukaryotic cell-cell fusion families. Curr Top Membr. 68:209–234. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD et al. . 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh CT, Brizzio V, Rose MD. 1997. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J Cell Biol. 139:1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc D, Nicholls R, Sargeaunt PG. 1989. Experimental production of new zymodemes of Entamoeba histolytica supports the hypothesis of genetic exchange. Trans R Soc Trop Med Hyg. 83:787–790. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Pezza RJ, Mazina OM, Voloshin ON, Camerini-Otero RD, Mazin AV. 2011. The resistance of DMC1 D-loops to dissociation may account for the DMC1 requirement in meiosis. Nat Struct Mol Biol. 18:56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter ML, Assaf ZJ, Gourguechon S, Cande WZ. 2012. Nuclear inheritance and genetic exchange without meiosis in the binucleate parasite Giardia intestinalis. J Cell Sci. 125(Pt 10):2523–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Fiore-Donno AM, Chao E, Kudryavtsev A, Berney C, Snell EA, Lewis R. 2015. Multigene phylogeny resolves deep branching of Amoebozoa. Mol Phylogenet Evol. 83:293–304. [DOI] [PubMed] [Google Scholar]
- Chen EH, Grote E, Mohler W, Vignery A. 2007. Cell-cell fusion. FEBS Lett. 581:2181–2193. [DOI] [PubMed] [Google Scholar]
- Chen YK, Leng CH, Olivares H, Lee MH, Chang YC, Kung WM, Ti SC, Lo YH, Wang AH, Chang CS et al. . 2004. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc Natl Acad Sci U S A. 101:10572–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepurnov VA, Mann DG, Sabbe K, Vyverman W. 2004. Experimental studies on sexual reproduction in diatoms. Int Rev Cytol. 237:91–154. [DOI] [PubMed] [Google Scholar]
- Chi J, Mahe F, Loidl J, Logsdon J, Dunthorn M. 2014a. Meiosis gene inventory of four ciliates reveals the prevalence of a synaptonemal complex-independent crossover pathway. Mol Biol Evol. 31:660–672. [DOI] [PubMed] [Google Scholar]
- Chi J, Parrow MW, Dunthorn M. 2014b. Cryptic sex in Symbiodinium (Alveolata, Dinoflagellata) is supported by an inventory of meiotic genes. J Eukaryot Microbiol. 61:322–327. [DOI] [PubMed] [Google Scholar]
- Cole ES, Cassidy-Hanley D, Fricke Pinello J, Zeng H, Hsueh M, Kolbin D, Ozzello C, Giddings T Jr, Winey M, Clark TG. 2014. Function of the male-gamete-specific fusion protein HAP2 in a seven-sexed ciliate. Curr Biol. 24:2168–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. 1994. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 13:2708–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdos GW, Raper KB, Vogen LK. 1973. Mating types and macrocyst formation in Dictyostelium discoideum. Proc Natl Acad Sci U S A. 70:1828–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdos GW, Raper KB, Vogen LK. 1975. Sexuality in cellular slime-mold Dictyostelium giganteum. Proc Natl Acad Sci U S A. 72:970–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erenpreisa J, Cragg MS, Salmina K, Hausmann M, Scherthan H. 2009. The role of meiotic cohesin REC8 in chromosome segregation in gamma irradiation-induced endopolyploid tumour cells. Exp Cell Res. 315:2593–2603. [DOI] [PubMed] [Google Scholar]
- Fedry J, Liu Y, Pehau-Arnaudet G, Pei J, Li W, Tortorici MA, Traincard F, Meola A, Bricogne G, Grishin NV et al. . 2017. The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell. 168:904–915.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller RS, Brake A, Thorner J. 1989. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci U S A. 86:1434–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg SG, Martin WF. 2016. Mitochondria, the cell cycle, and the origin of sex via a syncytial eukaryote common ancestor. Genome Biol Evol. 8:1950–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner G, Noegel AA. 2012. Comparative genomics in the Amoebozoa clade. Biol Rev. 88:215–225. [DOI] [PubMed] [Google Scholar]
- Goh PY, Surana U. 1999. Cdc4, a protein required for the onset of S phase, serves an essential function during G/M transition in Saccharomyces cerevisiae. Mol Cell Biol. 19:5512–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow LP, Belcher JH, Page FC. 1974. A light- and electron-microscopical study of Sappinia diploidea, a sexual amoeba. Protistologica. 2:207–216. [Google Scholar]
- Grant JR, Katz LA. 2014. Building a phylogenomic pipeline for the eukaryotic tree of life—addressing deep phylogenies with genome-scale data. PLoS Curr. 6,7. doi: 10.1371/currents.tol.c24b6054aebf3602748ac042ccc8f2e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel EHPA. 1866. Generelle morphologie der organismen. Allgemeine grundzüge der organischen formen-wissenschaft, mechanisch begründet durch die von Charles Darwin reformirte descendenztheorie. Berlin: G. Reimer. [Google Scholar]
- Hirai M, Arai M, Mori T, Miyagishima SY, Kawai S, Kita K, Kuroiwa T, Terenius O, Matsuoka H. 2008. Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr Biol. 18:607–613. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, Halsey C. 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9:1728–1739. [DOI] [PubMed] [Google Scholar]
