Abstract
A small cysteine-rich protein with antimicrobial activity was isolated from pokeweed (Phytolacca americana) seeds and purified to homogeneity. The protein inhibits the growth of several filamentous fungi and gram-positive bacteria. The protein was highly basic, with a pI higher than 10. The entire amino acid sequence of the protein was determined to be homologous to antimicrobial protein (AMP) from Mirabilis jalapa. The cDNA encoding the P. americana AMP (Pa-AMP-1) and chromosomal DNA containing the gene were cloned and sequenced. The deduced amino acid sequence shows the presence of a signal peptide at the amino terminus, suggesting that the protein is synthesized as a preprotein and secreted outside the cells. The chromosomal gene shows the presence of an intron located within the region encoding the signal peptide. Southern hybridization showed that there was small gene family encoding Pa-AMP. Immunoblotting showed that Pa-AMP-1 was only present in seeds, and was absent in roots, leaves, and stems. The Pa-AMP-1 protein was secreted into the environment of the seeds during germination, and may create an inhibitory zone against soil-borne microorganisms. The disulfide bridges of Pa-AMP-1 were identified. The three-dimensional modeling of Pa-AMP-1 indicates that the protein has a small cystine-knot folding, a positive patch, and a hydrophobic patch.
During evolution, plants have developed a variety of defense systems to protect themselves from potential pathogens. Plants often produce small Mr chemicals inhibitory to microbial growth. These chemicals are either induced as a result of activation of a group of genes encoding the enzymes of the synthetic pathway upon pathogen infection, such as phytoalexins (Smith, 1994), or are constitutive, such as saponins (Osborn, 1996). In recent years, it has been realized that proteins also play important roles in plant defense systems. Proteins including thionins (Bohlmann and Apel, 1991), plant defensins (Broekaert et al., 1995; Epple et al., 1997), and chitinases (Schumbaum et al., 1986) have been shown to play active roles against pathogen infections.
Seed germination is likely to be one of the most vulnerable periods for pathogen attack in a plant's life cycle because the rupture of the seed coat could allow invasion of pathogens into the seed storage tissues. Plants have developed defense systems such as defensins to prevent pathogen infection at this stage in their life cycle. The plant defensins are small, Cys-rich antifungal proteins present in many plants (Broekaert et al., 1995). Other proteins have also been shown to play an important role in plant defense systems. Cammue et al. (1992) isolated two small proteins with strong antimicrobial activity from Mirabilis jalapa seeds. These two proteins contained six Cys residues, all of which are involved in disulfide bond formation for stabilizing protein tertiary structure. The genes encoding these two proteins have been cloned and sequenced (De Bolle et al., 1995).
In searching for antifungal proteins from various sources, we found that pokeweed (Phytolacca americana) seeds contained strong activity against growth of some soil-borne fungal pathogens. We report the isolation and characterization of an active antimicrobial protein (AMP) from pokeweed seeds and molecular cloning of the gene encoding the protein.
MATERIALS AND METHODS
Purification of AMP from Pokeweed Seeds
Protein Purification
Pokeweed (Phytolacca americana) plants were grown in the greenhouse of Peking University. For isolation and purification of the AMP from pokeweed seeds (Pa-AMP-1), 20 g of seeds were collected. The seeds were ground into powder and suspended in approximately 20 mL of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (25 mm, pH 8.0) containing 1 mm NaCl. The solution was incubated at 25°C for about 1 h before it was filtered to remove undissolved materials. The filtrate was incubated at 90°C for 10 min, followed by a centrifugation at 15,000 rpm for 10 min. The supernatant was loaded onto a Sephadex-SP (Sigma, St Louis) column (2.5 × 30 cm). The column was washed with 5 bed volumes of buffer A (25 mm HEPES, pH 8.0, 10 mm NaCl, and 10 mm KCl) and eluted with a NaCl gradient from 10 to 500 mm in buffer A. The flow rate was 25 mL h−1, and the elution was monitored at 280 nm. The fractions with antimicrobial activities were pooled and concentrated by lyophilization. The active fractions were then loaded onto a Sephadex G-50 gel filtration column (2 × 45 cm) and eluted with buffer A at a flow rate of 15 mL h−1. The fractions with antimicrobial activity were pooled and concentrated by ultrafiltration with a 3-kD cutoff membrane (Amicon, Beverly, MA).
HPLC Analysis
A micro-HPLC system (model 173, Perkin Elmer-Applied Biosystems, Foster City, CA) was used for HPLC analysis of the purified protein. Solution A contains 0.1% (v/v) trifluoroacetic acid (TFA) and solution B contained 0.1% (v/v) TFA in 100% (v/v) acetonitrile. The column (0.5 mm × 15 cm) was equilibrated with 2% (v/v) solution B in solution A before sample injection. An elution gradient (2%–45% B from 0–75 min, then maintained at 45%) was employed to elute the protein. The elution was monitored at 210 nm.
Characterization of the AMP
Protein Sequencing
The N-terminal sequence of the purified protein was determined using an automatic protein sequencer (ABI491, Perkin Elmer-Applied Biosystems). The protein was first dissolved in 4 m guanidine and reduced with 10 mm diothiothreitol (DTT). The Cys residues were modified by incubating with 2 m acrylamide at 37°C for 1 h at pH 8.3. The derivatized protein was adsorbed to a polyvinylidene difluoride (PVDF) membrane using the Prosorb kit (Perkin Elmer-Applied Biosystems). Five microliters of Biobrene (Perkin Elmer-Applied Biosystems) was added to the membrane before it was subjected to sequencing.
