ABSTRACT
During immature capsid assembly in cells, human immunodeficiency virus type 1 (HIV-1) Gag co-opts a host RNA granule, forming a pathway of intracellular assembly intermediates containing host components, including two cellular facilitators of assembly, ABCE1 and DDX6. A similar assembly pathway has been observed for other primate lentiviruses. Here we asked whether feline immunodeficiency virus (FIV), a nonprimate lentivirus, also forms RNA granule-derived capsid assembly intermediates. First, we showed that the released FIV immature capsid and a large FIV Gag-containing intracellular complex are unstable during analysis, unlike for HIV-1. We identified harvest conditions, including in situ cross-linking, that overcame this problem, revealing a series of FIV Gag-containing complexes corresponding in size to HIV-1 assembly intermediates. Previously, we showed that assembly-defective HIV-1 Gag mutants are arrested at specific assembly intermediates; here we identified four assembly-defective FIV Gag mutants, including three not previously studied, and demonstrated that they appear to be arrested at the same intermediate as the cognate HIV-1 mutants. Further evidence that these FIV Gag-containing complexes correspond to assembly intermediates came from coimmunoprecipitations demonstrating that endogenous ABCE1 and the RNA granule protein DDX6 are associated with FIV Gag, as shown previously for HIV-1 Gag, but are not associated with a ribosomal protein, at steady state. Additionally, we showed that FIV Gag associates with another RNA granule protein, DCP2. Finally, we validated the FIV Gag-ABCE1 and FIV Gag-DCP2 interactions with proximity ligation assays demonstrating colocalization in situ. Together, these data support a model in which primate and nonprimate lentiviruses form intracellular capsid assembly intermediates derived from nontranslating host RNA granules.
IMPORTANCE Like HIV-1 Gag, FIV Gag assembles into immature capsids; however, it is not known whether FIV Gag progresses through a pathway of immature capsid assembly intermediates derived from host RNA granules, as shown for HIV-1 Gag. Here we showed that FIV Gag forms complexes that resemble HIV-1 capsid assembly intermediates in size and in their association with ABCE1 and DDX6, two host facilitators of HIV-1 immature capsid assembly that are found in HIV-1 assembly intermediates. Our studies also showed that known and novel assembly-defective FIV Gag mutants fail to progress past putative intermediates in a pattern resembling that observed for HIV-1 Gag mutants. Finally, we used imaging to demonstrate colocalization of FIV Gag with ABCE1 and with the RNA granule protein DCP2. Thus, we conclude that formation of assembly intermediates derived from host RNA granules is likely conserved between primate and nonprimate lentiviruses and could provide targets for future antiviral strategies.
KEYWORDS: ABCE1, DCP2, DDX6, FIV, Gag, HIV, immature capsid assembly
INTRODUCTION
Feline immunodeficiency virus (FIV) is one of the most common infectious diseases in domestic cats, with a prevalence in North America of ∼2% (1). The natural history of FIV infection follows a pattern similar to that of the human immunodeficiency virus type 1 (HIV-1), starting with an initial acute infection phase, followed by an asymptomatic phase of variable length, and a terminal phase that results in feline AIDS (reviewed in reference 1). A commercial FIV vaccine has been developed for cats, but it may not provide complete protection from circulating FIV strains (2, 3). In infected cats, when less aggressive management fails to prevent recurrent disease, antiretroviral chemotherapy can be initiated using antiretroviral drugs. The antiretroviral drug typically prescribed is the HIV-1 reverse transcriptase inhibitor zidovudine (AZT) (4). Although AZT can improve quality of life and extend life expectancy, resistance can develop quickly, and treatment can have serious side effects (reviewed in reference 5). However, because cats can live healthier lives with treatment, drugs that specifically and potently inhibit FIV replication could be of benefit. In addition, since FIV infection of cats can result in AIDS, it has been proposed that such infection could serve as a useful model system for studying new antiviral treatments and vaccines for HIV-1.
For FIV infection to be useful as a model system for the study of HIV-1 will require understanding how FIV is similar to and different from HIV-1, which has been defined for some stages of the two viral life cycles (reviewed in reference 6). FIV has some notable differences from HIV-1; for example, FIV encodes a dUTPase, which is not found in the primate lentiviruses, and encodes fewer accessory proteins than HIV-1. Additionally, FIV encodes OrfA, which may not have a direct HIV-1 ortholog. Here we were interested in addressing whether mechanisms involved in intracellular immature capsid assembly are conserved between HIV-1 and FIV. Like all exogenous retroviruses, HIV-1 and FIV encode three proteins that carry out essential steps common to all retroviral life cycles—Gag, Pol, and Env. Because Gag is the only viral protein required for immature capsid assembly, the domains of HIV-1 Gag required for this process have been studied extensively (reviewed in reference 7). The HIV-1 matrix domain (MA) confers plasma membrane (PM) targeting, and the HIV-1 capsid domain (CA), which contains an N-terminal and a C-terminal subdomain (CA-NTD and CA-CTD, respectively), provides important Gag-Gag contacts required for the immature capsid structure. The HIV-1 nucleocapsid domain (NC) mediates both specific interactions with viral genomic RNA and nonspecific RNA interactions, while the HIV-1 late domain (p6) recruits host factors important for budding of the virus from infected cells. FIV Gag encodes the same domains as HIV-1 Gag, and mutational studies have confirmed that the FIV Gag domains (MA, CA, NC, and late domain p2) function similarly to their HIV-1 counterparts (8–11). For example, the late domain of both viruses encodes a TSG101 binding motif to recruit TSG101 and ESCRT machinery to promote budding (11). Additionally, in both cases, full-length Gag in the immature virus undergoes maturation when the viral protease cleaves Gag into separate domains (MA, CA, p1, NC, and p2 for FIV; MA, CA, sp1, NC, sp2, and p6 for HIV-1) (12). Importantly, numerous assembly-defective mutants have been generated in structure-function analyses of HIV-1 Gag (reviewed in reference 13), but these have not been generated for FIV Gag. Thus, one goal of our study was to generate a diverse set of assembly-defective FIV Gag point mutants that could serve as tools for studying the mechanism of FIV Gag assembly.
Similarity between HIV-1 and FIV Gag proteins in sequence or structure could argue for shared intracellular mechanisms of assembly. At the level of overall amino acid sequence, conservation among different retroviral Gag proteins is low, with the major homology region of Gag being a notable exception (14–16). However, known atomic structures of CA subdomains from different retroviruses display a high degree of conservation (17–19), which raises the possibility of shared mechanisms for immature capsid assembly. This could include conservation of specific Gag-Gag interactions in immature capsids but also conservation of Gag-host interactions that may facilitate immature capsid assembly by common mechanisms within cells. To determine whether FIV and HIV-1 Gag assemble via similar mechanisms, here we asked whether intracellular events that occur during HIV-1 immature capsid assembly are conserved for FIV.
Studies have shown that HIV-1 immature capsid assembly proceeds via a sequential pathway of posttranslational assembly intermediates (20–22). At steady state, four major intermediates of increasing size can be identified in cells, starting with the first intermediate (∼10S) and followed by progression through the subsequent ∼80S, ∼150S, and ∼500S intermediates to the final completed immature capsid (∼750S) (20–22). Evidence that the complexes observed at steady state are bona fide assembly intermediates comes from pulse-chase studies showing that they are formed sequentially (20, 21) and mutational analyses showing that all assembly-defective HIV-1 Gag mutants studied to date are arrested at various steps in this assembly pathway, with each mutant accumulating only the intermediate at which the mutant is arrested and all preceding intermediates (22–25). Moreover, other components found in the released virus, such as the HIV-1 Vif protein, GagPol, and genomic RNA (gRNA), are also found in assembly intermediates (21, 22, 26).
Notably, host proteins have been shown to play a role in facilitating the HIV-1 capsid assembly pathway. Immunodepletion studies from cell extracts and dominant negative studies in cells established that the cellular enzyme ATP binding cassette protein E1 (ABCE1) facilitates HIV-1 immature capsid assembly (26). Although its exact mechanism of action has not been defined, endogenous ABCE1 is found associated with the ∼80S and larger assembly intermediates in human cells and is released before completion of the immature capsid (26). ABCE1 is conserved in archaebacteria and eukaryotes and is thought to be required for termination of translation and recycling of ribosomes (reviewed in reference 27). Another cellular protein that facilitates HIV-1 capsid assembly in cells is DEAD box helicase 6 (DDX6), which, like ABCE1, is associated with the ∼80S and larger HIV-1 assembly intermediates but is not in the final completed immature capsid (28). Knockdown of DDX6 in HIV-1-expressing cells resulted in the reduction of released immature capsids without affecting the steady-state Gag levels; moreover, the finding that virus production in DDX6-depleted cells could be rescued with wild-type (WT) DDX6, but not with an ATPase mutant of DDX6, established a role for the RNA helicase activity of DDX6 in promoting HIV-1 immature capsid assembly (28). In uninfected cells, DDX6 catalyzes remodeling of mRNAs as they transition between translation and a translationally silenced state (29). In keeping with our finding that ABCE1 and DDX6 are associated with Gag in immature capsid assembly intermediates and dissociate upon completion of the immature capsid (26, 28), a proteomics study found that ABCE1 and DDX6 are associated with full-length Gag in cells but are not present in the virus (30). Yet another cellular protein that may be associated with HIV-1 assembly intermediates is mRNA decapping enzyme 2 (DCP2). DCP2 is a component of the mRNA decapping complex and plays a role in the 5′-to-3′ RNA degradation pathway but catalyzes cleavage of the mRNA cap structure only when associated with activating factors (31). Previously, yellow fluorescent protein (YFP)-tagged DCP2 was found associated with both full-length HIV Gag and ABCE1 in COS-1 cells, raising the possibility that endogenous DCP2 may also be a component of the HIV-1 assembly intermediates (28).
Interestingly, DDX6 and DCP2 are well-studied enzymatic markers of cellular RNA granules, which are host complexes involved in the regulation and metabolism of RNA in the cytoplasm (reviewed in references 29 and 32). Large RNA granules that are easily visible by light microscopy are well studied; these include P bodies, stress granules, and neuronal granules (reviewed in reference 33). However, RNA granules are likely much more diverse in size. Indeed, when microscopically visible P bodies are disrupted, their functions are not lost, arguing that the resulting small RNA granules that are not easily visualized by light microscopy are functional (34, 35). DDX6 and DCP2 are found in P bodies; additionally, DDX6 appears to be in smaller foci that likely correspond to small RNA granules (28; B. C. Barajas, J. C. Reed, J. R. Lingappa, unpublished observations). Although ABCE1 was not described previously as an RNA granule protein, immunoprecipitation (IP) studies have shown that endogenous ABCE1 and DDX6 are associated in the absence of assembling Gag (28), suggesting that ABCE1 is also found in some DDX6-containing RNA granules. Studies to date support a model in which HIV-1 Gag co-opts a class of small RNA granules containing at least DDX6, Argonaute 2 (AGO2), and ABCE1 during assembly, thereby forming the ∼80S cytoplasmic assembly intermediate (22, 28). Gag then appears to target, along with ABCE1, DDX6, and AGO2 (and possibly the RNA granule protein DCP2), to the PM (22, 28), where Gag multimerizes. ABCE1, DDX6, and possibly other associated RNA granule proteins dissociate upon completion of the immature capsid, before virus budding and release (26, 28). Interestingly, the RNA granule proteins DDX6 and DCP2 have been shown to play a role in assembly and/or genome packaging of other retroviruses and even retroelements. Specifically, both DCP2 and the Saccharomyces cerevisiae DDX6 homolog, Dhh1, colocalize with assembling Gag of the Ty3 retrotransposon, an S. cerevisiae retroelement that is distantly related to HIV-1 (36). Moreover, DCP2 and the yeast DDX6 homolog are required for efficient Ty3 retrotransposition (36–38). Lastly, in cells infected with primate foamy virus, DDX6 helicase activity promotes the encapsidation of human foamy virus genomic RNA (39). Here we were interested in determining whether FIV, like HIV-1, co-opts small RNA granules during assembly. Such a finding would support the utility of the FIV animal model for validating small molecules that might inhibit FIV and HIV-1 assembly, as well as the assembly of other retroviruses, by interfering with a shared assembly pathway.
Other primate lentiviruses (e.g., HIV-2 and both the macaque isolate 239 and African green monkey isolate of simian immunodeficiency virus) form similar intermediates that are associated with ABCE1 (40), suggesting that co-opting a host RNA granule at an early stage of immature capsid assembly is a conserved feature among primate lentiviruses. To date, a role for ABCE1-containing RNA granules in assembly of other retroviruses, besides primate lentiviruses, has not been shown. The structural conservation of lentiviral Gags described above led us to ask whether nonprimate lentiviruses also co-opt host RNA granules to form immature capsid assembly intermediates. Here we showed that FIV WT Gag forms complexes in feline cells that are similar in size to the assembly intermediates formed by HIV-1. To confirm that these complexes behave like assembly intermediates, we first generated FIV Gag mutants that correspond to known assembly-defective HIV-1 Gag mutants. We then showed that these FIV Gag mutants are assembly defective and appear to be arrested at the same assembly intermediates as the corresponding HIV-1 Gag mutants. Additionally, we demonstrated that FIV Gag is associated with endogenous ABCE1 and DDX6 by coimmunoprecipitation (coIP) in feline cells, as would be expected if FIV Gag forms RNA granule-derived assembly intermediates similar to those formed by HIV-1. Moreover, we showed that endogenous ABCE1 colocalizes with assembly-competent FIV Gag, but not assembly-incompetent FIV Gag, in situ by using the proximity ligation assay (PLA). Lastly, we tested the hypothesis that FIV Gag is also associated with yet another protein found in RNA granules, DCP2. We demonstrated by coIP that both FIV WT Gag and an assembly-defective but oligomerization-competent FIV Gag mutant associate with endogenous DCP2 in feline cells, while an assembly-incompetent FIV Gag mutant does not; moreover, in situ colocalization of FIV Gag with endogenous DCP2 was confirmed by PLA. Based on these findings, we propose that FIV Gag forms complexes that likely correspond to RNA granule-derived intracellular assembly intermediates and that the assembly pathway defined previously for primate lentiviruses is conserved in a nonprimate lentivirus. Given the identification of small molecules that inhibit rabies virus replication by targeting ABCE1-containing assembly intermediates (41), the conservation of such intermediates has important implications for the development of new antiretroviral compounds.
(This article was submitted to an online preprint archive [42].)
