Abstract
The increasing interest in extracellular vesicles (EVs) research is fueled by reports indicating their unique role in intercellular communication and potential connection to the development of common human diseases. The unique role assumes unique protein and nucleic acid cargo. Unfortunately, accurate analysis of EVs cargo faces a challenge of EVs isolation. Generally used isolation techniques do not separate different subtypes of EVs and even more, poorly separate EVs from non-EVs contaminants. Further development of EVs isolation protocols urgently needs a quantitative method of EVs purity assessment. We report here that multiple reaction monitoring assay using internal standards carrying peptides for quantification of EVs and non-EVs proteins is a suitable approach to assess purity of EVs preparations. As a first step in potential standardization of EVs isolation, we have evaluated polymer-based precipitation techniques and compared them to traditional ultracentrifugation protocol.
Graphical abstract
Extracellular vesicles (EVs) are released from all cell types into the extracellular space and play an important role in intercellular communication through the transfer of nucleic acids, proteins, and lipids to recipient cells. EVs have been linked to tissue regeneration, immune response modulation, tumorigenesis, neurodegeneration and spread of viruses and pathogenic agents (reviewed in refs 1–3). In addition to being biomarkers of disease and therapeutic targets, EVs have potential as cargo carriers in drug and vaccine delivery applications.1–3
The most challenging aspect of EVs studies is their diversity. There are multiple EVs subtypes with different biogenesis, cargo, and physicochemical properties. Furthermore, EVs are isolated from different body fluids and in vitro cell culture media using a variety of general techniques (reviewed in ref 4), which do not well separate subtypes of EVs and typically result in partially purified preparations of EVs contaminated by nonvesicular components. The International Society for Extracellular Vesicles has emphasized the urgent need for standardization in EVs research on all levels, including special attention to the methods of isolation.5–7
In the present study, we evaluated the usefulness of so-called polymer-based precipitation method in isolation of EVs. Polymer-based precipitation is a broadly used method of EVs isolation.7–13 The basic procedure includes mixing of a polymer solution and the sample of interest, incubation, and low-speed centrifugation to collect precipitated EVs. Several commercial kits are available and EVs isolation can be performed with (Exospin kit from Cell Guidance Systems) and without (Total Exosome Isolation kit from Invitrogen) the subsequent spin-column step. The popularity of polymer-based precipitation results from less laborious protocols. Although precipitation techniques are rarely used without further isolation techniques,7 the rationale for using precipitation remains obscure in general.
We first generated 15N-labeled quantification concatamer (QconCAT)14,15 to quantify a pattern of targeted EVs and non-EVs proteins in multiple reaction monitoring assay (MRM).16,17 These quantifications were then used to compare purity of EVs preparations obtained from human serum by two protocols; the first one includes ultracentrifugation and size exclusion chromatography (SEC), the second one includes polymer-based precipitation techniques in-front of ultracentrifugation. The rationale for such comparison was (i) to demonstrate that MRM with QconCATs can be used for evaluation of EVs purity and (ii) to address the question whether precipitation techniques offer advantages for proteomic downstream applications.
