Abstract
The Steroidogenic acute regulatory protein (StAR) directs mitochondrial cholesterol uptake through a C-terminal cholesterol binding domain (CBD) and a 62 amino acid N-terminal regulatory domain (NTD) that contains an import sequence and conserved sites for inner membrane metalloproteases. Deletion of the NTD prevents mitochondrial import while maintaining steroidogenesis but with compromised cholesterol homeostasis. The rapid StAR-mediated cholesterol transfer in adrenal cells depends on concerted mRNA translation, p37 StAR phosphorylation and controlled NTD cleavage. The NTD controls this process with two cAMP-inducible modulators of, respectively, transcription and translation SIK1 and TIS11b/Znf36l1. High-resolution fluorescence in situ hybridization (HR-FISH) of StAR RNA resolves slow RNA splicing at the gene loci in cAMP-induced Y-1 cells and transfer of individual 3.5 kb mRNA molecules to mitochondria. StAR transcription depends on the CREB coactivator CRTC2 and PKA inhibition of the highly inducible suppressor kinase SIK1 and a basal counterpart SIK2. PKA-inducible TIS11b/Znf36l1 binds specifically to highly conserved elements in exon 7 thereby suppressing formation of mRNA and subsequent translation. Co-expression of SIK1, Znf36l1 with 3.5 kb StAR mRNA may limit responses to pulsatile signaling by ACTH while regulating the transition to more prolonged stress
Keywords: StAR, steroidogenesis, CRTC, SIK, TIS11B
1. StAR integrates inter-membrane cholesterol transfer with mitochondrial electron transfer processes
The steroidogenic acute regulatory protein (StAR) initiates steroidogenesis by transferring cholesterol from outside the mitochondria to cytochrome P450 11A1 (CYP11A1) in the inner mitochondrial membrane (IMM) (Artemenko et al., 2001; Caron et al., 1997; Clark et al., 1994; Kiriakidou et al., 1996). Even after adrenocorticotropic hormone (ACTH) stimulation, cholesterol metabolism by CYP11A1 in adrenal mitochondria can exceed StAR mediated transfer so that cholesterol normally does not accumulate. ACTH stimulated cholesterol accumulation is produced by the CYP11A1 inhibitor aminoglutethimide (AMG) resulting in up to 3–5 cholesterol molecules per CYP11A1. This stimulation is paralleled by cholesterol–CYP11A1 complex formation (Jefcoate et al., 1973), which has been reproduced in cultured bovine adrenal cells (DiBartolomeis and Jefcoate, 1984). Turnover of this pool of reactive cholesterol at CYP11A1 is driven by reduced nicotinamide adenine dinucleotide phosphate (NADPH) generated most effectively by succinate dehydrogenase and the ATP-dependent NADH/NADPH transhydrogenase (NNT) (Hanukoglu and Jefcoate, 1980; Yamazaki et al., 1995). This process competes with transfer to IMM Cyp11b1 as shown by the opposing effect of cholesterol accumulation at CYP11A1 (Yamazaki et al., 1993). Mitochondrial intermembrane 3 beta-hydroxysteroid dehydrogenase (Hsd3b2) may have activity integrated with StAR activity (Rajapaksha et al., 2016) to relieve product inhibition of CYP11A1.
2. StAR functions through C-terminal Cholesterol binding domain
StAR consists of two domains: the N-terminal domain (NTD), which includes about 62 amino acids, and the C-terminal domain (CBD), which forms cholesterol complexes and is the conserved core of the STARD family. The NTD has the typical positive charge characteristics of other mitochondrial import sequences in the initial N-terminal amino acids and additional sequences that provide an unusually appreciable helical content and unusual dual cleavage sites (Bose et al., 1999).
The crystal structures of the CBD of StAR/STARD1 and StARD3 are similar even though they have very different specialized NTD (Kang et al., 2010; Letourneau et al., 2015). Each complex has a single cholesterol molecule. The transgenic deletion of the StAR gene in mice reproduces the pathology of human adrenal lipidemic hyperplasia (ALH) (Bose et al., 2002; Parker et al., 1998). Mutations, which cause the human disease, concentrate in the cholesterol binding domain rather than the NTD (Sahakitrungruang et al., 2010). However, the R182 mutation retains full cholesterol exchange activity but does not stimulate activity at CYP11A1 (Baker et al., 2005; Barbar et al., 2009).
The StAR activity under hormonal control is mediated by phosphorylation at S194 by cAMP and protein kinase A (PKA) in fasciculata cells, and by Ca–dependent kinases in glomerulosa cells (Dyson et al., 2009; Elliott et al., 1993). A second phosphorylation by extracellular signal-regulated kinase (ERK) at S232 affects mitochondrial import (Duarte et al., 2014). A large number of cholesterol molecules transferred for each newly synthesized StAR protein (Artemenko et al., 2001). This high turnover suggests that cholesterol activation of the CBD directs receptor-like activity for StAR. The cholesterol induced conformational change in StAR, which delivers a more flexible structure matches this concept (Rajapaksha et al 2013). Such complexes are active on the mitochondrial outer membrane (OMM) where they may enrich cholesterol at sites proximal to the IMM mitochondrial permeability transition pore (mPTP). StAR transfer of cholesterol is inhibited by cholesterol sulfate with the consequence that cholesterol sulfatase can enhance activity (Sugawara and Fujimoto, 2004).
Protein cross-linking and many other studies like the yeast two-hybrid system suggest coordination with voltage-dependent anion channel (VDAC) proteins and translocator protein (TSPO)/peripheral benzodiazepine receptor (PBR) (Li et al., 2001; Prasad et al., 2015; Shanmughapriya et al., 2015). The systemic TSPO-ko mice retain full steroidogenic activity in the testis much as is seen in MA-10 cells (Tu et al., 2015). It is also seen for an SF1-cre directed conditional loss of TSPO in the testis (Tu et al., 2016). By contrast, this same mouse shows a loss of ACTH induced glucocorticoid synthesis in the adrenal cortex. There appears to be a difference in the StAR-mediated cholesterol transfer process in the testis compared to the adrenal cortex. We have emphasized here that adrenal cells can produce peak steroidogenesis at very low levels of StAR mRNA thus pointing to a much more efficient process. The integration of TSPO and the StAR NTD into a distinctive adrenal process provides one explanation for the difference. The established effect of TSPO on mitochondrial Ca-sensitive permeability channels also suggests that mitochondria may rapidly adapt with alternative controls over membrane contacts that enhance StAR activity.
Cholesterol transfer depends on the continuous translation of the 37kd StAR pre-protein with concomitant phosphorylation by PKA. Thus, Inhibition with cycloheximide (CHX) stops ACTH-stimulated steroidogenesis within 5 minutes, while causing an accumulation of cholesterol in the OMM that remains inaccessible to IMM CYP11A1 (Pon et al., 1986; Privalle et al., 1983). This cholesterol accumulation in intact mitochondria generated by CHX is comparable to that generated by inhibition of CYP11A1 by AMG. For CHX, the cholesterol is in the OMM and inaccessible to CYP11A1, whereas for AMG the pool forms cholesterol-CYP11A1 complexes and is completely converted in minutes to pregnenolone after removal of the inhibitor. This early data indicates that cholesterol can enter the OMM and any associated membranes without StAR. The CBD can function in COS1 cells as a TOM 20 chimera, thus demonstrating that activity on the OMM alone effects cholesterol transfer to the IMM (Bose et al., 2002). However, the rate and efficiency here are possibly 100 times lower that in adrenal cells suggesting that other factors including the NTD can enhance the turnover. This activity has been reconstituted with rat adrenal mitochondria and StAR CBD. Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) were shown to be co-activators of StAR activity (Lin et al., 2016).
The inter-membrane cholesterol barrier is readily breached by mild treatments such as elevated Ca or hypotonic media. This is an issue for many early assays for StAR cholesterol transfer activity. The metabolism of this artificially transferred cholesterol is supported by NADPH generated by isocitrate but not by succinate in conjunction with the nicotinamide nucleotide transhydrogenase (NNT) (Yamazaki et al., 1995). This precaution is realized in the SNARE assay by using succinate to support CYP11A1 activity.
