Abstract
Recent advances in deep learning have impacted various scientific and industrial fields. Due to the rapid application of deep learning in biomedical data, molecular imaging has also started to adopt this technique. In this regard, it is expected that deep learning will potentially affect the roles of molecular imaging experts as well as clinical decision making. This review firstly offers a basic overview of deep learning particularly for image data analysis to give knowledge to nuclear medicine physicians and researchers. Because of the unique characteristics and distinctive aims of various types of molecular imaging, deep learning applications can be different from other fields. In this context, the review deals with current perspectives of deep learning in molecular imaging particularly in terms of development of biomarkers. Finally, future challenges of deep learning application for molecular imaging and future roles of experts in molecular imaging will be discussed.
Keywords: Deep learning, Molecular imaging, Machine learning, Convolutional neural network, Precision medicine
Introduction
The amount of biomedical data including images and medical records as well as omics data is rapidly increasing [1]. However, the heterogeneous, complex, and multidimensional nature of biomedical data make it difficult to decipher clinical meaning [2]. Thus, machine learning (ML), a method set of artificial intelligence in which a computer captures patterns underlying data and utilizes them to help decision making, have been extensively and increasingly applied for handling biomedical data. Recently, deep learning (DL), a special type of machine learning methods, has emerged as a new area of machine learning research [3]. DL is a class of machine learning that automatically learns hierarchical features of data by multiple layers composed of simple and nonlinear modules. It transforms the data to representations which are important for discriminating the data [4]. Since this method was successfully applied and overwhelmingly beat other previous ML methods for visual recognition tasks at a competitive challenge in 2012, ImageNet [5, 6], it has dramatically improved tasks of various scientific and industrial fields including not only computer vision but speech recognition, drug discovery, and bioinformatics [4, 7, 8]. Subsequently, DL techniques have become the method of choice in computer vision and several image processing tasks. This success came from development of new modules enabling learning of deep structures and reducing overfitting as well as efficient use of hardware including general-purpose graphics processing units. Most of all, structured large datasets such as ImageNet data contribute to the success of this technique.
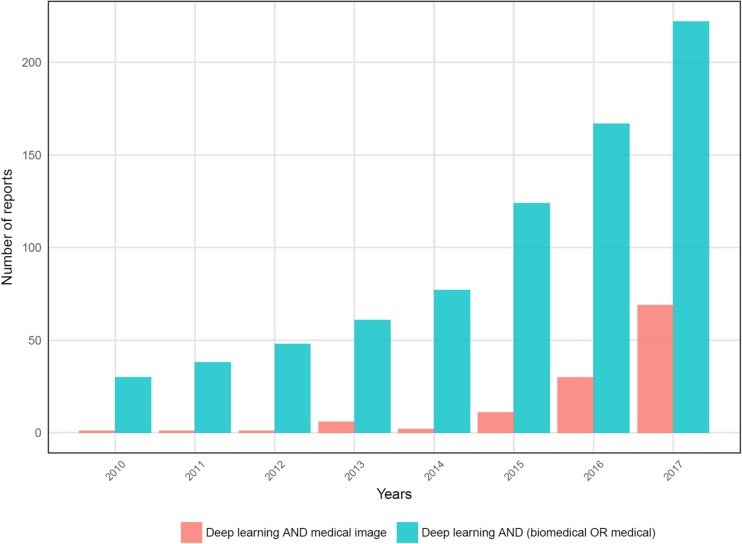
DL has been gradually applied to medical image data as medical image analyses are considerably similar to computer vision techniques. As well-structured and labeled medical data are relatively limited, applications of DL were relatively delayed. However, the number of reports regarding DL applications in medical data grew rapidly in 2016 (Fig. 1). The application areas are widening and range from anatomical segmentation to disease classification [9]. Although the many studies initially started from relatively small datasets by using a pretrained DL model as a feasibility study, a robust validation of the medical application has been required [10, 11]. In this regard, medical image big data have started to be collected to validate the feasibility of medical applications. For example, recently, Google researchers collected large data sets consisting of more than 120,000 retinal fundus images for diagnosing diabetic retinopathy and showed high sensitivity and specificity for the detection [12]. An automated skin lesion classification system comparable to dermatologists was recently developed using more than 120,000 clinical images [13]. These systems based on large datasets will further improve DL-based diagnosis and eventually make considerable changes in medical fields by supporting clinical decisions in the clinic in the near future.
Fig. 1.
Number of papers regarding deep learning applications in biomedical fields. Number of papers dealing with deep learning in biomedical fields are recently rapidly increasing. Bar graphs represent results of the number of PubMed paper searched by words: [Deep learning AND medical image] and [Deep learning AND (medical OR biomedical)], respectively
Due to the rapid application of DL in biomedical data, molecular imaging can also be influenced. The application of DL for molecular imaging can be ranged from disease diagnosis and classification to image processing. The potential impact of DL throughout the entire process of molecular imaging acquisition and interpretation requires nuclear medicine physicians and molecular imaging researchers to have knowledge about this technique. To this end, this review will give an overview of deep learning techniques particularly focusing on image recognition. In addition, the next section will introduce recent progress and current perspectives of medical application of DL focusing on molecular imaging. As the adaptation of DL in molecular imaging may inevitably happen, the direction of DL application in molecular imaging and the possible changes in the role of physicians will be discussed.
