Abstract
The factors predisposing toward the development of pulmonary nontuberculous mycobacterial (pNTM) disease and influencing disease progression remain unclear. Impaired immune responses have been reported in individuals with pNTM disease, but data are limited and inconsistent. In this study, we sought to use gene expression profiling to examine the host response to pNTM disease. Microarray analysis of whole-blood gene expression was performed on 25 subjects with pNTM disease and 27 uninfected control subjects with respiratory disease. Gene expression results were compared with phenotypic variables and survival data. Compared with uninfected control subjects, pNTM disease was associated with downregulation of 213 transcripts enriched for terms related to T cell signaling, including IFNG. Reduced IFNG expression was associated with more severe computed tomography changes and impaired lung function. Mortality was associated with the expression of transcripts related to the innate immune response and inflammation, whereas transcripts related to T and B cell function were associated with improved survival. These findings suggest that pNTM disease is associated with an aberrant immune response, which may reflect an underlying propensity to infection or result from NTM infection itself. There were important differences in the immune response associated with survival and mortality in pNTM disease.
Keywords: nontuberculous mycobacteria, gene expression profiling, interferon-γ
Clinical Relevance
These findings suggest that pulmonary nontuberculous mycobacterial disease is associated with an altered immune response that may contribute to both the development of disease as well as its severity. The expression of genes identified in this study warrant further investigation as potential markers of disease activity and suggest targets for potential therapeutic intervention.
Pulmonary nontuberculous mycobacterial (pNTM) disease is an increasingly common and challenging infection. Although ubiquitous in the environment, disease usually occurs in the context of an existing pulmonary disease, such as bronchiectasis or chronic obstructive pulmonary disease (COPD); however, in a subgroup of patients, disease occurs in the absence of any known risk factors. The disease is associated with a high mortality, ranging between 12.5% and 41.1% at 5 years (1–5). The clinical course of disease is variable and unpredictable; although some individuals develop progressive disease, some may remain stable without treatment (1, 6, 7). The factors governing the acquisition of disease and its subsequent clinical course are poorly understood.
Disseminated NTM disease occurs in the context of immunodeficiency, such as acquired immunodeficiency syndrome or inherited and acquired defects of the IFN-γ/IL-12 pathway, and several studies have also demonstrated functional defects in the host immune response of subjects with pNTM disease (8–13). A recent study employing exome sequencing revealed an excess of variants affecting genes implicated in the immune response in subjects with pNTM disease compared with both the control population and unaffected family members (14). These findings suggest that defective immune responses may play a role in predisposing individuals to pNTM disease. The aims of this study were to investigate the host response to pNTM disease using global gene expression profiling of peripheral blood and to explore the relationship between gene expression and clinical outcomes.
Methods
Individuals with pNTM disease were recruited from the Royal Brompton Hospital and Chelsea and Westminster Hospital between September 2012 and November 2013. All participants met American Thoracic Society 2007 criteria, and were unanimously deemed by two clinicians with expertise in pNTM disease (authors M.R.L. and R.W.) to have active disease requiring treatment. Control subjects with respiratory disease (bronchiectasis or COPD), but no radiological or microbiological evidence of mycobacterial disease, were recruited from the same departments. Written consent was obtained from all participants and the study was approved by the local research ethics committee (reference 12/LO/1034).
Participants underwent clinical assessment and pulmonary function testing. Peripheral blood was taken for RNA extraction and clinical testing, and spontaneously expectorated sputum was collected for phenol auramine microscopy and bacterial, mycobacterial, and fungal cultures. If not performed within the previous 6 months, a high-resolution computed tomography (CT) was performed. For pNTM cases, CT scoring was performed by a specialist radiologist blinded to clinical details, as described previously (15), and a composite score calculated by summing the scores for individual features. Further methods are provided in the data supplement.
Gene Expression Analysis
Peripheral blood was collected into PAXgene RNA tubes (Becton Dickinson Co.) and RNA extracted using the PAXgene Blood RNA Kit (PreAnalytiX). Extracted RNA was amplified to complementary DNA using the Ovation Pico WTA System V2 (NuGEN Technologies), then cDNA was fragmented and labeled using the Encore Biotin Module (NuGEN Technologies) and immediately hybridized to Affymetrix Human Gene 1.1 ST array plates and scanned using the GeneTitan instrument (Affymetrix). Samples were randomized before amplification and again before fragmentation, labeling, and hybridization.
Outliers were identified using the arrayQualityMetrics package (16) in the R environment version 3.1.2, and removed. Raw probe-level data from .CEL files passing quality control were summarized, quantile normalized, and log2 transformed using the Affymetrix Power Tools package. Expression data were reimported into R, where low-expressed and unannotated probes were removed and all downstream analyses performed.
Differentially expressed genes were identified using significance analysis of microarrays (SAM) (17) applying a false discovery rate (FDR) of 5% for significance. Gene ontology analyses were performed using the WebGestalt tool (http://www.webgestalt.org) using the Gene Ontology (GO) Consortium and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. All expressed genes were used as the reference set, and an adjusted P value of 0.05 was deemed significant.
Weighted gene coexpression gene network analysis (WGCNA) was performed using the WGCNA package (18) with the default-recommended parameters. All expressed genes were entered into the analysis; however, only genes with a connectivity in the highest tertile were used to construct the topological overlap matrix.
Correlation between gene (or gene module) expression and clinical variables was performed using the Pearson or Spearman correlation for continuous variables according to their distribution, and polyserial correlation for categorical variables. The t test and ANOVA were used to compare expression values between groups. The log-rank test was used to compare differences in survival curves.
Results
A total of 52 subjects (25 pNTM disease cases and 27 control subjects) matched for age, sex, and ethnicity was studied. Clinical characteristics of the study cohort are detailed in Table 1, with further details provided in Tables E2 and E3 of the data supplement. The proportion of subjects with a diagnosis of bronchiectasis was lower (although not significantly so) in the group of pNTM cases due to the inclusion of subjects with “Lady Windermere” syndrome. These individuals (n = 7) had no underlying lung disease before their NTM infection, although, at the time of enrolment, bronchiectasis was present in all subjects on high-resolution CT.
Table 1.
