Abstract
Detailed observations of transcriptional, translational, and post-translational events in the human brain are essential to improving our understanding of its development, function, and vulnerability to disease. Here, we exploited label-free quantitative tandem mass-spectrometry proteomics to create an in-depth proteomic survey of adult human brain regions. Integration of protein data with existing whole-transcriptome sequencing (RNA-seq) from the BrainSpan project revealed varied patterns of protein:RNA relationships with generally increased magnitudes of protein abundance differences between brain regions compared to RNA. Many of the differences amplified in protein data were reflective of cyto-architectural and functional variation between brain regions. Comparing structurally similar cortical regions revealed significant differences in the abundance of receptor-associated and resident plasma membrane proteins that were not readily observed in the RNA expression data.
Introduction
Work from converging fields is beginning to disentangle the immense complexity of the mammalian brain1, from the mapping of the connectome2–6 to ever deeper molecular characterisation of brain regions7–10 and cell-types11–15. Extensive maps of RNA abundance across the mammalian brain have been available since 201116. The focus of projects such as BrainSpan16, PsychENCODE17, and organisations such as the Allen Brain Institute (www.brain-map.org) is to increase the depth of these datasets, cataloguing cell types, coding and non-coding RNA expression, and epigenetic modifications17. Technological advances in microfluidics and nucleic acid sequencing have led to single cell and single cell type transcriptome analyses of increasingly high quality11–13,18. Automation of electron microscopy is providing unprecedented resolution with which to survey the connections of neurons in fixed tissue2, while optogenetics allows us to define functionally important circuits in rodent models6. Despite this huge increase in our collective ability to interrogate the mammalian nervous system, comprehensive protein-level data, particularly in human brains, is a notable exception. Given the likely poor correlation between mRNA and protein abundance19,20, and that protein levels are closer to the biosynthetic output of the cell21,22, a systematic survey of the human brain proteome is vital. Here we present a detailed proteomic analysis of seven major regions of the human brain across postnatal development from early infancy through adulthood.
Tandem liquid chromatography mass spectrometry (LC-MS/MS) is the gold standard method for obtaining unbiased, high-resolution proteomic data22,23. Current technology makes it possible to obtain quantitative observations of peptides derived from several thousand proteins per sample24–26. However, the depth of mass-spectrometry is limited by the complexity of the sample being investigated. In heterogeneous brain tissue, sensitivity to low abundance proteins is likely decreased due to the presence of a large number of spectra from high abundance proteins, such as actin or tubulin22. This effect is magnified when highly diverse samples are combined in an isobaric labelling experiment27,28. For this reason, we adopted a dual approach to analysing human brain samples similar to that of a recent high-quality study of the mouse brain proteome29. First was a discovery phase, designed to create a highly sensitive, heavily fractionated spectral library for each adult brain region. Then, to produce a quantitative analysis for each adult and postnatal development sample, ‘single shot’ label free LC-MS/MS runs were used to accurately quantify proteins based on detected precursor peptide ion mass.
To maximise compatibility with existing *omic profiles of the human brain, the post-mortem tissue samples used for quantitative proteomics were exact matches to those already profiled by mRNA-seq in the BrainSpan16 project (www.brainspan.org); thus these matched RNA and protein samples constitute a detailed, deeply integrated, and unique resource. Finally, the human discovery and quantitative proteomic data were compared to the mouse brain proteome29 to create a valuable point of reference for the neuroscience community.
Results
Powdered frozen tissue was obtained from post-natal samples previously profiled by RNA-seq as part of the Brainspan Project30 (Fig 1A). Of the 16 regions profiled in BrainSpan, 7 regions that showed large inter-regional differences in gene expression by RNA-seq were selected: cerebellum (CBC), striatum (STR), mediodorsal thalamic nucleus (MD), amygdala (AMY), hippocampus (HIP), primary visual cortex (V1C), and dorsolateral prefrontal cortex (DFC). Developmental samples spanned the period from early infancy (1 year post conception) to adulthood (42 years)16, and were derived from a near equal mix of male and female donors (Fig 1A; Table S1). Consistent and stringent quality control measures were applied at the level of tissue acquisition and dissection, sample preparation, and data processing (help.brain-map.org/download/attachments/3506181/Transcriptome_Profiling.pdf).
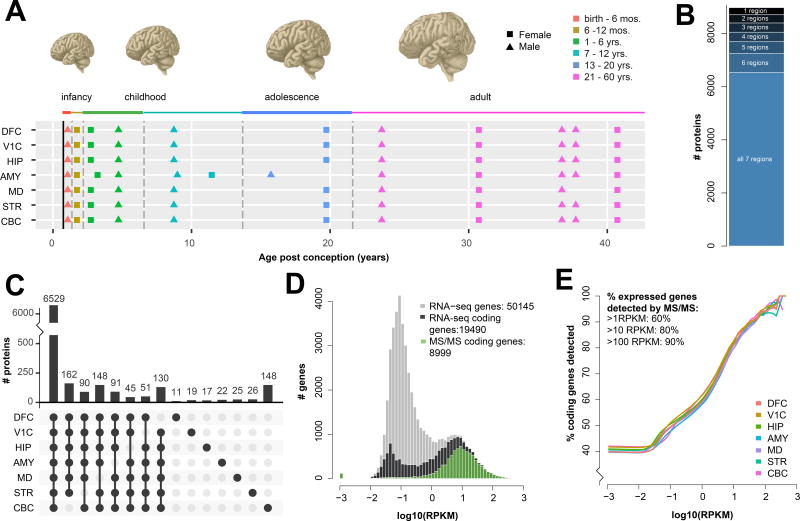
Figure 1. Resource overview and peptide library illustrate broad coverage of both the adult human brain and of expressed genes.
A) Individual brain regions (dorsolateral frontal cortex (DFC), primary visual cortex (V1C), hippocampus (HIP), amygdala (AMY), mediodorsal nucleus of the thalamus (MD), striatum (STR) and cerebellum (CBC)) and ages used in this study; all samples pooled for fractionation were derived from adult brains. B) On average 7,945 proteins were detected in each brain region, with 8,980 proteins detected in at least one region and 6,529 proteins detected in all 7 regions. C) Over two-thirds (6,529 of 8,980) of the proteins detected were consistently identified in all 7 brain regions. Counts of proteins (bars, top) detected in each combination of regions (black dots, below) are shown for all combinations indicating enrichment or depletion in a single region. Region-specific dropouts are far more frequent than region-specific detections; with the exception of 148 CBC-specific proteins, the largest groups are single region depletions that, together, total 717 proteins. D) Using the whole-brain distribution of gene expression (all genes; grey) defined by RNA-seq, peptides (green) correspond to the majority of higher abundance mRNAs (RNA coding genes, black). E) Sliding a lower threshold from the left to right of the histogram for each brain region, a robust pattern of increasing peptide coverage is observed with increasing RNA expression.
Peptide library of seven adult human brain regions
To create a peptide library for each region, we generated 7 region-specific collections of tissue homogenate pooled from adult subjects. Trypsin digested peptides from these pools were separated offline by high pH reverse phase chromatography into 15 fractions prior to online LC-MS/MS. Mass spectra were searched in MaxQuant31 against the Ensembl/Gencode proteome. A total of 111,456 peptides, corresponding to 8,980 proteins, were detected in at least one region, with an average of 75,303 peptides from 7,945 proteins per region (Fig 1B–C, Fig S1, 2, Table S2). On average, we detected over 9 peptides per protein (Fig S2) resulting in a mean-average coverage of 28% of the observed proteins (Table S3A–B). Data from several recent, large scale, proteomic projects suggests that the number of proteins detectable in a single biological sample tends to saturate at approximately 11,00024,25 and is in rough agreement with RNA analyses, which frequently report ~50% of protein-coding genes are expressed in a given tissue or cell-type32. In our data, the observed proteins constituted the majority of coding genes detected by RNA-seq, with a strong bias towards proteins corresponding to more highly expressed RNAs (Fig 1D). Over 60% of genes expressed at greater than 1 read per kilobase per million (RPKM) and over 80% of genes expressed at more than 10 RPKM were detected as proteins (Fig 1E). Peptides were detected for proteins across a wide range of molecular weights, from two 44 amino acid variants of beta thymosin (TMSB10 and TMSB4X) to fragments of TTN, the largest known protein. Analysis of the gene ontology category ‘integral membrane component’ showed no significant enrichment or depletion of these potentially difficult to extract proteins. Taken together, these data indicate a broad and detailed protein library.
