Summary
A systematic characterization of the genetic alterations driving ALCLs has not been performed. By integrating massive sequencing strategies, we provide a comprehensive characterization of driver genetic alterations (somatic point mutations, copy number alterations, and gene fusions) in ALK− ALCLs. We identified activating mutations of JAK1 and/or STAT3 genes in ∼20% of 155 ALK− ALCLs and demonstrated that 38% of systemic ALK− ALCLs displayed double lesions. Recurrent chimeras combining a transcription factor (NFkB2 or NCOR2) with a tyrosine kinase (ROS1 or TYK2) were also discovered in WT JAK1/STAT3 ALK− ALCL. All these aberrations lead to the constitutive activation of the JAK/STAT3 pathway, which was proved oncogenic. Consistently, JAK/STAT3 pathway inhibition impaired cell growth in vitro and in vivo.
Introduction
Peripheral T cell lymphomas (PTCLs) are a heterogeneous group of tumors derived from post-thymic lymphocytes (Swerdlow et al., 2008). They are orphan diseases, accounting for 12% to 15% of all non-Hodgkin's lymphoma in Western populations (Vose et al., 2008), and display great variability in their clinical, morphological, immunophenotypic, cytogenetic, and molecular features. The classification of anaplastic large cell lymphomas (ALCLs) has been revised several times, and ALCLs are nowadays designated as a distinct entity of systemic PTCL (Swerdlow et al., 2008). Meanwhile, cutaneous forms of ALCLs (cALCLs) are recognized as a different variant. Among systemic ALCLs, patients harboring translocations of anaplastic lymphoma kinase (ALK) generally have a more favorable clinical course (Vose et al., 2008), although aggressive outcomes exist (Grewal et al., 2007). In contrast, ALK− ALCL patients have high morbidity and mortality, and ALCL remains an incurable disease in ∼70% of patients (Savage et al., 2008).
The genetics of ALK+ ALCL is characterized by translocations of the ALK proto-oncogene leading to ALK fusion proteins. The ALK chimeras activate STAT3, whose deregulated program is required for the maintenance of the neoplastic phenotype in ALK+ ALCL (Chiarle et al., 2005). Conversely, the mechanisms of transformation and maintenance of the ALK− ALCLs remain elusive. Recurrent translocations and loss of TP53 and PRDM1/BLIMP1 have been proved to have a pathogenic role associated with less favorable outcomes (Boi et al., 2013; Parilla Castellar et al., 2014). Last, comparative genomic hybridization studies have shown that ALK+ ALCLs display a more stable genome than ALK− ALCL or cALCL (Boi et al., 2013).
There are several alternative mechanisms leading to hyperactive STAT signaling in human cancers. These include aberrant or chronic stimulation via cytokines and growth factors, constitutive engagement of wild-type (WT) and mutated RTK receptors, and deregulated activation of several G protein-coupled receptors. Likewise, STAT3 hyper-activation occurs within multiple elements of stromal compartment and/or host immune cells, making STAT3 a central actor for inflammation-induced cancers (Bournazou and Bromberg, 2013). Disrupting mutations controlling epigenetically endogenous regulators of STAT3 (Johnston and Grandis, 2011) and somatic mutations of STATs, detectable in rare solid tumors and selected lymphoproliferative disorders, have been described (Kiel et al., 2014; Koskela et al., 2012; Pilati et al., 2011). These data validate STAT3 as a valuable therapeutic target.
To characterize the spectrum of mutations in ALK− ALCL and to identify potential therapeutic targets, we used massive genomic sequencing of both RNA and DNA. We investigated the landscape of somatic point mutations, copy number alterations, and gene fusions and we infer the associated mutational mechanisms of disease along with a set of in vitro and in vivo models.
Results
Whole-Exome Sequencing Somatic Mutation Analyses Demonstrate the Presence of Recurrent Mutations in ALK− ALCL
The number of mutations per case varied markedly (mean of 36 non-synonymous somatic mutations, from 1 to 150) without any preferential chromosomal distribution (Figure 1A). Mutations were largely represented by single-nucleotide substitutions leading to amino acid changes, namely, missense mutations (n = 752 [90%]), but included insertions or deletions (n = 15 [1.8%]), nonsense mutations (n = 63 [7.6%]), and alterations in canonical splice sites (n = 1 [0.1%]) (Figure S1).
Figure 1. Somatic Mutation and STAT3 Expression in ALCL.
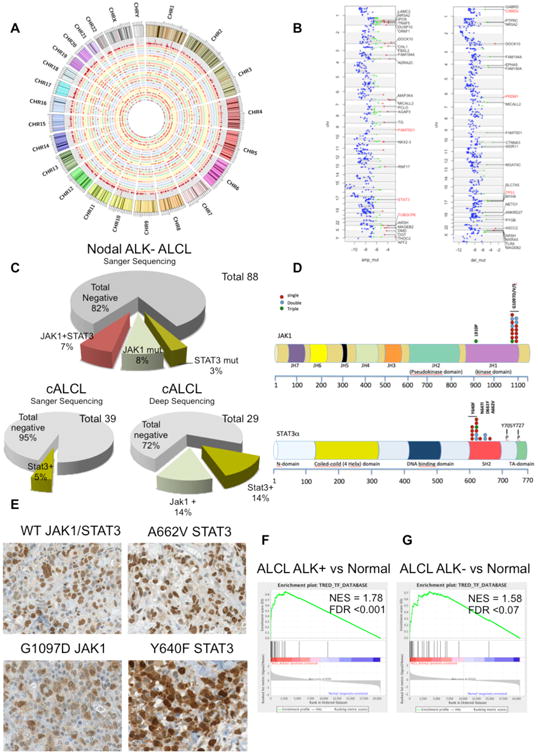
(A) Circos plot graphical representation of somatic synonymous and non-synonymous SNVs displays the mutational distribution across chromosomes (represented with different colors). Concentric circles are distinguished by different color background. Mutations are depicted as red points, and the outer circle depicts the histogram of the mutations per genomic position (red bars over gray background).
(B) Chromosome view of ALCL genes scoring at the top of mutated genes in regions of focal and recurrent amplifications/deletions (respectively, amp-mut and del-mut). Each color represents a different tier: red, 1; green, 2; and blue, 3.
(C) Prevalence of the JAK1, STAT3, and JAK1/STAT3 somatic mutations in systemic ALK− ALCL and cALCL by Sanger DNA sequencing.
(D) Schematic representation of human STAT3 and JAK1 proteins with their functional domains. Symbols depict distinct types of substitution mutations occurring as single (blue dots), dual (red dots), and triple (green dots) defects in systemic and cutaneous ALCL. Individual mutants were validated by Sanger DNA sequencing.
(E) Expression of STAT3 by immunohistochemistry in systemic ALK− ALCL. The black scale bar represents 50 mm and the red scale bar represents 20 μm.
(F) GSEA of STAT3 gene targets in ALK− ALCL patient samples versus normal resting and activated T cells (GSE6338, GSE14879, and GSE19069).
(G) GSEA of STAT3 gene targets in ALK− ALCL patient samples versus normal resting and activated T cells.
See also Figure S1 and Tables S1–S4.
Mutations were identified in PRDM1/BLIMP1, TP53, STAT3, JAK1, and BANK1 (Figure 1A). Integration of somatic mutations and focal copy number alterations highlighted PRDM1/BLIMP1, TP53, and CSMD2 as commonly mutated or deleted genes. TUBGCP6 and STAT3 genes were shown to be mutated or amplified (Figure 1B). Next we estimated the statistical significance of recurrent mutated genes and identified 13 putative candidate drivers on the basis of known functions and bio-informatics prediction (Figure S1, Tables S1 and S2, and Supplemental Information); those pathogenic roles require further functional studies.
Mutations of JAK1 and STAT3 Are Common in ALK− ALCL
JAK-STAT pathway genes (i.e., STAT3 and JAK1) were recurrently mutated in the discovery ALK− ALCL panel (Figure S1), suggesting that a STAT3-mediated oncogenic mechanism may be shared by all ALCLs, independent of ALK status. To define the mutation recurrence of JAK/STAT3 genes in ALK− ALCL, we analyzed by targeted re-sequencing the mutation hot spots of the STAT3 (i.e., the SH2 domain) and JAK1-3 (i.e., the kinase domain [KD]) in a validation panel of PTCL.