Clinical Characteristics of the Study Population
| Characteristics | pNTM Cases (n = 25) | Controls (n = 27) |
|---|---|---|
| Age, yr | 66.9 (±10.3) | 65.4 (±7.5) |
| Male sex, n (%) | 9 (36) | 10 (37) |
| Ex- or current smoker, n (%) | 14 (56) | 14 (52) |
| Underlying diagnosis, n (%) | ||
| COPD | 7 (28) | 7 (26) |
| Bronchiectasis | 11 (44) | 20 (74) |
| No underlying lung disease | 7 (28) | 0 |
| FEV1 % predicted | 48.64 (±31.42) | 71.27 (±25.98) |
| FVC % predicted | 73.78 (±39.2) | 102.25 (±20.87) |
| Prophylactic antibiotics, n (%) | 6 (24) | 8 (29) |
| Systemic corticosteroids, n (%) | 3 (12) | 2 (7) |
| Inhaled corticosteroids, n (%) | 12 (48) | 17 (63) |
| NTM treatment, n (%) | 9 (36) | — |
| NTM species, n (%) | ||
| MAC | 14 (56) | — |
| M. abscessus | 4 (16) | — |
| M. xenopi | 3 (12) | — |
| M. kansasii | 2 (8) | — |
| M. malmoense | 1 (4) | — |
| M. fortuitum | 1 (4) | — |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; FEV1 = forced expiratory volume in 1 second; FVC = forced vital capacity; M. = Mycobacterium; MAC = M. avium complex; NTM = nontuberculous mycobacterial; pNTM = pulmonary nontuberculous mycobacterial.
Continuous variables are stated as means (±SD).
Gene Expression and pNTM Disease
A total of 213 transcripts was identified as differentially expressed between cases and control subjects, with an FDR of 5% (for full list, see Table E4 and Figure E1). All transcripts were downregulated in pNTM disease. The top 25 genes with the highest fold change are detailed in Table 2.
Table 2.
The Top 25 Differentially Expressed Genes between Pulmonary Nontuberculous Mycobacterial Cases and Control Subjects
| Rank | Gene Name | Entrez ID | Fold Change | d-Statistic | Name |
|---|---|---|---|---|---|
| 1 | FLJ45825 | 100505530 | 1.657 | 2.646 | Uncharacterized LOC100505530 |
| 2 | GZMK | 3003 | 1.61 | 2.857 | Granzyme K (granzyme 3; tryptase II) |
| 3 | TARP | 445347 | 1.609 | 2.619 | TCR γ alternate reading frame protein |
| 4 | TARP | 445347 | 1.609 | 2.619 | TCR γ alternate reading frame protein |
| 5 | XCL2 | 6846 | 1.553 | 3.269 | Chemokine (C motif) ligand 2 |
| 6 | A2M | 2 | 1.507 | 2.293 | α-2-macroglobulin |
| 7 | CRTAM | 56253 | 1.465 | 3.57 | Cytotoxic and regulatory T cell molecule |
| 8 | PMS2P1 | 5379 | 1.453 | 4.053 | Postmeiotic segregation increased 2 pseudogene 1 |
| 9 | FCRL3 | 115352 | 1.446 | 3.26 | Fc receptor-like 3 |
| 10 | PZP | 5858 | 1.435 | 2.275 | Pregnancy-zone protein |
| 11 | TIGIT | 201633 | 1.4 | 3.333 | T cell immunoreceptor with Ig and ITIM domains |
| 12 | PPIH | 10465 | 1.393 | 2.897 | Peptidylprolyl isomerase H (cyclophilin H) |
| 13 | AK5 | 26289 | 1.39 | 2.515 | Adenylate kinase 5 |
| 14 | MUC12 | 10071 | 1.39 | 2.388 | Mucin 12, cell surface associated |
| 15 | IFNG | 3458 | 1.382 | 2.622 | IFN, γ |
| 16 | FAHD2A | 51011 | 1.364 | 3.021 | Fumarylacetoacetate hydrolase domain containing 2A |
| 17 | SMA4 | 11039 | 1.361 | 3.602 | Glucuronidase, β pseudogene |
| 18 | VSIG1 | 340547 | 1.356 | 2.784 | V-set and immunoglobulin domain containing 1 |
| 19 | SAMD3 | 154075 | 1.35 | 2.367 | Sterile α motif domain containing 3 |
| 20 | XCL1 | 6375 | 1.348 | 2.247 | Chemokine (C motif) ligand 1 |
| 21 | IL2RB | 3560 | 1.347 | 2.242 | IL-2 receptor, β |
| 22 | PSPH | 5723 | 1.339 | 2.232 | Phosphoserine phosphatase |
| 23 | LDHB | 3945 | 1.336 | 2.312 | Lactate dehydrogenase B |
| 24 | NELL2 | 4753 | 1.323 | 2.276 | NEL-like 2 (chicken) |
| 25 | NFATC2 | 4773 | 1.322 | 2.804 | Nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 2 |
Definition of abbreviations: ITIM = immunoreceptor tyrosine-based inhibition motif; TCR = T-cell receptor.
Genes are ranked by fold change; all displayed decreased expression in pulmonary nontuberculous mycobacterial cases.
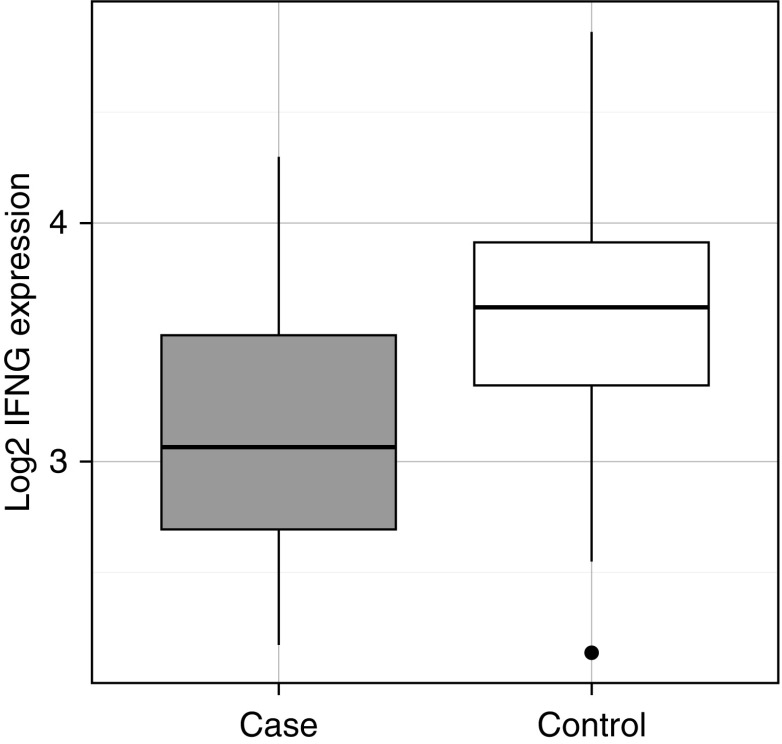
One of the top differentially expressed genes in pNTM disease cases compared with control subjects was IFN-γ (IFNG) (Figure 1). IFN-γ is the primary effector cytokine of the T-helper cell type 1 pathway, which plays a vital role in mycobacterial immunity. In light of this, it is notable that other differentially expressed genes included nuclear factor of activated T cells 2 (NFATC2), cytotoxic and regulatory T cell molecule (CRTAM), and X-C motif chemokine ligands 1 and 2 (XCL1, XCL2). NFATC2 encodes NFATc2, a member of the NFAT family of transcription factors. NFATc2 has been shown to be an in vitro regulator of both IFN-γ (19, 20) and TNF-α (21, 22) release. The product of CRTAM also promotes IFN-γ release from CD8+ T cells (23–27) and natural killer (NK) cells (28, 29). XCL2 encodes a chemokine that is chemotactic for CD8+ and CD4+ T cells (30, 31), and its homolog, XCL1, encodes a chemokine associated with the T-helper cell type 1 response, the silencing of which has been shown to lead to reduced IFN-γ production and disordered granuloma formation in tuberculosis-infected mice (32, 33).
