Abstract
Targeted covalent inhibitors have emerged as a powerful approach in the drug discovery pipeline. Key to this process is the identification of signaling pathways (or receptors) specific to (or overexpressed in) disease cells. In this context, fragment-based ligand discovery (FBLD) has significantly expanded our view of the ligandable proteome and affords tool compounds for biological inquiry. To date, such covalent ligand discovery has almost exclusively employed cysteine-reactive small-molecule fragments. However, functional cysteine residues in proteins are often redox-sensitive and can undergo oxidation in cells. Such reactions are particularly relevant in diseases, like cancer, which are linked to excessive production of reactive oxygen species. Once oxidized, the sulfur atom of cysteine is much less reactive toward electrophilic groups used in the traditional FBLD paradigm. To address this limitation, we recently developed a novel library of diverse carbon-based nucleophile fragments that react selectively with cysteine sulfenic acid formed in proteins via oxidation or hydrolysis reactions. Here, we report analysis of sulfenic acid-reactive C-nucleophile fragments screened against a colon cancer cell proteome. Covalent ligands were identified for >1280 S-sulfenylated cysteines present in “druggable” proteins and orphan targets, revealing disparate reactivity profiles and target preferences. Among the unique ligand–protein interactions identified was that of a pyrrolidinedione nucleophile that reacted preferentially with protein tyrosine phosphatases. Fragment-based covalent ligand discovery with C-nucleophiles affords an expansive snapshot of the ligandable “redoxome” with significant implications for covalent inhibitor pharmacology and also affords new chemical tools to investigate redox-regulation of protein function.
Graphical abstract
INTRODUCTION
In recent years, targeting of disease proteins and pathways with covalent inhibitors has emerged as a viable option to overcome drug resistance, mutations, and toxicity associated with non-covalent inhibitors.1,2 Advantages of targeted covalent inhibition (TCI) strategies include extended duration of action and the ability to target rare residues and shallow binding sites. The mechanism by which TCIs achieve desired inhibition necessitate two distinct steps—the first involves the reversible association of a high-affinity ligand with its biological target in such a manner that an electrophilic “warhead” on the ligand is brought into an appropriately proximate position to form a covalent adduct with nucleophilic residue on the protein (more often than not, a cysteine). Majority of TCIs are being developed to target kinases for their role in cancer.3 Indeed, FDA approval of drugs such as “Afatinib” and “Ibrutinib” validated this strategy of incorporating Michael acceptor (electrophilic in nature) functionality in small-molecule inhibitors to covalently target cysteine residue in the active sites of diseased proteins (Figure 1a). While extremely attractive, designing TCIs is difficult in practice because of the difficulties in striking the right balance between reactivity and selectivity. In other words, electrophilic warheads may suffer from chemoselectivity issues due to the presence of other nucleophilic species (such as lysine, histidine, etc.). The electrophiles could also be scavenged by ubiquitous low-molecular-weight nucleophiles such as glutathione (GSH). Moreover, current TCIs predominantly target protein cysteines, and among many nucleophiles present in the cellular system, cysteine thiols/thiolates (Cys-SH/S−) are primary targets for the redox-based modulation of protein activity. Oxidation of a protein cysteine thiol (Cys-SH) to sulfenic acid (Cys-SOH) by reactive oxygen species (ROS) (e.g., H2O2) is termed S-sulfenylation (Figure 1b). It is a reversible post-translational modification that plays a crucial role in regulating several protein functions.4–8 Over the past decade several groups have reported the role of Cys-SOH in regulation of proteins such as transcription factors, kinases, phosphatases, ion channels, peroxidases and cysteine proteases, sirtuins, human serum albumin, and many others.7,9–18 Aberrant Cys-SOH formations have been shown to be a biomarker of disease state as well.19,20 The aforementioned examples and many other reports have established protein S-sulfenylation as a global signaling mechanism and a potential drug target.21–23
Figure 1.
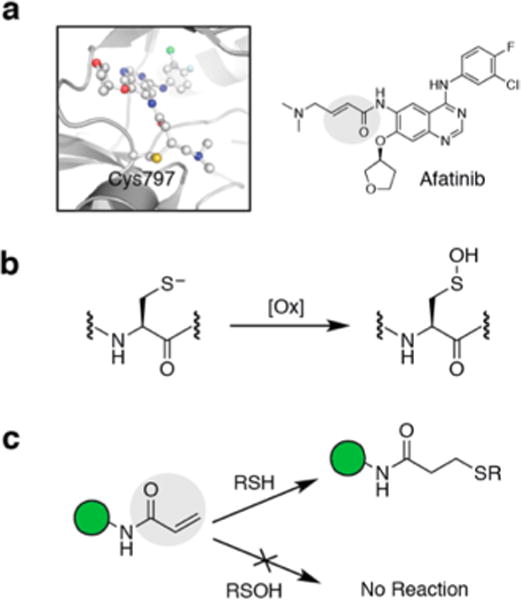
Targeting the cysteine thiol (Cys-SH) with covalent inhibitors. (a) FDA-approved Afatinib forms a covalent adduct with EGFR 797Cys-SH via Michael addition. (b) Oxidation of Cys-SH generates cysteine sulfenic acid (Cys-SOH). (c) Covalent inhibitors with electrophilic warheads (such as the Michael acceptor highlighted in gray) undergo facile reaction with Cys-SH but not with Cys-SOH under physiological conditions.
