Abstract
Background
Expression profiling of skin biopsies has established molecular features of the skin in atopic dermatitis (AD). Invasiveness of biopsies has prevented their use in defining individual level AD pathobiological mechanisms (endotypes) in large research studies.
Objective
To determine if minimally invasive skin tape strip transcriptome analysis identifies gene expression dysregulation in AD and molecular disease endotypes.
Methods
We sampled non-lesional and lesional skin tape strips and biopsies from adult Caucasian subjects AD patients (18 males, 12 females; age (Mean±SE) 36.3±2.2 yrs) and healthy controls (9 males, 16 females; age (Mean±SE) 34.8±2.2 yrs). Ampliseq whole transcriptome sequencing was performed on extracted RNA. Differential expression, clustering/pathway analyses, immunostaining of skin biopsies, and clinical trait correlations were performed.
Results
Skin tape expression profiles were distinct from skin biopsy profiles and better sampled epidermal differentiation complex genes. Skin tape expression of 29 immune and epidermis-related genes (FDR<5%) separated AD from healthy subjects. Agnostic gene set analyses and clustering revealed 50% of AD subjects exhibited a type 2 inflammatory signature (type 2-high endotype) characterized by differential expression of 656 genes including overexpression of IL13, IL4R, CCL22, CCR4 (log2FC=5.5, 2.0, 4.0, and 4.1, respectively), and at a pathway level by T-helper 2/dendritic cell activation. Both expression and immunostaining of skin biopsies indicated this type 2-high group was enriched for inflammatory, type 2-skewed dendritic cells expressing the high affinity IgE receptor (FcεRI). The type 2-high endotype group exhibited more severe disease by both EASI score and body surface area covered by lesions.
Conclusion
Minimally invasive expression profiling of non-lesional skin reveals stratification in AD molecular pathology by type 2 inflammation that correlates with disease severity.
Keywords: skin, atopic dermatitis, RNA sequencing, Type 2 immune response, inflammation
INTRODUCTION
Atopic dermatitis (AD) is the most common chronic inflammatory skin disease in the general population, with a prevalence of up to 25% in children and up to 7% in adults.1, 2 A large proportion of patients suffer from sleep disturbance and impaired quality of life. AD is caused by the complex interplay between epithelial and immune-driven abnormalities.3, 4 The role of genetics in epithelial dysfunction is clearly demonstrated by the robust association of AD development and fillaggrin null mutations, which disrupt healthy skin barrier function.5–7 Similarly, the contributions of immune dysfunction to AD have been substantiated by strong associations of disease status with genetic variants in the STAT3, IL13, TSLP, and IL1RL1 genes, as well as the HLA gene locus.8, 9 Moreover, the involvement of both immune and epithelial constituents in AD pathology is corroborated by the efficacy of both skin emollients and T-cell therapeutics, such as cyclosporine.10, 11 These genetic studies are also revealing by the small increase in disease risk estimated to be conferred by any one of these loci.8 This is in contrast to the high heritability of AD estimated from twin studies.12 Although there are many potential explanations for these observations, the existence of multiple AD subgroups stratified by pathobiological mechanisms, or endotypes, may explain this phenomenon.13 Further supporting this endotypic model of AD is recent drug trials of dupilumab, an IL-4 receptor antagonist, which produced clearing or almost clearing in AD skin disease in ~40% of patients.14, 15
An inability to easily stratify patients by individual disease profile confounds tests of significance in both genetics and clinical trial studies. Moreover, approval of duplimab and similar drugs creates an urgent need to identify endotype-specific treatment response groups. The precision medicine that AD endotyping would allow could have significant benefits to individual patients and the health care system in reducing expense, treatment failures, and risk of significant side effects. Gene expression profiling of RNA isolated from skin biopsies has revealed significant differences in the expression profiles of AD subjects, implicating type 2 immune pathways as well as dysregulation of the skin barrier through modification of cornification genes and lipid biosythesis.16–19 However, the requirement for large, scar-producing biopsies of lesional skin to obtain RNA amounts sufficient for expression profiling has impeded the study of AD disease endotypes in both clinical and research settings. Herein, we describe a minimally invasive method for sampling and gene expression profiling of non-lesional and lesional AD skin which we used to reveal molecular dysfunction of the skin and to identify molecular endotypic groups of AD subjects.
METHODS
Subject Recruitment
Skin samples were obtained from 30 adult Caucasian subjects with active AD (18 males, 12 females; age (Mean±SE) 36.3±2.2 yrs) and 25 non-atopic healthy individuals (9 males, 16 females; age (Mean±SE) 34.8±2.2 yrs) with neither personal nor family history of atopy and no skin disease. At the time of skin sample collection, Investigator’s Global Assessment scores (IGA) scores, eczema area and severity index (EASI) and information about the body surface area (BSA) % covered by lesions were recorded for all AD patients (IGA=2, mild – n=7; IGA=3, moderate – n=17; IGA=4, severe – n=6; EASI (Mean±SE) - 11.9±3.3; BSA % covered by lesions (Mean±SE) - 34±4%). AD study subjects had not received topical corticosteroids, topical calcineurin inhibitors, topical or oral antibiotics for one week prior to enrollment. Patients were not treated with systemic immunosuppressive medications for more than one month prior to enrollment into this study. The study was approved by the Institutional Review Board at National Jewish Health, Denver, CO. With over twenty five years of conducting clinical research studies, (both industry and investigator initiated) on atopic dermatitis at NJH we have a large database of research participants from which we pre-screen and select depending on the inclusion/exclusion criteria of the protocol. We estimate that 80% of those who were approached from that database and pre-screened, ended up qualifying for the protocol and then enrolling. Common reasons for exclusion included eczema that was not active or a disallowed medication like antibiotics and/or oral steroids. All subjects gave written informed consent prior to participation in the study.
Skin tape strip sampling was done for all study participants. In addition, two optional 2-mm skin punch biopsies were collected from the non-lesional skin area from the upper extremities. One of the biopsies was dissociated into epidermis and dermis after 1 h of digestion at 37°C in 5 U/ml dispase diluted in PBS (Corning, catalogue # 354235). Isolated epidermis and dermis were immediately submerged into RLT buffer (Qiagen, Valencia, CA) and frozen at −80°C for future RNA isolation. The second biopsy was preserved in 10% buffered formalin for immunofluorescent staining.
Tape Strip Collection and RNA Extraction
A total of 20 consecutive D-Squame® tape strips (22 mm diameter, CuDerm, Dallas, TX) were collected from the volar surface of the forearm. Previous skin biopsy studies of tape stripped sites had shown the depth of tape stripping included the upper half of the stratum granulosum, which includes viable cells containing RNA (Kim, Goleva and Leung, unpublished data). Both non-lesional and lesional skin from AD patients and skin from healthy control subjects was sampled. On application of the first tape disc, 4 marks were placed around the disc with a pen so that subsequent discs could be applied to the same location. Each tape disc was placed adhesive side up in a separate well of the two 12-well plates allocated for sample collection. Plates were kept on dry ice during the tape strip collection. Collected tape strips #11 through #20 were sequentially scraped into the RLT buffer (Qiagen) and frozen at −80°C. RNeasy Micro Kits (Qiagen) were used according to the manufacturer’s protocol to isolate RNA from skin tape strips, epidermis and dermis.
