Abstract
HLA-DRB1 is the major genetic risk factor for visceral leishmaniasis (VL). We used SNP2HLA to impute HLA DRB1 alleles and SNPTEST to carry out association analyses in 889 human cases and 977 controls from India. NetMHCIIpan2.1 was used to map epitopes and binding affinities across 49 Leishmania vaccine candidates, and across peptide epitopes captured from dendritic cells treated with crude Leishmania antigen and identified using mass spectrometry and alignment to a reference Leishmania genome. Cytokines were measured in peptide-stimulated whole blood from 26 cured VL cases and 8 endemic healthy controls. HLA-DRB1*1501 and DRB*1404/DRB1*1301 were the most significant protective versus risk alleles, respectively, with specific residues at amino acid positions 11 and 13 unique to protective alleles. We observed greater peptide promiscuity in sequence motifs for 9-mer core epitopes predicted to bind to risk (*1404/*1301) compared to protective (*1501) DRB1 alleles. There was a higher frequency of basic AAs in DRB1*1404-/*1301-specific epitopes, compared to hydrophobic and polar AAs in DRB1*1501-specific epitopes, at anchor residues P4 and P6 which interact with residues at DRB1 position 11 and 13. Cured VL patients made variable but robust interferon-γ, tumour necrosis factor and interleukin-10 responses to 20-mer peptides based on captured epitopes, with peptides based on DRB1*1501-captured epitopes resulting in a higher proportion (odds ratio 2.23; 95%CI 1.17-4.25; P=0.017) of patients with interferon-γ:IL10 ratios >2-fold compared to peptides based on DRB1*1301-captured epitopes. Our data provides insight into molecular mechanisms underpinning the association of HLA DRB1 alleles with risk versus protection in VL in humans.
Introduction
The importance of host genetic factors in determining outcome of infection with Leishmania species causing visceral leishmaniasis (VL) is indicated by familial clustering (1) and high sibling risk ratios (2). In mice, different genes control innate versus adaptive immunity (reviewed (3)), but the MHC class II region (H-2) stands out as one of the earliest identified (4), with the clearest effects of any locus in murine models of complex infectious disease. This attests to the importance of CD4+ T cell mediated immunity in determining disease outcome in leishmaniasis, with polarized antigen-specific T helper 1 (Th1) and Th2/T regulatory responses affecting favourable versus adverse outcomes, respectively, in murine models (5) and human disease (6). Clinical VL caused by L. donovani, in particular, has been associated with high Th2/Treg cytokine responses (6-10), while Th1-generated interferon-γ (IFN-γ) is higher in children infected with L. infantum chagasi that do not progress to clinical VL than in those who do (11). Nevertheless, HLA had not been highlighted as a putative susceptibility locus amongst a number of genome-wide linkage studies (12-15), and robust associations had not been observed in the few small studies in which HLA has been examined as a candidate gene region (reviewed (16)), likely because of low statistical power. As part of the Wellcome Trust Case Control Consortium 2 (WTCCC2), we undertook the first genome-wide association study (GWAS) of VL across major foci of disease caused by L. donovani in India and L. infantum chagasi in Brazil (17). Meta-analysis of discovery GWAS undertaken in 989 cases and 1089 controls from India and 357 cases in 308 families (1970 individuals) from Brazil identified the class II gene region HLA-DRB1-DQA1 as the major locus regulating susceptibility and resistance to clinical disease for L. donovani and L. infantum chagasi (Discovery Meta-P=1.6-×10−13), with replication in a second sample of 941 cases and 990 controls from India (Combined P=2.76×10−17, OR=1.41, 95%CI 1.30-1.52, across 3 cohorts). No other loci replicated at P<10−6 within India, and no non-MHC loci identified on the basis of the discovery GWAS in Indian samples were replicated in Brazil. The importance of this GWAS is the demonstration that HLA class II alleles are major genetic risk factors for VL that cross the epidemiological divides of geography and parasite species, renewing interest in research to understand the precise molecular basis to this and translate this knowledge to development of vaccines that will work even in genetically susceptible individuals.
Identification of peptides selected by MHC class II molecules during natural processing of proteins is fundamentally important to understanding the repertoire of CD4+ T cells regulating complex disease (18, 19). Improvements in mass spectrometry have facilitated examination of whole repertoires of peptides selected from natural processing, leading to key observations (reviewed (19)) on the nature of naturally processed peptides selected by class II: (i) variable in length (12-26 AA); (ii) families share a common 9 AA core; (iii) 9-mer binding core interacts with peptide binding groove, which consists of 4 major binding pockets P1, P4, P6 and P9; (iv) most allelic differences between class II molecules reside within the binding pocket, which determine allele-specific binding motifs; (v) flanking residues increase peptide-binding affinity; and (vi) heterogeneity in flanking residues reflects enzyme specific proteolysis in antigen processing. This has underpinned development of predictive algorithms (20) to analyse epitope selection in silico, which can be compared with those determined experimentally. Here we use a combination of in silico tools and experimental epitope capture to characterize the epitopes that bind to risk versus protective DRB1 alleles for VL. We show greater peptide promiscuity and sequence motifs for 9-mer core epitopes predicted to bind to the peptide binding groove of risk (*1404/*1301) compared to protective (*1501) DRB1 alleles. We find that cured VL patients make robust IFN-γ, tumour necrosis factor (TNF) and interleukin-10 (IL-10) responses to 20-mer peptides based on protein sequences for captured epitopes, with peptides based on DRB1*1501-captured epitopes resulting in a higher proportion (odds ratio 2.23; 95%CI 1.17-4.25; P=0.017) of patients with interferon-γ:IL10 ratios >2-fold compared to peptides based on DRB1*1301-captured epitopes.
Materials and Methods
Study subjects
For HLA typing and imputation studies, we used subjects from the Indian discovery GWAS recruited from Bihar state in northeast India, as previously described (17). Cases and controls were matched for self-reported age, sex, religion, caste and geographic region of recruitment. The post-quality control sample used in the original GWAS was 989 cases and 1089 controls. For HLA imputation (cf. below) we used data for 889 unrelated cases and 977 unrelated controls. For whole blood assays we recruited 26 cured VL cases (14 females; 12 males; mean age 29±13 years, range 9 to 52 years) and 8 healthy endemic controls (1 female; 7 males; mean age 41±8 years, range 26 to 55 years) from the same study area as the GWAS. Of the cured cases, 23 received single dose Ambisome, and 3 received a 30-day course of Amphotericin B. All cases had achieved complete recovery. Samples were collected 4 to 7 months after completion of treatment. Dendritic cells (cf. below) were prepared from a cryobank of peripheral blood mononuclear cells for HLA-typed de-identified donors held at Lonza Biologics plc (Cambridge, UK).
Ethical considerations
Ethical approvals for studies on Indian subjects were obtained from the ethical committee of the Institute of Medical Sciences, Banaras Hindu University (Varanasi, India). The study was carried out in accordance with the Declaration of Helsinki Principles, and each participant, or the parent/guardian of individuals <18 years old, signed informed consent forms to participate in the study and provide a blood sample.
HLA imputation and association analysis
Imputation of classical HLA alleles at 2- and 4-digit level, as well as amino acid variants, in the Indian discovery GWAS case-control cohort was performed in SNP2HLA (21) using extensive reference data for the T1DGC (22) panel (10450 haplotypes). Frequentist association tests for each allele were performed in SNPTEST (23) under an additive model, with the first 3 principal components for population genetic substructure as covariates. Odds ratios, associated P-values and frequency of the DRB1 alleles were determined.
In silico epitope predictions using NetMHCIIpan 2.1
We employed NetMHCIIpan 2.1 (20) to screen overlapping 20-mer peptides across 49 known vaccine candidate molecules, or across 12 proteins identified on the basis of epitope capture experiments (see results). This prediction tool outperforms other tools (24, 25) principally by implementation of NN-align, a neural network-based approach that combines the peptide sequence representation that was highly successful in predicting the binding specificity of HLA Class I molecules and now includes the representation of peptide flanking residues and peptide length applicable to HLA Class II molecules (25, 26). Binding affinities for each peptide are provided as nM IC50 values, with arbitrary cut-offs for strong and weak binding affinities set at IC50, 1-50nM (=High) and 51-500nM (=Low). The y-axis of all binding affinity plots is represented as relative binding affinity expressed as 1-log15,000 of the predicted nM binding affinity (20). Sequences for the 9-mer cores of 20-mer epitopes with binding affinities specific for risk or protective DRB1 alleles, or with binding affinity to both, were aligned using BlockLogo (27), and the resulting sequence motifs were visualized using WebLogo (28, 29). Peptides selected for whole blood assays are shown in Table S1 (cf. below). Note that reference to all binding affinities presented here are predicted, not measured, binding affinities.
