Abstract
Efficient and safe delivery of the CRISPR/Cas system is one of the key challenges for genome-editing applications in humans. Herein, we designed and synthesized a series of biodegradable lipidlike compounds containing ester groups for the delivery of mRNA-encoding Cas9. Two lead materials, termed N-methyl-1,3-propanediamine (MPA)-A and MPA-Ab, showed a tunable rate of biodegradation. MPA-A with linear ester chains was degraded dramatically faster than MPA-Ab with branched ester chains in the presence of esterase or in wild-type mice. Most importantly, MPA-A and MPA-Ab demonstrated efficient delivery of Cas9 mRNA both in vitro and in vivo. Consequently, these biodegradable lipidlike nanomaterials merit further development as genome-editing delivery tools for biological and therapeutic applications.
Keywords: biodegradable nanomaterials, lipidlike nanoparticles, CRISPR/Cas9, mRNA, genome editing
Graphical abstract
INTRODUCTION
CRISPR/Cas9-mediated genome editing has been widely used as a powerful tool for biological and therapeutic applications.1–5 Adeno-associated virus-derived vectors and DNA nanoclews were reported to deliver plasmid-encoding CRISPR/Cas in a wide variety of cells and animal models.6–10 Yet, potential off-target effects are significant safety issues if Cas9 is present for a long time.11 One alternative approach is to express Cas9 protein using Cas9 mRNA; however, efficient delivery of Cas9 mRNA is formidable due to its large size (up to 4.5 k nucleotides), high density of negative charges, and weak tolerance of enzymes in serum.7 Hence, new biomaterials are needed to overcome these obstacles and to enable broad applications of CRISPR/Cas9.
Nanomaterials have shown great promise for delivery of diverse therapeutic and diagnostic payloads.12–16 Particularly, lipid and lipidlike nanoparticles (LNPs and LLNs) are representative biomaterials for efficient delivery of RNAs, including siRNA, miRNA, and mRNA.17–24 Recently, a few LLN-based delivery systems demonstrated efficient delivery of mRNAs for expression of functional proteins, including factor IX, erythropoietin, and Cas9.17,20,25–28 Yet, many lipidlike compounds are not considered biodegradable, which may lead to potential side effects. Previous studies reported that biodegradable bonds enabled rapid elimination of lipids from plasma and tissues and substantially improved their tolerability in preclinical studies.22,29 For example, the biodegradable lipids reported previously were well-tolerated at a dose of 10 mg/kg in mice, whereas nonbiodegradable lipids caused acute mortality at a similar dose.30 Hence, it is essential to develop new materials with high delivery efficiency and compatible biodegradability.
RESULTS AND DISCUSSION
Design of Lipidlike Materials
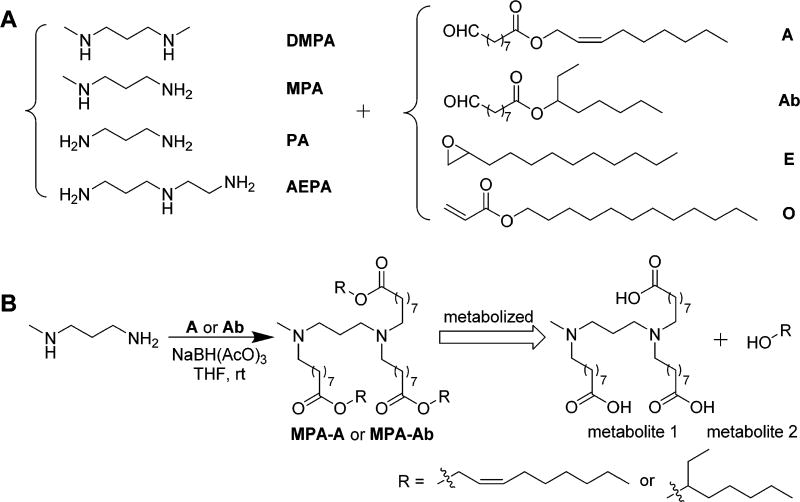
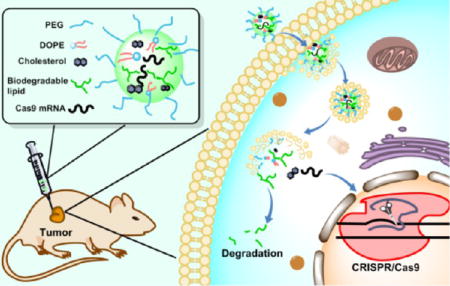
We report the design, synthesis, and biological evaluation of biodegradable amino-ester-derived LLNs for the delivery of Cas9 mRNA. As shown in Scheme 1, we installed four types of lipid chains, including two ester chains, one epoxide chain, and one acrylate chain on four amino cores, termed as N,N′-dimethyl-1,3-propanediamine (DMPA), N-methyl-1,3-propanediamine (MPA), 1,3-propanediamine (PA), and N-(2-aminoethyl)-1,3-propanediamine (AEPA). The linear and branched ester series are termed as 9-oxononanoic acid (Z)-non-2-en-yl ester (A) and 9-oxononanoic acid 2-ethyl-hexane-1-yl ester (Ab), respectively. The epoxide (E) and acrylate (O) series were reported previously for siRNA delivery,19,22 serving as control groups. We hypothesized that A and Ab series lipidlike compounds are biodegradable through interaction with esterases. Moreover, we are capable of tuning the hydrolysis rate by incorporation of functional groups with different steric effects (Scheme 1B).