Figure 1.
Boxplots of IFNG expression in pulmonary nontuberculous mycobacterial (pNTM) disease cases (gray) and control subjects (white). The central line, box boundaries, and whiskers represent the median, first and third quartiles, and the lowest and highest values lying within 1.5 times the interquartile range, respectively.
GO analysis of the set of differentially expressed transcripts revealed enrichment for one biological process, negative regulation of α-β T cell proliferation (enrichment ratio, 36.01; adjusted P = 0.049) and a further 10 GO cellular components. In addition, KEGG analysis revealed enrichment for the pathway T cell receptor signaling (enrichment ratio, 6.12; adjusted P < 0.001), with nine genes in the pathway (IFNG, AKT3, CD247, MAPK9, CBLB, NFATC2, ICOS, RASGRP1, and ITK) being significantly underexpressed in pNTM disease cases. Full details are given in Table E5.
To examine the influence of systemic steroid use and NTM treatment on the observed difference in gene expression, multiple linear modeling was performed using these as additional explanatory variables in addition to pNTM disease. The expression of 168 of the 213 genes (79%) remained significantly associated with pNTM disease (see Table E6).
The 213 transcripts were compared with the 380 gene “meta-signature” of active tuberculosis identified by Blankley and colleagues (34). There was an overlap of only nine genes (CDK5RAP2, EPHA4, FAIM3, FCRL3, GPR183, GZMK, KLF12, NELL2, and SMA4), most of which were also downregulated in tuberculosis, except CDK5RAP2 and SMA4, which showed increased expression. Comparison with studies of sarcoidosis, another granulomatous disease, revealed that 11 of the 213 transcripts (AKT3, CBLB, CCND2, CD247, ICOS, IFNG, IL2RB, ITK, MAPK9, NFATC2, and RASGRP1) are also present in the 31-gene sarcoid signature described by Zhou and colleagues (35), representing significant (P < 0.001) enrichment. However, none of the 20 genes in the experimentally derived “unbiased” signature from this study were present, and there was little or no overlap with other reported gene signatures in sarcoidosis (36, 37).
To identify modules associated with pNTM disease, WGCNA was performed using all 13,360 expressed transcripts from the 52 subjects, resulting in 10 modules being identified (detailed in Table E7 and E8). Three modules showed significantly (P ≤ 0.05) different expression in pNTM disease cases compared with control subjects. These were enriched for multiple GO terms, including those relating to T cell selection, differentiation, activation, and signaling, and others associated with IFN-γ activity, such as NK cell–mediated cytotoxicity, antigen processing and presentation, and major histocompatibility complex class II protein complex (Table 3). In all cases, the expression of genes within each module was lower in subjects with pNTM disease. Of the 213 genes identified to be differentially expressed in pNTM disease cases compared with control subjects, 114 (53.5%) were present in one of the three modules that showed association with pNTM disease.
Table 3.
Gene Modules Identified as Significantly Associated with Nontuberculous Mycobacterial Disease by Weighted Gene Coexpression Gene Network Analysis
| Module Name | No. of Genes | Correlation with pNTM Disease | P Value | Relevant Enriched GO Terms |
|---|---|---|---|---|
| Cyan | 1,139 | −0.31 | 0.02 | Thymic T cell selection |
| T cell receptor signaling pathway | ||||
| Antigen processing and presentation | ||||
| Greenyellow | 85 | −0.32 | 0.02 | Cellular defense response |
| Positive regulation of immune response | ||||
| T cell activation | ||||
| T cell receptor complex | ||||
| Natural killer cell mediated cytotoxicity | ||||
| CCR1 chemokine receptor binding | ||||
| Lightcyan | 37 | −0.27 | 0.05 | B cell activation |
| B cell receptor complex | ||||
| MHC class II protein complex |
Definition of abbreviations: GO = gene ontology; MHC = major histocompatibility complex; pNTM = pulmonary nontuberculous mycobacterial.
IFNG Expression and Clinical Outcomes
Given the essential role of IFN-γ in the immune response to mycobacteria, the relationship between IFNG expression in pNTM disease cases and clinical variables was explored further.
The expression of IFNG was significantly positively correlated with several measures of lung function, including forced expiratory volume in 1 second % predicted (Pearson’s r = 0.43, P = 0.05), forced vital capacity % predicted (Pearson’s r = 0.51, P = 0.019), and total lung capacity % predicted (Pearson’s r = 0.48, P = 0.016). No significant correlations, however, were seen for transfer factor for carbon monoxide or the transfer factor corrected for alveolar volume.
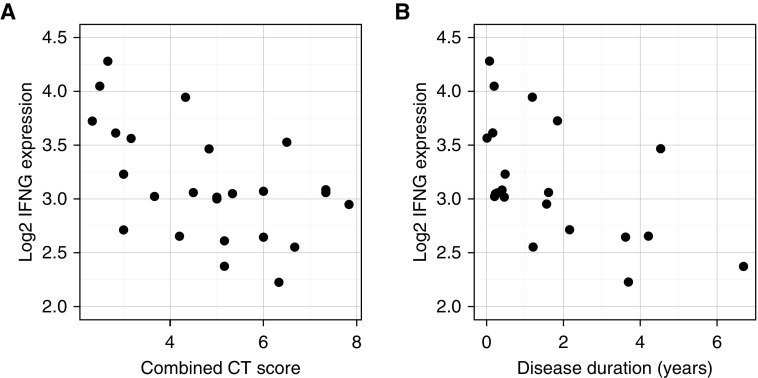
Expression of IFNG was strongly negatively correlated with combined CT score (Figure 2A; Pearson’s r = −0.53, P = 0.007). When individual CT features were examined (see Figure E2), IFNG expression was significantly lower in the presence of moderate or severely extensive bronchiectasis compared with a limited extent (P = 0.017 and P = 0.029, respectively).
Figure 2.
(A) Expression of IFNG and combined computed tomography (CT) score in pNTM cases. (B) Expression of IFNG and duration of pNTM disease.