RESULTS
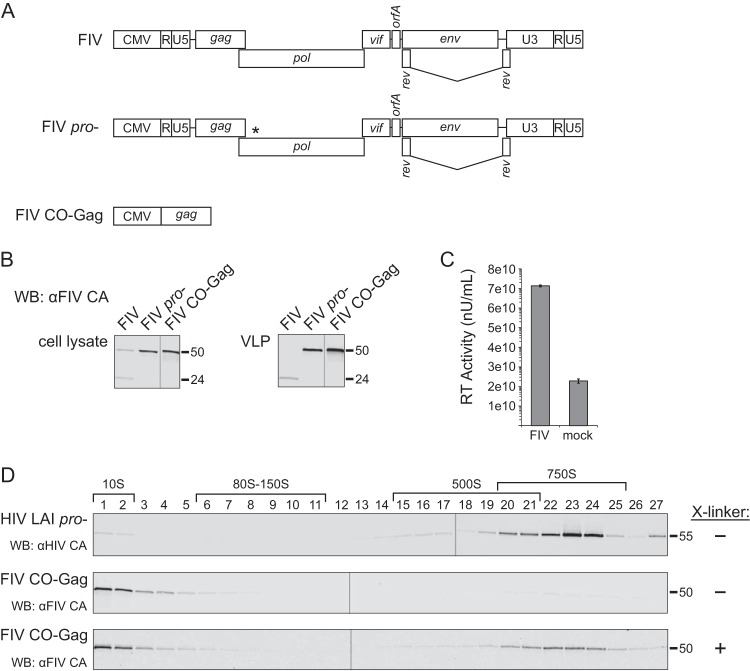
To study FIV immature capsid assembly, we generated three FIV Gag-expressing constructs (Fig. 1A). To generate the first construct (termed FIV), we introduced two previously described modifications into the FIV 34TF10 proviral clone (43): specifically, we restored OrfA expression (44, 45) and replaced the native FIV promoter with the cytomegalovirus (CMV) promoter to allow for expression in both feline and human cells (46). The second construct is an FIV proviral clone that contains the two modifications described above as well as an inactivating mutation in the viral protease gene (termed FIV pro−). This mutation was generated because preventing the protease-mediated cleavage of Gag following immature capsid assembly allows Gag to remain full length, which makes tracking and quantification by SDS-PAGE easier (Fig. 1A) (47). Finally, since Gag alone should be able to make virus-like particles (VLPs), we also codon-optimized FIV 34TF10 Gag and cloned it into an expression vector to make the third construct (termed FIV CO-Gag), which expresses FIV Gag in the absence of other viral proteins (Fig. 1A). To validate these three constructs, we transfected the feline astrocyte cell line G355-5, as described previously (48), and analyzed lysates of these cells for both steady-state expression of Gag and for VLP production by Western blotting (WB) with an antibody directed against FIV CA (αFIV CA). Transfection of the proviral constructs, FIV and FIV pro−, resulted in intracellular FIV Gag expression and production of either mature virus containing cleaved CA (∼24 kDa) or immature VLPs containing uncleaved Gag (∼50 kDa), respectively (Fig. 1B), both detected by αFIV CA. Expression of FIV CO-Gag alone resulted in high steady-state levels of Gag, as expected, and release of immature VLPs (Fig. 1B). To confirm the integrity of our infectious FIV construct, we demonstrated that virus released from G355-5 cells transfected with this construct displayed abundant reverse transcriptase (RT) activity (Fig. 1C). Moreover, when used to infect G355-5 cells, this virus stock caused a spreading infection, as indicated by increased reverse transcriptase activity and syncytium formation over time (D. T. B. Ressler and J. C. Reed, unpublished observations). To confirm that the noninfectious constructs assembled VLPs properly, we used equilibrium centrifugation to determine the density of immature VLPs produced from cells transfected with these constructs. In our experiments, the average density of FIV immature VLPs (with envelopes intact) ranged from 1.12 to 1.16 g/ml (N. Westergreen and J. C. Reed, unpublished observations), which is close to the published FIV VLP density estimate of 1.15 g/ml (49). Thus, measurements of RT activity and buoyant densities showed that all three of our constructs produced properly formed FIV VLPs in feline cells.
FIG 1.
FIV immature capsids are unstable when de-enveloped but stabilized by cross-linking. (A) Diagrams depicting gene maps of the FIV proviral construct (FIV), FIV proviral construct containing an inactivated protease gene (FIV pro−), and codon-optimized FIV Gag alone (FIV CO-Gag). (B) Each construct was expressed in feline G355-5 cells. Cell lysates and pelleted virus-like particles (VLPs) from medium were analyzed by WB with an antibody to FIV CA (αFIV CA). (C) Medium was collected from G355-5 cells transfected with FIV or mock transfected and examined for reverse transcriptase activity by an SG-PERT assay. Standards processed in parallel were used to determine reverse transcriptase activity in nU/ml. (D) Pelleted VLPs produced either by COS-1 cells transfected with HIV-1 LAI pro− or G355-5 cells transfected with FIV CO-Gag were de-enveloped and subjected to velocity sedimentation, followed by WB with an antibody to HIV-1 CA (αHIV CA) or with αFIV CA. Symbols (+ or −) on the right indicate whether VLPs were treated with cross-linker (X-linker) prior to removal of the viral envelope. Approximate S values are indicated with brackets above the WB of gradient fractions, and molecular size markers (55 kDa, 50 kDa, and 24 kDa) are shown to the right of blots. Data shown are from a single experiment and are representative of two independent repeats of the experiment.
Based on studies of HIV-1 assembly intermediates, we expect FIV assembly intermediates to be smaller than completed FIV immature capsids. For this reason, we first defined the migration of completed FIV immature capsids in velocity sedimentation gradients used previously to separate HIV-1 assembly intermediates (24). Immature VLPs released from cells expressing FIV CO-Gag or HIV-1 LAI pro− provirus were isolated from the culture medium, de-enveloped with nonionic detergent to release the immature capsid, and analyzed by velocity sedimentation. The HIV-1 immature capsids generated in this manner migrated at ∼750S in the velocity sedimentation gradient, as described previously (20, 24) (Fig. 1D, upper panel). However, when the FIV immature capsids generated in this manner were analyzed in parallel, FIV Gag was found only in the soluble (∼10S) region of the gradient (Fig. 1D, middle panel). Given that VLPs of the correct density were observed when envelopes were intact, as described above, the lack of intact immature capsids following de-envelopment suggested that Gag in FIV immature capsids dissociates following removal of the viral envelope. Consistent with this hypothesis, immature capsids produced by de-envelopment of VLPs from cells expressing the FIV pro− proviral clone were also unstable (N. Westergreen and J. C. Reed, unpublished observations). To examine whether FIV immature capsids are intrinsically less stable than HIV-1 immature capsids, we cross-linked FIV immature capsids with dithiobis(succinimidyl propionate) (DSP) to preserve their integrity prior to removal of the viral envelope. When velocity sedimentation was performed after cross-linking, de-enveloped FIV immature capsids migrated at ∼750S (Fig. 1D, lower panel). Thus, FIV immature capsids appear to have a sedimentation value (S value) similar to that of HIV-1 immature capsids but are more labile, requiring cross-linking for stability after de-envelopment.
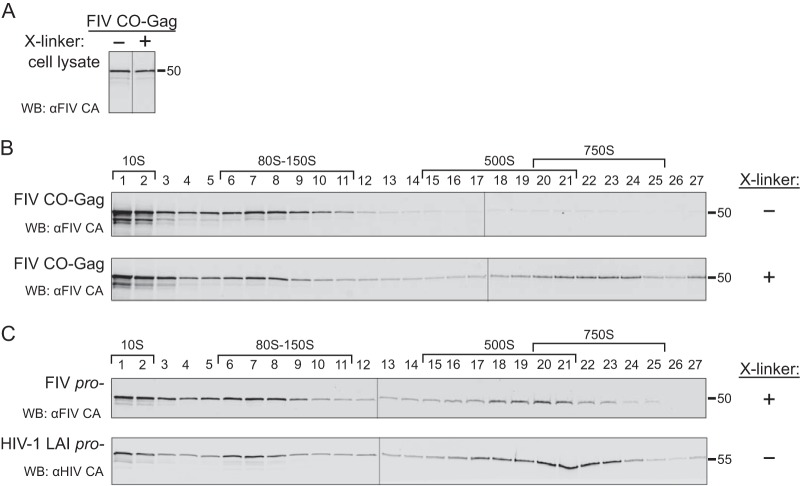
Having established the S value of the completed FIV immature capsid, we next asked whether we could identify putative intracellular FIV assembly intermediates, which should be present transiently in small quantities and would likely have S values similar to those of intracellular HIV-1 assembly intermediates, i.e., ∼10S, ∼80S, ∼150S, and ∼500S (21, 22, 26). Given the instability of the completed immature capsid, we hypothesized that one or more of the putative FIV assembly intermediates might also be unstable. To test this hypothesis, feline G355-5 cells were transfected to express FIV CO-Gag and prior to harvest were either mock treated or treated with DSP to cross-link putative FIV assembly intermediates. Harvested lysates were subjected to velocity sedimentation and analyzed by WB for FIV Gag. The velocity sedimentation conditions used here are expected to optimally separate ∼10S, ∼80S, and ∼500S complexes; under these conditions, peaks in the ∼80S and ∼150S regions comigrate and the ∼500S and ∼750S regions overlap. Intracellular steady-state Gag levels in cell lysates from mock-treated and DSP cross-linked cells were comparable (Fig. 2A). In mock-treated cell lysates, FIV Gag was found in complexes corresponding to the HIV-1 ∼10S and ∼80S assembly intermediates, with only faint FIV Gag bands observed in the ∼500S region (Fig. 2B, upper panel). In contrast, the DSP cross-linked cell lysates contained abundant FIV Gag in complexes corresponding to the HIV-1 ∼10S, ∼80S, and ∼500S assembly intermediates (Fig. 2B, lower panel). Taken together, these data suggest that FIV forms immature capsid assembly intermediates that are similar in size to previously described HIV-1 assembly intermediates but with the larger ∼500S FIV intermediate being less stable than its HIV-1 counterpart, as is the case with the released FIV immature capsid.
FIG 2.
FIV produces intracellular intermediates that are similar in size to HIV-1 assembly intermediates. (A) G355-5 cells transfected with FIV CO-Gag were either mock treated or treated with cross-linker (X-linker; 0.1 mM DSP) prior to harvest in a standard salt buffer. WB was performed with an antibody to FIV CA (αFIV CA), and bands were taken from a single exposure of film. (B) Cell lysates analyzed by WB in panel A were subjected to velocity sedimentation in parallel, and gradient fractions were analyzed by WB with αFIV CA. (C) G355-5 cells transfected with FIV pro− were cross-linked with 0.1 mM DSP and harvested in a standard salt buffer. Cell lysate was subjected to velocity sedimentation, and gradient fractions were analyzed by WB with αFIV CA. Separately, COS-1 cells transfected with HIV-1 LAI pro− were harvested in a standard salt buffer without cross-linking. Cell lysate was subjected to velocity sedimentation, and gradient fractions were analyzed by WB with an antibody to HIV Gag CA (αHIV CA). Symbols (+ or −) on the right indicate whether cells were treated with cross-linker (X-linker) prior to harvest. Approximate S values are indicated with brackets above the WB of gradient fractions, and expected migrations of full-length HIV-1 Gag (55 kDa) or FIV Gag (50 kDa) are shown on the right of the blots. Data shown are from a single experiment and are representative of two independent repeats of the experiment.
To determine if putative FIV assembly intermediates could also be detected in cells transfected to express an FIV provirus, we analyzed cross-linked G355-5 cells transfected to express FIV pro−, using velocity sedimentation and WB of gradient fractions for FIV Gag (Fig. 2C, upper panel). As with FIV CO-Gag, the ∼10S, ∼80S, and ∼500S putative FIV assembly intermediates were detected. In the case of FIV pro−, the putative ∼10S and ∼80S intermediates and the ∼500S completed immature capsids were abundant in the cross-linked lysates, with more of the late ∼500S putative assembly intermediate and less of the completed ∼750S capsid relative to that of FIV CO-Gag (Fig. 2B, lower panel). The difference in the amount of intracellular ∼750S completed immature capsids present at steady state could be explained by budding kinetics that may differ with constructs and cell types, as has been observed for HIV-1 (22, 25, 28). As a positive control, COS-1 cells transfected to express HIV-1 LAI pro− were also analyzed by velocity sedimentation without prior cross-linking. WB of gradient fractions for HIV-1 Gag revealed the ∼10S, ∼80S, and ∼500S HIV-1 assembly intermediates, as expected (Fig. 2C, lower panel). Moreover, a comparison of the results for HIV-1-expressing lysates confirmed that FIV Gag forms complexes that are similar in size to previously defined HIV-1 assembly intermediates, with the late ∼500S FIV complex being less stable than the corresponding HIV-1 late assembly intermediate.
Previously, rigorous pulse-chase studies demonstrated that HIV-1 Gag-containing complexes identified in cell lysates are part of a pathway of sequentially formed assembly intermediates; moreover, HIV-1 Gag mutants that are assembly defective are arrested at different steps of the assembly pathway and accumulate only the intermediate at which the mutant is arrested and all preceding intermediates, further confirming the sequential progression of Gag through the pathway (Fig. 3A) (21, 22, 24, 25). Thus, to further test the hypothesis that the FIV Gag-containing complexes that we identified in cells (Fig. 2B) are assembly intermediates, we generated five FIV Gag mutants that are predicted to be either budding defective or assembly defective and determined the pattern of FIV Gag complexes produced in cells transfected with each of these mutants. If key assembly-defective FIV Gag mutants are arrested at the same complex as the corresponding assembly-defective HIV-1 Gag mutant and form only the FIV Gag-containing complexes that precede the point of arrest, then FIV Gag-containing complexes are likely part of a pathway of sequential assembly intermediates analogous to the HIV-1 assembly pathway.
FIG 3.
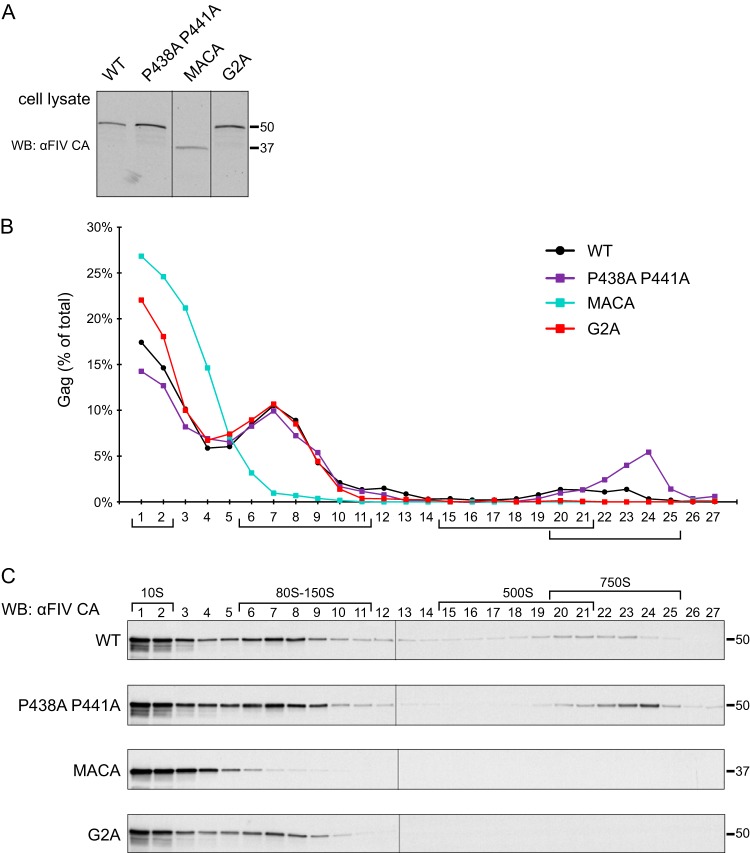
Defining VLP phenotypes of FIV Gag mutants. (A) Summary of results from a previous study demonstrating that HIV-1 Gag mutants are arrested in the assembly pathway (22). A diagram of the HIV-1 assembly pathway is shown at the top of the figure. HIV Gag mutants are listed below, with bars spanning all the complexes that are formed by each Gag mutant. The legend (boxed) shows colors and symbols representing HIV-1 Gag, genomic RNA (gRNA), DDX6, ABCE1, other granule proteins, and the plasma membrane. Point mutations are numbered relative to the start of HIV-1 Gag. (B) Diagram of WT FIV CO-Gag and FIV CO-Gag mutants used in this study, with point mutations indicated by red asterisks. Dark gray, MA; dark blue, CA-CTD; blue, CA-NTD; light blue, NC; light gray, spacer peptide (p1) and late domain (p2). FIV viral protease cleavage sites are diagrammed as vertical black lines, and the names of Gag domains are indicated above the diagrams. Mutated residues are numbered relative to the start of Gag. (C) Equivalent aliquots of cell lysates and pelleted virus-like particles (VLPs) from G355-5 cells transfected with WT FIV CO-Gag or the indicated FIV CO-Gag mutants were analyzed by WB with an antibody to FIV CA (αFIV CA). Expected migrations for full-length FIV Gag (50 kDa) and FIV Gag MACA (37 kDa) proteins are shown on the right. Data shown are from a single experiment and are representative of two independent repeats of the experiment.