Immunohistochemistry
Paraffin-embedded skin tissue samples were cut at 5 μm on frosted microscope slides. Using toluene and a series of ethanol washes, slides were deparaffinized and then rehydrated. Skin sections were then blocked with 5% BSA in Super Block (ScyTek Laboratories, Logan, UT) containing 10% non-immune donkey serum (Jackson Laboratories, West Grove, PA) for 60 min. Slides were stained with rabbit anti-Langerin (ab192027, Abcam, Cambridge, MA), rabbit-anti-IL-4R (ab131058, Abcam, Cambridge, MA) antibodies or control rabbit IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 4°C overnight. Slides were washed with PBS/Tween 0.05%, followed by incubation with a Cy3-conjugated donkey anti-rabbit IgG (Jackson Laboratories, West Grove, PA). Nuclei were visualized with DAPI and wheat germ agglutinin-conjugated fluorescein isothiocyanate (FITC) was used to stain the cytoskeleton.
Immunofluorescent staining was visualized with a fluorescent microscopy (Leica, Wetzlar Germany). All skin samples were evaluated by a blinded observer. Slides were evaluated under the 25x and 40x magnification oil immersion objectives. For IL-4R staining, a mean fluorescence intensity (MFI) was determined for the epidermal area. For Langerin staining, the number of the Langerin positive dendrites per 170μm of the granular layer of the epidermis were counted. Slidebook 6.0 software (Intelligent Imaging innovations, Denver, CO) was used for image acquisition and analysis.
RNA Transcriptome Gene Expression and Quality Control
RNA Ampliseq libraries were constructed and barcoded with the Ion Ampliseq™ Trasncriptome Human Gene Expression Kit and methods. For skin tape RNA since yields were less than the 10ng used as standard input for Ampliseq, we instead loaded the maximum volume of RNA (from a 12μl extraction) the reaction could accommodate for all samples (3.5μl). Average inputs for skin tape samples were 0.11±0.17ng. Yields were higher for skin biopsy epidermis and dermis but less than 10ng needed for standard input in most cases. Average inputs for epidermis and dermis samples were 3.55±3.24ng and 1.69±2.52ng, respectively. Barcoded RNA-seq libraries were pooled and sequenced on the Ion Torrent Proton sequencer using P1 chips.
Sequencing reads were mapped to Ampliseq transcriptome target regions with the torrent mapping alignment program (TMAP) and quantified with the Ion Torrent ampliSeqRNA plugin, using the uniquely mapping option. Duplicated sequences were removed from the FASTA file and incorrect amplicon locations were corrected as previously reported.20 Samples with total reads counts >8×106 were downsampled to 8×106 reads. The average total reads for skin tape, epidermis, and dermis samples were 7.84×106± 5.8×105, 7.95×106± 2.01×105, and 7.86×106± 4.87×105, respectively.
Normalization and Differential Expression Analysis of RNA-seq Data
The gene counts were normalized and differential expression performed using the DESeq2 R package21 (distributed via Bioconductor22). DESeq2 performs multiple testing correction by applying the Benjamini-Hochberg procedure at a false discovery rate (FDR) of 0.05.
Gene Set Enrichment
The Enrichr23 webtool was used to test for enrichment of gene ontology (GO) categories in the 500 most highly expressed genes from each sampling technique. Generally applicable gene set enrichment (GAGE)24 was used to perform functional enrichment analyses of GO categories. GSEA software25 was used to perform leading edge analysis of the significantly enriched GOs and identify the genes driving enrichment of each ontology category. QIAGEN’s Ingenuity Pathway Analysis (IPA*, QUIAGEN Redwood City, www.qiagen.com/ingenuity) was used to perform Canonical Pathway Analysis of the type 2-high versus type 2-low differential expression results.
Statistical Methods and Plot Generation
All statistical analyses outside of the bioinformatics programs were performed in the R statistical language. Hierarchical clustering was performed with the hclust function of the stats package. Heatmaps were generated with the heatmap3 package. Principal component analysis was performed using the prcomp function of the stats package. All other plots were generated using the ggplot2 package in concert with the reshape, ggrepel, lattice, grid and gridExtra packages.
Dendritic Cell Comparisons
Microarray data sets of dendritic cell subsets were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO26) using the Geoquery R package27 (distributed via Bioconductor22). Expression of monocyte-derived dendritic cells and Langerhans cells was obtained from the work of Széles et al., 201028 (available at accession GSE23618). Expression of plasmacytoid dendritic cells and Langerhans cells was obtained from the work of Hutter et al., 201229 (available at accession GSE35340). Differential expression of these cell types was performed using the limma R package29, (distributed via Bioconductor22) which implements a moderated t-test, and the resulting P values are adjusted using the Benjamini-Hochberg multiple testing procedure. Genes were considered differentially expressed with a false discovery rate (FDR) of 0.05 or less.
RESULTS
The skin tape transcriptome identifies a unique epidermal molecular profile in non-lesional AD skin
We collected non-lesional skin tape samples from 30 subjects with AD and 25 healthy controls. RNA extraction resulted in average total RNA yields of 260.33 pg (SD = 411.70 pg). Although these RNA amounts were too low for standard RNA-seq, we proceeded to generate Ampliseq Whole Transcriptome sequencing libraries for all samples. This method was based on the multiplexed amplification of cDNA amplicons for over 20,000 genes in a single reaction.20, 30 The cDNA amplicons were converted into an Ion Torrent sequencing library, with sequencing used to generate digital count data. This method has been previously used to robustly generate libraries in settings where RNA quantity and quality is limited. Sequencing data was successfully generated from non-lesional skin for 18 (60%) AD subjects and 13 (52%) healthy controls (p=0.35). We found an average of 14,520 and 13,201 genes expressed in AD and healthy control subjects, respectively. Among the top ten expressed genes (Figure 1A) were FLG, FLG2, SPRR2E, and KRT10, all of which are known to be highly expressed in the keratinized epidermal epithelium.
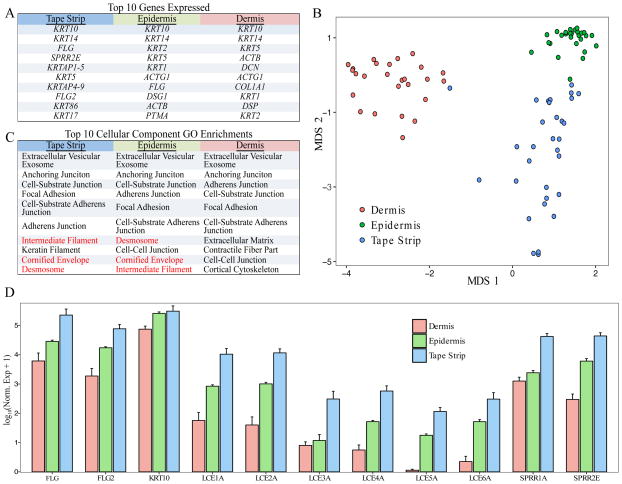
Figure 1. Non-lesional AD skin tape strip transcriptomes differ from skin biopsy transcriptomes and better assay the stratum corneum.