Scheme 1.
(A) Synthesis of Amino-Ester-Derived Lipidlike Compounds; (B) Rationale of Biodegradability for Newly Synthesized Lipidlike Compoundsa
aWe anticipate that MPA-A and MPA-Ab will be degraded to metabolites 1 and 2 by esterases.
Synthesis and Formulation of LLNs
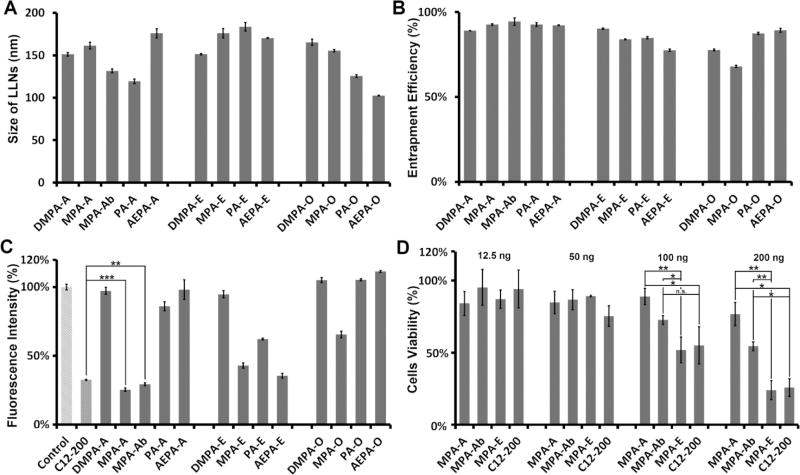
First, we installed lipid chains A and Ab on the four amines through a reductive amination reaction to afford amino-ester-derived lipidlike compounds (Scheme 1A). Epoxide (E) and acrylate (O) series were synthesized according to the procedures reported previously.19,22 The structures of the products were validated by 1H NMR spectroscopy and mass spectrometry (Supporting Information). According to the methods reported previously,17–22,31 newly synthesized amino-ester-derived lipidlike compounds were formulated with 2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), cholesterol (Chol), 1,2-dimyristoyl-sn-glycerol, methoxypoly(ethylene glycol) (DMG-PEG2000), and Cas9 mRNA in an optimized molar ratio (lipid/DOPE/Chol/DMG-PEG2000 = 20/30/40/0.75).17 Particle properties, including particle size, zeta potential, and entrapment efficiency of Cas9 mRNA, were measured using a Zetasizer Nano ZS and a ribogreen assay (Figures 1A,B and S1). The size of these particles ranged from 119 to 184 nm with PDI <0.3. Most materials exhibited moderately positive charges except PA-A and PA-O, which were close to neutral (Figure S1). We speculate that slight structural alternations may affect their interactions with mRNA and particle formulation. The highest entrapment efficiency of mRNA was 94%.
Figure 1.
Characterizations of amino-ester LLNs: (A) Particle size. (B) Entrapment efficiency of Cas9 mRNA. (C) Delivery of Cas9 mRNA using amino-ester LLNs in vitro. Control: phosphate-buffered saline (PBS). (D) Cell viability through a 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay in 293T cells (triplicate; *P < 0.05; **P < 0.01; ***P < 0.001; n.s., P > 0.05; t test, double-tailed).