Quantitative single shot proteomics of postnatal human brain regions
Single shot, label-free protein quantifications (LFQs) were obtained from the seven brain regions of 16 individuals spanning one year post conception to 42 years. LFQ was used to maximise the number of protein identifications per sample27,28,33, and also for comparability with the recent high quality study of the mouse brain29. As with the fractionated peptide atlas, mass spectral data from the single-shot runs were searched in MaxQuant against the Ensembl/Gencode proteome. To increase the number of protein identifications, single shot data were searched alongside the fractionated samples, using the “match between runs” feature in MaxQuant. A total of 63,478 peptides derived from 7,244 proteins were detected in one or more of the 77 single-shot samples analysed, detecting on average 18,835 peptides from 3,612 proteins per sample. The use of match between runs produced ~50% more protein identifications per sample than would have otherwise been detected (Fig S3). A large fraction (5,151; 71%) of the 7,244 total proteins were reliably quantified by LFQ (Table S4).
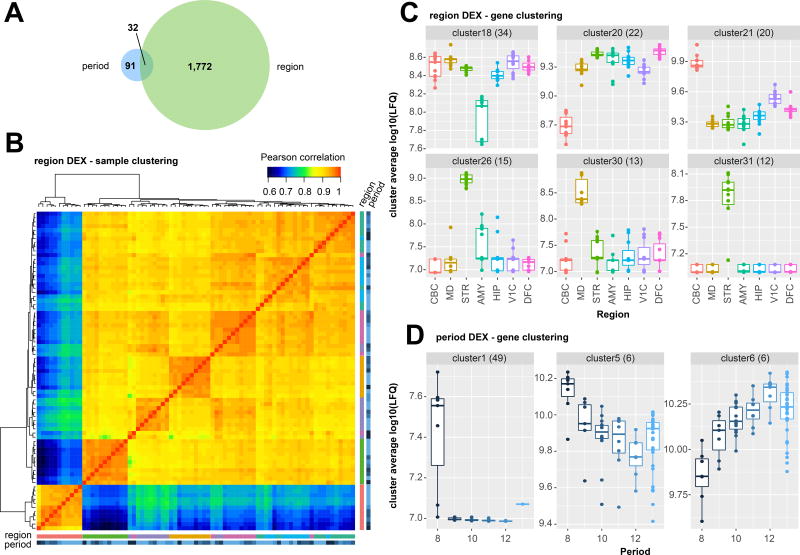
Single shot LFQs were filtered, normalised, and log transformed (see Methods, Fig S4) prior to differential expression (DEX) analysis, jointly modelling protein changes over time and between brain regions by regression and ANOVA. A total of 1,804 proteins were found to be significantly DEX between one or more brain region. Given the limited number of developmental samples available (6 time-points spanning from infancy to adulthood), we had comparatively low power to detect significant time-dependent changes, likely resulting in a small number of proteins significantly DEX across development. We detected 123 proteins significantly DEX across developmental periods (Bonferroni adjusted p-values < 0.05, Fig 2A; Table S5). Using the proteins found to be differentially expressed between brain regions, samples clustered in a predictable manner, with CBC a clear outlier (Fig 2B, Fig S5).
Figure 2. Differentially expressed proteins across human brain regions and post-natal development.
A) The majority of significantly differentially expressed genes were between brain regions, rather than over developmental periods (Two-way ANOVA, significant DEX defined as Bonferroni corrected p value < 0.05. Adjusted p values of n=5141 genes across 7 regions (6 DoF, Fig 1A) 6 timepoints (1 DoF, Fig 1A) in Table S5). B) Clustering all samples subjected to MS/MS using proteins significantly differentially expressed between brain regions revealed expected bulk differences between brain regions. Samples are defined by the same colour scheme used to depict regions and developmental period in Figs 2C and 2D (see also fully labelled zoomable version in Fig S5). Cerebellum (CBC) and Striatum (STR) are clear outliers (lower left), as they are by RNA-seq16, and the remaining samples cluster well by region with the exception of occasional outlying samples derived from the youngest subject, HSB139 (dark blue). C) Clustering all proteins significantly differentially expressed between regions reveals consistent patterns of expression that favour region-specific enrichment or region-specific depletion in abundance (all clusters Fig S6, dendrogram Fig S7). Variable y axes are used to best visualize the inter-regional differences within clusters. The center line indicates the median, limits indicate the interquartile range (IQR), and the whiskers either 1.5* the IQR or the min/max value if it falls within 1.5* the IQR. Each individual data point is shown as a dot. D) Clustering all proteins significantly differentially expressed over developmental period reveals proteins enriched shortly after birth (period 8) and proteins more gently increasing or decreasing in abundance over developmental period. Boxplots are defined the same as Fig 2C.
We performed cluster analysis, where DEX proteins were grouped according to their relative abundance across all single-shot samples. Proteins formed distinct groups that reflected the substantial differences in their abundance between brain regions. Clusters comprising proteins enriched or depleted in the major non-cortical structures, such as the CBC (for example Fig 2C, clusters 20 & 21) and STR (clusters 26 & 31), were the most striking (see Methods, Tables S5A, 5B, 5C, Fig S6, Fig S7). The largest CBC enriched cluster (cluster 7) contained a significant number of proteins annotated as involved in mRNA processing, such as RNPS1, SF3A2, SNRPA1, SRRM1, SRSF10, SRSF11, SRSF6, and SRSF9 (‘RNA processing’, adj. p-value = 9.6E-07, cellular component ‘nucleus’ adj. p-value = 1.85E-06)). This functional enrichment is reflective of many of the CBC enriched clusters, and presumably reflects the much higher density of nuclei in the cerebellar granular layer compared to the other brain regions. Several clusters highlighted the diversity of the other brain regions. For example, clusters 26 & 31 (Fig 2C), contain PDE10A, TH (cluster 26), and CHAT (cluster 31), proteins known to be functionally important in the STR34. This was confirmed by network analysis (STRING35) of clusters 26 & 31 combined, which showed a strong enrichment for established protein-protein interactions (adj. p-value = 1.55E-15) and for KEGG pathways involved in stimulant addiction (‘cocaine addiction’ adj. p-value = 2.55E-06, ‘amphetamine addiction’ adj. p value = 0.0006 Fig S8), and ‘dopaminergic synapse’, (adj. p-value = 0.001), well established processes relevant to the human striatum36.
Quantitative single shot proteomics of brain development
Several clusters of temporally co-expressed proteins highlighted increasing or decreasing abundance over postnatal brain development (Fig 2D, Fig S9, Table S5). A striking number of the time-DEX proteins appeared to be exclusively expressed during early infancy, a time characterised by corticocortical and long afferent axon reorganization, synaptogenesis, and spinogenesis. While there were no statistically enriched gene ontologies in this cluster (Cluster 1, Fig 2D), it included many candidates such as EFNB3, BZW2, CNTN3, CELSR2, NYAP2, TENM4, CSPG4, SEZ6, RAPH1, and LRRTM4, known to function in cell-cell adhesion and downstream signalling, a critical process for appropriate neuronal maturation37,38.
Cluster 6 was a small cluster of 6 proteins that increased in abundance across postnatal development and that may be linked to several neurological disorders. NFASC, an alternatively spliced protein present in cluster 6 and which anchors myelin to axons, is known to be disrupted in Multiple Sclerosis39,40. Decreased gene body methylation of TPPP, another member of cluster 6, has been linked to depression in children subject to early life stress41,42. TPPP may also be found in alpha-synuclein positive protein deposits in Parkinson’s and Lewy body dementia43.
Quantitative spatiotemporal comparison of mRNA and protein abundance
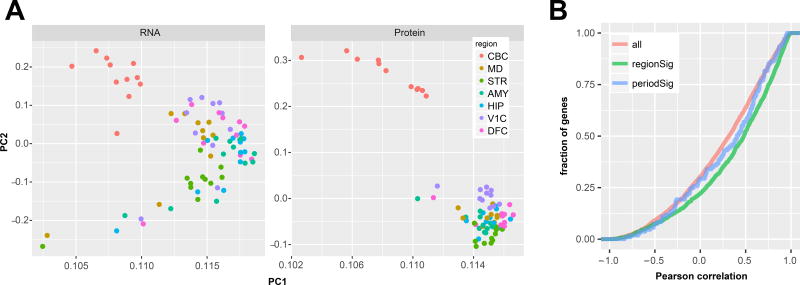
All the brains used in this study have been previously characterised by RNA-seq as part of the BrainSpan project10 (Table S6A,B), allowing the ready integration of gene-level RNA expression data with quantitative proteomic data (5,039 genes in total). This allowed us to disambiguate the protein-level expression of 264 genes (5%) where the peptide mapping was ambiguous (Tables S6A,B; see Methods). Principle component analysis of protein and RNA-level data showed a similar proportion of variance attributed to the first and second components (90.5% protein and 91.2 RNA; Fig 3A). In these components however, the CBC showed a greater separation from the other regions in the protein data, whilst also showing clearer grouping of the other regions compared to RNA. The correlation between absolute mRNA and protein abundance within one sample is typically lower than the correlation between mRNA and protein fold changes. We therefore directly compared interregional fold-changes between RNA and protein across all possible region pairs. This approach produced a median Pearson correlation coefficient of 0.32 (Fig 3B). Genes found to be significantly DEX between brain regions at the protein level were significantly skewed towards a more positive correlation (median correlation coefficient of 0.48; KS p-value < 1E-16; Fig 3B). A similar trend was observed for proteins DEX over the developmental time course (median correlation coefficient of 0.41), but there were too few to achieve significance. Region-level summary expression and differential expression results for both the RNA and protein data are provided in Table S7.