A total of 155 primary ALCL samples (88 ALK− and 23 ALK+ ALCLs and 44 cALCLs) and 74 PTCLs (29 angioimmunoblastic T cell lymphomas, 31 PTCLs not otherwise specified [PTCL-NOS], and 14 NK-T cell lymphomas) were sequenced. Non-synonymous somatic mutations of STAT3 and/or JAK1 were identified in 18% of systemic ALK− ALCLs and 5% of cALCLs (Figure 1C). Remarkably, 37.5% of the systemic ALK− ALCL cases harbored mutations of STAT3 and JAK1 (p < 0.0009, Fisher's exact test). Because Sanger DNA sequencing detects somatic mutations when mutated alleles are well represented (>10% of the total), we performed two deep sequencing strategies (deep sequencing analysis [DSA]; sensitivity down to 0.1%–1%) to detect variants restricted to a small fraction of the alleles (Figure S1). DSA excluded the occurrence of mutated subclones in systemic ALK− ALCL that lacked STAT3 and JAK1 mutations by Sanger sequencing. cALCLs, characterized by an indolent clinical course (Savage et al., 2008), were also analyzed by Sanger sequencing and DSA. By applying both strategies, cALCLs were found to harbor either JAK1 (6 of 29) or STAT3 (3 of 29) variants, but no co-occurrence of JAK1-STAT3 mutations. In systemic ALK− and cutaneous ALCLs, STAT3 mutations were substitutions targeting a hot spot in the SH2 domain, including recurrent (Y640F [Pilati et al., 2011], N647I, D661Y [Jerez et al., 2013], and A662V) missense substitutions (exon 21). JAK1 mutations were mostly selected to affect the 1097 (G1097D/S) codon within the KD of JAK1 (Figure 1D). A proline substitution of JAK1 in position 910 (L910P) was observed in a single case, harboring both G1097D/V JAK1 and Y640F STAT3 mutations (Figure 1D). JAK2 and JAK3 were in germline configuration in all samples.
Among PTCLs, JAK1 and STAT3 mutations were specific for ALK− ALCL, and no somatic mutations were present in either ALK+ ALCL or in PTCL-NOS, including 16 CD30-positive PTCL-NOS, which share morphological and phenotypic features with ALK− ALCL. STAT3 and JAK1 mutations were absent in highly purified CD3+CD4+, CD3+CD8+, and CD3−CD16+ normal cells derived from healthy individuals (DSA, ∼10−3).
Nuclear pSTAT3 expression, with a staining pattern similar to that of ALK+ ALCLs, was observed in a fraction of ALK− ALCLs (43% [27 of 63]; Figures 1E and S1). As predicted, all JAK/STAT3-mutated ALK− ALCLs were found to have strong nuclear pSTAT3 staining. Nonetheless, a subset of pSTAT3-positive ALK− ALCLs displayed no recurrent JAK/STAT3 mutations (74% [20 of 27]), suggesting alternative mechanisms for the constitutive activation of STAT3 (Figure 1E; Table S4). The role of the JAK/STAT pathway in ALCL was further strengthened by the gene set enrichment analysis (GSEA) of T cell-associated STAT3 genes (Piva et al., 2010) in both ALK−2+ and ALK− ALCL samples (GSEA false discovery rate [FDR] respectively q < 0.001 and q < 0.07; Figure 1F) and by the preferential expression of STAT3-regulated genes (∼30%; Figure S1), suggesting that ALK− ALCL may include a subset characterized by the constitutive activation of STAT3.
STAT3 Mutants Are Constitutively Phosphorylated and Oncogenic
To assess the properties of ALCL-related STAT3 mutants, two different STAT3−/− mouse embryonic fibroblast (MEF) cell lines were transduced with encoding WT or the mutated (Y640F, D661Y, and A662V, in brief as YF, DY, and AV) STAT3 viruses (Figures 2A and S2). The K658Y (in brief as KY) STAT3, aberrantly expressed in other human cancers, was included for comparison. Puromycin-selected cells were cultured in low serum with or without recombinant interleukin-6 (IL-6). As shown in Figure 2A, YF and KY STAT3 cells displayed high levels of Tyr705 pSTAT3 (and Ser727; Figure S2), which were further increased after IL-6 stimulation. Although the IL-6 receptor engagement led to detectable pSTAT3 in all conditions, YF and KY STAT3 cells displayed robust activation. Similar data were seen in both STAT3−/− MEF cell lines and replicated in human cell lines carrying WT STAT3 (human embryonic kidney 293T [HEK293T], lung [A549], and prostate [DU145]) and murine Lewis lung carcinoma lines (Figures 2B and S2). Conversely, the changes in the phosphorylation status of other signaling adaptors might be related to IL-6 exposure and/or the individual phenotypes (Figures 2A, 2B, and S2). Because lymphokine-mediated signaling plays a critical role in the maintenance of neoplastic phenotypes (Ngo et al., 2011; Zhang et al., 2012), we tested the phosphorylation status of WT and mutated STAT3 cells after a short IL-6 stimulation. As shown in Figure 2C, levels of WT pSTAT3 decreased over time; in contrast, YF pSTAT3 remained stable. Similarly, the mRNA transcript of SOCS3, a known STAT3-regulated gene, mirrored the phosphorylation of WT and YF STAT3 cells after IL-6 stimulation (Figure S2).
Figure 2. Signaling and Oncogenic Role of Mutated STAT3.
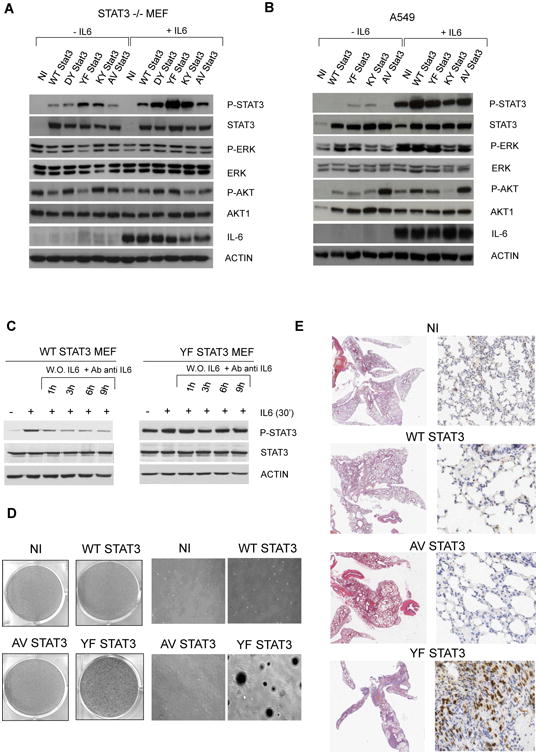
(A) Immunoblotting from whole-cell lysates of reconstituted STAT3−/− MEF cells.
(B) Immunoblotting from whole-cell lysates of reconstituted human lung adenocarcinoma A549 cells.
(C) Immunoblotting from whole-cell lysates of reconstituted STAT3−/− MEF with WT or YF STAT3 constructs after human IL-6 stimulation and its neutralization with specific antibodies against human IL-6. The relative ratio pSTAT3/STAT3 (P-S/S) is indicated.
(D) Colony assay in reconstituted STAT3−/− MEF cells.
(E) Soft agar colony assay in reconstituted STAT3−/− MEF cells.
(F) Histology and phospho-STAT3 expression by immunohistochemistry in mice injected with reconstituted STAT3−/− MEF cells. The red scale bar represents 1.5 mm, the green scale bar represents 10 μm, and the black scale bar represents 20 μm.
See also Figure S2.
Thereafter, we sought to demonstrate the oncogenic potential of mutated STAT3. YF STAT3 MEF displayed the largest number colonies (Figure 2D) and when injected via tail vein in NOD scid gamma (NSG) mice, produced lung metastases (Figure 2E), eventually killing the mice 5 to 6 weeks after implantation, whereas AV STAT3 and WT STAT3 or STAT3−/− control mice remained healthy at 9 weeks.
These findings demonstrated that the non-synonymous ALCL somatic mutations of STAT3 can sustain cell transformation and that the activation status of STAT3 is enhanced by lymphokine signaling activation.
JAK1 Mutants Lead to the Constitutive Phosphorylation of STAT and Synergize with STAT3 Mutants
Next we tested whether the ectopic expression of JAK1 mutants (G1097D and/or S and L910P, in brief GD, GS, and LP) could lead to constitutive pSTAT. ALCL-associated mutants of JAK1 were associated with high levels of pSTAT3 (Figure 3A). Once co-expressed with WT STAT3 in STAT3−/− MEF cells, a low amount of JAK1 mutants resulted in a stronger phosphorylation of STAT3 compared with WT JAK1 (Figure 3B). Moreover, higher levels of pJAK1 and pSTAT3 were seen in MEF ectopically co-expressing both JAK1 and STAT3 mutant proteins (Figures 3C and S3).
Figure 3. Signaling and Oncogenic Role of Mutated JAK1.