In Vitro Delivery Efficiency of LLNs
Next, we evaluated the delivery efficiency of Cas9 mRNA in 293T cells that stably express enhanced green fluorescent protein (eGFP) and the corresponding eGFP guide RNA. Effective delivery of Cas9 mRNA induces cleavage of eGFP gene and consequently reduces the eGFP signals in these cells. We treated these 293T cells with newly formulated LLNs at a Cas9 mRNA dose of 50 ng per well in a 24-well clear bottom plate. After treatment for 48 h, the eGFP signal was quantified using fluorescence-activated cell sorting (FACS) analysis. C12-200 was reported for delivery of Cas9 mRNA and showed effective gene editing, serving as a positive control.10 As shown in Figure 1C, the six types of LLNs displayed dramatic reduction of the eGFP signal, including MPA-A, MPA-Ab, MPA-E, PA-E, AEPA-E, and C12-200. Regarding lipid tails, the amino-ester and epoxide series are more favorable than the acrylate series. With respect to amino cores, LLNs based on MPA exhibited a strong reduction of the eGFP signal. Importantly, MPA-A and MPA-Ab decreased the eGFP signal by 74 and 70%, respectively, which showed a higher delivery efficiency compared to that of other LLNs including C12-200 (p < 0.01 or 0.001), a lead material reported previously.10 Meanwhile, we observed the dose-dependent reduction of the eGFP signal (Figure S2, Cas9 mRNA: 12.5, 25, 50, and 100 ng/well). MPA-A and MPA-Ab showed a significantly higher delivery efficiency compared to that of C12-200 and MPA-E (Figure S2). To study the applicability of MPA-A and MPA-Ab, we then formulated them with mRNA-encoding Firefly luciferase (Fluc) and treated the Hep3B cells accordingly. Both MPA-A and MPA-Ab LLNs showed a higher efficiency than C12-200 and MPA-E LLNs (Figure S3). Furthermore, we examined the delivery efficiency of these materials for mRNA-encoding eGFP in 293T cells. All four nanomaterials (MPA-A, MPA-Ab, C12-200, and MPA-E LLNs) induced eGFP expression in over 98% of cells (Figure S4). MPA-A and MPA-Ab showed significantly higher delivery efficiency compared to that of C12-200 and MPA-E (Figure S4). To evaluate their cytotoxicity, we then performed MTT assays at four doses (12.5, 50, 100, and 200 ng/well). At the dose of 200 ng/well, MPA-A and MPA-Ab LLNs displayed a lower toxicity than that of C12-200 and MPA-E (Figure 1D, p < 0.01 or 0.005) in 293T cells. A similar trend of cytotoxicity was detected in Hep3B cells (Figure S5). On the basis of these results, MPA-A and MPA-Ab were selected for further study.
In Vitro Biodegradability Study
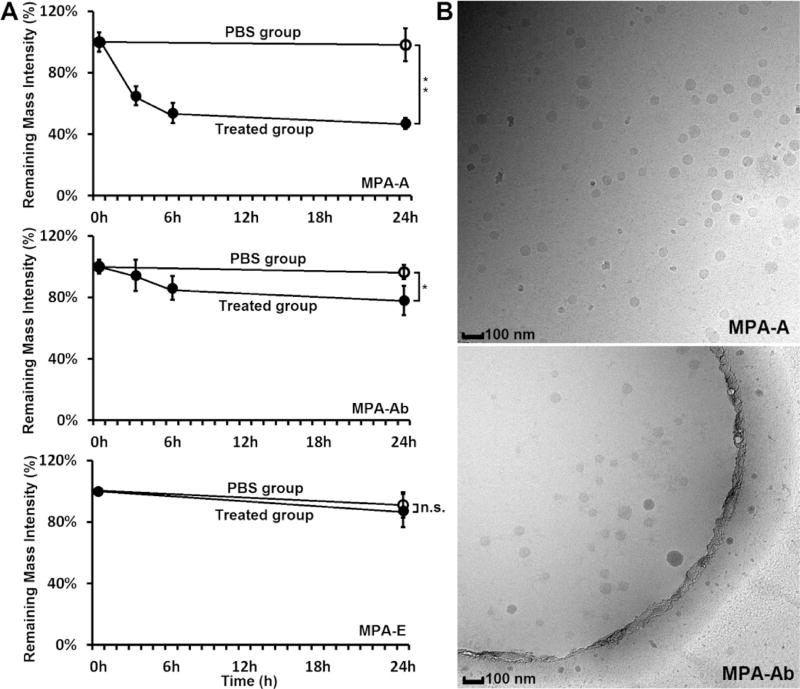
Next, we investigated the biodegradability of MPA-A and MPA-Ab using MPA-E as a control group. The degradation assay was carried out at 37 °C using esterase according to the method reported in the literature.32 The amount of each compound was quantified through mass spectra analysis. As shown in Figure 2A, over 53% of MPA-A was degraded 24 h after incubation, whereas 22% of MPA-Ab was degraded under the same conditions. In addition, MPA-E, a nonbiodegradable control, showed no significant change in mass spectra intensity after 24 h of treatment with the esterase. These results validated our design: that is, adjusting the ester chains with different steric effects is able to tune the degradation rate. Cryo-transmission electron microscopy (cryo-TEM) images revealed that both MPA-A and MPA-Ab LLNs were spherical, showing relatively homogenous particle sizes (Figure 2B).
