ABSTRACT
Adequate information on the precise molecular and biological composition of the viral strains that establish HIV infection in the human host will provide effective means of immunization against HIV infection. In an attempt to identify the transmitted founder (TF) virus and differentiate the biological properties and infectious potential of the TF virus from those of the population of the early transmitted viruses, 250 patient-derived gp120 envelope glycoproteins were cloned in pMN-K7-Luc-IRESs-NefΔgp120 to obtain chimeric viruses. Samples were obtained from eight infants who had recently become infected with HIV through mother-to-child transmission (MTCT) and two adults who acquired infection through the heterosexual route and were in the chronic stage of infection. Among the 250 clones tested, 65 chimeric viruses were infectious, and all belonged to HIV-1 subtype C. The 65 clones were analyzed for molecular features of the envelope, per-infectious-particle infectivity, coreceptor tropism, drug sensitivity, and sensitivity to broadly neutralizing antibodies. Based on genotypic and phenotypic analysis of the viral clones, we identified 10 TF viruses from the eight infants. The TF viruses were characterized by shorter V1V2 regions, a reduced number of potential N-linked glycosylation sites, and a higher infectivity titer compared to the virus variants from the adults in the chronic stage of infection. CXCR6 coreceptor usage, in addition to that of the CCR5 coreceptor, which was used by all 65 chimeric viruses, was identified in 13 viruses. The sensitivity of the TF variants to maraviroc and a standard panel of neutralizing monoclonal antibodies (VRC01, PG09, PG16, and PGT121) was found to be much lower than that of the virus variants from the adults in the chronic stage of infection.
IMPORTANCE Tremendous progress has been made during the last three and half decades of HIV research, but some significant gaps continue to exist. One of the frontier areas of HIV research which has not seen a breakthrough yet is vaccine research, which is because of the enormous genetic diversity of HIV-1 and the unique infectious fitness of the virus. Among the repertoire of viral variants, the virus that establishes successful infection (transmitted founder [TF] virus) has not been well characterized yet. An insight into the salient features of the TF virus would go a long way toward helping with the design of an effective vaccine against HIV. Here we studied the biological properties of recently transmitted viruses isolated from infants who acquired infection from the mother and have come up with unique characterizations for the TF virus that establishes infection in the human host.
KEYWORDS: transmitted founder virus, amino acid diversity, coreceptor tropism, human immunodeficiency virus, mother-to-child transmission, resistance to neutralization
INTRODUCTION
Since the discovery of HIV, understanding the mechanism of HIV transmission and preventing the spread of infection have been the focus of intense research. The ultimate aim is to develop a potential vaccine (1) that can act in a subtype-independent manner (2) or at least have the potential to control viral replication and prevent CD4+ T-cell loss and disease progression (3). A useful broadly neutralizing antibody (bNAb) response is required to overcome the genetic variability, structural complexity, and escape mechanisms of the virus. Unfortunately, attempts to develop an immunogen capable of eliciting such antibodies have not met with success to date (4, 5), and HIV has proved such a formidable foe to vaccine designers and the immune system alike.
Nearly half of the global HIV infections are caused by the subtype C strain (HIV-1C), which is the most predominant among the nine pure HIV-1 subtypes and numerous unique and circulating recombinant forms (6). In India, approximately 2.1 million people were reported to be living with HIV in the middle of 2017 (UNAIDS global AIDS update, 2017), with more than 99% of infections being caused by HIV-1 subtype C (HIV-1C) strains alone (7–11). Despite the unique characteristics of HIV-1C, there are considerable differences in the features of viruses within the quasispecies, especially in the transmission efficiency of the virus in vivo.
Among the repertoire of transmitted viral variants, it is well-known that only a small proportion of the viruses (commonly referred to as transmitted founder [TF] viruses) are successful in establishing infection. However, we are still limited in our understanding of the properties of these viruses. It is widely believed that the early immune response to HIV infection is likely to be an essential factor in determining the clinical course of the disease. Thus, a better understanding of the characteristics of TF viruses and their role in early infection will throw light on the features that bestow these variants with the unique advantage of successfully establishing infection and contribute significantly to the design and development of a protective HIV vaccine. The conformational changes in the viral envelope induced by the interaction between the HIV-1 gp120 and the CD4 receptor and coreceptor enable viral entry into the host cell (12, 13).
Therefore, this study aimed to construct recombinant gp120 clones from infants recently infected with HIV and to identify and characterize the TF variants for their unique biological properties, including coreceptor usage, infectivity potential, drug sensitivity, and sensitivity to neutralizing antibodies.
RESULTS
General features of the recombinant viruses.
Two hundred fifty representative clones (25 from each subject) were selected from the 10 individuals and subjected to plasmid DNA isolation, restriction digestion with the enzymes NgoMIV and MluI, and sequence analysis. A total of 150 clones were confirmed by both sequence analysis and digestion with a restriction endonuclease. Recombinant viruses were generated by transfection of 293T cells with the plasmids, and the viruses were tested for infectivity using TZM-bl reporter cells. Based on luciferase expression (number of relative light units [RLU]), 43% (65/150) of the recombinant viruses, belonging to nine individuals, were found to be infectious and were employed for further downstream analysis. Unfortunately, none of the clones obtained from one individual, IN_01, were found to be infectious. Table 1 provides the clinical and demographic profiles of the study participants.
TABLE 1.
Clinical and demographic profiles of study subjects and viral characteristics of the chimeric viral variantsa
| Sample | Age/sex | Mode of transmission | Yr of sampling | Diagnostic method | Viral load (no. of copies/ml) | CD4 T-cell count (no. of cells/μl) | No. of clones | No. of infectious clones | No. of TF viruses | Subtypeb | Predicted coreceptor determined byc: |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PhenoSeq | G2P | |||||||||||
| IN_01 | 34 yr/M | HET | 2013 | ELISA | 84,356 | 207 | 21 | 0 | 0 | C | CCR5 | CCR5 |
| IN_02 | 25 yr/F | HET | 2016 | ELISA | 148,000 | 352 | 21 | 05 | 0 | C | CCR5 | CCR5 |
| IN_03 | 45 days/F | MTCT | 2014 | DNA PCR | NA | 623 | 15 | 02 | 1 | C | CCR5 | CCR5 |
| IN_04 | 04 mo/M | MTCT | 2014 | DNA PCR | 459,494 | NA | 04 | 02 | 1 | C | CCR5 | CCR5 |
| IN_05 | 14 mo/M | MTCT | 2015 | DNA PCR | 4,723,382 | 224 | 07 | 05 | 1 | C | CCR5 | CCR5 |
| IN_06 | 04 mo/F | MTCT | 2014 | DNA PCR | 2,354,238 | 328 | 22 | 08 | 1 | C | CCR5 | CCR5 |
| IN_07 | 02 mo/M | MTCT | 2015 | DNA PCR | NA | 655 | 22 | 18 | 1 | C | CCR5 | CCR5 |
| IN_08 | 04 mo/M | MTCT | 2015 | DNA PCR | 2,991,258 | 925 | 16 | 14 | 2 | C | CCR5 | CCR5 |
| IN_09 | 06 mo/F | MTCT | 2014 | DNA PCR | 127,706 | 432 | 07 | 05 | 1 | C | CXCR4 | CCR5 |
| IN_10 | 13 mo/F | MTCT | 2014 | DNA PCR | 1,485,978 | NA | 15 | 06 | 2 | C | CCR5 | CCR5 |
F, female; M, male; HET, heterosexual route; MTCT, mother-to-child-transmission; NA, not available; G2P, Geno2Pheno algorithm.
Based on the ML phylogenetic tree and three online automated tools, RIP, jpHMM, and REGA.