Determination of the Disulfide Bridges of Pa-AMP-1
The purified Pa-AMP-1 at a concentration of 2 mg mL−1 was first cleaved chemically with N-bromosuccinimide (NBS). The protein solution was diluted 10-fold with (NH4)2CO3 buffer (100 mm), followed by tryptic digestion with the modified trypsin (Promega, Beijing) according to the instructions of the supplier. The digested products were analyzed by micro-HPLC. The C18 column (0.5 mm × 15 cm) was first equilibrated with 2% (v/v) buffer B before the sample was injected. The peptides were eluted with an isocratic gradient. The elutants were blotted directly on a PVDF membrane strip with the on-line blotter and sequenced to determine their N-terminal sequences. The results are summarized in Table II. The method by Zhang and Liang (1993) was used to determine which residues would produce dehydro-Ala (Δ-Ser). The monitoring wavelength of the protein sequencer detector was adjusted to 313 nm, where phenylhydantoin (PTH)-Δ-Ser has a strong absorption.
Table II.
N-Terminal sequences obtained from the peaks separated with micro-HPLC shown in Fig. 6
| Peak No. | Sequence |
|---|---|
| 4 | NGGR (6–9) |
| 8 | AGCI (1–5) |
| CCSS (19–22) | |
| GVCK (33–36) | |
| 10, 11 | CNASAG (10–15) |
| CFQIAG (24–29) | |
| 12 | NGGRCNASA (6–14) |
| CFQIAGQS (24–31) | |
| 13 | AGCIK (1–5) |
| 14 | AGCIK (1–5) |
| CCSS (19–22) |
Electrophoresis
SDS-electrophoretic analysis of the Pa-AMP-1 was performed using N-[2-hydroxy-1,1-Bis(hydroxymethyl)-ethyl]glycine (Tricine)-SDS gel according to the method of Schagger and von Jagow (1987). The gel was stained using a silver-staining kit (Bio-Rad, Hercules, CA). To determine the pI of the Pa-AMP-1, isoelectric focusing gel electrophoresis was performed according to the method of Zhou et al. (1998). The gel was stained with Coomassie Brilliant Blue.
Immunological Detection of Pa-AMP-1
To obtain polyclonal antibodies against Pa-AMP-1, a green fluorescence protein (GFP)-PA-AMP-1 fusion protein was first overproduced in Escherichia coli (C. Xu and J. Zhao, unpublished data). The rabbit polyclonal antibodies were raised according to the method of Harlow and Lane (1988). The antibodies were specific to Pa-AMP-1 and GFP. Immunoblotting after Tricine SDS-PAGE was performed using horseradish peroxidase-conjugated goat anti-rabbit IgG antibodies as secondary antibodies, as described in Zhou et al. (1998). To detect Pa-AMP-1 secretion from the pokeweed seeds during germination, seeds were first incubated in water for 12 h at 28°C, and then transferred onto a PVDF membrane, which was wet first before being placed on top of four layers of Whatman filter paper in a Petri dish. The seeds were incubated for various times at 28°C before removal. The Pa-AMP-1 released from each seed onto the PVDF membrane was detected using the antibodies against Pa-AMP-1 as described above.
Molecular Cloning of the Gene Encoding the AMP of Pokeweed Seeds
Genomic DNA from pokeweed leaves was isolated according to the method of Dellaporta et al. (1983). Total RNA from pokeweed seeds was isolated as described by De Vries et al. (1988). mRNA was isolated using the mRNA isolation kit from Promega according to the manufacturer's instructions. To isolate cDNA encoding the AMPs, reverse transcription PCR (RT-PCR) was performed. The mRNAs isolated from the seeds were first reverse-transcribed with reverse transcriptase using poly(T) oligonucleotides as the primer. The cDNAs encoding the mature AMP were amplified by PCR using Taq enzyme (Promega). The primers for amplification were 5′-CNGGNTG(C/T) AT(A/T/C) AA(A/G) AA(T/C) GG-3′, where N represents any nucleotide, and poly(T) primer. The PCR conditions were: 92°C for 1 min, 48°C for 1 min, and 72°C for 1.5 min, for 35 cycles. The amplified fragments were cloned into a T-vector from Promega and sequenced with an automatic DNA sequencer (model 477, Perkin Elmer-Applied Biosystems). To obtain the full-length cDNA encoding the AMP, the isolated mRNAs were subjected to a 5′RACE according to the method of Frohman et al. (1988). The primers used in the PCR were: (a) 5′-TTTTGCAAACACCATAGGAT- TGTCC-3′, which was designed based on the sequence encoding the mature protein; and (b) poly(T) oligonucleotides. The PCR conditions were the same as described above. The amplified fragment was cloned and sequenced as described above.
To amplify chromosomal DNA encoding the AMP, 2 primers were designed based on the sequence of the cDNA obtained above. The 5′ primer was 5′-ACGTTATCAATC- TCCGCCTTACC-3′ and the 3′ primer was CTTATTC- ATCATGATAGGGCC-3′. The amplification with PCR was performed in the presence of Pfu DNA polymerase as described by Cheng et al. (1994). A 1.0-kb fragment was obtained and cloned in pUC18. It was sequenced as described above.
Southern Hybridization
Genomic DNA was digested with EcoRI and XbaI followed by agarose (0.8%, w/v) electrophoresis. The separated DNA was transferred to a nitrocellulose membrane, and the membrane was baked at 80°C for 2 h under vacuum. Radioactive probe was first prepared with a random primer extension kit (Promega) with 32P-dATP using the cDNA encoding Pa-AMP-1 as a template. The hybridization was performed according to the method of Zhao et al. (1993).