EXPERIMENTAL SECTION
Materials
15N-Ammonium chloride (99.9%) was from Cambridge Isotope Laboratories (Andover, MA). The DC Protein Assay kit was from Bio-Rad Laboratories (Hercules, CA). EVs purification kit (Exo-02) was purchased from Cell Guidance Systems (St. Louis, MO). Total exosome isolation reagent (Invitrogen) was from ThermoFisher Scientific (Carlsbad, CA). Human serum was pooled from 10 individual male samples (Bioreclamation, Westbury, NY). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
15N-Labeled QconCAT Expression, Purification, and Characterization
The quantification concatamer (Qcon-CAT) is an artificial protein composed of concatenated tryptic peptides from targeted proteins and used as an internal standard for quantification of these targeted proteins by MRM mass spectrometry.14,15,18,19 The amino acid sequences of EXO1 and EXO2 QconCATs designed for quantification of EVs proteins are shown in Figure S1 (Supporting Information). The synthetic genes encoding these sequences were synthesized and incorporated into the pET21a expression vector, with codon optimization for E. coli (Biomatik, Cambridge, Ontario). The plasmid was transformed into One Shot BL21 (DE3) competent E. coli cells (Invitrogen, Grand Island, NY) and grown in M9 minimal media with 1 g/L 15NH4Cl (Cambridge Isotope Laboratories, Andover, MA) as the sole nitrogen source at 37 °C until the optical density reached 0.6 to 0.8 at 600 nm. Protein expression was induced by 0.5 mmol/L isopropyl β-d-1-thiogalactopyranoside. After 3 h of growth, the cells were harvested by centrifugation at 5000 g for 10 min. QconCATs were expressed as insoluble inclusion bodies. Collected cells were first washed with water and then disrupted by sonication in water. QconCATs were collected by centrifugation at 5000 g for 10 min and washed with 7 M urea in water. Final pellet, which was insoluble in urea, was collected by centrifugation at 5000 g for 10 min, dissolved in 2% SDS and separated using SDS polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were eluted from the gel at 100 mA for 40 min into 14 fractions using a Mini Whole Gel Eluter (Bio-Rad Laboratories, Hercules, CA). The fractions containing QconCATs as compared to molecular mass standards were collected and concentrated using Amicon Ultra-4 centrifugal filters with 10 K molecular weight cutoff. The QconCAT concentration was measured in the presence of 2% SDS using DC protein assay kit and bovine serum albumin as a standard. The final QconCATs were aliquoted and kept frozen at −80 °C.
The purity of QconCATs was considered ≥95% based on SDS-PAGE (Figure S2, Supporting Information) and no correction for protein concentration of QconCATs was made during data analysis. Optimal MRM transitions for Q-peptides were experimentally selected after tryptic digestion of QconCATs (Table S1, Supporting Information). These transitions were then used to determine the level of stable isotope incorporation into the QconCATs. The isotope incorporation was calculated as the percentile of the area of the labeled peak to the sum of the unlabeled and labeled peaks. Calculation based on three representative Q-peptides resulted in 99.66 ± 0.27% and 99.74 ± 0.27% isotope incorporation for EXO1 and EXO2 QconCATs, respectively (Table S2, Supporting Information). These values were accepted as a complete labeling and no correction for labeling efficiency was applied during data analysis.
Basic Protocol for EVs Isolation from Human Serum
Basic protocol for EVs isolation includes low-speed centrifugations, ultracentrifugation and SEC. All centrifugations were performed at 4 °C. The purification starts with low-speed centrifugations, first at 2000 g for 15 min, and then supernatant was further centrifuged at 10000 g for 30 min. The pellets from 2000 g and 10000 g centrifugations were discarded. The supernatant of 10000 g centrifugation was further centrifuged at 70000 rpm (average 208000 g) for 60 min using Beckman TLA-100.3 rotor and TL-100 ultracentrifuge. The pellet of this centrifugation was resuspended in 1 mL of phosphate buffered saline (PBS) and centrifuged again. The PBS-washed pellet was called EVs(208K).
For SEC, an AKTA FPLC (Amersham Biosciences, Piscataway, NJ, U.S.A.) was used to separate the EVs(208K) on a Superose 6 Increase 10/300 GL column in PBS. The flow rate was 0.4 mL/min and 0.5 mL fractions were collected. The fractions eluted in void volume were pooled and centrifuged at 70000 rpm (average 208000 g) for 60 min. The pellet of this centrifugation was called EVs(SEC).
Dynamic Light Scattering
A DynaPro NanoStar (556-DPN, WYATT Technology, U.S.A.) was used for the dynamic light scattering (DLS) measurements. The laser was set at λ = 661 nm with the detector angle at 90° for measurements. Both EVs samples, EVs(208K) and EVs(SEC) were adjusted with PBS to give concentration ranges from 0.2 to 2.0 mg/mL. PBS buffer was filtered through a Millipore Millex-GV 0.22 µm PVDF filter. DLS spectra were acquired in a disposable cyclic olefin copolymer cuvette at 25 °C. Each spectrum was collected over 5 runs consisting of 10 ten-second scans. The five runs were then averaged. Dynamics software (7.5.0.17, WYATT Technology, U.S.A.) was used to acquire and analyze the spectra. The regularization method was used to fit the autocorrelation functions.
