ABSTRACT
The mechanisms of RNA interference (RNAi) as a defense response against viruses remain unclear in many plant-pathogenic fungi. In this study, we used reverse genetics and virus-derived small RNA profiling to investigate the contributions of RNAi components to the antiviral response against Fusarium graminearum viruses 1 to 3 (FgV1, -2, and -3). Real-time reverse transcription-quantitative PCR (qRT-PCR) indicated that infection of Fusarium graminearum by FgV1, -2, or -3 differentially induces the gene expression of RNAi components in F. graminearum. Transcripts of the DICER-2 and AGO-1 genes of F. graminearum (FgDICER-2 and FgAGO-1) accumulated at lower levels following FgV1 infection than following FgV2 or FgV3 infection. We constructed gene disruption and overexpression mutants for each of the Argonaute and dicer genes and for two RNA-dependent RNA polymerase (RdRP) genes and generated virus-infected strains of each mutant. Interestingly, mycelial growth was significantly faster for the FgV1-infected FgAGO-1 overexpression mutant than for the FgV1-infected wild type, while neither FgV2 nor FgV3 infection altered the colony morphology of the gene deletion and overexpression mutants. FgV1 RNA accumulation was significantly decreased in the FgAGO-1 overexpression mutant. Furthermore, the levels of induction of FgAGO-1, FgDICER-2, and some of the FgRdRP genes caused by FgV2 and FgV3 infection were similar to those caused by hairpin RNA-induced gene silencing. Using small RNA sequencing analysis, we documented different patterns of virus-derived small interfering RNA (vsiRNA) production in strains infected with FgV1, -2, and -3. Our results suggest that the Argonaute protein encoded by FgAGO-1 is required for RNAi in F. graminearum, that FgAGO-1 induction differs in response to FgV1, -2, and -3, and that FgAGO-1 might contribute to the accumulation of vsiRNAs in FgV1-infected F. graminearum.
IMPORTANCE To increase our understanding of how RNAi components in Fusarium graminearum react to mycovirus infections, we characterized the role(s) of RNAi components involved in the antiviral defense response against Fusarium graminearum viruses (FgVs). We observed differences in the levels of induction of RNA silencing-related genes, including FgDICER-2 and FgAGO-1, in response to infection by three different FgVs. FgAGO-1 can efficiently induce a robust RNAi response against FgV1 infection, but FgDICER genes might be relatively redundant to FgAGO-1 with respect to antiviral defense. However, the contribution of this gene in the response to the other FgV infections might be small. Compared to previous studies of Cryphonectria parasitica, which showed dicer-like protein 2 and Argonaute-like protein 2 to be important in antiviral RNA silencing, our results showed that F. graminearum developed a more complex and robust RNA silencing system against mycoviruses and that FgDICER-1 and FgDICER-2 and FgAGO-1 and FgAGO-2 had redundant roles in antiviral RNA silencing.
KEYWORDS: Fusarium graminearum virus, mycovirus, Argonaute, RNA silencing, antiviral response
INTRODUCTION
Most eukaryotic organisms have evolved diverse defense mechanisms against pathogens, including viruses. One of the conserved host defense mechanisms is RNA interference (RNAi). The RNAi pathway uses small, noncoding RNAs to regulate endogenous genes and to defend against virus infection, transposons, and transgene expression (1–3). The canonical RNAi pathway requires a set of host cellular proteins that includes dicer, Argonaute (AGO), and RNA-dependent RNA polymerase (RdRP) proteins (2). Dicer proteins recognize double-stranded RNA (dsRNA) molecules, including viral dsRNAs, and cleave them to generate small interfering RNAs (siRNAs), microRNAs (miRNAs), and other short dsRNAs (2, 3). One of the strands from these small RNA (sRNA) duplexes is bound to an Argonaute protein and incorporated into a functional RNA-induced silencing complex (RISC) that can bind cRNA sequences and specifically cleave the target RNA (4, 5). In certain organisms, including plants, nematodes, and fungi, RdRP is required to convert single-stranded RNA (ssRNA) into dsRNA and to amplify the sRNA signals (4, 6).
Following the first report of RNAi-mediated posttranscriptional gene silencing (referred to as “quelling”) in the model fungus Neurospora crassa, diverse molecular mechanisms and quelling/RNAi components have been identified in many fungi, including Schizosaccharomyces pombe, Mucor circinelloides, and Cryptococcus neoformans (4, 7–9). Previous studies demonstrated that N. crassa has two RNA silencing pathways, including a quelling pathway and a pathway involving meiotic silencing by unpaired DNA (MSUD) (10, 11). The quelling pathway is accompanied by one of the dicer-like proteins (DCL-2), an Argonaute-like protein (QDE-2), and an RdRP (QDE-1) in the vegetative stage of N. crassa (12–14). MSUD only affects the expression of unpaired genes during meiosis, and it requires DCL-1, an Argonaute-like protein (SMS-2), and an RdRP (SAD-1) (4). However, detailed information regarding RNAi components related to antiviral defense responses in N. crassa is lacking, because mycoviruses have not been reported in this fungus (15).
The RNA silencing mechanisms associated with antiviral defense responses have been studied in many fungi, including Cryponectria parasitica, Aspergillus nidulans, and Rosellinia necatrix (16–19). The RNA silencing response against Cryponectria parasitica hypovirus 1 (CHV1) strain EP713 in C. parasitica has been well analyzed and involves the induction of dcl-2 and agl-2 transcripts and the production of hairpin RNA (hpRNA) (16, 20, 21). In addition, CHV1 encodes p29, which interferes with the upregulation of dcl-2 and agl-2 and thereby acts as an RNA silencing suppressor to counter the host's defense response (20, 22). Researchers recently reported that RNA silencing genes were differently upregulated in response to infection by five unrelated dsRNA mycoviruses in R. necatrix (19). Among these viruses, infection by Rosellinia necatrix mycoreovirus 3 (RnMyRv3) or Rosellinia necatrix megabirnavirus 1 (RnMBV1) resulted in upregulation of the expression of R. necatrix genes RnDCL-2, RnAGL-2, RnRdRP-1, and RnRdRP-2, but this upregulation was not triggered by the expression of exogenous dsRNA (19).
Fusarium graminearum is a homothallic, ascomycetous, phytopathogenic fungus associated with root rot and scab disease (Fusarium head blight) of small grains (23, 24). Infection by F. graminearum is a problem in many parts of the world because, in addition to reducing yields, the fungus causes mycotoxin contamination that is harmful to humans and animals (24–26). F. graminearum contains genes encoding two dicers (F. graminearum DICER-1 [FgDICER-1] and FgDICER-2), two Argonautes (FgAGO-1 and FgAGO-2), and five RdRPs (FgRdRP-1 to -5) (27, 28). Previous studies confirmed that F. graminearum has meiotic silencing and hpRNA-mediated gene silencing mechanisms (28, 29). Furthermore, Chen et al. (28) reported that FgAGO-1 and FgDICER-2 proteins play a critical role in hpRNA-mediated gene silencing and that FgDICER-2 is involved in miRNA-like small RNA (milRNA) generation in F. graminearum. A recent study showed that FgAGO-2 and FgDICER-1 primarily mediate a sex-specific RNAi pathway (30). Although the properties of sRNAs in Fusarium graminearum hypovirus 1 (FgHV1) and FgHV2 in F. graminearum were recently analyzed by deep sequencing (31), the detailed biological functions of RNAi components and the specific interaction between RNAi core components and mycovirus in F. graminearum are still unknown. We previously established a model system composed of F. graminearum strain PH-1 and four phylogenetically different mycoviruses, including Fusarium graminearum virus 1 (FgV1), FgV2, FgV3, and FgV4 (32). Based on analysis of the phenotypes induced in the host fungus by the four mycoviruses and on RNA sequencing-based genomewide transcriptome data, we confirmed that infections by FgV1 to -4 differentially up- or downregulate numerous host genes, including some RNAi components, in F. graminearum (32).
In this study, we used a reverse genetics strategy and analysis of virus-derived-sRNA profiles to investigate the role(s) of host genes related to the antiviral RNA silencing response in F. graminearum. We generated gene disruption and overexpression (OE) mutants for individual genes encoding Argonautes, dicers, and RdRPs in F. graminearum and compared the effects of gene disruption and overexpression mutations on infection by three different FgVs. Our results showed that the functions of two dicer genes, FgDICER-1 and FgDICER-2, in the response to infection by FgV1, -2, and -3 might be partially redundant and that FgAGO-1 might have an important role(s) in the antiviral responses associated with the RNA silencing pathway.
RESULTS
Differential levels of expression of RNAi-related genes in F. graminearum following FgV infections.
We previously used comprehensive genomewide transcriptome analysis to investigate the effects of the four Fusarium mycoviruses on the expression of F. graminearum genes responsible for RNA silencing (32). The data suggested that the expression of RNA silencing-related genes might be virus specific in F. graminearum. To confirm this analysis in the current study, we examined the transcript levels of RNA silencing-related genes in F. graminearum in response to FgV infections. Real-time reverse transcription-quantitative PCR (qRT-PCR) measurement showed that the expression of most RNA silencing-related genes was induced by infection but that the specific quantitative aspects of this increase in expression depended on the specific gene and virus (FgV1, FgV2, and FgV3) (Table 1). All dicer and Argonaute genes were upregulated by FgV2 infection, but the expression of FgDICER-1 (RefSeq accession number FGSG_09025) was only induced by FgV1 and FgV2 infection. The altered expression patterns of FgV1-responsive genes differed from those of FgV2- or FgV3-responsive genes. The expression levels of FgDICER-2 (RefSeq accession number FGSG_04408) and FgAGO-1 (RefSeq accession number FGSG_08752) were significantly increased by FgV2 and FgV3 infection but reduced by FgV1 infection. Among RdRP genes, the FgRdRP-1 (RefSeq accession number FGSG_06504), FgRdRP-3 (RefSeq accession number FGSG_01582), and FgRdRP-5 (RefSeq accession number FGSG_09076) transcript levels were reduced in response to FgV1 infection. The FgRdRP-3 (RefSeq accession number FGSG_01582), FgRdRP-4 (RefSeq accession number FGSG_04619), and FgRdRP-5 (RefSeq accession number FGSG_09076) transcript levels were increased by FgV2 or FgV3 infection, whereas FgRdRP-1 (RefSeq accession number FGSG_06504) and FgRdRP-2 (RefSeq accession number FGSG_08716) were not upregulated in response to FgV2 or FgV3 infection. These results indicate that the RNA silencing response in F. graminearum differs depending on which of three unrelated mycoviruses causes the infection.
TABLE 1.