Figure 3. Comparison of the proteome and transcriptome of the human brain.
A) Principal component analysis of RNA and protein data show a clear separation of the cerebellum from the other six regions in the first two PCs in both datasets. Clustering of samples by region is tighter in these components in the protein data compared to RNA. B) The cumulative frequency of Pearson correlations between each gene’s RNA and protein shows a modest median correlation of 0.32 (n=5039). However, when considering only genes significantly differentially expressed at the protein level between brain regions (n=1776), these correlations are significantly increased (median 0.5; K-S pVal < 10−16).
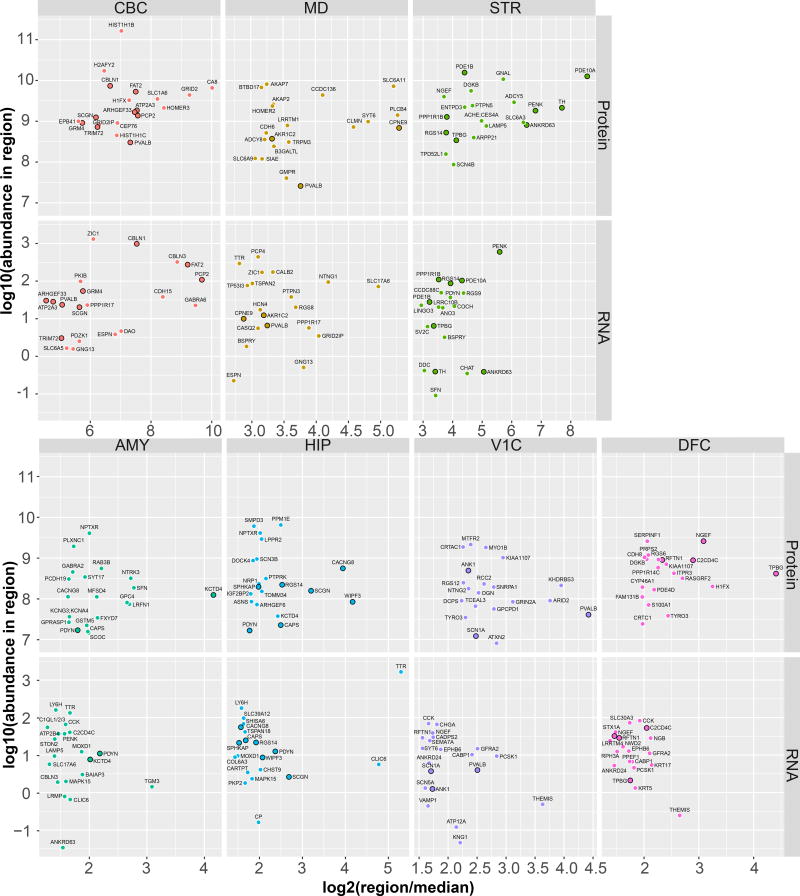
Using the region-averaged protein and RNA expression data, we identified the abundance and relative expression of the 20 most enriched proteins and RNAs in each brain region (Fig 4). In general, the top 20 genes were enriched to a greater degree in CBC or STR compared to neocortical regions by both RNA and protein. Furthermore, the fraction of genes that appeared in the top 20 for both protein and RNA (black outline) was greater in these non-cortical regions (~50% in the CBC and STR) than in neocortical regions (~20% in the V1C and DFC).
Figure 4. Abundance and enrichment of the 20 most enriched proteins and RNAs in each brain region.
Scatter plots of log10 protein and RNA abundance vs log2 fold enrichment (in each region over the median of all other regions) for the top 20 genes enriched in each region. Variable y axes are used to reflect the variation in abundance of the most enriched proteins between regions. Points representing genes enriched in a given region in both RNA and protein are slightly enlarged and highlighted with a black outline. Generally, the abundance and fold-enrichments of these gene products is highest in sub-cortical regions (CBC, STR) compared to neocortical regions (V1C, DFC).
For all possible region pairs, we categorised gene products based on their fold-change similarity between RNA and protein (Fig 5A; Fig S10A–G). Over all region-pairs, the majority of genes (62.5%) showed a significant but not substantial (≤2-fold) inter-regional abundance difference by both RNA and protein (grey, Fig 5A, B, all two region comparisons in Fig S11A), and were considered the ‘no change’ group. A further 17.4% showed larger (>2-fold) region-specific expression differences in the same direction in both RNA and protein (green and purple), and were considered the ‘agree’ and ‘partial agree’ (same direction of change, different magnitude of fold change) groups. However, we observed over twice as many genes ≥2-fold DEX by protein (blue ‘protein only’) but not RNA (11.5%) compared to ≥2-fold DEX by RNA (orange, ‘RNA only’) but not protein (5.2%). A small fraction (3.3%) of genes disagreed on the direction of change between RNA and protein (red), and these were defined as the ‘not agree’ category. Finally, the differences in a gene’s protein abundance between regions were generally greater than the difference in RNA abundance (Fig S11B).
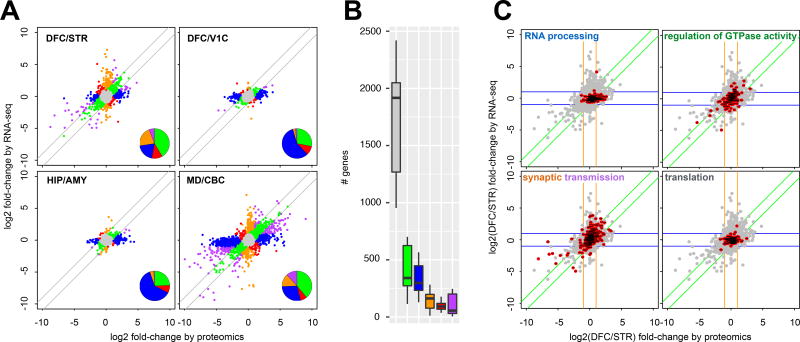
Figure 5. Ontological enrichments of inter-regional protein and RNA changes.
A) RNA and protein abundance differences between pairs of brain regions for genes significantly differentially expressed at the protein level. Genes are coloured based on their agreement or disagreement between the RNA and protein measurements; genes for which the protein variability between regions is <2-fold of that reported at the RNA-level were considered consistent (green and grey points). Purple coloured genes are those with consistent direction but variable magnitude of change (≥2-fold) between the regions at the protein and RNA level, while red genes disagree in the direction of change between RNA and protein. Blue genes vary between regions according to protein but not RNA, orange genes vary by RNA but not protein. Inset pie charts illustrate the relative dominance of genes in the green ‘agree’ and blue ‘protein-only’ categories. B) Distribution of the number of gene products annotated in each of the colour categories defined in A) across all unique pairs of brain regions. The center line indicates the median, limits indicate the IQR, and the whiskers either 1.5* the IQR or the min/max value if it falls within 1.5* the IQR. C) Scatter plots for the DFC/STR comparison show the position of gene products contained within ontological terms of interest. RNA processing genes are enriched by ‘protein-only’. Regulation of GTPase activity shows an enrichment in the ‘agree’ category. Synaptic transmission genes are found in both ‘RNA-only’ and ‘partial-agree’. Genes involved in translation are almost entirely in the ‘no change’ category.
Across all unique pairs of regions, genes found to be consistently differentially expressed (Table S8) by protein-only, i.e. DEX in protein but not RNA, were largely nuclear proteins (cellular component ‘nucleus’, adj. p-value = 8.6E-29, Table S9A,B) and within the enriched nuclear GO terms (Table S9A), there was a bias towards ‘RNA processing’ (adj. p-value = 3.3E-28). The enrichment of these categories of nuclear proteins as the major ‘protein-only’ DEX genes is likely due to their greater stability relative to that of their mRNAs19. While the strongest differences in nuclear proteins were between CBC and other regions, differences clearly also existed between all sub-cortical regions and regions with larger or more numerous nuclei.
