Abstract
Na+/Ca2+ exchangers are low-affinity high-capacity transporters that mediate Ca2+ extrusion by coupling Ca2+ efflux to the influx of Na+ ions. The Na+/Ca2+ exchangers form a super-family comprised of three branches each differing in ion-substrate selectivity: Na+/Ca2+ exchangers (NCX), Na+/Ca2+/K+ exchangers, and Ca2+/cation exchangers. Their primary function is to maintain Ca2+ homeostasis and play a particularly important role in excitable cells that experience transient Ca2+ fluxes. Research into the role and activity of Na+/Ca2+ exchangers has focused extensively on the cardio-vascular system, however, growing evidence suggests that Na+/Ca2+ exchangers play a key role in neuronal processes such as memory formation, learning, oligodendrocyte differentiation, neuroprotection during brain ischemia and axon guidance. They have also been implicated in pathologies such as Alzheimer’s disease, Parkinson’s disease, Multiple Sclerosis and Epilepsy, however, a clear understanding of their mechanism during disease is lacking. To date, there has never been a central resource or database for Na+/Ca2+ exchangers. With clear disease relevance and ever-increasing research on Na+/Ca2+ exchangers from both model and non-model species, a database that unifies the data on Na+/Ca2+ exchangers is needed for future research. NCX-DB is a publicly available database with a web interface that enables users to explore various Na+/Ca2+ exchangers, perform cross-species sequence comparison, identify new exchangers, and stay-up to date with recent literature. NCX-DB is available on the web via an interactive user interface with an intuitive design, which is applicable for the identification and comparison of Na+/Ca2+ exchanger proteins across diverse species.
Electronic supplementary material
The online version of this article (10.1186/s12868-018-0423-2) contains supplementary material, which is available to authorized users.
Keywords: NCX, NCKX, NCLX, Sodium calcium exchanger, Database, Antiporter
Background
The sodium calcium exchanger superfamily of antiporters function to maintain calcium homeostasis. This superfamily is composed of the Na+/Ca2+ exchangers (NCX) which couple the extrusion of 1 Ca2+ ion with the influx of 3 Na+ ions, the Na+/Ca2+/K+ exchangers (NCKX) which exchange 1 Ca2+ ion and 1K+ ion in exchange for 4 Na+ ions, and Ca2+/Cation exchangers (also called NCLX) which couple the extrusion of 1 Ca2+ ion with the influx of either 3 Na+ or 3 Li+ ions [1–11]. The Na+/Ca2+ exchanger super-family is highly conserved across species in both invertebrate and vertebrate organisms [12–16]. The activity of the exchanger and rate of calcium extrusion is largely dependent on intracellular calcium levels. Studies of NCX in retinal cells of Drosophila melanogaster revealed that forward mode activity is initiated when cytosolic calcium levels reach 500 nM [17]. During high intracellular Na+ concentrations, Na+/Ca2+ exchangers will reverse by initiating calcium influx [9, 18]. For instance, reverse-mode NCX plays a role in regulating glutamatergic glial transmission in rat cortical astrocytes [19]: during resting state in cortical astrocytes, the Na+/K+ ATPase pump regulates cellular Na+ homeostasis by extruding Na+ and the Plasma Membrane Ca2+ ATPase pump regulates cellular calcium homeostasis by extruding calcium [19]; however, upon mechanical stimulation, an increase in intracellular Na+ causes NCX to function in reverse mode, which mediates calcium entry into astrocytes, and increase in intracellular calcium stimulates glutamatergic gliotransmission between astrocytes and neurons [19]. Similarly in Purkinje neurons, an increase in intracellular sodium triggers NCX to function in the reverse mode [20]. Reverse mode NCX also plays a role in astrogliosis whereby astrocytes proliferate and migrate into areas of neuronal damage [21]. Consequently, attenuation of NCX activity via a pharmacological inhibitor results in decreased calcium transients within astrocytes following mechanical injury and stimulation [21]. Although they can facilitate bidirectional transport, under normal physiological conditions, Na+/Ca2+ exchangers function primarily in the forward direction by coupling the extrusion of calcium with the influx of sodium ions [22]. Under pathophysiological conditions is has been shown that NCX can play a neuroprotective role. Ischemic preconditioning is a neuroprotective mechanism in which a brief ischemia protects the brain from a subsequent lethal insult. During a non-injurious episode of brain ischemia it was shown that both NCX1 and NCX3 were up-regulated where they conferred neuroprotective roles, and knockdown of NCX1 and NCX3 reversed this neuroprotective effect [23, 24]. Furthermore, increased expression of NCX3 was observed after a prolonged harmful ischemic episode (ischemic post-conditioning), where again it was shown to confer a neuroprotective effect [23]. More recently it has been shown that sumoylation of the lysine at position 590 of NCX3 may be a key determinant for enhancing ischemic preconditioning-induced neuroprotection [25]. Thus, during both normal and pathophysiological conditions, NCX plays a central role in homeostasis by regulating Ca2+ exchange.
Genetic studies of NCX in the nervous system, have revealed a role for NCX3 in the expression of myelin marker genes: CNPase and myelin basic protein [26]. Moreover, ncx3−/− mice exhibit reduced spinal cord size, thereby implicating NCX3 in spinal cord formation [26]. NCX3 is also expressed in the CA1 region of the hippocampus [27]. In ncx3−/− knockout mice, basal intracellular calcium levels are elevated and presynaptic intracellular calcium levels exhibit a slower decline after depolarization [27]. The NCX type exchanger, NCX2, is also expressed in CA1 pyramidal neurons [28]. In ncx2−/− knockout mice, clearance of intracellular calcium is significantly delayed at the presynaptic site of CA1 neurons [28] and consequently, ncx2−/− knockout mice show significantly enhanced performance in learning and memory behaviors [28]. NCX1 has been shown to regulate neurite outgrowth in cortical neurons via modulation of ER calcium content and PI3K/Akt signaling [29], and more recently research in Caenorhabditis elegans has revealed a role for the NCLX-type exchanger NCX-9 in regulating asymmetric patterning of a motor circuit by engaging Netrin and heparan sulfate signaling effectors [30].
