Abstract
Light regulates various aspects of plant growth, and the photoreceptor phytochrome B (phyB) mediates many responses to red light. In a screen for Arabidopsis mutants with phenotypes similar to those of phyB mutants, we isolated two new elf3 mutants. One has weaker morphological phenotypes than previously identified elf3 alleles, but still abolishes circadian rhythms under continuous light. Like phyB mutants, elf3 mutants have elongated hypocotyls and petioles, flower early, and have defects in the red light response. However, we found that elf3 mutations have an additive interaction with a phyB null mutation, with phyA or hy4 null mutations, or with a PHYB overexpression construct, and that an elf3 mutation does not prevent nuclear localization of phyB. These results suggest that either there is substantial redundancy in phyB and elf3 function, or the two genes regulate distinct signaling pathways.
Plants adjust their growth and development according to diurnal, seasonal, and local variations in their light environment. Light can induce leaf formation, leaf expansion, and chloroplast differentiation; inhibit stem elongation; induce bending toward or away from light; and induce or repress flowering. Light can also phase the circadian rhythm. Several photoreceptors sense light, including red/far-red light receptors called phytochromes, blue light receptors called cryptochromes, the NPH1 photoreceptor required for phototropism, and hypothesized UV light receptors (Fankhauser and Chory, 1997; Deng and Quail, 1999).
Genetic analyses in Arabidopsis have been particularly helpful in dissecting the roles of the various photoreceptors. Arabidopsis has five phytochromes, phyA to phyE, and two cryptochromes, cry1 (also known as HY4) and cry2. Analyses of the effects of mutations in genes encoding PHYA, PHYB, PHYD, PHYE, CRY1, CRY2, and NPH1 and transgenic plants overexpressing PHYA, PHYB, PHYC, CRY1, or CRY2 have revealed the developmental functions and capabilities of each of these photoreceptors (Fankhauser and Chory, 1997; Deng and Quail, 1999). The various phytochromes and cryptochromes share some functions, but are also specialized to some degree. For example, different photoreceptors contribute to inhibition of hypocotyl elongation under different light conditions. In white light, phyB and cry1 play the largest roles and phyA, phyD, and cry2 play lesser roles (Reed et al., 1994; Aukerman et al., 1997; Smith et al., 1997; Lin et al., 1998). Signal transduction pathways downstream of these photoreceptors probably interact. For example, under some light conditions phyB and cry1 require each other's activity for maximum inhibition of hypocotyl elongation (Casal and Boccalandro, 1995; Casal and Mazzella, 1998). Conversely, whereas phyB normally inhibits flowering, phyA and cry2 each promote flowering under certain light conditions (Johnson et al., 1994; Reed et al., 1994; Guo et al., 1998). cry2 mutant plants flower later than wild-type plants in light containing both red and blue frequencies, and a phyB mutation suppresses this effect, indicating that cry2 antagonizes phyB-mediated inhibition of flowering (Guo et al., 1998; Mockler et al., 1999). Thus, signal transduction pathways downstream of different photoreceptors may reinforce or antagonize each other, depending on the response.
Phytochromes exist in two photointerconvertible forms called Pr and Pfr. Red light converts Pr to Pfr, which absorbs far-red light. Far-red light reconverts Pfr to Pr. For most responses it is thought that Pfr is the active form, because most phytochrome-mediated responses are induced by red light (Furuya, 1993; Quail et al., 1995). However, phyA mediates far-red light responses, and therefore it is possible that the Pr form of phyA is active (Shinomura et al., 2000). Recent biochemical results have shown that phytochromes act as kinases (Yeh et al., 1997; Yeh and Lagarias, 1998; for review, see Reed, 1999). Both phyA and phyB proteins localize to the nucleus under light conditions when they mediate light responses, suggesting that nuclear localization may be important for phytochrome signaling (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999).
Other recent work has aimed to identify downstream targets of phytochromes. Several mutations cause phenotypes similar to those caused by mutations in phytochrome genes (Whitelam et al., 1993; Ahmad and Cashmore, 1996; Barnes et al., 1996; Lin and Cheng, 1997; Wagner et al., 1997; Soh et al., 1998; Hudson et al., 1999) or confer hypersensitive red and/or far-red light responses (Genoud et al., 1998; Hoecker et al., 1998). These mutations may affect genes encoding immediate targets of phytochrome action or downstream regulators of phytochrome signaling. Other potential phytochrome signaling components have been identified in yeast two-hybrid screens. The PIF3 and PKS1 proteins can interact with both phyA and phyB, and NDPK2 interacts with phyA (Ni et al., 1998; Choi et al., 1999; Fankhauser et al., 1999). Studies with plants that overexpress or underexpress these genes suggest that PIF3 and NDPK2 activate phytochrome responses, whereas PKS1 may repress phytochrome responses (Ni et al., 1998; Choi et al., 1999; Fankhauser et al., 1999; Halliday et al., 1999). Given the complexity of light responses and the relatively subtle phenotypes of these transgenic plants, these proteins are probably just a subset of actual phytochrome signaling components.
The circadian system controls biological rhythms with a period of roughly 24 h (Lumsden and Millar, 1998). Circadian-regulated outputs in Arabidopsis include expression of many genes, leaf movements, and hypocotyl growth (Millar and Kay, 1991; Hicks et al., 1996; Dowson-Day and Millar, 1999). Both red and blue light signals control the phase, period, and amplitude of circadian rhythms in higher plants (Lumsden and Millar, 1998). In Arabidopsis, phyA, phyB, and cry1 have all been shown to participate in these responses of the circadian system (Millar et al., 1995; Anderson et al., 1997; Somers et al., 1998). Light regulation and circadian control may allow more flexible responses together than either does alone; the two modes of regulation are frequently associated, sometimes in a complex manner. The processes that are directly regulated by phyB, for example, overlap with those controlled by circadian rhythms. For CAB gene activation, the amplitude of the light response is modulated by the circadian clock (Millar and Kay, 1996; Anderson et al., 1997).
Arabidopsis phyB mutants have several defects in red light responses, including reduced seed germination, reduced induction of CAB gene expression, elongated hypocotyls and stems, and a longer circadian rhythm period, and they also flower early (Koornneef et al., 1980; Reed et al., 1993, 1994; Halliday et al., 1994; Shinomura et al., 1994; Somers et al., 1998). In a screen for mutants with phenotypes similar to those of the phyB mutants, we have discovered two alleles of a previously known locus called ELF3. elf3 mutants were first identified based on their early flowering phenotype, but also have elongated hypocotyls and lack circadian rhythms in constant light (Hicks et al., 1996; Zagotta et al., 1996; Dowson-Day and Millar, 1999). In constant darkness, elf3-1 plants retained a circadian rhythm, and it was proposed that ELF3 mediates an interaction between light and the circadian clock, rather than being a component of the clock itself (Hicks et al., 1996). We report genetic and physiological experiments that explore the relationship between ELF3 and phyB. We have compared several phenotypes of elf3 and phyB mutants, and have assayed light responsiveness of hypocotyl elongation in elf3 mutants and several double mutants between elf3 and photoreceptor mutations. These phenotypic analyses showed that elf3 mutants resemble phyB mutants in several respects, and we have therefore tested whether the elf3-1 null mutation affects either the ability of overexpressed phyB to confer a phenotype or phyB nuclear localization. Finally, we have examined the effects of a weak elf3 allele on circadian rhythms. We find that ELF3 and phyB can act independently to control a common set of phenotypes.