3. What is the role of the StAR NTD?
The sequence encompassing these NTD cleavage sites is highly conserved across species (Yamazaki et al., 2006). The cleavage pattern for bovine StAR in COS1 cells is similar to that for native StAR in MA-10 cells. Mutation of the conserved cleavage sites in bovine StAR singly and in combination show a complex additive cleavage process involving two separate processing pathways. The double mutation of these sites slowed but did not prevent NTD cleavage or decrease cholesterol metabolism in transfected COS1 cells. The IMM metalloproteases (MMP) that cleave the StAR NTD appear likely to interact with OMM VDAC1 each as participants in the mitochondrial permeability core complex (Shanmughapriya et al., 2015). This pore complex mediates the Ca inhibition of mitochondrial cholesterol metabolism, which is blocked by cyclosporine, a drug that binds to cyclophilin, a component of the pore complex (Kowluru et al., 1995). NTD-modulatory activity beyond the mitochondrial import function may involve the StAR 30–62 sequences, which are downstream of the first conserved MMP cleavage site. The MMP cleavage enzymes are located on the inner face of the IMM thus requiring transfer of p37 StAR to this point for cleavage. Recent evidence suggests that this cleavage facilitates interaction of StAR with the VDAC2, which then facilitates both cholesterol transfer and cleavage of this sequence (Prasad et al., 2015).
The role of IMM proteases in StAR activity is well demonstrated by the inhibition of cholesterol metabolism in Y-1 cells by o-phenanthroline (OP), a metalloprotease inhibitor (Artemenko et al., 2001). OP prevents NTD cleavage while inhibiting Br-cAMP-induced cholesterol metabolism but without any effect on the CYP11A1 cleavage of 20-hydroxy-cholesterol. When OP is washed out, the metabolism of cholesterol to pregnenolone continues as the cleavage of StAR ensues, but now without the need for the further synthesis of StAR. Four phosphorylated forms of p30-BAC/p32/rodent have been captured in 2D gels (Artemenko et al., 2001; Epstein and Orme-Johnson, 1991). A plausible model is that an IMM cleavage releases cleavage at NTD site A, which then allows IMM StAR to interact with OMM VDAC2 to enhance OMM-IMM contact and cholesterol transfer. The new and old concepts described here have been merged in the model shown in Figure 1.
Figure 1.
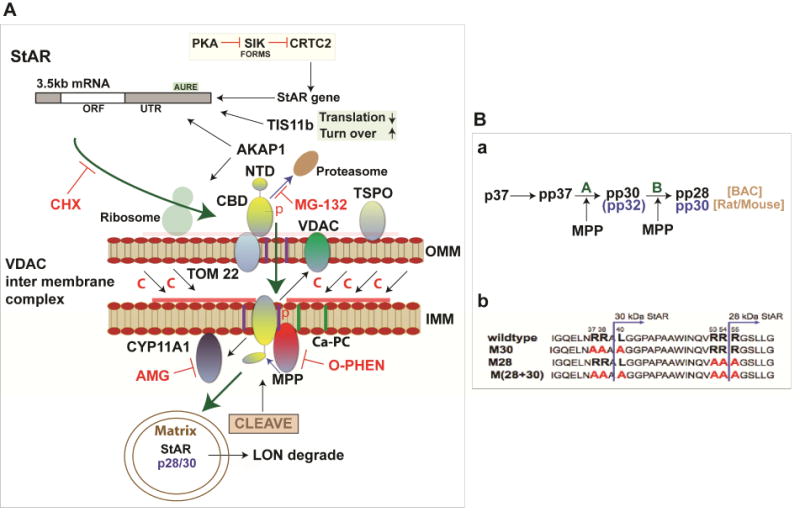
Model for high efficiency StAR-mediated adrenal cholesterol transfer to Cyp11a1 (A, B (a)) Step 1. StAR 3.5kb mRNA locates to OMM, where the cycloheximide (CHX) sensitivity of transfer implicates the need for continuous synthesis to offset inactivation after full import to the mitochondrial matrix (p37, progress to the matrix—green arrow). This process is not necessary for cholesterol transfer from lipid droplets, which are CHX-insensitive (DiBartolomeis and Jefcoate, 1984). Step 2. P37 StAR docks at TOM22 prior to reversible import to the IMM driven by the initial 30 AA of the NTD. In Step 1 or at this stage, PKA phosphorylated S194, and OMM ERK phosphorylates S232. In this phase, cholesterol enrichment in the region of the Ca-sensitive permeability channel (Ca-PC) is facilitated by TSPO/VDAC1 interaction (Rone et al., 2012). Step 3. NTD cleavage by IMM metalloprotease (MMP) at conserved site A (Blue arrows) exposes a conserved hydrophobic N-terminus that then associates with VDAC2 to enhance OMM-IMM contacts demonstrated by crosslinking (Prasad et al., 2015). These contacts enhance O-phenanthroline (OP) inhibition of site A cleavage and remove pp-30 while completely blocking cholesterol transfer. Removal of OP leads to recovery of cholesterol metabolism without renewal of p37 synthesis. Step 4. NTD cleavage at conserved site B inactivates the NTD and leads to further migration of p30/p28 into the matrix, where cholesterol transfer activity is lost. Turnover by the matrix protease LON is slow unless activated by oxidative stress (Bahat et al., 2015). A LON insensitive phase of mitochondrial importance corresponding to Step 3 has been identified. Step 5. Cholesterol transfer without NTD activity occurs at slower rates but produces deficiencies due to the cytoplasmic activity of CBD (Bahat et al., 2015). When persistent due to inhibited import (CCCP), p37 and other intermediates are removed by the proteasome degradation (MG132 inhibition). (B (b)) Mutation of conserved cleavage sites in N-terminal regulatory domain
GTP enhances mitochondrial uptake of cholesterol in partnership with Ca (Kowluru et al., 1995). GTPases such as mitochondrial fusion-associated protein 2 (MFN2) and opacity associated (OPA) proteins, which mediates the continuous dynamics of inter-mitochondrial fusion, may also play pivotal roles in the cAMP-stimulated cholesterol transfer (Duarte et al., 2012). Mitochondrial fusion with the ER through sigma receptor sites has also been implicated (Prasad et al., 2015). Each share affects on the inner membrane transition pore that may enhance inter-membrane contacts.
4. StAR NTD and cholesterol homeostasis
Deletion of the N-terminal mouse sequence (N-47 mouse) shows that this region is involved in the extra-mitochondrial activity, notably cholesterol homeostasis involving liver X receptor (LXR) forms and sterol regulatory element binding factor 1c and 2 (SREBP) (Sasaki et al., 2008). StAR functions from outside the mitochondria without the NTD to mediate linkage to lipid droplets (Arakane et al., 1996; Manna et al., 2002; Tu et al., 2014). This linkage of StAR activity to SNARE proteins, which function with lipid droplets has been reproduced in a reconstituted system using adrenal rat mitochondria (Lin et al., 2016). The appreciable potential for StAR to affect the cytoplasmic and nuclear cell compartments in multiple cell types has recently been highlighted, including through control of 27-hydroxy cholesterol, an LXR activator that also functions in the adrenal (Anuka et al., 2013; Manna et al., 2014).
TSPO also recruits PAP7/ACBD3 to these cholesterol-rich regions of the OMM. ACBD is an acronym for acyl-CoA binding domain, additionally suggesting participation in cholesterol esterification. ATP-binding cassette (ABCD3) has been now shown to be a suppressor of SREBP forms one that control both fatty acid synthesis genes (Chen et al., 2012).
5. Y-1 cells exhibit sufficient basal StAR mRNA to mediate a full acute steroidogenic response
Y-1 adrenal cells like their primary counterparts and in contrast to MA-10 cells exhibit sufficient basal mRNA to mediate maximum steroidogenesis within 15 minutes without an increase in mRNA. This basal expression has been addressed by a combination of q-PCR and high-resolution fluorescence in situ hybridization (HR-FISH) microscopy described in detail elsewhere (Lee et al., 2016a).
