ABSTRACT
Methylmercury (MeHg) production was compared among nine cultured methanogenic archaea that contain hgcAB, a gene pair that codes for mercury (Hg) methylation. The methanogens tested produced MeHg at inherently different rates, even when normalized to growth rate and Hg availability. Eight of the nine tested were capable of MeHg production greater than that of spent- and uninoculated-medium controls during batch culture growth. Methanococcoides methylutens, an hgcAB+ strain with a fused gene pair, was unable to produce more MeHg than controls. Maximal conversion of Hg to MeHg through a full batch culture growth cycle for each species (except M. methylutens) ranged from 2 to >50% of the added Hg(II) or between 0.2 and 17 pmol of MeHg/mg of protein. Three of the species produced >10% MeHg. The ability to produce MeHg was confirmed in several hgcAB+ methanogens that had not previously been tested (Methanocella paludicola SANAE, Methanocorpusculum bavaricum, Methanofollis liminatans GKZPZ, and Methanosphaerula palustris E1-9c). Maximal methylation was observed at low sulfide concentrations (<100 μM) and in the presence of 0.5 to 5 mM cysteine. For M. hollandica, the addition of up to 5 mM cysteine enhanced MeHg production and cell growth in a concentration-dependent manner. As observed for bacterial Hg methylators, sulfide inhibited MeHg production. An initial evaluation of sulfide and thiol impacts on bioavailability showed methanogens responding to Hg complexation in the same way as do Deltaproteobacteria. The mercury methylation rates of several methanogens rival those of the better-studied Hg-methylating sulfate- and iron-reducing Deltaproteobacteria.
KEYWORDS: Archaea, methylmercury, bioavailability, complexation, cysteine, hgcAB, mercury, methyltransferase, methylation, thiols
IMPORTANCE
Archaea, specifically methanogenic organisms, play a role in mercury methylation in nature, but their global importance to MeHg production and the subsequent risk to ecosystems are not known. Methanogenesis has been linked to Hg methylation in several natural habitats where methylmercury production incurs risk to people and ecosystems, including rice paddies and permafrost. In this study, we confirm that most methanogens carrying the hgcAB gene pair are capable of Hg methylation. We found that methylation rates vary inherently among hgcAB+ methanogens but that several species are capable of MeHg production at rates that rival those of the better-know Hg-methylating sulfate- and iron-reducing bacteria. Methanogens may need to be considered equally with sulfate and iron reducers in evaluations of MeHg production in nature.
INTRODUCTION
The discovery of the hgcAB gene pair led to the identification of several groups of microorganisms not previously identified as capable of mercury methylation, including methanogens. With the exception of a few syntrophic and fermentative Firmicutes (4), demonstrated mercury (Hg) methylators are all strictly anaerobic bacteria that use sulfate, iron, and carbon dioxide as terminal electron acceptors. Several hgcAB+ putative methylators have been identified in the phyla Bacteroidetes, Chloroflexi, and Nitrospirae (2) and other phyla, but none of these have been confirmed to produce methylmercury (MeHg) in culture. Overall, ~210 microbial species containing hgcAB have been identified among all of the bacteria and Archaea whose genomes have been sequenced, numbering >10,000.
The first list of hgcAB+ organisms was published in 2013 (3), with subsequent updates (2, 4). In all cases, the list has been based on in silico screening for both hgcA and hgcB by using the encoded amino acids to eliminate genomic variability such as codon wobble and species-specific nucleotide bias. Screening for hgcA included the presence of the distinctive conserved motif NVWCAAGK in the active methyltransferase site and predicted C-terminal transmembrane helices. The ferredoxin-encoding gene hgcB is generally, but not always, immediately downstream from hgcA. All but two of the hgcAB+ methanogens identified are in one class, Methanomicrobia. However, the overall importance of methanogens to MeHg production in nature is not well understood. Hg methylation has been confirmed in culture for only four organisms to date, Methanospirillum hungatei (5), Methanomethylovorans hollandica, Methanolobus tindarius (4), and Methanomassiliicoccus luminyensis (2). Here, we examined the Hg methylation rates of several strains to understand how their MeHg production compares to that of the better-studied methylators in the Deltaproteobacteria.
It is clear that there are several habitats, like rice paddies (1), some freshwater sediments (6), and periphyton (7, 8), where methanogens play an important role in MeHg production. However, their global importance to MeHg production and the subsequent risk to ecosystems are not known. A survey of publically available metagenomes showed that methanogens may also be dominant Hg methylators in termite hindguts, permafrost peat, Arctic sediments, and some bioreactors (2). Early work with clade-specific primers for hgcAB+ methanogens, Deltaproteobacteria, and Firmicutes suggested that archaeal Hg methylators are important in freshwater sediments from East Fork Poplar Creek, TN (9, 10).
In this study, several cultured representatives of hgcAB+ methanogens were used to understand the extent to which these organisms can produce MeHg and to begin to evaluate the biogeochemical controls on Hg methylation by methanogens. Specifically, we asked whether hgcAB+ methanogens have the ability to methylate significant portions of the available Hg, how methylation rates vary among species and metabolisms, and how the methylation rates of methanogens compare to those of Deltaproteobacteria and Firmicutes. We examined how the ability to methylate Hg is clustered within the Archaea, phylogenetically and metabolically.
Last, we began to evaluate the Hg complexes that are available for uptake and methylation by hgcAB+ methanogens. We wanted to know if the same types of Hg complexes that are available to the best-known group of Hg methylators, sulfate- and iron-reducing Deltaproteobacteria, are also available to Hg-methylating methanogens. Small thiols appear to be most bioavailable for methylation by sulfate-reducing bacteria (SRB) and iron-reducing bacteria (FeRB) (11, 12). Sulfide reduces the bioavailability of Hg to Hg-methylating Deltaproteobacteria via precipitation of mercuric sulfides that are less available for uptake (13–15). Dissolved organic matter (DOM) can slow the precipitation of nanoparticulate HgS (HgSnp) (16–18), enhancing bioavailability to SRB (13, 14). The characteristics of DOM, including its aromaticity (19), thiol content, and degree of sulfidization (20), all influence how DOM impacts Hg and HgSnp bioavailability to SRB. Understanding the range of methylation rates and the biogeochemical controls on Hg methylation by methanogens is a critical part of understanding the role of methanogens in MeHg production and risk in nature.
RESULTS
Methylation results.
We quantified and compared the MeHg production of 9 of the now 19 identified hgcAB+ methanogens, all Methanomicrobia (see Table S1 in the supplemental material). The species tested included previously untested hgcAB+ methanogens (Methanocella paludicola SANAE, Methanocorpusculum bavaricum, Methanofollis liminatans GKZPZ, Methanococcoides methylutens, and Methanosphaerula palustris E1-9c) plus previously reported methylators Methanospirillum hungatei JF-1 (5), Methanomethylovorans hollandica, Methanolobus tindarius (4). Additionally, the bioavailability of Hg sulfide and Hg thiol complexes to methanogens was tested for selected species. Full information is provided in Data Set S1.