Na+/Ca2+ exchangers are widely expressed across the nervous system (as well as many other tissues and cell types) where they function in development, learning, memory formation, and motor function. They have also been implicated in several disease states of the nervous system such as Alzheimer’s disease, Parkinson’s disease, Multiple Sclerosis and Epilepsy, however, the exact role and mechanism of Na+/Ca2+ exchangers during disease is still unclear [31–40]. The field of researchers that study Na+/Ca2+ exchangers encompass many different branches of biology and employ diverse model systems to address various aspects of their biology [41]. In order to facilitate the exchange of data and further the field of Na+/Ca2+ exchange biology, there is a clear need for a central database that unifies Na+/Ca2+ exchanger data across species to facilitate analysis and discovery. NCX-DB is the first database of Na+/Ca2+ exchangers, and represents a central resource for discovery, cross-species comparison, while also providing a portal to recent literature in the field of Na+/Ca2+ exchangers.
Construction and content
Data contained within NCX-DB was mined from UniProt (ver. June 2017) [42] as well as the animal model organism databases that comprise the Alliance of Genome Resources (http://www.alliancegenome.org/). These are: WormBase (ver. WS260) [43], XenBase (ver. 4.5.0) [44], FlyBase (FB2017_04) [45], the Zebrafish Information Network (ZFIN: https://zfin.org/), Mouse Genomics Informatics (http://www.informatics.jax.org/), and the Rat Genome Database (https://rgd.mcw.edu/). By including the Alliance Genomes databases we could include all the major model organisms, and we chose UniProt as it is considered the most comprehensive protein database because it includes literature based curation in its functional annotations. Candidate exchangers for NCX-DB BLAST database were mined using text-based queries that relied upon literature based annotations and also by using BlastP and psi-Blast searches of downloaded genomes from each database (Fig. 1 and Table 1). Candidate exchangers were then aligned using MUSCLE [46] and filtered through a Position Weighted Matrix (PWM) based upon known sodium calcium exchanger transport repeats (α1 and α2) [6, 47–52]. To design the PWM, a Position Probability Matrix (PPM) was determined from the residue frequency of each site of aligned known sodium calcium exchangers. The PPM (M) was then used to calculate a PWM, M’, which was generated by transforming the matrix with a background model, bi, that assumes equal residue frequency, as follows:
where, Mij is the probability of residue i at position j in M above.
Fig. 1.
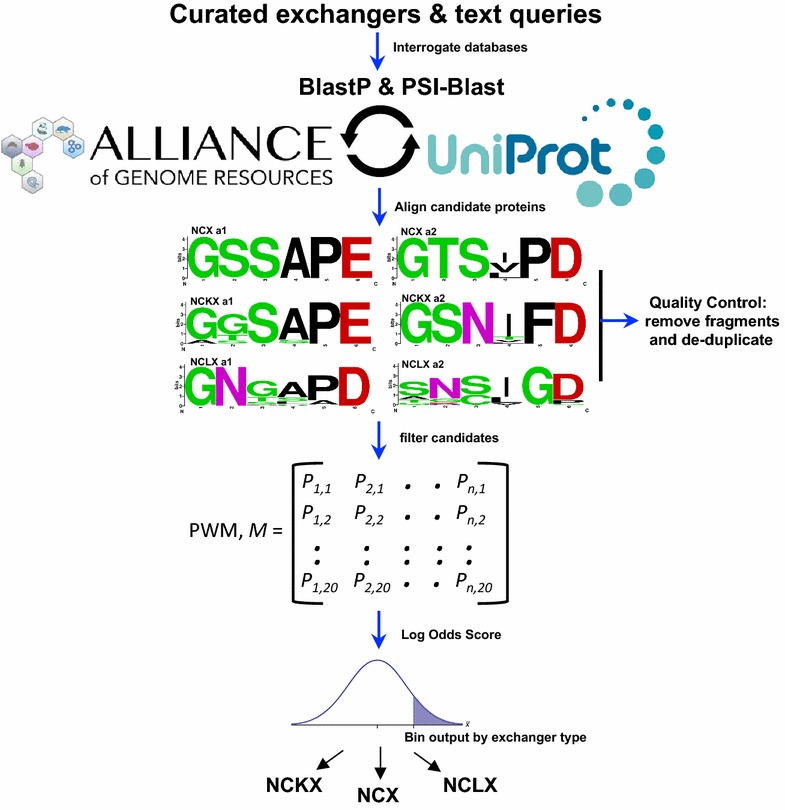
NCX-DB data workflow. Candidate exchangers were sourced from UniProt as well as the animal model organism databases that comprise the Alliance of Genome Resources. These model organism databases are: WormBase, XenBase, FlyBase, the Zebrafish Information Network, Mouse Genomics Informatics, and the Rat Genome Database. The resulting candidate exchangers were then aligned and filtered through a Position Weighted Matrix (PWM) to return a log odds score used to classify exchangers by subtype
Table 1.
Summary of NCX-DB BLAST database content
| Classification | Number of entries |
|---|---|
| Sodium Calcium Exchangers | 483 |
| Total amino-acids (a.a) | 348,524 |
| Average protein length (a.a) | 723 |
| Largest protein length (a.a) | 1283 |
| Smallest protein length (a.a) | 242 |
| Acidic residues | 56,677 |
| Basic residues | 35,288 |
| Neutral residues | 56,677 |
| Most common amino acid | Leucine (33,547) |
| Least common amino acid | Tryptophan (6031) |
Using this matrix, candidate exchangers were filtered by exchanger type based upon their Log Odds score. The alpha-repeats have been shown to form a diamond shaped transport vestibule which facilitates ion transport [53]. The filtered data was then converted to protein databases using BLAST [54] local executables (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/) and organized in various databases by taxonomy and exchanger subtype. BLAST searches are ran on the server-side using PHP scripts and served asynchronously using Ajax (http://api.jquery.com/jquery.ajax/) and jQuery (http://jquery.com/). FASTA formatted sequences can be retrieved at the NCX-DB BLAST page by using the NCBI Entrez Programming Utilities (https://www.ncbi.nlm.nih.gov/books/NBK25501/). Accession numbers can be entered as a comma separated list and retrieved remotely. The following parameters are implemented for BlastP searches using NCX-DB: initial word_size match is 3; the minimum threshold score to add a word to the BLAST lookup table is 11; the composition-based score adjustment is defined as per [55]; the heuristic value (in bits) for the final gapped alignment is 25; and the window size for multiple hits is set to 40.
The un-filtered exchanger data is also available at NCX-DB under the Browse link. The Browse data includes fragments, and is comprised of predicted exchangers collected into NoSQL tables and converted into searchable JavaScript object notation (JSON) objects. Data is visualized using jQuery table plugins. The user can select the number of entries to view per page (default is 25), and search the data by species (common or Latin name), or exchanger. The data can also be ordered into groups by clicking on the various table headers. Entries within the browsing tables also return URLs for each entry which link to various databases that were used to source the exchangers within NCX-DB.