Predicted coreceptor based on the V3 amino acid population sequence.
A maximum likelihood (ML) phylogenetic tree based on the amino acid sequence of the V1 to V3 region of Env was constructed using MEGA software (v7) (see Fig. S1 in the supplemental material). As expected, the V1 to V3 region of Env from an individual study subject formed a monophyletic cluster. On the basis of the amino acid sequence diversity, the number of potential N-linked glycosylation sites (PNGs) in the V1V2 region, and per-infectious-particle infectivity, we identified a total of 10 TF viruses from the eight infant samples (Table 1). No TF viruses were identified from the adult samples. The length of the V1 to V3 region and the PNG positions were defined on the basis of the strain HXB2 reference sequence. A vast variation in the length of the V1 region was identified. While the V1 region comprised 30 to 46 amino acids (average, 42 amino acids) and three to five PNGs in the amplicons derived from the adult samples, the recently transmitted virus isolates had short (25-amino-acid long) and very short (12-amino-acid long) V1 regions with a reduced number of PNGs (1 to 3) (Table 2). The length of variable regions V1 to V3, the number of PNGs, and the sequences in the different clones are provided in Table S1.
TABLE 2.
Characteristics of V1 to V3 region of infectious molecular clonesa
| Sample | Length of variable region (no. of amino acids) |
No. of PNGs |
||||
|---|---|---|---|---|---|---|
| V1 | V2 | V3 | V1 | V2 | V3 | |
| IN_02 | 41 (30–45) | 41 | 35 | 5 (3–5) | 2 | 1 |
| IN_03 | 21 | 44 | 35 | 2 (1–2) | 2 | 1 |
| IN_04 | 18 | 39 | 45 | 2 (1–2) | 2 | 1 |
| IN_05 | 13 | 41 | 35 | 2 (1–2) | 2 | 1 |
| IN_06 | 24 | 41 | 35 | 3 (1–3) | 2 | 1 |
| IN_07 | 25 (23–25) | 46 | 35 (34–35) | 3 (1–3) | 2 | 1 |
| IN_08 | 12 | 45 | 35 | 2 (1–2) | 3 (1–3) | 1 |
| IN_09 | 28 | 47 | 34 (34–35) | 2 (1–2) | 2 | 1 |
| IN_10 | 24 | 40 | 35 | 3 | 2 (1–2) | 1 |
Values given are for the average length of the variable region or number of PNGs; numbers in parentheses indicate ranges.
Identification of transmitted/founder virus variants and relative infectivity analysis.
Since blood samples were collected from infants during the acute phase of infection, we hypothesize that the viral variants obtained from the plasma RNA would very closely resemble that of the virus variants that were responsible for establishing infection in these individuals. The amino acid diversity in the variable region (V1 to V3) of the 65 infectious clones is provided in Fig. S2. It can be seen from the highlighter plot that the clones exhibited a substantial amount of sequence diversity and variation in the number of PNGs. Phylogenetic analysis using amino acid sequences identified a few high-diversity lineages among the intraindividual clones that differed by only a few amino acids. We measured the infectivity of each of the envelope clones to examine the impact of amino acid diversity in the V1 to V3 regions on infectivity of the virus. Interestingly, we found that clones having the highest diversity showed a high infectious potential (Fig. S3).
We performed a single-round-infection assay to determine the infectious potential of the recently transmitted viruses, TF viruses, and control viruses derived from chronically infected individuals (referred to here as chronic viruses) on a per-infectious-particle basis in TZM-bl cell lines. The number of RLU expressed was measured and used to calculate the infectious potential of a single viral particle. The results showed that the viral variants derived from four recently infected individuals had statistically significantly (P < 0.05) enhanced infectivity compared to the variants derived from chronically infected individuals (Fig. 1a). The median magnitude of per-infectious-particle infectivity of recently transmitted viral variants was found to be 1.2- to 3.7-fold higher than that of chronic viruses. In particular, the infectivity of viral variants from two individuals, IN_08 and IN_09, was found to be very high (average, 3.5-fold). Further, we checked the infectious potential of recently transmitted viruses, TF viruses, and chronic control viruses (Fig. 1b). Both TF viruses and recently transmitted viruses had statistically significantly higher infectious titers than chronic control viruses (P = 0.02 and P = 0.01, respectively). Interestingly, the TF viruses were found to have statistically significantly higher infectious titers than recently transmitted viruses (P = 0.02)
FIG 1.
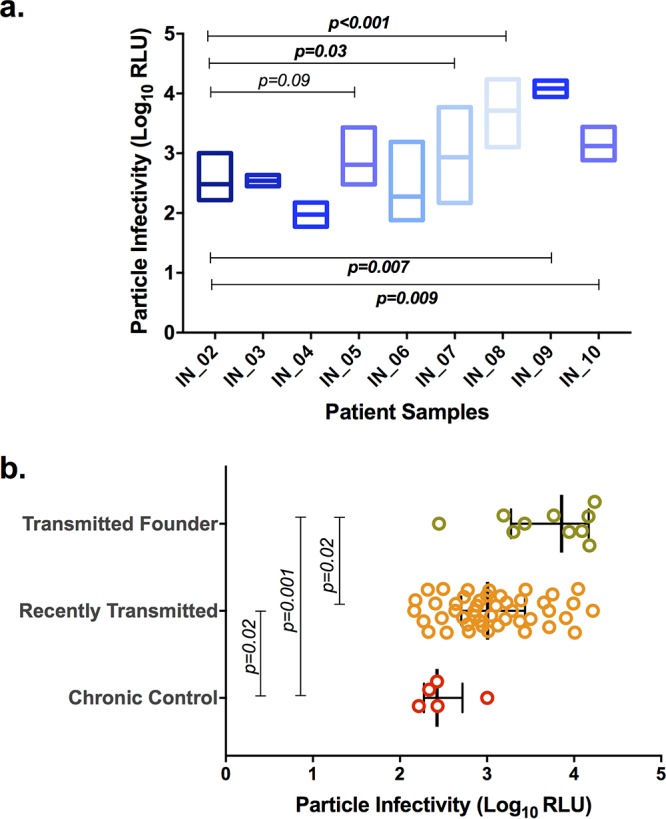
Relative infectious potential of chimeric viruses on a per-infectious-particle basis. Infectivity values for chronic and recently transmitted viruses are expressed in terms of the number of relative light units (RLU) as log10 values. Particle infectivity is defined by the infectious titer determined on TZM-bl cells. (a) Particle infectivity based on the number of RLU for each individual is presented. The box plot represents the infectivity of a single viral particle of multiple variants from each infected individual. Infections were performed in TZM-bl cells with virus at an MOI of 0.005 in the presence of DEAE-dextran. Pairwise comparisons were performed using the Mann-Whitney test, and a P value of <0.05 was considered statistically significant. (b) Particle infectivity of the chronic control, recently transmitted, and TF viruses. Pairwise comparisons were performed using the Mann-Whitney test, and a P value of <0.05 was considered statistically significant.
Coreceptor usage and tropism.