Under basal conditions, Y-1 cells show appreciable levels of labile primary RNA (p-RNA) and spliced transcripts (sp-RNA) indicative of active transcription and splicing. This basal expression has been shown to derive from a combination of PKA and Janus kinase 2 (Jak2) activity (Lefrancois-Martinez et al., 2011). Stimulation by Br-cAMP substantially increases initiation and elongation up to the beginning of exon 7. However, the absence of increases in either exon 7 or spliced transcripts for 15 minutes indicates a pause in pol2 elongation at this point, which is coupled to splicing. Very similar delays are apparent in both Y-1 and MA-10 cells (Lee et al., 2016a; Lee et al., 2015). This delay in elongation delivers spatially resolved clusters of unspliced and spliced transcripts that can be visualized at the loci with the Nikon’s structured illumination microscope (N-SIM) (Figure 2A). High resolution fluorescence in situ hybridization (HR-FISH) is achieved with hybridization of 30–40 fluorescent 20mers that bind head to tail synergistically. We have used this approach to resolve primary and spliced transcripts at StAR and CYP11A1 at the gene loci and mRNA in the cytoplasm. At the StAR loci, the p-RNA and sp-RNA are variable resolved by N-SIM to separate by 100–300nm in MA-10 cells.
Figure 2.
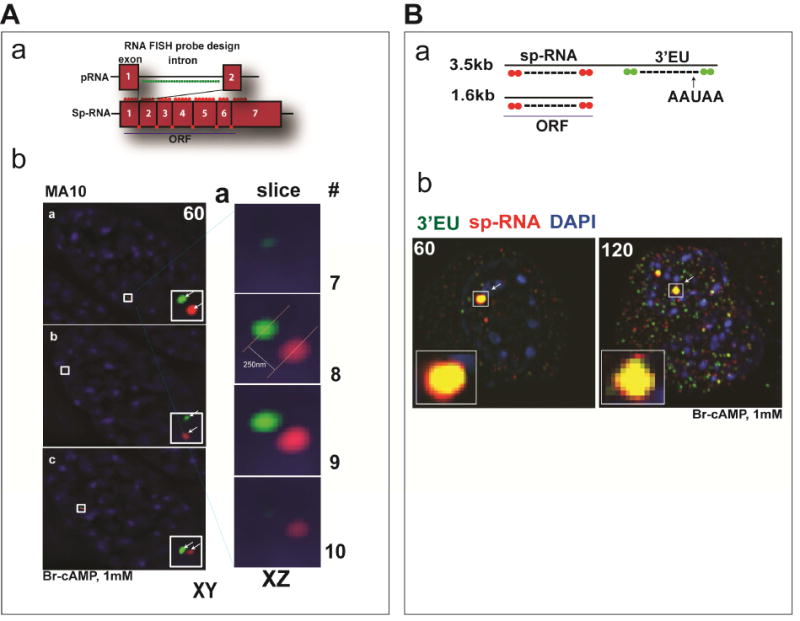
Time-dependent appearance of p-RNA, sp-RNA, and 3′UTR at loci after stimulation by Br-cAMP (A, a) Design of HR-FISH fluorescent probes consisting of 40 fluorescent probes (about 20 mers each) for StAR p-RNA and sp-RNA/mRNA. The RNA probe sets for StAR was generated by using the stellaris probe designer (http://www.biosearchtech.com/stellarisdesigner/). The p-RNA probes target introns while the sp-RNA probes selectively target spliced sequences by limiting the number of oligomers in each exon and exon-exon junction region. (A, b) The N-SIM microscope system (Nikon) was used to detect and visualize p-RNA and sp-RNA/mRNA. Left, the images show the analyses of loci in three dually labeled cells in the XY plane. The arrows indicate p-RNA (green) and sp-RNA (red) at the locus. Right, 3D-SIM image stacks were acquired with a Z-distance of 0.2μm, covering the entire thickness of the cell (about 10μm). Fifteen raw images per plane were acquired and computationally reconstructed using the reconstruction slice system from NIS-Elements software (Nikon). The locus (top, a) is examined by Z-slicing at 0.2um intervals. We labeled the slices spanning the one of the loci from the top to the bottom. p-RNA and sp-RNA are seen after 60 minutes stimulation with Br-cAMP (1mM) in MA-10 cells. (B, a) Design of HR-FISH fluorescent probes for StAR sp-RNA/mRNA and extended 3′UTR (3′EU). (B,b) Resolution of sp-RNA and 3′EU transcripts at StAR loci. The arrows indicate sp-RNA (red) and 3′EU (green) at the locus. Messages are seen after 60 and 120 minutes stimulation with Br-cAMP (1mM) in Y-1 cells.
The delayed splicing occurs after elongation up to the terminal intron 6. This pause should provide an accumulation of p-RNA approximating to the beginning of exon 7. A second accumulation of spliced sp-RNA generates a separate accumulation at the end of exon 7. We suppose that 3′ untranslated region (UTR) cleavage and polyadenylation ensues at this site. Additional oligomers that probe the terminal segment of the 3′UTR more closely overlap the sp-RNA at the locus. This separation at the locus is modeled (Figure 2B). q-PCR analyses show that elongation proceeds to the end of the 3′UTR. Thus there are minimal amounts of either p-RNA or sp-RNA that show short 3′UTR sequences in absence of terminal 3′UTR sequences. In Y-1 cells, the basal expression of StAR p-RNA and sp-RNA at the loci, individual mRNA/sp-RNA in the cytoplasm, and StAR protein in the inner mitochondria have been clearly resolved.
In Figure 3A, these features of a single Y-1 cell under basal conditions are presented as a 3D reconstruction of NSIM Z-stack slices, which we term the “cell in a box”. Consistent features are the midline nuclear positioning of the StAR loci with cytoplasmic mRNA and mitochondrial StAR protein between this position and the adherent cell membrane. The individual Z slices also indicate proximity of single mitochondria and single StAR mRNA (<0.5uM). This 1:1 pairing which we find in some cell locations with al least 50 percent incidence represents a dynamic association of 3.5 kb StAR mRNA with mitochondria. Cholesterol transfer and pregnenolone synthesis depend on translation of StAR mRNA, PKA phosphorylation, mitochondrial import and IMM cleavage of the NTD all occurring within 5 minutes (Artemenko et al., 2001). We assume that proteins that bind to the extended 3′UTR including A-kinase anchoring protein (Akap)1 (Grozdanov and Stocco, 2012) and Tis11b/Znf36l1 (Duan et al., 2009) target StAR mRNA to ribosomes associated with the OMM. This association is supported by the N-SIM images of mRNA–mitochondrial pairing. The stoichiometry is a surprise. The process is likely to proceed through translation and eventually dissociation of the mRNA to other non-mitochondrial sites that we detect close to the adherent cell membrane. The number of cycles of translation for each mRNA prior to dissociation is a key issue.We are presently counting the prevalence of this pairing in various activity situations. This may represent a first view of active StAR directed cholesterol metabolism (Figure 3B). The relationship to mitochondrial fusion and ER– mitochondrial associations that have each been linked to StAR activity are potential contributors.
Figure 3.
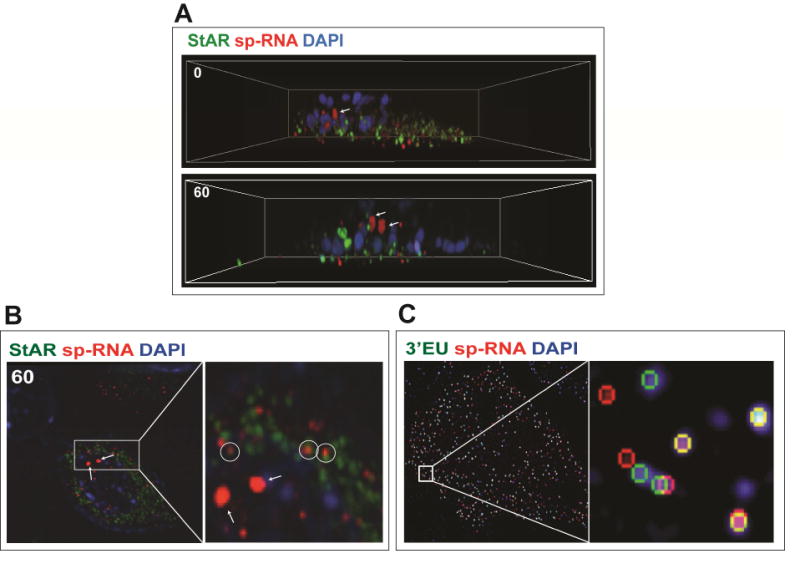
3D N-SIM image of Y1 cell separates loci, StAR protein (mitochondria), and mRNA. (A) HR-FISH of StAR mRNA (sp-RNA) and immunochemistry of StAR protein after 3D N-SIM image in XYZ axis. The arrows indicate the loci. sp-RNA/mRNA (red) and StAR proteins (green) are seen after 60 minutes stimulation with Br-cAMP (1mM) in Y-1 cells. (B) Enlarged region from 60-minute stimulation shows pairing of StAR mRNA and mitochondrial matrix StAR protein (circle). The arrows indicate the loci. (C) HR-FISH of StAR mRNA (sp-RNA) and 3′EU. Some messages show complete separation in the XY plane with N-SIM image processing.