Figure 2.
(A) Degradation rate in esterase solutions: treated group with esterase (solid circle) and PBS control without esterase (hollow circle). Over 53% of MPA-A was degraded after 24 h incubation, whereas 22% of MPA-Ab was degraded. No significant degradation was observed for MPA-E, a nonbiodegradable control group.(triplicate; *P < 0.05; **P < 0.01; t test, double-tailed). (B) Cryo-TEM images of MPA-A and MPA-Ab LLNs. Scale bar: 100 nm.
In Vivo Delivery Efficiency of LLNs
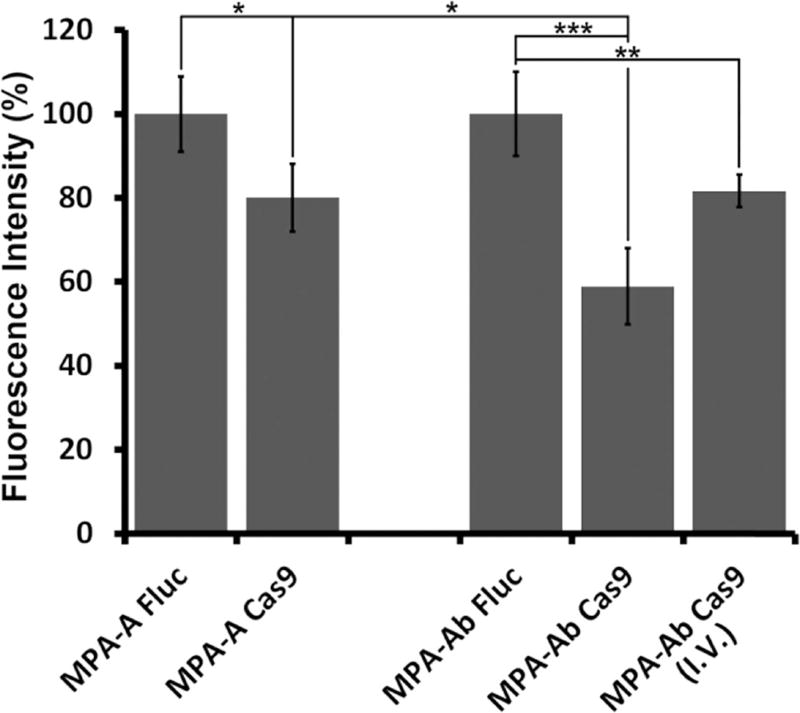
Given the promising in vitro results, we next examined MPA-A and MPA-Ab LLNs for their delivery efficiency of Cas9 mRNA in vivo. We established a mouse xenograft tumor model using the 293T cells mentioned above. Nude mice bearing xenograft tumors were randomly divided into five groups (n = 4), and four groups were injected intratumorally, with two treated groups (MPA-A/Cas9 LLNs and MPA-Ab/Cas9 LLNs) and two control groups (MPA-A/Fluc LLNs and MPA-Ab/Fluc LLNs), at a dose of 2.5 µg/100 mm3 tumor; for the last group, MPA-Ab/Cas9 LLNs were injected intravenously at a dose of 0.88 mg/kg. Five days after treatment, the tumors were dissected and lysed. As mentioned above, the eGFP signal was quantified using FACS analysis. As shown in Figure 3, both MPA-A LLNs and MPA-Ab LLNs showed significant suppression of the eGFP signal compared to that of the control groups (statistically significant), demonstrating efficient delivery of Cas9 mRNA in vivo. MPA-Ab LLNs (41% reduction of eGFP signal) were more potent than MPA-A LLNs (20% reduction of eGFP signal) when injected intratumorally. Moreover, MPA-Ab LLNs also showed reduction of the eGFP signal (18%, statistically significant) after intravenous injection.
Figure 3.
In vivo delivery of Cas9 mRNA. Both MPA-A and MPA-Ab LLNs encapsulating Cas9 mRNA significantly reduced the eGFP signal in a mouse xenograft model (n = 4; *P < 0.05; **P < 0.01; ***P < 0.001; t test, double-tailed).