To generate a budding-defective mutant (FIV CO-Gag P438A P441A), we altered the TSG101 binding motif in FIV p2 from PSAP to ASAA, which is known to reduce the production of FIV VLPs (Fig. 3B) (11). Given that the analogous PTAP mutation in HIV-1 inhibits virus budding but not Gag multimerization (50, 51), we expected FIV CO-Gag P438A P441A to be assembly competent and therefore form all the assembly intermediates but subsequently fail to complete virus budding and release. We also generated two FIV Gag mutants that are not expected to form immature capsids. One is truncated at the end of CA, resulting in expression of only MA and CA (FIV CO-Gag MACA) (Fig. 3B). We expected that like HIV-1 MACA (Fig. 3A) (22), FIV CO-Gag MACA will fail to produce VLPs, will be arrested as the ∼10S assembly intermediate, and will thereby serve as an assembly-incompetent mutant that marks the first step in the assembly pathway. Moreover, a similar FIV Gag truncation mutant, FIV MA-CA-p1, does not produce VLPs (9), further suggesting that FIV CO-Gag MACA is likely to be assembly defective. In the second FIV Gag mutant, the glycine at position 2 of FIV Gag was replaced with an alanine to generate FIV CO-Gag G2A (Fig. 3B); this mutation is known to reduce VLP production (8). The analogous HIV-1 Gag G2A mutant is competent for oligomerization (52) but fails to undergo myristoylation and subsequent targeting to the PM site of assembly and is arrested as a cytosolic ∼80S assembly intermediate (Fig. 3B) (22). Hence, we expected the assembly-defective but oligomerization-competent FIV CO-Gag G2A mutant to be arrested at the ∼80S assembly intermediate. To verify that these three constructs are expressed in cells and display the expected VLP phenotypes, we examined steady-state intracellular Gag levels as well as VLP production (Fig. 3C). The budding mutant, FIV CO-Gag P438A P441A, was expressed at a level comparable to that of FIV WT CO-Gag and displayed a 3-fold reduction in VLP release relative to the WT (Fig. 3C). Although this budding defect was not as profound as that observed previously (11), this may reflect differences in cell lines (53). For the two other FIV Gag mutants described thus far (G2A and MACA), almost no VLPs were released from transfected cells, despite intracellular Gag levels similar to those of FIV WT Gag (Fig. 3C), confirming that both are assembly defective, as expected.
Having established that these three FIV Gag mutants (P438A P441A, G2A, and MACA) display the expected VLP phenotypes, we expressed them in cells at similar steady-state levels (Fig. 4A) and asked whether they are arrested in the assembly pathway in a manner similar to the comparable HIV-1 Gag mutants (Fig. 4B and C). Velocity sedimentation analysis of cell lysates (harvested using the DSP cross-linking conditions described above) revealed that FIV CO-Gag produces the expected pattern of putative intracellular assembly intermediates, displaying a prominent ∼80S complex and a less prominent ∼500S complex, as well as the ∼750S completed immature capsid. As expected, the budding-defective P438A P441A mutant produced all the putative assembly intermediates formed by FIV CO-Gag, consistent with immature capsid assembly being unaffected by mutation of the TSG101 binding motif that is important for budding but not assembly. Interestingly, the budding-defective P438A P441A mutant accumulated more ∼750S completed immature capsids than WT FIV CO-Gag, consistent with the completely assembled immature capsids of the P438A P441A mutant being arrested at the PM before undergoing release. In contrast, FIV CO-Gag MACA produced only the ∼10S complex, and FIV CO-Gag G2A produced only the ∼10S and ∼80S complexes (Fig. 4B and C). Thus, the assembly-incompetent FIV CO-Gag MACA mutant and assembly-defective FIV CO-Gag G2A mutant were each arrested at the same Gag-containing complex as their HIV-1 counterparts (Fig. 3A) (22). Taken together, these data further support the model that intracellular FIV Gag assembles via a stepwise pathway of assembly intermediates, analogous to those described previously for HIV-1, and that these assembly intermediates can be identified at steady-state in cell lysates.
FIG 4.
Assembly-defective FIV Gag MACA and FIV Gag G2A appear to be arrested at putative assembly intermediates. (A) G355-5 cells transfected with FIV CO-Gag or the indicated constructs were treated with cross-linker (0.1 mM DSP) and harvested in a standard salt buffer. Equivalent aliquots of cell lysate were analyzed for steady-state FIV Gag expression by WB with an antibody to FIV CA (αFIV CA). (B and C) Lysates shown in panel A were also subjected to velocity sedimentation, and gradient fractions were analyzed by WB with αFIV CA. A graph displays quantification of the WB results from panel C, with the amount of Gag in each fraction shown as a percentage of total Gag in the gradient. The approximate S values of FIV Gag-containing complexes are indicated with brackets below the graph and above the gradient WB, and expected migrations for full-length FIV Gag (50 kDa) and FIV Gag MACA (37 kDa) proteins are shown on the right. Data shown are from a single experiment and are representative of two independent repeats of the experiment.
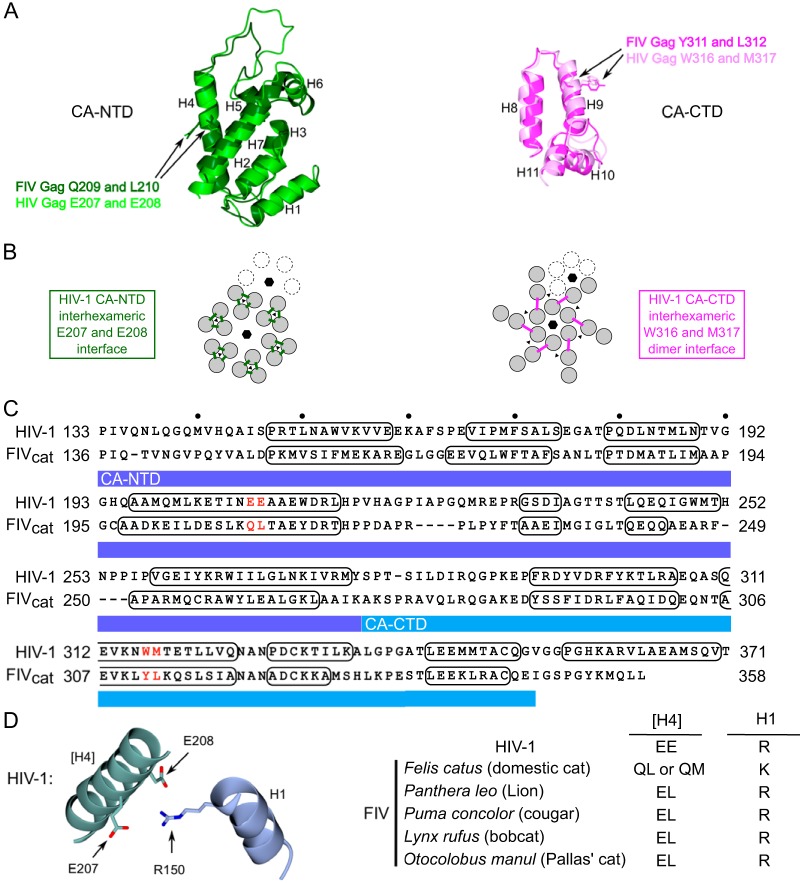
Given the important role of the CA domain in HIV-1 immature capsid assembly, we also wanted to identify point mutations in FIV CA that result in assembly defects and determine whether they also cause arrest of FIV Gag in the putative assembly pathway. While point mutations in CA-NTD and CA-CTD that impair HIV-1 immature capsid assembly have been defined previously (16, 24, 54–58), they have not yet been identified for FIV. Such FIV point mutants would allow us to compare arrest of FIV and HIV assembly-defective mutants, thereby providing additional insights into assembly mechanisms. Since high-resolution structures of the HIV-1 CA-NTD and CA-CTD are available, we sought to identify FIV residues that correspond to known HIV-1 assembly-critical residues by aligning the structure of the FIV CA subdomains with the structure of the HIV-1 CA subdomains (Fig. 5A). For CA-CTD, we aligned the published FIV CA-CTD structure (PDB accession number 5DCK [18]) with the HIV-1 CA-CTD domain from a published structure of HIV-1 CA (PDB accession number 5L93 [59]) (Fig. 5A). However, because a high-resolution structure of FIV CA-NTD had not been reported at the time we were doing our studies, we generated a model of the FIV CA-NTD structure, using SWISS-MODEL, with the published RELIK CA-NTD structure as a template (PDB accession number 2XGU [19]). The RELIK structure was selected as a template because it had the best predicted fit of relevant structures in the SWISS-MODEL ranking (see Materials and Methods for details). The FIV CA-NTD model generated using RELIK CA-NTD as a template was then aligned with the published HIV-1 CA-NTD structure (PDB accession number 5L93 [59]) (Fig. 5A).
FIG 5.
Structural homology modeling allows identification of candidate CA residues critical for FIV capsid assembly. (A) On the left is a superimposition of an HIV-1 CA-NTD structure (light green; PDB accession number 5L93 [59]) and a homology-modeled structure of FIV CA-NTD (dark green) generated using RELIK CA-NTD as a template (PDB accession number 2XGU [19]). On the right is a superimposition of an HIV-1 CA-CTD structure (light pink; PDB accession number 5L93) and an FIV CA-CTD structure (dark pink; PDB accession number 5DCK [18]). The side chains of FIV residues that were selected for mutation are indicated with arrows and are shown in the same color as the main chain, as are their HIV-1 counterparts. CA alpha-helices 1 to 11 are numbered (H1 to H11). (B) On the left is a diagram of the HIV-1 immature capsid structure depicting the 6-fold and 3-fold points of symmetry in the CA-NTD layer (indicated by black hexagons and black triangles, respectively). On the right is a diagram of the structure of the CA-CTD layer. Dotted subunits indicate an adjacent hexamer. The HIV-1 CA-NTD interhexameric interface analyzed in this study is shown by green lines but is shown only for the central hexamer. The HIV-1 CA-CTD interhexameric interface analyzed in this study is shown by pink lines but is shown only for the central hexamer. Interfaces were identified in a high-resolution structure of the completed immature capsid (59). (C) Shown is an amino acid sequence alignment of the HIV-1 CA and FIV CA domains based on a structural alignment of CA-NTD and CA-CTD from sources described for panel A above. Numbering on the right and left of each line is from the start of Gag. Residues forming alpha-helical structures are outlined in black. CA-NTD and CA-CTD are highlighted with dark blue and light blue bars, respectively. Red letters indicate residues chosen for mutation (FIV Gag Q209A and L210A and FIV Gag Y311A and L312A). (D) Diagram of the residues proposed to form a salt bridge in the HIV-1 immature capsid structure (HIV-1 Gag R150 in H1 with HIV-1 Gag E207 E208 in H4), generated using PDB accession number 5L93. Brackets indicate helix in neighboring 3-fold symmetry mate. The table lists consensus residues at these positions in helix 4 and helix 1 for HIV-1, FIV from Felis catus (domestic cat), FIV from Panthera leo (lion), FIV from Puma concolor (cougar), FIV from Lynx rufus (bobcat), and FIV from Otocolobus manul (Pallas' cat).
After aligning the FIV CA subdomain structures on an HIV-1 CA structure (Fig. 5A), we next asked which residues in the FIV structures most closely correspond to assembly-critical HIV CA residues. In CA-NTD, residues in helix 4 were of particular interest because helix 4 forms an exposed surface (57) and is critical for interhexameric CA-NTD contacts in a high-resolution cryoelectron microscopy (cryoEM) structure (Fig. 5B) (59). In keeping with this, the HIV-1 Gag helix 4 E207A E208A mutant (Fig. 5C) displays reduced VLP production (57) but forms all the assembly intermediates, including the late ∼500S assembly intermediate (22), indicating arrest just prior to formation of the ∼750S completed capsid (Fig. 3A). Moreover, a recent cryoEM structure of HIV-1 CA suggests that E207 and E208 in CA-NTD helix 4 may participate in a salt bridge with R150 in CA-NTD helix 1 of a neighboring 3-fold symmetry mate (59) (Fig. 5D). This interaction may contribute to stabilizing the immature HIV-1 capsid; thus, it is possible that the observed arrest of the E207 E208 mutant in the assembly pathway is explained by disruption of this salt bridge. CA-NTD helix 4 is also important for FIV immature capsid assembly since its deletion abrogated VLP production (9). Thus, we chose to mutate the FIV residues Q209 and L210 to alanine based on their alignment with E207 and E208 in the superimposed HIV-1 and FIV CA-NTD structure (Fig. 5A and C). Additionally, the side chains of Q209 and L210 residues had an orientation similar to the corresponding E207 and E208 residues in the HIV-1 CA structure (Fig. 5A), further supporting the selection of Q209 and L210 for mutation. Consistent with their similar alignment and orientation, QL (or QM) residues are highly conserved at this position in FIV variants from domestic cats (QL is present in 312 out of 322 FIV Gag sequences from domestic cats deposited into NCBI) (Fig. 5D; see Table S1 in the supplemental material). Notably, as described in Discussion, the finding that Q209 in FIV CA-NTD of domestic cats corresponds to E209 in HIV-1 CA-NTD based on alignment and orientation raises some interesting structural issues, since unlike glutamic acid, glutamine would not be expected to form a salt bridge in FIV Gag due to its lack of a negative charge.
To test whether the FIV Q209 and L210 residues are analogous to the assembly-critical HIV-1 E207 and E208 residues, we introduced Q209A and L210A mutations into FIV CO-Gag and examined the effect on VLP production and progression through the assembly pathway. We found that VLP production by FIV CO-Gag Q209A L210A was dramatically reduced despite detectable intracellular Gag (Fig. 3C), thus recapitulating the VLP defect observed for the corresponding HIV-1 mutant (22, 57). When expressed in cells at similar intracellular levels as FIV WT CO-Gag (Fig. 6A), FIV CO-Gag Q209A L210A formed the ∼10S, ∼80S, and ∼500S complexes but little to no ∼750S completed immature capsid, as indicated by velocity sedimentation analysis (Fig. 6B and C). From these data, we conclude that FIV CO-Gag Q209A L210A is arrested at the ∼500S assembly intermediate, since it forms the ∼500S intermediate (Fig. 6C) but fails to complete assembly and undergo release (Fig. 3C), as observed for the corresponding HIV-1 E207A E208A mutant (Fig. 3A) (22).
FIG 6.
Assembly-defective FIV Gag CA-NTD and CA-CTD mutants appear to be arrested at putative assembly intermediates. (A) G355-5 cells transfected with FIV CO-Gag or the indicated constructs were treated with cross-linker (0.1 mM DSP) and harvested in a standard salt buffer. Equivalent aliquots of cell lysate were analyzed for steady-state FIV Gag expression by WB with an antibody to FIV CA (αFIV CA). (B and C) Lysates shown in panel A were also subjected to velocity sedimentation, and gradient fractions were analyzed by WB with αFIV CA. A graph displays quantification of the WB results from panel C, with the amount of Gag in each fraction shown as a percentage of total Gag in the gradient. The approximate S values of FIV Gag-containing complexes are indicated with brackets below the graph and above the gradient WB, and the expected migration for full-length FIV Gag protein (50 kDa) is shown on the right. Data shown are from a single experiment and are representative of two independent repeats of the experiment.
Similarly, we also sought to identify residues in FIV CA-CTD that correspond to assembly-critical HIV-1 residues. In HIV-1 CA-CTD, CA helix 9 mediates an important interhexameric CA-CTD dimerization interface in the completed immature capsid (16) (Fig. 5B). We expected that FIV helix 9 also forms this critical CA-CTD dimer interface, especially given that FIV CA-CTD can functionally replace SIV CA-CTD, as shown with SIV-FIV chimeras (60). It was demonstrated previously that the W316 and M317 mutations within helix 9 of HIV-1 CA inhibit VLP production (22, 57) and arrest HIV-1 Gag assembly at the ∼80S intermediate (Fig. 3A) (22). To determine if FIV assembly also depends on this critical CA-CTD dimerization interface, FIV Gag residues Y311 and L312 were chosen for mutation based on their alignment with, and similar orientation to, residues W316 and M317 in HIV-1 helix 9 (Fig. 5A and C).
When expressed in cells, FIV CO-Gag Y311A L312A displayed dramatically reduced VLP production (Fig. 3C), despite detectable intracellular Gag, thus recapitulating the VLP defect observed for the analogous HIV-1 Gag W316A M317A (22, 57). Velocity sedimentation analysis revealed that FIV CO-Gag Y311A L312A produced only the ∼10S and ∼80S complexes, while FIV WT CO-Gag produced 10S, ∼80S, and ∼500S/750S intermediates when expressed at similar steady-state intracellular levels and analyzed in parallel (Fig. 6A to C). Thus, FIV CO-Gag Y311A L312A is arrested at the ∼80S putative assembly intermediate, as observed for the corresponding HIV-1 W316A M317A mutant (Fig. 3A) (22). Together, analysis by velocity sedimentation shows that each of the four assembly-defective Gag mutants that we examined appears to be arrested at the same assembly intermediate as its HIV-1 counterpart (Fig. 4 and 6; summarized in Fig. 7A).