Three-Dimensional Modeling of Pa-AMP-1
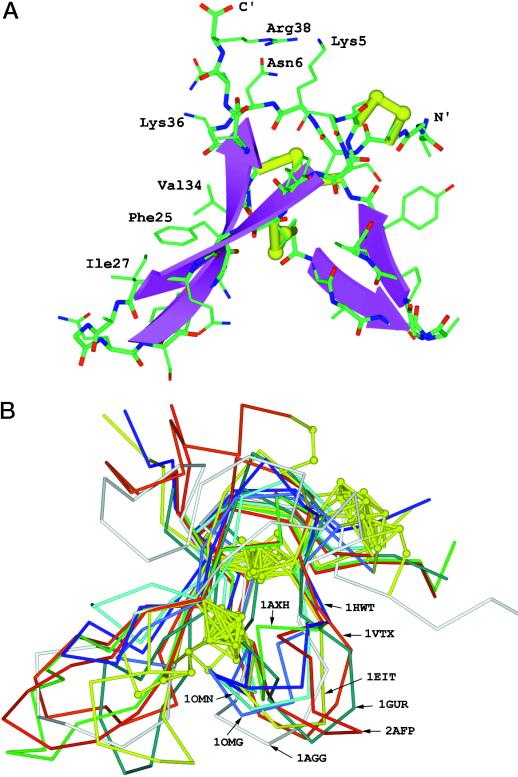
Model building was with the molecular modeling program Whatif (Vriend et al., 1998). The NMR coordinates of 1AXH were used to build up the backbone fragments. Loops were searched against the Whatif built-in loop fragment database. The modeled structure was refined geometrically with Whatif and energy minimized with the CHARM program to reduce side chain crash. Seven structure templates were taken from the Brookhaven Protein Data Bank (PDB) and used in the modeling work. The PDB codes are: 1AXH, 1AGG, 1EIT, 1VTX, 1OMN, 1OMG, and 1GUR. The structure of the Chinese bird spider toxin Huwentoxin-I (1HWT) was solved recently in our laboratory, but has not been deposited to the PDB. Sequence alignment in Figure 8 was performed taking into account that three disulfide bridges are conserved among all these peptides.
Figure 8.
A, Three-dimensional modeling of Pa-AMP-1. The NMR coordinates of 1AXH were used to build up the backbone fragments. Loops were searched against the Whatif built-in loop fragment database. The modeled structure was refined geometrically within Whatif and energy minimized with the CHARM program to reduce side chain crash. B, Superimposition of the constructed model onto eight template molecules shown in Figure 7.
Antimicrobial Activity Assays
For antifungal activity assays, Alternaria panax, Fusarium sp., and Rhizoctonia solani, all soil-borne pathogenic fungi, were first grown on potato-dextrose agar plates at 28°C until the diameters of the fungal colonies were around 3 cm. The antifungal activities of the samples were assayed using Oxford cups. Assays for anti-bacterial activity were performed according to the method of Vivanco et al. (1999) as follows. Bacillus megaterium and Staphyanococcus sp. cells were first grown in liquid medium overnight at 30°C. The cultures were diluted 100-fold and spread on solid medium plates. Whattman filter papers with a diameter of 0.5 cm were laid on top of the plates before various amount of the protein samples were added. The plates were then incubated for 24 h to observe the inhibition zone of the bacterial growth lawn around the filter papers. For determination of IC50 (concentration leading to 50% inhibition of growth rate, in micrograms per milliliter) of Pa-AMP-1 against B. megaterium, Staphyanococcus sp., and E. coli, the bacterial suspension in liquid medium was diluted to an A600 of 0.05. Pa-AMP-1 at various concentrations was added to 5-mL aliquots of the cultures. The cultures were incubated with shaking. The A600 was measured for monitoring bacterial growth.
RESULTS
Purification of Pa-AMP-1 and Determination of Its Amino Acid Residue Sequence
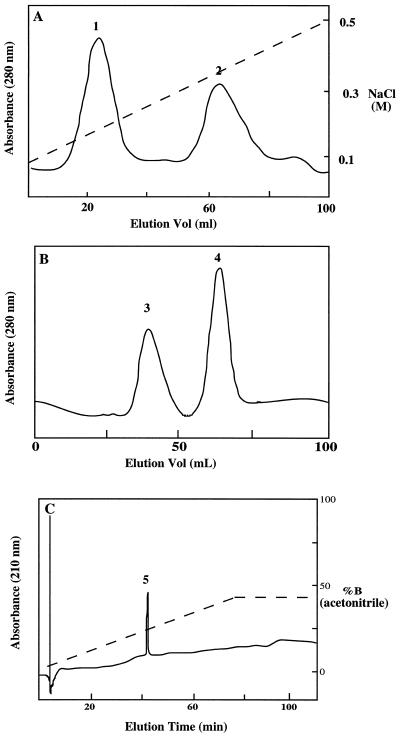
In searching for antifungal proteins from various sources, we found that the extracts from pokeweed seeds had strong activity against several soil-borne pathogenic fungi such as A. panax, Fusarium sp., and R. solani. The growth of R. solani was inhibited by the seed extract and purified protein (Fig. 1). The purified protein was also inhibitory to the growth of several gram-positive bacteria such as B. megaterium and Staphyanococcus sp., as shown in Table I. The IC50 of B. megaterium was approximately 8 μg mL−1 (Table I), which is comparable to that of Mj-AMP-1 (Cammue et al., 1992). The protein was not inhibitory to the growth of gram-negative bacteria such as E. coli (Table I). The antimicrobial activity was not affected by high-temperature (90°C) treatment of the extract. We took advantage of this heat-stable property of the protein in the purification procedure. The seed extract was first incubated at 90°C for 15 min before it was centrifuged to remove insoluble materials. The supernatant was loaded onto a cation exchange column and eluted with a NaCl gradient from 50 to 500 mm. Two major peaks were obtained (Fig. 2A), and the antimicrobial activity was associated with peak 2. Fractions of peak 2 were pooled and fractionated with a gel filtration column (Fig. 2B). Two major peaks (3 and 4) were obtained. The antimicrobial activity was assayed and found to be associated with peak 4. The purity of the peak D was evaluated with micro-HPLC equipped with a C18 column (15 cm × 0.5 mm). Only one peak (peak 5) was detected (Fig. 2C) and it contained antimicrobial activity. The peak eluted at approximately 30% (v/v) acetonitrile, indicating that this was a small protein molecule (Pa-AMP-1). Peak 3 shown in Figure 2B had no detectable antimicrobial activity.