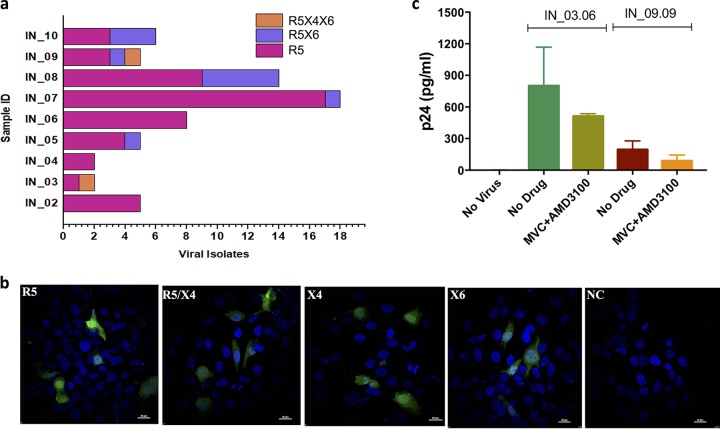
To determine the coreceptor usage profile of the viral clones generated, we used luciferase expression as well as green fluorescent protein (GFP) expression in GHOST(3) cells expressing CCR5 (R5), CXCR4 (X4), both CCR5 and CXCR4 (R5X4), and CXCR6 (X6) chemokine receptors (Fig. 2). Among the 65 infectious viruses tested, 80% (52/65) were found to use the CCR5 coreceptor for entry exclusively (R5-tropic). Interestingly, 3% of the clones (2/65) were dual tropic (R5X4) and also used the CXCR6 coreceptor. Twenty percent of the clones (13/65) used both the CCR5 and CXCR6 coreceptors (R5X6) (Fig. 2a). Among the eight recently infected individuals, six had at least one clone that exhibited CXCR6 tropism. Among the ten TF viruses, four were found to use both CCR5 and CXCR6. Interestingly, viruses that used CXCR4 or CXCR6 exclusively were not identified. The representative infection with virus that used CCR5, CXCR4, and CXCR6 (R5X4X6) in Bonzo-positive cells (R5, R5X4, X4, and X6) is presented as Fig. 2b. We next sought to determine whether the R5X4X6-tropic viruses can infect the CD8-depleted peripheral blood mononuclear cells (PBMCsCD8−) after blocking both the CCR5 and CXCR4 coreceptors with maraviroc (MVC) and AMD3100, respectively, by quantifying the p24 within the cells using the cell lysate. After the CCR5 and CXCR4 coreceptors were blocked, PBMCsCD8− showed evidence of infection with the two R5X4X6-tropic viruses (Fig. 2c). However, the data should be interpreted with caution. The ability of the viruses to infect the cells when the CCR5 and CXCR4 coreceptors were blocked could be due not only to the CXCR6 coreceptor, as coreceptors other than CXCR6 can be used in the absence of CCR5 and CXCR4.
FIG 2.
Coreceptor tropism and viral entry. The expression of luciferase upon infection with GHOST(3) cells (CXCR4+, CCR5+, CXCR4-CCR5+, and CXCR6+) by the recombinant viruses was measured and used for the identification of coreceptor usage. (a) The bar diagram represents the number of viral isolates that were identified as R5-, R5X4-, and R5X4X6-tropic based on the coreceptors used for viral entry. (b) Confocal microscopy images of green fluorescent protein expression by IN_09.09 viral isolates that use the R5, X4, and X6 coreceptors for viral entry. NC, no-drug control. (c) Quantification of p24 for detection of cellular entry by the two R5X4X6 chimeric viruses in CD8-depleted PBMCs. The experiments were performed in duplicate. The mean (SD) is presented.
Sensitivity to CCR5 antagonist.
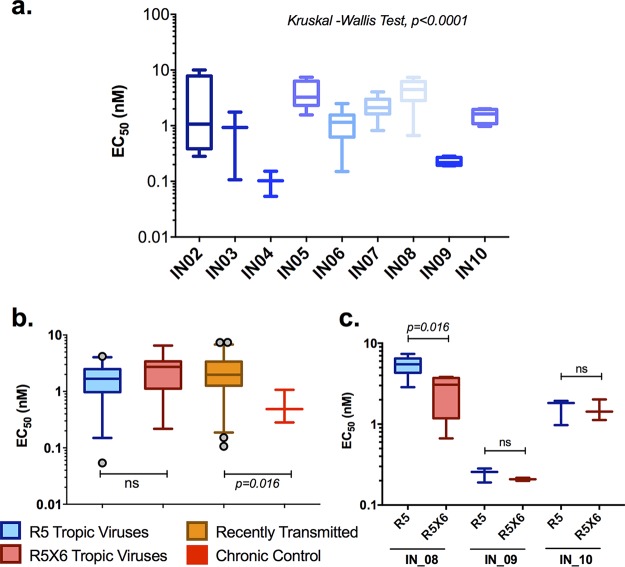
Next, we determined the sensitivity of the recombinant viruses to maraviroc (MVC), a CCR5 antagonist, in a single-round-infection assay at various concentrations starting from 1 × 104 nM and going to 1 × 10−3 nM in 10-fold serial dilutions. The median 50% effective concentrations (EC50) for the individual patients are presented in Fig. 3a. The EC50 were found to be greater for the recently transmitted viral variants than the chronic viral variants; the median EC50 for the chronic and recently transmitted variants was found to be 0.49 nM and 1.97 nM, respectively (P = 0.016) (Fig. 3b). The median half-maximal response to MVC for R5-tropic and R5X6-tropic viruses varied marginally (1.65 nM and 2.72 nM, respectively). It was interesting to note that when the R5- and R5X6-tropic viruses obtained from any given individual (IN_08, IN_09, and IN_10) were compared, the R5-tropic viruses had significantly higher (1.8-fold) median EC50 than the R5X6-tropic viruses in patient IN_08, while in the other two patients, the difference was 1.3-fold higher without any statistical significance (Fig. 3c).
FIG 3.
Half-maximal effective concentration (EC50) of a CCR5 antagonist for suppression of infection by chimeric viral variants. TZM-bl cells were infected with the chimeric virus at an MOI of 0.005 and cultured in the presence of the CCR5 antagonist maraviroc at concentrations ranging from 1 × 104 nM to 1 × 10−3 nM with 10-fold serial dilutions. The box plots represent the EC50 of maraviroc for each viral variant from the individuals (a), the susceptibilities of viral isolates derived from later and early stages of infection to maraviroc (b), and the susceptibility difference between R5- and R5X6-capable early-transmitted viruses (c). Inhibition data from replicates were plotted using GraphPad Prism software, and EC50 were determined using variable-slope nonlinear regression analysis. Experiments were carried out in triplicate in two biological replicates. The statistical comparisons were performed using the Kruskal-Wallis test (among the individual patients) and the Mann-Whitney test (between groups), and a P value of <0.05 was considered statistically significant. ns, not significant.
Sensitivity to neutralizing antibodies.