Epitope capture from dendritic cells
Dendritic cells (DCs; 4-5×106) were prepared essentially as described (30) from cryopreserved peripheral blood mononuclear cells (PBMC) from human donors homozygous for risk (*1301, homozygous *1404 donors were not available) or protective (*1501) DRB1 alleles. Briefly, PBMC were isolated from fresh human whole blood by density gradient centrifugation in under 8 hours and stored in vapour phase nitrogen according to the ethically approved protocol and donors informed consent. To generate dendritic cells, cryopreserved PBMC were thawed and monocytes isolated using negative magnetic bead isolation (Stem Cell Technologies, Cambridge, UK). Monocytes were then differentiated into dendritic cells with human GM-CSF and human IL-4 (Peprotech, London, UK) in a tissue culture flask (BD Biosciences, Oxford, UK) for 5 days at 37°C, 5% CO2. Crude L. donovani (strain MHOM/ET/67/HU3) antigen was prepared from stationary-phase promastigotes by resuspension in 5mM CaCl2, 10mM Tris, pH 7.4, 1:100 cOmplete™ EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich, Castle Hill, Australia), and freeze-thawing 3x over liquid nitrogen (31). After differentiation the crude antigen was added to the dendritic cells to a final concentration of 50 μg/mL, along with TNFα and IL-1β for 24 hours to mature the dendritic cells. After maturation the DC were transferred to tubes and washed with assay medium before being centrifuged at 300g for 10 mins to pellet the cells. Once pelleted the medium was removed and the dendritic cell pellets stored at −80°C until HLA-DR purification. Membrane fractions were prepared by re-suspension and gentle homogenization of the cell pellets in hypotonic lysis buffer containing 0.5% IGEPAL CA-630 (octylphenoxypolyethoxyethanol; Sigma-Aldrich, Castle Hill, NSW, Australia), 50mM Tris-HCl (pH 8.0), 150mM NaCl, 2.5mM EDTA and protease inhibitors (Complete Protease Inhibitor Cocktail Tablet; Roche, Sydney, NSW, Australia) at ~107 cells/mL, and rotated 1h at 4°C. Cellular debris was removed by ultracentrifugation at 180,000×gmax for 60min. Supernatants were retained, and the pellet resuspended and processed again through lysis buffer and ultracentrifugation. The combined clear supernatants were retained for purification of DRB1 molecules over CNBr-activated Sepharose 4 Fast Flow columns containing 5mg of L227 anti-human HLA-DR (1 mg antibody/mL of resin; L227 is specific for an HLA-DR monomorphic epitope – isotype IgG1) affinity matrix. The flow-through lysate was reloaded onto the Sepharose columns ×1. Columns were washed in 10 column volumes of buffer containing 50mM Tris, 150mM NaCl, and 0.5% IGEPAL CA-630 (final pH 8.0). Columns were sequentially washed by 5 column volumes of 50mM Tris and 150mM NaCl (final pH 8.0) followed by 5 column volumes of a high salt buffer, to remove non-specifically bound material (50mM Tris and 500mM NaCl (final pH 8.0)) and finally washed with 5 column volumes of 50mM Tris (pH 8.0). Bound HLA-DRB1*1301 and HLA-DRB1*1501 peptide complexes were eluted with 14mL of 10% acetic acid and eluates were collected as separate fractions of 2mL each. Western blotting was performed to determine (a) which fractions contained the eluted DRB1 molecules, and (b) to check for the ratio of αβ dimers to β-chain monomers indicative of the degree of dissociation of the DRB1 dimers and therefore of peptides from DRB1 molecules. DRB1-containing eluates were then filtered through 10kDa polyethersulfone ultracentrifugation filters (Sartorius, Dandenong South, Victoria, Australia) and frozen at −80°C. Samples were defrosted, 3.5mL H2O added, vortexed, frozen with liquid N2, and frozen samples freeze dried ready for mass spectrometry. Mass spectrometry was performed by Proteomics International Laboratories Ltd (Perth, Western Australia). Samples were re-suspended in 100μL of 2% CAN/0.05% TFA and 50μL injected onto the HPLC system. Peptides were analyzed by LC-MS analysis using the Shimadzu Prominence nano HPLC system (Shimadzu, Malaga, Western Australia) coupled to a 5600 TripleTOF mass spectrometer (AB Sciex, Mulgrave, Victoria, Australia). Peptides were loaded onto an Agilent Zorbax 300SB-C18, 3.5μM (Agilent Technologies, Singapore) and separated with a gradient of 2-40% acetonitrile (0.1% formic acid) over 100min. Spectral data for the samples were analyzed against a database comprising the TriTrypDB-6.0_LdonovaniBPK282A1_ AnnotatedProteins with SwissProt curated Human protein sequences. Hits were considered as true hits against a Leishmania protein when the same spectrum matched with higher confidence to the Leishmania predicted peptide than to Human or Bovine Serum Albumin (BSA) matches for the same spectrum.
Whole blood assays
Whole blood was collected into heparinised tubes and samples were diluted 1 in 8 in serum-free complete medium comprising RPMI 1640 medium supplemented with 2 mM L-glutamine, 100μg/mL of streptomycin, and 100 IU/mL of penicillin (Gibco, USA). Diluted blood (180μL/well) was plated into 96-well U-bottomed plates (Nunc, Rochester, USA) and antigen added in triplicate wells at a final concentration of 10 μg/mL soluble Leishmania antigen (32) prepared from an Indian strain of L. donovani, 5 μg/mL individual peptides, 5 μg/mL PHA, or no antigen control. Peptides for 20-mer amino acid epitopes based on the predicted affinity of binding to risk and/or protective HLA DRB1 alleles (Table S1) were synthesized commercially (Peptide2.0, Chantilly, VA, USA). They were initially solubilized in dimethylsulfoxide (final concentration in the well <0.1% DMSO), diluted in endotoxin-free phosphate-buffered saline at a final concentration of 50 μg/mL, and stored at −80°C. Cytokine release (IFN-γ, IL-10) was measured by ELISA in supernatants harvested at 24 h post antigen stimulation using matched antibody pairs (BD Pharmingen, Franklin Lakes, NJ, USA). The limit of detection for these ELISAs was 31 pg/mL. TNF was measured using the Human TNF-α ELISA MAX™ Set (Catalogue Number 430206; BioLegend, San Diego, CA, USA).
Statistical methods
Fisher’s Exact Test was used to determine whether the proportions of responders to non-responders differed between peptides of different affinities for risk versus protective DRB1 alleles.
Results
Genetic associations of classical HLA DRB1 alleles with VL
A forest plot demonstrating the association between VL and imputed HLA DRB1 alleles at the 2- and 4-digit level is presented in Figure 1. This concurs with our previous analysis (17) showing that HLA DRB1*16, DRB1*01 and DRB1*15 allele groups provide significant protection (OR 0.43 to 0.79) from VL, while DRB1*14, DRB1*11, and DRB1*13 are significant risk (OR 1.37 to 1.76) groups. More specifically, for further functional analyses designed to relate DRB1 molecular function to VL susceptibility, we selected DRB1*1501 (OR 0.71; 95%CI 0.59-0.86; P=3.01×10−4) as representative of protective alleles, and DRB1*1404 (OR 1.37; 95%CI 1.09-1.71; P=6.73×10−3) or DRB1*1301 (OR 1.48; 95%CI 1.10-2.00; P=0.01) as representative risk alleles. These alleles were individually the most frequent risk and protective HLA-DRB1 alleles in the Indian population (HLA-DRB1*1301: 0.05; *1404: 0.09; *1501: 0.14). The analysis of imputed alleles also showed that VL susceptibility is associated with variants at amino acid (AA) positions 9, 11, 13, and 37 (Figure 2) which contribute to forming the DRB1 binding groove pockets that interact with residues of the 9-mer core of the peptide (33) (Figure 3). For example, Proline and Leucine (pocket P6 of the binding groove) at DRB1 AA position 11, and Serine at AA position 37 (pocket P9 of the binding groove), are associated with protection (OR 0.67; 95%CI 0.58-0.77; P=1.48×10−8) and are unique to the protective allele groups DRB1*01/*15/*16. Alternative AA alleles (Serine, Valine, Asparagine) at DRB1 AA position 11 are associated with risk (OR 1.45; 95%CI 1.28-1.65; P=1.27×10−8) and are common to risk allele groups DRB1*03/*04/*09/*10/*11/*12/*13/*14. This evidence highlighting AA positions that influence structure and conformation of the peptide-binding groove of the DRB1 molecule provided the rationale for further functional analysis of epitope binding affinities.
FIGURE 1.
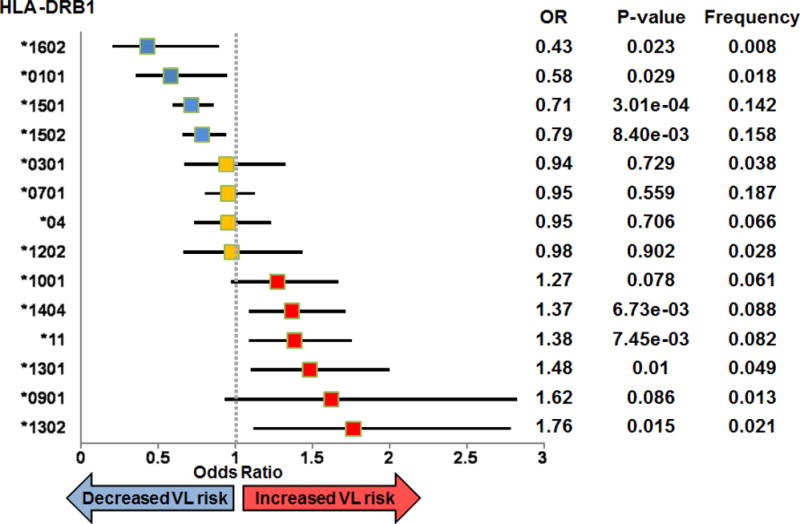
Forest plot showing associations between VL and imputed 2- or 4-digit classical HLA DRB1 alleles. The plot shows odds ratios (OR) and 95% confidence intervals for risk (OR>1; red symbols), neutral (OR~1; yellow symbols), and protective (OR<1; blue symbols) haplotypes. Information to the right of the plot shows values for the OR, the P-value for the association, and the allele frequency.