Due to overexpression, mutations, and constitutive activation in cancer cells, EGFR is targeted by a major class of covalent drugs that are aimed to covalently add at 797Cys-SH.1,3 However, 797Cys present in the catalytic pocket of EGFR is a target of signal-derived H2O2 oxidation which enhances the kinase activity of this enzyme.12,14,24 We recently showed that chronic oxidation result in an EGFR subpopulation that is refractory to covalent drugs such as Afatinib (Figure 1c).14 Inhibitors targeting 797Cys-SOH would be better suited for the purpose of preferential reactivity as well as enhanced selectivity. Another example of an enzyme class that is redox regulated is the protein tyrosine phosphatases (PTPs), including PTP1B for Type II diabetes and obesity, SHP2 for cancer, and others for rheumatoid arthritis, making them attractive therapeutic targets.25–29 In the past, we have reported redox-based trifunctional probes consisting of a warhead to covalently target Cys-SOH of PTPs, a ligand that directs the binding to the target, and a reporter tag used for the identification, purification or direct visualization of the labeled PTP. These redox-based probes showed low micromolar inhibitory activity against phosphatase YopH.30
Fragment-based ligand discovery (FBLD) and activity-based protein profiling (ABPP) approaches are powerful tools for the investigation of protein function and identification of drug leads for new therapeutics.31,32 Over the years, several FBLD and ABPP approaches have been reported that profile the reactivity of carbon electrophiles toward biological nucleophiles (such as Cys-SH) in cellular proteomes.32–34 However, ligands identified in these studies are incapable of reacting with cysteine sulfenic acid (Cys-SOH) under physiological conditions, and cysteine oxidation is expected to impact the pharmacology of inhibitors based on covalent targeting of Cys-SH.14 For this reason, we have proposed a complementary strategy that uses clickable carbon nucleophiles in a chemical proteomic assay and studied the Cys sulfenylome in RKO colon adenocarcinoma cells. Based on the results of this work, the sulfenyl form of therapeutically important proteins could be exploited to develop a new class of irreversible covalent inhibitors targeting this unique Cys oxoform.
RESULTS AND DISCUSSION
Current approaches to study protein sulfenylation are based on a β-dicarbonyl scaffold that shows moderate reactivity compared to other biologically relevant reactions (such as disulfide formation).8,12,35–38 Moreover, due to the lack of scaffold diversity among present Cys-SOH probes, current proteomics-based approaches (such as FBLD and ABPP) to identify small-molecule ligands targeting sulfenylated proteins could not be implemented effectively.13,36,39–41 To remedy the moderate reactivity of current nucleophile probes and to increase the structure diversity, we recently developed a library of cyclic and linear C-nucleophiles that showed diverse reactivity profiles toward Cys-SOH in a small molecule dipeptide model as well as in a protein model.42–44 The next logical step is to identify the biological target preferences of these newly developed nucleophiles. To do so, we first broadly divided the collection of ∼100 cyclic C-nucleophiles into eight distinct scaffolds - cyclohexane-1,3-diones (1), (thio)barbituric acids (2), different sized β-dicarbonyl rings (3), dihydrothiophen-3(2H)-one 1,1-dioxides/2-substituted isothiazolidin-4-one 1,1-dioxides (4) pyrrolidine-2,4-diones (5), 1,3-indandiones (6), piperidine-2,4-5(6H)-one (7), 1,2-thiazinan-5-one 1,1-dioxides/2H-1,2-thiazin-5(6H)-one 1,1-dioxides/1H-benzo-[c][1,2]thiazine-4(3H)-one 2,2-dioxides (8) (Figure 2a). Nucleophiles belonging to these scaffolds showed reaction rates that ranged from 2- to 150-fold higher than that of 1 (Figure 2a).42–44 Next, we chose cyclic C-nucleophiles from the above-described eight scaffolds based on differences in reactivity rates and structural diversity (Figure 2b, Figure S1). An alkyne was incorporated into the framework of these nucleophiles to provide a click chemistry handle for gel analysis, enrichment, and mass spectrometric analysis. DYn-2 is an established probe for Cys-SOH detection.12 Owing to the stability of resultant enolic carbanion (due to resonance stabilization), its reaction rate with Cys-SOH is modest (10 M−1 s−1).42 Minimum energy calculations show that this resonance stabilization also enables DYn-2 to toggle between half-chair conformers (Figure 2c). Thiazolidin-4-one 1,1-dioxide-based TD (kobs = 20 M−1 s−1) exists predominantly in a keto form with an envelope conformation. This is attributed to the sulfonamide S being out of the plane (due to sterics), thus distorting the tetrahedral geometry around the S atom (Figure 2c). In the case of pyrrolidine-2,4-dione-based PYD (kobs = 330 M−1 s−1), the keto−enol equilibrium favors the keto form.42 Minimum energy calculations for PYD showed an almost complete preference for planar geometry (Figure 2c). Piperidine-2,4-dione-based PRD (kobs = 1155 M−1 s−1) is structurally similar to DYn-2, but exhibits rate enhancement of more than 2 orders of magnitude, which is attributed to the instability of the carbanion in a keto–enol equilibrium that favors the keto form.42 Minimum energy calculation indicated that PRD preferentially exists in the boat conformation (Figure 2c). Benzo[c][1,2]thiazine-based BTD (kobs = 1700 M−1 s−1) showed the highest reaction rate toward Cys-SOH, which is attributed to predominance of the keto conformation and a distorted tetrahedron geometry around the S atom.42 Minimum energy calculation showed that BTD adopts a half-boat conformation with sulfone S out of the plane, similar to TD (Figure 2c).