Levels of Fusarium graminearum RNA silencing-related gene transcript accumulation in response to infection by Fusarium graminearum viruses 1, -2, and -3
| Protein family | Gene | Mean mRNA level ± SD ina: |
|||
|---|---|---|---|---|---|
| Virus-free PH-1 | PH-1 infected with: |
||||
| FgV1 | FgV2 | FgV3 | |||
| Dicer | FgDICER-1 | 0.90 ± 0.17 | 5.08 ± 2.71 B | 2.89 ± 0.7 B | 0.51 ± 0.15 |
| FgDICER-2 | 1.07 ± 0.1 | 0.41 ± 0.06 C | 42.13 ± 9.9 A | 17.77 ± 4.61 B | |
| Argonaute | FgAGO-1 | 0.89 ± 0.28 | 0.16 ± 0.05 C | 2.66 ± 0.51 B | 2.23 ± 0.54 B |
| FgAGO-2 | 0.95 ± 0.16 | 1.64 ± 0.3 C | 3.29 ± 1.0 B | 0.86 ± 0.35 | |
| RdRp | FgRdRP-1 | 1.05 ± 0.08 | 0.42 ± 0.07 B | 0.31 ± 0.11 B | 0.44 ± 0.11 B |
| FgRdRP-2 | 1.09 ± 0.33 | 0.99 ± 0.23 | 1.26 ± 0.29 | 0.56 ± 0.19 A | |
| FgRdRP-3 | 1.06 ± 0.08 | 0.32 ± 0.07 B | 27.18 ± 3.85 A | 27.66 ± 7.46 A | |
| FgRdRP-4 | 0.85 ± 0.19 | 1.21 ± 0.42 | 16.57 ± 5.14 A | 17.24 ± 3.71 A | |
| FgRdRP-5 | 1.01 ± 0.07 | 0.14 ± 0.04 B | 11.41 ± 6.37 A | 7.48 ± 1.81 A | |
mRNA levels of dicer, Argonaute, and RdRP gene transcripts in virus-infected F. graminearum PH-1 relative to their levels in virus-free PH-1 after 120 h of incubation were measured by real-time qRT-PCR. Values are from three independent experiments. Values in each row followed by different letters are significantly different from the mean value for virus-free PH-1 according to Tukey's test (P < 0.05).
Effects of FgDICER and FgAGO on phenotypic changes and viral RNA accumulation in response to FgV1 infection.
To identify the functions of the F. graminearum RNA silencing gene(s) in defending against FgV infections, single gene deletion mutants with deletions of DICER, AGO, and RdRP genes were generated by targeted gene replacement. Among the five RdRP genes, we selected FgRdRP-1 and FgRdRP-4, which are phylogenetically related to qde-1 in N. crassa. We also generated overexpression mutants for analysis of these genes. All gene deletion and overexpression mutants were further confirmed by Southern blotting and RT-PCR analysis, and three independent strains of each deletion and overexpression mutant were used for the analyses (Fig. 1A). Each fungal mutant strain was morphologically similar to the wild type (Fig. 1B), which is consistent with previous research demonstrating that these gene deletion mutations do not change the colony morphology of F. graminearum (28). We confirmed that the target gene expression level in each overexpression mutant strain was mostly higher than the virus-induced transcript level in the wild-type strain infected with FgV1, -2, or -3, except for the expression of FgDICER-2 during FgV2 infection, which was slightly higher than the level in the FgDICER-2 overexpression mutant (Table 2). We then used hyphal anastomosis to individually infect all of the mutant strains with FgV1 in order to understand the RNA silencing response to FgV1 infections.
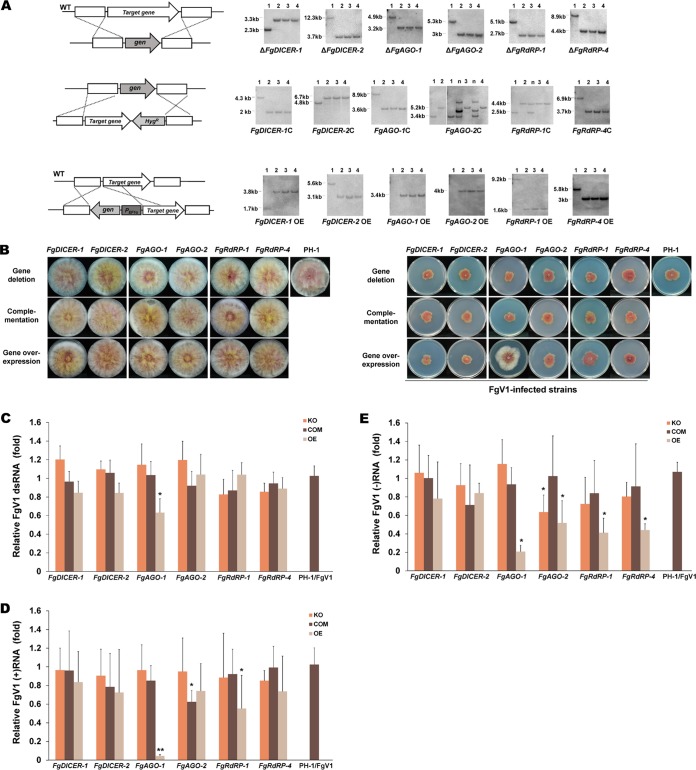
FIG 1.
Colony morphologies and FgV1 RNA accumulation levels of the Fusarium graminearum RNA silencing mutant strains. (A) Generation of mutants in F. graminearum. Gene deletion (top), complementation (middle), and overexpression (bottom) mutants were generated in this study. Schematic representations of the homologous gene recombination strategies used to generate the RNA silencing-related F. graminearum mutants (left) and results of Southern blot hybridization of the mutants (right) are shown. The ORFs of the target genes were fused with the hygromycin resistance cassette to generate complementary strains. The promoter was replaced with the elongation factor 1α promoter (PEF1α) in the overexpression strain. All DNA probes and restriction enzymes used for constructing each mutant strain are shown on the right, and the expected DNA sizes indicated. A 32P-labeled DNA fragment of the 5′-flanking or 3′-flanking region of the target gene was used as a probe for Southern blot hybridization. The sizes of the DNA bands are indicated to the left of the Southern blot images. Lanes 1, wild-type (WT) strain PH-1; lanes 2 to 4, different biological replicates of the indicated single gene deletion mutant used in this study; lane(s) n, nonhomologous (ectopic) insertion that was not selected. (B) Colony morphologies of virus-free (left) and Fusarium graminearum virus 1 (FgV1)-infected (right) RNA silencing gene deletion, complementation, and overexpression mutant strains. All cultures were photographed after 5 days on CM. (C) Semiquantification of viral dsRNA accumulation levels of the wild type and different mutant strains. Shown is the viral dsRNA accumulation level of each RNA silencing mutant relative to that of PH-1/FgV1, which was set at one as determined with ImageJ software. Values were calculated using the results from three independent biological replicates, and error bars indicate standard deviations (SD). (D and E) Quantification of plus-strand (D) and minus-strand (E) viral RNA accumulation at 120 h postinoculation (hpi) using real-time reverse transcription-quantitative RT-PCR (qRT-PCR). EF1α and UBH gene transcripts were used as internal controls. Mean values (± SD) from two biological replicates and at least three replicated experiments are shown. (C to E) Mean values with different numbers of asterisks are significantly different (P < 0.05) from each other based on Tukey's test. KO, gene deletion (knockout) mutant; COM, complementation mutant; OE, overexpression mutant.
TABLE 2.
Target gene expression levels in virus-free overexpression strains
| Protein family | OE target gene | Mean mRNA level ± SDa |
|---|---|---|
| Dicer | FgDICER-1 | 101.23 ± 22.56 |
| FgDICER-2 | 28.58 ± 7.36 | |
| Argonaute | FgAGO-1 | 6.20 ± 0.82 |
| FgAGO-2 | 1,113.71 ± 572.74 | |
| RdRp | FgRdRP-1 | 44.24 ± 6.17 |
| FgRdRP-4 | 21.08 ± 5.08 |
Target gene expression levels in virus-free overexpression mutant strains relative to their levels in virus-free PH-1 after 120 h of incubation were measured by real-time qRT-PCR. Values are from three independent experiments. All values are significantly different from the mean for virus-free PH-1 according to Tukey's test (P < 0.05).
The colony morphologies and mycelial growth rates of the FgV1-infected gene deletion mutants and the FgV1-infected wild type (PH-1/FgV1) were similar (Fig. 1B). The mycelial growth rates of the FgV1-infected overexpression mutants and the FgV1-infected wild type were also similar, except that the mycelial growth rates of the FgV1-infected FgAGO-1 overexpression mutants were much higher than those of the FgV1-infected wild type. We examined the dsRNA and ssRNA accumulation levels in virus-infected RNA silencing mutant strains (Fig. 1C to E). Only the FgV1-infected FgAGO-1 overexpression mutant showed a significantly reduced level of viral dsRNA accumulation compared to that of the FgV1-infected PH-1 strain. We also quantitated viral sense (plus)-strand and antisense (minus)-strand ssRNA accumulation levels in virus-infected RNA silencing gene deletion mutant strains using qRT-PCR. We observed substantial decreases in the ssRNA accumulation levels, especially for plus-strand ssRNA, in the FgAGO-1 OE mutant compared to the levels in the FgV1-infected wild type (Fig. 1D and E). In contrast, the ΔFgAGO-1 mutant only allowed slightly greater FgV1 accumulation, by ca. 1.2-fold, but this difference is within the experimental fluctuation. Neither the ΔFgDICER-1 nor the ΔFgDICER-2 mutant showed increased FgV1 RNA accumulation, nor did overexpression of these genes directly affect the accumulation of viral RNA. The FgV1 RNA accumulation levels did not differ significantly between FgRdRP-1 and FgRdRP-4 gene deletion strains; the viral ssRNA accumulation levels in overexpression strains, however, were slightly reduced compared to those in the FgV1-infected PH-1 strain. As noted earlier, the FgV1-infected FgAGO-1 overexpression mutants grew faster than the FgV1-infected PH-1 strain (Fig. 1B), indicating that low levels of viral RNA accumulation in FgAGO-1 overexpression strains might affect mycelial growth. These results suggest that FgAGO-1 overexpression greatly affects FgV1 RNA accumulation in F. graminearum.
