Abstract
Rhabdomyosarcoma is the most common soft-tissue sarcoma in childhood and histologically resembles developing skeletal muscle. Alveolar rhabdomyosarcoma (ARMS) is an aggressive subtype with a higher rate of metastasis and poorer prognosis. The majority of ARMS tumors (80%) harbor a PAX3-FOXO1 or less commonly a PAX7-FOXO1 fusion gene. The presence of either the PAX3-FOXO1 or PAX7-FOXO1 fusion gene foretells a poorer prognosis resulting in clinical re-classification as either fusion-positive (FP-RMS) or fusion-negative RMS (FN-RMS). The PAX3/7-FOXO1 fusion genes result in the production of a rogue transcription factors that drive FP-RMS pathogenesis and block myogenic differentiation. Despite knowing the molecular driver of FP-RMS, targeted therapies have yet to make an impact for patients, highlighting the need for a greater understanding of the molecular consequences of PAX3-FOXO1 and its target genes including microRNAs. Here we show FP-RMS patient-derived xenografts and cell lines display a distinct microRNA expression pattern. We utilized both loss- and gain-of function approaches in human cell lines with knockdown of PAX3-FOXO1 in FP-RMS cell lines and expression of PAX3-FOXO1 in human myoblasts and identified microRNAs both positively and negatively regulated by the PAX3-FOXO1 fusion protein. We demonstrate PAX3-FOXO1 represses miR-221/222 that functions as a tumor suppressing microRNA through the negative regulation of CCND2, CDK6, and ERBB3. In contrast, miR-486-5p is transcriptionally activated by PAX3-FOXO1 and promotes FP-RMS proliferation, invasion, and clonogenic growth. Inhibition of miR-486-5p in FP-RMS xenografts decreased tumor growth, illustrating a proof of principle for future therapeutic intervention. Therefore, PAX3-FOXO1 regulates key microRNAs that may represent novel therapeutic vulnerabilities in FP-RMS.
Introduction
Rhabdomyosarcoma (RMS), the most common soft-tissue sarcoma in children [1], is divided into two major histologic subclasses: embryonal RMS (ERMS) and alveolar RMS (ARMS) [2]. ARMS is notoriously more aggressive and most (~80%) ARMS tumors harbor chromosomal translocations t(2:13)(q35;q14) or t(1:13)(p36;q14) resulting in PAX3-FOXO1 or PAX7-FOXO1 gene fusions respectively [3–5]. The presence of the PAX3/7-FOXO1 fusion gene foretells a worse prognosis and is superior to histology in predicting survival [6–9]. ARMS patients without a PAX3/7-FOXO1 translocation have both molecular features and clinical outcome similar to ERMS [7, 10]. Therefore, molecular classification as fusion-positive RMS (FP-RMS) and fusion-negative RMS (FN-RMS) based on the presence or absence of the PAX3/7-FOXO1 fusion more accurately represents the clinical features and biology of RMS. Despite the recently expanded genomic understanding of RMS, patient survival and the treatment strategies have not improved [11–14].
In both PAX3-FOXO1 and PAX7-FOXO1 fusion proteins, the amino terminus of the paired box (PAX) protein including the DNA-binding domain is fused to the carboxy terminus of the forkhead box O1 (FOXO1) protein including the transcriptional transactivation domain. Thus, it is thought that the PAX3/7-FOXO1 fusion proteins function as aberrant transcription factors at PAX3 or PAX7 target genes [15]. FP-RMS cells are addicted to PAX3-FOXO1 expression making it therapeutically attractive [16, 17]. Given targeting transcription factors remains a challenge, many groups have focused on identifying and inhibiting PAX3-FOXO1 target genes [18]. However, little is known about the regulation of non-coding RNAs including microRNAs by PAX3-FOXO1.
MicroRNAs are single-stranded ~22 nucleotide non-coding RNAs that function to reduce gene expression [19]. Primarily through binding complementary sequences in the 3′ untranslated regions (UTR) of target mRNAs, microRNAs target transcripts for degradation or repress transcript translation [20]. Individual microRNAs can modulate the expression of hundreds of target genes allowing for regulation of complex biological processes. MicroRNAs have been implicated in normal development, differentiation, cellular homeostasis, and diseases including cancer [21–24]. Methods to block or replace microRNA function with either inhibitors or mimics are now in clinical trials opening the potential to leverage microRNA biology to target multiple nodes in signaling pathways or biological processes [22, 25].
Given the PAX3-FOXO1 fusion protein is thought to function as a rogue transcription factor; transcriptional regulation by PAX3-FOXO1 is thought to be central to FP-RMS pathogenesis. Nothing is known about how the PAX3-FOXO1 fusion protein controls microRNA expression. We hypothesize that microRNAs are transcriptionally regulated by the PAX3-FOXO1 fusion protein and actively participate in FP-RMS pathogenesis.
Results
Distinct microRNA expression in FP-RMS and FN-RMS
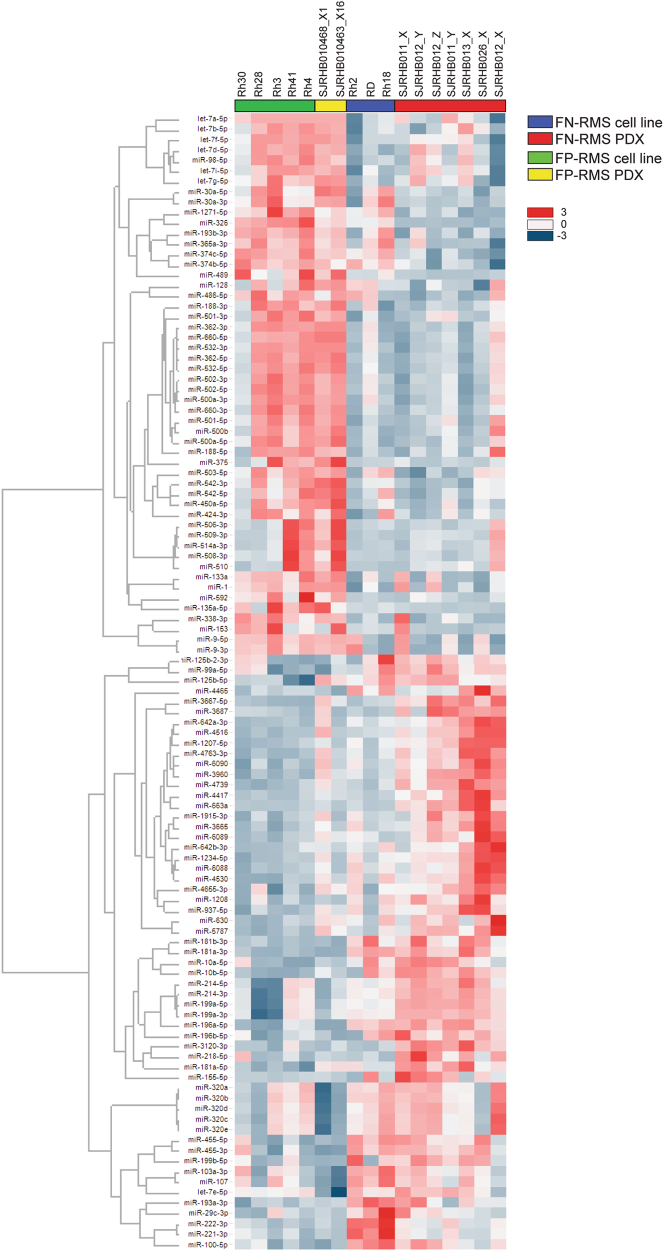
We hypothesized that if microRNAs participate in driving the clinical differences between FN- and FP-RMS microRNA expression patterns should be distinct. To interrogate microRNA expression in RMS, we assessed the microRNA expression profile in a panel of RMS cell lines as well as patient-derived xenografts (PDX) from the Childhood Solid Tumor Network [26]. Our panel included seven FN-RMS PDX samples, three FN-RMS cell lines, two FP-RMS PDX samples, and five FP-RMS cell lines. All of the FP-RMS cell lines and the SJRHB010463_X16 PDX harbor the PAX3-FOXO1 translocation; however, the SJRHB010468_X1 PDX contains the PAX7-FOXO1 translocation. Comparing the microRNA expression between FP-RMS and FN-RMS with greater than two-fold difference and P-value less than 0.05, we identified 57 microRNAs decreased and 52 with increased expression in FP-RMS compared to FN-RMS (Fig. 1). Because the PAX3-FOXO1 translocation is the driver of FP-RMS, we hypothesized that microRNAs differentially expressed in FP-RMS are transcriptionally regulated by the PAX3-FOXO1 fusion protein.
Fig. 1.