Figure 2.
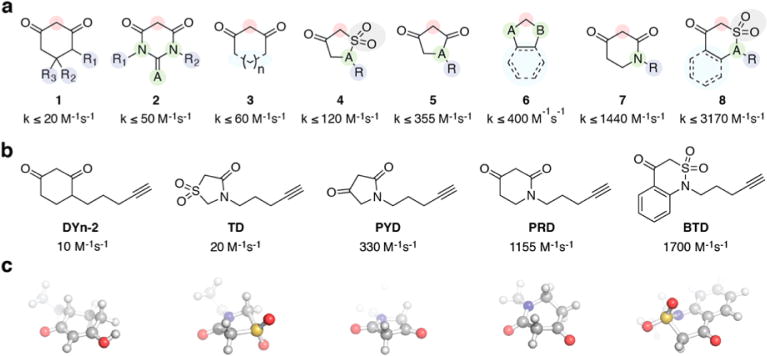
Novel classes of nucleophile probes to profile cysteine oxidation (a) Structural classes of cyclic C-nucleophiles (1–8). In each structure, the nucleophilic carbon is highlighted in the red circle. Rate constants from ref 42. (b) From the pool of cyclic C-nucleophiles, DYn-2 and four new C-nucleophiles were selected on the basis of a range of reaction rate constants and scaffold diversity. (c) Energy-minimized 3-dimensional representation of the probes.
Application of these nucleophilies to RKO colon adenocarcinoma cells under basal conditions, followed by click chemistry with a biotin azide reporter tag and visualization of labeled proteins by SDS-PAGE demonstrated that the panel of nucleophiles exhibited a range of reactivities consistent with their rate constants (Figure S2). As expected, the highest reactivity was observed for the BTD probe, which demonstrated substantial protein labeling at concentrations as low as 100 μM. The PRD and PYD probes showed moderate to high levels of labeling, while DYn-2 and TD displayed less extensive labeling at low concentrations (Figure S2). From these results, it was apparent that if the protein site accessibility is not limiting, the comparative reactivity of these probes correlates well with their reaction rate constants toward Cys-SOH.
Subsequently, we further examined the peptide labeling profiles for these probes by analyzing their proteome reactivities using a mass spectrometry platform and a workflow referred to as SulfenM to site-specifically map the S-sulfenylome in RKO cells. The selectivity, accuracy, and reproducibility of the workflow have been reported previously.13,39 The workflow consists of eight steps: lysing the RKO cells under native conditions (using lysis buffer containing non-ionic detergent and catalase) in the presence of C-nucelophile (Figure 3a), reducing the native disulfides with DTT, alkylating the reduced cysteines, digesting the labeled proteins, conjugating labeled peptides with a photocleavable azide biotin via “click” chemistry, capturing labeled peptides with streptavidin beads, photorelease by cleavage of the biotin linker with UV light, and MS-based proteomic analysis of the probe-labeled peptides (Figure S3). A total of 1283 S-sulfenylated sites on 761 proteins were successfully labeled by the five C-nucleophiles (Figure 3b,c). Interestingly, only nine proteins (ACLY, EEF2, GAPDH, HSPA8, PFN1, RPS20, RPS21, RPS27A, RPS3A) were labeled by all five nucleophiles. For example, in GAPDH, 152Cys-SOH (by DYn-2, PYD, PRD, BTD), 156Cys-SOH (by DYn-2, BTD), and 247Cys-SOH (TD, PYD, PRD, BTD) were successfully labeled. GAPDH is known as a major redox-sensitive protein, which has three Cys residues, one (152Cys) in the exposed active site and two (156Cys, 247Cys) that are buried (Figure 4a). While redox-sensitivity of 152Cys is well established, 156Cys and 247Cys were recently reported to be sensitive to storage-dependent oxidation.45 Even more interestingly, only two S-sulfenylated sites (17Cys-SOH on RPS21 and 128Cys-SOH on PFN1) were labeled by all five nucleophiles (Figure S4). An interesting target was PARK7 (also known as DJ-1), which is a highly conserved homodimeric small protein associated with Parkin-son’s disease.46 Studies have demonstrated that DJ-1 protects cells against oxidative stress-mediated apoptosis, the mechanism of which is largely unknown.7 DJ-1 is also known to regulate the activity of phosphatase and tensin homologue (PTEN) via transnitrosylation reaction.47 Structurally, DJ-1 has three potentially redox-active cysteines (46Cys, 53Cys, and 106Cys). However, 106Cys, which is highly conserved, was shown to be highly sensitive to oxidation and formed a stable Cys-SO2H.7 We showed that DJ-1 was labeled by DYn-2 and PRD at 106Cys, and BTD at 46Cys, respectively. The rank order of the number of distinct probe-modified sites identified by these chemoproteomic analyses was BTD ≫ DYn-2 > PYD > PRD > TD (Table 1). Another interesting target was CARM1, in which 421Cys was labeled by TD. CARM1 (also known as PRMT4) is a member of arginine-specific protein methyltransferases (PRMTs). While most PRMTs methylate Gly- and Arg-rich (GAR) motifs in their substrates, CARM1 displays unique substrate specificity by methylating Pro-, Gly-, and Metrich motifs (PGM motifs).48
Figure 3.