FIG 7.
FIV Gag, like HIV-1 Gag, is associated with both ABCE1 and DDX6 by coIP. (A) Summary of the results in the present study demonstrating that FIV Gag mutants appear to be arrested in the assembly pathway at the same points as their HIV-1 counterparts. A diagram of the HIV-1 assembly pathway is shown at the top of the figure. HIV and FIV Gag mutants are listed below, with bars spanning all the complexes that are formed by each Gag mutant. The legend (boxed) shows symbols representing Gag (FIV or HIV-1), genomic RNA (gRNA), DDX6, ABCE1, other granule proteins, and the plasma membrane. (B) G355-5 cells transfected with the indicated constructs were treated with cross-linker (0.1 mM DSP) and lysed in a standard salt buffer. Lysates were subjected to IP with an antibody to ABCE1 (αA) or an antibody to DDX6 (αD6) alongside a nonimmune (NI) control. IP eluates and an aliquot of input (In) were analyzed by WB using an antibody to FIV CA (αFIV CA). (C) G355-5 cells transfected with FIV CO-Gag were harvested in a low-salt buffer without cross-linking. Lysates were subjected to velocity sedimentation, and gradient fractions were analyzed by WB with an αFIV CA (top panel). Pooled gradient fractions from the 10S, 80S to 150S, and 500S regions were also subjected to IP with an antibody to DDX6 (αD6) alongside a nonimmune control (NI), with gradient fractions that were pooled indicated by brackets below the gradient fraction WB. IP eluates and an aliquot of pooled gradient fraction input (In) were analyzed by WB with αFIV CA. The approximate S values of FIV Gag-containing complexes are indicated with brackets above the gradient WB, and the expected migrations for full-length FIV Gag (50 kDa) and FIV Gag MACA (37 kDa) proteins are shown on the right of the blots. Data shown are from a single experiment and are representative of two independent repeats of the experiment.
To further confirm that the FIV Gag-containing complexes closely resemble assembly intermediates, we asked whether they contain the same cellular proteins found in HIV-1 assembly intermediates. The ∼80S and ∼500S HIV-1 capsid assembly intermediates contain several host proteins, of which the best studied are the enzymes ABCE1 (26) and DDX6 (28). Both ABCE1 and DDX6 facilitate events in HIV-1 assembly (26, 28) and serve as markers for assembly intermediates. Thus, we asked whether FIV Gag is also associated with endogenous ABCE1 and DDX6 in feline G355-5 cells. FIV WT CO-Gag was expressed in G355-5 cells, and cell lysates were subjected to IP with either antibody to human ABCE1 or antibody to human DDX6 alongside a nonimmune control, followed by WB with αFIV CA to detect associated FIV Gag. Because both ABCE1 and DDX6 are highly conserved between humans and domestic cats (99.8% identity) and the ABCE1 and DDX6 antibodies we used (described previously [26, 28]) are directed against peptides with 100% identity between humans and domestic cats, we expected both antibodies to recognize the feline homologs. Indeed, we found that FIV WT CO-Gag was associated with endogenous feline ABCE1 and DDX6 in unfractionated lysates by coIP (Fig. 7B). These data suggested that like HIV-1 Gag, FIV Gag co-opts an ABCE1- and DDX6-containing RNA granule.
The ∼10S HIV-1 assembly intermediate, which corresponds to soluble Gag, is not associated with ABCE1 and DDX6 (26, 28, 40), indicating that HIV-1 Gag likely associates with ABCE1 and DDX6 after it targets an RNA granule. If FIV forms RNA granule-derived assembly intermediates similar to those formed by HIV-1, then we would not expect the assembly-incompetent FIV Gag MACA mutant, which is arrested at the ∼10S assembly intermediate (Fig. 4C), to be associated with ABCE1 and DDX6. In contrast, we would expect all the other assembly-defective mutants studied here (FIV Gag G2A, FIV Gag Q209A L210A, and FIV Gag Y311A L312A) to associate with ABCE1 and DDX6, since they all appear to be arrested at either the ∼80S intermediate (FIV Gag G2A and FIV Gag Y311A L312A [Fig. 4C and 6C]) or the ∼500S intermediate (FIV Gag Q209A L210A [Fig. 6C]). Additionally, the assembly-competent FIV CO-Gag P438A P441A, which forms all the assembly intermediates (Fig. 4C), should also associate with ABCE1 and DDX6. Indeed, as expected, all the FIV Gag mutants we studied here, with the exception of FIV Gag MACA, were associated with endogenous ABCE1 and DDX6 by coIP in lysates of feline G355-5 cells that were subjected to DSP cross-linking before harvest (Fig. 7B). These findings support a model in which FIV Gag localizes to an RNA granule that contains ABCE1 and DDX6, as shown previously for HIV-1.
If FIV Gag co-opts RNA granules to form the putative ∼80S intermediate, we would expect FIV Gag to be associated with the RNA granule marker DDX6 in the ∼80S and ∼500S putative assembly intermediates but not with FIV Gag in the soluble ∼10S region of the gradient, as observed for HIV-1 (28). To test this hypothesis, we performed DDX6 IPs from gradient fractions. However, while DSP cross-linking had allowed us to detect the unstable late FIV putative assembly intermediate (Fig. 2B), cross-linking is also known to mask immunoreactive protein epitopes and can therefore interfere with IPs (61). For this reason, we tested an alternative to cross-linking, in which we harvested cells in a low-salt buffer (NaCl reduced from ∼150 mM to 10 mM). We hypothesized that such a low-stringency buffer would preserve the integrity of the putative ∼500S assembly intermediate and thereby eliminate the need for cross-linking. Consistent with this, we observed that DDX6 IPs were more effective from lysates treated with lower concentrations of DSP cross-linker (J. C. Reed and J. R. Lingappa, unpublished observations). Furthermore, when G355-5 cells transfected to express FIV WT CO-Gag were harvested in low-salt buffer without cross-linking and analyzed by velocity sedimentation, the ∼10S, ∼80S/150S, and ∼500S putative assembly intermediates were still detected, albeit with a slightly different migration of ∼80S/150S complex that could be explained by preservation of proteins loosely associated with complexes under the low-stringency harvest conditions (e.g., compare Fig. 6C to Fig. 7C). To determine if FIV Gag is associated with DDX6 in the putative RNA granule-derived assembly intermediates, pooled gradient fractions containing the ∼10S, ∼80S/150S, or ∼500S putative assembly intermediates were immunoprecipitated with a DDX6 antibody, followed by WB for FIV Gag. DDX6 was associated with FIV Gag in the ∼80S/150S and ∼500S putative assembly intermediates but not the ∼10S putative assembly intermediate (Fig. 7C). Thus, the ∼80S/150S and ∼500S putative FIV assembly intermediates contain an RNA granule marker, which suggests that they are derived from an RNA granule, unlike the ∼10S FIV assembly intermediate. The same pattern of DDX6-Gag association was observed for HIV-1 assembly intermediates (28), which were validated using pulse-chase experiments in cells (21), lending further support to these complexes being FIV assembly intermediates.
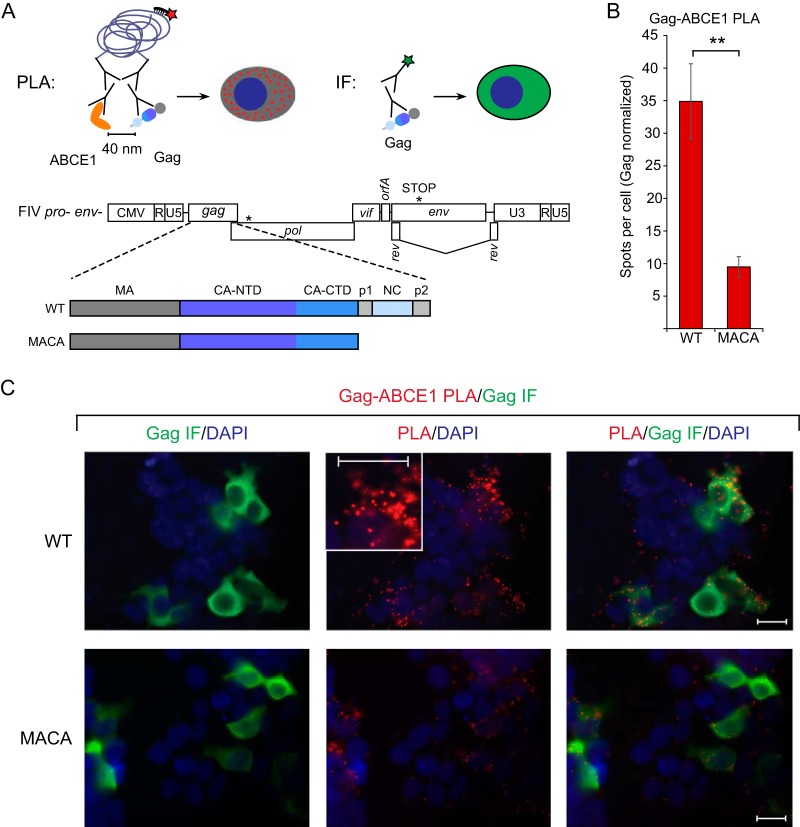
Given that our studies of HIV-1 Gag suggest that ABCE1 is a marker of assembly intermediates and having shown that ABCE1 associates with FIV Gag by coIP, we next asked if we could confirm the association of FIV Gag with ABCE1 by demonstrating Gag-ABCE1 colocalization in situ. For this purpose, we utilized the proximity ligation assay (PLA), which produces fluorescent spots only at sites where two proteins are within 40 nm of each other in situ. Briefly, proteins of interest (FIV Gag and ABCE1) are labeled with primary antibodies, which in turn are detected by a pair of secondary antibodies that are each conjugated to one of two complementary oligonucleotides; the two secondary antibodies are ligated via annealing between the conjugated complementary oligonucleotides only when the proteins of interest are within 40 nm of one another. Reagents added subsequently result in a rolling-circle amplification product that is recognized by a fluorophore-conjugated oligonucleotide, creating a punctate fluorescent signal wherever colocalization occurs (Fig. 8A) (62). Thus, if FIV Gag is in close proximity to ABCE1 within an ABCE1-containing RNA granule, cells expressing FIV WT Gag would be expected to display abundant Gag-ABCE1 PLA spots. In contrast, cells expressing the assembly-defective FIV Gag MACA mutant would be expected to display few PLA spots, since FIV Gag MACA is arrested at the ∼10S intermediate and does not associate with ABCE1 by coIP. To avoid FIV Env-induced cytopathicity (63) that might interfere with the PLA, we utilized proviral constructs that expressed only a truncated form of Env (FIV pro− env−; diagrammed in Fig. 8A). We found that cells transfected with an FIV pro− env− provirus encoding WT Gag (Fig. 8A, WT), when normalized by Gag immunofluorescence (IF) intensity, contained an average of 34.9 Gag-ABCE1 PLA spots per cell (Fig. 8B and C), and concurrent FIV Gag indirect IF staining showed that the vast majority of these PLA spots occurred in cells expressing FIV WT Gag at low or high levels (Fig. 8C). In contrast, cells transfected with FIV pro− env− encoding the assembly-defective Gag MACA mutant (Fig. 8A, MACA), when normalized by Gag IF intensity, contained nearly 4-fold fewer Gag-ABCE1 PLA spots per cell than FIV WT Gag (an average of 9.5 Gag-ABCE1 PLA spots per cell for FIV Gag MACA versus an average of 34.9 spots per cell for FIV WT Gag, P < 0.01) (Fig. 8B; see also Fig. 8C for representative images), despite abundant FIV Gag MACA staining by indirect immunofluorescence (Fig. 8C). Note that because ABCE1 as well as WT and mutant FIV Gag proteins are all present in the ∼10S region of velocity sedimentation gradients (Fig. 4C and 6C) (J. C. Reed and J. R. Lingappa, unpublished observations), FIV Gag and ABCE1 may be in proximity to some extent in the soluble fraction of the cytoplasm, outside of RNA granules. This might explain the presence of a background level of PLA spots for FIV Gag MACA, which was not coimmunoprecipitated by ABCE1 antibody to a detectable extent (Fig. 7B). Despite this, significantly more Gag-ABCE1 PLA spots were observed in FIV WT Gag-expressing cells than in FIV Gag MACA-expressing cells (Fig. 8B, see P value). Importantly, the number of PLA spots was negligible in cells that were negative for Gag by IF and were present in the same fields as Gag-expressing cells. Specifically, the average number of Gag-ABCE1 PLA spots per cell in Gag IF-negative cells was 14-fold lower than that in cells expressing FIV Gag MACA (0.62 spots per cell versus 8.59 spots per cell, respectively, P < 0.0001, without normalization for Gag expression; n = 25 Gag IF-negative cells and n = 37 FIV Gag MACA-positive cells). Thus, the near absence of nonspecific background PLA spots in Gag-negative cells confirms that the PLA spots represent colocalization of Gag and ABCE1. Moreover, the finding that MACA-expressing cells display a low but significant number of PLA spots is consistent with a low level of background PLA signal from nonspecific colocalization of soluble protein. Together, these data show that ABCE1 colocalizes in situ to a greater extent with FIV WT Gag, which is found in RNA granules, than with FIV Gag MACA, which is not found in RNA granules. Together with the coIP data, the PLA data further support a model in which assembling FIV Gag co-opts an ABCE1-containing RNA granule.
FIG 8.
PLA confirms colocalization of FIV Gag and ABCE1 in situ upon provirus expression. (A) Experimental schematic for PLA and concurrent immunofluorescence (IF). The PLA schematic shows primary antibodies directed against FIV Gag and ABCE1, with mouse and rabbit secondary antibodies attached to oligonucleotides that undergo ligation, leading to a rolling-circle amplification product that is detected by a red fluorophore (red star). Red PLA spots indicate regions in which Gag is within 40 nm of ABCE1 in situ. The concurrent IF schematic shows primary antibody directed against FIV Gag with mouse secondary antibody conjugated to a green fluorophore (green star), which allows for quantification of intracellular Gag levels. Shown below the experimental schematic is the gene map of the FIV pro− env− provirus utilized in this PLA experiment, with the point mutation in protease and the premature stop codon in env marked by asterisks. The proviral constructs encoded either FIV WT Gag or Gag MACA, as shown in the colored domain map below the provirus gene map. (B) Following transfection of these proviruses into 293T cells and processing for PLA, the average number of PLA spots per cell was determined for all cells IF positive for Gag in five randomly chosen fields and normalized to Gag levels as measured by IF signal intensity. Error bars indicate SEM. Double asterisks indicate a significant difference relative to the results for the WT (P ≤ 0.01 in a two-tailed, paired t test). (C) Panels show representative images from each transfection. Columns from left to right for each construct: column 1, Gag IF (green) with DAPI-stained nuclei (blue); column 2, FIV Gag-ABCE1 PLA signal (red) with DAPI-stained nuclei (blue); column 3, a merge of images from the first two columns. The middle PLA panel in the top row contains an inset showing a high-magnification view of the cell to the right of the inset. Scale bars, 5 μm. Data shown are from analysis of 35 to 37 cells taken from five fields for each condition from a single experiment and are representative of two independent repeats of the experiment.