In order to facilitate the discovery of novel exchangers or characterize predicted exchangers, NCX-DB contains a tool under the Predict link which searches a user supplied protein sequence for candidate sodium calcium exchanger transport repeats. To search for candidate exchanger motifs, NCX-DB will scan a user supplied sequence for a fuzzy match to the motif; if a match is found, NCX-DB then uses the PWM described above, to sum the corresponding log likelihood values at each residue to return a score for the candidate exchanger motif. The Predict tool provides a graphical sequence output highlighting the position of the alpha repeats within the candidate exchanger, and also a chart depicting residue frequency. The sequence graphical output was modified from the neXtProt protein sequence viewer available at biojs.io, and the chart is generated using a JavaScript function to convert the residue frequency into a JSON object for canvas.js (https://canvasjs.com/).
To examine the phylogenetic relationship between exchanger subtypes, the user can visit the Phylogeny page and view radial unrooted phylogenetic trees for each exchanger subtype (i.e. NCX, NCKX, and NCLX). Protein sequences for each tree was aligned using MUSCLE [46, 56] and the resulting alignments were used to generate Newick formatted trees using PhyML [57, 58]. Newick trees were converted to phyloXML (http://www.phyloxml.org/) and visualized using jsPhyloSVG [59]. XML files for each tree was edited to include clade and phylum specific features so as to make the experience more interactive. Each tip on the tree is also linked to other databases where the user can obtain more information and background on each exchanger.
Finally, NCX-DB also contains a Stats page which details the number of exchangers, exchanger sub-types, and residue counts contained within the database, as well as a Twitter feed for NCX-DB (Twitter handle: @ncxdb). The Twitter account for NCX-DB serves as an information service about all things related to sodium calcium exchangers (meetings, societies, etc.) while also providing data related to NCX-DB. In addition the Twitter account has been configured to serve as a twitterbot that captures the RSS feed on searches related to sodium calcium exchangers from PubMed (https://www.ncbi.nlm.nih.gov/pubmed) as well as the pre-print servers: bioRxiv (https://www.biorxiv.org/) and arXiv (https://arxiv.org/). Therefore, visitors can keep up-to-date with the literature related to sodium calcium exchangers by following @ncxdb or by visiting NCX-DB. The front-end for NCX-DB was written in JavaScript and uses Bootstrap (http://getbootstrap.com/) and jQuery (http://jquery.com/) to provide an intuitive graphical user interface (Fig. 2).
Fig. 2.
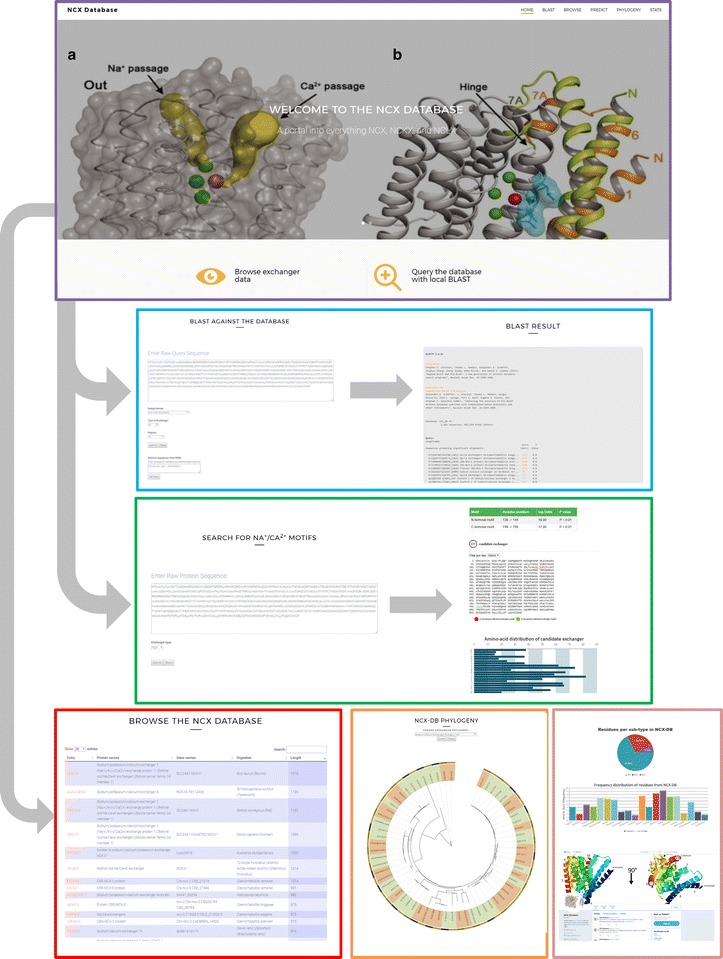
NCX-DB interface and features. The landing page of NCX-DB and various features are shown. From the landing page, users can navigate through various links to browse NCX-DB in table form, query NCX-DB via local BLAST executables, examine phylogenetic relationships, and characterize novel exchangers using the Predict tool. Users can view recent literature and developments in the field of Na+/Ca2+ exchange biology, and also link to other databases through NCX-DB. NCX-DB is designed with an interactive graphical user interface which is intuitive and provides seamless exploration and discovery options
Utility and discussion
NCX-DB represents the first centralized database for sodium calcium exchangers. Data contained within NCX-DB is mined from highly annotated databases so as to collate the most up to date and informative information for each exchanger across the main model organisms. On the landing page, the user can explore NCX-DB using the links along the top panel or by clicking on the graphical icons under the banner (Fig. 2). The same navigation menu along the top panel, can be viewed on all NCX-DB pages. The first link on the menu brings the user to the BLAST page for NCX-DB, where the user can input raw protein sequence data and perform BLAST searches of a query sequence against NCX-DB. Various drop-down menus permit taxonomic as well as exchanger specific BLAST searches. The user can also visualize the results in a variety of different formats, although the pairwise representation is set as the default. Sequences can also be remotely retrieved from The National Center for Biotechnology Information (NCBI) by simply entering an accession number in the textbox under Retrieve sequences from NCBI. Any number of accession numbers can be entered, however, for two or more, the accession numbers must be separated by a comma. To browse raw exchanger data, the user can click on the Browse link to view a table of predicted exchanger entries. The user can change the number of entries viewed per page in the table from 25 (default) to 100. By clicking on the table headers, the user can sort the data by heading for example, species, gene name, or exchanger length. Search terms can also be entered into the Search box at the top of the table to dynamically sort entries that contain search terms. As well as providing an overview of each entry, the Browsing tables also link to various databases which were used to source the exchangers within NCX-DB, therefore visitors can explore and learn more about specific exchangers by clicking on links in the first column of the browse table.