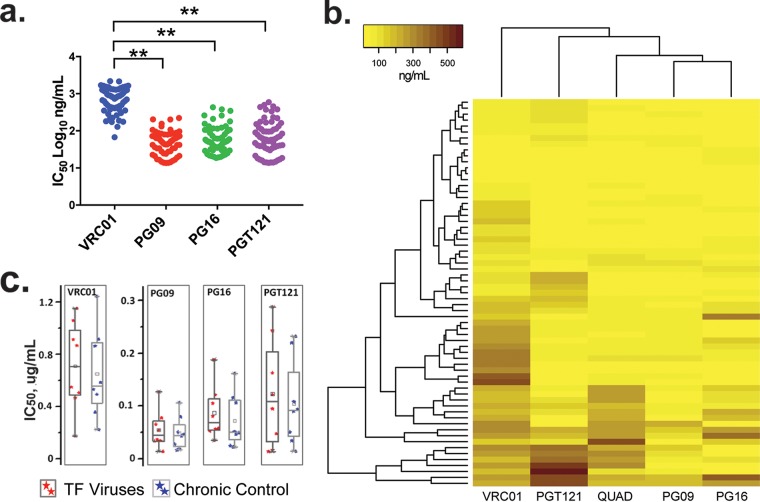
We used a panel of well-characterized broadly neutralizing antibodies (bNAbs), VRC01, PG09, PG16, and PGT121, to determine the neutralization sensitivity of the viral clones. Based on the percent neutralization, chimeric viruses were categorized into three groups (<30%, 30 to 60%, and >60% neutralization), and the neutralization sensitivity results are represented in Fig. S4. PG16 showed the broadest activity, neutralizing >60% of the viral variants at concentrations below 1 μg/ml. This was followed by the Quad monoclonal antibody (MAb), PGT121, and PG09, which neutralized 57%, 34%, and 32% of the viruses, respectively. VRC01, which targets the CD4 binding site by binding to the V1 loop of Env, neutralized merely 9% of the viruses, even when tested at a concentration of 5 μg/ml. We also determined the potency of the neutralization response by calculating 50% inhibitory concentrations (IC50) using a dose-response curve fit with a nonlinear function using GraphPad Prism (v5.0) software (Fig. 4a). We observed that the potency of neutralization of the viral variants was significantly lower for VRC01 than for PG09, PG16, and PGT121 (P < 0.001 in all cases). The results of the cluster analysis of neutralization sensitivity are shown in Fig. 4b. In general, the potency of neutralization of all the four MAbs for TF viruses was found to be lower than for chronic viruses (Fig. 4c). The neutralization potency of VRC01, PG16, and PGT121 for the TF viruses was 708 ng/ml, 68 ng/ml, and 106 ng/ml., respectively, while it was 582 ng/ml, 49 ng/ml, and 95 ng/ml, respectively, for the chronic viruses. On the other hand, the potency of neutralization of the PG09 MAb was similar for both TF and chronic viruses (mean IC50, 46 ng/ml and 48 ng/ml, respectively). It was interesting to observe that there was a difference in the neutralization sensitivity of TF and chronic viruses obtained from the same individual. The neutralization sensitivity of the viruses for the entire panel of monoclonal antibodies is shown in Fig. S5.
FIG 4.
Neutralizing potential of a panel of 4 monoclonal antibodies against the chimeric viruses. The figure represents the potency of neutralization of viral isolates by the VRC01, PG09, PG16, and PGT121 monoclonal antibodies. Twofold dilutions of all MAbs, except VRC01, starting from 1 μg/ml and going up to 8 serial dilutions, were tested for their potency against the 65 infectious viral variants; VRC01 was tested at a higher concentration of 5 μg/ml. Data from replicates were plotted using GraphPad Prism software, and IC50 were determined using variable-slope nonlinear regression analysis. Experiments were carried out in duplicate. (a) The IC50 of each dot represents the amount of MAbs required to neutralize half of the virus in each patient. (b) The difference in color intensity in the heatmap indicates the potency of neutralization. The darker and lighter colors indicate a stronger and a weaker neutralization potency of the virus, respectively. (c) The box plot represents the neutralization potency of MAbs against the TF and chronic viruses. The statistical comparisons were performed using the Mann-Whitney test (between groups), and a P value of <0.05 was considered statistically significant.
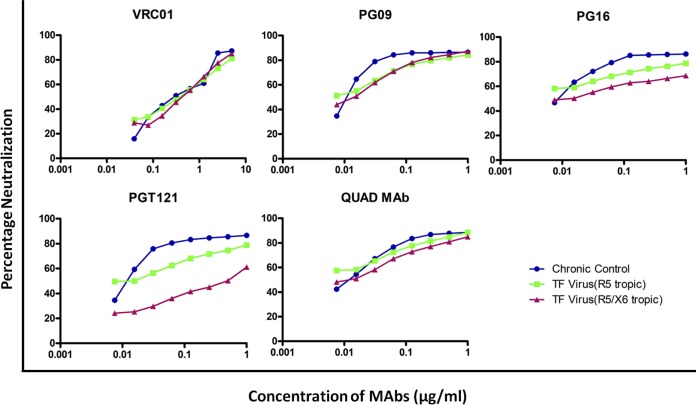
Correlation between tropism and neutralization response.
Though there was no significant difference in the neutralization profile of the chronic viruses and the R5- and R5X6-tropic TF viral clones with the VRC01, PG09, and Quad MAbs, we did find a unique pattern (Fig. 5). At all concentrations tested, the percent inhibition of R5-tropic chronic viruses, R5-tropic TF viruses, and R5X6-tropic TF viruses with VRC01 was more or less similar. On the other hand, there was a considerable difference in the neutralization sensitivity of the three groups of viruses with PG16 and PGT121 MAbs. The percent neutralization of the viruses at 1 ng/μl (the maximum concentration tested) was found to be 86% and 78% for the two groups of R5-tropic viruses but was much less (68%) for the R5X6-tropic viruses with PG16 MAb. Similarly, with the PGT121 MAb, the percent neutralization of the two groups of R5-tropic viruses was 86% and 78%, while that of R5X6-tropic viruses was 61%.
FIG 5.
Representation of neutralization profile in association with coreceptor tropism. The graph depicts the percent inhibition of chronic control R5-capable and recently transmitted (R5 and R5X6) viruses against neutralizing monoclonal antibodies.
DISCUSSION
In the present study, we developed replication-competent chimera viruses comprising the gp120 gene amplified from ten HIV-1C-infected individuals, two adults with chronic infection who acquired HIV-1 through sexual contact and eight infants who recently acquired the infection from the mother during pregnancy. The data obtained from the 65 infectious clones demonstrate that the viral variants from early transmission have a higher infectivity and reduced neutralization sensitivity. Interestingly, we also found that several recently transmitted viruses were capable of CXCR6-dependent cell entry, in addition to CCR5-dependent cell entry, indicating that coreceptor usage during the early event of transmission may differ.
There are no clear data on the number of viruses required for establishing an efficient clinical infection. It is well-known that all the viruses derived from an HIV-infected individual are not necessarily infectious or capable of establishing a productive infection (14) and that just a few viruses or even a single virus particle with enormous transmission fitness is sufficient for transmitting infection. On this account, it may be surprising that the infection caused by the single viral particle proceeds to a low multiplicity and limited evolution of the viral population in the early stages of infection (15). In an attempt to identify unique characteristics that distinguish the TF viruses from the chronic viruses, we examined the length of the variable regions of HIV-1 gp120, the number of PNGs present in the variable regions (V1 to V3), and the infectivity fitness of the chimeric viruses. We found a difference in the length of the V1V2 region, with TF viruses having a shorter V1V2 region than the viral variants obtained from the later stages of disease. A previous study also reported that the numbers of amino acids and PNGs in the gp120 variable regions are unique determinants that contribute to the successful establishment of early infection among recently transmitted viral isolates (16). In the present study, we noticed that envelopes from the same individual showed a significant variation in the number of amino acids and PNGs in the V1V2 region. It was interesting to note that envelope clones with a shorter V1V2 region and a reduced number of PNGs exhibited more infectious fitness than those with a longer V1V2 region and a higher number of PNGs. It has previously been reported that changes in the length of the variable regions might alter antibody recognition of both the V3 loop and the CD4 binding site under host selection pressure (16, 17). In contrast to the findings of an earlier study where TF viruses did not demonstrate increased viral fitness regarding particle infectivity in heterosexual transmission (18), our study showed an increased particle infectivity of the TF viruses as well as recently transmitted viruses (19, 20). It is thought that the TF viruses carry an excess of functional envelope spikes and that such an excess seems to provide an advantage to the virus during the very early stages of infection (20). The presence of viral variants with a higher infectious titer at or near the moment of infection ensures stable binding to the CD4 receptor and establishment of infection during the transmission event (21). It is to be noted that TF viruses can also establish de novo infection and resist the host antiviral activity of IFITMs (interferon-induced transmembrane protein 1) (22).