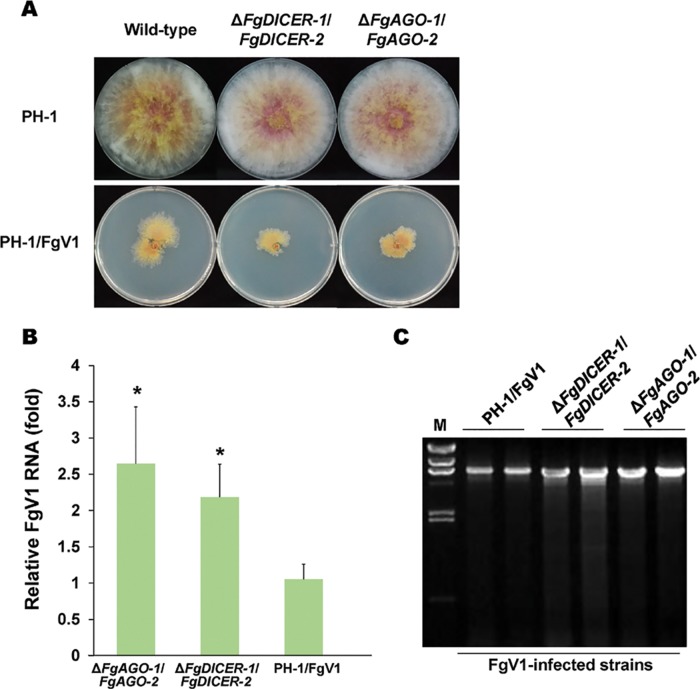
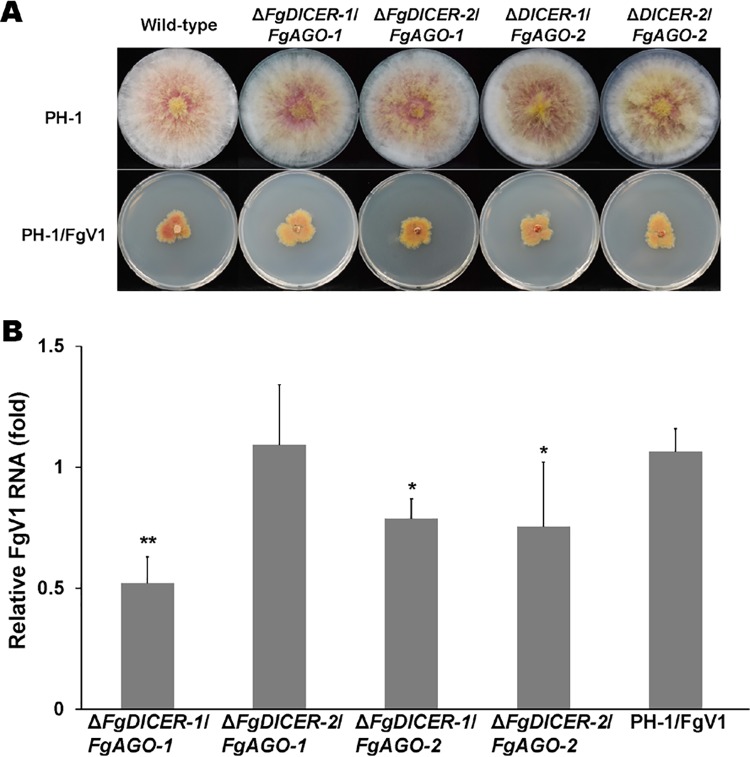
As mentioned earlier, F. graminearum contains multiple dicer and Argonaute genes. As shown by the data in Table 1, we confirmed that FgV2 infection induced all dicer and Argonaute genes, while FgV1 infection induced FgDICER-1 and FgAGO-2 and FgV3 infection induced FgDICER-2 and FgAGO-1. Moreover, the levels of accumulation of viral RNAs in a single gene deletion mutant might also affect the antiviral RNA silencing response to virus infection. To determine whether there is compensation between FgDICER-1 and FgDICER-2 and between FgAGO-1 and FgAGO-2, we generated ΔFgDICER-1/FgDICER-2 and ΔFgAGO-1/FgAGO-2 double gene deletion mutant strains (Fig. 2). These double deletion mutant strains also exhibited normal colony morphology (Fig. 2A). However, the FgV1-infected mutant strains showed growth retardation compared to the growth of the FgV1-infected wild-type strain or single gene deletion mutants. The FgV1 RNA accumulation levels increased significantly, by 2.1-fold in the ΔFgDICER-1/FgDICER-2 strain and by 2.6-fold in the ΔFgAGO-1/FgAGO-2 strain (Fig. 2B). The FgV1 dsRNA accumulation levels also increased in the ΔFgDICER-1/FgDICER-2 and ΔFgAGO-1/FgAGO-2 strains (Fig. 2C). Moreover, we generated ΔFgAGO-1 or -2/FgDICER-1 or -2 cross-double knockout gene deletion mutant strains to determine the redundant roles of Argonaute and dicer genes against virus infection in F. graminearum. These FgV1-infected cross-double deletion mutant strains showed a normal colony morphology in comparison to that of the FgV1-infected wild type (Fig. 3A). The FgV1 RNA accumulation levels were not significantly different in the ΔFgDICER-2/FgAGO-1 strain and the FgV1-infected wild type. The ΔFgDICER-1/FgAGO-1, ΔFgDICER-1/FgAGO-2, and ΔFgDICER-2/FgAGO-2 mutant strains showed slightly decreased levels of FgV1 accumulation (Fig. 3B). These observations indicate that dicer and Argonaute genes might have redundant role(s) in response to FgV1 infection in F. graminearum.
FIG 2.
Analysis of the double gene deletion mutants. (A) Colony morphologies of ΔFgDICER-1/FgDICER-2 and ΔFgAGO-1/FgAGO-2 double gene deletion virus-free and virus-infected mutants. Cultures were photographed after 5 days on CM. (B) Accumulation of FgV1 RNA in double gene deletion mutants at 120 hpi according to qRT-PCR. Means with an asterisk are significantly different (P < 0.05) from the mean of the virus-infected wild type (PH-1/FgV1) based on Tukey's test. (C) Agarose gel (1%) analysis of the dsRNA accumulation for PH-1/FgV1 and double gene deletion mutants. A 10-μg quantity of total RNA per sample was treated with DNaseI and S1 nuclease. The largest band in the FgV1-infected samples represents the full-length viral dsRNA (6.6 kb); smaller bands indicate internally deleted forms of viral dsRNA. Lane M, lambda DNA digested with HindIII.
FIG 3.
Colony morphologies and FgV1 RNA accumulation levels in cross-double gene deletion mutants of F. graminearum. (A) Colony morphologies of ΔFgDICER-1/FgAGO-1, ΔFgDICER-2/FgAGO-1, ΔFgDICER-1/FgAGO-2, and ΔFgDICER-2/FgAGO-2 mutants. Cultures were photographed after 5 days on CM. (B) Accumulation of FgV1 RNA in mutant strains at 120 hpi according to qRT-PCR. Mean values with different numbers of asterisks are significantly different (P < 0.05) from each other based on Tukey's test.
Involvement of FgAGO-1 in transcriptional induction of FgDICER genes in response to FgV1 infection.
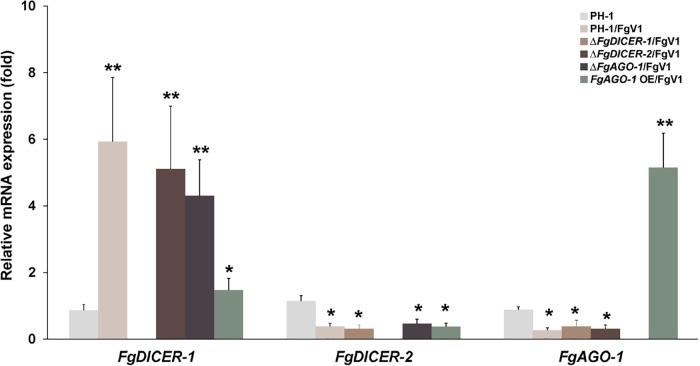
The expression levels of key RNAi components, such as the Argonaute protein QDE-2 and the dicer protein DCL-2, are elevated following the induction of dsRNA expression in N. crassa and C. parasitica (4, 21). Moreover, similar levels of induction of dcl-2 and agl-2 in C. parasitica were observed in response to infection by a mutant CHV1-EP713 hypovirus that lacked p29, which is a suppressor of RNA silencing (20, 21). Researchers have reported that agl-2 in C. parasitica is required for the induction of dcl-2 expression in response to CHV1 infection (21). To determine whether FgAGO-1 expression is correlated with FgDICER-2 expression, we examined the transcript levels of FgAGO-1, FgDICER-1, and FgDICER-2 in wild-type and mutant strains. The transcript levels of each Argonaute, dicer, and RdRP gene in the gene deletion mutants did not differ significantly from the levels in the virus-free wild-type PH-1 strain (data not shown). The transcript levels of FgAGO-1 and FgDICER-2 in the gene deletion mutant strains were also similar to the levels in the wild type following FgV1 infection (Fig. 4). The significantly induced gene expression of FgDICER-1 in the FgV1-infected strain was not accompanied by the induction of FgAGO-1. However, the transcript level of FgDICER-1 was decreased in the FgV1-infected FgAGO-1 overexpression strains. These data indicate that the induction or suppression of FgAGO-1 gene expression might not directly affect the expression of FgDICER genes.
FIG 4.
Accumulation of FgDICER-1, FgDICER-2, and FgAGO-1 gene transcripts in response to FgV1 infection. The transcript levels of FgDICER-1, FgDICER-2, and FgAGO-1 in the FgV1-infected wild type (PH-1/FgV1) and the FgV1-infected mutants were determined by qRT-PCR. Mean values (± SD) are shown. Mean values with different numbers of asterisks are significantly different (P < 0.05) from each other based on Tukey's test. OE, overexpression.
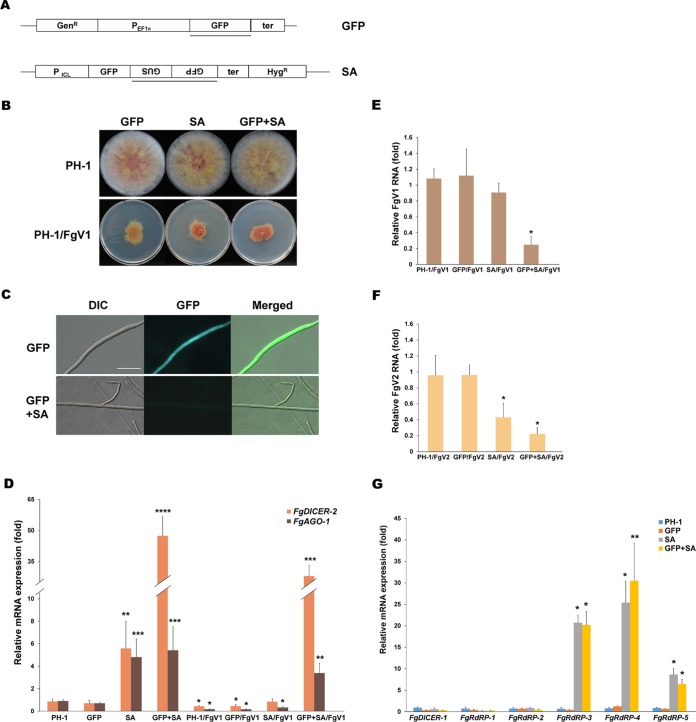
Comparison of levels of gene expression associated with FgV1-mediated gene silencing and hpRNA-induced gene silencing.