Differential microRNA expression in FN-RMS and FP-RMS. Heat map of differentially expressed microRNAs based on P < 0.05 and fold change >2 in FP-RMS cell lines (green, Rh3, Rh4, Rh28, Rh30, Rh41), FP-RMS patient-derived xenografts (PDX, yellow, SJRHB010468_X1, SJRHB010463_X16), FN-RMS cell lines (blue, RD, Rh2, Rh18), and FN-RMS PDX (red, SJRHB011_X, SJRHB011_Y, SJRHB012_x, SJRHB012_Y, SJRHB012_Z, SJRHB013_X, SJRHB026_X)
Knockdown of PAX3-FOXO1 reduces transformative properties and promotes differentiation of FP-RMS
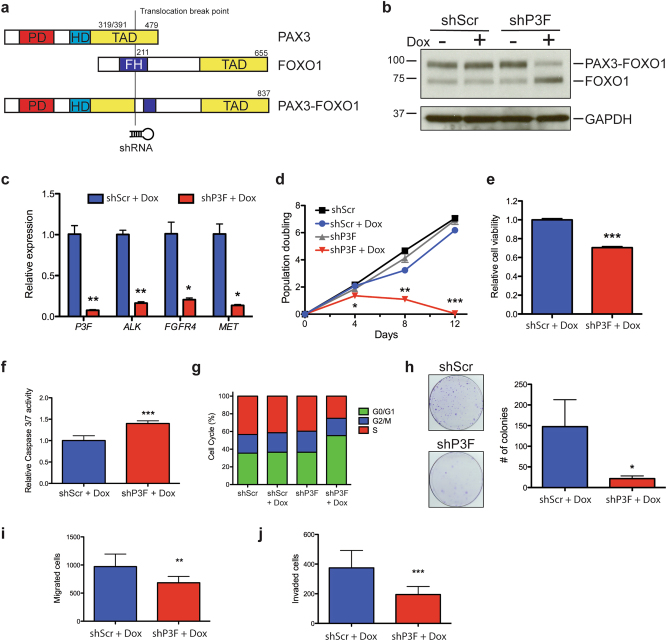
In order to identify microRNAs regulated by PAX3-FOXO1 in FP-RMS, we generated a doxycycline-inducible lentivirus expressing shRNA directed against PAX3-FOXO1 breakpoint or a scrambled shRNA control. The PAX3-FOXO1 shRNA (shP3F) targets the translocation breakpoint to minimally complement endogenous PAX3 or FOXO1 sequences (Fig. 2a). We utilized the broadly studied FP-RMS Rh30 and Rh41 cell lines harboring the PAX3-FOXO1 translocation to established stable cell lines that express doxycycline-inducible shP3F or scramble control (shScr) shRNA [27, 28]. Treatment with doxycycline in both Rh30 and Rh41 cells leads to a significant knockdown of PAX3-FOXO1 protein (Fig. 2b and Supplementary Figure S1a) and transcript levels (Fig. 2c and Supplementary Figure S1c). Reduced PAX3-FOXO1 protein correlated with a reduction of well-known PAX3-FOXO1 target genes ALK, FGFR4, and MET (Fig. 2c and Supplementary Figure S1c). FOXO1 and PAX3 proteins are not decreased with the shP3F illustrating specificity (Fig. 2b and Supplementary Figure S1a and b).
Fig. 2.
Inducible knockdown of PAX3-FOXO1 decreases proliferation, migration, and invasion of FP-RMS. a Schematic of PAX3-FOXO1 translocation and location of shRNA expressed by doxycycline-inducible Tet-pLKO lentivirus. b Expression of PAX3-FOXO1 by immunoblot with FOXO1 directed antibody in Rh41 cells stably transduced with Tet-pLKO-P3F or Tet-pLKO-Scr treated with and without doxycycline for 5 days. c Expression of PAX3-FOXO1 and transcriptional targets of PAX3-FOXO1 by qRT-PCR in Rh41 cells as in (b). d PAX3-FOXO1 knockdown decreases population doubling of Rh41 cells. e Cell Titer Glo and f Caspase 3/7 Glo assays in cells from (b) treated with doxycycline for 3 days. g Cell cycle analysis of cells from (b) after 5 days of doxycycline treatment. h Low-density colony formation: i cell migration and j invasion in Rh41 PAX3-FOXO1 knockdown cells. *P < 0.05, **P < 0.01, ***P < 0.001 for shScr+Dox vs shP3F+Dox
To confirm functional PAX3-FOXO1 knockdown in our system, we measured the phenotypic consequences in Rh30 and Rh41 cells. The PAX3-FOXO1 knockdown in Rh41 cells markedly decreased the population doubling of cells to the point that cells had senesced or died after 12 days in doxycycline (Fig. 2d). In contrast, PAX3-FOXO1 knockdown in Rh30 significantly reduced population doubling but permitted continued although attenuated growth (Supplementary Figure S1d). PAX3-FOXO1 knockdown reduced cell viability and increased apoptosis (Fig. 2e, f, Supplementary Figure S1e and f). Cell cycle analysis with FACS illustrated PAX3-FOXO1 knockdown results in a G1 arrest (Fig. 2g). PAX3-FOXO1 knockdown in both Rh41 and Rh30 reduced colony formation in clonogenic cell survival assays (Fig. 2h and Supplementary Figure S1g). Since FP-RMS has a higher propensity for metastasis, we assessed the effect of PAX3-FOXO1 knockdown on cell migration and invasion both of which were reduced with PAX3-FOXO1 knockdown (Fig. 2i, j). We noted that with PAX3-FOXO1 knockdown both Rh30 and Rh41 cells elongated and formed myosin heavy chain-positive multinucleated myotubes and expressed mature skeletal muscle gene CKM indicative of myogenic differentiation (Supplementary Figure S2a–f). These results illustrate that our inducible shP3F system reduces expression of known PAX3-FOXO1 target genes, reduces the oncogenic capacity and invasion potential while inducing myogenic differentiation of FP-RMS cell lines.
Identification of candidate microRNAs regulated by PAX3-FOXO1
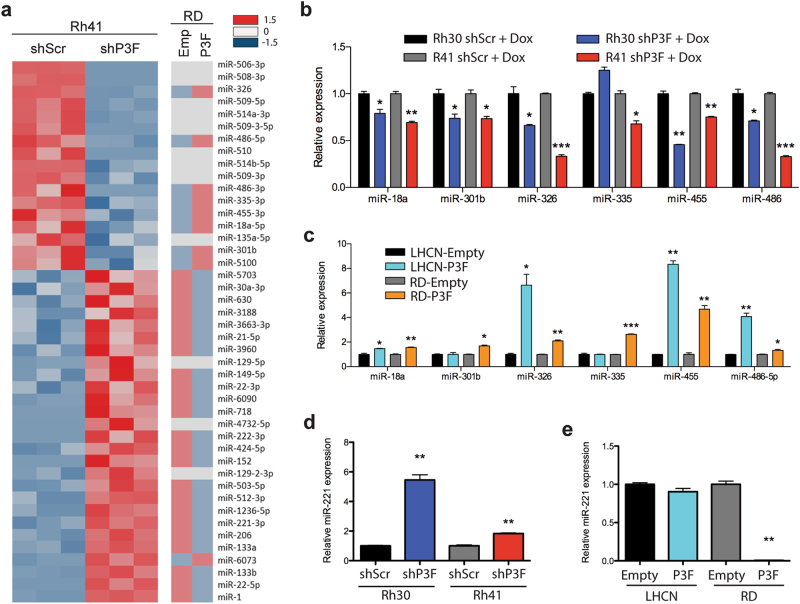
To identify microRNAs regulated by PAX3-FOXO1 in FP-RMS, we utilized both loss- and gain-of-function approaches. First, shP3F and shScr Rh41 knockdown and control cells respectively were treated with doxycycline for 5 days (before population doubling decreases) and RNA was prepared for microRNA expression profiling (Fig. 3a). To complement the loss-of-function PAX3-FOXO1 knockdown, PAX3-FOXO1 was stably expressed in the FN-RMS RD cell line with correspondingly increased expression of PAX3-FOXO1 target genes (Supplementary Figure S3a and b). We hypothesized that microRNAs transcriptionally activated by PAX3-FOXO1 would have decreased expression in the shP3F knockdown Rh41 cells and increased expression in the PAX3-FOXO1 forced expression RD cells. In contrast, microRNAs transcriptionally repressed by PAX3-FOXO1 would have increased expression in the shP3F Rh41 cells and decreased expression in the RD cells with PAX3-FOXO1 forced expression.
Fig. 3.