In Vivo Biodegradability and Histology Study
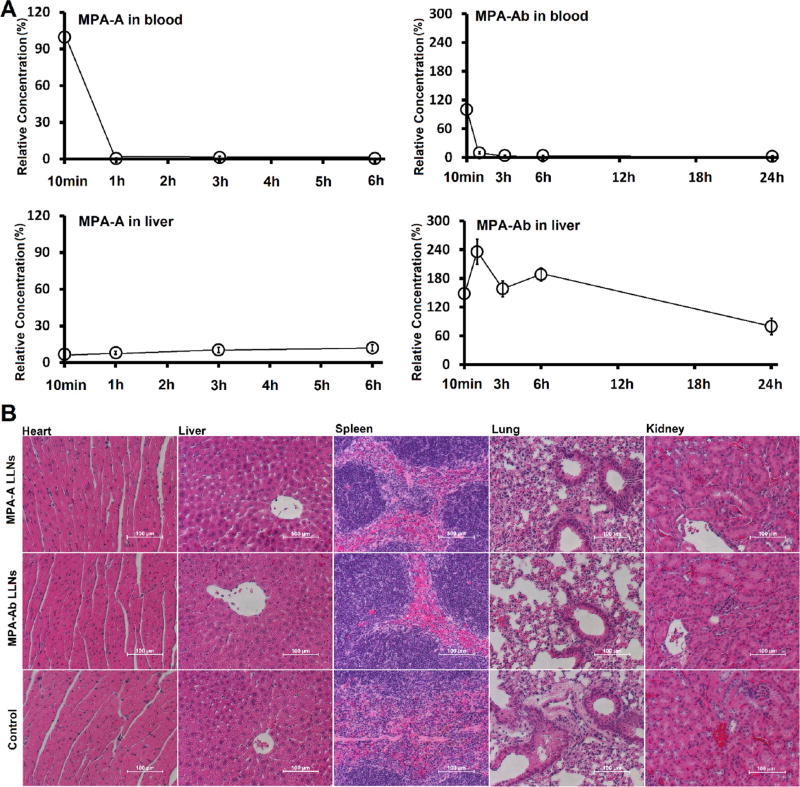
Finally, we evaluated the biodegradability of MPA-A and MPA-Ab in C57BL/6 mice as well as their histology. After MPA-A and MPA-Ab LLNs were administered intravenously, the lipid levels in the blood and liver were measured at various time points using mass spectrometry. The data were normalized as the percentage of the lipid level in blood at the first time point (10 min post injection) for each compound. As shown in Figure 4A, both MPA-A and MPA-Ab LLNs exhibited rapid clearance from blood in the first hour. In the liver, MPA-A showed minimum accumulation, whereas MPA-Ab reached the peak level at 1 h post injection and afterward displayed an evident clearance from 6 to 24 h. Moreover, we observed no evident changes in clinical appearance in the MPA-A- and MPA-Ab-treated groups compared to that in the untreated mice. Histopathology analysis indicated no apparent damage to the organs tested (Figure 4B).
Figure 4.
(A) Biodegradability of MPA-A and MPA-Ab LLNs through in vivo phamarcokinetic study. Relative concentration–time profiles of lipids in blood and liver (normalized with mass intensity in blood 10 mins post injection). (B) Histology images of hearts, livers, spleens, lungs, and kidneys from mice treated with MPA-A and MPA-Ab LLNs. Control: untreated mice.
CONCLUSIONS
In summary, we designed and synthesized a series of biodegradable amino-ester LLNs for Cas9 mRNA delivery. By tuning the functional groups on the ester chains, we were capable of controlling the rate of their biodegradability. The more the steric effects in the chains, the slower the degradation rate. Both MPA-A and MPA-Ab showed a higher delivery efficiency for Cas9 mRNA compared to that of the E & O series LLNs and C12-200, a lead material reported previously. Moreover, they were less toxic than C12-200 in the cells tested. In vivo studies further demonstrated that MPA-A and MPA-Ab LLNs exhibited efficient delivery of Cas9 mRNA. Taken together, these amino-ester LLNs offer promising genome-editing delivery tools for future biological and therapeutic applications.
EXPERIMENTAL SECTION
Synthesis of Nanomaterials
Ester chains A and Ab were synthesized according to the methods reported previously.33,34
General Method for the Synthesis of DMPA-A, MPA-A, MPA-Ab, PA-A, and AEPA-A
Excess amounts of A or Ab and 0.1 mmol corresponding amine were stirred in 5mL of anhydrous tetrahydrofuran at RT. After adding NaBH(OAc)3, the reaction mixture was stirred at RT for 12 h. After the solvent was evaporated, the residue was purified by column chromatography using a Combiflash Rf system with a gradient elution from 100% CH2Cl2 to CH2Cl2/MeOH/NH4OH (75/22/3 by volume) to give the products.