Precipitation-Based Protocols for EVs Purification from Human Serum
EVs from human serum were prepared with four protocols using two commercial kits, Exospin Blood from Cell Guidance Systems and Total Exosome Isolation (from serum) from Invitrogen. Similar to the basic protocol, all precipitation-based protocols started with two low-speed centifugations, at 2000 g for 15 min and then at 10000 g for 30 min. The supernatant of 10000 g centrifugation was further directed to Exospin buffer precipitation (protocol #1), Exospin buffer precipitation and column purification (protocol #2), Invitrogen buffer precipitation (protocol #3), and Invitrogen buffer precipitation with following column purification using columns from Exospin kit (protocol #4). All of these steps were performed in accordance with manufacturer’s recommendations. The pellets after buffer precipitation were redisolved in PBS, while samples after column purification were already eluted in PBS. All of these PBS samples were further centrifuged at 70000 rpm (average 208000 g) for 60 min using Beckman TLA-100.3 rotor and TL-100 ultracentrifuge. The pellet of this centrifugation was resuspended in 1 mL of PBS and centrifuged again. The PBS-washed pellets were used for MRM measurements of targeted proteins. A scheme depicting the basic protocol and all four precipitation-based protocols is shown in Figure S3 (Supporting Information).
Sample Processing for Mass Spectrometry
Purified EVs samples were dissolved in 100 µL of 50 mmol/L NH4HCO3/2% SDS and 10 µL was used for total protein concentration determination by DC assay. During the following processing, samples were supplemented with 3 pmol of each 15N-labeled internal standards and 10 mmol/L dithiothreitol (DTT). After incubation for 60 min at room temperature, samples were treated with 30 mmol/L iodoacetamide for another 60 min in the dark and precipitated with chloroform/methanol.20 The pellets obtained from precipitation were sonicated in 100 µL of 50 mmol/L NH4HCO3/0.1% RapiGest and treated with trypsin (1:5 w/w) for 15 h at 37 °C. After trypsinolysis, the samples were acidified with 0.5% trifluoroacetic acid (TFA) for 30 min at 37 °C and centrifugated at 16000 g to collect the supernatant. Finally, samples were dried in a vacuum centrifuge (Eppendorf AG, Hamburg, Germany), redissolved in H2O, and dried again.
LC-MS/MS Analysis
For LC-MS/MS analysis, dried peptides were reconstituted in 3% acetonitrile/0.1% formic acid (volume fraction). Separation was performed on an Agilent Zorbax Eclipse Plus C18 RRHD column (2.1 mm × 50 mm, 1.8 µm particle) and MRM analysis was performed on an Agilent 6490 iFunnel Triple Quadrupole LC/MS system (Santa Clara, CA). Peptides were eluted at a flow rate of 200 µL/min using the following gradient of solvent B in solvent A: 3% B for 3 min, 3% to 30% B in 30 min, 30% to 50% B in 5 min, and 50% to 3% B in 5 min. Solvent A was water containing 0.1% formic acid (volume fraction) and solvent B was acetonitrile containing 0.1% formic acid (volume fraction). The acquisition method used the following parameters in positive mode: fragmentor 380 V, collision energy 20 V, dwell time 100 ms, cell accelerator 4 V, electron multiplier 500 V, and capillary voltage 3500 V. MRM transitions for 2+ charge precursor ions and 1+ charge product ions were predicted using PinPoint software (Thermo Fisher Scientific, Waltham, MA).
Data Analysis
MRM peak area integration was performed using Skyline (3.7.0.10940) (University of Washington). Excel was used to calculate peak area ratios. Peak integration was manually inspected and adjusted, if necessary. The peak ratios from transitions were averaged to yield the peptide ratios. All experiments were performed in duplicate with three replicate injections to assess error and reproducibility. Data is represented as the mean ± SD.