A previous report indicated that FgAGO-1 and FgDICER-2 are important in the hpRNA-induced target gene silencing in F. graminearum, while FgAGO-2 and FgDICER-1 participate primarily in the sex-specific RNAi pathway (28, 30). In this study, we showed that the transcript levels of FgAGO-1 and FgDICER-2 increased in the FgV2- or FgV3-infected wild-type strain but not in the FgV1-infected wild-type strain (Table 1). To compare the Argonaute and dicer gene transcript accumulation levels resulting from FgV-induced and hpRNA-induced RNA silencing, we generated green fluorescent protein (GFP) gene-bearing hairpin silencing constructs in order to trigger the transcriptional GFP transgene-silencing response (Fig. 5A to C). The morphologies of the GFP-expressing strain (GFP), the GFP hairpin RNA-expressing strain (GFP spacer and antisense strand [SA]), and the GFP-expressing strain with a GFP hairpin RNA expression construct (GFP+SA) were similar to that of the wild-type PH-1 strain (Fig. 5B). The wild-type strain and three independent transformants were examined to determine the gene expression levels of FgAGO-1 and FgDICER-2. The FgAGO-1 and FgDICER-2 transcript levels were approximately 5- and 47-fold higher, respectively, in the GFP+SA strain than in the wild-type PH-1 strain (Fig. 5D). The FgAGO-1 and FgDICER-2 transcript levels were also significantly higher in the SA transformants than in the wild type. In contrast, the FgAGO-1 and FgDICER-2 transcript levels were decreased in FgV1-infected GFP and SA single transformants and GFP+SA double transformants compared to the levels in each virus-free mutant strain. We also observed that the accumulation level of FgV1 or FgV2 RNA in the FgV-infected GFP+SA double mutant was at least 60% less than the level in the FgV1- or FgV2-infected wild type (Fig. 5E and F). These results suggested that the induction of FgAGO-1 and FgDICER-2 negatively affected the viral RNA accumulation in F. graminearum and that FgV1 could interfere with the host's antiviral response by suppressing FgAGO-1 and FgDICER-2 expression.
FIG 5.
Functional analysis of dicer and Argonaute in hairpin RNA-induced gene silencing. (A) Diagram of transformation of plasmids used to study GFP silencing. The pSKGen vector was under the control of the EF1 alpha promoter, and the pGFP-SA construct was under the control of the isocitrate lyase (ICL) promoter. A HincII fragment of the β-glucuronidase (GUS) gene was used as a spacer. Segments used as probes in Southern blot hybridization analysis are indicated by bars. GFP, transformant that only had the pSKGen construct; SA, transformant with the pSA construct. (B) Colony morphologies of virus-free GFP silencing-related transformants and FgV1-infected mutant strains. Cultures were photographed after 5 days on CM. GFP+SA, GFP and pSA were transformed together into the wild type. (C) GFP expression in mutant strains. Mycelia of GFP transformants and GFP-silenced transformants (GFP+SA) were observed with a microscope at 2 days postincubation in CM. DIC, differential interference contrast. Scale bar = 20 μm. (D) Accumulation of FgDICER-2 and FgAGO-1 gene transcripts in GFP expression or hairpin dsRNA-producing mutants. Accumulation levels of FgDICER-2 and FgAGO-1 in all virus-free and FgV1-infected mutants relative to the levels in the virus-free wild type (PH-1), which were set to one, are shown. EF1α and UBH gene transcripts were used as the internal controls. (E and F) Accumulation of FgV1 (E) and FgV2 (F) RNA in mutant strains. qRT-PCR was used to quantify FgV1 and FgV2 RNA1 at 120 hpi. (G) Confirmation of accumulation of FgDICER-1 and FgRdRP1 to -5 in hairpin RNA-induced gene silencing mutants. Gene expression levels of FgDICER-1 and five FgRdRP genes were analyzed by qRT-PCR at 120 hpi. (D to G) Mean values (± SD) from at least two biological replicates and three independent experiments are shown. Mean values with different numbers of asterisks are significantly different (P < 0.05) from each other based on Tukey's test.
Unlike FgV1 infection, hairpin dsRNA production did not trigger upregulation of the FgDICER-1 gene. The levels of transcript accumulation of FgDICER-1 did not differ between the GFP-silenced strains and the wild type (Fig. 5G). Among the five FgRdRP genes, the transcript levels of FgRdRP-3, FgRdRP-4, and FgRdRP-5 were significantly increased in the hairpin dsRNA-producing strains (Fig. 5G). This result indicates that the induction of FgDICER-1 is involved in specific FgV-mediated responses but not in hpRNA-induced gene silencing in F. graminearum.
Effects of deletion and overexpression of RNAi-related genes on FgV2 or FgV3 infection.
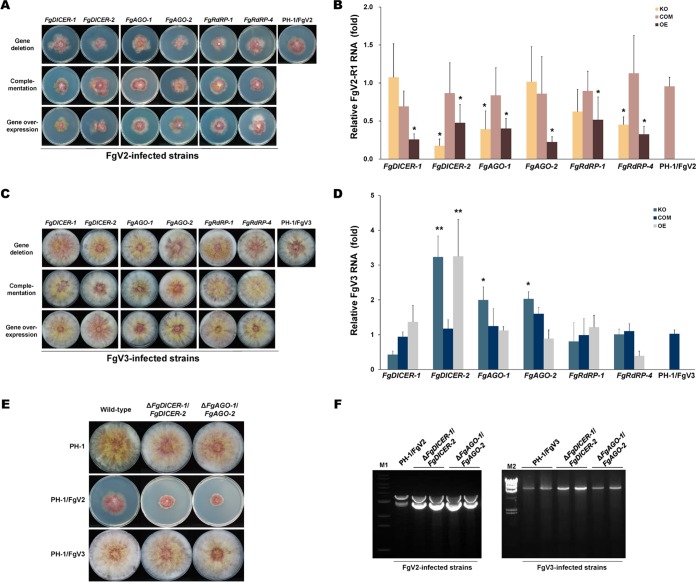
As shown by the results in Fig. 1 and 2, the deletion or overexpression of specific genes involved in RNAi affected the mycelial growth phenotype and FgV1 RNA accumulation. To compare the effects of the same mutations on the other FgV infections, we further investigated FgV2- and FgV3-infected mutants with mutations of RNAi-related genes. The colony morphologies were similar in the FgV2-infected single gene deletion mutants and the FgV2-infected wild type (PH-1/FgV2), i.e., colonies of both had reduced aerial mycelia, increased pigmentation, and irregular margins (Fig. 6A). The colony morphologies were also similar in the FgV3-infected mutant strains and the FgV3-infected wild type (PH-1/FgV3) (Fig. 6C).
FIG 6.
Accumulation of FgV RNA in virus-infected RNA silencing gene mutant strains of F. graminearum. (A) Colony morphologies of mutant strains infected with Fusarium graminearum virus 2 (FgV2). (B) Quantification of FgV2 RNA1 at 120 hpi using qRT-PCR. (C) Colony morphologies of mutant strains infected with FgV3. (D) Quantification of FgV3 RNA at 120 hpi using qRT-PCR. (A and C) All cultures were photographed after 5 days on CM. (B and D) Mean values (± SD) of two biological replicates and at least three replicated experiments are shown. EF1α and UBH gene transcripts were used as internal controls. Mean values with different numbers of asterisks are significantly different (P < 0.05) from each other based on Tukey's test. (E) Colony morphologies of ΔFgDICER-1/FgDICER-2 or ΔFgAGO-1/FgAGO-2 double gene deletion virus-free and virus-infected mutants. Cultures were photographed after 5 days on CM. (F) Agarose gel (1%) analysis of dsRNA accumulation in PH-1/FgV2, PH-1/FgV3, and double gene deletion mutants. A 20-μg quantity of total RNA per sample was treated with DNaseI and S1 nuclease. Lane M1, 1-kb ladder; lane M2, lambda DNA digested with HindIII.
The FgV2 RNA accumulation levels in mutant strains were determined using primer sets derived specifically for FgV2 RNA segment RNA1 (Fig. 6B). When the RNA1-specific primer set was used, the RNA1 accumulation levels in FgV2-infected ΔFgDICER-1, ΔFgAGO-2, and ΔFgRdRP-1 mutants were similar to the level in the FgV2-infected wild type, but the levels in FgV2-infected ΔFgDICER-2, ΔFgAGO-1, and ΔFgRdRP-4 mutants were lower than the level in the FgV2-infected wild type. Most of the single gene disruption mutations might not interfere directly with the antiviral response to FgV2 infection in F. graminearum. However, the overexpression of RNAi-related genes caused reductions of FgV2 RNA1 accumulation levels compared to the level in the wild type.
Among FgV3-infected strains, the viral RNA accumulation levels were approximately 2-fold higher in the ΔFgAGO-1 or ΔFgAGO-2 mutant than in the FgV3-infected wild type, but the accumulation levels were similar in the FgAGO-1 and FgAGO-2 overexpression strains and the wild type (Fig. 6D). For FgDICER-2, disruption and overexpression mutants showed increased FgV3 RNA accumulation levels compared to the levels in the FgV3-infected wild type, while the ΔFgDICER-1 strain showed a similar viral RNA accumulation level. As was the case in FgV1- and FgV2-infected mutant strains, the deletion or overexpression of FgRdRP-1 and FgRdRP-4 did not affect viral RNA accumulation in FgV3-infected mutant strains.
We also transferred both viruses to double gene deletion strains. The growth retardation was greater for the FgV2-infected than for the FgV1-infected double gene knockout strains (Fig. 6E). The accumulation of FgV2 and FgV3 double-stranded RNAs also increased compared to their levels in the wild type. Interestingly, FgV3 RNA accumulation was efficiently increased by double gene deletions of FgDICER-1 and FgDICER-2 (Fig. 6F). These results suggest that the two FgDICER and two FgAGO genes have redundant roles in response to FgV2 or FgV3 infection in F. graminearum.
FgV1-derived small RNAs in F. graminearum.