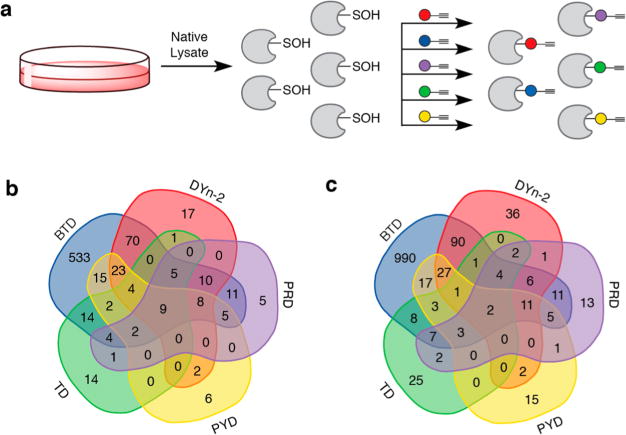
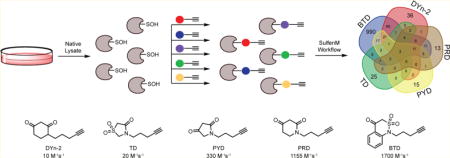
Application of new nucleophile probes for studying cellular sulfenylomes. (a) RKO whole cell lysates were prepared under nondenaturing conditions. Native lysate was divided into equal portions and incubated with one of five nucleophile probes (or vehicle) in separate reactions. Labeled proteins from each reaction were digested separately and the resulting peptides subjected to click reaction with UV-cleavable biotin-N3. Enrichment and photorelease afforded different probe-labeled peptides samples for proteomic analysis. (b) Protein Venn diagram of unique sulfenylated proteins. (c) Site Venn diagram of unique sulfenylated peptides.
Figure 4.
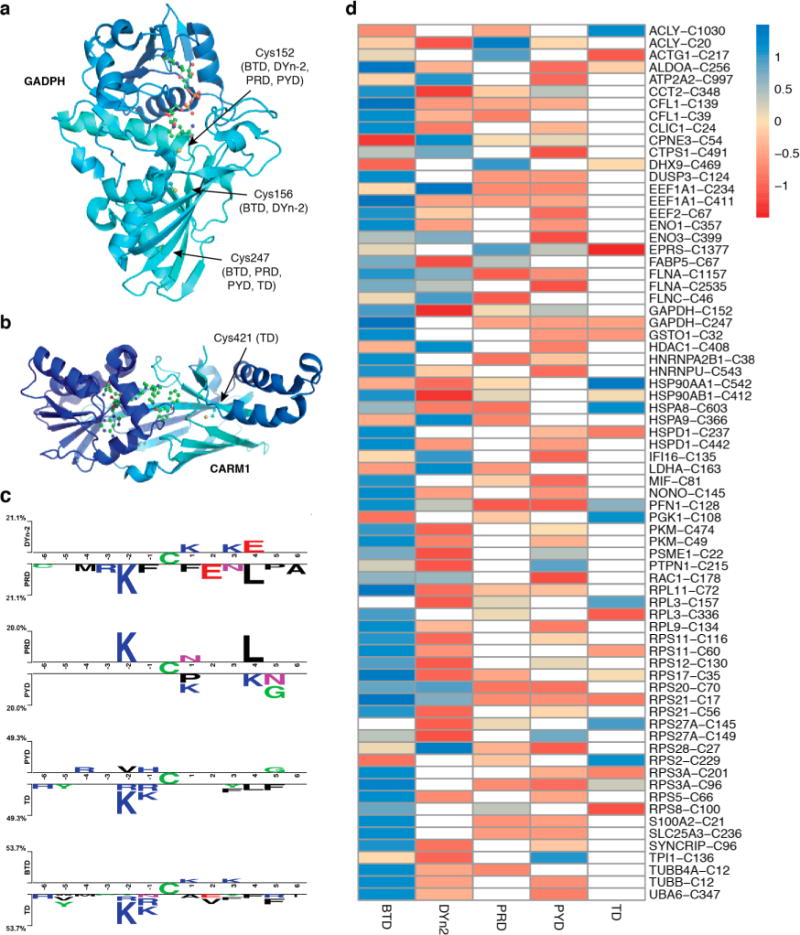
Chemoproteomic analysis of RKO cellular S-sulfenylome. (a) Crystal structures of GAPDH (PDB: 4WNC) showing Cys152, Cys156, and Cys247 labeled by different probes. (b) CARM1 (PDB: 5U4X) showing Cys421 that is labeled by TD. (c) Comparative sequence analysis of peptides labeled with different probes. (d) Heat map of Z-score normalized peak intensities of S-sulfenylated sites labeled with various probes. Values in each row direction have been mean centered and scaled.