NCX-DB also provides facilities to characterize and search for new exchangers. To characterize candidate exchangers, users can use the Predict tool under the Predict link in the header menu. Users can enter the raw protein sequence of a candidate exchanger and the Predict tool at NCX-DB will scan the user supplied sequence for matches to exchanger specific domains. The Predict tool at NCX-DB returns a tabulated output with the position (in residues) of the candidate motifs and their resulting log odds score that are determined by using the PWM described above. In addition, a graphical output of the user supplied input is returned that illustrates the N terminal and C terminal candidate exchanger motifs highlighted in red and green respectively, as well as a chart of the amino-acid frequency distribution for the user-inputted sequence. Using this tool, users can characterize candidate exchangers.
Another bioinformatics tool that users can use to learn about sodium calcium exchangers is available under the Phylogeny link. At this page, users can examine interactive phylogenentic trees of representative exchangers from each of the three exchanger subgroups. The user can select the subtype from a drop-down menu and click submit to view a tree for that specific exchanger subtype. The resulting tree exhibits markings for the Phylum and clades which are denoted by colored arcs around the radial tree, and each tip can be clicked to reveal more information and also links to external databases for each exchanger. This feature enables users to explore the evolutionary history of an exchanger subtype and identify closely related exchangers to exchangers of interest.
A utility of NCX-DB and the Predict tool within NCX-DB is its ability to identify candidate exchangers and filter these exchangers into the appropriate exchanger subtype. In order to benchmark the ability of NCX-DB and Predict to identify and filter candidates appropriately, we compared our training data with a randomized data set which was generated using a customized Perl script (Additional file 1). In our filtering algorithm, NCX-DB searches for alpha repeat structures and filters candidates based upon log odds scores obtained for each motif. We generated a training data set comprised of known sodium calcium exchangers from mice, human, rat, Drosophila melanogaster, and Caenorhabditis elegans. The training data was only comprised of exchangers for which there is experimental and literature based evidence of functional NCX, NCKX or NCLX annotation. For our random data set we generated 100,000 random alpha repeat motifs for each exchanger type (Fig. 3) and plotted the density of their log odds scores (e.g. see Fig. 3a for random NCX alpha 1 repeat). In all cases we observed log odds scores around zero, which is what we would expect if the motifs were generated by chance. The 95th percentile log odds ranged from 4.3 to 6.4 (Fig. 3). Next, we compared the randomized data set with our training data to see how well NCX-DB predicts the correct exchanger in a background of randomized data. From this analysis, the ROC curves revealed AUC values close to 1 or at 1 for each motif: NCX alpha 1 repeat AUC = 0.979 and random NCX alpha 1 repeat AUC = 0.4744 (Fig. 3b); the NCX alpha 2 repeat AUC = 1 and random NCX alpha 2 repeat AUC = 0.4491 (Fig. 3d); NCKX alpha 1 repeat AUC = 1 and random NCKX alpha 1 repeat AUC = 0.4622 (Fig. 3f); NCKX alpha 2 repeat AUC = 1 and random NCKX alpha 2 repeat AUC = 0.4896 (Fig. 3h); NCLX alpha 1 repeat AUC = 1 and random NCLX alpha 1 repeat AUC = 0.509 (Fig. 3j); NCLX alpha 2 repeat AUC = 0.9985 and random NCLX alpha 2 repeat AUC = 0.5458 (Fig. 3l). These data revealed that NCX-DB predicts the correct exchanger in almost 100% of cases as compared with background simulated data. However, these tests do not take into account real background data, and nor do they consider how many exchangers NCX-DB may miss (i.e. false negatives). To address these concerns, we chose to use NCX-DB to predict exchanger subtypes within the Gorilla genome (Gorilla gorilla). We chose Gorilla for two reasons: firstly, a search in PubMed for “Gorilla” + “NCX” returns zero hits (as per 02/2018); therefore, little is known about the sodium calcium exchanger super-family in Gorilla and this makes it a good test case for determining how well NCX-DB can report exchanger subtypes from an unknown sample; secondly, we chose Gorilla because of its close phylogenetic relationship to humans. There has been a lot of work done on NCX from humans and therefore, we can hypothesis that the Gorilla genome should have at least the same number of exchanger subtypes as humans, and this will allow us to estimate the false negative rate for NCX-DB. We downloaded the genome of Gorilla (release 91) from Ensembl (https://useast.ensembl.org/Gorilla_gorilla/Info/Index), and used NCX-DB to identify candidate exchangers and filter them by exchanger subtype. We then aligned candidate exchangers using MUSCLE [56], and determined the best model of evolution from Prottest [60] before reconstructing the phylogenetic relationship of the candidate Gorilla exchangers with Human exchangers by maximum-likelihood using PhyML [57] (Fig. 4). In total, we identified 23 candidate exchangers from Gorilla. Four are predicted isoforms of NCX1, 2 are predicted isoforms of NCX2, and 3 are predicted isoforms for NCX-3. We identified 5 isoforms of NCKX1, 2 candidate isoforms of NCKX2, 2 candidate isoforms of NCKX3, a single NCKX4 ortholog, and 2 candidate isoforms of NCKX5. Finally, we also identified 2 candidate isoforms of NCLX (Fig. 4). The canonical isoforms for each subtype grouped perfectly with their human counterparts, with NCLX (NCKX6) forming a sister group to the NCKX subtypes (Fig. 4). These data provide clear evidence that the algorithm employed by NCX-DB can identify all the main exchanger subtypes from a genome with no published NCX exchanger experiments, and although there may still be exchangers that were missed, our algorithm clearly identified multiple isoforms for each exchanger subtype, with the exception of NCKX4 which was represented by a single protein. Overall, this suggests that our pipeline used to identify exchangers in NCX-DB returns almost no false positives, and based on the many isoforms predicted from the Gorilla genome, we conclude that at worst, NCX-DB exhibits a very low false negative rate. Although, more data would be needed to determine this rate more accurately.
Fig. 3.