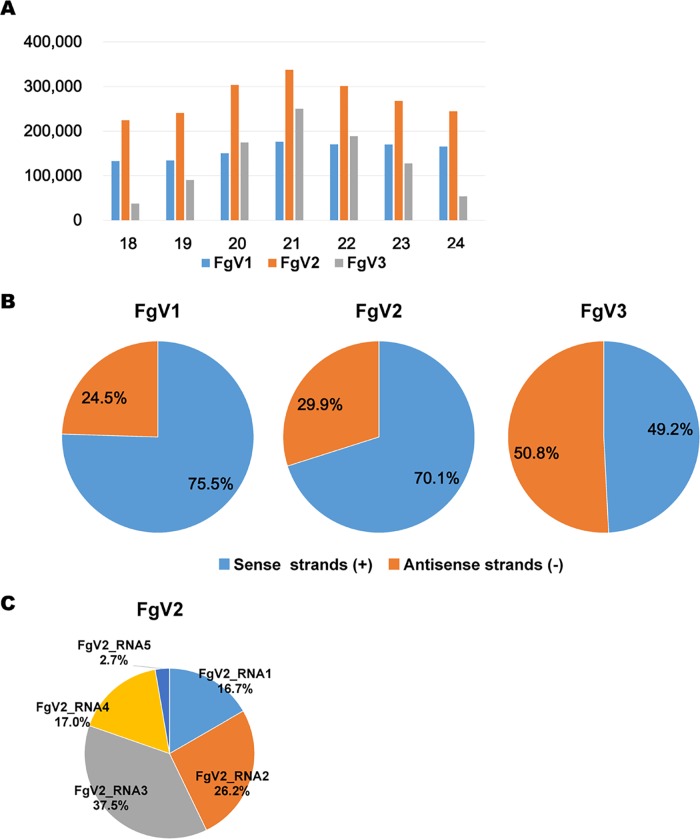
In general, the RNA silencing mechanism in many organisms is associated with the generation of small interfering RNAs (siRNAs). Several previous studies have reported that many fungal hosts produce virus-derived siRNAs (vsiRNAs) in response to diverse mycovirus infections (19, 20, 31, 33). These studies showed that dicer and Argonaute genes are involved in vsiRNA production and that the vsiRNAs generated function in the RNA silencing pathway (19, 20, 33). In addition to experiments using the reverse genetics approach, we conducted small RNA profiling to understand the RNA silencing mechanism for known mycoviruses in the model host F. graminearum. We conducted high-throughput sequencing for small RNA libraries obtained from F. graminearum PH-1 strains infected by one of three mycoviruses (FgV1, FgV2, or FgV3). The distributions of vsiRNAs on the individual virus genomes differed. Large numbers of vsiRNAs, ranging from approximately 40,000 to 3,900,000 reads, were obtained from the four libraries (data not shown). The vsiRNA profiling data indicate that vsiRNA abundances and distributions differ among the FgV1, -2, and -3 genomes even when these viruses are infecting the same fungal host. The proportion of FgV1- or FgV2-derived siRNA within the total siRNA reads was relatively higher than the proportion of FgV3 siRNA. The size distributions of the 18- to 24-nucleotide (nt) vsiRNA reads in strain PH-1 infected with each virus demonstrated that 20- to 22-nt-long vsiRNAs were dominant following FgV2 and FgV3 infection but that no length was clearly dominant following FgV1 infection (Fig. 7A). The strand polarities of vsiRNA reads differed depending on which virus infected the fungus. The percentage of vsiRNA reads was clearly greater for the sense strand than for the antisense strand following FgV1 or FgV2 infection. In the case of FgV3 infection, the percentages of vsiRNA reads were almost equal for both strands (Fig. 7B). For the segmented virus FgV2, the percentages of vsiRNA reads differed among the segments (Fig. 7C). Among the five segments of FgV2, the percentage on the dsRNA3 segment, which encodes an ORF of unknown function, was relatively large.
FIG 7.
Identification of vsiRNAs by small RNA sequencing. (A) Numbers of vsiRNAs identified (y axis) based on size (number of nucleotides; x axis). (B) Percentages of vsiRNAs located on sense and antisense strands of each virus. (C) Percentages of identified vsiRNAs located on individual RNA segments of FgV2.
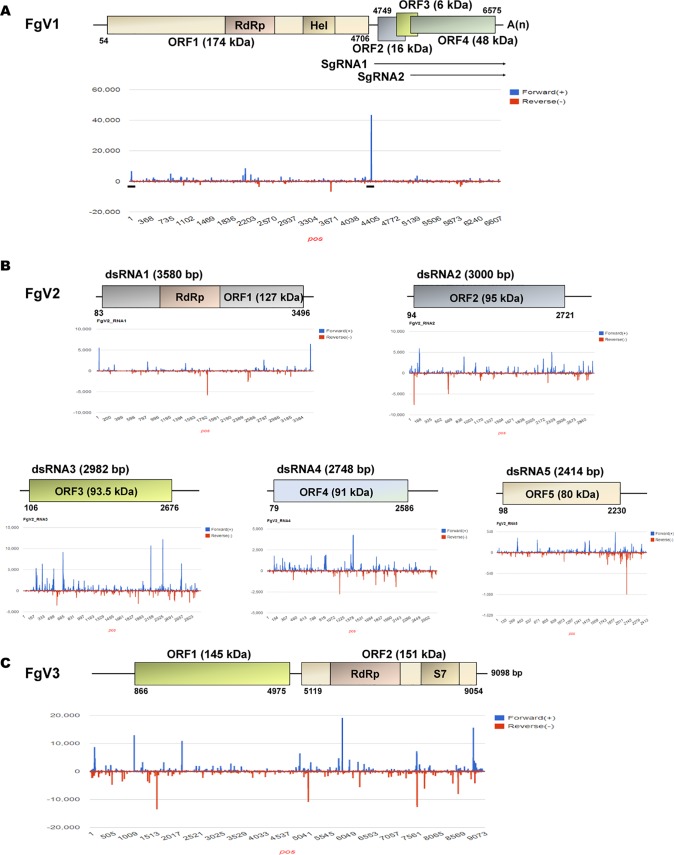
The vsiRNAs resulting from FgV1 infection had an asymmetric distribution on both strands. In FgV1, the numbers of vsiRNAs were highest on an internal region and were relatively high on the 5′-terminal region of the sense strand (Fig. 8A). In FgV2, the vsiRNAs identified appeared to be unevenly distributed among the five dsRNA segments (Fig. 8B). In the dsRNA1 segment of FgV2, the numbers of vsiRNAs were high in the 5′ and 3′ regions of the sense strand and in the internal region of the antisense strand. For dsRNA2, -4, and -5 of FgV2, peaks in numbers of vsiRNAs were scattered throughout each segment. In FgV2, the dsRNA3 segment had many more hotspots of vsiRNAs than the other four segments. This result indicates that dsRNA3 of FgV2 might be preferentially recognized and targeted by RNAi components. In FgV3, the sense- and antisense-strand vsiRNA peaks were distributed throughout the viral genome, and some of the higher sense-strand vsiRNA peaks were located in the 5′ and 3′ terminal regions (Fig. 8C). These results indicate the differences in the vsiRNA accumulation proportion and distribution patterns along each viral genome among different mycoviruses that showed different patterns of induction and suppression of RNA silencing-related genes upon infection by FgV1, -2, and -3.
FIG 8.
Profiling of viral small interfering RNAs (vsiRNAs) on FgV1, -2, and -3 genomes. The identified vsiRNAs induced by FgV1, FgV2, or FgV3 are mapped on the corresponding virus genome. (A to C) Distributions of vsiRNAs mapped on the FgV1, FgV2, and FgV3 genomes. FgV3 has only one RNA segment, while FgV2 has five RNA segments. The genome organization for each virus genome is depicted. Blue and red bars indicate vsiRNAs located on the sense strand (+) and the antisense strand (−) of the corresponding virus genome, as observed by stem-loop RT-PCR.
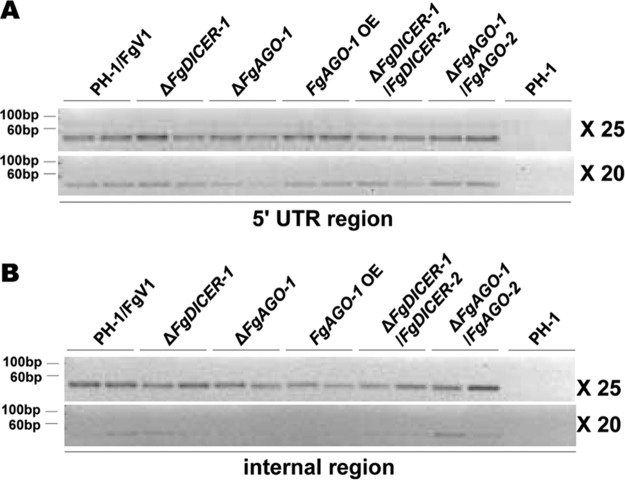
Based on the small RNA sequencing data, we conducted stem-loop RT-PCR to validate the accumulation of some specific vsiRNA peaks. First, we selected two candidate vsiRNA peaks on the sense strand of FgV1 in the 5′-terminal region and the internal region from nt 4304 to 4323, which had the highest numbers of vsiRNAs (Fig. 8A). In addition, we assessed the vsiRNA accumulation levels in ΔFgDICER-1 and ΔFgAGO-1 mutants to determine whether FgV1 RNA accumulation in these mutants was related to the accumulation of specific vsiRNAs. We confirmed the presence of vsiRNAs in the 5′-terminal and internal regions in FgV1-infected strains but not in the virus-free wild-type strain (Fig. 9). The accumulation of vsiRNAs in the ΔFgDICER-1/FgDICER-2 strain showed a decreased level, as expected. The vsiRNA accumulation level was lower in the FgV1-infected ΔFgAGO-1 strain than in the FgV1-infected wild-type strain, but not in the FgV1-infected ΔFgDICER-1 strain, although these two mutant strains showed similar increased FgV1 RNA accumulation levels. This result suggested that the increased accumulation level of viral RNA in the FgAGO-1 mutant was accompanied by a significant decrease in vsiRNA levels.
FIG 9.
Detection of FgV1 vsiRNAs. Stem-loop RT-PCR of vsiRNAs of FgV1 in the 5′-end untranslated region (UTR) (A) and internal region (B). A 10-ng quantity per cDNA sample was used for PCR amplification. The numbers of PCR cycles are indicated to the right. Three independent biological replicates were used for this experiment, and two samples from each were visualized on a 4% agarose gel.
DISCUSSION
In this study, we used a reverse genetics approach to investigate the antiviral role(s) of RNA silencing components in F. graminearum. To study the fungus's defense response against mycovirus infections, we used the established model system of F. graminearum PH-1 and three unrelated viruses, FgV1, FgV2, and FgV3. Although both FgV1 and FgV2 are hypovirulent, their genomic organizations differ. FgV3 did not cause any phenotypic change in F. graminearum. All three virus infections, however, induced certain RNA silencing-related genes (Table. 1). Following FgV2 and FgV3 infection, the expression levels of FgAGO-1 and FgDICER-2 increased significantly. The expression levels of FgRdRP-3, FgRdRP-4, and FgRdRP-5 were also greatly increased by FgV2 and FgV3 infection. Similar results were observed following FgHV2 infection, i.e., the expression levels of FgAGO-1, FgDICER-2, and FgRdRP-3 were significantly upregulated in an FgHV2-infected strain of F. graminearum (34). In C. parasitica, in addition to many other genes, dcl-2 and agl-2 were induced by CHV1 and mycoreovirus 1 (MyRV1) infections (16, 20). Our data indicate that FgAGO-1 and FgDICER-2 might be preferentially induced following virus infection and hairpin RNA production. These results suggest that F. graminearum has a dsRNA antiviral response similar to that in C. parasitica.