Table 1.
Total Number of Probe-Modified Proteins and Sites
| Probe | Total no. of labeled proteins | Total no. of labeled sites |
|---|---|---|
| DYn-2 | 150 | 183 |
| TD | 56 | 58 |
| PYD | 76 | 87 |
| PRD | 60 | 68 |
| BTD | 715 | 1186 |
Owing to its superior reaction rate toward Cys-SOH, BTD exhibited the highest level of S-sulfenylome reactivity. However, a very restricted reactivity profile was observed for other probes. Of particular interest was the relatively low number of sulfenylated proteins labeled by the PYD and PRD probes in RKO sulfenylome (Table 1), which contrasted sharply with the robust reactivity shown by these probes in native RKO cell lysate (Figure S2). Potential reversibility of thioether adducts under reducing conditions under the proteomics workup conditions could be responsible for unexpected labeling profile of PYD and PRD.44 To further explore structural features that may define the protein sulfenylation, we examined the flanking sequences of S-sulfenylated Cys residues modified by the five probes with the pLogo algorithm for the presence of consensus motifs (Figure S5).49 We found several consensus motifs for the nucleophile-labeled S-sulfenylome, such as KxC and KC. Of interest, Lys was significantly overrepresented at almost every adjacent position (from −6 to +6) of BTD-labeled Cys-SOH. This observed enrichment of Lys is intriguing and may indicate that positive charge in the vicinity of the reactive cysteine is critical for thiolate generation. However, if this were strictly the case, then Arg enrichment would also be observed. Another possibility is that due to its elevated reactivity, BTD was able to capture highly transient sulfenylated proteins. But the abundance of such BTD labeled peptides is low, and probe-tagged peptides containing Lys ionize better, thus enabling detection of these peptides above the noise. We also compared the flanking sequences of S-sulfenylated Cys residues modified by the five probes using Two Sample Logo (2logP) algorithm (Figure 4c).50 Structurally, DYn-2 and PRD differ by just one atom (C4 in DYn-2 vs N in PRD). 2LogP analyses indicated that PRD-labeled peptides show higher frequency of Lys (at −2) and Leu (at +4) residues compared to DYn-2 modified peptides (Figure 4c, panel 1). On the other hand, both PYD and PRD are β-ketolactams, but they differ in ring size, which leads to differences in conformation but which also resulted in higher frequency of Lys (at −2) and Leu (+4) in the peptides modified by PRD (Figure 4c, panel 2). KxC and CxxxL thus appeared to be the conserved sequence motifs in PRD modified peptides. Indeed, a pLogo analysis of PRD modified peptides show a conserved KxC sequence motif as well (Figure S5). Similarly, when sequences of peptides labeled by TD probe were compared with those modified by PYD (owing to both having 5-membered ring structure) and BTD (due to structural analogy), the KxC sequence motif was again found to be conserved (Figure 4c, panels 3 and 4), an observation which also was seen in pLogo sequence analysis of TD (Figure S5).
To further estimate the stoichiometries and selectivity of nucleophile labeling, we obtained peak intensities of all the detected S-sulfenylated peptides using MS1 filtering-based label-free quantification as previously described (Figure S6).13,39 The label-free quantification results from each nucleophile-labeling experiment were then standardized with Z-score normalization to enable further comparison. The quantitative comparison of peptide-nucleophile labeling is illustrated in a heat map (Figure 4d). These data suggested that the nucleophiles display different trends of S-sulfenylome reactivity and selectivity in complex proteomes. For example, 442Cys-SOH of HSPD1 (Hsp60) showed a clear preference for BTD over DYn-2 (Figure 4d). In some cases, S-sulfenylated sites on the same protein showed different probe preferences. DYn-2, PYD, PRD, and BTD labeled 152Cys-SOH of GAPDH with almost equal efficiencies, but BTD was more efficient in labeling surface-exposed 247Cys-SOH compared to TD, PYD, and PRD (Figure 4d). These results could be biased by differences in ionization efficiency among different peptide-probe adducts. To evaluate this possibility, we compared the ionization of Cbz-Cys-Val-OMe peptide labeled with different nucleophiles. Each probe was exposed to limiting amount of Cbz-Cys(SOH)-Val-OMe under standard buffer conditions (Figure S7a). Thioether products corresponding to each nucleophile were HPLC-purified, mixed together in equimolar quantities, and analyzed by LC/MS. The relative ionization intensities of these adducts differed by one-fold between the lowest and highest ionizing species, with lowest being Cbz-Cys(S-BTD)-Val-OMe and highest being Cbz-Cys(S-DYn-2)-Val-OMe, indicating limited effect of ionization efficiencies of peptide–probe adducts (Figure S7b).
