Abstract
Long interspersed element-1 (LINE-1 or L1) is a transposable element with the ability to self-mobilize throughout the human genome. The L1 elements found in the human brain is hypothesized to date back 56 million years ago and has survived evolution, currently accounting for 17% of the human genome. L1 retrotransposition has been theorized to contribute to somatic mosaicism. This review focuses on the presence of L1 in the healthy and diseased human brain, such as in autism spectrum disorders (ASD). Throughout this exploration, we will discuss the impact L1 has on neurological disorders that can occur throughout the human lifetime. With this, we hope to better understand the complex role of L1 in the human brain development and its implications to human cognition.
Keywords: LINE-1, Retrotransposition, Brain, Neurological Disorders, Autism
Introduction
The invention of the wheel, the landing on the moon, the ability to hypothesize a theory of evolution- is there a single component of the human brain that has allowed us to make all of these great feats? Although the human brain does have slightly larger physical characteristics such as size of cerebral cortex (Hofman 1988; Azevedo et al. 2009; Herculano-Houzel 2009, 2012) and surface area (Hofman 2014) when compared to other species of primates, a new wave of research has started exploring the genetic contribution to the uniqueness of the human brain. The evolution towards the present human brain has been a series of mutations and natural selection. This review will focus on the role of transposable elements, specifically L1 retrotransposons that contributed to this selection and its impact on the human brain, health, and disease throughout a single lifetime. Based on the current data, we postulate that L1 elements can affect the formation of brain networks, leading to alterations in cognition and behavior, such as observed in Autism Spectrum Disorders (ASD) or in late-onset conditions.
Transposable Elements in the Human Genome
Transposable elements (TEs) are colloquially known as “jumping genes” because of their ability to move, or transpose, within a genome. TEs were first discovered in maize by Barbara McClintock in the 1940s (McClintock 1956). At this time, the idea of mobile DNA was novel and much of the scientific community still believed DNA was stagnant, however, the progression towards the idea of a more dynamic genome has taken precedence and is widely accepted now. TEs are found in all living organisms, but the percentage they occupy within a genome and their ongoing activity vary. In humans specifically, TEs contribute to approximately 45% of the genome (Kazazian 2004).
TEs are categorized into two major classes: DNA transposons and retrotransposons (Goodier and Kazazian 2008). DNA transposons are found to be active in many plants and lower organisms, but the ability to transpose in humans was evolutionarily lost due to mutation. Indeed, DNA transposons only make up about 3% of the human genome (Lander et al. 2001). DNA transposons are often described to replicate through a “cut and paste” mechanism (Munoz-Lopez and Garcia-Perez 2010; Craig 2002). By encoding the enzyme transposase, DNA transposons are removed, or “cut”, from one location of the genome and reintegrated, or “pasted” into a new location. Retrotransposons instead work through a “copy and paste” mechanism, which undergoes an RNA intermediate in the process of amplification (Moran and Gilbert 2002). The majority of TEs found in humans and mice are categorized under retrotransposons. Retrotransposons can further be divided by the presence or absence of long terminal repeat sequences (LTR), falling either into the category of LTR or non-LTR retrotransposons respectively. LTR retrotransposons are also known as endogenous retroviruses (ERVs) due to similar structure to simple retroviruses (Rowe and Trono 2011; Maksakova et al. 2008). LTR retrotransposons account for 8% of the human genome (Mager and Stoye 2015), but their activity in our genome is uncertain and is still continuously researched and discussed.
The non-LTR retrotransposons, on the other hand, are known to be active within the human genome (Mills et al. 2007). Non-LTR retrotransposons can further be classified into non-autonomous or autonomous depending if their coding sequence has the enzymatic machinery to mobilize themselves throughout the genome (Lander et al. 2001). Alu and SVA retrotransposons, non-autonomous elements, comprise approximately 10% of the human genome, while long interspersed element-1 (LINE-1 or L1s) are autonomous transposons and contribute to approximately 17% (Mills et al. 2007). L1s are the only known class of autonomous retrotransposons to be active. In 2003, Brouha and colleagues conducted a study where they determined 44% out of 90 L1s with intact open reading frames (ORFs) were polymorphic across the human population. After determining their capability to mobilize through cultured cell assays, termed as retrotransposition, they determined there were six highly active L1s, termed “hot L1s”, that comprised 84% of the total retrotransposition capability observed. Five full-length L1s are known to be involved in disease-causing insertions (Divoky et al. 1996; Schwann et al. 1998; Brouha et al. 2002; Dombroski et al. 1991; Holmes et al. 1994). The authors discovered that these five L1s are very active in comparison to the cloned L1s with intact ORFs, suggesting that these hot L1s are responsible for the retrotransposition occurring in humans, possibly playing a role in predisposition to certain disorders.
Where LINE-1 Elements Impact Humans
The first reported observation of a L1 insertion was in 1988, when Haig Kazazian and colleagues investigated a patient with hemophilia A, who had no familial history of the disorder. Upon investigation, they observed a new exonic L1 insertion in X-linked gene factor VIII, suggesting new L1 insertions in cell types where the next generation can inherit genetic information (i.e. germ cells in early embryogenesis) (Kazazian et al. 1988; Kazazian and Moran 2017; Richardson et al. 2017). Since then, examples of human genetic disorders caused by de novo L1 insertions continue to accumulate, and more than 100 cases have been shown to cause heritable diseases, such as haemophilia, β-thalassaemia, Duchenne muscular dystrophy, cystic fibrosis, Apert syndrome, neurofibromatosis and cancer (Belancio et al. 2009; Cordaux and Batzer 2009; Chen et al. 2005). However, recent research has determined that retrotransposition is not only limited to the germline. L1 expression and activity has been observed in several human tumors (Iskow et al. 2010; Lee et al. 2012; Shukla et al. 2013; Solyom et al. 2012) and in selected tissues of the human brain (Baillie et al. 2011; Coufal et al. 2009; Evrony et al. 2012; Muotri et al. 2005; Evrony 2015; Upton et al. 2015) as well, concluding that somatic retrotransposition is more prevalent than previously anticipated. Intriguingly, a new retrotransposition insertion can not only be mutagenic by disrupting a coding sequence. L1 insertions can impact the expression of nearby genes by generating new splice sites, promoters, adenylation signals and transcription factor-binding sites that can reorganize gene expression. L1s can also contribute to genetic instability by generating target site deletions, insertions of flanking DNA, recombination with other retrotransposons, and the possible generation of chromosomal inversions and interchromosomal translocations (Goodier and Kazazian 2008; Beck et al. 2011; Kazazian 2004; Hancks and Kazazian 2012). In addition, inactive L1 elements (unable to mobilize) can contain mutations in their sequence and express truncated proteins causing varying levels of cell toxicity and DNA damage (Kines et al. 2014). In summary, new TE insertions can profoundly affect the human genome in multiple ways, thus this chapter will focus on the association between L1 activity and a complex tissue as the human brain.
In order to understand how L1 elements arrived to the human brain, we must first look at its presence in evolution. During the Eocene, the most ancient subfamilies of L1, HERV, and Alu were hypothesized to first appear in early prosimians, which had relatively small brains when compared to their body mass (Gilbert et al. 2005; Zilles et al. 1989; Linker et al. 2017). About 25 million years ago, additional L1 and Alu subfamilies appeared and SVAs first originated during the split between hominoids and cercopithecoids (Schrago and Russo 2003; Batzer and Deininger 2002; Khan et al. 2006; Wang et al. 2005). Also at this time, the frontal cortex increased in size (Linker et al. 2017). Approximately 5 million years ago, after the Pan-Homo split, the primate brain size dramatically increased (Semendeferi et al. 2002) and in Homo, the subfamilies L1Hs, AluYa5, AluYb8, and SVA were the most active, and are still the most active in the human genome today (Cordaux et al. 2006; Ewing and Kazazian 2010; Xing et al. 2009; Linker et al. 2017). Interestingly, human restriction factors have evolved in parallel with transposable elements in order to repress its activity (Castro-Diaz et al. 2014; Jacobs et al. 2014), however, these elements were still able to escape this repressive mechanism through mutation. Additionally, a study conducted by Ramsay and colleagues (2017) used an induced pluripotent stem cell (iPSC) model to generate RNA-seq data for four different primate species- human, chimpanzee, gorilla, and rhesus. The authors reported 30% of TE instances found in human iPSCs also had orthologous TE instances expressed in chimpanzee and gorilla (Ramsay et al. 2017). Although we have established L1 insertions in the evolution of the primate brain, retrotransposition in somatic cells is a random phenomenon and cannot be evolutionary mapped in the same way. However, these insertions can be studied in different times of a human individual’s life. First, we must look to understand the mechanism of how L1 can mobilize within a human.
Retrotransposition Mechanism of Active LINE-1 Elements
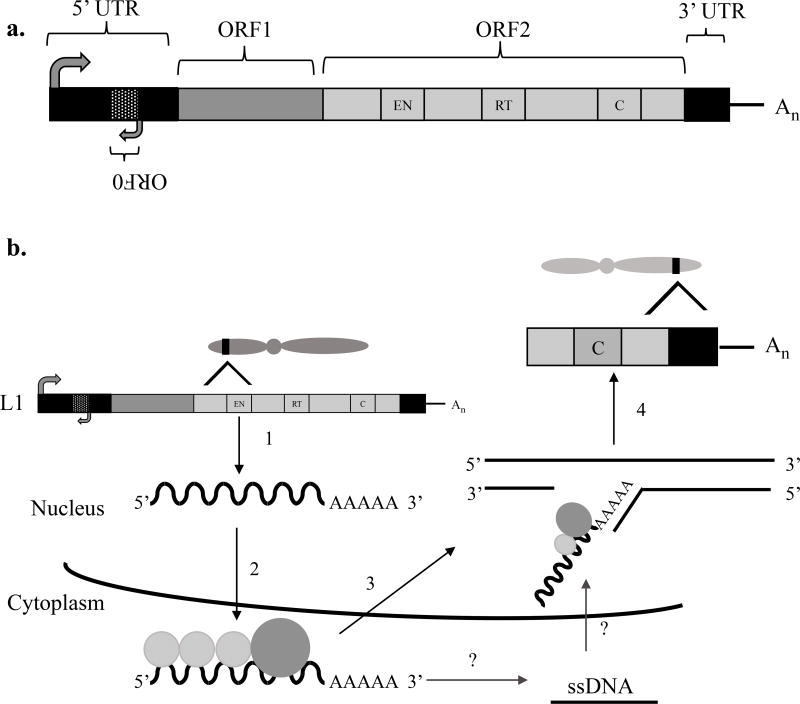
As mentioned previously, retrotransposons undergo a copy and paste mechanism in order to reintegrate into new genomic locations. With more than 500,000 L1 copies in the genome (Booth, Ready, & Smith, 1996; Lander et al., 2001), more than 99% are unable to move due to 5’ truncation, rearrangement, or mutation (Goodier and Kazazian 2008; Dombroski et al. 1991; Scott et al. 1987; Beck et al. 2010). However, the remaining 80–100 L1s that are mobile are called retrotransposition competent L1s (RC-L1s) (Beck et al. 2010; Brouha et al. 2003). These RC-L1s contain a promoter within its 5’ untranslated region (UTR) with sense and antisense activities (Swergold 1990; Speek 2001; Beck et al. 2011; Macia et al. 2011) and ends with a short 3’ UTR and a poly(A) tail (Figure 1a) (Dombroski et al. 1991; Scott et al. 1987). RNA can be transcribed and transported into the cytoplasm to encode the open reading frame-1 and open reading frame-2 proteins (ORF1p and ORF2p, respectively) (Moran et al. 1996) (Figure 1b). ORF1 encodes an RNA binding protein with nucleic acid chaperon activity (Hohjoh and Singer 1997b, a; Khazina and Weichenrieder 2009; Martin and Bushman 2001). ORF2 produces a protein with enzymatic activities, two of which include an apurinic/apyrimidinic (AP)- like endonuclease (EN) (Martin et al. 1995; Feng et al. 1996) and a reverse transcriptase (RT) (Mathias et al. 1991). With the exception of the 3’ UTR in certain cell types (Moran et al. 1996), all of these components are necessary in RC-L1 mobilization (Figure 1b). More recently, Denli and colleagues (2015) identified an ORF in the human antisense L1 5’ UTR. Because of its position before ORF1 and ORF2, it has been named ORF0 (Figure 1a). This primate-specific antisense ORF can be fully encoded by approximately 781 loci in the human genome (Denli et al. 2015; Macia et al. 2017a). Researchers observed an increase in L1 mobility when ORF0 was overexpressed in HEK293T cells and human embryonic stem cell (hESC)-derived neural progenitor cells (NPCs). This suggests that ORF0’s ability to enhance L1 activity could contribute to somatic variation (Denli et al. 2015).