Brief Overview of Deep Learning for Images
Basics of Neural Networks: Single Layer Perceptron to Deep Neural Networks
In this section, DL concepts particularly for image analysis are briefly introduced. The detailed basic concepts of DL are reviewed by LeCun et al. [4] and general applications of DL in biomedical fields are reviewed by Mamoshina et al. [14].
DL is a type of ML defined by models with many hierarchical layers of information processing compared with conventional ‘shallow’ learning. ML including both shallow and deep learning is a field of artificial intelligence in which learning occurs without explicit programming. Both methods capture patterns underlying complicated data and utilize them as discriminative features of data. DL has advantages in learning intricate patterns from high dimensional raw data while conventional shallow learning usually requires handcrafted features extracted from raw data. Regardless of depth of layers, ML is generally divided into two methods, supervised and unsupervised learning. Supervised learning typically predicts a target variable representing a specific class (classification) of data or specific continuous values (regression). The inputs are raw data or features paired with the target label or target variable. Thus, a supervised ML model can be simply summarized by a prediction model that predicts target values from the data. Parameters of the model are adjusted to accurately predict the target by training process. On the other hand, unsupervised learning is a type of ML that finds patterns of the data without target labels or variables. Data clustering is an example of unsupervised learning. In terms of applications of unsupervised learning, representations extracted by unsupervised learning can be used for the other supervised tasks as well as data clustering.
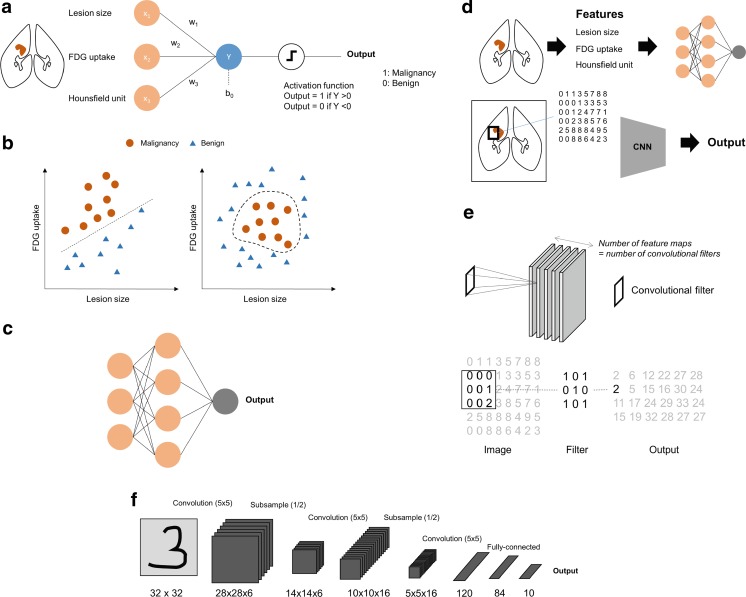
DL typically means deep neural networks. Thus, neural networks, a type of machine learning, are the basis of recent deep learning techniques. The perceptron is the earliest neural network model which consists of a single layer (Fig. 2a). A simple single layer model which aims at discriminating malignancy from benign lesion is exhibited in Fig. 2a. Inputs of the model are features of the lesion such as tumor size and FDG uptake. The model parameters, W = (w1, w2,…,wn), are multiplied to the inputs and then the activation function is applied. An example of simple activation function is that the output is 1 if the value is more than 0 and otherwise, the output is 0. The parameters of this simple model are optimized for the accurate discrimination. This process is defined as training. The training can be formulated as the minimization of the error between predicted outputs and real values. Practically, the gradient descent method which iteratively updates the model parameters according to gradient of the error function is used for the training [15]. This single layer perceptron has limitations in complicated nonlinear data patterns (Fig. 2b). As shown in the left plot of Fig. 2b, a linear discriminative line can differentiate malignancy from benign lesions while it cannot differentiate it for some nonlinear data distribution patterns such as the right plot. To overcome these certain issues, hidden layers between inputs and outputs are added. This type of neural network is a well-known traditional neural network, multi-layer perceptron (Fig. 2c). Although the application of hidden layer has improved performance of neural networks, it is difficult to train deep architectures. Deep neural networks have received attention after utilization of a new training method, pretraining [16, 17]. Efficient training was achieved by unsupervised feature extraction from input data followed by supervised learning.
Fig. 2.
Brief overview of neural networks. a An example of a single-layer perceptron is built for discriminating malignancy from benign lesion. Image features such as tumor size and FDG uptake are inputs of the model. Model parameters, weights, are multiplied to input variables and the output is predicted by a simple activation function. b A simple perceptron model has limitations in nonlinear problems. Data exhibited in the left plot are simply divided by a discriminative line while those in the right plot require a nonlinear and complex curve for the discrimination. c Multilayer perceptron models can be built by hierarchical composition of perceptron. Hidden layers hierarchically capture representations of data. d While conventional multilayer perceptron models use manual features as inputs, current convolutional neural network models directly use pixels/voxels. e An explanation of convolutional filter is exhibited. Convolutional layers of the network architectures have a certain number of convolutional filters as network model parameters. It produces feature maps as outputs by multiplying input pixels and filter values. f An example of a convolutional neural network represents multiple convolutional layers followed by pooling layers. Hierarchical feature maps are eventually connected to feature vectors and then connected to outputs
While the pretraining initiated the popularity of deep learning, current models particularly for image analysis fields can be summarized by simplified direct supervised training of specialized architecture such as convolutional neural networks (CNN). While previous network architecture used data with vector form, current models using CNN utilize structural information of neighboring pixels/voxels of the image. Besides, another currently popular architecture, recurrent neural network (RNN), considers information of the time-varying pattern of the data. In addition to these specialized network architectures, various techniques have improved performance by reducing overfitting and efficient training. They include special layers, such as dropout [18], batch normalization [19], and modified activation function such as rectified-linear unit (ReLU) [20]. Furthermore, network models trained by large datasets including ImageNet are used to extract general image features, so that they can transfer to another domain such as medical image classification as aforementioned pretraining [21]. Thus, current concepts of deep learning mean not only neural networks with multiple layers but also special architectures, such as CNN and RNN, combined with the various techniques improving the performance and training process. As CNNs are the most widely used for medical image analysis, the next section will additionally describe brief concepts of CNNs.