MATERIALS AND METHODS
Mutant Alleles and Double Mutants
The phyB-1 mutation has a stop codon in the PHYB gene and is in the Arabidopsis Landsberg erecta ecotype (Reed et al., 1993). All other mutants used in this study were in the Columbia ecotypic background. The elf3-1 mutation (Zagotta et al., 1996) and the elf3-9 mutation each create a stop codon and behave like null alleles of ELF3 (K. Hicks and D.R. Wagner, personal communication). elf3-1 also carries the gl1 mutation. elf3-7 has a splice site mutation (K. Hicks and D.R. Wagner, personal communication). elf3-9 was isolated in the phyA-211 background. phyA-211 has a rearrangement in the PHYA gene and is a presumed null mutation (Reed et al., 1994; C. Fankhauser, personal communication). The phyB-9 mutation has a stop codon and is also a presumed null mutation (Reed et al., 1993). hy4-101 was originally called blu1 and has not been sequenced, but lacks CRY1 protein (Liscum and Hangarter, 1991; Bagnall et al., 1996; Mockler et al., 1999). To construct double mutants between the linked elf3-1 (or elf3-7) and phyB-9 mutations, single mutants were crossed, and the resulting F1 double heterozygotes were crossed with the corresponding elf3 single mutant. Multiple tall F1 individuals (elf3/elf3) from this second cross were test-crossed with phyB-9 to identify individuals with a recombinant elf3 phyB-9 chromosome. Double mutants were then identified among the self progeny of plants carrying such a recombinant chromosome. Double mutants involving unlinked mutations were identified by phenotypic criteria and then confirmed by test crosses.
Growth Conditions and Light Response Experiments
Growth conditions and light response experiments were performed as described previously (Reed et al., 1998): Seeds were surface-sterilized and plated on Murashige and Skoog (MS)/agar plates containing 1× MS salts (Gibco, Grand Island, NY), 0.8% (w/v) Phytagar (Gibco), and 1× Gamborg's B5 vitamin mix (Sigma, St. Louis) with or without 2% (w/v) Suc, stored overnight at 4°C, and moved to the appropriate light condition. Red, far-red, and blue light sources were as described previously (Reed et al., 1998). For far-red and blue light experiments, results for only the highest fluence rates (approximately 50 μmol m−2 s−1 for far-red light and 10 μmol m−2 s−1 for blue light) are shown. For flowering time determinations, seedlings were grown on MS/agar/2% (w/v) Suc plates for 10 to 14 d, and then transplanted to soil (Pro-Mix BX, Hummert, St. Louis). Plants were grown in a Conviron (Winnipeg, Manitoba, Canada) growth chamber at 21°C. Light was provided on a 9:15 h day:night cycle from 12 fluorescent (F72T12/CW/VHO, 160 W) and six incandescent (60 W) bulbs, and had an intensity at plant height of 170 to 220 μmol m−2 s−1.
Circadian Rhythm Assays
Seeds for circadian rhythm experiments were grown in MS/agar medium with 3% (w/v) Suc, as described previously (Millar et al., 1995; Dowson-Day and Millar, 1999). Seed containing the CAB2::LUC reporter gene in the elf3-1 background have also been described previously (Hicks et al., 1996). The same reporter gene was introduced into the elf3-7 background in a similar manner, following a cross between a homozygous elf3-7 mutant and a line in which the CAB2::LUC reporter had been introgressed into the Columbia background. Rhythmic cotyledon movement and hypocotyl elongation were analyzed (Dowson-Day and Millar, 1999), and the percentage of arrhythmic plants was calculated (Hicks et al., 1996). Studies of circadian-regulated CAB gene expression were performed (Millar et al., 1995) and analyzed as for the morphological data (Dowson-Day and Millar, 1999). Unless indicated in the text, the data presented are representative of at least two experiments (CAB2::LUC rhythms).
Immunodetection of PHYB
Protein extracts were prepared from 10-d-old seedlings grown in constant light on MS/agar/2% (w/v) Suc plates or from young leaves of greenhouse-grown plants. To prepare the extracts, 100 mg of tissue was added to a microfuge tube containing about 80 to 100 mg of sand and ground with a pestle for 2 min in 75 μL of extraction buffer containing 100 mm Tris, pH 6.8, 4% (w/v) SDS, 20% (w/v) glycerol, 200 mm dithiothreitol (DTT), 5 mm phenylmethylsulfonyl fluoride (PMSF), and 100 μg/mL leupeptin at 95°C. The extracts were incubated at 95°C for 3 more min, then centrifuged at 14,000 rpm for 2 min. The supernatant was transferred to a fresh tube and the protein concentration was determined using a protein assay reagent (Bio-Rad Laboratories, Hercules, CA). The desired volume of total protein extract was mixed with one-tenth volume of bromphenol blue (0.05%, w/v), heated again at 95°C for 2 min, and fractionated on 12% (w/v) SDS-PAGE (Laemmli, 1970).
Proteins were transferred to nitrocellulose membrane (no. BA85, Schleicher & Schuell, Keene, NH) using transblot buffer (containing 3.02 g of Tris, 14.42 g of Gly, and 200 mL of methanol per liter) at 100 V for 30 min at 4°C. The membrane was blocked in TBST (20 mm Tris, pH 7.5, 150 mm NaCl, and 0.1% [w/v] Tween 20) containing 1% (w/v) nonfat dry milk at room temperature for at least 1 h. The antibodies were diluted in PBS (135 mm NaCl, 2.7 mm KCl, 1.5 mm KH2PO4, and 15 mm Na2HPO4, pH 7.5) containing 3% (w/v) bovine serum albumin. The primary antibody, anti-phyB monoclonal mBA2 (a gift from Akira Nagatani, Kyoto University, Kyoto) or anti-c-myc monoclonal from hybridoma line 9E10 (obtained from the Tissue Culture Facility at the University of North Carolina at Chapel Hill Lineberger Comprehensive Cancer Center), was used at a final concentration of 5 μg/mL at room temperature for 2 h. The membrane was washed with TBST three times for 10 min each. The secondary antibody was alkaline-phosphatase-conjugated anti-mouse IgG (Sigma) and was diluted 1:2,000. Proteins were visualized using the chromogenic reagents nitroblue tetrazolium chloride (Jersey Lab Supply, Livingston, NJ) and 5-bromo-4-chloro-3-indolyl phosphate toluidine salt (Jersey Lab Supply) as described in Sambrook et al. (1989).