Earlier studies have indicated that TF viruses from heterosexual transmission also possibly have enhanced virus entry (23), which could be due to the broader coreceptor usage, as relative fitness differs significantly among TF viruses (24). We also analyzed the coreceptor usage pattern of the viral variants using a phenotypic assay with reporter cell lines. Most of the infectious clones (52/65; 78%) generated in this study were found to exclusively use the CCR5 coreceptor, which is in line with the findings of previous studies (15, 20). Only 20% of the clones used both the CCR5 and CXCR6 coreceptors; two clones could use all three coreceptors, CCR5, CXCR4, and CXCR6. Further virus entry was observed in the CD8-depleted PBMCs. It has been reported earlier that TF viruses choose CCR5-dependent cell entry for establishing productive and persistent infection, as their genetic units are more conserved toward the R5 coreceptor (25). Although CXCR6 is considered a major coreceptor for HIV-2 and simian immunodeficiency virus but a minor coreceptor for HIV-1 (26), we found that some of our HIV-1 clones could use CXCR6 as a coreceptor (27). To the best of our knowledge, no study has reported on CXCR6 usage in TF viruses, and this is the first report on the presence of viral variants capable of using the CXCR6 coreceptor in recently infected infants. More importantly, four of the ten TF viruses identified in this study were capable of using both CCR5 and CXCR6 coreceptors. Various subsets of leukocytes express the chemokine receptor CXCR6 (28). RNA sequencing data show higher numbers of transcripts of CXCR6 per million (TPM) transcripts both in lymphoid tissue and in the placenta (29). Genome-wide association studies have identified polymorphisms within the CXCR6 gene (rs2234358) that correlate with HIV-1 control in long-term nonprogressors (30). A recent transcriptomics study reported that CXCR6 gene expression was significantly downregulated in elite controllers compared to viremic progressors (31). This could potentially point to a mechanism of increased HIV-1 replication control in HIV-1-positive elite controllers caused by a decreased susceptibility of T lymphocytes for HIV-1 entry. Our data thus indicate that coreceptor usage during the early event of transmission could be different and that the role of CXCR6 as a coreceptor in the initial stage of infection, transmission, and the establishment of the disease needs to be looked at in greater detail.
The effective inhibitory concentration of maraviroc, a CCR5 antagonist, was found to vary for viruses isolated from the same individual as well as viruses from different individuals. In general, the amount of drug required to inhibit half of the virus among the chronic virus population was found to be much less than that required to inhibit the TF virus. A unique property of HIV-1C strains is that a proportion of HIV-1C strains has broad coreceptor usage (32–34). An earlier study and our unpublished data showed that maraviroc could inhibit dual-tropic viruses (35). However, given the broader coreceptor usage, this could be expected as the viruses from recently infected individuals can also use the other coreceptor, besides CCR5, for entry. It was observed that within a given individual, the amount of drug needed to reduce half of the R5X6-tropic viruses was higher than that required to reduce half of the solely CCR5-dependent viruses. We therefore posit that, given the current guideline of test and treat, where the treatment is initiated at a very early phase of infection, maraviroc may not be a good option with which to start therapy. It has already been reported that viral escape from CCR5 antagonists can evolve by adopting CXCR4 or CXCR6 for entry (36). Our recent study also indicated that HIV-1C strains are less susceptible than non-subtype C strains to maraviroc (37).
We also analyzed the sensitivity of the viruses to neutralization by the well-characterized bNAbs, such as PG09, PG16, PGT121, and VRC01 (38). Overall, the recombinant viruses exhibited reduced sensitivity to neutralization by the CD4 binding site-directed MAb VRC01 compared to that by the N160 glycan binding site (V1V2 glycan) MAbs, PG09 and PG16, and the N332 glycan binding site (V3 glycan) MAb PGT121. Our findings are in agreement with previous reports that HIV-1 strains are evolving to become resistant to CD4 binding site-directed neutralizing activity (38) and suggest different neutralization mechanisms (39, 40). The neutralization sensitivity of chronic viruses to all the four MAbs was found to be higher than that of the TF viral variants. This increased resistance to neutralization by the TF virus could be due to the shorter length of the V1V2 region of their Env proteins and the smaller number of PNGs. Evidence from the literature also suggests that the epitopes in this region are conformationally masked (15). These factors might be responsible for the difference in neutralization sensitivity between the TF and chronic viruses. However, there is one contradictory report which says that a longer V1V2 region is associated with resistance to neutralization (41). Of note, site-specific glycosylation can also influence neutralizing antibody elicitation in a subtype-specific manner (42, 43). Although the HIV isolates had increased resistance to bNAbs at the population level, the newly identified bNAb PG16 displayed an extraordinary breadth and potency of neutralization with the chimeric viruses. In contrast, another glycan-dependent bNAb, PG09, exhibited a very limited breadth and potency of neutralization with the same viruses. This discrepancy in neutralization might be advantageous for TF virus in establishing successful infection early after transmission. Further, we observed that the potency of neutralization varied with respect to the number and type of coreceptors used by the viral variants. The level of inhibition by the neutralizing antibodies PG16 and PGT121 of the viral variants that could use the CXCR6 coreceptor for establishing infection was found to be minimal compared to that of the variants that could use CCR5 alone.
Our study has a few limitations that merit mention. First, we included viral clones from only one chronically infected control subject in our analyses. Second, we cloned only the gp120 portion of the HIV-1 Env and not the entire Env gp160, and the backbone used was of HIV-1 subtype B. Although the use of full-length infectious molecular clones (IMC) containing the complete HIV-1 genome has several advantages (20), the methodology for generating full-length infectious clones is difficult and makes it extremely hard to produce a large number of clones for such analysis. Third, after blocking the CCR5 and CXCR4 coreceptors, we found that R5X4X6 viruses could infect PBMCs. However, our study could not confirm that infection in these cases was exclusively through the CXCR6 coreceptor, as earlier studies have shown that HIV-1C strains can use a broad range of coreceptors to infect primary cells (32–34). Finally, as the vertical transmission from mother to child mostly involves transfer of a homogeneous population of viruses, the characteristics of the viruses transmitted through the sexual route or drug use may differ significantly. This aspect is under investigation in our laboratory at Karolinska Institute with primary HIV infections.
However, our study has revealed several interesting characteristics of viral variants found at the time of transmission, such as higher infectivity, reduced neutralization sensitivity, and broader coreceptor tropism compared to those for viruses present in the chronic stage of the disease. Further, discrepancies in the breadth and neutralization potency of different bNAbs for the viruses might be advantageous to TF virus for establishing successful infection early after transmission. The use of other coreceptors, like CXCR6, along with CCR5, at the early stage of infection points to a different cellular entry mechanism employed by TF viruses.
MATERIALS AND METHODS
Cell lines.
TZM-bl, GHOST(3) CXCR4+, GHOST(3) CCR5+ (Hi-5), and GHOST(3) CXCR6 cells were obtained from the NIH AIDS Reagent Program, NIH, USA. 293T cells were purchased from ATCC, USA. TZM-bl and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS) and 2 mM l-glutamine. The GHOST cell lines were maintained in high-glucose DMEM supplemented with 10% FBS.
Clinical specimens.
The study population comprised eight infants who acquired HIV infection from the mother during pregnancy and two adults (chronic controls [CC]) who got the infection through heterosexual transmission. Their HIV infection status was confirmed using an HIV-1 DNA PCR and enzyme-linked immunosorbent assay (ELISA), respectively. The detailed demographic profiles of the subjects, including their CD4 cell counts and viral loads, are shown in Table 1. Ethical clearance for the study was obtained from the Institutional Ethics Committee of the National Institute for Research in Tuberculosis (formerly known as Tuberculosis Research Centre ICMR; TRC-IEC 2009009).