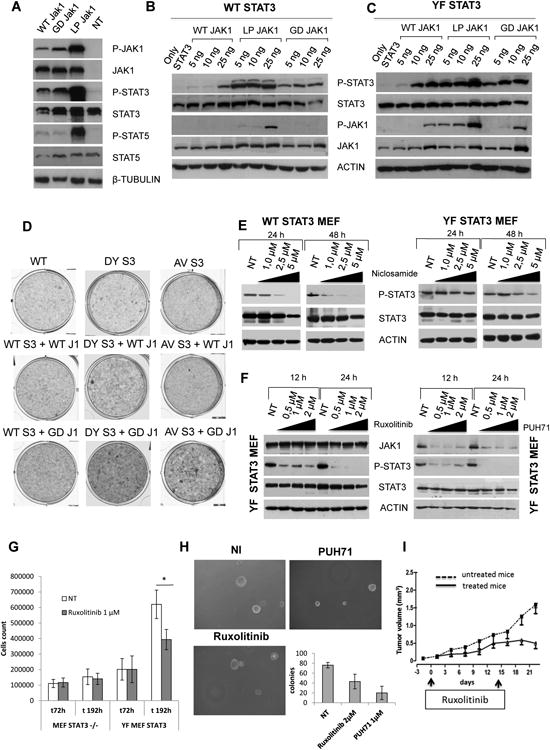
(A) Immunoblotting from whole-cell lysates of transfected HEK293T cells.
(B) Immunoblotting from whole-cell lysates of STAT3−/− MEF cells, cotransfected with WT STAT3 and mutated JAK1 cassettes. The absolute ratio pSTAT3/STAT3 (P-S/S) is indicated.
(C) Immunoblotting from whole-cell lysates of STAT3−/− MEF cells cotransfected with YF STAT3 and mutated JAK1 cassettes. The absolute ratio pSTAT3/STAT3 (P-S/S) is indicated.
(D) Colony assay in infected STAT3−/− MEF cells.
(E) Viability assay of SUP-M2 transduced cells with WT or mutated JAK1 and/or WT or mutated STAT3 treated with CEP28122 (50 nM, daily [with or without]) for 96 hr. All values were normalized to untreated control cells.
(F) Immunoblotting from whole-cell lysates of transduced STAT3−/− MEF cells treated with niclosamide
(G) Immunoblotting from whole-cell lysates of transduced STAT3−/− MEF cells treated with ruxolitinib and PUH71.
(H) Cell growth of reconstituted STAT3−/− MEF cells treated with ruxolitinib. The p value (*p < 0.01) is indicated. Values correspond to the mean ± SD.
(I) Tumor growth of mutated JAK1/STAT3 ALCL PDT treated in vivo with vehicle (12 tumors) or ruxolitinib (8 tumors, 25 mg/kg for 14 days). Treatment schedule is indicated. Values correspond to the mean ± SD.
See also Figure S3.
To test the oncogenic potential of double JAK/STAT mutants, we first performed an in vitro colony assay. Unexpectedly, the co-expression of LP JAK1 with YF or KY STAT3 (but not with WT) cassettes led to cell death (data not shown). Because GD JAK1 and DY STAT3 were most frequently detected in ALK− ALCLs, we focused on these associations. GD/S JAK1 and DY STAT3 double-mutant cells displayed higher number of colonies compared with GD/S JAK1 alone and/or JAK1 GD/S and WT STAT3, suggesting a synergistic role of these proteins (Figures 3D and S3). Because MEFs may not represent the best model to test mutations associated with neoplastic T cells, we forced the expression of single JAK1 or STAT3 in SUMP-M2, reasoning that ALK+ ALCLs are totally dependent on STAT3 signaling and are most related to ALK− ALCL (Piva et al., 2010). Control and transduced cells were treated with a selective ALK TKi and alive cells were enumerated. As shown in Figure 3E, the TKi exposure led to the total cell death of control and WT JAK1 or STAT3 cells. Conversely, both GD JAK1 and YF/DY STAT3 cells were partially rescued, while double-positive elements were salvaged. Interestingly, when an equal number of single transfected GD JAK1 and DY STAT3 were co-cultured, only 50% of them were alive at 96 hr, a value similar to that of DY STAT3. These later findings indicate that independent clones within the same micro-environment do not support each other. Last, the concomitant expression of GD JAK1 and DY STAT3 in lymphocytes from normal individuals was proved to favor the in vitro expansion as single mutants enhanced the cell growth of natural killer T (NK/T) cells in low interleukin-2 (IL-2) concentration (Figure S3).
Then, we evaluated the therapeutic feasibility of STAT3 or JAK1/2 inhibitors. We initially studied WT and YF STAT3 cells exposed to niclosamide, a molecule capable of inhibiting STAT3 signaling (Li et al., 2013). WT STAT3 cells treated with increasing amount of the drug displayed lower pSTAT3 levels, whereas after 48 hr of exposure, the highest non-toxic dose of niclosamide had a limited effect in YF STAT3 MEF (Figure 3F). Ruxolitinib, an inhibitor of JAK1/JAK2 molecules and the PUH71, a Hsp90 inhibitor, rapidly abrogated STAT3 phosphorylation (Figure 3G). As expected, the treatment with PUH71 led to JAK1 protein downregulation, consistent with an increased degradation of the active JAK/STAT complex. Drug exposure was associated with a decreased cell proliferation (Figure 3H) and a smaller number of colonies in agar (Figure S3) in the absence of apoptosis (data not shown). Last, to assess the efficacy of human dose equivalent of ruxolitinib, we took advantage of a patient-derived tumorgraft (PDT) line (ALCL-2 PDT) generated from a primary ALK− ALCL carrying both JAK1 and STAT3 mutations. Lymphoma-bearing mice treated for 14 consecutive days showed a significant tumor growth inhibition compared with vehicle-treated animals (Figure 3I). Overall, these data demonstrate that the pharmacological inhibition of JAK/STAT pathway represents a viable strategy in molecular stratified ALCL.
The Deregulated Expression of STAT3 Leads to the Upregulation of ATF3
To gain additional data on the mechanism(s) driving STAT3-mediated transformation, we analyzed the transcriptome of reconstituted STAT3 MEF−/− cells. An unsupervised clustering analysis showed that YF and KY STAT3 cells had similar signatures compared with AV and DY STAT3 or control MEF (Figure 4A). Canonical STAT3 genes were preferentially enriched in YF and KY STAT3 cells (Figure 4B), even in the absence of IL-6 stimulation (Table S5). GSEA showed a significant enrichment in YF STAT3 MEF (GSEA normalized enrichment score = 1.89, FDR < 0.001; Figure 4C), and a pathway enrichment analysis identified additional signals, some of which have been linked to STAT3 activation (Figure 4D). Among STAT3 genes in MEF, we found a set of transcripts modulated by the knock-down (kd) of ALK and/or STAT3 in NPM-ALK ALCL cells. These included members of the AP1 transcription factor family, co-activators and co-repressors (Figure S4). We focused on ATF3, which has a bifunctional role contributing to cell transformation (Yin et al., 2008). GSEA showed a statistical enrichment of ATF3-regulated genes (Table S6) in YF and KY STAT3 MEF as well as in ALK+ ALCL cells, as determined by a STAT3 kd (Figure 4E). These data were further confirmed by the statistical enrichment of ATF3 genes in NPM-ALK cells, documented by genomic or pharmacological kds of ALK (Figure 4E). The relationship between pSTAT3 and ATF3 protein expression was then documented in NPM-ALK Karpas 299 cells treated with an ALK inhibitor (CEP28122; Figure 4F) and after IL-2 withdrawal of NK/T cells (Figure 4G).
Figure 4. Gene Transcriptional Activation of Mutated STAT3.
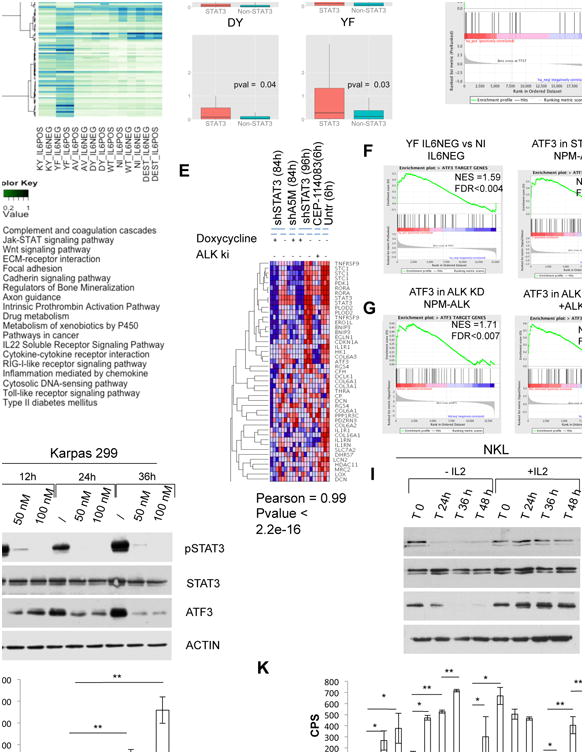
(A) Unsupervised hierarchical clustering of top 100 genes with higher variance of expression across the control and reconstitute STAT3−/− MEF cells: backbone vector (pLentiPGKpuroDEST, in brief DEST), WT, AV, DY, KY, and YF STAT3 cassettes. Control (nil) cells are included. Cells were cultured in low serum or in the presence of recombinant human IL-6 (50 ng/ml for 30 min).