Construction of Transgenic Plants
T-DNA constructs were introduced into plants by vacuum infiltration (Bechtold et al., 1993). The 35S::PHYB construct was a gift from R. Sharrock (Hirschfeld et al., 1998). An elf3-1 transformant that overexpressed PHYB was identified by western blot, crossed to wild-type Columbia, and ELF3/ELF3 and elf3-1/elf3-1 progeny of this cross used for the phenotypic analyses. The overexpressing line selected had more than one insertion element (data not shown), and therefore we performed western blots on the same seed analyzed for phenotype to confirm that the ELF3/ELF3 and elf3-1/elf3-1 lines contained similar amounts of PHYB protein. Deduced elf3 genotypes of these lines were confirmed by a cleaved-amplified polymorphic sequence assay for elf3-1 generously performed by Karen Hicks (Kenyon College, Gambier, OH).
For nuclear localization experiments, we engineered a c-myc epitope at the C-terminal end of a PHYB cDNA expressed behind its own promoter. We amplified the 3′ end of PHYB by PCR using the oligonucleotides PB7J (5′-TCTGTTTCTTGCAAATCCCGAGC-3′) and PB41 (5′CCT-CCCGGGACCATATGGCATCATCAGCATC-3′, SmaI site underlined), cut the product with EcoRI and SmaI, and ligated the resulting fragment into the corresponding sites of a PHYB cDNA clone in pBluescript (Elich and Chory, 1998). This step eliminated the stop codon and introduced a SmaI restriction site at the C-terminal end of the coding sequence. We then ligated a c-myc cassette derived from a DraI/SacI digestion of plasmid CD3-128 (Arabidopsis Biological Resource Center, Ohio State University, Columbus) into this SmaI site. We sequenced the resulting plasmid to confirm that the c-myc epitope tag was fused in-frame with PHYB at the C-terminal end. Finally, this PHYB-c-myc cDNA construct was fused at an internal SacI site to a genomic PHYB gene containing 2.3 kb of genomic DNA 5′ of the PHYB start codon (corresponding to the upstream sequences determined by Reed et al. [1993]), in a derivative of T-DNA binary vector pBI121 (CLONTECH, Palo Alto, CA) lacking the 35S::GUS gene. Columbia, phyB-1, and elf3-1 transformants carrying this construct and segregating approximately 3:1 for kanamycin resistance were used to generate homozygous lines for the immunolocalization experiments.
Immunolocalization
Tissue for immunolocalization experiments was obtained from leaves of greenhouse-grown (photoperiod artificially extended in winter to 16 h of light/8 h of dark) plants. Protoplasts were obtained as described in Staub et al. (1996), except that a 90-μm nylon mesh was used to filter the protoplasts. Ten to 25 μL of protoplasts were placed per chamber in several chambers of a 16-well glass slide (Lab-Tek Chamber slide system, Nunc, Napierville, IL), allowed to bind to the slide for 2 to 3 min, and extra wash solution was removed with a pipette. The protoplasts were fixed immediately with 50 μL of fixative (3.5% [w/v] paraformaldehyde, 50 mm potassium phosphate, pH 7.0, and 1 mm EGTA) per chamber at room temperature for 30 min. The fixative was discarded, and the protoplasts were allowed to air-dry for 45 min and then left at 4°C overnight.
Fixed protoplasts were treated with cold methanol for 10 min once followed by PBST (PBS + 0.5% [w/v] Tween 20) for 10 min twice to permeabilize the cells. For immunoreactions, the antibodies were diluted in PBST containing 200 μg bovine serum albumin/mL. Both the primary antibody (monoclonal from hybridoma line 9E10) and the secondary antibody fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Cappell, Durham, NC) were used at a dilution of 1:100. The protoplasts were incubated with the primary antibody for 2 h and with the secondary antibody for 1 h at room temperature. Washes were done with PBS (once for 5 min) followed by PBS plus 0.1% (w/v) Tween 20 (once for 5 min), and then PBS (twice for 5 min each). Nuclei were stained with 4′,6-diamidino-2-phenyl-indole (DAPI) at 10 μg/mL at room temperature for 20 min, followed by two washes of 5 min each with PBS. The samples were mounted in Mowiol (Calbiochem, San Diego) containing the antifade reagent p-phenylene diamine dihydrochloride (Polysciences, Warrington, PA), and the slides were left in the cold room overnight to harden. Immunofluorescence was observed using a fluorescence microscope (Nikon, Tokyo) equipped with an FITC filter (excitation 450–490, barrier 520) and a UV filter (excitation 360–370, barrier 400). For photographs, Kodak Select 400 films (Eastman Kodak, Rochester, NY) were used.
RESULTS
Isolation and Morphology of elf3 Mutants
In screens for long hypocotyl mutants, we isolated two mutants that had a strong resemblance to phyB mutants in having elongated hypocotyls and petioles and in flowering early. Mapping and complementation analyses established that the mutations in both of these were alleles of ELF3 (data not shown), and we have called them elf3-7 and elf3-9. The elf3-7 mutation alters a splice site junction (K. Hicks and D.R. Wagner, personal communication), and conferred slightly less extreme phenotypes than the elf3-1 null mutation (Zagotta et al., 1996; see below). The elf3-9 mutant had a stop codon in the coding sequence and is likely a null mutant (K. Hicks and D.R. Wagner, personal communication). For our phenotypic studies we have used the previously described elf3-1 null mutant and our weaker elf3-7 mutant.
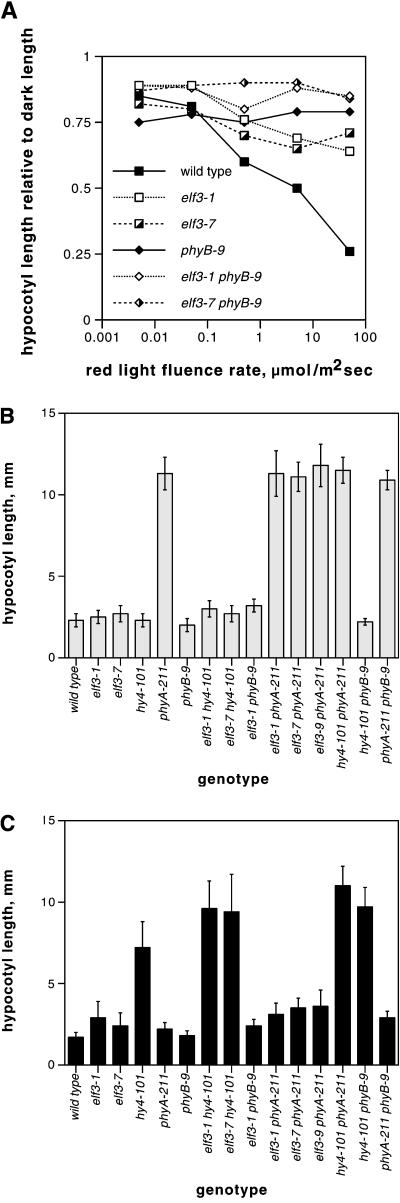
When grown under short-day conditions, elf3-1 and elf3-7 mutants each had long hypocotyls, as did the phyB-9 mutant (Fig. 1A; Table I). In addition, all three mutants had elongated petioles (Fig. 1B; Table I) and flowered earlier and with fewer leaves than wild type (Table I). For each of these phenotypes, the elf3-1 and elf3-7 mutants had more extreme phenotypes than the phyB-9 mutant. The relative effect of the two elf3 alleles depended on the assay. elf3-1 and elf3-7 mutants had similar petiole lengths, but the elf3-1 mutant had a slightly longer hypocotyl and flowered earlier than the elf3-7 mutant. As noted previously for elf3-1 (Zagotta et al., 1996), the elf3-7 long hypocotyl phenotype was less severe when the plants were grown under constant white light (data not shown).