(A) The top 10 expressed genes by tissue type are listed in rank order. (B) Multidimensional scaling (MDS) of the top 500 variant genes in non-lesional tape strip, epidermis, and dermis samples separates the samples by tissue. (C) The top 10 GO cellular component enrichments in non-lesional tape strips, skin biopsy separated epidermis, and skin biopsy dermis. GO categories highlighted in red were only observed in epidermis and skin tape strip samples. (D) Expression of canonical epidermal genes is consistently greatest in skin tape strip, next highest in epidermis, and lowest in dermis samples.
To compare the molecular information contained in the skin tape transcriptome to skin biopsies, we generated transcriptomes from the epidermis and dermis of separated non-lesional skin biopsies collected from the skin tape subjects, concurrent with skin taping at adjacent sites. Sample clustering was performed using multidimensional scaling (MDS) of the most variant genes across all three data sets. Samples separated based on epidermis, dermis, and skin tape sample types (Figure 1B). Gene ontology (GO) analysis of the most highly expressed genes among each sample type supported a greater similarity between the molecular profiles of skin tape and epidermis than skin tape and dermis (Figure 1C). Specifically, the cornified envelope, desmosome, and intermediate filament GO categories, characteristic of the granular layers of the keratinizing epidermis, were among the top ten GO enrichments for only the epidermis and skin tape. Since tape strip sampling collects the most superficial layers of the skin, we hypothesized that late cornification genes would be most highly expressed in the skin tape samples. Comparing expression of these late cornification envelope genes between the sample types we found the lowest expression of these genes in the dermis, high expression of these genes in the epidermis, and the highest expression in the skin tape samples (Figure 1D).
Non-lesional skin tape gene expression reflects AD disease status
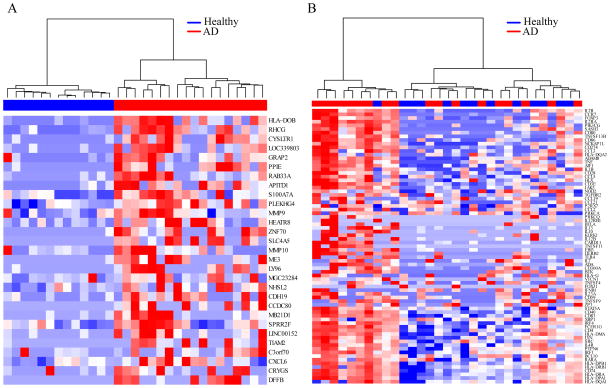
We then performed single gene differential expression between non-lesional AD (n=18) and healthy control (n=13) skin tape transcriptomes. We identified 29 differentially expressed genes (FDR < 0.05), all upregulated in the AD subjects. Hierarchical clustering of expression for these genes separated AD from healthy subjects (Figure 2A). Among the upregulated genes in AD was the cysteine leukotriene receptor 1 (CYSLTR1), a major mediator of hypersensitivity and allergy, which is expressed on mast cells, basophils and eosinophils31. We also identified the Small Proline Rich Protein 2F (SPRR2F), a gene that encodes a protein involved in cell membrane cross-linkage in keratinocytes32, and may have a role in antimicrobial responses. Also among these genes were two matrix metalloproteinase genes, MMP9 and MMP10. Another of these upregulated genes, coiled-coil domain containing 80 (CCDC80), has been identified in a recent AD GWAS analysis34. The top differentially expressed gene by FDR, HLA-DOB, encodes the beta chain of the human leukocyte antigen class II major histocompatibility DO complex. The HLA-DO complex is involved in inhibition of antigen loading and presentation by HLA-DM.36
Figure 2. Hierarchical clustering of non-lesional skin tape gene expression separates AD from healthy control samples and identifies immune-activated AD subjects.
(A) Expression heatmap and hierarchical clustering of AD and healthy control subjects using non-lesional skin tape expression of genes differentially expressed in AD subjects. (B) Expression heatmap and hierarchical clustering of the 75 most recurrent leading-edge genes from all immune-related gene ontology categories significantly enriched in AD subjects. A cluster of AD samples (immune-activated) is identified that demonstrates distinct upregulation of immune-related genes compared to other AD and healthy control subjects.
Gene set analysis reveals a subgroup of AD subjects with a strong immune activation signature in non-lesional AD skin
To identify functional groupings or pathways of genes altered in AD non-lesional skin tape samples, we utilized the GAGE package to perform gene set enrichment analysis of predefined GOs categories. In AD patients versus healthy controls, we identified 154 and 11 GO terms that were up- and down-regulated, respectively (STable 1). The top down-regulated GO term was detection of chemical stimulus involved in sensory perception (FDR=3.75×10−7), with 6 other highly related down-regulated terms, including sensory perception of smell (FDR=1.73×10−6), and olfactory receptor activity (FDR=1.07×10−7). Examination of the genes in this data set revealed most were olfactory receptor genes. The three other down-regulated GO terms all related to intermediate filaments, specifically, keratin filament (FDR=2.61×10−7), intermediate filament (FDR=3.26×10−7), and intermediate filament cytoskeleton (FDR=4.67×10−7). In contrast, the top upregulated GO categories were almost all related to the regulation and function of immune cells. For example, the top categories were lymphocyte activation (FDR=8.22×10−8), leukocyte cell-cell adhesion (FDR=8.22×10−8), and regulation of: (1) cell activation (FDR=8.22×10−8), (2) leukocyte activation (FDR=8.22×10−8), and (3) lymphocyte activation (FDR=1.21×10−7). To eliminate redundancy in this analysis we identified the genes contributing most to these gene set associations by performing GSEA on the significant GO categories. This resulted in 1455 significant leading edge genes across all GO categories (STable 2). We performed hierarchical clustering of all subjects based on the 75 most frequently enriched of the leading edge genes (Figure 2B). Interestingly, we found upregulation in these genes was driven by a subset (9 of 20) of the AD patients, the remainder of AD subjects had low expression of these genes similar to the healthy controls (Figure 2B). We defined this group of AD patients as patients with the evidence of immune cell activation.
The type 2-high AD endotype is defined by antigen presenting cell and T cell signaling and activation in non-lesional AD skin
To determine the molecular nature of AD in the immune-activated subjects, we performed differential expression analysis of non-lesional skin RNA transcripts between this group (n=9) and AD subjects not exhibiting this immune activation signature (n=9). We identified 656 differentiated expressed genes between these two AD groups, including 599 and 57 up and down regulated genes, respectively (STable 3). We performed canonical pathway analysis of the dataset with ingenuity pathway analysis (IPA) and observed the T-helper 2 (Th2) cell pathway was most significantly enriched (5.0×10−29, Figure 3A). Supporting this, the canonical type 2 cytokine, IL13, was highly upregulated in these subjects (log2FC=5.5, Figure 3B). Moreover, other well described type 2 pathway genes were upregulated including CCR4 (log2FC=4.1), IL4R (log2FC=2.0), and CCL22 (log2FC=4.0) (Figure 3B). Based on this expression pattern we labeled the immune-activated subjects, type 2-high, and the non-activated as type 2-low.
Figure 3. Immune activation signature in AD subjects reflects significant upregulation of type 2 inflammatory genes and presence of activated T helper 2 and dendritic cells.