(B) Log2 fold change of genes belonging to STAT3 target genes (up- and downregulated) and the remaining genes in DEST, AV, DY, and YF STAT3 reconstitute STAT3−/− MEF cells. All fold changes are computed versus empty vector STAT3−/−. The p values were calculated with the Wilcoxon rank sum test.
(C) GSEA of STAT3 gene targets in YF STAT3 cells versus control (nil, DEST) STAT3−/− MEF.
(D) Heatmap of overrepresented pathways among up- and downregulated genes in YF/KY STAT3 and AV/DY STAT3 MEF cells. Pathway enrichment is measured by hypergeometric p values.
(E) GSEA of ATF3 target genes: YF STAT3 mutant versus STAT3−/− MEF (upper left); non-induced versus STAT3 shRNA NPM-ALK (upper right); non-induced versus doxycycline-induced ALK shRNA NPM-ALK (lower left); untreated versus CEP14823 treated NPM-ALK cells (lower right).
(F) Immunoblotting from whole-cell lysates of NPM-ALK ALCL cells (Karpas 299) treated over time with an anti-ALK inhibitor (CEP28122).
(G) Immunoblotting from whole-cell lysates of NKL cells supplemented with (20 ng/ml) or without human recombinant IL-2.
(H and I) Luciferase expression of cells with single- or double-transfected STAT3−/− MEF cells. The p values (*p < 0.01 and **p < 0.001) are indicated. Values correspond to the mean ± SD.
See also Figure S4 and Tables S5 and S6.
Next, we proved that the transformation events associated with JAK/STAT3 mutants were mediated by a canonical STAT3 signaling pathway. As shown in Figure 4H, the forced expression of YF and KY STAT3 in STAT3−/− MEF cells led a robust transactivation of a STAT3-driven luciferase reporter construct. A dominant-negative form of STAT3 (Y795F) and a high amount of WT STAT3 could inhibit the transactivation of STAT3 mutants in a dose-dependent manner (Figure S4). Strong luciferase activation was also seen in transiently transfected STAT3−/− MEF cells with increasing amount of GD and LP JAK1 and STAT3 mutants (Figure 4I), suggesting a cooperative effect of JAK1 and STAT3 mutants.
ALK– ALCLs Express Dual Chimera Derived from Transcriptional Regulators and Tyrosine Kinases
To identify additional genetic defects in ALCL and to discover new mechanisms responsible for the constitutive activation of STAT3, we implemented a bioinformatics analysis of whole-transcriptome sequencing (RNA sequencing [RNA-seq]) of 23 ALK+/ALK− ALCLs. A total of 28 fusions were identified and validated (Figures 5A and S5), including four NPM-ALK and one TRAF1-ALK (Abate et al., 2014a) chimeras in the five ALK+ cases. Four of 18 ALK− ALCLs displayed fusion transcripts involving tyrosine kinases (TKs) known to play a pathogenetic role either in hematologic or solid neoplasms (Shaw et al., 2013) (Figure 5B), with two ALCLs bearing more than one fusion (Table S7) all in the absence of JAK1 or STAT3 mutations. Several samples shared the same kinase fusions, albeit fused to different partners, suggesting a non-random event (Figure 5B). All TKs underwent recombination with partners capable of providing homo- or hetero-dimerization domains. At their 5′-NH2 terminus, three chimeras showed sequences corresponding to transcription factors or transcriptional repressors (Figure 5C). These included samples expressing hybrid transcripts in which the coding region of the NFkB2 (exons 1–13/16) was fused to the intracytoplasmic domain of ROS1 (NFkB2-ROS1) or TYK2 (NFkB2-TYK2), respectively (Figures 5D and 5E). The 5′ end of both NFkB2 fusions predicted the expression of proteins with the RHD, NLS, and GRR domain, lacking all (13–24) or most (16–24) of the ankyrin region of the NFkB2 (Figure 5C). Loss of the NFkB2 ankyrin region is necessary for the translation of constitutively active NFkB-fusion proteins and for its oncogenic role (Neri et al., 1991). Previously reported NFkB2 translocations included ectopic genomic regions capable of stabilizing the expression of NFkB2 proteins but lacked alternative coding segments. Within the discovery panel, two additional samples displayed chimera involving either ROS1 or TYK2. In case GSP99, the 5′ region of NCOR2 was fused to the intracytoplasmic region of ROS1 (NCOR2-ROS1), while in case GSP50, PABPC4 was fused to the intra-cytoplasmic domain of TYK2 (PABPC4-TYK2) (Figure 5C). NCOR2 is a transcriptional repressor and aberrations altering the SMRT and N-CoR complex have been associated with metabolic defects and cancers (Privalsky, 2004).
Figure 5. Strategy and Discovery of Chimeric Fusion in ALK− ALCL.
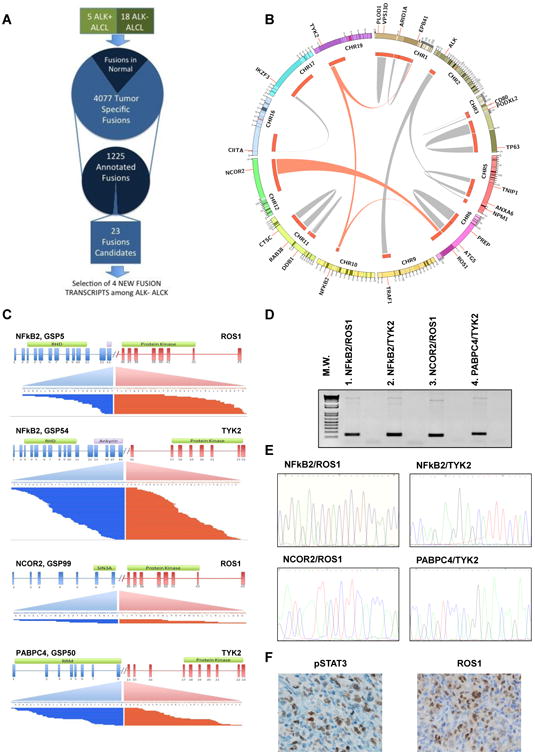
(A) Schematic representation of RNA-seq data gene fusion analysis.
(B) Circos plot showing validated intra- and inter-chromosomal gene fusions. Gene fusions undergoing functional validation are depicted with red links. Different gene fusions exchange gene partners, suggesting a non-random pattern.
(C) Gene fusions detected by paired-end massively parallel whole-transcriptome sequencing. Mate-pair sequences are shown, aligning on either side of the breakpoints. Intron-exon configuration is depicted: 5′ (blue) and 3′ (red) partners are indicated. Transcribed exonic sequences are shown as part of a transcript variant resulting from the fusions.
(D) PCR amplification products spanning the breakpoints of the fusion of ALK− ALCL samples (discovery samples).
(E) DNA sequencing electropherograms of chimeric fusion after PCR from ALK− ALCL samples (discovery samples).
(F) Immunohistochemistry on a representative sample carrying the NFkB2-ROS1 fusion, using specific antibodies against pSTAT3 and ROS1. The scale bar represents 25 μm.
See also Figure S5 and Table S7.
NFkB2-TK Fusions Are Oncogenic and Lead to the Constitutive Activation of STAT3
Because the N terminus of NFkB provides a domain capable of homo- or hetero-dimerization, we speculated that NFkB2-TK could dimerize and undergo trans-phosphorylation. To test this hypothesis, we first transfected the cDNA of NFkB2-ROS1 into HEK293T cells (Figure S5). A constitutive tyrosine phosphorylation was documented in NFkB2-ROS1 and NPM-ROS1 cells, but not in NFkB2-ROS1 dead mutant HEK293T cells. The NPM-ROS1 cassette encoded a control fusion protein in which the 5′-NPM provides a dimerization domain (Figures 6A and S5). The ectopic expression of NFkB2-ROS1 and NPM-ROS1 was associated with the phosphorylation of JAK (JAK2-3) and STAT3, but not of other signaling cascades (Figure S7). Similar data were obtained in cells transfected with NFkB2-TYK2, NCOR2-ROS1, and lung-associated ROS1 fusions (Figures 6B-6D). Genetic inhibition of the TK domain of these fusions through mutations of the ATP binding pocket (K to F) did not elicit the phosphorylation of NFkB2 fusions and their downstream adaptors/molecules (Figure 6).
Figure 6. Signaling, Cellular Distribution, and Transcriptional Activity of NFkB Chimeric Fusion in ALK− ALCL.
