Abstract
Campylobacter concisus is an oral bacterium that is associated with inflammatory bowel disease (IBD) including Crohn’s disease (CD) and ulcerative colitis (UC). C. concisus consists of two genomospecies (GS) and diverse strains. This study aimed to identify molecular markers to differentiate commensal and IBD-associated C. concisus strains. The genomes of 63 oral C. concisus strains isolated from patients with IBD and healthy controls were examined, of which 38 genomes were sequenced in this study. We identified a novel secreted enterotoxin B homologue, Csep1. The csep1 gene was found in 56% of GS2 C. concisus strains, presented in the plasmid pICON or the chromosome. A six-nucleotide insertion at the position 654–659 bp in csep1 (csep1-6bpi) was found. The presence of csep1-6bpi in oral C. concisus strains isolated from patients with active CD (47%, 7/15) was significantly higher than that in strains from healthy controls (0/29, P = 0.0002), and the prevalence of csep1-6bpi positive C. concisus strains was significantly higher in patients with active CD (67%, 4/6) as compared to healthy controls (0/23, P = 0.0006). Proteomics analysis detected the Csep1 protein. A csep1 gene hot spot in the chromosome of different C. concisus strains was found. The pICON plasmid was only found in GS2 strains isolated from the two relapsed CD patients with small bowel complications. This study reports a C. concisus molecular marker (csep1-6bpi) that is associated with active CD.
Introduction
Campylobacter concisus is a Gram-negative bacterium that is associated with inflammatory bowel disease (IBD), due to its significantly higher prevalence in the intestinal tissues of patients with IBD1–4. IBD is a chronic inflammatory condition of the gastrointestinal tract with Crohn’s disease (CD) and ulcerative colitis (UC) being the two major clinical forms5. In addition to IBD, C. concisus may also have a role in diarrhoeal disease as this bacterium was frequently isolated from the diarrhoeal stool samples6–9.
C. concisus is an oral bacterium, which is present in the oral cavity of nearly every individual including both patients with IBD and healthy controls10. Some individuals are colonised by multiple C. concisus strains in the oral cavity, which are more often seen in patients with active IBD11. There are no distinct oral or enteric C. concisus strain clusters and C. concisus strains in the intestinal tissues of patients with IBD were found to originate from oral C. concisus strains12. Some oral C. concisus strains were able to invade intestinal epithelial cells and induce epithelial production of IL-8, suggesting that translocation of these oral virulent C. concisus strains from the oral cavity into the intestinal tract may cause intestinal inflammation12–15.
C. concisus consists of two genomospecies (GS), which can be consistently divided based on the analysis of 23S rRNA gene, housekeeping genes and the core genome7,16–22. Both GS1 and GS2 contain diverse C. concisus strains23. Two C. concisus virulence factors have been characterised. Phospholipase A was shown to damage the membrane of mammalian cells; and prophage-encoded zonula occludens toxin (Zot) was found to cause prolonged damage to the intestinal epithelial barrier and enhance the responses of macrophages to other enteric bacterial species24–26. However, the prevalence of these virulence factors was not associated with IBD11. Currently, there are no available bacterial molecular markers that can differentiate commensal C. concisus strains from those that are associated with IBD; such markers were investigated in this study. Through genomic analysis, we identified a novel molecular marker in oral C. concisus strains that is associated with active CD.
Results
The genomes of 38 C. concisus strains sequenced in this study
The genomes of 63 oral C. concisus strains isolated from saliva samples of 19 patients with IBD (6 active CD, 6 active UC and 7 CD patients in remission) and 23 healthy controls were examined in this study (Table 1). Of the 63 genomes of oral C. concisus strains, 38 genomes were sequenced in this study and the remaining 25 genomes (5 GS1 and 20 GS2 strains) were obtained from National Center for Biotechnology Information (NCBI) database23.
Table 1.
C. concisus strains used in this study
| Strain id | Health status | Disease state | Age/sex | Montreal classification | Current treatment | GS | Csep1 P | Csep1 C | N50 | Genome size (bp) | No. of contigs | Coverage |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1CDO2 | CD | Active | 2/M | L2 and L4 | 2 | — | T | 80,888 | 1.99 | 36 | 67 | |
| P1CDO3 | 2 | — | Pa | 63,559 | 2.01 | 53 | 71 | |||||
| P2CDO3 | CD | Relapse, active | 19/M |
L3 and L4 Previous ileocolonic resection due to stricture |
2 | — | Pa | |||||
| P2CDO4 | 2 | Pa | Pa | 2.10 | 2 | 42 | ||||||
| P2CDO-S6 | 2 | — | Pa | |||||||||
| P20CDO-S1 | CD | Relapse, active | 22/M |
L1 Previous ileocolonic resection due to stricture |
2 | — | P | |||||
| P20CDO-S2 | 2 | Pa | — | |||||||||
| P20CDO-S3 | 2 | Pa | P | |||||||||
| P20CDO-S4 | 1 | — | — | |||||||||
| P21CDO-S1 | CD | Relapse, active | 21/M | L1 and L4 | 2 | — | — | |||||
| P21CDO-S2 | 2 | — | P | |||||||||
| P21CDO-S4 | 2 | — | T | |||||||||
| P10CDO-S1 | CD | New case, active | 19/M | L3 | 1 | — | — | 275,885 | 1.81 | 13 | 312 | |