HR-FISH shows variable levels of expression at the loci in each cell, representing different numbers of transcripts. Some cells have only one active locus, some none. At higher sensitivity mRNA is visible in the cytoplasm, even under basal conditions. This contrasts with MA-10 testis cells where expression under basal conditions is not detectable either at loci or in the cytoplasm. The ratio of locus sp-RNA expression to cytoplasmic mRNA is highly variable between individual cells. We suggest that each cell is at a different stage of endogenous stimulation as marked by this ratio. These cell differences are consistent with the asymmetric activation of StAR loci and asynchronous responses of individual cells previously reported for MA-10 cells (Lee et al., 2016a; Lee et al., 2015).
Stimulation for 15 minutes by Br-cAMP, which produces maximum steroidogenesis in these Y-1 cells, (Artemenko et al., 2001) increases primary transcription at the loci but without an increase in splicing or cytoplasmic mRNA. mRNA and protein are positioned below the loci. This mRNA is almost entirely 3.5 kb mRNA based on qPCR analyses. The HR-FISH probes show that half of these mRNA exhibit dual hybridization by sp-RNA and 3′EU probes that mostly show clear image resolution on single mRNA molecules (<0.1 microns) (Figure 3C).
6. Activation of transcription of StAR gene loci
We have used HR-FISH with Y-1 cells to show that Br-cAMP increases StAR transcripts at the gene loci asymmetrically. Thus, individual loci are activated on different time courses while single cells become engaged separately over the course of about 60 minutes(Lee et al., 2016a). Early and late onset processes have been distinguished in MA-10 cells. This is simplified by the absence of basal expression. The distinguishing features of the early onset process, which starts directly after PKA activation are the asymmetric activation of loci with delayed elongation and slow splicing. The late onset loci, which become active after 60 minutes exhibit an increased transcription rate but with highly coupled splicing. In Y-1 cells, the asymmetry and slow splicing of early onset locus activation superimpose on this basal expression. For the first 15 minutes of Br-cAMP stimulation, the number of engaged loci and mean locus expressions each increase but any increase in sp-RNA at the loci. This indicates an absence of splicing which is confirmed by parallel q-PCR analyses. Remarkably, this involves a 4-fold increase only in transcription of the first 6 introns and exons. Like early onset MA-10 transcription, there is a pol2 pause in elongation at the beginning of exon 7 while splicing remains at the low basal level. After 15 minutes, the level of p-RNA corresponding to the first 6 introns/exons remains constant over 6 h while transcripts corresponding to both complete elongation of exon 7 and full splicing increase linearly and in parallel to a new steady state which is about 5-fold higher. HR-FISH shows increased mRNA in the cytoplasm during this period. The number of engaged loci increases between 15 and 60 min.
It remains to be determined whether Y-1 cells have two types of StAR loci. In many respects, the basal turnover and 15 minutes stimulation, which is sufficient for maximum steroidogenesis, correspond to the MA-10 early onset process while the post-15-minute stimulation represents a transition to the late onset transcription with extensive coupling of transcription and splicing.
7. Sik1/CRTC2 intervention in Br-cAMP stimulation of StAR transcription
PKA surprisingly regulates StAR transcription by inactivating the salt inducible kinases (SIK1) (Lee et al., 2015) and (SIK2), which inactivate CREB-regulated transcription coactivator (CRTC) forms (CRTC2 in Y-1 cells) CRTC proteins are co-activators of CREB that aid in the recruitment of the histone acetyl transferase CBP. We suspect that this intervention occurs, also, to speed up shutdown of CREB-mediated transcription as cAMP levels fall. The SIK/CRTC combination is found extensively in conjunction with the metabolic regulation mediated by cAMP, including in the hypothalamus, pituitary, and liver, where rapid fluxes in stimulation are frequent (Altarejos and Montminy, 2011; Evans et al., 2013; Itoh et al., 2015).
The role of continuous translation in the StAR cholesterol transfer activity places particular demands on the transcription regulation. Modifications of SF1 (203 phosphorylation, K194 de-sumoylation), CREB and GATA4 (phosphorylation) play key roles in acute stimulation of StAR transcription by cAMP/PKA (Daems et al., 2015; Manna et al., 2009). CREB phosphorylation at S133 by PKA is effective in mediating PMA/Erk activation of StAR in the absence of any partnership with the CRTC co-activator since SIK forms are unaffected (Jefcoate et al., 2011). The PKA-mediated inhibition of SIK forms and subsequent phosphatase activities releases CRTC2 first from the cytoplasm (PP2B/from calcineurin) and then from nuclear speckles (PPI or PP4) to combine with CREB on the StAR promoter. CRTC forms also function at sites that facilitate other steps in transcription, notably splicing and elongation that is delayed at early onset StAR loci (Amelio et al., 2007).
SIK1 is induced extensively in the adrenal in vivo and in cultured Y-I cells by PKA before StAR expression (Jefcoate et al., 2011; Spiga et al., 2011). SIK1 nuclear location and inhibitory activity depend on phosphorylation at S182 by LKB1(Takemori et al., 2009). PKA inhibits SIK1 activity through S577 phosphorylation, which additionally shifts SIK1 out of the nucleus. The combination of kinase effects on SIK1 appears to keep the nuclear impact of SIK1 on CRTC2 relatively low at least until cAMP declines.
The stimulation of SIK1 by PKA shares several features of the StAR stimulation, which may facilitate their coordination. Br-cAMP stimulates SIK1 p-RNA to a steady state with a high copy number and a 15 minute delay before a rapid increase 4.4 kb mRNA. The delay before the appearance of this mRNA indicates a delay in splicing similar to that seen for StAR. This mRNA also contains an extended 3′UTR with multiple AU-rich regulatory elements at the 3′end. A steady state in stimulation is reached within 60 minutes indicating a rapid turnover of this 4.4 kb Sik1 mRNA.
8. Activation of StAR transcription by inhibition of SIK alone
SIK inhibition by staurosporine or the more specific HG-9-91-01 (HG-9) (Clark et al., 2012) alone activates the expression of StAR and most steroidogenic genes in Y-1 cells almost as effectively as cAMP (Jefcoate et al., 2011; Takemori et al., 2007). This inhibition of SIK forms replicates PKA stimulation of SIK1 transcription. This activation process implicates CRTC in SIK1 transcription and therefore a feedback control, perhaps to prevent excessive SIK1 induction. Staurosporine produces negligible phosphorylation of CREB in combination with nuclear translocation of CRTC2 and association with the StAR promoter Evidently CRTC2 associates with CREB without S133 phosphorylation. SIK2 has the high basal expression that responds only modestly to Br-cAMP. Inhibition of both SIK1 and SIK2 each contribute to this inhibitor response, although their respective contributions will depend on their relative expression (Lee et al., 2016b). We speculate that SIK2 mediates CRTC2 inactivation in the cytoplasm under the basal condition in Y-1 cells. The slower increase in StAR p-RNA response with staurosporine compared to Br-cAMP probably indicates a stimulatory contribution to initiation and elongation from CREB S133 phosphorylation or some other PKA target. Splicing of the StAR p-RNA following the stimulation by staurosporine in Y-1 cells occurs at the same time and rate as for Br-cAMP stimulation. Thus, while Br-cAMP can increase splicing factors (Schimmer and Cordova, 2015), an activation of CRTC shared by Br-cAMP and staurosporine is sufficient for this acute activation.
Staurosporine stimulation is not reproduced in MA-10 cells. Staurosporine is a very general kinase inhibitor although it is typically not as potent for other kinases as for SIK forms (Gani and Engh, 2010). In MA-10 cells, the stimulation of StAR by Br-cAMP was effectively inhibited by staurosporine. This inhibition indicates that other staurosporine-sensitive kinases contribute to activity in MA-10 cells but not Y-1 cells. Increased participation Dax1, an inhibitor of SF1, which is absent from Y-1 cells, is being explored as a possible discriminatory component. In Y-1 cells, we find that staurosporine at 5nM functions synergistically with Br-cAMP.