In addition, IFNG expression was associated with other markers of disease severity: it was positively correlated with serum albumin (Spearman’s ρ = 0.50, P = 0.011), and negatively correlated with C-reactive protein (Spearman’s ρ = −0.42, P = 0.034) and neutrophil count (Pearson’s r = −0.52, P = 0.007). Interestingly, IFNG was also negatively correlated with time since diagnosis (Figure 2B; Spearman’s ρ = −0.60, P = 0.005), although there was no correlation between the combined CT score and disease duration (Spearman’s ρ = 0.24, P = 0.287).
No significant differences were seen in mean IFNG expression between survivors and nonsurvivors, although, when subjects were dichotomized into low and high IFNG expression, there was a trend toward reduced survival in the low-expression group. This, however, did not reach significance (log-rank test P = 0.06).
For disease control subjects, no significant associations between any measures of lung function or markers of disease severity and IFNG expression were observed (Table E9), suggesting that the changes were specific to pNTM disease, and not merely due to disease severity in general.
Gene Expression and Survival in pNTM Disease
Survival data were available for 24 of the pNTM disease cases who had a mortality rate of 33% (eight deaths) during the study. No deaths were observed in the control group. Survival analysis was performed on these 24 cases using SAM with an FDR of 5%; 215 genes were found to show association with decreased survival and 1,131 genes with increased survival. The top enriched GO terms in relation to survival are given in Table 4 (for full results of the analysis, please see Tables E10–E12 and Figure E3).
Table 4.
The Top Five Most Significantly Enriched Gene Ontology Terms in Genes Associated with Decreased and Increased Survival in Nontuberculous Mycobacterial Disease
| Top GO Terms | GO Accession Number | Enrichment Ratio | Adjusted P Value |
|---|---|---|---|
| Associated with decreased survival | |||
| Response to other organism | GO:0051707 | 3.16 | 0.001 |
| Response to biotic stimulus | GO:0009607 | 3.00 | 0.001 |
| Activation of innate immune response | GO:0002218 | 5.56 | 0.001 |
| Detection of molecule of bacterial origin | GO:0032490 | 30.71 | 0.001 |
| Regulation of innate immune response | GO:0045088 | 4.23 | 0.001 |
| Associated with increased survival | |||
| T cell selection | GO:0045058 | 6.19 | 1 × 104 |
| Lymphocyte differentiation | GO:0030098 | 2.37 | 6 × 104 |
| Thymic T cell selection | GO:0045061 | 6.97 | 6 × 104 |
| Immune system development | GO:0002520 | 1.74 | 0.002 |
| Leukocyte differentiation | GO:0002521 | 2.01 | 0.002 |
Definition of abbreviation: GO = gene ontology.
The top genes most strongly associated with reduced survival included genes forming part of the inflammasomes (absent in melanoma 2 [AIM2], caspase 1 [CASP1], and NLR family caspase recruitment domain [CARD] domain containing 4 [NLRC4]), or related to them (caspase 4 [CASP4] and CARD family member 16 and 17 [CARD16, CARD17]). The set of genes associated with reduced survival was enriched for 50 GO terms, many of which were related to the innate immune response and inflammation, including the ice protease-activating factor and AIM2 inflammasome complexes. In addition, the genes were enriched for six KEGG pathways, including Toll-like receptor (TLR) signaling pathway.
The top genes most strongly associated with increased survival included CD4, the T-helper cell coreceptor; CD28, a coreceptor which mediates T cell activation; and IL2RB, the β subunit of the receptor for IL-2, which plays a key role in the regulation of lymphocyte proliferation and activation, and is also of importance in NK cell function through its roles as a subunit of the IL-15 receptor. The set of genes associated with improved survival was enriched for 26 GO terms, the majority of which were related to T cell and B cell function, and four KEGG pathways, including “antigen processing and presentation.”
WGCNA was repeated focusing only on the pNTM disease cases to investigate disease survival. Nine modules were identified, three of which showed significant differential expression in nonsurvivors compared with survivors (Table 5 and Tables E13 and E14). Of the 215 genes associated with reduced survival by SAM, 74 (34.6%) were present in either the “white” or “grey60” modules, whereas 666 (58.9%) of the 1,131 genes associated with increased survival were present in the “darkorange” module.
Table 5.
Gene Modules Identified as Significantly Associated with Decreased and Increased Survival in Nontuberculous Mycobacterial Disease
| Module Name | No. of Genes | Correlation with Mortality | P Value | Relevant Enriched GO Terms |
|---|---|---|---|---|
| darkorange | 1,413 | −0.5 | 0.01 | T cell receptor signaling pathway Thymic T cell selection |
| grey60 | 490 | 0.55 | 0.004 | RAGE receptor binding |
| White | 71 | 0.46 | 0.02 | None |
Definition of abbreviations: GO = gene ontology; RAGE = receptor for advanced glycation endproducts.
Of note is the enrichment of the grey60 module for the GO term “RAGE receptor binding.” RAGE (the receptor for advanced glycation endproducts) is a pattern recognition receptor that has been implicated in neutrophilic inflammation in tuberculosis (38, 39). Although grey60 module itself was not significantly correlated with neutrophil count (Pearson’s r = 0.29, P = 0.152), many of the genes associated with RAGE were found to be significantly correlated with neutrophilia (HMBG2: Pearson’s r = 0.5, P = 0.011; S100A8: Pearson’s r = 0.56, P = 0.003; S100A9: Pearson’s r = 0.56, P = 0.003; S100A12: Pearson’s r = 0.53, P = 0.006).
Discussion
In this study, we have shown that pNTM disease is associated with the differential expression of over 200 genes. These were characterized by the reduced expression of many genes associated with cellular immunity, some of which have previously been identified to be important in the host response to mycobacterial infection. One of the top differentially expressed genes was IFNG, which plays an essential role in antimycobacterial immunity. Not only was IFNG expression lower in subjects with pNTM disease, but, within this group, reduced IFNG expression was significantly associated with more severe radiological disease and impaired lung function. There was also a trend toward an increased mortality in subjects with lower IFNG expression.
These data are consistent with experimental evidence from in vitro studies, which have demonstrated diminished type 1 cytokine responses in patients with pNTM disease, showing reduced IL-12 (8–10) and IFN-γ (8–13) release after stimulation with both mitogen and NTM. These studies have employed varying methodologies (different cell types, stimuli, and read-outs), and not all such studies have detected a difference in the immune response in subjects with pNTM disease (40–42). The evidence from the present study therefore provides valuable, unbiased evidence for an impaired IFN-γ response in subjects with pNTM disease, and is the first study to link this impairment with adverse clinical features.