We next investigated whether the S-sulfenylomes labeled by the five probes had distinct structural features. Based on the calculations resulting from 20 proteins with structure files available in Protein Data Bank (PDB) that contained both buried and exposed Cys residues, we observed that DYn-2 and PRD prefer to label S-sulfenylated residues on protein surfaces. In contrast, PYD and TD display substantial reactivity toward the buried S-sulfenylated sites (Table 2). Of particular interest was the distinct reactivity of the PYD probe with the S-sulfenylated sites within the buried catalytic domains of several classical protein tyrosine phosphatase (PTPs) such as PTPN1 (PTP1B), PTPN7, PTPN11, and DUSP23 (Figure 5a, Figure S8). The preferential reactivity of PYD toward PTP1B was also verified by treatment of recombinant, oxidized phosphatase with each nucleophile followed by click chemistry-mediated conjugation to an biotin-azide reporter tag and visualization by Strepavidin-HRP Western blot (Figure 5b). Molecular docking (using AutoDock Vina)51 of PYD into the catalytic pocket of PTP1B showed a good fit with the catalytic pocket, ostensibly owing to the almost planar conformation of this C-nucleophile (Figure 5c). The CX5R sequence is the signature motif of the PTP superfamily and forms the critical phosphate-binding loop (also known as the P-loop) in the active site (Figure 5d). Irrespective of the variations in the X5 segment, conformation of the P-loop is conserved (to keep catalytic Cys and Arg in close proximity) and can be easily superimposed among different PTP structures. PTPs are established targets of ROS because the signature motif CX5R of this family contains an invariant cysteine that has an extremely low pKa, which promotes its function as a nucleophile in catalysis and renders it highly susceptible to oxidative inactivation.29
Table 2.
Surface Distribution Analysis of Probe-Labeled Proteinsa
| Probe | E (%) | B (%) |
|---|---|---|
| DYn-2 | 35.3 | 52.9 |
| TD | 0.0 | 80.0 |
| PYD | 21.4 | 71.4 |
| PRD | 33.3 | 55.6 |
| BTD | 25.0 | 62.5 |
| UM | 19.6 | 58.7 |
Calculations were based on 20 proteins with 3D structure files found in the Protein Data Bank (PDB) that contained both buried and exposed cysteine residues. Abbreviations: E, exposed (RSA > 25%); B, buried (RSA < 10%); UM, unmodified cysteine residues.
Figure 5.
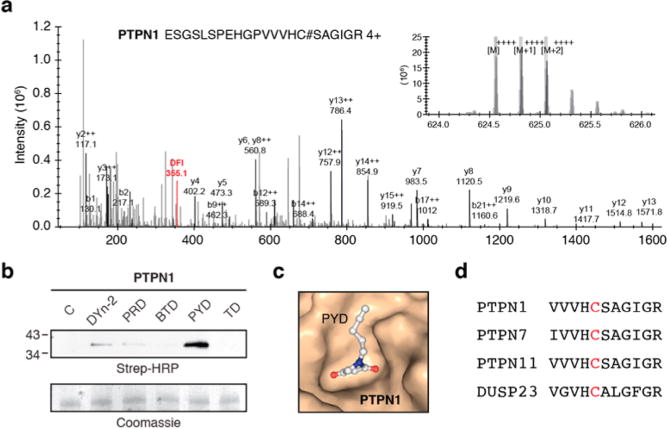
PTPs as privileged targets of PYD. (a) Representative MS/MS spectra of PTPN1 (PTP1B) labeled by PYD probe. (b) PYD probe preferentially modifies the recombinant oxidized form of PTP1B. (c) Molecular docking (using AutoDock Vina) of PYD into catalytic pocket of PTPN1 (PDB: 1OET). (d) Abbreviated sequence alignment of PTPs that harbor a Cys-SOH labeled by PYD probe.
CONCLUSION
In order to develop inhibitors that effectively target redox-active cysteines in therapeutically important proteins, it is imperative to complement the structural knowledge of S-sulfenylated sites with an understanding of warhead reactivity. With suitable chemical tools, modern chemical proteomics approaches such as FBLD and ABPP present a way to achieve this task in a systematic and efficient manner. Not only do such approaches provide the technology that help identify sites of covalent modification, but they also differentiate between selective versus promiscuous covalent modifiers and thus aid future drug design and development. Herein, we utilized a novel multi-probe approach and a highly efficient chemoproteomics platform to globally profile the reactivity of five C-nucleophile probes, including four novel molecules, in the RKO S-sulfenylome under normal physiological conditions. These five C-nucleophiles covalently labeled 1283 redox-sensitive Cys residues on 761 proteins that are endogenously S-sulfenylated. We show that distinct structural classes of C-nucleophiles targeted discrete protein populations and S-sulfenylated residues with little overlap. Whereas BTD labeled the greatest numbers of S-sulfenylated sites by far, the smaller size of the five-membered rings of TD and PYD appears to facilitate internal site labeling. In addition to physical size, the conformations of five-membered (envelope for TD and almost planar for PYD) versus six-membered (chair and boat) probes also differ, which also would favor buried site labeling by TD and PYD. Insights from this proteomics study have important implications in the development of covalent inhibitors that target cysteines within the thiol “redoxome”. For example, PTPs, including PTP1B for Type II diabetes and obesity, SHP2 for cancer, and others for rheumatoid arthritis, are attractive therapeutic targets.25–28 Our study has identified the catalytic Cys in PTP1B (215Cys), PTPN7 (396Cys), PTPN11 (459Cys), and DUSP23 (95Cys), PTPs as favored targets of pyrrolidine-2,4-dione-based nucleophile, PYD. The enrichment of PYD-labeled PTPs suggests that PYD could be coupled to binding elements that capture interactions with adjacent pockets surrounding the active site to develop covalent inhibitors for this class of proteins. This strategy can be expanded and applied to other attractive targets. Our expanding library of novel warheads that target S-sulfenylated cysteines will enable high-throughput profiling to pair distinct C-nucleophilic scaffolds with various redox-regulated proteins in a combined FBLD/ABPP approach to develop effective covalent irreversible/ reversible inhibitors-based therapeutics.23,32,52 Finally, these probes can be used to study how oxidative cysteine modifications impact both cell signaling and drug pharmacology.