FIGURE 2.
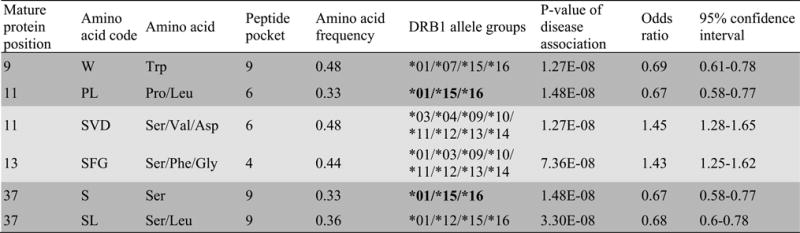
SNPTEST results for association between Indian VL and imputed AA variants within the HLA-DRB1 binding groove. Imputation was carried out using SNP2HLA and reference data from T1DGC, as referenced in the main text. Dark grey shading indicates protective alleles (odds ratio <1), light grey shading risk alleles (odd ratio >1). Bold lettering highlights AA at positions 11 and 37 that are unique to protective allele groups, as discussed in the text.
FIGURE 3.
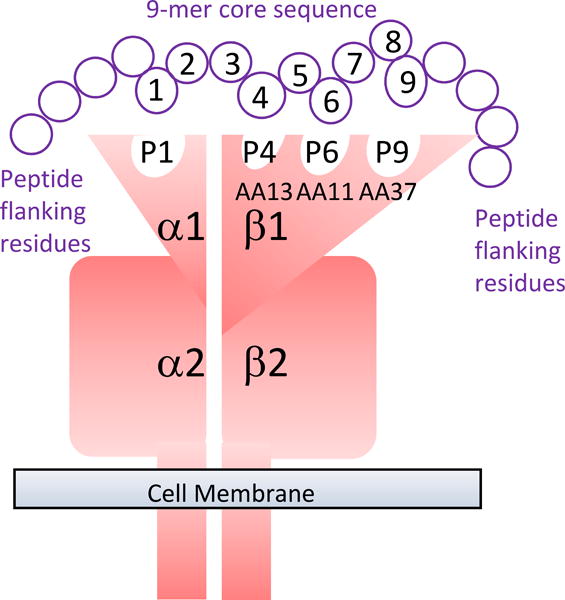
Diagrammatic representation of the DRA/DRB alpha/beta dimer to show how specific amino acids in the binding groove of DRB1 form the pockets that interact with different positions in the 9-mer core epitope of the antigen being presented.
In silico epitope predictions for candidate Leishmania vaccines
To determine whether risk versus protective 4-digit HLA-DRB1 alleles differ in epitope binding affinities and peptide repertoire for Leishmania antigens we selected a subset of 49 known diagnostic and vaccine candidate proteins for in silico analysis (Table I). Using NetMHCIIpan 2.1 (20), a sliding window of 1-mer overlapping 20-mer peptides based on the full-length amino acid sequences of the 49 antigens were screened in order to identify peptides with strong and weak binding affinities (arbitrarily IC50, 1-50nM=strong; 51-500nM=weak). Figure S1 provides plots mapping binding affinities for epitopes across Leishmania proteins predicted to bind to risk DRB1*1404 compared to protective DRB1*1501 4-digit alleles. Of interest, some well-studied vaccine candidates (40, 59) were almost devoid of epitopes that would be predicted to bind to risk or protective DRB1 alleles (see, for example, A2 and HASPB1 in Figure S1). To gain a better understanding of the nature of epitopes binding to different DRB1 alleles, we selected the 9-mer core for the 20-mer with the top binding affinity to either HLA-DRB1*1404 or HLA-DRB1*1501, or with equal binding affinity to both. For the analysis of DRB1*1404- versus DRB1*1501-specific epitopes, preferential binders were defined as those with ≤500nM affinity for one DRB1 allele and ≥1000nM for the alternative allele. Table S1 gives examples of the NetMHCIIpan 2.1 output and how these 9-mer cores were selected. Across all proteins, the total number of unique epitopes with preferential binding to DRB1*1404 was 97 (mean±SD binding affinity for *1404 = 350±93; for *1501 = 1231±260), with 82 unique epitopes preferentially binding to DRB1*1501 (mean±SD binding affinity for *1501 = 356±105; for *1404 = 1277±469). There were 188 unique epitopes with peak binding affinities <100nM common to both DRB1 alleles. WebLogo plots (Figure 4) show that the sequence motif for 9-mer core peptides that bind preferentially to DRB1*1501 differs from those of DRB1*1404-preferential or common binders. In addition to greater promiscuity of amino acid usage in DRB1*1404-preferential 9-mer core peptides, the sequence motif plots show that the anchor residues interacting with the HLA-DR binding pockets 4 (P4) and P6 differ between the two alleles in terms of the nature and frequencies of amino acids at these positions. While hydrophobic residues are generally preferred for P1 for both alleles, P4 preferred residues are polar-hydrophilic (specifically, serine and threonine) or basic for DRB1*1404 but mainly non-polar hydrophilic (specifically, glycine) or hydrophobic in DRB1*1501. Similarly, residues that interact with P6 of the HLA-DR molecule are generally basic or polar-hydrophilic (specifically, serine and threonine) for the risk DRB1*1404 allele but non-polar hydrophilic (specifically, glycine) or hydrophobic for the protective DRB1*1501 allele. These differences are of specific interest given that the anchor residues 4 and 6 interact with the HLA-DRB1 residues at positions 11 and 13, respectively (Figure 3). The latter are unique to the protective HLA-DRB1*15 allele groups that our HLA imputation studies demonstrate are associated with VL (Figure 2). Except for residues at position 1, which are similarly conserved across all 9-mer core peptides, motifs of 9-mer core peptides that are common to both DRB1 alleles (Figure 4) exhibit a diverse sequence pattern with lower sequence conservation at P4 and P6 compared to allele-specific motifs. The differences in amino acid usage for motifs across the 20-mer peptides from which 9-mer cores were selected also reflect the differences apparent in the 9-mer core epitope motifs (Figure S2).
Table I.
Details of 49 proteins selected for in silico analysis of epitope binding affinity predictions using NetMHCIIpan2.1
| Protein name | Product | GeneDB/GenBank Accession | Reference |
|---|---|---|---|
| A2 LINJ220670 | hypothetical protein | UniProtKB - A4HZU7_LEIIN | (34) |
| CPL2 | cathepsin L-like protease | LdBPK_080960.1 | (35) |
| CPA/CBA | cysteine peptidase A | LdBPK_191460.1 | (36) |
| CPB | cysteine peptidase B | LdBPK_070600.1 | (36) |
| CPC | cysteine peptidase C | LdBPK_290860.1 | (37) |
| gp46/PSA/M-2 | surface membrane protein gp46 | LdBPK_311490.1 | (38) |
| gp63/MSP | GP63, leishmanolysin | GenBank: ACT31401.1 | (39) |
| HASPB1/K26 | hydrophilic acylated surface protein B | GenBank: CAA09789.1 | (40) |
| HbR | hemoglobin receptor | LdBPK_210300.1; | (41) |
| Hsp70 | HSP70-like protein | LdBPK_283000.1 | (39) |
| J41 | hypothetical protein | LdBPK_331620.1 | (42) |
| J89 | hypothetical protein | LdBPK_260990.1 | (42) |
| KMP11 | kinetoplastid membrane prot-1 | LdBPK_352260.1 | (43) |
| L15 | anaphase promoting complex subunit | LdBPK_354570.1 | (42) |
| L21 | nucleolar protein family a member | LdBPK_343810.1 | (42) |
| lmd29 | hypothetical protein | LdBPK_180600.1 | (42) |
| L302.06 | hypothetical protein, conserved | LdBPK_040220.1 | (42) |
| L31/584C | 60S ribosomal subunit L31 | LdBPK_353330.1 | (42) |
| L37 | 60S ribosomal L37 | LdBPK_342620.1 | (42) |
| L5 | 60S ribosomal L5 | LdBPK_351870.1 | (44) |
| L3 | ribosomal protein L3-like | LdBPK_323320.1 | (44) |
| LACK/p36 | activated protein kinase c receptor | LdBPK_282970.1 | (45) |
| LeIF | eukaryotic initiation factor 4a | LdBPK_010790.1 | (46) |
| Lepp12 | phosphoprotein lepp12 | LdBPK_365970.1 | (47) |
| LmSTI1 | stress-induced protein sti1 | LdBPK_081020.1 | (48) |
| LPG3 | lipophosphoglycan biosynthetic | LdBPK_290790.1 | (49) |
| M18 | amastin-like protein | LdBPK_080720.1 | (42) |
| N52 | vacuolar ATP synthase subunit | LdBPK_120480.1 | (42) |
| NH36 | nonspecific nucleoside hydrolase | LdBPK_181570.1 | (50) |
| PABP1 | polyadenylate-binding protein 1 | LdBPK_355360.1 | (51) |
| PABP2 | polyadenylate-binding protein 2 | LdBPK_354200.1 | (51) |
| PABP3 | poly(A)-binding protein, putative | LdBPK_250080.1 | (51) |
| Pxn4 | peroxidoxin_4 | LdBPK_230050.1 | (52) |
| PO | 60S acidic ribosomal protein P0 | LdBPK_072480.1 | (53) |
| P25 | hypothetical protein | LdBPK_354800.1 | (42) |
| P31 | hypothetical protein, conserved | LdBPK_342300.1 | (42) |
| PRP-2 | paraflagellar rod protein 2C | LdBPK_161510.1 | (54) |
| Q24 | dynein light chain, flagellar outer arm | LdBPK_320240.1 | (42) |
| Q51 | hypothetical protein | LdBPK_301750.1 | (42) |
| R32 | hypothetical protein | LdBPK_333110.1 | (42) |
| R71 | 60S ribosomal protein L22 | LdBPK_363430.1 | (42) |
| S4 | 40S ribosomal protein S4 | LdBPK_131120.1 | (55) |
| S33 | eukaryotic initiation factor 5a | LdBPK_250740.1 | (42) |
| SOD B1 | iron superoxide dismutase | LdBPK_321910.1 | (56) |
| SMT | sterol 24-c-methyltransferase | LdBPK_362520.1 | (57) |
| TSA/TRYP | tryparedoxin peroxidase | LdBPK_151140.1 | (31) |
| TUZIN1 | Tuzin, putative (62.3 kDa) | LdBPK_342640.1 | (58) |
| TUZIN2 | Tuzin, putative (58.7 kDa) | LdBPK_080730.1 | (58) |
| TUZIN3 | Tuzin, putative (70.2 kDa) | LdBPK_080740.1 | (58) |
Except for A2, all protein sequences were based on L. donovani sequences, as represented in GeneDB or GenBank. References to vaccine candidacy are not comprehensive, especially where multiple studies have been undertaken. Where possible we quote a reference where pre-clinical vaccine studies were undertaken in a model of visceral leishmaniasis.