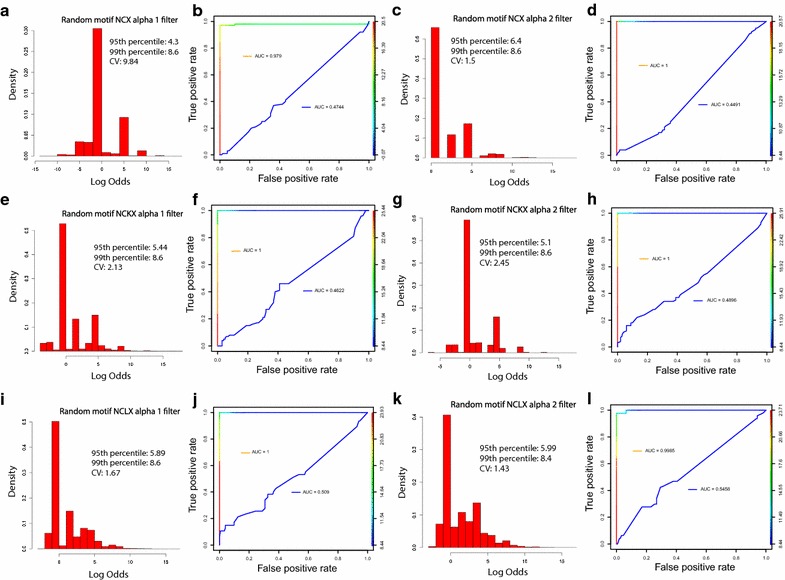
Comparison of NCX-DB exchanger classification to simulated random data. The classification feature of NCX-DB was benchmarked against a simulated dataset. In the NCX-DB filtering algorithm, NCX-DB searches for alpha repeat structures and filters candidates based upon log odds scores obtained for each motif. To benchmark this filtering algorithm, we compared the prediction accuracy of NCX-DB for a training dataset of known sodium calcium exchangers to a randomized data set. 100, 000 randomized alpha repeats were generated using a custom Perl script (Additional file 1). The density of log odds scores for each randomized alpha repeat is shown as histograms. a NCX alpha 1 repeat randomized data. 95th percentile = 4.3, 99th percentile = 8.6 coefficient of variation (CV) = 9.84; c NCX alpha 2 repeat randomized data. 95th percentile = 6.4, 99th percentile = 8.6, CV = 1.5; e NCKX alpha 1 repeat randomized data. 95th percentile = 5.44, 99th percentile = 8.6, CV = 2.13; g NCKX alpha repeat 2 randomized data. 95th percentile = 5.1, 99th percentile = 8.6, CV = 2.45; i NCLX alpha repeat 1 randomized data. 95th percentile = 5.89, 99th percentile = 8.6, CV = 1.67; k NCLX alpha repeat 2 randomized data. 95th percentile = 5.99, 99th percentile = 8.4, CV = 1.43. In all cases we observed the highest density of log odds scores close to zero. The 95th percentile log odds ranged from 4.3 to 6.4, and 99th percentile log odds ranged from 8.4 to 8.6. We then used this data to examine how well NCX-DB predicts the correct exchanger in a background of randomized data. The ROC curves in each case revealed AUC values close or at 1 for each motif: b NCX alpha 1 repeat AUC = 0.979 and random NCX alpha 1 repeat AUC = 0.4744; d the NCX alpha 2 repeat AUC = 1 and random NCX alpha 2 repeat AUC = 0.4491; f NCKX alpha 1 repeat AUC = 1 and random NCKX alpha 1 repeat AUC = 0.4622; h NCKX alpha 2 repeat AUC = 1 and random NCKX alpha 2 repeat AUC = 0.4896; j NCLX alpha 1 repeat AUC = 1 and random NCLX alpha 1 repeat AUC = 0.509; l NCLX alpha 2 repeat AUC = 0.9985 and random NCLX alpha 2 repeat AUC = 0.5458. Histograms were generated using the hist(x,…) function in R. ROC curves were made in R using the ROCR package (https://rocr.bioinf.mpi-sb.mpg.de/) using the perf ← performance(pred,”tpr”,”fpr”) to plot true positive rate against false positive rate. Control randomized data was added (add = TRUE) to the experimental colorized data
Fig. 4.
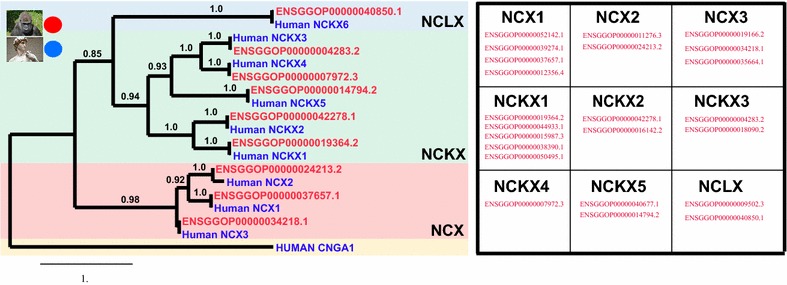
Identification and classification of sodium calcium exchangers from the Gorilla gorilla genome using NCX-DB. Phylogenetic tree constructed using PhyML [57] of the predicted canonical sodium calcium exchangers from Gorilla gorilla and human. The human cyclic nucleotide gated channel subunit, CNGA1, is used as an outgroup. 23 candidate sodium calcium exchangers isoforms were identified from Gorilla: 4 NCX1; 2 NCX2; 3 NCX3; 5 NCKX1; 2 NCKX2; 2 NCKX3, 1 NCKX4; 2 NCKX5; and 2 NCLX
Finally, when designing NCX-DB, we wanted to create a facility for researchers to stay up to date with new developments and literature as well as news in the field of sodium calcium exchangers. To accomplish this, we added a Stats page which provides a breakdown of data contained within NCX-DB and also a Twitter feed from a Twitter account which we set up for NCX-DB (Twitter handle @ncxdb). This Twitter account serves as an information center for everything related to sodium calcium exchangers and also a Twitterbot which automatically Tweets about new literature related to sodium calcium exchangers that come from PubMed (https://www.ncbi.nlm.nih.gov/pubmed), as well as the preprint servers: arXiv (https://arxiv.org/) and bioRxiv (https://www.biorxiv.org/). Twitter users can follow @ncxdb on Twitter or visit NCX-DB to stay up to date on new literature related to sodium calcium exchangers.