RESULTS AND DISCUSSION
Basic Protocol for EVs Isolation
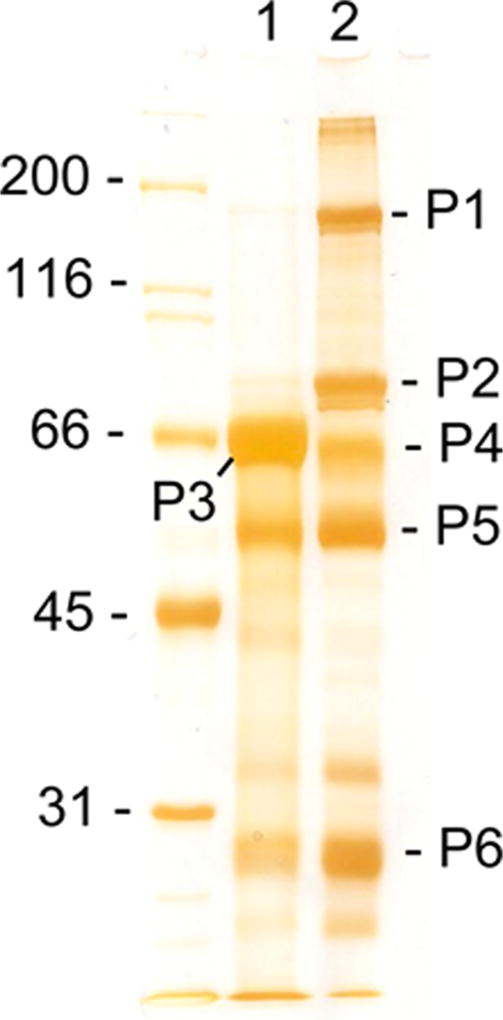
Ultracentrifugation remains the most common technique for EVs isolation7 and was selected as a first step for the basic protocol used in this study. Figure 1 shows the side-by-side comparison of abundant proteins in two samples, serum before ultracentrifugation and EVs(208K) after ultracentrifugation. The identification of proteins after in-gel digestion is summarized in Table S3, Supporting Information. The serum pattern includes three major bands, albumin (P3) and two chains of IgG, heavy (P5) and light (P6). The EVs(208K) pattern includes five major bands, α-2-macroglobulin (P1) and its fragment (P2), IgM heavy chain (P4), and both chains of IgG (P5 and P6). From one side, there is no visually abundant albumin in the EVs(208K) and it means that ultracentifugation efficiently removes some serum proteins. From another side, ultracentrifugation obviously enriched EVs(208K) with some other serum proteins, such as α-2-macroglobulin and IgM heavy chain. This observation prevents us from normalizing future MRM measurements of targeted proteins to the level of albumin as a measure of EVs purity. This also was a clear sign that ultracentrifugation itself may generate EVs that are still heavily contaminated with serum proteins.
Figure 1.
SDS-PAGE (9%) of serum (1) and EVs(208K) (2). Silver-stained bands used for in-gel digestion and protein identification are marked from P1 to P6. The molecular mass standards in kDa are shown on the left.
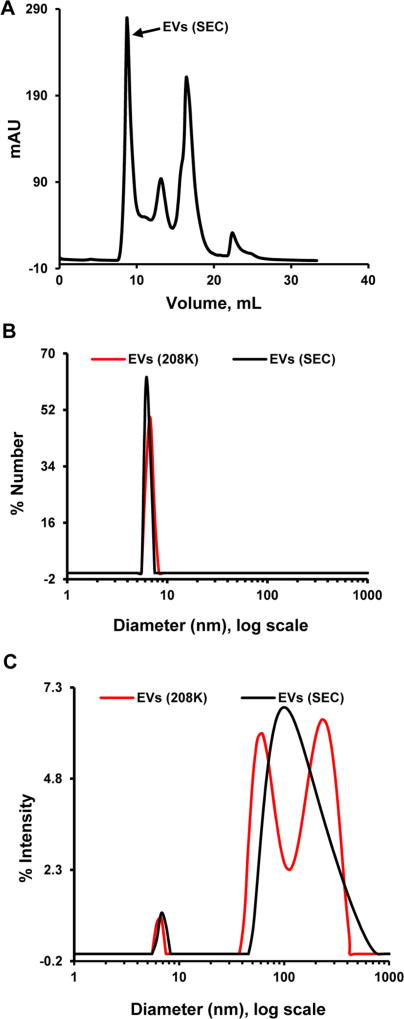
Since we are trying to separate vesicles from serum proteins, size exclusion chromatography (SEC) was selected as a second step for the basic protocol used in this study. Figure 2A shows EVs(208K) separation on Superose 6 Increase 10/300 GL column. Based on UV detection, the material eluted in the void volume and called EVs(SEC) represents less than 40% of the loaded EVs(208K). This points to approximately 2-fold purification of EVs preparation in this SEC application. EVs(208K) and EVs(SEC) were also compared by DLS. Figure 2B shows that the major component of both preparations is the same with hydrodynamic diameter near 8 nm. This is too small for EVs and most likely represents either protein(s) such as IgG21 or lipoprotein complexes such as high-density lipoprotein,22 which coisolate with EVs. The bigger components (not visible by number in Figure 2B) are present based on intensity of scattering and their pattern change after SEC (Figure 2C). The broad peak in EVs(SEC) with maximum at 103 nm matches well to expected EVs size, but has high polydispersity suggesting the presence of multiple species. Overall, we concluded that although SEC provides for further EVs purification, the final preparation remains heavily contaminated with non-EVs proteins.