Identification of microRNAs regulated by PAX3-FOXO1. a Heat map comparing differential expression of microRNAs in stably transduced Rh41 cells with Tet-pLKO-P3F or Tet-pLKO-Scr and treated with doxycycline for 5 days, and RD cells stably transduced with PAX3-FOXO1 or empty control vector. Downregulated genes indicated in blue (−1.5 SD) and upregulated genes indicated in red (1.5 SD) with FDR < 0.05. b Validation of microarray by qRT-PCR for microRNAs downregulated with PAX3-FOXO1 knockdown in Rh30 and Rh41 Tet-pLKO cells treated with doxycycline for 5 days. c Expression of PAX3-FOXO1 regulated microRNAs by qRT-PCR in RD and LHCN myoblasts stably transduced with control empty vector or PAX3-FOXO1 overexpression. d Expression of miR-221 by qRT-PCR in PAX3-FOXO1 knockdown cells treated with doxycycline for 5 days or e stably transduced RD or LHCN myoblasts. *P < 0.05, **P < 0.01, ***P < 0.001 for shScr+Dox vs shP3F+Dox or empty vs PAX3-FOXO1
MicroRNA microarray analysis identified a total of 44 microRNAs with greater than two-fold expression difference with PAX3-FOXO1 knockdown (Fig. 3a). Seventeen microRNAs had decreased expression and 27 had increased expression with PAX3-FOXO1 knockdown. To further define microRNAs specifically regulated by the PAX3-FOXO1 fusion protein, we compared the microRNA expression in the RD cells expressing PAX3-FOXO1. Of the 17 microRNAs with decreased expression with shP3F, 8 microRNAs (miR-18a-5p, miR-301b, miR-326, miR-335-3p, miR-455-3p, miR-486-5p, miR-486-3p, and miR-5100) increased expression in RD cells with PAX3-FOXO1 overexpression. We hypothesized that these microRNAs are transcriptional target genes of PAX3-FOXO1. In contrast, 23 of the 28 microRNAs with increased expression with PAX3-FOXO1 knockdown had decreased expression in RD cells with PAX3-FOXO1 expression, suggesting they are repressed by PAX3-FOXO1.
We validated six of the eight microRNAs with decreased expression with PAX3-FOXO1 knockdown in FP-RMS cells and concordant increased expression with forced expression of PAX3-FOXO1 in the FN-RMS, RD cell line. We excluded miR-5100 as it is poorly conserved. Both miR-486-5p and miR-486-3p had decreased expression with PAX3-FOXO1 knockdown on the microarray; however, we restricted our experiments to miR-486-5p as it is the dominantly expressed mature microRNA [29]. By qRT-PCR we validated that all six microRNAs have decreased expression with PAX3-FOXO1 knockdown in Rh41 cells and all but miR-335 are decreased in Rh30 cells (Fig. 3b). Similar results were observed with transient transfection of siRNA to PAX3-FOXO1 (Supplementary Figure S3c–e). PAX3-FOXO1 expression in RD cells resulted in increased expression of all six microRNAs. As an additional control, PAX3-FOXO1 was stably expressed in the LHCN-M2 human myoblast cell line. PAX3-FOXO1 protein level in LHCN-M2 cells was lower than that in Rh30 and RH41 cells; however, PAX3-FOXO1 target genes were upregulated (Supplementary Figure 3a and b). All of the regulated microRNAs except miR-301b and miR-335 were upregulated by PAX3-FOXO1 in LHCN-M2 cells (Fig. 3c).
Interestingly, more microRNAs had increased rather than decreased expression with PAX3-FOXO1 knockdown and were decreased by PAX3-FOXO1 overexpression, suggesting that PAX3-FOXO1 may also function as a transcriptional repressor. Among the microRNAs that increase expression with PAX3-FOX01 knockdown were miR-1, miR-206, and miR-133b that are associated with myogenic differentiation. The increased expression of these myogenic microRNAs may be secondary to the enhanced muscle differentiation induced with PAX3-FOXO1 knockdown. Interestingly, miR-221 and miR-222 were some of the most downregulated microRNAs in FP-RMS compared to FN-RMS (Fig. 1) and had increased expression with PAX3-FOXO1 knockdown and decreased with PAX3-FOXO1 overexpression (Fig. 3a). MiR-221 and miR-222 are co-expressed as a cluster on the X chromosome and share an identical seed sequence, thus share the same target mRNAs. Since they are transcriptionally co-regulated and target the same mRNAs, we focused on miR-221 and validated it to be inversely correlated with PAX3-FOXO1 expression by qRT-PCR (Fig. 3d, e, Supplementary Figure S3e). Thus based on its low expression level in FP-RMS, known potential role in myogenic differentiation [30], and unknown role in FP-RMS we decided to interrogate the possible tumor suppressive role of miR-221 in FP-RMS.
miR-221 decreases proliferation and invasion by regulating CCND2, CDK6, and ERBB3
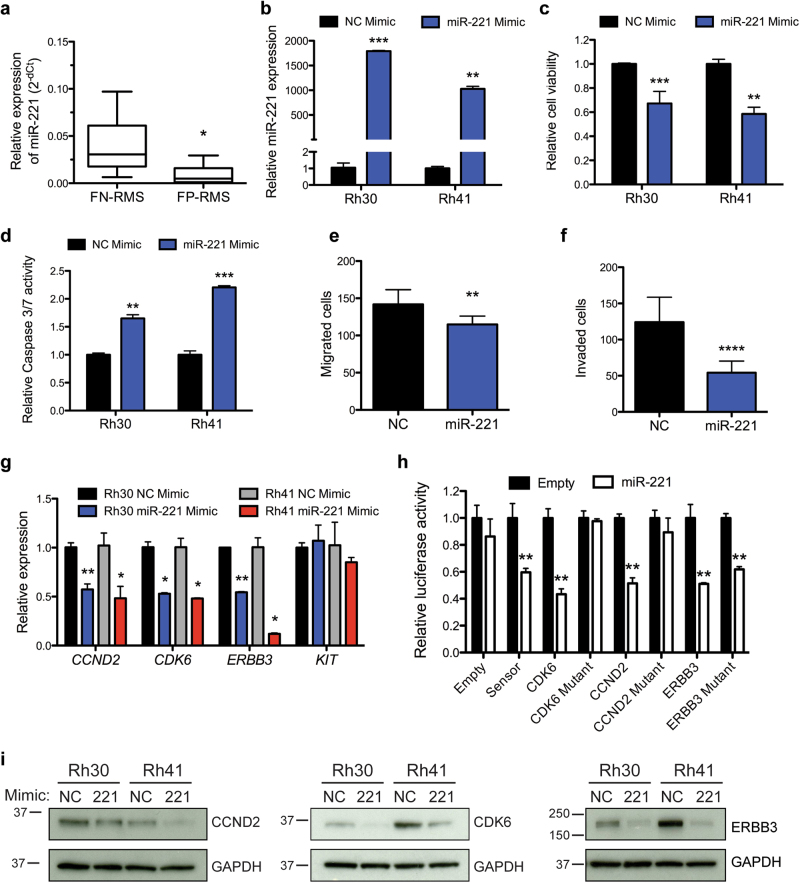
To further explore the role of miR-221 in FP-RMS, we first validated its expression level in an expanded cohort of PDX samples consisting of four FP-RMS PDX samples with two PAX3-FOXO1 fusions (SJRHB010463_X16 and SJRHB013759_X1) and two PAX7-FOXO1 fusions (SJRHB010468_X1 and SJRHB013757_X2) as well as 13 FN-RMS from the Childhood Solid Tumor Network [26]. MiR-221 is significantly reduced in FP-RMS compared to FN-RMS (Fig. 4a) consistent with a recent study in 64 RMS patients [31]. Transient transfection of miR-221 mimic led to overexpression as evaluated by qRT-PCR, reduction of cell viability, and an increase in apoptosis in both Rh30 and Rh41 cells (Fig. 4b–d). We next determined if miR-221 overexpression could contribute to FP-RMS pathogenesis beyond proliferation and found a striking reduction in the migratory capacity and invasive potential of the cells (Fig. 4e, f). Thus, miR-221 expression decreased proliferative, survival and metastatic potential of FP-RMS.
Fig. 4.
miR-221 functions as a tumor suppressor in FP-RMS and targets several known oncogenes. a Expression of miR-221 by qRT-PCR in patient-derived xenografts from the Childhood Solid Tumor Network (CSTN), including FN-RMS (n = 13, SJRHB011_X, SJRHB011_Y, SJRHB012_Y, SJRHB012_Z, SJRHB013_X, SJRHB026_X, SJRH010928_X1, SJRHB013758_X2, SJRHB015721_X1, SJRHB013758_X1, SJRHB012405_X1, SJRHB010927_X1, and SJRHB015720_X1) and FP-RMS (n = 4, SJRHB010463_X16, SJRHB013759_X1, SJRHB010468_X1 and SJRHB013757_X2). b Relative expression of miR-221 in Rh41 and Rh30 cells transfected with NC or miR-221 mimic. c Transfection of miR-221 mimic decreases cell viability and d increases apoptosis, e cell migration, and f invasion. g Expression of predicted miR-221 targets by qRT-PCR in Rh30 and Rh41 cells transfected with NC or miR-221 mimic. h Luciferase activity in C2C12 cells co-transfected with miR-221 or control vector and wild type or mutant miR-221 site 3′UTRs. Luciferase activity represented as mean S.E.M. (n = 4), the Renilla/Firefly luciferase ratio normalized to empty reporter (no miR-221). i Expression of miR-221 targets CCND2, CDK6, and ERBB3 by immunoblot analysis in cells as in (g). *P < 0.05, **P < 0.01, ***P < 0.001, **** P < 0.0001 for FN-RMS vs FP-RMS or miR-221 mimic vs NC
Given that overexpression of miR-221 decreased proliferation and the invasiveness of FP-RMS cells, we sought to identify the downstream targets of miR-221 responsible for the observed phenotypes. We interrogated the microRNA target prediction algorithms TargetScan [32] as well as previously published miR-221 targets with a focus on known oncogenes in RMS [33–36]. Thus we identified CCND2, CDK6, KIT, and ERBB3 as potential miR-221 targets for further analysis. MiR-221 overexpression in Rh30 and Rh41 cells decreased the expression of all the selected targets except KIT by qRT-PCR, protein levels on immunoblots, and we confirmed direct miR-221 regulation of CCND2, CKD6, and ERBB3 with 3′UTR luciferase reporter assays in C2C12 cells (Fig. 4g–i). Importantly, luciferase activity was rescued following mutation of the miR-221-binding site, although the ERBB3 rescue was not complete suggesting additional non-predicted sites in the 3′UTR (Supplementary Figure S4). Collectively these data suggest that miR-221 is negatively regulated by PAX3-FOXO1 and functions as a tumor suppressor in FP-RMS partially through targeting CCND2, CDK6, and ERBB3 decreasing proliferation and migration/invasion.