Supplementary Material
Acknowledgments
Part of this work is supported by the National Institutes of Health under awards R01 GM102187 and R01 CA174864 to K.S.C. J.Y. acknowledges support from the National Key Research and Development Program (No. 2016YFA0501303), the National Natural Science Foundation of China (No. 31500666 and No. 81573395), the Beijing Natural Science Foundation (No. 5162009), and Beijing Nova Program. V.G. thanks Dr. Mauro Lo Conte (Department of Chemistry, Scripps Florida) for biotin-N3 reagent, and Dr. Peng Wu (Department of Chemistry, Scripps La Jolla) and Dr. Hua Wang (Department of Chemistry, Scripps La Jolla) for providing BTTP “click” ligand used in the click chemistry for WB analysis.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b01791.
Experimental details and protocols, including Schemes S1–S4 and Figures S1–S8, and NMR spectra (PDF)
Data summary for S-sulfenylated peptides and proteins identified in RKO cells using BTD probe (XLSX)
Data summary for S-sulfenylated peptides and proteins identified in RKO cells using DYn-2 probe (XLSX)
Data summary for S-sulfenylated peptides and proteins identified in RKO cells using PRD probe (XLSX)
Data summary for S-sulfenylated peptides and proteins identified in RKO cells using PYD probe (XLSX)
Data summary for S-sulfenylated peptides and proteins identified in RKO cells using TD probe (XLSX)
Data for 1283 S-sulfenylated sites on 761 proteins identified by using multiple probe-labeling strategy (XLSX)
Data used to generate Venn diagrams (XLSX)
Data used to generate heat map of Z-score normalized peak intensities of S-sulfenylated peptides labeled with various probes (XLSX)
Sequence data used for generating 2LogP and pLogO plots (XLSX)
ORCID
Jing Yang: 0000-0001-8486-273X
Daniel C. Liebler: 0000-0002-7873-3031
Kate S. Carroll: 0000-0002-7624-9617
Notes
The authors declare no competing financial interest.
References
- 1.Singh J, Petter RC, Baillie TA, Whitty A. Nat Rev Drug Discovery. 2011;10:307. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 2.Baillie TA. Angew Chem, Int Ed. 2016;55:13408. doi: 10.1002/anie.201601091. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, Gray NS. Chem Biol. 2013;20:146. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulsen CE, Carroll KS. Chem Rev. 2013;113:4633. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddie KG, Carroll KS. Curr Opin Chem Biol. 2008;12:746. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 6.Paulsen CE, Carroll KS. ACS Chem Biol. 2010;5:47. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo Conte M, Carroll KS. J Biol Chem. 2013;288:26480. doi: 10.1074/jbc.R113.467738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta V, Carroll KS. Biochim Biophys Acta, Gen Subj. 2014;1840:847. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denu JM, Tanner KG. Biochemistry. 1998;37:5633. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 10.Svoboda LK, Reddie KG, Zhang L, Vesely ED, Williams ES, Schumacher SM, O’Connell RP, Shaw R, Day SM, Anumonwo JM, Carroll KS, Martens JR. Circ Res. 2012;111:842. doi: 10.1161/CIRCRESAHA.111.263525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulathu Y, Garcia FJ, Mevissen TE, Busch M, Arnaudo N, Carroll KS, Barford D, Komander D. Nat Commun. 2013;4:1569. doi: 10.1038/ncomms2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Nat Chem Biol. 2011;8:57. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Gupta V, Carroll KS, Liebler DC. Nat Commun. 2014;5:4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truong TH, Ung PM, Palde PB, Paulsen CE, Schlessinger A, Carroll KS. Cell Chem Biol. 2016;23:837. doi: 10.1016/j.chembiol.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett TJ, Pattison DI, Leonard SE, Carroll KS, Davies MJ, Hawkins CL. Free Radical Biol Med. 2012;52:1075. doi: 10.1016/j.freeradbiomed.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alili L, Sack M, von Montfort C, Giri S, Das S, Carroll KS, Zanger K, Seal S, Brenneisen P. Antioxid Redox Signaling. 2013;19:765. doi: 10.1089/ars.2012.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D, Vetrivelan R, Clish CB, Robinson AJ, Gygi SP, Spiegelman BM. Nature. 2016;532:112. doi: 10.1038/nature17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hourihan JM, Moronetti Mazzeo LE, Fernandez-Cardenas LP, Blackwell TK. Mol Cell. 2016;63:553. doi: 10.1016/j.molcel.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zito E, Hansen HG, Yeo GS, Fujii J, Ron D. Mol Cell. 2012;48:39. doi: 10.1016/j.molcel.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo YH, Carroll KS. Proc Natl Acad Sci U S A. 2009;106:16163. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Truong TH, Carroll KS. Biochemistry. 