Method for the Biodegradation Assay
For the in vitro assay, MPA-A, MPA-Ab, and MPA-E were dissolved with acetone to obtain 3.24 mol/L stock solutions. The stock solution (15 µL) was then added into 485 µL of PBS with or without esterase (0.8 mg/mL) and bile salt (5 mg/mL) in a 1.5 mL sealed tube and shaken at 37 °C for fixed times, such as 0, 3, 6, and 24 h. The degradation was stopped by adding 1mL of dichloromethane. The organic phase was used for mass spectrometry analysis (Bruker amaZon ETD). The degradation percentage was calculated by the intensity of the molecular ion peak using Compass OpenAccess mass spectrometry software. For the in vivo assay, the freshly prepared LLNs were injected intravenously at a mRNA dose of 1 mg/kg. Three mice were used for each treatment group at each time point. The blood and liver were harvested at 10 min, 1, 3, 6, and 24 h after injection. Samples were then prepared immediately for mass spectral analysis. Blood (20 µL) or liver (20 mg) was added to 1 mL of MeOH and 0.5 mL of CH2Cl2 and shaken vigorously for 5 min. After filtration, the solutions were used for mass spectrometry analysis (Bruker amaZon ETD).
Formulation of mRNA-Loaded LLNs
LLNs were formulated with DOPE, Chol, DMG-PEG2000 (molar ratio 20/30/40/0.75), and Cas9 mRNA, Fluc mRNA, or eGFP mRNA via pipetting for in vitro studies or via a microfluidic-based mixing device (Precision NanoSystems) for in vivo studies. For in vivo studies, the freshly prepared LLNs were dialyzed in PBS buffer using Slide-A-Lyzer dialysis cassettes (3.5 K MWCO; Life Technologies, Grand Island, NY). The particle size and zeta potential of LLNs were measured using a Nano ZS Zetasizer (Malvern, Worcestershire, U.K.) at a scattering angle of 173° and a temperature of 25 °C. The entrapment efficiency of LLNs was determined using the Ribogreen assay reported previously.19,35
Cryo-TEM
Cryo-TEM specimens were prepared by thin-film plunge freezing.36 A small aliquot (2.5 µL) of MPA-A or MPA-Ab LLNs was applied to a C-flat TEM grid (EMS, Hatfield, PA) inside a FEI Mark IV Vitrobot (FEI, Hillsboro, OR). After the blotting process using filter papers, the grid was immediately plunged into liquid ethane to achieve vitrified electron-transparent thin films. The grid was transferred under liquid nitrogen in a cryotransfer station to a Gatan 626.DH cryo-TEM holder (Gatan, Pleasanton, CA). The cryo-TEM holder was loaded into a Tecnai F20 S/TEM (FEI, Hillsboro, OR), and the specimen temperature was maintained at <170 °C. Cryo-TEM images were recorded using a Gatan Ultrascan 4 K CCD camera under low-dose conditions.
MTT Assay
Cytotoxicity assays were performed in triplicate, and the cell viability of each treatment group was normalized to untreated cells. For the 293T and Hep3B cell lines, the cells were grown in 96-well plates, 24 h prior to treatment, at a density of 1 × 104 cells/well. After incubation with MPA-A, MPA-Ab, C12-200, or MPA-E LLNs at four mRNA doses of 12.5, 50, 100, and 200 ng for 48 h, MTT was added to each well and the cells were then incubated for 4 h at 37 °C. The medium was then removed, and 150 µL of dimethyl sulfoxide was added. After shaking for 10 min, the absorbance at a wavelength of 570 nm was measured on a SpectraMax M5 microplate reader.
In Vitro Delivery of Cas9 mRNA
293T cells that stably express eGFP and corresponding gRNA were seeded in 24-well clear bottom plates (2 × 104 cells/500 µL). After 24 h incubation, 5 µL LLNs containing Cas9 mRNA were added in test groups (50 ng/well) and PBS was added in control groups. After 48 h of treatment, the cells were collected and the eGFP signal was quantified with a BD LSR II flow cytometry analyzer (BD Biosciences, San Jose, CA).
In Vivo Studies of MPA-A and MPA-Ab LLNs for Cas9 mRNA Delivery
All procedures used in animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the Ohio State University and were also consistent with local, state, and federal regulations as applicable. NU/J, homozygous female mice (9 weeks old) were purchased from the Jackson Laboratory. To establish the xenograft model, 100 µL of 293T cells (3 × 107/mL) that stably express eGFP and the corresponding gRNA were suspended in PBS and subcutaneously injected into the right flank of the mice. When the subcutaneous nodule reached approximately 800 mm3 in size, the mice were euthanized and subcutaneous tumors were harvested. The tumorlike bulge was carefully isolated to remove fat and necrotic tissues and cut into small pieces (approximately 2 mm3 in diameter) under aseptic conditions. Each individual piece was inserted into the right incision of each receptor mouse made by surgical scissors. The incision site was immediately closed with one drop of 3M Vetbond tissue adhesive. Three weeks later, the mice inoculated with tumorlike tissue were randomly divided into five groups (n = 4) and injected intratumorally for four groups with lipid particles, MPA-A/Cas9 and MPA-A/Fluc (control) or MPA-Ab/Cas9 and MPA-A/Fluc (control) (0.05 mg/mL, 50 µL/100mm3); for the last group, MPA-Ab/Cas9 LLNs were injected intravenously at a mRNA dose of 0.88 mg/kg. Five days after treatment, the tumorlike bulges were isolated and dissociated with a Tumor Dissociation Kit_human (Miltenyi Biotec). The eGFP signal was measured with a BD LSR II flow cytometry analyzer (BD Biosciences, San Jose, CA).