Figure 1.
Morphology of elf3, phyB, and elf3 phyB double mutant plants grown under short-day conditions. A, Appearance of seedlings after 8 d of growth under short-day conditions; B, appearance after 3 weeks of growth under short-day conditions.
Table I.
Measurements of wild type, elf3, phyB, and elf3 phyB double mutant plants grown under short-day conditions ± sd
| Genotype
|
||||||
|---|---|---|---|---|---|---|
| Phenotype | Wild type | elf3-1 | elf3-7 | phyB-9 | elf3-1 phyB-9 | elf3-7 phyB-9 |
| Hypocotyl length, mm | 1.8 ± 0.1 | 6.8 ± 0.6 | 4.9 ± 0.3 | 4.0 ± 0.3 | 8.2 ± 0.5 | |
| Petiole length, mm | 5.6 ± 0.7 | 9.3 ± 1.4 | 9.1 ± 0.7 | 7.4 ± 0.9 | 12.9 ± 0.7 | 12.4 ± 1.4 |
| Leaf blade length, mm | 5.6 ± 0.6 | 4.3 ± 0.5 | 4.1 ± 0.8 | 3.8 ± 0.6 | 4.2 ± 0.5 | 4.1 ± 0.6 |
| Leaf blade width, mm | 4.3 ± 0.5 | 3.1 ± 0.2 | 3.0 ± 0.3 | 3.2 ± 0.4 | 3.0 ± 0.2 | 2.8 ± 0.2 |
| Days to flower | 38.1 ± 4.2 | 24.6 ± 0.6 | 30.6 ± 2.2 | 34.4 ± 6.7 | 23.3 ± 0.5 | 25.0 ± 0.8 |
| Leaf number upon flowering | 31.9 ± 6.3 | 8.4 ± 0.7 | 16.8 ± 2.9 | 20.2 ± 8.6 | 6.9 ± 0.7 | 7.6 ± 1.1 |
Hypocotyl lengths are means of between 12 and 20 measurements of 8-d-old seedlings. Other data are means of 12 to 14 measurements. Leaf dimensions are for the first pair of true leaves of 3-week-old plants.
Effects of elf3-7 on Circadian Rhythms
The elf3-1 mutant is aperiodic in constant light for several circadian rhythm outputs, including CAB::LUC expression, hypocotyl elongation, and cotyledon movements (Hicks et al., 1996; Dowson-Day and Millar, 1999). We tested the elf3-7 mutant for each of these circadian rhythm outputs. As shown in Figure 2, in constant light the elf3-7 mutant was arrhythmic for all three outputs (Fig. 2, A and C), just as has been found previously for the elf3-1 mutant (Hicks et al., 1996). For example, when the criteria of Hicks et al. (1996) are applied to our data, only 12% of elf3-7 plants (n = 26 from five experiments) showed a robust circadian rhythm of cotyledon movement, compared with 93% of wild-type plants (n = 27 from 10 experiments) and 8% of elf3-1 plants (n = 24 from nine experiments). The pattern of CAB::LUC expression was also similar in elf3-1 and elf3-7 plants tested under a light-dark cycle followed by constant darkness (Fig. 2B). The phase advance of the first peak in darkness is less pronounced in elf3-7 (approximately 2 h before the wild type) than in elf3-1 (approximately 4 h before the wild type). The rapid damping of CAB::LUC gene expression in prolonged darkness prevented accurate scoring of later peaks.
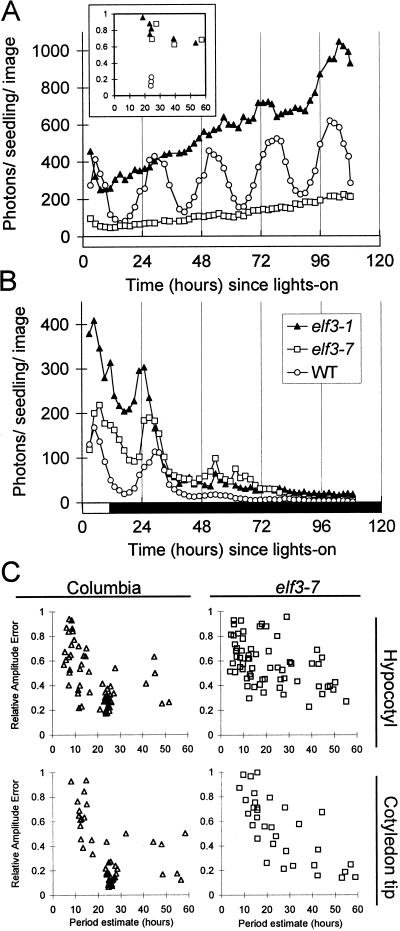
Figure 2.
Circadian rhythm defects in elf3-7. The activity of the CAB::LUC reporter transgene was assayed in seedlings grown under light-dark cycles for 7 d and transferred to constant light at time 0 (A) or constant darkness at time 12 (B). Inset, Rhythmicity of wild-type CAB2::LUC (○; n = 3), elf3-1 (▴; n = 6), and elf3-7 (□; n = 5) plants in the experiment depicted in A was assessed by FFT-NLLS analysis, as in C. Mean expression levels in elf3-7 were variable among experiments, falling within approximately a 4-fold range on either side of the wild-type mean (compare A and B). Black box on time axis, Dark interval; white box, light interval. C, Circadian rhythms of hypocotyl elongation (upper panels) and cotyledon movement (lower panels) were assayed in seeds germinated under two light-dark cycles and transferred to constant dim light. Rhythmicity was assessed by FFT-NLLS analysis (Dowson-Day and Millar, 1999). The clustering of data points with approximately 24-h period and low relative amplitude error (indicating robust rhythmicity) reflect the strong circadian rhythms of wild-type seedlings. elf3-7 seedlings show a uniform scatter of periods, almost all of which have high relative amplitude error (indicating weak rhythms or noise).
Effects of elf3 Mutations on Light Responses
The similar morphological phenotypes of elf3 and phyB mutants suggested that ELF3 may regulate signaling by phyB or other photoreceptors. To determine whether elf3 mutations might preferentially affect responses to any particular photoreceptor, we tested the effect of elf3 mutations on inhibition of hypocotyl elongation by continuous red, far-red, or blue light. phyB mutants have a decreased hypocotyl inhibition response to red light compared with wild type, but a normal response to far-red light and only a subtle decrease in response to blue light. As shown in Figure 3, elf3 mutants also had a decreased response to red light. This decrease was most obvious at high fluence rates, just as for the phyB mutant (Fig. 3A). The magnitude of the effect was similar for elf3-1 and elf3-7 seedlings, each of which showed a slightly greater responsiveness than phyB-9 seedlings.