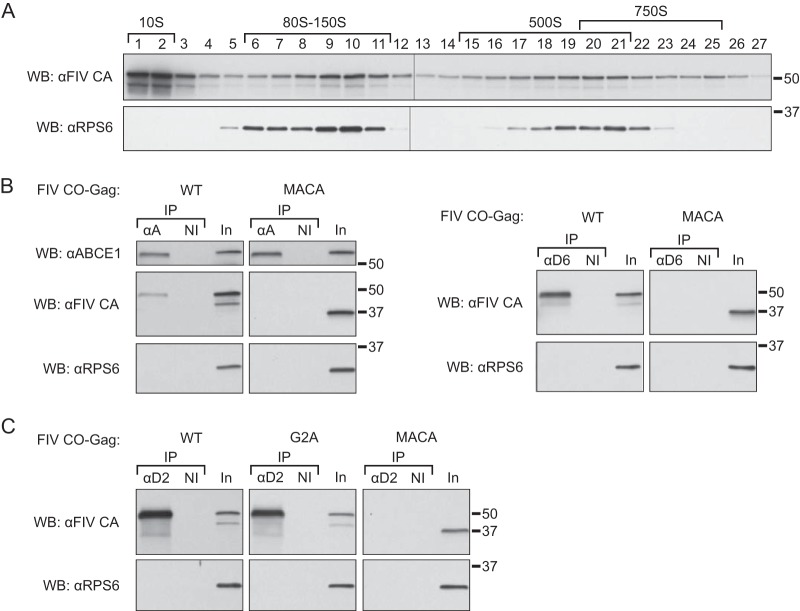
Although ABCE1 is a marker for HIV-1 assembly intermediates, it is also critical for termination of translation leading to ribosome recycling (reviewed in reference 27). Thus, one might expect that ABCE1 would be associated, perhaps to different extents, with two functionally different complexes: ribosomes undergoing translation termination and assembly intermediates. Therefore, among other possibilities, complexes immunoprecipitated by ABCE1 antibody might consist of (i) both assembly intermediates and ribosomal complexes, (ii) mostly ribosomal complexes, or (iii) mostly assembly intermediates. Previously, we have shown that HIV-1 Gag remains associated with ABCE1 in ∼80S/150S and ∼500S complexes when ribosomes are dissociated into the 40S and 60S subunits with 10 mM EDTA, arguing that ABCE1 is associated with Gag in assembly intermediates independent of ABCE1 association with translating ribosomes (40). Here we asked whether FIV assembly intermediates are distinct from ribosome-associated complexes in feline cells by using a different strategy in which we determined whether a well-studied ribosomal protein is present in the putative ABCE1-containing assembly intermediates. For this purpose, we examined whether antibody to ABCE1 or DDX6, both of which coimmunoprecipitate FIV Gag by WB, also coimmunoprecipitate the small ribosomal protein S6 (RPS6) by WB. If RPS6 is associated with ABCE1-containing complexes under conditions in which ABCE1 coimmunoprecipitates FIV Gag in G355-5 cells, then this would indicate that the ABCE1 antibody precipitates both ribosomal complexes and assembly intermediates, or mainly ribosomal complexes. For this purpose, we utilized a well-studied commercial antibody to RPS6 (64, 65) and validated the antibody using lysates from G355-5 cells transfected to express FIV WT CO-Gag, harvested in low-salt buffer, and analyzed by velocity sedimentation. WB of individual gradient fractions demonstrated that the RPS6 antibody detects a single ∼32-kDa protein that migrates in fractions that should contain monosomes (80S), disomes (160S), and polysomes (>480S), as expected (Fig. 9A). Moreover, RPS6 was not present in the soluble (∼10S) region of the gradient, as also expected (Fig. 9A). Additionally, when the RPS6 WB was reprobed with FIV CA antibody, FIV Gag was found in the ∼10S, ∼80/150S, and ∼500S fractions (Fig. 9A), as observed earlier (Fig. 2, 4, 6, and 7).
FIG 9.
ABCE1, DDX6, and DCP2 are associated with FIV WT Gag but not with the small ribosomal protein RPS6. (A) Lysates from cells transfected to express FIV WT CO-Gag were harvested in low-salt buffer without cross-linking and subjected to velocity sedimentation. Gradient fractions were analyzed by WB with an antibody to ribosomal protein S6 (αRPS6), followed by a WB reprobe with antibody to FIV CA (αFIV CA). (B) Lysates of G355-5 cells transfected with the indicated constructs were subjected to IP with an antibody to ABCE1 (αA) or an antibody to DDX6 (αD6) alongside a nonimmune (NI) control. IP eluates were analyzed by WB with αRPS6, followed by a WB reprobe with αFIV CA, with 1% of IP input analyzed in parallel (In). Subsequently, ABCE1 IP eluates were also WB reprobed with an antibody to ABCE1 (αABCE1) to confirm the ABCE1 direct IP. (C) G355-5 cells transfected with the indicated constructs were harvested in low-salt buffer without cross-linking. Cell lysates were subjected to IP with an antibody to DCP2 (αD2), and IP elutes were analyzed by WB with αRPS6, followed by a WB reprobe with αFIV CA, with 1% IP input analyzed in parallel (In). Direct IPs were not shown for DDX6 and DCP2 because both migrate close to the heavy chain; however, the specificity of both of these IP antibodies was confirmed by direct IP of YFP-tagged DDX6 and DCP2, respectively (28; J. C. Reed and J. R. Lingappa, unpublished observations). The approximate S values of FIV Gag-containing complexes are indicated with brackets above the gradient WB, and molecular size markers (50 kDa and 37 kDa) are shown on the right of the blots. Data shown are from a single experiment and are representative of two independent repeats of the experiment.
Having validated the RPS6 antibody, we next asked whether ABCE1 is associated with RPS6 by coIP at steady state in FIV Gag-expressing cells. Association of RPS6 with ABCE1 would be consistent with the presence of ribosomal complexes comigrating with our assembly intermediates; conversely, the absence of RPS6 in ABCE1 IPs would suggest that the association of ABCE1 with ribosomal complexes may be very transient at steady state under the conditions used to study FIV Gag assembly in cells. Lysates from G355-5 cells transfected to express FIV WT CO-Gag or assembly-incompetent FIV CO-Gag MACA were subjected to IP with an antibody to ABCE1 (with rabbit nonimmune controls and 1% of input cell lysate analyzed in parallel on the same blot), and IP eluates were probed for RPS6 by WB, followed by a reprobe for FIV CA by WB. Here we utilized the low-salt harvest conditions without cross-linking, described above, to preserve both immunoreactive epitopes and the integrity of the putative FIV assembly intermediates; thus, negative coIP data could not be ascribed to epitope masking by the cross-linker (or disassociation due to a high-stringency buffer). Interestingly, we did not detect RPS6 associated with ABCE1 in these IPs, despite the presence of RPS6 in lysate inputs analyzed on the same blot (Fig. 9B) and comigration of RPS6 and FIV Gag in gradient fractions from cells harvested in the same manner (Fig. 9A). Notably, a subsequent reprobe of the ABCE1 IP blots for Gag revealed coIP of FIV WT Gag but not assembly-incompetent FIV Gag MACA (Fig. 9B). An additional reprobe of the same blots with ABCE1 antibody showed that the 68-kDa ABCE1 protein was indeed immunoprecipitated by the ABCE1 antibody, as expected (Fig. 9B). Taken together, these data indicate that at steady state and under the harvest conditions used here, most endogenous ABCE1 in FIV-expressing feline cells is not in ribosome-containing complexes, at least to the limit of our detection. As discussed below, these findings might be explained by ABCE1 being associated with ribosomal complexes very transiently during translation termination or by alternate harvest conditions being required to trap the ABCE1-ribosome association. Either way, these data suggest that at steady state, the association of FIV Gag with ABCE1 is independent of ribosomes. Moreover, these data support our previous studies demonstrating that HIV-1 Gag is largely present in ABCE1-containing assembly intermediates, rather than ABCE1-associated translating ribosomes, as indicated by the insensitivity of those complexes to treatment with 10 mM EDTA (40) or puromycin followed by high salt (M. Tanaka, B. C. Barajas, J. C. Reed, and J. R. Lingappa, unpublished observations).
To further confirm that the complexes identified here containing FIV-1 Gag, ABCE1, and DDX6 are independent of ribosomes, we also analyzed DDX6 IPs for RPS6. Like ABCE1, DDX6 was also not associated with RPS6 (Fig. 9B), as expected given that DDX6 is not directly involved in translation (29). A subsequent reprobe of the DDX6 IP blots for Gag revealed coIP of FIV WT Gag but not assembly-incompetent FIV Gag MACA, as expected (Fig. 9B). Thus, our finding that FIV Gag is associated with ABCE1 and DDX6, but not with the ribosome marker RPS6 (to the limit of our detection), supports a model in which most FIV Gag at steady state in feline cells is associated with ABCE1 and DDX6 in an RNA granule that is not involved in translation or translation termination. Moreover, our mutational studies show that this FIV Gag association with ABCE1 and DDX6 holds true for WT Gag and oligomerization-competent Gag mutants, but not assembly-incompetent Gag, consistent with these nontranslating, FIV Gag-containing complexes being involved in assembly.
Previously, we have shown that HIV-1 Gag is also associated with a cotransfected tagged version of yet another RNA granule protein, DCP2, in cell lysates (28). We were interested in DCP2 because it has been found associated with assembling Ty3 Gag in RNA granule-derived complexes in yeast and facilitates Ty3 retrotransposition (36, 38). To test whether FIV Gag is associated with DCP2, lysates from G355-5 transfected to express either FIV WT CO-Gag, FIV CO-Gag G2A, or FIV CO-Gag MACA were harvested in low-salt buffer and subjected to IP with a DCP2 antibody, followed first by WB with RPS6 and then a WB reprobe for FIV Gag. DCP2 was not associated with RPS6 by coIP regardless of whether FIV WT Gag, FIV Gag G2A, or FIV Gag MACA was expressed (Fig. 9C). However, notably, DCP2 was associated with FIV WT Gag and FIV Gag G2A but not assembly-incompetent FIV Gag MACA (Fig. 9C). Thus, under the harvest conditions used here, DCP2, like ABCE1 and DDX6, does not appear to be associated with ribosomes to the limit of our detection but is associated with assembling FIV Gag.
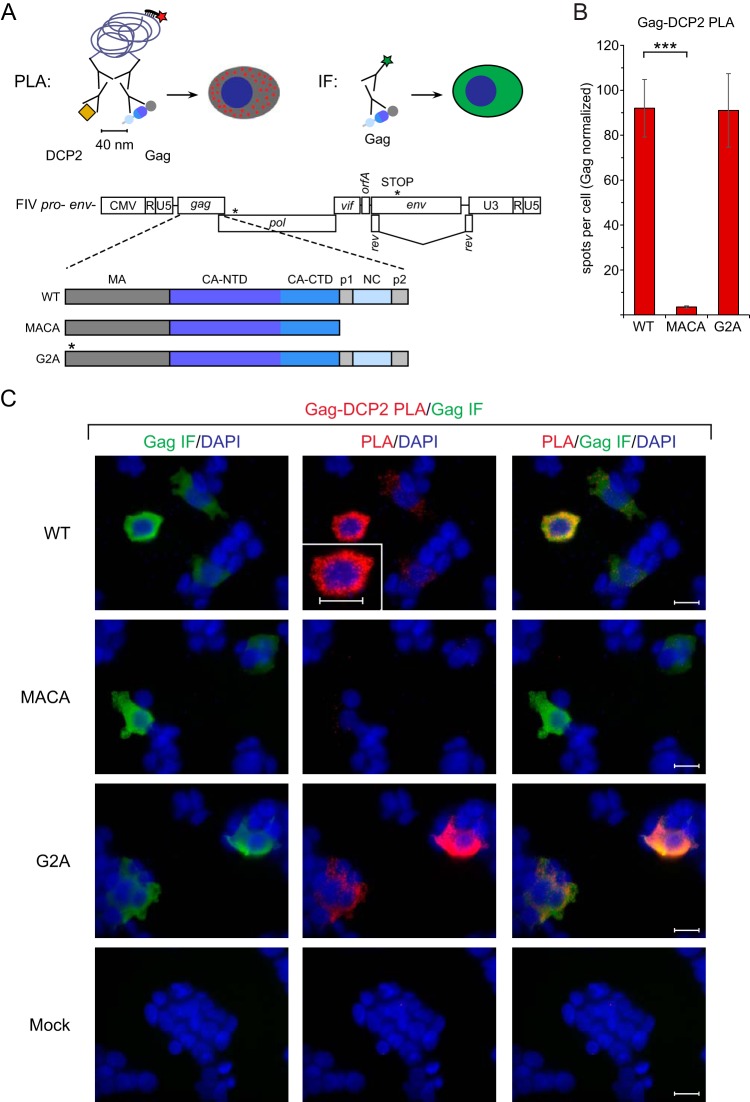
Having confirmed the in situ colocalization of FIV Gag with ABCE1, we also wanted to confirm colocalization of FIV Gag with one of the two canonical RNA granule proteins that associated with FIV Gag by coIP, DDX6 or DCP2. We chose to use DCP2 for this purpose, because a commercially available antibody against DCP2 was listed as validated for PLA. 293T cells transfected with FIV pro− env− encoding WT Gag (Fig. 10A, WT), when normalized by Gag IF intensity, contained an average of 92.0 Gag-DCP2 PLA spots per cell (Fig. 10B and C), and concurrent indirect immunofluorescence staining of FIV Gag showed that the majority of these PLA spots occurred in cells expressing FIV Gag at low or high levels (Fig. 10C). In contrast, cells transfected with FIV pro− env− encoding the assembly-defective Gag MACA mutant (Fig. 10A, MACA), when normalized by Gag IF intensity, contained an average of 3.5 Gag-DCP2 PLA spots per cell (Fig. 10B and C), despite FIV Gag MACA staining by indirect immunofluorescence comparable to that of FIV WT Gag (Fig. 10C). Because DCP2 is associated with FIV Gag G2A by IP, we also examined the localization of FIV Gag G2A with DCP2 by PLA. We found that cells transfected with FIV pro− env− encoding the Gag G2A mutant (Fig. 10A, G2A), when normalized by Gag IF intensity, contained an average of 91.0 Gag-DCP2 PLA spots per cell (Fig. 10B and C), similar to what we observed for FIV WT Gag. Lastly, as an additional control, we performed a Gag-DCP2 PLA on mock-transfected cells (Fig. 10C). These data are not included in Fig. 10B, which shows the PLA spot number normalized to Gag levels; however, as expected, the spot number was extremely low in mock-transfected controls. Specifically, the average number of Gag-DCP2 PLA spots per cell in mock-transfected cells was 53-fold lower than that in cells expressing FIV Gag MACA (0.08 spots per cell versus 4.21 spots per cell, respectively, P < 0.0003, without normalization for Gag expression; n = 25 mock-transfected cells and n = 15 FIV Gag MACA-positive cells). Thus, the near absence of nonspecific background PLA spots in mock-transfected cells confirmed that the PLA spots are specific to Gag-expressing cells. Moreover, the finding that MACA cells display a low but significant number of PLA spots is consistent with a low level of background PLA signal from nonspecific colocalization of soluble protein. In conclusion, the RNA granule protein DCP2 colocalizes in situ to a much greater extent with the FIV WT Gag and the partially assembling FIV Gag G2A mutant than with the assembly-incompetent FIV Gag MACA and associates with assembling FIV Gag proteins by coIP in feline cells. Together, the data support a model in which assembling FIV Gag, like HIV Gag, co-opts RNA granule proteins to form complexes that likely correspond to assembly intermediates.
FIG 10.
PLA confirms colocalization of FIV Gag and DCP2 in situ upon provirus expression. (A) Experimental schematic for PLA and concurrent immunofluorescence (IF). The PLA schematic shows primary antibodies directed against FIV Gag and DCP2, with mouse and rabbit secondary antibodies attached to oligonucleotides that undergo ligation, leading to a rolling-circle amplification product that is detected by a red fluorophore (red star). Red PLA spots indicate regions in which Gag is within 40 nm of DCP2 in situ. The concurrent IF schematic shows primary antibody directed against FIV Gag, with mouse secondary antibody conjugated to a green fluorophore (green star), which allows for quantification of intracellular Gag levels. Shown below the experimental schematic is a gene map of the FIV pro− env− proviral construct utilized in this PLA experiment, with the point mutation in protease and the premature stop codon in env marked by asterisks. The proviral constructs encoded either FIV WT Gag, Gag MACA, or Gag G2A, as shown in the colored domain map below the provirus gene map. The point mutation in Gag G2A is indicated by an asterisk. Cells were also mock transfected and analyzed in parallel. (B) Following transfection of these proviruses (or mock transfection) into 293T cells and processing for PLA, the average number of PLA spots per cell was determined for all cells positive for Gag IF in five randomly chosen fields and normalized to Gag levels as measured by IF signal intensity. Error bars indicate SEM. Triple asterisks indicate a significant difference between MACA results and WT results (P ≤ 0.001 in a two-tailed, paired t test). (C) Panels show representative images from each transfection. Columns from left to right for each construct: column 1, Gag IF (green) with DAPI-stained nuclei (blue); column 2, FIV Gag-DPC2 PLA signal (red) with DAPI-stained nuclei (blue); column 3, a merge of images from the first two columns. The middle PLA panel in the top row contains an inset showing a high-magnification view of the cell above the inset. Scale bars, 5 μM. Data shown are from analysis of 15 to 21 cells taken from five fields for each condition from a single experiment and are representative of two independent repeats of the experiment.