Unlike staurosporine, the more selective SIK inhibitor HG-9-91-01 effectively stimulates StAR expression in MA-10 cells. However, despite a Kd of only 10nM, stimulation was only achieved at 10uM. Unlike the normal direct stimulant, these increases depend on the extent of inhibition of SIK forms and their activity on CRTC forms. The improved selectivity of HG-9-91-01 may diminish the off-target inhibitory effect. However, a need for near complete SIK inhibition will shift the dose response curve to higher concentrations.
9. S577A-SIK1-GFP as a mechanistic probe of SIK1 activity
Many biological processes that involve SIK forms involve the regulation of CREB/CRTC directed transcription. However, there are many other activities, including some that may occur in adrenal cells. Notable examples include the original Na regulation of Na/K-ATPase (Popov et al., 2011), the control of Type2 histone deacetylase inhibitors (HDACs) (Berdeaux et al., 2007; Takemori et al., 2009) and most recently the activation of the cholesterol transporter, scavenger receptor class B (SR-B1) (Hu et al., 2015). Br-cAMP effects S577 phosphorylation of SIK1 and the equivalent S587 of SIK2 and mediates the sequestration of each by 14-3-3 forms in the cytoplasm. This protein functions as a homodimer to sequester proteins including SIK1, SIK2, CRTC forms and Tis11b/Znf36l1 through sites where serine is phosphorylated (Al-Hakim et al., 2005; Screaton et al., 2004). This phosphorylation prevents kinase activity but also the nuclear entry of SIK1. When mutated as S577A, SIK1 remains in nuclear speckles both with and without PKA stimulation (Lee et al., 2015). Thus, only nuclear activities of the SIK forms are prevented. This should, however, include inhibition of all CRTC activity but not other non-genomic cytoplasmic effects of SIK forms.
Transfection of cells with S577A-SIK1-GFP compared to GFP alone has been combined with HR-FISH to test the effects in transfected cells. SIK1 and CRTC2 then co-localize in the same speckle structures. Stimulation of Y-1 cells by Br-cAMP is completely prevented in cells that express SIK1-S577-GFP whereas full expression is maintained in neighboring cells lacking the active SIK1 chimera.
10. 3.5 kb StAR mRNA is selectively targeted by TIS 11b in the 3′UTR
Br-cAMP also stimulates the transcription of Znf36l1/Tis11b, a Zn finger protein (Figure 4A), which limits StAR activity in a different way. A complex homodimer format AU-rich TATTTATT elements in the extended 3′UTR. This complex interacts with proteins that bind the polyA tail and recruits ribonucleases selectively to degrade the StAR 3.5 kb mRNA (Baou et al., 2009). The 3.5 kb form is less stable than the 1.6 kb form in multiple cell types and then is selectively destabilized by further expression of Tis 11b (Duan et al., 2009).
Figure 4.
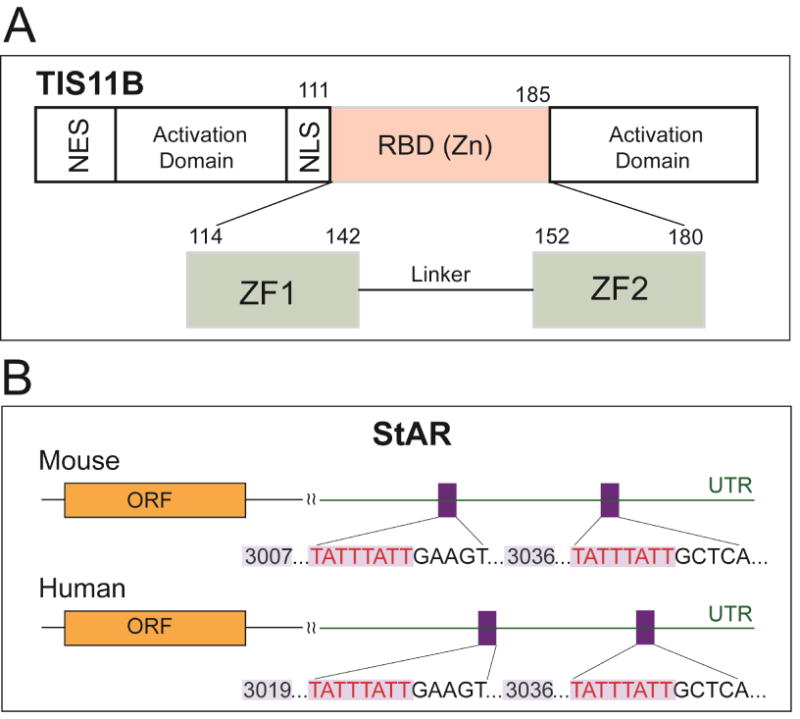
Selective targeting of extended StAR 3′UTR by transfected TIS11b/Znf36l1. (A) The zinc finger protein TIS11B has been documented as a post-transcriptional regulator of gene expression. Activation domains, NLS (nuclear localization signal), NES (nuclear export sequence), and RBD (RNA-binding domain) are shown. (B) Tis11b targeted sequences conserved in mouse and human StAR 3′UTR each with a conserved relationship to the terminal cleavage/polyadenylation site.
Znf36l1/Tis11b is one of three members of the TTP/Znf36 family of Zinc-finger regulators. The best-known form TTP/Znf36 restricts inflammatory responses by targeting elements in the 3′UTR of cytokines such as Tnf. The most notable target is the VEGF, which also functions in adrenal cortex cells. Over production of vascular endothelial growth factor (VEGF) results in the lack of viability for Znf36l1/Tis11b deficient mice due to aberrant vascular development at around e8.5 (Baou et al., 2009).
These TIS11b sites are conserved in the extended 3′UTR sequences of mouse and human StAR despite low 3′UTR conservation in surrounding sequences (Figure 4B). Br-cAMP stimulates expression in both cell types in parallel with StAR increases. The additional expression stimulated by Br-cAMP occurs with expression in both the nuclei and cytoplasm. Znf36l1/Tis11b is phosphorylated at multiple sites, including by p38 stress-activated kinases, which have been linked to cytoplasmic distribution. Interestingly, binding occurs in cytoplasmic 14-3-3 forms that also bind phosphorylated CRTC2 and SIK forms (Al-Hakim et al., 2005; Screaton et al., 2004).
Removal of Tis11b in Y-1 and MA-10 cell lines through the high expression of shRNA enhances the potency of Br-cAMP as a stimulant of StAR expression. These lines show a similar elevation of both forms, whereas Tis11b siRNA selectively targets the 3.5 kb form (Duan et al., 2009). Evidently there are differences in the impact on Tis11b activity. The long-term suppression by sh-RNA is more effective in suppressing Tis11b expression. Due to the slow splicing, transcription of the full 3′UTR precedes cleavage and polyadenylation to generate the 1.6 kb form. There is also no evidence of premature cessation of elongation to generate shortened 3′UTR p-RNA or sp-RNA. Thus, Tis11b may also function at the StAR loci on the p-RNA to affect splicing, mRNA processing or translation. Previous work has shown that proteins in this Znf36 family play a role in redirecting mRNA to P-bodies and stress particles (Chang and Tarn, 2009). We have seen in HR-FISH images that a proportion of StAR mRNA is not associated with mitochondria.
11. StAR Responses to episodic in vivo ACTH signaling
In rodent adrenals, in vivo ACTH stimulates cAMP through activation of the MC2R receptor. The cAMP generation is enhanced by the cAMP-inducible MC2R co-activating protein MRAP. ACTH circulates as low-level pulses of about 60 minutes duration, which are released from the pituitary under control of hypothalamic corticotropin-releasing hormone (CRH). These pulses superimpose on a fivefold StAR circadian fluctuation, in part due to changes in ultradian amplitude (Liu et al., 2013). Restraint stress increases ACTH over 30 minutes followed by a decline back to basal levels, which is matched by a single high dose ACTH administration. This stimulus delivers parallel changes corticosterone and StAR p-RNA with a delay of 30 minutes before the increase in sp-RNA/mRNA. A large dose ACTH bolus provides similar kinetics. In these same adrenals, SIK1 rises to high levels over 30 minutes. Overall these restraint responses closely match the changes seen in Y-1 cells with Br-cAMP.