Construction of gp120 molecular clones.
HIV RNA was extracted, using a QIAamp viral RNA minikit (Qiagen, USA), from 140 μl of the patients' plasma. cDNA was synthesized using the SuperScript III reverse transcriptase enzyme (Invitrogen, Life Technologies, MA, USA) and an oligo(dT)18 primer (Thermo Scientific). The first round of PCR was performed using high-fidelity Kapa HiFi HotStart Ready mix (2×; Kapa Biosystems, MA, USA) with primers 5550F (5′-AGARGAYAGATGGAACAAGCCCCAG-3′; HXB2 coordinates 5550 → 5574) and 9555R (5′-TCTACCTAGAGAGACCCAGTACA-3′; HXB2 coordinates 9555 → 9533), followed by a second round with primers 6433F (5′-CYACCAACGCGTGTGTACCCACAGA-3′; HXB2 coordinates 6433 → 6457) and 8329R (5′-CCCTGCCGGCCTCTATTYAYTATAGAAA-3′; HXB2 coordinates 8356 → 8329). The primers contained the NgoMIV and MluI restriction sites, respectively, for cloning. The resultant fragments were gel purified using the QIAquick gel extraction kit (Qiagen, USA) and cloned into the pMN-K7-Luc-IRESs-NefΔgp120 plasmid (44) (kindly gifted by Thomas Klimkait, University of Basel, Basel, Switzerland).
Generation of virus stocks and determination of infectivity.
Virus stocks were produced from the infectious molecular clones (IMC) by transfection of 293T cells as described previously (45). In brief, 1 × 105 293T cells were seeded in each well of 12-well plates on the day before transfection. Cells were transfected with 2.5 μg of DNA from the pMN-gp120 molecular clone using the FuGENE HD (Promega, USA) reagent following the manufacturer's protocol. The supernatant was harvested after 48 h and clarified by centrifugation, aliquoted, and stored at −80°C until use.
The 50% tissue culture infective dose (TCID50) of the generated virus stocks was determined as previously described (45). In brief, 1 × 105 TZM-bl cells per well in 96-well plates were infected with a serial dilution of the virus stock in sextuplicate, starting with a 1/10 dilution in the presence of 10 μg/ml DEAE-dextran (Sigma). The luciferase expressed by the TZM-bl cells was measured, using a Bright-Glo luciferase assay system (Promega, USA), at 48 h postinfection on an Infinite M200Pro plate reader (Tecan, Switzerland) and expressed as relative light units (RLU).
Sequence alignment and analysis.
Genotyping was performed using the method described previously (10). In brief, the gp120 sequence data were aligned using SeqScape software (v2.5) with the prescribed default parameters. The V1, V2, and V3 regions of the envelope were defined on the basis of the corresponding sequences in the HXB2 envelope as nucleotides 391 to 471, 472 to 585, and 886 to 993, respectively. The phylogenetic tree was constructed from amino acid sequence alignments on the basis of the maximum likelihood method with the Poisson correction model using MEGA (v7) software. HIV-1 subtyping was performed using the online subtyping and recombinant detection tools RIP (www.hiv.lanl.gov/content/sequence/RIP/RIP.html, accessed in December 2017), jpHMM (http://jphmm.gobics.de/submission_hiv.html, accessed in December 2017), and REGA (v3.0; http://dbpartners.stanford.edu:8080/RegaSubtyping/stanford-hiv/typingtool/, accessed in December 2017). Coreceptor usage was predicted on the basis of the V3 loop sequence using the online tools geno2pheno (http://coreceptor.bioinf.mpi-inf.mpg.de/index.php, accessed in December 2017) and phenoseq (http://tools.burnet.edu.au/phenoseq/, accessed in December 2017) and also on the basis of the net charge of the V3 region. The amino acid sequence deviation and PNG positions were analyzed using the highlighter tool provided in the Los Alamos National Laboratory database (https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html/, accessed in December 2017). The consensus sequence of the amplicons from an individual was generated before performing highlighter analysis.
Determination of viral coreceptor tropism.
GHOST(3) CXCR4+, X4R5+, Hi5 (CCR5+), and Bonzo (CXCR6)-positive cells were seeded in 96-well plates (10,000 cells per well) 24 h before infection. The cells were inoculated with a fixed dose of harvested virus at a multiplicity of infection (MOI) of 1 and a 10-μg/ml final concentration of DEAE in DMEM. Coreceptor tropism was determined by measuring the luciferase expression as described above. In addition to luciferase expression, green fluorescent protein (GFP) expression was also captured using confocal microscopy (Olympus FluoView, v2.0b). For this purpose, GHOST(3) cells were seeded onto glass coverslips placed in each well of a 24-well tissue culture plate, as described above. The monolayers were then inoculated with chimeric virus at a multiplicity of infection (MOI) of 1. At 48 h postinfection, the cells in the coverslips were fixed with 4% paraformaldehyde (PFA) for 1 h and examined using a Nikon Optiphot microscope attached to a Bio-Rad MRC 500 confocal system (LaserSharp). Images were acquired with a 60× oil-immersion objective and processed in an Olympus FluoView (v2.0b) microscope.
Determination of infection through the CXCR6 coreceptor in PBMCsCD8− after blocking CCR5 and CXCR4 coreceptors.
Peripheral blood mononuclear cells (PBMCs) were isolated from healthy individuals. To avoid CD8-mediated killing, CD8+ T cells were depleted from the healthy PBMCs using a RosetteSep human CD8 depletion cocktail (Stem Cell Technologies UK Ltd., UK), and the purity was checked by flow cytometry. The infection of PBMCsCD8− was carried out using a modified protocol as described previously (46). PBMCsCD8− were activated with phytohemagglutinin (20 μg/ml) in the presence of interleukin-2 (20 U/ml) in complete RPMI 1640 medium supplemented with 10% FBS. At 3 days poststimulation, cells were incubated with a CCR5 antagonist (maraviroc, 15 μM) and a CXCR4 antagonist (AMD3100; 15 μM; Sigma) in combination (1:1) for 2 h, followed by spinoculation (1,200 × g for 90 min) with R5X4X6 chimeric viruses in the presence of DEAE-dextran (10 μg/ml). The plates were incubated at 37°C with 5% CO2 for 24 h. On the following day, the cells were washed thrice with 1× phosphate-buffered saline, replenished with fresh complete RPMI 1640, and incubated further for 3 days. After 72 h, cells were lysed with radioimmunoprecipitation assay buffer without Triton X-100 and p24 levels were quantified using an electrochemiluminescence immunoassay (ECLIA) with a Cobas 8000 analyzer (Roche Diagnostics, USA).
Determination of infectious potential of chimeric viruses by per infectious viral particle.
TZM-bl cells seeded in 96-well plates were infected with 50 particles of each virus. Luciferase expression was measured after 48 h of culture and expressed as a measure of the infectious potential per 50 viral particles. The infectivity of the single viral particle was determined by dividing the number of RLU by 50 and calculating the logarithmic values.
Determination of neutralization sensitivity.