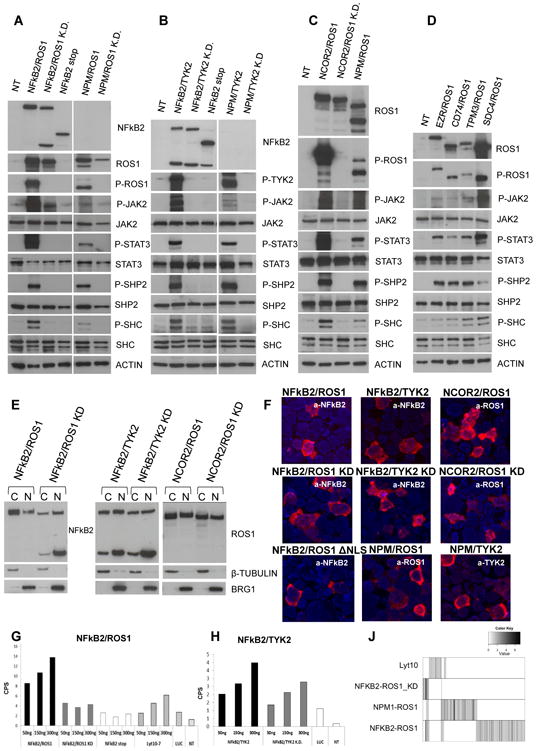
(A–D) Immunoblotting from whole-cell lysates of transfected HEK293T cells.
(E) Cellular localization of NFkB2-kinase fusions in transfected HEK293T cells after cell fraction.
(F) Immunofluorescence of chimeric fusions in transfected HEK293T cells using PE-labeled antibodies. The scale bar represents 15 mm.
(G and H) Luciferase expression of transfected HEK293T cells. Values correspond to the mean ± SD.
(I) Heatmap of genes differentially expressed in NFkB2-ROS1, NPM-ROS1, NFKB2-ROS1, and NFKB2 Lyt10 construct versus normal T cells.
See also Figure S6.
We next treated EZR-, CD74-, TPM3-, and SDC4-ROS1 cells with either crizotinib or TAE684, known to inhibit lung-associated ROS1 fusions, or an anti-ROS1 (JNJ-ROS1i-A) small molecule (see Supplemental Information) demonstrating a robust inhibition of phosphorylation of these fusions (Figure S7). Unexpectedly, crizotinib and TAE684 did not inhibit NFKB2-ROS1 or NCOR2-ROS1, whose phosphorylation was however abrogated by the JNJ-ROS1i-A inhibitor (> 250 nM; Figure S7).
To gain further information of the properties of these new fusions, we studied the half-lives of NFkB2-ROS1 and TYK2 proteins (>36 hr) demonstrating that these fusions could undergo proteasome independent proteolitic cleavage (data not shown). Notably, the inhibition of the tyrosine activity of NFkB2-ROS1 enhanced this phenomenon, suggesting that the kinase activity contributes to the NFkB2-ROS1 protein stability (Figure S7).
NFkB2-ROS1 and NFkB2-TYK2 Are Chimeric Transcription Factors
Using a cell fractionation assay, we demonstrated that NFkB2 and NCOR2 fusions localized into the nucleus and cytoplasm and that NCOR2-ROS1 and NFkB2-TYK2 proteins had a similar distribution (Figure 6E). These findings were corroborated using an immunofluorescence staining. Last, we showed a restricted expression in the cytoplasm of an arginine/lysine-rich NLSs mutant form of NFkB2-ROS1 (ΔNLS-NFkB2-ROS1) (Figures 6F and S6), which indicates that an importin-mediated transfer is required for NFkB2-ROS1 to entry into the nucleus.
Cotransfection of HEK293T with NFkB2-ROS1, NFkB2-ROS1 kd and a known activated form of NFkB2 (Lyt-10-7) (Neri et al., 1991) vectors in the presence of an IgK-HIV-kB-driven luciferase reporter construct led to a dose-dependent luciferase expression with values higher than those seen in Lyt-10-7 transfected cells, whereas catalytic dead mutants were less active. This suggests that the kinase activity of the fusions can contribute to their transcriptional activity (Figures 6G–6H). We then proved that Rel-B could increase NFKB2-ROS1-mediated transcription, whereas p50 or p65 had no major effects (data not shown).
Last, knowing that Rel/NFkB transcription factors lead to the selective expression of different target genes, we studied the transcription of Lyt10, NFkB2-ROS1, NFkB2-ROS1 kd, and NPM-ROS1 in HEK293T cells. Only a fraction of NFkB2-ROS1 upregulated genes were overlapping to NPM-ROS1 or NFkB2-ROS1 kd; meanwhile, NFkB2-ROS1 cells displayed a unique set of genes (Figure 6J). To elucidate the contribution of additional pathways, we performed a C2-GSEA demonstrating that NFkB2-ROS1 fusion elicits the expression of genes involved in membrane trafficking, replication, protein export, TNF signaling, T cell activation (Figure S6). This result suggests that NFkB2-ROS1 signature is not merely due to the individual contribution of NFkB2 and ROS1 signaling.
Last, to infer of the relative frequency of NFkB2-ROS1 fusions, we stained a set of ALCLs with an antibody detecting a formalin-resistant epitope of ROS1. ROS1-positive ALCL cells (2 of 100 ALCLs known to carry ROS1 fusions; Figure 5F) displayed nuclear staining for pSTAT3. Expression data confirmed these data (Figure S5), in agreement with a recent study (Velusamy et al., 2014). Nevertheless, molecular and fluorescence in situ hybridization analyses on large PTCL patient cohorts are required to conclusively assess the frequency of these fusions.
NFkB2-TK and NCOR2-ROS1 Are Tumorigenic and Require STAT3 Signaling
To test the oncogenic activity in vitro of ALK− ALCL-associated fusions, we transfected mouse 3T3 fibroblasts. Within 2 weeks, NFkB2-ROS1, NFkB2-TYK2, and NCOR2-ROS cells had established a larger number of colonies compared with KD-matched constructs or untrasfected cells (Figures 7A and S7). However NPM-ALK cells had the highest transformation potential. Once NFkB2-ROS1 and NFkB2-ROS1 kd NIH 3T3 cells were injected subcutaneously (s.c.) into NSG mice, they produced tumors, although NFkB2-ROS1 cells grew faster and reached larger masses overtime. No mice injected with parental cells developed tumors (∼4-weeks after injection; Figure 7B). To dissect the tumorigenic role and the dual function of the NFkB2-ROS1 chimera in vivo, NFkB2-ROS1 NIH 3T3-bearing NSG mice were treated with JNJ-ROS1i-A, showing a growth inhibition of NFkB2-ROS1 cells (Figure 7C).
Figure 7. Oncogenic Role of NFkB2-ROS1 Fusion.
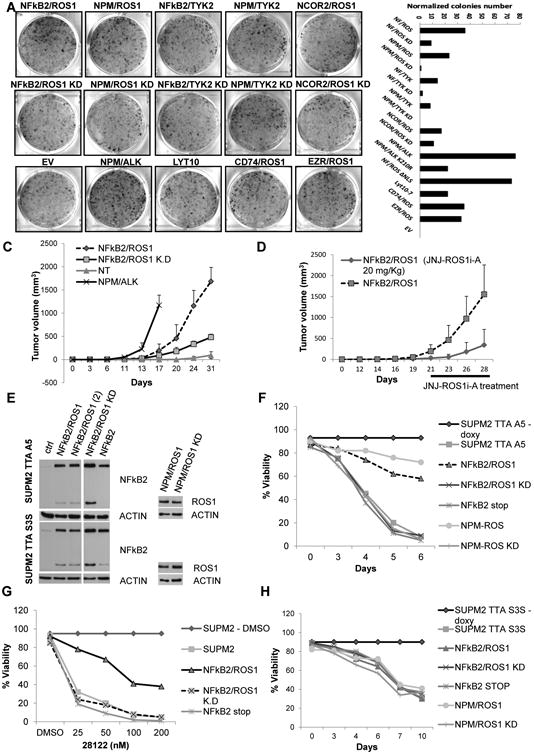
(A) Colony assay of NIH 3T3 cells after transfection with fusion and control cassettes. Number of colonies at day 14 after transfection are depicted. Values correspond to the mean ± SD.
(B) Tumor growth of transfected NIH 3T3 in NSG mice. Values correspond to the mean ± SD.
(C) Tumor growth of transduced NIH 3T3 with NFkB2/ROS1 in NSG mice (vehicle and anti-ROS, 20 mg/kg; six tumors per group). Time schedule is indicated. Values correspond to the mean ± SD.
(D) Viability of inducible shRNA ALK engineered NPM-ALK cells. Cells were infected with different lentiviral preparations, coding for control and ROS1 fusions, and then after puromycin selection were treated with doxycycline.
(E) Viability of inducible shRNA STAT3 engineered NPM-ALK cells. Cells were transduced with different lentiviral preparations and then after puromycin selection induced with doxycycline.
(F) Immunoblotting from whole-cell lysates of transduced cells with shRNA ALK (TTA A5) and shRNA STAT3 (TTA S3S) lentiviral cassettes.