The ultradian pulsatile changes correspond to much lower fluctuations of ACTH, which nevertheless, produce 60-minutes fluctuations of corticosterone that reach nearly 50 percent of the levels produced by restraint stress. Corticosterone is rapidly metabolized by 11-dehydrogenases such that the steroid pulses track after the ACTH pulses with only a short delay. StAR p-RNA and p-CREB show parallel fluctuations but with no increase in sp-RNA/mRNA. These low ultradian increases in ACTH appear to function through direct effects of PKA on the basal level of StAR mRNA through the rapid translation-dependent process shown in figure 1 that involves mitochondrial import and IMM metalloprotease cleavage. This process has been shown to provide a cholesterol transfer cycle in Y-1 cells of about 5 minutes, which is fully compatible with the modeling carried out for ultradian responses (Spiga et al., 2015). The rapid modulation provided by processing of the NTD can play a role in synchronizing ACTH fluctuations with mitochondrial cholesterol transfer and with changes in the cytoplasmic availability of cholesterol.
The slow splicing of StAR p-RNA that we have described has the effect of minimizing increases in StAR mRNA during the rapid low-level pulses, except perhaps with stress clusters multiple pulses. The rapid NTD modulation of CBD activity effectively uncouples ultradian pulses from the much slower diurnal regulation derived from the hypothalamus both through the autonomic stimulation of the adrenal medulla and from ACTH pulse variation. These diurnal changes closely correlate with changes in adrenal transcription factors (Spiga et al., 2015).
A non-genomic suppression of steroid synthesis is detected in the latter part of the ultradian pulse. Direct intra-adrenal glucocorticoid feedback has been suggested (Kalafatakis et al., 2016; Spiga et al., 2015). However, the elevations of SIK1 and TIS11b and changes in their nuclear/cytoplasmic distribution, which have been introduced here occur within 5 minutes and, therefore, function on the time scale of ultradian pulses. Their capacity to suppress transcription and translation may be expected to affect the partnership between mitochondria and StAR mRNA. The kinase activity delivered by SIK, for which mRNA increases tenfold within 30 minutes can extensively, suppress StAR transcription when cAMP levels fall. This inhibition is most rapidly produced through direct CRTC2 phosphorylation by SIK1 within the nucleus prior to any translocation to the cytoplasm.
HR-FISH has allowed us to show a remarkable pairing of single StAR mRNA with mitochondria. This pairing is likely to be affected by the suppression effects of TIS11b on translation and possibly on the mitochondrial location. During the 60 minutes window of ultradian cAMP fluctuations, other rapid dynamic processes including mitochondrial and endoplasmic reticulum (ER) membrane fusion changes may be critical. These more rapid spatiotemporal effects are the focus of present efforts with HR-FISH. While stress biology has naturally focused almost entirely on the brain, the remarkable plasticity of these adrenal pathways suggests that individuals may vary substantially in their adrenal glucocorticoid control (Kalafatakis et al., 2016).
Acknowledgments
MA-10 cell line was the generous gifts from Professor Mario Ascoli. Y-1 cell line was the generous gifts from Professor Bernard Schimmer. This study was financially supported by NIH (RO1 DK074819, RO1 DK090249).
Abbreviations
- 3′EU
extended 3′UTR
- ACTH
adrenocorticotropic hormone
- AMG
aminoglutethimide
- CBP
CREB binding protein
- CREB
cAMP responsive element binding protein 1
- CRTC
CREB regulated transcription coactivator
- CBD
C-terminal cholesterol binding domain
- CHX
cycloheximide
- FISH
fluorescence in situ hybridization
- HR-FISH
high resolution fluorescence in situ hybridization
- IMM
inner mitochondrial membrane
- MMP
metalloproteases
- NNT
NADH/NADPH transhydrogenase
- N-SIM
Nikon’s Structured Illumination Microscope
- NNT
nicotinamide nucleotide transhydrogenase
- NTD
N-terminal regulatory domain
- OP
o-phenanthroline
- PKA
Protein kinase A
- p-RNA
primary RNA
- SIK1
salt inducible kinase 1
- sp-RNA
spliced primary RNA
- StAR
steroidogenic acute regulatory protein
- TSPO
translocator protein
- VDAC
voltage-dependent anion channel
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Hakim AK, Goransson O, Deak M, Toth R, Campbell DG, Morrice NA, Prescott AR, Alessi DR. 14-3-3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J Cell Sci. 2005;118:5661–5673. doi: 10.1242/jcs.02670. [DOI] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio AL, Miraglia LJ, Conkright JJ, Mercer BA, Batalov S, Cavett V, Orth AP, Busby J, Hogenesch JB, Conkright MD. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20314–20319. doi: 10.1073/pnas.0707999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anuka E, Gal M, Stocco DM, Orly J. Expression and roles of steroidogenic acute regulatory (StAR) protein in ‘non-classical’, extra-adrenal and extra-gonadal cells and tissues. Molecular and cellular endocrinology. 2013;371:47–61. doi: 10.1016/j.mce.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Arakane F, Sugawara T, Nishino H, Liu Z, Holt JA, Pain D, Stocco DM, Miller WL, Strauss JF., 3rd Steroidogenic acute regulatory protein (StAR) retains activity in the absence of its mitochondrial import sequence: implications for the mechanism of StAR action. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13731–13736. doi: 10.1073/pnas.93.24.13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemenko IP, Zhao D, Hales DB, Hales KH, Jefcoate CR. Mitochondrial processing of newly synthesized steroidogenic acute regulatory protein (StAR), but not total StAR, mediates cholesterol transfer to cytochrome P450 side chain cleavage enzyme in adrenal cells. The Journal of biological chemistry. 2001;276:46583–46596. doi: 10.1074/jbc.M107815200. [DOI] [PubMed] [Google Scholar]
- Bahat A, Perlberg S, Melamed-Book N, Isaac S, Eden A, Lauria I, Langer T, Orly J. Transcriptional activation of LON Gene by a new form of mitochondrial stress: A role for the nuclear respiratory factor 2 in StAR overload response (SOR) Molecular and cellular endocrinology. 2015;408:62–72. doi: 10.1016/j.mce.2015.02.022. [DOI] [PubMed] [Google Scholar]
- Baker BY, Yaworsky DC, Miller WL. A pH-dependent molten globule transition is required for activity of the steroidogenic acute regulatory protein, StAR. The Journal of biological chemistry. 2005;280:41753–41760. doi: 10.1074/jbc.M510241200. [DOI] [PubMed] [Google Scholar]
- Baou M, Jewell A, Murphy JJ. TIS11 family proteins and their roles in posttranscriptional gene regulation. Journal of biomedicine & biotechnology. 2009;2009:634520. doi: 10.1155/2009/634520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbar E, Lehoux JG, Lavigne P. Toward the NMR structure of StAR. Molecular and cellular endocrinology. 2009;300:89–93. doi: 10.1016/j.mce.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Berdeaux R, Goebel N, Banaszynski L, Takemori H, Wandless T, Shelton GD, Montminy M. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- Bose HS, Lingappa VR, Miller WL. Rapid regulation of steroidogenesis by mitochondrial protein import. Nature. 2002;417:87–91. doi: 10.1038/417087a. [DOI] [PubMed] [Google Scholar]
- Bose HS, Whittal RM, Baldwin MA, Miller WL. The active form of the steroidogenic acute regulatory protein, StAR, appears to be a molten globule. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:7250–7255. doi: 10.1073/pnas.96.13.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron KM, Soo SC, Wetsel WC, Stocco DM, Clark BJ, Parker KL. Targeted disruption of the mouse gene encoding steroidogenic acute regulatory protein provides insights into congenital lipoid adrenal hyperplasia. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:11540–11545. doi: 10.1073/pnas.94.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WL, Tarn WY. A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucleic acids research. 2009;37:6600–6612. doi: 10.1093/nar/gkp717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Patel V, Bang S, Cohen N, Millar J, Kim SF. Maturation and activity of sterol regulatory element binding protein 1 is inhibited by acyl-CoA binding domain containing 3. PLoS One. 2012;7:e49906. doi: 10.1371/journal.pone.0049906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BJ, Wells J, King SR, Stocco DM. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR) The Journal of biological chemistry. 1994;269:28314–28322. [PubMed] [Google Scholar]