There is evidence that immunotherapy with IFN-γ may be beneficial in mycobacterial disease. Recombinant IFN-γ has been shown to improve sputum culture conversion in pulmonary tuberculosis infection (43), and has been used in refractory cerebral tuberculosis (21). Case reports have reported benefit from IFN-γ administration in NTM infection both in the presence (23, 25–27) and absence (28) of immunodeficiency. Two randomized, controlled trials have been conducted in pNTM disease, with one finding no benefit with inhaled IFN-γ (30) and the other finding a significant increase in treatment response with the addition of intramuscular IFN-γ (32).
These results were supported by the networks analysis, which identified gene modules significantly underexpressed in pNTM disease that were enriched for several terms linked to IFN-γ activity. In light of reports of immunodeficiency due to anti-cytokine autoantibodies (44), it is also interesting to note that one module was enriched for genes regulating thymic T cell selection.
The 215 genes associated with reduced survival were significantly enriched for multiple terms relating to innate immunity and pattern recognition receptors, including TLR signaling. Although TLR2 may have a protective role in murine models of NTM infection (45–49), and has been reported to display reduced expression in humans with pNTM disease (8), the proinflammatory response triggered by TLR2 may also be deleterious in NTM infection. The “rough” variant of Mycobacterium abscessus has been shown to trigger a hyperinflammatory response via TLR2 signaling (50), which may play a role in the virulence associated with this phenotype (51).
Interestingly, genes associated with inflammasome formation and pyroptosis were also overrepresented in the genes associated with reduced survival. Inflammasome activation has been reported in tuberculosis (52, 53), and has been proposed as a mechanism for the immune reconstitution inflammatory syndrome seen in human immunodeficiency virus (54). To date, only one report has demonstrated inflammasome activation by NTM (55), and little is known about its role in NTM infection.
Network analysis identified an association between mortality and the expression of genes related to RAGE, including the proteins S100A8 and S100A9 (which, together, form calprotectin, an important proinflammatory mediator) (56). In a murine model, expression of RAGE in lung tissue has been shown to increase in response to tuberculosis infection, and gene knockout caused an increase in weight loss and mortality (57). The expression of both S100A8 and S100A9 have been associated with neutrophilic inflammation in tuberculosis (38, 39), with the expression of S100A9 being suggested as a potential tuberculosis biomarker (58).
Improved survival was associated with 1,131 genes, which were enriched for several GO terms predominantly relating to T cell differentiation and activation. Despite the trend toward higher expression in surviving pNTM cases, IFNG was not identified by SAM as being significantly associated with survival, although it was related to many of the enriched functional terms. Again, the results of network analysis were also supportive of these findings and identified a gene module associated with survival that was enriched for terms associated with T cell development and signaling.
Taken together, these findings suggest that pNTM disease is associated with the downregulation of genes involved in the T cell response, including IFNG, which is consistent with the results of in vitro studies in subjects with NTM infection. In subjects in whom disease has developed, the expression of genes involved in the T cell response is associated with a better prognosis, perhaps reflecting the preservation of an appropriate response to NTM infection. Conversely, a poor prognosis is associated with an upregulation of genes involved in innate immunity and inflammation, which may potentially mediate tissue damage and, thus, the progressive radiological changes observed.
Infection with Mycobacterium tuberculosis has been reported to downregulate in vitro IFN-γ release (59); thus, it is not clear whether the differences in gene expression represent a predisposing defect in host response or are a result of immunomodulation after NTM infection. The reduction in IFNG seen as disease duration increases may point toward the latter. Similarly, the question remains as to whether the upregulation of the innate immune response associated with poor survival is in some way contributory to pathogenesis, perhaps due to excessive inflammation and tissue damage, or is merely a response to an increased burden of mycobacterial infection in those with severe disease.
Pulmonary NTM disease is a heterogeneous condition containing a variety of clinical phenotypes, the most clinically relevant distinction being between fibrocavitary and nodular-bronchiectatic disease. It is likely that the transcriptional response differs between these groups. Given the small sample size, no comparison was made between different radiological patterns; however, it is notable that IFNG expression strongly correlated with composite CT score, and was lower (although not significantly so) in individuals with severe cavitation. Another notable subgroup of individuals with pNTM disease is that which demonstrates stable, nonprogressive disease in the absence of treatment (1, 6, 7), and it is also likely that the gene expression profiles of those subjects differ from those seen in the individuals with active infection included in the present cohort.
Several studies have been conducted examining gene expression in tuberculosis, and, recently, Blankley and colleagues (34) characterized a 380-gene “meta-signature” of active tuberculosis showing consistent differential expression in at least 9 datasets. There was little overlap between this signature and the genes associated with pNTM disease in the current study, and, in contrast to pNTM disease gene expression in tuberculosis, was characterized by an upregulation of IFN-γ–regulated genes (although not IFNG itself). This difference may relate to the infecting organism, or reflect a more appropriate host response to tuberculosis in a generally younger population with fewer comorbidities, as opposed to the potentially aberrant response seen in individuals with pNTM disease. There was, however, a point of similarity in that genes related to T cells were consistently downregulated in both tuberculosis and pNTM disease.
There was no overlap with experimentally derived expression signatures of sarcoidosis (35–37). However, the 213 transcripts associated with pNTM disease were significantly enriched for genes in the 31-gene sarcoid signature described by Zhou and colleagues (35). These genes were chosen a priori, as they are part of the T cell receptor, Janus kinase-signal transducer and activator of transcription, and cytokine–cytokine receptor signaling pathways implicated in sarcoidosis, and were able to differentiate sarcoidosis from control subjects with 82.2% accuracy. In the current study, 11 (35%) of these genes were also associated with pNTM disease, all of which were regulated in the same direction (down) in sarcoidosis. This is particularly interesting, given the hypothesis that sarcoidosis may be driven by an aberrant immune response to mycobacterial antigens (60).
This study has some limitations. The selection of subjects with definite active disease was necessary to provide a cohort with a well defined phenotype, but has limited the sample size, and therefore the statistical power, of the study. This may therefore limit the ability to generalize these findings to the wider NTM population. Even with such selection, the pNTM study population remains highly heterogeneous, further limiting the power to detect differences in expression. This may explain the relatively modest differences in expression seen between groups in contrast to studies of more homogenous populations, such as those seen in tuberculosis. The use of inhaled or systemic steroids may have an effect on gene expression; however, the proportions were similar between pNTM cases and control subjects, and the exclusion of these subjects would necessitate the exclusion of most subjects with COPD, an important group at risk of pNTM disease, and would further limit the ability to generalize the findings.