Candidate oncogenic microRNAs contribute to FP-RMS pathogenesis
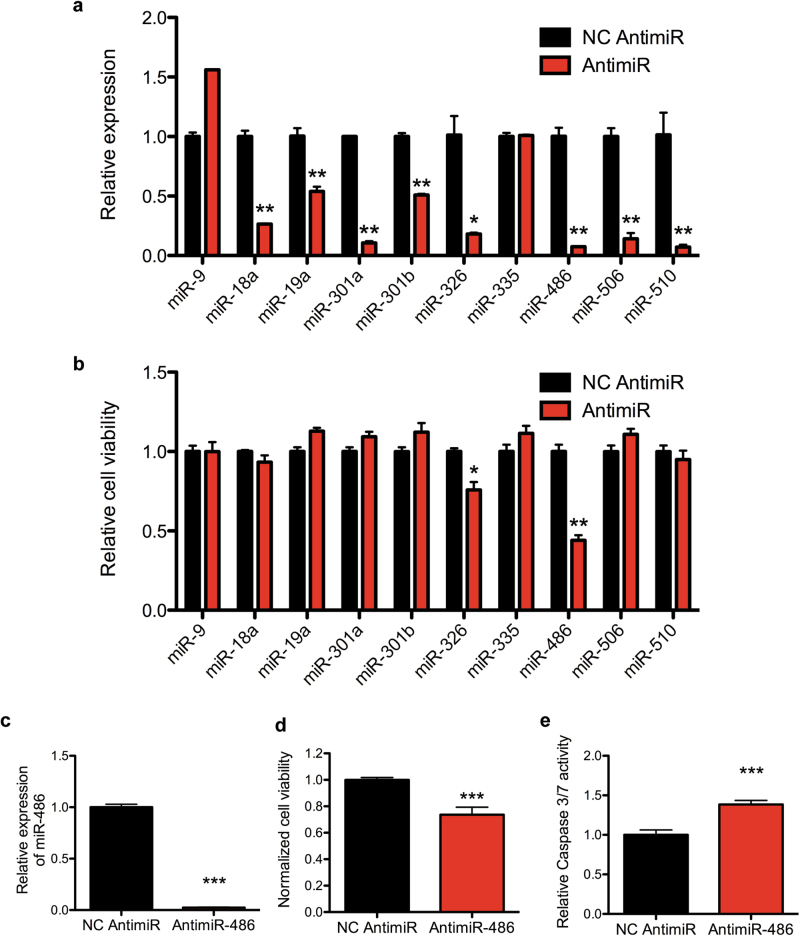
We hypothesized that microRNAs positively regulated by PAX3-FOXO1 could contribute to FP-RMS pathogenesis functioning as oncogenes. To this end, we assessed cell viability and apoptosis after transfection with antimiRs for microRNAs positively regulated by PAX3-FOXO1 and upregulated in FP-RMS cell lines and patient-derived xenografts (Supplementary Table S1). We included miR-506 and miR-510 given they had the largest expression change with PAX3-FOXO1 knockdown although there was no expression change with PAX3-FOXO1 expression in RD cells (Fig. 3a). We also included miR-9 given it is uniformly increased in FP-RMS cell lines and PDXs although not regulated by PAX3-FOXO1 knockdown (Fig. 1). We excluded miR-455 since it is enriched in FN-RMS compared to FP-RMS (Fig. 1). AntimiRs transiently transfected in the Rh41 cells resulted in a significant decrease in microRNA level with the exception of antimiR-9 and antimiR-335 (Fig. 5a). Since antimiRs could function through sequestering the mature microRNA without degradation, we continued with phenotypic analysis despite the lack of decreased miR-9 and miR-335 expression. However, only miR-486-5p and miR-326 inhibition reduced cell viability (Fig. 5b) while none of the antimiRs increased apoptosis in Rh41 cells (Supplementary Figure S5). Because miR-486-5p had the largest effect, we validated the findings in Rh30 cells, revealing similar knockdown of miR-486-5p, significant reduced cell viability, and increased apoptosis (Fig. 5c–e).
Fig. 5.
Contribution of candidate oncogenic microRNAs to FP-RMS. a qRT-PCR microRNA expression in Rh41 cells 3 days after transfection with indicated negative control (NC) or antimiR. b Cell Titer Glo cell viability assay in Rh41 cells as in (a). c qRT-PCR in Rh30 cells transfected with antimiR-486-5p or NC 3 days after transfection. d Cell Titer Glo cell viability and e Caspase 3/7 Glo apoptosis assay in Rh30 cells as in (c). *P < 0.05, **P < 0.01, ***P < 0.001 for antimiRs vs NC
miR-486-5p is a direct transcriptional target of PAX3-FOXO1
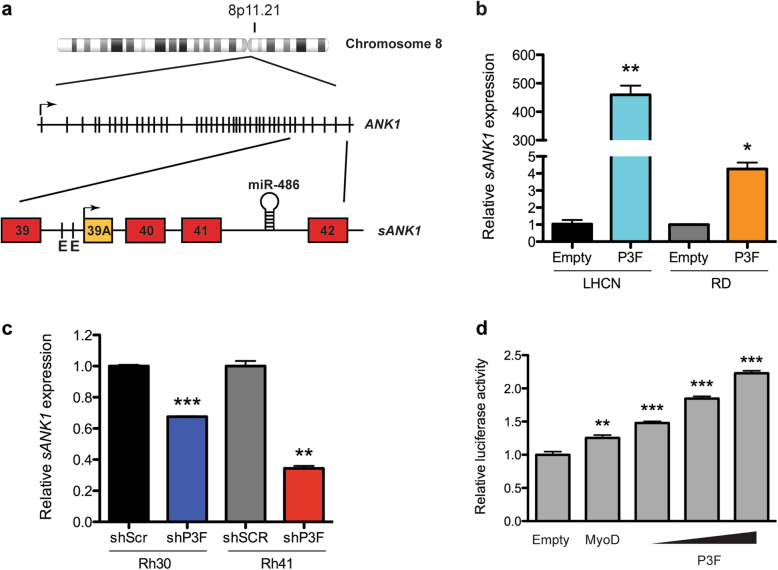
We focused on miR-486-5p as it is a muscle-enriched microRNA and antimiR-486-5p treatment in Rh41 cells had a strong antiproliferative phenotype. miR-486-5p is transcribed from intron 41 of ANK1 (Fig. 6a). ANK1 is an erythroid-specific ankyrin-repeat protein linking the plasma membrane and the cytoskeleton. A muscle-specific promoter upstream of an alternative exon 39a drives the expression of a muscle-specific ANK1 protein called small ANK1 (sANK1) that links the sarcoplasmic reticulum to the sarcomere [37]. The promoter region preceding sANK1 contains multiple E-Box binding sites for the myogenic transcription factor MYOD1 [38]. Comparable to miR-486-5p, sANK1 expression increases with PAX3-FOXO1 expression in LHCN-M2 and RD cells and decreases with PAX3-FOXO1 knockdown in FP-RMS cells (Fig. 6b, c). To determine if PAX3-FOXO1 transcriptionally regulates sANK1 and miR-486 we cloned the 3 kb promoter region 5′ of ANK1 exon 39a containing the MYOD1 responsive E-boxes upstream of luciferase for reporter assays. Increasing PAX3-FOXO1 overexpression led to a dose-dependent increase in sANK1 promoter-driven luciferase activity comparable to luciferase activity with MyoD (Fig. 6d). Thus, both sANK1 and miR-486-5p are transcriptionally regulated by PAX3-FOXO1 through the sANK1 promoter.
Fig. 6.