Conclusions
NCX-DB is the first database of sodium calcium exchangers and provides a unified platform and suite of tools for researchers to compare exchangers across species while also discovering and characterizing new exchangers. To construct NCX-DB, large numbers of exchangers were sourced from several highly curated databases so as to provide the most up-to-date information of annotated exchangers in diverse species. We will test for updates each time one of the sourced databases issues a new release, and then provide updates to NCX-DB accordingly; these updates will be announced through Twitter and online. NCX-DB also contains a literature portal, where researchers can stay informed of recent publications and developments in the field of sodium calcium exchangers. In conclusion, NCX-DB will serve as a useful platform for exploration and discovery in future experiments on Na+/Ca2+ exchange biology.
Availability of data
Project name: NCX-DB
Project home page: http://www.ncx-db.net
Operating system(s): Platform independent
Programming language: JavaScript, PHP, HTML, CSS
Other requirements: Modern Browser
License: GNU General Public License version 2, June 1991
Any restrictions to use by non-academics: None
Additional file
Additional file 1. Random Peptide. A Perl script used to generate random peptides for histograms in Fig. 3.
Authors’ contributions
DO’H conceived the idea for NCX-DB and designed the website. DO’H and KB designed the database and wrote the manuscript. KB performed testing of NCX-DB. Both authors read and approved the final manuscript.
Acknowledgements
I thank members of the O’Halloran lab for critical reading of the manuscript.
Competing interests
The authors declare no competing interests.
Availability of data and materials
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
Funding for this study was provided by The George Washington University (GWU) Columbian College of Arts and Sciences, GWU Office of the Vice-President for Research, and the GWU Department of Biological Sciences.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- NCX-DB
sodium calcium exchanger database
- NCX
sodium calcium exchanger
- NCKX
sodium calcium potassium exchanger
- NCLX
sodium calcium lithium exchanger
- CCX
calcium cation exchanger
- JSON
JavaScript object notation
- SQL
structured query language
- PWM
position weight matrix
- PPM
position probability matrix
- NCBI
National Center for Biotechnology Information
- BLAST
basic local alignment search tool
- RSS
rich site summary
- ROC
receiver operating characteristic
- AUC
area under the curve
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12868-018-0423-2) contains supplementary material, which is available to authorized users.
Contributor Information
Katrin Bode, Email: kb1988@gwmail.gwu.edu.
Damien M. O’Halloran, Phone: 202-994-8955, Email: damienoh@gwu.edu
References
- 1.Visser F, Lytton J. K + -dependent Na+/Ca2+ exchangers: key contributors to Ca2+ signaling. Physiology (Bethesda) 2007;22:185–192. doi: 10.1152/physiol.00001.2007. [DOI] [PubMed] [Google Scholar]
- 2.Lytton J, Li XF, Dong H, Kraev A. K+ -dependent Na+/Ca2+ exchangers in the brain. Ann N Y Acad Sci. 2002;976:382–393. doi: 10.1111/j.1749-6632.2002.tb04765.x. [DOI] [PubMed] [Google Scholar]
- 3.Kraev A, Quednau BD, Leach S, Li XF, Dong H, Winkfein R, Perizzolo M, Cai X, Yang R, Philipson KD, Lytton J. Molecular cloning of a third member of the potassium-dependent sodium-calcium exchanger gene family, NCKX3. J Biol Chem. 2001;276(25):23161–23172. doi: 10.1074/jbc.M102314200. [DOI] [PubMed] [Google Scholar]
- 4.Lytton J. Na+/Ca2+ exchangers: three mammalian gene families control Ca2+ transport. Biochem J. 2007;406(3):365–382. doi: 10.1042/BJ20070619. [DOI] [PubMed] [Google Scholar]
- 5.Haug-Collet K, Pearson B, Webel R, Szerencsei RT, Winkfein RJ, Schnetkamp PP, Colley NJ. Cloning and characterization of a potassium-dependent sodium/calcium exchanger in Drosophila. J Cell Biol. 1999;147(3):659–670. doi: 10.1083/jcb.147.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altimimi HF, Szerencsei RT, Schnetkamp PP. Functional and structural properties of the NCKX2 Na(+)-Ca (2 +)/K (+) exchanger: a comparison with the NCX1 Na (+)/Ca (2 +) exchanger. Adv Exp Med Biol. 2013;961:81–94. doi: 10.1007/978-1-4614-4756-6_8. [DOI] [PubMed] [Google Scholar]
- 7.Winkfein RJ, Pearson B, Ward R, Szerencsei RT, Colley NJ, Schnetkamp PP. Molecular characterization, functional expression and tissue distribution of a second NCKX Na+/Ca2+ -K+ exchanger from Drosophila. Cell Calcium. 2004;36(2):147–155. doi: 10.1016/j.ceca.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 8.Webel R, Haug-Collet K, Pearson B, Szerencsei RT, Winkfein RJ, Schnetkamp PP, Colley NJ. Potassium-dependent sodium-calcium exchange through the eye of the fly. Ann N Y Acad Sci. 2002;976:300–314. doi: 10.1111/j.1749-6632.2002.tb04753.x. [DOI] [PubMed] [Google Scholar]
- 9.Blaustein MP, Lederer WJ. Sodium/calcium exchange: its physiological implications. Physiol Rev. 1999;79(3):763–854. doi: 10.1152/physrev.1999.79.3.763. [DOI] [PubMed] [Google Scholar]
- 10.Palty R, Hershfinkel M, Sekler I. Molecular identity and functional properties of the mitochondrial Na+/Ca2+ exchanger. J Biol Chem. 2012;287(38):31650–31657. doi: 10.1074/jbc.R112.355867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palty R, Silverman WF, Hershfinkel M, Caporale T, Sensi SL, Parnis J, Nolte C, Fishman D, Shoshan-Barmatz V, Herrmann S, Khananshvili D, Sekler I. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc Natl Acad Sci USA. 2010;107(1):436–441. doi: 10.1073/pnas.0908099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai X, Clapham DE. Ancestral Ca2 + signaling machinery in early animal and fungal evolution. Mol Biol Evol. 2012;29(1):91–100. doi: 10.1093/molbev/msr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai X, Lytton J. The cation/Ca(2 +) exchanger superfamily: phylogenetic analysis and structural implications. Mol Biol Evol. 2004;21(9):1692–1703. doi: 10.1093/molbev/msh177. [DOI] [PubMed] [Google Scholar]