Figure 3.
Hypocotyl lengths of elf3 mutants and double mutants in different constant light conditions. A, Hypocotyl lengths under various fluence rates of constant red light, normalized to hypocotyl length in the dark. B and C, Hypocotyl lengths under constant far-red (B) and blue light (C). Between 14 and 25 seedlings were measured for each data point. sds in A are omitted for clarity, but were generally about 20% of the mean or less, with higher relative sd at higher fluence rates.
We performed similar experiments under constant far-red light and under constant blue light. In Arabidopsis, phyA mediates far-red light inhibition of hypocotyl elongation (Nagatani et al., 1993; Parks and Quail, 1993; Whitelam et al., 1993), and multiple photoreceptors including cry1, cry2, phyA, phyB, and possibly other phytochromes mediate blue light inhibition of hypocotyl elongation (Koornneef et al., 1980; Young et al., 1992; Ahmad and Cashmore, 1993; Casal and Mazzella, 1998; Lin et al., 1998; Neff and Chory, 1998). As shown in Figure 3B, elf3 seedlings had similar hypocotyl lengths as wild-type seedlings under far-red light. These data suggest that ELF3 plays little role in phyA signaling. Under blue light, elf3-1 and elf3-7 mutant seedlings had slightly longer hypocotyls than wild-type seedlings (Fig. 3C). However, this phenotype was far less dramatic than that of hy4 mutant seedlings (Fig. 3C), suggesting that ELF3 plays a lesser role than cry1 does in this response.
The elf3 seedlings also resembled phyB seedlings in their response to end-of-day far-red light. When given far-red light at the end of each day, hypocotyls of wild-type plants elongate more than if not given far-red light. This response is attributed to persistence in wild-type plants of phyB (and possibly other stable phytochromes) in the active Pfr form at night. When this population is converted to the inactive Pr form by a pulse of far-red light at the end of the day, the hypocotyls elongate more. Since phyB mutant plants lack phyB, they elongate constitutively and thus show only a slight response to end-of-day far-red treatments (Robson et al., 1993). As shown in Figure 4, elf3-1 and elf3-7 mutants had a reduced response, very similar to that of the phyB-9 mutant.
Figure 4.
End-of-day far-red hypocotyl elongation response. Seedlings were grown under short-day conditions, given either no extra light or a saturating pulse of far-red light at the end of each of the 3rd, 4th, 5th, and 6th d, and the hypocotyl lengths measured on the 7th d (±sd). Between 19 and 27 seedlings were measured for each data point.
Genetic Interactions between elf3 Mutations and Photoreceptor Mutations
These hypocotyl elongation results suggested that ELF3 may participate in light responses. To explore the relations among ELF3 and photoreceptors in more detail, we constructed double mutants between elf3-1 and elf3-7 and each of the photoreceptor mutations hy4-101, phyA-211, and phyB-9. Each of these photoreceptor mutations is a putative null allele based on phenotype and biochemical data (see “Materials and Methods”).
In constant red light, the elf3-1 phyB-9 and elf3-7 phyB-9 double mutants had longer hypocotyls than the corresponding single mutants (Fig. 3A). The elf3-1 phyB-9 and elf3-7 phyB-9 double mutants also had more elongated petioles than the corresponding single mutants, and flowered earlier (Fig. 1). These additive phenotypes show that, to some degree, ELF3 and phyB act independently of each other. The elf3-1 phyB-9 double mutant retained a small end-of-day far-red response, as did each of the single mutants (Fig. 4). In constant blue light, the elf3-1 phyB-9 double mutant had a hypocotyl length similar to that of the elf3-1 single mutant (Fig. 3C).
The elf3-1 phyA-211 and elf3-7 phyA-211 double mutants had the same hypocotyl lengths as the phyA-211 single mutant under constant far-red light (Fig. 3B), which is consistent with phyA mediating the entire hypocotyl inhibition response to far-red light (Whitelam et al., 1993) and with the lack of phenotype of elf3 single mutants in far-red light. Under constant blue light, the elf3-1 phyA-211 and elf3-7 phyA-211 double mutants were as tall or slightly taller than the corresponding single mutants (Fig. 3C; data not shown).
The elf3-1 hy4-101 and elf3-7 hy4-101 double mutants were taller than the corresponding elf3 and hy4-101 single mutants in constant blue light (Fig. 3C). The hy4-101 phyB-9 double mutant was also significantly taller than the hy4-101 single mutant. These results show that both ELF3 and phyB can affect blue light responses in a manner that does not depend on cry1. None of these double mutants was as tall in blue light as the hy4-101 phyA-211 double mutant (Fig. 3C). Consistent with these data, previous studies have concluded that phyA and cry1 contribute more to the blue light response than does phyB (Whitelam et al., 1993; Neff and Chory, 1998).
An elf3 Null Mutation Does Not Eliminate phyB-Mediated Hypocotyl Elongation Control
The additive phenotypes in the elf3 phyA, elf3 phyB, and elf3 hy4 double mutants reveal no functional interaction between ELF3 and these photoreceptors. However, phytochromes and cryptochromes act redundantly with each other, and this redundancy may have masked such functional interactions. As the morphological and light response phenotypes of elf3 mutants most closely resembled those of phyB mutants, we performed two experiments to test more directly whether ELF3 is required for phyB function. First, we examined whether an elf3 null mutation could suppress the effect of overexpression of phyB. Second, we tested whether an elf3 mutation affected nuclear localization of phyB.
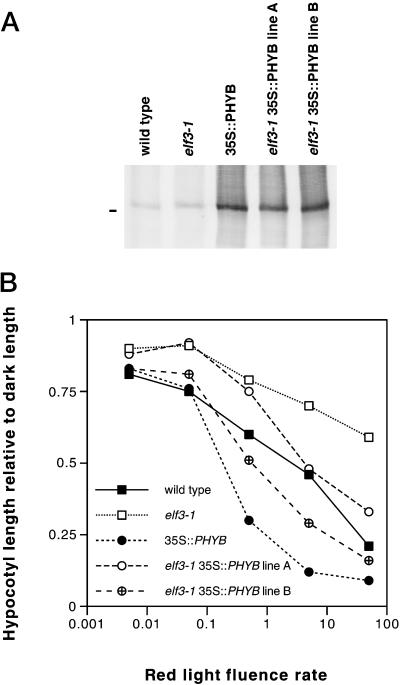
To test the effect of elf3 on phenotypes caused by overexpression of PHYB, we introduced a construct that drives overexpression of PHYB (35S::PHYB; Hirschfeld et al., 1998) into elf3-1 plants (see “Materials and Methods”). We crossed an elf3-1 transgenic plant carrying this construct with a wild-type (ELF3) plant, and from among the progeny selected cousin ELF3 and elf3-1 lines expressing similar levels of PHYB protein for phenotypic analysis (Fig. 5A). PHYB overexpression in a wild-type ELF3 background markedly enhanced the red light response (Fig. 5B), as described previously (Wagner et al., 1991; Wester et al., 1994; Hirschfeld et al., 1998). Similarly, overexpression of PHYB in the elf3-1 background increased the red light response, although these seedlings were still taller than ELF3 35S::PHYB seedlings (Fig. 5B). Thus, the elf3-1 mutation and the overexpressed PHYB appeared to interact additively.