Based on phylogenetic analysis, FgAGO-2 and FgDICER-1 are closely related to sms-2 and dcl-1, respectively, in N. crassa (27). Previous research suggested that only FgSMS2 (FgAGO-2) controls meiosis and the subsequent developmental pathways in F. graminearum (35). Recently, it was reported that FgDICER-1 and FgAGO-2 mainly mediated the sex-specific RNAi pathway in F. graminearum (30). Although we expected that FgAGO-2 and FgDICER-1 would function in the meiotic silencing by unpaired DNA (MSUD) pathway in F. graminearum, the present study showed that expression of FgAGO-2 and FgDICER-1 was induced by FgV1 and FgV2 infection but not by FgV3 infection. Perhaps the induction of FgAGO-2 or FgDICER-1 by FgV1 or FgV2 infection is related to the functional redundancy of dicers and Argonautes in F. graminearum. We showed that the transcription level of FgDICER-1 was not changed by GFP-expressing mutant and GFP hairpin RNA-expressing mutant strains that did not show any phenotypic change (Fig. 5G). In addition, the transcription level of FgDICER-1 was decreased in the FgV1-infected FgAGO-1 OE mutant, which showed mild symptoms and low accumulation of FgV1 RNA relative to those in FgV1-infected wild-type PH-1 train (Fig. 4). As mentioned above, among five unrelated dsRNA viruses in R. necatrix, only infection by RnMyRv3 and RnMBV1, which affect mycelial growth and virulence in the fungal host, caused the upregulation of RnDCL-2, RnAGL-2, RnRdRP-1, and RnRdRP-2 expression (19). These observations suggest that FgDICER-1 and FgAGO-2 can be induced by hypovirulence-associated phenotypic changes, e.g., reductions in conidiation and virulence or defects in perithecium development. Further studies are required to clarify how dicer and Argonaute genes are transcriptionally upregulated by mycovirus infection in F. graminearum.
Research has clearly indicated that host cellular RdRPs may function in the synthesis of virus-derived siRNAs and in the antiviral defense response in Arabidopsis and rice (36, 37). The roles of RdRPs have also been characterized in the RNA silencing pathway in some fungi. In N. crassa, RdRP contributes to transgene-induced silencing, meiotic silencing, and the production of DNA damage-induced QDE-2-interacting sRNAs (qiRNAs) (12, 38, 39). Two RdRP genes in M. circinelloides participate in different steps of the same transgene-induced RNA silencing pathway (40). Rdp1 also has a role in sex-induced silencing in C. neoformans (9). For some fungi, however, Rdp genes do not seem to be required for the antiviral RNA silencing response (41). In this study, we examined the effects of gene deletion and overexpression of FgRdRP-1 and FgRdRP-4 on the antiviral RNA silencing responses of F. graminearum. These two genes are phylogenetically closely related to qde-1, whose function is related to quelling pathway and dsRNA-triggered gene silencing in N. crassa (27, 42). We assumed that FgRdRP-1 and FgRdRP-4 might have functional role(s) in the antiviral RNA silencing response because the virus-mediated transcriptional activation response is similar to the hairpin dsRNA-induced transcriptional response in C. parasitica (21). As shown by the results in Table 1, FgV1 infection did not induce any RdRP genes, but FgV2 and FgV3 infection did induce FgRdRP-3, -4, and -5. However, neither the phenotype nor the viral RNA accumulation levels were greatly altered in the FgRdRP-1 and FgRdRP-4 gene deletion and overexpression mutants. Although neither the FgRdRP-1 nor the FgRdRP-4 gene alone seems to have a distinct role against FgV infections, these genes might have a role in the RNA silencing pathway. On the other hand, given that RdRP initiates or amplifies the RNA silencing signal, there might be an alternative pathway for RNA silencing. Researchers previously demonstrated that five C. parasitica Rdr genes do not play a significant role in the RNA silencing antiviral defense response or in viral RNA recombination, although the Rdr3 and Rdr4 gene expression levels were increased following CHV1/EP713 infection (41); the researchers concluded that an antiviral RNA silencing response in C. parasitica may be triggered by viral dsRNA or hairpin dsRNA molecules without the participation of RdRPs if sufficient dsRNA is generated by other means (41). Although FgRdRP-2 is required for meiotic silencing in F. graminearum (29), the physiological and biological functions of FgRdRP genes remain largely unclear because five predicted RdRP proteins in the F. graminearum genome may function redundantly. We found that FgRdRP-3, -4, and -5 were induced by hairpin RNA production, but the functions of other FgRdRP genes in the antiviral RNA silencing response and in other cellular processes remain unclear and require further study.
In N. crassa, dcl-1 can compensate for dcl-2 in the quelling pathway when dcl-2 is disrupted (13, 27). Although the expression levels of some genes were increased by virus infection in the current study, most of the single gene deletion mutants did not show alterations in colony morphology or viral RNA accumulation. We confirmed that FgV1 and FgV2 replicated better in ΔFgDICER-1/FgDICER-2 and ΔFgAGO-1/FgAGO-2 double knockout mutants; however, there was no significant increase in the accumulation of FgV1 in any of the ΔFgDICER/FgAGO cross-double knockout mutant strains (Fig. 2 and 3). In addition, the deletion of single dicer/Argonaute genes did not significantly affect the expression levels of the other genes (data not shown). These results suggest that antiviral-response-related genes, such as FgDICER-1 or -2 and FgAGO-1 or -2, might compensate for the loss of gene function in F. graminearum. Although these genes have functional redundancy, FgAGO-1 has a major role in the antiviral RNA silencing response against FgV1 infection.
In profiling the vsiRNAs in F. graminearum strains infected with FgV1, -2, and -3, we observed that the abundances and the percentages of vsiRNAs on sense versus antisense strands differed among three different viruses. Moreover, the distribution of vsiRNAs across the viral genomes differed even among the segments of the same virus. The formation of vsiRNA hot spots is related to secondary structures that are optimal for dicer binding and processing and for vsiRNA stability (43, 44). Therefore, these differences might be associated with the different characteristics of each virus, such as viral genome structures, organization, replication, and strategies for overcoming antiviral host defense responses. Our analysis focused on sRNAs specific to FgV1, -2, and -3, and to look at differences in small RNA metabolism between different virus infections, a more comprehensive analysis of viral and host small RNA profiling, including a combined analysis of the transcripts and small RNAs, is still required. In plants, AGOs associated with vsiRNA or virus-activated siRNAs (vasiRNAs) can directly target complementary viral RNAs or host genes for degradation and, thus, confer antiviral activity or regulate host gene expression (45, 46). Further study is required to explore aspects of virus-host interaction for these small regulatory RNAs.
We confirmed that overexpression of FgAGO-1 reduces FgV1 accumulation. In addition, we observed specific FgV1-derived vsiRNAs in the 5′ and internal regions, and the accumulation of these vsiRNAs was affected by FgAGO-1 deletion (Fig. 9). In contrast to the ΔFgDICER-1 strain, ΔFgAGO-1 strains showed decreased vsiRNA levels in both regions. These results suggest that FgAGO-1 is important for the accumulation of vsiRNAs. The vsiRNA accumulation levels in the FgV1-infected FgAGO-1 OE and ΔFgAGO-1 strains differed in the 5′ and internal regions. This could be explained by the specificity of the selected regions. Moreover, the vsiRNA accumulation levels in both regions in the FgV1-infected ΔFgDICER-1 strain appeared similar to those in the FgV1-infected wild type. In contrast, the FgV1-infected ΔFgDICER-1/FgDICER-2 strain showed significantly decreased levels of specific vsiRNAs. These results suggest that FgDICER-1 and FgDICER-2 have redundant roles in antiviral RNA silencing responses. In addition, it is possible that vsiRNAs can be generated in F. graminearum via an alternative, DICER-independent route that has not yet been demonstrated. Therefore, FgAGO-1 might be required for the accumulation of vsiRNAs during antiviral RNA silencing responses against FgV1 infection, but FgAGO-1 alone might not be sufficient to defend against mycovirus infection.
FgV1 might have the ability to counter the antiviral defenses of F. graminearum. Unlike FgV2 or FgV3 infection, FgV1 infection did not induce RNA silencing genes other than FgDICER-1 and FgAGO-2 (Table 1). Nevertheless, significant upregulation of FgDICER-2 and FgAGO-1 can negatively affect FgV accumulation, although this transcriptional induction occurs in response to GFP silencing (Fig. 5E and F). Similarly, a previous study reported that the inhibition of Rosellinia necatrix victorivirus 1 (RnVV1) replication is associated with transcriptional induction of dcl2 and agl2 in C. parasitica by the infection of CHV1-Δp69 or MyRV1 or by transgenic expression of hairpin dsRNA (47). We also confirmed that, in an hpRNA-mediated GFP silencing mutant in which the transcript levels of these genes were upregulated in the virus-free mutant, the transcript levels of FgAGO-1 and FgDICER-2 genes were reduced after FgV1 infection. It is not clear how FgAGO-1 and FgDICER-2 are coregulated by FgV1 infection. We assume that FgV1 has an effective strategy for avoiding the antiviral defense mechanism of the host. One possibility is that FgV1 can suppress RNA silencing by interfering with the antiviral function of FgAGO-1. Previous research determined that CHV1 suppresses RNA silencing via p29, a suppressor protein that inhibits the expression of dcl-2 and agl-2 in C. parasitica (20, 21). Additional research is needed to identify a possible suppressor(s) of RNA silencing in FgV1.
MATERIALS AND METHODS
Fungal strains and growth conditions.
Virus-free and FgV1-infected F. graminearum PH-1 isolates were stored in 20% (vol/vol) glycerol at −80°C and were reactivated on potato dextrose agar (PDA) at 25°C with a 12-h-light/12-h-dark cycle. F. graminearum cultures used for the extraction of RNA or genomic DNA were grown as described previously (32). Freshly grown mycelia from plates with complete medium (CM) were added to 50 ml of CM broth, and the cultures were incubated at 25°C for 120 h on an orbital shaker (150 rpm). After hyphae were collected by filtering through 3MM paper, they were washed with distilled water, dried by blotting with paper towels, and frozen at −80°C. All fungal strains used in this study are listed in Table S1 in the supplemental material.
Generation of gene deletion, complementation, and overexpression mutants.