PAX3-FOXO1 regulates miR-486-5p expression. a Map of the genomic location of miR-486-5p within the ANK1 gene downstream of the alternative exon 39A of the sANK1 isoform. b Expression of sANK1 by qRT-PCR in RD or LHCN cells stably transduced with empty control vector or PAX3-FOXO1. c Expression of sANK1 by qRT-PCR in Rh30 or Rh41 shRNA knockdown cells after doxycycline treatment for 5 days. d Luciferase activity in 293T cells co-transfected with the pGL3-sANK1-Promoter reporter vector and either Empty control, MyoD, or increasing dose of PAX3-FOXO1 expression vectors. *P < 0.05, **P < 0.01, ***P < 0.001 shSCR vs shP3F, or Empty vs P3F
miR-486-5p promotes invasion of FP-RMS cell lines
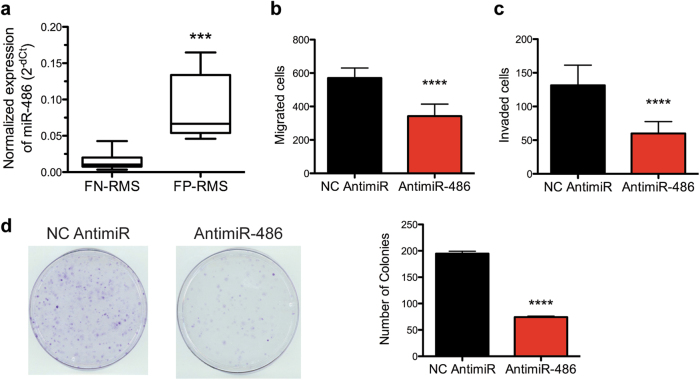
We confirmed miR-486-5p overexpression in FP-RMS compared to FN-RMS in patient-derived xenografts (Fig. 7a). Because miR-486-5p expression is positively regulated by PAX3-FOXO1 and upregulated in FP-RMS PDXs, we hypothesized that miR-486-5p could contribute to phenotypes of FP-RMS cells other than promoting proliferation. Similar to PAX3-FOXO1 knockdown, Rh41 cells with antimiR-486-5p transfection reduced both cell migration and invasion in transwell assays (Fig. 7b, c). In addition, miR-486-5p inhibition reduced clonogenic colony formation capacity of Rh41 cells (Fig. 7d). Thus, inhibition of miR-486-5p phenocopies PAX3-FOXO1 knockdown illustrating that miR-486-5p contributes to cell proliferation, cell migration and invasion, and clonogenic growth in FP-RMS.
Fig. 7.
Oncogenic miR-486-5p contributes to FP-RMS pathogenesis. a Expression of miR-486-5p by qRT-PCR in patient-derived xenografts from the CSTN consisting of 13 FN-RMS and 4 FP-RMS samples. b Transwell migration and c invasion assays in Rh41 cells transfected with NC or antimiR-486-5p. d Clonogenic colony formation assay in Rh41 cells transfected with NC or antimiR-486-5p. ***P < 0.001, ****P < 0.0001 for FP-RMS vs FN-RMS or antimiR-486 vs NC antimiR
To elucidate the molecular mechanisms of miR-486-5p in FP-RMS, we sought to explore the miR-486-5p target genes in FP-RMS. First, we identified 2755 putative target genes of miR-486-5p using the target prediction algorithm TargetScan irrespective of target site conservation. To narrow the list of putative target genes, we compared the putative target genes from the TargetScan algorithm to that of genes with increased expression with PAX3-FOXO1 knockdown (GSE73483) [39]. We identified 35 miR-486-5p putative target genes with increased expression with PAX3-FOXO1 knockdown (Supplementary Figure S6a). Of the 35 putative miR-486-5p target genes, 5 have tumor suppressive roles in cancer, including ARID1A, CYLD, GRIN20, PNPLA4, and SOCS2. We interrogated miR-486-5p regulation by transfecting Rh41 cells with NC or antimiR-486-5p and performing qRT-PCR (Supplementary Figure S6b). However, none of these five genes were upregulated with miR-486-5p inhibition. We also assessed previously identified miR-486-5p targets, including ARID4B, EMP, FOXO1, PIK3AP1, PTEN, SMARCD2, TOB1, and TWF1. Interestingly, only subtle but significant changes in gene expression were detected for several predicted target genes, including ARID4B, PIK3AP1, SMARCD2, and TWF1 (Supplementary Figure S6b). Despite the known role of miR-486-5p in regulating the PI3-kinase/AKT pathways in skeletal muscle and lung cancer, miR-486-5p inhibition did not result in increased expression of previously identified target genes PTEN, FOXO1, or PIK3R1 or activation of the PI3K/AKT pathway (Supplementary Figure S6b and c) [38, 40, 41]. One of the top predicted targets of miR-486-5p is Twinfillin1 (TWF1). TWF1 is an actin monomer binding protein that contributes to motility of cells through actin cytoskeleton rearrangement. Inhibition of miR-486-5p also led to an increase in TWF1 protein levels in both Rh30 and Rh41 cells (Supplementary Figure S6d). We confirmed that miR-486-5p directly regulates TWF1 using a luciferase reporter with the 3′UTR of TWF1 (Supplementary Figure S6e and f).
miR-486-5p inhibition reduces tumor growth in mouse xenografts
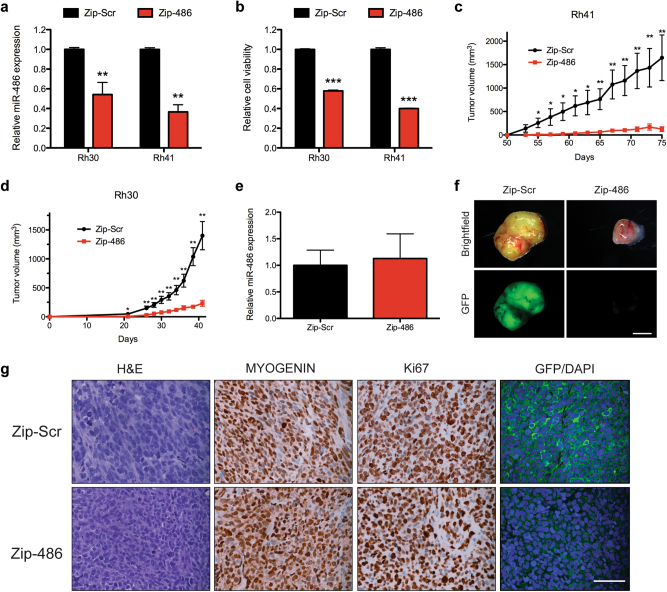
Given that miR-486-5p inhibition in FP-RMS cell lines reduces proliferation, we sought to determine the effect of long-term miR-486-5p inhibition in vitro as well as in vivo tumor growth. We inhibited miR-486-5p with transduction of lentivirus expressing miRZip-486-5p (lenti-miRZip-486-5p) that stably express single-stranded anti-microRNAs that bind the endogenous microRNA target, thus inhibiting its function. Transduction of Rh30 and Rh41 with lenti-miRZip-486-5p significantly decreased miR-486-5p expression and cell viability (Fig. 8a, b). Transduction efficiency was evaluated by GFP fluorescence expressed from the lenti-miRZip and greater than 80% of the cells were GFP positive 48 h after transduction (Supplementary Figure S7a). Transduction of additional FP-RM cell lines, including Rh3, Rh4, and Rh28 led to similar transduction efficiencies and decreases in miR-486-5p expression and cell viability (Supplementary Figure S7a–c).
Fig. 8.
miR-486-5p inhibition reduces in vivo tumor growth. a qRT-PCR expression of miR-486-5p in Rh30 and Rh41 cells 48 h following transduction with miRZip-Scr or miRZip-486-5p. b Cell Titer Glo cell viability assay in cells as in (a). Subcutaneous tumor growth in Rh41 cells (c) and Rh30 cells (d) transduced with miRZip-Scr or miRZip-486-5p (n = 10 mice per group). e qRT-PCR expression of miR-486-5p in resulting Rh30 xenografts from (c) (P = 0.227). f Whole-mount brightfield and GFP imaging of Rh30 xenografts. Scale bar, 4 mm. g H&E staining and IHC staining for MYOGENIN, Ki67, and GFP/DAPI in Rh30 xenografts. Scale bar 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001 for miRZip-Scr vs miRZip-486-5p
Rh30 and Rh41 cells were transduced with lenti-miRZip-486 or lenti-miRZip-Scr control and subcutaneously injected into immunocompromised SCID-Beige mice 48 h after transduction. A striking decrease in xenograft growth was observed in both Rh30 and Rh41 xenografts transduced with miRZip-486-5p (Fig. 8c, d, and Supplementary Figure S8a and b). In fact, measurable tumors developed in only 4 of 10 mice in Rh41 and 6 of 10 mice in Rh30 cells with miRZip-486-5p, compared to 9 of 10 mice with miRZip-Scr transduced cells. The expression of miR-486-5p was assessed in the resulting xenografts by qRT-PCR. No significant decreased miR-486-5p expression was observed suggesting a negative selection for untransduced cells or for cells with low expression of the miRZip-486-5p (Fig. 8e and Supplementary Figure S8c). Indeed, whole-mount imaging of the xenografts illustrates that miRZip-486-5p-transduced xenografts expressed significantly less GFP (Fig. 8f and Supplementary Figure S8d). The histology and immunohistochemistry (IHC) staining of the miRZip-Scr and miRZip-486 were indistinguishable in regard to MYOGENIN and Ki67 staining (Fig. 8g and Supplementary Figure S8e). However, the GFP staining on tumor sections was consistent with the whole-mount imaging with remarkably less GFP staining in the miRZip-486-5p-transduced xenografts. Taken together, miR-486-5p promotes FP-RMS tumorigenesis with a strong selection to maintain high expression of miR-486-5p.