FIGURE 4.
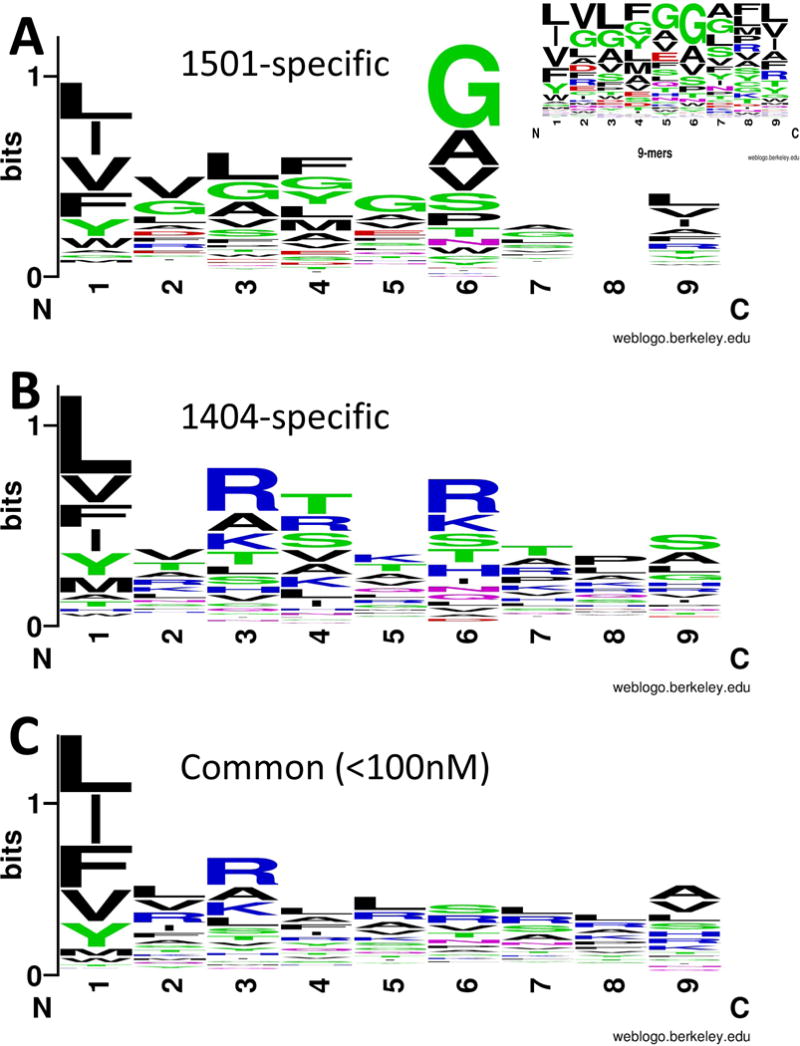
Sequence motifs that characterise 9-mer core epitopes for 20-mer peptides predicted to bind preferentially to (A) DRB1*1501 or (B) DRB1*1404 alleles, or (C) predicted to bind to both DRB1 alleles at <100nM. Preferential binders were defined as those with ≤500nM affinity for one DRB1 allele and ≥1000nM for the alternative allele (see Table S1 for examples). In silico predictions for 20-mer peptides were generated across 49 Leishmania vaccine candidate proteins using NetMHCIIpan 2.1. Sequences were aligned using BlockLogo and plotted using WebLogo, as referenced in the main text. The plots are based on 9-mer cores for 20-mer epitopes that spanned the peaks as plotted in Figure S1. Only one 9-mer core was selected to represent each epitope peak. The colours of the amino acids (AA) correspond to their chemical properties: polar AA are shown in green (G,S,T,Y,C; note G is non-polar when it forms a bond with other AA) or purple (Q,N); basic AA in blue (K,R,H); acidic AA in red (D,E); hydrophobic AA in black (A,V,L,I,P,W,F,M). The size of the letter relates to the frequency with which the AA is found in this position of core 9-mers, the overall peak height on the y-axis indicates the degree of conservation for specific epitopes at this location. Position 8 for DRB1*1501 showed no AA conservation; frequencies of AA at this position are shown in the insert figure.
Epitope capture from dendritic cells
In silico analysis provided evidence for different characteristics of peptide motifs binding to 9-mer cores for epitopes predicted to bind to risk versus protective DRB1 4-digit alleles. To determine whether similar differences occur for naturally processed and presented leishmanial antigens, we eluted peptides from DRB1 molecules purified from dendritic cells prepared from donors homozygous for risk (DRB1*1301) or protective (DRB1*1501) 4-digit alleles. In this experiment, 428 unique peptides were captured from DRB1*1501 human dendritic cells and 482 from DRB1*1301. Of these, 12 peptides (range 8- to 28-mers) mapped definitively back to the Leishmania genome alone, one in common between DRB1*1301 and DRB1*1501 dendritic cells, 6-specific to DRB1*1301, and 5 specific to DRB1*1501 (Figure 5). One peptide (not shown) had identical spectral matches to elongation factor 1-alpha against both human and Leishmania databases. This could represent a genuine auto-antigen or could be a captured Leishmania epitope.
FIGURE 5.
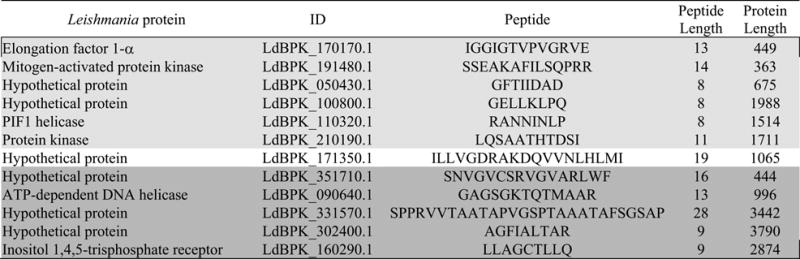
Leishmania peptides eluted from human dendritic cells treated with crude Leishmania antigen. ID, is for the matching proteins in the TriTrypDB-6.0_LdonovaniBPK282A1_AnnotatedProteins database. Light grey shading indicates epitopes captured from the risk DRB1*1301 Class II molecules, dark grey shading from the protective DRB1*1501. No shading indicates the epitope common to both allele groups. Mean length of proteins = 1625AA.
In silico epitope binding affinity predictions for captured Leishmania antigens
As for diagnostic/vaccine antigens, NetMHCIIpan2.1 was used to map epitopes across the full-length amino acid sequences for Leishmania proteins (Figure 5) from which epitopes had been captured. Figure S3 provides epitope binding affinity plots for 1-mer overlapping 20-mers predicted to bind to risk DRB1*1301 compared to protective DRB1*1501 4-digit alleles, with arrows indicating the AA positions matching the captured epitopes. Proteins from which epitopes were captured were generally large (Figure 5; 363AA to 3790AA in length), with multiple epitopes predicted to bind to one or both DRB1 alleles. Importantly, peak binding 20-mers incorporating captured peptides for all except one eluted peptide (from hypothetical protein LdBPK_331570.1; Table S1) were predicted to bind at <500nM, generally with stronger predicted binding affinity for the DRB1 allele from which they were captured. Based on 9-mer cores for the 20-mer (incorporating the captured peptides) with the strongest DRB1 allele-specific binding affinity (Figure 6) there was, as in Figure 4, a bias towards hydrophobic and polar AAs in DRB1*1501-specific peptides at 4 and 6, pointing again to the potential importance of these anchor residues interacting with the DRB1 residues at position 11 and 13 (Figure 3), shown to be associated with VL (Figure 2).