Figure 2.
Isolation of EVs(SEC) and its comparison to EVs(208K). (A) Sample of EVs(208K) was separated on a Superose 6 Increase 10/300 GL column in PBS. The material eluted in void volume was named EVs(SEC). (B and C) DLS for EVs(208K) and EVs(SEC). Distribution by number and by intensity is shown in (B) and (C), respectively.
The purpose of this study was not to obtain pure EVs, but rather to develop a quantitative MRM assay for assessing gradual increase in EVs purity. For this purpose, we were in need of a basic well-characterized protocol, which can be later used as a reference for evaluation of efficiency in EVs isolation by other protocols. The basic, two-step protocol described herein was adequate for our goal.
Internal Standards for MRM Assay
The initial step in developing the MRM assay was designing QconCATs, internal standards for quantification of targeted proteins. The QconCATs included two groups of targeted proteins: non-EVs contaminants and EVs markers. For tracking of major non-EVs contaminants, peptides from abundant serum proteins, albumin and α-2-macroglobulin, were included. For quantification of EVs, peptides from protein markers unique to EVs needed to be used. However, selecting appropriate EVs markers was more complicated than initially expected. Numerous proteomic studies were performed on EVs from various sources and the data were annotated in several databases, such as Vesiclepedia (http://microvesicles.org/#), EVpedia (http://student4.postech.ac.kr/evpedia2_xe/xe/index.php?mid=Home), and ExoCarta (http://exocarta.org/index.html). In total, thousands of proteins are annotated as EVs proteins, including the extended pattern of the same high abundance serum and tissue proteins, which are typically detected by proteomic analysis in virtually any biological sample analyzed. This makes the outlining of EVs-specific markers unclear. Another layer of complexity in designing QconCATs came from the MRM method itself, which has some preferences for tryptic peptides used for quantification. These peptides should preferably not carry cysteine and methionine residues and be approximately 9–20 amino acid residues in length. For example, out of four tetraspanins (CD9, CD63, CD81, and CD82), which are classically used as EVs markers, only CD82 has tryptic peptides suitable for MRM quantification. Fortunately, the above-mentioned databases also have a list of top 100 proteins that are often identified in EVs preparations. Since we purified EVs from human serum, which is not supposed to have membrane proteins other than those present in EVs, we designed the first QconCAT (EXO1) based on membrane proteins from the list of top 100 EVs proteins. EXO1 includes tryptic peptides from eight membrane proteins: L1CAM (single-pass), ADAM10 (single-pass), CD82 (multiple-pass), glypican-1 (lipid-anchor, GPI-anchor), TSG101 (peripheral), flotillin-1 (peripheral), EHD4 (peripheral), and moesin (peripheral). EXO1 also accommodates tryptic peptides from two abundant serum proteins mentioned above, albumin and α-2-macroglobulin. The second QconCAT (EXO2) includes tryptic peptides from six molecular chaperones: DNAJA1 (hDj-2), DNAJB1 (Hsp40), HSP90AA1, HSP90AB1, HSPA8 (Hsc70), and PTGES3 (p23). Molecular chaperones are not necessarily strictly EVs proteins. However, molecular chaperones are predisposed to strong protein–protein interactions and commonly found in EVs preparations (ExoCarta, http://exocarta.org/index.html).
The design of QconCATs included six amino acid long extensions from their natural sequences on both sides of the peptides.15,18,19 To prevent oxidation and disulfide bond formation, the methionine and cysteine residues in the extension sequences were replaced with isoleucine and alanine residues, respectively. The amino acid sequences of EXO1 and EXO2 QconCATs are shown in Figure S1 (Supporting Information). 15N-Labeled EXO1 and EXO2 were expressed in E. coli with higher than 99% isotope incorporation (Table S2, Supporting Information) and purity of final QconCAT preparations was considered higher than 95% based on SDS-PAGE (Figure S2, Supporting Information). These preparations were used as internal standards in MRM assay.