Discussion
In this study, we found distinct expression of microRNAs in FP-RMS and FN-RMS and interrogated the role of microRNAs regulated by PAX3-FOXO1 in FP-RMS. Using microRNA microarrays, we found many microRNAs positively and negatively correlated with PAX3-FOXO1 in patient-derived xenografts and cell lines as well as gain- and loss-of-function RMS cells. We found miR-221/222 and miR-486-5p as novel candidates of tumor suppressive and oncogenic microRNAs, respectively, in FP-RMS.
We found miR-221/222 among the most downregulated microRNAs in FP-RMS. These microRNAs, clustered on the X chromosome, are expressed as a single pri-microRNA and contain identical seed sequences [42]. One of the first studies on miR-221/222 found that they function as tumor suppressors through the modulation of the KIT receptor in erythroleukemia cells and acute myeloid leukemia [42, 43]. In addition, miR-221 is downregulated in cholangiocarcinoma [44], gastrointestinal stromal tumors [45, 46], and pancreatic cancer where it targets IRF2 and SOCS3 [47, 48]. In endothelial cells, miR-221/222 inhibit proliferation, migration, and angiogenesis [49]. Consistent with our findings, miR-221/222 are downregulated specifically in FP-RMS cell lines and patients [31, 50]. Interestingly miR-221/222 expression correlates with overall survival and progression-free survival of RMS [31]. In contrast, miR-221/222 are overexpressed and function as oncogenes in breast cancer, hepatocellular carcinoma, glioblastoma, and others [51, 52]. Furthermore, miR-221/222 are highly expressed in myoblasts promoting proliferation and delaying differentiation [30, 53]. Thus the function of miR-221/222 is highly context dependent on the expression and regulation of its targets [54]. The context-dependent oncogenic roles of miR-221/222 abrogates the temptation to proceed with miR-221/222 replacement therapy in RMS.
In FP-RMS, we identified CCND2, CDK6, and ERBB3 as novel targets of miR-221. Interestingly, CCND2 is the predominant D-Type Cyclin expressed in RMS and its downregulation by miR-1/206 decreases RMS proliferation [34, 55, 56]. Furthermore, Cyclin D and CDK4/6 activity is tightly regulated in myoblast proliferation and differentiation [57, 58] and overexpressed/hyperactive in cancer and RMS [33]. ERBB3 is significantly upregulated in FP-RMS, promotes RMS, and may represent a therapeutic target [35, 59]. Interestingly, ERBB3 upregulation may contribute to MEK inhibition resistance in RMS [60]. However, the context-dependent function of miR-221/222 suggests that therapeutic manipulation with replacement or inhibition of miR-221 needs to be rigorously evaluated since it can function as both an oncogene and tumor suppressor.
In evaluating the phenotypic effect of inhibition of 13 microRNAs positively regulated by PAX3-FOXO1, we found miR-486-5p to have the strongest proliferative phenotype. miR-486-5p is overexpressed and can function as an oncogene in KRAS-mutated colorectal cancer [61], pancreatic cancer [62], gliomas [63], Sezary syndrome lymphoma [64], and leukemias [29, 65, 66]. In contrast, miR-486-5p is downregulated in gastric cancer [67], lung cancer [40], and hepatocellular carcinoma [68]. The tumor suppressing phenotype is primarily through downregulation of IGF signaling pathway components. Similar to the promiscuous roles of miR-221 in cancer, the varied functions of miR-486-5p in different tumors highlight the context dependence of microRNA function likely secondary to differential target gene regulation in different cell types.
miR-486-5p is a muscle-enriched microRNA that is upregulated by MRTFs and can promote myogenic differentiation [38, 69]. miR-486-5p also induces proliferation and increases myotube size in primary mouse myoblasts [70]. Furthermore, miR-486-5p induces skeletal muscle hypertrophy in Myostatin knockout mice and ameliorates muscular dystrophy through Pten regulation of Akt signaling [41, 71]. In addition, miR-486-5p can reduce skeletal muscle atrophy in chronic kidney disease [72]. Consistent with increased proliferation in myoblasts overexpressing miR-486-5p [70], we find that inhibition of miR-486-5p reduces proliferation and xenograft growth of FP-RMS. In addition, we find that miR-486-5p inhibition decreases migration and invasion of FP-RMS, indicating that miR-486-5p contributes to the increased metastatic potential of FP-RMS. We were unable to identify a single specific miR-486-5p target gene in our model system responsible for the phenotypes observed with miR-486-5p inhibition. This is consistent with idea that the “1 microRNA-1 target” model is insufficient to explain the complex phenotypes regulated by microRNAs. Modest regulation of many target genes collectively are likely responsible for the complex phenotypes induced by microRNAs. The subtle gene expression changes we report here subsequent to inhibition of predicted miR-486-5p target genes are consistent with this model.
Despite knowing the molecular driver of FP-RMS, targeted therapies have yet to make an impact for patients, highlighting the need for a greater understanding of the molecular consequences of PAX3-FOXO1 expression. Our data suggest that PAX3-FOXO1 regulates the expression of key microRNAs involved in FP-RMS pathogenesis. The hypothesis that microRNAs can target hundreds of mRNAs and therefore be more therapeutically valuable than agents against a single molecule or pathway suggest that identifying crucial microRNAs may lead to therapeutics to improve patient outcomes. miR-221/222 and miR-486-5p may represent novel therapeutic vulnerabilities in FP-RMS with inhibition of miR-486-5p and replacement of miR-221/222.
Materials and methods
Cell lines
Cells were from the following sources: RD (CCL-136) and Rh30 (CRL-2061) from ATCC (Manassas, VA, USA), Rh3, RH4, Rh28, and Rh41 (Gerard Grosveld, St. Jude), 293T (Martine F. Roussel, St. Jude), and LHCN-M2 (Woodring Wright, University of Texas Southwestern Medical Center) [73]. Cells were authenticated with short tandem repeat profiling (Supplementary Table S2) [56].
RNA and gene expression
Total RNA was isolated, reverse transcribed, and quantitative real-time PCR (qRT-PCR) performed as previously described using primers and probes in Supplementary Table S3 [56]. For microRNA microarrays, total RNAs were labeled and hybridized to The Human microRNA Microarray Release 19.0, 8 × 60k (G4872A-046064; Agilent, Santa Clara, CA, USA). Normalized signal data were log2 transformed and each probeset was compared by unequal variance t-test (Partek Genomics Suite 6.6, St. Louis, MO, USA). The microarray data have been deposited to GEO (GSE97553). Differential microRNA expression was considered significant if P < 0.05 and the fold change was greater than 2. MicroRNA targets predicted based on TargetScan (Release 7.1) [32].
Molecular cloning, viral transduction, and luciferase assays
Doxycycline-inducible shRNAs (shP3F and shScr) were generated by ligating annealed overlapping oligonucleotides for PAX3-FOXO1 or Scrambled to AgeI and EcoRI digested Tet-pLKO-Puro (gift from Dmitri Wiederschain, Addgene plasmid #21915) [74]. Primers used in cloning are described in Supplementary Table S4. pBabe-PAX3-FOXO1-puro was generated by PCR amplifying PAX3-FOXO1 from pcDNA-PAX3-FOXO1 (gift from Rene Galindo) digesting with BamHI and SalI (New England Biolabs (NEB), Ipswich, MA, USA) and ligating to pBabe-puro (gift from Jay Morgenstern and Hartmut Land Addgene plasmid #1764) [75]. Retrovirus was packaged as described previously [24]. Cells were selected and maintained in 0.5 μg/mL puromycin. shRNA expression was induced with 50 ng/mL doxycycline. miRZip-486-5p (MZIP486-5p-PA-1; System Bioscience Inc., Palo Alto, CA, USA) and miRZip-Scr (MZIP000-PA-1) were packaged following manufacturers’ instructions. Transduction efficiency of miRZip was determined by counting the number of GFP-positive cells divided by the total number of cells in four random fields for each condition.