(A) IPA canonical pathway analysis was performed using the non-lesional skin tape differentially expressed genes between immune-activated AD vs. non-activated AD subjects. The 15 most significantly enriched canonical pathways indicate upregulation of T helper 2 and dendritic cell activity in immune-activated AD subjects. (B) Expression of canonical type 2 inflammatory genes is upregulated in the skin tape samples of immune-activated AD subjects. (C) Graphical display of the canonical IPA “Th2 Pathway,” showing genes overlaid on an antigen presenting cell and a T cell. Differentially expressed genes are colored by log2 fold change.
Examining the top 15 most significant canonical pathways in type 2-high samples, we found many centered on the activation of and signaling between T-helper and antigen presenting cells (APC) (Figure 3C). For example, 6 APC MHC class II receptor genes (HLA-DOB, -DOA, -DMA, -DPA1, -DPB1, -DRA, and -DRB1) were upregulated in the type 2-high group as were the APC co-stimulatory receptors CD80, CD86, and CD40. In line with APC-driven activation of T-helper cells, we found upregulation of CD28, CD3, ICOS, IL2R, and IL25R.
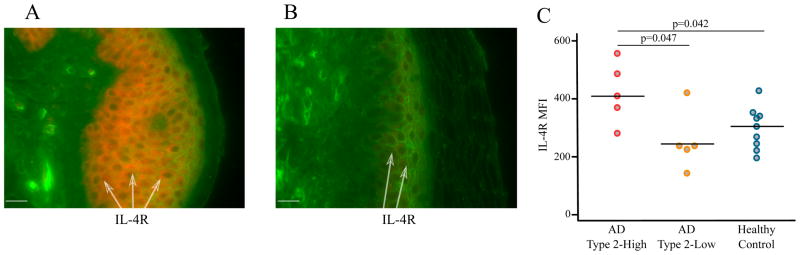
Our IL4R result was confirmed at a protein level in the granular epidermal layers by immunohistochemistry: we found significantly higher IL4-R staining by mean fluorescence intensity (MFI) in type 2-high AD subjects versus either healthy controls or type 2-low AD subjects (Figure 4). In total, these results are supportive of active, enhanced APC-T cell signaling in the superficial skin layers of type 2-high AD subjects.
Figure 4. IL-4R protein expression is increased in the non-lesional skin of type 2-high AD subjects.
Skin biopsies were stained for IL-4R with fluorescent immunohistochemistry. IL-4R staining in (A) type 2-high and (B) type 2-low subjects. IL-4R (cy3, red), wheat germ agglutinin (FITC, green), magnification 400x, bar=20μm. The arrows point to IL-4R expressing keratinocytes in the epidermis. (C) Quantification of IL-4R staining revealed significantly greater lL-4R levels in the non-lesional skin of type 2-high than type 2-low AD subjects.
The non-lesional skin of type 2-high AD subjects is characterized by upregulation of inflammatory DCs
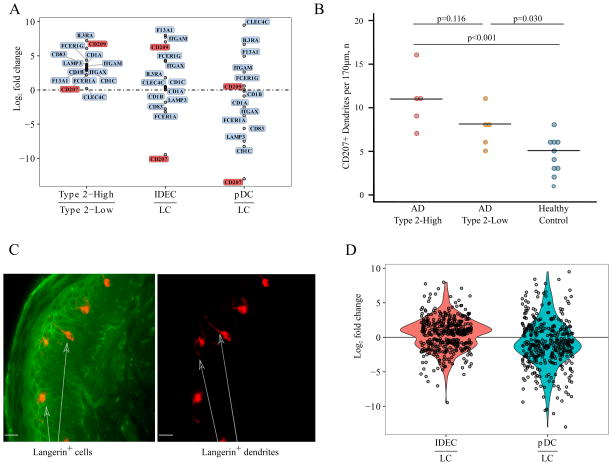
Given the strong APC signal among the type 2-high AD subjects and that dendritic cells are a common APC population in the skin, we investigated whether these APC signals reflected specific dendritic cell (DC) gene expression patterns. Specifically, we considered resident epidermal Langerhans DCs (LCs), but also two populations of peripherally derived DC’s that have been described to infiltrate AD skin: inflammatory, monocyte derived DCs (IDECs) and plasmacytoid dendritic cells (pDCs). We found upregulated expression for multiple markers of all three DC subsets in our type 2-high vs. type 2-low subjects (Figure 5A).
Figure 5. Non-lesional skin of type 2-high AD subjects is characterized by inflammatory dendritic cell signatures.
(A) Gene expression log2 fold changes for dendritic cell (DC) markers between non-lesional skin of type 2-high versus type 2-low AD subjects is plotted. Gene expression log2 fold changes for these DC genes in published inflammatory dendritic epidermal cells (IDEC or monocyte-derived DCs) versus Langerhans DCs (Széles et al. 2010)28, and plasmacytoid DC (pDC) versus Langerhans DCs (Hutter et al., 2012)29. (B) Immunohistochemistry of skin biopsies reveals numbers of CD207+ (Langerin) DCs are greatest in type 2-high AD patients, followed by type 2-low AD, and healthy control subjects. (C) CD207 (Langerin) staining of skin biopsies from type 2-high subjects reveals extension of DC dendrites into the superficial layers of the epidermis. Langerin (cy3, red), wheat germ agglutinin (FITC, green); magnification, 400x, bar=20μm. The arrows point to Langerin+ cells and dendrites in the upper granular layer of the epidermis. FITC/cy3 channels or cy3 channel only are shown. (D) Genes upregulated in the non-lesional skin of type 2-high versus type 2-low AD subjects are plotted using the log2 fold changes of differential expressions from the IDECs vs. Langerhans DCs (Szeles et al.)28 and pDC versus Langerhans DCs (Hutter et al.)29 generated from published gene expression data. Log2 fold changes of the type 2-high-upregulated genes indicate a skew toward IDECs. The plotted genes were filtered using a significance threshold of FDR <0.05.
To better refine the potential DC subset source of these markers, we generated relative expression plots between LCs and both IDECs and pDCs, using previously reported DC expression profiling data (Széles et al., 201028 Hutter et al., 201229) (Figure 5A). The more general markers of DCs, CD1A, CD1B, CD1C were all upregulated in type 2-high AD subjects. The CD83 gene, which we find highly enriched in LCs, was also upregulated in type 2-high AD subjects. Moreover, examining expression of CD207, the gene that encodes the Langerin protein and marker of LCs, we found significant upregulation in type 2-high AD subjects (FC=2.2, FDR=0.018, Figure 5A). Supporting this, immunohistochemistry of the granular epidermal layers revealed progressively higher numbers of Langerin positive dendrites in healthy control, type 2-low AD and type 2-high AD subjects. (Figure 5B,C). The FCER1A (enriched in LCs) and FCER1G (enriched in IDECs) genes combine to produce the trimeric (αγ2) form of the high-affinity IgE receptor (FcεRI), which has been observed in LCs and IDECs in AD lesional skin. Moreover, these IgE Fc receptor genes have previously been shown to be upregulated by type 2 cytokines, including IL-13. In contrast, the β subunit (MS4A2), which combines with the α and γ subunits to produce the tetrameric form (αγβ2) of the FcεRI receptor and is expressed by mast cells and basophils37, was not expressed in our subjects. In general we found the genes upregulated in type 2-high AD subjects were skewed toward higher expression in IDECs versus LCs (Figure 5D). In fact, CD209 (DC-SIGN), CD11C (ITGAX), and CD11B (ITGAM), markers that strongly differentiate IDECs from LCs, were strongly upregulated in type 2-high AD subjects. Finally, we also observed the IL3RA gene (CD123), a marker of plasmacytoid DCs was the most strongly upregulated gene in the AD type 2-high subjects. Together, this data supports strong upregulation of resident and infiltrating inflammatory DC sets in the non-lesional skin of AD type 2-high subjects.