- Clark K, MacKenzie KF, Petkevicius K, Kristariyanto Y, Zhang J, Choi HG, Peggie M, Plater L, Pedrioli PG, McIver E, et al. Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16986–16991. doi: 10.1073/pnas.1215450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daems C, Di-Luoffo M, Paradis E, Tremblay JJ. MEF2 Cooperates With Forskolin/cAMP and GATA4 to Regulate Star Gene Expression in Mouse MA-10 Leydig Cells. Endocrinology. 2015;156:2693–2703. doi: 10.1210/en.2014-1964. [DOI] [PubMed] [Google Scholar]
- DiBartolomeis MJ, Jefcoate CR. Characterization of the acute stimulation of steroidogenesis in primary bovine adrenal cortical cell cultures. The Journal of biological chemistry. 1984;259:10159–10167. [PubMed] [Google Scholar]
- Duan H, Cherradi N, Feige JJ, Jefcoate C. cAMP-dependent posttranscriptional regulation of steroidogenic acute regulatory (STAR) protein by the zinc finger protein ZFP36L1/TIS11b. Mol Endocrinol. 2009;23:497–509. doi: 10.1210/me.2008-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A, Castillo AF, Podesta EJ, Poderoso C. Mitochondrial fusion and ERK activity regulate steroidogenic acute regulatory protein localization in mitochondria. PLoS One. 2014;9:e100387. doi: 10.1371/journal.pone.0100387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A, Poderoso C, Cooke M, Soria G, Cornejo Maciel F, Gottifredi V, Podesta EJ. Mitochondrial fusion is essential for steroid biosynthesis. PLoS One. 2012;7:e45829. doi: 10.1371/journal.pone.0045829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson MT, Kowalewski MP, Manna PR, Stocco DM. The differential regulation of steroidogenic acute regulatory protein-mediated steroidogenesis by type I and type II PKA in MA-10 cells. Molecular and cellular endocrinology. 2009;300:94–103. doi: 10.1016/j.mce.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott ME, Goodfriend TL, Jefcoate CR. Bovine adrenal glomerulosa and fasciculata cells exhibit 28.5-kilodalton proteins sensitive to angiotensin, other agonists, and atrial natriuretic peptide. Endocrinology. 1993;133:1669–1677. doi: 10.1210/endo.133.4.8404608. [DOI] [PubMed] [Google Scholar]
- Epstein LF, Orme-Johnson NR. Regulation of steroid hormone biosynthesis. Identification of precursors of a phosphoprotein targeted to the mitochondrion in stimulated rat adrenal cortex cells. The Journal of biological chemistry. 1991;266:19739–19745. [PubMed] [Google Scholar]
- Evans AN, Liu Y, Macgregor R, Huang V, Aguilera G. Regulation of hypothalamic corticotropin-releasing hormone transcription by elevated glucocorticoids. Mol Endocrinol. 2013;27:1796–1807. doi: 10.1210/me.2013-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gani OA, Engh RA. Protein kinase inhibition of clinically important staurosporine analogues. Natural product reports. 2010;27:489–498. doi: 10.1039/b923848b. [DOI] [PubMed] [Google Scholar]
- Grozdanov PN, Stocco DM. Short RNA molecules with high binding affinity to the KH motif of A-kinase anchoring protein 1 (AKAP1): implications for the regulation of steroidogenesis. Mol Endocrinol. 2012;26:2104–2117. doi: 10.1210/me.2012-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanukoglu I, Jefcoate CR. Mitochondrial cytochrome P-450sec. Mechanism of electron transport by adrenodoxin. The Journal of biological chemistry. 1980;255:3057–3061. doi: 10.1016/S0021-9258(19)85851-9. [DOI] [PubMed] [Google Scholar]
- Hu Z, Hu J, Shen WJ, Kraemer FB, Azhar S. A Novel Role of Salt-Inducible Kinase 1 (SIK1) in the Post-Translational Regulation of Scavenger Receptor Class B Type 1 Activity. Biochemistry. 2015;54:6917–6930. doi: 10.1021/acs.biochem.5b00147. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Sanosaka M, Fuchino H, Yahara Y, Kumagai A, Takemoto D, Kagawa M, Doi J, Ohta M, Tsumaki N, et al. Salt-inducible Kinase 3 Signaling Is Important for the Gluconeogenic Programs in Mouse Hepatocytes. The Journal of biological chemistry. 2015;290:17879–17893. doi: 10.1074/jbc.M115.640821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefcoate CR, Lee J, Cherradi N, Takemori H, Duan H. cAMP stimulation of StAR expression and cholesterol metabolism is modulated by co-expression of labile suppressors of transcription and mRNA turnover. Molecular and cellular endocrinology. 2011;336:53–62. doi: 10.1016/j.mce.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefcoate CR, Simpson ER, Boyd GS, Brownie AC, Orme-Johnson WH. The detection of different states of the P-450 cytochromes in adrenal mitochondria: changes induced by ACTH. Ann N Y Acad Sci. 1973;212:243–261. doi: 10.1111/j.1749-6632.1973.tb47600.x. [DOI] [PubMed] [Google Scholar]
- Kalafatakis K, Russell GM, Zarros A, Lightman SL. Temporal control of glucocorticoid neurodynamics and its relevance for brain homeostasis, neuropathology and glucocorticoid-based therapeutics. Neuroscience and biobehavioral reviews. 2016;61:12–25. doi: 10.1016/j.neubiorev.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Kang HW, Wei J, Cohen DE. PC-TP/StARD2: Of membranes and metabolism. Trends Endocrinol Metab. 2010;21:449–456. doi: 10.1016/j.tem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, McAllister JM, Sugawara T, Strauss JF., 3rd Expression of steroidogenic acute regulatory protein (StAR) in the human ovary. J Clin Endocrinol Metab. 1996;81:4122–4128. doi: 10.1210/jcem.81.11.8923870. [DOI] [PubMed] [Google Scholar]
- Kowluru R, Yamazaki T, McNamara BC, Jefcoate CR. Metabolism of exogenous cholesterol by rat adrenal mitochondria is stimulated equally by physiological levels of free Ca2+ and by GTP. Molecular and cellular endocrinology. 1995;107:181–188. doi: 10.1016/0303-7207(94)03441-u. [DOI] [PubMed] [Google Scholar]
- Lee J, Foong YH, Musaitif I, Tong T, Jefcoate C. Analysis of specific RNA in cultured cells through quantitative integration of q-PCR and N-SIM single cell FISH images: Application to hormonal stimulation of StAR transcription. Molecular and cellular endocrinology. 2016a doi: 10.1016/j.mce.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Tong T, Duan H, Foong YH, Musaitif I, Yamazaki T, Jefcoate C. Regulation of StAR by the N-terminal Domain and Coinduction of SIK1 and TIS11b/Znf36l1 in Single Cells. Frontiers in endocrinology. 2016b doi: 10.3389/fendo.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Tong T, Takemori H, Jefcoate C. Stimulation of StAR expression by cAMP is controlled by inhibition of highly inducible SIK1 via CRTC2, a co-activator of CREB. Molecular and cellular endocrinology. 2015;408:80–89. doi: 10.1016/j.mce.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois-Martinez AM, Blondet-Trichard A, Binart N, Val P, Chambon C, Sahut-Barnola I, Pointud JC, Martinez A. Transcriptional control of adrenal steroidogenesis: novel connection between Janus kinase (JAK) 2 protein and protein kinase A (PKA) through stabilization of cAMP response element-binding protein (CREB) transcription factor. The Journal of biological chemistry. 2011;286:32976–32985. doi: 10.1074/jbc.M111.218016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau D, Lefebvre A, Lavigne P, LeHoux JG. The binding site specificity of STARD4 subfamily: Breaking the cholesterol paradigm. Molecular and cellular endocrinology. 2015;408:53–61. doi: 10.1016/j.mce.