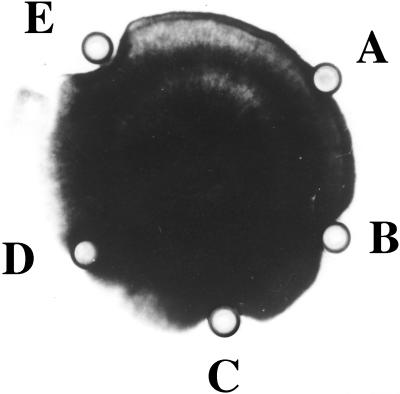
Figure 1.
Inhibition of R. solani growth by pokeweed seed extract and purified AMP. A, Bovine serum albumin in 25 mm HEPES buffer, pH 7.0 (control); B, seed extract; C and D, supernatant and pellet after the treatment of the seed extract at 90°C and centrifuging, respectively; E, purified Pa-AMP-1. Approximately 50 μg of protein was added to the Oxford cup, except cup E, which contained approximately 20 μg of protein.
Table I.
Effect of Pa-AMP-1 on bacterial growth
| Species | IC50 |
|---|---|
| μg mL−1 | |
| B. megaterium | 8 |
| Staphyanococcus sp. | 11 |
| E. coli | >300 |
Serial dilutions of purified Pa-AMP-1 were added to cultures of the bacteria listed. The growth rates of the microorganisms were determined by monitoring the A600, and the IC50 values are given.
Figure 2.
Purification of Pa-AMP-1 from pokeweed seeds. The basic and heat-stable protein in supernatant after heat treatment and centrifugation was the starting material for cation exchange chromatographic purification (A). Ten milliliters of supernatant was loaded onto a CM-Sephadex (Sigma) column (2.5 × 30 cm) previously equilibrated with elution buffer. The flow rate was 25 mL h−1. Fractions in 2 mL were collected and assayed for antimicrobial activity. The fractions containing antimicrobial activity (peak 2) were pooled and concentrated before being loaded onto a gel filtration column (B). The flow rate for the gel filtration was 15 mL h−1, and fractions in 2 mL were collected and assayed for antimicrobial activity. Peak 4 contained antifungal activity. C, Micro-HPLC analysis of the purified Pa-AMP-1. Conditions for micro-HPLC are described in “Materials and Methods.”
The pI of Pa-AMP-1 was estimated with isoelectric focusing gel electrophoresis and found to be above 10 (data not shown) using egg white lysozyme as the reference.
To determine its amino acid residue sequence, 100 μg of Pa-AMP-1 was first reduced with DTT and its Cys residues derivatized. The modified protein (2 μg) was adsorbed to a PVDF membrane and sequenced with an automatic sequencer. The entire amino acid sequence was: AGCIK NGGRC NASAG PPYCC SSYCF QIAGQ SYGVC KNR. The amino acid sequence of Pa-AMP-1 shows that it is rich in Cys residues and highly basic. There are several basic amino acid residues, while no acidic residue is present in the protein.
Molecular Cloning of the Gene Encoding Pa-AMP-1
To clone the gene encoding Pa-AMP-1, total mRNA from pokeweed was first isolated from the seeds. RT-PCR was performed using poly-T oligonucleotides and a degenerate oligonucleotide designed based on the amino acid sequence of Pa-AMP-1. A 350-bp fragment was obtained by RT-PCR. This fragment was cloned into the plasmid pGEM-T and its nucleotide sequence determined. The sequence revealed that the cloned fragment indeed contained a open reading frame with a deduced amino acid sequence identical to that of Pa-AMP-1. To obtain the full length of the cDNA of Pa-AMP-1, 5′-RACE was performed. A 280-bp fragment was obtained after amplification of the desired cDNA with PCR. This fragment was cloned as above and sequenced. The complete cDNA sequence of the gene encoding Pa-AMP-1 contains an open reading frame of 65 residues (Fig. 3A). The C-terminal portion of the deduced protein from the 28th to the 65th residue was identical to the amino acid sequence determined for the Pa-AMP-1 protein. The first 27 residues had characteristic features of a transit peptide found in excreted proteins (Nakai and Kanehisa, 1992). The presence of a transit peptide suggests that Pa-AMP-1 is synthesized as a precursor and processed into its mature form in the process of secretion (Von Heijne, 1986). The 3′-untranslated region of the gene is 185 nucleotides in length.
Figure 3.
Nucleotide sequences of Pa-AMP genes. A, cDNA sequence of the gene encoding Pa-AMP-1. The position of an intron (GenBank accession no. GI2939456) is indicated by the arrow; B, sequence alignment of Pa-AMP-1 and Pa-AMP-2 with other AMPs, Mj-AMP-1 and Mj-AMP-2 (De Bolle et al., 1995) and Mc-AMP-1 (GenBank accession no. AF069321). All Cys residues are conserved and are in bold. Identical residues are highlighted in gray. The Pa-AMP-1 and Pa-AMP-2 gene sequences have the database accession numbers AF048745 and AF209857, respectively.
Based on the cDNA sequence of Pa-AMP-1, a chromosomal DNA fragment containing the Pa-AMP-1 gene was amplified with PCR in the presence of Pfu DNA polymerase (Li et al., 1998) to ensure high fidelity in PCR amplification. The chromosomal gene of Pa-AMP-1 was nearly 900 bp in length and was cloned and sequenced. We found one intron located within the transit peptide region.