AD clinical severity measures are strongly related to non-lesional skin type 2-high gene signature
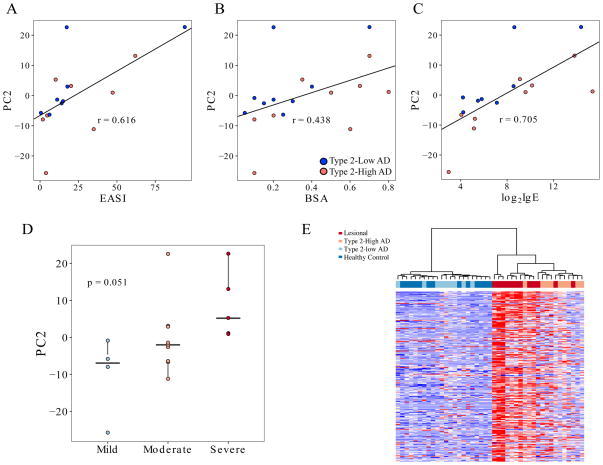
Examining the severity classification of AD subjects based on IGA scores, we observed 4 of 9 type 2-high subjects were severe (IGA score=4) versus only 1 of the 11 type 2-low AD subjects. Therefore, we explored the relationship between expression of the type 2-high signature genes and several clinical measures of AD severity. To accomplish this, we performed principle component analysis (PCA) of the 656 genes associated with type 2-high status. The PCs generated were tested for correlation with the eczema area and severity index (EASI) severity scores for all AD subjects. We found PC2 exhibited a strong correlation (ρ= 0.616, p=9.79×10−3) with EASI score (Figure 6A). Relatedly, PC2 was also strongly correlated with the body surface area (BSA) covered with lesions (ρ= 0.438, p=1.45×10−2) (Figure 6B). The PC2 values also exhibited a striking correlation with lgE levels (ρ= 0.705, p=6.96×10−5), reflecting the highly allergic nature of these subjects and this gene set (Figure 6C). We also note increased correlation of serum IgE and BSA with PC2 when only the type 2-high subjects were analyzed (ρ= 0.811 and .643, respectively). Supporting these quantitative relationships, PC2 values were strongly associated with severity status with a median value of −6.94 for mild (IGA score=2), −2.00 for moderate (IGA score=3), and 5.19 for severe (IGA score=4) subjects (p=5.12×10−2), (Figure 6D).
Figure 6. Non-lesional skin type 2 inflammatory gene expression patterns are reflective of disease severity and lesional skin expression profile in AD.
(A) Principal component analysis was performed on non-lesional skin tape strip expression from 656 differentially expressed genes between type 2-high and type 2-low AD samples. The second principal component was strongly correlated (Pearson) with clinical measurements of AD. PC2 positively correlated with EASI score (ρ = 0.616, p=1.11×10−2), (B) % body surface area covered with lesions (ρ = 0.438, p=1.89×10−2), and (C) log2 serum IgE levels (ρ = 0.705, p=1.56×10−3). Type two-low samples are denoted with blue points, and type 2-high samples are plotted in red. (D) PC2 values displayed a positive significant trend (via Cochran-Armitage trend test) across AD severity levels (p=5.12×10−2). (E) The top 250 differentially expressed genes between AD lesional and healthy control tape strips agnostically group non-lesional type 2-high samples with lesional tape strips, and type 2-low with healthy control tape strips.
Considering the strong relationship between non-lesional type 2-high expression signature and AD severity, we hypothesized the lesional skin expression profile would be similarly upregulated in the non-lesional skin of type 2-high but not type 2-low subjects. Therefore we generated lesional skin tape transcriptomes on 12 subjects. We clustered the non-lesional and lesional skin samples on the top 250 genes differentially expressed in lesional skin vs. healthy control skin (STable 4). We found non-lesional skin from type 2-high subjects clustered with lesional skin samples, while healthy control and non-lesional type 2-low skin clustered separately (Figure 6E).
DISCUSSION
Over a decade of genomic profiling studies, examining skin punch biopsy samples from AD subjects, has revealed significant heterogeneity in disease mechanisms. Various studies have implicated permutations of excessive Th2, Th22, and, to a lesser extent, Th17 activation and inflammation in AD skin.17, 38, 39 Some of these and other studies have also implicated deficiencies in either or both lipid synthesis and terminal skin differentiation genes.5, 17, 19, 40, 41 The heterogeneity in disease mechanisms across the AD patient population, coupled with the development of targeted biological therapeutics, has created an urgent need for methods which allow query of the AD skin transcriptome endotype. Recent studies in this Journal have highlighted the importance of epithelial abnormalities and immune acrtivation in the pathogenesis of AD.3, 4 Skin transcriptome measurements could allow stratification of patients to endotype groups for large research studies (e.g. genetic), assignment to targeted biologic therapy, and facilitate biological measures of treatment response with repeat sampling. Although skin punch biopsy genomic profiling has elucidated disease mechanisms well, its widespread adoption is impeded by the invasive, scarring nature of the sampling.
Our paper details the use of a much less invasive, non-scarring method of tape stripping to collect AD patient skin. In line with the small tissue quantities collected using this technique, the yield of RNA from the tape strips were significantly less than from skin punch biopsies. Therefore we adopted a well-described20, 30, 42, transcript amplicon enrichment method (AmpliSeq) to generate full transcriptome sequencing data from sub-nanogram total RNA levels. This method is also robust to RNA degradation since only short transcript fragments of 100–200 bp are required. We have previously used this method successfully for endotyping of asthmatics using RNA isolated from sputum and nasal airway epithelial cells.30 This sampling method not only collects less skin, but assays a more superficial epidermal layer.
This was made clear by the distinct clustering of the skin tape samples by transcriptome in comparison to the separated epidermal and dermal biopsy layers. In line with the superficial nature of the sample, the skin tape transcriptome was most closely related to the epidermal samples, but with even higher expression late cornification genes, including FLG, FLG2, and LCE & SPRR family genes.
We observed a set of 29 significantly upregulated genes in AD tape strips which exhibited sufficient differences to separate the AD from healthy subjects by hierarchial clustering. This included multiple genes previously observed to be upregulated in non-lesional AD skin, including MMP9, MMP10, and S100A7A and is supportive of previous work linking MMP function and disruption of barrier function in AD17, 19, 33, 38. One of the most consistently high expressed genes across subjects was the receptor for cysteinyl leukotrienes, CYSLTR1. Expression of this gene is induced by type 2 cytokines and tightly linked with hypersensitivity in multiple diseases.43, 44 Also of high interest was identification of the CCDC80 gene as upregulated in AD. The CCDC80 gene is one of only 8 loci identified in an AD GWAS of Japanese subjects.34 The CCDC80 protein has been shown to function in control of energy homeostasis, with its deletion in mice resulting in obesity.45 Additionally, CCDC80 is strongly upregulated in both mouse and rat models of pulmonary arterial hypertension (PAH), as well as among human PAH disease.35 Although it is unclear how CCDC80 might function in AD, the independent genome-wide significant genetic and genomic signals for this gene suggest extensive study of CCDC80 in AD is warranted. We did not observe widespread downregulation of terminal differentiation genes as reported by the Suarez-Farinas et al non-lesional skin punch biopsy study17. These differences may be a function of the less severe nature of our subjects, which included mild patients. Additionally, we note the Cole et al non-lesional AD skin study did not find this widespread terminal differentiation gene downregulation, but instead detected increases in filaggrin expression in non-FLG mutation AD carriers vs. controls19. Finally, we suggest that the superficial epidermis sampled by tape strips provides a more like-to-like comparison of skin between healthy and AD subjects than skin punch biopsies would allow, since AD disease dramatically alters the epidermal and dermal layer thicknesses. These alterations in layer thickness could dramatically influence the expression levels observed when comparing whole biopsies between AD and healthy skin.