DISCUSSION
Here we present evidence that, in feline cells, the nonprimate lentivirus FIV Gag likely forms immature capsids via a pathway of RNA granule-derived assembly intermediates, as shown for HIV-1 and other primate lentiviruses (20–22, 26, 28, 40). Support for this conclusion comes from four findings: first, FIV Gag forms complexes in cells that resemble well-studied HIV-1 immature capsid assembly intermediates in size and shape (Fig. 2); second, assembly-defective FIV Gag mutants are arrested in the putative assembly pathway and the points of arrest are the same as for the analogous HIV-1 Gag assembly-defective mutants (Fig. 4 and 6); third, FIV Gag is associated with the RNA granule proteins ABCE1, DDX6, and DCP2 by coIP (Fig. 7B and C and 9B and C), as observed for HIV-1 Gag in assembly intermediates; and fourth, assembling FIV Gag colocalizes with ABCE1 and DCP2 in situ, while an assembly-defective mutant does not (Fig. 8 and 10). Taken together, these data argue that the ∼80S and ∼500S FIV Gag-containing complexes likely correspond to early and late immature capsid assembly intermediates, respectively, and that the immature capsid assembly pathway defined by studies of HIV-1 is conserved between primate and nonprimate lentiviruses (Fig. 7A). Notably, this evolutionary conservation could extend further, given that the assembling Gag protein of the S. cerevisiae retrotransposon Ty3 is also associated with DCP2 and the S. cerevisiae DDX6 homolog in RNA granule-derived complexes (36). Because of the similarity between RNA granules involved in Ty3 assembly, which have been termed retrosomes (66), and RNA granule-derived retroviral assembly intermediates, we here propose the term “assemblysome” to collectively refer to RNA granule-derived capsid assembly intermediates of different sizes (e.g., the ∼80S and ∼500S HIV-1 and FIV assembly intermediates).
While a pulse-chase analysis would be required to fully confirm that these complexes are assembly intermediates, the numerous similarities between the FIV Gag-containing complexes identified here and HIV-1 assembly intermediates identified previously through pulse-chase analysis strongly suggest that these complexes correspond to FIV assembly intermediates. Additionally, our study directly addressed the alternate possibility that these FIV Gag-containing complexes correspond not to assembly intermediates but to translating complexes (i.e., monosomes, disomes, and polysomes), which migrate at similar S values. Previously, we showed that HIV-1 assembly intermediates are resistant to EDTA (40) and puromycin–high-salt treatments (M. Tanaka, B. C. Barajas, J. C. Reed, and J. R. Lingappa, unpublished observations), which disrupt polysomes. Additionally, the failure to find Gag MACA associated with ABCE1 by coIP also argues that the complexes we are studying are likely not translating complexes (Fig. 7B and 9B and C). Here we took yet another approach to this question in which we showed that the well-studied small ribosomal subunit protein RPS6 is not associated with ABCE1, DDX6, or DCP2 at steady state, to the extent that we could detect using an IP-WB approach, in feline cells expressing FIV Gag. Thus, under the conditions of our harvests, we did not find a significant association between any of these three proteins and RPS6, even though our controls confirm that RPS6 appears to be in ribosomes. DDX6 and DCP2 would not be expected to be associated with ribosomes, so those results are not surprising. In contrast, ABCE1 is critical for translation termination in archaebacteria and eukaryotes (reviewed in reference 27) and would be expected to be associated with ribosomes, albeit perhaps only at a very low level. ABCE1 has been found associated with ribosomal proteins in tandem affinity purification experiments in yeast (67, 68) and with initiation factors in vertebrate cells (69); however, to our knowledge, ABCE1 has not been shown to associate with ribosomal proteins by coIP in any vertebrate cell type. Our findings suggest that the ABCE1-ribosome association may be very transient or is unstable under our harvest conditions. In either case, our findings reveal that ABCE1 associates with assembling FIV Gag in complexes that appear to be ribosome independent and are likely derived from RNA granules, with PLA demonstrating the expected colocalization in situ. Future studies will be needed to clarify whether ABCE1 is associated with ribosomal proteins to an extent detectable by coIP in the absence of retroviral Gag expression, by using other harvest conditions, or by using other WB antibodies.
Our finding that both primate (HIV-1, HIV-2, SIV) and nonprimate (FIV) lentiviruses appear to form assemblysomes by co-opting a similar subclass of RNA granules suggests that many or all Gag proteins have evolved to utilize this cellular complex. While many questions about these RNA granules remain unanswered, this conservation argues that these granules play an important role in the retroviral life cycle. Other types of RNA granules, such as P bodies and stress granules, transiently store nontranslating cellular mRNA (reviewed in reference 70); thus, it is possible that these ABCE1-, DDX6-, and DCP2-containing complexes could also be sites for storing nontranslating cellular mRNA. Moreover, because FIV and HIV-1 unspliced RNAs resemble cellular mRNA in that they are capped and polyadenylated, one might expect unspliced FIV and HIV-1 viral RNA to also be stored in these ABCE1- and DDX6-containg granules. Consistent with this possibility, unspliced HIV-1 RNA is associated with ABCE1 in these small RNA granules (22). Thus, targeting of assembling retroviral Gag proteins to these small RNA granules might be advantageous to the virus for a number of reasons. First, assembling Gag could utilize cellular facilitators of immature capsid assembly present in RNA granules, given studies showing that ABCE1 and DDX6 facilitate HIV-1 capsid assembly (22, 25, 26, 28). Second, these RNA granules could provide a site for concentrating Gag, thereby facilitating Gag-Gag interactions. Third, RNA granule localization could bring assembling Gag into proximity with granule-associated unspliced retroviral RNA, thereby promoting packaging of the RNA genome into the assembling virus. Finally, these RNA granules could also shield retroviral RNA or other retroviral components from host innate immune sensing.
Many viruses are known to utilize RNA granules at various stages of their life cycles (71). Canonical RNA granules include stress granules and P bodies, both of which are much larger than the ABCE1- and DDX6-containing RNA granule from which assembly intermediates are derived. For example, P bodies range from 100 to 300 nm in size (72), whereas our ∼80S assembly intermediate is expected to be approximately the same size as an ∼80S ribosome (∼25 nm) (73). Others have reported that HIV-1 Gag is associated with RNA granule proteins, including Staufen1 (74, 75), AGO2 (76), and MOV10 (77). HIV-1 Gag is not found in stress granules (75), whose formation is induced by stress; nevertheless, Gag has been shown to modulate the formation of stress granules (75, 78, 79). Similarly, a recent study reported that HIV-1 Gag does not colocalize with P bodies by fluorescence microscopy (80). Consistent with that report (80), our recent PLA studies show that HIV-1 Gag is not typically colocalized with P bodies, although it colocalizes with the P body protein DDX6 in much smaller granular foci (B. C. Barajas and J. R. Lingappa, unpublished observations). Thus, P bodies, stress granules, and assemblysomes likely represent RNA granules of different sizes and functions; however, more work is needed to understand how they are related to each other and to the HIV-1 life cycle. One hypothesis consistent with published data is that the small RNA granules that are co-opted by HIV-1 Gag, and possibly FIV Gag, are subunits of P bodies (and possibly of stress granules) that can also exist independently of larger RNA granules. In support of this hypothesis, the granules that appear to be co-opted by HIV-1 Gag share three components with P bodies (DDX6, AGO2, and DCP2) (28), one of which (DDX6) is also found in stress granules (reviewed in reference 81). Thus, this subunit hypothesis would explain why Gag is associated with DDX6 and ABCE1 but does not colocalize with P bodies or stress granules and why P bodies and stress granules can be impacted when Gag is expressed. While future studies will be needed to test these possibilities, the current study adds to a large body of evidence indicating that a common feature of Gag proteins is their ability to co-opt a small ABCE1-, DDX6-, and DCP2-containing RNA granule during immature capsid assembly.
The association of HIV-1 Gag with endogenous host enzymes DDX6, AGO2, and ABCE1 in assemblysomes was shown previously using coIP and immunoelectron microscopy (21, 28); additionally, previous studies of a tagged RNA granule protein DCP2 suggested that it too may be present in HIV-1 assemblysomes (28). Here we showed that like HIV-1 Gag, FIV Gag is associated with endogenous ABCE1 and DDX6 by coIP; additionally, for the first time, we demonstrated the association of a retroviral Gag protein with endogenous DCP2 by coIP. Moreover, for the first time, we utilized PLA to demonstrate colocalization in situ of a retroviral Gag with ABCE1 and, separately, with DCP2. Unlike direct or indirect fluorescence microscopy, PLA results in fluorescent spots only when two proteins are in close proximity, thus allowing colocalization to be quantified even when proteins are present diffusely throughout the cytoplasm. Taken together, two different in situ approaches—PLA and immunoelectron microscopy—support the finding that assembling Gag proteins of various lentiviruses associate with ABCE1 and RNA granule proteins, while assembly-incompetent Gag MACA mutants do not. Moreover, our PLA results suggest that PLA may be a promising approach for in situ confirmation of other retrovirus-host protein associations.
Since DCP2 acts enzymatically to remove the 5′ cap that is found on cellular mRNAs as well as on retroviral RNAs, it may seem puzzling that DCP2 is present in RNA granules that are thought to be co-opted during retroviral assembly. However, DCP2 is not enzymatically active in the absence of activating proteins such as DCP1 and LSM1 (reviewed in reference 31). Further studies are required to determine if these activating proteins are absent from putative FIV assemblysomes and the small RNA granules from which they are derived. However, previously, tagged LSM1 was not found associated with ABCE1 in HIV-1-expressing cells, in contrast to tagged DCP2 which was found associated with ABCE1 (28). Moreover, once again the parallels with Ty3 retrosomes are instructive: Ty3 retrosomes in yeast also contain endogenous DCP2 as well as the exonuclease XRN1, even though intact Ty3 RNA is found in these retrosomes and retrosomes are strongly implicated as sites of Ty3 packaging and assembly (36). Together these data fit with the model described above, in which these small RNA granules serve as subunits of P bodies; as such, they may lack some components that are found in P bodies, such as activators of decapping and RNA degradation. Further studies are required to test this hypothesis and also to determine if DCP2 functions in FIV or HIV assembly; however, if these host complexes constitute a type of RNA granule that lacks activators required for DCP2-mediated RNA degradation but contains enzymes that promote RNA remodeling, they would offer important advantages to viruses for packaging and assembly.
Although our study found numerous similarities between FIV and HIV-1, we did find one significant difference, namely, that the late ∼500S FIV assembly intermediate and completed FIV immature capsids were relatively unstable compared to the corresponding HIV-1 complexes. Notably, our study demonstrated that two approaches can be used to overcome this problem: in situ cross-linking, which stabilizes these complexes, or harvest in low-salt buffers, which appears to keep these labile complexes intact. While the first approach is useful for demonstrating that the complexes were present before cell lysis, the latter approach has the advantage of not masking epitopes important for detection by IP. In keeping with this, low-salt buffers were also used to identify the association of DDX6 with HIV-1 Gag assembly intermediates (28). Thus, these two approaches for maintaining the stability of assemblysomes should allow assemblysomes for other viruses to be isolated and analyzed intact in the future.
The present study also advances our understanding of residues and interfaces critical for assembly of FIV immature capsids and may have identified an example of coevolution that could have structural implications. Previously, it has been shown that E207 and E208 in helix 4 of HIV-1 CA-NTD are required for assembly (57) and for progression past the ∼500S step in the assembly pathway (22), consistent with a study showing that these residues are critical for an interhexameric interface formed between helix 4 and helix 1 of a neighboring 3-fold symmetry mate (59). Here we showed that Q209 and L210 in FIV CA-NTD helix 4 function in a manner analogous to E207 and E208 in helix 4 of HIV-1 CA-NTD, in that Q209 and L210 are important for VLP production and for progression past the ∼500S Gag-containing complex (Fig. 6B and C). However, we also noted a difference in the HIV-1 and FIV residues that refines our understanding of this interface. Others have proposed that a salt bridge may form between the negatively charged E207 and E208 in helix 4 of HIV-1 CA-NTD and the positively charged R150 in helix 1 of HIV-1 Gag CA-NTD (E-R salt bridge) (59), but this salt bridge is unlikely to form between the comparable FIV Gag residues, Q209 and L210 (neither of which is negatively charged), and K152, which is analogous to HIV-1 R150. Our finding that FIV Gag Q209 and L210 are required for progression past the ∼500S putative assembly intermediate and may serve a function similar to that of E207 and E208 raises the possibility that a Q209-K152 hydrogen bond (Q-K hydrogen bond) in the FIV immature capsid structure could substitute for the proposed E-R salt bridge in the HIV-1 immature capsid structure and be sufficient for formation of this critical interhexameric helix 1-helix 4 CA-NTD interface. If this is the case, then the FIV Q-K hydrogen bond in place of the E-R salt bridge at the interhexameric CA-NTD interface could contribute to the marked instability of completed FIV immature capsids and the late (but not the early) FIV assembly intermediates relative to their HIV-1 counterparts, as shown above (Fig. 1D and 2B). While Q209 and K152 are well conserved in FIV Gag sequences from isolates obtained from domestic cats (Felis catus) (Fig. 5D), they are not present in FIV isolates from felines other than domestic cats, including isolates from lions, cougars, bobcats, and Pallas' cats (Panthera leo, Puma concolor, Lynx rufus, and Otocolobus manul, respectively), which more closely resemble HIV-1 at the relevant positions (Fig. 5D). This observation suggests that the E-R salt bridge could form in FIV Gag isolates from lions, cougars, bobcats, or Pallas' cats and that the immature FIV capsids formed by those isolates may be more stable than those from isolates found in domestic cats. Additionally, this might be an example of coevolved substitutions (18), since helix 1 contains an arginine whenever helix 4 contains a glutamic acid (e.g., in HIV-1 and FIV isolates from species besides the domestic cat), while helix 1 contains a lysine whenever helix 4 contains a glutamine (e.g., in most FIV isolates from domestic cats). Notably, arginine forms bonds in more directions and is therefore thought to be more stabilizing than lysine (82), thus providing another reason, in addition to the salt bridge, for why the E-R pair may be more stabilizing than the Q-K pair. Future mutational studies could address whether the instability of FIV immature capsid and the ∼500S assembly intermediate results from having a Q-K pair in place of the E-R pair at the interhexameric CA-NTD interface and whether residues that reduce immature capsid stability also reduce the quantity of infectious virus produced.
Another important HIV-1 interface is the CA-CTD dimer interface, formed by residues W316 and M317 in helix 9 of HIV CA (16, 59). Our finding that mutation of the corresponding two hydrophobic residues (Y311 and L312) in FIV CA helix 9 inhibited VLP production and arrested the assembly pathway at the same assembly intermediate as observed with the corresponding HIV-1 mutant argues that helix 9 in CA-CTD plays a similar role in FIV and HIV-1 immature capsid assembly and likely forms the CA-CTD dimer interface in both cases. Although a previous study found that deletion of FIV CA helix 9 and part of helix 10 did not inhibit VLP production (9), very different phenotypes can result from deletion versus point mutation.
Finally, a key implication of our findings is that compounds that target the ABCE1-, DDX6-, and DCP2-containing RNA granules co-opted by lentiviral Gag proteins could have broad-acting antiretroviral activity, inhibiting both primate and nonprimate lentiviruses and possibly other retroviruses. Notably, small molecules that target ABCE1-containing complexes are known to effectively inhibit rabies virus assembly in vitro, thereby providing proof of principle that ABCE1-containing assemblysomes are druggable (41). The finding that rabies virus, HIV-1, other primate lentiviruses, and the nonprimate lentivirus FIV all form ABCE1-containing assemblysomes also suggests a much broader conservation of this mechanism of assembly.