- 14.He C, O’Halloran DM. Analysis of the Na+/Ca2+ exchanger gene family within the phylum Nematoda. PLoS ONE. 2014;9(11):e112841. doi: 10.1371/journal.pone.0112841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma V, He C, Sacca-Schaeffer J, Brzozowski E, Herranz DM, Mendelowitz Z, Fitzpatrick DA, O’Halloran DM. Insight into the family of Na+/Ca2+ exchangers of Caenorhabditis elegans. Genetics 2013. [DOI] [PMC free article] [PubMed]
- 16.Sharma V, O’Halloran DM. Nematode sodium calcium exchangers: a surprising lack of transport. Bioinform Biol Insights. 2016;10:1–4. doi: 10.4137/BBI.S37130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Xu H, Oberwinkler J, Gu Y, Hardie RC, Montell C. Light activation, adaptation, and cell survival functions of the Na+/Ca2+ exchanger CalX. Neuron. 2005;45(3):367–378. doi: 10.1016/j.neuron.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 18.Baker PF, Blaustein MP, Hodgkin AL, Steinhardt RA. The influence of calcium on sodium efflux in squid axons. J Physiol. 1969;200(2):431–458. doi: 10.1113/jphysiol.1969.sp008702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes RC, Verkhratsky A, Parpura V. Plasmalemmal Na+/Ca2+ exchanger modulates Ca2 + -dependent exocytotic release of glutamate from rat cortical astrocytes. ASN Neuro. 2012 doi: 10.1042/AN20110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roome CJ, Knopfel T, Empson RM. Functional contributions of the plasma membrane calcium ATPase and the sodium-calcium exchanger at mouse parallel fibre to Purkinje neuron synapses. Pflugers Arch. 2013;465(2):319–331. doi: 10.1007/s00424-012-1172-1. [DOI] [PubMed] [Google Scholar]
- 21.Pappalardo LW, Samad OA, Black JA, Waxman SG. Voltage-gated sodium channel Nav 1.5 contributes to astrogliosis in an in vitro model of glial injury via reverse Na+/Ca2+ exchange. Glia. 2014;62(7):1162–1175. doi: 10.1002/glia.22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilgemann DW, Lin MJ, Fine M, Frazier G, Wang HR. Toward an understanding of the complete NCX1 Lifetime in the cardiac sarcolemma. Adv Exp Med Biol. 2013;961:345–352. doi: 10.1007/978-1-4614-4756-6_29. [DOI] [PubMed] [Google Scholar]
- 23.Pignataro G, Cuomo O, Vinciguerra A, Sirabella R, Esposito E, Boscia F, Di Renzo G, Annunziato L. NCX as a key player in the neuroprotection exerted by ischemic preconditioning and postconditioning. Adv Exp Med Biol. 2013;961:223–240. doi: 10.1007/978-1-4614-4756-6_19. [DOI] [PubMed] [Google Scholar]
- 24.Boscia F, Gala R, Pignataro G, de Bartolomeis A, Cicale M, Ambesi-Impiombato A, Di Renzo G, Annunziato L. Permanent focal brain ischemia induces isoform-dependent changes in the pattern of Na+/Ca2+ exchanger gene expression in the ischemic core, periinfarct area, and intact brain regions. J Cereb Blood Flow Metab. 2006;26(4):502–517. doi: 10.1038/sj.jcbfm.9600207. [DOI] [PubMed] [Google Scholar]
- 25.Cuomo O, Pignataro G, Sirabella R, Molinaro P, Anzilotti S, Scorziello A, Sisalli MJ, Di Renzo G, Annunziato L. Sumoylation of LYS590 of NCX3 f-Loop by SUMO1 participates in brain neuroprotection induced by ischemic preconditioning. Stroke. 2016;47(4):1085–1093. doi: 10.1161/STROKEAHA.115.012514. [DOI] [PubMed] [Google Scholar]
- 26.Boscia F, D’Avanzo C, Pannaccione A, Secondo A, Casamassa A, Formisano L, Guida N, Sokolow S, Herchuelz A, Annunziato L. Silencing or knocking out the Na(+)/Ca(2 +) exchanger-3 (NCX3) impairs oligodendrocyte differentiation. Cell Death Differ. 2012;19(4):562–572. doi: 10.1038/cdd.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molinaro P, Viggiano D, Nistico R, Sirabella R, Secondo A, Boscia F, Pannaccione A, Scorziello A, Mehdawy B, Sokolow S, Herchuelz A, Di Renzo GF, Annunziato L. Na+–Ca2+ exchanger (NCX3) knock-out mice display an impairment in hippocampal long-term potentiation and spatial learning and memory. J Neurosci. 2011;31(20):7312–7321. doi: 10.1523/JNEUROSCI.6296-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon D, Yang YM, Jeong MJ, Philipson KD, Rhim H, Shin HS. Enhanced learning and memory in mice lacking Na+/Ca2+ exchanger 2. Neuron. 2003;38(6):965–976. doi: 10.1016/S0896-6273(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 29.Secondo A, Esposito A, Sirabella R, Boscia F, Pannaccione A, Molinaro P, Cantile M, Ciccone R, Sisalli MJ, Scorziello A, Di Renzo G, Annunziato L. Involvement of the Na+/Ca2+ exchanger isoform 1 (NCX1) in neuronal growth factor (NGF)-induced neuronal differentiation through Ca2 + -dependent Akt phosphorylation. J Biol Chem. 2015;290(3):1319–1331. doi: 10.1074/jbc.M114.555516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma V, Roy S, Sekler I, O’Halloran DM. The NCLX-type Na+/Ca2+ exchanger NCX-9 is required for patterning of neural circuits in caenorhabditis elegans. J Biol Chem. 2017;292(13):5364–5377. doi: 10.1074/jbc.M116.758953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pannaccione A, Secondo A, Molinaro P, D’Avanzo C, Cantile M, Esposito A, Boscia F, Scorziello A, Sirabella R, Sokolow S, Herchuelz A, Di Renzo G, Annunziato L. A new concept: Abeta1-42 generates a hyperfunctional proteolytic NCX3 fragment that delays caspase-12 activation and neuronal death. J Neurosci. 2012;32(31):10609–10617. doi: 10.1523/JNEUROSCI.6429-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim YA, Rhein V, Baysang G, Meier F, Poljak A, Raftery MJ, Guilhaus M, Ittner LM, Eckert A, Gotz J. Abeta and human amylin share a common toxicity pathway via mitochondrial dysfunction. Proteomics. 2010;10(8):1621–1633. doi: 10.1002/pmic.200900651. [DOI] [PubMed] [Google Scholar]