Figure 1.
a. Structure of active human L1 element. The relative positions of the 5’ untranslated region (5’ UTR), the open reading frames: ORF0, ORF1, and ORF2, 3’ untranslated region (3’ UTR), and poly A tail are shown. EN denotes endonuclease, RT denotes reverse transcriptase, and C denotes cysteine-rich domain (all encoded by ORF2). The arrows located on the 5’ UTR symbolize the sense and antisense promoters. More details provided in the main text. b. Model of the retrotransposition cycle. 1) Generation of full-length L1 mRNA using the sense promoter in 5’ UTR. 2) mRNA is transported to the cytoplasm, where ORF1p and ORF2p are translated. ORF1p and ORF2p bind to the L1 mRNA to form a ribonucleotide particle (RNP) through a process called cis-preference. 3) Return to the nucleus (Mechanism is still undetermined). Once in the nucleus, the ORF2 EN activity will make a break in the DNA, exposing a free 3’OH in the bottom strand, serving as a template for the RT to generate the first cDNA strand linked to the genome. 4) This mechanism called target-primed reverse transcription (TPRT) will allow the integration of a new L1 copy. More detail provided in the main text.
Retrotransposition first begins with the generation of a full-length L1 mRNA using the sense internal promoter within its 5’ UTR (Figure 1b). The RNA is transported into the cytoplasm where translation occurs to encode ORF1p and ORF2p (Alisch et al. 2006; Dmitriev et al. 2007). Once translated, the L1-encoded proteins bind to the L1-encoding mRNA through a process known as cis-preference (Wei et al. 2001). It is believed that this binding forms a ribonucleoprotein particle (RNP) and serves as an essential intermediate throughout the retrotransposition process (Figure 1b) (Doucet et al. 2010; Goodier et al. 2007; Kulpa and Moran 2005; Martin 1991). The L1-RNP is imported back into the nucleus where a new L1 copy is integrated through target-primed reverse transcription (TPRT) (Luan et al. 1993; Cost et al. 2002). During TPRT, the EN of the L1-ORF2p cleaves the bottom strand of the DNA at a degenerate consensus sequence (5’-TTTT/A-3’) (Jurka 1997). With this nick, a 3’ OH is exposed (Luan et al. 1993; Cost et al. 2002) and used as a primer for the RT of L1-ORF2p (Monot et al. 2013), where the L1 mRNA serves as a template (Luan et al. 1993) for the generation of cDNA (Figure 1b). The TPRT model may have alternative mechanisms with the possibility of RT activity occurring also in the cytoplasm (Telesnitsky and Goff 1997; Thomas et al. 2017; Garcia Perez and Alarcon-Riquelme 2017). Existing research has shown that the configuration of four T nucleotides at the end of the target DNA is the sequence that will give rise to the highest efficiency of initiation of reverse transcription process (Monot et al. 2013). Although unclear, a process similar to the one just described occurs with the top strand of DNA, generating yet another L1 insertion in the genome. While it seems as though many L1 insertions are produced, the majority are 5’ truncated (Grimaldi et al. 1984; Goodier and Kazazian 2008). This truncation prevents the formation of a functional RNP intermediate and is therefore incapable of mobilization. Although L1s are major drivers of genome evolution, these L1-driven mutagenic events can also cause genetic disorders, thus host restriction mechanisms that control the accumulation of new insertions are essential on the stability of mammalian genomes.
Regulation of L1 activity in the mammalian genome
The L1 retrotransposition cycle as described is very complex and entails many crucial steps, however, mammalian evolution has tried to prevent the occurrence of retrotransposition at different points of the cycle. Methylation is one host factor that can inhibit L1 expression. L1 promoters include a CpG island which has the possibility of being methylated, consequently inhibiting expression (Coufal et al. 2009; Bourc'his and Bestor 2004; Thayer et al. 1993). DNA methyltransferase-3 like (DNMT3L) plays a role in DNA-methylation of certain retroelements in primordial germ cells (Bourc'his and Bestor 2004; Macia et al. 2015). The early embryogenesis stage is associated with hypomethylation, possibly relating to the accumulation of L1 copies during this period (Munoz-Lopez et al. 2011). This hypomethylation of L1 promoters serves as a characteristic of mouse, human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs) (Garcia-Perez et al. 2007; Munoz-Lopez et al. 2011; Shen et al. 2006; Wissing et al. 2012; Macia et al. 2011). Further along in the L1 retrotransposition cycle, splicing has been shown to regulate L1 mobilization in somatic tissues and cell cultures by generating non-functional LINE-1 transcripts (Belancio et al., 2006; Perepelitsa-Belancio and Deininger, 2003). In addition, PIWI proteins, which are highly conserved, interact with PIWI-interacting small RNAs (piRNAs) for TE RNA degradation (Heras et al. 2014). piRNAs are predicted to work in unison with DNA methylation, with the piRNA pathway generating DNA methylation patterns on repeated elements in mice (Aravin et al. 2007; Aravin et al. 2008; Newkirk et al. 2017; Heras et al. 2014; Xiol et al. 2014; Brennecke et al. 2007). Also, the Microprocessor (Drosha-DGCR8), which is required for microRNA biogenesis, has been shown to also regulate L1 mRNAs (Heras et al. 2013). Finally, post-translational control mechanisms have been proposed to restrict the ability of L1 to mobilize. Host factors like ribonuclease L (RNaseL), described to degrade L1 RNA produced by the sense and antisense L1 promoters (Yang and Kazazian 2006; Zhang et al. 2014); TEX19.1, which can directly interact with the L1-ORF1p in mouse embryonic stem cells, stimulating its degradation (MacLennan et al. 2017); APOBEC3 (A3), a cytidine deaminase enzyme, capable of limiting L1 retrotransposition through deaminating and editing L1 sequences (Schumann 2007; Wissing et al. 2011; Marchetto et al. 2013; Richardson et al. 2014b), are a few examples shown to interfere with later steps of the retrotransposition cycle. The majority of host factors that control L1 retrotransposition is yet to be discovered, however research in this area continues in order to better understand the mechanisms of regulation and the coevolution with humans. L1 retrotransposition can impact the developing human brain at different stages of human life (infancy, childhood, adulthood, aging) in health and disease (McConnell et al. 2017).
L1, brain and disease
L1 mobilization is active in healthy human brain (Muotri et al. 2005; Coufal et al. 2009; Baillie et al. 2011; Evrony et al. 2012; Goodier 2014). Indeed, several studies in healthy post-mortem brains and hESC-derived neural cells have provided evidence of accumulation of L1 insertions. There is currently an array of techniques in the field to detect L1 expression in neuronal lineage. The first method, used by Moran and colleagues (1996), utilized an active engineered L1 element tagged with a reporter cassette that was only activated after a complete round of retrotransposition. Since then, retrotransposition assays have often been used as a reliable method to detect L1 retrotransposition (Garcia-Perez et al. 2007, 2010; Wissing et al. 2011). A cell culture-based retrotransposition assay consists of a complete human RC-L1 tagged with a reporter cassette containing an antisense copy of a specific marker, which is disrupted by an intron. The RC-L1 and the intron are in the same transcriptional orientation, but the reporter cassette is in the opposite transcriptional orientation (Moran et al. 1996). The reporter gene is flanked by a heterologous promoter and a polyadenylation signal. This arrangement allows L1 transcripts to splice the intron, and due to the antisense orientation, expression of the marker can only be detected when the transcript is reverse transcribed, integrated, and expressed by the heterologous promoter (Moran et al. 1996). The different reporter cassettes used in the detection of L1 mobilization are diverse, from fluorescent protein, luciferase or antibiotic selection genes. For example, L1-EGFP (enhanced green fluorescent protein) indicator cassettes have also been used to visually detect L1 retrotransposition through EGFP-positive cells in adult rat and human NPCs (Muotri et al. 2005), hESCs (Garcia-Perez et al. 2007) and in transgenic L1-EGFP mice (Muotri et al. 2005), where the EGFP-positive cells are only seen after L1 retrotransposition is completed. In 2009, Coufal and colleagues isolated NPCs from human fetal brain stem cells and additionally derived NPCs from hESCs. Using these two cell types, the authors performed retrotransposition assays and detected L1 retrotransposition events in both conditions (Coufal et al. 2009). Furthermore, through quantitative polymerase chain reaction (qPCR), the authors observed a significantly higher amount of L1 ORF2 copy number in various regions of healthy adult human brain sample when compared to the liver and heart, indeed suggesting more L1 activity in the brain. Notably, Coufal and colleagues consistently noticed higher levels of L1 ORF2 copy number in the adult hippocampus when compared to other regions of the brain, providing evidence that L1 elements could be active as early as the formation of the central nervous system. Although the specific L1 integration sites within the genome cannot be accurately determined (Kempen et al. 2017), the striking difference between the hippocampus and other areas of the brain and body demonstrates the contribution L1 has in neuronal diversity and in somatic mosaicism in general (Coufal et al. 2009; Richardson et al. 2014a). In 2017, Macia and colleagues explored L1’s ability to retrotranspose in somatic cells as mesenchymal stem cells (MSCs), hematopoietic stem cells (HSCs) and NPCs, as well as in mature human neuronal cells. The authors were able to detect higher levels of L1 mRNA and L1 ORF1p in NPCs when compared to other somatic cells (Macia et al. 2017b; Garcia-Perez et al. 2016). The majority of the cells in the human brain are mature neuronal cells (Tang et al. 2001). Thus, the researchers then focused on L1 retrotransposition in mature nondividing cells. In order to do this, a plasmid containing an RC-L1 (Brouha et al. 2002) was transfected into differentiating NPCs at day 31(Macia and Muotri 2017), showing efficient L1 retrotransposition in these nondividing mature neuronal cells (Macia et al. 2017b). Although these L1-fluorescent reporter assays are convenient, Kempen and colleagues (2017) point out that the fairly large EGFP cassette must be reverse transcribed for fluorescence to ensue, making this method variable. Additionally, there is also the possibility of the EGFP promoter being silenced by the host genome (Garcia-Perez et al, 2010). Also, the use of retrotransposition assays in this field does not necessarily conclude that endogenous elements retrotranspose in the cell type or tissue, rather it allows researchers to determine if a particular cell type or tissue can support retrotransposition.
More recently, to subdue these disadvantages, studies have been utilizing high-throughput DNA sequencing to both detect and locate endogenous L1 variants, providing stronger evidence of L1’s role in somatic mosaicism. In 2011, Baillie and colleagues described a high-throughput method, named RC-seq, in order to detect L1 insertions in hippocampal and caudate nucleus samples of three healthy individuals. With the use of RC-seq, the authors identified somatic L1 insertions within protein-coding genes differentially expressed and active in the brain (Baillie et al. 2011). Researchers validated only a portion of their somatic candidate insertions using 5’ end nested PCR. Out of their 850 found full-length L1 insertions, they selected 14 L1 examples to validate, and all were confirmed. In 2012, Evrony and colleagues used single-neuron genome-wide sequencing to detect full-length L1 insertions. 300 single neurons originating from the cerebral cortex and caudate nucleus of three normal individuals were profiled, and although somatic L1 insertions were observed, only about 1.1 somatic insertions were found per neuron, and less than 0.6 unique somatic insertions were discovered per neuron. These results contradict the Coufal et al. (2009) study and instead suggest L1 does not contribute to neuronal diversity between the cortex and caudate (Evrony et al. 2012). However, out of the 81 insertions found, only five were validated. On the other hand, in 2015, Upton and colleagues utilized RC-seq for single cell analysis of hippocampal neuron and glial cell. The authors estimated 13.7 and 6.5 somatic L1 insertions in each cell type respectively (Upton et al. 2015), which displays a drastically different result from Evrony et al. (2012). Upon further investigation of the hippocampal L1 insertions, the authors observed enrichment in transcribed neuronal stem cell enhancers and hippocampus genes, suggesting retrotransposition in the brain and diversity of genomes between individual neurons (Upton et al. 2015). However, again validation of the L1 insertions was limited. In 2016, Erwin and colleagues performed single-cell and bulk sequencing methods to capture Somatic L1 Associated Variants (SLAVs) in frontal cortex and hippocampus samples from three healthy individuals. Single nuclei were isolated and amplified by multiple displacement amplification (MDA). Similar to Upton et al. (2015), the results of this study are in favor of L1 as a contributor to somatic mosaicism. In order to isolate somatic insertions from false positives, the authors did not take into account previously identified known non-reference germline loci and private germline insertion loci. Whole genome amplification often occurs near known L1 loci (Evrony et al. 2012; Upton et al. 2015), and the machine-learning approach used cannot detect proximity to know L1 loci, therefore the researchers did not consider candidates within 10 kb of known non-reference germline loci, germline L1HS, and PA2 and PA3, both of which are subfamilies of L1s. Chimeric DNA rearrangements, often generated when utilizing MDA (Zhang et al. 2006), have been shown infrequent for segments more than 10 kb apart, however SLAVs are still difficult to differentiate from a MDA chimera unless found and validated in more than one MDA amplified cell or in bulk (Lasken and Stockwell 2007). Indeed, confirmation of somatic L1 retrotransposition was found in neurons, glia, and progenitor cells and was extensively validated through PCR and Sanger sequencing. For validation, the authors used a PCR assay confirming the 3’ end of the L1 candidate using complementary primers to both the 3’ end of L1 and the flanking genomic sequence. They also performed another PCR assay using primers complementary to the 5’ and 3’ flanking sequences. This was important for validation because chimera formation often occurs when a displaced 3’ end primes to new templates (Lasken and Stockwell 2007). SLAVs not containing target site duplications were found and deemed somatic deletions that were independent of retrotransposition. The authors hypothesized for these SLAVs to occur primarily at germline cells. By overexpressing L1 in human hippocampal neural differentiation, researchers confirmed an increase in dsDNA damage at genomic L1 loci suggesting these genomic regions to be predisposed to somatic copy number variants (CNVs) in the brain and thus a heritable genetic contributor to somatic mosaicism (Erwin et al. 2016). Additionally, recent studies have suggested that some polymorphic L1 copies (Philippe et al. 2016) and some Alu elements (Payer et al. 2017) within individuals might be associated with an increased risk to certain human disorders. Although somatic mutations are not inherited, donor L1s, which cause de novo L1 insertions within somatic cells, can be passed through the germline and be a source of both germline and somatic mosaicism (Faulkner and Garcia-Perez 2017; Richardson et al. 2017; Scott et al. 2016). It has been shown that active donor L1s in somatic cells have the capability of predisposing an individual to specific diseases and cancers (Scott et al. 2016; Singer et al. 2010; Miki et al. 1992), but this is dependent on their region of mobilization (Brouha et al. 2003; Evrony 2015; Faulkner and Garcia-Perez 2017). In 2016, Carreira and colleagues utilized RC-seq and investigated L1 mutations in 14 glioblastoma multiforme (GBM) tumors. In four of these tumors, the authors discovered an endonuclease-independent polymorphic L1 insertion, two L1-associated rearrangements and an Alu-Alu recombination event in close proximity to an L1, however after sequencing the L1 integration sites, there was no found tumor-specific, endonuclease-dependent L1 insertions (Carreira et al. 2016). In sum, the study of L1 activity with new technologies has allowed us to find L1 activity in the healthy brain, but is also well studied among brain disorders that span throughout a human’s lifetime.