Quantification of Targeted Proteins in EVs(208K) and EVs(SEC)
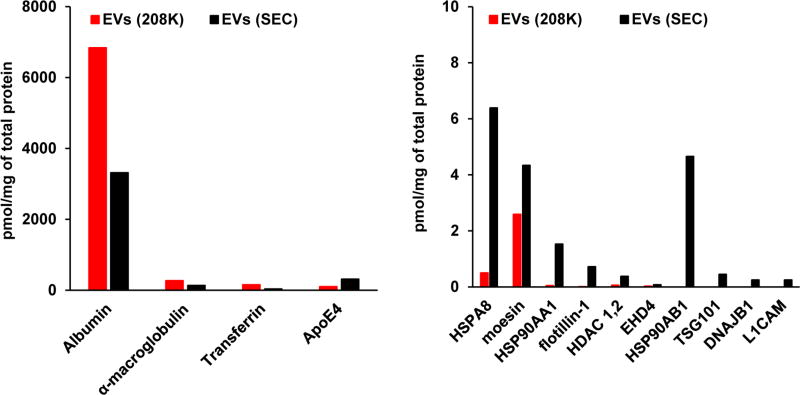
In addition to proteins covered by EXO1 and EXO2 internal standards, we also measured transferrin, apolipoprotein E (apoE) and histone deacetylase isoform 1 and 2 (HDAC 1,2). The generation of 15N-labeled internal standards and development of MRM assay for these three proteins was described before.23–25 These proteins were included because transferrin is a typical abundant serum protein, apoE was reported as an EVs-associated regulatory protein,26 and there are reports supporting the presence of histones in EVs.27,28 The data in pmol of targeted protein per mg of the total protein in EVs(208K) and EVs(SEC) representative preparations are shown in Figure 3. Since the dynamic range of targeted proteins is approximately 105, the data for a representative experiment are shown in two separate groups with different y-axis scales. It can be obviously seen that the presence of abundant serum proteins, such as albumin, α-2-macroglobulin, and transferrin, decreased in EVs(SEC) in comparison to EVs(208K). At the same time, the presence of EVs proteins (apoE, HSPA8, moesin, HSP90AA1, flotillin-1, HDAC 1,2, and EHD4) increased. Some proteins (HSP90AB1, TSG101, DNAJB1, and L1CAM), for which quantification in EVs(208K) was not possible, show reasonable measurements in EVs(SEC).
Figure 3.
Quantification of targeted proteins in EVs(208K) and EVs(SEC). Since the dynamic range protein concentrations covers 105, the data for a representative experiment are shown in two bar graphs with different y-axis scale.
Comparison of Precipitation-Based Protocols with Basic Protocol
Table 1 summarizes the quantitative data received by MRM comparison of EVs(208K) to EVs obtained by four different precipitation protocols. The scheme depicting all of these protocols is shown in Figure S3 (Supporting Information) and details are described in the Experimental Section. Here, it is important to emphasize that all of four precipitation protocols includes final ultracentrifugation at 208 K. It means that comparing data in Table 1 addresses the basic question whether precipitation protocols in-front of ultracentrifugation improve purity of the EVs preparation or not.
Table 1.