| P10CDO-S2 | 1 | — | — | 428,977 | 1.92 | 12 | 298 | |||||
| P11CDO-S1b | CD | New case, active | 33/M | L3 | 2 | — | Pa | 136,390 | 2.02 | 27 | 295 | |
| P3UCO1 | UC | New case, active | 23/M | Extensive, S1 | 1 | — | — | |||||
| P7UCO-S2 | UC | New case, active | 65/M | Left sided, S2 | 2 | — | P | 226,478 | 1.97 | 26 | 327 | |
| P13UCO-S1 | UC | New case, active | 22/M | Extensive, S1 | 2 | — | P | 130,673 | 2.00 | 34 | 324 | |
| P13UCO-S3 | 2 | — | P | |||||||||
| P15UCO-S2 | UC | New case, active | 39/M | Extensive, S1 | 2 | — | — | |||||
| P16UCO-S1 | UC | New case, active | 67/M | Extensive, S1 | 2 | — | — | 181,794 | 1.92 | 24 | 89 | |
| P16UCO-S2 | 2 | — | P | 341,483 | 1.98 | 11 | 122 | |||||
| P26UCO-S1 | UC | New case, active | 34/M | Extensive, S1 | 2 | — | Pa | 122,849 | 2.08 | 41 | 290 | |
| P26UCO-S2 | 1 | — | — | 125,041 | 1.87 | 28 | 345 | |||||
| P6CDO1 | CD | Remission | 13/M | Mesalazine | 2 | — | T | 348,922 | 2.03 | 21 | 125 | |
| P18CDO-S1 | CD | Remission | 14/F | Azathioprine | 2 | — | P | 32,428 | 1.91 | 94 | 93 | |
| P19CDO-S1 | CD | Remission | 9/M | Mesalazine, azathioprine and iron supplements | 1 | — | — | 1,174,594 | 1.81 | 9 | 150 | |
| P24CDO-S2 | CD | Remission | 20/F | Azathioprine | 2 | — | NCS | |||||
| P24CDO-S3 | 2 | — | NCS | |||||||||
| P24CDO-S4 | 2 | — | NCS | |||||||||
| P25CDO-S3 | CD | Remission | 71/F | Azathioprine | 1 | — | — | 273,413 | 1.95 | 18 | 417 | |
| P27CDO-S1 | CD | Remission | 16/F | Azathioprine | 2 | — | P | 205,442 | 2.00 | 29 | 312 | |
| P27CDO-S2 | 1 | — | — | 201,601 | 1.83 | 18 | 257 | |||||
| P28CDO-S1 | CD | Remission | 17/M | Cotrimoxazole, tacrolimus, calcium and fish oil | 1 | — | — | 1,034,549 | 1.96 | 10 | 71 | |
| H1O1 | Healthy | 23/F | 1 | — | — | |||||||
| H3O1 | Healthy | 58/M | 2 | — | P | 211,732 | 1.94 | 22 | 191 | |||
| H7O-S1 | Healthy | 4/M | 2 | — | T | 346,163 | 1.94 | 17 | 127 | |||
| H9O-S1 | Healthy | 27/F | 2 | — | P | 203,101 | 2.09 | 19 | 154 | |||
| H9O-S2 | 2 | — | P | |||||||||
| H10O-S1 | Healthy | 18/M | 1 | — | — | 267,337 | 1.84 | 13 | 77 | |||
| H11O-S1 | Healthy | 21/F | 2 | — | T | 128,646 | 1.98 | 26 | 81 | |||
| H11O-S2 | 2 | — | P | 200,644 | 1.95 | 18 | 142 | |||||
| H12O-S1 | Healthy | 16/M | 1 | — | — | 687,684 | 1.92 | 11 | 211 | |||
| H14O-S1 | Healthy | 41/F | 2 | — | — | |||||||
| H15O-S1 | Healthy | 60/F | 1 | — | — | 194,730 | 1.84 | 14 | 574 | |||
| H16O-S1 | Healthy | 23/F | 2 | — | — | 262,245 | 1.96 | 19 | 123 | |||
| H17O-S1 | Healthy | 12/M | 1 | — | — | |||||||
| H19O-S1 | Healthy | 22/F | 2 | — | NCS | 197,598 | 2.01 | 31 | 342 | |||
| H20O-S1 | Healthy | 22/F | 2 | — | P | 131,516 | 2.00 | 26 | 174 | |||
| H21O-S1 | Healthy | 25/F | 2 | — | P | |||||||
| H21O-S2 | 2 | — | — | |||||||||
| H21O-S3 | 1 | — | — | |||||||||
| H21O-S5 | 2 | — | T | |||||||||
| H22O-S1 | Healthy | 65/M | 2 | — | NCS | |||||||
| H23O-S1 | Healthy | 62/F | 2 | — | T | |||||||
| H24O-S1 | Healthy | 23/F | 1 | — | — | 39,412 | 1.76 | 88 | 150 | |||
| H25O-S1 | Healthy | 9/F | 1 | — | — | 180,040 | 1.91 | 21 | 205 | |||
| H26O-S1 | Healthy | 16/M | 1 | — | — | 1,025,414 | 1.84 | 6 | 493 | |||
| H27O-S1 | Healthy | 5/M | 1 | — | — | 1,209,207 | 1.88 | 10 | 239 | |||
| H28O-S1 | Healthy | 67/F | 1 | — | — | 91,334 | 1.82 | 30 | 267 | |||
| H28O-S2 | 1 | — | — | 360,820 | 1.82 | 14 | 334 | |||||
| H29O-S1 | Healthy | 13/M | 2 | — | P | 64,541 | 2.00 | 47 | 253 | |||
| H30O-S1 | Healthy | 16/M | 1 | — | — | 96,450 | 1.82 | 31 | 315 |
The 63 C. concisus strains in Table 1 were examined for the presence of pICON plasmid, csep1P, csep1C and csep1C2 genes. The details of the draft genomes of 37 C. concisus strains and the complete genome of strain P2CDO4 sequenced in this study were listed. The remaining strains have had their draft genomes sequenced in our previous study23. Letters P and H in strain ID indicate strains isolated from patients with inflammatory bowel disease and healthy controls, respectively
Csep1P pICON plasmid-encoded csep1 gene, Csep1C chromosomally encoded csep1 gene, Csep1C2 second copy of chromosomally encoded csep1 gene, CD Crohn’s disease, UC ulcerative colitis, P present, T truncated protein, NCS non-coding sequence, — negative for pICON, csep1P, csep1C or csep1C2
aCsep1-6bpi
bThe csep1C2 gene was only found in strain P11CDO-S1
The sizes of the draft genome of the 37 C. concisus strains sequenced using MiSeq method ranged between 1.76 and 2.09 Mb and all draft genomes had more than 50 folds coverage (range 67 to 574). The complete genome of strain P2CDO4, which was sequenced using PacBio method, had a genome coverage of 42 with the genome size being 2.10 Mbp. The details of the C. concisus genomes sequenced in this study are summarised in Table 1.
The genomospecies of the 63 oral C. concisus strains
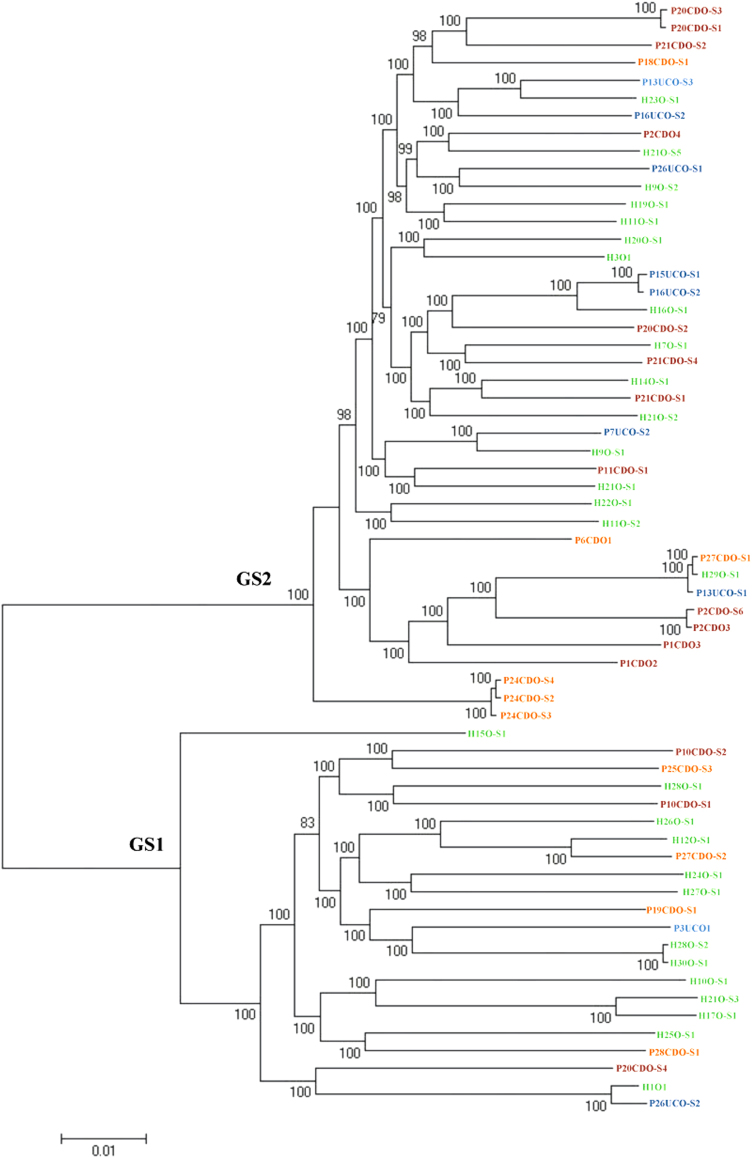
The 63 oral C. concisus strains examined in this study were consistently divided into GS1 and GS2 based on the core genome (Fig. 1) and the 23S rRNA gene (Supplementary Figure S1), of which 22 strains belonged to GS1 and 41 strains belonged to GS2 (Fig. 1). The core genome of these 63 strains contained 589 genes that contributed to 29% (589/2077) of the genes present in C. concisus strain P2CDO4. The core genomes of GS1 and GS2 C. concisus strains consisted of 1014 and 1109 genes, respectively.