In sequencing the cloned PCR fragments amplified with primers of poly-T and the degenerate oligonucleotides based on the N-terminal amino acid sequence, we found a second cDNA and named it Pa-AMP-2. The deduced amino acid sequence of Pa-AMP-2 is similar to that of Pa-AMP-1 (Fig. 3B), only seven amino acids of Pa-AMP-1 was conservatively replaced in Pa-AMP-2.
A database search showed that both Pa-AMP-1 and Pa-AMP-2 are homologous to the two AMPs isolated from M. jalapa seeds, Mj-AMP-1 and Mj-AMP-2 (Cammue et al., 1992), and an AMP from Mesembryanthemum crystallinum, Mc-AMP-1 (GenBank accession no. AF069321) (Fig. 3B). The most striking features of these proteins are that they all contain six Cys residues, are all conserved in their positions in the primary sequences, and are all rich in basic amino acid residues.
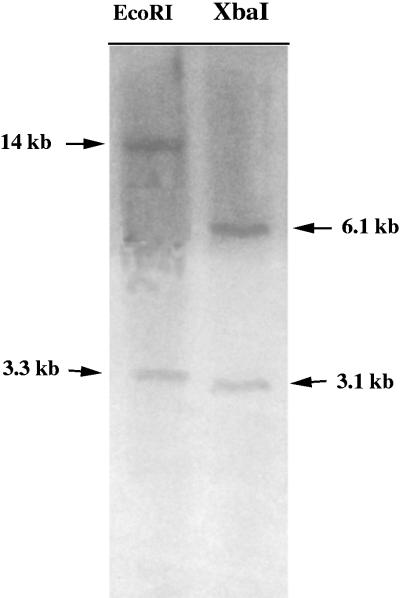
Southern hybridization was performed to determine how many copies of the Pa-AMP genes were present in the pokeweed genome, and the results are shown in Figure 4. Two bands could be detected when total DNA was digested with two different restriction enzymes, suggesting that two copies of the genes are present in pokeweed.
Figure 4.
Fluorogram of Southern blot hybridized with the 0.5-kb cDNA gene encoding Pa-AMP-1 of pokeweed. Ten micrograms of total DNA from pokeweed was digested with EcoRI (lane 1) or XbaI (lane 2) before being subjected to electrophoretic separation. The sizes of the hybridization bands are indicated with arrows.
Localization of Pa-AMPs
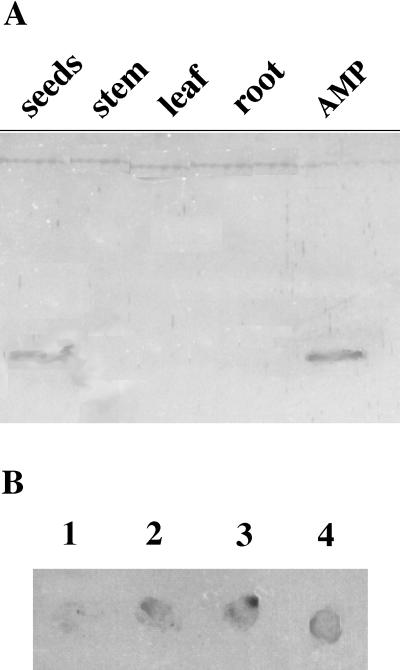
To investigate the spatial pattern of the Pa-AMP protein distribution in pokeweed plant, we performed immunoblotting using polyclonal antibodies against Pa-AMP-1 (Fig. 5A). Proteins showing cross-reaction with the antibodies could only be detected in mature seeds, not in leaves, stems, and roots. This result shows that Pa-AMPs are seed specific.
Figure 5.
Localization of Pa-AMPs in pokeweed by immunoblotting. A, Immunoblotting with total cell extracts from seeds (lane 1), leaves (lane 2), roots (lane 3), and stems (lane 4). Lane 5 contains purified Pa-AMP-1. B, Release of Pa-AMP-1 during seed germination. Seeds were soaked in water overnight before being placed on a PVDF membrane for 1 (spot 1), 24 (spot 2), and 48 h (spot 3). The level of Pa-AMPs released from each seed was measured by immunodetection. About 5 μg of purified Pa-AMP-1 was spotted at spot 4 as a control.
Analysis of the transit peptide of Pa-AMP-1 using the PSORT program (Nakai and Kanehisa, 1992) suggests that the protein could either be secreted out of the cells or into vacuoles. Immunoblotting was performed to investigate the location of the protein during seed germination. Pokeweed seeds were first soaked in water for 8 h before each individual seed was planted on top of a PVDF membrane. The germinating seeds were removed at various times from the PVDF membrane, and the Pa-AMPs adsorbed onto the PVDF membrane were detected with immunoblotting (Fig. 5B). The Pa-AMPs could be detected after the rupture of the pokeweed seeds, which usually occurred approximately 18 to 24 h after soaking in water, indicating that the Pa-AMPs are released into the environment of the germinating seed (Fig. 5B). It is possible that not all Pa-AMPs released from each seed were detected by this method because the binding of the Pa-AMPs to PVDF membranes could be weak due to their small size and highly hydrophilic nature. Some Pa-AMPs could diffuse out of the PVDF membrane.