Histology Study
Freshly prepared LLNs were injected intravenously at a dose of 1 mg/kg. After treatment for 6 h, the organs, including the heart, liver, spleen, lung, and kidney, were harvested. After being fixed in 10% formaldehyde solution for 12 h, these organs were stored in 70% ethanol. Tissues were then embedded into paraffin, sectioned, and imaged on an ECLIPSE Ti inverted fluorescence microscope (Nikon, Japan).
Supplementary Material
Acknowledgments
The cryo-TEM data were obtained at the Liquid Crystal Institute Characterization Facility, Kent State University. Histology slides were obtained at the Comparative Pathology and Mouse Phenotyping Shared Resource (CPMPSR), College of Veterinary Medicine at the Ohio State University, supported in part by NCI grant P30 CA016058. We thank Dr. Wen Xue for providing 293T cells that stably expressed eGFP and the corresponding eGFP guide RNA. This work was supported by the Early Career Investigator Award from the Bayer Hemophilia Awards Program, Research Awards from the National PKU Alliance, New Investigator Grant from the AAPS Foundation, Maximizing Investigators’ Research Award 1R35GM119679 from the National Institute of General Medical Sciences, and the startup fund from the College of Pharmacy at the Ohio State University.
Footnotes
ASSOCIATED CONTENT
- Materials, syntheses, and 1H NMR spectra of the compounds (PDF)
The authors declare no competing financial interest.
References
- 1.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–72. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, Kornepati AV, Sousa A, Collins MA, Jayaram H, Cullen BR, Bumcrot D. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu XB, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–U98. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platt RJ, Chen SD, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng GP, Sharp PA, Zhang F. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun WJ, Ji WY, Hall JM, Hu QY, Wang C, Beisel CL, Gu Z. Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR-Cas9 for Genome Editing. Angew. Chem., Int. Ed. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin H, Song CQ, Dorkin JR, Zhu LHJ, Li YX, Wu QQ, Park A, Yang J, Suresh S, Bizhanova A, Gupta A, Bolukbasi MF, Walsh S, Bogorad RL, Gao GP, Weng ZP, Dong YZ, Koteliansky V, Wolfe SA, Langer R, Xue W, Anderson DG. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Cheng H, Peng HS, Zhou H, Li PY, Langer R. Non-genetic engineering of cells for drug delivery and cell-based therapy. Adv. Drug Delivery Rev. 2015;91:125–140. doi: 10.1016/j.addr.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen TX, Huang L, Gauthier M, Yang G, Wang Q. Recent advances in liposome surface modification for oral drug delivery. Nanomedicine. 2016;11:1169–1185. doi: 10.2217/nnm.16.9. [DOI] [PubMed] [Google Scholar]
- 14.Lu Y, Aimetti AA, Langer R, Gu Z. Bioresponsive materials. Nat. Rev. Mater. 2017;2(16075) [Google Scholar]
- 15.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitragotri S, Burke PA, Langer R. Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat. Rev. Drug Discovery. 2014;13:655–672. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Luo X, Deng BB, Wang JF, McComb DW, Shi YM, Gaensler KML, Tan X, Dunn AL, Kerlin BA, Dong YZ. An Orthogonal Array Optimization of Lipid-like Nanoparticles for mRNA Delivery in Vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang YL, Chen DL, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang WH, Schroeder A, Lytton-Jean AKR, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin JN, Querbes W, Koteliansky V, Langer R, Anderson DG. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DWY, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenton OS, Kauffman KJ, McClellan RL, Appel EA, Dorkin JR, Tibbitt MW, Heartlein MW, DeRosa F, Langer R, Anderson DG. Bioinspired Alkenyl Amino Alcohol Ionizable Lipid Materials for Highly Potent In Vivo mRNA Delivery. Adv. Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 22.Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang YL, Olejnik KT, Yesilyurt V, Chen DL, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 2014;5(4277) doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li MH, Yu H, Wang TF, Chang ND, Zhang JQ, Du D, Liu MF, Sun SL, Wang R, Tao HQ, Geng SL, Shen ZY, Wang Q, Peng HS. Tamoxifen embedded in lipid bilayer improves the oncotarget of liposomal daunorubicin in vivo. J. Mater. Chem. B. 2014;2:1619–1625. doi: 10.1039/c3tb21423k. [DOI] [PubMed] [Google Scholar]