FIGURE 6.

Sequence motifs that characterise 9-mer core epitopes for 20-mer peptides incorporating peptide sequences as captured from dendritic cells from donors homozygous for risk (*1301) or protective (*1501) DRB1 alleles. Plots generated as in Figure 2, except that only the relative frequencies of AA are shown at each location in the 9-mer core of the 20-mer epitope since there were too few captured epitopes to determine degree of sequence conservation.
Cytokine responses to peptides based on captured epitopes
To determine whether epitopes captured from dendritic cells treated with crude Leishmania antigen in vitro were representative of antigens processed and presented to naturally exposed individuals, we had peptides synthesized based on captured epitopes (Table S1) for use in whole blood assays (60, 61). The peptides were based on the 20-mer with the strongest predicted binding that overlapped the captured epitopes. Cytokine responses to the 12 peptides were studied in cured patients and endemic healthy controls. Analysis focused on comparing cytokine responses for peptides of differing binding affinities for risk versus protective alleles. Cured VL patients made robust but individually variable IFN-γ, and IL-10 responses to 20-mer peptides based on captured epitopes (Figure 7). Of interest, peptides based on DRB1*1501-captured epitopes resulted in a higher proportion (odds ratio 2.23; 95%CI 1.17-4.25; P=0.017) of patients with IFN-γ:IL10 ratios >2-fold compared to peptides based on DRB1*1301-captured epitopes (Figure 7D). TNF measured in a subset of cured patients (Figure 8A) again showed robust but individually variable responses across all peptides based on captured epitopes. Figure 8 (B to D) shows plots of cytokine responses to all 12 peptides across endemic healthy controls. Very few endemic healthy controls showed IL-10 or TNF responses to peptides or SLA. Individual IFN-γ responsiveness to peptides or SLA among these individuals can be accounted for by endemic exposure to the parasite, as asymptomatic infections based on modified quantiferon positivity (61) are common in the population (62).
FIGURE 7.
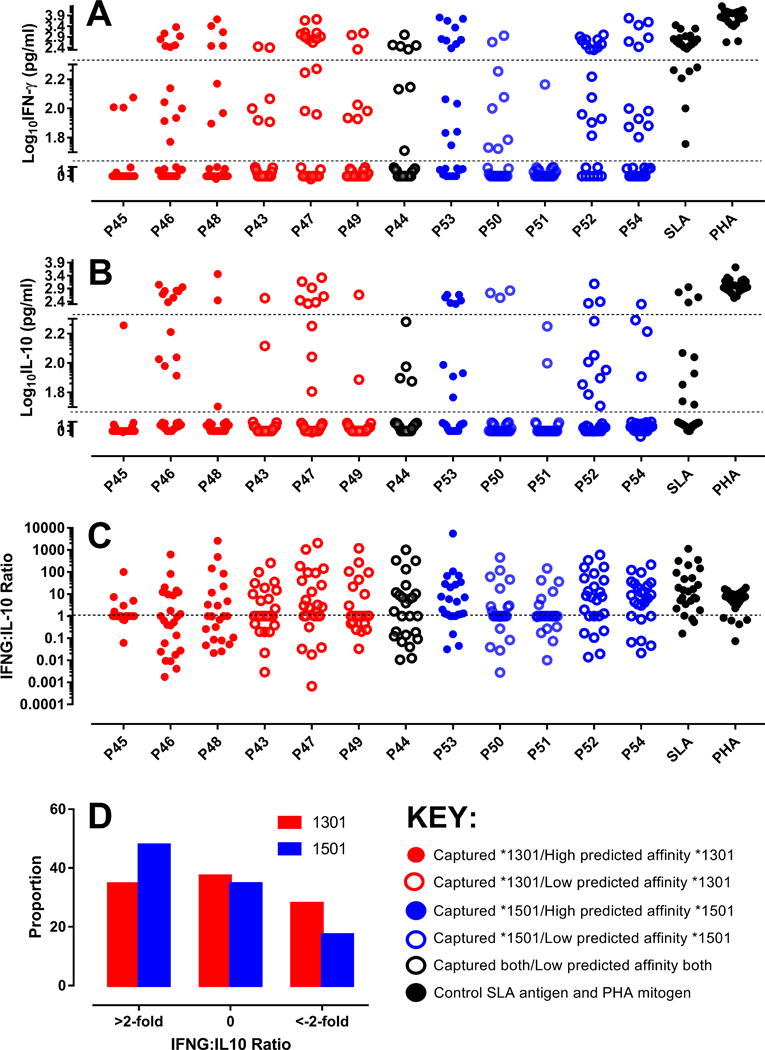
(A) IFN-γ and (B) IL-10 responses in cured patients. The dotted lines represent cut-offs for high-responders (>200 pg/ml), responders (50-200 pg/ml) and non-responders (<50 pg/ml). All values are after subtraction of the NIL antigen control. The y-axes are split to give visual emphasis to the cut-offs for responder status. They are numerically contiguous; all values appear on the graph. TNF responses measured in a subset of these individuals are presented in Figure 5. (C) provides IFN-γ:IL-10 ratios. (D) shows the proportions of individuals with IFN-γ:IL-10 ratios >2-fold, between −2-fold and +2-fold, and <−2-fold. The key indicates which DRB1 allele peptides were captured from, and what their predicted affinity of binding was to that DRB1 allele.
FIGURE 8.
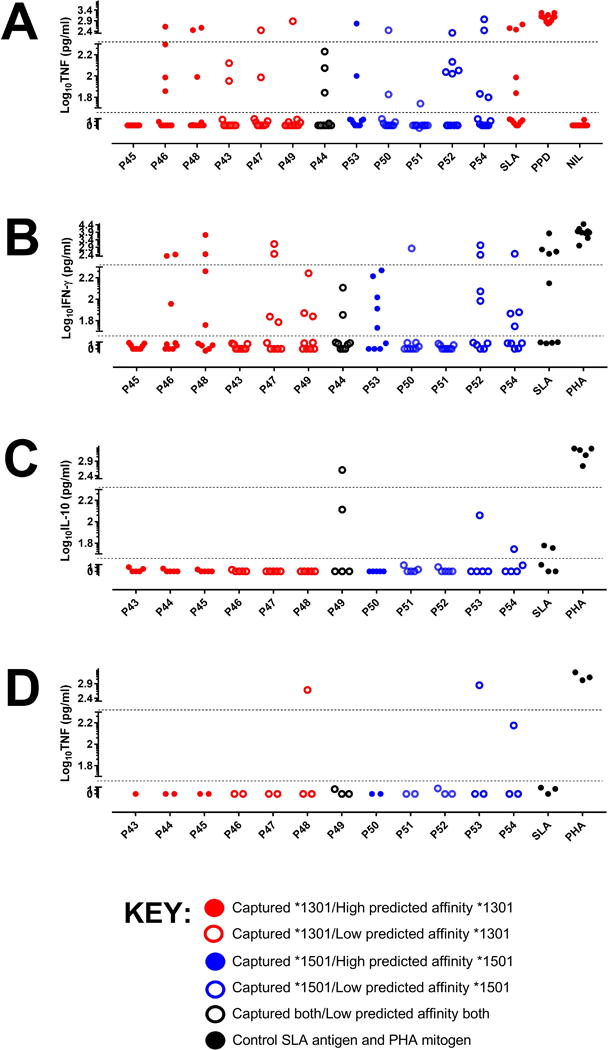
(A) TNF responses for a subset of cured cases for whom IFN-γ and IL-10 were measured (see Figure 4). (B), (C) and (D) are cytokine responses in endemic healthy control donors; (B) IFN-γ, (C) IL-10, and (D) TNF. The dotted lines represent cut-offs for high-responders (>200 pg/ml), responders (50-200 pg/ml) and non-responders (<50 pg/ml). All values are after subtraction of the NIL antigen control. The y-axes are split to give visual emphasis to the cut-offs for responder status. They are numerically contiguous; all values appear on the graph.
DISCUSSION
Previously we identified the class II gene region HLA-DRB1-DQA1 as the major locus regulating susceptibility and resistance to clinical disease for L. donovani in India and L. infantum chagasi in Brazil (17), indicating that HLA class II alleles provide genetic risk factors for VL that cross the epidemiological divides of geography and parasite species. Here we used HLA imputation to further refine genetic risk factors for VL in India at the level of 4-digit classical HLA-DRB1 alleles. We also demonstrate that the association maps to specific amino acids encoded in exon 2 that determine the interaction with amino acids at positions P4 and P6 of the 9-mer cores of foreign epitopes binding to the groove created by DRA1/DRB1 alpha/beta dimers. Based on these findings we used in silico analysis of both known Leishmania vaccine candidate antigens, as well as epitopes captured from dendritic cells of homozygous HLA DRB1 donors treated with crude Leishmania antigen, to demonstrate major differences in sequence motifs for 9-mer cores of Leishmania epitopes that bind preferentially to risk compared to those that bind preferentially to protective DRB1 alleles. Further, individuals drug-cured from VL showed strong recall T cell responses to peptides based on captured epitopes, indicating that these captured epitopes were representative of antigens processed and presented to antigen-specific CD4 T cells in naturally exposed individuals.