Analysis of the genes found to be consistently represented in the ‘RNA-only’ DEX category showed enrichment for ontological terms that describe proteins that may be transported away from the nucleus to the synapse and distal regions of neurons. Thus while the majority of the mRNA in a cell is found in the cell body within the region measured, synaptic proteins are often transported along projections typically into a different region. Enriched GO terms (Table S9A) in the ‘RNA-only’ category include ‘ion transmembrane transport’ (adj. p-value = 3.2E-06), ‘signalling’ (adj. p-value = 5.9E-07), and ‘synaptic transmission’ (adj. p-value = 3E-04). Synaptic transmission was also significantly enriched in the ‘partial agree’ category, where RNA and protein changes occurred in the same direction within a region, but to a different magnitude.
Looking more closely at regional comparisons, DFC and STR are well established as functionally linked regions44, but different with respect to their cyto-architecture and developmental origin. In gene products differentially expressed between these two regions, there was still a protein-only enrichment of nuclear proteins in DFC (Fig 5C, GO term ‘RNA processing’ adj. p-value = 0.03). ‘Synaptic transmission’ was found in the ‘RNA-only’ category, and also in the ‘partial-agree’ group reflecting the distal transport of some of these proteins out of the region containing the cell bodies (Fig 5C, adj. p-values = 2.5E-04, 6.8E-09). In the ‘agree’ category, many GO terms reflected axonal proteins and development (Table S9C,D), although signalling terms such as ‘regulation of GTPase activity’ (Fig 5C, adj. p-value = 3.2E-05) were also found in this category. As in all two region comparisons (Fig 5B), the largest number of gene products in the DFC/STR comparison were found in the ‘no change’ category (grey, Fig S11A). These included proteins with stable mRNA and proteins with conserved functions19 across all cells, such as ‘translation’ (Fig 5C, adj. p-value 1.8E-09) and ‘cellular respiration’ (Fig 5C, adj. p-value = 3.7E-08).
Comparing the two regions in our dataset with the highest developmental and cyto-architectural similarity, the neocortical areas DFC and V1C, we observed much more pronounced protein-level differential expression compared to RNA (254 protein-only DEX genes vs 11 RNA-only; Fig 5A, Fig S12A). The protein-only DEX genes are enriched for protein protein interactions (adj. p-value = 0.0016, Figure S12B) and include CNR1 (CB1 receptor),GRM2 & 3 (mGluR2 & 3 receptors) and GRIA1 (GluR1 receptor). To validate these findings with a different molecular technique, we chose 5 of the proteins from this DFC enriched PPI network that had well established antibodies, and immunoblotted the DFC and V1C samples. Four of these proteins showed clear enrichment in DFC (Fig S12C,D, paired Student’s t-test). NTRK3 was not found to be enriched by immunoblotting.
Integration with mouse protein abundance from the Mouse Brain Proteome project
Given wide ranging interest in the use of mouse models for human disorders, we compared the human protein data with a recent study on mouse brain regions29. No temporal data from whole tissue was available from the mouse; however, 5 brain regions matched those in our study: CBC, STR, thalamus, HIP, and pre-frontal cortex. Of the 16,217 1:1 human-mouse orthologs annotated by Ensembl, 4,052 were detected by LC-MS/MS in both the mouse and human datasets. Mouse and human protein expression showed a similar correlation to that of human RNA/protein (median Pearson correlation = 0.3); however, considering only proteins differentially expressed between human regions, this correlation was significantly increased (median Pearson correlation = 0.65, KS p-value < 10−16; Fig 6A). This generally positive correlation with, presumably, PMI-controlled mouse tissue was encouraging given the potential for non-tryptic protease action during the post-mortem interval in human tissue45.
Figure 6. Comparison of the human and mouse brain proteome.
A) The cumulative frequency of Pearson correlations for each 1:1 ortholog protein between human and mouse shows a median correlation of 0.3 (n=4052). When considering only genes significantly differentially expressed at the protein level between human brain regions (n=1517), these correlations are significantly increased (median 0.65; K-S pVal < 10−16). B) Human and mouse protein abundance differences between two example brain regions, PFC and STR, shows a lower overall degree of consistency between organisms compared to between human RNA and protein (see Fig 5A). As before, genes are coloured based on their agreement or disagreement between human and mouse; genes for which the human variability between regions was <2-fold of that reported for mouse were considered consistent (green and grey points). Purple coloured genes are those with consistent direction, but variable magnitude of change between the regions of human and mouse, while red genes disagree even in the direction of change. Blue and orange genes vary between regions according to human but not mouse and vice-versa. C) Distribution of the number of genes annotated in each of the colour categories defined in B) across all unique pairs of brain regions. The center line indicates the median, limits indicate the IQR, and the whiskers either 1.5* the IQR or the min/max value if it falls within 1.5* the IQR. D) Genes with poor correlation between human and mouse regions tend to have more sequence differences in their coding sequence compared to those genes with greater correlation (red vs. green (p = 0.04) or grey (p-value = 8.97E-06)). Box plots are defined as in 6C, with the individual points representing outliers that fall 1.5* below the IQR.
Fold-changes were computed for the human proteins and their mouse orthologues between the 5 shared regions (Fig 6B DFC/STR; Fig S13 summary counts for all regions, Fig S14A–E scatters of all other regions, Table S10). No substantial region specific differences (≤2-fold) were detected for 47.5% of genes in both human and mouse (Fig 6C, grey). A further 27.2% of genes showed large (>2-fold) region specific expression differences, in the same direction, in both human and mouse protein (green and purple). Around half as many proteins appeared to vary substantially in the mouse but not human (5.6% of genes, orange) compared to those that vary in human but not mouse (11.6%, blue). Compared to the human RNA vs protein comparison, there were more genes that completely disagreed on their direction of change in human (8.1%, red). There was a weak but significant relationship between the conservation of protein sequence between human and mouse and the correlation of the human and mouse inter-regional fold changes (Fig 6D). Those proteins with a fold change greater than 2 in human only (blue) had a significantly lower percent sequence identity than those genes exhibiting no substantial regional difference (grey; p-value 8.97E-06) or those genes in high agreement (green; p-value 0.04) between the two organisms (Fig 6D).
Discussion
This study constitutes the largest human brain proteomic dataset yet completed. We anticipate this resource will be of great use to the neuroscience community to investigate proteins of interest, their expression patterns, and conservation with mouse models. Overall, we directly observed 111,456 peptides, derived from 8,980 proteins and obtained reliable quantifications for 5,151 of these proteins. A large fraction (~35%) of the quantified proteins (1804) were found to be differentially expressed between brain regions with much fewer (123) being identified to change over the postnatal developmental time-course. The small number of significantly DEX proteins between infancy and adult is likely a result of low power. A large proportion of the differential expression was driven by differences between other regions and the CBC. There were also very clear STR enriched clusters of genes, which were reflected in the strikingly robust sample clustering of CBC and STR. We observed fewer developmental changes, which is likely a reflection of the fact that the most dramatic changes observed in gene expression occur at earlier timepoints during prenatal development1,8,16.
Integration of protein data with existing whole-transcriptome sequencing revealed generally increased magnitudes of protein abundance differences between brain regions compared to RNA. While many of these differences reflect cyto-architectural, developmental, and functional differences between brain regions, comparison of the more similar neocortical regions in our study revealed the presence of potential ‘protein only’ region markers. Comparison of human and mouse data showed a generally positive correlation between protein orthologues, which was significantly increased in those proteins found to be DEX between human regions.
In complex tissues like the brain, protein abundance is highly dependent on the sites of synthesis and localisation. It is also the case that different proteins and, to a lesser extent, mRNAs possess vastly different half-lives19,21. To be able to accurately assess these relationships between RNA and protein therefore, it is a great advantage to measure gene product abundance in the same exact biological samples, as we have done here. We have shown that although a majority of genes had similar magnitudes of change in their mRNA and protein abundance between brain regions, there were a large number that showed consistent differences in relative extent and direction of change. As such, mRNA expression is an incomplete measure of protein abundance, requiring direct proteomic assessment to generate an overview of gene expression and regulation in different regions of the brain.
Genes identified as differentially expressed between regions by ‘protein-only’ tended to be enriched for nuclear functions with a strong bias toward ‘RNA-processing’ over other ontological terms, such as ‘chromatin modification’. RNA-processing genes have been reported to have stable protein but unstable RNA and would therefore be expected to be more enriched compared to genes related to chromatin modification, which have both unstable RNA and unstable protein19. The stability of proteins involved in aspects of nuclear function likely contributed strongly to the significant differences between regions that arise from the number, relative density, and size of nuclei. The most extreme example is the cerebellum, which contains approximately 68 billion of the 86 billion neurons estimated to make up the human brain46, and this high density of nuclei was reflected in the highly significant enrichment for nuclear GO terms seen in many CBC enriched clusters.