The targeted gene deletion, complementation, and overexpression DNA constructs were generated by the double-joint (DJ) PCR method as described previously, with modifications (48). In brief, to generate the FgDICER-1 gene deletion mutant, the 5′- and 3′-flanking regions of the gene were amplified from F. graminearum PH-1 using the primer pairs fgsg09025-5F/-5R and fgsg09025-3F/-3R, respectively. A Geneticin resistance cassette (gen), which was used as a selectable marker, was amplified from vector pII99 using primer pairs GenF/GenR (49). The three amplicons were mixed at a 1:3:1 molar ratio and fused by DJ PCR under previously described PCR conditions (49). The final fusion construct was amplified with the nested primers. The same strategy was used to generate the FgDICER-2, FgAGO-1, FgAGO-2, FgRdRP-1, and FgRdRP-4 gene deletion mutants. Double gene deletion mutant strains were generated with the same strategy, using the Geneticin resistance cassette and the hygromycin phosphotransferase (Hyg) gene resistance cassette. We amplified the fusion constructs for the ΔFgDICER-2 and ΔFgAGO-1 mutations with the Hyg resistance cassette by DJ PCR and used the constructs for protoplast transformation of the fungus as described below. Conidia from the ΔFgDICER-1 and ΔFgAGO-2 strains were used to prepare protoplasts. For complementation, the DNA fragments carrying the native promoter and located upstream from the FgDICER-1 and FgDICER-1 open reading frames (ORFs) and from the 3′-flanking regions were amplified with fgsg09025-5F/fgsg09025 com-5F Rv and fgsg09025 com-3F Fw/-3F Rv, respectively. The Hyg resistance cassette was amplified with pBCATPH hph-fw/-rv primers from pBCATPH. The hygromycin B phosphotransferase (HPH) cassette and both flanking regions were fused by DJ PCR, and the final constructs were amplified with the nested primer set. The same strategy was used to complement the FgDICER-2, FgAGO-1, FgAGO-2, FgRdRP-1, and FgRdRP-4 gene deletion mutants. To generate FgDICER-1 overexpression mutants, the 5′ end and 3′ end of the FgDICER-1 region were amplified by primer sets fgsg09025-5F/fgsg09025 OE-5F rv primer and fgsg09025 OE-3F fw/fgsg09025-3R, respectively. The Gen-PEF1α construct was amplified with Gen_EF1 F/EF1 pro R primers using pSKGEN, which contains the Geneticin resistance cassette and the elongation factor 1α promoter (PEF1α) from F. verticillioides (50). Amplicons were joined as described above, and a final PCR product was generated using fgsg09025 nested F/fgsg09025 OE nested rv primers. The same strategy was used to generate FgDICER-2, FgAGO-1, FgAGO-2, FgRdRP-1, and FgRdRP-4 overexpression mutants. All final DNA constructs were used for fungal protoplast transformation. Conidia were harvested 5 days after inoculation of CMC medium and were added to 50 ml of YPG (1% yeast extract, 1% peptone, 2% glucose) medium for 13 h at 25°C. Protoplasts were prepared with mycelia by treating them with NH4Cl containing 10 mg/ml of Driselase (Sigma-Aldrich, St. Louis, MO) and incubating for 4 h at 30°C. Fungal protoplasts were treated with polyethylene glycol (PEG) for transformation as previously described (48). Transformants were selected on PDA supplemented with 50 μg/ml of hygromycin or Geneticin for further study. After all transgenic strains were confirmed by Southern blotting hybridization (described below), FgV1, -2, or -3 was introduced into the virus-free transformant strains through hyphal anastomosis. Viral infection was detected by RT-PCR using virus-specific primer pairs. All primer sets used in this study are available upon request.
DNA extraction and Southern blot hybridization.
After the fungal strains were incubated for 5 days in CM broth, mycelia were collected by filtration through Whatman 3MM filter paper (GE Healthcare, Uppsala, Sweden). The mycelia were washed with distilled water, pressed onto paper towels to remove the excess water, and stored at −80°C. Genomic DNA was extracted as previously described (32). For Southern blot hybridization of PH-1 and transgenic mutants, 10 μg of genomic DNA was digested with the appropriate enzyme. The digest was loaded on an 0.8% agarose gel and then subjected to gel electrophoresis, capillary blotting, radiolabeling of DNA probes, and hybridization as previously described (32). The hybridized blots were exposed to phosphoimaging screens (BAS-IP MS 2040; Fuji Photo Film, Japan) and were visualized using a BAS-2500 image analysis system (Fuji Photo Film Co.).
Construction of GFP-silencing vectors.
pSA was constructed by inserting the spacer and antisense strand (SA) of the GFP gene into the NotI site of the pIGPAPA vector (51). The HincII fragment of the β-glucuronidase (GUS) gene in pMDC139 was isolated to use as a spacer. The GUS spacer and GFP gene were fused by PCR amplification and then inserted into the NotI site of the pIGPAPA vector. The orientation of this pSA clone was confirmed by PCR analysis and sequencing. A 10-μg quantity of pSA plasmid DNA was introduced into the pSKGen-induced GFP expression strain (Geneticin resistant) by fungal protoplast transformation. Candidate transformants were selected by exposure to Geneticin and hygromycin for secondary screening. All GFP expression strains and GFP-silenced strains were confirmed by observing GFP fluorescence and by Southern blotting.
Preparation of total-RNA or ssRNA samples and cDNA synthesis for RT-PCR.
For nucleic acid extraction, frozen mycelia were pulverized using liquid nitrogen and a mortar and pestle. Total RNAs were extracted with Iso-RNA lysis reagent (5 Prime, Hamburg, Germany). Extracted total RNA was treated with DNase I (TaKaRa Bio, Shiga, Japan) to remove genomic DNA according to the manufacturer's instructions. These total-RNA samples were precipitated with ethanol and resuspended in DEPC-treated water. Next, 5 μg of total RNA of each sample was used to synthesize first-strand cDNA with an oligo(dT)18 primer and Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, WI) according to the manufacturer's protocols. All synthesized cDNAs were diluted 1:10 with nuclease-free water for RT-PCR. To isolate the ssRNA fraction, total-RNA extracts were precipitated with LiCl to a final concentration of 2 M. Samples were precipitated after incubation at 4°C for 2 h. ssRNA pellets were washed in 75% ethanol and dissolved in RNase-free water. First-strand cDNA synthesis was conducted with the GoScript reverse transcription system (Promega, Madison, WI) using virus strand-specific primers (FsRT Fw and FsRT Rv for minus and plus strands, respectively) and oligo(dT)18 primer, using 2 μg of ssRNA.
Viral dsRNA semiquantification.
Amounts of 3 μg of total RNA from all virus-infected mutants were loaded into 1% agarose gels for analysis of viral dsRNA accumulation. After separation on the agarose gels, ethidium bromide-stained gels were visualized in a UV transilluminator. Gel images were measured using ImageJ software to determine the relative band intensities of viral dsRNA. The intensities of viral dsRNA bands were normalized to that of 18S rRNA in the samples.
Real-time RT-PCR analysis.
Real-time reverse transcription-quantitative PCR (qRT-PCR) was performed with a Bio-Rad CFX384 real-time PCR system using gene-specific internal primers. Each reaction mixture (10 μl) consisted of 25 ng of total cDNA, 5 μl of 2× iQSYBR green supermix (Bio-Rad, Hercules, CA), and 10 pmol of each primer. The thermal profile was as follows: 3 min at 95°C and 40 cycles of 10 s at 95°C and 30 s at 60°C, with melting curve data obtained by increasing the temperature from 55 to 95°C. Two endogenous reference genes, UBH, encoding ubiquitin C-terminal hydrolase (RefSeq accession number FGSG_01231) (52), and EF1α, encoding elongation factor 1α (RefSeq accession number FGSG_08811) (48), were used as internal controls to normalize qRT-PCR results. Data were analyzed using Bio-Rad CFX Manager software, version 1.6.541.1028 (Bio-Rad, Hercules, CA). RNA samples were extracted from at least two independent, biologically replicated experiments, and each PCR product was evaluated in at least three independent experiments, including three technical replicates.
Small RNA library preparation and deep sequencing.
Total RNAs extracted from mycelia were used for small RNA preparation. At least three individual samples for each condition were subjected to total RNA extraction in order to minimize bias between samples. We generated four small RNA libraries, including F. graminearum PH-1 infected by FgV1, FgV2, FgV3, and FgV4, using the TruSeq small RNA sample prep kit version 2 (Illumina, San Diego, CA) and following the manufacturer's instructions. To purify small RNAs (fewer than 30 nt), polyacrylamide gel electrophoresis (PAGE) gel was used. The 3′ and 5′ adapters were added to the purified small RNAs, and cDNA was then synthesized by RT-PCR using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). A linear PCR step was used to amplify the DNA fragments. Libraries were multiplexed using 3′ PCR primers containing unique 6-nucleotide sequences. The prepared libraries were single-end sequenced with the HiSeq 2000 system in the National Instrumentation Center for Environmental Management (NICEM, Seoul, South Korea).
Identification of vsiRNAs by bioinformatics analysis.
Adapter sequences and poor-quality sequences from the raw data were trimmed using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). In addition, noncoding RNAs, including tRNA, rRNA, snRNA, and snoRNA, were deleted by using the BWA program with the following parameters (mismatch ≤ 2, gap open = 0, and e value ≤ 0.05) (53) against the RFAM database (http://www.sanger.ac.uk/software/Rfam). Only clean reads of small RNA sequences were used for further analysis. To identify vsiRNAs, small RNA sequences (18 to 24 nt) from each library were mapped on the corresponding virus genome using the BWA program.
Validation of small RNA sequencing results using stem-loop RT-PCR.
To confirm the data for vsiRNA accumulation in response to FgV1 infection, we selected vsiRNAs in the regions from nt 26 to 46 and nt 4304 to 4325 on the sense strand. The low-molecular-weight (LMW) and total-RNA samples were used for the stem-loop RT reaction. The LMW RNA sample was prepared using the mirVana miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer's protocol, and stem-loop pulsed RT and PCR amplification were conducted as previously described (54, 55). In brief, 500 ng of each LMW RNA sample was denatured at 65°C, and the pulsed RT reaction was carried out using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) with the following conditions: 30 min at 16°C, followed by 60 cycles of 30 s at 30°C, 30 s at 42°C, and 1 s at 50°C, and a final incubation at 85°C for 5 min; the reaction mixture was then held at 4°C. A 10-ng quantity of the diluted RT mixture was amplified with Ex Taq polymerase (TaKaRa Bio, Shiga, Japan) with the following PCR conditions: 5 min at 94°C, followed by 20 or 25 cycles at 94°C for 15 s and 60°C for 1 min; the reaction mixture was then held at 4°C. FgV1 vsiRNA-specific primers for RT and primer sets for PCR were used (primer information is available upon request). The final PCR products were visualized on a 4% agarose gel that was stained with ethidium bromide.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by grants from the National Research Foundation (grant no. NRF-2016R1D1A1B03936370), funded by the Ministry of Education, Science, and Technology (MEST), the Vegetable Breeding Research Center (grant no. 710011-3), through the Agriculture, Food and Rural Affairs Research Center Support Program of the Ministry of Agriculture, Food and Rural Affairs, and the Next-Generation BioGreen 21 Program (grant no. PJ013225), Rural Development Administration (RDA), Republic of Korea. W.K.C. was supported by a research fellowship from the Brain Korea 21 Plus Project.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01756-17.