1- The human infant brain
L1 and Aicardi-Goutieres Syndrome
Aicardi-Goutieres Syndrome (AGS) is a severe neuroinflammatory disorder that is inherited in an autosomal recessive pattern with an onset in early infancy (Crow et al. 2015) (Figure 2). AGS often arises when three-prime repair exonuclease 1 (TREX1) becomes mutated (Crow and Manel 2015). TREX1 is an anti-viral DNase that breaks down nucleic acids in the cytosol (Mazur and Perrino 1999; Richards et al. 2007). With this function, TREX1 prevents accumulation of single-stranded and double stranded DNA, in turn preventing type I interferon (IFN) associated inflammatory response (Crow and Rehwinkel 2009; Yang et al. 2007). With TREX1 function compromised, abnormal amounts of nucleic acid species in the cytosol can result in an unintended immune response.
Figure 2.
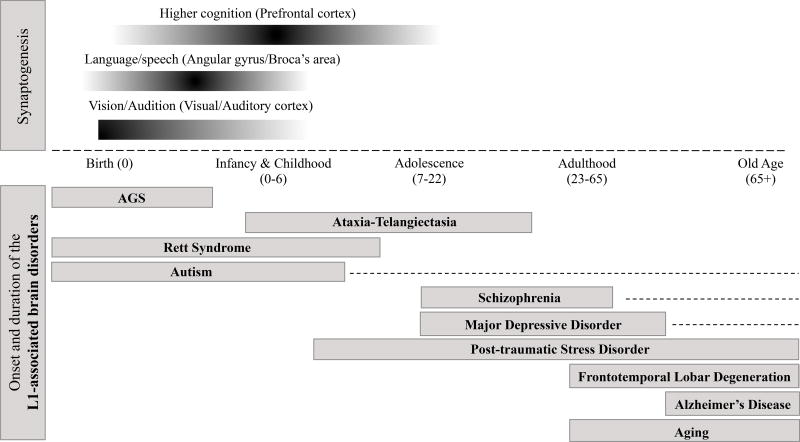
Timeline of impact of L1 in the human brain during a single lifetime. The top curves represent a timeline of major events occurring in brain development from conception to adulthood (adapted from Thompson and Nelson (2001)). The L1-associated neurodevelopmental, neurodegenerative and neurological disorders discussed in the main text are graphed with respect to the age of impact in a human lifetime.
In 2008, Stetson and colleagues were the first to recognize the relationship between TREX1 and L1 retrotransposons (Stetson et al. 2008). Curious about the source of the TREX1 DNA substrates that activate the interferon-stimulatory DNA (ISD) pathway, researchers purified, amplified, and cloned the endogenous TREX1 DNA substrates from TREX-1 knock out (KO) mice. They then compared the DNA recovered from control and TREX1-deficient mice hearts. The DNA collected from TREX1-deficient hearts was more abundant (32-fold increase), suggesting the KO heart extracts had substantially more DNA that the wild-type (WT) extracts. Additionally, DNA fragments derived from endogenous retroelements were highly detected in the deficient hearts when compared to wild-type. The authors recovered 25 retroelement sequences (consisting of LINE, LTR, and short interspersed elements (SINE)) from knockout samples, 12 of which were derived from L1 retrotransposons. Mapping between the ssDNA fragments to consensus L1 sequences showed that L1-derived DNA fragments mapped preferentially to the 3’ end of the consensus sequence, alluding to the majority of L1 copies that are 5’ truncated. The authors also analyzed L1 retrotransposition by co-transfecting an engineered active L1 element containing a neomycin resistance cassette (neor) in reverse orientation to the retroviral coding sequence, with and without a TREX1 expression vector. They used a human L1 element and a LTR-containing murine intracisternal type A particle (IAP) as their marked retroelements. When the marked element was transcribed into mRNA, reverse transcribed, and integrated into the genome, the neor gene became active. As a control, HeLa cells were transfected with IAP and formed neomycin-resistant colonies, with each colony representing a single retrotransposition event. The authors observed a reduction in IAP retrotransposition efficiency when they co-transfected with a TREX1 expression vector. An 80% reduction in L1 retrotransposition was also seen with an expression vector for TREX1. Lastly, the authors also focused on AGS-causing TREX1 mutations and their effect on IAP and L1 retrotransposition. Loss of function is observed in TREX1 R114H and TREX1 V201D mutations. The mutations, both which cause recessive AGS, did not affect IAP or L1 retrotransposition. When the TREX1 D200N catalytic mutant, which causes autosomal dominant AGS, was co-transfected, an increase in IAP retrotransposition efficiency was observed, but not for L1 retrotransposition. This was the first suggestion that DNA fragments derived from endogenous retroelements accumulate in TREX1-deficient samples.
A study conducted by Pokatayev and colleagues (2016) modeled AGS in mice by exploring the associated RNase H2 mutation. RNase H2 is the main contributor to ribonuclease H activity. It plays a role in removing ribonucleotides accidentally incorporated by DNA polymerases within DNA-DNA duplexes (Nick McElhinny et al. 2010; Reijns et al. 2012; Chon et al. 2013). A mutation in the catalytic A subunit results in a more severe disease phenotype when compared to the other subunits. In the presence of a mutation in the conserved glycine (G37S) on the RNase H2A subunit, an early onset of AGS occurs (Crow et al. 2006). When Glycine 37 is substituted with Serine in RNase H2, a reduction in RNase activity is observed (Rohman et al. 2008; Chon et al. 2009; Crow et al. 2006; Coffin et al. 2011). Researchers produced a mouse model that reflected AGS patients by knocking in both RNase H2 copies with G37S mutation, resulting in homozygous Rnaseh2aG37s/G37s mice, causing interferon stimulated gene (ISG) activation. Through measurement of ISG expression levels in mutated and control embryos, researchers determined the G37S mutation activates a DNA-sensing cGAS-STING pathway. When the protein cyclic GMP-AMP Synthase (cGAS) detects cytosolic DNA, it produces cGAMP leading to activation of the STING pathway. Additionally, the authors measured L1 DNA levels in the mice from cytosolic extract. They observed high levels of L1 DNA in the G37S-deficient embryos suggesting this mutation increases L1 DNA and consequently activates the cGAS-STING pathway.
Recently, Thomas and colleagues (2017) also investigated the relationship between L1 elements and type I IFN inflammation, but rather, in a human developmental model. Using the CRIPSR/Cas9 genome-editing system, authors generated three different cell lines, each with a distinct TREX1 mutation, and differentiated each cell line into NPCs, neurons, cortical organoids, and astrocytes. The authors also utilized two reverse-transcriptase inhibitors (RTis): Lamivudine (3TC) and Stavudine (d4T) that have been previously shown to inhibit L1 retrotransposition (Dai et al. 2011). Researchers extracted and sequenced extrachromosomal DNA from a TREX-1 deficient NPC line, its isogenic control, and the TREX-1 deficient line treated with 3TC/d4T. After sequencing, the authors detected an abundant amount of repetitive elements, more so, the TREX-1 deficient line contained an increase in L1 elements when compared to control. Particularly, the TREX-1 deficient line contained 70% more human-specific L1 (L1 Hs) when compared to control, but with 3TC/d4T treatment, the L1 and L1 Hs levels returned to near normal. Throughout the neuronal differentiation process of the TREX1-deficient lines, the authors observed a morphology relating to cell sickness and a downregulation of canonical neuronal markers not seen in control lines. By measuring an increase in cleaved caspase-3 (CC3) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cells, they determined the TREX1-deficient cells were experiencing neurotoxicity. 3TC/d4T treated neurons and organoids revealed a healthier appearance with lower CC3 and TUNEL-positive cells, suggesting the RTis rescued neurotoxicity. Similarly, the 3TC/d4T treatment also reduced expression of IFNb and IFNa13 levels in TREX-1 deficient astrocytes to near control, suggesting that L1 retroelements contribute a significant amount to intracellular DNA species and consequently stimulate type I IFN expression. Treatment with 3TC/d4T also reduced phosphorylated IRF3 levels to control and reduced three out of the six STING-dependent ISGS that were upregulated in TREX1-deficient astrocytes. Ultimately, these data propose IRF3 activation from cytosolic DNA induces both type I IFN and ISG upregulation in TREX1-deficient astrocytes.
AGS is also associated with SAMHD1 mutations (Dale et al. 2010; Rice et al. 2009; Thiele et al. 2010; Xin et al. 2011). SAMHD1 is a known inhibitor against certain retroviruses, including HIV and simian immunodeficiency virus (SIV) (Hrecka et al. 2011; Laguette et al. 2011). In 2013, Zhao and colleagues demonstrated that the SAMHD1 protein inhibits L1 retrotransposition using an L1-EGFP reporter assay in HEK293T cells. While examining AGS patients with SAMHD1 mutations, the authors observed a significant reduction of L1 inhibition in these individuals (Zhao et al. 2013). The researchers suggest that nuclear localization is necessary for SAMHD1-mediated L1 inhibition because the nucleus is where both L1 reverse transcription occurs and SAMHD1 localizes (Zhao et al. 2013). Through the use of qPCR and a L1 element amplification protocol (LEAP) assay, which assesses L1 ORF2p’s ability to reverse transcribe L1 RNA in vitro (Kopera et al. 2016; Kulpa and Moran 2006), the authors determined that SAMHD1 reduced ORF2p expression and ORF2-mediated endogenous reverse transcription of L1 RNA by 62% and 83%, respectively (Zhao et al. 2013). This provides evidence identifying L1 ORF2p and its reverse transcriptase as a target of SAMHD1.
L1 and Ataxia-Telangiectasia
Ataxia-telangiectasia (AT) is a progressive neurodegenerative disease with an onset between the ages of one and four, and a life expectancy typically into early adulthood (Gatti and Perlman 1993) (Figure 2). AT is caused by mutation in the ataxia telangiectasia mutated (ATM) gene, which encodes a serine/threonine kinase that is activated by double-stranded DNA breaks and senses cellular DNA damage (Shiloh 2001). Upon activation, DNA damage checkpoint and cell cycle arrest occurs. The DNA is either repaired or p53-mediated apoptosis ensues for the cell (Shiloh 2001). In 2011, Coufal and colleagues investigated the relationship between L1 retrotransposition and ATM deficiency using four different approaches: a mouse model, an embryonic/neuronal stem cell model, a cancer cell line model, and an examination of postmortem human brain tissue (Coufal et al. 2011). Researchers generated ATM KO mice containing an L1-EGFP transgene and observed an increase in the number of EGFP-positive cells in ATM KO mice when compared to controls, suggesting the ATM KO mice have an increase in retrotransposition events. The most striking increase was seen in the hippocampus. Thereafter, hESC-derived NPCs were generated with shRNAs knocking down ATM expression and subsequently reducing protein levels. Researchers observed a twofold increase in L1 retrotransposition in ATM-deficient NPCs. Turning to a parental human HCT116 colorectal cancer cell line and its isogenic derivatives that lacked the p53 gene or genes necessary for nonhomologous end-joining (NHEJ), the authors explored the role of ATM in L1 mobilization in a non-embryonic/non-neuronal cell type. They observed L1 retrotransposition in both p53-proficient and –deficient HCT116 cells, but reported a twofold increase in ATM knockdown p53-deficient HCT116 cells. Lastly, authors used qPCR to analyze the copy number of endogenous, human specific L1s in hippocampal sections from AT patients. An increase in ORF2 copy numbers in AT neurons was observed when compared to age- and sex-matched control patients, thereby again suggesting an increase in L1 retrotransposition in AT patients.