Data on the cultures and controls used in this study. Download DATA SET S1, XLSX file, 0.03 MB (29.2KB, xlsx) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Methanogen strains used in this study. Download TABLE S1, PDF file, 0.04 MB (39.6KB, pdf) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
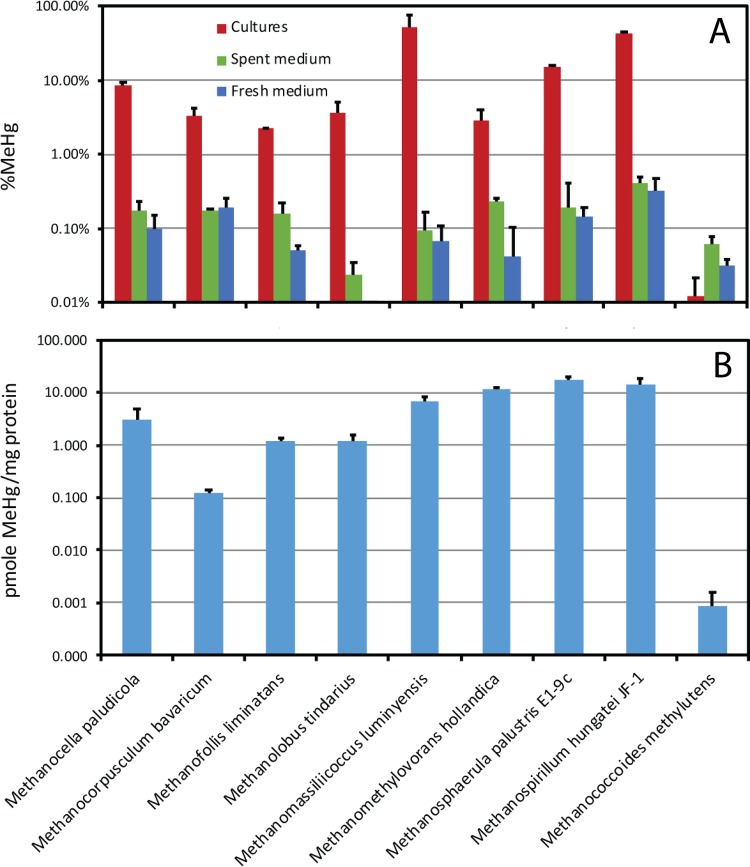
Figure 1 (top) provides a comparison of the maximum measured MeHg production levels of all nine of the hgcAB+ methanogens tested. Methylation was assayed in batch cultures. MeHg concentrations were measured once cultures entered stationary phase, and percentage of MeHg formation was calculated on the basis of the total measured Hg concentration in the culture medium at the same time. Controls included MeHg production in fresh and spent media for each strain. Several organisms were tested with different concentrations of sulfide or cysteine in culture medium; the data in Fig. 1 are methylation rates under the most favorable observed conditions, usually between 0.5 and 4 mM cysteine and 10 and 100 μM sulfide.
FIG 1 .
Maximal conversion of inorganic Hg to MeHg during batch culture growth by each hgcAB+ methanogen tested, expressed two ways. (Top) MeHg as a percentage of the total measured Hg in the culture medium (both unfiltered) at the end of log growth. Green and blue bars show noncellular control values. (Bottom) MeHg normalized to protein measured at the end of the log growth phase. Note the log scales. Bars are averages of triplicate separate cultures or controls. Methylation was measured from a 1 nM 201Hg(II) spike. Error bars are based on the standard deviation of MeHg only. M. luminyensis data are from reference 2.
Eight of the nine species tested were capable of more MeHg production than controls during batch culture growth. Maximal conversion of Hg to MeHg for each strain through a full batch culture growth cycle ranged from 2 to ~65%. Three of the species produced >10% MeHg, M. luminyensis, M. palustris, and M. hungatei. Using lower sulfide concentrations, M. tindarius and M. hollandica produced somewhat higher percentages of MeHg than previously reported. MeHg production by M. hungatei (43%) was similar to that reported by Yu et al. (5). Methanococcoides methylutens, a species with a fused hgcAB gene pair, did not produce more MeHg than controls in triplicate assays repeated three times.
There was significant loss of Hg from the aqueous phase in many of the methylation assays. Hg losses were comparable in controls (spent and fresh media) and cultures (see Data Set S1). Differences in Hg loss appear to be related to medium chemistry, with greater losses often observed in higher-sulfide media. However, no systematic tests of medium chemistry effects on Hg losses was done, nor were the mechanisms of loss (bottle sorption versus reduction to Hg°) systematically evaluated.
Although the percentage of MeHg is an easily understandable way to evaluate the magnitude of MeHg production by cultures, a better way to report and compare rates among organisms and conditions is methylation normalized to growth (Fig. 1, bottom). Protein-normalized MeHg production by the Hg-methylating methanogens ranged from roughly 1 to 20 pmol of MeHg/mg of protein, similar to that in prior assays of M. tindarius and M. hollandica. Yu et al. (5) reported about 100 pmol of MeHg/mg of protein for M. hungatei during log-phase growth, which was reduced to <10 pmol of MeHg/mg of protein in the presence of a sulfide reductant.
Protein-normalized MeHg production by Deltaproteobacteria and Firmicutes, measured by the same batch culture approach, ranged from about 1 to 600 and 1 to 50 pmol of MeHg/mg of protein, respectively, with Geobacter bemidjiensis and Desulfovibrio sp. strain ND132 the highest producers to date (4). A survey of eight hgcAB+ Desulfovibrio species produced between about 20 and 250 pmol of MeHg/mg of protein in short-term (3 h) washed cell assays with 500 μM cysteine and no added sulfide (21).
Methylation by methanogenic Archaea in comparison to Bacteria.
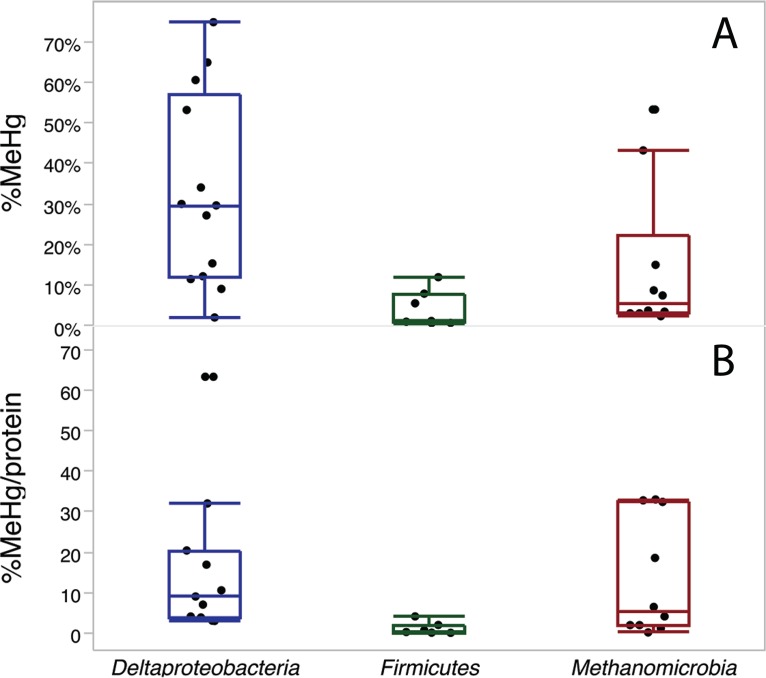
To compare methylation abilities among clades and across studies with different growth conditions and Hg levels, we compared absolute conversion of Hg to MeHg (percentage of MeHg; Fig. 2, top) and also normalized MeHg production for each species to both protein and Hg (Fig. 2, bottom). In a past study, we found that methylation rates in washed-cell assays were proportional to the inorganic Hg(II) concentration (21). For this comparison, we chose the highest measured methylation rate across all of the conditions tested for each species measured, usually between 0.5 and 4 mM cysteine and 10 and 100 μM sulfide. Figure 2 compiles data from this study, our prior batch culture assays of all three clades (4), and washed-cell assays of Desulfovibrio species (21). We did not correct the methylation rates for time, although batch cultures were often incubated for several days while washed cell assays ran 3 h. Our observation has been that most MeHg production in cultures occurs in the first hours of incubation with active cells (12, 21). Nevertheless, the differences in timing, culture conditions, and Hg concentrations among assays add uncertainty to this comparison.