- 24.Du D, Chang ND, Sun SL, Li MH, Yu H, Liu MF, Liu XY, Wang GT, Li HC, Liu XP, Geng SL, Wang Q, Peng HS. The role of glucose transporters in the distribution of p-aminophenyl-alpha-D-mannopyranoside modified liposomes within mice brain. J. Controlled Release. 2014;182:99–110. doi: 10.1016/j.jconrel.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Karikó K, Muramatsu H, Keller JM, Weissman D. Increased Erythropoiesis in Mice Injected With Submicrogram Quantities of Pseudouridine-containing mRNA Encoding Erythropoietin. Mol. Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong Y, Dorkin JR, Wang WH, Chang PH, Webber MJ, Tang BC, Yang J, Abutbul-Ionita I, Danino D, DeRosa F, Heartlein M, Langer R, Anderson DG. Poly(glycoamidoamine) Brushes Formulated Nanomaterials for Systemic siRNA and mRNA Delivery in Vivo. Nano Lett. 2016;16:842–848. doi: 10.1021/acs.nanolett.5b02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kauffman KJ, Dorkin JR, Yang JH, Heartlein MW, DeRosa F, Mir FF, Fenton OS, Anderson DG. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 28.Miller JB, Zhang SY, Kos P, Xiong H, Zhou KJ, Perelman SS, Zhu H, Siegwart DJ. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem., Int. Ed. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier MA, Jayaraman M, Matsuda S, Liu J, Barros S, Querbes W, Tam YK, Ansell SM, Kumar V, Qin J, Zhang XM, Wang QF, Panesar S, Hutabarat R, Carioto M, Hettinger J, Kandasamy P, Butler D, Rajeev KG, Pang B, Charisse K, Fitzgerald K, Mui BL, Du XY, Cullis P, Madden TD, Hope MJ, Manoharan M, Akinc A. Biodegradable Lipids Enabling Rapidly Eliminated Lipid Nanoparticles for Systemic Delivery of RNAi Therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barros SA, Gollob JA. Safety profile of RNAi nanomedicines. Adv. Drug Delivery Rev. 2012;64:1730–1737. doi: 10.1016/j.addr.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Dong Y, Eltoukhy AA, Alabi CA, Khan OF, Veiseh O, Dorkin JR, Sirirungruang S, Yin H, Tang BC, Pelet JM, Chen DL, Gu Z, Xue Y, Langer R, Anderson DG. Lipid-Like Nanomaterials for Simultaneous Gene Expression and Silencing In Vivo. Adv. Healthcare Mater. 2014;3:1392–1397. doi: 10.1002/adhm.201400054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oda H, Yamashita H, Kosahara K, Kuroki S, Nakayama F. Esterified and Total 7-Alpha-Hydroxycholesterol in Human Serum as an Indicator for Hepatic Bile-Acid Synthesis. J. Lipid Res. 1990;31:2209–2218. [PubMed] [Google Scholar]
- 33.Rajabi M, Lanfranchi M, Campo F, Panza L. Synthesis of a Series of Hydroxycarboxylic Acids as Standards for Oxidation of Nonanoic Acid. Synth. Commun. 2014;44:1149–1154. [Google Scholar]
- 34.Woodcock SR, Marwitz AJV, Bruno P, Branchaud BP. Synthesis of nitrolipids. All four possible diastereomers of nitrooleic acids: (E)- and (Z)-, 9- and 10-nitro-octadec-9-enoic acids. Org. Lett. 2006;8:3931–3934. doi: 10.1021/ol0613463. [DOI] [PubMed] [Google Scholar]
- 35.Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong YZ, Whitehead KA, Anderson DG. Rapid Discovery of Potent siRNA-Containing Lipid Nanoparticles Enabled by Controlled Microfluidic Formulation. J. Am. Chem. Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 36.Gao M, Kim YK, Zhang CY, Borshch V, Zhou S, Park HS, Jakli A, Lavrentovich OD, Tamba MG, Kohlmeier A, Mehl GH, Weissflog W, Studer D, Zuber B, Gnagi H, Lin F. Direct Observation of Liquid Crystals Using Cryo-TEM: Specimen Preparation and Low-Dose Imaging. Microsc. Res. Tech. 2014;77:754–772. doi: 10.1002/jemt.22397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.