Convolutional Neural Networks
Traditional neural networks and general machine learning models use inputs as features with vector form. As aforementioned example, to build a machine learning model to differentiate malignancy from benign lesion, we select features such as tumor size and FDG uptake from images. Instead of these features, CNN generally directly uses pixels/voxels as inputs (Fig. 2d). To use structural information of neighboring pixels, convolutional layers are used instead of densely connected perceptron layers. A convolutional layer consists of a specific number of convolutional filters. The example of a 3 × 3 convolutional filter application to an image matrix is exhibited in Fig. 2e. For convolutional layers, values of convolutional filters correspond to weight parameters of the fully-connected neural network layers. Thus, convolutional filter values are adjusted during the training process. As a convolutional filter generated an output feature image, the number of convolutional filters are the number of feature images. In general, after the convolutional layer, consisting of a certain set of convolutional filters, an activation layer follows. Pooling layers are usually added to convolutional layers to subsample feature maps to reduce the amount of learning parameters of the model. These types of layers are hierarchically connected to the output. An example of a simple CNN model is represented in Fig. 2f [22]. The recent model is similar with this figure, however, it has deeper layers and a larger number of convolutional kernels. The dramatic improvement of image recognition in 2012 was achieved by AlexNet, which consists of five convolutional layers and combines modified activation function, ReLU [6]. After 2012, novel architectures improve the performance by using deeper architectures. In 2014, a 22-layer network named Inception [23] was the winner of ImageNet and in 2015, 152-layer network named ResNet further improved performance of image recognition [24]. These architectures and trained models were applied to other domains including medical images. For example, the diabetic retinopathy detection model [12] was based on fine-tuning of Inception model. The application of DL has widened becuase of its flexibility. For example, combining CNN with RNN produced interesting results such as automatic image captioning [25] and various CNN models were also used for object detection [26, 27] and object segmentation [28, 29] from images. These techniques have also been applied to medical images for diagnosis, lesion detection, and segmentation.
Perspectives of Deep Learning in Nuclear Medicine Molecular Imaging
The Distinct Characteristics of Molecular Imaging in DL Application
Even though DL applications in medical images rapidly grow, molecular imaging has distinct characteristics particularly in terms of its purposes and application. Firstly, the aim of the application of deep learning to molecular imaging is substantially different from computer vision fields. While DL models in image recognition aimed at identifying the ground-truth class, the ground-truth of DL is ambiguous in medical fields. Although recent diagnostic applications of DL to medical images generally used clinical or pathologic diagnosis as a ground-truth label, diagnosis can be varied according to pathologists as well as clinicians [30]. Furthermore, in many cases, various disorders are defined as a deviation from the normal population spectrum rather than clear-cut discriminative states. Thus, the application of DL to molecular imaging needs to aim at developing a good biomarker which can define the spectrum of disorders, monitor the status and reflect patients’ outcome instead of simple classification. Besides, molecular imaging aims at representing and visualizing the biological process with regard to physiologic and pathologic changes. The biological process which molecular imaging aims at already reflects the subjects’ outcome as well as surrogate marker of disease pathophysiology. To this end, the application of DL should focus on the discovery of crucial and latent information from imaging data. It should make the best use of molecular imaging information and discover a key pathophysiologic process by summarizing high-dimensional data to a few characteristic and discriminative parameters. The importance of this purpose of DL application in molecular imaging can be inferred from the reason maximum standardized uptake value (SUV) is widely used as the representative parameter for cancer PET imaging in spite of its limitations [31]. Maximum SUV is a single representative value for characterizing a lesion which consists of more than thousands of voxel data. This is a one-dimensional value that quantitatively summarizes an image as a set of high dimensional data to explain tumor glucose metabolism associated with patients’ outcome. Going beyond such a conventional imaging parameter, DL can be a method to capture the most important and discriminative information from the multidimensional and multimodal data of molecular imaging.
Secondly, in terms of data structure and size, molecular imaging is different from natural images. Even more, compared with other medical images such as simple X-ray, it is more difficult to collect large datasets for molecular imaging. As the spectrum of biological targets is very broad, there are numerous types of molecular imaging. In addition, several modalities, e.g., PET, SPECT, optical imaging, and MRI, increase their variety. As it substantially requires case-by-case applications of DL, training with small-sized samples is inevitable. Thus, a challenging issue is to apply deep learning to small medical image datasets. To overcome this issue, transfer learning has been widely used for the application of DL. The models trained by ImageNet, such as AlexNet and ResNet, were used for medical image feature extraction which can be further used for classification, segmentation, and lesion detection. For example, Wang H et al. used AlexNet as a feature extractor for differentiating mediastinal lymph node metastasis of lung cancer from FDG PET/CT [32]. Another method to overcome the limited number of images was to use image patches instead of full-size images. It was commonly used for anatomical segmentation in medical images [33–36]. For segmentation tasks, because classes are designated for each voxel, it is effective to use patch-based learning. Though data augmentation is generally used for natural images, medical images are cautious because manually transformed images could have different clinical information. Nonetheless, to overcome the data size issue, some studies have tried image rotation and flipping for data augmentation [32, 37, 38]. Furthermore, molecular imaging is sometimes used for detecting rare conditions, which require appropriate training methods to solve class imbalance. While the commonly used DL models were developed by data with uniform classes, the training for molecular imaging is usually performed by uneven diagnostic classes. Segmentation and detection also requires identifying small sized lesions in large image matrices. These issues of unique characteristics of molecular imaging require an appropriately adjusted neural networks model.