In all, these studies have highlighted disorders that show the impact L1 insertions have on the human infant brain, nevertheless the adult brain is still susceptible to L1-associated brain disorders.
L1, Autism and Rett Syndrome
Autism and the autism spectrum disorders (ASD) are the most severe developmental disabilities prevalent in today’s society (American Psychological Association, 2017). Affecting 1 in 68 children (Center for Disease Control, 2017), ASD affects all racial, ethnic, and socioeconomic groups. According to the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders, ASD is characterized by two main symptoms: deficits in both verbal and nonverbal social communication and repetitive behaviors. Criteria for diagnosis states that these symptoms must have occurred early in development and are measured in severity by the amount of support the individual needs. Autism, along with other early onset childhood neurological disorders, have been the focus of research and discussion within the scientific community (Figure 2).
The genesis of ASD still remains unknown, although studies suggests that autism develops from a combination of genetic and non-genetic, or environmental influences (Grabrucker 2012). Phenotypic characteristics of ASD are often shown to be heritable through family and twin studies (Sandin et al. 2014; Hallmayer et al. 2011; Folstein and Rutter 1977; Bailey et al. 1995; Ronald and Hoekstra 2011). With these genetic implications, genome-wide analyses have become popular in this research field to determine genetic variants. Among the genetic variants found, the most noteworthy discovery is the identification of protein-disrupting de novo copy-number variants (CNVs) (Szatmari et al. 2007; Sebat et al. 2007; Ronald and Hoekstra 2011; Pinto et al. 2010; Marshall et al. 2008; Levy et al. 2011; Itsara et al. 2010; Glessner et al. 2009; Bucan et al. 2009). The genetic variants were only detected in the affected child, not in the healthy parents, suggesting their appearance occurred in the germline. The most recognized CNVs in autism research are 16p11.2 deletions and 15q11-q13 duplications (Sanders et al. 2015). A recent study conducted in China performed a genome-wide CNV analysis in 546 subjects (343 parent and child trios) with ASD (Guo et al. 2017). Existing genome-wide studies have mainly occurred in primary European samples, making this particular study novel. In all, they detected an overall higher number of CNVs in patients with ASD when compared to controls. They confirmed the reoccurrence of the 15q11-13 de novo duplication CNV among their subjects and actually reported a higher frequency of occurrence when compared to primary European samples. Another study analyzed 5,193 individuals, 2,620 of those individuals were diagnosed with ASD, and the remaining were relatives (RK et al. 2017). The study identified 61 ASD risk-genes, reporting 54 de novo loss-of-function (LOF) mutations/missense mutations and seven maternally inherited LOF mutations, including MECP2, AFF2, and PCDH11X. In total, approximately 4% of the study’s subjects carried these identified mutations. 43 out of the 61 ASD risk-genes mentioned have previously been detected in earlier CNV studies. With CNVs possibly leading to the development of sporadic disorders including ASD, research has also focused on what contributes to CNV instability. Intriguingly, transposable elements have been associated with this. One source of CNVs is non-allelic homologous recombination (NAHR) between low-copy repeat (LCR) genome sequences (Stankiewicz and Lupski 2002). High-copy repeats, which contribute to a larger portion of the human genome when compared to LCRs, can also contribute to CNV instability. Among high-copy repeats are interspersed repeats, which make up 44% of the human genome (Lander et al. 2001). Existing research has shown these transposable elements to be associated with supporting NAHR and the formation of CNVs (Sasaki et al. 2010; Matejas et al. 2006; Gu et al. 2008; Erez et al. 2009; de Smith et al. 2008). Indeed, L1 retrotransposons in particular, have been associated with the autism spectrum disorders (Muotri et al. 2010; Shpyleva et al. 2017).
Rett syndrome (RTT), once considered a part of the ASD, is a progressive neurological disorder associated with the mutation of the X-linked gene methyl CpG binding protein 2 (MeCP2) (Amir et al. 1999). MeCP2 epigenetically regulates its target genes by binding to methylated CpG dinucleotides within promoters, acting as a transcriptional repressor (Chahrour et al. 2008; Yasui et al. 2007). L1 5’ UTR sequences are included among MeCP2 targets resulting in methylation-dependent repression (Yu et al. 2001; Klose et al. 2005; Muotri et al. 2010). RTT patients seem to have normal development until 6–18 months of age (Figure 2), however it is followed by impaired motor function, regression of developmental skills, and autistic characteristics and behaviors (Neul et al. 2010). Muotri and colleagues (2010) showed MeCP2 regulation of L1 expression and retrotransposition in mouse and human samples. Muotri et al. compared wild-type and MeCP2 KO brains of L1-EGFP transgenic mice and observed a higher number of EGFP-positive cells in the MeCP2 KO mice brains when compared to wild-type, suggesting an increase in retrotransposition events in the KO mice. Similarly, a two-fold increase of EGFP-positive cells were observed in RTT NPCs in comparison to controls, suggesting the LOF mutation of MeCP2 (as in RTT patients) correlates with an accumulation of L1 insertions. L1 ORF2 sequences in human brain were also analyzed using qPCR. The brains of RTT patients showed a significantly higher number of ORF2 sequences when compared to age- and gender-matched controls. Thus, for the first time, investigators showed that disease-related genetic mutations could influence the frequency of neuronal L1 retrotransposition.
Another study recently examined the levels of L1 ORF1 and ORF2 transcripts in idiopathic autism postmortem brain, looking at four brain regions: BA9 (frontal cortex), BA24 (anterior cingulate), BA22 (auditory processing), and cerebellum (Shpyleva et al. 2017). In this study, Sphyleva and colleagues observed a significant increase in total ORF1 and ORF2 RNA and mRNA in the autism cerebellum compared to control, however there was no significant difference in overall L1 copies. Correspondingly to Muotri et al., the authors used chromatin immunoprecipitation (ChIP) technology to measure MeCP2 binding to the 5’ promoter region. Although no significant difference of binding was observed between autism and control samples, authors detected a negative association between MeCP2 binding to 5’ UTR and ORF1 expression in autism brains not found in control, proposing that reduced MeCP2 binding contributes to the increase in ORF1 expression seen in the autism cerebellum. To investigate L1 expression in the presence of epigenetic alterations, researchers used methylated DNA immunoprecipitation (MeDIP) methodology. They reported no differences in DNA methylation density in 5’ UTR, ORF1, and ORF2 sequences between autism and control samples, however, trimethylation of histone H3 lysine 9 (H3K9me3), which prevents L1 activation, was significantly reduced at ORF1 and ORF2 sequences. This suggests that reduced H3K9me3 levels, and consequently an increase in chromatin accessibility, contributes to the overexpression of ORF1 and ORF2 in the autism cerebellum. Thus, it is intriguing that L1 expression might be upregulated in a fraction of the ASD brain, generating local mosaicism upon retrotransposition. Future work will focus on the mapping of neurons and circuits that could be affected by this mechanism. It is tempting to propose that the L1 mosaicism in early development could affect certain brain regions but not others, contributing to the heterogeneity of the ASD behavior later in life, including some of the “savant” skills observed in certain individuals with ASD.
2- The young adult brain
L1 and Schizophrenia
According to the Diagnostic and Statistical Manual of Mental Disorders, schizophrenia is the most common psychosis with symptoms including hallucinations, delusions, disorganized thinking, abnormal motor behavior, and negative symptoms. Symptoms typically develop in early adulthood (Figure 2). Schizophrenia is considered a neuropsychiatric disorder associated with both genetic and environmental factors (Keshavan et al. 2011). Many schizophrenia studies performed have created a strong association between microdeletions in chromosome 22q11.2 and schizophrenia (Karayiorgou et al. 1995; Xu et al. 2008). The 22q11.2 deletion syndrome has an incidence of 1 in 2000–4000 live births (Robin and Shprintzen 2005; Kobrynski and Sullivan 2007; Botto et al. 2003). Children with this phenotype have consistently revealed language delays (Moss et al. 1999; Gerdes et al. 1999; Scherer et al. 1999; Solot et al. 2000; Solot et al. 2001) and impairments in attention and executive function (Woodin et al. 2001; Sobin et al. 2004). Also, verbal memory is constantly reported stronger than visual-spatial memory (Bearden et al. 2001; Lajiness-O'Neill et al. 2005; Sobin et al. 2005). These individuals are typically diagnosed first with attention-deficit hyperactivity disorder (ADHD), generalized anxiety disorder, obsessive-compulsive disorder or ASD (Arnold et al. 2001; Feinstein et al. 2002; Antshel et al. 2006; Antshel et al. 2007; Vorstman et al. 2006). By early adulthood, up to one-third of these individuals develop schizophrenia or schizoaffective disorder (Pulver et al. 1994; Murphy et al. 1999; Gothelf et al. 2007; Green et al. 2009). The 22q11.2 deletion accounts for 1–2% of schizophrenia cases and is a recurrent structural mutation responsible for sporadic cases of schizophrenia (Karayiorgou et al. 1995; Xu et al. 2008; Consortium 2008; Stefansson et al. 2008). In a recent study, Guffanti and colleagues sequenced a family of six using next generation sequencing. Out of the six family members, four are affected (the mother and three children). The authors identified many non-reference L1s, which are L1s that are either present or absent in individuals and are not annotated in the current release of the genome (Guffanti et al. 2016). More than one third of the non-reference L1s detected were located in the open reading frame of protein-coding genes implicated in schizophrenia (Guffanti et al. 2016), suggesting that L1 retrotransposition contributes to the pathogenesis of schizophrenia.
In 2014, Bundo and colleagues observed a high L1 copy number in patients with schizophrenia. This increase was apparent in the neurons from the prefrontal cortex of patients and in iPSC-derived neurons with a 22q11 deletion (Bundo et al. 2014). Through whole-genome sequencing, the authors determined that the L1 insertions were concentrated in gene areas related to schizophrenia and synaptic function. Recently, Doyle and colleagues analyzed 36 postmortem brains of individuals diagnosed with schizophrenia, specifically looking at dorsolateral prefrontal cortex neurons. The researchers reported a drastic increase in intragenic novel L1s in the schizophrenia brains when compared to age-matched controls, and many L1 insertions were found within genes related to “cell projection” and “postsynaptic membrane” (Doyle et al. 2017). With a different approach, Misiak and colleagues instead investigated L1 methylation in peripheral blood leukocytes from 48 patients with first-episode schizophrenia. Upon analysis, the authors observed significantly lower L1 methylation in patients with schizophrenia who had a history of childhood trauma compared to schizophrenia patients without childhood trauma or healthy controls (Misiak et al. 2015). Existing research has shown low L1 methylation is also seen in other mental disorders, including major depressive disorder and post-traumatic stress disorder.
L1 and Major Depressive Disorder
Major Depressive Disorder (MDD) is a common disorder consisting of episodes of depressed mood that most typically affects individuals ages 25–44 (Kessler et al. 2003; Kupfer et al. 2012) (Figure 2). Both genetic and environmental factors have been associated with MDD (Mill and Petronis 2007; Tsankova et al. 2007), and L1 and other mobile elements are considered environmental stress sensors in the brain (Erwin et al. 2014; Muotri et al. 2009). In 2016, Liu and colleagues obtained peripheral blood of 105 MDD patients, and using qPCR, an increase in L1 copy number was seen in MDD patients when compared to controls (Liu et al. 2016). Additionally, authors reported lower methylation levels of L1 5’ UTR in MDD patients (Liu et al. 2016), suggesting the increase in L1 activity resulted from hypomethylation.
L1 and Post-Traumatic Stress Disorder
Post-traumatic stress disorder (PTSD) is an anxiety disorder that has no particular age of onset (Figure 2), but typically follows a psychologically traumatic event (Shalev 2009). Although the molecular mechanism is still to be determined, epigenetic factors are being investigated in this field of research. In 2013, Rusiecki and colleagues obtained blood samples from 75 US military service members with a post-deployment diagnosis of PTSD and DNA methylation patterns were analyzed. The researchers reported that L1 was hypomethylated in the PTSD cases when compared to controls (Rusiecki et al. 2012; Macia et al. 2017a). The above studies have provided evidence of L1 activity mental illness in the young adult brain, however in the following section, we will explore the element’s presence in a mature, adult brain affected by different neurodegenerative diseases.
3- The adult brain
L1 and Frontotemporal Lobar Degeneration
Frontotemporal lobar degeneration (FTLD) is a form of dementia characterized by a progressive decline in behavior and language (Baralle et al. 2013; Cruts et al. 2013; Rohrer et al. 2015), with an onset age of 65 (Onyike 2013) (Figure 2). This disorder is associated with the accumulation of TAR DNA-binding protein 43 (TDP-43) containing cytoplasmic inclusions (Cohen et al. 2011). In 2012, a study demonstrated binding between TDP-43 and L1-derived RNA transcripts (Li et al. 2012). Specifically, in FTLD patients with a mutated TDP-43, this binding is reduced, which results in an elevation of L1 transcripts (Li et al. 2012). Amyotrophic lateral sclerosis (ALS) is a neuromuscular disease that is also associated with mutations in TDP-43. Recent research has suggested activation of HERV-K, another transposable element, in sporadic ALS patients (Li et al. 2015). These studies demonstrate how L1 and other transposable elements could possibly contribute to TDP-43 related and other neurodegenerative diseases.