Previous analyses of the mRNA from the brain samples used revealed very subtle expression differences between the regions of the neocortex16. Thus, the 11 neocortical regions were so similar by mRNA-seq that they were collapsed when compared to the sub-cortical regions and over the developmental time-course16, a phenomenon that is even more apparent in pre-adolescence8. Meanwhile, total-RNA analysis of the neocortex, which is not restricted to measuring only coding RNAs, suggests that it is lower-abundance non-coding RNAs that are largely responsible for the differences between neocortical regions. However, our proteomic analysis of the DFC and V1C found many more differentially expressed proteins between these neocortical regions than we found differentially expressed mRNAs. Furthermore, we found little evidence of nuclear enrichment in these protein-only DEX genes, likely due to the similar cytoarchitecture of these two regions. Instead we observed significant enrichments for genes implicated in ‘receptor activity’ or ‘localisation to the plasma membrane’. It is therefore plausible to hypothesise that coding genes substantially contribute to the diversity and molecular differentiation of the neocortical regions, but that these differences are being magnified in their protein abundance compared to their mRNA expression.
In neurons, the extent of localized (non cell body) protein translation is unclear. As high throughput techniques for measuring the impact of local translation improve47, it seems likely that production of protein from mRNA transcripts occurs primarily in the cell body, with a subset of functionally important proteins being synthesized in the dendritic and potentially axonal compartments48. It is therefore possible that differences in mRNA and protein abundance are driven by highly mobile proteins and mRNAs localized far from their brain region of origin. For example, we observed an enrichment of synaptic proteins in the ‘RNA only’ category, suggesting that while the mRNA would largely be present in the source region containing the cell soma but not in the axonal target region, protein that is subsequently trafficked between the brain regions would be detected in both. Such genes would only be differentially expressed between the regions in the ‘RNA only’ data. Therefore, while it is important to understand where and when a gene is expressed, this may not accurately reflect the abundance of the final protein product.
Despite inherent problems working with the post-mortem human brain, notably the highly variable post-mortem intervals that limit observability of PTMs and affect non-tryptic protease digestion45,49,50, the data presented here are a rich source of information. For example, the database of observable peptides in each brain region (Table S2) may be used to design targeted assays as a replacement for low-throughput methods such as immunoblotting. Peptides observed in the single shot data can be used as a high-confidence subset for quantitation and the protein quantifications themselves (Table S4) can be used to estimate stoichiometry for molecular modelling of these brain regions. In cases where the mRNA and protein are highly correlated (Table S7,8), RNA measurement is likely preferable in terms of ease, cost, and replicability. It is also straightforward to compare regional protein abundance in the mouse vs human (Table S10), an important consideration for animal models of disease and neuropharmacology. These data have the potential to become an invaluable resource for neuroscientists and the approaches used are a step towards broader adoption of proteomic techniques by the neuroscience community.
Online methods
Tissue procurement
This study was conducted using frozen post-mortem human brain specimens from a number of tissue collections facilitated by the Brainspan and psychENCODE consortia - see Kang et al16 for specific sample handling and preservation details. Briefly, tissue was collected after obtaining parental or next of kin consent and with approval by the institutional review boards at the Yale School of Medicine, the National Institutes of Health (NIH), and at each institution from which tissue specimens were obtained. Tissue was handled in accordance with ethical guidelines and regulations for the research use of human brain tissue set forth by the NIH (http://bioethics.od.nih.gov/humantissue.html) and the WMA Declaration of Helsinki (http://www.wma.net/en/30publications/10policies/b3/index.html). Appropriate informed consent was obtained and all available non-identifying information was recorded for each specimen. Specimens range in age from one year post conception to 40 years. The postmortem interval (PMI) was defined as hours between time of death and time when tissue samples were frozen. To ensure appropriate representation of the dissected sample region, frozen samples were pulverized in liquid nitrogen using a ceramic mortar and pestle.
Of the 16 brain regions profiled in Brainspan, seven regions that showed the largest interregional differences in gene expression by RNA-seq were selected: cerebellum (CBC), striatum (STR), mediodorsal thalamic nucleus (MD), amygdala (AMY), hippocampus (HIP), primary visual cortex (V1C), and dorsolateral prefrontal cortex (DFC). Developmental samples spanned early infancy (1 year post conception; developmental period 8) to adulthood (42 years; developmental period 13)16; the subjects were a near equal mix of males and females (Fig 1A; Table S1). No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in Sharma et al29.
Sample dissections
Please see the supplement of Kang et al16 for more detailed descriptions of the dissections of the regional samples. Briefly, the dissections were as follows:
-
-
DFC was sampled from approximate border between the anterior and middle third of the medial frontal gyrus. DFC corresponds approximately to Brodmann areas (BA) 9 and 46.
-
-
V1C was sampled from the area surrounding the calcarine fissure. Only samples in which the stria of Gennari could be recognized were included. V1C corresponds to BA 17 or the primary visual cortex.
-
–
HIP was sampled from the middle third of the retrocommissural hippocampal formation, located on the medial side of the temporal lobe. Sampled areas always contained dentate gyrus and the cornu ammonis.
-
-
AMY - the whole amygdala was dissected.
-
-
STR includes the head of the caudate nucleus and the putamen, separated by the internal capsule and ventrally connected to the nucleus accumbens.
-
-
MD - the whole mediodorsal nucleus of the thalamus (MD) was sampled from the dorsal and medial thalamus. Small quantities of surrounding thalamic nuclei may be present in the samples.
-
-
CBC was sampled from the lateral part of the posterior lobe. The sampled area contained all three layers of cerebellar cortex and underlying white matter but not the deep cerebellar nuclei.
Sample preparation
Frozen powdered brain samples were weighed and added to lysis buffer (8 M Urea, 0.4 M ammonium bicarbonate, Complete Protease inhibitor (Roche)) at 1:10 weight:volume. Samples were homogenised by sonication, and cleared by centrifugation at 14,000 rpm, 4°C, 10 min on a desktop centrifuge. Lysates were quantified by BCA assay, and adjusted to 100 µg protein in 50 µL 8 M Urea, 0.4 M ammonium bicarbonate. pH was confirmed to be ~8. Dithiothreitol (45 mM at 1/10th sample volume) was added to lysates for 30 min at 37°C, followed by addition of iodoacetamide (100 mM at 1/10th sample volume) for 30 min in the dark at room temperature. Samples were diluted to 2 M urea with deionised water, before addition of trypsin at 1:20 trypsin:protein ratio. Proteins were digested for 16 hours at 37°C. The digestion was quenched by adjusting the pH to below 3 by the addition of 20% trifluoroacetic acid, and desalted using C18 Macro Spin Columns (Nest Group) according to the manufacturer’s instructions. Peptides eluted from the column were dried in a SpeedVac and stored at −20°C. Dried peptides were dissolved in 3.5% formic acid (FA) / 0.1% trifluoroacetic acid (TFA), and peptide concentrations were estimated from A280 absorbance using a Thermo Scientific Nanodrop 2000. Aliquots were diluted accordingly with additional 0.1% TFA to a final concentration of 0.04 µg/µl, with 0.2 µg loaded on column for mass spectrometric analysis. This procedure was used for both fractionated and single-shot runs.
Fractionation of samples
Master region pools were produced by adding 40 µg of homogenate from each of the adult (period 13) subjects, to make 200 µg total per region. Pooled lysates were processed for proteomic analysis as described above. For the fractionated samples, peptides were first dissolved in 53 µL 0.1% TFA and injected onto a Waters ACQUITY UPLC (BEH) C18 column (130 Angstrom, 1.7 µm, 2.1 mm id × 100 mm) at a flow rate of 0.4 mL/min. An orthogonal ‘high pH reverse phase separation was carried out (using buffers A:100% water with 10 mM ammonium acetate, pH 10; & B: 90% water/10% acetonitrile (ACN) with 10 mM ammonium acetate, pH 10) with a gradient 0.0 min - 2% B, 2.19 min - 2% B, 19.83 min −37% B, 28.65 min - 75% B, 33.06 min - 98% B, 34.53 min - 98% B, 37.47 min - 2% B, and 40 min - 2% B to separate the peptides. 48 fractions were collected from each brain region and were then pooled based on their estimated concentration from analysis of the chromatogram into 15 pools. Each pool was subsequently analysed individually by LC MS/MS.
Mass-spectrometry analysis (LC MS/MS)
For both the pooled fractionated and single shot quantitative runs, MS analyses were performed on a Q Exactive Plus mass spectrometer (ThermoFisher Scientific) coupled online to a Waters nanoAcquity UPLC in ‘low pH’ condition. Peptides were separated over a 180-min gradient run using a Waters Symmetry C18 trap (1.7 µm, 180 µm × 20 mm) and a ACQUITY UPLC PST (BEH) C18 column (130 Angstrom, 1.7 µm × 75 µm id × 250 mm) at 37°C. Trapping was carried out for 3 min at 5 µL/min, 99% Buffer A (99% water, 0.1% FA) and 1% Buffer B (0.1% FA in ACN) prior to eluting with linear gradients that reached 0.0 min – 1% B, 140 min - 30% B, 155 min - 40% B and 160 min - 85% B with a flow rate of 300 nL/min. MS1 (300 to 1500 m/z, target value 3E6, maximum ion injection times 45 ms) were acquired and followed by higher energy collisional dissociation based fragmentation (normalised collision energy 28). A resolution of 70,000 at m/z 200 was used for MS1 scans, and up to 20 dynamically chosen, most abundant, precursor ions were fragmented (isolation window 1.7 m/z). The MS2 scans were acquired at a resolution of 17,500 at m/z 200 (target value 1E5, maximum ion injection times 100 ms). Samples were run in regional blocks, with control samples interspersed throughout to allow for correction of batch effects.