Briefly, application of DL for molecular imaging needs to focus on the development of good biomarkers rather than simple image classification. As molecular imaging itself provides biologically important information, DL has roles in discovering a crucial biomarker from high-dimensional data and maximum utilization of image information instead of simply replacing human’s visual cognition. To achieve this goal, DL should be properly applied to molecular imaging considering its unique characteristics compared to natural images. Thus, the next sections will discuss recent DL application results and directions in the light of biomarker development. It will be discussed by dividing the applications of DL into two categories: biomarker acquisition supported by DL and discovery of DL-based biomarker (Table 1).
Table 1.
Directions of deep learning applications for molecular imaging
| Indirect | Direct | |
|---|---|---|
| Explanation | Imaging biomarker acquisition supported by DL | DL-based imaging biomarker |
| Examples | ✓ Segmentation helps measurement of tracer uptake in target regions | ✓DL output scores for diagnostic classification |
| ✓ Low dose acquisition and image reconstruction | ✓DL-based image features | |
| ✓Predicting CT images for attenuation correction | ✓Combining multimodality data |
Acquisition of Imaging Biomarker Supported by Deep Learning
DL application of medical images could indirectly improve well-known biomarkers provided by molecular imaging. Because recent clinically applicable molecular imaging studies, e.g., PET/CT and SPECT/CT, are combined with anatomical imaging, the application of DL to anatomical imaging for lesion delineation and organ segmentation can support quantification of coregistered molecular imaging. Furthermore, DL can be used for improvement of image processing and acquisition which support robust imaging biomarkers by producing better images.
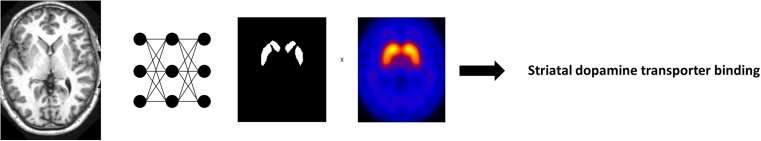
The most common subject of DL application in medical image is segmentation using anatomical CT and MR images. It is ranged from normal anatomical structure segmentation such as whole brain extraction [34, 39], brain substructure, heart and airway segmentation [36, 40–43] to pathological lesion segmentation [44–46]. Before the DL applications, several methods have been reported for the segmentation. However, DL-based medical image segmentation shows faster results as well as better performance. For example, CNN-based striatum segmentation took only a few seconds and showed better accuracy than conventional methods, while FreeSurfer, a widely used brain image processing tool, required several hours [36]. Because most of DL-based segmentation methods are fully automated, quantitative analysis of combined functional images can be accurately and easily performed by utilizing them. For example, using tumor segmentation techniques based on DL, we can simply obtain well-known parameters which require tumor volumes such as mean SUV or total lesion glycolysis. Utilization of this technique may offer a good solution for the issue of variability in tumor volume parameters associated with tumor delineation [47, 48]. Using automated DL-based brain structure segmentation, brain molecular imaging such as amyloid PET and dopamine transporter SPECT/PET can be accurately and easily quantified without complicated processing such as spatial normalization (Fig. 3).
Fig. 3.
An example of imaging biomarker acquisition supported by deep learning. Using automated deep learning-based brain structure segmentation, a well-known brain imaging biomarker such as dopamine transporter SPECT/PET can be easily and accurately quantitated
Another application area for improving imaging biomarker is related to image acquisition and processing. DL has markedly improved image processing fields as well as image recognition. An example is super-resolution, which predicts a high resolution image from low resolution information [49, 50]. This kind of concept could be extended to the PET images estimating standard images from low-dose PET images [51]. Recent studies suggest DL-based reconstruction of sinograms which can outperform computational time and show comparability with conventional iterative algorithms [52, 53]. Even though this kind of technique requires evidence of clinical equivalence, it can be expected that they support reducing radiation exposure as well as scan and reconstruction time.
As one of various application fields of DL, specific neural network models can be used for generating images of different modality [54]. Some neural network models were developed for predicting pseudo-CT images from MR images [55, 56]. These models could produce realistic CT images corresponding to MR images though synthesized CT images could hardly apply to clinical decision making such as lesion detection and characterization. Nonetheless, this method can be used for solving CT-less attenuation correction particularly for PET/MR [57]. Moreover, as this method is bidirectional, various ways of translation are feasible. As a future work, pseudo-image generation can be combined with conventional image processing and quantification methods such as partial volume correction for SPECT and PET images.