L1 and Alzheimer’s disease
Alzheimer’s disease (AD) is a neurodegenerative disorder consisting of progressive impairments in memory and cognitive abilities (Blennow et al. 2006), with the majority of diagnosed individuals age 65 and older (Figure 2). Existing research has suggested epigenetic changes, particularly DNA methylation, as a contributor to late onset AD. In 2011, Bollati and colleagues used bisulfite-PCR pyrosequencing to evaluate methylation of L1 in about 40 AD patients and healthy controls. The authors reported an increase of L1 in AD patients when compared to controls (Bollati et al. 2011). Additionally, a mini mental state examination was performed on AD patients and those with high levels of L1 methylation performed the best (Bollati et al. 2011). On the other hand, in 2014, Hernandez and colleagues measured L1 DNA methylation levels in peripheral blood samples from about 30 patients with late-onset AD. Using a methylation-sensitive high-resolution melting quantitative assay, the authors detected no difference in L1 methylation between AD patients and controls (Hernandez et al. 2014). Due to the conflicting results of these two studies, further investigation is needed to understand the role of L1 methylation in AD pathogenesis. Recently, in another interesting research study relating L1 elements and memory formation, it was shown that L1 expression in adult mice hippocampus enabled long-term memory formation suggesting that genomic mosaicism originating from L1 retrotransposition is involved in cognitive processes (Bachiller et al. 2017).
L1 and Aging
Although the previous topics have discussed L1 in association with infancy, childhood, and adulthood disorders, there has been recent research exploring the role of L1 in aging. Aging is typically correlated with the accumulation of DNA damage and ineffective DNA repair mechanisms (Nijnik et al. 2007; Wilson et al. 2008; Li et al. 2008), and L1 activity has been hypothesized to be a source of DNA damage (Wallace et al. 2008). Wallace and colleagues (2008) investigated the individual roles of the endonuclease and reverse transcriptase of the L1 ORF2p with regard to cell viability, hypothesizing that mutations in each domain would decrease the deleterious effect on L1 expression. The authors generated the mutations using QuikChange Site-Directed Mutagenesis Kit, introducing expression constructs with the desired mutation to the cell lines. In HeLa and MCF7 cells, a mutated ORF2p EN domain significantly increased the number of viable cells, thought to be due to the reduction of DNA double-stranded breaks that accumulate with normal EN activity. Likewise, a mutated ORF2 RT domain also increased the amount of viable cells, however not as significant as the mutated EN. When both the EN and RT domains were mutated, the number of viable cells reached empty vector control levels. Additionally, authors performed an assay measuring expression of senescence specific ®-galactosidase to determine if L1 and ORF2 expression can induce a senescence-like state. Indeed, both L1 expression and ORF2 expression resulted in significantly higher ®-galactosidase levels than the control and is responsible for the loss of viable cells.
In 2013, Li and colleagues reported the activity of transposable elements in Drosophila brain during aging. By comparing TE transcripts in head tissues of flies at different ages using qPCR, the authors discovered a significant increase of expression of R2 (a LINE-like element) and gypsy (an LTR element) with age (Li et al. 2013). With the generation of a gypsy-TRAP reporter system, the authors had a way to measure de novo gypsy through GFP expression in the neurons of the mushroom body. As a result, they observed no labeled neurons in 2–4 day old animals, but saw a significant increase in GFP-labeled cells at later ages. dAGO2 is a protein necessary for TE silencing in somatic tissues (Czech and Hannon 2011). Authors used dAGo2 mutants to disrupt the somatic TE control mechanism and detected higher expression levels of R2 and gypsy in young dAgo2 mutants, with 2–4 day old mutant animals acquiring the same transcript levels as 28-day old wild-type animals. Similarly, long-term memory performance assays were conducted displaying a reduction in memory of dAgo2 mutants at 2–4 days old, which could be rescued with neuronal expression of dAgo2. dAgo2 mutants also exhibited significantly shorter lifespans when compared to wildtype. In all, the authors suggest that the increase in TE activation with age contributes to neuronal decline. These results can possibly give us insight into TE activation with age in humans.
More recently, the activation of L1s in somatic tissues during aging has been explored. Sirtuin 6 (SIRT6) is a protein deacetylase and mono-ADP ribosyltransferase that interacts with KAP1 (KRAB-associated protein 1) to silence chromatin and facilitate DNA repair (Van Meter et al. 2014). Previously shown, SIRT6 KO mice exhibited premature aging syndrome (Mostoslavsky et al. 2006), while mice overexpressing SIRT6 displayed extended lifespans (Kanfi et al. 2012). In 2014, Van Meter et al. showed the importance of SIRT6 in repressing L1 activity. Using qPCR, the authors detected higher genomic L1 DNA in SIRT6 KO mouse embryonic fibroblasts when compared to WT. Furthermore, SIRT6KO cells contained hypomethylated CpG islands in the L1 5’ UTR and depleted levels of H3K9me3, MeCP2, and Kap1, suggesting the vital role of SIRT6 in packaging L1 loci into transcriptionally repressive heterochromatin. SIRT6 binds to L1 loci to repress transcription, however with an accumulation in DNA damage as is the case with aging, SIRT6 will vacate those sites to perform DNA repair, consequently allowing increased L1 activity. These data can contribute to understanding SIRT6 activity in human aging.
Conclusions
The studies mentioned in this review all contribute to make the cogent case that L1 retrotransposition indeed impacts the human brain. On two different time scales, L1 can be seen throughout the evolution of the primate brain and within a human individual lifetime. At the individual level, retrotransposition during early neurodevelopment could contribute to the phenotypes associated with ASD and even late onset disorders. This novel concept might help to explain, for example, the lack of heritable genetic factors in some ASD individuals as well as specific networks responsible for the “savant” phenotype. Since the discovery of transposable elements in the 1940s, this scientific field has also made great feats. From understanding the mobilization mechanism to causing disease, our knowledge on how L1 impacts the human genome has grown. However, many questions still remain for future studies to answer. How has LINE-1 contributed to the evolution of the human brain? Can we prevent retrotransposition and therefore disease? What are the environmental factors that affect neuronal L1 retrotransposition? With the existing knowledge of L1s, future research can now develop and design new technologies, approaches, and experiments to better understand the role of L1s in the human brain.
Acknowledgments
This work was supported by grants from the National Institutes of Health through the NIH R01MH094753. The work was also supported by the California Institute for Regenerative Medicine (CIRM) award DISC1-08825 and a UCSD CTRI pilot grant to Dr. Muotri. Dr. Macia is supported by a NARSAD Young Investigator grant.
Footnotes
Authors declare no conflict of interest
References
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes & development. 2006;20(2):210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature genetics. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Antshel KM, Aneja A, Strunge L, Peebles J, Fremont WP, Stallone K, Abdulsabur N, Higgins AM, Shprintzen RJ, Kates WR. Autistic spectrum disorders in velo-cardio facial syndrome (22q11.2 deletion) Journal of autism and developmental disorders. 2007;37(9):1776–1786. doi: 10.1007/s10803-006-0308-6. [DOI] [PubMed] [Google Scholar]
- Antshel KM, Fremont W, Roizen NJ, Shprintzen R, Higgins AM, Dhamoon A, Kates WR. ADHD, major depressive disorder, and simple phobias are prevalent psychiatric conditions in youth with velocardiofacial syndrome. Journal of the American Academy of Child and Adolescent Psychiatry. 2006;45(5):596–603. doi: 10.1097/01.chi.0000205703.25453.5a. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science (New York, NY) 2007;318(5851):761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold PD, Siegel-Bartelt J, Cytrynbaum C, Teshima I, Schachar R. Velo-cardio-facial syndrome: Implications of microdeletion 22q11 for schizophrenia and mood disorders. American journal of medical genetics. 2001;105(4):354–362. doi: 10.1002/ajmg.1359. [DOI] [PubMed] [Google Scholar]
- Azevedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE, Jacob Filho W, Lent R, Herculano-Houzel S. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. The Journal of comparative neurology. 2009;513(5):532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- Bachiller S, del-Pozo-Martín Y, Carrión ÁM. L1 retrotransposition alters the hippocampal genomic landscape enabling memory formation. Brain, behavior, and immunity. 2017;64(Supplement C):65–70. doi: 10.1016/j.bbi.2016.12.018. doi: https://doi.org/10.1016/j.bbi.2016.12.018. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychological medicine. 1995;25(1):63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, Talbot RT, Gustincich S, Freeman TC, Mattick JS, Hume DA, Heutink P, Carninci P, Jeddeloh JA, Faulkner GJ. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479(7374):534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle M, Buratti E, Baralle FE. The role of TDP-43 in the pathogenesis of ALS and FTLD. Biochemical Society transactions. 2013;41(6):1536–1540. doi: 10.1042/bst20130186. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nature reviews Genetics. 2002;3(5):370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Bearden CE, Woodin MF, Wang PP, Moss E, McDonald-McGinn D, Zackai E, Emannuel B, Cannon TD. The neurocognitive phenotype of the 22q11.2 deletion syndrome: selective deficit in visual-spatial memory. Journal of clinical and experimental neuropsychology. 2001;23(4):447–464. doi: 10.1076/jcen.23.4.447.1228. [DOI] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141(7):1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annual review of genomics and human genetics. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: transposable elements and disease. Genome medicine. 2009;1(10):97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. Lancet (London, England) 2006;368(9533):387–403. doi: 10.1016/s0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- Bollati V, Galimberti D, Pergoli L, Dalla Valle E, Barretta F, Cortini F, Scarpini E, Bertazzi P, Baccarelli A. DNA methylation in Repetitive Elements and Alzheimer disease. Brain, behavior, and immunity. 2011;25(6):1078–1083. doi: 10.1016/j.bbi.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, May K, Fernhoff PM, Correa A, Coleman K, Rasmussen SA, Merritt RK, O'Leary LA, Wong LY, Elixson EM, Mahle WT, Campbell RM. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics. 2003;112(1 Pt 1):101–107. doi: 10.1542/peds.112.1.101. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in <em>Drosophila</em>. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian HH., Jr Evidence consistent with human L1 retrotransposition in maternal meiosis I. American journal of human genetics. 2002;71(2):327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(9):5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, Kim C, Gidaya NB, Lindquist I, Hutman T, Sigman M, Kustanovich V, Lajonchere CM, Singleton A, Kim J, Wassink TH, McMahon WM, Owley T, Sweeney JA, Coon H, Nurnberger JI, Li M, Cantor RM, Minshew NJ, Sutcliffe JS, Cook EH, Dawson G, Buxbaum JD, Grant SF, Schellenberg GD, Geschwind DH, Hakonarson H. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS genetics. 2009;5(6):e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, Sunaga F, Toritsuka M, Ikawa D, Kakita A, Kato M, Kasai K, Kishimoto T, Nawa H, Okano H, Yoshikawa T, Kato T, Iwamoto K. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81(2):306–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- Carreira PE, Ewing AD, Li G, Schauer SN, Upton KR, Fagg AC, Morell S, Kindlova M, Gerdes P, Richardson SR, Li B, Gerhardt DJ, Wang J, Brennan PM, Faulkner GJ. Evidence for L1-associated DNA rearrangements and negligible L1 retrotransposition in glioblastoma multiforme. Mob DNA. 2016;7 doi: 10.1186/s13100-016-0076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes & development. 2014;28(13):1397–1409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science (New York, NY) 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Human genetics. 2005;117(5):411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, Cerritelli SM. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic acids research. 2013;41(5):3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic acids research. 2009;37(1):96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin SR, Hollis T, Perrino FW. Functional consequences of the RNase H2A subunit mutations that cause Aicardi-Goutieres syndrome. The Journal of biological chemistry. 2011;286(19):16984–16991. doi: 10.1074/jbc.M111.228833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Lee VM, Trojanowski JQ. TDP-43 functions and pathogenic mechanisms implicated in TDP-43 proteinopathies. Trends in molecular medicine. 2011;17(11):659–667. doi: 10.1016/j.molmed.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nature reviews Genetics. 2009;10(10):691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. The EMBO Journal. 2002;21(21):5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MC, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(51):20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460(7259):1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL. Mobile DNA II. ASM Press; Washington, D.C.: 2002. [Google Scholar]
- Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, Abdel-Hamid MS, Abdel-Salam GM, Ackroyd S, Aeby A, Agosta G, Albin C, Allon-Shalev S, Arellano M, Ariaudo G, Aswani V, Babul-Hirji R, Baildam EM, Bahi-Buisson N, Bailey KM, Barnerias C, Barth M, Battini R, Beresford MW, Bernard G, Bianchi M, Billette de Villemeur T, Blair EM, Bloom M, Burlina AB, Carpanelli ML, Carvalho DR, Castro-Gago M, Cavallini A, Cereda C, Chandler KE, Chitayat DA, Collins AE, Sierra Corcoles C, Cordeiro NJ, Crichiutti G, Dabydeen L, Dale RC, D'Arrigo S, De Goede CG, De Laet C, De Waele LM, Denzler I, Desguerre I, Devriendt K, Di Rocco M, Fahey MC, Fazzi E, Ferrie CD, Figueiredo A, Gener B, Goizet C, Gowrinathan NR, Gowrishankar K, Hanrahan D, Isidor B, Kara B, Khan N, King MD, Kirk EP, Kumar R, Lagae L, Landrieu P, Lauffer H, Laugel V, La Piana R, Lim MJ, Lin JP, Linnankivi T, Mackay MT, Marom DR, Marques Lourenco C, McKee SA, Moroni I, Morton JE, Moutard ML, Murray K, Nabbout R, Nampoothiri S, Nunez-Enamorado N, Oades PJ, Olivieri I, Ostergaard JR, Perez-Duenas B, Prendiville JS, Ramesh V, Rasmussen M, Regal L, Ricci F, Rio M, Rodriguez D, Roubertie A, Salvatici E, Segers KA, Sinha GP, Soler D, Spiegel R, Stodberg TI, Straussberg R, Swoboda KJ, Suri M, Tacke U, Tan TY, te Water Naude J, Wee Teik K, Thomas MM, Till M, Tonduti D, Valente EM, Van Coster RN, van der Knaap MS, Vassallo G, Vijzelaar R, Vogt J, Wallace GB, Wassmer E, Webb HJ, Whitehouse WP, Whitney RN, Zaki MS, Zuberi SM, Livingston JH, Rozenberg F, Lebon P, Vanderver A, Orcesi S, Rice GI. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. American journal of medical genetics Part A. 2015;167a(2):296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nature genetics. 2006;38(8):910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Manel N. Aicardi-Goutieres syndrome and the type I interferonopathies. Nature reviews Immunology. 2015;15(7):429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Human molecular genetics. 2009;18(R2):R130–136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, Van Langenhove T, van der Zee J, Van Broeckhoven C. Current insights into the C9orf72 repeat expansion diseases of the FTLD/ALS spectrum. Trends in neurosciences. 2013;36(8):450–459. doi: 10.1016/j.tins.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nature reviews Genetics. 2011;12(1):19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Huang Q, Boeke JD. Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochemistry. 2011;12(1):18. doi: 10.1186/1471-2091-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale RC, Gornall H, Singh-Grewal D, Alcausin M, Rice GI, Crow YJ. Familial Aicardi-Goutieres syndrome due to SAMHD1 mutations is associated with chronic arthropathy and contractures. American journal of medical genetics Part A. 2010;152a(4):938–942. doi: 10.1002/ajmg.a.33359. [DOI] [PubMed] [Google Scholar]
- de Smith AJ, Walters RG, Coin LJM, Steinfeld I, Yakhini Z, Sladek R, Froguel P, Blakemore AIF. Small Deletion Variants Have Stable Breakpoints Commonly Associated with Alu Elements. PLoS ONE. 2008;3(8) doi: 10.1371/journal.pone.0003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, Diedrich JK, Aslanian A, Ma J, Moresco JJ, Moore L, Hunter T, Saghatelian A, Gage FH. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. 2015;163(3):583–593. doi: 10.1016/j.cell.2015.09.025. [DOI] [PubMed] [Google Scholar]
- Divoky V, Indrak K, Mrug M, Brabec V, Huisman THJ, Prchal JT. A novel mechanism of beta thalassemia: The insertion of L1 retrotransposable element into beta globin IVS II. Blood. 1996;88:580–580. [Google Scholar]
- Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, Merrick WC, Shatsky IN. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5' untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Molecular and Cellular Biology. 2007;27(13):4685–4697. doi: 10.1128/mcb.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science (New York, NY) 1991;254(5039):1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, Gilbert N. Characterization of LINE-1 ribonucleoprotein particles. PLoS genetics. 2010;6(10) doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle GA, Crist RC, Karatas ET, Hammond MJ, Ewing AD, Ferraro TN, Hahn CG, Berrettini WH. Analysis of LINE-1 Elements in DNA from Postmortem Brains of Individuals with Schizophrenia. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2017 doi: 10.1038/npp.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez A, Patel AJ, Wang X, Xia Z, Bhatt SS, Craigen W, Cheung SW, Lewis RA, Fang P, Davenport SL, Stankiewicz P, Lalani SR. Alu-specific microhomology-mediated deletions in CDKL5 in females with early-onset seizure disorder. Neurogenetics. 2009;10(4):363–369. doi: 10.1007/s10048-009-0195-z. [DOI] [PubMed] [Google Scholar]
- Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nature reviews Neuroscience. 2014;15(8):497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Paquola ACM, Singer T, Gallina I, Novotny M, Quayle C, Bedrosian T, Ivanio F, Butcher CR, Herdy JR, Sarkar A, Lasken RS, Muotri AR, Gage FH. L1-Associated Genomic Regions are Deleted in Somatic Cells of the Healthy Human Brain. Nature neuroscience. 2016;19(12):1583–1591. doi: 10.1038/nn.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD. Cell lineage analysis in human brain using endogenous. 2015;85(1):49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, Park PJ, Walsh CA. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151(3):483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome research. 2010;20(9):1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Garcia-Perez JL. L1 Mosaicism in Mammals: Extent, Effects, and Evolution. Trends in Genetics. 2017 doi: 10.1016/j.tig.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Feinstein C, Eliez S, Blasey C, Reiss AL. Psychiatric disorders and behavioral problems in children with velocardiofacial syndrome: usefulness as phenotypic indicators of schizophrenia risk. Biological psychiatry. 2002;51(4):312–318. doi: 10.1016/s0006-3223(01)01231-8. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Genetic influences and infantile autism. Nature. 1977;265(5596):726–728. doi: 10.1038/265726a0. [DOI] [PubMed] [Google Scholar]
- Garcia Perez JL, Alarcon-Riquelme ME. The TREX1 Dinosaur Bites the Brain through the LINE. Cell stem cell. 2017;21(3):287–288. doi: 10.1016/j.stem.2017.08.010. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Human molecular genetics. 2007;16(13):1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, Hammer GD, Collins KL, O'Shea KS, Menendez P, Moran JV. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466(7307):769–773. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Widmann TJ, Adams IR. The impact of transposable elements on mammalian development. Development. 2016;143(22):4101–4114. doi: 10.1242/dev.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti R, Perlman S. In: Ataxia-Telangiectasia. Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews(R); Seattle (WA): 1993. [Google Scholar]
- Gerdes M, Solot C, Wang PP, Moss E, LaRossa D, Randall P, Goldmuntz E, Clark BJ, 3rd, Driscoll DA, Jawad A, Emanuel BS, McDonald-McGinn DM, Batshaw ML, Zackai EH. Cognitive and behavior profile of preschool children with chromosome 22q11.2 deletion. American journal of medical genetics. 1999;85(2):127–133. [PubMed] [Google Scholar]
- Gilbert SL, Dobyns WB, Lahn BT. Genetic links between brain development and brain evolution. Nature reviews Genetics. 2005;6(7):581–590. doi: 10.1038/nrg1634. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459(7246):569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL. Retrotransposition in tumors and brains. Mobile DNA. 2014;5(1):11. doi: 10.1186/1759-8753-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135(1):23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Molecular and Cellular Biology. 2007;27(18):6469–6483. doi: 10.1128/mcb.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothelf D, Feinstein C, Thompson T, Gu E, Penniman L, Van Stone E, Kwon H, Eliez S, Reiss AL. Risk factors for the emergence of psychotic disorders in adolescents with 22q11.2 deletion syndrome. The American journal of psychiatry. 2007;164(4):663–669. doi: 10.1176/ajp.2007.164.4.663. [DOI] [PubMed] [Google Scholar]
- Grabrucker AM. Environmental Factors in Autism. Frontiers in Psychiatry. 2012;3 doi: 10.3389/fpsyt.2012.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T, Gothelf D, Glaser B, Debbane M, Frisch A, Kotler M, Weizman A, Eliez S. Psychiatric disorders and intellectual functioning throughout development in velocardiofacial (22q11.2 deletion) syndrome. Journal of the American Academy of Child and Adolescent Psychiatry. 2009;48(11):1060–1068. doi: 10.1097/CHI.0b013e3181b76683. [DOI] [PubMed] [Google Scholar]
- Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of KpnI family segments. The EMBO Journal. 1984;3(8):1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. PathoGenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti G, Gaudi S, Klengel T, Fallon JH, Mangalam H, Madduri R, Rodriguez A, DeCrescenzo P, Glovienka E, Sobell J, Klengel C, Pato M, Ressler KJ, Pato C, Macciardi F. LINE1 insertions as a genomic risk factor for schizophrenia: Preliminary evidence from an affected family. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2016;171(4):534–545. doi: 10.1002/ajmg.b.32437. [DOI] [PubMed] [Google Scholar]
- Guo H, Peng Y, Hu Z, Li Y, Xun G, Ou J, Sun L, Xiong Z, Liu Y, Wang T, Chen J, Xia L, Bai T, Shen Y, Tian Q, Hu Y, Shen L, Zhao R, Zhang X, Zhang F, Zhao J, Zou X, Xia K. Genome-wide copy number variation analysis in a Chinese autism spectrum disorder cohort. Scientific reports. 2017;7:44155. doi: 10.1038/srep44155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, Lotspeich L, Croen LA, Ozonoff S, Lajonchere C, Grether JK, Risch N. Genetic heritability and shared environmental factors among twin pairs with autism. Archives of general psychiatry. 2011;68(11):1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH. Active human retrotransposons: variation and disease. Review Curr Opin Genet Dev. 2012;22 doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras SR, Macias S, Caceres JF, Garcia-Perez JL. Control of mammalian retrotransposons by cellular RNA processing activities. Mobile genetic elements. 2014;4:e28439. doi: 10.4161/mge.28439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Caceres JF. The Microprocessor controls the activity of mammalian retrotransposons. Nature structural & molecular biology. 2013;20(10):1173–1181. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. The human brain in numbers: a linearly scaled-up primate brain. Frontiers in human neuroscience. 2009;3:31. doi: 10.3389/neuro.09.031.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. Neuronal scaling rules for primate brains: the primate advantage. Progress in brain research. 2012;195:325–340. doi: 10.1016/b978-0-444-53860-4.00015-5. [DOI] [PubMed] [Google Scholar]
- Hernandez HG, Mahecha MF, Mejia A, Arboleda H, Forero DA. Global long interspersed nuclear element 1 DNA methylation in a Colombian sample of patients with late-onset Alzheimer's disease. American journal of Alzheimer's disease and other dementias. 2014;29(1):50–53. doi: 10.1177/1533317513505132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman MA. Size and shape of the cerebral cortex in mammals. II. The cortical volume. Brain, behavior and evolution. 1988;32(1):17–26. doi: 10.1159/000116529. [DOI] [PubMed] [Google Scholar]
- Hofman MA. Evolution of the human brain: when bigger is better. Frontiers in Neuroanatomy. 2014;8 doi: 10.3389/fnana.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. Journal of molecular biology. 1997a;271(1):7–12. doi: 10.1006/jmbi.1997.1159. [DOI] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. The EMBO Journal. 1997b;16(19):6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH., Jr A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nature genetics. 1994;7(2):143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141(7):1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, Eichler EE. De novo rates and selection of large copy number variation. Genome research. 2010;20(11):1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516(7530):242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proceedings of the National Academy of Sciences. 1997;94(5):1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(17):7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science (New York, NY) 2004;303(5664):1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Moran JV. Mobile DNA in Health and Disease. The New England journal of medicine. 2017;377(4):361–370. doi: 10.1056/NEJMra1510092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332(6160):164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Kempen M-JHC, Bodea GO, Faulkner GJ. Neuronal Genome Plasticity: Retrotransposons, Environment and Disease. In: Cristofari G, editor. Human Retrotransposons in Health and Disease. Cham: Springer International Publishing; 2017. pp. 107–125. [Google Scholar]