Determination of the Disulfide Bridges and Three-Dimensional Modeling of Pa-AMP-1
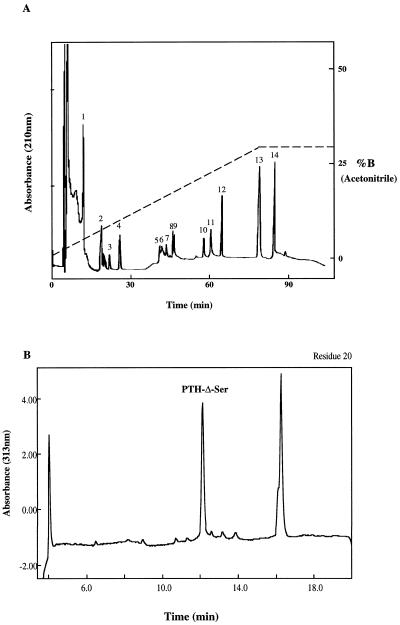
It has been shown that all Cys residues are involved in disulfide bridge formation in Mj-AMPs (Cammue et al., 1992), even though the exact disulfide bridges have not been determined. The fact that all six Cys residues in these proteins are conserved (Fig. 3) suggests that they play an important role in their structures. To investigate the pattern of disulfide bonding within Pa-AMP-1, the protein was fragmented first chemically with NBS before digestion with trypsin. NBS specifically cleaves at the peptide bonds formed by Tyr and Trp. Since there is no Trp residue in the protein, the only peptide bonds cleaved were at the tyrosal bonds. The chemical fragmentation was needed because Pa-AMP-1 is resistant to digestion by trypsin, as are Mj-AMPs (Cammue et al., 1992). The digested products were separated with micro-HPLC (Fig. 6A), and the fragments sequenced to determine their N-terminal sequences.
Figure 6.
Micro-HPLC peptide mapping of Pa-AMP-1. The protein was first cleaved chemically with NBS before digestion with trypsin. The peptides were separated with micro-HPLC (A) as described in “Materials and Methods.” B, Detection of a Δ-Ser at the 20th residue in Pa-AMP-1 sequencing . The protein was loaded to the sequencer without reduction. The first three Cys residues were linked to other Cys residues through disulfide bonds and could not be detected. Based on the results shown in Table I, the detection of the Δ-Ser at the 20th residue (the fourth Cys) shows that there is a disulfide bond between Cys-3 and Cys-20.
The relevant sequence information is summarized in Table II. The sequence information shown in peaks 10, 11, and 12 suggests that there is a disulfide bond between Cys-10 and Cys-24. The N-terminal sequences shown in peak 8 suggest that there are two disulfide bonds among the three peptides: a pair of disulfide bonds either between Cys-3 to Cys-19 and Cys-20 to Cys-35 or between Cys-3 to Cys-20 and Cys-19 to Cys-35. To determine which pattern is correct in Pa-AMP-1, N-terminal sequencing was performed in the absence of DTT in the step of PTH conversion. The PTH-dehydro-Ala, which is formed from a Cys residue or Ser residue and has characteristic A313, was detected as described by Zhang and Liang (1993), and the result is shown in Figure 6B. Because PTH-dehydro-Ala was detected in the twentieth cycle of sequencing, the first Cys can only be linked to the fourth Cys (the 20th amino acid residue). Based on the above information, the disulfide bonds of Pa-AMP-1 are Cys-3 to Cys-20, 10 to 24, and 19 to 35.
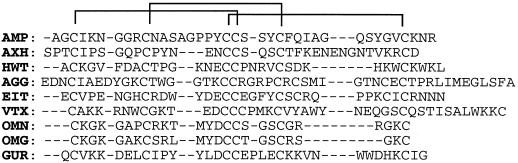
A database search showed that the disulfide bridge pattern found in Pa-AMP-1 was also present in some small peptide toxins (Fig. 7). The most obvious feature in all of the sequences is the pattern of distribution of the six Cys and disulfide bridges, even though they have a low homology in their primary amino acid sequences. They all have a pattern of X(0–3) CX(5–7) CX(5–8) CCX(3–4) CX(4–13) CX(0–12), i.e. they all have a disulfide bridge pattern of 1 to 4, 2 to 5, and 3 to 6.
Figure 7.
Sequence alignment of Pa-AMP-1 with eight proteins of known three-dimensional structure. AXH, Atracotoxin-HVI, a Blue Mountains funnel-web spider toxin; HWT, huwentoxin-I, a Chinese bird spider (Selenoicosmia huwena) toxin; AGG, ω-agatoxin-IVb, a funnel-web spider toxin that is a P-type calcium channel antagonist; EIT, μ-atatoxin-I, a Blue Mountains funnel-web toxin that is a sodium channel blocker; OMN, ω-conotoxin-MVIIc, a magus cone toxin that is a P-type calcium channel antagonist; OMG, ω-conotoxin-MVIIa, a Magus cone toxin that is a P-type calcium channel antagonist; GUR, gurmarin, a sweet-taste-suppressing protein from Gymnema sykvestre. The structures of all listed proteins can be accessed in the PDB with the three letter codes.
The functions and three-dimensional structures of the small peptide toxins are known and are shown in Figure 7. Structurally, these toxins belong to the cystine-knot superfamily (Pallaghy et al., 1994). Comparison of Pa-AMP-1 with the toxins shown in Figure 7 suggests that Pa-AMP-1 may also belong to this folding group. To help understand the mechanism of Pa-AMP-1 inhibition of microbial growth and to reveal the three-dimensional structure of Pa-AMP-1, computer modeling of Pa-AMP-1 was performed (Fig. 8A). The model shows that the Pa-AMP-1 molecule has a characteristic feature of cystine-knot folding: disulfide bridges 1 to 4 and 2 to 5, together with the backbone of the protein, form a ring and the third disulfide bridge crosses it. Another key feature in this model is the anti-parallel β-sheets that were found in all template molecules. The position of the anti-parallel sheets and the disulfide bridges of Pa-AMP-1 shows that the molecule has a β-cross-folding (Pallaghy et al., 1994). The side chains of three basic residues, Lys-5, Lys-36, and Arg-38, form a positive patch at one side (top right in Fig. 8A) of the molecule. On the left side of the molecule, the side chains of three hydrophobic residues, Phe-25, Ile-27, and Val-34, form a hydrophobic surface.