Immunological studies have long indicated the importance of CD4+ T cell mediated immunity in determining disease outcome in leishmaniasis, with polarized antigen-specific T helper 1 (Th1) and Th2/T regulatory responses affecting favourable versus adverse outcomes, respectively, in murine models (5) and human disease (6). The question of how the MHC affects the differentiation of Th1 compared to Th2/T regulatory populations in the context of a complex infection has been a long-term interest of immunologists (63-65). Early work demonstrating that peptides of highest affinity for a given MHC class II molecule elicited a shift towards the Th1 subset [e.g. (66)], led to theories of immuno-dominance of pathogen molecules and a role for affinity (i.e. the strength of a single bond) or avidity (i.e. the combined strength of multiple bond interactions) in dictating Th1 versus Th2 immunity (63). In support of this, specific HLA class II genotypes have been linked to Th1- or Th2-like responses against defined antigens in malaria and leprosy (64, 65). A growing body of studies in human and avian models of infection has explored how peptide promiscuity and sequence motif relates to functional avidity and disease susceptibility (67-71). Peptide-binding repertoires differ across MHC class I and II alleles (70, 71) with the structure of the binding groove influencing peptide-binding promiscuity and explaining resistance to viral infection (67). In humans, HLA-DRB1 variants linked with low viremia in HIV promiscuously present a larger breadth of peptides with lower functional avidity when compared to DRB1 variants linked with high viremia (71). Similarly, dominant and highly promiscuous epitopes characterize the CD4+ T helper cell response of spontaneously controlled Hepatitis C virus infection (69). We therefore hypothesised that sequence motifs of peptides predicted to bind preferentially to protective versus risk alleles could highlight important residues and help to explain differences in CD4+ T cell cytokine responses. When we tested this in 49 known vaccine candidates we observed differences in sequence motifs and greater promiscuity of peptide motifs for 9-mer core epitopes that were predicted to bind to the risk alleles DRB1*1404 compared to the strongest protective DRB1*1501 allele. This perhaps seems counter-intuitive relative to the earlier HIV (71) and hepatitis (69) studies. However, strong pro-inflammatory responses characteristic of low viremia in the latter cases may contribute to disease pathogenesis in clinical VL (72). Ultimately it is the balance between macrophage-activating Th1 cytokines IFN-γ/TNF and regulatory IL-10 that is important in determining a curative response, and is also a hallmark of vaccine-induced cure in pre-clinical models of disease (45). The critical question in our study was whether the same pattern of differences between sequence motifs for the 9-mer cores of epitopes binding to risk versus protective DRB1 alleles would emerge if naturally processed and presented antigens were identified and studied in endemic human disease cases. Our study of epitopes captured from dendritic cells indicated that this was, indeed, the case.
To reflect the functional role of cytokine responses in determining disease outcome we examined IFN-γ, TNF and IL-10 responses to peptides based on captured epitopes in drug-cured VL patients and endemic healthy controls. Robust but variable responses for all three cytokines were observed across all peptides, indicating that these captured epitopes were representative of antigens processed and presented during natural infections. Of note, there were no obvious differences in cytokine responses related to whether peptides were predicted to bind with strong or weak affinity to the risk versus protective DRB1 alleles from which epitopes were captured. However, as to be expected given the strong association between VL and HLA-DRB1 (17), only 3 of 26 cases studied were heterozygous for the single nucleotide polymorphic variant that tags the protective DRB1*1501 allele (data not shown). Hence, it was not possible to differentiate cured case donors as carrying risk versus protective alleles when studying immune responses to peptides based on captured antigens. Nevertheless, it was of interest that there was a bias towards higher IFN-γ:IL-10 ratios in cured patients in response to peptides based on antigens eluted from the dendritic cell donor homozygous for the protective DRB1*1501 allele. Further in-depth analysis of responses to these captured antigens in asympotomatic individuals identified as quantiferon-positive (61) in endemic areas will help to determine whether these specific antigens preferentially constitute useful candidates for future vaccines.
In summary, our study has refined the association between HLA Class II molecules and susceptibility to VL in this Indian population, and demonstrated clear differences in sequence motifs for 9-mer cores epitopes that bind to risk versus protective DRB1 alleles. These findings contribute to further understanding of the molecular interaction between DRB1 and CD4 T cells that determine the outcome of VL disease caused by L. donovani, providing novel naturally processed and presented candidate antigens that could contribute to future vaccine design.
Supplementary Material
Acknowledgments
We would like to thank the hospital staffs at Kala–azar Medical Research Centre, Muzaffarpur for their assistance in the collection of blood samples and all research scholars of Infectious Disease Research Laboratory, Banaras Hindu University for their kind help during the study.
This work was supported by the NIH as part of a Tropical Medicine Research Centre award.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
Statement about ethics
Ethical approvals for studies on Indian subjects were obtained from the ethical committee of the Institute of Medical Sciences, Banaras Hindu University (Varanasi, India). The study was carried out in accordance with the Declaration of Helsinki Principles, and each participant, or the parent/guardian of individuals <18 years old, signed informed consent forms to participate in the study and provide a blood sample.
Author contributions
Conceived and designed the experiments: TS MF NS JMB. Performed the experiments: TS JO NS APS AKrS. Analyzed the data: TS MF JMB. Supervised the clinical workup and experimental studies in patient donors and endemic healthy controls: JC. Wrote the paper: JMB with assistance from TS MF. Supervised the research in India: SS. Supervised the research in Australia and Cambridge: JMB. All authors read and approved the manuscript.
References
- 1.Cabello PH, Lima AM, Azevedo ES, Kriger H. Familial aggregation of Leishmnaia chagasi infection in northeastern Brazil. Am J Trop Med Hyg. 1995;52:364–365. doi: 10.4269/ajtmh.1995.52.364. [DOI] [PubMed] [Google Scholar]
- 2.Peacock CS, Collins A, Shaw MA, Silveira F, Costa J, Coste CH, Nascimento MD, Siddiqui R, Shaw JJ, Blackwell JM. Genetic epidemiology of visceral leishmaniasis in northeastern Brazil. Genet Epidemiol. 2001;20:383–396. doi: 10.1002/gepi.8. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell JM. Genetic susceptibility to leishmanial infections: studies in mice and man. Parasitology. 1996;112:S67–S74. [PubMed] [Google Scholar]
- 4.Blackwell J, Freeman J, Bradley D. Influence of H-2 complex on acquired resistance to Leishmania donovani infection in mice. Nature. 1980;283:72–74. doi: 10.1038/283072a0. [DOI] [PubMed] [Google Scholar]
- 5.Locksley RM, Scott P. Helper T-cell subsets in mouse leishmaniasis induction, expansion and effector function. Immunol Today. 1991;12:A58–A61. doi: 10.1016/S0167-5699(05)80017-9. [DOI] [PubMed] [Google Scholar]
- 6.Sundar S, Reed SG, Sharma S, Mehrotra A, Murray HW. Circulating T helper 1 (Th1) cell- and Th2 cell-associated cytokines in Indian patients with visceral leishmaniasis. Am J Trop Med Hyg. 1997;56:522–525. doi: 10.4269/ajtmh.1997.56.522. [DOI] [PubMed] [Google Scholar]
- 7.Karp CL, el-Safi SH, Wynn TA, Satti MM, Kordofani AM, Hashim FA, Hag-Ali M, Neva FA, Nutman TB, Sacks DL. In vivo cytokine profiles in patients with kala-azar. Marked elevation of both interleukin-10 and interferon-gamma. J Clin Invest. 1993;91:1644–1648. doi: 10.1172/JCI116372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenney RT, Sacks DL, Gam AA, Murray HW, Sundar S. Splenic cytokine responses in Indian kala-azar before and after treatment. J Infect Dis. 1998;177:815–818. doi: 10.1086/517817. [DOI] [PubMed] [Google Scholar]