- Keshavan MS, Nasrallah HA, Tandon R. Schizophrenia, "Just the Facts" 6. Moving ahead with the schizophrenia concept: from the elephant to the mouse. Schizophrenia research. 2011;127(1–3):3–13. doi: 10.1016/j.schres.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, Rush AJ, Walters EE, Wang PS. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) Jama. 2003;289(23):3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome research. 2006;16(1):78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proceedings of the National Academy of Sciences. 2009;106(3):731–736. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kines KJ, Sokolowski M, deHaro DL, Christian CM, Belancio VP. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic acids research. 2014;42(16):10488–10502. doi: 10.1093/nar/gku687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Molecular cell. 2005;19(5):667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11.2 deletion syndromes. Lancet (London, England) 2007;370(9596):1443–1452. doi: 10.1016/s0140-6736(07)61601-8. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Human molecular genetics. 2005;14(21):3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nature structural & molecular biology. 2006;13(7):655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Kupfer DJ, Frank E, Phillips ML. Major depressive disorder: new clinical, neurobiological, and treatment perspectives. Lancet (London, England) 2012;379(9820):1045–1055. doi: 10.1016/s0140-6736(11)60602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajiness-O'Neill RR, Beaulieu I, Titus JB, Asamoah A, Bigler ED, Bawle EV, Pollack R. Memory and learning in children with 22q11.2 deletion syndrome: evidence for ventral and dorsal stream disruption? Child neuropsychology : a journal on normal and abnormal development in childhood and adolescence. 2005;11(1):55–71. doi: 10.1080/09297040590911202. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lasken RS, Stockwell TB. Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnology. 2007;7(1):19. doi: 10.1186/1472-6750-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, Wheeler DA, Gibbs RA, Kucherlapati R, Lee C, Kharchenko PV, Park PJ. Landscape of somatic retrotransposition in human cancers. Science (New York, NY) 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, Buja A, Krieger A, Yoon S, Troge J, Rodgers L, Iossifov I, Wigler M. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70(5):886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Li H, Mitchell JR, Hasty P. DNA double-strand breaks: a potential causative factor for mammalian aging? Mechanisms of ageing and development. 2008;129(7–8):416–424. doi: 10.1016/j.mad.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Jin Y, Prazak L, Hammell M, Dubnau J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS ONE. 2012;7(9):e44099. doi: 10.1371/journal.pone.0044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lee M-H, Henderson L, Tyagi R, Bachani M, Steiner J, Campanac E, Hoffman DA, von Geldern G, Johnson K, Maric D, Morris HD, Lentz M, Pak K, Mammen A, Ostrow L, Rothstein J, Nath A. Human endogenous retrovirus-K contributes to motor neuron disease. Science Translational Medicine. 2015;7(307):307ra153–307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Prazak L, Chatterjee N, Gruninger S, Krug L, Theodorou D, Dubnau J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nature neuroscience. 2013;16(5):529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker SB, Marchetto MC, Narvaiza I, Denli AM, Gage FH. Examining non-LTR retrotransposons in the context of the evolving primate brain. BMC Biology. 2017;15 doi: 10.1186/s12915-017-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Du T, Liu Z, Shen Y, Xiu J, Xu Q. Inverse changes in L1 retrotransposons between blood and brain in major depressive disorder. Scientific reports. 2016;6:37530. doi: 10.1038/srep37530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72(4):595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Macia A, Blanco-Jimenez E, Garcia-Perez JL. Retrotransposons in pluripotent cells: Impact and new roles in cellular plasticity. Biochimica et biophysica acta. 2015;1849(4):417–426. doi: 10.1016/j.bbagrm.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Macia A, Munoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, Marchal JA, Badge RM, Garcia-Perez JL. Epigenetic control of retrotransposon expression in human embryonic stem cells. Molecular and Cellular Biology. 2011;31(2):300–316. doi: 10.1128/mcb.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia A, Muotri AR. Analysis of LINE-1 Retrotransposition in Neural Progenitor Cells and Neurons. In: Frade JM, Gage FH, editors. Genomic Mosaicism in Neurons and Other Cell Types. Springer New York; New York, NY: 2017. pp. 189–208. [DOI] [Google Scholar]
- Macia A, Tejwani L, Mesci P, Muotri A, Garcia-Perez JL. Activity of Retrotransposons in Stem Cells and Differentiated Cells. In: Cristofari G, editor. Human Retrotransposons in Health and Disease. Springer International Publishing; Cham: 2017a. pp. 127–156. [DOI] [Google Scholar]
- Macia A, Widmann TJ, Heras SR, Ayllon V, Sanchez L, Benkaddour-Boumzaouad M, Munoz-Lopez M, Rubio A, Amador-Cubero S, Blanco-Jimenez E, Garcia-Castro J, Menendez P, Ng P, Muotri AR, Goodier JL, Garcia-Perez JL. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome research. 2017b;27(3):335–348. doi: 10.1101/gr.206805.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan M, Garcia-Canadas M, Reichmann J, Khazina E, Wagner G, Playfoot CJ, Salvador-Palomeque C, Mann AR, Peressini P, Sanchez L, Dobie K, Read D, Hung CC, Eskeland R, Meehan RR, Weichenrieder O, Garcia-Perez JL, Adams IR. Mobilization of LINE-1 retrotransposons is restricted by Tex19.1 in mouse embryonic stem cells. eLife. 2017;6 doi: 10.7554/eLife.26152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Stoye JP. Mammalian Endogenous Retroviruses. Microbiology Spectrum. 2015;3(1) doi: 10.1128/microbiolspec.MDNA3-0009-2014. doi: [DOI] [PubMed] [Google Scholar]
- Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cellular and molecular life sciences : CMLS. 2008;65(21):3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MCN, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola ACM, Desai KN, Herai RH, Weitzman MD, Yeo GW, Muotri AR, Gage FH. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503(7477):525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. American journal of human genetics. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Maranon C, Olivares M, Alonso C, Lopez MC. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. Journal of molecular biology. 1995;247(1):49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Molecular and Cellular Biology. 1991;11(9):4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Molecular and Cellular Biology. 2001;21(2):467–475. doi: 10.1128/mcb.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matejas V, Huehne K, Thiel C, Sommer C, Jakubiczka S, Rautenstrauss B. Identification of Alu elements mediating a partial PMP22 deletion. Neurogenetics. 2006;7(2):119–126. doi: 10.1007/s10048-006-0030-8. [DOI] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science (New York, NY) 1991;254(5039):1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3'-->5' exonucleases. The Journal of biological chemistry. 1999;274(28):19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- McClintock B. Controlling elements and the gene. Cold Spring Harbor symposia on quantitative biology. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, Erwin JA, Fasching L, Flasch DA, Freed D, Ganz J, Jaffe AE, Kwan KY, Kwon M, Lodato MA, Mills RE, Paquola ACM, Rodin RE, Rosenbluh C, Sestan N, Sherman MA, Shin JH, Song S, Straub RE, Thorpe J, Weinberger DR, Urban AE, Zhou B, Gage FH, Lehner T, Senthil G, Walsh CA, Chess A, Courchesne E, Gleeson JG, Kidd JM, Park PJ, Pevsner J, Vaccarino FM, Brain Somatic Mosaicism N. Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science (New York, NY) 2017;356(6336) doi: 10.1126/science.aal1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52(3):643–645. [PubMed] [Google Scholar]
- Mill J, Petronis A. Molecular studies of major depressive disorder: the epigenetic perspective. Molecular psychiatry. 2007;12(9):799–814. doi: 10.1038/sj.mp.4001992. [DOI] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends in genetics : TIG. 2007;23(4):183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Misiak B, Szmida E, Karpinski P, Loska O, Sasiadek MM, Frydecka D. Lower LINE-1 methylation in first-episode schizophrenia patients with the history of childhood trauma. Epigenomics. 2015;7(8):1275–1285. doi: 10.2217/epi.15.68. [DOI] [PubMed] [Google Scholar]
- Monot C, Kuciak M, Viollet S, Mir AA, Gabus C, Darlix JL, Cristofari G. The specificity and flexibility of l1 reverse transcription priming at imperfect T-tracts. PLoS genetics. 2013;9(5):e1003499. doi: 10.1371/journal.pgen.1003499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, Gilbert N. Mobile DNA II. American Society of Microbiology; 2002. Mammalian LINE-1 retrotransposons and related elements; pp. 836–869. [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Moss EM, Batshaw ML, Solot CB, Gerdes M, McDonald-McGinn DM, Driscoll DA, Emanuel BS, Zackai EH, Wang PP. Psychoeducational profile of the 22q11.2 microdeletion: A complex pattern. The Journal of pediatrics. 1999;134(2):193–198. doi: 10.1016/s0022-3476(99)70415-4. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Munoz-Lopez M, Garcia-Perez JL. DNA transposons: nature and applications in genomics. Current genomics. 2010;11(2):115–128. doi: 10.2174/138920210790886871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Lopez M, Macia A, Garcia-Canadas M, Badge RM, Garcia-Perez JL. An epi [c] genetic battle: LINE-1 retrotransposons and intragenomic conflict in humans. Mobile genetic elements. 2011;1(2):122–127. doi: 10.4161/mge.1.2.16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468(7322):443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Zhao C, Marchetto MCN, Gage FH. Environmental Influence on L1 Retrotransposons in the Adult Hippocampus. Hippocampus. 2009;19(10):1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Archives of general psychiatry. 1999;56(10):940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- Neul JL, Kaufmann WE, Glaze DG, Christodoulou J, Clarke AJ, Bahi-Buisson N, Leonard H, Bailey ME, Schanen NC, Zappella M, Renieri A, Huppke P, Percy AK. Rett syndrome: revised diagnostic criteria and nomenclature. Annals of neurology. 2010;68(6):944–950. doi: 10.1002/ana.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newkirk SJ, Lee S, Grandi FC, Gaysinskaya V, Rosser JM, Vanden Berg N, Hogarth CA, Marchetto MCN, Muotri AR, Griswold MD, Ye P, Bortvin A, Gage FH, Boeke JD, An W. Intact piRNA pathway prevents L1 mobilization in male meiosis. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(28):E5635–e5644. doi: 10.1073/pnas.1701069114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(11):4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, Enver T, Bell JI, Slijepcevic P, Goodnow CC, Jeggo PA, Cornall RJ. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447(7145):686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- Onyike CU. The Epidemiology of Frontotemporal Dementia. 2013;25(2):130–137. doi: 10.3109/09540261.2013.776523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer LM, Steranka JP, Yang WR, Kryatova M, Medabalimi S, Ardeljan D, Liu C, Boeke JD, Avramopoulos D, Burns KH. Structural variants caused by Alu insertions are associated with risks for many human diseases. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(20):E3984–e3992. doi: 10.1073/pnas.1704117114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C, Vargas-Landin DB, Doucet AJ, van Essen D, Vera-Otarola J, Kuciak M, Corbin A, Nigumann P, Cristofari G. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. eLife. 2016;5:e13926. doi: 10.7554/eLife.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, Piven J, Ponting CP, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Sequeira AF, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stein O, Sykes N, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Webber C, Weksberg R, Wing K, Wittemeyer K, Wood S, Wu J, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Devlin B, Ennis S, Gallagher L, Geschwind DH, Gill M, Haines JL, Hallmayer J, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Scherer SW, Sutcliffe JS, Betancur C. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver AE, Nestadt G, Goldberg R, Shprintzen RJ, Lamacz M, Wolyniec PS, Morrow B, Karayiorgou M, Antonarakis SE, Housman D, et al. Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. The Journal of nervous and mental disease. 1994;182(8):476–478. doi: 10.1097/00005053-199408000-00010. [DOI] [PubMed] [Google Scholar]
- Ramsay LA, Marchetto MC, Caron M, Chen SH, Busche S, Kwan T, Pastinen T, Gage FH, Bourque G. Conserved expression of transposon-derived non-coding transcripts in primate stem cells. BMC Genomics. 2017;18 doi: 10.1186/s12864-017-3568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149(5):1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nature genetics. 2009;41(7):829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A, van den Maagdenberg AM, Jen JC, Kavanagh D, Bertram P, Spitzer D, Liszewski MK, Barilla-Labarca ML, Terwindt GM, Kasai Y, McLellan M, Grand MG, Vanmolkot KR, de Vries B, Wan J, Kane MJ, Mamsa H, Schafer R, Stam AH, Haan J, de Jong PT, Storimans CW, van Schooneveld MJ, Oosterhuis JA, Gschwendter A, Dichgans M, Kotschet KE, Hodgkinson S, Hardy TA, Delatycki MB, Hajj-Ali RA, Kothari PH, Nelson SF, Frants RR, Baloh RW, Ferrari MD, Atkinson JP. C-terminal truncations in human 3'-5' DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nature genetics. 2007;39(9):1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- Richardson SR, Gerdes P, Gerhardt DJ, Sanchez-Luque FJ, Bodea GO, Munoz-Lopez M, Jesuadian JS, Kempen MHC, Carreira PE, Jeddeloh JA, Garcia-Perez JL, Kazazian HH, Jr, Ewing AD, Faulkner GJ. Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome research. 2017;27(8):1395–1405. doi: 10.1101/gr.219022.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Morell S, Faulkner GJ. L1 Retrotransposons and Somatic Mosaicism in the Brain. Annual Review of Genetics. 2014a;48(1):1–27. doi: 10.1146/annurev-genet-120213-092412. [DOI] [PubMed] [Google Scholar]
- Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. eLife. 2014b;3:e02008. doi: 10.7554/eLife.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RK CY, Merico D, Bookman M, J LH, Thiruvahindrapuram B, Patel RV, Whitney J, Deflaux N, Bingham J, Wang Z, Pellecchia G, Buchanan JA, Walker S, Marshall CR, Uddin M, Zarrei M, Deneault E, D'Abate L, Chan AJ, Koyanagi S, Paton T, Pereira SL, Hoang N, Engchuan W, Higginbotham EJ, Ho K, Lamoureux S, Li W, MacDonald JR, Nalpathamkalam T, Sung WW, Tsoi FJ, Wei J, Xu L, Tasse AM, Kirby E, Van Etten W, Twigger S, Roberts W, Drmic I, Jilderda S, Modi BM, Kellam B, Szego M, Cytrynbaum C, Weksberg R, Zwaigenbaum L, Woodbury-Smith M, Brian J, Senman L, Iaboni A, Doyle-Thomas K, Thompson A, Chrysler C, Leef J, Savion-Lemieux T, Smith IM, Liu X, Nicolson R, Seifer V, Fedele A, Cook EH, Dager S, Estes A, Gallagher L, Malow BA, Parr JR, Spence SJ, Vorstman J, Frey BJ, Robinson JT, Strug LJ, Fernandez BA, Elsabbagh M, Carter MT, Hallmayer J, Knoppers BM, Anagnostou E, Szatmari P, Ring RH, Glazer D, Pletcher MT, Scherer SW. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nature neuroscience. 2017;20(4):602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin NH, Shprintzen RJ. Defining the clinical spectrum of deletion 22q11.2. The Journal of pediatrics. 2005;147(1):90–96. doi: 10.1016/j.jpeds.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Rohman MS, Koga Y, Takano K, Chon H, Crouch RJ, Kanaya S. Effect of the disease-causing mutations identified in human ribonuclease (RNase) H2 on the activities and stabilities of yeast RNase H2 and archaeal RNase HII. Febs j. 2008;275(19):4836–4849. doi: 10.1111/j.1742-4658.2008.06622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JD, Isaacs AM, Mizielinska S, Mead S, Lashley T, Wray S, Sidle K, Fratta P, Orrell RW, Hardy J, Holton J, Revesz T, Rossor MN, Warren JD. C9orf72 expansions in frontotemporal dementia and amyotrophic lateral sclerosis. The Lancet Neurology. 2015;14(3):291–301. doi: 10.1016/s1474-4422(14)70233-9. [DOI] [PubMed] [Google Scholar]
- Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2011;156b(3):255–274. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411(2):273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Rusiecki JA, Chen L, Srikantan V, Zhang L, Yan L, Polin ML, Baccarelli A. DNA methylation in repetitive elements and post-traumatic stress disorder: a case–control study of US military service members. Epigenomics. 2012;4(1) doi: 10.2217/epi.11.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, Goldberg AP, Jinlu C, Keaney JF, 3rd, Klei L, Mandell JD, Moreno-De-Luca D, Poultney CS, Robinson EB, Smith L, Solli-Nowlan T, Su MY, Teran NA, Walker MF, Werling DM, Beaudet AL, Cantor RM, Fombonne E, Geschwind DH, Grice DE, Lord C, Lowe JK, Mane SM, Martin DM, Morrow EM, Talkowski ME, Sutcliffe JS, Walsh CA, Yu TW, Ledbetter DH, Martin CL, Cook EH, Buxbaum JD, Daly MJ, Devlin B, Roeder K, State MW. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87(6):1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A. The familial risk of autism. Jama. 2014;311(17):1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germline. Nature reviews Molecular cell biology. 2010;11(3):182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer NJ, D'Antonio LL, Kalbfleisch JH. Early speech and language development in children with velocardiofacial syndrome. American journal of medical genetics. 1999;88(6):714–723. [PubMed] [Google Scholar]
- Schrago CG, Russo CA. Timing the origin of New World monkeys. Molecular biology and evolution. 2003;20(10):1620–1625. doi: 10.1093/molbev/msg172. [DOI] [PubMed] [Google Scholar]
- Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochemical Society transactions. 2007;35(Pt 3):637–642. doi: 10.1042/bst0350637. [DOI] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, Pinckers AJ, Fundele R, Rosenthal A, Cremers FP, Ropers HH, Berger W. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nature genetics. 1998;19(4):327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1(2):113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome research. 2016;26(6):745–755. doi: 10.1101/gr.201814.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science (New York, NY) 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semendeferi K, Lu A, Schenker N, Damasio H. Humans and great apes share a large frontal cortex. Nature neuroscience. 2002;5(3):272–276. doi: 10.1038/nn814. [DOI] [PubMed] [Google Scholar]
- Shalev AY. Posttraumatic Stress Disorder (PTSD) and Stress Related Disorders. The Psychiatric clinics of North America. 2009;32(3):687–704. doi: 10.1016/j.psc.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Chow J, Wang Z, Fan G. Abnormal CpG island methylation occurs during in vitro differentiation of human embryonic stem cells. Human molecular genetics. 2006;15(17):2623–2635. doi: 10.1093/hmg/ddl188. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM (ataxia telangiectasia mutated): expanding roles in the DNA damage response and cellular homeostasis. Biochemical Society transactions. 2001;29(Pt 6):661–666. doi: 10.1042/0300-5127:0290661. [DOI] [PubMed] [Google Scholar]
- Shpyleva S, Melnyk S, Pavliv O, Pogribny I, Jill James S. Overexpression of LINE-1 Retrotransposons in Autism Brain. Molecular neurobiology. 2017 doi: 10.1007/s12035-017-0421-x. [DOI] [PubMed] [Google Scholar]
- Shukla R, Upton KR, Munoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, Sinha S, Iannelli F, Radaelli E, Dos Santos A, Rapoud D, Guettier C, Samuel D, Natoli G, Carninci P, Ciccarelli FD, Garcia-Perez JL, Faivre J, Faulkner GJ. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153(1):101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends in neurosciences. 2010;33(8):345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin C, Kiley-Brabeck K, Daniels S, Blundell M, Anyane-Yeboa K, Karayiorgou M. Networks of attention in children with the 22q11 deletion syndrome. Developmental neuropsychology. 2004;26(2):611–626. doi: 10.1207/s15326942dn2602_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin C, Kiley-Brabeck K, Daniels S, Khuri J, Taylor L, Blundell M, Anyane-Yeboa K, Karayiorgou M. Neuropsychological characteristics of children with the 22q11 Deletion Syndrome: a descriptive analysis. Child neuropsychology : a journal on normal and abnormal development in childhood and adolescence. 2005;11(1):39–53. doi: 10.1080/09297040590911167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski M, Chynces M, deHaro D, Christian CM, Belancio VP. Truncated ORF1 proteins can suppress LINE-1 retrotransposition in trans. Nucleic acids research. 2017;45(9):5294–5308. doi: 10.1093/nar/gkx211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solot CB, Gerdes M, Kirschner RE, McDonald-McGinn DM, Moss E, Woodin M, Aleman D, Zackai EH, Wang PP. Communication issues in 22q11.2 deletion syndrome: children at risk. Genetics in medicine : official journal of the American College of Medical Genetics. 2001;3(1):67–71. doi: 10.1097/00125817-200101000-00015. [DOI] [PubMed] [Google Scholar]
- Solot CB, Knightly C, Handler SD, Gerdes M, McDonald-McGinn DM, Moss E, Wang P, Cohen M, Randall P, Larossa D, Driscoll DA. Communication disorders in the 22Q11.2 microdeletion syndrome. Journal of communication disorders. 2000;33(3):187–203. doi: 10.1016/s0021-9924(00)00018-6. quiz 203-184. [DOI] [PubMed] [Google Scholar]
- Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B, Wheelan S, Upton KR, Shukla R, Faulkner GJ, Largaespada DA, Kazazian HH., Jr Extensive somatic L1 retrotransposition in colorectal tumors. Genome research. 2012;22(12):2328–2338. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek M. Antisense Promoter of Human L1 Retrotransposon Drives Transcription of Adjacent Cellular Genes. Molecular and Cellular Biology. 2001;21(6):1973–1985. doi: 10.1128/mcb.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends in genetics : TIG. 2002;18(2):74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Molecular and Cellular Biology. 1990;10(12):6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu CE, Roge B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bolte S, Feineis-Matthews S, Herbrecht E, Schmotzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, Van Engeland H, de Jonge M, Kemner C, Koop F, Langemeijer M, Hijmans C, Staal WG, Baird G, Bolton PF, Rutter ML, Weisblatt E, Green J, Aldred C, Wilkinson JA, Pickles A, Le Couteur A, Berney T, McConachie H, Bailey AJ, Francis K, Honeyman G, Hutchinson A, Parr JR, Wallace S, Monaco AP, Barnby G, Kobayashi K, Lamb JA, Sousa I, Sykes N, Cook EH, Guter SJ, Leventhal BL, Salt J, Lord C, Corsello C, Hus V, Weeks DE, Volkmar F, Tauber M, Fombonne E, Shih A, Meyer KJ. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nature genetics. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Nyengaard JR, De Groot DM, Gundersen HJ. Total regional and global number of synapses in the human brain neocortex. Synapse (New York, NY) 2001;41(3):258–273. doi: 10.1002/syn.1083. [DOI] [PubMed] [Google Scholar]
- Telesnitsky A, Goff SP. Reverse Transcriptase and the Generation of Retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 1997. [PubMed] [Google Scholar]
- Thayer RE, Singer MF, Fanning TG. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene. 1993;133(2):273–277. doi: 10.1016/0378-1119(93)90651-i. [DOI] [PubMed] [Google Scholar]
- Thiele H, du Moulin M, Barczyk K, George C, Schwindt W, Nurnberg G, Frosch M, Kurlemann G, Roth J, Nurnberg P, Rutsch F. Cerebral arterial stenoses and stroke: novel features of Aicardi-Goutieres syndrome caused by the Arg164X mutation in SAMHD1 are associated with altered cytokine expression. Human mutation. 2010;31(11):E1836–1850. doi: 10.1002/humu.21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CA, Tejwani L, Trujillo CA, Negraes PD, Herai RH, Mesci P, Macia A, Crow YJ, Muotri AR. Modeling of TREX1-Dependent Autoimmune Disease using Human Stem Cells Highlights L1 Accumulation as a Source of Neuroinflammation. Cell stem cell. 2017 doi: 10.1016/j.stem.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RA, Nelson CA. Developmental science and the media. Early brain development. The American psychologist. 2001;56(1):5–15. doi: 10.1037/0003-066x.56.1.5. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nature reviews Neuroscience. 2007;8(5):355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, Vanderver A, Faulkner GJ. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161(2):228–239. doi: 10.1016/j.cell.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nature communications. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorstman JA, Morcus ME, Duijff SN, Klaassen PW, Heineman-de Boer JA, Beemer FA, Swaab H, Kahn RS, van Engeland H. The 22q11.2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. Journal of the American Academy of Child and Adolescent Psychiatry. 2006;45(9):1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419(1–2):75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. Journal of molecular biology. 2005;354(4):994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Molecular and Cellular Biology. 2001;21(4):1429–1439. doi: 10.1128/mcb.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Bohr VA, McKinnon PJ. DNA damage, DNA repair, ageing and age-related disease. Mechanisms of ageing and development. 2008;129(7–8):349–352. doi: 10.1016/j.mad.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. The Journal of biological chemistry. 2011;286(42):36427–36437. doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Munoz-Lopez M, Macia A, Yang Z, Montano M, Collins W, Garcia-Perez JL, Moran JV, Greene WC. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Human molecular genetics. 2012;21(1):208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodin M, Wang PP, Aleman D, McDonald-McGinn D, Zackai E, Moss E. Neuropsychological profile of children and adolescents with the 22q11.2 microdeletion. Genetics in medicine : official journal of the American College of Medical Genetics. 2001;3(1):34–39. doi: 10.1097/00125817-200101000-00008. 10.109700125817-200101000-00008. [DOI] [PubMed] [Google Scholar]
- Xin B, Jones S, Puffenberger EG, Hinze C, Bright A, Tan H, Zhou A, Wu G, Vargus-Adams J, Agamanolis D, Wang H. Homozygous mutation in SAMHD1 gene causes cerebral vasculopathy and early onset stroke. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(13):5372–5377. doi: 10.1073/pnas.1014265108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, Jorde LB. Mobile elements create structural variation: analysis of a complete human genome. Genome research. 2009;19(9):1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Spinelli P, Laussmann Maike A, Homolka D, Yang Z, Cora E, Couté Y, Conn S, Kadlec J, Sachidanandam R, Kaksonen M, Cusack S, Ephrussi A, Pillai Ramesh S. RNA Clamping by Vasa Assembles a piRNA Amplifier Complex on Transposon Transcripts. Cell. 2014;157(7):1698–1711. doi: 10.1016/j.cell.2014.05.018. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nature genetics. 2008;40(7):880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nature structural & molecular biology. 2006;13(9):763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131(5):873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(49):19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Zingler N, Schumann G, Stratling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic acids research. 2001;29(21):4493–4501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Dong B, Doucet AJ, Moldovan JB, Moran JV, Silverman RH. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic acids research. 2014;42(6):3803–3820. doi: 10.1093/nar/gkt1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Sequencing genomes from single cells by polymerase cloning. Nat Biotech. 2006;24(6):680–686. doi: 10.1038/nbt1214. doi: http://www.nature.com/nbt/journal/v24/n6/suppinfo/nbt1214_S1.html. [DOI] [PubMed] [Google Scholar]
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, Cheung LE, Wang G, Kazazian HH, Yu XF. Modulation of LINE-1 and Alu/SVA Retrotransposition by Aicardi-Goutières Syndrome-Related SAMHD1. Cell reports. 2013;4(6):1108–1115. doi: 10.1016/j.celrep.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Armstrong E, Moser KH, Schleicher A, Stephan H. Gyrification in the cerebral cortex of primates. Brain, behavior and evolution. 1989;34(3):143–150. doi: 10.1159/000116500. [DOI] [PubMed] [Google Scholar]