Data analysis
Data collection and analysis were not performed blind to the conditions of the experiments. No samples were excluded from the analysis. Mass spectra were processed using MaxQuant31 (v1.5.2.1). Spectra were searched against the full set of human protein sequences annotated in Gencode51 (version 21; hg38) using the Andromeda search engine52. This search included a fixed modification, cysteine carbamidomethylation, and two variable modifications, N-acetylation and methionine oxidation. Peptides shorter than 7 amino-acids were not considered for further analysis due to lack of uniqueness and a 1% false-discovery rate (FDR) was used to filter poor identifications at both the peptide and protein level. Where possible, peptide identification information was matched between runs of the fractionated and single-shot samples within MaxQuant31. This exploited the accurate mass and retention times across liquid chromatography (LC)-MS runs to infer the identity of a peptide in a particular run in which the precursor ion was detected but was not selected for identification by MS2.
For the single shot spatio-temporal analysis of human brain samples, mass spectra were matched between the fractionated brain-region-specific samples (Table S2) and the adult single shot runs. Due to the number of extra protein IDs gained using this feature, we endeavoured to estimate the likelihood of protein misidentification by the match between runs (MBR) feature. We took the 148 proteins observed only in the CBC fractionated data (“CBC specific”), and counted how many times, and by what means these same proteins were identified in the adult single shot samples (Table S11). Only 49 of these “CBC-specific” proteins were identified by any means in the single shot samples, a reflection of the substantially increased depth of fractionated vs single shot proteomics. As would likely be expected in this small subset of proteins, there were more identifications by all methods in the CBC single shot samples. 3.4% of possible observations from non-CBC single shot samples occurred only at the MS1 match level. These observations may reflect a false identification of an isobaric “imposter” peptide by the MBR feature, or may simply reflect a peptide missed by the fractionated proteomic profiling of the other regions. Although it is not possible to confidently differentiate between these two possibilities, we estimate an upper limit of 3.4% for the protein misidentification rate, and a 1.8% rate at which proteins were undetected in the non-CBC fractionated runs.
To be included in the region-specific dataset, peptides were required to have at least 2 MS/MS scans. Protein identification required at least one unique or razor peptide per protein group. Quantification in MaxQuant was performed using the label free quantification (maxLFQ) algorithm53. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD005445.
Sample preparation - RNA extraction, library preparation & sequencing
Details of tissue handling, RNA sample preparation, library construction, and sequencing can be found in the white paper at: http://help.brain-map.org/download/attachments/3506181/Transcriptome_Profiling.pdf
RNA sequencing alignment and expression quantification
RNA-seq reads were re-aligned and re-quantified using a custom analysis pipeline. This involved filtering reads against known laboratory contaminant sequences in UniVec, before further filtering by explicit alignment to the human 5.8S and 45S rRNA sequences. Reads aligned to these sequences were removed. Reads that did not map to UniVec or rRNA were aligned against the human genome (hg38) and known splice junctions (gencode v21) using the STAR aligner (v2.4.2a), using default parameters with the exception of the following:
-
-
quantMode: TranscriptomeSAM
-
-
outFilterMismatchNoverLmax: 0.1
-
-
outFilterMultimapNmax: 50
Gene expression values (Reads Per Kilobase of exon model per Million mapped; RPKM) were computed from the aligned reads using RSEQTools54. The exon model used was the composite (union of all overlapping exons) of all transcripts for each gene; a description of this composite model is available both in the RSEQTools paper and in the whitepaper above.
Downstream analysis
All downstream analysis was performed using R/Bioconductor55.
Normalisation / pre-processing
Human mass spectrometry data were obtained from the ‘proteinGroups.txt’ and ‘peptides.txt’ tables output from MaxQuant. As spectra were mapped to gencode isoforms, Ensembl transcript IDs were generalised to their corresponding Ensembl gene IDs. Entries corresponding only to reverse DB hits or to common mass-spectrometry contaminants were removed. Label Free Quantitation (LFQ) for the single shot samples were extracted and duplicated rows (redundant gene entries) were summed. Sample LFQ distributions were log10 transformed, scaled by their 75th percentile, and batch corrected using ComBat (Figure S4)56. Batch correction was employed as the single shot samples were prepared and run in two discrete batches due to sample availability. These final, normalised, LFQ data are available in Table S4.
Human mRNA-seq RPKM distributions were log10 transformed, quantile normalised, and batch corrected, again using ComBat56. Expression data were further filtered to remove nuisance high abundance small-RNA biotypes (miRNA, misc_RNA, snRNA, snoRNA, rRNA, Mt_rRNA, Mt_tRNA). This was only important for the RNA/protein coverage comparison in Figure 1 as these nuisance biotypes led to a large skew to the RNA expression distribution that is not relevant to the analysis presented here. For the RNA-seq and protein comparison, RNA-seq expression data were used to resolve genes for which the peptide alignment was ambiguous. Briefly, considering a set of peptides that map equally well to multiple different genes, if the RNA-seq could rule out one or more of these genes based on low abundance (<10% of the sum of the RNA abundance over all of these genes), then it was removed from the protein identifier. Subsequently, if the remaining ambiguous genes were also roughly equally expressed by RNA, then the RNA-seq data for these genes was summed to match the protein entry.
Region averaged mouse mass-spectrometry data were obtained from the supplement of Sharma et al29. Human:Mouse 1:1 orthologues were obtained from BioMart (Ensembl Genes 85). Ortholog gene pairs were defined by matching gene-symbols in the human with those in the mouse dataset.
Statistics
Normalised LFQ human protein and RNA data (5141 proteins, 77 samples from 17 individuals) were subjected to a gene-wise linear model to compute the coefficients of development period (continuous variable, 6 periods) and brain region (discrete variable, 7 regions). Data distribution was assumed to be normal but this was not formally tested. Statistical significance was computed by two-way ANOVA and p-values were corrected for multiple hypothesis tests using Bonferroni’s method. A threshold of p<0.05 was used to define significantly differentially expressed (DEX) genes across either developmental period or brain region, with an additional criteria of a minimum fold change of 2 for genes included in GO analysis.
DEX genes over developmental period or brain region were clustered using the R package ‘dynamicTreeCut’57. Euclidean distances between genes were clustered using the ‘hclust’ function (method=‘average’) prior to cluster discretisation using a dynamic tree cut (method=‘hybrid’, deepSplit=T, pamStage=T, maxPamDist=0). The minimum cluster size was varied between the different gene sets to produce more easily interpretable results; for period-DEX proteins the minimum cluster membership was set to 2, for region-DEX proteins the minimum cluster membership was set to 10.
For the protein/RNA comparison, KS tests were performed to assess whether RNA:protein correlation was altered in regionally DEX genes (n=1776) versus all detected genes (n=5039). For the human/mouse comparison, only those data from the 5 regions (CBC, STR, MD, HIP, and frontal cortex: dorsolateral PFC in primate, PFC in mouse) and 4052 genes represented in both datasets were used (1517 genes were region significant). Human differential expression analysis was performed on this subset of genes and regions in the same manner as before. For the mouse, equivalent replicate-level data were unavailable from the supplement of Sharma et al.29, so instead human-equivalent coefficients were computed from the region-averaged data available in their Supplementary Information. This simply involved a re-scaling of the reported abundances from log2 to log10.
In order to assess gene-set enrichments in the regionally differentially expressed proteins and transcripts, we downloaded gene symbol to gene ontology terms and accessions for each of the three GO domains using the biomaRt package in R. All ontological analysis was performed using the R package topGO (http://www.mpi-sb.mpg.de/alexa) and all p-values corrected for multiple tests using the Benjamini Hochberg method58. Briefly, genes were classified based on their RNA vs protein fold-change agreement in each region pair, visually summarised in Figure 5. We sought to identify enriched ontological terms in genes that either agree, partially agree, or disagree in the fold-change reported by RNA-seq and proteomics, including genes that appear highly differentially expressed in one of these assays but not the other. Only significantly differentially expressed genes were input (background) and the classification based on +/− 2-fold changes between region pairs.