(G) Western blot analysis of HEK293T cotransfected with NFkB2/TYK2 or NPM/TYK2 cassettes and a specific shRNA expression vector.
(H) Western blot analysis of SUP-M2 transduced with NFkB2/TYK2, NFkB2/TYK2 KD, and control viruses.
See also Figure S7.
To investigate a potential role of STAT3, we first forced the expression of NFkB2-ROS1, NFkB2-TYK2 and NCOR-ROS1 in STAT3−/− MEF cells. Contrary to WT STAT3 MEF cells, ROS1 and TYK2 fusions in STAT3−/− MEF cells lea only to a slightly larger number of colony compared to control (Figure S7), suggesting a strict requirement for STAT3-mediated transformation, likewise as for ALK chimera (Chiarle et al., 2005).
Although in vitro models have been instrumental to elucidate the tumorigenic properties of oncogenes, these assays do not fully recapitulate the oncogenic properties of specific lesions operating in different lineages. Because ROS1 fusion expression in activated human T cells led to senescence (data not shown), we tested whether NFkB2-ROS1 could rescue the cell death of ALK+ ALCL cells after ALK kd signaling. Taking advantage of the resistance of ROS1 fusions to crizotinib and CEP28122 ALK Ki, we infected SUP-M2, SU-DHL-1, and JB-6 NPM-ALK ALCL cells with NFkB2-ROS1 or control vectors (Figure S7). The kd of ALK signaling using CEP28122 or an inducible ALK RNAi was associated with growth inhibition and loss of viability, changes largely overcome by the forced expression NFKB2-ROS1 but not in NFkB2-ROS1kd (Figures 7D and S7).
We then tested the role of STAT3 in NFkB2-ROS1 signaling in these models. NFkB2-ROS1, NFkB2-ROS1 kd or NPM-ROS1 were introduced in S3S SUP-M2-TS cells, in which STAT3 mRNA levels are controlled by a doxycycline-inducible RNAi system. As expected, loss of STAT3 in control cells led to cell death (>120 hr), a phenotype that could not be rescued by NFkB2-ROS1 signaling. Overall, these findings demonstrated that NFkB2-ROS1 can rescue ALK oncogenic addiction and that the STAT3 signaling is required for NFkB2-ROS1-mediated rescue (Figures 7D and 7E).
Last, we investigated the signaling pathway of NFkB2-TYK2. We demonstrated that its ectopic expression led to the robust activation of pSTAT1 and/or/3/5 in Jurkat, HEK293T, and MEF cells (Figures 7G and S7) and pSTAT could be inhibited by a selective JAK/TYK2 inhibitor (Chrencik et al., 2010; Sanda et al., 2013) and by a specific small hairpin RNA (shRNA) (Figures 7G and S7). Conversely, NFkB2-TYK2 in ALK+ ALCL cells did not modulate either pSTAT1 or pSTAT5 levels (Figure 7H).
Discussion
We provide molecular and biological data demonstrating that a subset of systemic ALK− ALCLs and cutaneous ALCLs display the constitutive activation of the JAK/STAT3 pathway via multiple and alternative genomic mechanisms. Mutually exclusive oncogenic mutations of JAK (Bellanger et al., 2014) and STAT have been reported in several human cancers (Odejide et al., 2014; Pilati et al., 2011) and in some CD30-positive lymphoproliferative disorders (Ohgami et al., 2013). Our study provides a paradigm, demonstrating the presence of concomitant mutations on the JAK/STAT3 pathway in human cancers. Functional tests of JAK1 and STAT3 mutants showed that individual aberrations could cooperate when co-expressed. Treatment with a JAK1/2 and Hsp90 inhibitors was associated with growth inhibition of reconstitute YF STAT3 MEF cells. An in vivo PDT model derived from a double JAK/STAT3 mutant ALK− ALCL patient corroborated this finding. We have also identified TK fusions derived from the juxtaposition of transcription/repressor factors to TKs, whose oncogenic properties rely on the STAT3 signaling pathway. Interestingly, rare fusions were described in human cancers (Shern et al., 2014) and in a subset of CD30 lymphoproliferative disorders of the skin (Velusamy et al., 2014). A comprehensive map of the different lesions found by next-generation sequencing demonstrate that the majority of the ALK− ALCL bear driver mutations, but no bona fide drivers could be recognized in some tumors.
The deregulated expression of STAT3 is commonly seen in human cancer, and pro-oncogenic STAT3 mutations have been described in some neoplasms and are required for the maintenance of the ALK+ ALCL phenotype (Chiarle et al., 2005). We found that in ALK∼ ALCL, JAK1 and STAT3 mutations most commonly occurred in hot spots, with an exception represented by the mutation of JAK1 in L910P. When different STAT3 mutants are coexpressed with L910P JAK1 cells undergo cell growth arrest and ultimately senescence, suggesting that a non-random association of specific mutations may occur. In the case of JAK1, mutations have been reported to arise in amino acids facing the interface in which the pseudokinase and kinase interact, resulting in increased kinase activity (Lupardus et al., 2014). However, the residue G1097, which most frequently mutated in ALCL, is sitting on the surface of the molecule, and it might modulate protein-protein interactions. In the case of STAT3, mutations commonly occur within the SH2 domain. The residue in position Y640 of STAT3 is near the TAD domain, which allows protein and DNA interaction. Changes in the polarity of this residue may stabilize STAT3 dimers, consistent with the prolonged phosphorylation status of Y640F STAT3.
The association between convergent and recurrent point mutations in genes coding for two interacting proteins is unexpected. First, the distribution of mutations in hot spots points to gain-of-function mutations in both proteins. Second, and more surprisingly, the concurrence of these alterations indicates that these double mutants are strongly selected beyond single mutants. If mutations are randomly distributed along the genome, and the probability of a single mutation is small, the probability of double mutations should theoretically be the square of the probability of having a single mutation. This argues that these double mutants might undergo very strong selection. Notably, the possibility that JAK1 and STAT3 mutants could be present in two independent subclones within the double-positive samples cannot be conclusively ruled out. However, the deep sequencing of single double-positive samples and its derived PDT confirmed that these aberrations were present in heterozygosity in both primary and PDT derived tissue samples, strongly suggesting that they co-exist.
Our findings showed that mutated STAT3 proteins are potent transcriptional activators and the constitutive activation of the JAK1/STAT3 pathway leads to the modulation of a set of genes in different cells (MEF, ALK+ ALCL, and NKL) and can protect ALK+ ALCL cells when treated with ALK TKi. This suggests that STAT3 mediated transformation requires the modulation of a critical number of genes, including ATF3 and its regulated targets. Deregulation of ATF3 genes has been linked to cell transformation (Yin et al., 2008) but never associated with STAT3 activation in human cancers. Although these data were largely generated in MEF cells, we have proven that cells derived from different lineages (MEF, HEK293T, NK and T cells) co-share STAT3- and/or ATF3-regulated genes. These data must be confirmed in more reliable ALK− ALCL models, which are currently lacking. The use of PDT derived from different human PTCLs may overcome in part this limitation.
It is now known that the host plays a critical role in the maintenance of the neoplastic phenotype, and IL-6-mediated stimulation by intratumoral infiltrating lymphocytes has been proved to be critical in liver disorders (Pilati et al., 2011; Rebouissou et al., 2009). Here, we have shown that cells carrying mutated STAT3 displayed high and prolonged levels of pSTAT3 after IL-6 stimulation, and this activation is repressed, inhibiting JAK1/2, suggesting that pSTAT3 of mutated forms requires JAK1/2. In lymphoma, high levels of lymphokines can be provided through an autocrine loop or via host cells, a scenario also seen in lymphoproliferative disorders (Anderson, 2007). High expression levels of IL-2 and interleukin-22 can occur as after the loss of the transcriptional repressor (i.e., BLIMP1), which is often lost in many ALK− ALCLs (Boi et al., 2013).
The RNA-seq data have provided an additional layer of complexity for the constitutive activation of STAT3. We had originally shown that both ALK+ and ALK− ALCLs share a common signature (Piva et al., 2010). Now we provide data confirming that ALK− ALCLs can display a constitutive pSTAT3 in the absence of either JAK/STAT3 mutations, as a result of NFkB2-ROS1 and NFkB2-TYK2 fusions. Interestingly, we demonstrated that NFkB2-TYK2 can lead to pSTAT1/3/5 and that individual STATs can be differentially activated in different cells. In ALK chimera, the ALK partners contribute by providing only dimerization domains, lacking oncogenic activities, with TRAF-1 being the exception (Abate et al., 2014b). The scenario is different for NFkB2-ROS1 and TYK2 fusions, because they are transcriptional active and their transcriptional activity can be enhanced by the enzymatic activity of the kinases. This may be due in part to the protein stability of NFkB2-ROS1. However, an alternative scenario may be taking place, as a result of their translocation into the nucleus leading to the aberrant phosphorylation of proteins not otherwise accessible to the native kinases (i.e., transcriptional holocomplexes). This is partially supported by the unique signature of NFkB2-ROS1, which has minimal overlaps to either NFkB2 or ROS1 profiles.