- Howard-Till RA, Lukaszewicz A, Novatchkova M, Loidl J. 2013. A single cohesin complex performs mitotic and meiotic functions in the protist tetrahymena. PLoS Genet. 9:e1003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianzini F, Kosmacek EA, Nelson ES, Napoli E, Erenpreisa J, Kalejs M, Mackey MA. 2009. Activation of meiosis-specific genes is associated with depolyploidization of human tumor cells following radiation-induced mitotic catastrophe. Cancer Res. 69:2296–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA. 1991. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 113:539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 88:375–384. [DOI] [PubMed] [Google Scholar]
- Kim J, Jeon S, Yang YS, Kim J. 2004. Posttranscriptional regulation of the karyogamy gene by Kem1p/Xrn1p exoribonuclease and Rok1p RNA helicase of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 321:1032–1039. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. 1997. Evolutionary genetics of life cycles. Ann Rev Ecol Syst. 28:391–435. [Google Scholar]
- Kudryavtsev A, Bernhard D, Schlegel M, Chao EEY, Cavalier-Smith T. 2005. 18S ribosomal RNA gene sequences of Cochliopodium (Himatismenida) and the phylogeny of Amoebozoa. Protist. 156:215–224. [DOI] [PubMed] [Google Scholar]
- Kudryavtsev A, Wylezich C, Pawlowski J. 2011. Ovalopodium desertum n. sp. and the phylogenetic relationships of Cochliopodiidae (Amoebozoa). Protist. 162:571–589. [DOI] [PubMed] [Google Scholar]
- Lahr DJ, Grant J, Nguyen T, Lin JH, Katz LA. 2011. Comprehensive phylogenetic reconstruction of amoebozoa based on concatenated analyses of SSU-rDNA and actin genes. PLoS One. 6:e22780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr DJ, Parfrey LW, Mitchell EA, Katz LA, Lara E. 2011. The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms. Proc Biol Sci. 278:2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tewari R, Ning J, Blagborough AM, Garbom S, Pei J, Grishin NV, Steele RE, Sinden RE, Snell WJ, Billker O. 2008. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 22:1051–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Howard-Till RA, Loidl J. 2013. Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex. Nucleic Acids Res. 41:9296–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM JR. 2008. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One. 3:e2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. 1978. The evolution of sex. Cambridge: Cambridge University Press. [Google Scholar]
- Melloy P, Shen S, White E, Rose MD. 2009. Distinct roles for key karyogamy proteins during yeast nuclear fusion. Mol Biol Cell. 20:3773–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 62:1191–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L, Dudin O, Martin SG. 2013. Mate and fuse: how yeast cells do it. Open Biol. 3:130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 91:35–45. [DOI] [PubMed] [Google Scholar]
- Michel R, Smirnov AV. 1999. The genus Flamella Schaeffer, 1926 (Lobosea, Gymnamoebia), with description of two new species. Eur J Protistol. 35: 400–410. [Google Scholar]
- Mignot J-P, Raikov IB. 1992. Evidence for meiosis in the testate amoeba Arcella. J Eurkaryot Microbiol. 39:287–289. [Google Scholar]
- Mihake A. 1996. Fertilization and sexuality in ciliates. In: Hausmann K, Bradbury PC, editors. Ciliates: cells as organisms. Stuttgart: Gustav Fischer; p. 243–290. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). (2010 November 4; New Orleans, LA: ). p. 1–8. [Google Scholar]
- Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J et al. . 2013. Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 27:1198–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant KT, Chen C, Shinohara M, Shinohara A, Alani E. 2010. Genetic analysis of baker’s yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability. PLoS Genet. 6:e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JE, Ross-Macdonald PB, Roeder GS. 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics. 158:1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Yamada L, Fujisaki Y, Bloomfield G, Yoshida K, Kuwayama H, Sawada H, Mori T, Urushihara H. 2016. Two HAP2-GCS1 homologs responsible for gamete interactions in the cellular slime mold with multiple mating types: Implication for common mechanisms of sexual reproduction shared by plants and protozoa and for male-female differentiation. Dev Biol. 415:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page FC. 1971. Two marine species of Flabellula (Amoebida, Mayorellidae). J Protozool. 18:37–44. [Google Scholar]
- Parfrey LW, Lahr DJ, Katz LA. 2008. The dynamic nature of eukaryotic genomes. Mol Biol Evol. 25:787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S, Moeys S, Von Dassow P, Huysman MJ, Mapleson D, De Veylder L, Sanges R, Vyverman W, Montresor M, Ferrante MI. 2015. Identification of the meiotic toolkit in diatoms and exploration of meiosis-specific SPO11 and RAD51 homologs in the sexual species Pseudo-nitzschia multistriata and Seminavis robusta. BMC Genomics. 16:930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Vargas J, Krey T, Valansi C, Avinoam O, Haouz A, Jamin M, Raveh-Barak H, Podbilewicz B, Rey FA. 2014. Structural basis of eukaryotic cell-cell fusion. Cell. 157:407–419. [DOI] [PubMed] [Google Scholar]