Comparison of Precipitation-Based Protocols with Basic Protocol
| protein (pmol/mg total protein)a | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| protein | Exospin (protocol #1) | Exospin (protocol #2) | total Exo (protocol #3) | total Exo (protocol #4) | EVs (208 K) | EVs (SEC) |
| albumin | 3163.0 ± 817.5 | 1592.0 ± 284.8 | 1119.0 ± 1.4 | 603.0 ± 222.5 | 6839.0 ± 177.4 | 3316.0 ± 40.3 |
| transferrin | 109.6 ± 15.3 | 22.9 ± 1.9 | 25.4 ± 4.9 | 32.8 ± 5.7 | 154.2 ± 0.01 | 35.1 ± 0.1 |
| α-2-macroglobulin | 4038.0 ± 254.5 | 6529.0 ± 504.2 | 1132.0 ± 56.6 | 676.0 ± 119.3 | 270.0 ± 18.0 | 133.5 ± 7.9 |
| apoE | 79.2 ± 5.8 | 121.9 ± 7.4 | 81.8 ± 7.0 | 480.9 ± 28.9 | 102.8 ± 0.03 | 314.0 ± 0.1 |
| HSPA8 | 0.12 ± 0.05 | 0.11 ± 0.03 | 0.08 ± 0.02 | 0.09 ± 0.05 | 0.50 ± 0.02 | 6.39 ± 2.4 |
| moesin | 0.67 ± 0.20 | 0.76 ± 0.16 | 0.49 ± 0.17 | 0.54 ± 0.31 | 2.59 ± 0.12 | 4.34 ± 0.10 |
| HSP90AA1 | 0.05 ± 0.07 | 1.53 ± 0.05 | ||||
| EHD4 | 0.03 ± 0.04 | 0.08 ± 0.01 | ||||
| flotillin-1 | 0.02 ± 0.05 | 0.72 ± 0.02 | ||||
| HDAC 1,2 | 0.06 ± 0.03 | 0.38 ± 0.03 | ||||
| HSP90AB1 | 4.66 ± 0.11 | |||||
| TSG101 | 0.39 ± 0.01 | |||||
| DNAJB1 | 0.25 ± 0.20 | |||||
| L1CAM | 0.25 ± 0.13 | |||||
MRM measurements were normalized to the total protein in each sample. Exospin Blood kit was from Cell Guidance Systems and total exosome isolation (from serum) kit was from Invitrogen. Protocols #1 through #4 are summarized in Figure S3 (Supporting Information). All experiments were performed in duplicate with three replicate injections to assess error and reproducibility. Data is represented as the mean ± SD.
For two abundant serum proteins, albumin and transferrin, precipitation protocols in-front of ultracentrifugation provides for better EVs preparations than ultracentrifugation alone. Albumin levels range from 603 to 3163 pmol/mg with precipitation and show a high level (6839 pmol/mg) without precipitation. However, data for α-2-macroglobulin shows the opposite tendency and ultracentrifugation alone results in better purity EVs preparation with 270 pmol/mg versus higher values measured in all four protocols that include precipitation. This observation points to the fact that precipitation is double-acting to serum proteins. It can remove some serum proteins from EVs preparation while simultaneously enriching EVs preparation with other ones. In terms of EVs proteins, inclusion of precipitation in-front of ultracentrifugation did not result in noticeable improvement compared to the basic protocol. Levels for those proteins, which we were able to quantify in precipitation protocols (apoE, HSPA8, moesin, and HSP90AB1) were lower than in ultracentrifugation alone. Other proteins measured in centrifugation alone (EHD4, flotillin-1, and HDAC 1,2) were not detected in protocols with precipitation. It can possibly be explained by the lower yield of EVs in protocols with precipitation and/or by interference of residual polymer with mass spectrometry analysis of low abundance proteins. Table 1 also includes data for EVs(SEC) from Figure 3, which obviously favor a combination of ultracentrifugation with SEC over combination of ultracentrifugation with precipitation.
CONCLUSIONS
We have demonstrated that QconCATs comprised of peptides for quantification of EVs and non-EVs proteins are suitable internal standards for assessing purity of EVs preparations in MRM assay. QconCAT standards can be expanded to include additional EVs and non-EVs proteins for more complete characterization. Additionally, differential comparison of various EVs subtypes could be performed using this MRM assay. This assay can be used for potential round robin interlaboratory comparison of EVs isolation techniques aiming to develop best practice recommendations. As a first step, we have evaluated use of polymer-based precipitation techniques and did not find a convincing rationale for their applications in EVs isolation.
Supplementary Material
Acknowledgments
The authors thank Dr. Meiyao Wang for expression and purification of 15N-labeled QconCAT for transferrin and full-size apoE4. Certain commercial materials, instruments, and equipment are identified in this manuscript in order to specify the experimental procedure as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that the materials, instruments, or equipment identified are necessarily the best available for the purpose.
Footnotes
ASSOCIATED CONTENT
- Additional information as noted in text (PDF).
The authors declare no competing financial interest.