We also classified each gene based on its consistency of RNA/protein fold-change agreement across all pairs of regions. For each gene, regions with fold-changes < 2 in both RNA and protein were discarded, and genes appearing in the same category (i.e. protein-only DEX) in more than 50% of the remaining region pairs were annotated as consistent, in this example, as ‘proteinOnly’. Ontological enrichment analysis was performed as before with the individual region pairs.
Finally, we sought to increase the scope and relevance of this enrichment analysis by including all hallmark, positional, BioCarta, KEGG, reactome, miRNAtargets, and TFtargets available in the Molecular Signatures Database (v5.2)59. Enrichments here were computed using Fisher’s Exact Tests and p-values again corrected for multiple comparisons using the Benjamini Hochberg method. Exact Ns for each functional enrichment test can be found in the “Annotated” and “Significant” columns of Figure S9.
To generate the network figures in S7 and S12 STRING criteria were set at “medium” stringency (0.4) and included all sources of interactions (Text mining, experiments, databases, co-expression, neighbourhood, gene fusion & co-occurrence). For the STR figure (S8), the p value survives when stringency is increased to “highest” (0.9). All enrichment analyses were performed using a background list of genes that were significantly differentially expressed as RNA or protein (this same list was used to generate the scatter plots).
Immunoblotting
Lysates prepared in 8 M Urea were separated by SDS-PAGE (4–20% tris-glycine gels, Life Technologies). Proteins were transferred onto 0.2 µM Nitrocellulose (Biorad). Primary antibodies used were anti mGluR2/3 (GRM2/3, EMD Millipore, 06–676, 1:1000), anti CB1 (CNR1, Cell Signalling, D5N5C, 1:1000), anti PDE4D (Millipore ABS22, 1:1000), anti TrkC (NTRK3, Cell Signalling, C44H5, 1:1000) and anti GAPDH (Calbiochem CB1001, 1:5000). The antibodies used have been validated for this assay in other species, but not in human brain. Comprehensive validation of antibodies for use in human brain tissue is extremely difficult, as discussed by the Antibody Validation Working group60, but given that for all antibodies except TrKC the antibodies show a comparable pattern of expression to the LC-MS/MS data, and bands of the appropriate size, we can presume these antibodies are working appropriately. In TrkC’s case, the disagreement may depend on the epitope used by the antibody, drop out of peptides due to post translational modification, or a lack of sensitivity by the antibodies for a relatively modest change. The primary antibodies were visualized using anti mouse or rabbit HRP (Vector Laboratories, PI-2000 Ms, PI-1000 Rb 1:3000) and a ChemiDoc Imager (Biorad), or in the case of GAPDH, the Licor IRDye 800 anti-mouse secondary antibody (Rockland 610-102-041, 1:15,000) and a Licor Odyssey Infra Red Scanner. Bands were quantified using ImageJ, normalized within lane to GAPDH, and paired Student’s T-tests were performed using Prism 7 (n= 5 biological replicates per group, all data shown on blots/graphs).
A summary of important reproducibility related information from these methods can be found in the accompanying “Life Sciences Reporting Summary.”
Data Availability
The Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE61 partner repository with the data identifier PXD005445.
Code availability
Analysis code and the required input tables have been provided as a zipped Supplementary Software file. This large supplementary file (>600 MB unzipped) contains a readme file with system specific instructions for code running.
Accession Codes
PXD005445: https://www.ebi.ac.uk/pride/archive/
Supplementary Material
Table S1 | Metadata for all 77 Brainspan samples subjected to MS/MS for this study.
Table S2 | Peptide-level data obtained from heavily fractionated per-region MS/MS
Table S3 | Protein-level summary of the fractionated per-region and single-shot MS/MS. A) Number of peptides observed per protein, per sample. B) Percent of the coding sequence represented by these observed peptides, per protein, per sample.
Table S4 | Label free protein quantification (LFQ) of all single shot samples Log10 LFQ protein data for each single shot sample, ordered alphabetically by gene symbol and corresponding Ensembl gene ID. Samples are ordered by subject.
Table S5 | Results of the proteomic spatiotemporal differential expression analysis. This excel table includes averaged LFQ data for each brain region, and cluster membership for each DEX gene. Individual tabs show data ordered for easy inspection of region and period DEX clusters
Table S6 | Protein and RNA expression data for genes expressed in both datasets. A) Protein: log10 LFQ data for all samples. B) RNA: log10 RPKM data for all samples.
Table S7 | Inter-regional protein and RNA abundance and differential expression summary. Regional differential expression pValues, regional abundance averages (A) and intra-regional abundance standard deviations (B).
Table S8 | Summary of the RNA vs protein differential consistency of each gene in accordance with the definitions introduced in Figure 5.
Proteins are assigned as consistent to the categories defined in Fig 5 if they fall into a single category in >50% of the all region comparisons – i.e. if they appear in the protein only category in 11 of the 21 total comparisons, they are consistently protein only.
Table S9 | Complete ontology and gene-set enrichment analysis results consistent with the definitions introduced in Fig 5.
Multi tab Excel table showing GO enrichments for genes consistently found in the protein only, RNA only and agree categories. There were no significant GO enrichments in other categories. Further tabs show data from the DFC/STR comparison in all categories.
Table S10 | Inter-regional human and mouse protein abundance summary.
Protein conservation, log10 LFQ data from comparable mouse and human regions.
Table S11 | Identification methods of “CBC specific” proteins observed in the adult single shot samples, expressed as a percentage of total possible observations. The majority of “CBC specific” proteins were not observed in the single shot samples. From this analysis we estimate an upper limit of 3.4% for the protein misidentification rate. 1.8% of proteins were identified in the non-CBC single shot runs by either MS2 (0.75%) or a mix of MS2 and MBR (1.05%), and thus may represent proteins that were unidentified in the non-CBC fractionated runs.
Acknowledgments
We would like to thank S. Leslie and D. Li for helpful discussions. Data were generated as part of the PsychENCODE Consortium, supported by: U01MH103339, U01MH103365, U01MH103392, U01MH103340, U01MH103346, R01MH105472, R01MH094714, R01MH105898, R21MH102791, R21MH105881, R21MH103877, and P50MH106934 awarded to: S. Akbarian (Icahn School of Medicine at Mount Sinai), G. Crawford (Duke), S. Dracheva (Icahn School of Medicine at Mount Sinai), P. Farnham (USC), M. Gerstein (Yale), D. Geschwind (UCLA), T. M. Hyde (LIBD), A. Jaffe (LIBD), J. A. Knowles (USC), C. Liu (UIC), D. Pinto (Icahn School of Medicine at Mount Sinai), N. Sestan (Yale), P. Sklar (Icahn School of Medicine at Mount Sinai), M. State (UCSF), P. Sullivan (UNC), F. Vaccarino (Yale), S. Weissman (Yale), K. White (UChicago) and P. Zandi (JHU). This work was supported by the Yale/NIDA Neuroproteomics Centre (DA018343-12), by NIA grant AG047270-02, by NIMH grant MH110926, by NIH SIG grants 1S10OD019967-0 & 1S10ODOD018034-01, and by the State of Connecticut, Department of Mental Health & Addiction Services. B.C.C. was supported by a 2014 NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation.
Footnotes
Author Contributions: B.C.C. designed the experiments, performed the experiments, analysed the data and wrote the manuscript. R.R.K. designed the experiments, analysed the data and wrote the manuscript. J.E.K. performed the experiments. E.Z.V performed the experiments. M.P. contributed to tissue and sample processing. A.M.M.S. contributed to tissue and sample processing. T.T.L. designed the experiments and wrote the manuscript. M.B.G. contributed to RNA-seq data generation and provided computational resources. N.S. designed the experiments, contributed to tissue and sample processing, contributed to RNA-seq data generation and wrote the manuscript. A.C.N. designed the experiments, and wrote the manuscript.
Competing Financial Interests Statement: The authors have no competing financial interests.