Fig. 1. The phylogenetic tree generated based on C. concisus core genome.
The phylogenetic tree based on the core genome of 63 oral C. concisus strains (the genomes of 38 strains were sequenced in this study) was generated to show the GS1 and GS2 strains. The phylogenetic tree was generated using Roary. Strains from active CD, active UC, remission CD and healthy controls were coloured in red, blue, orange and green, respectively. Bootstrap values were generated from 1000 replicates. Bootstrap values of more than 70 were indicated. GS genomospecies
The 63 oral C. concisus strains included in this study are individual strains; the sequences of their core genome genes were not identical, confirming that they are individual strains (Fig. 1).
Identification of a novel plasmid pICON in oral C. concisus strains isolated from relapsed CD patients with previous ileocecal resection
By comparing the draft genomes of the 63 C. concisus strains, we found a highly similar genomic fragment in the draft genomes of strains P2CDO4 (contig 6), P20CDO-S2 (contig 8 and 9) and P20CDO-S3 (contig 9) (Supplementary Figure S2A), which were oral C. concisus strains isolated from the two relapsed CD patients with previous ileocecal resection due to small bowel stricture (Table 1). The complete genome of strain P2CDO4 sequenced using the PacBio method confirmed that this fragment was a plasmid (Fig. 2a; Supplementary Figure S2B). The origin of replication (ori) site was found at the nucleotide positions between 100,021 and 100,675 bp (655 bp), and contained three dnaA boxes including TTATACCCA, TTATATACA and TTATACAAA, and three AT-rich repeats (Fig. 2b). Furthermore, a plasmid-encoded replication initiation protein (CCS77_2118) was found at 110,672–111,694 bp (Fig. 2a). These molecular features were also present in the genomic fragment of strains P20CDO-S2 and P20CDO-S3. Collectively, using previously published criteria for defining a plasmid, these findings confirm that the genomic fragment found in strains P2CDO4, P20CDO-S2 and P20CDO-S3 is a plasmid27. We named this plasmid pICON.
Fig. 2. The pICON plasmid.
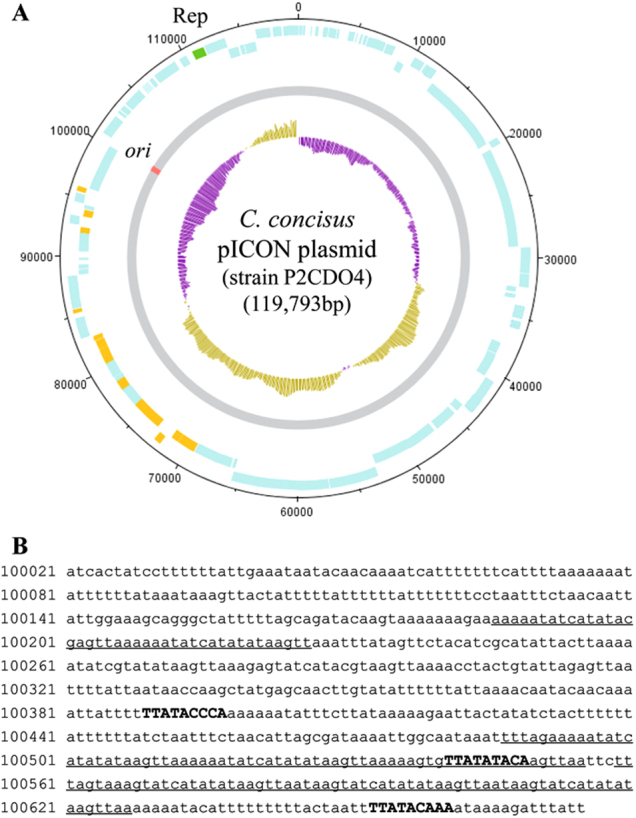
a Circularised diagram of the pICON plasmid in C. concisus strain P2CDO4. The outer black circle indicated genome base positions around the plasmid, and the outer blue circles represented CDSs on both forward and reverse strands. The plasmid replication initiation protein (Rep) was coloured in green. Predicted secreted proteins were coloured in orange. The grey circle represented the plasmid nucleotide sequence with predicted origin of replication (ori) site coloured in red. The inner circle was G+C content plot, G+C content below and above average were coloured in purple and brown, respectively. b Position and sequence of the ori site. Ori was predicted at the position with lowest G+C content. AT-rich regions containing sequence repeats were underlined. DnaA boxes were shown in upper case and bold
Comparison of the nucleotide sequences of the pICON plasmid with the known plasmids in NCBI bacterial genome database did not identify similar plasmids, showing that pICON is a novel plasmid.
Of the 63 oral C. concisus strains, only three strains, including P2CDO4, P20CDO-S2 and P20CDO-S3, carried the pICON plasmid, which was consistent in the genome search and PCR detection of pICON plasmid. All the three strains were GS2 C. concisus (Table 1). The prevalence of pICON plasmid in patients with active CD was significantly higher than that in healthy controls (2/6 vs. 0/23, P = 0.037).
The Csep1 protein
We compared the proteins encoded by the pICON plasmid in strains P2CDO4, P20CDO-S2 and P20CDO-S3 with known bacterial virulence proteins and found that the protein encoded by gene CCS77_2074 in the pICON plasmid was homologous to Staphylococcus aureus enterotoxin B (E = 0.04) and predicted to be secreted (Supplementary Table S1 and S2). We named it C. concisus-secreted protein 1 (Csep1). We found another Csep1 protein encoded by gene CCS77_0139 in the chromosome of C. concisus strain P2CDO4, which had 85% amino acids identical to the Csep1 protein encoded by gene CCS77_2074 in the pICON plasmid. We used Csep1P and Csep1C to differentiate the pICON plasmid-encoded and chromosomally encoded Csep1 proteins.
Except for Csep1P, all proteins encoded by the pICON plasmid had an amino acid identity of <40% as compared to proteins encoded by the chromosome of strain P2CDO4, showing that the csep1 gene in the chromosome was not due to the integration of pICON plasmid into the chromosome.
The csep1 gene in different oral C. concisus strains and their prevalence in patients with IBD and controls
The csep1 gene in different C. concisus strains was identified by genome search and then confirmed using various PCR methods, which showed consistent results.
The csep1P gene was found in the three C. concisus strains containing pICON plasmid (P2CDO4, P20CDO-S2 and P20CDO-S3). The csep1C gene was found in 22 C. concisus strains, all contained one copy of the csep1C gene except for strain P11CDO-S1, which contained two copies of the csep1C gene (csep1C and csep1C2). Strains P2CDO4 and P20CDO-S3 contained both csep1P and csep1C, strain P20CDO-S2 had csep1P but no csep1C.