Although multiple immune-related genes were among the 29 detected in the differential expression analysis, pathway analysis identified a much stronger pattern of immune activation and inflammation in AD subjects. Hierchial clustering of subjects based on these immune acitivation pathways revealed the striking stratification in gene expression among AD subjects, explaining the lack of association in the single gene analysis among the entire group. Examination of this immune-activated group of AD subjects found over 600 differentially expressed genes. A strong upregulation of type 2 inflammatory signatures (high IL13, IL4R, CCL22) differentiated the immune-activated from other AD subjects who exhibited type 2 gene expression levels similar to healthy controls. This type 2-high group exhibited a strong signature of T-helper 2 cell activation including upregulated iCOS and CD28 signaling, replicating signals observed in both AD lesional and non-lesional skin from a recent AD transcriptome meta-analysis.16 These findings are also in line with two prior immunohistochemistry studies which found increased numbers of T cells in the non-lesional skin of AD vs. healthy control skin.18, 46
An equally prominent component of the type 2-high AD subjects was the activated dendritic cell signature. We observed increased expression of CD207 and other markers of resident Langerhans DCs. We confirmed an increased presence of these Langerhans DCs in the stratum corneum of type 2-high vs. healthy control subjects and the extension of the Langerin positive dendrites upwards in the upper granular layer of the non-lesional skin of type 2-high subjects. Related, recent work found activated Langerhans cells elongate their dendrites to penetrate keratinocyte tight junctions and survey the environment located outside of the tight junction barrier, likely capturing antigens, a process which was significantly enhanced in AD lesional skin47. Examination of the type 2-high AD gene signature for Langerhans DCs versus monocyte-derived inflammatory infiltrating DCs (IDEC) indicated the expression pattern among the type 2-high subjects was more enriched for the IDECs. Additionally, we found specific markers of pDCs, IL3RA and CLEC4C, were highly upregulated in type 2-high subjects, with IL3RA being the top upregulated gene in the type 2-high group. Finally, we found upregulated expression of the high-affinity IgE receptor (FceRI) subunits, expressed by DCs (FCER1A and FCER1G) in the type 2-high subjects. We note prior studies have found increased CD11C+ DCs in non-lesional AD skin and another study which found increased FceRI staining in non-lesional skin DCs for AD patients48, 49. Taken together our data is supportive of increased penetration into the skin strateum corneum of multiple highly activated DCs subsets among type 2-high subjects.
Interestingly, we found the non-lesional type 2-high skin endotype was strongly related to disease severity as judged by multiple measures, including EASI score and body surface area covered by lesions, revealing the clinical value of this endotype. Our observations of heterogeneity in the non-lesional skin type 2 inflammation and its relationship with clinical severity and lesional skin expression profile is in agreement with the Suarez-Farinas et al skin biopsy transcriptome study17. This study found complete separation of lesional skin from the skin of healthy subjects by transcriptome pattern, with the non-lesional AD skin occupying a continuum from healthy to lesional AD skin. In this study, inflammatory gene expression (mostly type 2) in the non-lesional skin was also strongly correlated with SCORAD measured disease severity. Likewise, our lesional data on skin tape strips indicates a much more uniform pattern of type 2 inflammation than the dichotomous nature of the non-lesional samples. Taken together, these results suggest that although most exacerbations of AD involve type 2 inflammation, the type 2 inflammatory state of the visibly unaffected skin associates with severity. We notice the parallel between our results and the type 2-high endotype observed in both the baseline bronchial and nasal airway of asthmatic subjects, where disease severity also associates with type 2-high endotype. In fact, the observed frequency of the type 2-high airway endotype is ~50% across several studies.30, 50 We speculate the baseline type 2 inflammation observed in both airway and skin may involve the same systemic immune system skew and drive development of the respective type 2-high endotypes of these diseases. This mechanism is consistent with the strong association between AD, asthma, and other allergic diseases.
The ease of our technique has potential for paired use in the development and eventual clinical administration of multiple biologics under development, most notably, type 2 cytokine pathway inhibitors. Dupilumab, a IL4Rα blocker, has now been shown to improve nearly all severity measures in 2 different Phase 3 clinical trials for moderate-severe AD14, 15. Despite its success, only ~40% of subjects experienced success for the primary outcome (complete clearing or almost clear AD) in these trials. We speculate that these subjects could be identified a priori by non-lesional skin tape transcriptome analysis. We note both IL4Rα gene expression and protein staining was increased in type 2-high AD subjects. The use of skin transcriptomics as a suitable biomarker for response is also supported by earlier Dupilumab study which found transcriptomic profiles for lesional skin improved with treatment and correlated with clinical response51.
We acknowledge several limitations to our current study. First, the transcriptome generation failed in ~40% of AD non-lesional skin samples due to lack of RNA. We acknowledge the possibility that this failure rate could have skewed the relative percentages of subjects in the skin endotype groups. However, in comparing the severity measures and transcriptomes (SFigure 1) of the failed-to-successful samples, we do not see any relationship. This high failure rate could also impede deployment of non-lesional skin tape profiling in research and clinical practice. However, we are currently working to improve the sampling and extraction procedure. In fact, we resampled 11 of the subjects that failed to generate a successful skin tape transcriptome on the first attempt reported in this paper and we were able to generate successful libraries on all subjects with repeat sampling. The non-scarring nature of the sample collection make patients much more likely to provide a sample initially and repeatedly. In fact, in this study we find the consent rate for skin tape stripping was 100% in contrast to only 40% for skin biopsies, when biopsy sampling was suggested as optional. Lastly, we note the success rate for lesional AD skin tape transcriptome analysis was 86%. This higher rate is likely due to the increased immune cell load in lesions, with resultant higher quality RNA. The lesional skin analysis could serve as an important tool to judge reduction in inflammatory and other immune characteristics in clinical trials for all subjects, as has previously been shown. Furthermore, the high failure rate in non-lesional AD skin may reflect a low level of immune activation and cell infiltration in a subset of subjects. We draw attention to the concept that barrier properties that differ between AD and healthy controls, or between type 2-high and type 2-low subjects could result in differential skin sampling depth. These depth differences could drive broad expression profile differences given the multilayered nature of the skin. This clearly is not the case for AD vs. healthy controls in our current study, as only a limited number of genes were differentially expressed, and these genes were not relegated to only skin differentiation states or layers. Likewise, this does not appear to be the case for the type 2-high vs. type 2-low AD comparison. Most genes differentially expressed here were immune (rather than skin layer) related. If anything, our skin tape results for AD and type 2-high differential expression show considerably less variation in skin layer and barrier-related genes than prior skin biopsy transcriptome studies, where the portion of the biopsy originating from different skin layers (epidermis vs. dermis) may have indeed played a role in results. An interesting aspect of the study is the immune signature readout in such superficial skin collected by tape stripping. However, as noted in our histology and others’ results, infiltration of inflammatory immune cells into the superficial skin layers is a real phenomenon. Clearly, this is also seen in our lesional skin results, where it was more expected. In summary we believe our endotypes truly identify type 2 inflamed subjects. We recoginze that the participation of this study was limited to adult caucasions, and the scope of this work may not translate to pediatric atopic dermatitis. Further study of children is needed, and would be facilitated by our methods. Finally, we also acknowledge the cross-sectional, single sampling nature of our data. Future studies will need to be conducted which perform repeat sampling varying time and body location. These studies would allow determination of persistance for this endotype and its potential uniformity across body location.