FIG 2 .
Comparison of maximal MeHg production among the three major clades of Hg-methylating bacteria. (Top) Maximal conversion of inorganic Hg to MeHg by 13 Deltaproteobacteria, 7 Firmicutes, and 10 Methanomicrobia species. (Bottom) MeHg production normalized to both the measured Hg and protein concentrations at the end of methylation assays, for the same organisms and assays, as (MeHg/Hg)/(mg of protein/liter). Hg spike levels ranged from 1 to 50 nM. Data combine assays done over the course of batch culture growth in this study, in the study described in reference 4, and during 3-h washed-cell assays (21). In these bar-and-whisker plots, the average value for each species is represented by a dot; the centerline, top, and bottom of each bar are the median and upper and lower quartiles, respectively, across all species; and the whiskers are drawn to the furthest data point within 1.5× the quartile value (thus excluding outliers).
On average, across all of the species tested, Deltaproteobacteria produced the highest average percentage of MeHg and the highest percentage of MeHg normalized to protein. There are few hgcAB+ Deltaproteobacteria that convert <10% of Hg(II) to MeHg under ideal conditions (moderate thiol levels, low sulfide levels) during a few hours of log-phase growth. Of 13 species tested, 4 are able to convert more than half of the available Hg(II) to MeHg. However, normalized to protein and Hg, MeHg production by methanogens rivals that of Deltaproteobacteria. While methanogens tended to convert a smaller fraction of Hg(II) to MeHg during batch culture growth, their cell densities and growth rates in culture are usually lower than those of sulfate and iron reducers. Expressed as the percentage of MeHg per milligram of protein, several methanogens exhibited methylation rates equal to those of the best Deltaproteobacteria. Including this study, the methylation rates of about the same number of Deltaproteobacteria (13) and Methanomicrobia (9) have been tested quantitatively in culture. SRB and FeRB have been tested in several labs (4, 12, 15, 21–37), but only one hgcAB+ methanogen has been tested in culture outside our group (5).
About 40 hgcAB+ Firmicutes have been identified to date, but only 7 have been tested for methylation in culture so far (4). Almost all produced <10% MeHg in culture and a uniformly low percentage of MeHg per milligram of protein (Fig. 2). The majority of these are sulfite and sulfate reducers.
Influence of cysteine on methylation by methanogens.
The presence of small thiols often enhances the bioavailability of Hg for methylation to Deltaproteobacteria. We evaluated Hg methylation by M. hollandica and M. tindarius across cysteine gradients to begin to assess the bioavailability of Hg thiols to hgcAB+ methanogens. Methanogens often require a source of reduced S for growth. Addition of cysteine, other thiols, or sulfide can stimulate growth, complicating the interpretation of Hg methylation data. In this study we used methionine, which is not a strong ligand for Hg, to provide reduced sulfur for growth. The amino acid methionine contains S in a thioether side chain but not a free thiol. By providing an alternative reduced-S source, we hoped to test the influence of cysteine on Hg bioavailability, separately from its influence on cell growth.
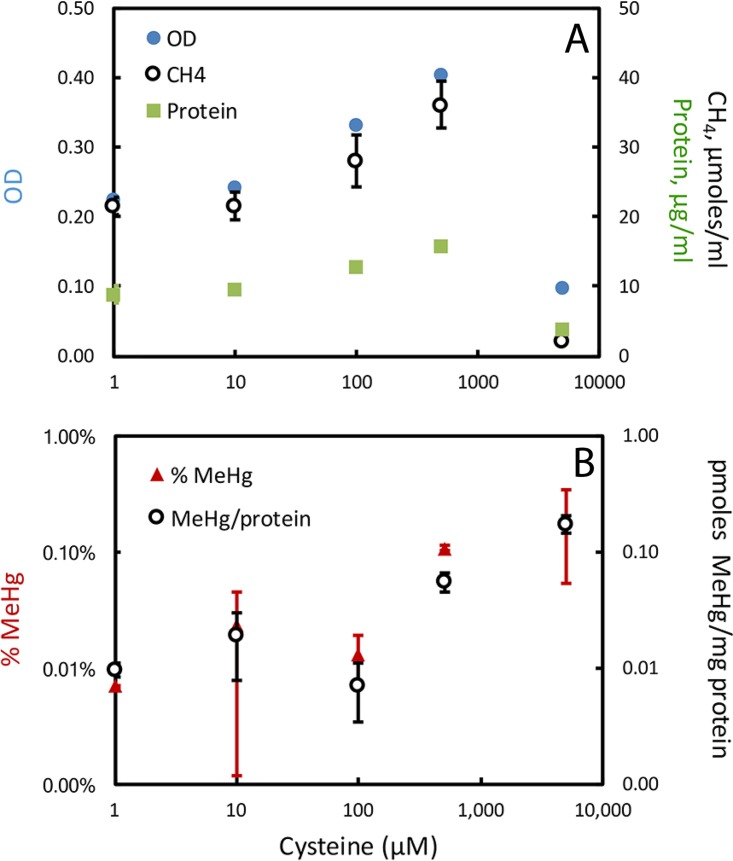
Cysteine enhanced the production of MeHg by both cultures. For M. hollandica, MeHg production was tested in batch culture across a cysteine gradient of 1 to 5,000 μM, with 1 mM methionine added to the medium. However, even when methionine was provided, cysteine up to 500 μM enhanced cell growth in a concentration-dependent manner (Fig. 3). Growth was inhibited at 5 mM cysteine. Methionine (1 mM) as the primary reduced-S source produced less growth than 500 μM cysteine (in comparison with a prior assay [4]). In prior experiments, M. hollandica grown in batch culture methylation assays with 500 μM cysteine, 50 μM sulfide, and 10 nM Hg produced ~1% MeHg and ~5 pmol of MeHg/mg of protein. Under those conditions, cultures grew well, achieving an optical density (OD) of ~0.5 and ~22 μg of protein/ml. Cysteine may be a more available form of reduced sulfur than methionine and also serves as an additional reductant (although all media were reduced with TiNTA (titanium nitrilotriacetic acid) and monitored visually with resazurin). Corrected for its effects on growth, cysteine enhanced the bioavailability of Hg for methylation by M. hollandica. The absolute amount of MeHg produced increased with the cysteine concentration in the medium, as did MeHg production normalized to cell protein, with the most MeHg production in the 5 mM cysteine cultures, despite their relatively weak growth.