MATERIALS AND METHODS
Construction of proviral and codon-optimized FIV Gag plasmids.
The proviral clone FIV 34TF10 was obtained from the NIH AIDS Reagent Program (catalog no. 1236; GenBank accession no. M25381.1). The previously described premature stop codon (TGA) in orfA within the proviral clone (44) was reverted to tryptophan (TGG) by overlap extension PCR. This construct was used to generate the construct termed FIV, in which the FIV promoter was replaced with the CMV promoter using a previously described strategy (46). To make the construct termed FIV pro−, a nonoverlapping site-directed mutagenesis approach was used to introduce the D30N mutation in the FIV protease domain (numbered according to the start of protease) (83). FIV pro− env− was made by introducing two stop codons in env at position 7175 (numbering according to reference 43) to generate an Env truncation mutant similar to that described previously (84). The G2A mutation was introduced into FIV pro− env− by overlap extension PCR. FIV Gag MACA pro− env− was made by introducing two stop codons after L357 at the end of CA by overlap extension PCR. FIV CO-Gag was ordered as a gene block (Integrated DNA Technologies) and cloned into pcDNA3.1/Neo(+) using HindIII and XbaI. FIV CO-Gag G2A, FIV CO-Gag P438A P441A, FIV CO-Gag Q209A L210A, and FIV CO-Gag Y311A L312A were made by overlapping site-directed mutagenesis. FIV CO-Gag MACA was made by introducing a stop codon after L357 at the end of CA by overlapping site-directed mutagenesis. HIV-1 experiments utilized an HIV-1 LAI provirus that was also pro− env− vpr− and contained the puromycin gene in place of nef. To generate this construct, XhoI and XbaI were utilized to insert the puromycin gene in place of nef starting with HIV-GFP env− (28, 85), resulting in HIV env− puro. The vpr mutation was made by introducing two stop codons at the NcoI site in vpr by overlap extension PCR, resulting in HIV env− vpr− puro. The protease mutations (D25K, G49W, I50W) were introduced in two steps by overlap extension PCR, resulting in HIV env− vpr− pro− puro. The FIV CO-Gag sequence and the sequences of the oligonucleotides used to generate each mutation are available upon request.
Cells and transfection.
The feline astrocyte cell line G355-5 was obtained from ATCC (CRL-2033) and maintained in McCoy 5A (modified) medium (Life Technologies; 16600-108) supplemented with 10% fetal bovine serum (Thermo Fisher; 26140079). Cells were transfected with 1 to 6 μg DNA in 6-cm dishes using X-tremeGENE 9 (Roche; 6365787001). The COS-1 cell line was obtained from ATCC (CRL-1650) and maintained in Dulbecco modified Eagle medium (DMEM) (Life Technologies; 10566-024) supplemented with 10% fetal bovine serum (Thermo Fisher; 26140079). COS-1 cells were transfected with 2 μg DNA in 6-cm dishes using polyethyleneimine (PEI) as described previously (25). The 293T/17 cell line was obtained from ATCC (CRL-11268) and maintained in DMEM (Life Technologies; 10566-024) supplemented with 10% fetal bovine serum (Thermo Fisher; 26140079). Transfection of 293T/17 cells for the proximity ligation assay (PLA) is described below.
FIV virus production and RT assay.
G355-5 cells were transfected as described above but in 6-well plates. Briefly, each well was transfected with 2 μg per well of FIV or was mock transfected. At 24 h posttransfection, cells were washed with medium twice and medium was replaced with 4 ml of complete McCoy 5A. At 144 h posttransfection, medium was filtered with a 0.45-μm syringe filter, aliquoted, and stored at −80°C. Reverse transcriptase activity in medium from FIV- or mock-transfected cells was estimated using a SYBR green I-based product-enhanced reverse transcriptase (SG-PERT) assay (86). A set of recombinant RT standards (EMD Millipore catalog no. 382129) ranging from 5.12e−3 nU/ml to 5.12e−9 nU/ml was used to estimate the activity (as nU/ml) of reverse transcriptase activity in the FIV stock.
Cell lysis and DSP cross-linking.
For all experiments, cells were harvested 24 to 48 h posttransfection. When harvested without cross-linking, cells were washed twice with Dulbecco's phosphate-buffered saline (DPBS) with no calcium and no magnesium (Thermo Fisher; 14190250) and lysates were collected in the presence of the protease inhibitor cocktail (Sigma-Aldrich; P8340), Ribolock RNase inhibitor (Thermo Fisher; E00381) in standard salt buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 50 mM KCl, 0.625% NP-40). Where indicated, cells were harvested in a low-salt lysis buffer (10 mM Tris [pH 7.4], 10 mM NaCl, 1 mM MgCl2, 0.2 mM EDTA, 0.35% Triton X-100). When harvested with cross-linking versus mock treatment, cells were washed twice with wash buffer and treated for 30 min either with wash buffer containing DSP at the indicated concentration (Thermo Fisher; 22586) in 0.5% dimethyl sulfoxide (DMSO) or with wash buffer with 0.5% DMSO alone (mock). DSP solution or mock treatment was removed from the cells, the reaction was quenched with standard salt buffer without detergent for 15 min, and cells were subsequently harvested as described for cells without DSP cross-linking. Following harvest, in standard salt buffer, cells were sheared 20 times with an 18-gauge syringe and clarified by centrifugation at low speed in a Beckman F241.5 rotor at 200 × g for 10 min at 4°C, followed by a second clarification by centrifugation in a Beckman F241.5 rotor at 18,000 × g for 1 min at 4°C. Cells harvested in low-salt lysis buffer were processed the same way, except that the second clarification step was increased from 1 min to 10 min in a F241.5 rotor at 18,000 × g and 4°C.
Virus-like particle harvest.
Culture supernatants were collected 24 to 48 h posttransfection and filtered with a 0.45-μm PES syringe filter to remove any remaining cells. Virus-like particles were centrifuged through a 30% sucrose cushion in a Beckman SW55Ti rotor at 130,000 × g for 30 min at 4°C. For WB, the virus pellet was harvested directly in loading buffer for analysis by WB with an antibody to FIV CA (NIH AIDS Reagent Program; 4814). For velocity sedimentation, virus pellets were resuspended in the presence of the protease inhibitor cocktail (Sigma-Aldrich; P8340), Ribolock RNase inhibitor (Thermo Fisher; E00381) in phosphate-buffered saline (PBS). Harvested virus pellets were agitated at 1,000 rpm for 20 min at room temperature to further resuspend VLPs. Resuspended VLPs were clarified by centrifugation in a Beckman F241.5 rotor at 18,000 × g for 1 min at 4°C. Where indicated, resuspended virus pellets were cross-linked for 30 min at room temperature (prior to de-enveloping the pelleted virus) by the addition of DSP to a final concentration of 0.125 mM. The cross-linking reaction was quenched by the addition of Tris, pH 7.4, to a final concentration of 20 mM. Virus was de-enveloped by adding detergent buffer (to a final concentration of 10 mM Tris [pH 7.4], 100 mM NaCl, 50 mM KCl, 0.625% NP-40), followed by incubation on ice for 15 min. De-enveloped virus was clarified by centrifugation in a Beckman F241.5 rotor at 18,000 × g for 1 min at 4°C.
Velocity sedimentation.
Harvested virus pellet or cell lysate was layered on a 5-ml step gradient containing equivalent layers (10%, 15%, 20%, 40%, 50%, 66%, and 80% sucrose in the same lysis buffer used to harvest the cells) and subjected to velocity sedimentation in a Beckman MLS-50 rotor at 217,000 × g for 45 min at 4°C. Gradients were fractionated from top to bottom, and pelleted material was harvested for WB. Aliquots of fractions and pellet were analyzed by SDS-PAGE, followed by WB with either an antibody to FIV CA (NIH AIDs Reagent Program catalog no. 4814) or antibody to HIV CA (NIH AIDs Reagent Program catalog no. 1513), as indicated in the figure legends. Gag in pellet, which likely represents denatured Gag, was not included in the quantification of the velocity sedimentation gradients and represented less than 10% of total Gag signal in the gradient. The method for estimating the migration of particles with different S values in gradients has been described previously (20).
Immunoprecipitation and Western blotting.
Cell lysates or 2-fold-diluted pooled gradient fractions were subjected to IP with 1 to 2 μg of an antibody to ABCE1 (26), DDX6 (Bethyl; A300-461A), DCP2 (Novus Biologicals; NB100-56198), or rabbit IgG antibody (Bethyl; P120-101) by incubating antibody with lysate for 30 min to overnight at 4°C with rotation, followed by incubation with protein G-coupled Dynabeads (Life Technologies; 10004D) for 1 to 4 h at 4°C with rotation. The beads were then washed with the same lysis buffer used for harvesting the cells, twice with buffer containing detergent and once with buffer without detergent. IP eluates were analyzed by WB, with antibodies to the following: FIV CA (NIH AIDs Reagent Program; 4814), RPS6 (Cell Signaling Technology; 2317), or ABCE1 (26). WB signals were detected by using secondary antibodies conjugated to horseradish peroxidase (HRP) and Pierce ECL substrate (Thermo Fisher Scientific) with Carestream Kodak Biomax Light film or by using secondary antibodies conjugated to infrared dyes (LI-COR, Lincoln, NE). Quantification of Gag bands on film was performed using Image Studio software (LI-COR).
PLA.
For the proximity ligation assay (PLA), 293T/17 cells were plated into 6-well dishes containing poly-l-lysine-coated coverslips with Grace Biolabs CultureWell silicone chambers (Sigma-Aldrich) attached to create four chambers on each coverslip. Cells were transfected with 2 to 3 μg of plasmid per well (mock-transfected cells received no DNA) using PEI as described previously (25) and 16.5 h later were fixed for 15 min in 4% paraformaldehyde in PBS (pH 7.4), permeabilized in 0.3% Triton X-100 in PBS (pH 7.4) for 10 min, and blocked in Duolink blocking solution (Sigma-Aldrich) at 37°C for 30 min. Cells were incubated in primary antibody (described in “Immunoprecipitation and Western blotting”), followed by the Duolink reagents (Sigma-Aldrich) oligonucleotide-linked secondary antibody, ligation mix, and red amplification/detection mix, with washes in between, per the Duolink protocol. For concurrent immunofluorescence (IF), cells were incubated for 15 min at room temperature with 1:1,000 Alexa Fluor 488 anti-mouse secondary antibody after the final buffer B PLA washes. Coverslips were mounted onto glass slides using Duolink in situ mounting medium with DAPI (4′,6-diamidino-2-phenylindole), sealed to the glass slides with clear nail polish, allowed to dry for 24 h at room temperature, and stored at −20°C. Imaging was performed with a Zeiss Axiovert 200M deconvolution microscope using a Zeiss Plan-Apochromat lens (63× aperture, 1.4 objective with oil immersion), with AxioVision Rel. 4.8 software. For quantification, five fields containing at least two Gag IF-positive cells were chosen at random and imaged using identical exposure times for the red and green channels (for ABCE1 PLA, red and green exposures were 1 s and 2.5 s, respectively; for DCP2 PLA, red and green exposures were 25 ms and 2 s, respectively). In the case of the Gag-ABCE1 PLA, the five fields selected for analysis from the FIV WT Gag group contained 35 cells, and the five selected fields from the FIV Gag MACA group contained 37 cells. In the case of the Gag-DCP2 PLA, the five fields selected for analysis from the FIV WT Gag group contained 18 cells, the five fields selected from the FIV Gag MACA contained 15 cells, and the five fields selected from the FIV Gag G2A group contained 21 cells. Images were captured as a Z-stack of five 1-μm slices centered on the focal point for the PLA. Images were deconvolved using the AxioVision software, the Z-stack images were then exported as tif files, and Image J was used to outline Gag IF-positive cells in the central Z-stack image from each field. Within those Gag IF-positive cells, this same central image was used to count the number of PLA “spots” and to quantify Gag IF intensity where indicated by using Image J. The PLA spot number for each field was then normalized to the average Gag IF intensity within that field, and the results were plotted with error bars representing the standard errors of the mean (SEM) for five fields. For the Gag IF-negative cells within WT Gag-transfected fields (Fig. 8), ImageJ was used to subtract PLA and DAPI signal contributed by all Gag IF-positive cells in the same images used for WT PLA spot counting, five IF-negative cells were then chosen per field, the boundaries were manually outlined, and the PLA spots were counted within these boundaries. Mock-transfected cells (Fig. 10) were quantified in the same way (but without subtraction of Gag IF-positive cells) using the central Z-stack image from each field in the mock-transfected condition. The PLA spot count for these negative cells was then compared to the PLA spot number for Gag IF-positive cells under each condition, without normalization to Gag IF. Prior to export of the images used in the figures, the red channel gain for all conditions was adjusted to 2 (for all Gag-DCP2 PLA images) or 3 (for all Gag-ABCE1 PLA images) in the AxioVision software to allow spots to be easily seen by eye. Gain was not adjusted for the blue or green channels. Images were then imported in 8-bit color into Inkscape to create the final figure layout, without further adjustments for color balance or gamma correction. Each of the images shown in Fig. 8 and 10 is an image from the center slice of a Z-stack that is representative of the mean. PLA experiments were repeated once, with similar results.
Structure alignment and sequence analysis.
FIV 34TF10 Gag CA-NTD was submitted to SWISS-MODEL (87–90), which uses BLAST and HHBlits to identify evolutionarily related structures matching the submitted target sequence. The top-scoring template was RELIK CA-NTD (PDB accession number 2XGU [19]), which was then selected for homology modeling using SWISS-MODEL. The resulting FIV CA-NTD homology model had a global quality estimation score (GMQE score) of 0.73 (with a perfect score being 1.0), a QMEAN score of −0.17, and a normalized QMEAN4 Z-score of <1 in SWISS-MODEL. Local quality estimates of the model were largely >0.6. The FIV CA-NTD model obtained from SWISS-MODEL and the FIV CA-CTD crystal structure (PDB accession number 5DCK [18]) were separately aligned with HIV-1 CA structure (PDB accession number 5L93 [59]) using FATCAT (91, 92), resulting in optimized root mean square deviations (RMSD) of 2.75 Å for the NTD domains and 1.94 Å for the CTD domains. While this report was in the final stages of review, a structure of the full FIV CA protein was published (PDB accession number 5NA2 [93]). Our RELIK-based model for FIV CA-NTD fits well with the newly published FIV CA model, with alignment of the RELIK-based CA-NTD structure and the newly published FIV CA-NTD structure resulting in an RMSD of 2.67 Å using FATCAT. Note that the sequence alignments from FATCAT are shown in Fig. 5A and the superposed domains, rendered by CCP4MG (94), are shown in Fig. 5B.
For pairwise sequence alignments of human ABCE1 and DDX6 (GenBank accession numbers NP_001035809.1 and NP_001244120.1, respectively) with feline ABE1 and DDX6 (GenBank accession numbers XP_003985024.1 and XP_011284692.1, respectively), EMBOSS Needle was used with default settings (95).
For alignments of FIV Gag from different feline species, FIV Gag sequences were obtained from the NCBI protein database using the search terms “Feline immunodeficiency virus Gag,” resulting in a return of 618 records. These records were then filtered to select only records that contained at least Gag sequence that spanned the residues of interest (i.e., position 152 in helix 1 to position 210 in helix 4, based on the FIV Gag sequence shown in Fig. 5), reducing the data set from 618 to 405 sequences. These records were annotated for species (see Table S1 in the supplemental material for accession numbers and species annotation) and then aligned using Clustal Omega (95).
Supplementary Material
ACKNOWLEDGMENTS
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: FIV 34TF10 from John Elder, anti-FIV p24 monoclonal antibody (PAK3-2C1) from DAIDS, NIAID (Custom Monoclonal Antibodies, Inc.), anti-HIV-1 p24 hybridoma antibody (183-H12-5C) from Bruce Chesebro and Hardy Chen.
Jaisri R. Lingappa and Jonathan C. Reed have shares or stock options in Prosetta Biosciences, and Jaisri R. Lingappa receives some unrestricted research funding from Prosetta Biosciences. The studies reported here likely do not pose a conflict of interest, given that they do not relate directly to any of Prosetta's antiviral compounds.