Discovering of Deep Learning-Based Biomarker
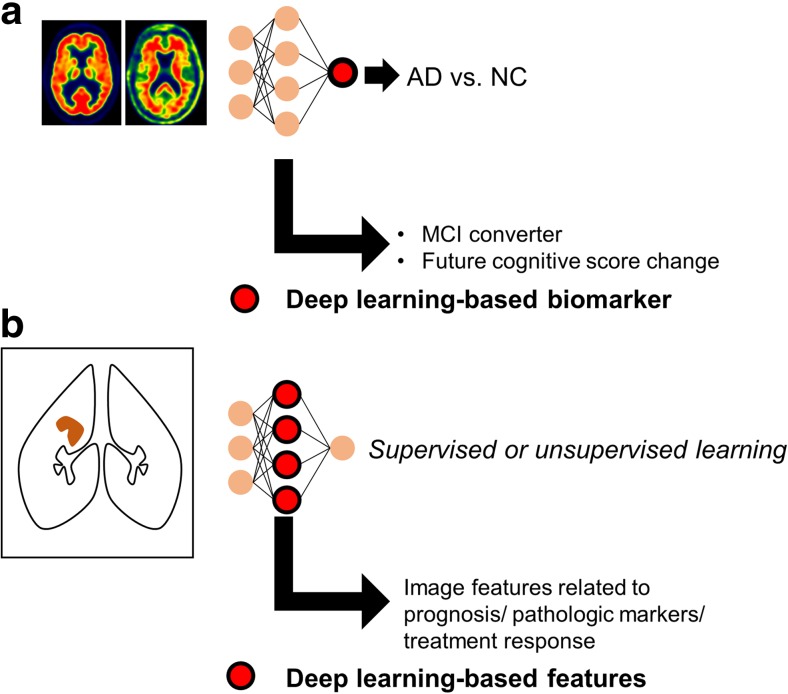
DL can be directly applied to molecular imaging to obtain reliable biomarkers. Deep CNN models for classifying each subject’s image is a representative direct application of DL as a supervised learning. A study of deep CNN application to mediastinal metastatic lymphadenopathy detection using FDG PET was built by supervised training [32]. Multimodal images including PET and MRI were used as inputs of DL models for the disease classification problems. A report used manual image features of PET and MRI to discriminate Alzheimer’s disease from normal controls [58]. Another recent study suggested a deep 3-dimensional CNN model that used a pair of FDG and florbetapir PET images as input and discriminated Alzheimer’s disease from normal controls [59]. In terms of biomarker development, a quantitative marker representing patients’ outcome based on image data has been required instead of simple diagnostic classification of images. The CNN model using FDG and florbetapir PET images was directly transferred to mild cognitive impairment patients to identify who would rapidly convert into full-blown dementia [59]. In result, the final output of the model could be used for future cognitive score decline. Thus, the final output of deep neural network could be used for a biomarker to predict future cognitive decline (Fig. 4a). Another directly extracted biomarker using DL was reported in dopamine transporter imaging [60]. The model trained for discriminating FP-CIT SPECT images of Parkinson’s disease from normal was applied to scans without evidence of dopaminergic deficit (SWEDD) patients determined by human reading. The DL-based model revealed that a number of SWEDD patients already had abnormal imaging patterns at baseline, which suggested the output of DL could be used for refinement of diagnosis for Parkinson’s disease and SWEDD. Briefly, the role of DL in such functional imaging is not limited to image classification but extended to provide a quantitative output value summarizing multidimensional and multimodal image data, which can be used for the predictive marker.
Fig. 4.
Two types of deep learning-based imaging biomarkers. a Outputs of deep learning model can be used as a biomarker. A deep convolutional neural network model for discriminating Alzheimer’s disease could be directly transferred into identification of mild cognitive impairment patients who would rapidly convert to dementia. Additionally, the output score of the model was well correlated with future decline of cognitive score. b As deep learning is an automated discriminative feature learning, hierarchical features of neural networks can be used as multidimensional biomarkers. Image features related to prognosis or treatment response can be selected after the training of deep learning models
Since DL is a representation learning, DL-based features automatically extracted from images can be used for imaging biomarkers. Some recent studies define these features as ‘DL-based radiomics features’ [61, 62]. ‘Radiomics’ is originally an emerging concept that represents a series of quantitative analysis for identifying high-dimensional image features which can provide predictive and prognostic information [63, 64]. So far, the features have been manually extracted, which included texture features as well as simple parameters such as mean and maximum SUV. On the other hand, as DL is a feature learning method, the hierarchical features of hidden layers can be used as radiomics features (Fig. 4b). A study suggested DL-based image features could be related to patients’ prognosis from chest CT images [61]. Another study using MR images showed that DL model trained for tumor segmentation can be used as a feature extractor, which could identify predictive features for IDH1 mutation in low-grade glioma [62]. DL-based features can be obtained by supervised learning as well as unsupervised learning. A study identified autism-related functional connectivity patterns using an autoencoder, a type of unsupervised learning, for brain network analysis [65]. Though DL-based features are optimized for image discrimination and effectively summarize high-dimensional image data, their usage as a clinically reliable biomarker should be validated in large, independent cohorts.
The direct extraction of DL-based image biomarkers may facilitate utilization of molecular imaging in precision medicine. The key to successful precision medicine relies on the development of good biomarkers that define individual variability as the basic concept of precision medicine to plan prevention and treatment strategies taking individual variability into account [66]. DL can play a role in translating high-dimensional and complex images into quantitative and objective values. Though visual analysis and manual feature extraction such as SUV also provide information on molecular imaging, application of DL enriches the quantitative information by capturing the most discriminative features of images.