FIG 3 .
Impact of cysteine on M. hollandica growth and MeHg production. (Top) Growth assessed by OD and methane production. (Bottom) MeHg as a percentage of the total Hg in the culture medium and MeHg normalized to protein. All measurements were made once all cultures reached stationary phase (312 h). Methionine at 1 mM was included in all culture media. Sulfide was added to all at 10 μM, but the concentration was to 2 to 4 μM at the end of log-phase growth. Note the log scales. Error bars are standard deviations of triplicate cultures.
Cysteine also enhanced methylation by M. tindarius. M. tindarius was grown across the same cysteine gradient in medium with or without the addition of 100 μM sulfide and 500 μM methionine (Fig. 4). Cysteine enhanced growth more strongly in the medium without added sulfide or methionine. In medium without added sulfide, MeHg as a fraction of the Hg in the medium (at the end of incubation) and MeHg production normalized to cell protein both increased up to about 100 μM cysteine but declined as the cysteine concentration increased further. Sulfide had a strong inhibitory effect on methylation. MeHg production was lower in medium with added sulfide, and the addition of cysteine did not increase MeHg production, even though growth was much stronger. The sulfide concentration in cultures without added sulfide averaged 0.3 μM at the end of log-phase growth; cultures with 100 μM added sulfide averaged 1.4 μM. In medium with or without sulfide, cysteine held Hg in solution in the culture medium in a concentration-dependent way, based on measurement of the Hg remaining in the aqueous medium (including cells) at the end of growth (Fig. S1). Interestingly, in the absence of added sulfide, cysteine increased methylation even when normalized to the final Hg concentration, suggesting that the mechanism for cysteine enhancement is not just its ability to enhance Hg solubility, but that the Hg-cysteine complex is more available than other forms. This is consistent with findings on Deltaproteobacteria (11, 21).
FIG 4 .
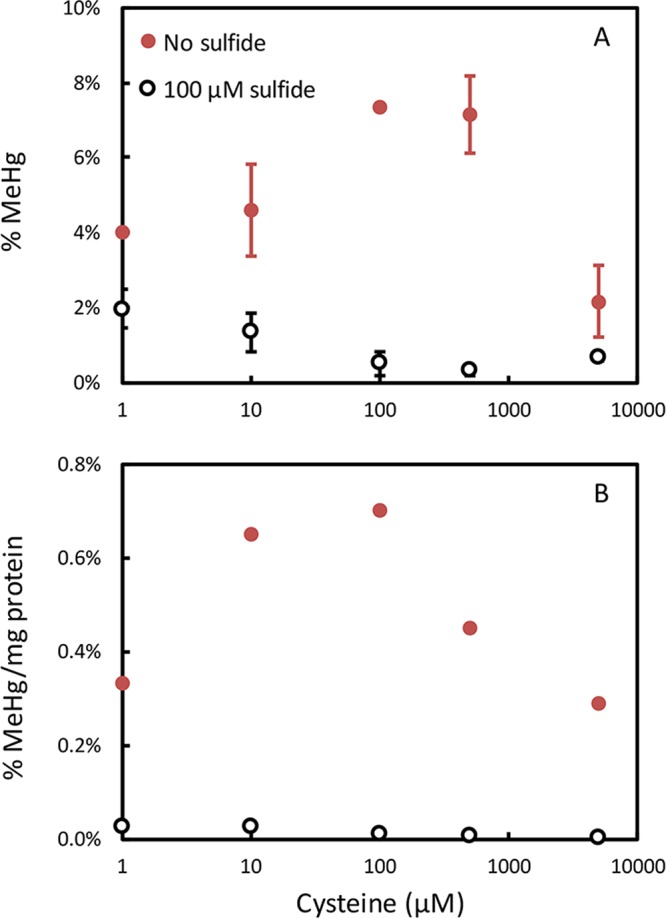
Impact of cysteine on MeHg production by M. tindarius in medium without (red) and with (black) 100 μM sulfide and 500 μM methionine. (Top) MeHg as a percentage of the total Hg in the culture medium. (Bottom) MeHg production normalized to both the measured Hg and protein concentrations at the end of methylation assays, as (MeHg/Hg)/(mg of protein/liter). All measurements were made once all cultures reached stationary phase. Error bars are standard deviations of triplicate cultures. Growth data are in Fig. S1.
Impact of cysteine on M. tindarius growth and MeHg production. Download FIG S1, PDF file, 0.05 MB (52.9KB, pdf) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Influence of sulfide on Hg methylation by methanogens.
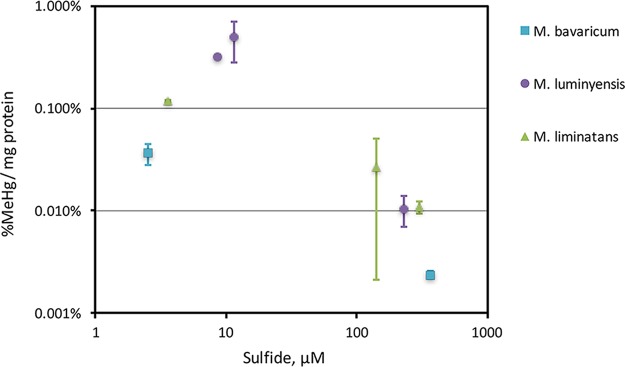
Sulfide reduces the bioavailability of Hg to Hg-methylating Deltaproteobacteria via precipitation of mercuric sulfides that are less available for uptake (13–15). Sulfide also inhibited MeHg production by three hgcAB+ methanogens grown in medium with sulfide additions between 10 and 1,000 μM (Fig. 5). Sulfide additions resulted in loss of Hg from medium to either bottle walls or reduction (see Fig. S1 for an example). Even so, sulfide reduced MeHg production normalized to the final measured Hg concentration in the culture medium.
FIG 5 .
Impact of sulfide on MeHg production by three methanogens based on values measured at the end of log growth in triplicate batch cultures. Sulfide was added to all cultures (between 10 and 1,000 μM); the sulfide concentration shown was measured at the end of the log phase of growth. MeHg production is normalized to both the measured Hg and protein concentrations at the end of methylation assays, as (MeHg/Hg)/(mg of protein/liter). All cultures were grown with 4 mM cysteine. Error bars are standard deviations of triplicate cultures. All measurements were made once all cultures reached stationary phase.
DISCUSSION
Newly identified Hg-methylating methanogens.
This is the first confirmation of methylation ability in culture of hgcAB+ Methanocella paludicola SANAE, Methanocorpusculum bavaricum, Methanofollis liminatans GKZPZ, and Methanosphaerula palustris E1-9c. Methanocella paludicola SANAE is the first confirmed methylator in the order Methanocellales. We identified a second organism with a fused hgcAB gene pair that is unable to produce MeHg under the conditions tested, Methanococcoides methylutens; the first was Pyrococcus furiosus (2).