These studies were funded by Prosetta Biosciences and by NIAID RO1 grant AI106397 to J.R.L.
Neither of the funders (Prosetta or NIH) had a role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01761-17.
REFERENCES
- 1.Hartmann K. 2012. Clinical aspects of feline retroviruses: a review. Viruses 4:2684–2710. doi: 10.3390/v4112684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhl EW, Martin M, Coleman JK, Yamamoto JK. 2008. Advances in FIV vaccine technology. Vet Immunol Immunopathol 123:65–80. doi: 10.1016/j.vetimm.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunham SP, Bruce J, MacKay S, Golder M, Jarrett O, Neil JC. 2006. Limited efficacy of an inactivated feline immunodeficiency virus vaccine. Vet Rec 158:561–562. doi: 10.1136/vr.158.16.561. [DOI] [PubMed] [Google Scholar]
- 4.Levy J, Crawford C, Hartmann K, Hofmann-Lehmann R, Little S, Sundahl E, Thayer V. 2008. 2008 American Association of Feline Practitioners' feline retrovirus management guidelines. J Feline Med Surg 10:300–316. doi: 10.1016/j.jfms.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann K, Wooding A, Bergmann M. 2015. Efficacy of antiviral drugs against feline immunodeficiency virus. Vet Sci 2:456–476. doi: 10.3390/vetsci2040456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienzle D. 2014. FIV in cats—a useful model of HIV in people? Vet Immunol Immunopathol 159:171–179. doi: 10.1016/j.vetimm.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Freed EO. 2015. HIV-1 assembly, release and maturation. Nat Rev Microbiol 13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manrique ML, Celma CC, Gonzalez SA, Affranchino JL. 2001. Mutational analysis of the feline immunodeficiency virus matrix protein. Virus Res 76:103–113. doi: 10.1016/S0168-1702(01)00249-0. [DOI] [PubMed] [Google Scholar]
- 9.Abdusetir Cerfoglio JC, Gonzalez SA, Affranchino JL. 2014. Structural elements in the Gag polyprotein of feline immunodeficiency virus involved in Gag self-association and assembly. J Gen Virol 95:2050–2059. doi: 10.1099/vir.0.065151-0. [DOI] [PubMed] [Google Scholar]
- 10.Manrique ML, Rauddi ML, Gonzalez SA, Affranchino JL. 2004. Functional domains in the feline immunodeficiency virus nucleocapsid protein. Virology 327:83–92. doi: 10.1016/j.virol.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 11.Luttge BG, Shehu-Xhilaga M, Demirov DG, Adamson CS, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. 2008. Molecular characterization of feline immunodeficiency virus budding. J Virol 82:2106–2119. doi: 10.1128/JVI.02337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elder JH, Schnolzer M, Hasselkus-Light CS, Henson M, Lerner DA, Phillips TR, Wagaman PC, Kent SB. 1993. Identification of proteolytic processing sites within the Gag and Pol polyproteins of feline immunodeficiency virus. J Virol 67:1869–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lingappa JR, Reed JC, Tanaka M, Chutiraka K, Robinson BA. 2014. How HIV-1 Gag assembles in cells: putting together pieces of the puzzle. Virus Res 193:89–107. doi: 10.1016/j.virusres.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patarca R, Haseltine WA. 1985. A major retroviral core protein related to EPA and TIMP. Nature 318:390. doi: 10.1038/318390a0. [DOI] [PubMed] [Google Scholar]
- 15.Orlinsky KJ, Gu J, Hoyt M, Sandmeyer S, Menees TM. 1996. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J Virol 70:3440–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 17.Schur FK, Hagen WJ, Rumlova M, Ruml T, Muller B, Krausslich HG, Briggs JA. 2015. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 18.Khwaja A, Galilee M, Marx A, Alian A. 2016. Structure of FIV capsid C-terminal domain demonstrates lentiviral evasion of genetic fragility by coevolved substitutions. Sci Rep 6:24957. doi: 10.1038/srep24957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstone DC, Yap MW, Robertson LE, Haire LF, Taylor WR, Katzourakis A, Stoye JP, Taylor IA. 2010. Structural and functional analysis of prehistoric lentiviruses uncovers an ancient molecular interface. Cell Host Microbe 8:248–259. doi: 10.1016/j.chom.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Lingappa JR, Hill RL, Wong ML, Hegde RS. 1997. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol 136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dooher JE, Schneider BL, Reed JC, Lingappa JR. 2007. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic 8:195–211. doi: 10.1111/j.1600-0854.2006.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson BA, Reed JC, Geary CD, Swain JV, Lingappa JR. 2014. A temporospatial map that defines specific steps at which critical surfaces in the Gag MA and CA domains act during immature HIV-1 capsid assembly in cells. J Virol 88:5718–5741. doi: 10.1128/JVI.03609-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingappa JR, Dooher JE, Newman MA, Kiser PK, Klein KC. 2006. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J Biol Chem 281:3773–3784. doi: 10.1074/jbc.M507255200. [DOI] [PubMed] [Google Scholar]
- 24.Klein KC, Reed JC, Tanaka M, Nguyen VT, Giri S, Lingappa JR. 2011. HIV Gag-leucine zipper chimeras form ABCE1-containing intermediates and RNase-resistant immature capsids similar to those formed by wild-type HIV-1 Gag. J Virol 85:7419–7435. doi: 10.1128/JVI.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka M, Robinson BA, Chutiraka K, Geary CD, Reed JC, Lingappa JR. 2015. Mutations of conserved residues in the major homology region arrest assembling HIV-1 Gag as a membrane-targeted intermediate containing genomic RNA and cellular proteins. J Virol 90:1944–1963. doi: 10.1128/JVI.02698-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmerman C, Klein KC, Kiser PK, Singh ARS, Firestein BL, Riba SC, Lingappa JR. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]
- 27.Schuller AP, Green R. 2017. The ABC(E1)s of ribosome recycling and reinitiation. Mol Cell 66:578–580. doi: 10.1016/j.molcel.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Reed JC, Molter B, Geary CD, McNevin J, McElrath J, Giri S, Klein KC, Lingappa JR. 2012. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J Cell Biol 198:439–456. doi: 10.1083/jcb.201111012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostareck DH, Naarmann-de Vries IS, Ostareck-Lederer A. 2014. DDX6 and its orthologs as modulators of cellular and viral RNA expression. Wiley Interdiscip Rev RNA 5:659–678. doi: 10.1002/wrna.1237. [DOI] [PubMed] [Google Scholar]
- 30.Engeland CE, Brown NP, Borner K, Schumann M, Krause E, Kaderali L, Muller GA, Krausslich HG. 2014. Proteome analysis of the HIV-1 Gag interactome. Virology 460-461:194–206. doi: 10.1016/j.virol.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 31.Grudzien-Nogalska E, Kiledjian M. 17 July 2017. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip Rev RNA doi: 10.1002/wrna.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kedersha N, Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol 431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 33.Buchan JR. 2014. mRNP granules. Assembly, function, and connections with disease. RNA Biol 11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balagopal V, Parker R. 2009. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol 21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beliakova-Bethell N, Beckham C, Giddings TH Jr, Winey M, Parker R, Sandmeyer S. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, Dou Y, Liou W, Sandmeyer S. 2005. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res 15:641–654. doi: 10.1101/gr.3739005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bilanchone V, Clemens K, Kaake R, Dawson AR, Matheos D, Nagashima K, Sitlani P, Patterson K, Chang I, Huang L, Sandmeyer S. 2015. Ty3 retrotransposon hijacks mating yeast RNA processing bodies to infect new genomes. PLoS Genet 11:e1005528. doi: 10.1371/journal.pgen.1005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu S, Lujan P, Jackson D, Emerman M, Linial M. 2011. The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog 7:e1002303. doi: 10.1371/journal.ppat.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dooher JE, Lingappa JR. 2004. Conservation of a step-wise, energy-sensitive pathway involving HP68 for assembly of primate lentiviral capsids in cells. J Virol 78:1645–1656. doi: 10.1128/JVI.78.4.1645-1656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lingappa UF, Wu X, Macieik A, Yu SF, Atuegbu A, Corpuz M, Francis J, Nichols C, Calayag A, Shi H, Ellison JA, Harrell EK, Asundi V, Lingappa JR, Prasad MD, Lipkin WI, Dey D, Hurt CR, Lingappa VR, Hansen WJ, Rupprecht CE. 2013. Host-rabies virus protein-protein interactions as druggable antiviral targets. Proc Natl Acad Sci U S A 110:E861–E868. doi: 10.1073/pnas.1210198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed JC, Westergreen N, Barajas BC, Ressler D, Phuong DJ, Swain JV, Lingappa VR, Lingappa JR. 22 October 2017. The ABCE1 capsid assembly pathway is conserved between primate lentiviruses and the non-primate lentivirus feline immunodeficiency virus. BioRxiv doi: 10.1101/183848. [DOI] [PMC free article] [PubMed]
- 43.Talbott RL, Sparger EE, Lovelace KM, Fitch WM, Pedersen NC, Luciw PA, Elder JH. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci U S A 86:5743–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters AK, De Parseval AP, Lerner DL, Neil JC, Thompson FJ, Elder JH. 1996. Influence of ORF2 on host cell tropism of feline immunodeficiency virus. Virology 215:10–16. doi: 10.1006/viro.1996.0002. [DOI] [PubMed] [Google Scholar]
- 45.Fadel HJ, Saenz DT, Poeschla EM. 2012. Construction and testing of orfA +/− FIV reporter viruses. Viruses 4:184–199. doi: 10.3390/v4010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poeschla EM, Wong-Staal F, Looney DJ. 1998. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat Med 4:354–357. doi: 10.1038/nm0398-354. [DOI] [PubMed] [Google Scholar]
- 47.Lin YC, Brik A, de Parseval A, Tam K, Torbett BE, Wong CH, Elder JH. 2006. Altered gag polyprotein cleavage specificity of feline immunodeficiency virus/human immunodeficiency virus mutant proteases as demonstrated in a cell-based expression system. J Virol 80:7832–7843. doi: 10.1128/JVI.00374-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips TR, Talbott RL, Lamont C, Muir S, Lovelace K, Elder JH. 1990. Comparison of two host cell range variants of feline immunodeficiency virus. J Virol 64:4605–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen N, Ho E, Brown M, Yamamoto J. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- 50.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A 88:3195–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang M, Orenstein JM, Martin MA, Freed EO. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol 69:6810–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kutluay SB, Bieniasz PD. 2010. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog 6:e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demirov DG, Orenstein JM, Freed EO. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol 76:105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu HH, Chang YF, Wang CT. 2006. Mutations in the alpha-helix directly C-terminal to the major homology region of human immunodeficiency virus type 1 capsid protein disrupt Gag multimerization and markedly impair virus particle production. J Biomed Sci 13:645–656. doi: 10.1007/s11373-006-9094-6. [DOI] [PubMed] [Google Scholar]
- 55.Joshi A, Nagashima K, Freed EO. 2006. Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation. J Virol 80:7939–7951. doi: 10.1128/JVI.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ono A, Waheed AA, Joshi A, Freed EO. 2005. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J Virol 79:14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol 77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grover JR, Llewellyn GN, Soheilian F, Nagashima K, Veatch SL, Ono A. 2013. Roles played by capsid-dependent induction of membrane curvature and Gag-ESCRT interactions in tetherin recruitment to HIV-1 assembly sites. J Virol 87:4650–4664. doi: 10.1128/JVI.03526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schur FK, Obr M, Hagen WJ, Wan W, Jakobi AJ, Kirkpatrick JM, Sachse C, Krausslich HG, Briggs JA. 2016. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 60.Esteva MJ, Affranchino JL, Gonzalez SA. 2014. Lentiviral Gag assembly analyzed through the functional characterization of chimeric simian immunodeficiency viruses expressing different domains of the feline immunodeficiency virus capsid protein. PLoS One 9:e114299. doi: 10.1371/journal.pone.0114299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vani K, Bogen SA, Sompuram SR. 2006. A high throughput combinatorial library technique for identifying formalin-sensitive epitopes. J Immunol Methods 317:80–89. doi: 10.1016/j.jim.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 63.Poeschla EM, Looney DJ. 1998. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J Virol 72:6858–6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schipany K, Rosner M, Ionce L, Hengstschlager M, Kovacic B. 2015. eIF3 controls cell size independently of S6K1-activity. Oncotarget 6:24361–24375. doi: 10.18632/oncotarget.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geissler R, Golbik RP, Behrens SE. 2012. The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes. Nucleic Acids Res 40:4998–5011. doi: 10.1093/nar/gks070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sandmeyer SB, Clemens KA. 2010. Function of a retrotransposon nucleocapsid protein. RNA Biol 7:642–654. doi: 10.4161/rna.7.6.14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC. 2005. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J 24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janaky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, Lill R. 2005. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J 24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen ZQ, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M. 2006. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J Biol Chem 281:7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- 70.Anderson P, Kedersha N. 2006. RNA granules. J Cell Biol 172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eulalio A, Behm-Ansmant I, Izaurralde E. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 73.Verschoor A, Warner JR, Srivastava S, Grassucci RA, Frank J. 1998. Three-dimensional structure of the yeast ribosome. Nucleic Acids Res 26:655–661. doi: 10.1093/nar/26.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. 2004. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol 24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clement JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, Boccaccio G, Mouland AJ. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci 123:369–383. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- 76.Bouttier M, Saumet A, Peter M, Courgnaud V, Schmidt U, Cazevieille C, Bertrand E, Lecellier CH. 2012. Retroviral GAG proteins recruit AGO2 on viral RNAs without affecting RNA accumulation and translation. Nucleic Acids Res 40:775–786. doi: 10.1093/nar/gkr762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Izumi T, Burdick R, Shigemi M, Plisov S, Hu WS, Pathak VK. 2013. Mov10 and APOBEC3G localization to processing bodies is not required for virion incorporation and antiviral activity. J Virol 87:11047–11062. doi: 10.1128/JVI.02070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cinti A, Le Sage V, Ghanem M, Mouland AJ. 2016. HIV-1 Gag blocks selenite-induced stress granule assembly by altering the mRNA cap-binding complex. mBio 7:e00329. doi: 10.1128/mBio.00329-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Valiente-Echeverria F, Melnychuk L, Vyboh K, Ajamian L, Gallouzi IE, Bernard N, Mouland AJ. 2014. eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat Commun 5:4819. doi: 10.1038/ncomms5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Phalora PK, Sherer NM, Wolinsky SM, Swanson CM, Malim MH. 2012. HIV-1 replication and APOBEC3 antiviral activity are not regulated by P bodies. J Virol 86:11712–11724. doi: 10.1128/JVI.00595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Decker CJ, Parker R. 2012. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sokalingam S, Raghunathan G, Soundrarajan N, Lee SG. 2012. A study on the effect of surface lysine to arginine mutagenesis on protein stability and structure using green fluorescent protein. PLoS One 7:e40410. doi: 10.1371/journal.pone.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Laco GS, Schalk-Hihi C, Lubkowski J, Morris G, Zdanov A, Olson A, Elder JH, Wlodawer A, Gustchina A. 1997. Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor. Biochemistry 36:10696–10708. doi: 10.1021/bi9707436. [DOI] [PubMed] [Google Scholar]
- 84.Kemler I, Barraza R, Poeschla EM. 2002. Mapping the encapsidation determinants of feline immunodeficiency virus. J Virol 76:11889–11903. doi: 10.1128/JVI.76.23.11889-11903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamashita M, Emerman M. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol 78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, Taghon T, Pizzato M, Verhasselt B. 2012. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One 7:e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 90.Guex N, Peitsch MC, Schwede T. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 91.Ye Y, Godzik A. 2003. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19(Suppl 2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 92.Ye Y, Godzik A. 2004. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res 32:W582–W585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Folio C, Sierra N, Dujardin M, Alvarez G, Guillon C. 2017. Crystal structure of the full-length feline immunodeficiency virus capsid protein shows an N-terminal beta-hairpin in the absence of N-terminal proline. Viruses 9:E335. doi: 10.3390/v9110335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.