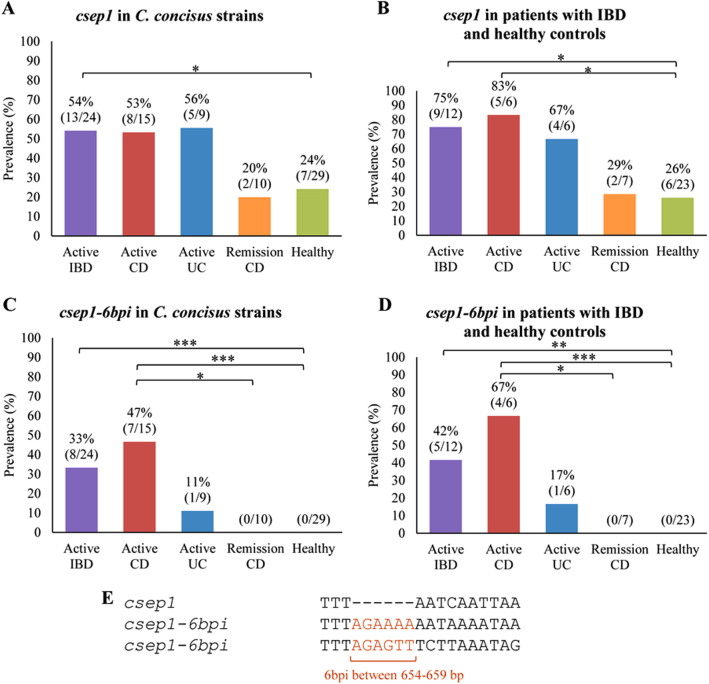
The csep1 gene (either csep1P or csep1C) was found in GS2 C. concisus strains (56%, 23/41) and in none of the GS1 strains. More than half of the oral C. concisus strains isolated from patients with IBD (54%, 13/24) contained the csep1 gene, which was significantly higher than that in the oral C. concisus strains isolated from healthy controls (24%, 7/29, P = 0.045) (Fig. 3a). The prevalence of csep1-positive C. concisus strain was significantly higher in patients with active CD (83%, 5/6) as compared to healthy controls (26%, 6/23, P = 0.019) (Fig. 3b).
Fig. 3. The prevalence of csep1 and csep1-6bpi genes in patients with IBD and healthy controls.
a The prevalence of the csep1 gene in C. concisus strains isolated from active IBD was significantly higher than that from healthy controls (P = 0.045). b The prevalence of csep1-positive C. concisus strains from active CD was significantly higher than that from healthy controls (P = 0.019). c A six bp insertion (6bpi) at position 654–659 bp was mainly found in the csep1 genes (csep1-6bpi) from C. concisus strains isolated from active CD, not from remission CD and healthy controls. (P = 0.02 and P = 0.0002, respectively). d The prevalence of csep1-6bpi positive C. concisus strains in patients with active CD was significantly higher than that in remission CD and health controls (P = 0.021 and P = 0.0006, respectively). e Majority of the csep1-6bpi contained AGAAAA between 654 and 659 bp, while only one contained AGAGTT. *Indicates statistical significance (*P < 0.05, **P < 0.01 and ***P < 0.001). CD Crohn’s disease, UC ulcerative colitis
A six bp insertion in the csep1 gene is strongly associated with active CD
The sequences of the csep1 gene in different C. concisus strains were compared (Supplementary Figure S3). The csep1 gene in different C. concisus strains had sizes ranging between 651 and 672 bp, encoding proteins of 216–223 amino acids. All Csep1 proteins were predicted to be secreted proteins, containing a signal peptide (Supplementary Figure S4). In addition, 12 strains had truncated csep1C or non-coding csep1C genes. The truncated csep1C genes had stop codons at various positions within the gene and the non-coding csep1C genes were gene fragments without a start codon or very short gene fragments. The truncated and non-coding csep1C genes and their flanking genes were in the same contig, their presence therefore was not due to assembly. The truncated and non-coding csep1C genes were also confirmed by the PCR method targeting the flanking sequences.
Nucleotide insertions were found at six positions in the csep1 gene in different C. concisus strains (Supplementary Figure S3). The six bp insertion at the nucleotide 654–659 bp of the csep1 gene (csep1-6bpi) was found in seven oral C. concisus strains isolated from patients with active CD, one strain from a patient with active UC and none of the strains from CD patients in remission and healthy controls (Supplementary Figure S3). The presence of csep1-6bpi gene in oral C. concisus strains isolated from patients with active CD (47%, 7/15) was significantly higher than that in oral strains from healthy controls (0/29, P = 0.0002) and patients with CD in remission (0/10, P = 0.02) (Fig. 3c). The prevalence of csep1-6bpi positive C. concisus strains was significantly higher in patients with active CD (67%, 4/6) as compared to healthy controls (0/23, P = 0.0006) and CD patients in remission (0/7, P = 0.021) (Fig. 3d). When comparing the prevalence of csep1-6bpi positive C. concisus strain in patients and healthy controls, if an individual is colonised by multiple csep1-6bpi positive strains, the positivity was counted only once.
Of the eight strains that had the csep1-6bpi, the six bp insertion sequences were AGAAAA in seven strains and AGAGTT in one strain (Fig. 3e). Both Csep1P and Csep1C from strain P2CDO4 contained AGAAAA.
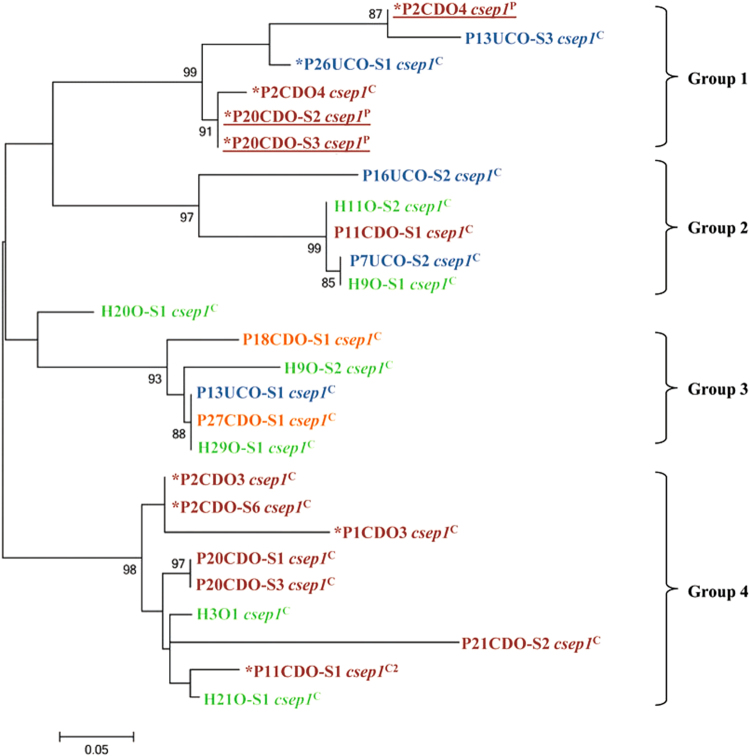
Phylogenetic analysis of the csep1 gene in different oral C. concisus strains
The phylogenetic tree generated based on the csep1 gene in different oral C. concisus strains formed four groups (groups 1–4, Fig. 4). The csep1P and csep1C genes did not form distinct groups. Five of the csep1-6bpi genes (AGAAAA) were in group 1 and the remaining four csep1-6bpi genes (three AGAAAA and one AGAGTT) were in group 4. The phylogenetic clustering of the csep1 genes was not consistent with the phylogenetic grouping of the C. concisus strains based on their core genomes.
Fig. 4. The phylogenetic tree generated based on csep1 genes in C. concisus strains.
Maximum likelihood method was used to generate the phylogenetic tree. Bootstrap values were generated from 1000 replicates. Bootstrap values of more than 70 were indicated. The csep1 genes from C. concisus strains isolated from patients with active CD, active UC, remission CD and healthy controls were coloured in red, blue, orange and green, respectively. The csep1P genes were underlined. *The csep1-6bpi gene
The csep1 gene insertion sites in the C. concisus genome
To understand the insertion patterns of csep1P, csep1C and csep1C2 in the C. concisus genome, the upstream and downstream flanking genes were compared.