Future Directions and Challenges
DL has received great attention in the medical imaging domain because of its success in various scientific and industrial fields. Due to the unique features of molecular imaging, DL should be appropriately modified for molecular imaging analysis. Most of all, the aim of DL application in molecular imaging is different from previous common usage of DL. These images represent physiologic and pathologic processes which require discovering crucial and latent biological information instead of simple diagnostic classification. Thus, DL in molecular imaging should focus on extracting good biomarkers from multimodal and high-dimensional image data.
Although DL has only recently begun to be applied to molecular imaging fields, there are some challenges to be solved in terms of biomarker discovery based on DL. Firstly, as the aim of DL application is to capture good prognostic and predictive information, the target of DL training needs to be diversified. So far, most of the medical application of DL has been aimed at diagnostic classification. However, the DL-based biomarker is aimed at predicting individual subjects’ outcome and treatment response. A good example of another clinically feasible training target is the usage of specialized regression for survival data. Recent studies suggested a prognostic stratification model based on genomic data and DL, which was trained by survival time and events [67, 68]. This DL-based survival analysis can provide a robust prognostic score from the patients’ data. This type of method can be applied to the patients’ molecular imaging data for risk stratification. Secondly, clinical application of DL-based molecular imaging diagnostics requires diagnostic uncertainty and diagnosis of multiple domains. Current DL models are optimized for classification and regression without suggestion of decision reliability. However, the uncertainty measure of the DL-based decision is critical in the clinical setting. Cases where it is difficult to determine diagnosis or produce quantitative value should be identified as they need additional diagnostic tests. Recent studies have attempted to combine Bayesian approximation with DL for uncertainty measurement [69], which will be used in clinical application of DL. Third, application of unsupervised learning will be increased to exploit unlabeled image data and clinical implication. The large dataset of pairs of image data and confirmed diagnosis is limited in spite of the large amount of raw image data. Unsupervised learning is used for image feature extraction, which can be applied to supervised learning with relatively small-sized datasets. Additionally, clustering of data, a result of unsupervised learning, may give additional insights of subgroups of disorders. A clinically permeated example of unsupervised clustering is breast cancer subtyping based on gene expression data [70]. In this context, we expect patients subtyping based on DL-based molecular imaging biomarkers. DL-based molecular imaging biomarker may support individualized risk stratification and personalized treatment by playing an important role in precision medicine.
Roles as an Expert in Nuclear Medicine and Molecular Imaging
Due to the recent development of DL, a myth that the practice of visual tasks in medicine including nuclear medicine image reading will be replaced by machines has emerged. Even though it is difficult to predict the future medical environment, it will be inevitable that some specific tasks are replaced by automated systems. For example, conventional image interpretation tasks such as detecting lesions and simple diagnostic classification may be considerably replaced. Although the clinical value of DL-based conventional nuclear image interpretation has not yet been fully proved, it is evident that DL-based interpretation has the great advantage of the absence of interobserver variability as reported in the DL-based model for FP-CIT SPECT interpretation [60]. Furthermore, the model showed higher accuracy than visual reading for discriminating Parkinson’s disease [60]. For common radiologic images such as mammography, DL-based interpretation showed comparable diagnostic classification to experts’ reading for specific tasks [71]. Nonetheless, the key to molecular imaging is to provide good biomarkers reflecting biological processes. The role of DL in molecular imaging will be capturing maximum information from images by generating predictive biomarkers, which is not related to replacement of human tasks. In this regard, the role of nuclear medicine physicians will be changed to translate and integrate biomarkers automatically extracted from image data for clinical decision making. Various DL applications such as anatomical segmentation and image processing will indirectly help to obtain and augment well-known biomarkers. Image features and biomarkers directly obtained by DL will be used to predict clinical outcomes as well as select individualized treatment options. Thus, for nuclear medicine physicians, knowledge of multimodal data and analytic methods will be needed for the translation of molecular imaging data to clinical implications.
DL applications in molecular imaging will eventually facilitate precision medicine. It will help to make the most of information in molecular imaging. To accelerate proper DL application in molecular imaging, more emphasis should be placed on the intent of each molecular imaging as DL aims at developing good biomarkers. Furthermore, large and open databases are needed to advance the techniques. The innovation of DL in computer vision was initiated by a publicly available large dataset, ImageNet. Large genomic databases such as The Cancer Genome Atlas have accelerated the understanding of cancer and development of novel biomarkers based on big omics data analysis. Recently, data sharing has started to be seriously discussed for raising integrity and accelerating medical research [72]. In medical imaging fields, large datasets in common modalities such as chest X-ray data have started to open [73]. Accordingly, it is urgently required to establish large databases of molecular imaging data as well. It will be the starting point for molecular imaging as an important part of future data-driven precision medicine.
Conclusion
Medical decisions are made by a comprehensive interpretation of all relevant patient data including symptoms, signs, laboratory tests, and imaging. As reviewed in this article, DL has advantages in automatically extracting discriminative features in high-dimensional data. Thus, DL will largely impact medical field tasks particularly related to quantitative analysis. As molecular imaging is a set of image data which provides quantitative information, various DL techniques will be rapidly applied to molecular imaging. Even though some current tasks may be partly replaced by automatic systems, DL may enhance usage of molecular imaging as it can maximally extract information obtained by imaging data to develop biomarkers. As experts in translating molecular imaging information to clinical decision making, we need to focus on exploiting DL for developing biomarkers from molecular imaging to be an important part of the precision medicine era.
Compliance with Ethical Standards
Conflict of Interest
Hongyoon Choi declares no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
For this study formal consent is not required.