However, the oncogenic contribution of NFkB is unclear, because STAT3 is critical for the NFkB2-ROS1-mediated phenotypein MEFand in ALK+ ALCL cells. Although the role of NFkB in B cell neoplasia is well established, its contribution in T cell disorders is less defined. Toward this end, the activation of NFkB can be seen in both ALK− and in ALK+ ALCLs, suggesting that this pathway can contribute to the lymphoma phenotype (Abate et al., 2014b).
Many innovative protocols have been explored to improve the clinical outcome of ALK− ALCL or refractory ALK+ ALCL (overall survival of 35%–40% at 5 years) with limited success. No upfront biomarkers for refractory or relapsing patients exist, and refractoriness-driving lesions remain unknown. The recurrent aberrations (Boi et al., 2013; Parilla Castellar et al., 2014) need to be tested in prospective cohorts to discern their prognostic clinical significance. Nevertheless, the discovery of targetable pathogenetic defects provides the rationale for the triage and selective treatment of molecularly defined ALK− ALCL patients. In fact, the discovery that selective compounds targeting either JAK or STAT3 can inhibit cell growth offers alternative therapeutic avenues. As selective STAT3 inhibitors have not been approved, JAK inhibitors have entered the clinical arena (Sonbol et al., 2013) and could be tested in molecularly stratified ALCL patients, as suggested by our pre-clinical ALCL PDT model. The central role of STAT3 in these and in many other types of hematological malignancies urges the execution of new drug discovery programs and the design of innovative protocols.
Experimental Procedures
Tissue Samples
Fresh and viable cryopreserved samples from primary lymphoma were obtained at the time of diagnosis or at relapse after chemotherapy. Diagnoses were assigned according to the World Health Organization classification (Table S3). Tissues used for NGS analyses were selected for their high tumor cell content (>50%). All studies were approved by the institutional human ethics review board, and patients provided written informed consent in accordance with the Declaration of Helsinki.
Immunohistochemistry and Immunofluorescence
Immunohistochemistry stains of primary lymphoma were implemented on formalin-fixed paraffin-embedded sections with specific antibodies (Supplemental Experimental Procedures). The immunofluorescence was completed as described (Voena et al., 2013).
RNA Preparation, Quantitative Real-Time PCR, and Microarray Analysis
Total RNAs were isolated using Trizol reagent (Ambion). Quantitative realtime PCR analysis was performed using the iQ SYBR Green Real-Time PCR Supermix (BioRad). Transcript levels were normalized to the a-actin or GAPDH level. A list of specific primers is available in Supplemental Experimental Procedures.
Gene expression profiling data were analyzed as previously described (Boi et al., 2013).
Whole-Exome and RNA-Seq
Whole-exome sequencing and RNA-seq were performed as previously described (Palomero et al., 2014; Abate et al., 2014b)
Bioinformatics Analysis
Single-nucleotide variants (SNVs) were determined by means of the Statistical Algorithm for Variant Identification. Copy number variations and SNVs were integrated with the MutComFocal algorithm, and chimeric transcripts were detected by deFuse and Chimeracan. Pegasus was used to assign fusion annotation and functional selection (Supplemental Information). mRNA quantification in RNA-seq data analyses were determined by the Cufflinks package and GSEA by GSEA pre-ranked tool. Gene expression data had been previously reported (accession numbers GSE6184, GSE6338, GSE14879, and GSE19069).
DSA
Purified PCR DNA fragments were obtained after genomic DNA amplification with primer specific for spanning hot spot mutations/SH2 domains along with 10-bp multiplex identifier tag sequences and high-fidelity Taq polymerase (FastStart High Fidelity PCR System; Roche Diagnostics). Purified DNA fragments were subjected to ultra-deep NGS on the Genome Sequencer Junior instrument (454 Life Sciences).
Multiparametric Assays
Cells were culture in standard conditions. Multiparametric assays were conducted in 96-well microtiter plates (40,000 cells/well). Cell viability and Renilla-based luciferase expression were measured using CellTiter-Glo Luminescent Cell Viability Assay and the Dual-Luciferase Reporter system (Promega).
Mice
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were injected s.c. or intravenously with 1 × 106 cancer cells. For experimental metastasis assays, mice were sacrificed after 4 to 8 weeks after intravenous injection. PDTs were established by implanting fresh tissue fragments s.c. PDT mice were treated with ruxolitinib (100 mg/kg twice daily) and NFKB2-ROS1 NIH 3T3-bearing mice with the JNJ-ROS1i-A small molecule (20 mg/kg twice daily). The Animal and Bioethical Committee of the University of Torino (TO-0072468) and the Institutional Animal Care and Use Committee of Weill Cornell Medical College (2014-0024) approved the animal protocols.
Supplementary Material
Highlights.
Co-occurring somatic mutations of JAK1 and STAT3 promote oncogenesis
Novel chimera fuse transcription/repressor domains to tyrosine kinases (TKs)
Dual functional TK fusions are oncogenic and mediate STAT3 transformation
JAK/STAT3 inhibitors have therapeutic efficacy in pre-clinical ALCL models
In Brief.
Crescenzo et al. provide a comprehensive characterization of driver genetic alterations in ALK− anaplastic large cell lymphomas (ALCLs) and uncover mechanisms leading to the constitutive activation of STAT3 in ALK− ALCL. JAK/STAT3 inhibitors have therapeutic efficacy in pre-clinical ALCL models.
Significance.
The JAK/STAT3 signaling pathway is often deregulated in hematopoietic disorders. We describe two mechanisms leading to the constitutive activation of STAT3 in ALK− ALCL. Oncogenic JAK1 or STAT3 mutations are associated with hyperactive pSTAT3 that regulat canonical STAT3 and ATF3 genes. Moreover, synergizing JAK1 and STAT3 mutants sustain the neoplastic growth, which can be efficiently controlled in vitro and in an ALCL patient-derived tumorgraft model by JAK1/2 inhibitors. We discovered that chimera, displaying concomitant transcriptional and kinase activities, are power oncogenes capable to sustain via STAT3 the ALCL phenotype. The pharmacological inhibition of JAK/STAT3 represents an alternative strategy for the treatment of molecularly stratified ALCL.
Acknowledgments
We thank Drs. L Cerchietti, J. Wang, H. Khiabanian, and P. Kyriakides for discussion and suggestions on the manuscript. We are grateful to S.A. Fasteris and the Epigenomics Core Facility of Weill Cornell Medical College for their technical support. We are grateful to Drs. B. Ruggeri and W. Chan (ALK inhibitors); V. Poli, F. Bussolino, and A. Bardelli (cell lines); R. Doebele (ROS1 fusion); R. Dalla Favera (Lyt10 and IgK-HIV-kB); and J. Bromberg (dominant-negative STAT3) for their reagents. We would like to thank Drs. Olivier Elemento, Neel S. Madhukar, and Peter L Nagy, for their support in the bioinformatics analyses. We also thank E. Mereu for her technical support. We show gratitude to the technical staff of the Immunopathology Laboratory at Weill Cornell Medical College. G.I. is supported by the Italian Association for Cancer Research (5×1000 No. 10007); Regione Piemonte (ONCOPROT, CIPE 25/2005); and ImmOnc (BIO F.E.S.R. 2007/13), the Oncology Program of Compagnia di San Paolo, Torino, and Ricerca Finalizzata, Ministero della Salute. RP is supported by AIRC (IG-13358) and Compagnia di San Paolo (TO_Call2_2012_0061). LDS is supported by NIH Cancer Core grant CA034196. R.R. is supported by National Institutes of Health grants R01 CA185486, R01 CA179044, and U54 CA121852 and the Stewart Foundation. F.B. is supported by Oncosuisse KLS-02403-02-2009, Anna Lisa Stiftung, and the Nelia and Amadeo Barletta Foundation.
Footnotes
Accession Numbers: Sequencing data are accessible at Sequence Read Achieve (SRA; http://www.ncbi.nlm.nih.gov/sra/) (BioProject PRJNA255877, SRA identifier SRP044708). SNP array data have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE50253 and gene expression data under accession numbers GSE6338, GSE14879, and GSE19069.
Supplemental Information: Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.ccell.2015.03.006.