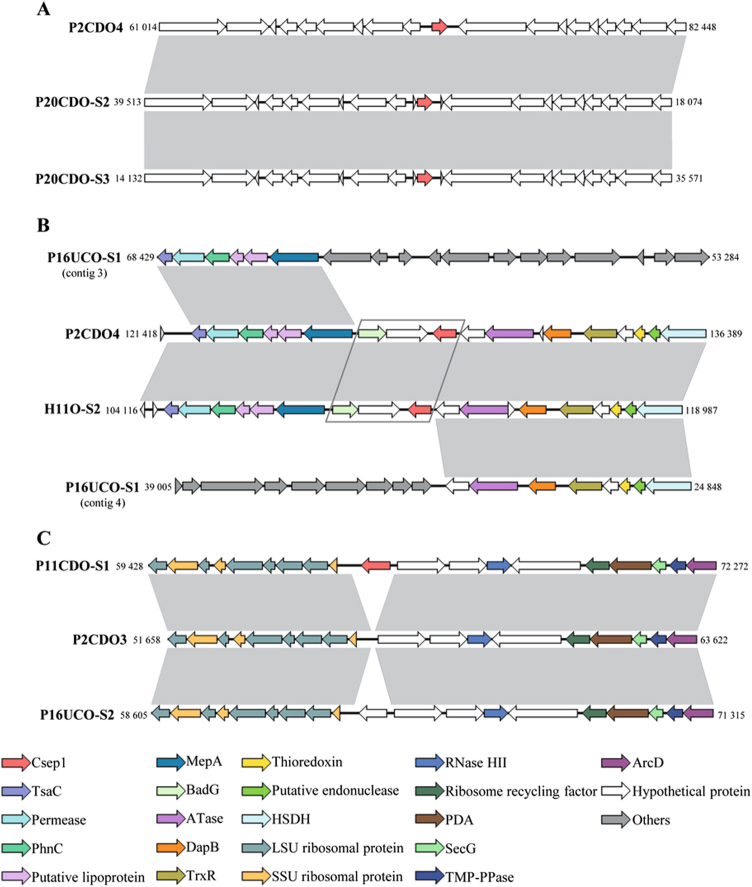
The csep1P was located between 72,094 and 72,762 bp in the pICON plasmid of strain P2CDO4. All three pICON plasmids identified in this study contained the csep1P gene and the flanking genes were almost identical (with more than 80% of nucleotide identity), showing that the csep1P gene was inserted at the same location in the pICON plasmid in different C. concisus strains (Fig. 5a).
Fig. 5. Insertion sites of csep1 genes in C. concisus genomes.
a The flanking genes of the csep1P gene in the pICON plasmid of strain P2CDO4, P20CDO-S2 and P20CDO-S3 were almost identical, indicating the csep1P was inserted at the same location in the pICON plasmid of different strains. b The flanking genes of the csep1C gene in all the csep1C-positive strains shared similar patterns, strains P2CDO4 and H11O-S2 are shown here as examples. The two genes (boxed) immediately upstream of the csep1C gene were absent in all csep1P-negative strains, while other flanking genes were present, although distantly located, strain P16UCO-S1 is shown here as an example. c The csep1C2 gene was inserted at a rare spot within the chromosome, in which only appeared in strain P11CDO-S1. In the remaining strains with similar flanking gene arrangements, there was either no gene insertion, such as strain P2CDO3; or insertion of other genes such as strain P16UCO-S2. Nucleotide sequences sharing more than 80% identity were shaded in grey. Nucleotide positions referred to the positions within the contig, except strain P2CDO4 which had the genome sequenced without gap. Gene sizes in strains between a, b and c were not on scale. HSDH homoserine dehydrogenase, PDA polysaccharide deacetylase
The csep1C in the chromosome was located between 128,877 and 129,542 bp in strain P2CDO4 (Fig. 5b). Of the 23 copies of the csep1 gene in the chromosome carried by 22 C. concisus strains, 22 copies of the csep1C gene were in the same position, demonstrated by their flanking genes which were almost identical (strains P2CDO4 and H11O-S2 were used as examples to show the location of the csep1C gene). Most of the flanking genes encode for bacterial enzymes. The flanking genes were also present in the csep1C-negative strains, but were distantly located (strain P16UCO-S1 was used to show the distantly located flanking genes in Fig. 5b), indicating that gene rearrangement has occurred. Furthermore, the two genes immediately upstream of the csep1C gene were absent in all csep1C negative strains, these two genes encode for a hypothetical protein and benzoyl-CoA reductase subunit BadG (boxed in Fig. 5b).
P11CDO-S1 is the only strain that carried a second copy of the csep1 in the chromosome (csep1C2) and most of the flanking genes encoded ribosomal proteins and bacterial enzymes. Among the remaining 62 csep1C2-negative strains, 33 strains had other genes inserted at the same position encoding for putative type-IIS restriction/modification enzyme or hypothetical proteins; 26 strains had no insertion; two strains had the flanking genes located distantly; and one strain had contigs ended at the insertion site, thus information regarding gene insertion was unavailable. Strains P11CD-S1, P2CDO3 and P16UCO-S2 were used to show the insertion site of csep1C2 and the flanking genes (Fig. 5c).
Detection of Csep1 protein expression in C. concisus culture supernatant
Csep1 proteins, including both Csep1P and Csep1C, were predicted to be secreted proteins (Supplementary Table S2; Supplementary Figure S4). Using mass spectrometry analysis, both Csep1P and Csep1C were detected in the bacterial culture supernatant of C. concisus strain P2CDO4. Unique peptides containing amino acids specific to Csep1P or Csep1C were detected (Csep1P: LIEINTRPISTDNAK and NDIDNKTIK; Csep1C: NIPAIDLIK; specific amino acids were underlined), common peptides shared between Csep1P and Csep1C were also detected (MLEYGCNELK and TIPEYCYDKK).
The prevalence of pICON plasmid and csep1 in other C. concisus strains in the public databases
There are genomes of 125 other C. concisus strains available from the public databases including 42 GS1 and 83 GS2 C. concisus strains. Most of these strains were enteric strains isolated from the stool samples and intestinal biopsies of patients with diarrhoea, CD, UC or healthy individuals. There were 16 oral strains isolated from 5 patients with UC, 3 patients with CD, 1 patient with gingivitis and 4 healthy individuals23,28–31. It was not clear whether patients with IBD had active disease and whether they were receiving IBD treatment at the time of C. concisus strain isolation, these isolates are therefore not suitable for analysing the prevalence of csep1-6bpi, as we had previously shown that the drugs such as azathioprine and mercaptopurine used for IBD treatment could inhibit the growth of C. concisus32.
We examined the presence csep1 gene and pICON plasmid in these strains. We found that the five oral strains isolated from four healthy individuals were all negative for the csep1-6bpi gene and the pICON plasmid. There were only two strains had genomic fragments similar to the pICON plasmid, and these two strains were isolated by Kirk et al.31 from a patient with UC. However, the contigs of the oral strain from this patient were really short, and the contigs did not cover the full length of the csep1 gene (Supplementary Figure S5A). The enteric strain had longer contigs (Supplementary Figure S5B), and this strain contained the csep1-6bpi gene with the flanking genes similar to csep1P. Overall, these data suggest that these two strains have the pICON plasmid. Interestingly, these strains are GS1 strains, suggesting that pICON plasmid can be transmitted between GS1 and GS2 C. concisus strains. However, the genomes sequenced by Kirk et al. were not complete genomes without gaps; therefore, we cannot carry further analysis of the pICON plasmid in their strains.
Discussion
In this study, we analysed the genomes of 63 oral C. concisus strains isolated from patients with IBD and controls and the genomes of 38 C. concisus strains were sequenced in this study. We identified a novel bacterial biomarker that is associated with active CD, and this marker was confirmed by PCR methods.