References
- 1.Weber GM, Mandl KD, Kohane IS. Finding the missing link for big biomedical data. JAMA. 2014;311:2479–2480. doi: 10.1001/jama.2014.4228. [DOI] [PubMed] [Google Scholar]
- 2.Kuo M-H, Sahama T, Kushniruk AW, Borycki EM, Grunwell DK. Health big data analytics: current perspectives, challenges and potential solutions. Int J Big Data Intell. 2014;1:114–126. doi: 10.1504/IJBDI.2014.063835. [DOI] [Google Scholar]
- 3.Bengio Y. Learning deep architectures for AI. Foundations and trends® in. Mach Learn. 2009;2:1–127. doi: 10.1561/2200000006. [DOI] [Google Scholar]
- 4.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 5.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 6.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Adv Neural Inf Proces Syst. 2012;25:1090–8.
- 7.Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA-and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning–based sequence model. Nat Methods. 2015;12:931. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M et al. A survey on deep learning in medical image analysis. arXiv:170205747. 2017. [DOI] [PubMed]
- 10.Xu Y, Mo T, Feng Q, Zhong P, Lai M, Eric I et al., editors. Deep learning of feature representation with multiple instance learning for medical image analysis. Acoustics, Speech and Signal Processing (ICASSP), 2014 I.E. International Conference; 2014.
- 11.Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35:1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 12.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 13.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of deep learning in biomedicine. Mol Pharm. 2016;13:1445–1454. doi: 10.1021/acs.molpharmaceut.5b00982. [DOI] [PubMed] [Google Scholar]
- 15.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Cogn Model. 1988;5:1. [Google Scholar]
- 16.Bengio Y, Lamblin P, Popovici D, Larochelle H. Greedy layer-wise training of deep networks. Adv Neural Inf Proces Syst. 2006;19:153–160.
- 17.Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- 19.Ioffe S, Szegedy C, editors. Batch normalization: accelerating deep network training by reducing internal covariate shift. Proceedings of the 32nd International Conference on Machine Learning; 2015.
- 20.Nair V, Hinton GE, editors. Rectified linear units improve restricted boltzmann machines. Proceedings of the 27th International Conference on Machine Learning (ICML-10); 2010.
- 21.Oquab M, Bottou L, Laptev I, Sivic J, editors. Learning and transferring mid-level image representations using convolutional neural networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2014.
- 22.LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86:2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 23.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D et al., editors. Going deeper with convolutions. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015.
- 24.He K, Zhang X, Ren S, Sun J, editors. Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016.
- 25.Vinyals O, Toshev A, Bengio S, Erhan D, editors. Show and tell: a neural image caption generator. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015.
- 26.Girshick R, editor. Fast r-cnn. Proceedings of the IEEE International Conference on Computer Vision; 2015.
- 27.Ren S, He K, Girshick R, Sun J. Faster R-CNN: Towards real-time object detection with region proposal networks. Adv Neural Inf Proces Syst. 2015;28:91–9. [DOI] [PubMed]
- 28.Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. arXiv:160600915. 2016. [DOI] [PubMed]
- 29.Badrinarayanan V, Kendall A, Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. arXiv:151100561. 2015. [DOI] [PubMed]
- 30.Elmore JG, Longton GM, Carney PA, Geller BM, Onega T, Tosteson AN, et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA. 2015;313:1122–1132. doi: 10.1001/jama.2015.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keyes JW., Jr SUV: standard uptake or silly useless value? J Nucl Med. 1995;36:1836–1839. [PubMed] [Google Scholar]
- 32.Wang H, Zhou Z, Li Y, Chen Z, Lu P, Wang W, et al. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18 F-FDG PET/CT images. EJNMMI Res. 2017;7:11. doi: 10.1186/s13550-017-0260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, et al. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Li R, Deng H, Wang L, Lin W, Ji S, et al. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage. 2015;108:214–224. doi: 10.1016/j.neuroimage.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth HR, Lu L, Farag A, Shin H-C, Liu J, Turkbey EB et al., editors. Deeporgan: multi-level deep convolutional networks for automated pancreas segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention.Berlin: Springer; 2015.
- 36.Choi H, Jin KH. Fast and robust segmentation of the striatum using deep convolutional neural networks. J Neurosci Methods. 2016;274:146–153. doi: 10.1016/j.jneumeth.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Shen W, Zhou M, Yang F, Yang C, Tian J, editors. Multi-scale convolutional neural networks for lung nodule classification. International Conference on Information Processing in Medical Imaging. Berlin: Springer; 2015. [DOI] [PubMed]
- 38.Setio AAA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel SJ, et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging. 2016;35:1160–1169. doi: 10.1109/TMI.2016.2536809. [DOI] [PubMed] [Google Scholar]
- 39.Kleesiek J, Urban G, Hubert A, Schwarz D, Maier-Hein K, Bendszus M, et al. Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. NeuroImage. 2016;129:460–469. doi: 10.1016/j.neuroimage.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 40.de Brebisson A, Montana G, editors. Deep neural networks for anatomical brain segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops; 2015.