Author Contributions: All authors contributed as a team to experimental design and interpretation of data of manuscript. R.C., E.L., E.S., F.T., F.D.G., M.B., M.T., B.P., M.G., A. Rinaldi, E.E., C.C., and F.B. performed experiments and data analysis. R.C., E.L., R.P. and G.I. crossvalidated the TK fusions and determine their relative frequency in an extended ALCL panel. F.A. and R.R. completed the bioinformatics analyses. D.N., T.T., A. Rosenwald, D.W., J.C., L.K., L.C., M.A.P., S.P., E.T., B.F., and G.I. diagnosed and stratified pathological samples. LM. and J.E.V. identified and selected the JNJ-ROS1i-A compound. N.C., F.T., and M.G. performed DNA Sanger sequencing and/or immunohistochemistry stains. F.T. and M.G. created the pathological and clinical database. G.I., R.C., and F.A. wrote the manuscript.
References
- Abate F, Todaro M, van der Krogt J, Boi M, Landra I, Machiorlatti R, Tabbo F, Messana K, Barreca A, Novero D, et al. A novel patient derived tumorgraft model with TRAF1-ALK anaplastic large cell lymphoma translocation. Leukemia Published online December. 2014a;23:2014. doi: 10.1038/leu.2014.347. http://dx.doi.org/10.1038/leu.2014.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abate F, Zairis S, Ficarra E, Acquaviva A, Wiggins CH, Frattini V, Lasorella A, Iavarone A, Inghirami G, Rabadan R. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol. 2014b;8:97. doi: 10.1186/s12918-014-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KC. Targeted therapy of multiple myeloma based upon tumor-microenvironmental interactions. Exp Hematol. 2007;35(4,Suppl. 1):155–162. doi: 10.1016/j.exphem.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Bellanger D, Jacquemin V, Chopin M, Pierron G, Bernard OA, Ghysdael J, Stern MH. Recurrent JAK1 and JAK3 somatic mutations in T-cell prolymphocytic leukemia. Leukemia. 2014;28:417–419. doi: 10.1038/leu.2013.271. [DOI] [PubMed] [Google Scholar]
- Boi M, Rinaldi A, Kwee I, Bonetti P, Todaro M, Tabbò F, Piva R, Rancoita PM, Matolcsy A, Timar B, et al. PRDM1/BLIMP1 is commonly inactivated in anaplastic large T-cell lymphoma. Blood. 2013;122:2683–2693. doi: 10.1182/blood-2013-04-497933. [DOI] [PubMed] [Google Scholar]
- Bournazou E, Bromberg J. Targeting the tumor microenvironment: JAK-STAT3 signaling. JAK-STAT. 2013;2:e23828. doi: 10.4161/jkst.23828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, Karras JG, Levy DE, Inghirami G. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- Chrencik JE, Patny A, Leung IK, Korniski B, Emmons TL, Hall T, Weinberg RA, Gormley JA, Williams JM, Day JE, et al. Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6. J Mol Biol. 2010;400:413–433. doi: 10.1016/j.jmb.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Grewal JS, Smith LB, Winegarden JD, 3rd, Krauss JC, Tworek JA, Schnitzer B. Highly aggressive ALK-positive anaplastic large cell lymphoma with a leukemic phase and multi-organ involvement: a report of three cases and a review of the literature. Ann Hematol. 2007;86:499–508. doi: 10.1007/s00277-007-0289-3. [DOI] [PubMed] [Google Scholar]
- Jerez A, Clemente MJ, Makishima H, Rajala H, Gómez-Seguí I, Olson T, McGraw K, Przychodzen B, Kulasekararaj A, Afable M, et al. STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood. 2013;122:2453–2459. doi: 10.1182/blood-2013-04-494930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston PA, Grandis JR. STAT3 signaling: anticancer strategies and challenges. Mol Interv. 2011;11:18–26. doi: 10.1124/mi.11.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Velusamy T, Rolland D, Sahasrabuddhe AA, Chung F, Bailey NG, Schrader A, Li B, Li JZ, Ozel AB, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood. 2014;124:1460–1472. doi: 10.1182/blood-2014-03-559542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmäki H, Andersson EI, Lagström S, Clemente MJ, Olson T, Jalkanen SE, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366:1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, You S, Hu Z, Chen ZG, Sica GL, Khuri FR, Curran WJ, Shin DM, Deng X. Inhibition of STAT3 by niclosamide synergizes with erlotinib against head and neck cancer. PLoS ONE. 2013;8:e74670. doi: 10.1371/journal.pone.0074670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupardus PJ, Ultsch M, Wallweber H, Bir Kohli P, Johnson AR, Eigenbrot C. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci U S A. 2014;111:8025–8030. doi: 10.1073/pnas.1401180111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri A, Chang CC, Lombardi L, Salina M, Corradini P, Maiolo AT, Chaganti RS, Dalla-Favera R. Bcell lymphoma-associated chromosomal translocation involves candidate oncogene lyt-10, homologous to NF-kappa B p50. Cell. 1991;67:1075–1087. doi: 10.1016/0092-8674(91)90285-7. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, Kim S, van Bodegom D, Bolla S, Schatz JH, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgami RS, Ma L, Merker JD, Martinez B, Zehnder JL, Arber DA. STAT3 mutations are frequent in CD30+ T-cell lymphomas and T-cell large granular lymphocytic leukemia. Leukemia. 2013;27:2244–2247. doi: 10.1038/leu.2013.104. [DOI] [PubMed] [Google Scholar]
- Palomero T, Couronné L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, Carpenter Z, Abate F, Allegretta M, Haydu JE, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parilla Castellar ER, Jaffe ES, Said JW, Swerdlow SH, Ketterling RP, Knudson RA, Sidhu JS, Hsi ED, Karikehalli S, Jiang L, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. 2014;124:1473–1480. doi: 10.1182/blood-2014-04-571091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati C, Amessou M, Bihl MP, Balabaud C, Nhieu JT, Paradis V, Nault JC, Izard T, Bioulac-Sage P, Couchy G, et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J Exp Med. 2011;208:1359–1366. doi: 10.1084/jem.20110283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva R, Agnelli L, Pellegrino E, Todoerti K, Grosso V, Tamagno I, Fornari A, Martinoglio B, Medico E, Zamò A, et al. Gene expression profiling uncovers molecular classifiers for the recognition of anaplastic large-cell lymphoma within peripheral T-cell neoplasms. J Clin Oncol. 2010;28:1583–1590. doi: 10.1200/JCO.2008.20.9759. [DOI] [PubMed] [Google Scholar]
- Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Rebouissou S, Amessou M, Couchy G, Poussin K, Imbeaud S, Pilati C, Izard T, Balabaud C, Bioulac-Sage P, Zucman-Rossi J. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2009;457:200–204. doi: 10.1038/nature07475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda T, Tyner JW, Gutierrez A, Ngo VN, Glover J, Chang BH, Yost A, Ma W, Fleischman AG, Zhou W, et al. TYK2-STAT1-BCL2 pathway dependence in T-cell acute lymphoblastic leukemia. Cancer Discov. 2013;3:564–577. doi: 10.1158/2159-8290.CD-12-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage KJ, Harris NL, Vose JM, Ullrich F, Jaffe ES, Connors JM, Rimsza L, Pileri SA, Chhanabhai M, Gascoyne RD, et al. International Peripheral T-Cell Lymphoma Project. ALK- anaplastic large-cell lymphoma is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International Peripheral T-Cell Lymphoma Project. Blood. 2008;111:5496–5504. doi: 10.1182/blood-2008-01-134270. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonbol MB, Firwana B, Zarzour A, Morad M, Rana V, Tiu RV. Comprehensive review of JAK inhibitors in myeloproliferative neoplasms. Ther Adv Hematol. 2013;4:15–35. doi: 10.1177/2040620712461047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Haris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumors of Haemotolopoietic and Lymphoid Tissues, Fourth Edition. Sterling: Stylus Publishing; 2008. [Google Scholar]
- Velusamy T, Kiel MJ, Sahasrabuddhe AA, Rolland D, Dixon CA, Bailey NG, Betz BL, Brown NA, Hristov AC, Wilcox RA, et al. A novel recurrent NPM1-TYK2 gene fusion in cutaneous CD30-positive lymphoproliferative disorders. Blood. 2014;124:3768–3771. doi: 10.1182/blood-2014-07-588434. [DOI] [PubMed] [Google Scholar]
- Voena C, Di Giacomo F, Panizza E, D'Amico L, Boccalatte FE, Pellegrino E, Todaro M, Recupero D, Tabbò F, Ambrogio C, et al. The EGFR family members sustain the neoplastic phenotype of ALK+ lung adenocarcinoma via EGR1. Oncogenesis. 2013;2:e43. doi: 10.1038/oncsis.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vose J, Armitage J, Weisenburger D International T-Cell Lymphoma Project. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26:4124–4130. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- Yin X, Dewille JW, Hai T. A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene. 2008;27:2118–2127. doi: 10.1038/sj.onc.1210861. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yang J, Qian J, Li H, Romaguera JE, Kwak LW, Wang M, Yi Q. Role of the microenvironment in mantle cell lymphoma: IL-6 is an important survival factor for the tumor cells. Blood. 2012;120:3783–3792. doi: 10.1182/blood-2012-04-424630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