We identified the C. concisus Csep1 protein, which is homologous to enterotoxin B encoded by S. aureus. Staphylococcal enterotoxin B has multiple pathogenic effects such as inducing diarrhoea and acting as a human superantigen that non-specifically activates T cells to produce a large amount of proinflammatory cytokines33. Further analysis found nucleotide insertions in the csep1 gene in different C. concisus strains and the csep1-6bpi insertion at the position 654–659 bp was only found in oral C. concisus strains isolated from patients with active IBD particularly in CD. The prevalence of csep1-6bpi positive C. concisus strains in patients with active CD was significantly higher than that in the healthy controls (P = 0.0006). Future studies are needed to assess the effects of C. concisus Csep1 protein encoded by the csep1-6bpi gene on human gastrointestinal epithelial cells and the mucosal immune system, which will provide information regarding whether this protein has a role in the development or pathogenesis of CD. The csep1 gene was located in the chromosome (csep1C) or the pICON plasmid (csep1p). The csep1C in the majority of the C. concisus strains were at the position 128,877–129,542 bp (nucleotide position in strain P2CDO4), showing that this is a csep hot spot. One strain (P11CDO-S1) had a second copy of the csep1 gene (csep1C2), which was identified at the location between 1,819,244 and 1,820,490 bp (nucleotide position in strain P2CDO4). The Csep1 was predicted to be a secreted protein, containing a signal peptide (Supplementary Table S2). Proteomics analysis indeed detected Csep1 proteins encoded by the csep1 gene in both pICON plasmid and the chromosome from the culture supernatant of C. concisus strain P2CDO4.
Phylogenetic analysis of the csep1 gene from different C. concisus strains identified four groups. Csep1P and csep1C did not form distinct groups, showing that they were from the same ancestor. We also compared the flanking genes of the csep1 gene in both pICON and the chromosome (Fig. 5). The flanking genes of csep1P in the pICON plasmid in different strains were nearly identical, suggesting the csep1P was transmitted by the plasmid between the strains. The csep1C appeared not stable, in 12 GS2 C. concisus strains, truncated or non-coded csep1C gene was found, implying that the csep1C genes in these C. concisus strains have undergone mutations (Table 1).
A novel and rare C. concisus plasmid, the pICON plasmid, is reported for the first time in this study. Of 63 oral C. concisus strains examined in this study, only 3 GS2 strains isolated from 2 relapsed CD patients contained the pICON plasmid. These 2 patients were not related, and their saliva samples were collected from different hospitals. Interestingly, both patients had previous ileocecal resection due to small bowel restriction within 2 years of their diagnosis of CD, suggesting that CD patients colonised by pICON plasmid-positive GS2 C. concisus strains may be more likely to develop complications, which should be further investigated.
C. concisus consists of two GS23. In comparison with the GS1 strains, GS2 strains are better adapted to the human gastrointestinal tract. More GS2 strains were isolated from the saliva samples of patients with IBD as compared to healthy controls and previous studies showed that GS2 C. concisus strains were more invasive to human intestinal epithelial cell lines as compared to GS1 strains34. Each C. concisus GS contained diverse strains, as shown by the number of genes in the GS core genome. We found that csep1 were present in the chromosome of 56% of oral GS2 C. concisus strains, showing that it is possible to further divide GS2 strains into CD-associated strains and the other strains based on this gene.
The csep1-6bpi-positive C. concisus strains were not detected in the seven CD patients in remission. These patients were receiving IBD treatment at the time of sample collection. We previously showed that immunosuppressive drugs used to treat IBD such as azathioprine and mercaptopurine inhibited the growth of C. concisus strains under laboratory conditions32. It is possible that IBD treatment drugs have inhibited the growth of csep1-6bpi-positive C. concisus strains in these patients.
In conclusion, we report an active CD-associated C. concisus molecular marker (csep1-6bpi), which is present in the bacterial chromosome and the novel pICON plasmid. The pathogenic role of the protein encoded by the csep1-6bpi gene requires further investigation.
Materials and methods
Oral C. concisus strains used in this study
C. concisus strains sequenced in this study were isolated in our previous studies, under the ethics approval granted by the Ethics Committees of the University of New South Wales and the South East Sydney Area Health Service, Australia (HREC 09237/SESIAHS 09/078 and HREC08335/SESIAHS (CHN)07/48)3,10–12. Patients and healthy controls were recruited from Sydney, Australia. For the saliva sample from each patient or healthy individual, 12 putative C. concisus isolates were collected. The putative C. concisus isolates were subjected to a C. concisus-specific PCR to confirm the identity of C. concisus and then subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) for whole-cell protein profile analysis to define the strains. Isolates with identical SDS-PAGE pattern were defined as the same strain. Some individuals were colonised by multiple oral C. concisus strains and these strains have been named accordingly11. The details of each C. concisus strains are listed in Table 1.
C. concisus culture and bacterial DNA extraction
C. concisus strains were grown on horse blood agar (HBA) plates under anaerobic conditions supplemented with 5% H2 as described previously35. Bacterial DNA used for genome sequencing through the MiSeq method was extracted using Gentra Puregene Yeast/Bacteria Kit (Qiagen, Australia) according to manufacturer’s instructions. Bacterial DNA used for genome sequencing through the PacBio method was extracted with phenol-chloroform, followed by purification with Agencourt AMPure XP beads (A63881, Beckman Coulter, UK)36. The quality of DNA was determined using Nanodrop and Qubit Fluorometer.
Genome sequencing, assembly and annotation
The genomes of 37 C. concisus strains were sequenced using the MiSeq method at the University of Western Australia, WA, Australia. Bacterial genomic libraries were prepared according to Nextera XT protocol (Ver. May 2012). Libraries were prepared using Nextera XT V2 on MiSeq Personal Sequencer (Illumina Inc., San Diego, CA, USA) running version MiSeq Control Software 1.1.1 to obtain 250 bp paired-end reads. Reagent contamination was controlled by barcoding all DNA samples and primers. The quality of reads was assessed by the Phred quality score and the reads mapping fold coverage was determined with qualimap_v2.2.137. The raw reads were assembled as described previously23. Contigs <1000 bp and with coverage <10× were removed. Gene annotation was performed by the Rapid Annotations software at Subsystems Technology server (RAST, Ver. 2.0)38.
The draft genome of C. concisus strain P2CDO4 has been sequenced in our previous study using the MiSeq method. To confirm the identity of a novel genomic fragment, the DNA extracted from this strain was re-sequenced in this study using the PacBio method to obtain the complete genome. Large insert libraries (20 kb) were constructed and sequenced using the PacBio RS II platform (Ramaciotti Centre for Genomics, University of New South Wales, Australia). The PacBio reads were assembled into contigs using CANU v 1.339. The assembly was rearranged using Circlator to produce accurate linear representations of circular sequences40. The assembly was then subjected to polishing using Quiver, followed by polishing with Illumina reads obtained from our previous study using Pilon23,41,42.
We ensured that all genomes sequenced using the MiSeq or PacBio methods had fold coverage of at least 50× or 20× respectively, which were shown to be adequate for genome characterisation39,43.
Determination of C. concisus genomospecies
The genomospecies status of the 37 C. concisus strains sequenced using the MiSeq method in this study was determined by phylogenetic analysis of the core genome and the 23S rRNA gene. The 23S rRNA gene phylogenetic tree was generated using the maximum likelihood method implemented in MEGA644. The core genome phylogenetic tree was generated by Roary45.
Plasmid identification
By comparing the draft genomes of 63 C. concisus strains generated using the MiSeq method, we found a genomic fragment that was only present in the draft genomes of strains P2CDO4, P20CDO-S2 and P20CDO-S3, which were oral strains isolated from the two relapsed CD patients with previous ileocecal resection due to small bowel stricture (Table 1). The fragment from these three strains were aligned using Mauve46. We re-sequenced the genome of strain P2CDO4 using PacBio method, which generated two contigs with the large contig being the chromosome, and the small contig that corresponds to the genome fragment being the plasmid. The plasmid was also consistently identified by plasmidSPAdes47. Plasmid identification was performed using bioinformatics tools according to previously described criteria27. The criteria defining a plasmid include the presence of ori site containing AT-rich repetitive sequences, dnaA box sequences and plasmid-encoded replication initiation protein27. The ori and the dnaA box sequences were predicted using Ori-Finder, and AT-rich repetitive sequences were identified using Tandem Repeats Finder48,49. DNAPlotter was used to visualise the plasmid genome50.