- 9.Nylen S, Maurya R, Eidsmo L, Manandhar KD, Sundar S, Sacks D. Splenic accumulation of IL-10 mRNA in T cells distinct from CD4+CD25+ (Foxp3) regulatory T cells in human visceral leishmaniasis. J Exp Med. 2007;204:805–817. doi: 10.1084/jem.20061141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nylen S, Sacks D. Interleukin-10 and the pathogenesis of human visceral leishmaniasis. Trends Immunol. 2007;28:378–384. doi: 10.1016/j.it.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Carvalho EM, Barral A, Pedral-Sampaio D, Barral-Netto M, Badaro R, Rocha H, Johnson WD., Jr Immunologic markers of clinical evolution in children recently infected with Leishmania donovani chagasi. J Infect Dis. 1992;165:535–540. doi: 10.1093/infdis/165.3.535. [DOI] [PubMed] [Google Scholar]
- 12.Jamieson SE, Miller EN, Peacock CS, Fakiola M, Wilson ME, Bales-Holst A, Shaw MA, Silveira F, Shaw JJ, Jeronimo SM, Blackwell JM. Genome-wide scan for visceral leishmaniasis susceptibility genes in Brazil. Genes Immun. 2007;8:84–90. doi: 10.1038/sj.gene.6364357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeronimo SM, Duggal P, Ettinger NA, Nascimento ET, Monteiro GR, Cabral AP, Pontes NN, Lacerda HG, Queiroz PV, Gomes CE, Pearson RD, Blackwell JM, Beaty TH, Wilson ME. Genetic predisposition to self-curing infection with the protozoan Leishmania chagasi: a genomewide scan. J Infect Dis. 2007;196:1261–1269. doi: 10.1086/521682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller EN, Fadl M, Mohamed HS, El Zein A, Jamieson SE, Cordell HJ, Peacock CS, Fakiola M, Raju M, Khalil EA, El Hassan AM, Ibrahim ME, Blackwell JM. Y chromosome lineage- and village-specific genes on chromosomes 1p22 and 6q27 that control visceral leishmaniasis in The Sudan. PLoS Genet. 2007;3:679–688. doi: 10.1371/journal.pgen.0030071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bucheton B, Abel L, El-Safi S, Kheir MM, Pavek S, Lemainque A, Dessein AJ. A major susceptibility locus on chromosome 22q12 plays a critical role in the control of kala-azar. Am J Hum Genet. 2003;73:1052–1060. doi: 10.1086/379084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clinical microbiology reviews. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fakiola M, Strange A, Cordell HJ, Miller EN, Pirinen M, Su Z, Mishra A, Mehrotra S, Monteiro GR, Band G, Bellenguez C, Dronov S, Edkins S, Freeman C, Giannoulatou E, Gray E, Hunt SE, Lacerda HG, Langford C, Pearson R, Pontes NN, Rai M, Singh SP, Smith L, Sousa O, Vukcevic D, Bramon E, Brown MA, Casas JP, Corvin A, Duncanson A, Jankowski J, Markus HS, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Wilson ME, Deloukas P, Peltonen L, Christiansen F, Witt C, Jeronimo SM, Sundar S, Spencer CC, Blackwell JM, Donnelly P. Common variants in the HLA-DRB1-HLA-DQA1 HLA class II region are associated with susceptibility to visceral leishmaniasis. Nat Genet. 2013;45:208–213. doi: 10.1038/ng.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suri A, Lovitch SB, Unanue ER. The wide diversity and complexity of peptides bound to class II MHC molecules. Curr Opin Immunol. 2006;18:70–77. doi: 10.1016/j.coi.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Unanue ER. From antigen processing to peptide-MHC binding. Nat Immunol. 2006;7:1277–1279. doi: 10.1038/ni1206-1277. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen M, Justesen S, Lund O, Lundegaard C, Buus S. NetMHCIIpan-2.0 - Improved pan-specific HLA-DR predictions using a novel concurrent alignment and weight optimization training procedure. Immunome Res. 2010;6:9. doi: 10.1186/1745-7580-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, Raychaudhuri S, de Bakker PI. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rich SS, Concannon P, Erlich H, Julier C, Morahan G, Nerup J, Pociot F, Todd JA. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 23.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimitrov I, Garnev P, Flower DR, Doytchinova I. MHC Class II Binding Prediction-A Little Help from a Friend. J Biomed Biotechnol. 2010;2010:705821. doi: 10.1155/2010/705821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen LR, Kudahl UJ, Simon C, Sun J, Schonbach C, Reinherz EL, Zhang GL, Brusic V. BlockLogo: visualization of peptide and sequence motif conservation. J Immunol Methods. 2013;400-401:37–44. doi: 10.1016/j.jim.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rombach-Riegraf V, Karle AC, Wolf B, Sorde L, Koepke S, Gottlieb S, Krieg J, Djidja MC, Baban A, Spindeldreher S, Koulov AV, Kiessling A. Aggregation of human recombinant monoclonal antibodies influences the capacity of dendritic cells to stimulate adaptive T-cell responses in vitro. PLoS ONE. 2014;9:e86322. doi: 10.1371/journal.pone.0086322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stober C, Lange U, Roberts MTM, Alcami A, Blackwell JM. IL-10 from regulatory T cells determines vaccine efficacy in murine Leishmania major infection. J Immunol. 2005;175:2517–2514. doi: 10.4049/jimmunol.175.4.2517. [DOI] [PubMed] [Google Scholar]
- 32.Scott P, Pearce E, Natovitz P, Sher A. Vaccination against cutaneous leishmaniasis in a murine model. I. Induction of protective immunity with a soluble extract of promastigotes. J Immunol. 1987;139:221–227. [PubMed] [Google Scholar]
- 33.Bondinas GP, Moustakas AK, Papadopoulos GK. The spectrum of HLA-DQ and HLA-DR alleles, 2006: a listing correlating sequence and structure with function. Immunogenetics. 2007;59:539–553. doi: 10.1007/s00251-007-0224-8. [DOI] [PubMed] [Google Scholar]
- 34.Charest H, Matlashewski G. Developmental gene expression in Leishmania donovani: differential cloning and analysis of an amastigote-stage-specific gene. Mol Cell Biol. 1994;14:2975–2984. doi: 10.1128/mcb.14.5.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selzer PM, Pingel S, Hsieh I, Ugele B, Chan VJ, Engel JC, Bogyo M, Russell DG, Sakanari JA, McKerrow JH. Cysteine protease inhibitors as chemotherapy: lessons from a parasite target. Proc Natl Acad Sci U S A. 1999;96:11015–11022. doi: 10.1073/pnas.96.20.11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rafati S, Zahedifard F, Nazgouee F. Prime-boost vaccination using cysteine proteinases type I and II of Leishmania infantum confers protective immunity in murine visceral leishmaniasis. Vaccine. 2006;24:2169–2175. doi: 10.1016/j.vaccine.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 37.Khoshgoo N, Zahedifard F, Azizi H, Taslimi Y, Alonso MJ, Rafati S. Cysteine proteinase type III is protective against Leishmania infantum infection in BALB/c mice and highly antigenic in visceral leishmaniasis individuals. Vaccine. 2008;26:5822–5829. doi: 10.1016/j.vaccine.2008.08.065. [DOI] [PubMed] [Google Scholar]
- 38.Handman E, Symons FM, Baldwin TM, Curtis JM, Scheerlinck JP. Protective vaccination with promastigote surface antigen 2 from Leishmania major is mediated by a TH1 type of immune response. Infect Immun. 1995;63:4261–4267. doi: 10.1128/iai.63.11.4261-4267.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaur S, Kaur T, Joshi J. Immunogenicity and protective efficacy of DNA vaccine against visceral leishmaniasis in BALB/c mice. J Biomed Res. 2016;30:304–313. doi: 10.7555/JBR.30.20150125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stager S, Smith DF, Kaye PM. Immunization with a recombinant stage-regulated surface protein from Leishmania donovani induces protection against visceral leishmaniasis. J Immunol. 2000;165:7064–7071. doi: 10.4049/jimmunol.165.12.7064. [DOI] [PubMed] [Google Scholar]
- 41.Guha R, Gupta D, Rastogi R, Vikram R, Krishnamurthy G, Bimal S, Roy S, Mukhopadhyay A. Vaccination with leishmania hemoglobin receptor-encoding DNA protects against visceral leishmaniasis. Sci Transl Med. 2013;5:202ra121. doi: 10.1126/scitranslmed.3006406. [DOI] [PubMed] [Google Scholar]
- 42.Stober C, Lange U, Roberts MT, Gilbmartin B, Francis R, Almeida R, Peacock CS, McCann S, Blackwell JM. From genome to vaccines for leishmaniasis: Screening 100 novel vaccine candidates against murine Leishmania major infection. Vaccine. 2006;24:2602–2614. doi: 10.1016/j.vaccine.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Bhaumik S, Basu R, Sen S, Naskar K, Roy S. KMP-11 DNA immunization significantly protects against L. donovani infection but requires exogenous IL-12 as an adjuvant for comparable protection against L. major. Vaccine. 2009;27:1306–1316. doi: 10.1016/j.vaccine.2008.12.053. [DOI] [PubMed] [Google Scholar]