Rarity of hgcAB among methanogens.
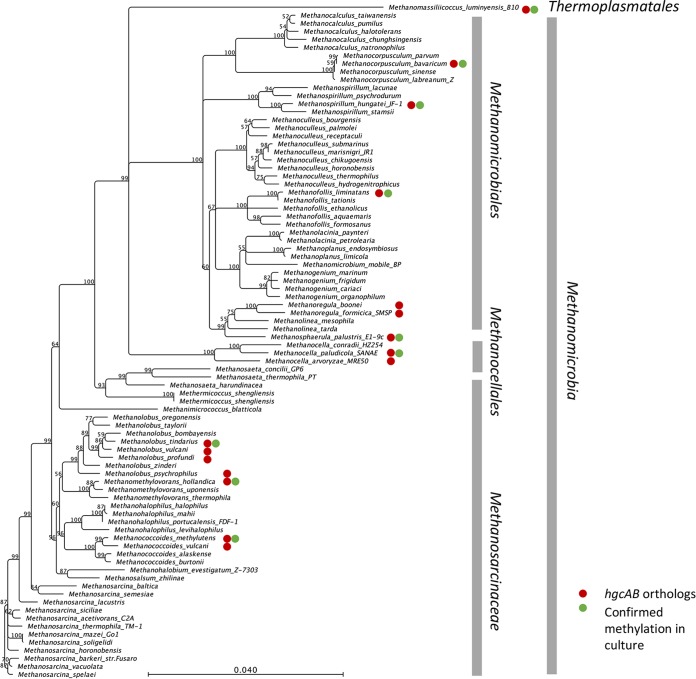
To date, hgcAB orthologs have been identified in only 18 sequenced Euryarchaeota genomes, among several dozen sequenced genomes (Fig. 6). Methylation in culture has been confirmed for eight of those. All but two of the identified Hg methylators are in one class, Methanomicrobia (Methanomassiliicoccus has tentatively been placed in the class Thermoplasmata [38]). There are no identified hgcAB+ organisms in the other four classes of methanogens, including the Methanobacteria/Methanococci superclass, Methanopyri, or the newly identified WSA2 class (“Candidatus Methanofastidiosa”) (39). Among the members of the class Methanomicrobia, most of the hgcAB+ strains are in the two larger orders Methanomicrobiales and Methanosarcinaceae. Hg methylators are most common in the genera Methanolobus, Methanoregula, and Methanocella but, aside from these small clusters, are scattered throughout a 16S rRNA gene-based phylogeny. In the two methanogenic orders that contain hgcAB+ organisms, Methanomicrobia and Thermoplasmata, there are currently about 85 sequenced genomes. About 20% of those species are hgcAB+.
FIG 6 .
16S rRNA gene distance phylogeny of the Methanomicrobia/Thermoplasmata superclass (excluding the Halobacteria) within the methanogenic Euryarchaeota, showing all of the available sequenced genomes. Red circles show organisms with hgcAB orthologs. Green circles are organisms confirmed to methylate Hg in culture. Branch points supported with bootstrap values of ≥50% are shown. The scale bar indicates 0.04 substitution per nucleotide position.
In comparison, hgcAB orthologs are found in 5 of the 10 orders of Deltaproteobacteria (Desulfarculales, Desulfobacterales, Desulfovibrionales, Desulfuromonadales, and Syntrophobacterales). Overall, there are ~110 hgcAB+ species among the >1,000 identified species in these five orders, or roughly 10% of the species identified. Known Hg methylators are concentrated in the genus Desulfovibrio in the order Desulfovibrionales (26 species, or about 25% of the named species) and in the genus Geobacter in the order Desulfuromonadales (19 species). Almost all of the members of the genus Geobacter whose genomes have been sequenced contain hgcAB. In the large order Desulfobacterales, there are 22 identified species with hgcAB orthologs (Hg methylation has been demonstrated in 7 of them) spread among several genera and >250 identified species.
Carbon metabolism of Hg-methylating methanogens.
Substrates for methanogenesis include H2-CO2, acetate, and methylated compounds. Among the methanogens whose genomes have been sequenced, Hg methylators were found among the methylotrophic and hydrogenotrophic methanogens but not among the small group of described aceticlastic methanogens. None of the hydrogenotrophic Hg methylators are class I methanogens (40). This distribution was also observed in a broad query of available microbial metagenomes (2).
Several methylotrophic hgcAB+ methanogens were identified in the order Methanosarcinales, the only methanogens that contain cytochromes and can metabolize methylated compounds via a membrane-bound electron transport chain. Most of the Hg-methylating members of the order Methanosarcinales are Methanolobus species, which are all methylotrophic and somewhat salt tolerant. Substrates for some of the species include methanol (MeOH), methylamines, and methyl sulfides. We tested Hg methylation by Methanolobus tindarius growing on MeOH under a N2-CO2 atmosphere. The other hgcAB+ Methanosarcinales member is M. hollandica, which is also obligately methylotrophic. We tested its methylation ability on trimethylamine-MeOH under a N2-CO2 atmosphere.
Other methylotrophic methanogens are obligately H2 dependent. Methanomassiliicoccus is a recently identified genus of obligately methylotrophic and H2-dependent methanogens isolated from the intestinal tracts of humans, ruminants, and termites; anaerobic digesters; and soils. Along with Methanoplasma, Methanogranum, and Methanomethylophilus, these organisms have been classified as a new, seventh order of methanogens, Methanomassiliicoccales, in the class Thermoplasmatales, phylogenetically distant from other methanogenic orders (41). The order lacks the pathway for CO2 reduction to methyl coenzyme M and produces methane by H2-dependent reduction of MeOH or methylamines. The two known species of Methanomassiliicoccus both contain hgcAB. Methanomassiliicoccus luminyensis B10 produced MeHg in cultures growing on MeOH supplemented with rumen fluid (2).
The remaining hgcAB+ methanogens are obligately hydrogenotrophic organisms in the orders Methanomicrobiales and Methanocellales. The order Methanomicrobiales is a highly polyphyletic group of hydrogenotrophic methanogens, but with a narrow substrate range, generally utilizing only CO2, with either formate or H2 as the reducing agent. We identified eight species with hgcAB orthologs in four families. Most were in the family Methanoregulaceae, formerly the E1/E2 and R10 groups or “fen cluster” phylotypes (42). Five Methanoregulaceae species in three genera, Methanoregula, Methanosphaerula, and Methanolinea, have been sequenced (42). Neither of the two Methanolinea strains contain hgcAB, but both Methanoregula species (M. boonei and M. formicica) and the single Methanosphaerula species (M. palustris) do. Both M. boonei and M. palustris were isolated from oligotrophic peat environments (43, 44). M. formicica was isolated from an anaerobic sludge reactor treating brewery effluent (45). The unclassified Methanoregulaceae archaeon JGI M3C4D3-001-G22, whose genome has recently been sequenced, is also hgcAB+. Hg methylation was tested in Methanosphaerula palustris E1-9c cultures growing in H2-CO2 supplemented with acetate.