- Pinello JF, Lai AL, Millet JK, Cassidy-Hanley D, Freed JH, Clark TG. 2017. Structure-function studies link class II viral fusogens with the ancestral gamete fusion protein HAP2. Curr Biol. 27:651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poxleitner MK, Carpenter ML, Mancuso JJ, Wang CJ, Dawson SC, Cande WZ. 2008. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science. 319:1530–1533. [DOI] [PubMed] [Google Scholar]
- Raikov IB. 1982. The protozoan nucleus: morphology and evolution. Wien: Springer-Verlag. [Google Scholar]
- Raikov IB. 1994. The diversity of forms of mitosis in protozoa: a comparative review. Eur J Protistol. 30:253–269. [Google Scholar]
- Ramesh MA, Malik S-B, Logsdon JM. 2005. A phylogenomic inventory of meiotic genes: evidence for sex in Giardia and an early eukaryotic origin of meiosis. 15:185. [DOI] [PubMed] [Google Scholar]
- Rose MD. 1996. Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu Rev Cell Dev Biol. 12:663–695. [DOI] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. 1989. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 57:1211–1221. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. 2002. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 4:626–631. [DOI] [PubMed] [Google Scholar]
- Schaudinn F. 1899. Untersuchungen über den generationswechsel von Trichosphaerium sieboldii. Schn Abh Konigl Preuss Akad Wiss (Berlin). 1–93. [Google Scholar]
- Schurko AM, Logsdon JM Jr. 2008. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. Bioessays. 30:579–589. [DOI] [PubMed] [Google Scholar]
- Seravin LN, Goodkov AV. 1984. Possible forms of agamic genetic interactions in protists and ways of establishment of the sexual process. Tsitologiya. 26:1224–1236. [Google Scholar]
- Shodhan A, Lukaszewicz A, Novatchkova M, Loidl J. 2014. Msh4 and Msh5 function in SC-independent chiasma formation during the streamlined meiosis of Tetrahymena. Genetics. 198:983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Bhattacharya A, Bhattacharya S. 2013. Homologous recombination occurs in Entamoeba and is enhanced during growth stress and stage conversion. PLoS One. 8:e74465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speijer D, Lukes J, Elias M. 2015. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc Natl Acad Sci U S A. 112:8827–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 57:758–771. [DOI] [PubMed] [Google Scholar]
- Stearns T, Hoyt MA, Botstein D. 1990. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics. 124:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekle YI, Anderson OR, Katz LA, Maurer-Alcala XX, Romero MA, Molestina R. 2016. Phylogenomics of ‘Discosea’: a new molecular phylogenetic perspective on Amoebozoa with flat body forms. Mol Phylogenet Evol. 99:144–154. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Anderson OR, Lecky AF. 2014. Evidence of parasexual activity in “asexual amoebae” Cochliopodium spp. (Amoebozoa): extensive cellular and nuclear fusion. Protist. 165:676–687. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Anderson OR, Lecky AF, Kelly SD. 2013. A new freshwater amoeba: Cochliopodium pentatrifurcatum n. sp. (Amoebozoa, Amorphea). J Eukaryot Microbiol. 60:342–349. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Gorfu LA, Anderson OR. 2015. Cochliopodium arabianum n. sp. (Amorphea, Amoebozoa). J Eukaryot Microbiol. 62:623–628. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Grant J, Anderson OR, Nerad TA, Cole JC, Patterson DJ, Katz LA. 2008. Phylogenetic placement of diverse amoebae inferred from multigene analyses and assessment of clade stability within ‘Amoebozoa’ upon removal of varying rate classes of SSU-rDNA. Mol Phylogenet Evol. 47:339–352. [DOI] [PubMed] [Google Scholar]
- Tekle YI, Wood FC, Katz LA, Cerón-Romero MA, Gorfu LA. 2017. Amoebozoans are secretly but ancestrally sexual: evidence for sex genes and potential novel crossover pathways in diverse groups of amoebae. Genome Biol Evol. 9:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walen KH. 2010. Mitosis is not the only distributor of mutated cells: non-mitotic endopolyploid cells produce reproductive genome-reduced cells. Cell Biol Int. 34:867–872. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 400:461–464. [DOI] [PubMed] [Google Scholar]
- Wenrich DH. 1954. Sex in protozoa: a comparative review. Washington, DC: AAAS. [Google Scholar]
- Wong JL, Johnson MA. 2010. Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol. 20:134–141. [DOI] [PubMed] [Google Scholar]
- Xu H, Beasley M, Verschoor S, Inselman A, Handel MA, Mckay MJ. 2004. A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep. 5:378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomes used in this study have been deposited at the National Center for Biotechnology Information (NCBI) under BioProject 394890.