458 bp of the human genomic DNA sequence containing miR-221 was synthesized (gene block; Integrated DNA Technologies (IDT), Coralville, IA, USA), digested with EcoRI and SalI and ligated to pCMV6. 337 bp of the human genomic DNA sequence containing miR-486 was PCR amplified using primers detailed in Supplementary Table S4, digested with EcoRI and SalI and ligated to pCMV6. The psiCHECK2-miR-221-sensor and psiCHECK2-miR-486-5p-sensor were constructed by annealing oligonucleotides with the reverse complement sequence of the mature microRNA with 5′ XhoI and 3′ NotI overhangs (IDT) and ligating to psiCHECK2 reporter. CCND2 and TWF1 3′UTRs were described previously [56]. Gene blocks (IDT) for wild type and miR-221 site mutant 3′UTRs (Supplementary Table S5) for CDK6 (6643-7109 bp) and ERBB3 (1-478 bp) were digested with XhoI and NotI and ligated to psiCHECK2 reporter. The miR-221 or miR-486-5p recognition sites in CCND2 and TWF1 were mutated with Quick Change II Site Directed Mutagenesis (200524; Agilent Technologies, Santa Clara, CA, USA). Luciferase reporter assays performed as previously described [56].
The sANK1 promoter was PCR amplified from LHCN-M2 myoblast genomic DNA digested with KpnI and NheI and ligated to pGL3-Promoter vector (Promega). MYOD1 cDNA (MHS6278-202832821; GE Dharmacon Open Biosystems, Lafayette, CO, USA) was amplified, PCR digested with EcoRI and SalI, and ligated to pCMV6. PAX3-FOXO1 cDNA was amplified by PCR, digested with SalI and BamHI, and ligated to pCMV6. Promoter assays performed by co-transfecting 293T cells with 50 ng of pGL3-promoter empty or sANK1 promoter, 400 ng of pCMV6-Empty, pCMV6-MYOD1, or pCMV6-PAX3-FOXO1, and 5 ng of pCMV6-LacZ. Luciferase assays performed using the Luciferase Assay System (E1501; Promega) and the FluoReporter LacZ/Galactosidase Quantification Kit (F-2905; Thermo Fisher Scientific, Waltham, MA, USA). Luciferase activity was normalized to β-galactosidase activity relative to empty control.
Phenotypic assays
For assessment of cell viability, cells were treated as indicated and the Cell Titer Glo assay (G7570, Promega) or Caspase 3/7 Glo Assay (G8090, Promega) was performed measuring luminescence with BioTek Synergy 2. Population doubling was determined as previously described [24]. Cell cycle FACS analysis was performed with propidium iodide in cells treated with 50 ng/mL doxycycline for 5 days. Clonogenic low-density colony formation was assessed by seeding 400 cells per 6 cm dish. Colonies were fixed and stained with a solution of 0.05% crystal violet, 1% formaldehyde, 1% methanol, phosphate-buffered saline, and counted. Transwell migration (3422; Costar, Corning, NY, USA) and matrigel invasion assays (354480, Costar) were performed as described previously [76].
Immunoblotting
Immunoblotting performed as described previously [77]. Primary antibodies detailed in Supplementary Table S6.
Transfection of siRNA, microRNA mimics and inhibitors
Cells were transfected with 30 nM miRVana microRNA antimiRs or mimics (Thermo Fisher Scientific, Waltham, MA, USA) with inhibitors and mimics (Supplementary Table S7) as described previously [56]. The NC Dicer-substrate siRNA (51-01-14-03, IDT) and PAX3-FOXO1 siRNA (detailed in Supplementary Table S4) were transfected with 3 nM duplex as above.
In vivo tumorigenesis
Rh30 and Rh41 cells were transduced with miRZip-Scr or miRZip-486-5p, 48 h after infection. 1 × 106 Rh41 cells or 7.5 × 105 were subcutaneously injected in the flank of 8-week-old age-matched, randomized female SCID-Beige mice (n = 10 per group). Tumor growth measured every other day blinded to treatment once palpable tumors formed. Tumor volume calculated as previously described [24]. The tumor volume at each time point was compared using an unpaired Student’s t-test. All studies involving animal studies were reviewed and approved by the St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee.
Immunohistochemistry
Histology and IHC performed following standard protocols using antibodies detailed in Supplementary Table S6 [24].
Statistical analysis
All results expressed as the mean ± SEM. All pair-wise comparisons utilized a two-tailed, unpaired Student’s t-test using GraphPad Prism Version 5 (Graph Pad Software, Inc., San Diego, CA, USA). P-values <0.05 considered significant. The number of mice required (n = 10 per group) for in vivo xenograft experiments was determined by power analysis using G Power 3.1 [78] with two sided, 90% power, Type I error of 1%.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the views of the NIH.
Electronic supplementary material
Acknowledgements
This work was supported by the National Cancer Institute of the National Institutes of Health under award numbers R01CA216344 and K08CA151649 to M.E.H. and 5R25CA23944 to D.M. and J.R. The Hatley lab is also supported by the V foundation for Cancer Research, Hyundai Hope on Wheels, and Alex’s Lemonade Stand Foundation and by the American Lebanese Syrian Associated Charities.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
The online version of this article (10.1038/s41388-017-0081-3) contains supplementary material, which is available to authorized users.
References
- 1.Sultan I, Qaddoumi I, Yaser S, Rodriguez-Galindo C, Ferrari A. Comparing adult and pediatric rhabdomyosarcoma in the surveillance, epidemiology and end results program, 1973 to 2005: an analysis of 2,600 patients. J Clin Oncol. 2009;27:3391–7. doi: 10.1200/JCO.2008.19.7483. [DOI] [PubMed] [Google Scholar]
- 2.Parham DM, Barr FG. Alveolar rhabdomyosarcoma. In: Fletcher CDM, Bridge JA, Hogendoorn P, Mertens F, editors. WHO classification of tumours of soft tissue and bone. 4th ed. Lyon, France: IARC; 2013. pp. 130–2. [Google Scholar]
- 3.Shapiro DN, Sublett JE, Li B, Downing JR, Naeve CW. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 1993;53:5108–12. [PubMed] [Google Scholar]
- 4.Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, III, Emanuel BS, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–5. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 5.Davis RJ, D’Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–72. [PubMed] [Google Scholar]
- 6.Missiaglia E, Williamson D, Chisholm J, Wirapati P, Pierron G, Petel F, et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol. 2012;30:1670–7. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 7.Skapek SX, Anderson J, Barr FG, Bridge JA, Gastier-Foster JM, Parham DM, et al. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a children’s oncology group report. Pediatr Blood Cancer. 2013;60:1411–7. doi: 10.1002/pbc.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan F, Smith LM, Gustafson DM, Zhang C, Dunlevy MJ, Gastier-Foster JM, et al. Genomic and clinical analysis of fusion gene amplification in rhabdomyosarcoma: a report from the Children’s Oncology Group. Genes Chromosomes Cancer. 2012;51:662–74. doi: 10.1002/gcc.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children’s oncology group. J Clin Oncol. 2002;20:2672–9. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 10.Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28:2151–8. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Shern JF, Wei JS, Yohe ME, Song YK, Hurd L, et al. Clonality and evolutionary history of rhabdomyosarcoma. PLoS Genet. 2015;11:e1005075. doi: 10.1371/journal.pgen.1005075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24:710–24. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–31. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashi VP, Hatley ME, Galindo RL. Probing for a deeper understanding of rhabdomyosarcoma: insights from complementary model systems. Nat Rev Cancer. 2015;15:426–39. doi: 10.1038/nrc3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahajan P, Leavey PJ, Galindo RL. PAX genes in childhood oncogenesis: developmental biology gone awry? Oncogene. 2015;34:2681–9. doi: 10.1038/onc.2014.209. [DOI] [PubMed] [Google Scholar]
- 16.Ayyanathan K, Fredericks WJ, Berking C, Herlyn M, Balakrishnan C, Gunther E, et al. Hormone-dependent tumor regression in vivo by an inducible transcriptional repressor directed at the PAX3-FKHR oncogene. Cancer Res. 2000;60:5803–14. [PubMed] [Google Scholar]
- 17.Bernasconi M, Remppis A, Fredericks WJ, Rauscher FJ, III, Schafer BW. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc Natl Acad Sci USA. 1996;93:13164–9. doi: 10.1073/pnas.93.23.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall AD, Grosveld GC. Alveolar rhabdomyosarcoma—the molecular drivers of PAX3/7-FOXO1-induced tumorigenesis. Skelet Muscle. 2012;2:25. doi: 10.1186/2044-5040-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions—beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 21.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11:860–72. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 24.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–22. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 26.Stewart E, Federico S, Karlstrom A, Shelat A, Sablauer A, Pappo A, et al. The Childhood Solid Tumor Network: a new resource for the developmental biology and oncology research communities. Dev Biol. 2016;411:287–93. doi: 10.1016/j.ydbio.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douglass EC, Valentine M, Etcubanas E, Parham D, Webber BL, Houghton PJ, et al. A specific chromosomal abnormality in rhabdomyosarcoma. Cytogenet Cell Genet. 1987;45:148–55. doi: 10.1159/000132446. [DOI] [PubMed] [Google Scholar]
- 28.Petak I, Douglas L, Tillman DM, Vernes R, Houghton JA. Pediatric rhabdomyosarcoma cell lines are resistant to Fas-induced apoptosis and highly sensitive to TRAIL-induced apoptosis. Clin Cancer Res. 2000;6:4119–27. [PubMed] [Google Scholar]