Two other methylators in the order Methanomicrobiales were identified. Methanospirillum hungatei is the only organism in the family Methanospirillaceae whose genome has been sequenced. Various phylogenies place M. hungatei within or just outside Methanoregula (46). Isolated from sewage sludge, it was one of the earliest methanogens characterized (47) and the first confirmed Hg-methylating methanogen (5). We grew this organism in H2-CO2 supplemented with acetate and formate for Hg methylation testing. Methanocorpusculum bavaricum (family Methanocorpusculaceae) was isolated from a sugar wastewater pond. In addition to H2-CO2 methanogenesis, it can utilize secondary alcohols as electron donors to reduce CO2 and requires rumen fluid in culture. To test Hg methylation, this organism was cultured under H2-CO2 in medium supplemented with acetate, formate, rumen fluid, and fatty acids.
To summarize, the hgcAB gene pair appears to be confined to two classes of methanogens, Methanomicrobia and Thermoplasmata, but is found in organisms with a wide variety of pathways for methanogenesis. Methanosarcina may metabolize one-carbon compounds like MeOH, acetate, and methylamines via a membrane-bound electron transport chain. Other methylotrophic methanogens containing hgcAB require H2. Our methylation assays of several previously untested hgcAB+ Methanomicrobia species lead us to believe that these methanogens should be considered equally with sulfate and iron reducers in evaluations of potential Hg-methylating biogeochemical conditions in nature. Our understanding of the distribution and activity of hgcAB+ organisms in nature and of the controls on hgcAB expression remains in its infancy.
MATERIALS AND METHODS
Methanogen strains evaluated.
The methanogen strains used in this study are listed in Table S1 along with the sources of the cultures and the growth media used. Species purity was checked through 16S rRNA gene nucleotide sequencing.
Hg methylation assays.
MeHg production was evaluated by measuring the production of Me201Hg from a 1 or 10 nM inorganic >98% enriched 201Hg spike during triplicate batch culture growth (4). The culture media and growth conditions used for methylation assays are described in Table S2. Several cultures were tested under a range of added thiol and sulfide levels. Controls included fresh and spent culture media (filtrate of mature cultures) under matched redox conditions. Spent medium was prepared by anaerobic filtration of late-log- or stationary-phase cultures. Organisms without hgcAB were used as negative-control strains. All assays and controls were conducted in separate, triplicate bottles. Preparation of medium and spent-medium controls was done under O2-free N2 or other reduced headspace gases, as appropriate for each culture. Several cultures were tested multiple times (Data Set S1).
Culture media and growth conditions used in this study. Download TABLE S2, PDF file, 0.1 MB (67.2KB, pdf) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
During batch culture growth and Hg methylation assays, cell density, protein concentration, sulfide, and headspace methane were measured to assess growth and monitor redox and medium chemistry (4). Some culture media contained added sulfide; in other media, cells produced sulfide from added cysteine. Culture media were amended with additional cysteine, methionine, or sulfide as noted for methylation assays. For most assays, medium recipes were amended with 500 μM cysteine. The Hg-Cys complex is highly bioavailable to Hg-methylating Deltaproteobacteria (11, 12, 21).
Methylation was calculated as the concentration of excess Me201Hg in unfiltered culture medium at the end of log-phase growth divided by the measured concentration of excess total 201Hg measured in the same sample. This provides a maximal estimate of methylation, given the loss of Hg(II) from culture medium to bottle walls and potentially to reduction during growth. Cultures were considered to be methylators when they produced significantly more MeHg than matched fresh- and spent-medium controls. Since small amounts of MeHg can be produced chemically (48), it is important to assess MeHg production in appropriate controls.
Hg and MeHg analysis.
All measurements were made by isotope dilution inductively coupled plasma mass spectrometry (ICP-MS) as described previously (4). Briefly, the total Hg in filtered or unfiltered samples was measured after digestion, SnCl2 reduction, purging, and trapping. MeHg was determined following aqueous phase distillation and ethylation. Me199Hg isotope dilution standards were synthesized in house from 199HgCl2 (Oak Ridge National Laboratory; 91.95% enriched) (49). All measurements were made with Brooks Rand MERX automated systems (Brooks Rand Instruments, Seattle, WA) interfaced with a PerkinElmer Elan DRC II ICP-MS apparatus (PerkinElmer Inc., Shelton, CT). The detection limit for Me201Hg and total 201Hg averaged 0.9 and 27 pM, respectively, in the dilutions used for analysis. The average MeHg percentage in controls was 0.7% of the aqueous 201Hg.
16S rRNA gene phylogeny.
A distance scaffold and supporting multiple-sequence alignment were constructed with CLC Sequence Viewer 7 (Qiagen) from 16S rRNA gene sequences obtained from GenBank and JGI. The phylogeny used a neighbor-joining algorithm with Jukes-Cantor nucleotide distance measurement and 1,000 bootstrap runs.
Genome accession numbers, phylogeny, culture source, and references for original isolation of the methanogens used in this study. Download TABLE S3, PDF file, 0.1 MB (77.5KB, pdf) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
This research was sponsored by the Office of Biological and Environmental Research, Office of Science of the U.S. Department of Energy (DOE), as part of the Mercury Science Focus Area at Oak Ridge National Laboratory, which is managed by UT-Battelle LLC for the DOE under contract DE-AC05-00OR22725, and by NIEHS R01ES024284-01 to Upal Ghosh (University of Maryland Baltimore County), C.C.G., and D.A.E.
We thank S. Sakai (JAMSTEC) for the gift of Methanocella paludicola SANAE, H. Cadillo-Quiroz (Arizona State University) for Methanosphaerula palustris E1-9c and Methanospirillum hungatei JF-1, and Kevin Sowers (University of Maryland Institute of Marine and Environmental Technology) for Methanococcoides methylutens.
C.C.G., D.A.E., and M.P. designed the research. A.L.B. and A.M. performed the experiments. C.G. wrote the paper with input from D.A.E., M.P., and A.L.B. All of us read and approved the paper.
Footnotes
Citation Gilmour CC, Bullock AL, McBurney A, Podar M, Elias DA. 2018. Robust mercury methylation across diverse methanogenic Archaea. mBio 9:e02403-17. https://doi.org/10.1128/mBio.02403-17.