A panel of four bNAbs targeting different epitopes on HIV-1 gp120 were tested for their ability to neutralize the viral variants and prevent infection of target cells. Antibodies were tested in 2-fold serial dilutions (up to eight dilutions) in duplicate, starting at the highest concentration indicated, unless otherwise stated: CD4bs MAb VRC01 (5 μg/ml), anti-V1V2 apex MAbs PG9 and PG16 (1 μg/ml each), and anti-V3 MAb PGT121 (1 μg/ml). Pooled MAbs consisting of the Quad MAb (VRC01, PG9, PG16, and PGT121 in a 1:1:1:1 [wt/vol] ratio) were also tested for neutralization efficiency. For the neutralization assay, the viral supernatant was normalized to TCID50 values, and 200 TCID50 of the virus was used to infect 1.25 × 105 TZM-bl cells seeded in flat-bottom 96-well plates. The viral supernatant was preincubated with an equal volume of medium containing the MAb at 37°C for 2 h before infection of the TZM-bl cells. At 40 to 48 h postinfection, cells were lysed with 1× passive lysis buffer, and luciferase expression was measured as the number of relative light units using a Bright-Glo luciferase assay system. The IC50 of each MAb was calculated using GraphPad Prism software. In cases where the 50% inhibitory concentration was higher or lower than the concentrations tested, the amount was reported as the maximum or minimum concentration/dilution, respectively.
Determination of sensitivity to maraviroc.
TZM-bl cells were plated in a 96-well format (1 × 104 cells/well) for 24 h before infection. Cells were preincubated with different dilutions of maraviroc (MVC; 10-fold dilutions starting from 1 × 104 nM and going to 1 × 10−3 nM) for 2 h at 37°C before inoculation with chimeric env luciferase reporter virus (50 viral particles/well, based on the TCID50 value). All assays were performed in triplicate. The level of inhibition by MVC with respect to the EC50 was measured as described above.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by Swedish Research Council Establishment grant 2017-01330 to U.N. and by Stockholm County Council grant ALF 20160074. M.A. acknowledges the HIV Research Trust, United Kingdom, for providing a fellowship to carry out this study at the Karolinska Institute, Stockholm, Sweden. Confocal imaging for this study was performed at the Live Cell Imaging facility, Karolinska Institute, Stockholm, Sweden, which is supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Research Council, the Centre for Innovative Medicine, and the Jonasson Center at the Royal Institute of Technology, Sweden.
We thank Thomas Klimkait, University of Basel, Basel, Switzerland, for providing the pMN-K7-Luc-IRESs-NefΔgp120 plasmid. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc. and GHOST(3) cells (CXCR4+, CCR5+ [Hi-5], and CXCR6+) from Vineet N. Kewal Ramani and Dan R. Littman. We also thank Wang Zhang, Karolinska Institute, for his assistance with confocal imaging.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00063-18.
REFERENCES
- 1.Keele BF. 2010. Identifying and characterizing recently transmitted viruses. Curr Opin HIV AIDS 5:327–334. doi: 10.1097/COH.0b013e32833a0b9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, Mulenga J, Allen SA, Pinter A, Shaw GM, Hunter E, Robinson JE, Gnanakaran S, Derdeyn CA. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog 5:e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferre AL, Hunt PW, Critchfield JW, Young DH, Morris MM, Garcia JC, Pollard RB, Yee HF Jr, Martin JN, Deeks SG, Shacklett BL. 2009. Mucosal immune responses to HIV-1 in elite controllers: a potential correlate of immune control. Blood 113:3978–3989. doi: 10.1182/blood-2008-10-182709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascola JR, Montefiori DC. 2010. The role of antibodies in HIV vaccines. Annu Rev Immunol 28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 6.Hemelaar J. 2012. The origin and diversity of the HIV-1 pandemic. Trends Mol Med 18:182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri RP, Neogi U, Rao SD, Shet A. 2014. Genetic factors associated with slow progression of HIV among perinatally-infected Indian children. Indian Pediatr 51:801–803. doi: 10.1007/s13312-014-0505-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neogi U, Palchaudhuri R, Bommana S, Shet A. 2013. Genetic architecture of HIV type 1 Nef and Tat from HLA-B57-typed long-term survivors in an Indian cohort of perinatally HIV-infected children. AIDS Res Hum Retroviruses 29:1613–1616. doi: 10.1089/aid.2013.0195. [DOI] [PubMed] [Google Scholar]
- 9.Neogi U, Sharma Y, Sood V, Wanchu A, Banerjea AC. 2010. Diversity of HIV type 1 long terminal repeat (LTR) sequences following mother-to-child transmission in North India. AIDS Res Hum Retroviruses 26:1299–1305. doi: 10.1089/aid.2010.0128. [DOI] [PubMed] [Google Scholar]
- 10.Ashokkumar M, Nesakumar M, Cheedarla N, Vidyavijayan KK, Babu H, Tripathy SP, Hanna LE. 2017. Molecular characteristics of the envelope of vertically transmitted HIV-1 strains from infants with HIV infection. AIDS Res Hum Retroviruses 33:796–806. doi: 10.1089/aid.2016.0260. [DOI] [PubMed] [Google Scholar]
- 11.Neogi U, Bontell I, Shet A, De Costa A, Gupta S, Diwan V, Laishram RS, Wanchu A, Ranga U, Banerjea AC, Sonnerborg A. 2012. Molecular epidemiology of HIV-1 subtypes in India: origin and evolutionary history of the predominant subtype C. PLoS One 7:e39819. doi: 10.1371/journal.pone.0039819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tilton JC, Doms RW. 2010. Entry inhibitors in the treatment of HIV-1 infection. Antiviral Res 85:91–100. doi: 10.1016/j.antiviral.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Roche M, Jakobsen MR, Sterjovski J, Ellett A, Posta F, Lee B, Jubb B, Westby M, Lewin SR, Ramsland PA, Churchill MJ, Gorry PR. 2011. HIV-1 escape from the CCR5 antagonist maraviroc associated with an altered and less-efficient mechanism of gp120-CCR5 engagement that attenuates macrophage tropism. J Virol 85:4330–4342. doi: 10.1128/JVI.00106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gnanadurai CW, Pandrea I, Parrish NF, Kraus MH, Learn GH, Salazar MG, Sauermann U, Topfer K, Gautam R, Munch J, Stahl-Hennig C, Apetrei C, Hahn BH, Kirchhoff F. 2010. Genetic identity and biological phenotype of a transmitted/founder virus representative of nonpathogenic simian immunodeficiency virus infection in African green monkeys. J Virol 84:12245–12254. doi: 10.1128/JVI.01603-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashokkumar M, Tripathy SP, Hanna LE. 2016. Variability in V1V2 and PNGs in pediatric HIV-1 viral variants transmitted through vertical route. AIDS Res Hum Retroviruses 32:942–943. doi: 10.1089/aid.2016.0093. [DOI] [PubMed] [Google Scholar]