Figure 5.
Effect of overexpressed PHYB protein on red light inhibition of hypocotyl elongation in wild-type and elf3-1 seedlings. A, PHYB protein levels in 1-week-old seedlings of wild-type Columbia, elf3-1, Columbia 35S::PHYB, and two different elf3-1 35S::PHYB populations. Forty micrograms of total protein was loaded for wild-type and elf3-1 lanes, and 10 μg of total protein was loaded for each of the 35S::PHYB lanes. The bar to the left of the blot marks the location of the 118-kD molecular mass marker. B, Hypocotyl length after growth of the same populations under different fluence rates of constant red light for 4 d. Between 11 and 24 seedlings were measured for each data point. sds are omitted for clarity, but were generally about 20% of the mean or less, with higher relative sd at higher fluence rates.
Effect of ELF3 on Localization of PHYB
Western blots showed that wild-type and elf3-1 mutant plants had similar steady-state levels of PHYB protein (Fig. 5A). Moreover, the elf3-1 mutation still caused a decreased red light response in transgenic lines having similar levels of overexpressed PHYB (Fig. 5). These results suggest that ELF3 does not affect light responses by regulating expression or stability of PHYB, but rather affects either some downstream aspect of phyB signaling or some phyB-independent regulatory pathway.
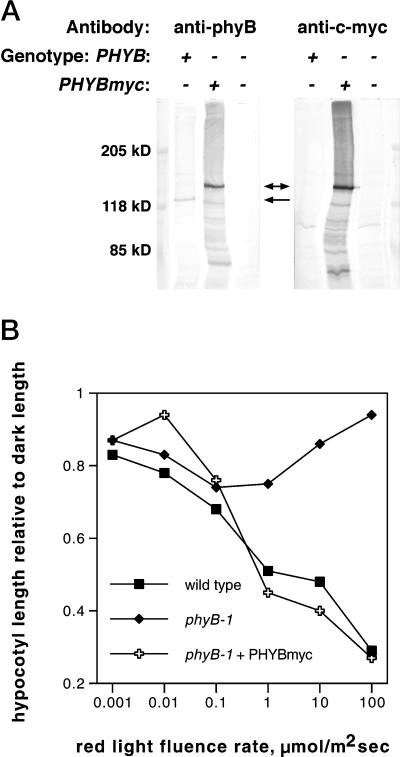
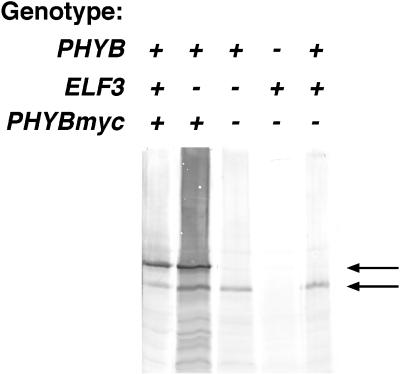
phyB has been shown to localize to the nucleus in light-grown plants (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999). To test whether ELF3 is required for phyB nuclear localization, we constructed a c-myc epitope-tagged version of PHYB and introduced this into ELF3 and elf3-1 transgenic plants. This PHYBmyc construct directed synthesis of a protein slightly larger than native phyB (Figs. 6A and 7) and complemented the elongated hypocotyl phenotype of a phyB-1 mutant (Fig. 6B), indicating that the protein was biologically active.
Figure 6.
Rescue of a phyB-1 mutant by PHYBmyc. A, Western blots of total proteins from wild-type, phyB-1 PHYBmyc transgenic, and phyB-1 mutant plants. Duplicate blots were probed with antibody against phyB (left panel) or c-myc (right panel). Arrows indicate PHYBmyc protein (double headed arrow) or phyB protein (single-headed arrow). Equal volumes of protein extracts were loaded in each lane. B, Hypocotyl lengths relative to dark lengths of plants of the same genotypes as in A under different fluence rates of continuous red light. Between 18 and 29 seedlings were measured for each data point. sds were generally about 20% of the mean or less, with higher relative sd at higher fluence rates, and are omitted for clarity.
Figure 7.
PHYBmyc protein in transgenic lines. Extracts of transgenic and non-transgenic lines of the indicated genotypes were probed with anti-phyB monoclonal antibody BA2. Arrows indicate positions of PHYBmyc (top arrow) and native phyB (bottom arrow). Equal volumes of protein extracts were loaded in each lane.
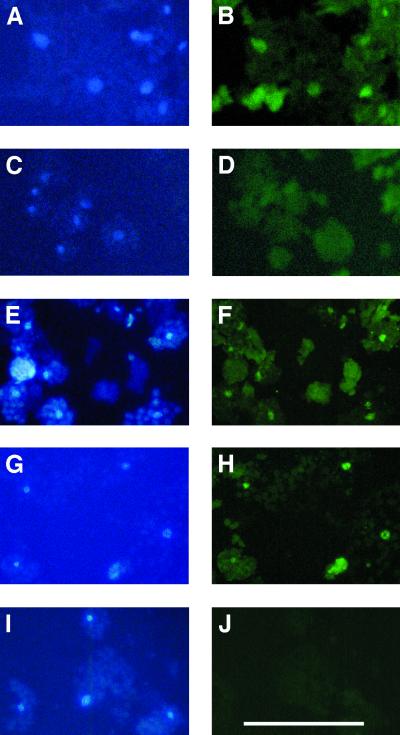
We localized PHYBmyc protein in fixed protoplasts of light-grown transgenic plants by indirect immunofluorescence. As shown in Figure 8, PHYBmyc localized to the nucleus in protoplasts of phyB-1 or wild-type plants carrying the PHYBmyc construct (Fig. 8, A, B, E, and F). In the elf3-1 plants, PHYBmyc also localized to the nucleus (Fig. 8, G and H). Control experiments with wild-type (non-transgenic) plants showed only a very faint background band in western blots probed with anti-myc antibody (Fig. 6A), and no nuclear fluorescence in immunolocalization experiments (Fig. 8, C and D), indicating that the anti-c-myc antibody specifically recognized nuclear PHYBmyc in the transgenic plants. The nuclear fluorescence also depended on the presence of the anti-c-myc antibody (Fig. 8, I and J). Therefore, these experiments revealed no effect of the elf3-1 mutation on nuclear localization of phyB.
Figure 8.