REFERENCES
- 1.Rothenberg SE, Anders M, Ajami NJ, Petrosino JF, Balogh E. 2016. Water management impacts rice methylmercury and the soil microbiome. Sci Total Environ 572:608–617. doi: 10.1016/j.scitotenv.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Podar M, Gilmour CC, Brandt CC, Soren A, Brown SD, Crable BR, Palumbo AV, Somenahally AC, Elias DA. 2015. Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci Adv 1:e1500675. doi: 10.1126/sciadv.1500675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parks JM, Johs A, Podar M, Bridou R, Hurt RA Jr, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, Palumbo AV, Smith JC, Wall JD, Elias DA, Liang L. 2013. The genetic basis for bacterial mercury methylation. Science 339:1332–1335. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 4.Gilmour CC, Podar M, Bullock AL, Graham AM, Brown SD, Somenahally AC, Johs A, Hurt RA Jr, Bailey KL, Elias DA. 2013. Mercury methylation by novel microorganisms from new environments. Environ Sci Technol 47:11810–11820. doi: 10.1021/es403075t. [DOI] [PubMed] [Google Scholar]
- 5.Yu RQ, Reinfelder JR, Hines ME, Barkay T. 2013. Mercury methylation by the methanogen Methanospirillum hungatei. Appl Environ Microbiol 79:6325–6330. doi: 10.1128/AEM.01556-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avramescu ML, Yumvihoze E, Hintelmann H, Ridal J, Fortin D, Lean DRS. 2011. Biogeochemical factors influencing net mercury methylation in contaminated freshwater sediments from the St. Lawrence River in Cornwall, Ontario, Canada. Sci Total Environ 409:968–978. doi: 10.1016/j.scitotenv.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Correia RRS, Miranda MR, Guimarães JRD. 2012. Mercury methylation and the microbial consortium in periphyton of tropical macrophytes: effect of different inhibitors. Environ Res 112:86–91. doi: 10.1016/j.envres.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Hamelin S, Amyot M, Barkay T, Wang YP, Planas D. 2011. Methanogens: principal methylators of mercury in lake periphyton. Environ Sci Technol 45:7693–7700. doi: 10.1021/es2010072. [DOI] [PubMed] [Google Scholar]
- 9.Christensen GA, Wymore AM, King AJ, Podar M, Hurt RA, Santillan EU, Soren A, Brandt CC, Brown SD, Palumbo AV, Wall JD, Gilmour CC, Elias DA. 2016. Development and validation of broad-range qualitative and clade-specific quantitative molecular probes for assessing mercury methylation in the environment. Appl Environ Microbiol 82:6068–6078. doi: 10.1128/AEM.01271-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen GA, Somenahally AC, Moberly JG, Miller CM, King AJ, Gilmour CC, Brown SD, Podar M, Brandt CC, Brooks SC, Palumbo AV, Wall JD, Elias DA. 2018. Carbon amendments alter microbial community structure and net mercury methylation potential in sediments. Appl Environ Microbiol 84:e01049-17. doi: 10.1128/AEM.01049-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaefer JK, Morel FMM. 2009. High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens. Nat Geosci 2:123–126. doi: 10.1038/ngeo412. [DOI] [Google Scholar]
- 12.Gilmour CC, Elias DA, Kucken AM, Brown SD, Palumbo AV, Schadt CW, Wall JD. 2011. Sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Appl Environ Microbiol 77:3938–3951. doi: 10.1128/AEM.02993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T, Kim B, Levard C, Reinsch BC, Lowry GV, Deshusses MA, Hsu-Kim H. 2012. Methylation of mercury by bacteria exposed to dissolved, nanoparticulate, and microparticulate mercuric sulfides. Environ Sci Technol 46:6950–6958. doi: 10.1021/es203181m. [DOI] [PubMed] [Google Scholar]
- 14.Graham AM, Aiken GR, Gilmour CC. 2012. Dissolved organic matter enhances microbial mercury methylation under sulfidic conditions. Environ Sci Technol 46:2715–2723. doi: 10.1021/es203658f. [DOI] [PubMed] [Google Scholar]
- 15.Benoit JM, Gilmour CC, Mason RP. 2001. The influence of sulfide on solid phase mercury bioavailability for methylation by pure cultures of Desulfobulbus propionicus (1pr3). Environ Sci Technol 35:127–132. doi: 10.1021/es001415n. [DOI] [PubMed] [Google Scholar]
- 16.Ravichandran M, Aiken GR, Ryan JN, Reddy MM. 1999. Inhibition of precipitation and aggregation of metacinnabar (mercuric sulfide) by dissolved organic matter isolated from the Florida Everglades. Environ Sci Technol 33:1418–1423. doi: 10.1021/es9811187. [DOI] [Google Scholar]
- 17.Deonarine A, Hsu-Kim H. 2009. Precipitation of mercuric sulfide nanoparticles in NOM-containing water: implications for the natural environment. Environ Sci Technol 43:2368–2373. doi: 10.1021/es803130h. [DOI] [PubMed] [Google Scholar]
- 18.Slowey AJ. 2010. Rate of formation and dissolution of mercury sulfide nanoparticles: the dual role of natural organic matter. Geochim Cosmochim Acta 74:4693–4708. doi: 10.1016/j.gca.2010.05.012. [DOI] [Google Scholar]
- 19.Graham AM, Aiken GR, Gilmour CC. 2013. Effect of dissolved organic matter source and character on microbial hg methylation in Hg-S-DOM solutions. Environ Sci Technol 47:5746–5754. doi: 10.1021/es400414a. [DOI] [PubMed] [Google Scholar]
- 20.Graham AM, Cameron-Burr K, Hajic H, Lee C, Msekela D, Gilmour CC. 2017. Sulfurization of dissolved organic matter increases hg-sulfide-DOM bioavailability to a Hg-methylating bacterium. Environ Sci Technol 51:9080–9088. doi: 10.1021/acs.est.7b02781. [DOI] [PubMed] [Google Scholar]
- 21.Graham AM, Bullock AL, Maizel AC, Elias DA, Gilmour CC. 2012. Detailed assessment of the kinetics of Hg-cell association, Hg methylation, and methylmercury degradation in several Desulfovibrio species. Appl Environ Microbiol 78:7337–7346. doi: 10.1128/AEM.01792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malcolm EG, Schaefer JK, Ekstrom EB, Tuit CB, Jayakumar A, Park H, Ward BB, Morel FMM. 2010. Mercury methylation in oxygen deficient zones of the oceans: no evidence for the predominance of anaerobes. Mar Chem 122:11–19. doi: 10.1016/j.marchem.2010.08.004. [DOI] [Google Scholar]
- 23.King JK, Kostka JE, Frischer ME, Saunders FM. 2000. Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl Environ Microbiol 66:2430–2437. doi: 10.1128/AEM.66.6.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekstrom EB, Morel FMM, Benoit JM. 2003. Mercury methylation independent of the acetyl-coenzyme A pathway in sulfate-reducing bacteria. Appl Environ Microbiol 69:5414–5422. doi: 10.1128/AEM.69.9.5414-5422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benoit JM, Gilmour CC, Mason RP. 2001. Aspects of bioavailability of mercury for methylation in pure cultures of Desulfobulbus propionicus (1pr3). Appl Environ Microbiol 67:51–58. doi: 10.1128/AEM.67.1.51-58.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benoit JM, Gilmour CC, Mason RP, Heyes A. 1999. Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediment pore waters. Environ Sci Technol 33:951–957. doi: 10.1021/es9808200. [DOI] [Google Scholar]