- 17.Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 18.Deymier MJ, Ende Z, Fenton-May AE, Dilernia DA, Kilembe W, Allen SA, Borrow P, Hunter E. 2015. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-alpha resistance. PLoS Pathog 11:e1005154. doi: 10.1371/journal.ppat.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Rola M, West JT, Tully DC, Kubis P, He J, Kankasa C, Wood C. 2010. Functional properties of the HIV-1 subtype C envelope glycoprotein associated with mother-to-child transmission. Virology 400:164–174. doi: 10.1016/j.virol.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachrach E, Dreja H, Lin YL, Mettling C, Pinet V, Corbeau P, Piechaczyk M. 2005. Effects of virion surface gp120 density on infection by HIV-1 and viral production by infected cells. Virology 332:418–429. doi: 10.1016/j.virol.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 22.Foster TL, Wilson H, Iyer SS, Coss K, Doores K, Smith S, Kellam P, Finzi A, Borrow P, Hahn BH, Neil SJD. 2016. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe 20:429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, Hopper J, Hora B, Kumar A, Mahlokozera T, Yuan S, Coleman C, Vermeulen M, Ding H, Ochsenbauer C, Tilton JC, Permar SR, Kappes JC, Betts MR, Busch MP, Gao F, Montefiori D, Haynes BF, Shaw GM, Hahn BH, Doms RW. 2012. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathog 8:e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song H, Hora B, Giorgi EE, Kumar A, Cai F, Bhattacharya T, Perelson AS, Gao F. 2016. Transmission of multiple HIV-1 subtype C transmitted/founder viruses into the same recipients was not determined by modest phenotypic differences. Sci Rep 6:38130. doi: 10.1038/srep38130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naidoo VL, Mann JK, Noble C, Adland E, Carlson JM, Thomas J, Brumme CJ, Thobakgale-Tshabalala CF, Brumme ZL, Brockman MA, Goulder PJR, Ndung'u T. 2017. Mother-to-child HIV transmission bottleneck selects for consensus virus with lower Gag-protease-driven replication capacity. J Virol 91:e00518-17. doi: 10.1128/JVI.00518-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao F, Alkhatib G, Peden KW, Sharma G, Berger EA, Farber JM. 1997. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med 185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohlmann S, Krumbiegel M, Kirchhoff F. 1999. Coreceptor usage of BOB/GPR15 and Bonzo/STRL33 by primary isolates of human immunodeficiency virus type 1. J Gen Virol 80(Pt 5):1241–1251. [DOI] [PubMed] [Google Scholar]
- 28.Wilbanks A, Zondlo SC, Murphy K, Mak S, Soler D, Langdon P, Andrew DP, Wu L, Briskin M. 2001. Expression cloning of the STRL33/BONZO/TYMSTR ligand reveals elements of CC, CXC, and CX3C chemokines. J Immunol 166:5145–5154. doi: 10.4049/jimmunol.166.8.5145. [DOI] [PubMed] [Google Scholar]
- 29.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, Backstrom A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson A, Sjostedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Ponten F, von Feilitzen K, Lilley KS, Uhlen M, Lundberg E. 2017. A subcellular map of the human proteome. Science 356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 30.Limou S, Coulonges C, Herbeck JT, van Manen D, An P, Le Clerc S, Delaneau O, Diop G, Taing L, Montes M, van't Wout AB, Gottlieb GS, Therwath A, Rouzioux C, Delfraissy JF, Lelievre JD, Levy Y, Hercberg S, Dina C, Phair J, Donfield S, Goedert JJ, Buchbinder S, Estaquier J, Schachter F, Gut I, Froguel P, Mullins JI, Schuitemaker H, Winkler C, Zagury JF. 2010. Multiple-cohort genetic association study reveals CXCR6 as a new chemokine receptor involved in long-term nonprogression to AIDS. J Infect Dis 202:908–915. doi: 10.1086/655782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Ambikan AT, Sperk M, van Domselaar R, Nowak P, Noyan K, Russom A, Sonnerborg A, Neogi U. 2017. Transcriptomics and targeted proteomics analysis to gain insights into the immune-control mechanisms of HIV-1 infected elite controllers. EBioMedicine 27:40–50. doi: 10.1016/j.ebiom.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isaacman-Beck J, Hermann EA, Yi Y, Ratcliffe SJ, Mulenga J, Allen S, Hunter E, Derdeyn CA, Collman RG. 2009. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J Virol 83:8208–8220. doi: 10.1128/JVI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dash PK, Siddappa NB, Mangaiarkarasi A, Mahendarkar AV, Roshan P, Anand KK, Mahadevan A, Satishchandra P, Shankar SK, Prasad VR, Ranga U. 2008. Exceptional molecular and coreceptor-requirement properties of molecular clones isolated from an human immunodeficiency virus type-1 subtype C infection. Retrovirology 5:25. doi: 10.1186/1742-4690-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cashin K, Jakobsen MR, Sterjovski J, Roche M, Ellett A, Flynn JK, Borm K, Gouillou M, Churchill MJ, Gorry PR. 2013. Linkages between HIV-1 specificity for CCR5 or CXCR4 and in vitro usage of alternative coreceptors during progressive HIV-1 subtype C infection. Retrovirology 10:98. doi: 10.1186/1742-4690-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Symons J, van Lelyveld SF, Hoepelman AI, van Ham PM, de Jong D, Wensing AM, Nijhuis M. 2011. Maraviroc is able to inhibit dual-R5 viruses in a dual/mixed HIV-1-infected patient. J Antimicrob Chemother 66:890–895. doi: 10.1093/jac/dkq535. [DOI] [PubMed] [Google Scholar]
- 36.Tilton JC, Wilen CB, Didigu CA, Sinha R, Harrison JE, Agrawal-Gamse C, Henning EA, Bushman FD, Martin JN, Deeks SG, Doms RW. 2010. A maraviroc-resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists depends on both N-terminal and extracellular loop domains of drug-bound CCR5. J Virol 84:10863–10876. doi: 10.1128/JVI.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddik AB, Haas A, Rahman MS, Aralaguppe SG, Amogne W, Bader J, Klimkait T, Neogi U. 2018. Phenotypic co-receptor tropism and maraviroc sensitivity in HIV-1 subtype C from East Africa. Sci Rep 8:2363. doi: 10.1038/s41598-018-20814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Euler Z, Bunnik EM, Burger JA, Boeser-Nunnink BD, Grijsen ML, Prins JM, Schuitemaker H. 2011. Activity of broadly neutralizing antibodies, including PG9, PG16, and VRC01, against recently transmitted subtype B HIV-1 variants from early and late in the epidemic. J Virol 85:7236–7245. doi: 10.1128/JVI.00196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bunnik EM, Euler Z, Welkers MR, Boeser-Nunnink BD, Grijsen ML, Prins JM, Schuitemaker H. 2010. Adaptation of HIV-1 envelope gp120 to humoral immunity at a population level. Nat Med 16:995–997. doi: 10.1038/nm.2203. [DOI] [PubMed] [Google Scholar]
- 42.Smith SA, Burton SL, Kilembe W, Lakhi S, Karita E, Price M, Allen S, Hunter E, Derdeyn CA. 2016. Diversification in the HIV-1 envelope hyper-variable domains V2, V4, and V5 and higher probability of transmitted/founder envelope glycosylation favor the development of heterologous neutralization breadth. PLoS Pathog 12:e1005989. doi: 10.1371/journal.ppat.1005989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chikere K, Webb NE, Chou T, Borm K, Sterjovski J, Gorry PR, Lee B. 2014. Distinct HIV-1 entry phenotypes are associated with transmission, subtype specificity, and resistance to broadly neutralizing antibodies. Retrovirology 11:48. doi: 10.1186/1742-4690-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edwards S, Stucki H, Bader J, Vidal V, Kaiser R, Battegay M, Klimkait T. 2015. A diagnostic HIV-1 tropism system based on sequence relatedness. J Clin Microbiol 53:597–610. doi: 10.1128/JCM.02762-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozaki DA, Gao H, Todd CA, Greene KM, Montefiori DC, Sarzotti-Kelsoe M. 2012. International technology transfer of a GCLP-compliant HIV-1 neutralizing antibody assay for human clinical trials. PLoS One 7:e30963. doi: 10.1371/journal.pone.0030963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wetzel KS, Yi Y, Elliott ST, Romero D, Jacquelin B, Hahn BH, Muller-Trutwin M, Apetrei C, Pandrea I, Collman RG. 2017. CXCR6-mediated simian immunodeficiency virus SIVagmSab entry into Sabaeus African green monkey lymphocytes implicates widespread use of non-CCR5 pathways in natural host infections. J Virol 91:e01626-16. doi: 10.1128/JVI.01626-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.