- 33.Molinaro P, Cataldi M, Cuomo O, Viggiano D, Pignataro G, Sirabella R, Secondo A, Boscia F, Pannaccione A, Scorziello A, Sokolow S, Herchuelz A, Di Renzo G, Annunziato L. Genetically modified mice as a strategy to unravel the role played by the Na(+)/Ca (2 +) exchanger in brain ischemia and in spatial learning and memory deficits. Adv Exp Med Biol. 2013;961:213–222. doi: 10.1007/978-1-4614-4756-6_18. [DOI] [PubMed] [Google Scholar]
- 34.Lim YA, Ittner LM, Lim YL, Gotz J. Human but not rat amylin shares neurotoxic properties with Abeta42 in long-term hippocampal and cortical cultures. FEBS Lett. 2008;582(15):2188–2194. doi: 10.1016/j.febslet.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Fitzpatrick AL, Kuller LH, Lopez OL, Diehr P, O’Meara ES, Longstreth WT, Jr, Luchsinger JA. Midlife and late-life obesity and the risk of dementia: cardiovascular health study. Arch Neurol. 2009;66(3):336–342. doi: 10.1001/archneurol.2008.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovacs R, Kardos J, Heinemann U, Kann O. Mitochondrial calcium ion and membrane potential transients follow the pattern of epileptiform discharges in hippocampal slice cultures. J Neurosci. 2005;25(17):4260–4269. doi: 10.1523/JNEUROSCI.4000-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaiswal MK, Zech WD, Goos M, Leutbecher C, Ferri A, Zippelius A, Carri MT, Nau R, Keller BU. Impairment of mitochondrial calcium handling in a mtSOD1 cell culture model of motoneuron disease. BMC Neurosci. 2009;10:64. doi: 10.1186/1471-2202-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Black JA, Newcombe J, Trapp BD, Waxman SG. Sodium channel expression within chronic multiple sclerosis plaques. J Neuropathol Exp Neurol. 2007;66(9):828–837. doi: 10.1097/nen.0b013e3181462841. [DOI] [PubMed] [Google Scholar]
- 39.Ago Y, Kawasaki T, Nashida T, Ota Y, Cong Y, Kitamoto M, Takahashi T, Takuma K, Matsuda T. SEA0400, a specific Na+/Ca2+ exchange inhibitor, prevents dopaminergic neurotoxicity in an MPTP mouse model of Parkinson’s disease. Neuropharmacology. 2011;61(8):1441–1451. doi: 10.1016/j.neuropharm.2011.08.041. [DOI] [PubMed] [Google Scholar]
- 40.Wood-Kaczmar A, Deas E, Wood NW, Abramov AY. The role of the mitochondrial NCX in the mechanism of neurodegeneration in Parkinson’s disease. Adv Exp Med Biol. 2013;961:241–249. doi: 10.1007/978-1-4614-4756-6_20. [DOI] [PubMed] [Google Scholar]
- 41.Sharma V, O’Halloran D. Recent structural and functional insights into the family of sodium calcium exchangers. Genetics. 2014;52(2):93. doi: 10.1002/dvg.22735. [DOI] [PubMed] [Google Scholar]
- 42.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LS. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2004, 32(Database issue):D115-9. [DOI] [PMC free article] [PubMed]
- 43.Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, Done J, Grove C, Howe K, Kishore R, Lee R, Li Y, Muller HM, Nakamura C, Ozersky P, Paulini M, Raciti D, Schindelman G, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Wong JD, Yook K, Schedl T, Hodgkin J, Berriman M, Kersey P, Spieth J, Stein L, Sternberg PW: WormBase 2014: new views of curated biology. Nucleic Acids Res 2014, 42(Database issue):D789-93. [DOI] [PMC free article] [PubMed]
- 44.James-Zorn C, Ponferrada VG, Burns KA, Fortriede JD, Lotay VS, Liu Y, Brad Karpinka J, Karimi K, Zorn AM, Vize PD. Xenbase: core features, data acquisition, and data processing. Genesis. 2015;53(8):486–497. doi: 10.1002/dvg.22873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P. The FlyBase Consortium: FlyBase at 25: looking to the future. Nucleic Acids Res. 2017;45(D1):D663–D671. doi: 10.1093/nar/gkw1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palty R, Hershfinkel M, Yagev O, Saar D, Barkalifa R, Khananshvili D, Peretz A, Grossman Y, Sekler I. Single alpha-domain constructs of the Na+/Ca2+ exchanger, NCLX, oligomerize to form a functional exchanger. Biochemistry. 2006;45(39):11856–11866. doi: 10.1021/bi060633b. [DOI] [PubMed] [Google Scholar]
- 48.Liao J, Li H, Zeng W, Sauer DB, Belmares R, Jiang Y. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335(6069):686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 49.Giladi M, Khananshvili D. Molecular determinants of allosteric regulation in NCX proteins. Adv Exp Med Biol. 2013;961:35–48. doi: 10.1007/978-1-4614-4756-6_4. [DOI] [PubMed] [Google Scholar]
- 50.Giladi M, Tal I, Khananshvili D. Structural features of ion transport and allosteric regulation in sodium-calcium exchanger (NCX) proteins. Front Physiol. 2016;7:30. doi: 10.3389/fphys.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giladi M, Shor R, Lisnyansky M, Khananshvili D. Structure-functional basis of ion transport in sodium-calcium exchanger (NCX) proteins. Int J Mol Sci. 2016;17(11):E1949. doi: 10.3390/ijms17111949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winkfein RJ, Szerencsei RT, Kinjo TG, Kang K, Perizzolo M, Eisner L, Schnetkamp PP. Scanning mutagenesis of the alpha repeats and of the transmembrane acidic residues of the human retinal cone Na/Ca-K exchanger. Biochemistry. 2003;42(2):543–552. doi: 10.1021/bi026982x. [DOI] [PubMed] [Google Scholar]
- 53.Liao J, Li H, Zeng W, Sauer DB, Belmares R, Jiang Y. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335(6069):686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 55.Yu YK, Altschul SF. The construction of amino acid substitution matrices for the comparison of proteins with non-standard compositions. Bioinformatics. 2005;21(7):902–911. doi: 10.1093/bioinformatics/bti070. [DOI] [PubMed] [Google Scholar]
- 56.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 58.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 59.Smits SA, Ouverney CC. jsPhyloSVG: a javascript library for visualizing interactive and vector-based phylogenetic trees on the web. PLoS ONE. 2010;5(8):e12267. doi: 10.1371/journal.pone.0012267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Random Peptide. A Perl script used to generate random peptides for histograms in Fig. 3.