Although being matched for basic demographics, important phenotypic differences still remained between cases and control subjects, such as the high mortality seen in cases, systemic steroid use, and the treatment given for NTM. These differences may potentially contribute to the observed differences in gene expression. However, intervening to reduce the differences between groups by altering treatment would not have been feasible on ethical grounds, and the mortality in our cohort is in keeping with the known high mortality associated with pNTM disease reported in other cohorts (1–5). Furthermore, the expression of the majority of the differentially expressed transcripts (including IFNG) was significantly associated with pNTM disease, independent of systemic steroid use or NTM treatment. Similarly, several other factors were observed to be associated with survival (such as the presence of mycetoma), and the association seen between gene expression and survival may therefore be confounded by these factors. Unfortunately, the sample size precludes multivariate analyses to investigate them further.
Conclusions
This is the first study of gene expression associated with NTM infection. The expression of many genes mediating cellular immunity was depressed in pNTM disease, including IFNG, which correlated negatively with markers of disease severity. Although requiring further validation in independent cohorts, these findings are mechanistically plausible and consistent with previous experimental data. Mortality in pNTM disease was associated with reduced expression of genes related to cellular immunity and increased expression of genes related to innate immunity, suggesting that disease progression may be driven by immune dysregulation. To our knowledge, this is the first time the immune response to NTM has been linked to differences in disease outcomes. The expression of genes identified in this study warrants further investigation as potential markers of disease activity, and suggests targets for potential therapeutic intervention.
Acknowledgments
Acknowledgment
The authors thank the staff of the Royal Brompton Hospital Respiratory Clinical Research Facility for their help in the collection of samples and phenotypic data. We would like to thank Dr. Edward Chan (National Jewish Health, Denver, CO) for alerting us to a typograhical error in the original ahead-of-print version of this article.
Footnotes
This work was supported by a grant from the Welton Foundation (S.A.C. and M.R.L.); additional funding was provided by the Asmarley Trust and the Wellcome Trust. Support was also provided by the National Institute for Health Research Respiratory Disease Biomedical Research Unit at the Royal Brompton and Harefield National Health Service Foundation Trust and Imperial College London. D.M.H. is the recipient of a National Institute of Health Research Senior Investigator Award.
Author Contributions: S.A.C. collected samples and phenotypic information, performed the gene expression experiments and subsequent data analysis, and composed the manuscript; R.W. treated the patients and contributed phenotypic information and critical input; J.J. and D.M.H. scored the computed tomography scans; W.O.C.C. and M.F.M. advised on experimental and study design, and provided expertise in gene expression and data analysis; P.K. helped analyze immunological data; M.R.L. treated the patients, contributed phenotypic information, and conceived and designed the study; all authors contributed to the final manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2017-0230OC on December 5, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Gochi M, Takayanagi N, Kanauchi T, Ishiguro T, Yanagisawa T, Sugita Y. Retrospective study of the predictors of mortality and radiographic deterioration in 782 patients with nodular/bronchiectatic Mycobacterium avium complex lung disease. BMJ Open. 2015;5:e008058. doi: 10.1136/bmjopen-2015-008058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayashi M, Takayanagi N, Kanauchi T, Miyahara Y, Yanagisawa T, Sugita Y. Prognostic factors of 634 HIV-negative patients with Mycobacterium avium complex lung disease. Am J Respir Crit Care Med. 2012;185:575–583. doi: 10.1164/rccm.201107-1203OC. [DOI] [PubMed] [Google Scholar]
- 3.Andréjak C, Thomsen VØ, Johansen IS, Riis A, Benfield TL, Duhaut P, et al. Nontuberculous pulmonary mycobacteriosis in Denmark: incidence and prognostic factors. Am J Respir Crit Care Med. 2010;181:514–521. doi: 10.1164/rccm.200905-0778OC. [DOI] [PubMed] [Google Scholar]
- 4.Fleshner M, Olivier KN, Shaw PA, Adjemian J, Strollo S, Claypool RJ, et al. Mortality among patients with pulmonary non-tuberculous mycobacteria disease. Int J Tuberc Lung Dis. 2016;20:582–587. doi: 10.5588/ijtld.15.0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gommans EPAT, Even P, Linssen CFM, van Dessel H, van Haren E, de Vries GJ, et al. Risk factors for mortality in patients with pulmonary infections with non-tuberculous mycobacteria: a retrospective cohort study. Respir Med. 2015;109:137–145. doi: 10.1016/j.rmed.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki Y, Kubo K, Takamizawa A, Yamamoto H, Honda T, Sone S. Markers indicating deterioration of pulmonary Mycobacterium avium–intracellulare infection. Am J Respir Crit Care Med. 1999;160:1851–1855. doi: 10.1164/ajrccm.160.6.9902019. [DOI] [PubMed] [Google Scholar]
- 7.Lee G, Kim HS, Lee KS, Koh W-J, Jeon K, Jeong B-H, et al. Serial CT findings of nodular bronchiectatic Mycobacterium avium complex pulmonary disease with antibiotic treatment. AJR Am J Roentgenol. 2013;201:764–772. doi: 10.2214/AJR.12.9897. [DOI] [PubMed] [Google Scholar]
- 8.Ryu YJ, Kim EJ, Lee S-H, Kim SY, Suh GY, Chung MP, et al. Impaired expression of Toll-like receptor 2 in nontuberculous mycobacterial lung disease. Eur Respir J. 2007;30:736–742. doi: 10.1183/09031936.00039507. [DOI] [PubMed] [Google Scholar]
- 9.Vankayalapati R, Wizel B, Samten B, Griffith DE, Shams H, Galland MR, et al. Cytokine profiles in immunocompetent persons infected with Mycobacterium avium complex. J Infect Dis. 2001;183:478–484. doi: 10.1086/318087. [DOI] [PubMed] [Google Scholar]
- 10.Kwon Y-S, Kim EJ, Lee S-H, Suh GY, Chung MP, Kim H, et al. Decreased cytokine production in patients with nontuberculous mycobacterial lung disease. Hai. 2007;185:337–341. doi: 10.1007/s00408-007-9040-z. [DOI] [PubMed] [Google Scholar]