Immunolocalization of PHYBmyc. Protoplasts from different plant lines were fixed and stained with anti-myc primary antibody and FITC-conjugated anti-mouse secondary antibody, and stained with DAPI. A, DAPI fluorescence of phyB-1 PHYBmyc protoplasts. B, FITC fluorescence of phyB-1 PHYBmyc protoplasts stained with primary and secondary antibodies. C, DAPI fluorescence of wild-type (non-transgenic) protoplasts. D, FITC fluorescence of wild-type protoplasts. E, DAPI fluorescence of Columbia PHYBmyc protoplasts. F, FITC fluorescence of Columbia PHYBmyc protoplasts. G, DAPI fluorescence of elf3-1 PHYBmyc protoplasts. H, FITC fluorescence of elf3-1 PHYBmyc protoplasts. I, DAPI fluorescence of elf3-1 PHYBmyc protoplasts. J, FITC fluorescence of elf3-1 PHYBmyc protoplasts stained with secondary antibody only. Scale bar in J = 100 μm.
DISCUSSION
The similar phenotypes of elf3 and phyB mutants show that ELF3 and phyB control an overlapping set of responses. However, they have a different relative importance for each response. Thus, phyB mutations decreased red-light-induced inhibition of hypocotyl elongation, as well as the “acute” induction of CAB in dark-grown seedlings, more than elf3 mutations (Anderson et al., 1997). In contrast, elf3 mutant plants had longer hypocotyls in blue light, and under short-day conditions had longer petioles and flowered earlier than phyB mutant plants. Lastly, elf3 mutations eliminate circadian rhythm outputs in constant light (Hicks et al., 1996; Dowson-Day and Millar, 1999), whereas phyB mutations have quantitative effects on amplitude and period without eliminating rhythmicity (Anderson et al., 1997; Somers et al., 1998).
Our analyses of inhibition of elf3 mutant hypocotyl elongation by different light qualities agree broadly with those of Zagotta et al. (1996), except that those investigators found a larger effect of the elf3-1 mutation on blue light response than we did. They also found that the elf3-1 deficiency in blue light was as severe as that of the hy4-101 mutant, whereas in our study the hy4-101 mutant was substantially taller than the elf3 mutants. Different light conditions or the different ages of the seedlings measured (2 d in the previous work versus 5 d in this work) may explain this quantitative discrepancy. As discussed below, our results suggest that ELF3 cannot be considered simply as a unique phytochrome or cryptochrome signaling component. ELF3 may affect red and blue light responses by changing phytochrome or cryptochrome signaling, or by altering circadian rhythms. Altered phytochrome signaling might account for both red and blue light phenotypes, as several groups have reported effects of phyB mutations on blue light responses, and have seen synergistic effects between phyB and cry1 (Casal and Boccalandro, 1995; Casal and Mazzella, 1998; Neff and Chory, 1998; see also the double mutant data in Fig. 3).
Under short-day conditions, the hypocotyl length and flowering time phenotypes of elf3-7 seedlings were intermediate between those of the wild type and the elf3-1 null mutant. The phase of CAB::LUC gene expression was also intermediate in the elf3-7 mutant grown in photoperiods of 12 h or less. While we do not know the precise effect of the elf3-7 mutation on ELF3 protein amount or structure, the mutation probably causes a partial loss of function. Therefore, these results suggest that the absolute level of ELF3 activity may determine the severity of phenotype, at least under short-day conditions. Consistent with this idea, we have observed partial dominance of both elf3-1 and elf3-7 mutations for flowering time under short-day conditions (K.S. Solomon and J.W. Reed, unpublished results). It appears paradoxical that these phenotypes are more severe under short-day conditions, whereas the circadian rhythms of elf3 plants are more aberrant in constant light. However, a photoperiodic response rhythm might also have an altered phase in the elf3 mutants under short photoperiods, and the phase change, rather than arrhythmia, might affect flowering time and hypocotyl elongation. Further work may resolve this question.
Experiments to determine whether the phenotypic similarities between elf3 and phyB-9 mutants reflect ELF3 participation in phyB signaling revealed no functional interaction. Thus, elf3 and phyB-9 mutations had additive effects on hypocotyl elongation, petiole length, and flowering time; and the elf3-1 mutation also had an additive effect on hypocotyl length in combination with the 35S::PHYB construct. Moreover, the elf3-1 mutation did not prevent phyB from moving to the nucleus. Although it remains possible that ELF3 and phyB signal in a common pathway, if this were the case, then both would have to act redundantly with other proteins. phyB acts redundantly with other phytochromes to control hypocotyl elongation (phyA, phyD), petiole elongation (phyD, phyE), and flowering (phyD, phyE) (Reed et al., 1994; Aukerman et al., 1997; Devlin et al., 1998), suggesting that ELF3 could mediate signals from multiple phytochromes. The multiplicity of putative direct phytochrome signaling targets recently identified also suggests that there may be considerable redundancy downstream of phyB (Ni et al., 1998; Choi et al., 1999; Fankhauser et al., 1999).
An alternative model is that the circadian rhythm regulates many of the same outputs as phyB does, and the phenotypes of elf3 mutants arise from a defective circadian rhythm. This would be consistent with the additive effects of the mutations and with the distinct effects of elf3 and phyB mutations on circadian rhythm outputs. Red light and the circadian rhythm each control hypocotyl elongation (Dowson-Day and Millar, 1999), CAB gene expression (Karlin-Neumann et al., 1988; Millar and Kay, 1991), and flowering time (Smith, 1994; Lumsden and Millar, 1998).
Evaluation of this model is complicated by interactions between light and circadian signaling (Lumsden and Millar, 1998). Although phyB contributes light input signals to the circadian system, a single photoreceptor mutation has little or no effect on circadian rhythms in white light (Millar et al., 1995; Somers et al., 1998), so the phenotypes of phyB mutants are unlikely to arise as secondary effects of a circadian defect. However, the circadian system may rhythmically regulate (or “gate”) phyB activity, or a downstream component of phyB signaling. The effectiveness of light to induce CAB depends on the phase of the circadian rhythm (Millar and Kay, 1996), indicating that such a model is plausible. As the acute activation of CAB by phytochrome is intact in elf3-1 plants (Anderson et al., 1997), it is unlikely that elf3 mutations uniformly suppress all phyB functions and phenocopy the phyB mutation as a secondary consequence of their circadian defect. These results also suggest that ELF3 and phyB act independently.
Molecular regulation at two levels may provide further insight into the mechanisms of this signaling network. First, rhythmic expression patterns have recently been discovered for the PHYB (Kozma-Bognar et al., 1999) and ELF3 genes (K. Hicks and D.R. Wagner, personal communication). Second, phyB moves to the nucleus in the light. Although the elf3-1 mutation did not prevent phyB nuclear localization, it remains possible that this localization is normally circadian. Experiments are currently under way to determine the effect of elf3 mutations on the temporal pattern of PHYB gene expression and phyB nuclear localization.
ACKNOWLEDGMENTS
We thank A. Nagatani for providing the BA2 antibody, R. Sharrock for the 35S::PHYB clone, L. Krall for help with western blots, K. Hicks for sequencing elf3 mutant alleles and performing cleaved-amplified polymorphic sequence assays on elf3-1 transgenic lines, and S. Kay for providing the CAB2::LUC (Columbia) introgression line.