- 44.Ramirez L, Corvo L, Duarte MC, Chavez-Fumagalli MA, Valadares DG, Santos DM, de Oliveira CI, Escutia MR, Alonso C, Bonay P, Tavares CA, Coelho EA, Soto M. Cross-protective effect of a combined L5 plus L3 Leishmania major ribosomal protein based vaccine combined with a Th1 adjuvant in murine cutaneous and visceral leishmaniasis. Parasit Vectors. 2014;7:3. doi: 10.1186/1756-3305-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dondji B, Perez-Jimenez E, Goldsmith-Pestana K, Esteban M, McMahon-Pratt D. Prime-boost vaccination using the LACK antigen protects against murine visceral leishmaniasis. Infect Immun. 2005;73:5286–5289. doi: 10.1128/IAI.73.8.5286-5289.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coler RN, Goto Y, Bogatzki L, Raman V, Reed SG. Leish-111f, a recombinant polyprotein vaccine that protects against visceral leishmaniasis by the elicitation of CD4+ T cells. Infect Immun. 2007 doi: 10.1128/IAI.00394-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fragaki K, Suffia I, Ferrua B, Rousseau D, Le Fichoux Y, Kubar J. Immunisation with DNA encoding Leishmania infantum protein papLe22 decreases the frequency of parasitemic episodes in infected hamsters. Vaccine. 2001;19:1701–1709. doi: 10.1016/s0264-410x(00)00398-4. [DOI] [PubMed] [Google Scholar]
- 48.Skeiky YA, Coler RN, Brannon M, Stromberg E, Greeson K, Crane RT, Campos-Neto A, Reed SG. Protective efficacy of a tandemly linked, multi-subunit recombinant leishmanial vaccine (Leish-111f) formulated in MPL adjuvant. Vaccine. 2002;20:3292–3303. doi: 10.1016/s0264-410x(02)00302-x. [DOI] [PubMed] [Google Scholar]
- 49.Pirdel L, Zavaran Hosseini A. Immune response to recombinant Leishmania infantum lipophosphoglycan 3 plus CpG oligodeoxynucleotides in BALB/c mice. Parasite Immunol. 2017;39 doi: 10.1111/pim.12345. [DOI] [PubMed] [Google Scholar]
- 50.Aguilar-Be I, da Silva Zardo R, Paraguai de Souza E, Borja-Cabrera GP, Rosado-Vallado M, Mut-Martin M, Garcia-Miss Mdel R, Palatnik de Sousa CB, Dumonteil E. Cross-protective efficacy of a prophylactic Leishmania donovani DNA vaccine against visceral and cutaneous murine leishmaniasis. Infect Immun. 2005;73:812–819. doi: 10.1128/IAI.73.2.812-819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soto M, Corvo L, Garde E, Ramirez L, Iniesta V, Bonay P, Gomez-Nieto C, Gonzalez VM, Martin ME, Alonso C, Coelho EA, Barral A, Barral-Netto M, Iborra S. Coadministration of the Three Antigenic Leishmania infantum Poly (A) Binding Proteins as a DNA Vaccine Induces Protection against Leishmania major Infection in BALB/c Mice. PLoS Negl Trop Dis. 2015;9:e0003751. doi: 10.1371/journal.pntd.0003751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daifalla NS, Bayih AG, Gedamu L. Immunogenicity of Leishmania donovani iron superoxide dismutase B1 and peroxidoxin 4 in BALB/c mice: the contribution of Toll-like receptor agonists as adjuvant. Exp Parasitol. 2011;129:292–298. doi: 10.1016/j.exppara.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Iborra S, Soto M, Carrion J, Nieto A, Fernandez E, Alonso C, Requena JM. The Leishmania infantum acidic ribosomal protein P0 administered as a DNA vaccine confers protective immunity to Leishmania major infection in BALB/c mice. Infect Immun. 2003;71:6562–6572. doi: 10.1128/IAI.71.11.6562-6572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saravia NG, Hazbon MH, Osorio Y, Valderrama L, Walker J, Santrich C, Cortazar T, Lebowitz JH, Travi BL. Protective immunogenicity of the paraflagellar rod protein 2 of Leishmania mexicana. Vaccine. 2005;23:984–995. doi: 10.1016/j.vaccine.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 55.Ramirez L, Santos DM, Souza AP, Coelho EA, Barral A, Alonso C, Escutia MR, Bonay P, de Oliveira CI, Soto M. Evaluation of immune responses and analysis of the effect of vaccination of the Leishmania major recombinant ribosomal proteins L3 or L5 in two different murine models of cutaneous leishmaniasis. Vaccine. 2013;31:1312–1319. doi: 10.1016/j.vaccine.2012.12.071. [DOI] [PubMed] [Google Scholar]
- 56.Campos BL, Silva TN, Ribeiro SP, Carvalho KI, Kallas EG, Laurenti MD, Passero LF. Analysis of iron superoxide dismutase-encoding DNA vaccine on the evolution of the Leishmania amazonensis experimental infection. Parasite Immunol. 2015;37:407–416. doi: 10.1111/pim.12206. [DOI] [PubMed] [Google Scholar]
- 57.Kumar R, Goto Y, Gidwani K, Cowgill KD, Sundar S, Reed SG. Evaluation of ex vivo human immune response against candidate antigens for a visceral leishmaniasis vaccine. Am J Trop Med Hyg. 2010;82:808–813. doi: 10.4269/ajtmh.2010.09-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lakshmi BS, Wang R, Madhubala R. Leishmania genome analysis and high-throughput immunological screening identifies tuzin as a novel vaccine candidate against visceral leishmaniasis. Vaccine. 2014;32:3816–3822. doi: 10.1016/j.vaccine.2014.04.088. [DOI] [PubMed] [Google Scholar]
- 59.Coelho EA, Tavares CA, Carvalho FA, Chaves KF, Teixeira KN, Rodrigues RC, Charest H, Matlashewski G, Gazzinelli RT, Fernandes AP. Immune responses induced by the Leishmania (Leishmania) donovani A2 antigen, but not by the LACK antigen, are protective against experimental Leishmania (Leishmania) amazonensis infection. Infect Immun. 2003;71:3988–3994. doi: 10.1128/IAI.71.7.3988-3994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh OP, Stober CB, Singh AK, Blackwell JM, Sundar S. Cytokine responses to novel antigens in an Indian population living in an area endemic for visceral leishmaniasis. PLoS Negl Trop Dis. 2012;6:e1874. doi: 10.1371/journal.pntd.0001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh OP, Sundar S. Whole blood assay and visceral leishmaniasis: Challenges and promises. Immunobiology. 2014;219:323–328. doi: 10.1016/j.imbio.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh OP, Hasker E, Sacks D, Boelaert M, Sundar S. Asymptomatic Leishmania infection: a new challenge for Leishmania control. Clin Infect Dis. 2014;58:1424–1429. doi: 10.1093/cid/ciu102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray JS. How the MHC selects Th1/Th2 immunity. Immunol Today. 1998;19:157–163. doi: 10.1016/s0167-5699(97)01237-1. [DOI] [PubMed] [Google Scholar]
- 64.Stephens HA, Brown AE, Chandanayingyong D, Webster HK, Sirikong M, Longta P, Vangseratthana R, Gordon DM, Lekmak S, Rungruang E. The presence of the HLA class II allele DPB1*0501 in ethnic Thais correlates with an enhanced vaccine-induced antibody response to a malaria sporozoite antigen. Eur J Immunol. 1995;25:3142–3147. doi: 10.1002/eji.1830251123. [DOI] [PubMed] [Google Scholar]
- 65.Mutis T, Cornelisse YE, Datema G, van den Elsen PJ, Ottenhoff TH, de Vries RR. Definition of a human suppressor T-cell epitope. Proc Natl Acad Sci U S A. 1994;91:9456–9460. doi: 10.1073/pnas.91.20.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar V, Bhardwaj V, Soares L, Alexander J, Sette A, Sercarz E. Major histocompatibility complex binding affinity of an antigenic determinant is crucial for the differential secretion of interleukin 4/5 or interferon gamma by T cells. Proc Natl Acad Sci U S A. 1995;92:9510–9514. doi: 10.1073/pnas.92.21.9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koch M, Camp S, Collen T, Avila D, Salomonsen J, Wallny HJ, van Hateren A, Hunt L, Jacob JP, Johnston F, Marston DA, Shaw I, Dunbar PR, Cerundolo V, Jones EY, Kaufman J. Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity. 2007;27:885–899. doi: 10.1016/j.immuni.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 68.Ranasinghe S, Flanders M, Cutler S, Soghoian DZ, Ghebremichael M, Davis I, Lindqvist M, Pereyra F, Walker BD, Heckerman D, Streeck H. HIV-specific CD4 T cell responses to different viral proteins have discordant associations with viral load and clinical outcome. J Virol. 2012;86:277–283. doi: 10.1128/JVI.05577-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schulze zur Wiesch J, Lauer GM, Day CL, Kim AY, Ouchi K, Duncan JE, Wurcel AG, Timm J, Jones AM, Mothe B, Allen TM, McGovern B, Lewis-Ximenez L, Sidney J, Sette A, Chung RT, Walker BD. Broad repertoire of the CD4+ Th cell response in spontaneously controlled hepatitis C virus infection includes dominant and highly promiscuous epitopes. J Immunol. 2005;175:3603–3613. doi: 10.4049/jimmunol.175.6.3603. [DOI] [PubMed] [Google Scholar]
- 70.Paul S, Weiskopf D, Angelo MA, Sidney J, Peters B, Sette A. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J Immunol. 2013;191:5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ranasinghe S, Cutler S, Davis I, Lu R, Soghoian DZ, Qi Y, Sidney J, Kranias G, Flanders MD, Lindqvist M, Kuhl B, Alter G, Deeks SG, Walker BD, Gao X, Sette A, Carrington M, Streeck H. Association of HLA-DRB1-restricted CD4(+) T cell responses with HIV immune control. Nat Med. 2013;19:930–933. doi: 10.1038/nm.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barral-Netto M, Badaro R, Barral A, Almeida RP, Santos SB, Badaro F, Pedral-Sampaio D, Carvalho EM, Falcoff E, Falcoff R. Tumor necrosis factor (cachectin) in human visceral leishmaniasis. J Infect Dis. 1991;163:853–857. doi: 10.1093/infdis/163.4.853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