- 11.Kartalija M, Ovrutsky AR, Bryan CL, Pott GB, Fantuzzi G, Thomas J, et al. Patients with nontuberculous mycobacterial lung disease exhibit unique body and immune phenotypes. Am J Respir Crit Care Med. 2013;187:197–205. doi: 10.1164/rccm.201206-1035OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greinert U, Schlaak M, Rüsch-Gerdes S, Flad HD, Ernst M. Low in vitro production of interferon-γ and tumor necrosis factor-α in HIV-seronegative patients with pulmonary disease caused by nontuberculous mycobacteria. J Clin Immunol. 2000;20:445–452. doi: 10.1023/a:1026407815946. [DOI] [PubMed] [Google Scholar]
- 13.Safdar A, Armstrong D, Murray HW. A novel defect in interferon-γ secretion in patients with refractory nontuberculous pulmonary mycobacteriosis. Ann Intern Med. 2003;138:521. doi: 10.7326/0003-4819-138-6-200303180-00030. [DOI] [PubMed] [Google Scholar]
- 14.Szymanski EP, Leung JM, Fowler CJ, Haney C, Hsu AP, Chen F, et al. Pulmonary nontuberculous mycobacterial infection: a multisystem, multigenic disease. Am J Respir Crit Care Med. 2015;192:618–628. doi: 10.1164/rccm.201502-0387OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoumot Z, Boutou AK, Gill SS, van Zeller M, Hansell DM, Wells AU, et al. Mycobacterium avium complex infection in non-cystic fibrosis bronchiectasis. Respirology. 2014;19:714–722. doi: 10.1111/resp.12287. [DOI] [PubMed] [Google Scholar]
- 16.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiani A, García-Cózar FJ, Habermann I, Laforsch S, Aebischer T, Ehninger G, et al. Regulation of interferon-γ gene expression by nuclear factor of activated T cells. Blood. 2001;98:1480–1488. doi: 10.1182/blood.v98.5.1480. [DOI] [PubMed] [Google Scholar]
- 20.Erb KJ, Twardzik T, Palmetshofer A, Wohlleben G, Tatsch U, Serfling E. Mice deficient in nuclear factor of activated T-cell transcription factor c2 mount increased Th2 responses after infection with Nippostrongylus brasiliensis and decreased Th1 responses after mycobacterial infection. Infect Immun. 2003;71:6641–6647. doi: 10.1128/IAI.71.11.6641-6647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J-Y, Yim J-J, Yoon B-W. Adjuvant interferon-γ treatment in two cases of refractory tuberculosis of the brain. Clin Neurol Neurosurg. 2012;114:732–734. doi: 10.1016/j.clineuro.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Kaminuma O, Kitamura F, Kitamura N, Hiroi T, Miyoshi H, Miyawaki A, et al. Differential contribution of NFATc2 and NFATc1 to TNF-α gene expression in T cells. J Immunol. 2008;180:319–326. doi: 10.4049/jimmunol.180.1.319. [DOI] [PubMed] [Google Scholar]
- 23.Squires KE, Murphy WF, Madoff LC, Murray HW. Interferon-γ and Mycobacterium avium–intracellulare infection. J Infect Dis. 1989;159:599–600. doi: 10.1093/infdis/159.3.599. [DOI] [PubMed] [Google Scholar]
- 24.Valle-Rios R, Patiño-Lopez G, Medina-Contreras O, Canche-Pool E, Recillas-Targa F, Lopez-Bayghen E, et al. Characterization of CRTAM gene promoter: AP-1 transcription factor control its expression in human T CD8 lymphocytes. Mol Immunol. 2009;46:3379–3387. doi: 10.1016/j.molimm.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Squires KE, Brown ST, Armstrong D, Murphy WF, Murray HW. Interferon-γ treatment for Mycobacterium avium–intracellular complex bacillemia in patients with AIDS. J Infect Dis. 1992;166:686–687. doi: 10.1093/infdis/166.3.686. [DOI] [PubMed] [Google Scholar]
- 26.Holland SM, Eisenstein EM, Kuhns DB, Turner ML, Fleisher TA, Strober W, et al. Treatment of refractory disseminated nontuberculous mycobacterial infection with interferon γ: a preliminary report. N Engl J Med. 1994;330:1348–1355. doi: 10.1056/NEJM199405123301904. [DOI] [PubMed] [Google Scholar]
- 27.Hallstrand TS, Ochs HD, Zhu Q, Liles WC. Inhaled IFN-γ for persistent nontuberculous mycobacterial pulmonary disease due to functional IFN-γ deficiency. Eur Respir J. 2004;24:367–370. doi: 10.1183/09031936.04.00036704. [DOI] [PubMed] [Google Scholar]
- 28.Chatte G, Panteix G, Perrin-Fayolle M, Pacheco Y. Aerosolized interferon γ for Mycobacterium avium–complex lung disease. Am J Respir Crit Care Med. 1995;152:1094–1096. doi: 10.1164/ajrccm.152.3.7663788. [DOI] [PubMed] [Google Scholar]
- 29.Mathew PA, Chuang SS, Vaidya SV, Kumaresan PR, Boles KS, Pham H-TK. The LLT1 receptor induces IFN-γ production by human natural killer cells. Mol Immunol. 2004;40:1157–1163. doi: 10.1016/j.molimm.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 30.Lam PK, Griffith DE, Aksamit TR, Ruoss SJ, Garay SM, Daley CL, et al. Factors related to response to intermittent treatment of Mycobacterium avium complex lung disease. Am J Respir Crit Care Med. 2006;173:1283–1289. doi: 10.1164/rccm.200509-1531OC. [DOI] [PubMed] [Google Scholar]
- 31.Cao W, Tang S, Yuan H, Wang H, Zhao X, Lu H. Mycobacterium tuberculosis antigen Wag31 induces expression of C-chemokine XCL2 in macrophages. Curr Microbiol. 2008;57:189–194. doi: 10.1007/s00284-008-9172-2. [DOI] [PubMed] [Google Scholar]
- 32.Milanés-Virelles MT, García-García I, Santos-Herrera Y, Valdés-Quintana M, Valenzuela-Silva CM, Jiménez-Madrigal G, et al. MACGAM Study Group. Adjuvant interferon γ in patients with pulmonary atypical mycobacteriosis: a randomized, double-blind, placebo-controlled study. BMC Infect Dis. 2008;8:17. doi: 10.1186/1471-2334-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei Y, Takahama Y. XCL1 and XCR1 in the immune system. Microbes Infect. 2012;14:262–267. doi: 10.1016/j.micinf.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Blankley S, Graham CM, Levin J, Turner J, Berry MPR, Bloom CI, et al. A 380-gene meta-signature of active tuberculosis compared with healthy controls. Eur Respir J. 2016;47:1873–1876. doi: 10.1183/13993003.02121-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou T, Zhang W, Sweiss NJ, Chen ES, Moller DR, Knox KS, et al. Peripheral blood gene expression as a novel genomic biomarker in complicated sarcoidosis. PLoS One. 2012;7:e44818. doi: 10.1371/journal.pone.0044818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koth LL, Solberg OD, Peng JC, Bhakta NR, Nguyen CP, Woodruff PG. Sarcoidosis blood transcriptome reflects lung inflammation and overlaps with tuberculosis. Am J Respir Crit Care Med. 2011;184:1153–1163. doi: 10.1164/rccm.201106-1143OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monast CS, Li K, Judson MA, Baughman RP, Wadman E, Watt R, et al. Sarcoidosis extent relates to molecular variability. Clin Exp Immunol. 