To examine whether the plasmid identified in C. concisus strains P2CDO4, P20CDO-S2 and P20CDO-S3 shared similarities with known plasmids, the nucleotide sequence of the identified plasmid was compared with the bacterial genomes (Taxonomy ID for bacteria: 2) available in NCBI genome database using BLASTn.
These approaches led to the identification of a novel C. concisus plasmid, the pICON plasmid (see results section).
Detection of the pICON plasmid in C. concisus strains isolated from patients with IBD and controls
The presence of the pICON plasmid in the 63 oral C. concisus strains isolated from patients with IBD and healthy controls was firstly examined by genome search using BLASTn51, then confirmed using two PCR methods targeting genes CCS77_2029 and CCS77_2093, which are genes exclusively present in the pICON plasmids not the chromosomes. The primers and thermocycling conditions were listed in Supplementary Figure S6A. The prevalence rates of pICON plasmid in C. concisus strains isolated from patients with IBD and controls were compared.
Prediction of secreted proteins and identification of putative virulence factors in C. concisus pICON plasmid
Secreted proteins were predicted using SignalP version 4.0, which identifies signal peptides in queried proteins52.
Virulence Factors Database (VFDB) was used for identification of putative virulence factors in the pICON plasmid53. The plasmid proteins were queried against the virulence factors in the VFDB core dataset using BLASTp with a cut-off E-value of 0.0554.
Detection and comparison of csep1 genes and Csep1 proteins in C. concisus strains isolated from patients with IBD and controls
The presence of the csep1 gene in the 63 oral C. concisus strains isolated from patients with IBD and controls was first examined by genome search using BLASTn. The sequences of the csep1 gene and Csep1 protein in different C. concisus strains were compared using Muscle55.
The presence of csep1 gene in the 63 oral C. concisus strains was then confirmed using PCR methods. PCR primers targeting the conserved regions upstream and downstream of csep1P (Pfla_F and Pfla_R), csep1C (Cfla_F and Cfla_R) and csep1C2 (C2fla_F and C2fla_R) were designed using Primer-BLAST (Supplementary Figure S6B)56. Strains that were negative for csep1 genes in the above PCR reactions were subjected to an additional PCR detection targeting the conserved regions within the csep1 genes (csep1_F and csep1_R), which amplifies all three copies of the csep1 genes (Supplementary Figure S6C). All positive PCR products were sequenced from both ends using BigDye v 3.1 reagents (Applied Biosystems, Foster City, CA) and analysed on an ABI Capillary DNA Sequencer ABI3730 (Applied Biosystems) at Ramaciotti Centre for Genomics.
As mentioned above, the csep1 gene was found at three different positions within the genome: one in the pICON plasmid and two in the chromosome. To investigate whether specific genomic structures are associated with the insertion site of the csep1 gene, the flanking genes of csep1-negative and positive C. concisus strains were compared using BLASTn and visualised using EasyFig51,57.
Phylogenetic analysis of the csep1 genes in different C. concisus strains
The phylogenetic tree of the 26 csep1 genes from the 63 oral C. concisus strains was generated using the maximum likelihood method implemented in MEGA644.
Detection of the expressed Csep1 proteins
C. concisus P2CDO4, which contains the csep1 gene in both the plasmid pICON and the chromosome, was used to examine the Csep1 protein expression. The strain was cultured on HBA plates for 48 h. Following cultivation, bacteria were collected from the plates and resuspended in 20 ml of heart infusion broth (HIB) (OxoidTM, Australia) to a final OD600 of 0.1, and further incubated for 24 h with rotation at 200 rpm35.
Following incubation in HIB, both C. concisus bacteria and supernatant were collected by centrifugation. The whole-cell lysates were prepared by three freeze-thaw cycles of the bacterial cells. The protein concentrations were determined using a BCA assay kit (Thermo Fisher Scientific, USA), and 20 µg of proteins were loaded onto SDS-polyacrylamide gel and separated by electrophoresis. The culture supernatant from bacteria cultured using HIB was filtered through a 0.22 μm MILLEX GP filter (Merck Millipore Ltd, Ireland) to remove any remaining bacteria. Supernatant was concentrated using Amicon® Ultra 3 K columns (Merck Millipore Ltd, Ireland), which was then loaded onto SDS-polyacrylamide gel and separated by electrophoresis. Protein bands were excised from Coomassie Blue stained polyacrylamide gels and digested with trypsin. Digested peptides were separated by liquid chromatography and analysed using a LTQ-FT Ultra mass spectrometer (Thermo Electron, Bremen, Germany) as previously described12. All MS/MS spectra were searched against the NCBI database using MASCOT (version 2.5.1) and then Scaffold Q+ (v.4.7.3, Proteome software, OR, US) was used to validate peptide and protein identities against the proteins encoded on the pICON plasmid of C. concisus strain P2CDO458. Mass spectrometry was conducted at the Bioanalytical Mass Spectrometry Facility, University of New South Wales, Australia.
Genbank sequence submission
The annotated complete genome of C. concisus strain P2CDO4 including its pICON plasmid and chromosome was submitted to Genbank genome assembly database (Biosample ID: SAMN07160232; Bioproject ID: PRJNA388128; accession number: CP021642 and CP021643 for chromosome and pICON plasmid respectively). The assembled genomes of the remaining 37 C. concisus strains sequenced using the MiSeq method were submitted to Genbank under the Bioproject ID PRJNA388128.
The presence of csep1 gene in the genome of other C. concisus strains
Currently, there are a further 125 C. concisus strains’ genomes available in public databases. We examined the presence of pICON and csep1 gene in these strains by genome search and comparison of the flanking genes.
Statistical analysis
Fisher’s exact test (two-tailed) was used to compare the prevalence of pICON plasmid, csep1 gene in C. concisus strains isolated from patients with IBD and healthy controls. Statistical analyses were performed using GraphPad Prism 6 software (San Diego, CA). P values <0.05 were considered as statistically significant.
Electronic supplementary material
Acknowledgements
This work is supported by Faculty Research Grant awarded to L.Z. from the University of New South Wales (Grant No: PS35329).
Authors’ contributions
F.L. conducted bacterial cultivation, DNA extraction and PCR, and had a major role in performing bioinformatics analysis. R.M. and F.L. performed proteomics analysis. S.O. participated in genome assembly. M.C.G., S.M.R., R.W.L. and S.C. provided important feedback on clinical aspect. R.L., S.O., M.M.T., C.Y.A.T. and H.K.L.C. provided important feedback on bioinformatics analysis and data presentation. L.Z. and F.L. conceived the project. F.L. and L.Z. had a major role in writing the manuscript. All authors have read the manuscript and provided feedback. All authors have approved the final version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0065-6).