- 29.Wang LS, Li L, Li L, Chu S, Shiang KD, Li M, et al. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood. 2015;125:1302–13. doi: 10.1182/blood-2014-06-581926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS ONE. 2009;4:e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Missiaglia E, Shepherd CJ, Aladowicz E, Olmos D, Selfe J, Pierron G, et al. MicroRNA and gene co-expression networks characterize biological and clinical behavior of rhabdomyosarcomas. Cancer Lett. 2017;385:251–60. doi: 10.1016/j.canlet.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saab R, Bills JL, Miceli AP, Anderson CM, Khoury JD, Fry DW, et al. Pharmacologic inhibition of cyclin-dependent kinase 4/6 activity arrests proliferation in myoblasts and rhabdomyosarcoma-derived cells. Mol Cancer Ther. 2006;5:1299–308. doi: 10.1158/1535-7163.MCT-05-0383. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Hu S, Schofield DE, Sorensen PH, Triche TJ. Selective usage of D-type cyclins by Ewing’s tumors and rhabdomyosarcomas. Cancer Res. 2004;64:6026–34. doi: 10.1158/0008-5472.CAN-03-2594. [DOI] [PubMed] [Google Scholar]
- 35.Nordberg J, Mpindi JP, Iljin K, Pulliainen AT, Kallajoki M, Kallioniemi O, et al. Systemic analysis of gene expression profiles identifies ErbB3 as a potential drug target in pediatric alveolar rhabdomyosarcoma. PLoS ONE. 2012;7:e50819. doi: 10.1371/journal.pone.0050819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang QE, Racicot KE, Kaucher AV, Oatley MJ, Oatley JM. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development. 2013;140:280–90. doi: 10.1242/dev.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou D, Birkenmeier CS, Williams MW, Sharp JJ, Barker JE, Bloch RJ. Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J Cell Biol. 1997;136:621–31. doi: 10.1083/jcb.136.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Small EM, O’Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA. 2010;107:4218–23. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebauer M, Wachtel M, Niggli FK, Schafer BW. Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene. 2007;26:7267–81. doi: 10.1038/sj.onc.1210525. [DOI] [PubMed] [Google Scholar]
- 40.Peng Y, Dai Y, Hitchcock C, Yang X, Kassis ES, Liu L, et al. Insulin growth factor signaling is regulated by microRNA-486, an underexpressed microRNA in lung cancer. Proc Natl Acad Sci USA. 2013;110:15043–8. doi: 10.1073/pnas.1307107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexander MS, Casar JC, Motohashi N, Vieira NM, Eisenberg I, Marshall JL, et al. MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J Clin Invest. 2014;124:2651–67. doi: 10.1172/JCI73579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–6. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brioschi M, Fischer J, Cairoli R, Rossetti S, Pezzetti L, Nichelatti M, et al. Down-regulation of microRNAs 222/221 in acute myelogenous leukemia with deranged core-binding factor subunits. Neoplasia. 2010;12:866–76. doi: 10.1593/neo.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52:297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 45.Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, et al. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–26. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- 46.Koelz M, Lense J, Wrba F, Scheffler M, Dienes HP, Odenthal M. Down-regulation of miR-221 and miR-222 correlates with pronounced Kit expression in gastrointestinal stromal tumors. Int J Oncol. 2011;38:503–11. doi: 10.3892/ijo.2010.857. [DOI] [PubMed] [Google Scholar]
- 47.Kneitz B, Krebs M, Kalogirou C, Schubert M, Joniau S, van Poppel H, et al. Survival in patients with high-risk prostate cancer is predicted by miR-221, which regulates proliferation, apoptosis, and invasion of prostate cancer cells by inhibiting IRF2 and SOCS3. Cancer Res. 2014;74:2591–603. doi: 10.1158/0008-5472.CAN-13-1606. [DOI] [PubMed] [Google Scholar]
- 48.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 49.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–71. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 50.Ciarapica R, Russo G, Verginelli F, Raimondi L, Donfrancesco A, Rota R, et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8:172–5. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 51.Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuzaki J, Suzuki H. Role of MicroRNAs-221/222 in digestive systems. J Clin Med. 2015;4:1566–77. doi: 10.3390/jcm4081566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cardinali B, Cappella M, Provenzano C, Garcia-Manteiga JM, Lazarevic D, Cittaro D, et al. MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis. 2016;7:e2086. doi: 10.1038/cddis.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor Suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76:3666–70. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L, Sarver AL, Alamgir S, Subramanian S. Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab Invest. 2012;92:571–83. doi: 10.1038/labinvest.2012.10. [DOI] [PubMed] [Google Scholar]
- 56.Hanna JA, Garcia MR, Go JC, Finkelstein D, Kodali K, Pagala V, et al. PAX7 is a required target for microRNA-206-induced differentiation of fusion-negative rhabdomyosarcoma. Cell Death Dis. 2016;7:e2256. doi: 10.1038/cddis.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao SS, Chu C, Kohtz DS. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix regulators. Mol Cell Biol. 1994;14:5259–67. doi: 10.1128/MCB.14.8.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skapek SX, Rhee J, Spicer DB, Lassar AB. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science. 1995;267:1022–4. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 59.Ricci C, Landuzzi L, Rossi I, De Giovanni C, Nicoletti G, Astolfi A, et al. Expression of HER/erbB family of receptor tyrosine kinases and induction of differentiation by glial growth factor 2 in human rhabdomyosarcoma cells. Int J Cancer. 2000;87:29–36. doi: 10.1002/1097-0215(20000701)87:1<29::AID-IJC5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 60.Bersani F, Lingua MF, Morena D, Foglizzo V, Miretti S, Lanzetti L, et al. Deep sequencing reveals a novel miR-22 regulatory network with therapeutic potential in rhabdomyosarcoma. Cancer Res. 2016;76:6095–106. doi: 10.1158/0008-5472.CAN-16-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mosakhani N, Sarhadi VK, Borze I, Karjalainen-Lindsberg ML, Sundstrom J, Ristamaki R, et al. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer. 2012;51:1–9. doi: 10.1002/gcc.20925. [DOI] [PubMed] [Google Scholar]
- 62.Omura N, Mizuma M, MacGregor A, Hong SM, Ayars M, Almario JA, et al. Overexpression of ankyrin1 promotes pancreatic cancer cell growth. Oncotarget. 2016;7:34977–87. doi: 10.18632/oncotarget.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song L, Lin C, Gong H, Wang C, Liu L, Wu J, et al. miR-486 sustains NF-kappaB activity by disrupting multiple NF-kappaB-negative feedback loops. Cell Res. 2013;23:274–89. doi: 10.1038/cr.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, et al. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sezary syndrome. Cell Death Dis. 2011;2:e151. doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seyyedi SS, Soleimani M, Yaghmaie M, Ajami M, Ajami M, Pourbeyranvand S, et al. Deregulation of miR-1, miR486, and let-7a in cytogenetically normal acute myeloid leukemia: association with NPM1 and FLT3 mutation and clinical characteristics. Tumour Biol. 2016;37:4841–7. doi: 10.1007/s13277-015-4289-y. [DOI] [PubMed] [Google Scholar]
- 66.Shaham L, Vendramini E, Ge Y, Goren Y, Birger Y, Tijssen MR, et al. MicroRNA-486-5p is an erythroid oncomiR of the myeloid leukemias of Down syndrome. Blood. 2015;125:1292–301. doi: 10.1182/blood-2014-06-581892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan IB, et al. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res. 2011;17:2657–67. doi: 10.1158/1078-0432.CCR-10-3152. [DOI] [PubMed] [Google Scholar]
- 68.Youness RA, El-Tayebi HM, Assal RA, Hosny K, Esmat G, Abdelaziz AI. MicroRNA-486-5p enhances hepatocellular carcinoma tumor suppression through repression of IGF-1R and its downstream mTOR, STAT3 and c-Myc. Oncol Lett. 2016;12:2567–73. doi: 10.3892/ol.2016.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–14. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alexander MS, Casar JC, Motohashi N, Myers JA, Eisenberg I, Gonzalez RT, et al. Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet Muscle. 2011;1:27. doi: 10.1186/2044-5040-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hitachi K, Nakatani M, Tsuchida K. Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. Int J Biochem Cell Biol. 2014;47:93–103. doi: 10.1016/j.biocel.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 72.Xu J, Li R, Workeneh B, Dong Y, Wang X, Hu Z. Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney Int. 2012;82:401–11. doi: 10.1038/ki.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu CH, Mouly V, Cooper RN, Mamchaoui K, Bigot A, Shay JW, et al. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515–23. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 74.Wiederschain D, Wee S, Chen L, Loo A, Yang G, Huang A, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 75.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–96. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Machado E, White-Gilbertson S, van de Vlekkert D, Janke L, Moshiach S, Campos Y, et al. Regulated lysosomal exocytosis mediates cancer progression. Sci Adv. 2015;1:e1500603. doi: 10.1126/sciadv.1500603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N, et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell. 2012;22:536–46. doi: 10.1016/j.ccr.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–91. doi: 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.