2012;51:9954. doi: 10.1021/bi301441e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truong TH, Carroll KS. Crit Rev Biochem Mol Biol. 2013;48:332. doi: 10.3109/10409238.2013.790873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visscher M, Arkin MR, Dansen TB. Curr Opin Chem Biol. 2016;30:61. doi: 10.1016/j.cbpa.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heppner DE, Hristova M, Dustin CM, Danyal K, Habibovic A, van der Vliet A. J Biol Chem. 2016;291:23282. doi: 10.1074/jbc.M116.749028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan CC, Ramachandran C, Gresser MJ, Tremblay ML, Kennedy BP. Science. 1999;283:1544. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 26.Saltiel AR, Kahn CR. Nature. 2001;414:799. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 27.He RJ, Yu ZH, Zhang RY, Zhang ZY. Acta Pharmacol Sin. 2014;35:1227. doi: 10.1038/aps.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barr AJ. Future Med Chem. 2010;2:1563. doi: 10.4155/fmc.10.241. [DOI] [PubMed] [Google Scholar]
- 29.Tonks NK. Nat Rev Mol Cell Biol. 2006;7:833. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 30.Leonard SE, Garcia FJ, Goodsell DS, Carroll KS. Angew Chem, Int Ed. 2011;50:4423. doi: 10.1002/anie.201007871. [DOI] [PubMed] [Google Scholar]
- 31.Murray CW, Rees DC. Nat Chem. 2009;1:187. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- 32.Backus KM, Correia BE, Lum KM, Forli S, Horning BD, Gonzalez-Paez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, Wolan DW, Cravatt BF. Nature. 2016;534:570. doi: 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weerapana E, Simon GM, Cravatt BF. Nat Chem Biol. 2008;4:405. doi: 10.1038/nchembio.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kathman SG, Xu Z, Statsyuk AV. J Med Chem. 2014;57:4969. doi: 10.1021/jm500345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddie KG, Seo YH, Muse WB, III, Leonard SE, Carroll KS. Mol BioSyst. 2008;4:521. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leonard SE, Reddie KG, Carroll KS. ACS Chem Biol. 2009;4:783. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 37.Qian J, Klomsiri C, Wright MW, King SB, Tsang AW, Poole LB, Furdui CM. Chem Commun. 2011;47:9203. doi: 10.1039/c1cc12127h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian J, Wani R, Klomsiri C, Poole LB, Tsang AW, Furdui CM. Chem Commun. 2012;48:4091. doi: 10.1039/c2cc17868k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Gupta V, Tallman KA, Porter NA, Carroll KS, Liebler DC. Nat Protoc. 2015;10:1022. doi: 10.1038/nprot.2015.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gould NS, Evans P, Martinez-Acedo P, Marino SM, Gladyshev VN, Carroll KS, Ischiropoulos H. Chem Biol. 2015;22:965. doi: 10.1016/j.chembiol.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akter S, Huang J, Bodra N, De Smet B, Wahni K, Rombaut D, Pauwels J, Gevaert K, Carroll K, Van Breusegem F, Messens J. Mol Cell Proteomics. 2015;14:1183. doi: 10.1074/mcp.M114.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta V, Carroll KS. Chem Sci. 2016;7:400. doi: 10.1039/c5sc02569a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta V, Carroll KS. Chem Commun (Cambridge, U K) 2016;52:3414. doi: 10.1039/c6cc00228e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta V, Paritala H, Carroll KS. Bioconjugate Chem. 2016;27:1411. doi: 10.1021/acs.bioconjchem.6b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reisz JA, Wither MJ, Dzieciatkowska M, Nemkov T, Issaian A, Yoshida T, Dunham AJ, Hill RC, Hansen KC, D’Alessandro A. Blood. 2016;128:e32. doi: 10.1182/blood-2016-05-714816. [DOI] [PubMed] [Google Scholar]
- 46.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Science. 2003;299:256. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 47.Choi MS, Nakamura T, Cho SJ, Han XM, Holland EA, Qu J, Petsko GA, Yates JR, Liddington RC, Lipton SA. J Neurosci. 2014;34:15123. doi: 10.1523/JNEUROSCI.4751-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Bedford MT. Nat Rev Cancer. 2013;13:37. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 49.O’Shea JP, Chou MF, Quader SA, Ryan JK, Church GM, Schwartz D. Nat Methods. 2013;10:1211. doi: 10.1038/nmeth.2646. [DOI] [PubMed] [Google Scholar]
- 50.Vacic V, Iakoucheva LM, Radivojac P. Bioinformatics. 2006;22:1536. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- 51.Trott O, Olson AJ. J Comput Chem. 2010;31:455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gruner BM, Schulze CJ, Yang D, Ogasawara D, Dix MM, Rogers ZN, Chuang CH, McFarland CD, Chiou SH, Brown JM, Cravatt BF, Bogyo M, Winslow MM. Nat Methods. 2016;13:883. doi: 10.1038/nmeth.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