The constructed model was superimposed onto eight template molecules (Fig. 8B), illustrating the structural similarity between Pa-AMP-1 and all templates. Although the mechanism of Pa-AMP-1 action against microbial growth is not yet known, the structure of Pa-AMP-1 shows that it belongs to the inhibitor cystine-knot group (Harrison and Sternberg, 1996).
DISCUSSION
Seed germination of plants occurs in an environment rich in pathogens. Many plant seeds contain antifungal and anti-bacterial chemicals. The pokeweed seed Pa-AMP protein reported here is another example of a plant defense system employed against pathogen infection. AMPs in pokeweed, like AMP in M. jalapa, are only synthesized in mature seeds (Fig. 5A). The cDNA sequence of the Pa-AMP-1 gene shows the presence of a transit peptide in the protein, suggesting that it is synthesized as a precursor and processed into its mature form in secretion. The protein is released outside the seed coat during germination (Fig. 5B). The release of the mature Pa-AMP into seed germination environment can create an inhibitory zone that prevents infection of the germinating seeds by pathogens. It has been shown that the mRNA of AMPs in M. jalapa (De Bolle et al., 1995) accumulated only in mature seeds, implying that the proteins are synthesized not only in the process of seed maturation, but also during germination.
The presence of a second AMP in pokeweed shows that, as in case of M. jalapa, the AMP genes are also present in a gene family. Southern hybridization (Fig. 4) showed that the Pa-AMP gene family has a low complexity. The promoters for these genes are apparently seed specific. It is interesting that the protein is missing in roots in both pokeweed, as shown in Figure 5A, and M. jalapa, as shown previously by northern analysis (De Bolle et al., 1995). This phenomenon could result from the fact that the AMPs are potent agents against microorganisms. If they were present in plant roots, they would interfere with root-microbe interactions crucial to plant growth.
There are several basic amino acid residues and no acidic amino acid residues in the primary sequence of the mature protein (Fig. 3A), showing that it is a basic protein. The isolation of Pa-AMP-1 with a cation exchange column (Fig. 2) also shows that the Pa-AMP-1 is a basic protein. The pI of Pa-AMP-1 was estimated to be above 10, higher than the value predicted by computer analysis based on its primary sequence. The difference could be due the special folding of the protein, which could result in a more basic pI of Pa-AMP-1. The basic amino acid residues are conserved in different AMPs (Fig. 3B). De Samblanx et al. (1997) showed that replacing Val-39 with the basic Arg increased the potency of antifungal protein from radish seeds (Rs-AFP1) (Terras et al., 1995) against fungi, while replacing Lys-44 with a neutral amino acid residue decreased the potency. It is likely that the basic residues are important to the functions of these small, Cys-rich AMPs. We have recently overproduced an active Pa-AMP-1 protein in E. coli (Y. Liu and J. Zhao, unpublished data). Site-specific mutagenesis of the gene should provide more information about what roles the positive residues have in antimicrobial activities.
Recently, the three-dimensional structures of several antifungal proteins, including the plant defensin from radish seeds (Fant et al., 1998) and γ-thionin from sorghum (Bloch et al., 1998), have been determined. Both Rs-AFP1 and γ-thionin have one α-helix and triple-stranded β-sheets. Although the mechanism of their antifungal activity is still unknown, the importance of a hydrophobic surface and a positive patch has been suggested (Bloch et al., 1998). The biochemical basis of the antimicrobial activity of Pa-AMP-1 has not been revealed. We have not found any effect of Pa-AMP-1 on α-amylase and subtilisin activity (Y. Liu and J. Zhao, unpublished data). Cammue et al. (1992) showed that all six Cys residues are involved in disulfide bridges in Mj-AMP. Sequence comparison of the AMPs (Fig. 4) shows that all six Cys residues are conserved, indicating that the formation of these intramolecular disulfide bonds is important to structure and function.
In the present study, we have shown that the three intramolecular disulfide bridges have a pattern of 1 to 4, 2 to 5, and 3 to 6, a folding pattern found in many toxin proteins (Harrison and Sternberg, 1996). The three-dimensional model suggests that Pa-AMP-1 has a different structure from both the antifungal protein of radish seeds (Fant et al., 1998) and the γ-thionin (Bloch et al., 1998). Our three-dimensional modeling of Pa-AMP-1 suggests that it belongs to the inhibitor cystine-knot group, one of the most compact and stable protein-folding motifs (Harrison and Sternberg, 1996). Stable folding is very critical to seed protein, since most of the seeds desiccate during maturation, which could denature most of unstable proteins. The model also suggests the presence of a hydrophobic surface and a positive patch on Pa-AMP-1 (Fig. 8). It is possible that Pa-AMP-1 could interact with the phospholipids of cell membranes, resulting in inhibition of fungal growth. Since Pa-AMP-1 is only inhibitory toward filamentous fungi and gram-positive bacteria, and has no inhibitory effect on gram-negative bacteria or yeast (Table I), it is likely that AMPs may specifically interact with membrane receptors rather than non-specifically binding to cell membranes (Thevissen et al., 1997). The positive patch of AMPs could provide the specific site for the interaction through ionic interactions. This suggestion is supported by the fact that the potency of Mj-AMPs was drastically reduced under high ionic strength (Cammue et al., 1992).
So far, AMPs have only been found in the seeds of pokeweed, M. jalapa (De Bolle et al., 1995), and M. crystallium (GenBank accession no. AF069321), and these three plants are not closely related phylogenetically. The presence of a small gene family in both M. jalapa and pokeweed, and the fact that the chromosomal genes from M. jalapa and pokeweed contain one intron located similarly in the signal peptide region suggest the possibility that these AMP genes were possibly conserved during evolution and may be present in some other plant seeds as well. We are currently testing this possibility using immunoblotting.
ACKNOWLEDGMENTS
The authors thank Profs. X. Wu and P. Xiao for their suggestions in this research. The technical assistance of W. Shen is appreciated.