- 41.Chen H, Dou Q, Yu L, Heng P-A. Voxresnet: deep voxelwise residual networks for volumetric brain segmentation. arXiv:160805895. 2016. [DOI] [PubMed]
- 42.Avendi M, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2016;30:108–119. doi: 10.1016/j.media.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Charbonnier J-P, Van Rikxoort EM, Setio AA, Schaefer-Prokop CM, van Ginneken B, Ciompi F. Improving airway segmentation in computed tomography using leak detection with convolutional networks. Med Image Anal. 2017;36:52–60. doi: 10.1016/j.media.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35:1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 45.Ghafoorian M, Karssemeijer N, Heskes T, van Uden IW, Sanchez CI, Litjens G, et al. Location sensitive deep convolutional neural networks for segmentation of white matter hyperintensities. Sci Rep. 2017;7. 10.1038/s41598-017-05300-5. [DOI] [PMC free article] [PubMed]
- 46.Trebeschi S, van Griethuysen JJ, Lambregts DM, Lahaye MJ, Parmer C, Bakers FC, et al. Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci Rep. 2017;7 [DOI] [PMC free article] [PubMed]
- 47.Han D, Yu J, Yu Y, Zhang G, Zhong X, Lu J, et al. Comparison of 18 F-fluorothymidine and 18 F-fluorodeoxyglucose PET/CT in delineating gross tumor volume by optimal threshold in patients with squamous cell carcinoma of thoracic esophagus. Int J Radiat Oncol Biol Phys. 2010;76:1235–1241. doi: 10.1016/j.ijrobp.2009.07.1681. [DOI] [PubMed] [Google Scholar]
- 48.Tylski P, Stute S, Grotus N, Doyeux K, Hapdey S, Gardin I, et al. Comparative assessment of methods for estimating tumor volume and standardized uptake value in 18F-FDG PET. J Nucl Med. 2010;51:268–276. doi: 10.2967/jnumed.109.066241. [DOI] [PubMed] [Google Scholar]
- 49.Dong C, Loy CC, He K, Tang X. Image super-resolution using deep convolutional networks. IEEE Trans Pattern Anal Mach Intell. 2016;38:295–307. doi: 10.1109/TPAMI.2015.2439281. [DOI] [PubMed] [Google Scholar]
- 50.Kim J, Kwon Lee J, Mu Lee K, editors. Accurate image super-resolution using very deep convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016.
- 51.Xiang L, Qiao Y, Nie D, An L, Wang Q, Shen D. Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing. 2017. 10.1016/j.neucom.2017.06.048. [DOI] [PMC free article] [PubMed]
- 52.Jiao J, Ourselin S. Fast PET reconstruction using Multi-scale Fully Convolutional Neural Networks. arXiv:170407244. 2017.
- 53.Jin KH, McCann MT, Froustey E, Unser M. Deep convolutional neural network for inverse problems in imaging. IEEE Trans Image Process. 2017;26:4509–4522. doi: 10.1109/TIP.2017.2713099. [DOI] [PubMed] [Google Scholar]
- 54.Isola P, Zhu J-Y, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. arXiv:161107004. 2016.
- 55.Nie D, Trullo R, Petitjean C, Ruan S, Shen D. Medical Image Synthesis with Context-Aware Generative Adversarial Networks. arXiv:161205362. 2016. [DOI] [PMC free article] [PubMed]
- 56.Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. 2017;44:1408–1419. doi: 10.1002/mp.12155. [DOI] [PubMed] [Google Scholar]
- 57.Bezrukov I, Mantlik F, Schmidt H, Schölkopf B, Pichler BJ, editors. MR-based PET attenuation correction for PET/MR imaging. Seminars in nuclear medicine. Amsterdam: Elsevier; 2013. [DOI] [PubMed]
- 58.Suk H-I, Lee S-W, Shen D. Initiative AsDN. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct Funct. 2015;220:841–859. doi: 10.1007/s00429-013-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi H, Jin KH. Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. arXiv:170406033. 2017. [DOI] [PubMed]
- 60.Choi H, Ha S, Im HJ, Paek SH, Lee DS. Refining diagnosis of Parkinson's disease with deep learning-based interpretation of dopamine transporter imaging. Neuroimage Clin. 2017. 10.1016/j.nicl.2017.09.010. [DOI] [PMC free article] [PubMed]
- 61.Oakden-Rayner L, Carneiro G, Bessen T, Nascimento JC, Bradley AP, Palmer LJ. Precision radiology: predicting longevity using feature engineering and deep learning methods in a radiomics framework. Sci Rep. 2017;7:1648. doi: 10.1038/s41598-017-01931-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep. 2017;7 [DOI] [PMC free article] [PubMed]
- 63.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Cavalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5 [DOI] [PMC free article] [PubMed]
- 64.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi H. Functional connectivity patterns of autism spectrum disorder identified by deep feature learning. arXiv:170707932. 2017.
- 66.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaudhary K, Poirion OB, Lu L, Garmire L. Deep Learning based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2017. 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed]
- 68.Choi H, Na KJ. A risk stratification model for lung cancer based on gene coexpression network. bioRxiv. 2017. 10.1101/179770. [DOI] [PMC free article] [PubMed]
- 69.Gal Y, Ghahramani Z, editors. Dropout as a Bayesian approximation: representing model uncertainty in deep learning. International Conference on Machine Learning; 2016.
- 70.Van De Vijver MJ, He YD, Van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 71.Carneiro G, Nascimento J, Bradley AP, editors. Unregistered multiview mammogram analysis with pre-trained deep learning models. International Conference on Medical Image Computing and Computer-Assisted Intervention. Berllin: Springer; 2015.
- 72.Warren E. Strengthening research through data sharing. N Engl J Med. 2016;375:401–403. doi: 10.1056/NEJMp1607282. [DOI] [PubMed] [Google Scholar]
- 73.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. arXiv:170502315. 2017.