In summary, we detail a novel method for molecular analysis of skin from AD patients which is non-scarring and minimally invasive. Our method indicates an endotypic subgroup of AD patients exists with high immune activation, positively associated with severity, and dominated by type 2 inflammation. With modifications to improve robustness, our method may have clinical application in precision medicine and allow, for the first time, birth cohorts and longitudinal disease evolution studies which repeatedly sample and genomically evaluate the skin. These studies will likely be required to truly reveal the mechanisms which underlie both the development and clinical course of AD disease.
Supplementary Material
STable 1. Gene Ontology enrichments of differentially expressed genes between the tape strips collected from nonlesional AD skin and skin of healthy control subjects Gene Ontologies Upregulated in AD Tape Strips
STable 2. Leading edge genes identified from the gene ontologies significantly enriched in the nonlesional AD skin tape strip samples vs healthy control skin samples.
STable 3. Significantly differentially expressed genes between type 2-high AD and type 2-low AD skin tape strip groups
Stable 4. The top 250 significantly differentially expressed genes between lesional AD and healthy skin tape strip groups
Multidimensional scaling (MDS) of the top 500 variant genes in non-lesional epidermis and dermis samples separates the samples by tissue. The MDS plot indicates no visual similarity (proximity) among samples by the success of sequencing their corresponding skin tape strip RNA-seq.
Clinical Implications.
The availability of minimally invasive expression profiling of skin tape strips will contribute to the development of precision medicine in various skin diseases.
Acknowledgments
Funding Sources: The project was funded in part by NIH/NIAMS grant R01 AR41256, NIH/NIAID grant U19 AI117673, NIH/NCRR grant UL1 RR025780 and The Edelstein Family Chair of Pediatric Allergy-Immunology at National Jewish Health.
Abbreviations
- AD
atopic dermatitis
- APC
antigen presenting cell
- [p]DC
[plasmacytoid] dendritic cell
- DEG
differentially expressed gene
- GAGE
generally applicable gene-set enrichment
- GO
gene ontology
- GSEA
gene set enrichment analysis
- IDEC
inflammatory dendritic epidermal cell
- LC
Langerhans cell
- IGA
Investigator’s Global Assessment scores
- EASI
eczema area and severity index
- BSA
body surface area
- FITC
fluorescein isothiocyanate
- MFI
mean fluorescence intensity
- TMAP
torrent mapping alignment program
- FDR
false discovery rate
- MDS
multidimensional scaling
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanifin JM, Reed ML, Eczema P Impact Working G. A population-based survey of eczema prevalence in the United States. Dermatitis. 2007;18:82–91. doi: 10.2310/6620.2007.06034. [DOI] [PubMed] [Google Scholar]
- 2.Silverberg JI, Simpson EL. Associations of childhood eczema severity: a US population-based study. Dermatitis. 2014;25:107–14. doi: 10.1097/DER.0000000000000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung DY, Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: shifting paradigms in treatment approaches. J Allergy Clin Immunol. 2014;134:769–79. doi: 10.1016/j.jaci.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunner PM, Guttman-Yassky E, Leung DY. The immunology of atopic dermatitis and its reversibility with broad-spectrum and targeted therapies. J Allergy Clin Immunol. 2017;139:S65–S76. doi: 10.1016/j.jaci.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 6.Weidinger S, Illig T, Baurecht H, Irvine AD, Rodriguez E, Diaz-Lacava A, et al. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J Allergy Clin Immunol. 2006;118:214–9. doi: 10.1016/j.jaci.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Gao PS, Rafaels NM, Hand T, Murray T, Boguniewicz M, Hata T, et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 2009;124:507–13. 13 e1–7. doi: 10.1016/j.jaci.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–56. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao PS, Rafaels NM, Mu D, Hand T, Murray T, Boguniewicz M, et al. Genetic variants in thymic stromal lymphopoietin are associated with atopic dermatitis and eczema herpeticum. J Allergy Clin Immunol. 2010;125:1403–7. e4. doi: 10.1016/j.jaci.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khattri S, Shemer A, Rozenblit M, Dhingra N, Czarnowicki T, Finney R, et al. Cyclosporine in patients with atopic dermatitis modulates activated inflammatory pathways and reverses epidermal pathology. J Allergy Clin Immunol. 2014;133:1626–34. doi: 10.1016/j.jaci.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson EL, Chalmers JR, Hanifin JM, Thomas KS, Cork MJ, McLean WH, et al. Emollient enhancement of the skin barrier from birth offers effective atopic dermatitis prevention. J Allergy Clin Immunol. 2014;134:818–23. doi: 10.1016/j.jaci.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elmose C, Thomsen SF. Twin Studies of Atopic Dermatitis: Interpretations and Applications in the Filaggrin Era. J Allergy (Cairo) 2015;2015:902359. doi: 10.1155/2015/902359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bieber T, D’Erme AM, Akdis CA, Traidl-Hoffmann C, Lauener R, Schappi G, et al. Clinical phenotypes and endophenotypes of atopic dermatitis: Where are we, and where should we go? J Allergy Clin Immunol. 2017;139:S58–S64. doi: 10.1016/j.jaci.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Beck LA, Thaci D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med. 2014;371:130–9. doi: 10.1056/NEJMoa1314768. [DOI] [PubMed] [Google Scholar]
- 15.Simpson EL, Bieber T, Guttman-Yassky E, Beck LA, Blauvelt A, Cork MJ, et al. Two Phase 3 Trials of Dupilumab versus Placebo in Atopic Dermatitis. N Engl J Med. 2016;375:2335–48. doi: 10.1056/NEJMoa1610020. [DOI] [PubMed] [Google Scholar]
- 16.Ewald DA, Malajian D, Krueger JG, Workman CT, Wang T, Tian S, et al. Meta-analysis derived atopic dermatitis (MADAD) transcriptome defines a robust AD signature highlighting the involvement of atherosclerosis and lipid metabolism pathways. BMC Med Genomics. 2015;8:60. doi: 10.1186/s12920-015-0133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suarez-Farinas M, Ungar B, Correa da Rosa J, Ewald DA, Rozenblit M, Gonzalez J, et al. RNA sequencing atopic dermatitis transcriptome profiling provides insights into novel disease mechanisms with potential therapeutic implications. J Allergy Clin Immunol. 2015;135:1218–27. doi: 10.1016/j.jaci.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Suarez-Farinas M, Tintle SJ, Shemer A, Chiricozzi A, Nograles K, Cardinale I, et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127:954–64. e1–4. doi: 10.1016/j.jaci.2010.12.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole C, Kroboth K, Schurch NJ, Sandilands A, Sherstnev A, O’Regan GM, et al. Filaggrin-stratified transcriptomic analysis of pediatric skin identifies mechanistic pathways in patients with atopic dermatitis. J Allergy Clin Immunol. 