References
- 1.Van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Pharmacol. Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 2.Raposo G, Stoorvogel W. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andaloussi SEL, Mager I, Breakefield XO, Wood MJA. Nat. Rev. Drug Discovery. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 4.Abramowicz A, Widlak P, Pietrowska M. Mol. BioSyst. 2016;12:1407–1419. doi: 10.1039/c6mb00082g. [DOI] [PubMed] [Google Scholar]
- 5.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-‘t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, Hochberg F. J. Extracell. Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Thery C. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardiner C, Di Vizio D, Sahoo S, Thery C, Witwer KW, Wauben M, Hill AF. J. Extracell. Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor DD, Zacharias W, Gercel-Taylor C. Methods Mol. Biol. 2011;728:235–246. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez ML. Methods Mol. Biol. 2014;1182:145–170. doi: 10.1007/978-1-4939-1062-5_13. [DOI] [PubMed] [Google Scholar]
- 10.Rekker K, Saare M, Roost AM, Kubo AL, Zarovni N, Chiesi A, Salumets A, Peters M. Clin. Biochem. 2014;47:135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Zlotogorski-Hurvitz A, Dayan D, Chaushu G, Korvala J, Salo T, Sormunen R, Vered M. J. Histochem. Cytochem. 2015;63:181–189. doi: 10.1369/0022155414564219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanchi Ravi R, Khosroheidari M, DiStefano JK. J. Visualized Exp. 2015;95:51158. doi: 10.3791/51158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deregibus MC, Figliolini F, D’Antico S, Manzini PM, Pasquino C, De Lena M, Tetta C, Brizzi MF, Camussi G. Int. J. Mol. Med. 2016;38:1359–1366. doi: 10.3892/ijmm.2016.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pratt JM, Simpson DM, Doherty MK, Rivers J, Gaskell SJ, Beynon RJ. Nat. Protoc. 2006;1:1029–1043. doi: 10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Turko IV. TrAC, Trends Anal. Chem. 2014;57:1–5. [Google Scholar]
- 16.Anderson L, Hunter CI. Mol. Cell. Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Liebler DC, Zimmerman LJ. Biochemistry. 2013;52:3797–3806. doi: 10.1021/bi400110b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung CSF, Anderson KW, Wang M, Turko IV. Anal. Chem. 2015;87:1097–1102. doi: 10.1021/ac503697j. [DOI] [PubMed] [Google Scholar]
- 19.Bauer Scott K, Turko IV, Phinney KW. Anal. Chem. 2015;87:4429–4435. doi: 10.1021/acs.analchem.5b00331. [DOI] [PubMed] [Google Scholar]
- 20.Liao W-L, Turko IV. Anal. Biochem. 2008;377:55–61. doi: 10.1016/j.ab.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 21.Cheung CSF, Anderson KW, Patel PM, Cade KL, Phinney KW, Turko IV. Sci. Rep. 2017;7:42497. doi: 10.1038/srep42497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuana Y, Levels J, Grootemaat A, Sturk A, Nieuwland R. J. Extracell. Vesicles. 2014;3:23262. doi: 10.3402/jev.v3.23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmerman TA, Wang M, Lowenthal MS, Turko IV, Phinney KW. Anal. Chem. 2013;85:10362–10368. doi: 10.1021/ac402326v. [DOI] [PubMed] [Google Scholar]
- 24.Wang M, Chen J, Turko IV. Anal. Chem. 2012;84:8340–8344. doi: 10.1021/ac3018873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson KW, Chen J, Wang M, Mast N, Pikuleva IA, Turko IV. PLoS One. 2015;10:e126592. doi: 10.1371/journal.pone.0126592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Niel G, Bergam P, Di Cicco A, Hurbain I, Lo Cicero A, Dingli F, Palmulli R, Fort C, Potier MC, Schurgers LJ, Loew D, Levy D, Raposo G. Cell Rep. 2015;13:43–51. doi: 10.1016/j.celrep.2015.08.057. [DOI] [PubMed] [Google Scholar]
- 27.Geis-Asteggiante L, Dhabaria A, Edwards N, Ostrand-Rosenberg S, Fenselau C. Int. J. Mass Spectrom. 2015;378:264–269. doi: 10.1016/j.ijms.2014.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiera G, Di Liegro CM, Pileo V, Colletta O, Fricano A, Cancemi P, Di Gara G, Di Liegro I. Int. J. Oncol. 2016;49:1807–1814. doi: 10.3892/ijo.2016.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.