Footnotes
This work was supported by the National Institutes of Health (grant no. R29–GM52456 to J.W.R.) and by the Biotechnology and Biological Science Research Council (BBSRC: grant no. G08667 to A.J.M.). K.S.S. was supported in part by the James Henley Thompson and Evelyn Barnett Thompson Undergraduate Research Fund. The imaging facilities at the University of Warwick are funded by the Gatsby Charitable Foundation, by the BBSRC (grant no. BI11209), and by the Royal Society.
LITERATURE CITED
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Cashmore AR. The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. Plant J. 1996;10:1103–1110. doi: 10.1046/j.1365-313x.1996.10061103.x. [DOI] [PubMed] [Google Scholar]
- Anderson SL, Somers DE, Millar AJ, Hanson K, Chory J, Kay SA. Attenuation of phytochrome A and B signaling pathways by the Arabidopsis circadian clock. Plant Cell. 1997;9:1727–1743. doi: 10.1105/tpc.9.10.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall DJ, King RW, Hangarter RP. Blue-light promotion of flowering is absent in hy4 mutants. Planta. 1996;200:278–280. doi: 10.1007/BF00208319. [DOI] [PubMed] [Google Scholar]
- Barnes SA, Quaggio RB, Whitelam GC, Chua N-H. fhy1 defines a branch point in phytochrome A signal transduction pathways for gene expression. Plant J. 1996;10:1155–1161. doi: 10.1046/j.1365-313x.1996.10061155.x. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Casal JJ, Boccalandro H. Co-action between phytochrome B and HY4 in Arabidopsis thaliana. Planta. 1995;197:213–218. doi: 10.1007/BF00202639. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Mazzella MA. Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiol. 1998;118:19–25. doi: 10.1104/pp.118.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G, Yi H, Kwon Y-K, Soh MS, Shin B, Luka Z, Hahn T-R, Song P-S. Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature. 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Quail PH. Signaling in light-controlled development. Semin Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 1999;17:63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- Elich TD, Chory J. Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell. 1998;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Yeh K-C, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Furuya M. Phytochromes: their molecular species, gene families, and functions. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:617–645. [Google Scholar]
- Genoud T, Millar AJ, Nishizawa N, Kay SA, Schäfer E, Nagatani A, Chua N-H. An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell. 1998;10:889–904. doi: 10.1105/tpc.10.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler T, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Hudson M, Ni M, Qin M, Quail PH. poc1: an Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc Natl Acad Sci USA. 1999;96:5832–5837. doi: 10.1073/pnas.96.10.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Koornneef M, Whitelam GC. Phytochrome B, and at least one other phytochrome, mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiol. 1994;104:1311–1315. doi: 10.1104/pp.104.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA. Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics. 1998;149:523–535. doi: 10.1093/genetics/149.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC. Photoresponses of light-grown phyA mutants of Arabidopsis: phytochrome A is required for the perception of daylength extensions. Plant Physiol. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin-Neumann GA, Sun L, Tobin EM. Expression of light-harvesting chlorophyll a/b-protein genes is phytochrome-regulated in etiolated Arabidopsis thaliana seedlings. Plant Physiol. 1988;88:1323–1331. doi: 10.1104/pp.88.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schäfer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochromes A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- Kozma-Bognar L, Hall A, Adam E, Thain SC, Nagy F, Millar AJ. The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc Natl Acad Sci USA. 1999;96:14652–14657. doi: 10.1073/pnas.96.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light photoreceptor cryptochrome 2. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cheng C-L. A chlorate-resistant mutant defective in the regulation of nitrate reductase gene expression in Arabidopsis defines a new HY locus. Plant Cell. 1997;9:21–35. doi: 10.1105/tpc.9.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Hangarter RP. Arabidopsis mutants lacking blue light-dependent inhibition of hypocotyl elongation. Plant Cell. 1991;3:685–694. doi: 10.1105/tpc.3.7.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden PJ, Millar AJ. Biological Rhythms and Photoperiodism in Plants. Oxford: BIOS Scientific Publishers; 1998. [Google Scholar]
- Millar AJ, Kay SA. Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell. 1991;3:541–550. doi: 10.1105/tpc.3.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua N-H, Kay SA. The regulation of circadian period by phototransduction pathways in Arabidopsis. Science. 1995;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development. 1999;126:2073–2082. doi: 10.1242/dev.126.10.2073. [DOI] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J. Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 1993;102:269–277. doi: 10.1104/pp.102.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Chory J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998;118:27–36. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH. hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell. 1993;5:39–48. doi: 10.1105/tpc.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Reed JW. Phytochromes are Pr-ipatetic kinases. Curr Opin Plant Biol. 1999;2:393–397. doi: 10.1016/s1369-5266(99)00011-4. [DOI] [PubMed] [Google Scholar]
- Reed JW, Elumalai RP, Chory J. Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signalling and hypocotyl elongation. Genetics. 1998;148:1295–1310. doi: 10.1093/genetics/148.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PRH, Whitelam GC, Smith H. Selected components of the shade avoidance syndrome are displayed in a normal manner in mutants of Arabidopsis thaliana and Brassica rapa deficient in phytochrome B. Plant Physiol. 1993;102:1179–1184. doi: 10.1104/pp.102.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M. The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B, and secondarily by phytochrome A. Plant Physiol. 1994;104:363–371. doi: 10.1104/pp.104.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Uchida K, Furuya M. Elementary responses of photoperception by phytochrome A for high irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. Sensing the light environment: the functions of the phytochrome family. In: Kendrick R, Kronenberg G, editors. Photomorphogenesis in Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 377–416. [Google Scholar]
- Smith H, Xu Y, Quail PH. Antagonistic but complementary actions of phytochromes A and B allow optimum seedling de-etiolation. Plant Physiol. 1997;114:637–641. doi: 10.1104/pp.114.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J. 1998;16:411–419. doi: 10.1046/j.1365-313x.1998.00307.x. [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- Staub JM, Wei N, Deng X-W. Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell. 1996;8:2047–2056. doi: 10.1105/tpc.8.11.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Hoecker U, Quail PH. RED1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant Cell. 1997;9:731–743. doi: 10.1105/tpc.9.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Tepperman JM, Quail PH. Overexpression of phytochrome B induces a short hypocotyl phenotype in transgenic Arabidopsis. Plant Cell. 1991;3:1275–1288. doi: 10.1105/tpc.3.12.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester L, Somers D, Clack T, Sharrock R. Transgenic complementation of the hy3 phytochrome B mutation and response to PHYB gene copy number in Arabidopsis. Plant J. 1994;5:261–272. doi: 10.1046/j.1365-313x.1994.05020261.x. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Wu S-H, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- Young JC, Liscum E, Hangarter RP. Spectral dependence of light-inhibited hypocotyl elongation in photomorphogenic mutants of Arabidopsis: evidence for a UV-A photosensor. Planta. 1992;188:106–114. doi: 10.1007/BF00198946. [DOI] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 1996;10:691–702. doi: 10.1046/j.1365-313x.1996.10040691.x. [DOI] [PubMed] [Google Scholar]