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Li H, Degenhardt B, Tobin D, Yao ZX, Tasken K, Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor- and PKA (RIalpha)-associated protein. Mol Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- Lin Y, Hou X, Shen WJ, Hanssen R, Khor VK, Cortez Y, Roseman AN, Azhar S, Kraemer FB. SNARE-Mediated Cholesterol Movement to Mitochondria Supports Steroidogenesis in Rodent Cells. Mol Endocrinol. 2016;30:234–247. doi: 10.1210/me.2015-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Smith LI, Huang V, Poon V, Coello A, Olah M, Spiga F, Lightman SL, Aguilera G. Transcriptional regulation of episodic glucocorticoid secretion. Molecular and cellular endocrinology. 2013;371:62–70. doi: 10.1016/j.mce.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna PR, Dyson MT, Jo Y, Stocco DM. Role of dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X chromosome, gene 1 in protein kinase A- and protein kinase C-mediated regulation of the steroidogenic acute regulatory protein expression in mouse Leydig tumor cells: mechanism of action. Endocrinology. 2009;150:187–199. doi: 10.1210/en.2008-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna PR, Huhtaniemi IT, Wang XJ, Eubank DW, Stocco DM. Mechanisms of epidermal growth factor signaling: regulation of steroid biosynthesis and the steroidogenic acute regulatory protein in mouse Leydig tumor cells. Biol Reprod. 2002;67:1393–1404. doi: 10.1095/biolreprod.102.007179. [DOI] [PubMed] [Google Scholar]
- Manna PR, Slominski AT, King SR, Stetson CL, Stocco DM. Synergistic activation of steroidogenic acute regulatory protein expression and steroid biosynthesis by retinoids: involvement of cAMP/PKA signaling. Endocrinology. 2014;155:576–591. doi: 10.1210/en.2013-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Jhala US, Radhakrishnan I, Yaffe MB, Reyes C, Shulman AI, Cantley LC, Wright PE, Montminy M. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- Pon LA, Hartigan JA, Orme-Johnson NR. Acute ACTH regulation of adrenal corticosteroid biosynthesis. Rapid accumulation of a phosphoprotein. The Journal of biological chemistry. 1986;261:13309–13316. [PubMed] [Google Scholar]
- Popov S, Silveira A, Wagsater D, Takemori H, Oguro R, Matsumoto S, Sugimoto K, Kamide K, Hirose T, Satoh M, et al. Salt-inducible kinase 1 influences Na(+),K(+)-ATPase activity in vascular smooth muscle cells and associates with variations in blood pressure. J Hypertens. 2011;29:2395–2403. doi: 10.1097/HJH.0b013e32834d3d55. [DOI] [PubMed] [Google Scholar]
- Prasad M, Kaur J, Pawlak KJ, Bose M, Whittal RM, Bose HS. Mitochondria-associated endoplasmic reticulum membrane (MAM) regulates steroidogenic activity via steroidogenic acute regulatory protein (StAR)-voltage-dependent anion channel 2 (VDAC2) interaction. The Journal of biological chemistry. 2015;290:2604–2616. doi: 10.1074/jbc.M114.605808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalle CT, Crivello JF, Jefcoate CR. Regulation of intramitochondrial cholesterol transfer to side-chain cleavage cytochrome P-450 in rat adrenal gland. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:702–706. doi: 10.1073/pnas.80.3.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapaksha M, Kaur J, Prasad M, Pawlak KJ, Marshall B, Perry EW, Whittal RM, Bose HS. An Outer Mitochondrial Translocase, Tom22, Is Crucial for Inner Mitochondrial Steroidogenic Regulation in Adrenal and Gonadal Tissues. Mol Cell Biol. 2016;36:1032–1047. doi: 10.1128/MCB.01107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rone MB, Midzak AS, Issop L, Rammouz G, Jagannathan S, Fan J, Ye X, Blonder J, Veenstra T, Papadopoulos V. Identification of a dynamic mitochondrial protein complex driving cholesterol import, trafficking, and metabolism to steroid hormones. Mol Endocrinol. 2012;26:1868–1882. doi: 10.1210/me.2012-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakitrungruang T, Soccio RE, Lang-Muritano M, Walker JM, Achermann JC, Miller WL. Clinical, genetic, and functional characterization of four patients carrying partial loss-of-function mutations in the steroidogenic acute regulatory protein (StAR) J Clin Endocrinol Metab. 2010;95:3352–3359. doi: 10.1210/jc.2010-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki G, Ishii T, Jeyasuria P, Jo Y, Bahat A, Orly J, Hasegawa T, Parker KL. Complex role of the mitochondrial targeting signal in the function of steroidogenic acute regulatory protein revealed by bacterial artificial chromosome transgenesis in vivo. Mol Endocrinol. 2008;22:951–964. doi: 10.1210/me.2007-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmer BP, Cordova M. Corticotropin (ACTH) regulates alternative RNA splicing in Y1 mouse adrenocortical tumor cells. Molecular and cellular endocrinology. 2015;408:5–11. doi: 10.1016/j.mce.2014.09.026. [DOI] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Shanmughapriya S, Rajan S, Hoffman NE, Higgins AM, Tomar D, Nemani N, Hines KJ, Smith DJ, Eguchi A, Vallem S, et al. SPG7 Is an Essential and Conserved Component of the Mitochondrial Permeability Transition Pore. Mol Cell. 2015;60:47–62. doi: 10.1016/j.molcel.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiga F, Liu Y, Aguilera G, Lightman SL. Temporal effect of adrenocorticotrophic hormone on adrenal glucocorticoid steroidogenesis: involvement of the transducer of regulated cyclic AMP-response element-binding protein activity. J Neuroendocrinol. 2011;23:136–142. doi: 10.1111/j.1365-2826.2010.02096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiga F, Walker JJ, Gupta R, Terry JR, Lightman SL. 60 YEARS OF NEUROENDOCRINOLOGY: Glucocorticoid dynamics: insights from mathematical, experimental and clinical studies. The Journal of endocrinology. 2015;226:T55–66. doi: 10.1530/JOE-15-0132. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Fujimoto S. The potential function of steroid sulphatase activity in steroid production and steroidogenic acute regulatory protein expression. Biochem J. 2004;380:153–160. doi: 10.1042/BJ20031379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemori H, Kanematsu M, Kajimura J, Hatano O, Katoh Y, Lin XZ, Min L, Yamazaki T, Doi J, Okamoto M. Dephosphorylation of TORC initiates expression of the StAR gene. Molecular and cellular endocrinology. 2007;265–266:196–204. doi: 10.1016/j.mce.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Takemori H, Katoh Hashimoto Y, Nakae J, Olson EN, Okamoto M. Inactivation of HDAC5 by SIK1 in AICAR-treated C2C12 myoblasts. Endocr J. 2009;56:121–130. doi: 10.1507/endocrj.k08e-173. [DOI] [PubMed] [Google Scholar]
- Tu LN, Morohaku K, Manna PR, Pelton SH, Butler WR, Stocco DM, Selvaraj V. Peripheral benzodiazepine receptor/translocator protein global knock-out mice are viable with no effects on steroid hormone biosynthesis. The Journal of biological chemistry. 2014;289:27444–27454. doi: 10.1074/jbc.M114.578286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu LN, Zhao AH, Hussein M, Stocco DM, Selvaraj V. Translocator Protein (TSPO) Affects Mitochondrial Fatty Acid Oxidation in Steroidogenic Cells. Endocrinology. 2016;157:1110–1121. doi: 10.1210/en.2015-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu LN, Zhao AH, Stocco DM, Selvaraj V. PK11195 effect on steroidogenesis is not mediated through the translocator protein (TSPO) Endocrinology. 2015;156:1033–1039. doi: 10.1210/en.2014-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Kowluru R, McNamara BC, Jefcoate CR. P450scc-dependent cholesterol metabolism in rat adrenal mitochondria is inhibited by low concentrations of matrix Ca2+ Arch Biochem Biophys. 1995;318:131–139. doi: 10.1006/abbi.1995.1213. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Matsuoka C, Gendou M, Izumi S, Zhao D, Artemenko I, Jefcoate CR, Kominami S. Mitochondrial processing of bovine adrenal steroidogenic acute regulatory protein. Biochim Biophys Acta. 2006;1764:1561–1567. doi: 10.1016/j.bbapap.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, McNamara BC, Jefcoate CR. Competition for electron transfer between cytochromes P450scc and P45011 beta in rat adrenal mitochondria. Molecular and cellular endocrinology. 1993;95:1–11. doi: 10.1016/0303-7207(93)90023-d. [DOI] [PubMed] [Google Scholar]