References
- 1.Silbereis JC, Pochareddy S, Zhu Y, Li M, Sestan N. The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron. 2016;89:248–268. doi: 10.1016/j.neuron.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasthuri N, et al. Saturated Reconstruction of a Volume of Neocortex. Cell. 2015;162:648–61. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- 3.Swanson LW, Lichtman JW. From Cajal to Connectome and Beyond. Annu. Rev. Neurosci. 2016;39:197–216. doi: 10.1146/annurev-neuro-071714-033954. [DOI] [PubMed] [Google Scholar]
- 4.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–14. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouyang A, et al. Spatial mapping of structural and connectional imaging data for the developing human brain with diffusion tensor imaging. Methods. 2015;73:27–37. doi: 10.1016/j.ymeth.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lerner TN, Ye L, Deisseroth K. Communication in Neural Circuits: Tools, Opportunities, and Challenges. Cell. 2016;164:1136–50. doi: 10.1016/j.cell.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fertuzinhos S, et al. Laminar and temporal expression dynamics of coding and noncoding RNAs in the mouse neocortex. Cell Rep. 2014;6:938–50. doi: 10.1016/j.celrep.2014.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pletikos M, et al. Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron. 2014;81:321–32. doi: 10.1016/j.neuron.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakken TE, et al. A comprehensive transcriptional map of primate brain development. Nature. 2016;535:367–75. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consortium B, et al. Technical White Paper: Transcriptome Profiling by RNA sequencing and exon microarray [Google Scholar]
- 11.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–9. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–42. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 14.Krishnaswami SR, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc. 2016;11:499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–48. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.PsychENCODE Consortium et al. The PsychENCODE project. Nat. Neurosci. 2015;18:1707–12. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgess DJ. Technology: A drop in single-cell challenges. Nat. Rev. Genet. 2015;16:376–377. doi: 10.1038/nrg3972. [DOI] [PubMed] [Google Scholar]
- 19.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 20.Li JJ, Bickel PJ, Biggin MD. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ. 2014;2:e270. doi: 10.7717/peerj.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwanhäusser B, Wolf J, Selbach M, Busse D. Synthesis and degradation jointly determine the responsiveness of the cellular proteome. Bioessays. 2013;35:597–601. doi: 10.1002/bies.201300017. [DOI] [PubMed] [Google Scholar]
- 22.Kitchen RR, Rozowsky JS, Gerstein MB, Nairn AC. Decoding neuroproteomics: integrating the genome, translatome and functional anatomy. Nat. Neurosci. 2014;17:1491–9. doi: 10.1038/nn.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann M, Kulak NA, Nagaraj N, Cox J. The coming age of complete, accurate, and ubiquitous proteomes. Mol. Cell. 2013;49:583–90. doi: 10.1016/j.molcel.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 24.Nagaraj N, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beck M, et al. The quantitative proteome of a human cell line. Mol. Syst. Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Alvarez S, Hicks LM. Comprehensive Comparison of iTRAQ and Label-free LC-Based Quantitative Proteomics Approaches Using Two Chlamydomonas reinhardtii Strains of Interest for Biofuels Engineering. J. Proteome Res. 2012;11:487–501. doi: 10.1021/pr2008225. [DOI] [PubMed] [Google Scholar]
- 28.Latosinska A, et al. Comparative Analysis of Label-Free and 8-Plex iTRAQ Approach for Quantitative Tissue Proteomic Analysis. PLoS One. 2015;10:e0137048. doi: 10.1371/journal.pone.0137048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma K, et al. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 2015;18:1819–31. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brainspan. BrainSpan Atlas of the Developing Human Brain. [Google Scholar]
- 31.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 32.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, et al. Systematic Comparison of Label-Free, Metabolic Labeling, and Isobaric Chemical Labeling for Quantitative Proteomics on LTQ Orbitrap Velos. J. Proteome Res. 2012;11:1582–1590. doi: 10.1021/pr200748h. [DOI] [PubMed] [Google Scholar]
- 34.Handbook of Basal Ganglia Structure and Function. Steiner H, Tseng KY, editors. Handbook of Behavioral Neuroscience. 2017;24:1–1012. [Google Scholar]
- 35.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volkow ND, Morales M. The Brain on Drugs: From Reward to Addiction. Cell. 2015;162:712–25. doi: 10.1016/j.cell.2015.07.046. [DOI] [PubMed] [Google Scholar]
- 37.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–33. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 38.Scheiffele P. Cell-cell signaling during synapse formation in the CNS. Annu. Rev. Neurosci. 2003;26:485–508. doi: 10.1146/annurev.neuro.26.043002.094940. [DOI] [PubMed] [Google Scholar]
- 39.Lindner M, Ng JKM, Hochmeister S, Meinl E, Linington C. Neurofascin 186 specific autoantibodies induce axonal injury and exacerbate disease severity in experimental autoimmune encephalomyelitis. Exp. Neurol. 2013;247:259–66. doi: 10.1016/j.expneurol.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Mathey EK, et al. Neurofascin as a novel target for autoantibody-mediated axonal injury. J. Exp. Med. 2007;204:2363–72. doi: 10.1084/jem.20071053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weder N, et al. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J. Am. Acad. Child Adolesc. Psychiatry. 2014;53:417–24.e5. doi: 10.1016/j.jaac.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montalvo-Ortiz JL, et al. The role of genes involved in stress, neural plasticity, and brain circuitry in depressive phenotypes: Convergent findings in a mouse model of neglect. Behav. Brain Res. 2016;315:71–4. doi: 10.1016/j.bbr.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovács GG, et al. Natively unfolded tubulin polymerization promoting protein TPPP/p25 is a common marker of alpha-synucleinopathies. Neurobiol. Dis. 2004;17:155–62. doi: 10.1016/j.nbd.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Hintiryan H, et al. The mouse cortico-striatal projectome. Nat. Neurosci. 2016;19:1100–1114. doi: 10.1038/nn.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seyfried NT, et al. Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards. J. Proteome Res. 2012;11:2721–38. doi: 10.1021/pr2010814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llinas RR, Walton KD, Lang EJ. Cerebellum. The Synaptic Organization of the Brain. 2003:271–310. doi: 10.1093/acprof:oso/9780195159561.003.0007. [DOI] [Google Scholar]
- 47.Namjoshi SV, Raab-Graham KF. Screening the Molecular Framework Underlying Local Dendritic mRNA Translation. Front. Mol. Neurosci. 2017;10:45. doi: 10.3389/fnmol.2017.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holt CE, Schuman EM. The Central Dogma Decentralized: New Perspectives on RNA Function and Local Translation in Neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dammer EB, et al. Neuron enriched nuclear proteome isolated from human brain. J. Proteome Res. 2013;12:3193–206. doi: 10.1021/pr400246t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tagawa K, et al. Comprehensive phosphoproteome analysis unravels the core signaling network that initiates the earliest synapse pathology in preclinical Alzheimer’s disease brain. Hum. Mol. Genet. 2015;24:540–558. doi: 10.1093/hmg/ddu475. [DOI] [PubMed] [Google Scholar]
- 51.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox J, et al. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 53.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–26. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Habegger L, et al. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–3. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 57.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 58.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing on JSTOR. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 59.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uhlen M, et al. A proposal for validation of antibodies. Nature Methods. 2016;13:823–827. doi: 10.1038/nmeth.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vizcaino JE. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 | Metadata for all 77 Brainspan samples subjected to MS/MS for this study.
Table S2 | Peptide-level data obtained from heavily fractionated per-region MS/MS
Table S3 | Protein-level summary of the fractionated per-region and single-shot MS/MS. A) Number of peptides observed per protein, per sample. B) Percent of the coding sequence represented by these observed peptides, per protein, per sample.
Table S4 | Label free protein quantification (LFQ) of all single shot samples Log10 LFQ protein data for each single shot sample, ordered alphabetically by gene symbol and corresponding Ensembl gene ID. Samples are ordered by subject.
Table S5 | Results of the proteomic spatiotemporal differential expression analysis. This excel table includes averaged LFQ data for each brain region, and cluster membership for each DEX gene. Individual tabs show data ordered for easy inspection of region and period DEX clusters
Table S6 | Protein and RNA expression data for genes expressed in both datasets. A) Protein: log10 LFQ data for all samples. B) RNA: log10 RPKM data for all samples.
Table S7 | Inter-regional protein and RNA abundance and differential expression summary. Regional differential expression pValues, regional abundance averages (A) and intra-regional abundance standard deviations (B).
Table S8 | Summary of the RNA vs protein differential consistency of each gene in accordance with the definitions introduced in Figure 5.
Proteins are assigned as consistent to the categories defined in Fig 5 if they fall into a single category in >50% of the all region comparisons – i.e. if they appear in the protein only category in 11 of the 21 total comparisons, they are consistently protein only.
Table S9 | Complete ontology and gene-set enrichment analysis results consistent with the definitions introduced in Fig 5.
Multi tab Excel table showing GO enrichments for genes consistently found in the protein only, RNA only and agree categories. There were no significant GO enrichments in other categories. Further tabs show data from the DFC/STR comparison in all categories.
Table S10 | Inter-regional human and mouse protein abundance summary.
Protein conservation, log10 LFQ data from comparable mouse and human regions.
Table S11 | Identification methods of “CBC specific” proteins observed in the adult single shot samples, expressed as a percentage of total possible observations. The majority of “CBC specific” proteins were not observed in the single shot samples. From this analysis we estimate an upper limit of 3.4% for the protein misidentification rate. 1.8% of proteins were identified in the non-CBC single shot runs by either MS2 (0.75%) or a mix of MS2 and MBR (1.05%), and thus may represent proteins that were unidentified in the non-CBC fractionated runs.
Data Availability Statement
The Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE61 partner repository with the data identifier PXD005445.