2017;188:444–454. doi: 10.1111/cei.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marais S, Wilkinson KA, Lesosky M, Coussens AK, Deffur A, Pepper DJ, et al. Neutrophil-associated central nervous system inflammation in tuberculous meningitis immune reconstitution inflammatory syndrome. Clin Infect Dis. 2014;59:1638–1647. doi: 10.1093/cid/ciu641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gopal R, Monin L, Torres D, Slight S, Mehra S, McKenna KC, et al. S100A8/A9 proteins mediate neutrophilic inflammation and lung pathology during tuberculosis. Am J Respir Crit Care Med. 2013;188:1137–1146. doi: 10.1164/rccm.201304-0803OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Jong E, Lim A, Waterer G, Price P. Monocyte-derived macrophages do not explain susceptibility to pulmonary non-tuberculous mycobacterial disease. Clin Transl Immunology. 2012;1:e2. doi: 10.1038/cti.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim A, Allison C, Price P, Waterer G. Susceptibility to pulmonary disease due to Mycobacterium avium–intracellulare complex may reflect low IL-17 and high IL-10 responses rather than Th1 deficiency. Clin Immunol. 2010;137:296–302. doi: 10.1016/j.clim.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Kim RD, Greenberg DE, Ehrmantraut ME, Guide SV, Ding L, Shea Y, et al. Pulmonary nontuberculous mycobacterial disease: prospective study of a distinct preexisting syndrome. Am J Respir Crit Care Med. 2008;178:1066–1074. doi: 10.1164/rccm.200805-686OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawson R, Condos R, Tse D, Huie ML, Ress S, Tseng C-H, et al. Immunomodulation with recombinant interferon-γ1b in pulmonary tuberculosis. PLoS One. 2009;4:e6984. doi: 10.1371/journal.pone.0006984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browne SK, Holland SM MD SKB. Anticytokine autoantibodies in infectious diseases: pathogenesis and mechanisms. Lancet Infect Dis. 2010;10:875–885. doi: 10.1016/S1473-3099(10)70196-1. [DOI] [PubMed] [Google Scholar]
- 45.Sampaio EP, Elloumi HZ, Zelazny A, Ding L, Paulson ML, Sher A, et al. Mycobacterium abscessus and M. avium trigger Toll-like receptor 2 and distinct cytokine response in human cells. Am J Respir Cell Mol Biol. 2008;39:431–439. doi: 10.1165/rcmb.2007-0413OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng CG, Scanga CA, Collazo-Custodio CM, Cheever AW, Hieny S, Caspar P, et al. Mice lacking myeloid differentiation factor 88 display profound defects in host resistance and immune responses to Mycobacterium avium infection not exhibited by Toll-like receptor 2 (TLR2)- and TLR4-deficient animals. J Immunol. 2003;171:4758–4764. doi: 10.4049/jimmunol.171.9.4758. [DOI] [PubMed] [Google Scholar]
- 47.Gomes MS, Sousa Fernandes S, Cordeiro JV, Silva Gomes S, Vieira A, Appelberg R. Engagement of Toll-like receptor 2 in mouse macrophages infected with Mycobacterium avium induces non-oxidative and TNF-independent anti-mycobacterial activity. Eur J Immunol. 2008;38:2180–2189. doi: 10.1002/eji.200737954. [DOI] [PubMed] [Google Scholar]
- 48.Carvalho NB, Oliveira FS, Durães FV, de Almeida LA, Flórido M, Prata LO, et al. Toll-like receptor 9 is required for full host resistance to Mycobacterium avium infection but plays no role in induction of Th1 responses. Infect Immun. 2011;79:1638–1646. doi: 10.1128/IAI.01030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin D-M, Yang C-S, Yuk J-M, Lee J-Y, Kim KH, Shin SJ, et al. Mycobacterium abscessus activates the macrophage innate immune response via a physical and functional interaction between TLR2 and dectin-1. Cell Microbiol. 2008;10:1608–1621. doi: 10.1111/j.1462-5822.2008.01151.x. [DOI] [PubMed] [Google Scholar]
- 50.Roux A-L, Ray A, Pawlik A, Medjahed H, Etienne G, Rottman M, et al. Overexpression of proinflammatory TLR-2–signalling lipoproteins in hypervirulent mycobacterial variants. Cell Microbiol. 2011;13:692–704. doi: 10.1111/j.1462-5822.2010.01565.x. [DOI] [PubMed] [Google Scholar]
- 51.Catherinot E, Clarissou J, Etienne G, Ripoll F, Emile J-F, Daffé M, et al. Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect Immun. 2007;75:1055–1058. doi: 10.1128/IAI.00835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hagar JA, Miao EA. Detection of cytosolic bacteria by inflammatory caspases. Curr Opin Microbiol. 2014;17:61–66. doi: 10.1016/j.mib.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Man SM, Karki R, Kanneganti T-D. AIM2 inflammasome in infection, cancer, and autoimmunity: role in DNA sensing, inflammation, and innate immunity. Eur J Immunol. 2016;46:269–280. doi: 10.1002/eji.201545839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan HY, Yong YK, Shankar EM, Paukovics G, Ellegård R, Larsson M, et al. Aberrant inflammasome activation characterizes tuberculosis-associated immune reconstitution inflammatory syndrome. J Immunol. 2016;196:4052–4063. doi: 10.4049/jimmunol.1502203. [DOI] [PubMed] [Google Scholar]
- 55.Lee H-M, Yuk J-M, Kim KH, Jang J, Kang G, Park JB, et al. Mycobacterium abscessus activates the NLRP3 inflammasome via Dectin-1-Syk and p62/SQSTM1. Immunol Cell Biol. 2012;90:601–610. doi: 10.1038/icb.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrchen JM, Sunderkötter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 57.van Zoelen MAD, Wieland CW, van der Windt GJW, Florquin S, Nawroth PP, Bierhaus A, et al. Receptor for advanced glycation end products is protective during murine tuberculosis. Mol Immunol. 2012;52:183–189. doi: 10.1016/j.molimm.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 58.Xu D, Li Y, Li X, Wei L-L, Pan Z, Jiang T-T, et al. Serum protein S100A9, SOD3, and MMP9 as new diagnostic biomarkers for pulmonary tuberculosis by iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics. 2015;15:58–67. doi: 10.1002/pmic.201400366. [DOI] [PubMed] [Google Scholar]
- 59.O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MPR. The immune response in tuberculosis. Annu Rev Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 60.Brownell I, Ramírez-Valle F, Sanchez M, Prystowsky S. Evidence for mycobacteria in sarcoidosis. Am J Respir Cell Mol Biol. 2011;45:899–905. doi: 10.1165/rcmb.2010-0433TR. [DOI] [PMC free article] [PubMed] [Google Scholar]