- 27.Rodríguez-González P, Epov VN, Bridou R, Tessier E, Guyoneaud R, Monperrus M, Amouroux D. 2009. Species-specific stable isotope fractionation of mercury during Hg(II) methylation by an anaerobic bacteria [sic] (Desulfobulbus propionicus) under dark conditions. Environ Sci Technol 43:9183–9188. doi: 10.1021/es902206j. [DOI] [PubMed] [Google Scholar]
- 28.Bridou R, Monperrus M, Gonzalez PR, Guyoneaud R, Amouroux D. 2011. Simultaneous determination of mercury methylation and demethylation capacities of various sulfate-reducing bacteria using species-specific isotopic tracers. Environ Toxicol Chem 30:337–344. doi: 10.1002/etc.395. [DOI] [PubMed] [Google Scholar]
- 29.Ranchou-Peyruse M, Monperrus M, Bridou R, Duran R, Amouroux D, Salvado JC, Guyoneaud R. 2009. Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: implications for environmental studies. Geomicrobiol J 26:1–8. doi: 10.1080/01490450802599227. [DOI] [Google Scholar]
- 30.Moberly JG, Miller CL, Brown SD, Biswas A, Brandt CC, Palumbo AV, Elias DA. 2012. Role of morphological growth state and gene expression in Desulfovibrio africanus strain Walvis Bay mercury methylation. Environ Sci Technol 46:4926–4932. doi: 10.1021/es3000933. [DOI] [PubMed] [Google Scholar]
- 31.Compeau GC, Bartha R. 1985. Sulfate-reducing bacteria—principal methylators of mercury in anoxic estuarine sediment. Appl Environ Microbiol 50:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin CC, Jay JA. 2007. Mercury methylation by planktonic and biofilm cultures of Desulfovibrio desulfuricans. Environ Sci Technol 41:6691–6697. doi: 10.1021/es062304c. [DOI] [PubMed] [Google Scholar]
- 33.Jay JA, Murray KJ, Gilmour CC, Mason RP, Morel FMM, Roberts AL, Hemond HF. 2002. Mercury methylation by Desulfovibrio desulfuricans ND132 in the presence of polysulfides. Appl Environ Microbiol 68:5741–5745. doi: 10.1128/AEM.68.11.5741-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerin EJ, Gilmour CC, Roden E, Suzuki MT, Coates JD, Mason RP. 2006. Mercury methylation by dissimilatory iron-reducing bacteria. Appl Environ Microbiol 72:7919–7921. doi: 10.1128/AEM.01602-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleming EJ, Mack EE, Green PG, Nelson DC. 2006. Mercury methylation from unexpected sources: molybdate-inhibited freshwater sediments and an iron-reducing bacterium. Appl Environ Microbiol 72:457–464. doi: 10.1128/AEM.72.1.457-464.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssen SE, Schaefer JK, Barkay T, Reinfelder JR. 2016. Fractionation of mercury stable isotopes during microbial methylmercury production by iron- and sulfate-reducing bacteria. Environ Sci Technol 50:8077–8083. doi: 10.1021/acs.est.6b00854. [DOI] [PubMed] [Google Scholar]
- 37.Kaschak E, Knopf B, Petersen JH, Bings NH, König H. 2014. Biotic methylation of mercury by intestinal and sulfate-reducing bacteria and their potential role in mercury accumulation in the tissue of the soil-living Eisenia foetida. Soil Biol Biochem 69:202–211. doi: 10.1016/j.soilbio.2013.11.004. [DOI] [Google Scholar]
- 38.Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. 2012. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62:1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- 39.Nobu MK, Narihiro T, Kuroda K, Mei R, Liu WT. 2016. Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J 10:2478–2487. doi: 10.1038/ismej.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson I, Ulrich LE, Lupa B, Susanti D, Porat I, Hooper SD, Lykidis A, Sieprawska-Lupa M, Dharmarajan L, Goltsman E, Lapidus A, Saunders E, Han C, Land M, Lucas S, Mukhopadhyay B, Whitman WB, Woese C, Bristow J, Kyrpides N. 2009. Genomic characterization of Methanomicrobiales reveals three classes of methanogens. PLoS One 4:e5797. doi: 10.1371/journal.pone.0005797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borrel G, Parisot N, Harris HM, Peyretaillade E, Gaci N, Tottey W, Bardot O, Raymann K, Gribaldo S, Peyret P, O’Toole PW, Brugère JF. 2014. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 15:679. doi: 10.1186/1471-2164-15-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imachi H, Sakai S. 2016. Methanoregulaceae, p 1–4. In Bergey DH, Harrison FC, Breed RS, Hammer BW, Huntoon FM (ed), Bergey’s manual of systematics of Archaea and Bacteria. Wiley, New York, NY. [Google Scholar]
- 43.Bräuer SL, Cadillo-Quiroz H, Ward RJ, Yavitt JB, Zinder SH. 2011. Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog. Int J Syst Evol Microbiol 61:45–52. doi: 10.1099/ijs.0.021782-0. [DOI] [PubMed] [Google Scholar]
- 44.Cadillo-Quiroz H, Yavitt JB, Zinder SH. 2009. Methanosphaerula palustris gen. nov., sp. nov., a hydrogenotrophic methanogen isolated from a minerotrophic fen peatland. Int J Syst Evol Microbiol 59:928–935. doi: 10.1099/ijs.0.006890-0. [DOI] [PubMed] [Google Scholar]
- 45.Yashiro Y, Sakai S, Ehara M, Miyazaki M, Yamaguchi T, Imachi H. 2011. Methanoregula formicica sp. nov., a methane-producing archaeon isolated from methanogenic sludge. Int J Syst Evol Microbiol 61:53–59. doi: 10.1099/ijs.0.014811-0. [DOI] [PubMed] [Google Scholar]
- 46.Browne P, Tamaki H, Kyrpides N, Woyke T, Goodwin L, Imachi H, Bräuer S, Yavitt JB, Liu WT, Zinder S, Cadillo-Quiroz H. 2017. Genomic composition and dynamics among Methanomicrobiales predict adaptation to contrasting environments. ISME J 11:87–99. doi: 10.1038/ismej.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferry JG, Smith PH, Wolfe R. 1974. Methanospirillum, a new genus of methanogenic bacteria, and characterization of Methanospirillum hungatii sp. nov. Int J Syst Evol Microbiol 24:465–469. [Google Scholar]
- 48.Bloom NS, Colman JA, Barber L. 1997. Artifact formation of methyl mercury during aqueous distillation and alternative techniques for the extraction of methyl mercury from environmental samples. Fresenius J Anal Chem 358:371–377. doi: 10.1007/s002160050432. [DOI] [Google Scholar]
- 49.Bancon-Montigny C, Yang L, Sturgeon RE, Colombini V, Mester Z. 2004. High-yield synthesis of milligram amounts of isotopically enriched methylmercury (CH3198HgCl). Appl Organomet Chem 18:57–64. doi: 10.1002/aoc.572. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data on the cultures and controls used in this study. Download DATA SET S1, XLSX file, 0.03 MB (29.2KB, xlsx) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Methanogen strains used in this study. Download TABLE S1, PDF file, 0.04 MB (39.6KB, pdf) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Impact of cysteine on M. tindarius growth and MeHg production. Download FIG S1, PDF file, 0.05 MB (52.9KB, pdf) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Culture media and growth conditions used in this study. Download TABLE S2, PDF file, 0.1 MB (67.2KB, pdf) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome accession numbers, phylogeny, culture source, and references for original isolation of the methanogens used in this study. Download TABLE S3, PDF file, 0.1 MB (77.5KB, pdf) .
Copyright © 2018 Gilmour et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.