2014;134:82–91. doi: 10.1016/j.jaci.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds SD, Rios C, Wesolowska-Andersen A, Zhuang Y, Pinter M, Happoldt C, et al. Airway Progenitor Clone Formation Is Enhanced by Y-27632-Dependent Changes in the Transcriptome. Am J Respir Cell Mol Biol. 2016;55:323–36. doi: 10.1165/rcmb.2015-0274MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 26.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–7. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 28.Szeles L, Poliska S, Nagy G, Szatmari I, Szanto A, Pap A, et al. Research resource: transcriptome profiling of genes regulated by RXR and its permissive and nonpermissive partners in differentiating monocyte-derived dendritic cells. Mol Endocrinol. 2010;24:2218–31. doi: 10.1210/me.2010-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutter C, Kauer M, Simonitsch-Klupp I, Jug G, Schwentner R, Leitner J, et al. Notch is active in Langerhans cell histiocytosis and confers pathognomonic features on dendritic cells. Blood. 2012;120:5199–208. doi: 10.1182/blood-2012-02-410241. [DOI] [PubMed] [Google Scholar]
- 30.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014;133:670–8. e12. doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson MD, Capra V, Clunes MT, Rovati GE, Stankova J, Maj MC, et al. Cysteinyl Leukotrienes Pathway Genes, Atopic Asthma and Drug Response: From Population Isolates to Large Genome-Wide Association Studies. Front Pharmacol. 2016;7:299. doi: 10.3389/fphar.2016.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–40. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 33.Harper JI, Godwin H, Green A, Wilkes LE, Holden NJ, Moffatt M, et al. A study of matrix metalloproteinase expression and activity in atopic dermatitis using a novel skin wash sampling assay for functional biomarker analysis. Br J Dermatol. 2010;162:397–403. doi: 10.1111/j.1365-2133.2009.09467.x. [DOI] [PubMed] [Google Scholar]
- 34.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–6. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 35.Sasagawa S, Nishimura Y, Sawada H, Zhang E, Okabe S, Murakami S, et al. Comparative Transcriptome Analysis Identifies CCDC80 as a Novel Gene Associated with Pulmonary Arterial Hypertension. Front Pharmacol. 2016;7:142. doi: 10.3389/fphar.2016.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denzin LK, Sant’Angelo DB, Hammond C, Surman MJ, Cresswell P. Negative regulation by HLA-DO of MHC class II-restricted antigen processing. Science. 1997;278:106–9. doi: 10.1126/science.278.5335.106. [DOI] [PubMed] [Google Scholar]
- 37.Kraft S, Kinet JP. New developments in FcepsilonRI regulation, function and inhibition. Nat Rev Immunol. 2007;7:365–78. doi: 10.1038/nri2072. [DOI] [PubMed] [Google Scholar]
- 38.Esaki H, Ewald DA, Ungar B, Rozenblit M, Zheng X, Xu H, et al. Identification of novel immune and barrier genes in atopic dermatitis by means of laser capture microdissection. J Allergy Clin Immunol. 2015;135:153–63. doi: 10.1016/j.jaci.2014.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol. 2003;112:1195–202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 40.Howell MD, Gallo RL, Boguniewicz M, Jones JF, Wong C, Streib JE, et al. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–8. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 41.Li S, Villarreal M, Stewart S, Choi J, Indra G, Babineau DC, et al. Altered composition of epidermal lipids correlates with Staphylococcus aureus colonization status in Atopic Dermatitis. Br J Dermatol. 2017 doi: 10.1111/bjd.15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Turner A, Aggarwal P, Matter A, Storvick E, Arnett DK, et al. Comprehensive evaluation of AmpliSeq transcriptome, a novel targeted whole transcriptome RNA sequencing methodology for global gene expression analysis. BMC Genomics. 2015;16:1069. doi: 10.1186/s12864-015-2270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh RK, Gupta S, Dastidar S, Ray A. Cysteinyl leukotrienes and their receptors: molecular and functional characteristics. Pharmacology. 2010;85:336–49. doi: 10.1159/000312669. [DOI] [PubMed] [Google Scholar]
- 44.Liu M, Yokomizo T. The role of leukotrienes in allergic diseases. Allergol Int. 2015;64:17–26. doi: 10.1016/j.alit.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Grill JI, Neumann J, Herbst A, Ofner A, Hiltwein F, Marschall MK, et al. Loss of DRO1/CCDC80 results in obesity and promotes adipocyte differentiation. Mol Cell Endocrinol. 2017;439:286–96. doi: 10.1016/j.mce.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 46.Brunner PM, Emerson RO, Tipton C, Garcet S, Khattri S, Coats I, et al. Non-lesional atopic dermatitis skin shares similar T-cell clones with lesional tissues. Allergy. 2017 doi: 10.1111/all.13223. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida K, Kubo A, Fujita H, Yokouchi M, Ishii K, Kawasaki H, et al. Distinct behavior of human Langerhans cells and inflammatory dendritic epidermal cells at tight junctions in patients with atopic dermatitis. J Allergy Clin Immunol. 2014;134:856–64. doi: 10.1016/j.jaci.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Bieber T, Novak N, Herrmann N, Koch S. Role of dendritic cells in atopic dermatitis: an update. Clin Rev Allergy Immunol. 2011;41:254–8. doi: 10.1007/s12016-010-8224-0. [DOI] [PubMed] [Google Scholar]
- 49.Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, Whynot J, Novitskaya I, Cardinale I, et al. Major differences in inflammatory dendritic cells and their products distinguish atopic dermatitis from psoriasis. J Allergy Clin Immunol. 2007;119:1210–7. doi: 10.1016/j.jaci.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–95. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamilton JD, Suarez-Farinas M, Dhingra N, Cardinale I, Li X, Kostic A, et al. Dupilumab improves the molecular signature in skin of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol. 2014;134:1293–300. doi: 10.1016/j.jaci.2014.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
STable 1. Gene Ontology enrichments of differentially expressed genes between the tape strips collected from nonlesional AD skin and skin of healthy control subjects Gene Ontologies Upregulated in AD Tape Strips
STable 2. Leading edge genes identified from the gene ontologies significantly enriched in the nonlesional AD skin tape strip samples vs healthy control skin samples.
STable 3. Significantly differentially expressed genes between type 2-high AD and type 2-low AD skin tape strip groups
Stable 4. The top 250 significantly differentially expressed genes between lesional AD and healthy skin tape strip groups
Multidimensional scaling (MDS) of the top 500 variant genes in non-lesional epidermis and dermis samples separates the samples by tissue. The MDS plot indicates no visual similarity (proximity) among samples by the success of sequencing their corresponding skin tape strip RNA-seq.