References
- 1.Zhang L, et al. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn’s disease. J. Clin. Microbiol. 2009;47:453–455. doi: 10.1128/JCM.01949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukhopadhya I, et al. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS ONE. 2011;6:e21490. doi: 10.1371/journal.pone.0021490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahendran V, et al. Prevalence of Campylobacter species in adult Crohn’s disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS ONE. 2011;6:e25417. doi: 10.1371/journal.pone.0025417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirk KF, et al. Optimized cultivation of Campylobacter concisus from gut mucosal biopsies in inflammatory bowel disease. Gut Pathog. 2016;8:27. doi: 10.1186/s13099-016-0111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartor RB, Mazmanian SK. Intestinal microbes in inflammatory bowel diseases. Am. J. Gastroenterol. 2012;Suppl 1:15–21. doi: 10.1038/ajgsup.2012.4. [DOI] [Google Scholar]
- 6.Lindblom G, et al. Campylobacter upsaliensis, C. sputorum sputorum and C. concisus as common causes of diarrhoea in Swedish children. Scand. J. Infect. Dis. 1995;27:187–188. doi: 10.3109/00365549509019006. [DOI] [PubMed] [Google Scholar]
- 7.Kalischuk L, Inglis G. Comparative genotypic and pathogenic examination of Campylobacter concisus isolates from diarrheic and non-diarrheic humans. Bmc. Microbiol. 2011;11:53. doi: 10.1186/1471-2180-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen, H. et al. High incidence of Campylobacter concisus in gastroenteritis in North Jutland, Denmark: a population-based study. Clin. Microbiol. Infect.19, 445–450 (2013). [DOI] [PubMed]
- 9.Lastovica AJ, Roux E. Efficient isolation of Campylobacteria from stools. J. Clin. Microbiol. 2000;38:2798–2800. doi: 10.1128/jcm.38.7.2798-2799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, et al. Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J. Clin. Microbiol. 2010;48:2965–2967. doi: 10.1128/JCM.02391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahendran V, et al. The prevalence and polymorphisms of zonula occluden toxin gene in multiple Campylobacter concisus strains isolated from saliva of patients with inflammatory bowel disease and controls. PLoS ONE. 2013;8:e75525. doi: 10.1371/journal.pone.0075525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ismail Y, et al. Investigation of the enteric pathogenic potential of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease. PLoS ONE. 2012;7:e38217. doi: 10.1371/journal.pone.0038217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, et al. Campylobacter concisus and inflammatory bowel disease. World J. Gastroenterol. 2014;20:1259–1267. doi: 10.3748/wjg.v20.i5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L. Oral Campylobacter species: Initiators of a subgroup of inflammatory bowel disease? World J. Gastroenterol. 2015;21:9239–9244. doi: 10.3748/wjg.v21.i31.9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ismail Y, et al. The effects of oral and enteric Campylobacter concisus strains on expression of TLR4, MD-2, TLR2, TLR5 and COX-2 in HT-29 cells. PLoS ONE. 2013;8:e56888. doi: 10.1371/journal.pone.0056888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aabenhus R, et al. Delineation of Campylobacter concisus genomospecies by amplified fragment length polymorphism analysis and correlation of results with clinical data. J. Clin. Microbiol. 2005;43:5091–5096. doi: 10.1128/JCM.43.10.5091-5096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller WG, et al. Multilocus sequence typing methods for the emerging Campylobacter Species C. hyointestinalis, C. lanienae, C. sputorum, C. concisus, and C. curvus. Front. Cell Infect. Microbiol. 2012;2:45. doi: 10.3389/fcimb.2012.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahendran V, et al. Delineation of genetic relatedness and population structure of oral and enteric Campylobacter concisus strains by analysis of housekeeping genes. Microbiology. 2015;161:1600–1612. doi: 10.1099/mic.0.000112. [DOI] [PubMed] [Google Scholar]
- 19.Engberg J, et al. Campylobacter concisus: an evaluation of certain phenotypic and genotypic characteristics. Clin. Microbiol. Infect. 2005;11:288–295. doi: 10.1111/j.1469-0691.2005.01111.x. [DOI] [PubMed] [Google Scholar]
- 20.Istivan T. Molecular Characterisation of Campylobacter concisus: A Potential Etiological Agent of Gastroenteritis in Children (School of Applied Sciences, RMIT University, Melbourne, 2005).
- 21.On S, et al. Characterisation of Campylobacter concisus strains from South Africa using amplified fragment length polymorphism (AFLP) profiling and a genomospecies-specific polymerase chain reaction (PCR) assay: identification of novel genomospecies and correlation with clinical data. Afr. J. Microbiol. Res. 2013;7:1845–1851. doi: 10.5897/AJMR12.2182. [DOI] [Google Scholar]
- 22.Nielsen HL, Nielsen H, Torpdahl M. Multilocus sequence typing of Campylobacter concisus from Danish diarrheic patients. Gut Pathog. 2016;8:44. doi: 10.1186/s13099-016-0126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung HK, et al. Genome analysis of Campylobacter concisus strains from patients with inflammatory bowel disease and gastroenteritis provides new insights into pathogenicity. Sci. Rep. 2016;6:38442. doi: 10.1038/srep38442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Istivan TS, et al. Characterization of a haemolytic phospholipase A(2) activity in clinical isolates of Campylobacter concisus. J. Med. Microbiol. 2004;53:483–493. doi: 10.1099/jmm.0.45554-0. [DOI] [PubMed] [Google Scholar]
- 25.Mahendran V, et al. Examination of the effects of Campylobacter concisus zonula occludens toxin on intestinal epithelial cells and macrophages. Gut Pathog. 2016;8:18. doi: 10.1186/s13099-016-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, et al. Zonula occludens toxins and their prophages in Campylobacter species. Gut Pathog. 2016;8:43. doi: 10.1186/s13099-016-0125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Solar G, et al. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshpande NP, et al. Comparative genomics of Campylobacter concisus isolates reveals genetic diversity and provides insights into disease association. BMC Genom. 2013;14:585. doi: 10.1186/1471-2164-14-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huq M, et al. The ribosomal RNA operon (rrn) of Campylobacter concisus supports molecular typing to genomospecies level. Gene Rep. 2017;6:8–14. doi: 10.1016/j.genrep.2016.10.008. [DOI] [Google Scholar]
- 30.Cornelius AJ, et al. Complete genome sequence of Campylobacter concisus ATCC 33237T and draft genome sequences for an additional eight well-characterized C. concisus strains. Genome Announc. 2017;5:e00711–e00717. doi: 10.1128/genomeA.00711-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirk KF, et al. Molecular epidemiology and comparative genomics of Campylobacter concisus strains from saliva, faeces and gut mucosal biopsies in inflammatory bowel disease. Sci. Rep. 2018;8:1902. doi: 10.1038/s41598-018-20135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu F, et al. Azathioprine, mercaptopurine, and 5-aminosalicylic acid affect the growth of IBD-associated Campylobacter species and other enteric microbes. Front. Microbiol. 2017;8:527. doi: 10.3389/fmicb.2017.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fries, B. C. & Varshney, A. K. Bacterial toxins—Staphylococcal enterotoxin B. Microbiol. Spectr.1, AID-0002-2012 (2013). [DOI] [PMC free article] [PubMed]
- 34.Wang Y, et al. Campylobacter concisus genomospecies 2 is better adapted to the human gastrointestinal tract as compared with Campylobacter concisus genomospecies 1. Front. Physiol. 2017;8:543. doi: 10.3389/fphys.2017.00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H, et al. Examination of the anaerobic growth of Campylobacter concisus strains. Int. J. Microbiol. 2014;2014:476047. doi: 10.1155/2014/476047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol.Chapter 2, Unit 2.4 (2001). [DOI] [PubMed]
- 37.Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2015;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koren S, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunt M, et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 43.Desai A, et al. Identification of optimum sequencing depth especially for de novo genome assembly of small genomes using next generation sequencing data. PLoS ONE. 2013;8:e60204. doi: 10.1371/journal.pone.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darling AC, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antipov, D. et al. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics32, 3380–3387 (2016). [DOI] [PubMed]
- 48.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao F, Zhang CT. Ori-Finder: a web-based system for finding oriC s in unannotated bacterial genomes. BMC Bioinformatics. 2008;9:79. doi: 10.1186/1471-2105-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carver T, et al. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen TN, et al. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 53.Chen L, et al. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weston J, et al. Semi-supervised protein classification using cluster kernels. Bioinformatics. 2005;21:3241–3247. doi: 10.1093/bioinformatics/bti497. [DOI] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye J, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS‐based proteomic studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.