REFERENCES
- 1.Ding S-W, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang C, Wu Z, Li Y, Wu J. 2015. Biogenesis, function, and applications of virus-derived small RNAs in plants. Front Microbiol 6:1237. doi: 10.3389/fmicb.2015.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee K, Campos H, Kolaczkowski B. 2013. Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol 30:627–641. doi: 10.1093/molbev/mss263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang S-S, Zhang Z, Liu Y. 2012. RNA interference pathways in fungi: mechanisms and functions. Annu Rev Microbiol 66:305–323. doi: 10.1146/annurev-micro-092611-150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutvagner G, Simard MJ. 2008. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 6.Nicolas FE, Torres-Martínez S, Ruiz-Vazquez RM. 2013. Loss and retention of RNA interference in fungi and parasites. PLoS Pathog 9:e1003089. doi: 10.1371/journal.ppat.1003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 8.Torres-Martínez S, Ruiz-Vázquez RM. 2016. RNAi pathways in Mucor: a tale of proteins, small RNAs and functional diversity. Fungal Genet Biol 90:44–52. doi: 10.1016/j.fgb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Hsueh Y-P, Li W, Floyd A, Skalsky R, Heitman J. 2010. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev 24:2566–2582. doi: 10.1101/gad.1970910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romano N, Macino G. 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol 6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 11.Shiu PK, Metzenberg RL. 2002. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics 161:1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cogoni C, Macino G. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 13.Catalanotto C, Pallotta M, ReFalo P, Sachs MS, Vayssie L, Macino G, Cogoni C. 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol 24:2536–2545. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catalanotto C, Azzalin G, Macino G, Cogoni C. 2000. Transcription: gene silencing in worms and fungi. Nature 404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- 15.Nuss DL. 2011. Mycoviruses, RNA silencing, and viral RNA recombination. Adv Virus Res 80:25–48. doi: 10.1016/B978-0-12-385987-7.00002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci U S A 104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond T, Andrewski M, Roossinck M, Keller N. 2008. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot Cell 7:350–357. doi: 10.1128/EC.00356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himeno M, Maejima K, Komatsu K, Ozeki J, Hashimoto M, Kagiwada S, Yamaji Y, Namba S. 2010. Significantly low level of small RNA accumulation derived from an encapsidated mycovirus with dsRNA genome. Virology 396:69–75. doi: 10.1016/j.virol.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Yaegashi H, Shimizu T, Ito T, Kanematsu S. 2016. Differential inductions of RNA silencing among encapsidated double-stranded RNA mycoviruses in the white root rot fungus Rosellinia necatrix. J Virol 90:5677–5692. doi: 10.1128/JVI.02951-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Segers GC, Sun Q, Deng F, Nuss DL. 2008. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J Virol 82:2613–2619. doi: 10.1128/JVI.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Q, Choi GH, Nuss DL. 2009. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc Natl Acad Sci U S A 106:17927–17932. doi: 10.1073/pnas.0907552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segers GC, van Wezel R, Zhang X, Hong Y, Nuss DL. 2006. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot Cell 5:896–904. doi: 10.1128/EC.00373-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goswami RS, Kistler HC. 2004. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol 5:515–525. doi: 10.1111/j.1364-3703.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 24.O'Donnell K, Ward TJ, Geiser DM, Kistler HC, Aoki T. 2004. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet Biol 41:600–623. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Bai G, Shaner G. 2004. Management and resistance in wheat and barley to Fusarium head blight. Annu Rev Phytopathol 42:135–161. doi: 10.1146/annurev.phyto.42.040803.140340. [DOI] [PubMed] [Google Scholar]
- 26.McMullen M, Jones R, Gallenberg D. 1997. Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Dis 81:1340–1348. doi: 10.1094/PDIS.1997.81.12.1340. [DOI] [PubMed] [Google Scholar]
- 27.Nakayashiki H. 2005. RNA silencing in fungi: mechanisms and applications. FEBS Lett 579:5950–5957. doi: 10.1016/j.febslet.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Gao Q, Huang M, Liu Y, Liu Z, Liu X, Ma Z. 2015. Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci Rep 5:12500. doi: 10.1038/srep12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Son H, Min K, Lee J, Raju NB, Lee Y-W. 2011. Meiotic silencing in the homothallic fungus Gibberella zeae. Fungal Biol 115:1290–1302. doi: 10.1016/j.funbio.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Son H, Park AR, Lim JY, Shin C, Lee Y-W. 2017. Genome-wide exonic small interference RNA-mediated gene silencing regulates sexual reproduction in the homothallic fungus Fusarium graminearum. PLoS Genet 13:e1006595. doi: 10.1371/journal.pgen.1006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Li P, Zhang J, Qiu D, Guo L. 2016. Generation of a high resolution map of sRNAs from Fusarium graminearum and analysis of responses to viral infection. Sci Rep 6:26151. doi: 10.1038/srep26151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K-M, Cho WK, Yu J, Son M, Choi H, Min K, Lee Y-W, Kim K-H. 2014. A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses. PLoS One 9:e100989. doi: 10.1371/journal.pone.0100989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campo S, Gilbert KB, Carrington JC. 2016. Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum. PLoS Pathog 12:e1005640. doi: 10.1371/journal.ppat.1005640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li P, Zhang H, Chen X, Qiu D, Guo L. 2015. Molecular characterization of a novel hypovirus from the plant pathogenic fungus Fusarium graminearum. Virology 481:151–160. doi: 10.1016/j.virol.2015.02.047. [DOI] [PubMed] [Google Scholar]
- 35.Kim H-K, Jo S-M, Kim G-Y, Kim D-W, Kim Y-K, Yun S-H. 2015. A large-scale functional analysis of putative target genes of mating-type loci provides insight into the regulation of sexual development of the cereal pathogen Fusarium graminearum. PLoS Genet 11:e1005486. doi: 10.1371/journal.pgen.1005486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X-B, Wu Q, Ito T, Cillo F, Li W- X, Chen X, Yu J-L, Ding S-W. 2010. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci U S A 107:484–489. doi: 10.1073/pnas.0904086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong W, Qian D, Sun R, Jiang L, Wang Y, Wei C, Zhang Z, Li Y. 2015. OsRDR6 plays role in host defense against double-stranded RNA virus, Rice dwarf phytoreovirus. Sci Rep 5:11324. doi: 10.1038/srep11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H-C, Chang S-S, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. 2009. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shiu PK, Raju NB, Zickler D, Metzenberg RL. 2001. Meiotic silencing by unpaired DNA. Cell 107:905–916. doi: 10.1016/S0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 40.Calo S, Nicolás FE, Vila A, Torres-Martínez S, Ruiz-Vázquez RM. 2012. Two distinct RNA-dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides. Mol Microbiol 83:379–394. doi: 10.1111/j.1365-2958.2011.07939.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang D-X, Spiering MJ, Nuss DL. 2014. Characterizing the roles of Cryphonectria parasitica RNA-dependent RNA polymerase-like genes in antiviral defense, viral recombination and transposon transcript accumulation. PLoS One 9:e108653. doi: 10.1371/journal.pone.0108653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres-Martínez S, Ruiz-Vázquez RM. 2017. The RNAi universe in fungi: a varied landscape of small RNAs and biological functions. Annu Rev Microbiol 71:371–391. doi: 10.1146/annurev-micro-090816-093352. [DOI] [PubMed] [Google Scholar]
- 43.Blevins T, Rajeswaran R, Aregger M, Borah BK, Schepetilnikov M, Baerlocher L, Farinelli L, Meins F, Hohn T, Pooggin MM. 2011. Massive production of small RNAs from a non-coding region of cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res 39:5003–5014. doi: 10.1093/nar/gkr119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donaire L, Wang Y, Gonzalez-Ibeas D, Mayer KF, Aranda MA, Llave C. 2009. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392:203–214. doi: 10.1016/j.virol.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Cao M, Du P, Wang X, Yu Y-Q, Qiu Y-H, Li W, Gal-On A, Zhou C, Li Y, Ding S-W. 2014. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc Natl Acad Sci U S A 111:14613–14618. doi: 10.1073/pnas.1407131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carbonell A, Carrington JC. 2015. Antiviral roles of plant ARGONAUTES. Curr Opin Plant Biol 27:111–117. doi: 10.1016/j.pbi.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiba S, Suzuki N. 2015. Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated virus. Proc Natl Acad Sci U S A 112:4911–4918. doi: 10.1073/pnas.1509151112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Son M, Lee K-M, Yu J, Kang M, Park JM, Kwon S-J, Kim K-H. 2013. The HEX1 gene of Fusarium graminearum is required for fungal asexual reproduction and pathogenesis and for efficient viral RNA accumulation of Fusarium graminearum virus 1. J Virol 87:10356–10367. doi: 10.1128/JVI.01026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu J-H, Hamari Z, Han K-H, Seo J-A, Reyes-Domínguez Y, Scazzocchio C. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Lee S, Son H, Lee J, Min K, Choi GJ, Kim J-C, Lee Y-W. 2011. Functional analyses of two acetyl coenzyme A synthetases in the ascomycete Gibberella zeae. Eukaryot Cell 10:1043–1052. doi: 10.1128/EC.05071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Y, Son H, Lee J, Min K, Choi GJ, Kim J-C, Lee Y-W. 2011. A putative transcription factor MYT1 is required for female fertility in the ascomycete Gibberella zeae. PLoS One 6:e25586. doi: 10.1371/journal.pone.0025586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim H-K, Yun S-H. 2011. Evaluation of potential reference genes for quantitative RT-PCR analysis in Fusarium graminearum under different culture conditions. Plant Pathol J 27:301–309. doi: 10.5423/PPJ.2011.27.4.301. [DOI] [Google Scholar]
- 53.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. 2007. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Serio F, Schöb H, Iglesias A, Tarina C, Bouldoires E, Meins F. 2001. Sense-and antisense-mediated gene silencing in tobacco is inhibited by the same viral suppressors and is associated with accumulation of small RNAs. Proc Natl Acad Sci U S A 98:6506–6510. doi: 10.1073/pnas.111423098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.