Footnotes
This research was supported by the Chinese National Natural Science Foundation (grant no. 39535002) and by the Department of Science and Technology (grant no. J99–A–032).
LITERATURE CITED
- Bloch C, Patel SU, Baud F, Zvelebil MJ, Carr MD, Sadler PJ, Thornton JM. 1H NMR structure of an antifungal gamma-thionin protein Sialpha1: similarity to scorpion toxins. Proteins. 1998;15:334–349. doi: 10.1002/(sici)1097-0134(19980815)32:3<334::aid-prot9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Bohlmann H, Apel K. Thionins. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:227–240. [Google Scholar]
- Broekaert WF, Terras FRG, Cammue BPA, Osborn RW. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Phyisol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammue BPA, De Bolle MFC, Terras FRG, Van Damme PPJ, Rees SB, Vanderleyden J, Broekaert WF. Isolation and characterization of a novel class of plant antimicrobial peptides from Mirabilis jalapa L. seeds. J Biol Chem. 1992;267:2228–2233. [PubMed] [Google Scholar]
- Cheng S, Fockler C, Barnes WM, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bolle MFC, Eggermont K, Duncan RE, Osborn RW, Terras FRG, Broekaert WF. Cloning and characterization of two cDNA clones encoding seed-specific antimicrobial peptides from Mirabilis jalapa L. Plant Mol Biol. 1995;28:713–721. doi: 10.1007/BF00021195. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- De Samblanx GW, Goderis IJ, Thevissen K, Raemaekers R, Fant F, Borremans F, Acland DP, Osborn RW, Patel S, Broekaert WF. Mutational analysis of a plant defensin from radish (Raphanus sativus L.) reveals two adjacent sites important for antifungal activity. J Biol Chem. 1997;272:1171–1179. doi: 10.1074/jbc.272.2.1171. [DOI] [PubMed] [Google Scholar]
- De Vries S, Hoge H, Bisseling T. Isolation of total and polysomal RNA from plant tissues. In: Gelvin SB, Schilperoort RA, Verma DPS, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. B6/1–B6/13. [Google Scholar]
- Epple P, Apel K, Bohlmann H. ESTs reveal a multigene family for plant defensins in Arabidopsis thaliana. FEBS Lett. 1997;400:168–172. doi: 10.1016/s0014-5793(96)01378-6. [DOI] [PubMed] [Google Scholar]
- Fant F, Vranken W, Broekaert W, Borremans F. Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1H NMR. J Mol Biol. 1998;279:257–270. doi: 10.1006/jmbi.1998.1767. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies, a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Harrison PM, Sternberg MJ. The disulphide beta-cross: from cystine geometry and clustering to classification of small disulphide-rich protein folds. J Mol Biol. 1996;6:603–623. doi: 10.1006/jmbi.1996.0664. [DOI] [PubMed] [Google Scholar]
- Li B, Gu C, Zhao J. Expression of Pfu DNA polymerase gene in Escherichia coli and its application in long-distance PCR. Chin Sci Bull. 1998;43:863–867. [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AE. Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell. 1996;8:1821–1831. doi: 10.1105/tpc.8.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallaghy PK, Nielsen KJ, Craik DJ, Norton RS. A common structural motif incorporating a cystine knot and a triple-stranded beta-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994;3:1833–1839. doi: 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schumbaum A, Mauch F, Vogeli U, Boller T. Plant chitinases are potent inhibitors of fungal growth. Nature. 1986;324:365–367. [Google Scholar]
- Smith C. Signal transduction in elicitation of phytoalexin synthesis. J Biochem Soc Trans. 1994;22:414–419. doi: 10.1042/bst0220414. [DOI] [PubMed] [Google Scholar]
- Terras FRG, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Cammue BPA, Broekaert WF. Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell. 1995;7:573–588. doi: 10.1105/tpc.7.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevissen K, Osborn RW, Acland DP, Broekaert WF. Specific, high affinity binding sites for an antifungal plant defensin on Neurospora crassa hyphae and microsomal membranes. J Biol Chem. 1997;272:32176–32181. doi: 10.1074/jbc.272.51.32176. [DOI] [PubMed] [Google Scholar]
- Vivanco JM, Savary BJ, Flores HE. Characterization of two novel type I ribosome-inactivating proteins from the storage roots of the Andean crop Mirabilis expansa. Plant Physiol. 1999;119:1447–1456. doi: 10.1104/pp.119.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta. 1986;974:307–333. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]
- Vriend G, Berendsen HJC, van den Burg B, Venema G, Eijsink VGH. Early steps in the unfolding of thermolysin-like proteases. J Biol Chem. 1998;273:35074–35077. doi: 10.1074/jbc.273.52.35074. [DOI] [PubMed] [Google Scholar]
- Zhang D, Liang S. Assignment of the three disulfide bridges of huwentoxin-I, a neurotoxin from the spider Selenocosmia huwena. J Protein Chem. 1993;12:735–740. doi: 10.1007/BF01024931. [DOI] [PubMed] [Google Scholar]
- Zhao J, Snyder WB, Muhlenhoff U, Rhiel E, Warren PV, Golbeck JH, Bryant DA. Cloning and characterization of the psaE gene of the cyanobacterium Synechococcus sp. PCC 7002: characterization of a psaE mutant and overproduction of the protein in Escherichia coli. Mol Microbiol. 1993;9:183–194. doi: 10.1111/j.1365-2958.1993.tb01680.x. [DOI] [PubMed] [Google Scholar]
- Zhou R, Wei X, Jiang N, Li H, Dong Y, Hsi KL, Zhao J. Evidence that HetR protein is an unusual serine-type protease. Proc Natl Acad Sci USA. 1998;95:4959–4963. doi: 10.1073/pnas.95.9.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]